Abstract
Current approaches to block KRAS oncogene function focus on inhibition of K-Ras downstream effector signaling. We evaluated the anti-tumor activity of selumetinib (AZD6244, ARRY-142886), a potent and selective MEK1/2 inhibitor, on a panel of colorectal carcinoma (CRC) cells and found no inhibition of KRAS mutant CRC cell anchorage-independent growth. While AKT activity was elevated in KRAS mutant cells, and PI3K inhibition did impair the growth of MEK inhibitor-insensitive CRC cell lines, concurrent treatment with selumetinib did not provide additional anti-tumor activity. Therefore, we speculated that inhibition of the Ral guanine exchange factor (RalGEF) effector pathway may be a more effective approach for blocking CRC growth. RalGEFs are activators of the related RalA and RalB small GTPases and we found activation of both in CRC cell lines and patient tumors. Interfering RNA stable suppression of RalA expression reduced CRC tumor cell anchorage-independent growth, but surprisingly, stable suppression of RalB greatly enhanced soft agar colony size and formation frequency. Despite their opposing activities, both RalA and RalB regulation of anchorage-independent growth required interaction with RalBP1/RLIP76 and components of the exocyst complex. Interestingly, RalA interaction with the Exo84 but not Sec5 exocyst component was necessary for supporting anchorage-independent growth, whereas RalB interaction with Sec5 but not Exo84 was necessary for inhibition of anchorage-independent growth. We suggest that anti-RalA-selective therapies may provide an effective approach for KRAS mutant CRC.
Keywords: Ras, MEK, AKT, RalBP1/RLIP76, exocyst
Introduction
KRAS is the most frequently mutated oncogene in colorectal cancer (CRC) (1, 2) and required for the maintenance of CRC cell growth (3–5). Therefore, it is believed that K-Ras inhibition will provide an effective therapeutic strategy for CRC. Current strategies have focused on inhibitors of Ras effector signaling (6). The best studied Ras effector pathways are the Raf-MEK-ERK mitogen-activated protein kinase and phosphotidylinositol 3-kinase (PI3K)-AKT serine/threonine protein kinase effector pathways (7), with inhibitors of components of both pathways currently under clinical evaluation 4. Further support for the functional role of these effectors in cancer growth comes from the identification of mutationally activated B-Raf (8, 9) or the p110α catalytic subunit of PI3K (10) in CRC. However, essential roles for other effectors in Ras-mediated oncogenesis have also been demonstrated: Ral (RalGDS) and Rac (Tiam1) guanine nucleotide exchange factors and PLCε (11–13). Tiam1 (14) or PLCε (15) deficiency impaired colon tumorigenesis in APC mutant Min mice. Tiam1 overexpression has also been implicated in colon cancer metastatic growth (13). These observations raise the question of whether inhibition of Raf or PI3K effector signaling, or another effector pathway, will be the most effective approach for treatment of KRAS mutant CRC.
Recent studies established that Ral small GTPases, activated by RalGEFs, are critical drivers of human oncogenesis (16). Our recent studies found more frequent activation of Ral, rather than the Raf or PI3K effector pathways, in KRAS mutant pancreatic ductal adenocarcinoma (PDAC) tissue and cell lines (17, 18). Ral has also been found to contribute to the growth of cancer types where RAS mutations are seen infrequently (19–21).
Mutated Ras stimulates RalGEF activation, which in turn stimulates formation of the active, Ral-GTP which then binds downstream effectors (16). One of the surprising findings regarding Ral GTPases and cancer is the striking, sometimes opposing functions of the otherwise highly related RalA and RalB (82% amino acid identity) proteins (19). Distinct roles for RalA and RalB were also described for prostate (20) and bladder (21), and our studies showed that suppression of RalA but not RalB expression impaired KRAS mutant PDAC anchorage-independent and tumorigenic growth (18).
Less clear is the role of specific effectors in Ral-mediated oncogenesis. Perhaps the best validated effector in tumor cell growth is the Sec5 subunit of the exocyst complex that regulates vesicle trafficking, where Sec5 activation of the TBK1 atypical IkappaB kinase is essential for tumor but not normal cell survival (22). The second best studied Ral effector is RalBP1/RLIP76 that functions as a GAP for Rho small GTPases (23) and RalBP1 inhibition suppressed tumor xenograft tumor growth in mice (24).
A key unresolved issue is which effector pathway(s) should be targeted for effective K-Ras inhibition in CRC. We utilized pharmacologic inhibition of Raf and PI3K and found variable effectiveness in blocking KRAS mutant CRC anchorage-independent growth. We explored a role for Ral GTPases and found that RalA was required for CRC growth, but surprisingly, suppression of RalB enhanced growth. Since we previously found that RalB was dispensable for pancreatic tumor cell anchorage-independent growth, our results demonstrate a striking difference in RalB function in two distinct mutant KRAS-driven cancers.
Materials and Methods
Cell culture
CRC cell lines were obtained from ATCC and maintained in either DMEM-H or RPMI-1640 supplemented with 10% fetal calf serum, and frozen down to maintain limited passage history. Cell lines were treated with either selumetinib (provided by AstraZeneca) or LY294002 for 24 h for inhibition of ERK or AKT, and soft agar analyses were done as described previously (25), with colony formation quantitated after 14 days. Mutation status for KRAS, BRAF and PIK3CA was derived from the COSMIC database5.
Plasmids
Short hairpin RNA (shRNA) sequences for human RalA or RalB were subcloned into pSuper.retro.blast (provided by Dr. John Minna) and pSuper.retro puro (Oligoengine), pBabe-puro retrovirus expression vectors encoding wild-type human RalA and RalB and RNAi-insensitive Ral cDNA sequences for RalA or RalB have been described previously (18), and were used to generate cDNA sequences encoding effector binding mutants. shRNA sequences for human Exo84 were cloned into pSuper.retro puro and the sequences are as follows: shExo84 1 – GGTGCCACTTTACTCTATA and shExo84 2 – ACAATATAATTTGAATGGCTAA. RalBP1 shRNA has been described previously (42). RNAi-insensitive RalBP1 was cloned into pBabeHAII puro with the following silent mutations (in italics): GTAGAACGTACGATGATGT. pLKO.1 puro lentiviral vectors encoding Sec5 shRNA (TRCN0000116102-TRCN0000116106) were obtained from OpenBiosystems TRC shRNA library and pLKO.1 puro NS shRNA (CAACAAGATGAAGAGCACCAA) was obtained from Sigma.
Immunoblotting
Blot analyses were done with antibodies for RalA (BD Laboratories), RalB (Millipore), β-actin and vinculin (Sigma), GAPDH and RalBP1 (Abcam), phosphorylated ERK1 and ERK2 T202/Y204, phospho-AKT S473, and total AKT (Cell Signaling), and total ERK1/2 (Santa Cruz Biotechnology), Exo84 (Orbigen) and Sec5 (Dr. Charles Yeaman, University of Iowa). Activated Ral-GTP was determined by pull down analyses as described previously (18). β-actin and vinculin blotting was done to verify equivalent total protein.
Statistical analysis
The Fisher’s exact test was used to analyze associations between two variables and the Pearson Chi-square test was used to analyze association between more than two variables.
Results
KRAS mutant CRC cell lines are insensitive to growth inhibition by blocking MEK
Our previous studies with two MEK1/2 inhibitors, U0126 and CI-1040 (PD184352), found that KRAS mutation status and ERK1/2 activation (pERK) did not correlate strongly with MEK inhibitor sensitivity (26). However, both inhibitors possess off-target activities. Therefore, we extended these analyses using a more potent and selective MEK1/2 inhibitor, selumetinib, that is currently undergoing extensive clinical trial analyses. The majority of previous studies evaluated MEK1/2 inhibitor activity against tumor cell lines grown as two-dimensional anchorage-dependent cultures (27–30). When compared, it was found that tumor cell growth in three-dimensional suspension cultures was more resistant to MEK inhibitor treatment (31, 32). Therefore, we evaluated colony formation in soft agar. Since previous studies determined that selumetinib inhibited pERK with a half-maximal inhibition (IC50) at a dose of <40 nM (26) or ranging from 10 to 100 nM (27), we treated cells with one set high concentration (200 nM) to assess sensitivity, determined the degree of pERK reduction and evaluated its effect on growth.
As we described previously (25), elevated pERK levels did not correlate with KRAS or BRAF mutation status (Figs. 1A–C). Similar to our previous observations (25), there was wide variation in selumetinib treatment reduction in pERK, with some cell lines showing high sensitivity and others with relative insensitivity when evaluated in adherent cultures. Interestingly, selumentinib treatment caused limited to no inhibition of growth in all six KRAS mutant CRC cell lines (p=0.031) (Fig. 1D). This pattern was distinct from what we observed with U0126 and CI-1040, where KRAS mutant SW480 growth was sensitive to both U0126 and CI-1040 (25). Four of five BRAF mutant CRC lines were inhibited by selumetinib, with only NCI-H508 showing insensitivity. NCI-H508 cells were also insensitive to U0126 and CI-1040 treatment (25). In contrast to other MEK inhibitors, selumetinib does not exhibit inhibition of MEK5 (26, 27). Nevertheless, despite inhibitor-specific differences in sensitivity, we reached the same conclusion with all three MEK inhibitors, that neither elevated pERK levels nor the degree of pERK inhibition was predictive of sensitivity to selumetinib growth inhibition.
Figure 1.
A–C, Steady state levels of activated pERK and pAKT, total ERK1/2 or AKT were determined by western blot analysis in vehicle- or selumetinib-treated cells. D, Colony forming activity was determined for vehicle- or selumetinib-treated CRC cell lines, with activity normalized to 1.0 for vehicle-treated cultures. E, PI3K inhibition reduces the growth of MEK inhibitor-insensitive cell lines. Mutation status for KRAS or BRAF or PIK3CA is indicated; WT indicates no mutation for KRAS or BRAF. Data shown are representative of two or three independent experiments.
A recent study with selumetinib and CRC cell lines found an association between inhibitor resistance and high pAKT levels (27). Therefore, we determined whether MEK inhibitor refractory CRC cells lines corresponded to those with PIK3CA mutation and/or AKT phosphorylation and activation (pAKT). While all five PIK3CA mutant CRC lines showed elevated pAKT levels, elevated pAKT was also seen in PIK3CA wild type cell lines (Figs. 1A–C) and PIK3CA mutation status did not correlate with selumitinib resistance (p=0.580). In contrast to our previous findings with KRAS mutant PDAC cell lines (25), we found elevated pAKT in all KRAS mutant CRC cell lines, while high pAKT levels was seen in only a subset of BRAF/KRAS WT or BRAF mutant CRC lines. High pAKT levels correlated weakly with selumetinib insensitivity (p=0.095). Finally, we determined if three selumitinib insensitive KRAS mutant cell lines were sensitive to PI3K inhibition. LY294002 treatment inhibited the growth of HCT-116, SW480, and T84 cells (~50–70%). However, concurrent treatment with both LY294002 and selumetinib did not result in further inhibitory activity (Fig. 1E). These results suggested that the inhibition of other Ras effector pathways, either alone or together with MEK and PI3K, may be required to effectively block the growth of KRAS mutant CRC cells. Since we recently identified a role for a third Ras effector pathway, leading to Ral GTPase activation, for pancreatic cancer growth (18), we focused on validating a role for Ral GTPases in CRC growth.
RalA and RalB are activated CRC cell lines
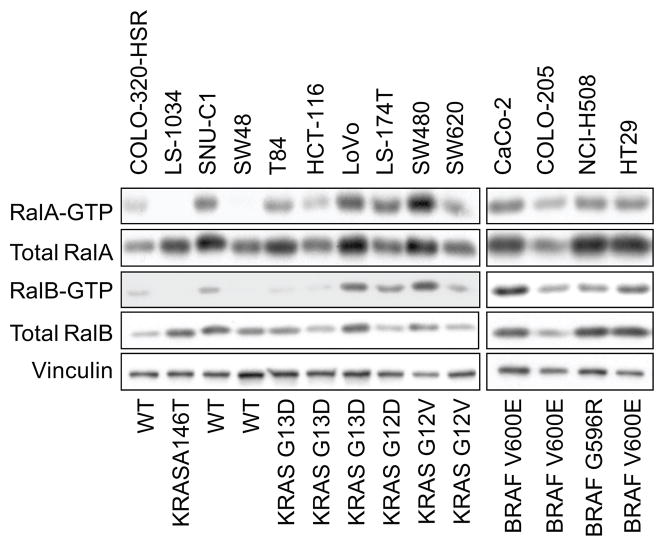
In our analyses of KRAS mutation positive PDAC cell lines and patient tumors, we determined that elevated steady-state levels of Ral GTPases, and not pERK or pAKT, were associated with the majority of cell lines and tumors (17, 18). Similarly, we found that ERK activation did not correlate with the KRAS mutation status of CRC cell lines. Instead, we determined that RalA and/or RalB are activated persistently in a majority CRC cell lines (Fig. 2A). Similar to PDAC cell lines, Ral activation did not correlate strictly with KRAS mutation status. Finally, we also detected activated GTP-bound RalA and RalB in CRC patient tumors (supplementary data Fig. 1). Although across all tumor stages, there was not a consistent difference when compared to matched normal tissue, tumors that were node positive (98-0265, 99-0148, 99-0296) had elevated levels of RalB-GTP, total RalA, and RalA-GTP (99-0148 and 99-0296) compared to the normal adjacent mucosa.
Figure 2.
Activated RalA and RalB expression in CRC cell lines. Pull down analysis with GST-RalBP1-RBD was done, followed by blot analyses with RalA-or RalB-specific antisera. Blot analysis of total cell lysate with anti-RalA or –RalB antiserum for total Ral protein.
RalA and RalB exhibit opposing activities in regulation of CRC cell line anchorage-independent growth
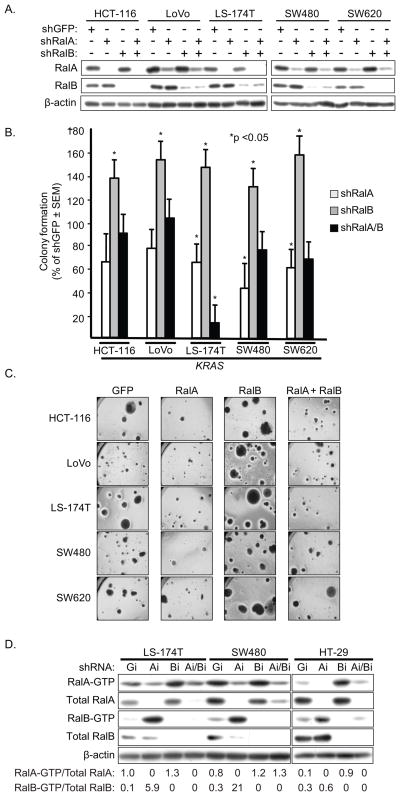
We found previously that sustained shRNA depletion of RalA but not RalB reduced PDAC anchorage-independent growth (18). To determine if these two related isoforms also served similar roles in CRC anchorage-independent growth, we evaluated KRAS or BRAF mutant or KRAS/BRAF WT CRC cell lines and additionally two PDAC cell lines from our previous study (18). Mass populations of PDAC and CRC cell lines stably-infected with each shRNA vector were characterized by western blot analyses to verify steady-state reduction in endogenous RalA or RalB protein (Fig. 3A and data not shown).
Figure 3.
RalA and RalB activity show opposing roles in regulation of CRC anchorage-independent growth. A, Stable suppression of endogenous RalA or RalB protein expression. CRC cell lines were stably-infected with pSuper.retro retrovirus vectors expressing shRNA for GFP (nonspecific control), RalA or RalB. Mass populations of drug-resistant cells were established and blot analyses were done to verify reduction in steady-state RalA or RalB protein expression, and for vinculin to verify equivalent total protein. B, Suppression of RalA reduces whereas suppression of RalB enhances colony formation in soft agar. Data shown are representative of two independent experiments. The mutation status of KRAS and BRAF is indicated with WT indicating wild type for KRAS and BRAF (COSMIC). C, Suppression of RalB enhances colony size. Representative fields of colonies from analyses quantitated in panel B.
As we observed previously, suppression of RalA but not RalB reduced the soft agar growth of the two PDAC cell lines (PANC-1 and T3M4; data not shown). Similarly, we found that RalA suppression reduced colony formation efficiency of eight of eight CRC cell lines, independent of KRAS mutation status (Fig. 3B). Surprisingly, suppression of RalB caused a dramatic increase in colony formation efficiency for all eight cell lines, with a two- to three-fold increase in colonies numbers for T84 and CaCo-2 cells. A significant increase in colony size was also seen (Fig. 4C).
Figure 4.
RalA activity is dominant over RalB in regulation of CRC anchorage-independent growth. A, Stable suppression of endogenous RalA and/or RalB protein expression. CRC cell lines were stably-infected with pSuper.retro retrovirus vectors expressing shRNA for GFP (nonspecific control), RalA or RalB, or both RalA and RalB, and established and characterized as described in Fig. 2A for RalA and RalB expression. B, C, Coordinate suppression of RalA reverses the colony formation enhancement caused by RalB suppression. (panel B) and representative fields of colonies were photographed (panel C). Data shown are normalized to % of shGFP and are representative of two independent experiments. D, Sustained Ral GTPase suppression is associated with increased activity of the related Ral isoforms. CRC cell lines with stable shRNA suppression of RalA or RalB expression were established and characterized as described in Fig. 2A. Numbers indicate the amount of Ral-GTP/total Ral protein for RalA and RalB respectively. Data shown are representative of two independent experiments.
RalA and RalB have been shown to have distinct functions in a number of cellular processes or biological activities (19, 33, 34). However, when concurrently suppressed, the phenotype associated with RalA is typically dominant over that of RalB (19, 21, 35). To address this possibility, we extended our analyses to a total of five KRAS mutant CRC cell lines with concurrent shRNA suppression of RalA and RalB (Fig. 4A and supplementary data Fig. 3 for LS-1034) and then evaluated colony formation in soft agar (Fig. 4B and supplementary data Fig. 2 for LS-1034). For five of six cell lines, colony formation was similar to that of the negative control scramble shRNA. However, for one cell line (LS174-T), concurrent suppression caused a more significant reduction in colony formation than was seen with suppression of RalA alone. Thus, it appears that co-depletion of RalA and RalB reverses the RalB-depletion phenotype to a level similar to that of control shGFP.
Our observation that suppression of RalB enhanced CRC anchorage-independent growth was unexpected, since it was reported previously that transient siRNA suppression of RalB induced apoptosis in the SW480 CRC cell line (19) as well as in KRAS mutant lung tumor cell lines (36). Our rationale for using sustained shRNA suppression was that we wanted to assess the consequences of prolonged antagonism of Ral, to more accurately model the situation that would be seen for therapeutic treatment of cancer. However, prolonged suppression may also allow time for compensatory mechanisms to arise to offset the acute consequences of RalB suppression. Since one compensatory mechanism may involve an alteration in the activity of the Ral isoform that is not targeted, we determined the expression and activation of one Ral isoform when the other isoform is suppressed by shRNA. Surprisingly, we found that shRNA suppression of RalA was associated with a 59- to 70-fold increase in RalB-GTP levels in the two KRAS mutant cell lines (Fig. 4D). Thus, the reduced soft agar growth caused by RalA suppression may be mediated by the concurrent loss of RalA function together with increased RalB activation. Conversely, suppression of RalB in KRAS mutant cell lines was associated with a modest 1.3- to 1.5-fold increased RalA-GTP that may contribute to the observed increased colony formation. For the BRAF mutant HT29 cells, a converse result was seen, where RalA suppression caused only a 2.0-fold increase in RalB-GTP formation, whereas RalB suppression caused a greater 9-fold increase in RalA-GTP formation.
RalA and RalB both utilize RalBP1, but distinct exocyst subunits, to regulate CRC anchorage-independent growth
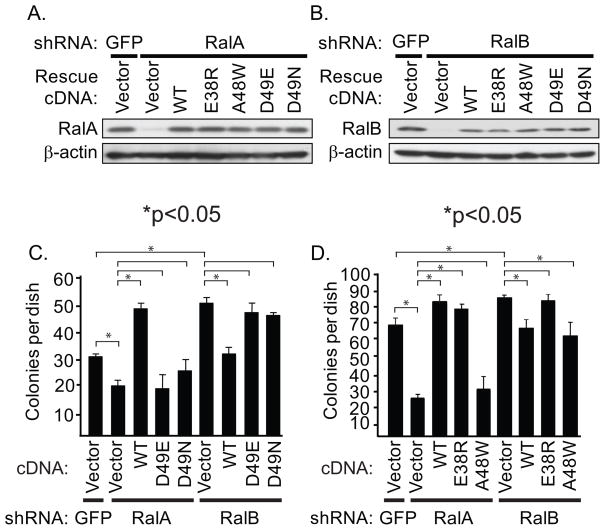
The opposing activities of RalA and RalB seen in CRC anchorage-independent growth suggests that these related isoforms may utilize different effectors in CRC cells. To address this possibility, we utilized effector domain mutants of Ral with differential impairment in effector binding. We first evaluated the activities of the D49E and D49N missense mutants, which are impaired in exocyst and RalBP1/RLIP76 effector binding, respectively (37–39). It is also possible that these mutants are defective in binding to unknown or recently described Ral effectors such as ZONAB. Using SW480 cells, we compared the ability of ectopic expression of WT or effector binding mutant RalA or RalB to rescue the growth effects caused by shRNA-mediated loss of the endogenous protein (Fig. 5A and 5B). The reduced soft agar growth caused by RalA shRNA was reversed and further enhanced by ectopic expression of WT RalA when expressed from an shRNA-insensitive cDNA expression vector (Fig. 5C). In contrast expression of either the D49E or D49N mutant of RalA did not restore colony formation activity, suggesting that both the exocyst and RalBP1 contribute to RalA promotion of CRC soft agar growth.
Figure 5.
RalA and RalB require interaction with RalBP1 but distinct exocyst subunits for regulation of anchorage-independent growth. A, RalA shRNA was used to suppress endogenous RalA expression in SW480 cells. The cells were then infected with pBabe retrovirus vectors encoding WT or effector binding mutant RalA proteins encoded by an shRNA-insensitive cDNA sequence. B, RalB shRNA was used to suppress endogenous RalB expression in SW480 cells. The cells were then infected with pBabe retrovirus vectors encoding WT or effector binding mutant RalB proteins encoded by an shRNA-insensitive cDNA sequence. Blot analyses were done with anti-RalA, anti-RalB and β-actin, to verify equivalent total protein loading. C, RalA and RalB require RalBP1 and the exocyst effector interactions to mediate their roles in CRC cell anchorage-independent growth. D, RalA requires Exo84 and RalB requires Sec5 effector interactions to regulate CRC cell anchorage-independent growth. Data shown are representative of two independent experiments.
We next utilized a second set of effector binding mutants, E38R and A48W (40), to assess which exocyst component was required for RalA activity. The E38R retains the ability to bind RalBP1 and Exo84, but not Sec5. The A48W mutant also retains the ability to bind RalBP1 and Sec5, but is impaired in Exo84 binding. RalA E38R but not A48W expression restored soft agar colony forming activity, indicating that Exo84 binding is important for RalA promotion of anchorage-independent growth.
Extending these analyses to RalB, we found that RalB shRNA enhancement of soft agar growth was reversed by ectopic expression of WT RalB expressed from an shRNA-resistant cDNA expression vector (Fig. 5D). However, neither ectopic expression of the D49E or D49N mutant of RalB was able to suppress soft agar colony formation activity, indicating that both effectors are required for RalB suppression. To further delineate the role of each exocyst component, we found that A48W but not E38R suppressed soft agar colony formation, indicating that RalB required Sec5 binding to suppress CRC anchorage-independent growth. Thus, RalA and RalB utilize different exocyst subunits to regulate their opposing actions on CRC anchorage-independent growth.
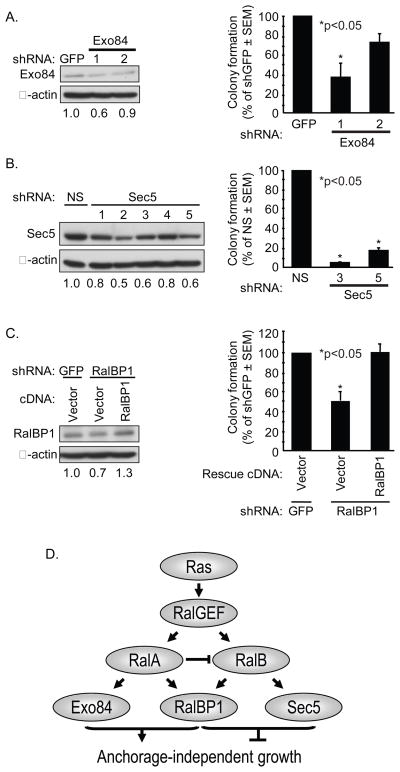
Finally, to directly evaluate a role for Ral effectors in CRC growth, we stably suppressed endogenous expression in SW480 cells (Fig. 6). As expected, since both Exo84 and RalBP1 binding were required for RalA support of anchorage-independent growth, suppression of Exo84 and RalBP1 reduced colony formation. However, surprisingly, since Sec5 binding was required for RalB suppression of anchorage-independent growth, Sec5 reduction reduced, rather than enhanced, soft agar growth. This may be a consequence of Ral-independent functions of Sec5.
Figure 6.
Ral effectors are required for SW480 CRC anchorage-independent growth. A, RalBP1 is required for the anchorage-independent growth of CRC cells. RalBP1 shRNA was used to stably suppress endogenous RalBP1 expression in SW480 cells and blot analyses were performed with anti-RalBP1 (left panel). Numbers indicate amount of RalBP1 expression compared to β-actin. Colony formation in soft agar was quantitated after 14 days (right panel). The cells were then infected with pBabe retrovirus vectors encoding shRNA-insensitive RalBP1 cDNA to verify shRNA specificity. B, Exo84 is required for SW480 anchorage-independent growth. SW480 cells were infected with nonspecific GFP shRNA or two independent Exo84 shRNA sequences to stably suppress endogenous Exo84 expression (left panel) and to evaluate colony formation in soft agar (right panel). C, Sec5 is required for SW480 anchorage-independent growth. SW480 cells were infected with nonspecific shRNA or five independent Sec5 shRNA sequences to stably suppress endogenous Sec5 expression (left panel) and to evaluate colony formation (right panel). Numbers indicate amount of Sec5 expression compared to β-actin. Data shown are representative of two independent experiments. E, Model for Ral in CRC anchorage-independent growth. Based on Ral effector binding mutant analyses only, since the shRNA suppression analyses of effector expression may be complicated by Ral-independent functions.
Discussion
Currently, the most vigorously pursued anti-Ras approaches are inhibitors of the Raf-MEK-ERK or PI3K-AKT effector signaling (6). However, these efforts are complicated by the likelihood that Ras-mediated oncogenesis involves these and other effector pathways. In this study, we extended our previous evaluation of MEK inhibitors (25) and concluded that KRAS mutation status but not pERK activity may be a marker to define selumitinib resistance in CRC. Although, pAKT activity was weakly associated with inhibitor insensitivity, PIK3CA mutation status was not. We also found Ral activation in CRC cell lines and tumors. However, in contrast to our observations in KRAS mutant PDAC, where RalA but not RalB promoted PDAC anchorage-independent and tumorigenic growth, we found that RalA and RalB exhibited opposing roles for CRC anchorage-independent growth. These results reveal the striking cell context functional differences that these GTPases may have in KRAS mutant cancers.
Our analyses with selumetinib reached the same conclusion as we did with other MEK1/2-selective inhibitors (25); pERK activation did not reliably predict MEK inhibitor sensitivity. However, we did find a different pattern of sensitivity to selumetinib when compared to U0126 and CI-1040. Whereas we found previously that a subset of KRAS mutant CRC cells did exhibit sensitivity to U0126 and CI-1040, we saw that all KRAS mutant CRC lines were resistant to treatment with selumetinib. Perhaps this different activity reflects the more specific nature of this MEK1/2 inhibitor and different off-target activities of the other inhibitors (e.g., MEK5-ERK5 inhibition). Also, one potential caveat to our analyses is that MEK inhibitory activity was determined on adherent cultures, whereas growth inhibitory activity was determined in nonadherent three-dimensional colonies. One recent study found that KRAS or BRAF mutation status did not correlate with selumetinib sensitivity, but did find that inhibitor resistance correlated with weak ERK and/or strong AKT activity (27). Consistent with their findings, we did find elevated pAKT in all KRAS mutant CRC cell lines and a weak association of elevated pAKT with selumitinib resistance. Although KRAS mutant cell lines showed partial sensitivity to PI3K inhibition, we found that concurrent PI3K inhibition did not further enhance MEK inhibitor sensitivity. Our results are consistent with another recent study where selumetinib response did not correlate with RAS mutation or PI3K activation (41). Our results support the need to assess the importance of other effectors in RAS mutant cancers.
We previously observed a striking requirement for RalA but not RalB for the anchorage-independent and tumorigenic growth of PDAC cell lines (18). In the present study, we found that RalA was also necessary for CRC anchorage-independent growth for both KRAS and BRAF mutant cell lines. Surprisingly, stable suppression of RalB caused a significant enhancement of soft agar colony size and colony forming efficiency. These results extend previous findings of striking functional differences with the related RalA and RalB isoforms (19, 21, 35), and additionally reveal a significant RalB functional difference in KRAS mutant tumor cells that arise from different tissues. While we do not have a mechanistic explanation for this cell context difference, it may reflect differences in RalB subcellular localization or posttranslational modifications, leading to different activation of effectors, in each tumor type.
The different functional roles of RalA and RalB in the growth of different tumor types complicate the issue of whether isoform-selective or pan-Ral therapeutic approaches will be the most effective. For five of six KRAS mutant CRC cell lines, we found that concurrent suppression of both RalA and RalB resulted in statistically insignificant reduction in colony formation when compared to the control shGFP cells. These results contrast with previous studies in different cancer types where the phenotype of RalA is dominant over that of RalB (19, 21, 35). These observations argue that a RalA-selective therapeutic approach may be the best approach for inhibiting the growth of CRC and PDAC cells. However, we also found that RalB was necessary for PDAC Matrigel invasion and lung colonization metastasis (18). Whether RalB loss will promote CRC invasion and metastasis will need to be established to better understand the consequences of RalA and RalB ablation for tumor growth in the CRC patient.
Our results with sustained RalB suppression differ from previous studies where transient RalB suppression caused CRC apoptotic cell death (19, 22, 36). When we evaluated transient RalB inactivation, we also observed cell death (data not shown). We suspect that with sustained suppression of RalB, compensatory events occur to offset the initial deleterious consequences of RalB loss. Consistent with this possibility, we observed a modest 1.3- to 1.5-fold increase in the steady-state level of RalA-GTP was increased by RalB suppression in KRAS mutant CRC lines that may contribute to the enhancement of growth. However, we suspect that additional more significant compensatory events must also contribute. In contrast, we observed a 59- to 70-fold increase in RalB-GTP levels by RalA suppression in KRAS mutant cells. Our observation that steady-state expression of constitutively activated RalB impaired CRC growth (data not shown) argues that this increase contributes to RalA suppression-associated growth inhibition. Since it is likely that targeted therapies focused on signal transduction molecules will require chronic therapy to maintain persistent suppression of target activity, we believe that our observations with sustained Ral suppression are relevant and important for understanding the potential consequences of Ral targeted therapies for CRC treatment.
In light of our observed opposing functions of sustained RalA and RalB depletion in CRC anchorage-independent growth, we were surprised to find that both RalA and RalB activities were dependent on RalBP1 binding. Since RalA and RalB exhibit different subcellular localizations, perhaps each GTPase engages RalBP1 in spatially-distinct locations, leading to distinct cellular outcomes. Interestingly, suppression of RalBP1 also reduced soft agar growth, indicating that its role in RalA function is dominant over its role in RalB function. In any case, our implication of RalBP1 in Ral-dependent oncogenesis contrasts with other studies where RalBP1 has not been involved. Furthermore, while both RalA and RalB required association with exocyst components to regulate CRC growth, RalA required association with Exo84 but not Sec5 whereas RalB required Sec5 but not Exo84 binding. One possible explanation for this result is that the differential requirements for Sec5 and Exo84 are unrelated to exocyst function. Certainly for Sec5, one exocyst independent function involves the TBK1 protein kinase (22). Similarly, it was suggested that Exo84 also exhibits an exocyst-independent function required for growth transformation (42). That suppression of Exo84 or Sec5 expression both reduced soft agar growth may reflect both Ral-dependent and –independent functions.
In summary, our results, while supporting the value of targeting Ral GTPases for KRAS mutant CRC, also indicate that Ral targeted therapies may need to be tailored differently for different cancers. For example, since we found that RalB was important for PDAC invasion and metastasis, a RalB-selective therapy may be ideally suited for advanced PDAC. In contrast, a RalB-selective therapy may enhance CRC tumor growth. Future studies with genetic ablation of RalA or RalB in KRAS-driven mouse models of PDAC and CRC will provide a more comprehensive understanding of the most effective approach for Ral inhibition for cancer treatment.
Supplementary Material
Acknowledgments
We thank Paul Smith (AstraZeneca) for providing selumetinib, Christopher Counter (Duke) for shRNA vectors for RalA and RalB and for shRNA-resistant cDNA sequences, the UNC Tissue Procurement Facility for providing human samples. We thank Lanika DeGraffenreid and Erin Hill for assistance in manuscript preparation.
Our research was supported by grants from the NIH to CJD (CA042978 and CA106991) and JJY (CA106991) and from the Emerald Foundation to JJY.
Footnotes
Disclosure of Potential Conflicts of Interest: none
References
- 1.Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–13. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 2.Sjoblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 3.Shirasawa S, Furuse M, Yokoyama N, Sasazuki T. Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science. 1993;260:85–8. doi: 10.1126/science.8465203. [DOI] [PubMed] [Google Scholar]
- 4.Scholl C, Frohling S, Dunn IF, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137:821–34. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 5.Luo J, Emanuele MJ, Li D, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–48. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeh JJ, Madigan JP, Campbell PM, DeGraffenreid L, Der CJ. The Handbook of Cell Signaling. 2. Academic Press; 2009. Targeting Ras for anti-cancer drug discovery; pp. 2837–57. [Google Scholar]
- 7.Repasky GA, Chenette EJ, Der CJ. Renewing the conspiracy theory debate: does Raf function alone to mediate Ras oncogenesis? Trends Cell Biol. 2004;14:639–47. doi: 10.1016/j.tcb.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 8.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 9.Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 10.Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez-Garcia A, Pritchard CA, Paterson HF, Mavria G, Stamp G, Marshall CJ. RalGDS is required for tumor formation in a model of skin carcinogenesis. Cancer Cell. 2005;7:219–26. doi: 10.1016/j.ccr.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 12.Malliri A, van der Kammen RA, Clark K, van der Valk M, Michiels F, Collard JG. Mice deficient in the Rac activator Tiam1 are resistant to Ras-induced skin tumours. Nature. 2002;417:867–71. doi: 10.1038/nature00848. [DOI] [PubMed] [Google Scholar]
- 13.Bai Y, Edamatsu H, Maeda S, et al. Crucial role of phospholipase Cepsilon in chemical carcinogen-induced skin tumor development. Cancer Res. 2004;64:8808–10. doi: 10.1158/0008-5472.CAN-04-3143. [DOI] [PubMed] [Google Scholar]
- 14.Malliri A, Rygiel TP, van der Kammen RA, et al. The rac activator Tiam1 is a Wnt-responsive gene that modifies intestinal tumor development. J Biol Chem. 2006;281:543–8. doi: 10.1074/jbc.M507582200. [DOI] [PubMed] [Google Scholar]
- 15.Li M, Edamatsu H, Kitazawa R, Kitazawa S, Kataoka T. Phospholipase Cepsilon promotes intestinal tumorigenesis of Apc(Min/+) mice through augmentation of inflammation and angiogenesis. Carcinogenesis. 2009;30:1424–32. doi: 10.1093/carcin/bgp125. [DOI] [PubMed] [Google Scholar]
- 16.Bodemann BO, White MA. Ral GTPases and cancer: linchpin support of the tumorigenic platform. Nat Rev Cancer. 2008;8:133–40. doi: 10.1038/nrc2296. [DOI] [PubMed] [Google Scholar]
- 17.Lim KH, Baines AT, Fiordalisi JJ, et al. Activation of RalA is critical for Ras-induced tumorigenesis of human cells. Cancer Cell. 2005;7:533–45. doi: 10.1016/j.ccr.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 18.Lim KH, O’Hayer K, Adam SJ, et al. Divergent roles for RalA and RalB in malignant growth of human pancreatic carcinoma cells. Curr Biol. 2006;16:2385–94. doi: 10.1016/j.cub.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 19.Chien Y, White MA. RAL GTPases are linchpin modulators of human tumour-cell proliferation and survival. EMBO Rep. 2003;4:800–6. doi: 10.1038/sj.embor.embor899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin J, Pollock C, Tracy K, et al. Activation of the RalGEF/Ral pathway promotes prostate cancer metastasis to bone. Mol Cell Biol. 2007;27:7538–50. doi: 10.1128/MCB.00955-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oxford G, Owens CR, Titus BJ, et al. RalA and RalB: antagonistic relatives in cancer cell migration. Cancer Res. 2005;65:7111–20. doi: 10.1158/0008-5472.CAN-04-1957. [DOI] [PubMed] [Google Scholar]
- 22.Chien Y, Kim S, Bumeister R, et al. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell. 2006;127:157–70. doi: 10.1016/j.cell.2006.08.034. [DOI] [PubMed] [Google Scholar]
- 23.Feig LA. Ral-GTPases: approaching their 15 minutes of fame. Trends Cell Biol. 2003;13:419–25. doi: 10.1016/s0962-8924(03)00152-1. [DOI] [PubMed] [Google Scholar]
- 24.Vatsyayan R, Lelsani PC, Awasthi S, Singhal SS. RLIP76: A versatile transporter and an emerging target for cancer therapy. Biochem Pharmacol. doi: 10.1016/j.bcp.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeh JJ, Routh ED, Rubinas T, et al. KRAS/BRAF mutation status and ERK1/2 activation as biomarkers for MEK1/2 inhibitor therapy in colorectal cancer. Mol Cancer Ther. 2009;8:834–43. doi: 10.1158/1535-7163.MCT-08-0972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeh TC, Marsh V, Bernat BA, et al. Biological characterization of ARRY-142886 (AZD6244), a potent, highly selective mitogen-activated protein kinase kinase 1/2 inhibitor. Clin Cancer Res. 2007;13:1576–83. doi: 10.1158/1078-0432.CCR-06-1150. [DOI] [PubMed] [Google Scholar]
- 27.Balmanno K, Chell SD, Gillings AS, Hayat S, Cook SJ. Intrinsic resistance to the MEK1/2 inhibitor AZD6244 (ARRY-142886) is associated with weak ERK1/2 signalling and/or strong PI3K signalling in colorectal cancer cell lines. Int J Cancer. 2009;125:2332–41. doi: 10.1002/ijc.24604. [DOI] [PubMed] [Google Scholar]
- 28.Solit DB, Garraway LA, Pratilas CA, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–62. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leboeuf R, Baumgartner JE, Benezra M, et al. BRAFV600E mutation is associated with preferential sensitivity to mitogen-activated protein kinase kinase inhibition in thyroid cancer cell lines. J Clin Endocrinol Metab. 2008;93:2194–201. doi: 10.1210/jc.2007-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pratilas CA, Hanrahan AJ, Halilovic E, et al. Genetic predictors of MEK dependence in non-small cell lung cancer. Cancer Res. 2008;68:9375–83. doi: 10.1158/0008-5472.CAN-08-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smalley KS, Haass NK, Brafford PA, Lioni M, Flaherty KT, Herlyn M. Multiple signaling pathways must be targeted to overcome drug resistance in cell lines derived from melanoma metastases. Mol Cancer Ther. 2006;5:1136–44. doi: 10.1158/1535-7163.MCT-06-0084. [DOI] [PubMed] [Google Scholar]
- 32.Haass NK, Sproesser K, Nguyen TK, et al. The mitogen-activated protein/extracellular signal-regulated kinase kinase inhibitor AZD6244 (ARRY-142886) induces growth arrest in melanoma cells and tumor regression when combined with docetaxel. Clin Cancer Res. 2008;14:230–9. doi: 10.1158/1078-0432.CCR-07-1440. [DOI] [PubMed] [Google Scholar]
- 33.Shipitsin M, Feig LA. RalA but not RalB enhances polarized delivery of membrane proteins to the basolateral surface of epithelial cells. Mol Cell Biol. 2004;24:5746–56. doi: 10.1128/MCB.24.13.5746-5756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosse C, Hatzoglou A, Parrini MC, White MA, Chavrier P, Camonis J. RalB mobilizes the exocyst to drive cell migration. Mol Cell Biol. 2006;26:727–34. doi: 10.1128/MCB.26.2.727-734.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamad NM, Elconin JH, Karnoub AE, et al. Distinct requirements for Ras oncogenesis in human versus mouse cells. Genes Dev. 2002;16:2045–57. doi: 10.1101/gad.993902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barbie DA, Tamayo P, Boehm JS, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–12. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cantor SB, Urano T, Feig LA. Identification and characterization of Ral-binding protein 1, a potential downstream target of Ral GTPases. Mol Cell Biol. 1995;15:4578–84. doi: 10.1128/mcb.15.8.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moskalenko S, Henry DO, Rosse C, Mirey G, Camonis JH, White MA. The exocyst is a Ral effector complex. Nat Cell Biol. 2002;4:66–72. doi: 10.1038/ncb728. [DOI] [PubMed] [Google Scholar]
- 39.Moskalenko S, Tong C, Rosse C, et al. Ral GTPases regulate exocyst assembly through dual subunit interactions. J Biol Chem. 2003;278:51743–8. doi: 10.1074/jbc.M308702200. [DOI] [PubMed] [Google Scholar]
- 40.Cascone I, Selimoglu R, Ozdemir C, et al. Distinct roles of RalA and RalB in the progression of cytokinesis are supported by distinct RalGEFs. EMBO J. 2008;27:2375–87. doi: 10.1038/emboj.2008.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dry JR, Pavey S, Pratilas CA, et al. Transcriptional Pathway Signatures Predict MEK Addiction and Response to Selumetinib (AZD6244) Cancer Res. doi: 10.1158/0008-5472.CAN-09-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Issaq SH, Lim KH, Counter CM. Sec5 and Exo84 foster oncogenic ras-mediated tumorigenesis. Mol Cancer Res. 8:223–31. doi: 10.1158/1541-7786.MCR-09-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.