Abstract
Two recent genome-wide association studies reported that single nucleotide polymorphisms (SNPs) in (or near) TERT (5p15), CCDC26 (8q24), CDKN2A/B (9p21), PHLDB1 (11q23), and RTEL1 (20q13) are associated with infiltrating glioma. From these reports it was not clear if the SNP associations predispose to glioma in general or whether they are specific to certain glioma grades or morphologic subtypes. To identify hypothesized associations between susceptibility loci and tumor subtype, we genotyped two case/control groups composed of the spectrum of infiltrating glioma subtypes, and stratified the analyses by type. We report that specific germline polymorphisms are associated with different glioma subtypes. CCDC26 (8q24) region polymorphisms are strongly associated with oligodendroglial tumor risk (rs4295627, OR=2.05, p=8.3*10−11), but not glioblastoma risk. The opposite is true of RTEL (20q13) region polymorphisms which are significantly associated with glioblastoma (rs2297440, OR = 0.56, p= 4.6*10−10) but not oligodendroglial tumor. The SNPs in or near CCDC26 (8q24) are associated with oligodendroglial tumors regardless of combined 1p and 19q deletion status; however, the association is greatest for those with combined deletion (rs4295627, OR=2.77, p=2.6*10-9). These observations generate hypotheses concerning the possible mechanisms by which specific SNPs (or alterations in linkage disequilibrium with such SNPs) are associated with glioma development.
Keywords: Glioma, Genetic Association, 1p/19q deletion
1. Introduction
Gliomas cause significant morbidity and mortality. Approximately 18,500 people in the U.S. are diagnosed with glioma each year. Because most gliomas are biologically aggressive, approximately 12,800 people in the U.S. succumb to these tumors every year[1]. Families with Li Fraumeni, Turcot, and Familial Melanoma/Glioblastoma syndromes [2] have a significantly increased risk of gliomas. However, such family cancer syndromes account for, at most, 5% of all gliomas [2]. Like many diseases, glioma development is hypothesized to be associated with relatively common germline alterations, each with limited penetrance. Two genome-wide association studies (GWAS) reported that single nucleotide polymorphisms (SNPs) in (or near) TERT (5p15), CCDC26 (8q24), CDKN2A/B (9p21), PHLDB1 (11q23), and RTEL1 (20q13) are associated with glioma development [3, 4]. Shete and colleagues analyzed gliomas of a variety of subtypes and identified all five regions [3]. Our group analyzed glioblastoma (GBM) and anaplastic astrocytoma (AA) and identified the CDKN2A/B and RTEL1 associations, and observed the TERT association during the discovery phase but not the replication phase [4]. We hypothesized that the different results were primarily due to varied glioma case mixes in the two studies. Differential histological associations of risk loci have been reported in lung cancer [5], ovarian cancer [6] and myeloproliferative disorders [7]. To test our hypothesis, we genotyped two case/control groups composed of a spectrum of glioma subtypes.
Using 582 cases and 532 controls from the Mayo Clinic for discovery and 864 cases and 602 controls from UCSF for confirmation, we examined the top 13 SNPs in the 5 associated regions from the two published GWAS [3, 4]. Data regarding patients with oligodendroglial tumors (n=390) and grade II astrocytoma (n=103) has not been previously published.
2. Materials and Methods
2.1 Study Populations
Mayo Clinic Case-Control Study
The Mayo Clinic case group included 582 individuals with glioma newly diagnosed between 2005 and 2009. Cases were identified within 24 hours of diagnosis, except for those who had their initial diagnosis elsewhere, followed by verification at the Mayo Clinic. Pathologic diagnosis was confirmed by review of the primary surgical material for all cases by two Mayo Clinic neuropathologists, C.Giannini and B. Scheithauer, based on surgically resected material. The control group consisted of consented individuals who had a general medical exam at the Mayo Clinic and matched to cases based on sex, date of birth (within two and one half years), self-identified race (Hispanic white, non-Hispanic white, American Indian, African American, Asian, Pacific Islander, Other) and residence. Geographic region of residence was matched in three zones based on the distance to the Mayo Clinic Rochester: Olmsted County; the rest of Minnesota, Wisconsin, Iowa, North Dakota and South Dakota; and the rest of the United States and Canada. Excluded were individuals under the age of 18 and those with a prior history of brain tumor. 1p/19q deletion data was obtained from the medical record, or if that was not available, by routine fluorescence in situ hybridization (FISH) analysis [8]. The Mayo Clinic case and control enrollment research protocol was approved by Mayo Institutional Review Board. Mayo Clinic subject characteristics are summarized in Supplemental Table 1.
UCSF Case-Control Study
UCSF cases and controls were taken from the San Francisco Bay Area Adult Glioma Study (AGS). Details of subject recruitment for AGS have been provided previously [9, 10]. Briefly, cases aged 20 or older diagnosed with histologically confirmed incident gliomas (International Classification of Diseases for Oncology, morphology codes 9380–9481) were recruited from the local population-based registry, the Northern California Rapid Case Ascertainment program and the University of California, San Francisco Neuro-oncology clinic between 1991 and 2009. Regarding pathology review, all 191 UCSF cases with oligodendroglial features, were either diagnosed or reviewed by a UCSF neuropathologist or, for one case whose materials were unavailable for review, the pathology report indicated deletion of 1p/19q. 1p/19q deletion data was obtained from the medical record, or if that was not available, by FISH analysis [8]. For the other histologies, 454/520 (87% of) glioblastomas (GBM), 89/95 (94% of) anaplastic astrocytoma, and 56/58 (97% of) grade 2 astrocytoma were either diagnosed or reviewed by a UCSF neuropathologist. Neuropathologists Kenneth Aldape (now at MD Anderson) and Tarik Tihan reviewed most cases used in this study, 49% and 23%, respectively. AGS controls aged 20 years or older from the same residential area as cases were identified using random digit dialing and were frequency matched to cases based on age, sex and ancestry. Consenting participants provided blood and/or buccal specimens and information during an in-person or telephone interview. UCSF subject characteristics are summarized in Supplemental Table 1.
2.2 Genotyping
SNP genotyping was performed on all Mayo Clinic cases and controls, 191 UCSF AGS cases with an oligodendroglial component, and 192 AGS controls using the custom GoldenGate™ Illumina genotyping platform (Illumina, San Diego CA). GoldenGate assays were developed for all 13 SNPs of most interest from the prior two GWAS studies [3, 4] as well as 89 non-redundant SNPS across the associated 8q24 region. Each GoldenGate assay batch included appropriate within-run and between-run controls as well as a large number (86) of additional SNPs. Excluding occasional single SNP failures, all within-run and between-run comparisons were completely concordant. We previously described SNP genotyping and quality control measures for UCSF GBM, anaplastic astrocytoma and astrocytoma grade 2 cases and 602 controls using the Illumina 370duo array panel [4].
2.3 Quality Control and Statistical Methods
Concordance in inter-plate, intra-plate, and overall subject replicates were summarized to investigate potential genotyping error. Subject level call rates were calculated and those subjects (N=12/1497, 0.8%) with call rates <0.9 were excluded from further analysis. Individual SNP call rates were summarized and SNPs (N=4/192, 2%) with call rates <0.9 were excluded from the analysis. The minor allele frequency (MAF) was calculated for each SNP and SNPs (N=3/192, 1.6%) with MAF <0.01 were excluded from further analysis. The above analyses were done on the complete set of data and each analysis was repeated separately for each plate to investigate any potential plate effects. The genotype frequency distribution for each SNP in the controls was tested against the Hardy-Weinberg equilibrium (HWE) using a chi-square test. SNPs with HWE p-values <0.001 were excluded from the analysis, as these may be SNPs with possible genotyping errors. Identity by descent estimates were calculated for all pairs of individuals to assess for relationships among subjects, as completed in PLINK [11]. Assessment of population stratification was completed using STRUCTURE software [12] based on the SNPs on the custom Illumina array, with no apparent population stratification observed. Therefore, association analysis was completed using all subjects. However, it should be noted that the SNPs genotyped on the Illumina array had limited information regarding ancestry. Assessment of each SNP with disease status was computed using logistic regression under a log-additive genetic model with genotypes coded as 0, 1, or 2 copies of the minor allele. Analysis of the association of glioma risk was performed for UCSF and Mayo separately as well as for UCSF and Mayo together. Separate histology and grade specific stratified analyses comparing gliomas to controls were performed for the following glioma groups: grade 2 astrocytoma, anaplastic astrocytoma, GBM, oligodendroglioma, mixed oligo-astrocytoma, oligodendroglioma/mixed oligo-astrocytoma, grade 2 oligodendroglioma/mixed oligo-astrocytoma, grade 3 oligodendroglioma/mixed oligo-astrocytoma, 1p/19q deleted tumors, and 1p/19q intact tumors. 1p/19q deleted versus 1p/19q intact case/case comparisons were also completed. These analyses included both age and gender as covariates in the logistic model. Assessment of genetic heterogeneity between sites was also assessed by including a site by SNP interaction term in the pooled analysis. No significant genetic heterogeneity in SNP effects was detected between the two sites. To adjust for multiple comparisons a Bonferroni approach was used with p-values <4.17*10−3 considered statistically significant.
2.4 Ethics
These studies were approved by the Mayo Clinic Office for Human Research Protection and the University of California San Francisco Committee on Human Research. Informed consent was obtained from all study participants.
3. Results
3.1 Stratification of Germline Risk Alleles by Oligodendroglial components
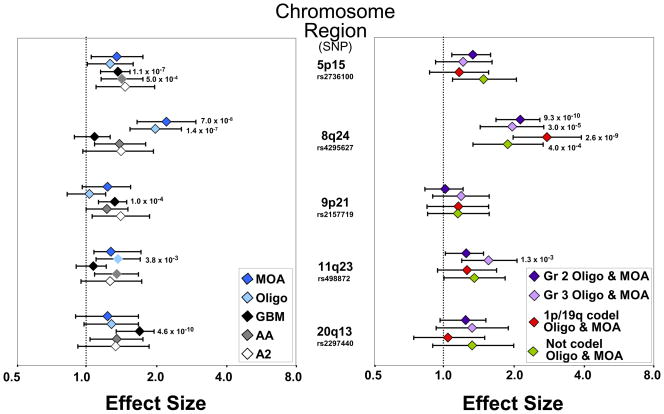
Of the 5 chromosomal regions examined, only 8q24 (CCDC26) was significantly associated with oligodendroglioma or oligoastrocytoma (Figure 1 and Supplemental Table 2). All three of the previously reported 8q24 SNPs were highly significant (e.g. for rs4295627, OR =2.05, 95%CI =1.65–2.54, p= 8.3*10−11). These associations were found in both case/control groups, and in tumors with either oligodendroglioma or mixed oligoastrocytoma (MOA) morphology (Supplemental Table 2) 8q24 polymorphisms were associated with oligodendroglial tumors irrespective of grade or 1p/19q deletion status, although the effect size was highest for co-deleted tumors (for rs4295627 OR=2.77; 95%CI=1.98–3.88). GBM was not associated with 8q24 polymorphisms, while non-GBM astrocytoma showed an association that did not reach statistical significance in our sample (Figure 1 and Supplemental Table 3).
Figure 1. Glioma Risk Stratified by Chromosomal Region and Morphology Type, Oligodendroglioma Grade and 1p/19q Codeletion Status.
The single SNP within each region that was most strongly associated is illustrated. For comparative purposes, some SNP-associated risks (and their 95% confidence limits) have been inverted (see Supplemental Tables 2 and 3). P-values are given for case-control comparisons significant after correction for multiple testing. (MOA=mixed oligoastrocytoma, Oligo=oligodendroglioma, GBM=glioblastoma, AA=anaplastic astrocytoma, A2=astrocytoma grade 2, codel=codeleted)
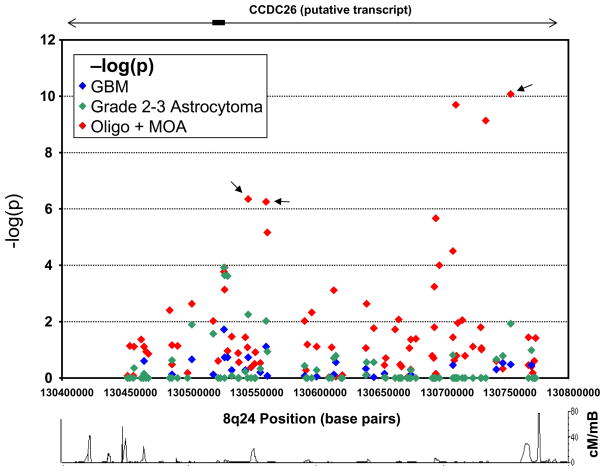
Figure 2 shows fine mapping of loci within the 8q24 region by tumor histologic type. Two peaks of association were noted for oligodendroglial tumors, but not GBM nor lower grade astrocytoma. Potential explanations for the distinct peaks include independent risk loci and/or a recombination event within the at-risk haplotype.
Figure 2. Fine mapping of loci within the 8q24 region by tumor histologic type.
−Log10(p) values for 89 SNPs across the 8q24 region stratified by glioma morphologic subtype. Combined Mayo Clinic and UCSF data are shown. Mayo Clinic glioma cases (N=582) and controls (N=532), and UCSF oligodendroglioma (oligo) and mixed oligoastrocytoma (MOA) cases (N=191) and controls (N=192) were genotyped using the Illumina VeraCode platform. UCSF astrocytic glioma cases (N=673) and controls (602) were evaluated by the Illumina 370duo platform. For these latter gliomas, only 28 SNPs are illustrated. Arrows indicate the SNPs reported by Shete et al[3]. The recombination rate (in cM/mbase) across the region from HapMap release 27 (http://hapmap.ncbi.nlm.nih.gov/) is also shown.
3.2 Stratification of Germline Risk Alleles by Astrocytoma Grade
The other four glioma risk regions demonstrated less specificity than that seen for 8q24, although RTEL region (20q13) risk estimates were strongest for GBM; e.g. for rs2297440, OR = 0.56, 95%CI = 0.47–0.68, p = 4.6*10−10. (Figure 1 and Supplemental Tables 2 and 3). Risk estimates were similar across glioma subtypes for the 5p15 region, although only GBM and AA associations were significant after adjusting for multiple testing. While the 9p21 region was strongly associated with GBM and null for oligodendroglioma, risk intervals for all histologic types overlap. Although the 11q23 glioma risk SNP rs498872 was significantly associated with oligodendroglioma, similar risk estimates, though not statistically significant, were observed for other non-GBM gliomas (Figure 1).
4. Discussion
We show that 8q24 polymorphisms were associated with gliomas containing an oligodendroglial component but not with GBM. Conversely, the 5p15, 9p21 and 20q13 regions were associated with GBM risk, but were not strongly associated with oligodendroglial tumors. This pattern supports the current model of glioma initiation and progression. For example, since 8q24 associations are seen for risk of oligodendroglioma regardless of 1p/19q deletion, as well as potentially with non-GBM astrocytoma, we predict 8q24 SNPs may be associated with gliomas containing IDH1/2 mutations [13]. Indeed, it is plausible to hypothesize that 8q24 alterations may facilitate the acquisition of IDH mutations or interact with IDH mutation to facilitate tumorigenesis [14]. We also predict that the 8q24 SNPs will be associated with secondary GBM and/or the CpG island methylator phenotype [15]. Unfortunately, we had limited case numbers and therefore insufficient statistical power to generate meaningful data for grade 2 astrocytoma and secondary GBM.
The associated 8q24 region in glioma is proximal to the regions near cMYC associated with the development of other cancers [16, 17]. It is unknown if the other tumors with 8q24 association are of different morphologic subtype or grade. Our glioma results suggest that 8q24 associations with risk in the other tumors may also be linked to lower grade and/or less aggressive behavior. The 8q24 region – including cMYC – is frequently gained or amplified in many cancers (including gliomas); the amplicon often includes CCDC26 [17]. It is not known whether the SNPs within 8q24 are associated with somatic gain of 8q24.
SNPs within or near the TERT (5p15), CDKN2A/B (9p21), and RTEL1 (20q13) regions were most strongly associated with GBMs. TERT is a component of the telomerase system, which maintains telomere length. RTEL1 has been hypothesized to be involved in the resolution of D-loops that occur during homologous recombination [18] and, like TERT, has a role in regulating telomere length [19]. Other genes in the 20q13 region near RTEL1 may also predispose to aneusomy. STMN3, which facilitates tubulin depolymerization and is regulated during mitosis [20] is adjacent to RTEL1. STMN3 levels are high in cancer and in brain [21, 22]. Germline alterations in STMN3 might affect tubulin binding and thus affect mitotic segregation. The CDKN2A/B region contains several tumor suppressor genes; deletions in this region are seen in hereditary glioma and melanoma syndromes [2].
In summary, we show for the first time that specific germline alterations predispose to gliomas with different morphologic subtypes, histologic grades, and somatic genetic alterations. Further studies will be necessary to determine if the known or additional genomic risk regions are associated with other morphologic or molecular genetic subgroups of glioma
Supplementary Material
Acknowledgments
We thank the University of Georgia Genome Center and the Mayo Clinic Genotyping Core for performing the custom SNP analyses. uIn addition to C. Gianinni, we thank B. Scheithauer for his careful histological review of all the gliomas collected at the Mayo Clinic. The San Francisco Adult Glioma Study thanks the Northern California Cancer Center for glioma patient case finding; we also thank Kenneth Aldape for pathology review and the pathology departments of Alexian, Alta Bates, Brookside, California Pacific, Doctors Pinole, Eden, El Camino, Good Samaritan, Highland, John Muir, Kaiser Redwood City, Kaiser San Francisco, Kaiser Santa Teresa, Los Gatos, Los Medanos, Marin General, Merrithew, Mills Peninsula, Mt. Diablo Hospital, Mt. Zion, Naval Hospital, O’Connor, Ralph K Davies, Saint Louise, San Francisco General, San Jose, San Leandro, San Mateo County, San Ramon Valley, Santa Clara Valley, Sequoia, Seton, St. Francis, St. Luke’s, St. Rose, Stanford, Summit, UC San Francisco, Valley Livermore, Veterans Palo Alto, Veterans SF, and Washington Hospitals and Medical Centers for providing tumor specimens for review. We used HapMap data to generate the recombination frequencies surrounding the associated 8q24 region. We thank the International HapMap Consortium (A second generation human haplotype map of over 3.1 million SNPs. Nature 449, 851–861. 2007.), and we thank the people in the Utah CEPH community who allowed the samples they donated earlier to be used by the International HapMap Consortium.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Central Brain Tumor Registry of the United States. 2010 [cited 2010 05/25/2010]; Available from: http://www.cbtrus.org/reports/reports.html.
- 2.Bahuau M, Vidaud D, Jenkins RB, Bieche I, Kimmel DW, Assouline B, et al. Germ-line deletion involving the INK4 locus in familial proneness to melanoma and nervous system tumors. Cancer Res. 1998;58(11):2298–303. [PubMed] [Google Scholar]
- 3.Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41(8):899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wrensch M, Jenkins RB, Chang JS, Yeh RF, Xiao Y, Decker PA, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41(8):905–8. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Landi MT, Chatterjee N, Yu K, Goldin LR, Goldstein AM, Rotunno M, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85(5):679–91. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song H, Ramus SJ, Tyrer J, Bolton KL, Gentry-Maharaj A, Wozniak E, et al. A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2. Nat Genet. 2009;41(9):996–1000. doi: 10.1038/ng.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldin LR, Bjorkholm M, Kristinsson SY, Samuelsson J, Landgren O. Germline and somatic JAK2 mutations and susceptibility to chronic myeloproliferative neoplasms. Genome Med. 2009;1(5):55. doi: 10.1186/gm55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith JS, Perry A, Borell TJ, Lee HK, O’Fallon J, Hosek SM, et al. Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J Clin Oncol. 2000;18(3):636–45. doi: 10.1200/JCO.2000.18.3.636. [DOI] [PubMed] [Google Scholar]
- 9.Felini MJ, Olshan AF, Schroeder JC, Carozza SE, Miike R, Rice T, et al. Reproductive factors and hormone use and risk of adult gliomas. Cancer Causes Control. 2009;20(1):87–96. doi: 10.1007/s10552-008-9220-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wrensch M, McMillan A, Wiencke J, Wiemels J, Kelsey K, Patoka J, et al. Nonsynonymous coding single-nucleotide polymorphisms spanning the genome in relation to glioblastoma survival and age at diagnosis. Clin Cancer Res. 2007;13(1):197–205. doi: 10.1158/1078-0432.CCR-06-1199. [DOI] [PubMed] [Google Scholar]
- 11.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–59. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan H, Bigner DD, Velculescu V, Parsons DW. Mutant metabolic enzymes are at the origin of gliomas. Cancer Res. 2009;69(24):9157–9. doi: 10.1158/0008-5472.CAN-09-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174(4):1149–53. doi: 10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–22. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiemeney LA, Thorlacius S, Sulem P, Geller F, Aben KK, Stacey SN, et al. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat Genet. 2008;40(11):1307–12. doi: 10.1038/ng.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsuchiya N, Slezak JM, Lieber MM, Bergstralh EJ, Jenkins RB. Clinical significance of alterations of chromosome 8 detected by fluorescence in situ hybridization analysis in pathologic organ-confined prostate cancer. Genes Chromosomes Cancer. 2002;34(4):363–71. doi: 10.1002/gcc.10064. [DOI] [PubMed] [Google Scholar]
- 18.Barber LJ, Youds JL, Ward JD, McIlwraith MJ, O’Neil NJ, Petalcorin MI, et al. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008;135(2):261–71. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, et al. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138(1):90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charbaut E, Curmi PA, Ozon S, Lachkar S, Redeker V, Sobel A. Stathmin family proteins display specific molecular and tubulin binding properties. J Biol Chem. 2001;276(19):16146–54. doi: 10.1074/jbc.M010637200. [DOI] [PubMed] [Google Scholar]
- 21.Ozon S, Byk T, Sobel A. SCLIP: a novel SCG10-like protein of the stathmin family expressed in the nervous system. J Neurochem. 1998;70(6):2386–96. doi: 10.1046/j.1471-4159.1998.70062386.x. [DOI] [PubMed] [Google Scholar]
- 22.Singer S, Malz M, Herpel E, Warth A, Bissinger M, Keith M, et al. Coordinated expression of stathmin family members by far upstream sequence element-binding protein-1 increases motility in non-small cell lung cancer. Cancer Res. 2009;69(6):2234–43. doi: 10.1158/0008-5472.CAN-08-3338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.