Abstract
The epidermal growth factor receptor (EGFR) signaling pathway has emerged as a promising target for cancer therapy. EGFR tyrosine kinase inhibitors (TKIs) such as erlotinib have been approved for cancer treatment but have demonstrated very limited activity in breast cancer patients. Clarifying the molecular mechanism underlying resistance to EGFR-TKIs could lead to more effective treatment against breast cancer. We previously reported that the sensitivity of breast cancer cells to erlotinib is partially dependent on p27 and that cytoplasmic localization of p27 is associated with erlotinib resistance. In the present study, we found that erlotinib induces p27 phosphorylation at serine (S) 10, and S10 p27 phosphorylation leads to erlotinib resistance in EGFR-expressing breast cancer. Inhibiting S10 phosphorylation of p27 by knocking down human kinase interacting stathmin (KIS), a nuclear protein that can phosphorylate p27 at S10, led to p27 accumulation in the nucleus and enhanced erlotinib-mediated cytotoxicity. Further, in vivo KIS gene silencing enhanced the antitumor activity of erlotinib in an orthotopic breast cancer xenograft model. KIS depletion also enhanced erlotinib sensitivity in erlotinib-resistant EGFR-expressing triple-negative breast cancer cells. Our study provides a rationale for the development of combinations of erlotinib with KIS inhibition to overcome EGFR-TKI resistance in EGFR-expressing breast cancer.
Keywords: EGFR, p27, KIS, erlotinib, breast cancer
Introduction
Epidermal growth factor receptor (EGFR), a member of the ErbB receptor tyrosine kinase family, is frequently overexpressed in human malignant tumors and is known to drive tumor growth, progression, and metastasis (1–4). EGFR overexpression is associated with poor prognosis and reduced overall survival in patients with lung cancer (5, 6). Therefore, the EGFR signaling pathway has emerged as a promising target for cancer therapy, in particular lung cancer. A number of tyrosine kinase inhibitors (TKIs) that target EGFR have been developed and used successfully to treat cancer patients. For example, erlotinib (also known as OSI-774 or Tarceva, Fig. 1A), a small-molecule EGFR TKI that targets the ATP-binding site of EGFR, is used to treat non–small-cell lung cancer and pancreatic cancer (7, 8).
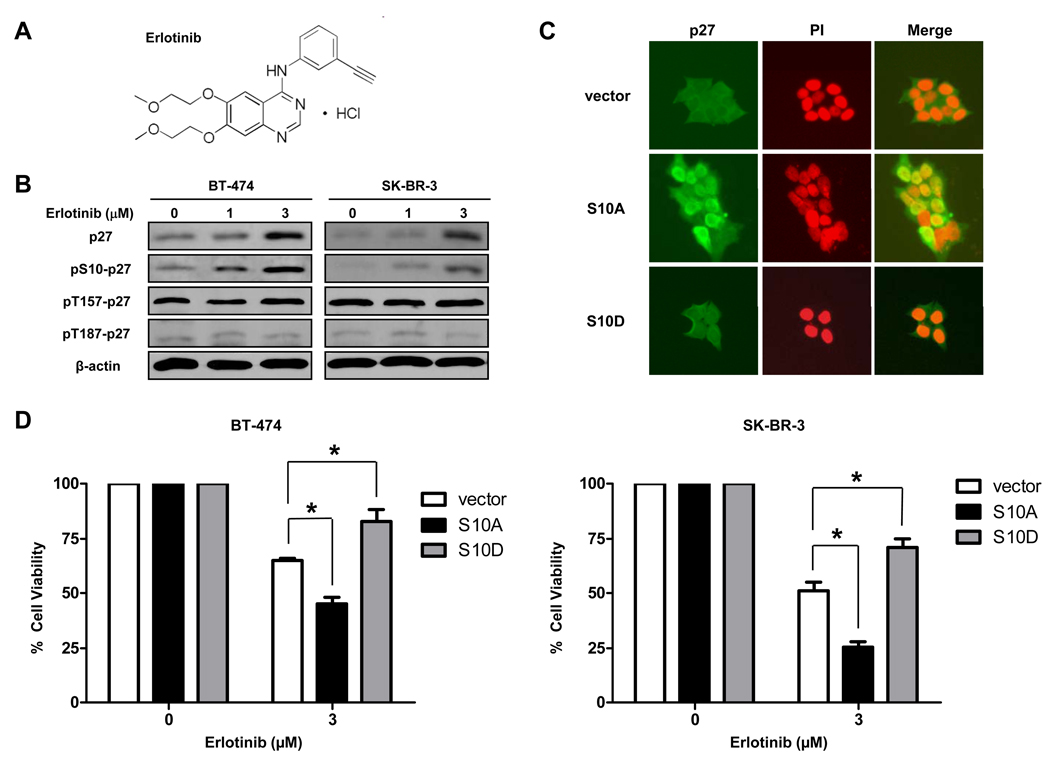
Figure 1.
S10 p27 phosphorylation leads to erlotinib resistance. A, the chemical structure of erlotinib. B, BT-474 and SK-BR-3 cells were treated with erlotinib at 0, 1, or 3 µM for 72 h, after which expression of total p27, p27 phosphorylated at S10 (pS10-p27), pT157-p27, and pT187-p27 was assessed by western blot analysis. β-actin was used as a loading control. C, BT-474 cells were transfected with pcDNA3 empty vector (vector), S10A-p27 (S10A) or S10D-p27 (S10D) mutants for 48 h. Immunofluorescent staining of p27 (green) was performed to detect the localization of p27. Nuclei were labeled by propidium iodide (PI, red). D, BT-474 and SK-BR-3 cells were transfected with empty vector, S10A-p27 or S10D-p27 for 48 h, then treated with erlotinib for another 72 h. Growth-inhibitory effects of erlotinib on these cells were studied by WST-1 assay. *, P < 0.01.
Although EGFR is overexpressed in 20% to 80% of breast cancers (3, 9, 10), erlotinib induces clinical responses in fewer than 10% of women with breast cancer. The molecular mechanism underlying resistance to EGFR TKIs in breast cancer must be better understood if EGFR is to be used successfully as a molecular target for breast cancer treatment.
p27Kip1 (p27), a major downstream molecule in EGFR signaling pathways, is a cyclin-dependent kinase (CDK) inhibitor that negatively regulates cellular proliferation by inhibiting progression through the cell cycle (11). The functions of p27 depend on the expression levels of p27 and its subcellular localization. In breast cancer, low p27 expression seems to be a marker of poor prognosis (12). The phosphorylation status of p27 is known to affect its nuclear-cytoplasmic localization (13–16), which itself is a critical determinant of growth arrest. p27 is phosphorylated at serine (S) 10, threonine (T) 157, T187, and T198. Phosphorylation of p27 at T187 is mediated by cyclin E-CDK2 (17), whereas phosphorylation at T157 and T198 is mediated by Akt (14–16, 18).
Phosphorylation of p27 at S10 is an important event in the regulation of the tumor suppressor function of p27 (19). Dephosphorylation of p27 at S10 inhibits p27 nuclear export and promotes its assembly into cyclin–CDK complexes, thereby inhibiting cell proliferation (13, 20–22). Phosphorylation of p27 at S10 takes place upon mitogenic stimulation by the nuclear protein human kinase interacting stathmin (KIS). KIS binds the C-terminal domain of p27 and phosphorylates it at S10 in vitro and in vivo, resulting in export of p27 from the nucleus to the cytoplasm and thereby regulating cell cycle progression (13, 23). KIS induces proliferation and cell cycle progression through the phosphorylation of p27 in leukemia cells (24). Overexpression of KIS can overcome p27-induced growth arrest, whereas KIS siRNA knockdown can enhance growth arrest by inhibiting S10 p27 phosphorylation (13).
We previously reported that the sensitivity of breast cancer cells to erlotinib is partially dependent on p27 and that cytoplasmic localization of p27 is associated with erlotinib resistance (25). In the present study, we showed that S10 p27 phosphorylation plays an important role in erlotinib sensitivity. Depletion of KIS by siRNA knockdown led to translocation of p27 to the nucleus and enhanced erlotinib’s cytotoxicity in breast cancer cells. More importantly, we found that the combination of erlotinib with KIS gene silencing had more significant antitumor effect than KIS siRNA knockdown alone or erlotinib alone in an orthotopic breast cancer xenograft model.
Materials and Methods
Cell lines, plasmid, and reagents
The breast cancer cell lines BT-474, SK-BR-3, MDA-MB-231 and MDA-MB-468 were purchased from American Type Culture Collection and were grown in Dulbecco’s modified Eagle’s medium/F12 medium supplemented with 10% fetal bovine serum (FBS, GIBCO/BRL), in a humidified atmosphere containing 5% CO2 at 37°C. The inflammatory breast cancer (IBC) cell line SUM149 was obtained from the University of Michigan. Another IBC cell line SUM190 was purchased from Asterand. The SUM149 and SUM190 cells were grown in Ham's F12 medium supplemented with 5% FBS, 5 µg/ml insulin, and 1 µg/ml hydrocortisone. SUM149 cells have been genotyped to ensure their identity by Link Genomics, Inc., Japan. Cells used for experiments were grown in culture for less than 2 months after resuscitation. All cell lines were tested to be mycoplasma free using a MycoAlert Mycoplasma Detection Kit (Lonza Cologne AG). pcDNA3-S10A-p27 (phosphoinhibitory S10A-p27 mutant generated by substituting an alanine at the S10 phosphorylation site) and pcDNA3-S10D-p27 (phosphomimetic S10D-p27 mutant generated by substituting an aspartic acid at the S10 phosphorylation site) plasmids were kindly provided by Dr. Keiichi I. Nakayama (Kyushu University, Japan) (22).
Erlotinib was kindly provided by OSI Pharmaceuticals, Inc. Erlotinib was dissolved in dimethyl sulfoxide (DMSO) as a 5 mM stock solution for in vitro experiments. Erlotinib was suspended in 0.5% methyl cellulose for oral gavage for in vivo animal work. Lapatinib was synthesized and dissolved in DMSO as a 10 mM stock solution as previously described (26).
Immunofluorescence analyses
Immunofluorescence analyses were performed as previously described (27). Rabbit anti-p27 antibody (Santa Cruz Biotechnology) was used as the primary antibody. Fluorescein isothiocyanate-conjugated antibodies (Biosource) were used as secondary antibodies. Cells were counterstained with propidium iodide before being mounted under glass coverslips and analyzed by confocal microscopy (FV300, Olympus).
Western blot analysis
Western blot were performed as previously described (25, 26). The antibodies used were rabbit anti-p27 antibody (Santa Cruz Biotechnology), rabbit anti-phospho-p27 (S10) antibody (Santa Cruz Biotechnology), rabbit anti-phospho-p27 (T157) antibody (R&D Systems), rabbit anti-phospho-p27 (T187) antibody (Santa Cruz Biotechnology), and mouse anti-β-actin antibody (Sigma-Aldrich).
WST-1 cell proliferation assay
WST-1 reagent (Roche Applied Science) was used to perform the WST-1 assay. A cell suspension of 4,000 cells/90 µl was seeded into each well of a 96-well plate and cultured overnight, after which 10 µl of erlotinib (or lapatinib) with various concentration was added to the individual wells. After 3 days of erlotinib (or lapatinib) treatment, 10 µl of the ready-to-use WST-1 reagent was added directly into the medium, the plates were incubated at 37°C for 1 h, and absorbance was measured on a plate reader at 450 nm. All experiments were done in triplicate. The percentage cell viability was calculated as (the absorbance of treated well minus the absorbance of cell-free control) / (absorbance of untreated control minus the absorbance of cell-free control) × 100. Median inhibitory concentrations were determined from these calculations.
Quantitative real-time reverse transcriptase-polymerase chain reaction (qRT-PCR)
RNA was extracted by using RNeasy kit (Qiagen). qRT-PCR was performed as described in detail elsewhere (28). The KIS primers used were as follows: upper primer: AAT CCT GGC AGA GGA CAA GTC TT, lower primer: GTA GAA TGT AGC CAC AAC AAA CTT CC.
siRNA knockdown
KIS siRNA (5’-AAGCAGTTCTTG CCGCCAGGA-3’) and nontargeting control siRNA were purchased from Dharmacon Research Inc. RNA interference assay was performed according to the manufacturer’s protocol (Dharmacon Research). Briefly, cells were seeded in 6-well culture plates at 30% confluence in culture medium supplemented with fetal bovine serum. The next day, cells were transfected with siRNA at a final concentration of 100 nM by using Oligofectamine (Invitrogen).
Flow cytometry analysis
For flow cytometry analysis, cells were plated in 60-mm dishes, cultured overnight, and then treated with or without erlotinib. After 48 h, floating and adherent cells were collected by trypsinization, fixed overnight in 70% ethanol, and resuspended in propidium iodide (25 µg/mL) supplemented with 0.1% RNase A. DNA content was measured with a FACScan flow cytometer (BD Biosciences). These experiments were repeated three times independently. Student’s t test was performed to compare the groups with respect to percentage of cells in G1 phase.
Anchorage-independent growth assay
Anchorage-independent growth assay was performed as previously described (29). In brief, cells were treated with control siRNA or KIS siRNA for 48 h. After that, 5000 cells were cultured on a plate containing 0.8% base agar and 0.4% top agar in medium containing 1 µM erlotinib and incubated at 37°C for 21 days. Plates were stained with 0.005% crystal violet for 1 h. Colonies were counted by use of a dissecting microscope. These experiments were done in triplicate.
Breast cancer xenograft model
A total volume of 0.15 mL of BT-474 cell suspension containing 5 × 106 cells with 50% Matrigel was injected into the bottom left mammary fat pad of 8-week-old athymic female nu/nu mice. When well-established tumors measured an average volume of 60 mm3, mice (8 per group) were randomly assigned to six groups to receive the following treatments for 2 weeks: group 1, vehicle control (0.5% methyl cellulose); group 2, control siRNA; group 3, KIS siRNA; group 4, erlotinib; group 5, erlotinib combined with control siRNA; group 6, erlotinib combined with KIS siRNA. Erlotinib (100 mg/kg of body weight) was given orally once a day. For siRNA delivery, intravenous injections were given twice a week. Alexa 555 siRNA was purchased from Qiagen. siRNA for in vivo delivery was incorporated into 1,2-dioleoyl-sn-glycero-3-phosphatidylcholine (DOPC; Avanti Lipids) liposomes. The mixture was hydrated with normal (0.9%) saline at a concentration of 15 µg/mL to achieve the desired dose in 100 µL/injection. Tumor volume was determined weekly by externally measuring the tumors in 2 dimensions using a caliper. Volume (V) was determined by the following equation, where L is length and W is width of the tumor: V = (L × W2) × 0.5. Two-sample t tests were used to compare tumor volumes between the groups. Tumor growth inhibition (%) was calculated as: 1- (the tumor volume change of treatment group / the tumor volume change of vehicle control group).
Statistical analyses
Statistical analyses were performed using commercially available software (Statview, version 5.0, SAS Institute). The data obtained were analyzed by t test. Statistical significance was defined as P < 0.05.
Results
S10 p27 phosphorylation leads to erlotinib resistance
We previously tested erlotinib sensitivity in breast cancer cell lines expressing various levels of EGFR and found that cytoplasmic localization of p27 is associated with erlotinib resistance in breast cancer cells (25). Among them, EGFR-expressing BT-474 and SK-BR-3 cells have limited sensitivity to erlotinib (IC50, 5.01 µM for BT-474 and 3.98 µM for SK-BR-3) (25). To determine the role of p27 localization in erlotinib cytotoxicity, we tested the expression levels of total p27 and phosphorylated p27 after erlotinib treatment in BT-474 and SK-BR-3 cells. Expression of both total p27 and p27 phosphorylated at S10 was significantly increased in these cells in response to erlotinib (Fig. 1B). Erlotinib did not, however, affect levels of p27 phosphorylated at T157 or T187, suggesting that phosphorylation of p27 at S10 may be the critical signal to export nuclear p27 to the cytoplasm in response to erlotinib (Fig. 1B).
Because S10 p27 phosphorylation leads to p27 cytoplasmic localization, we hypothesized that S10 p27 phosphorylation also leads to erlotinib resistance. To test this hypothesis, we transfected phosphoinhibitory S10A-p27 mutant or phosphomimetic S10D-p27 mutant to BT-474 and SK-BR-3 cells and then tested their sensitivity to erlotinib. We found that in BT-474 cells, p27 translocated to the nucleus after S10A-p27 transfection but remained in the cytoplasm after S10D-p27 transfection by immunofluorescence staining (Fig. 1C). p27 translocation to the nucleus was also detected in SK-BR-3 cells after S10A-p27 transfection (data not shown). We then treated cells with erlotinib and found that S10A-p27-transfected cells were more sensitive to erlotinib and S10D-p27-transfected cells were more resistant to erlotinib than empty vector-transfected cells (Fig. 1D), suggesting that phosphorylation of p27 at S10 leads to erlotinib resistance.
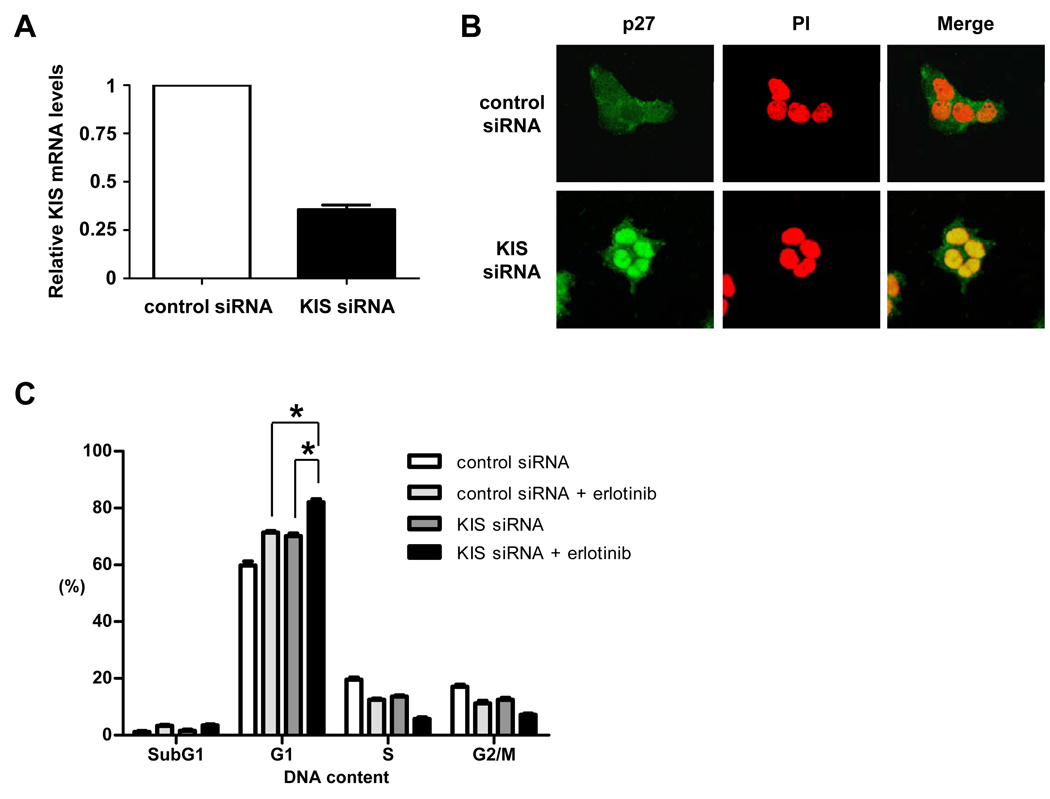
Inhibition of p27 phosphorylation at S10 by KIS siRNA knockdown leads to p27 nucleus accumulation and enhances erlotinib-induced G1 arrest
Because KIS is known to phosphorylate p27 at S10, we depleted KIS by KIS siRNA knockdown to inhibit S10 phosphorylation of p27 in BT-474 cells (Fig. 2A). KIS depletion led to strong accumulation of p27 in the nucleus (Fig. 2B). We then performed FACSan analysis to quantify the cell cycle distribution in control and KIS siRNA knockdown cells with and without erlotinib treatment. Even though both types of cells showed G1 cell cycle arrest after erlotinib treatment, more G1-phase cells were detected in KIS siRNA knockdown cells (82.1 ± 3.24%) than in control siRNA knockdown cells (71.4 ± 0.82%) after erlotinib treatment, indicating that KIS siRNA knockdown enhanced erlotinib-induced G1 cell cycle arrest (Fig. 2C).
Figure 2.
Dephosphorylation of p27 at S10 by KIS siRNA knockdown leads to p27 nucleus accumulation and enhances erlotinib-induced G1 arrest. A, BT-474 cells were treated with control or KIS siRNA knockdown for 48 h. qRT-PCR was then performed to quantify the expression levels of KIS. B, Immunofluorescent staining of p27 (green) at 48 h after control siRNA or KIS siRNA knockdown was performed in BT-474 cells. Nuclei were labeled by propidium iodide (PI, red). C, BT-474 cells were treated with control siRNA or KIS siRNA for 48 h, then treated with erlotinib at 0 or 3 µM for another 72 h. Flow cytometry analysis was performed to measure the cell cycle distribution. *, P < 0.01.
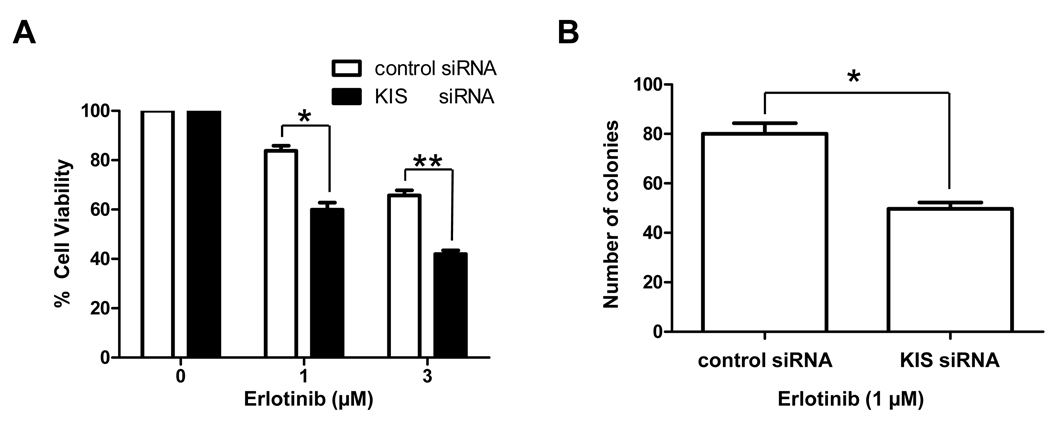
KIS depletion enhances the inhibitory effects of erlotinib on proliferation and anchorage-independent growth in breast cancer cells in vitro
We then tested whether inhibition of S10 phosphorylation of p27 enhances the anti-proliferative effects of erlotinib. BT-474 cells that were treated with KIS siRNA knockdown were more sensitive to erlotinib than were cells treated with control siRNA knockdown (Fig. 3A). To test whether the phosphorylation status of p27 at S10 plays a role in anchorage-independent growth in response to erlotinib, which may reflect in vivo tumorigenicity, we performed soft agar colony formation assay in BT-474 cells by treating them with KIS siRNA and erlotinib. We found that KIS siRNA-treated cells formed fewer colonies (i.e., were less tumorigenic) than control siRNA-treated cells in response to erlotinib (Fig. 3B). Similar results were obtained with SK-BR-3 cells (data not shown).
Figure 3.
KIS depletion enhances erlotinib’s cytotoxicity. A, Beginning 48 h after control or KIS siRNA knockdown, BT-474 cells were treated with the indicated concentrations of erlotinib for another 72 h, and then the growth-inhibitory effects of erlotinib were quantified by WST-1 assay. *, P < 0.01; **, P < 0.001. B, BT-474 cells were treated with control or KIS siRNA for 48 h, and then cells were plated in soft agar with 1 µM erlotinib to determine the anchorage-independent growth. Data shown are the colony numbers 21 days later. *, P < 0.01.
Since RNA interference can have off-target effects, we also transfected another siRNA targeting a different region of KIS into breast cancer cells. Depletion of KIS by KIS siRNA #2 also induced p27 translocation to the nucleus and enhanced erlotonib’s anti-proliferative effect in BT-474 cells (Supplementary Fig. S1).
Combination of erlotinib with KIS siRNA knockdown has more significant antitumor effect in an orthotopic breast cancer xenograft model
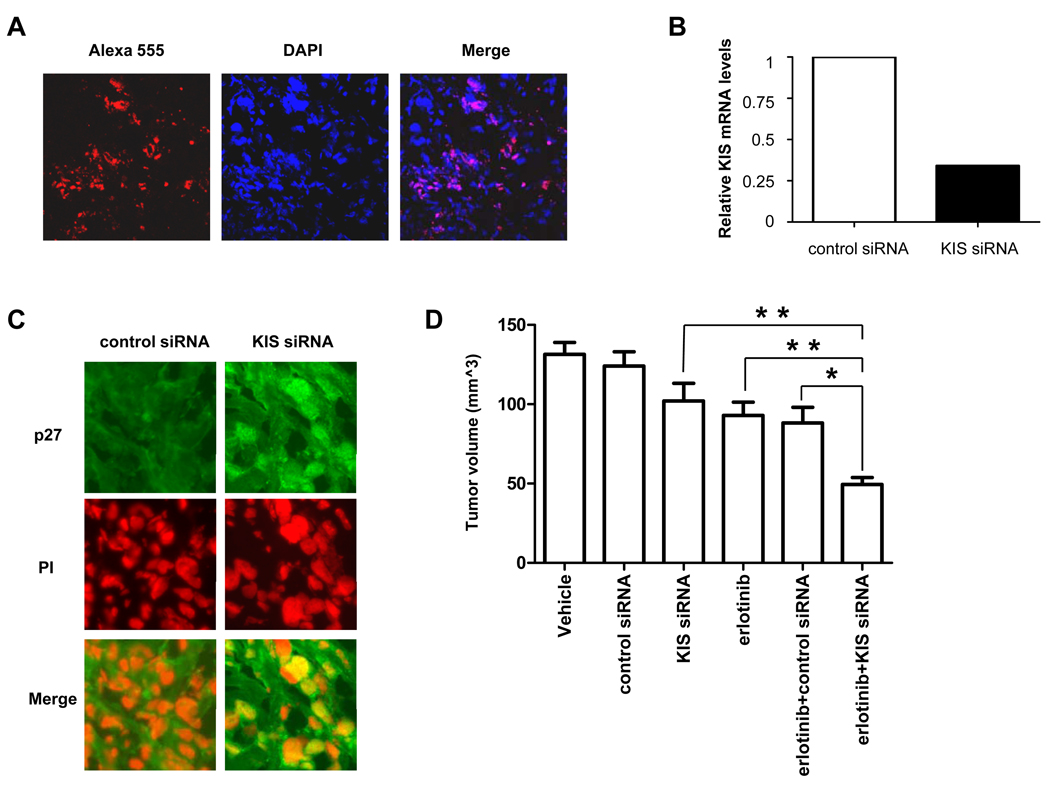
Because inhibiting the phosphorylation of p27 at S10 enhanced erlotinib’s cytotoxicity in vitro, we hypothesized that the combination of erlotinib with KIS siRNA knockdown exhibits a stronger antitumor effect than either treatment alone on breast cancer in vivo. To test this hypothesis, we examined the therapeutic efficacy of combining erlotinib with KIS siRNA knockdown in an orthotopic breast cancer xenograft model established by implanting BT-474 cells in the mammary fat pads of nude mice. We used a well-established method to deliver KIS siRNA to tumors. This is a highly efficient in vivo gene silencing method using neutral liposome (DOPC)–encapsulated siRNA (30, 31). To confirm the efficacy of siRNA-DOPC delivery, we injected Alexa 555–DOPC, a nonsilencing siRNA tagged with the fluorochrome Alexa 555, into a mouse bearing BT-474 tumor and found that Alexa 555–DOPC was taken up deeply by the tumor (Fig. 4A). qRT-PCR showed that tumor treated with KIS siRNA had significantly lower KIS expression than tumor treated with control siRNA (Fig. 4B). Furthermore, we found that p27 accumulated to the nuclei of the tumor cells after KIS siRNA administration, indicating that KIS siRNA administration is functional in vivo (Fig. 4C).
Figure 4.
KIS siRNA knockdown enhances the antitumor activity of erlotinib in an orthotopic breast cancer xenograft model. A, Alexa 555 control siRNA was injected into a BT-474-tumor-bearing mouse. Three days later, immunofluorescent staining was performed to detect Alexa 555 control siRNA (red) in the BT-474 xenograft. The nucleus was labeled by 4′,6-diamidino-2-phenylindole (DAPI, blue). B, mRNA levels of KIS in BT-474 xenografts were analyzed by qRT-PCR 3 days after control or KIS siRNA injection. C, BT-474-tumor-bearing mice were treated with control or KIS siRNA for 3 days. Immunofluorescent staining was performed to detect p27 in BT-474 xenografts. Nuclei were labeled by propidium iodide. D, Effect of erlotinib and KIS siRNA knockdown on tumor growth of BT-474 tumor xenograft model. Mice with established tumors were separated randomly and treated as described in Materials and Methods. Tumor volumes after treatment were measured weekly. Tumor volumes in 6 groups of mice were shown. Mean tumor volumes (mm3) were shown. Bars, SD. *, P < 0.01; **, P < 0.001.
At day 14 after treatment, tumors from vehicle-treated mice grew to an average size of 131 ± 22 mm3. Tumors from mice treated with KIS siRNA alone or erlotinib alone grew to an average size of 102 ± 33 mm3 and 93 ± 23 mm3, respectively, corresponding to a tumor growth inhibition of 37% and 53%, respectively. Compared with these single treatments, the combination of erlotinib with KIS siRNA knockdown was more efficient at inhibiting tumor growth: tumors from mice treated with the combination grew to an average size of 49 ± 13 mm3, corresponding to a 114% inhibition of tumor growth (Fig. 4D). The mice were treated for a total of 4 weeks and similar results were also found at different time points. At days 21 and 28 after treatment, combination of KIS siRNA knockdown and erlotinib exhibited enhanced antitumor activity than erlotinib alone or KIS siRNA knockdown alone, suggesting that KIS depletion enhanced erlotinib’s cytotoxicity in vivo (data not shown).
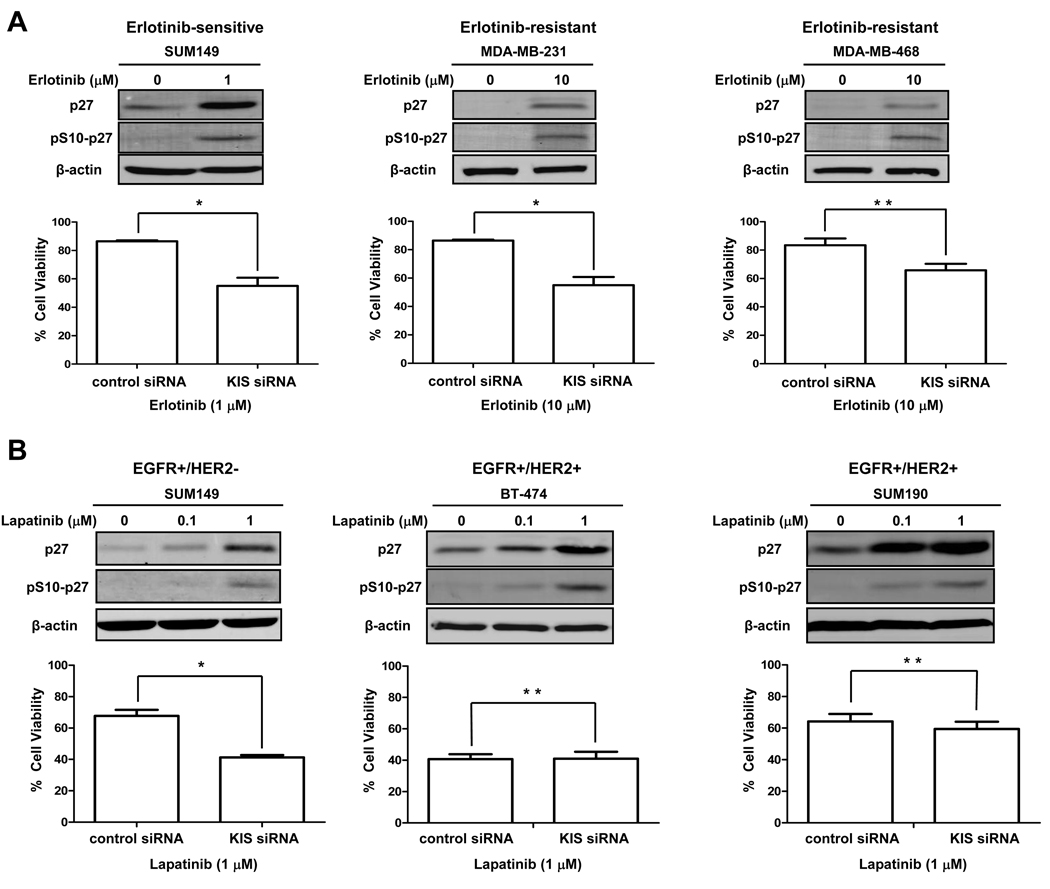
KIS depletion enhances the cytotoxicity of EGFR TKIs in both erlotinib-sensitive and erlotinib-resistant breast cancer cells
To determine whether the impact of phosphorylation of p27 at S10 on erlotinib sensitivity is a universal phenomenon in breast cancer, we performed similar experiments in other EGFR-expressing breast cancer cells: SUM149, an inflammatory breast cancer cell line that is sensitive to erlotinib (IC50 = 0.9 µM) (32), and MDA-MB-231 and MDA-MB-468, triple-negative breast cancer cell lines that are very resistant to erlotinib (IC50 > 20 µM for both) (25). In all three cell lines, we found the induction of pS10-p27 by erlotinib and found that KIS depletion enhanced erlotinib’s cytotoxicity, indicating that this effect is not restricted to a few cell lines in breast cancer (Fig. 5A).
Figure 5.
KIS depletion enhances cytotoxicity of EGFR TKIs in both erlotinib-sensitive and erlotinib-resistant breast cancer cells. A, SUM149, MDA-MB-231, and MDA-MB-468 cells were treated without or with erlotinib for 48 h. The expression levels of total p27 and pS10-p27 were detected by western blot analysis. Beginning 48 h after control or KIS siRNA knockdown, cells were treated with indicated concentrations of erlotinib for another 72 h, and then the growth-inhibitory effects of erlotinib were quantified by WST-1 assay. *, P < 0.01; **, P < 0.05. B, SUM149, BT-474, and SUM190 cells were treated without or with lapatinib for 48 h. The expression levels of total p27 and pS10-p27 were detected by western blot analysis. Beginning 48 h after control or KIS siRNA knockdown, cells were treated with indicated concentrations of lapatinib for another 72 h, and then the growth-inhibitory effects of lapatinib were quantified by WST-1 assay. *, P < 0.001; **, P > 0.05.
We also studied the effect of KIS depletion on the activity of another TKI targeting EGFR. Lapatinib, a dual EGFR/HER2 TKI, was used to treat breast cancer cells with or without KIS siRNA knockdown. First, we tested the anti-EGFR activity of lapatinib in SUM149 cells that express EGFR but not HER2. Use of these cells allowed us to focus on the EGFR-blocking activity of lapatinib. In SUM149 cells, we found that lapatinib treatment increased the expression level of S10-phosphorylated p27 and KIS siRNA knockdown enhanced the anti-proliferative activity of lapatinib, suggesting that other EGFR TKIs can produce the same effects as erlotinib (Fig. 5B). Similar experiments were then performed in breast cancer cell lines that express both EGFR and HER2: BT-474 and SUM190. Interestingly, even though the expression level of S10-phosphorylated p27 increased after lapatinib treatment in both cell lines, KIS siRNA knockdown did not enhance lapatinib’s anti-proliferative activity in either lapatinib-sensitive BT-474 cells (IC50 = 0.046 µM) or lapatinib-resistant SUM190 cells (IC50 = 2.9 µM) (Fig. 5B).
Discussion
In this study, we provided evidence that p27 cytoplasmic localization is associated with erlotinib resistance in breast cancer. Inhibition of S10 phosphorylation of p27 led to p27 nucleus accumulation and enhanced the anti-proliferative effect of erlotinib against breast cancer. Our findings elucidated the molecular mechanisms that explain why EGFR TKIs have minimum activity in breast cancer. Here we report for the first time to use KIS in vivo siRNA delivery technique in combination with erlotinib to enhance the EGFR TKI sensitivity.
p27 is an important negative regulator of the mammalian cell cycle. The activity of p27 depends on the subcellular localization of p27. Even though phosphorylation status of p27 at T157, T187, and S10 plays a role in p27 localization, we found that only S10 p27 phosphorylation increased in response to erlotinib treatment, indicating that S10 phosphorylation is involved in erlotinib resistance in breast cancer. The effects produced by KIS siRNA knockdown on growth and anchorage-independent growth are only partial. Therefore, other mechanisms may contribute to erlotinib resistance. We speculate that other types of phosphorylated p27 may play roles in erlotinib resistance. Our current study involved testing only three phosphorylation sites of p27 (S10, T157, and T187), but other phosphorylation sites of p27 may be important for regulating p27 localization and stability. For example, p27 can be phosphorylated by the oncogenic kinase Src at tyrosine (Y) 74 and Y88, and this phosphorylation reduces the p27-mediated inhibition of cyclin E-CDK2 (33). Phosphorylation of p27 at T198 by Akt leads to p27 cytoplasmic mislocalization (18). Therefore, currently we are also focusing on other phosphorylation sites on p27 to further elucidate the molecular mechanism of EGFR-TKI resistance.
HER2/ErbB2 is another member of the ErbB family of receptor tyrosine kinases. Overactivation of HER2 plays a critical role in the progression of human breast cancer (34). Targeting of HER2 by anti-HER2 antibodies such as trastuzumab is now widely used in the treatment of patients with HER2-positive breast cancer (35). Even though trastuzumab showed relevant clinical activity against HER2-positive breast cancer, the overall rate of response rates to trastuzumab as a single agent is only 15% to 30% (36). Resistance to trastuzumab therapy remains a significant clinical problem. p27 is also a downstream molecule of the HER2 pathway and is involved in trastuzumab resistance. It was reported recently that activation of MET receptor tyrosine kinase contributes to trastuzumab resistance, as Met protects cells against trastuzumab by abrogating p27 induction (37). By inhibiting Akt, trastuzumab can inhibit KIS expression and therefore inhibit KIS-induced nuclear export of p27 (21). Therefore, our study suggests that one way to overcome trastuzumab resistance is to modulate S10 phosphorylation and subcellular localization of p27.
Lapatinib is a dual TKI that inhibits both EGFR and HER2. We previously reported that the major activity of lapatinib in EGFR/HER2-positive breast cancer cells is through its anti-HER2 activity and not through anti-EGFR activity (26). In this study, our results provided further evidence to support this finding. When we used lapatinib to treat SUM149 cells, which express EGFR but not HER2, we found that EGFR blockage by lapatinib was enhanced by KIS depletion. In contrast, KIS depletion did not enhance lapatinib’s activity in cells that expressed both EGFR and HER2, suggesting that the activity of lapatinib in such cells is mainly through HER2.
In the current study, we tested the role of S10 phosphorylation of p27 in erlotinib sensitivity not only in cells that overexpress both EGFR and HER2, but also in cells that overexpress EGFR but not HER2. EGFR is reported to be overexpressed in > 60% of basal-like breast cancers (38, 39), and EGFR gene amplification is also found in a subset of basal-like tumors (40). Basal-like breast cancer often has a so-called triple-negative phenotype—in other words, it lacks expression of estrogen receptor, progesterone receptor, and HER2 (41, 42). Triple-negative breast cancer is the most aggressive form of primary breast cancer, and the majority of these tumors cannot be managed effectively with existing targeted treatments (trastuzumab and hormonal treatments) (43, 44). EGFR expression is associated with early relapse and poor survival in triple-negative breast cancer, suggesting that EGFR might be a promising target in this type of disease (45). Several studies have reported the use of cetuximab, a humanized monoclonal antibody against EGFR, in treatment of the basal-like tumor type (46). Another EGFR TKI, gefitinib, enhanced response to other chemotherapeutic drugs such as carboplatin and docetaxel (47). Therefore, even though erlotinib induces clinical responses in only a small proportion of breast cancer patients, there is still reason to believe that inhibition of the EGFR pathway can have substantial activity against triple-negative breast cancer. Identifying the molecules involved in and necessary for resistance to EGFR TKIs will enable us to develop clinically relevant therapeutic approaches by making EGFR a relevant target for breast cancer. In this study we tested the impact of p27 phosphorylation on erlotinib’s activity in 2 triple-negative EGFR-expressing breast cancer cells MDA-MB-231 and MDA-MB-468. We found that inhibiting S10 phosphorylation of p27 enhanced the sensitivity of erlotinib to these cells, suggesting that this effect is not restricted to a few cell lines but is a universal phenomenon.
It is still unclear whether there is some relationship between pS10-p27 expression and erlotinib sensitivity in breast cancer cells. To study whether erlotinib sensitivity depends on p27 phosphorylation in breast cancer cells, we tested the basal expression levels of pS10-p27 in a panel of breast cancer cell lines including erlotinib-sensitive cell lines SUM149 and KPL-4 (32), erlotinib-moderate-sensitive cell lines SK-BR-3 and BT-474 (25), and erlotinib-resistant cell lines MDA-MB-231 and MDA-MB-468 (25). The basal expression level of pS10-p27 is low in most of the cell lines and is not correlates with erlotinib sensitivity (data not shown). The underlying mechanism linking EGFR and KIS is also not clear. To investigate whether the KIS expression level is regulated by EGFR activation, we activated EGFR by EGF stimulation in BT-474 and SK-BR-3 cells and then tested KIS expression. We found that KIS expression did not change significantly after EGFR activation (Supplementary Fig. S2). In a future study, we will focus on detecting whether KIS can regulate the EGFR pathway.
Acquired resistance to EGFR TKIs in non–small-cell lung cancer commonly occurs after continuous drug administration. It is reported that MET amplification is involved in this acquired resistance (48). Therefore, targeting of MET may enhance the sensitivity of non–small cell lung cancer to EGFR TKIs. However, even though MET was found to be highly activated in cetuximab-resistant cells, inhibition of MET activity did not sensitize cetuximab-resistant cells to cetuximab. The reason why MET inhibition does not restore cetuximab sensitivity is still unknown. Because MET’s ligand hepatocyte growth factor induces cell cycle progression in medulloblastoma cells in a p27- and Cdk2-dependent manner (49), we speculate that modulating p27 directly may be more powerful than MET inhibition to abrogate the acquired resistance to EGFR TKIs and is thus worthy of prospective clinical investigation.
Overall, our study demonstrated that p27 phosphorylation at S10 plays a critical role in breast cancer sensitivity to erlotinib. Combining EGFR TKIs with siRNA knockdown of KIS, which leads to inhibition of S10 phosphorylation of p27, enhanced erlotinib activity both in vitro and in vivo in breast cancer. We expect this combination therapy to be potentially translatable to clinical use, where it may ultimately improve the efficacy of EGFR-TKIs for women with advanced breast cancer that is resistant to EGFR-TKIs.
Supplementary Material
Acknowledgements
We thank OSI Pharmaceuticals, Inc. for providing erlotinib, Dr. Keiichi I. Nakayama (Kyushu University, Japan) for providing pcDNA3-S10A-p27 and pcDNA3-S10D-p27 plasmids, Dr. Elizabeth G. Nabel (National Institutes of Health, Bethesda) for helpful suggestions, Ms. Ping Liu and Mr. Roland Bassett (Department of Biostatistics, The University of Texas M. D. Anderson Cancer Center) for statistical analysis, and Ms. Stephanie P. Deming (Department of Scientific Publications, The University of Texas M. D. Anderson Cancer Center) for editorial assistance.
Grant Support: NIH grant CA123318-01A1 (Naoto T. Ueno) and Susan G. Komen Postdoctoral Fellowship KG091192 (Dongwei Zhang).
Abbreviations List
- KIS
kinase interacting stathmin
- EGFR
epidermal growth factor receptor
- TKI
tyrosine kinase inhibitor
- siRNA
small-interfering RNA
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References
- 1.Berger MS, Gullick WJ, Greenfield C, Evans S, Addis BJ, Waterfield MD. Epidermal growth factor receptors in lung tumours. The Journal of pathology. 1987;152:297–307. doi: 10.1002/path.1711520408. [DOI] [PubMed] [Google Scholar]
- 2.Olayioye MA, Neve RM, Lane HA, Hynes NE. The ErbB signaling network: receptor heterodimerization in development and cancer. Embo J. 2000;19:3159–3167. doi: 10.1093/emboj/19.13.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seshadri R, McLeay WR, Horsfall DJ, McCaul K. Prospective study of the prognostic significance of epidermal growth factor receptor in primary breast cancer. Int J Cancer. 1996;69:23–27. doi: 10.1002/(SICI)1097-0215(19960220)69:1<23::AID-IJC5>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 4.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nature reviews. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 5.Cox G, Jones JL, O'Byrne KJ. Matrix metalloproteinase 9 and the epidermal growth factor signal pathway in operable non-small cell lung cancer. Clin Cancer Res. 2000;6:2349–2355. [PubMed] [Google Scholar]
- 6.Ohsaki Y, Tanno S, Fujita Y, et al. Epidermal growth factor receptor expression correlates with poor prognosis in non-small cell lung cancer patients with p53 overexpression. Oncology reports. 2000;7:603–607. doi: 10.3892/or.7.3.603. [DOI] [PubMed] [Google Scholar]
- 7.Moore MJ, Goldstein D, Hamm J, et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2007;25:1960–1966. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- 8.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 9.Cerra M, Cecco L, Montella M, Tuccillo F, Bonelli P, Botti G. Epidermal growth factor receptor in human breast cancer comparison with steroid receptors and other prognostic factors. Int J Biol Markers. 1995;10:136–142. doi: 10.1177/172460089501000302. [DOI] [PubMed] [Google Scholar]
- 10.Dabbs DJ. Correlations of morphology, proliferation indices, and oncogene activation in ductal breast carcinoma: nuclear grade, S-phase, proliferating cell nuclear antigen, p53, epidermal growth factor receptor, and c-erb-B-2. Mod Pathol. 1995;8:637–642. [PubMed] [Google Scholar]
- 11.Yakes FM, Chinratanalab W, Ritter CA, King W, Seelig S, Arteaga CL. Herceptin-induced inhibition of phosphatidylinositol-3 kinase and Akt Is required for antibody-mediated effects on p27, cyclin D1, and antitumor action. Cancer Res. 2002;62:4132–4141. [PubMed] [Google Scholar]
- 12.Porter PL, Malone KE, Heagerty PJ, et al. Expression of cell-cycle regulators p27Kip1 and cyclin E, alone and in combination, correlate with survival in young breast cancer patients. Nat Med. 1997;3:222–225. doi: 10.1038/nm0297-222. [DOI] [PubMed] [Google Scholar]
- 13.Boehm M, Yoshimoto T, Crook MF, et al. A growth factor-dependent nuclear kinase phosphorylates p27(Kip1) and regulates cell cycle progression. Embo J. 2002;21:3390–3401. doi: 10.1093/emboj/cdf343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang J, Zubovitz J, Petrocelli T, et al. PKB/Akt phosphorylates p27, impairs nuclear import of p27 and opposes p27-mediated G1 arrest. Nat Med. 2002;8:1153–1160. doi: 10.1038/nm761. [DOI] [PubMed] [Google Scholar]
- 15.Shin I, Yakes FM, Rojo F, et al. PKB/Akt mediates cell-cycle progression by phosphorylation of p27(Kip1) at threonine 157 and modulation of its cellular localization. Nat Med. 2002;8:1145–1152. doi: 10.1038/nm759. [DOI] [PubMed] [Google Scholar]
- 16.Viglietto G, Motti ML, Bruni P, et al. Cytoplasmic relocalization and inhibition of the cyclin-dependent kinase inhibitor p27(Kip1) by PKB/Akt-mediated phosphorylation in breast cancer. Nat Med. 2002;8:1136–1144. doi: 10.1038/nm762. [DOI] [PubMed] [Google Scholar]
- 17.Sheaff RJ, Groudine M, Gordon M, Roberts JM, Clurman BE. Cyclin E-CDK2 is a regulator of p27Kip1. Genes Dev. 1997;11:1464–1478. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]
- 18.Motti ML, De Marco C, Califano D, Fusco A, Viglietto G. Akt-dependent T198 phosphorylation of cyclin-dependent kinase inhibitor p27kip1 in breast cancer. Cell Cycle. 2004;3:1074–1080. [PubMed] [Google Scholar]
- 19.Besson A, Gurian-West M, Chen X, Kelly-Spratt KS, Kemp CJ, Roberts JM. A pathway in quiescent cells that controls p27Kip1 stability, subcellular localization, and tumor suppression. Genes Dev. 2006;20:47–64. doi: 10.1101/gad.1384406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodier G, Montagnoli A, Di Marcotullio L, et al. p27 cytoplasmic localization is regulated by phosphorylation on Ser10 and is not a prerequisite for its proteolysis. Embo J. 2001;20:6672–6682. doi: 10.1093/emboj/20.23.6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le XF, Pruefer F, Bast RC., Jr HER2-targeting antibodies modulate the cyclin-dependent kinase inhibitor p27Kip1 via multiple signaling pathways. Cell Cycle. 2005;4:87–95. doi: 10.4161/cc.4.1.1360. [DOI] [PubMed] [Google Scholar]
- 22.Ishida N, Hara T, Kamura T, Yoshida M, Nakayama K, Nakayama KI. Phosphorylation of p27Kip1 on serine 10 is required for its binding to CRM1 and nuclear export. J Biol Chem. 2002;277:14355–14358. doi: 10.1074/jbc.C100762200. [DOI] [PubMed] [Google Scholar]
- 23.Petrovic V, Costa RH, Lau LF, Raychaudhuri P, Tyner AL. FoxM1 regulates growth factor-induced expression of kinase-interacting stathmin (KIS) to promote cell cycle progression. J Biol Chem. 2008;283:453–460. doi: 10.1074/jbc.M705792200. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura S, Okinaka K, Hirano I, et al. KIS induces proliferation and the cell cycle progression through the phosphorylation of p27Kip1 in leukemia cells. Leuk Res. 2008;32:1358–1365. doi: 10.1016/j.leukres.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 25.Yamasaki F, Zhang D, Bartholomeusz C, et al. Sensitivity of breast cancer cells to erlotinib depends on cyclin-dependent kinase 2 activity. Mol Cancer Ther. 2007;6:2168–2177. doi: 10.1158/1535-7163.MCT-06-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang D, Pal A, Bornmann WG, et al. Activity of lapatinib is independent of EGFR expression level in HER2-overexpressing breast cancer cells. Mol Cancer Ther. 2008;7:1846–1850. doi: 10.1158/1535-7163.MCT-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang D, Hirota T, Marumoto T, et al. Cre-loxP-controlled periodic Aurora-A overexpression induces mitotic abnormalities and hyperplasia in mammary glands of mouse models. Oncogene. 2004;23:8720–8730. doi: 10.1038/sj.onc.1208153. [DOI] [PubMed] [Google Scholar]
- 28.Bieche I, Manceau V, Curmi PA, et al. Quantitative RT-PCR reveals a ubiquitous but preferentially neural expression of the KIS gene in rat and human. Brain Res Mol Brain Res. 2003;114:55–64. doi: 10.1016/s0169-328x(03)00132-3. [DOI] [PubMed] [Google Scholar]
- 29.Ueno NT, Yu D, Hung MC. Chemosensitization of HER-2/neu-overexpressing human breast cancer cells to paclitaxel (Taxol) by adenovirus type 5 E1A. Oncogene. 1997;15:953–960. doi: 10.1038/sj.onc.1201250. [DOI] [PubMed] [Google Scholar]
- 30.Landen CN, Jr, Chavez-Reyes A, Bucana C, et al. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 2005;65:6910–6918. doi: 10.1158/0008-5472.CAN-05-0530. [DOI] [PubMed] [Google Scholar]
- 31.Merritt WM, Lin YG, Spannuth WA, et al. Effect of interleukin-8 gene silencing with liposome-encapsulated small interfering RNA on ovarian cancer cell growth. J Natl Cancer Inst. 2008;100:359–372. doi: 10.1093/jnci/djn024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang D, Lafortune TA, Krishnamurthy S, et al. Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor Reverses Mesenchymal to Epithelial Phenotype and Inhibits Metastasis in Inflammatory Breast Cancer. Clin Cancer Res. 2009 doi: 10.1158/1078-0432.CCR-09-0951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chu I, Sun J, Arnaout A, et al. p27 phosphorylation by Src regulates inhibition of cyclin E-Cdk2. Cell. 2007;128:281–294. doi: 10.1016/j.cell.2006.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 35.Stebbing J, Copson E, O'Reilly S. Herceptin (trastuzamab) in advanced breast cancer. Cancer Treat Rev. 2000;26:287–290. doi: 10.1053/ctrv.2000.0182. [DOI] [PubMed] [Google Scholar]
- 36.Montemurro F, Aglietta M. Incorporating trastuzumab into the neoadjuvant treatment of HER2-overexpressing breast cancer. Clinical breast cancer. 2005;6:77–80. doi: 10.3816/CBC.2005.n.011. [DOI] [PubMed] [Google Scholar]
- 37.Shattuck DL, Miller JK, Carraway KL, 3rd, Sweeney C. Met receptor contributes to trastuzumab resistance of Her2-overexpressing breast cancer cells. Cancer Res. 2008;68:1471–1477. doi: 10.1158/0008-5472.CAN-07-5962. [DOI] [PubMed] [Google Scholar]
- 38.Nielsen TO, Hsu FD, Jensen K, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 39.Rakha EA, El-Sayed ME, Green AR, Paish EC, Lee AH, Ellis IO. Breast carcinoma with basal differentiation: a proposal for pathology definition based on basal cytokeratin expression. Histopathology. 2007;50:434–438. doi: 10.1111/j.1365-2559.2007.02638.x. [DOI] [PubMed] [Google Scholar]
- 40.Reis-Filho JS, Pinheiro C, Lambros MB, et al. EGFR amplification and lack of activating mutations in metaplastic breast carcinomas. The Journal of pathology. 2006;209:445–453. doi: 10.1002/path.2004. [DOI] [PubMed] [Google Scholar]
- 41.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 42.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishimura R, Arima N. Is triple negative a prognostic factor in breast cancer? Breast cancer (Tokyo, Japan) 2008;15:303–308. doi: 10.1007/s12282-008-0042-3. [DOI] [PubMed] [Google Scholar]
- 44.Razzak AR, Lin NU, Winer EP. Heterogeneity of breast cancer and implications of adjuvant chemotherapy. Breast cancer (Tokyo, Japan) 2008;15:31–34. doi: 10.1007/s12282-007-0007-y. [DOI] [PubMed] [Google Scholar]
- 45.Nogi H, Kobayashi T, Suzuki M, et al. EGFR as paradoxical predictor of chemosensitivity and outcome among triple-negative breast cancer. Oncology reports. 2009;21:413–417. [PubMed] [Google Scholar]
- 46.Oliveras-Ferraros C, Vazquez-Martin A, Lopez-Bonet E, et al. Growth and molecular interactions of the anti-EGFR antibody cetuximab and the DNA cross-linking agent cisplatin in gefitinib-resistant MDA-MB-468 cells: new prospects in the treatment of triple-negative/basal-like breast cancer. Int J Oncol. 2008;33:1165–1176. [PubMed] [Google Scholar]
- 47.Corkery B, Crown J, Clynes M, O'Donovan N. Epidermal growth factor receptor as a potential therapeutic target in triple-negative breast cancer. Ann Oncol. 2009 doi: 10.1093/annonc/mdn710. [DOI] [PubMed] [Google Scholar]
- 48.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 49.Li Y, Lal B, Kwon S, et al. The scatter factor/hepatocyte growth factor: c-met pathway in human embryonal central nervous system tumor malignancy. Cancer Res. 2005;65:9355–9362. doi: 10.1158/0008-5472.CAN-05-1946. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.