Abstract
Background
Sequential treatment with azacitidine can induce re-expression of epigenetically silenced genes through genomic DNA hypomethylation and reverse carboplatin resistance of epithelial ovarian cancer cells. We initiated a phase Ib-IIa clinical trial of this sequential combination of azacitidine and carboplatin in platinum-resistant or refractory epithelial ovarian cancer.
Methods
Patients with pathologically confirmed intermediate- or high-grade epithelial ovarian cancer who had disease progression within 6 months (resistant, n = 18) or during a platinum-based therapy (refractory, n = 12) were eligible. All patients had measurable disease.
Results
Thirty patients received a total of 163 cycles of treatment. This regimen produced 1 CR, 3 PR (ORR: 13.8%), and 10 SD among 29 evaluable patients. For those who achieved clinical benefits, the median duration of the treatment was 7.5 months. The median PFS and OS for all patients were 3.7 months and 14 months, respectively. Patients with platinum resistant disease achieved an ORR of 22%, with a median PFS of 5.6 months and a median OS of 23 months. The predominant toxicities were fatigue and myelosuppression. Correlative studies showed that DR4 methylation in peripheral blood leukocytes was decreased during treatment in 3 of 4 objective responders (75%), but in only 5 of 13 non-responders (38%).
Conclusions
To our knowledge, this study provides the first clinical evidence that a hypomethylating agent may partially reverse platinum resistance in ovarian cancer. Further clinical evaluation of hypomethylating agents in combination with carboplatin is warranted.
DNA methylation plays an essential role in regulating normal biologic processes as well as carcinogenesis1. Methylation of DNA is a heritable, DNA methyltransferase-induced modification of DNA structure that does not alter the specific sequence of base pairs responsible for encoding the genome, but can directly inhibit gene expression2. Two patterns of DNA methylation have been observed in cancer cells3: global hypomethylation across the genome, and localized hypermethylation at specific CpG islands within the gene promoter regions of certain genes. Decreased methylation due to global hypomethylation may permit the expression of previously quiescent proto-oncogenes and pro-metastatic genes and promote tumor progression. Alternatively, an aberrant increase in methylation patterns at previously unmethylated sites, such as the promoter regions of tumor suppressor genes, may result in transcriptional silencing and an inability to control tumor development4-8.
Methylation microarray analyses of late-stage ovarian cancers identified two distinct groups based on tumor methylation levels which appeared to have prognostic significance9. Progression-free survival after chemotherapy was significantly shorter for patients with higher levels of methylation (8 months or less) than those with lower levels (12 months or greater; p< 0.001), suggesting that a higher degree of CpG island methylation is associated with early recurrence and/or chemotherapy resistance.
Azacitidine is a hypomethylating agent that has been shown to induce the re-expression of hMLH1 in platinum-resistant ovarian cancer cell lines, leading to re-sensitization of treated cells to carboplatin10. Studies with clonogenic assays as well as human tumor xenografts have shown that treatment of platinum resistant ovarian cancer cells with hypomethylating agents such as decitabine or azacitidine increases sensitivity to platinum compounds, including carboplatin and cisplatin, in addition to other chemotherapeutic agents11, 12. Our preclinical studies revealed that in platinum resistant ovarian cancer cells, sequential treatment with azacitidine followed by carboplatin produced synergistic cytotoxicity. Additionally, azacitidine enhanced the sensitivity of platinum-resistant ovarian cancer cells to carboplatin associated with a DR4-mediated caspase 8-dependent apoptosis13. In this trial, we have assessed the safety and efficacy of a sequential regimen, in which azacitidine has been used to reverse resistance to carboplatin in patients with platinum resistant or refractory epithelial ovarian cancer. Choice of this design is based on the fact that platinum is the most effective chemotherapeutic agent in the treatment of human epithelial ovarian cancer14-17, on previous clinical experience that reversal of epigenetic changes can overcome chemotherapy resistance in other tumor types18, 19 and on our own preclinical data with ovarian cancer cell lines.
MATERIALS AND METHODS
Eligibility Criteria
Patients were eligible to participate in this trial if they had a pathologically confirmed diagnosis of intermediate- or high-grade epithelial cancers of the ovary, fallopian tube or peritoneum, which were considered platinum refractory (progression on or persistent disease following platinum-based therapy) or platinum resistant (progression within 6 months of a platinum-based regimen). Patients were at least 18 years of age and had measurable disease by imaging studies that had progressed within 3 months of study entry. All participants had an ECOG performance status of 2 or better. Additional eligibility criteria included adequate bone marrow function (absolute neutrophil count greater than 1,500 /μl, hemoglobin greater than 9.0 g/dL and platelet count greater than 75,000 /μl), renal function (serum creatinine less than 1.5 mg/dL or a calculated creatinine clearance of at least 60 mL/minute), and hepatic function (serum total bilirubin less than 2.0 mg/dL, and ALT less than 3 fold of the upper limit of normal). Patients were excluded if they had advanced hepatic metastases that occupied greater than 75% of the hepatic parenchyma or if they had undergone high dose chemotherapy for ovarian cancer.
Study Design
This study was a prospective open-label phase Ib-IIa clinical trial of a sequential regimen using azacitidine to reverse resistance to carboplatin in patients with platinum resistant or refractory epithelial ovarian cancer. The study was conducted at M D Anderson Cancer Center after the approval by the Institutional Review Board (IRB). All patients had to give written informed consent prior to study entry using forms approved by the IRB. This study included a dose-escalation phase based on the classical “3 + 3” design and an expansion phase at the recommended phase 2 dosages. The primary goal of this study was to define safety and clinical responses of this regimen.
Treatment Plan
Treatment was administered on an outpatient basis at M D Anderson Cancer Center. Patients received azacitidine at 75 mg/m2 subcutaneously daily for 5 days20 and carboplatin at either AUC 4 or AUC 5 intravenously over 1 hour on day 2 every 28 days. Chemotherapeutic agents were reconstituted according to the manufacturers’ manuals. All patients received antiemetics according to best local practice. Patients with evidence of response or without disease progression after the first 8-week treatment were eligible to receive further treatment until prohibitive toxicity or tumor progression.
Dose limiting toxicity (DLT) was defined as platelets less than 20,000 /μl, absolute neutrophil count less than 500 /μl for more than 7 days, neutropenic fever, or more than 7 days of delay in initiation of the next cycle at 100% dosage because of inadequate hematological parameters, as well as any grade 3 or greater non-hematological toxicity other than nausea, vomiting, or fatigue occurring during the first cycle of treatment. Carboplatin dosage was calculated using the Calvert formula = AUC × (GFR +25), in which glomerular filtration rate (GFR) was calculated by stable serum creatinine based on the Cockcroft-Gault equation21, 22.
Safety and Efficacy Evaluation
All patients underwent evaluation including complete medical history, physical examination, and laboratory tests. Radiographic imaging studies (helical CT or MRI scan) were obtained to assess measurable disease while serum CA125 was obtained to assess biochemical responses. Prior to each treatment, laboratory assessment, physical examination, functional status, toxicities and concomitant medications were documented. The severity of adverse events was graded according to the Common Terminology Criteria for Adverse Events v3.0.
Efficacy end points included response rate, progression-free survival (PFS), and overall survival (OS). All patients were observed until June, 2008 when this protocol was closed, or until death. The WHO criteria of complete response (CR), partial response (PR), stable disease (SD) and progressive disease (PD) were used to characterize tumor responses. Tumor size was determined by the product of two perpendicular diameters of marker lesions applied at the widest portion of the tumor. All of the measurable lesions were evaluated every two cycles. Serum CA125 levels were measured at baseline and before each cycle. The CA125 response criteria of 50% (four samples) and 75% (three samples) reduction were used as a supplementary assessment of possible antitumor activity. In order to be evaluable for treatment response, a patient must have received at least 2 cycles of treatment.
DNA extraction and methylation analysis
Blood samples from patients were collected into EDTA-vials at enrollment and before each cycle of treatment. Plasma was separated by centrifugation and white cells were separated on Ficoll-Hypaque gradients and stored at −20°C until analyzed.
DNA was extracted from peripheral blood mononuclear cells (PBMC) obtained from patients at different time points before and after treatment using standard phenol-chloroform extraction. It should be noted that this will represent DNA from lysed normal blood cells as well as tumor cells23. Methylation analysis was done using a methylation kit (EZ-96 gold; Zymo Research, Orange, CA). MethPrimer software was used for the prediction of CpG island of DR4 (ACCESSION EF064713; GI: 117606477) and design of methylation specific primers. The sequence of primers for methylated DR4 promoter was forward, TTGGAGCGTAATGGTTTTATTTC; reverse, AATACCTATAATCCCAACCACTCG, and that for unmethylated DR4 promoter was forward, GTTGGAGTGTAATGGTTTTATTTTG; reverse, AATACCTATAATCCCAACCACTCAA. The Methylation specific PCR (MSP) conditions were 94°C for 5 minutes with hot start, then 94°C for 30 seconds, 58°C or 60°C for 30 seconds, and 72°C for 1 minute repeated for 40 cycles. Universal methylated and unmethylated control DNAs were used for the positive control (Chemicon International). All MSP PCRs were repeated twice separately. Methylated DR4 was normalized by both unmethylated DR4. Image analysis (Scion Image for Windows) was used for semi-quantitative measurement of methylated and unmethylated DR4. We defined at least 10% of DR4 methylation changes as a cut-off value.
The hMLH1 methylation status in plasma DNA was determined by methylation specific PCR using a CPGWIZ hMLH1 amplification kit (Chemicon International) based on the manufacture’s instruction.
Statistical Considerations
This study is divided into 2 parts: dose-escalation was based on the standard 3+3 design and dose expansion was based on the Simon’s optimal two stage designs to enroll 27 patients assuming p0=20%, p1=35%, α=0.1 and β=0.1. Descriptive summary statistics were described as estimated proportions with 95% confidence intervals. Continuous variables not meeting the assumptions of normality (Shapiro-Wilk test), as well as, non-parametric data, were compared using the Fisher’s exact test. The intent-to-treat population was defined as all patients who were enrolled and received at least one partial dose of therapy while the evaluable population was defined as patients who completed at least two cycles of therapy. WHO criteria for efficacy were used to estimate overall response rate (ORR = complete response + partial response). All efficacy and safety analyses were conducted on the intent-to-treat (ITT) basis. Progression-free survival (PFS) and overall survival (OS) were estimated using the Kaplan-Meier method and log-rank test. PFS was defined as the interval from the first day of treatment with study drug to documented disease progression or death due to any cause while the patient was on study or during the long-term follow-up period. OS was defined as the time period from the first day of treatment with study drug to death. The association between the DR4 methylation in responders and non-responders was analyzed by using χ2 test.
RESULTS
Patient Characteristics
A total of 30 patients (median age, 63 years; range, 37 to 73 years) who met the inclusion and exclusion criteria were recruited onto this study. Among these patients, 7 patients were enrolled during the dose escalation phase with 1 early withdrawal, and 23 patients during the expansion phase. Twenty-nine patients had received 2 or more cycles of therapy, while one patient voluntarily withdrew from the study during dose escalation and was replaced. She was evaluable for toxicity since she completed one cycle of therapy. Characteristics of these patients are listed in Table 1. It was noted that 18 patients had platinum resistant and 12 had platinum refractory ovarian cancer. Twenty patients had received 3 or more types of systemic chemotherapy.
Table 1.
Patients’ Characteristics
| Number of patients (%) | |
|---|---|
| Age | |
| 63 yo (37-73) | |
| Pathology | |
| Serous | 18 (60) |
| Clear cell | 1 (3.3) |
| Endometrioid | 1 (3.3) |
| Mucinous | 1 (3.3) |
| Transitional | 1 (3.3) |
| Undifferentiated | 1 (3.3) |
| Mixed | 7 (23.3) |
| Initial Stage | |
| IB | 1 (3.3) |
| IIC | 1 (3.3) |
| IIIC | 21 (70) |
| IV | 7 (3.3) |
| Prior Treatment | |
| 1 regimen | 2 (6.6) |
| 2 regimen | 8 (26.7) |
| 3 regimen | 6 (20) |
| 4 regimen | 7 (23.3) |
| 5 regimen | 4 (13.3) |
| 6 regimen | 1 (3.3) |
| 8 regimen | 2 (6.6) |
| Platinum Sensitivity | |
| Resistant | 18 (60) |
| Refractory | 12 (40) |
Antitumor Activity
Of 29 evaluable patients who received 2 or more cycles of treatment, 11 received 6 or more cycles of treatment and 6 patients received 10 or more cycles of treatment. One patient discontinued treatment due to intermittent rectal bleeding caused by tumor necrosis after having completed 21 cycles. The ORR was 13.8% (4/29; 95% CI, 10.1%-17.5%): 1 patient achieved a clinical CR, 3 patients achieved clinical PR, and 10 patients had stable disease as shown in Table 2. For those who achieved clinical benefits, the median duration of the treatment was 7.5 months. The median progression-free survival (PFS) was 3.7 months while the median overall survival (OS) was 14 months (Figure 1). Among 27 patients who were eligible for CA125 response evaluation, 5 patients achieved a complete response, 6 patients achieved a partial response and 10 patients had stable disease. Though not pre-planned, subgroup analyses revealed that patients with platinum resistant disease achieved the ORR of 22%, the median PFS of 5.6 months and the median OS of 23 months, where patients with platinum refractory disease achieved the ORR of 0%, the median PFS of 1.9 months and the median OS of 10 months.
Table 2.
Major Characteristics of Individual Patients
| Patient Access |
Age | Platinum Resistant or Refractory |
Platinum free interval (months) |
Treatment free interval (months) |
Best Response by WHO |
Best Response by CA125 |
Progression free survival (Month) |
Survival (Month) |
|---|---|---|---|---|---|---|---|---|
| 1 | 51 | Resistant | 11 | 5 | PR | CR | 10 | 23 |
| 2 | 55 | Refractory | 3 | 1 | NE | NE | 1 | 3 |
| 3 | 45 | Resistant | 10 | 5 | SD | NE | 6 | 41 |
| 4 | 68 | Resistant | 13 | 3 | PD | SD | 2 | 7 |
| 5 | 62 | Resistant | 12 | 3 | SD | PR | 6 | 26 |
| 6 | 48 | Refractory | 1 | 1 | PD | PD | 2 | 5 |
| 7 | 44 | Refractory | 10 | 1 | PD | PR | 2 | 13 |
| 8 | 61 | Resistant | 14 | 5 | SD | CR | 10 | 21+ |
| 9 | 61 | Resistant | 9 | 4 | SD | PR | 5 | 24 |
| 10 | 69 | Resistant | 11 | 6 | PD | SD | 2 | 10 |
| 11 | 52 | Refractory | 10 | 1 | PD | SD | 2 | 18 |
| 12 | 69 | Resistant | 6 | 1 | SD | SD | 6 | 25+ |
| 13 | 63 | Refractory | 6 | 1 | PD | SD | 2 | 10 |
| 14 | 68 | Resistant | 32 | 4 | SD | PR | 4 | 37+ |
| 15 | 63 | Refractory | 10 | 1 | PD | SD | 4 | 5 |
| 16 | 66 | Resistant | 8 | 4 | PR | CR | 10 | 36 |
| 17 | 57 | Refractory | 6 | 1 | PD | SD | 2 | 14 |
| 18 | 62 | Resistant | 15 | 3 | CR | CR | 21 | 30 |
| 19 | 55 | Resistant | 31 | 3 | PR | CR | 16 | 36+ |
| 20 | 37 | Resistant | 7 | 1 | PD | SD | 2 | 4 |
| 21 | 64 | Resistant | 8 | 2 | PD | PD | 4 | 6 |
| 22 | 65 | Refractory | 7 | 1 | PD | PD | 2 | 3 |
| 23 | 51 | Refractory | 6 | 1 | PD | PD | 2 | 8 |
| 24 | 55 | Resistant | 6 | 2 | PD | PD | 2 | 11 |
| 25 | 58 | Resistant | 12 | 1 | SD | PD | 7 | 14 |
| 26 | 65 | Resistant | 14 | 1 | SD | PR | 14 | 32+ |
| 27 | 41 | Refractory | 6 | 3 | PD | PR | 2 | 2 |
| 28 | 73 | Resistant | 23 | 5 | SD | SD | 2 | 6 |
| 29 | 65 | Refractory | 1 | 1 | PD | SD | 2 | 6 |
| 30 | 48 | Refractory | 4 | 1 | SD | NE | 9 | 14 |
Abbreviation: CR: complete remission; PR: partial remission; SD: stable disease; PR: progressive disease; NE: not evaluable; and + means that patients were alive upon the last follow up.
Figure 1.
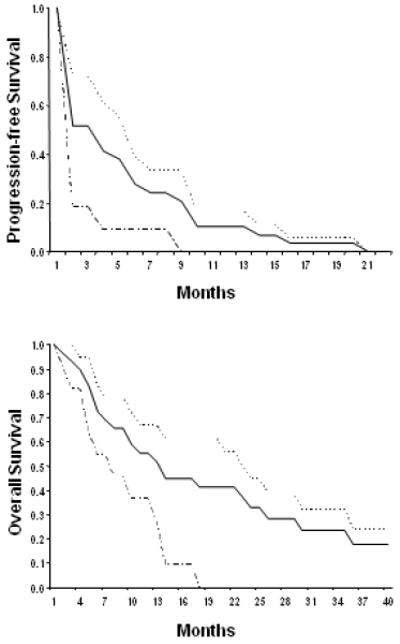
Progression-free survival and overall survival for ovarian cancer patients treated with azacitidine and carboplatin (Kaplan-Meier curves and log-rank test). Three groups of patients were analyzed: — indicated all patients (n=29), ------ indicated platinum-resistant patients (n=18), and ------ indicated platinum-refractory patients (n=11).
Toxicity
All 30 patients were evaluable for toxicity. No DLT or treatment-related deaths were observed. The most common adverse events included fatigue and myelosuppression. Grade 2 or higher toxicities are summarized in Table 3. Side effects included neutropenia, anemia, fatigue, nausea, and pain/irritation at the injection sites. No greater toxicities were observed with sequentially administered azacitidine and carboplatin than would be expected from single-agent carboplatin based on historical experiences in similar cohort of patients.
Table 3.
Grade 2 or Greater Toxicity Profile observed (No DLTs were observed)
| Toxicities | Grade 2 | Grade 3 | Grade 4 |
|---|---|---|---|
| Leukopenia | 16.7% | 20.0% | 0 |
| Neutropenia | 20.0% | 13.3% | 3.3% |
| Thrombocytopenia | 10.0% | 6.7% | 3.3% |
| Anemia | 33.3% | 0 | 0 |
| Nausea/Vomiting | 30.0% | 6.7% | 0 |
| Constipation | 20.0% | 3.3% | 0 |
| Fatigue | 33.3% | 30.0% | 0 |
| Pain | 43.3% | 13.3% | 0 |
| Neuropathy | 3.3% | 0 | 0 |
| Infection | 0 | 3.3% | 0 |
| Metabolic | 3.3% | 0 | 0 |
| Hepatic enzymes | 0 | 3.3% | 0 |
| Alopecia | 20.0% | 0 | 0 |
| Mucositis | 3.3% | 0 | 0 |
| Rash | 3.3% | 0 | 0 |
DR4 methylation in PBMC and hMLH1 in Plasma DNA
To define changes in DNA methylation, DR4 and hMLH1 methylation levels were determined using MSP of DNA extracted from PBMC sampled from patients before and after carboplatin and azacitidine treatment or plasma DNA from patients before carboplatin and azacitidine treatment.. When the dynamic changes of DR4 methylation in PBMC were analyzed in an objective responder (Figure 2), we found that DR4 methylation decreased slightly during the first cycle, reached a nadir during the second cycle by about 50%, and then increased slightly after several cycles of treatment. In contrast, no difference in hMLH1 methylation of plasma DNA was observed between objective responders and non-responders (Figure 3). In these heavily pretreated patients, hMLH methylation was found in 11 patients (3, 5, 7, 9, 10, 12, 13, 18, 20, 23, and 30) out of 26 patients examined (42%), supporting the hypothesis that chemotherapy increased hMLH1 methylation23.
Figure 2.
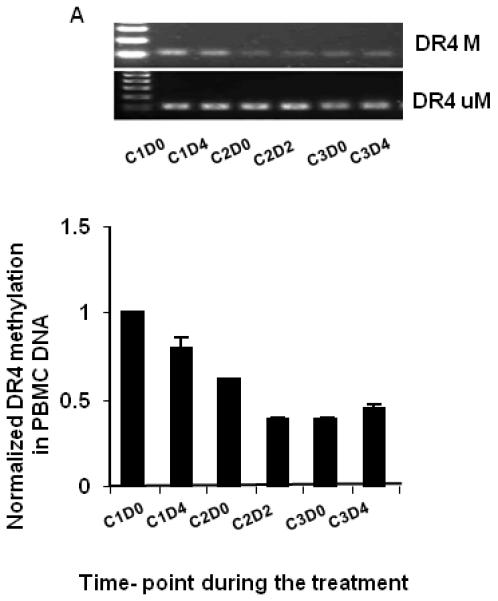
Dynamic changes in DR4 methylation in PBMC DNA from one objective responder (patient 18) before and after azacitidine and carboplatin detected by methylation specific PCR (A) and semi-quantitative analysis with NIH image tool (B). C: cycle; D: day. M: methylated DR4; U: un-methylated DR4.
Figure 3.
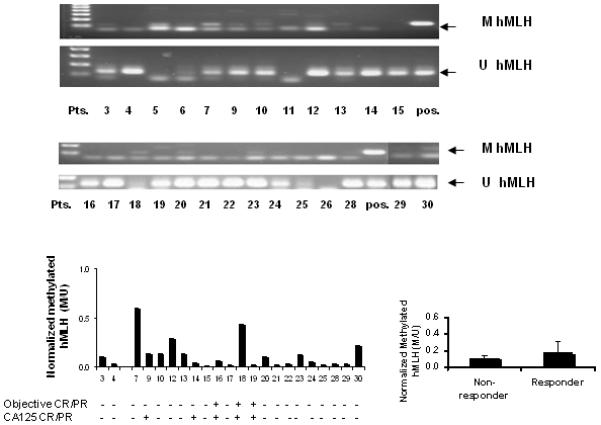
Basal level of hMLH methylation in plasma DNA from ovarian cancer patients before treatment with azacitidine and carboplatin. A: Methylation PSR; B and C: Semi-quantitative measurement of normalized methylated hMLH.
Among the 4 objective responders, 3 patients (75%) had a mean decrease of 40% (range 23-62%) in DR4 DNA methylation relative to baseline levels as detected by image analysis of methylated and unmethylated DR4 (Figure 4). Only 5 of 13 non-responders (38%) displayed similar decreases in the DR4 methylation levels, suggesting that DR4 hypomethylation is more frequently seen in the patients who responded to azacitidine followed by carboplatin.
Figure 4.
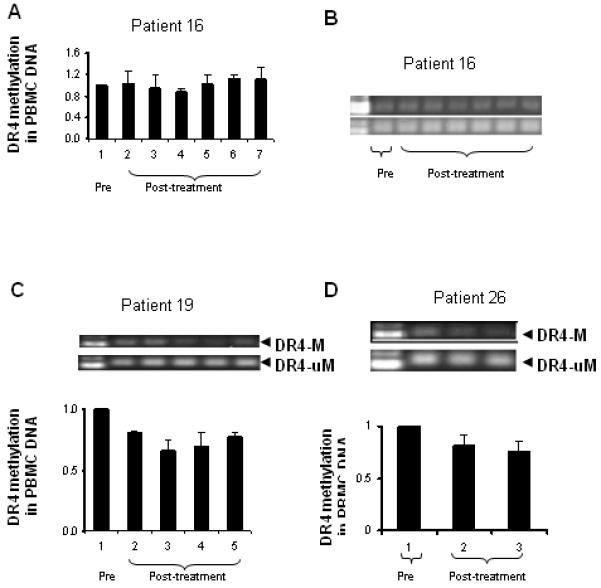
Dynamic changes in DR4 methylation in PBMC DNA from other three objective responder (patients 16, 19 and 26) before and after azacitidine and carboplatin detected by methylation specific PCR. A and B: patient 16; C: patient 19; and D: patient 26.
DISCUSSION
This trial provides the first clinical evidence that a hypomethylating agent may be able to partially reverse platinum resistance in ovarian cancer. In addition, this study includes several interesting observations. First, several cycles of sequential therapy were required in order to reverse carboplatin resistance, which was supported by our observation that patients with platinum refractory epithelial ovarian cancer achieved no clinical response and were removed from the study after 2 cycles of treatment for tumor progression. Second, patients who achieved clinical benefit displayed mixed responses, suggesting tumor and response heterogeneity to the hypomethylating agent. The third observation is that hepatic metastases generally failed to respond to this sequential regimen, which might be caused by hepatic inactivation of azacitidine. Finally, no additional chemotherapy-related toxicities were observed in this small cohort of patients indicating that the low-dose of azacitidine sufficient to induce target hypomethylation did not augment the toxicity profile of chemotherapeutic agents when used in combination.
Platinum resistant or refractory epithelial ovarian cancer was chosen as a model for the following 4 reasons: advanced epithelial ovarian cancer displays multiple hypermethylation phenotypes involving known tumor suppressor genes; carboplatin has significant efficacy in treating ovarian cancer; however, single-agent carboplatin induces minimal to no clinical response in relapsed platinum resistant and refractory ovarian cancer patients; and pretreatment with hypomethylating agents is able to reverse platinum resistance in platinum-resistant epithelial ovarian cancer cell models24-28.
The first retrospective study conducted at M. D. Anderson Cancer Center revealed that carboplatin retreatment resulted in a 21% partial response rate in 33 platinum resistant epithelial ovarian cancer patients (79% had a platinum-free interval of at least 12 months and 2 patients were potentially sensitive to platinum). No patients with a platinum-free interval of shorter than 12 months had an objective response29. Interestingly, a recent retrospective study at the same center identified 34 similar patients who received carboplatin retreatment. The median platinum-free interval from the time platinum was last received to re-treatment with carboplatin was 15.2 months. Only 2 patients achieved a partial response (5.9%) while 21 patients achieved stable disease (61.7%)30. Another retrospective study from Memorial Sloan-Kettering Cancer Center identified 30 platinum-resistant ovarian cancer patients with median platinum-free interval of 16.2 months from July 1997 to June 2001. At the time of platinum retreatment, 20 patients received single-agent platinum and 10 received platinum combinations with newer agents. Only 2 out of 21 patients achieved partial responses (9.5%) based on RECIST criteria by CT scans31. Moreover, in a phase 1 trial of bortezomib and carboplatin in patients with platinum resistant ovarian cancer, 8 of 18 patients had stable disease while 10 patients had progression disease32. Taken together, these data suggest that retreatment with carboplatin induces a very small fraction of objective response (less than 10%). In the current study, treatment with azacitidine and carboplatin achieved 22% objective responses based on the WHO criteria in patients with platinum resistant ovarian cancer while no objective responses were observed in patients with platinum refractory ovarian cancer. It was noted that 2 out of 4 responders had a platinum-free interval of less than 12 months.
The pathogenesis of ovarian carcinoma is poorly defined partly due to a lack of a tumor progression model as well as heterogeneous histological types33, 34. Gene expression profiling of malignant ovarian cancer demonstrated that a significant number of hypermethylated genes were down regulated35. A novel microarray system to assess gene expression, DNA methylation and histone acetylation in parallel, and to dissect the complex hierarchy of epigenetic changes has been developed using human ovarian cancer cell lines36. Aberrant CpG island methylation was found to be a major pathway leading to the inactivation of tumor suppressor genes and development of cancer37-39. Further studies revealed that changes in DNA methylation were cumulative with disease progression8, 40.
It is important to note that agents inducing hypomethylation do not benefit all patients with ovarian cancer. Therefore, additional identification of predictive and surrogate biomarkers is needed for better management of this disease. Given our previous in vitro data that azacitidine enhanced the sensitivity of platinum-resistant ovarian cancer cells to carboplatin through reactivation of DR4 and induction of caspase 8-mediated apoptosis13, we tested whether a similar mechanism might exist in a patient population. Although the number of patients studied is small and differences did not achieve statistical significance, combining azacitidine with carboplatin decreased DR4 methylation more frequently in patients who responded to this treatment than in those who did not, consistent with the possibility that reactivating silenced genes may be required for response. The basal methylation levels of the DR4 and hMLH1 gene promoters seem not to be critical for predicting the response to this treatment strategy27, 41.
This study suggests that a hypomethylating agent may enhance response to platinum in platinum-resistant ovarian cancers. The impact of epigenetic therapy might be further enhanced by concomitant or sequential use of a hypomethylating agent and a histone deacetylase inhibitor prior to platinum treatment. Alternatively, it may be possible to maximize the therapeutic potential of epigenetic treatments though prolonged exposure to epigenetic agents, while concurrently administering a series of chemotherapeutic or biological agents24. Further larger studies of combined azacitidine and carboplatin in patients with platinum resistant ovarian cancer are warranted.
Acknowledgements
The authors thank Carla Moore and Kristine Dice in the Department of Gynecological Medical Oncology at M. D. Anderson Cancer Center for patient care, De-yu Shen for the correlative study, Michael Worley in the Department of Scientific Publications at M. D. Anderson Cancer Center for editing our manuscript and financial support from Pharmion Corporation for the clinical and correlative studies. The study utilized shared resources supported by NCI CCSG grant CA16672.
REFERENCES
- 1.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8(4):286–98. doi: 10.1038/nrg2005. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17339880. [DOI] [PubMed] [Google Scholar]
- 2.Das PM, Singal R. DNA methylation and cancer. J Clin Oncol. 2004;22(22):4632–42. doi: 10.1200/JCO.2004.07.151. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15542813. [DOI] [PubMed] [Google Scholar]
- 3.Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009;1(2):239–59. doi: 10.2217/epi.09.33. Available from http://www.futuremedicine.com/doi/abs/10.2217/epi.09.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matei DE, Nephew KP. Epigenetic therapies for chemoresensitization of epithelial ovarian cancer. Gynecol Oncol. 2010;116(2):195–201. doi: 10.1016/j.ygyno.2009.09.043. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19854495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barton CA, Hacker NF, Clark SJ, O’Brien PM. DNA methylation changes in ovarian cancer: implications for early diagnosis, prognosis and treatment. Gynecol Oncol. 2008;109(1):129–39. doi: 10.1016/j.ygyno.2007.12.017. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18234305. [DOI] [PubMed] [Google Scholar]
- 6.Feng Q, Deftereos G, Hawes SE, Stern JE, Willner JB, Swisher EM, et al. DNA hypermethylation, Her-2/neu overexpression and p53 mutations in ovarian carcinoma. Gynecol Oncol. 2008;111(2):320–9. doi: 10.1016/j.ygyno.2008.07.036. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18757082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graham JS, Kaye SB, Brown R. The promises and pitfalls of epigenetic therapies in solid tumours. Eur J Cancer. 2009;45(7):1129–36. doi: 10.1016/j.ejca.2009.01.003. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19211243. [DOI] [PubMed] [Google Scholar]
- 8.Watts GS, Futscher BW, Holtan N, Degeest K, Domann FE, Rose SL. DNA methylation changes in ovarian cancer are cumulative with disease progression and identify tumor stage. BMC Med Genomics. 2008;1:47. doi: 10.1186/1755-8794-1-47. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18826610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei SH, Chen CM, Strathdee G, Harnsomburana J, Shyu CR, Rahmatpanah F, et al. Methylation microarray analysis of late-stage ovarian carcinomas distinguishes progression-free survival in patients and identifies candidate epigenetic markers. Clin Cancer Res. 2002;8(7):2246–52. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12114427. [PubMed] [Google Scholar]
- 10.Watanabe Y, Ueda H, Etoh T, Koike E, Fujinami N, Mitsuhashi A, et al. A change in promoter methylation of hMLH1 is a cause of acquired resistance to platinum-based chemotherapy in epithelial ovarian cancer. Anticancer Res. 2007;27(3B):1449–52. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17595760. [PubMed] [Google Scholar]
- 11.Plumb JA, Strathdee G, Sludden J, Kaye SB, Brown R. Reversal of drug resistance in human tumor xenografts by 2′-deoxy-5-azacytidine-induced demethylation of the hMLH1 gene promoter. Cancer Res. 2000;60(21):6039–44. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11085525. [PubMed] [Google Scholar]
- 12.Balch C, Yan P, Craft T, Young S, Skalnik DG, Huang TH, et al. Antimitogenic and chemosensitizing effects of the methylation inhibitor zebularine in ovarian cancer. Mol Cancer Ther. 2005;4(10):1505–14. doi: 10.1158/1535-7163.MCT-05-0216. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16227399. [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Hu W, Shen DY, Kavanagh JJ, Fu S. Azacitidine enhances sensitivity of platinum-resistant ovarian cancer cells to carboplatin through induction of apoptosis. Am J Obstet Gynecol. 2009;200(2):177, e1–9. doi: 10.1016/j.ajog.2008.08.030. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19110234. [DOI] [PubMed] [Google Scholar]
- 14.Markman M. Pharmaceutical management of ovarian cancer : current status. Drugs. 2008;68(6):771–89. doi: 10.2165/00003495-200868060-00004. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18416585. [DOI] [PubMed] [Google Scholar]
- 15.Morgan RJ, Jr., Alvarez RD, Armstrong DK, Boston B, Chen LM, Copeland L, et al. Ovarian cancer. Clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2008;6(8):766–94. doi: 10.6004/jnccn.2008.0058. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18926089. [DOI] [PubMed] [Google Scholar]
- 16.Markman M. Second-line therapy for ovarian cancer. Clin Adv Hematol Oncol. 2008;6(6):421–2. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18567987. [PubMed] [Google Scholar]
- 17.Bukowski RM, Ozols RF, Markman M. The management of recurrent ovarian cancer. Semin Oncol. 2007;34(2 Suppl 2):S1–15. doi: 10.1053/j.seminoncol.2007.03.012. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17512352. [DOI] [PubMed] [Google Scholar]
- 18.Candelaria M, Gallardo-Rincon D, Arce C, Cetina L, Aguilar-Ponce JL, Arrieta O, et al. A phase II study of epigenetic therapy with hydralazine and magnesium valproate to overcome chemotherapy resistance in refractory solid tumors. Ann Oncol. 2007;18(9):1529–38. doi: 10.1093/annonc/mdm204. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17761710. [DOI] [PubMed] [Google Scholar]
- 19.Appleton K, Mackay HJ, Judson I, Plumb JA, McCormick C, Strathdee G, et al. Phase I and pharmacodynamic trial of the DNA methyltransferase inhibitor decitabine and carboplatin in solid tumors. J Clin Oncol. 2007;25(29):4603–9. doi: 10.1200/JCO.2007.10.8688. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17925555. [DOI] [PubMed] [Google Scholar]
- 20.Lyons RM, Cosgriff TM, Modi SS, Gersh RH, Hainsworth JD, Cohn AL, et al. Hematologic response to three alternative dosing schedules of azacitidine in patients with myelodysplastic syndromes. J Clin Oncol. 2009;27(11):1850–6. doi: 10.1200/JCO.2008.17.1058. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19255328. [DOI] [PubMed] [Google Scholar]
- 21.Calvert AH, Newell DR, Gumbrell LA, O’Reilly S, Burnell M, Boxall FE, et al. Carboplatin dosage: prospective evaluation of a simple formula based on renal function. J Clin Oncol. 1989;7(11):1748–56. doi: 10.1200/JCO.1989.7.11.1748. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=2681557. [DOI] [PubMed] [Google Scholar]
- 22.Cockcroft DW, Gault MH. Prediction of creatinine clearance from serum creatinine. Nephron. 1976;16(1):31–41. doi: 10.1159/000180580. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=1244564. [DOI] [PubMed] [Google Scholar]
- 23.Gifford G, Paul J, Vasey PA, Kaye SB, Brown R. The acquisition of hMLH1 methylation in plasma DNA after chemotherapy predicts poor survival for ovarian cancer patients. Clin Cancer Res. 2004;10(13):4420–6. doi: 10.1158/1078-0432.CCR-03-0732. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15240532. [DOI] [PubMed] [Google Scholar]
- 24.Balch C, Huang TH, Brown R, Nephew KP. The epigenetics of ovarian cancer drug resistance and resensitization. Am J Obstet Gynecol. 2004;191(5):1552–72. doi: 10.1016/j.ajog.2004.05.025. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15547525. [DOI] [PubMed] [Google Scholar]
- 25.Gras E, Catasus L, Arguelles R, Moreno-Bueno G, Palacios J, Gamallo C, et al. Microsatellite instability, MLH-1 promoter hypermethylation, and frameshift mutations at coding mononucleotide repeat microsatellites in ovarian tumors. Cancer. 2001;92(11):2829–36. doi: 10.1002/1097-0142(20011201)92:11<2829::aid-cncr10094>3.0.co;2-3. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11753956. [DOI] [PubMed] [Google Scholar]
- 26.Strathdee G, MacKean MJ, Illand M, Brown R. A role for methylation of the hMLH1 promoter in loss of hMLH1 expression and drug resistance in ovarian cancer. Oncogene. 1999;18(14):2335–41. doi: 10.1038/sj.onc.1202540. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10327053. [DOI] [PubMed] [Google Scholar]
- 27.Horak P, Pils D, Haller G, Pribill I, Roessler M, Tomek S, et al. Contribution of epigenetic silencing of tumor necrosis factor-related apoptosis inducing ligand receptor 1 (DR4) to TRAIL resistance and ovarian cancer. Mol Cancer Res. 2005;3(6):335–43. doi: 10.1158/1541-7786.MCR-04-0136. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15972852. [DOI] [PubMed] [Google Scholar]
- 28.Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416(6880):552–6. doi: 10.1038/416552a. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11932749. [DOI] [PubMed] [Google Scholar]
- 29.Kavanagh J, Tresukosol D, Edwards C, Freedman R, de Leon C Gonzalez, Fishman A, et al. Carboplatin reinduction after taxane in patients with platinum-refractory epithelial ovarian cancer. J Clin Oncol. 1995;13(7):1584–8. doi: 10.1200/JCO.1995.13.7.1584. Available fromhttp://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=7602347. [DOI] [PubMed] [Google Scholar]
- 30.See HT, Freedman RS, Kudelka AP, Burke TW, Gershenson DM, Tangjitgamol S, et al. Retrospective review: re-treatment of patients with ovarian cancer with carboplatin after platinum resistance. Int J Gynecol Cancer. 2005;15(2):209–16. doi: 10.1111/j.1525-1438.2005.15205.x. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15823101. [DOI] [PubMed] [Google Scholar]
- 31.Leitao MM, Jr., Hummer A, Dizon DS, Aghajanian C, Hensley M, Sabbatini P, et al. Platinum retreatment of platinum-resistant ovarian cancer after nonplatinum therapy. Gynecol Oncol. 2003;91(1):123–9. doi: 10.1016/s0090-8258(03)00464-5. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14529671. [DOI] [PubMed] [Google Scholar]
- 32.Ramirez PT, Landen CN, Jr., Coleman RL, Milam MR, Levenback C, Johnston TA, et al. Phase I trial of the proteasome inhibitor bortezomib in combination with carboplatin in patients with platinum- and taxane-resistant ovarian cancer. Gynecol Oncol. 2008;108(1):68–71. doi: 10.1016/j.ygyno.2007.08.071. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17905421. [DOI] [PubMed] [Google Scholar]
- 33.Willner J, Wurz K, Allison KH, Galic V, Garcia RL, Goff BA, et al. Alternate molecular genetic pathways in ovarian carcinomas of common histological types. Hum Pathol. 2007;38(4):607–13. doi: 10.1016/j.humpath.2006.10.007. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17258789. [DOI] [PubMed] [Google Scholar]
- 34.Hennessy BT, Coleman RL, Markman M. Ovarian cancer. Lancet. 2009 doi: 10.1016/S0140-6736(09)61338-6. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19793610. [DOI] [PubMed] [Google Scholar]
- 35.Melnikov A, Scholtens D, Godwin A, Levenson V. Differential methylation profile of ovarian cancer in tissues and plasma. J Mol Diagn. 2009;11(1):60–5. doi: 10.2353/jmoldx.2009.080072. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19074590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi H, Wei SH, Leu YW, Rahmatpanah F, Liu JC, Yan PS, et al. Triple analysis of the cancer epigenome: an integrated microarray system for assessing gene expression, DNA methylation, and histone acetylation. Cancer Res. 2003;63(9):2164–71. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12727835. [PubMed] [Google Scholar]
- 37.Imura M, Yamashita S, Cai LY, Furuta J, Wakabayashi M, Yasugi T, et al. Methylation and expression analysis of 15 genes and three normally-methylated genes in 13 Ovarian cancer cell lines. Cancer Lett. 2006;241(2):213–20. doi: 10.1016/j.canlet.2005.10.010. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16303245. [DOI] [PubMed] [Google Scholar]
- 38.Menendez L, Walker D, Matyunina LV, Dickerson EB, Bowen NJ, Polavarapu N, et al. Identification of candidate methylation-responsive genes in ovarian cancer. Mol Cancer. 2007;6:10. doi: 10.1186/1476-4598-6-10. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17254359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strathdee G, Appleton K, Illand M, Millan DW, Sargent J, Paul J, et al. Primary ovarian carcinomas display multiple methylator phenotypes involving known tumor suppressor genes. Am J Pathol. 2001;158(3):1121–7. doi: 10.1016/S0002-9440(10)64059-X. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11238060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Makarla PB, Saboorian MH, Ashfaq R, Toyooka KO, Toyooka S, Minna JD, et al. Promoter hypermethylation profile of ovarian epithelial neoplasms. Clin Cancer Res. 2005;11(15):5365–9. doi: 10.1158/1078-0432.CCR-04-2455. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16061849. [DOI] [PubMed] [Google Scholar]
- 41.Shivapurkar N, Toyooka S, Toyooka KO, Reddy J, Miyajima K, Suzuki M, et al. Aberrant methylation of trail decoy receptor genes is frequent in multiple tumor types. Int J Cancer. 2004;109(5):786–92. doi: 10.1002/ijc.20041. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14999791. [DOI] [PubMed] [Google Scholar]


