SUMMARY
The chemical structures of cobamides (cobalamin [Cbl]-like compounds) are the same, except for the lower ligand, which in adenosylcobalamin (AdoCbl) is 5,6-dimethylbenzimidazole (DMB), and in adenosylpseudocobalamin (AdopseudoCbl) is adenine. Why the lower ligand of cobamides varies and what the mechanism of lower ligand replacement is are long-standing questions in the field of B12 biosynthesis. Work reported here uncovers the strategy used by the photosynthetic α-proteobacterium Rhodobacter sphaeroides to procure the cobamide it needs to grow on acetate as a carbon and energy source. On the basis of genetic and biochemical evidence we conclude that, in R. sphaeroides, the activity of the cobyric acid-producing amidohydrolase CbiZ enzyme is essential for the conversion of AdopseudoCbl into AdoCbl, the cobamide needed for the catabolism of acetate. The CbiZ enzyme uses AdopseudoCbl as a substrate, but not AdoCbl. Implications of these findings for cobamide remodeling in R. sphaeroides and in other CbiZ-containing microorganisms are discussed.
INTRODUCTION
Cobamides are complex cobalt-containing tetrapyrrole cofactors whose synthesis by a variety of bacteria and archaea requires substantial genetic information (Escalante-Semerena & Warren, 2008). Cobamides contain upper (β) and lower (α) axial ligands that interact with the cobalt ion of the corrin ring. The upper ligand of cobamides varies, but the one found in the coenzymic form of the cofactor is 5’-deoxyadenosine (Ado), the attachment of which is catalyzed by corrinoid adenosyltransferases found in all domains of life (Dobson et al., 2002, Buan et al., 2004, Buan et al., 2006, Johnson et al., 2004, Sanishvili et al., 2004). In AdoCbl, the lower ligand is 5,6-dimethylbenzimidazole (DMB) (Fig. 1A). Pseudocobalamin (pseudoCbl) is a commonly occurring cobamide whose lower ligand is N7-linked adenine (Fig. 1B) (Friedmann & Fyfe, 1969). Natural microbial communities convert Cbl into other cobamides (Girard et al., 2009, Allen & Stabler, 2008), but the mechanism by which lower ligand bases are exchanged is not known.
Figure 1. Structure of corrinoids used in this study.
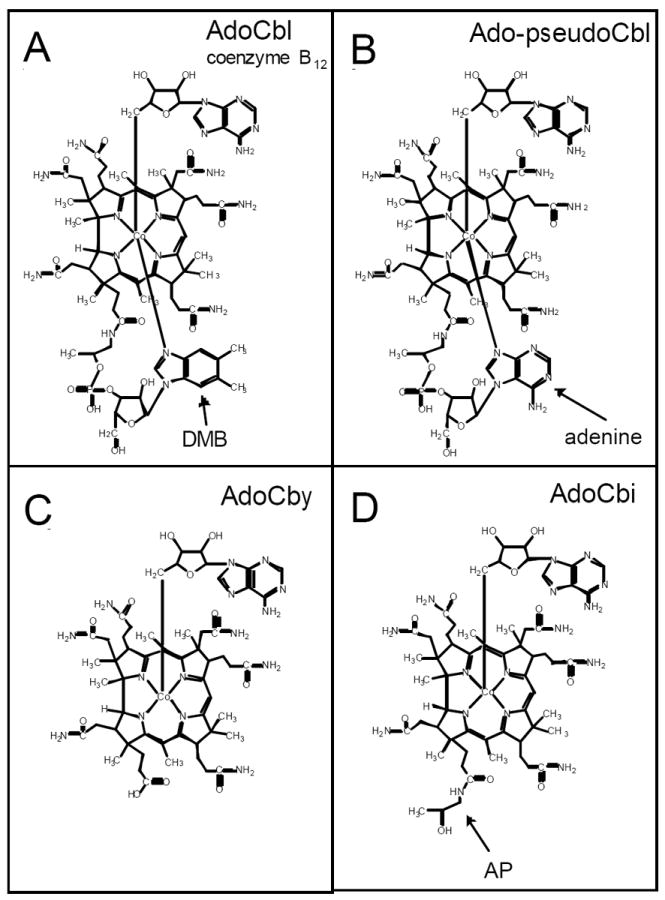
A. Adenosylcobalamin (AdoCbl). The 5,6-dimethylbenzimidazole (DMB) lower ligand is indicated. B. Adenosyl-pseudocobalamin (Ado-pseudoCbl). The N7-linked adenine lower ligand is indicated. C. Adenosylcobinamide (AdoCbi). The 1-amino-2-propanol (AP) moiety is indicated. D. Adenosylcobyric acid (AdoCby).
Many microorganisms salvage cobamide precursors that lack the lower ligand, such as cobinamide (Cbi) (Fig. 1C), from their environment. These precursors are used to synthesize complete cobamides. In bacteria, Cbi salvaging depends on the activity of a bifunctional adenosyl-Cbi (AdoCbi) kinase (EC 2.7.7.62) / AdoCbi:GTP guanylyltransferase (EC 2.7.1.156) (called CobP in Pseudomonas denitrificans or CobU in Salmonella enterica) (O’Toole & Escalante-Semerena, 1995, Blanche et al., 1991). To date, CobP/CobU homologs have not been reported in any archaeon. Archaea use a different strategy for Cbi salvaging, which depends on an AdoCbi amidohydrolase (called CbiZ), an enzyme that converts AdoCbi into 1-amino-2-propanol and adenosyl-cobyric acid (AdoCby) (Woodson et al., 2003, Woodson & Escalante-Semerena, 2004). Archaea and bacteria convert AdoCby in to AdoCbi-P via the activity of the AdoCbi-P synthetase (called CbiB in S. enterica and archaea, CobD in P. denitrificans) (Woodson et al., 2003, Zayas et al., 2007)
We recently identified a subset of bacteria whose genomes encode copies of both CobP and CbiZ (Gray et al., 2008). We have also demonstrated that, in the photosynthetic α-proteobacterium Rhodobacter sphaeroides, CobP and CbiZ are needed to salvage Cbi (Gray et al., 2008, Gray & Escalante-Semerena, 2009). These results raised the question of what the physiological role of CbiZ might be, particularly in organisms that synthesize two apparently redundant Cbi salvaging systems.
Here we report genetic and biochemical evidence that shows that growth of R. sphaeroides on acetate depends on AdoCbl (containing DMB as the lower ligand), and that the physiological role of CbiZ in R. sphaeroides is to remove the lower ligand of AdopseudoCbl so that the cell can replace the lower ligand with DMB, generating AdoCbl. We propose that CbiZ is a key enzyme used by microorganisms to remodel cobamides containing lower ligand bases that block the use of the cobamide as a coenzyme.
RESULTS
The combined results from in vivo and in vitro experiments described below support three conclusions: i) R. sphaeroides requires Cbl to grow on acetate; ii) CbiZ is required for the conversion of pseudoCbl to Cbl in this organism; and iii) CbiZ is an amidohydrolase with specificity for AdopseudoCbl.
CbiZ is required for the growth of R. sphaeroides on acetate with pseudoCbl
Results from control experiments for these studies are shown in column A. A cobB strain, which cannot synthesize the corrin ring, did not grow aerobically on acetate in the absence of exogenous corrinoids, but grew well when provided with dicyanocobinamide ([CN]2Cbi), cyanopseudocobalamin (CNpseudoCbl), or cyanocobalamin (CNCbl), regardless of whether or not DMB was present in the medium.
The ΔcobB ΔcbiZ strain carrying an empty cloning vector grew well on acetate in the presence of (CN)2Cbi or CNCbl (Fig. 2, column B, rows 2 and 6, black squares), but surprisingly, grew poorly in the presence of CNpseudoCbl (Fig. 2, column B, row 4, black squares). The presence of DMB in the medium did not affect growth of the ΔcobB ΔcbiZ strain (Fig. 2, column B, rows 3 and 5), but when a plasmid encoding the wild-type cbiZ gene was introduced into the ΔcobB ΔcbiZ strain, cells grew in response to CNpseudoCbl (Fig. 2, column B, rows 4 and 5, open triangles), indicating that the growth defect of the ΔcobB ΔcbiZ strain was due to the absence of cbiZ function.
Figure 2. Growth of R. sphaeroides on acetate is Cbl-dependent.

Corrinoid-dependent aerobic growth of R. sphaeroides strain JE10754 (ΔcobB) (column A), JE10755 (ΔcobB ΔcbiZ) (column B), JE11661 (ΔcobB bluB∷aadA) (column C), and JE11662 (ΔcobB ΔcbiZ bluB∷aadA) (column D) derivatives in mSistrom’s medium containing acetate (30 mM) and tetracycline (1 μg ml-1). OD650 was measured for 48 h at 30°C. Corrinoids were added at 15 nM. Plasmids used were: vector-only, pRK404 (black squares); pcbiZ+, pRsCBIZ6 (open triangles); pbluB+, pBLUB19 (open circles). Growth curves were obtained in triplicate using a PowerWave XS microplate reader (Bio-Tek Instruments), and error bars of one standard deviation are indicated.
DMB is required for the utilization of Cbi or pseudoCbl by R. sphaeroides
The above results suggest that growth of R. sphaeroides on acetate requires Cbl, and that this bacterium has a CbiZ-dependent mechanism for the conversion of pseudoCbl into Cbl. The lower ligand is the only difference between these molecules; adenine in pseudoCbl, DMB in Cbl (Fig. 1). To confirm that the presence of adenine as the lower ligand caused the growth defect of the ΔcobB ΔcbiZ strain, we constructed a ΔcobB bluB∷aadA strain, in which bluB, the gene encoding aerobic DMB synthase (Taga et al., 2007, Gray & Escalante-Semerena, 2007), was replaced with a spectinomycin-resistance cassette. Growth of the ΔcobB bluB∷aadA strain on acetate was poor in medium supplemented with (CN)2Cbi or CNpseudoCbl (Fig. 2, column C, rows 2 and 4, black squares). In contrast, the ΔcobB bluB∷aadA strain grew well on acetate when CNCbl was present in the medium (Fig. 2, column C, row 6). Inclusion of DMB in the medium (Fig. 2, column C, rows 3 and 5, black squares) or introduction of a plasmid encoding the wild-type allele of bluB restored growth of the ΔcobB bluB∷aadA strain (Fig. 2, column C, rows 2 and 4, open circles) on (CN)2Cbi and CNpseudoCbl. Together, the above results support the conclusion that growth of R. sphaeroides on acetate requires Cbl, and that pseudoCbl cannot substitute for Cbl.
CbiZ is required for the conversion of pseudoCbl to Cbl
We hypothesized that the phenotypes of the ΔcobB ΔcbiZ and ΔcobB bluB∷aadA strains would be explained if CbiZ were required for the removal of the lower ligand of pseudoCbl, so that the corrin ring of pseudoCbl could be recycled into Cbl by iuncorporation of DMB as the lower ligand. If this were the case, we predicted that the addition of pseudoCbl to a R. sphaeroides cbiZ+ strain would result in the accumulation of Cbl, and that the deletion of cbiZ would prevent this conversion.
To test this hypothesis, we used a combination of HPLC and a quantitative bioassay procedure (Gray & Escalante-Semerena, 2009) to determine the identity and quantity of cobamides accumulated by ΔcobB and ΔcobB ΔcbiZ strains (Table 1, Supplemental Figure S1). Stationary phase cultures of R. sphaeroides strains were grown aerobically with succinate as a sole carbon and energy source in the presence of exogenous corrinoids (50 nM). Cells were harvested by centrifugation and corrinoids were extracted, converted to their cyano forms, and resolved by reverse-phase (RP)-HPLC. The presence of substantial amounts of pigments in R. sphaeroides extracts masked the corrinoid peaks, thus fractions containing cobamides were identified using the abovementioned bioassay. The identification was based on the growth response of Salmonella enterica sv. Typhimurium strain JE8248 (ΔcobS), which lacks Cbl-5’-P synthase activity, and is therefore a cobamide auxotroph (O’Toole et al., 1993). At concentrations between 50 and 250 pM cobamide, the growth rate of strain JE8248 was directly proportional to the concentration of cobamide in the medium (Gray & Escalante-Semerena, 2009), and we used the growth rates of strain JE8248 in medium supplemented with fractionated R. sphaeroides extracts to calculate the amount of cobamide present in each fraction. The results were normalized to colony forming units (CFU) of R. sphaeroides to estimate the amount of cobamide accumulated per cell under the growth conditions tested. HPLC traces and a graphical representation of the bioassay data are shown in Figure S1.
Table 1.
Effect of cbiZ and bluB mutations on cobamide accumulation. a
| Genotype | Corrinoid added b | Percent total cobamide (in 2-min fractions) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 20 | 22 | 24 | 26 | 28 | 30 | 32 | 34 | 36 | 38 | ||
| ΔcobB | (CN)2Cbi | 1 | 2 | 14 | 60 | 23 | |||||
| ΔcobB | CNpseudoCbl | 2 | 25 | 63 | 10 | ||||||
| ΔcobB ΔcbiZ | CNpseudoCbl | 100 | |||||||||
| ΔcobB | (CN)2Cbi | 84 | 16 | ||||||||
| bluB∷aadA | |||||||||||
| Elution profiles of known cobamide standards c | |||||||||||
| Cobamide | 20 | 22 | 24 | 26 | 28 | 30 | 32 | 34 | 36 | 38 | |
| CNCbl | 3 | 30 | 61 | 5 | 1 | ||||||
| CNpseudoCbl | 95 | 5 | |||||||||
Cobamides accumulated by R. sphaeroides strains grown aerobically in mSistrom’s medium supplemented with succinate (30 mM) and the indicated corrinoids were separated by RP-HPLC; 2 min fractions were collected starting at the indicated times. The cobamide content in each fraction was determined by bioassay and normalized to the percentage of the total cobamide detected in the sample.
Corrinoids were added at 50 nM.
Authentic CNCbl and CNpseudoCbl standards were separated by RP-HPLC. The cobamide content in fractions was determined by bioassay and normalized to the percentage of the total corrinoid injected in the known sample.
The results of these experiments are summarized in Table 1. In the presence of (CN)2Cbi, the ΔcobB strain accumulated approximately 670 molecules of cobamide per CFU, with 60% of the detected cobamides eluting in the fraction starting 30 min after injection. This elution profile was consistent with the one obtained with authentic CNCbl (61% of detected cobamides in the fraction starting 30 min after injection). Under these HPLC conditions, CNCbl consistently elutes as a broad peak between 26 and 34 minutes. At present, we do not know why the Cbl peak is so broad. In the presence of CNpseudoCbl, the ΔcobB strain also accumulated cobamides (approximately 420 molecules per CFU) with 63% of the detected cobamides eluting in the fraction starting 30 min after injection, consistent with the conversion of pseudoCbl into Cbl by this strain. Notably, the ΔcobB ΔcbiZ strain did not convert pseudoCbl into Cbl. Instead, it accumulated approximately 200 molecules per CFU of a cobamide only detectable in the fraction starting 24 min after injection, a retention time that was consistent with the elution profile of authentic CNpseudoCbl (95% in the fraction starting 24 min after injection). This result indicated that cbiZ function was required to convert pseudoCbl into Cbl, and the fact that pseudoCbl accumulated in the ΔcobB ΔcbiZ strain showed that the growth defect of this strain on acetate (Fig. 2, column B, row 4, black squares) was not caused by a defect in pseudoCbl transport.
A bluB strain of R. sphaeroides cannot synthesize AdoCbl
We also used the quantitative bioassay method to investigate whether or not the ΔcobB bluB∷aadA strain synthesized Cbl. For this purpose we grew the ΔcobB bluB∷aadA strain aerobically on succinate in the presence of (CN)2Cbi. Under these conditions the ΔcobB bluB∷aadA strain accumulated approximately 30 molecules of cobamide per CFU, 84% of which eluted 24 min after injection (Table 1, Fig. S1), the retention time of authentic CNpseudoCbl. We did not detect any cobamide with retention times similar to that for authentic Cbl (61% in the fraction starting 30 min after injection). This result confirmed that bluB was required for Cbl biosynthesis in R. sphaeroides.
Purified CbiZ cleaves AdopseudoCbl, but not AdoCbl, to AdoCby in vitro
The CbiZ protein was isolated as a fusion protein with N-terminal maltose-binding protein (MBP) and His6 tags (hereafter referred to as MBP-CbiZ) (Rocco et al., 2008). Attempts to remove the tags with TEV protease resulted in insoluble, inactive protein, so the CbiZ activity data were obtained using MBP-CbiZ protein. Purified MBP-CbiZ protein (Fig. S2) was incubated with corrinoid substrates and the formation of Cby was monitored by HPLC.
Optimization of MBP-CbiZ function was performed using AdoCbi as substrate. Product was not formed (< 0.1 nmol Cby min-1 mg-1 protein) in the absence of DTT (1 mM) (data not shown); at 30°C, the highest specific activity was observed at pH 10 (Fig. 3A). We also determined MBP-CbiZ specific activities at pH 10 across a range of temperatures, observing the highest activity at 50°C (Fig. 3B). At pH 10 and 50°C, product formation was linear over time (up to 15 min) with 1 or 2 μg of MBP-CbiZ in the reaction mixture (Fig. 3C, triangles, squares). When 3 μg of MBP-CbiZ were used, the rate of Cby formation decreased after 10 min (Fig. 3C, diamonds). Subsequent determinations of MBP-CbiZ specific activity were performed with 2 μg of MBP-CbiZ, at pH 10, 50°C, for 10 min.
Figure 3. Purified MBP-CbiZ cleaves Cbi and pseudoCbl, but not Cbl.

Amidohydrolase activity of MBP-CbiZ (expressed as nanomoles of Cby min-1 mg-1 protein) is shown. Error bars of one standard deviation are indicated. Unless indicated, reactions contained CHES buffer (50 mM, pH 10), DTT (1 mM), MBP-CbiZ (2 μg), and AdoCbi (30 μM) and were incubated for 15 min at 50°C.
A. Activity of MBP-CbiZ as a function of pH at 30°C. B. Activity of MBP-CbiZ as a function of temperature at pH 10. C. Activity of MBP-CbiZ as a function of time and enzyme concentration. D. Activity of MBP-CbiZ with different corrinoid substrates (30 μM). Reactions were incubated for 10 min.
The specificity of MBP-CbiZ for its substrate was also investigated (Fig. 3D). AdoCbi was hydrolyzed to Cby at a rate of 20 ± 2 nmol min-1 mg-1 protein. AdopseudoCbl was hydrolyzed to Cby at a rate of 70 ± 4 nmol min-1 mg-1 protein. We did not observe any cleavage of AdoCbl into Cby. Significantly lower rates of product formation were observed with cyanated substrates: 2.4 ± 0.6 nmol Cby min-1 mg-1 protein for (CN)2Cbi and 4 ± 0.6 nmol Cby min-1 mg-1 protein for CNpseudoCbl. Hydrolysis of CNCbl into Cby was not observed. The high specific activity of MBP-CbiZ for AdopseudoCbl, combined with its inability to hydrolyze AdoCbl, supports the hypothesis that the physiological role of MBP-CbiZ in R. sphaeroides is to remove the lower ligand of alternate cobamides so that they can be converted to AdoCbl, the preferred cobamide in this organism.
DISCUSSION
CbiZ is a critical component of the cobamide remodeling system of R. sphaeroides
CbiZ homologs are found in a variety of distantly related bacteria whose genomes also encode AdoCbi kinases (Gray et al., 2008). Since the latter have been implicated in Cbi salvaging (for a review see (Escalante-Semerena, 2007)), it is unlikely that organisms that synthesize both CbiZ and AdoCbi kinases do so to ensure that Cbi is salvaged. Rather, we propose that, in cells that synthesize both CbiZ and AdoCbi kinases, CbiZ is a critical component of the mechanism that ensures that the correct lower ligand is incorporated into the end product of the pathway. In contrast, in organisms where AdoCbi kinases are absent (e.g., Methanocaldococcus jannaschii, Methanopyrus kandleri, Halobacterium, etc), we propose that the CbiZ enzyme is dedicated to salvaging Cbi from the environment (Woodson et al., 2003, Woodson & Escalante-Semerena, 2004).
Different organisms synthesize cobamides with different lower ligands (Renz, 1999), and we predict that CbiZ enzymes from prokaryotes that use cobamides other than Cbl may have evolved to acquire the substrate specificities needed to synthesize the correct cobamide. In support of this idea, we note that the specific activity of the CbiZ homolog from Methanopyrus kandleri was high when Cbl was the substrate (Woodson & Escalante-Semerena, 2006), while the CbiZ enzyme from R. sphaeroides reported in this work did not use Cbl as substrate at all. The lower ligand of the cobamide produced by M. kandleri has not been reported, but methanogens commonly synthesize 5-hydroxybenzimidazolyl-cobamide or pseudoCbl (Stupperich & Kräutler, 1988). Based on the results reported here and elsewhere, we predict that the CbiZ enzyme of M. kandleri (known as CbiS because it is fused to another cobamide biosynthetic enzyme) will cleave Cbl but not pseudoCbl, precisely the opposite of the specificity of the R. sphaeroides CbiZ enzyme.
How is the substrate specificity of CbiZ enzymes determined?
CbiZ does not contain any known corrinoid-binding motifs, and therefore probably represents a novel corrinoid-binding protein fold. Efforts to obtain a crystal structure of CbiZ are underway.
How does CbiZ interact with the corrinoid transport system?
In many bacteria, including R. sphaeroides, cbiZ homologs are found in operons containing homologs of the genes encoding the corrinoid-specific ABC transport system (Btu) (Gray et al., 2008). This observation suggests a specific role for CbiZ during the uptake of corrinoids from the environment. Maintenance of cbiZ in this organism suggests that corrinoid salvaging is physiologically important to this bacterium. R. sphaeroides is commonly found in freshwater environments, and although very little is known about the nature of cobamides found in this environment, other bacteria found in R. sphaeroides’ environment (e.g., cyanobacteria) synthesize substantial amounts of pseudoCbl (Watanabe et al., 1999, Watanabe et al., 2006, Miyamoto et al., 2006, Watanabe et al., 2007), suggesting that cyanobacteria may be a source of pseudoCbl that R. sphaeroides can salvage using CbiZ.
The Cby-forming amidohydrolase activity of CbiZ is sensitive to the identity of both the upper and lower ligands (Fig. 4D) (Woodson & Escalante-Semerena, 2006, Woodson & Escalante-Semerena, 2004). Activity of the bacterial Cbi-salvaging enzyme CobP requires an adenosylated substrate (O’Toole & Escalante-Semerena, 1995, Blanche et al., 1991). These observations are consistent with data in the literature, which shows that adenosylation of exogenous corrinoids occurs before entering the salvaging process (Escalante-Semerena et al., 1990).
Figure 4. The acetate catabolism pathway of R. sphaeroides.
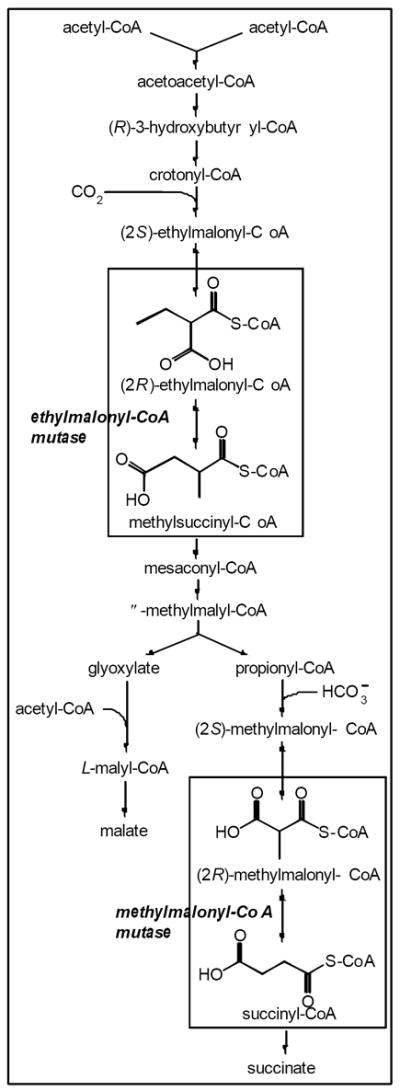
Modified from Erb et al (Erb et al., 2008).
Why does the lower ligand of cobamides vary?
A long-standing question in the field of B12 biosynthesis is why the lower ligand of cobamides varies. Naturally occurring cobamides containing lower ligands derived from benzimidazole, purine bases, and phenolic compounds have been described from a variety of different environments (Renz, 1999, Allen & Stabler, 2008), but the rationale for this variability is unclear. E. coli appears to be able to use cobamides with diverse lower ligands in vivo (Perlman & Barrett, 1958, Burkholder, 1951), and methylmalonyl-CoA mutase from Propionibacterium shermanii is only slightly more active in vitro with Cbl than with pseudoCbl or benzimidazolyl-cobamide (Lengyel et al., 1960). However, a number of enzymes have been described that differ substantially in their activity with different cobamides. These include the human methylmalonyl-CoA mutase (at least 1000-fold more active with Cbl than with pseudoCbl (Lengyel et al., 1960)), the glutamate mutase from Clostridium tetanomorphum (approximately 70-fold more active with benzimidazolyl-cobamide than with Cbl (Barker et al., 1960)), and the reductive dehalogenase from Sulfurospirillum (formerly Dehalospirillum) multivorans (50-fold more active with nor-pseudoCbl than with Cbl (Kräutler et al., 2003)). Whether this is due to differences in catalytic properties or binding affinities is unclear, and may vary from enzyme to enzyme.
Since only approximately 7% of bacterial genomes sequenced to date contain CbiZ homologs (Gray et al., 2008), the presence of CbiZ may serve as an identifier of bacteria that contain cobamide-dependent enzymes with specific lower ligand preferences. The presence of CbiZ could also reflect the presence of cobamides in the environment that could be modified to suit the physiological needs of the organism. The identification and characterization of these enzymes could lead to greater understanding of the reasons for the diversity of the lower ligand.
Which enzymes in R. sphaeroides require Cbl, and why?
In R. sphaeroides, acetate is catabolized via the ethylmalonyl-CoA pathway, a pathway that contains two cobamide-dependent enzymes: ethylmalonyl-CoA mutase and methylmalonyl-CoA mutase (EC 5.4.99.2) (Fig. 4) (Alber et al., 2006, Erb et al., 2007). The fact that growth of R. sphaeroides on acetate depends on Cbl strongly implies that one or both of these mutases can function only when the cofactor is AdoCbl, a cobamide with a DMB lower ligand. Both of these enzymes are predicted to bind coenzyme B12 in a “base-off” configuration (Mancia et al., 1996, Erb et al., 2008), in which the lower ligand is not coordinated to the cobalt in the active site of the enzyme. In studies using Cbl, the lower ligand nucleotide was reported to be important for the organization of the active site of methylmalonyl-CoA mutase from P. shermanii, but to have only a modest effect on the strength of cofactor binding (Chowdhury & Banerjee, 1999). Further experiments are needed to clarify the role of and specificity for the lower ligand in the R. sphaeroides mutases.
EXPERIMENTAL PROCEDURES
Bacterial strains and growth conditions
All strains and plasmids used in this study are listed in Table 2. All R. sphaeroides strains were derived from wild-type strain 2.4.1 (NC_007493, NC_007494). R. sphaeroides was grown at 30°C in Sistrom’s medium A (Sistrom, 1960) lacking glutamate and aspartate, and supplemented with CoCl2 (1 mg / liter); hereafter this modified Sistrom’s medium is referred to as mSistrom’s medium) (Gray & Escalante-Semerena, 2009); potassium succinate (30 mM) or potassium acetate (30 mM) was added as carbon source, as indicated. E. coli was grown at 37°C in lysogenic broth (LB) (Difco) (Bertani, 1951, Bertani, 2004). All Salmonella enterica sv. Typhimurium LT2 (hereafter S. enterica) strains were derived from strain TR6583 (metE205 ara-9). S. enterica was grown at 37°C in nutrient broth (NB) (Difco) or no-carbon essential (NCE) minimal medium (Berkowitz et al., 1968) containing MgSO4 (1 mM), glycerol (22 mM), and trace minerals (Balch & Wolfe, 1976).
Table 2.
Strains and plasmids used in this study.a
| Strain or plasmid | Marker(s)b | Relevant genotype | Reference or source |
|---|---|---|---|
| R. sphaeroides strains: | |||
| JE8777 | 2.4.1 wild-type | Lab collection | |
| JE10754 | ΔcobB | (Gray & Escalante-Semerena, 2009) | |
| JE10755 | ΔcobB ΔcbiZ | (Gray & Escalante-Semerena, 2009) | |
| JE10778 | TcR | ΔcobB / pRK404 | |
| JE10779 | TcR | ΔcobB ΔcbiZ / pRK404 | |
| JE10781 | TcR | ΔcobB ΔcbiZ / pRsCBIZ6 | |
| JE11661 | SpR SmR | ΔcobB bluB∷aadA | |
| JE11662 | SpR SmR | ΔcobB ΔcbiZ bluB∷aadA | |
| JE11805 | SpR SmR TcR | ΔcobB bluB∷aadA / pRK404 | |
| JE11806 | SpR SmR TcR | ΔcobB bluB∷aadA / pBLUB19 | |
| JE11807 | SpR SmR TcR | ΔcobB ΔcbiZ bluB∷aadA / pRK404 | |
| JE11808 | SpR SmR TcR | ΔcobB ΔcbiZ bluB∷aadA / pBLUB19 | |
| JE11821 | SpR SmR TcR | ΔcobB ΔcbiZ bluB∷aadA / pRsCBIZ6 | |
| S. enterica strains: | |||
| TR6583 | metE205 ara-9 derivative of strain LT2 | K. Sanderson via J.R. Roth | |
| JE8248 | metE205 ara-9 cobS1313 | lab collection | |
| E. coli strains: | |||
| DH5α/F’ | F’ / endA1 hsdR17(rk-mk+) glnV44 thi-1 recA1 gyrA(NalR) relA1 Δ(lacIZYA-argF)U169 deoR (ϕ80dlacΔ(lacZ)M15) | (Woodcock et al., 1989, Raleigh et al., 1989) | |
| S17-1 | recA pro hsdR RP4-2-Tc∷Mu-Km∷Tn7 | (Simonet al., 1983) | |
| BL21(DE3) | F-ompT gal dcm lon hsdSB(rB-mB-) λ(DE3 [lacl lacUV5-T7 gene 1 ind1 sam7 nin5]) | Novagen | |
| Plasmids: | |||
| pGEM®-T Easy | ApR | TA cloning vector | Promega |
| pK18mobsacB | KmR SucS | suicide vector for allelic exchange | (Schafer et al., 1994) |
| pRK404 | TcR | shuttle vector with lac promoter | (Ditta et al., 1985) |
| pSRA2 | ApR SpR SmR | aadA+ | (Frigaard et al., 2004) |
| pKLD66 | ApR | TEV protease-cleavable maltose-binding protein / His6 tag overexpression vector | (Rocco et al., 2008) |
| pRsCBIZ3 | ApR | R. sphaeroides cbiZ+ translational fusion to maltose-binding protein / His6 tag for protein purification | |
| pRsCBIZ6 | TcR | R. sphaeroides cbiZ+ | |
| pBLUB19 | TcR | R. sphaeroides bluB+ | |
| pBLUB20 | ApR | R. sphaeroides bluB+ region | |
| pBLUB21 | ApR | ΔbluB for R. sphaeroides | |
| pBLUB23 | ApR SpR SmR | bluB∷aadA for R. sphaeroides | |
| pBLUB24 | KmR SucS SpR SmR | bluB∷aadA for R. sphaeroides |
Unless otherwise indicated, all strains and plasmids were constructed during the course of these studies.
TcR, tetracycline resistance; SpR, spectinomycin resistance; SmR, streptomycin resistance; ApR, ampicillin resistance; KnR, kanamycin resistance; SucS, sucrose sensitivity.
For growth curves of R. sphaeroides, starter cultures were grown aerobically for two days in mSistrom’s medium containing succinate, CNCbl (1 nM), and appropriate antibiotics. Cells were washed twice with sterile mSistrom’s medium containing no carbon, and were used to inoculate fresh medium (5% v/v inoculum). For growth curves of S. enterica, starter cultures were grown aerobically overnight in NB + antibiotic, and were used to inoculate fresh medium (0.5% v/v inoculum). Growth behavior was analyzed under oxic conditions using a PowerWave XS Microplate Reader (Bio-Tek Instruments). When used, ampicillin was at 100 μg ml-1, kanamycin at 10 μg ml-1, tetracycline was at 12.5 μg ml-1 for E. coli and S. enterica or 1 μg ml-1 for R. sphaeroides, and spectinomycin was at 100 μg ml-1 for E. coli, or 50 μg ml-1 for R. sphaeroides. (CN)2Cby was a gift from Paul Renz (Institut für Biologische Chemie und Ernahrungswissenschaft, Universität-Hohenheim, Stuttgart, Germany). PseudoCbl was prepared as described (Gray & Escalante-Semerena, 2009). Adenosylated corrins were prepared as described (Thomas & Escalante-Semerena, 2000). All other chemicals were purchased from Sigma.
Genetic and molecular techniques
DNA manipulations were performed using described methods (Ausubel, 1989). Restriction and modification enzymes were purchased from Fermentas or Promega, and were used according to the manufacturer’s instructions. All DNA manipulations were performed in E. coli DH5α (Raleigh et al., 1989, Woodcock et al., 1989). Plasmid DNA was isolated with the Wizard Plus SV Plasmid Miniprep kit (Promega). PCR products were purified with the Wizard SV Gel and PCR Clean-Up System kit (Promega). DNA sequencing reactions were performed using nonradioactive BigDye® protocols (Applied Biosystems) and resolved at the Biotechnology Center of the University of Wisconsin, Madison. Vectors derived from plasmids pRK404 (Ditta et al., 1985) and pK18mobsacB (Schafer et al., 1994) were conjugated into R. sphaeroides as described (Liang et al., 1991). All primers used in this study are shown in Table 3.
Table 3.
Primers used in this study
| Primer Name | Sequence |
|---|---|
| Rs_ΔbluB_EcoRI_5’ | AGA ATT CTG TGT GGC CGC AAG AAG C |
| Rs_ΔbluB_XbaI_3’ | ATT CTA GAA AAA TCC GGC ATG GGA TC |
| Rs_ΔbluB_XbaI_5’ | ATT CTA GAC TGA TCA TGG AGG AAG GCT G |
| Rs_ΔbluB_SalI_3’ | ATG GTC GAC AGG AGC CGA TGA TCG CCG |
| Rs_bluB_seq_F | CTG TGA CGG GGA TGC TCC |
| Rs_bluB_seq_R | GGC GGT TGC GGA CCC TGC |
| Rs_bluB_HindIII_5’ | ATG AAG CTT CCA AGA GAC CTG CCT GCG |
| Rs_bluB_HindIII_3’ | ATG AAG CTT TCA CAA AGC GCG GTC G |
| Rs_cbiZ_HindIII_5’ | ACG GTT AAG CTT ATG ATC GAG GTC GCG CTG |
| Rs_cbiZ_BamHI_3’ | AGC GAA GGA TCC ATG TCA ACC CAT TGT CCC G |
| Rs_cbiZ_KpnI_5’ | ACG TTT GGT ACC ATG ATC GAG GTC GCG CTG GA |
| Rs_cbiZ_SalI_3’ | AGC TTT GTC GAC TCA CAG GAG GTC TCA GCC C |
| pRK404_seq_F | TAC CTG TCC GCC TTT CTC |
| pRK404_seq_R | GAG TTA GCT CAC TCA TTA |
| pKLD116_seq_F | CGA GCG GAA CCG CCT CG |
| pKLD37_seq_R | CCA TTC GCC AAT CCG GAT |
Construction of the R. sphaeroides bluB∷aadA strain
Primers Rs_ΔbluB_EcoRI_5’ and Rs_ΔbluB_SalI_3’ were used to amplify a 3,657-bp fragment of R. sphaeroides chromosomal DNA containing bluB, 1,461 bp of upstream and 1,503 bp of downstream sequence. This fragment was cloned into plasmid pGEM®-T Easy (Promega) according to the manufacturer’s instructions to yield plasmid pBLUB20. Primers Rs_ΔbluB_XbaI_5’ and Rs_ΔbluB_XbaI_3’ were used with pBLUB20 as a template to amplify a 6.1-kbp fragment, which was cut with XbaI and ligated to yield plasmid pBLUB21, which contained an in-frame deletion of bluB in which a 6-bp XbaI restriction site replaced bases 13 through 654. To construct an insertional replacement of bluB, pSRA2 (Frigaard et al., 2004) was cut with XbaI, and the aadA+ gene encoding spectinomycin resistance was gel-purified (Qiagen) and cloned into the XbaI site of pBLUB21 to yield pBLUB23. pBLUB23 was cut with EcoRI, and the 3.8-kbp fragment containing bluB∷aadA was ligated into the EcoRI site of pK18mobsacB to yield plasmid pBLUB24.
Plasmid pBLUB24 was conjugated into ΔcobB or ΔcobB ΔcbiZ R. sphaeroides strains, the resulting mixed cultures were inoculated into 150 ml of mSistrom’s medium supplemented with CNCbl (50 nM), sucrose (10% w/v), and spectinomycin, and incubated anoxically 3 days at 30°C with light. These cultures were diluted and plated on mSistrom’s agar containing CNCbl (15 nM) and spectinomycin, and incubated 3 days at 30°C. Colonies exposed to sucrose were screened for spectinomycin-resistant, kanamycin-sensitive variants on mSistrom’s agar plates containing CNCbl (15 nM). We confirmed the replacement of bluB by aadA by amplifying and sequencing the bluB region using primers Rs_bluB_seq_F and Rs_bluB_seq_R.
Construction of bluB+ plasmid
The R. sphaeroides bluB+ coding sequence plus 350 bp of 3’ sequence was amplified using primers Rs_bluB_HindIII_5’ and Rs_bluB_HindIII_3’, and the resulting product was cloned into the HindIII site of plasmid pRK404 to yield plasmid pBLUB19. The identity of the insert was confirmed by sequencing with primers pRK404_seq_F and pRK404_seq_R.
Construction of cbiZ+ plasmids
The R. sphaeroides cbiZ+ allele was amplified from the chromosome using primers Rs_cbiZ_KpnI_5’ and Rs_cbiZ_SalI_’3’, and the resulting product was cloned into the KpnI and SalI sites of plasmid pKLD66 (Rocco et al., 2008) to yield plasmid pRsCBIZ3. The identity of the insert was confirmed by sequencing with primers pKLD116_seq_F and pKLD37_seq_R. The R. sphaeroides cbiZ+ coding sequence plus 40 bp of 3’ sequence was amplified using primers Rs_cbiZ_HindIII_5’ and Rs_cbiZ_BamHI_3’, and the resulting product was cloned into the HindIII and BamHI sites of plasmid pRK404 to yield plasmid pRsCBIZ6. The identity of the insert was confirmed by sequencing with primers pRK404_seq_F and pRK404_seq_R.
Extraction and purification of corrinoids
R. sphaeroides 2.4.1 was grown aerobically for 3 days in mSistrom’s broth with potassium succinate (30 mM) and (CN)2Cbi or CNpseudoCbl (50 nM). Colony-forming units were quantified by plating serial dilutions of cultures in triplicate on mSistrom’s agar containing potassium succinate (30 mM) and CNCbl (15 nM). Cells were harvested by centrifugation (15 min at 5000 × g at 4°C). Corrinoids were extracted by re-suspending cell pellets in two volumes of methanol and shaking (200 rpm) for 2 h at 55°C (Woodson et al., 2005). Cell debris was removed by centrifugation (2 h at 40,000 × g at 4°C), supernatants were dried under vacuum, and re-suspended in 1 ml of potassium phosphate buffer (0.1 M, pH 6.5) containing KCN (10 mM). Cyano-corrinoids were obtained after exposure to bright light for 30 min, followed by filtration using Corning 0.2 μm Spin-X centrifuge filters.
Corrinoids were resolved using a Beckman Coulter System Gold® 126 HPLC system equipped with a Beckman Coulter System Gold® 508 autosampler and 150 × 4.6 mm Alltima HP C18 AQ column (Alltech). The column was developed with a modified System I of Blanche et al. (Blanche et al., 1990); corrinoids were detected with a photodiode array detector. The column was equilibrated at 1 ml min-1 with 98% solvent A [potassium phosphate buffer (0.1 M, pH 6.5) containing KCN (10 mM)] and 2% (v/v) solvent B [potassium phosphate buffer (50 mM, pH 8) containing KCN (5 mM) and acetonitrile (50%, v/v)]. The column was developed starting 5 min after injection with a 55 min linear gradient to 100% B. Fractions were collected, dried under vacuum, and re-suspended in 250 μl of ddH2O.
Analysis of corrinoids
The identity and quantity of cobamides produced by R. sphaeroides was assessed by a bioassay using S. enterica strain JE8248 (ΔcobS), as described (Gray & Escalante-Semerena, 2009). In strain JE8248, the last step of Cbl synthesis is blocked, making growth dependent on the presence of exogenous cobamides (O’Toole et al., 1993). Growth of strain JE8248 indicated the presence of Cbl or another cobamide in the extract. Growth kinetics under oxic conditions at 37°C were monitored at 650 nm, and the slopes of the growth curves between 6 and 8 h (early exponential phase) were proportional to the concentrations of CNCbl in the medium (25-250 pM) (Gray & Escalante-Semerena, 2009). The concentrations of cobamides in HPLC fractions were calculated using the equation of this standard curve. The limit of detection of the quantitative bioassay was 5 fmol of (CN)2Cbl.
Purification of MBP-CbiZ protein
R. sphaeroides CbiZ protein fused to N-terminal maltose-binding protein (MBP) and His6 tags (MBP-CbiZ) was overproduced using plasmid pRsCBIZ3 in the E. coli BL21 strain (Stratagene). A sample of an overnight culture (160 ml) of this strain grown in LB broth containing ampicillin (100 μg ml-1) was sub-cultured into eight liters of fresh medium, and was incubated for 1 h at 37°C with shaking at 180 rpm. Isopropyl-β-D-thiogalactopyranoside (IPTG) was added to a final concentration of 0.3 mM, and the culture was incubated for an additional 20 h at 15°C with shaking at 180 rpm. Cells were harvested by centrifugation (15 min at 5000 × g) and re-suspended in 30 ml of sodium phosphate (20 mM, pH 7.4) containing NaCl (0.5 M), imidazole (20 mM), and deoxyribonuclease I (0.01 g) (Sigma). One ml of protease inhibitor cocktail for use in purification of histidine-tagged proteins (Sigma) was added and cells were broken by two passages through a French press at 8.3 MPa. The cell lysate was clarified by centrifugation (30 min at 40,000 × g at 4°C) and filtered through a 0.2-μm pore size syringe filter (Nalgene). The cleared lysate was applied onto a 1-ml HisTrap HP column (Amersham Biosciences) at a rate of 0.3 ml min-1 using an ÄKTA Purifier FPLC system (Amersham Pharmacia), rinsed with 40 ml of sodium phosphate (20 mM, pH 7.4) containing NaCl (0.5 M), and imidazole (20 mM). MBP-CbiZ protein was eluted with a 30-ml linear gradient to 100% sodium phosphate (20 mM, pH 7.4) containing NaCl (0.5 M), and imidazole (500 mM). Purity was monitored using SDS / PAGE (Laemmli, 1970) and Coomassie blue staining (Sasse, 1991). Fractions containing MBP-CbiZ were pooled and dialyzed (molecular weight cut-off = 10,000 membrane; Pierce) at 4°C against two liters of Tris-HCl (10 mM, pH 7.9 at 4°C) containing ethylenediaminetetraacetic acid (EDTA, 10 mM) and NaCl (0.5 M), followed by dialysis against two liters of Tris-HCl (10 mM, pH 7.9 at 4°C) containing NaCl (0.5 M) and glycerol (10% v/v) with buffer changes (×3) every 4 h each. Protein purity was assessed using the TotalLab software package (Nonlinear Dynamics). Protein was flash frozen in liquid N2, and stored at -80°C until used.
Amidohydrolase (cobyric-acid producing) activity assays
MBP-CbiZ activity was assayed in 200-μl reaction mixtures containing 1 to 3 μg of MBP-CbiZ in 2-(N-cyclohexylamino)ethanesulfonic acid (CHES) buffer (50 mM, pH 10), dithiothreitiol (DTT, 1 mM), and corrinoid substrate (30 μM), unless otherwise indicated. Buffers (50 mM) used to assess pH optimum (other than at pH 10) were as follows: Tris-HCl for pH 7, 8, and 9; N-cyclohexyl-3-aminopropanesulfonic acid (CAPS) for pH 11. Reactions were incubated in dim light and stopped by incubation at 95°C for 10 min. Potassium cyanide was added to 0.1 M, and corrinoids were derivatized to their cyano forms by incubation under bright light for 15 min at room temperature (Fonseca & Escalante-Semerena, 2001). Precipitated protein was removed by centrifugation (1 min at 16,100 × g), samples were filtered with Spin-X filtration columns (2 μm pore size) (Costar), and product formation was monitored by HPLC.
HPLC analysis of the products of the amidohydrolase reaction
The product of the MBP-CbiZ reaction was resolved using a modification of the HPLC separation protocol described above. The column was equilibrated with 80% buffer A and 20% buffer B. Five minutes after injection, the column was developed for 30 min with a linear gradient to 40% buffer B, then developed for 5 min with a linear gradient to 100% buffer B. After 5 min, a final linear gradient returned the column to a composition of 80% buffer A / 20% buffer B over five min. Authentic (CN)2Cby, (CN)2Cbi, CNpseudoCbl, and CNCbl were used as standards, eluting at 8.8, 21.8, 17.8 and 31.5 min, respectively. Corrinoids were quantified by comparison to a standard curve of (CN)2Cbi. The limit of detection was 1 pmol of (CN)2Cbi.
DNA sequence analysis
Computer analyses of DNA and protein sequences were done using the Integrated Microbial Genomes system (Markowitz et al., 2006) and tools available from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) (accessed August 11, 2009).
Supplementary Material
Acknowledgments
This work was supported by PHS grant R01-GM40313 from the National Institute of General Medical Sciences (to J.C.E.-S.). M.J.G. was supported in part by the University of Wisconsin- Madison College of Agricultural and Life Sciences Louis and Elsa Thomsen Wisconsin Distinguished Graduate Fellowship.
References
- Alber BE, Spanheimer R, Ebenau-Jehle C, Fuchs G. Study of an alternate glyoxylate cycle for acetate assimilation by Rhodobacter sphaeroides. Mol Microbiol. 2006;61:297–309. doi: 10.1111/j.1365-2958.2006.05238.x. [DOI] [PubMed] [Google Scholar]
- Allen RH, Stabler SP. Identification and quantitation of cobalamin and cobalamin analogues in human feces. Am J Clin Nutr. 2008;87:1324–1335. doi: 10.1093/ajcn/87.5.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FA, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. Greene Publishing Associates & Wiley Interscience; New York, N.Y.: 1989. [Google Scholar]
- Balch WE, Wolfe RS. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressurized atmosphere. Appl Environ Microbiol. 1976;32:781–791. doi: 10.1128/aem.32.6.781-791.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker HA, Smyth RD, Weissbach H, Toohey JI, Ladd JN, Volcani BE. Isolation and properties of crystalline cobamide coenzymes containing benzimidazole or 5, 6-dimethylbenzimidazole. J Biol Chem. 1960;235:480–488. [PubMed] [Google Scholar]
- Berkowitz D, Hushon JM, Whitfield HJ, Jr, Roth J, Ames BN. Procedure for identifying nonsense mutations. J Bacteriol. 1968;96:215–220. doi: 10.1128/jb.96.1.215-220.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani G. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J Bacteriol. 2004;186:595–600. doi: 10.1128/JB.186.3.595-600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanche F, Debussche L, Famechon A, Thibaut D, Cameron B, Crouzet J. A bifunctional protein from Pseudomonas denitrificans carries cobinamide kinase and cobinamide phosphate guanylyltransferase activities. J Bacteriol. 1991;173:6052–6057. doi: 10.1128/jb.173.19.6052-6057.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanche F, Thibaut D, Couder M, Muller JC. Identification and quantitation of corrinoid precursors of cobalamin from Pseudomonas denitrificans by high-performance liquid chromatography. Anal Biochem. 1990;189:24–29. doi: 10.1016/0003-2697(90)90038-b. [DOI] [PubMed] [Google Scholar]
- Buan NR, Rehfeld K, Escalante-Semerena JC. Studies of the CobA-type ATP:Co(I)rrinoid adenosyltransferase enzyme of Methanosarcina mazei strain Gö1. J Bacteriol. 2006;188:3543–3550. doi: 10.1128/JB.188.10.3543-3550.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buan NR, Suh SJ, Escalante-Semerena JC. The eutT gene of Salmonella enterica encodes an oxygen-labile, metal-containing ATP:corrinoid adenosyltransferase enzyme. J Bacteriol. 2004;186:5708–5714. doi: 10.1128/JB.186.17.5708-5714.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkholder PR. Determination of vitamin B12 with a mutant strain of Escherichia coli. Science. 1951;114:459–460. doi: 10.1126/science.114.2966.459. [DOI] [PubMed] [Google Scholar]
- Chowdhury S, Banerjee R. Role of the Dimethylbenzimidazole Tail in the Reaction Catalyzed by Coenzyme B(12)-Dependent Methylmalonyl-CoA Mutase. Biochemistry. 1999;38:15287–15294. doi: 10.1021/bi9914762. [DOI] [PubMed] [Google Scholar]
- Ditta G, Schmidhauser T, Yakobson E, Lu P, Liang XW, Finlay DR, Guiney D, Helinski DR. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid. 1985;13:149–153. doi: 10.1016/0147-619x(85)90068-x. [DOI] [PubMed] [Google Scholar]
- Dobson CM, Wai T, Leclerc D, Kadir H, Narang M, Lerner-Ellis JP, Hudson TJ, Rosenblatt DS, Gravel RA. Identification of the gene responsible for the cblB complementation group of vitamin B12-dependent methylmalonic aciduria. Hum Mol Genet. 2002;11:3361–3369. doi: 10.1093/hmg/11.26.3361. [DOI] [PubMed] [Google Scholar]
- Erb TJ, Berg IA, Brecht V, Muller M, Fuchs G, Alber BE. Synthesis of C5-dicarboxylic acids from C2-units involving crotonyl-CoA carboxylase/reductase: the ethylmalonyl-CoA pathway. Proc Natl Acad Sci U S A. 2007;104:10631–10636. doi: 10.1073/pnas.0702791104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb TJ, Retey J, Fuchs G, Alber BE. Ethylmalonyl-CoA mutase from Rhodobacter sphaeroides defines a new subclade of coenzyme B12-dependent acyl-CoA mutases. J Biol Chem. 2008;283:32283–32293. doi: 10.1074/jbc.M805527200. [DOI] [PubMed] [Google Scholar]
- Escalante-Semerena JC. Conversion of cobinamide into adenosylcobamide in bacteria and archaea. J Bacteriol. 2007;189:4555–4560. doi: 10.1128/JB.00503-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante-Semerena JC, Suh SJ, Roth JR. cobA function is required for both de novo cobalamin biosynthesis and assimilation of exogenous corrinoids in Salmonella typhimurium. J Bacteriol. 1990;172:273–280. doi: 10.1128/jb.172.1.273-280.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante-Semerena JC, Warren MJ. Biosynthesis and Use of Cobalamin (B12) In: Böck A, Curtiss R III, Kaper JB, Karp PD, Neidhardt FC, Nyström T, Slauch JM, Squires CL, editors. EcoSal - Escherichia coli and Salmonella: cellular and molecular biology. Washington, D. C.: ASM Press; 2008. [Google Scholar]
- Fonseca MV, Escalante-Semerena JC. An in vitro reducing system for the enzymic conversion of cobalamin to adenosylcobalamin. J Biol Chem. 2001;276:32101–32108. doi: 10.1074/jbc.M102510200. [DOI] [PubMed] [Google Scholar]
- Friedmann HC, Fyfe JA. Pseudovitamin B12 biosynthesis. Enzymatic formation of a new adenylic acid, 7-α-D-ribofuranosyladenine 5’-phosphate. J Biol Chem. 1969;244:1667–1671. [PubMed] [Google Scholar]
- Frigaard NU, Li H, Milks KJ, Bryant DA. Nine mutants of Chlorobium tepidum each unable to synthesize a different chlorosome protein still assemble functional chlorosomes. J Bacteriol. 2004;186:646–653. doi: 10.1128/JB.186.3.646-653.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard CL, Santschi DE, Stabler SP, Allen RH. Apparent ruminal synthesis and intestinal disappearance of vitamin B12 and its analogs in dairy cows. J Dairy Sci. 2009;92:4524–4529. doi: 10.3168/jds.2009-2049. [DOI] [PubMed] [Google Scholar]
- Gray MJ, Escalante-Semerena JC. Single-enzyme conversion of FMNH2 to 5,6-dimethylbenzimidazole, the lower ligand of B12. Proc Natl Acad Sci U S A. 2007;104:2921–2926. doi: 10.1073/pnas.0609270104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MJ, Escalante-Semerena JC. In vivo analysis of cobinamide salvaging in Rhodobacter sphaeroides strain 2.4.1. J Bacteriol. 2009;191:3842–3851. doi: 10.1128/JB.00230-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MJ, Tavares NK, Escalante-Semerena JC. The genome of Rhodobacter sphaeroides strain 2.4.1 encodes functional cobinamide salvaging systems of archaeal and bacterial origins. Mol Microbiol. 2008;70:824–836. doi: 10.1111/j.1365-2958.2008.06437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CL, Buszko ML, Bobik TA. Purification and initial characterization of the Salmonella enterica PduO ATP:Cob(I)alamin adenosyltransferase. J Bacteriol. 2004;186:7881–7887. doi: 10.1128/JB.186.23.7881-7887.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kräutler B, Fieber W, Osterman S, Fasching M, Ongania K-H, Gruber K, Kratky C, Mikl C, Siebert A, Diekert G. The cofactor of tetrachloroethene reductive dehalogenase of Dehalospirillum multivorans is Norpseudo-B12, a new type of natural corrinoid. Helvetica Chimica Acta. 2003;86:3698–3716. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lengyel P, Mazumder R, Ochoa S. Mammalian Methylmalonyl Isomerase and Vitamin B(12) Coenzymes. Proc Natl Acad Sci U S A. 1960;46:1312–1318. doi: 10.1073/pnas.46.10.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang JH, Nielsen GM, Lies DP, Burris RH, Roberts GP, Ludden PW. Mutations in the draT and draG genes of Rhodospirillum rubrum result in loss of regulation of nitrogenase by reversible ADP-ribosylation. J Bacteriol. 1991;173:6903–6909. doi: 10.1128/jb.173.21.6903-6909.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancia F, Keep NH, Nakagawa A, Leadlay PF, McSweeney S, Rasmussen B, Bosecke P, Diat O, Evans PR. How coenzyme B12 radicals are generated: the crystal structure of methylmalonyl-coenzyme A mutase at 2Å resolution. Structure. 1996;4:339–350. doi: 10.1016/s0969-2126(96)00037-8. [DOI] [PubMed] [Google Scholar]
- Markowitz VM, Korzeniewski F, Palaniappan K, Szeto E, Werner G, Padki A, Zhao X, Dubchak I, Hugenholtz P, Anderson I, Lykidis A, Mavromatis K, Ivanova N, Kyrpides NC. The integrated microbial genomes (IMG) system. Nucleic Acids Res. 2006;34:D344–348. doi: 10.1093/nar/gkj024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto E, Tanioka Y, Nakao T, Barla F, Inui H, Fujita T, Watanabe F, Nakano Y. Purification and characterization of a corrinoid-compound in an edible cyanobacterium Aphanizomenon flos-aquae as a nutritional supplementary food. J Agric Food Chem. 2006;54:9604–9607. doi: 10.1021/jf062300r. [DOI] [PubMed] [Google Scholar]
- O’Toole GA, Escalante-Semerena JC. Purification and characterization of the bifunctional CobU enzyme of Salmonella typhimurium LT2. Evidence for a CobU-GMP intermediate. J Biol Chem. 1995;270:23560–23569. doi: 10.1074/jbc.270.40.23560. [DOI] [PubMed] [Google Scholar]
- O’Toole GA, Rondon MR, Escalante-Semerena JC. Analysis of mutants of defective in the synthesis of the nucleotide loop of cobalamin. J Bacteriol. 1993;175:3317–3326. doi: 10.1128/jb.175.11.3317-3326.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman D, Barrett JM. Biosynthesis of cobalamin analogues by Propionibacterium arabinosum. Can J Microbiol. 1958;4:9–15. doi: 10.1139/m58-002. [DOI] [PubMed] [Google Scholar]
- Raleigh EA, Lech K, Brent R. Selected topics from classical bacterial genetics. In: Ausubel FA, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley Interscience; 1989. p. 1.4. [DOI] [PubMed] [Google Scholar]
- Renz P. Biosynthesis of the 5,6-dimethylbenzimidazole moiety of cobalamin and of other bases found in natural corrinoids. In: Banerjee R, editor. Chemistry and Biochemistry of B12. New York: John Wiley & Sons, Inc.; 1999. pp. 557–575. [Google Scholar]
- Rocco CJ, Dennison KL, Klenchin VA, Rayment I, Escalante-Semerena JC. Construction and use of new cloning vectors for the rapid isolation of recombinant proteins from Escherichia coli. Plasmid. 2008;59:231–237. doi: 10.1016/j.plasmid.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanishvili R, Pennycooke M, Gu J, Xu X, Joachimiak A, Edwards AM, Christendat D. Crystal structure of the hypothetical protein TA1238 from Thermoplasma acidophilum: a new type of helical super-bundle. J Struct Funct Genomics. 2004;5:231–240. doi: 10.1007/s10969-005-3789-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse J. Detection of proteins. In: Ausubel FA, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley Interscience; 1991. pp. 10.16.11–10.16.18. [Google Scholar]
- Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Puhler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- Sistrom WR. A requirement for sodium in the growth of Rhodopseudomonas spheroides. J Gen Microbiol. 1960;22:778–785. doi: 10.1099/00221287-22-3-778. [DOI] [PubMed] [Google Scholar]
- Stupperich E, Kräutler B. Pseudo vitamin B12 or 5-hydroxybenzimidazolyl-cobamide are the corrinoids found in methanogenic bacteria. Arch Microbiol. 1988;149:213–217. [Google Scholar]
- Taga ME, Larsen NA, Howard-Jones AR, Walsh CT, Walker GC. BluB cannibalizes flavin to form the lower ligand of vitamin B12. Nature. 2007;446:449–453. doi: 10.1038/nature05611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MG, Escalante-Semerena JC. Identification of an alternative nucleoside triphosphate: 5’- deoxyadenosylcobinamide phosphate nucleotidyltransferase in Methanobacterium thermoautotrophicum ΔH. J Bacteriol. 2000;182:4227–4233. doi: 10.1128/jb.182.15.4227-4233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe F, Katsura H, Takenaka S, Fujita T, Abe K, Tamura Y, Nakatsuka T, Nakano Y. Pseudovitamin B(12) is the predominant cobamide of an algal health food, spirulina tablets. J Agric Food Chem. 1999;47:4736–4741. doi: 10.1021/jf990541b. [DOI] [PubMed] [Google Scholar]
- Watanabe F, Miyamoto E, Fujita T, Tanioka Y, Nakano Y. Characterization of a corrinoid compound in the edible (blue-green) alga, Suizenji-nori. Biosci Biotechnol Biochem. 2006;70:3066–3068. doi: 10.1271/bbb.60395. [DOI] [PubMed] [Google Scholar]
- Watanabe F, Tanioka Y, Miyamoto E, Fujita T, Takenaka H, Nakano Y. Purification and characterization of corrinoid-compounds from the dried powder of an edible cyanobacterium, Nostoc commune (Ishikurage) J Nutr Sci Vitaminol (Tokyo) 2007;53:183–186. doi: 10.3177/jnsv.53.183. [DOI] [PubMed] [Google Scholar]
- Woodcock DM, Crowther PJ, Doherty J, Jefferson S, De Cruz E, Noyer-Weidner M, Smith SS, Michael MZ, Graham MW. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucl Acids Res. 1989;17:3469–3478. doi: 10.1093/nar/17.9.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Escalante-Semerena JC. CbiZ, an amidohydrolase enzyme required for salvaging the coenzyme B12 precursor cobinamide in archaea. Proc Natl Acad Sci USA. 2004;101:3591–3596. doi: 10.1073/pnas.0305939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Escalante-Semerena JC. The cbiS gene of the archaeon Methanopyrus kandleri AV19 encodes a bifunctional enzyme with adenosylcobinamide amidohydrolase and alpha-ribazole-phosphate phosphatase activities. J Bacteriol. 2006;188:4227–4235. doi: 10.1128/JB.00227-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Reynolds AA, Escalante-Semerena JC. ABC transporter for corrinoids in Halobacterium sp. strain NRC-1. J Bacteriol. 2005;187:5901–5909. doi: 10.1128/JB.187.17.5901-5909.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Zayas CL, Escalante-Semerena JC. A new pathway for salvaging the coenzyme B12 precursor cobinamide in archaea requires cobinamide-phosphate synthase (CbiB) enzyme activity. J Bacteriol. 2003;185:7193–7201. doi: 10.1128/JB.185.24.7193-7201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayas CL, Claas K, Escalante-Semerena JC. The CbiB protein of Salmonella enterica is an integral membrane protein involved in the last step of the de novo corrin ring biosynthetic pathway. J Bacteriol. 2007;189:7697–7708. doi: 10.1128/JB.01090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


