Abstract
Objective
Adipose triglyceride lipase (ATGL) catalyzes the first step in adipocyte and muscle triglyceride hydrolysis, and Comparative Gene Identification-58 (CGI-58) is an essential cofactor. We studied the expression of ATGL and CGI-58 in human adipose and muscle, and examined correlations with markers of muscle fatty acid oxidation.
Materials/Methods
Non diabetic volunteers were studied. Subjects with impaired glucose tolerance were treated with pioglitazone or metformin for 10 weeks. Normal glucose tolerant subjects underwent a 12 week training program. We examined changes in ATGL and CGI-58 with obesity and insulin resistance, and effects of exercise and pioglitazone.
Results
ATGL mRNA expression showed no correlation with either body mass index (BMI) or insulin sensitivity (SI) in either adipose or muscle. However, adipose ATGL protein levels were inversely correlated with BMI (r=−0.64, p<0.02), and positively correlated with SI (r=0.67, p<0.02). In muscle, ATGL mRNA demonstrated a strong positive relationship with carnitine palmitoyltransferase I mRNA (r=0.82, p<0.0001), and the adiponectin receptors AdipoR1 mRNA (r=0.71, p<0.0001), and AdipoR2 mRNA (r=0.74, p<0.0001). Muscle CGI-58 mRNA was inversely correlated with intramyocellular triglyceride in both type 1 (r=−0.35, p<0.05) and type 2 (r=−0.40, p<0.05) fibers. Exercise training resulted in increased muscle ATGL and pioglitazone increased adipose ATGL by 31% (p<0.05). Pioglitazone also increased ATGL in adipocytes.
Conclusions
Adipose ATGL protein is decreased with insulin resistance and obesity, and muscle ATGL mRNA is associated with markers of fatty acid oxidation in muscle, as is CGI-58. The regulation of ATGL and CGI-58 have important implications for the control of lipotoxicity.
Keywords: CGI-58, muscle fatty acid oxidation, lipotoxicity, exercise, PPARγ
INTRODUCTION
The disruption of the balance among lipid synthesis, uptake and utilization is critical in the development of metabolic diseases such as insulin resistance and type 2 diabetes (T2D) [1,2]. Adipose tissue in mammals is the main source for energy storage and retrieval in the form of triacylglycerol (TAG). Hydrolysis of TAG by activation of lipolytic enzymes results in free fatty acid (FFA) release into the circulation for use as an energy substrate, primarily by muscle. Muscle is an important target tissue in insulin resistance, and an important defect in the muscle of insulin resistant subjects is the accumulation of triglyceride, due to either increased uptake or decreased oxidation, leading to the generation of diacylglycerol, ceramide, and other factors that impair insulin action.
Adipose triglyceride lipase (ATGL) catalyzes the first, rate-limiting step in TAG lipolysis, followed by additional lipolytic action by hormone sensitive lipase [3,4] and the release of FFA was decreased in 3T3-L1 cells treated with siRNA against ATGL [5]. Deletion of ATGL in mice caused weight gain and increased adipose mass, along with increased muscle glucose uptake due to an inability to use muscle lipid as fuel [6,7].
Full ATGL activity requires interaction with the activator protein CGI-58 [3,8-10]. CGI-58 knockout mice demonstrate an impaired lipolysis phenotype just like the ATGL knockout mouse [11]. The regulation of ATGL at the level of transcription is not well characterized, although ATGL mRNA expression is regulated by some nutritional and hormonal factors such as fasting, glucocorticoids, insulin, and leptin [7]. Several studies have shown that ATGL mRNA is increased by rosiglitazone in adipose tissue of rodents [12,13] as well as in 3T3-L1 adipocytes [14]. PPARγ-specific antagonist and siRNA-mediated inhibition of PPARγ inhibited rosiglitazone induced ATGL mRNA expressions, indicating a role for ATGL in triglyceride metabolism mediated by PPARγ [14].
In addition to the role of ATGL in adipose tissue lipolysis, this enzyme likely plays an important role in muscle. ATGL deficient mice showed significant TAG accumulation in skeletal muscle and less fatty acid oxidation measured with respiratory quotient, along with a shift to carbohydrate over fat as an energy source [5]. In addition, ATGL expression was decreased in skeletal muscle of obese, insulin resistant mice and rats [15], and ATGL overexpression in myotubes increased fatty acid oxidation and ceramide contents, suggesting a critical role of ATGL in triglyceride metabolism and storage in muscle. As suggested by Zimmermann et al [3], FFA is likely taken up by muscle and initially re-esterified into TAG, and the lipolysis of the intramyocellular TAG stores is a critical step in supplying muscle with the FFA fuel that is needed for oxidation and energy needs.
The precise role and degree of regulation of ATGL and CGI-58 in humans is not well known. There are several studies involving ATGL in humans and variable results have been obtained. Depending on the study, adipose ATGL mRNA was either decreased, increased, or unchanged with obesity, and ATGL protein either unchanged or decreased [16-19]. Studies in human muscle found that ATGL was associated with type 1 muscle fibers [20], and may be upregulated by endurance exercise training [20]. There are no data that related human ATGL and CGI-58 to insulin sensitivity, or examine the relationship between ATGL and CGI-58. Here, we report the relationship of muscle and adipose ATGL and CGI-58 with obesity, insulin sensitivity, and markers of fatty acid oxidation. To further understand the regulation of ATGL and CGI-58, we examined the influence of pioglitazone and exercise training on gene expression.
METHODS
Human subjects and tissues
The adipose tissue and muscle samples were obtained from three different groups of subjects. For each group, all subjects signed consent forms that were approved by the local Institutional Review Board.
Group 1: Adipose tissue and muscle biopsies
Subcutaneous adipose tissue (SAT) and muscle tissues were performed on generally healthy subjects on the University of Arkansas for Medical Sciences/Central Arkansas Veterans Healthcare System General Clinical Research Center and were recruited through local advertisement. Subjects were excluded if they were taking medications affecting adipocyte or lipid metabolism (e.g. fibrates, angiotensin converting enzyme-inhibitors). All subjects had fasting glucose levels under 110 mg/dl, and the 2h post-challenge glucose under 200 mg/dl, determined by a 75g oral glucose tolerance test (OGTT). A total of 99 subjects were recruited (85 women and 14 men, 21-61 years old), 37 subjects had impaired glucose tolerance on the OGTT, and the study group had a wide range of BMI (19-40 Kg/m2) and SI [0.21-13.64 × 10−4 × min−1/(μU/ml)], determined using a frequently sampled IV glucose tolerance test (FSIGT, described below). Fasting subjects underwent abdominal SAT and vastus lateralis muscle biopsies under local anesthesia. Subjects with IGT were treated with either pioglitazone 30 mg for 2 weeks and 45 mg daily for 8 weeks, or metformin 1000 mg for 2 weeks and 2000 mg daily for 8 weeks, followed by repeat biopsies. Previous studies have reported data from these subjects [21].
Group 2: Surgical adipose tissue
Paired visceral adipose tissue (VAT) and SAT were obtained from 14 patients undergoing elective abdominal surgery, including cholecystectomy, abdominal hysterectomy, hernia repair, and other routine procedures at the University of Maryland, and were generously provided by Dr. Susan Fried (Boston University). The subjects' age ranged from 24 to 62, and BMI ranged from 23 to 60 kg/m2. These patients were generally healthy, non-diabetic, and free of inflammatory disease by medical history. These subjects have been described previously [22].
Group 3: Exercise Training and muscle biopsies
Five female and one male subject (age: 53±1 years old; BMI: 33±2 kg/m2) were recruited to study the effects of exercise and weight loss on muscle gene expression, using methods described previously [23]. These subjects were non-smokers, sedentary, non-diabetic, and generally healthy, and were not taking any medications that would affect carbohydrate or lipid metabolism. Following medical screening procedures, all subjects underwent supervised training at 50% of VO2peak for 12 weeks, and performed approximately 2500 kcal/week of aerobic exercise on a cycle ergometer. All subjects in this group were placed on strict dietary control aimed at the maintenance of baseline caloric consumption such that weight loss would likely occur through the increased caloric expenditure due to exercise training.
Adipose Tissue Fractions and Cells
In group 1, adipocyte and stromal vascular fractions (SVF) were isolated from fresh adipose tissue by centrifugation after collagenase digestion as described previously [24].
An adipose cell line, derived from the stromal vascular fraction of an infant with Simpson-Golabi-Behmel syndrome (SGBS), was cultured and differentiated into adipocytes according to published methods [25]. Human primary adipose cells were generated from adult derived human adipocyte stem cells (ADHASC) isolated from discarded adipose tissue from subjects who underwent liposuction, as described previously [24,26]. Differentiation of both cell lines was determined by oil Red O staining and adipocyte-specific gene expression. For pioglitazone treatment, differentiated ADHASC cells were maintained in medium without any thiazolidinediones (TZD) for 4 days. The cells were then treated with pioglitazone (1.5 μM) or DMSO vehicle control for 48h.
Insulin Sensitivity Measurement
In group 1, peripheral insulin sensitivity was measured by an insulin-modified FSIGT using 11.4 g/m2 of glucose and 0.04 units/kg of insulin [27]. Plasma insulin was determined using a chemoluminescent assay (Molecular Light Technology Research, Ltd, Cardiff, Wales, UK), and plasma glucose was measured by a glucose oxidase assay. Insulin sensitivity was determined according to the insulin and glucose data using the MINMOD Millennium program [28].
RNA isolation and real time RT-PCR
Total RNA from human adipose tissue was isolated using an RNAeasy Lipid Tissue Mini kit from Qiagen (Valencia, CA). Total RNA from muscle biopsies and cell culture was isolated using an Ultraspec RNA Isolation System kit (Biotex, Houston, TX). Total RNA from cell culture was isolated using RNAqueous kit (Ambion, Austin, TX). The quantity and quality of the isolated RNA were determined by Agilent 2100 Bioanalyser (Palo Alto, CA). Real-time RT-PCR and the primer sequences of 18S was performed as described previously [24]. In brief, standard curves were generated using pooled RNA from the samples assayed, such that the data represent arbitrary units which accurately compare each set of samples to each other, but do not necessarily accurately compare samples between different assays. All data were expressed in relation to 18S RNA, and all samples were analyzed twice with and without reverse transcriptase, and no amplification was seen in the samples in the absence of reverse transcriptase. The primer sequences of ATGL, CGI-58, CPT-1, AdipoR1, and AdipoR2 are listed in Table 1.
Table 1.
Primer sequences
| Gene | Sequence (forward) | Sequence (reverse) |
|---|---|---|
| ATGL | ACCAGCATCCAGTTCAACCT | ATCCCTGCTTGCACATCTCT |
| CGI-58 | TGCAGACTCCAAGTGGTGAG | TGTCAGGGTGCATTTTACCA |
| CPT-1 | CTTTGGCCCTGTAGCAGATGA | TCGTCTCTGAGCTTGAGAACTT |
| AdipoR1 | TTCTTCCTCATGGCTGTGATGT | AAGAAGCGCTCAGGAATTCG |
| AdipoR2 | ATAGGGCAGATAGGCTGGTTGA | GGATCCGGGCAGCATACA |
Measurement of intramyocellular lipid (IMCL)
Skeletal muscle biopsies group 1 were immediately processed for fiber typing and lipid content analysis, using Oil Red O staining and imaging as described previously [21]. IMCL values were expressed as a percentage of lipid content in the muscle fiber by dividing the total area of lipid droplets in a muscle fiber by the total area of the same muscle fiber.
Measurement of ATGL protein
ATGL and β-actin protein levels in adipose tissue was measured by Western blot with a polyclonal antibody to human ATGL (Santa Cruz Biotechnology INC, Santa Cruz, CA). Frozen adipose tissue (150 mg) was lysed in 500 μl of M-PER mammalian protein extraction reagent (Pierce, Rockford, IL) containing protease inhibitors, and protein was loaded on a 10% polyacrylamide gel, transferred onto a nitrocellulose membrane, and blotted with anti-ATGL (1:1000), followed by horseradish peroxidase antirabbit IgG. The blot was analyzed using Lumi-Light Western blotting substrate (Roche, Indianapolis, IN). Densitometric analyses were carried out with Quantity One Digital Image software (Bio-Red, Hercules, CA).
Statistical Analysis
Student's two-sample t-tests were used to compare groups with respect to continuous variables. Paired t-tests were used to compare baseline and treatment measurements within a group. Pearson's correlation coefficients were used to describe the linear association between variables. All data from samples were expressed as mean ± SEM.
RESULTS
ATGL and CGI-58 mRNA expression in human adipose tissue and cell fractions
ATGL and CGI-58 mRNA expression was determined in human adipose tissue, isolated human adipocytes and the SVF from adipose tissue, and in skeletal muscle, and cultured human adipocytes. As shown in Table 2, ATGL was expressed in the adipocyte fraction was 23-fold higher than in the SVF. ATGL was expressed in skeletal muscle, but to a lesser extent compare to adipose tissue. CGI-58 was also expressed in adipose tissue; however there was no significant difference between adipocytes and the SVF. Both ATGL and CGI-58 were expressed at a higher level in SGBS adipocytes when compared to preadipocytes. LPL and leptin expression was measured in these samples as a control, and as expected, both were associated with the adipocyte fraction. To examine the expression of ATGL and CGI-58 in different adipose depots, ATGL and CGI-58 expression were measured in paired SAT and VAT samples obtained from 14 subjects, as described in the Methods. As shown in Table 2, ATGL expression was significantly higher in SAT than in VAT, while there was no difference in CGI-58 expression in the two depots. Leptin expression has previously been demonstrated to be higher in SAT.
Table 2.
ATGL and CGI-58 expression in human adipose tissue and culture
| ATGL* | CGI-58* | LPL* | Leptin* | |
|---|---|---|---|---|
| Surgical adipose tissue depots (n=14) | ||||
| SAT | 2.45 ± 0.41 | 2.68±0.43 | 1.35±0.21 | 1.74±0.21 |
| VAT | 1.36 ± 0.28 a | 3.12±0.32 | 0.75±0.21a | 0.68±0.12a |
|
| ||||
| Cell (n=2)/tissue (n=3) | ||||
| Whole adipose tissue | 2.24±0.79 | 1.40±0.22 | 5.09±1.84 | 3.65±1.11 |
| Adipocytes from adipose tissue | 4.14±0.52 | 1.33±0.12 | 5.00±0.65 | 3.44±0.58 |
| Stromal fraction from adipose tissue | 0.18±0.04 | 1.12±0.17 | 1.01±0.27 | 0.20±0.05 |
| Adipocytes from SGBS preadipocytes | 4.53±0.32 | 2.22±0.14 | 7.44±0.10 | 0.16±0.01 |
| Cultured SGBS preadipocytes | 0.13±0.03 | 0.35±0.07 | NE | NE |
| Skeletal muscle | 0.33±0.03 | 0.46±0.09 | 0.22±0.08 | NE |
Data are mean ± SE.
The RNA was pooled from all the cDNA samples to generate a standard curve. Therefore, the data are expressed relative to each other.
p<0.02 vs. SAT. NE: no expression.
ATGL gene expression in relation to obesity and insulin resistance
ATGL deficient mice had increased insulin sensitivity and glucose tolerance due to the decreased availability of FFAs for oxidation [5]. To determine whether ATGL was linked to obesity and/or insulin resistance, we measured ATGL mRNA levels in adipose tissue (n=86) and muscle (n=74) in Group 1 subjects using real-time RT-PCR. As described in Methods, the subjects covered a wide range of BMI and SI and the relationships between ATGL, BMI, insulin sensitivity (SI), and other parameters were studied using Pearson's correlation coefficients. As shown in Table 3, ATGL mRNA was not associated with BMI or SI in either adipose tissue or muscle. In addition, there was no significant association between ATGL mRNA expression in VAT and BMI (n=14)(data not shown).
Table 3.
Correlation coefficient of adipose and muscle of ATGL mRNA with different variables in plasma, muscle and adipose tissue
| Variable (n) | Adipose ATGL mRNA (n=86) |
Muscle ATGL mRNA (n=74) |
|---|---|---|
| BMI (86) | 0.21 | 0.14 |
| SI (86) | −0.18 | 0.19 |
| Plasma adiponectin (57) | −0.27 | 0.20 |
| Plasma TNFα (57) | 0.18 | −0.12 |
| IMCL type 1 fiber (62) | 0.09 | 0.14 |
| IMCL type 2 fiber (62) | 0.24 | 0.14 |
| Adipose CGI-58 mRNA (86) | 0.70* | NA |
| Muscle CGI-58 mRNA (74) | NA | 0.49* |
| Muscle CPT-1 mRNA (74) | NA | 0.82* |
| Muscle AdipoR1 mRNA (74) | NA | 0.71* |
| Muscle AdipoR2 mRNA (74) | NA | 0.74* |
p<0.001. NA, not applicable
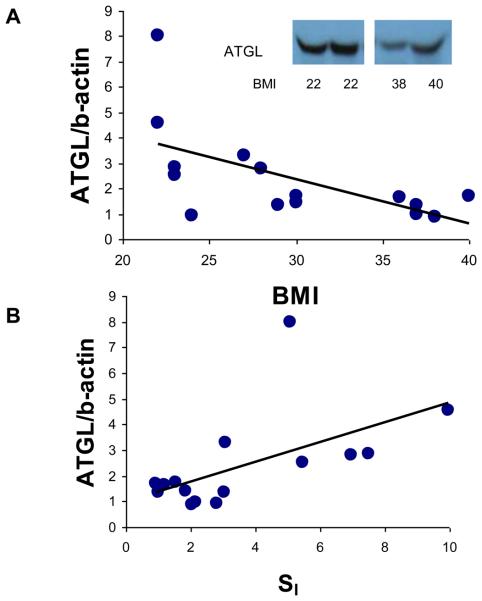
To further examine ATGL in obesity, we measured adipose ATGL protein using Western blot in 15 subjects who were representative of the larger group, ranging in BMI from 22 to 40 kg/m2. The protein level was determined with densitometry analysis and normalized with β-actin protein level (Figure 1). In contrast to adipose ATGL mRNA, adipose ATGL protein was negatively correlated with BMI (p<0.02) (Figure 1A) and positively with SI (p<0.02)(Figure 1B). Thus, adipose tissue ATGL protein is higher in lean, insulin sensitive subjects, although this association was not reflected at the mRNA level.
Figure 1.
ATGL protein levels in relation to obesity and insulin resistance. Human adipose ATGL protein levels were determined from the adipose tissue of 15 subjects as described in the Methods. ATGL protein level was expressed as its density ratio to actin. A. Relationship between adipose tissue ATGL protein level and BMI (r=−0.64, p<0.02). Inset: representative Western blot of two lean and two obese subjects. B. Relationship between adipose tissue ATGL and SI (r=0.67, p<0.02).
Muscle ATGL expression is associated with muscle fatty acid oxidation
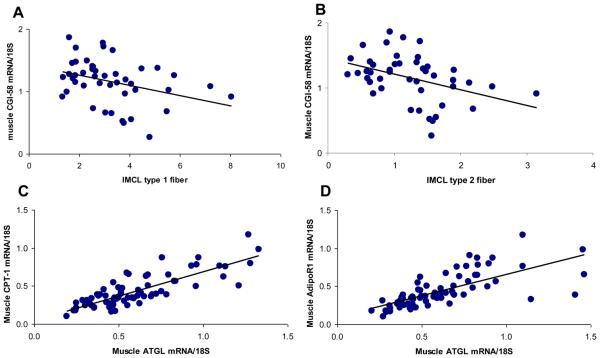
Because ATGL is associated with muscle fatty acid oxidation in mice, we further examined the relationship between ATGL and CGI-58 gene expression and muscle triglyceride content and markers of muscle triglyceride metabolism. ATGL mRNA expression was not associated with IMCL (Table 3). However, the expression of muscle CGI-58 was negatively associated with IMCL in both type 1 (r=−0.35, p<0.02) and type 2 fibers (r=−0.40, p<0.01) (Figure 2). ATGL mRNA level and CGI-58 mRNA levels were highly correlated with each other (Table 3), both in adipose tissue (r=0.71, p<0.001) and in muscle (r=0.49. p<0.001). In rodents, ATGL has been demonstrated to enhance muscle triglyceride oxidation, which is an important determinant of IMCL [5].
Figure 2.
Muscle CGI-58 and muscle ATGL mRNA expression are related to muscle fatty acid oxidation. A. Muscle CGI-58 was inversely related to IMCL in type 1 fibers (r=−0.35, p<0.02, n=45).B. Muscle CGI-58 was inversely related to IMCL in type 2 fibers (r=−0.40, p<0.01, n=45). C. Muscle ATGL mRNA was significantly associated with CPT-1 mRNA (r=0.82, p<0.0001, n=74). D. Muscle ATGL mRNA was significantly associated with AdipoR1 (r=0.71, p<0.001, n=74).
Hence, we examined the relationship between ATGL and other genes related to fatty acid oxidation. As shown in Figure 2, ATGL gene expression in muscle was strongly associated with fatty acid oxidation genes, such as muscle adiponectin receptor 1 (AdipoR1, r=0.71, p<0.001), adiponectin receptor 2 (AdipoR2, r=0.74, p<0.001), and with carnitine-palmitoyl transferase 1 (CPT-1, r=0.82, p<0.001). CGI-58 mRNA levels also were correlated with the muscle adiponectin receptors 1 (r=0.35, n=74, p<0.01), adiponectin receptor 2 (r=0.42, n=74, p<0.01), and CPT1 (r=0.35, n=74, p<0.01), but these relationships were not as strong as with ATGL.
Effects of exercise training on ATGL
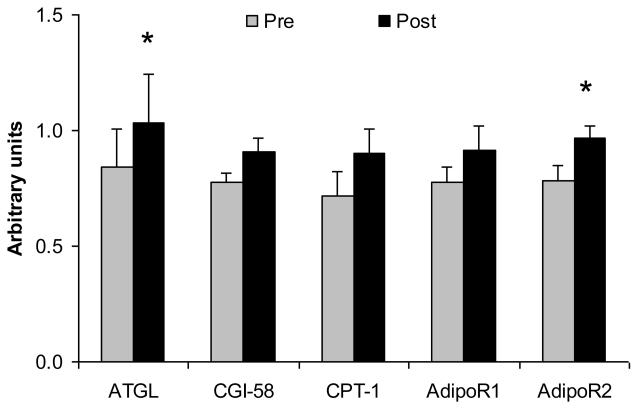
To further study the physiologic regulation of ATGL, we measured muscle ATGL in subjects before and after a 12 week period of exercise training with weight loss. As described in Methods, six subjects completed a 12 week training program of bicycle ergometer exercise under strict dietary control. These subjects lost 6±1 kg during this period (about 7% of initial weight). Muscle biopsies were performed before the training period, and following training and after caloric balance had been reestablished. As shown in Figure 3, ATGL and AdipoR2 gene expression were both significantly increased following training (p<0.05). Measurements of CPT-1, CGI-58, and AdipoR1 expression were also made, but these differences did not reach statistical significance. Thus, these data indicate that increased muscle ATGL expression occurs in association with the increased muscle oxidative requirements during training.
Figure 3.
Effects of exercise training and weight loss on ATGL and muscle oxidative enzymes. As described in Methods, muscle biopsies were performed in six subjects before and after 12 weeks of exercise training and modest weight loss. The gene expression of ATGL, CGI-58, CPT-1, AdipoR1, and AdipoR2 were determined by real-time RT-PCR and were normalized with 18S. Pre and post refers to before and after the training/weight loss. Data are mean ± SE, n=5. *p<0.05.
Pioglitazone increased adipose ATGL expression
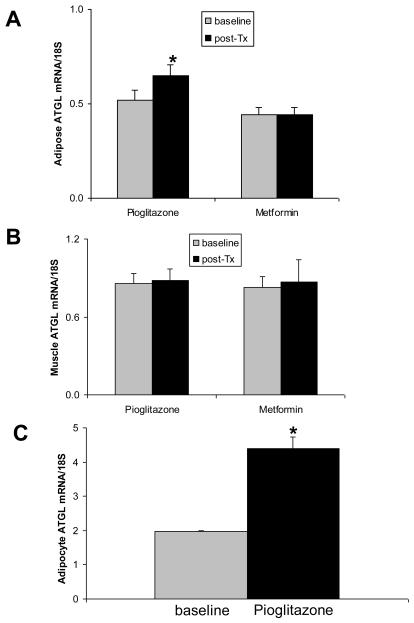
Previous studies showed that ATGL was regulated by PPARγ activation in rodent adipose tissue and cell culture. To study the effect of insulin sensitizers on ATGL and CGI-58 in humans, IGT subjects were randomized to treatment with either pioglitazone or metformin, as described in Methods. Metformin treatment did not affect SI, whereas pioglitazone resulted in an increase in SI from 1.44 ± 0.13 × 10−5 to 2.21 ± 0.21 × 10%−5 min%−1/(μU/ml), p<0.05. As shown in Figure 4, pioglitazone treatment resulted in a 31% increase of adipose ATGL expression (p<0.03), while there was no significant change in muscle ATGL. There was no change in CGI-58 expression in adipose tissue or muscle, and metformin had no effect on either ATGL or CGI-58 (data not shown). To determine the effect of pioglitazone on ATGL in vitro, human adipocytes were derived through the differentiation of stem cells, as described in methods, and were then treated with pioglitazone (1.5μM) for 48 hours, followed by measurement of ATGL mRNA. As shown in Figure 4C, ATGL expression was increased more than 2 fold after treatment with pioglitazone, indicating a direct effect on ATGL expression in adipose tissue.
Figure 4.
Effects of pioglitazone on ATGL expression. Human subjects' adipose tissue (n=18) and muscle (n=14) were obtained through biopsy before and after 10 weeks of pioglitazone or metformin treatment. ATGL expression in adipose tissue and muscle was determined by real-time RT-PCR. A. Adipose tissue. B. Muscle. C. Cultured human adipocytes. The cells of differentiated ADHASC were treated with 1.5 μM pioglitazone for 48h, followed by RNA isolation and measurement of ATGL expression by real-time RT-PCR. Data are mean ± SE, *p<0.05.
Discussion
Both adipose tissue and muscle contain lipid droplets, and the initial step in the hydrolysis of triglyceride involves ATGL, which hydrolyses FFA from triglyceride, leaving the diacylglycerol for subsequent hydrolysis by hormone sensitive lipase [29]. Subsequent studies of ATGL have suggested very different roles in adipose tissue and muscle. Based on observations in the ATGL deficient mouse, adipocytes are dependent on ATGL for lipolysis, and the absence of ATGL results in obesity and low plasma FFA levels. Absence of ATGL in muscle, however, results in muscle triglyceride accumulation, decreased use FFA for energy and decreased circulating FFA, followed by an increased dependence on carbohydrates as fuel, leading to increased glucose utilization [5,30]. CGI-58 is an essential cofactor for ATGL-mediated triglyceride hydrolysis [8], and mutations of both ATGL and CGI-58 have been found in humans with neutral lipid storage disease, suggesting that these proteins are also important in human tissues for the release of neutral lipid [31,32].
In this study, we examined ATGL expression profiles in different tissues and cells. ATGL expression was significantly higher in SAT compared to VAT, yet there was no difference in CGI-58 expression between depots. ATGL was highly expressed in adipocytes from either adipose tissue or cultured adipocytes, with lower expression in the stromal fraction and cultured preadipocytes, and ATGL expression was induced after adipocyte differentiation [3,7,33]. In contrast, CGI-58 expression in the adipocyte fraction was similar to the stromal fraction, which contains preadipocytes and other cell types. CGI-58 may have other functions, in addition to the activation of ATGL, as suggested by the different human syndromes caused by ATGL and CGI-58 deficiency [31]. Whereas ATGL knockout mice become obese, CGI-58 knockout mice die within 16 hr of birth with severe hepatic steatosis and dehydration from severe impairment of their skin permeability barrier [11]. Indeed, patients with CGI-58 mutations have prominent ichthyosis, perhaps suggesting some other role of CGI-58 in the dermis, perhaps to act as a cofactor for another neutral lipase [11].
ATGL mRNA levels in adipose tissue were not significantly associated with BMI or SI, although ATGL protein levels were lower with obesity and insulin resistance. Several studies have examined the physiologic regulation of ATGL or CGI-58 in human adipose tissue, and variable relationships with obesity or insulin resistance have been found [16-19,34]. Differences in degree of extraction of ATGL from the lipid droplet could explain some of these differences, although this is hard to discern, since every study involving extraction of ATGL from adipose tissue used different methods. With obesity or insulin resistance, several studies noted either an increase or minimal change in ATGL mRNA yet a decrease in ATGL protein [17,18], suggesting some level of posttranscriptional relationship. We also found no relationship between CGI-58 mRNA and obesity, which corresponds to a previous study [18]. Some of these inconsistent findings could be due to different study populations and different methods for obtaining adipose tissue. Our primary study group (group 1) consisted of subjects with a wide range of BMI and insulin sensitivities and biopsies were obtained by incision, whereas some other studies concentrated on obese subjects, or used surgical fat. Based on these data, the primary regulatory forces behind ATGL are not clear. Insulin inhibits ATGL mRNA levels in 3T3 L1 adipocytes [7], and obese subjects are hyperinsulinemic and insulin resistant. On the other hand, the obese environment is associated with adipose tissue macrophages [24], and TNFα decreases ATGL mRNA levels in adipocytes [35,36]. Since our data and others find evidence for posttranscriptional regulation, however, the regulation of ATGL is likely more complex and further studies are needed.
Previous studies in mice have demonstrated an important role for ATGL in muscle TAG lipolysis, fatty acid oxidation, and prevention of lipotoxicity [5,15]. In mice, treadmill exercise training did not affect ATGL mRNA or protein levels, however ATGL knockout mice demonstrated a reduced exercise capacity [37]. Fewer studies have been performed in human muscle, with one study demonstrating an increase in ATGL (but not CGI-58) following a period of training [38]. Although there was no association with BMI or SI, there were significant correlations relating either ATGL or CGI-58 expression to different elements of muscle fatty acid oxidation. There was a significant negative correlation between muscle CGI-58 expression and IMCL content in both type 1 and type 2 fibers. ATGL proteins have been shown to be exclusively expressed in human slow-twitch oxidative type 1 muscle fibers [20]. In line with those findings, we found a strong correlation between muscle ATGL expression and genes involved with fatty acid oxidation, including CPT-1, AdipoR1, and AdipoR2. Furthermore, we observed that muscle ATGL gene expression along with other oxidative genes was increased in subjects following weight loss with exercise training, where the need for muscle fatty acid oxidation increases. Thus, these data are consistent with the mouse data that suggest an important role of ATGL-CGI-58 mediated intramyocellular TAG lipolysis, which then leads to muscle fatty acid oxidation. It is not clear why muscle ATGL does not increase with exercise in mice [37], but could relate to other compensatory mechanisms for provision of FFA in mice, or perhaps the need for weight loss in addition to exercise. This increase in muscle lipid oxidation helps minimize muscle lipotoxicity, and is part of the response to weight loss and increased physical activity.
It is noteworthy that elements of muscle fatty acid oxidation (e.g. CPT-1, AdipoR1, AdipoR2) were associated with ATGL, but IMCL was associated with CGI-58. To our knowledge, no other study has identified regulation of muscle CGI-58 in humans. The precise mechanism for the interaction between ATGL and CGI-58 is yet to be understood. In adipocytes, CGI-58 binds to perilipin A and is released following hormonal stimulation to activate ATGL [39,40], however it is not known whether this precise mechanism is operative in muscle.
In previous studies, PPARγ agonists has been shown to directly increase ATGL mRNA and protein levels in pre-adipocytes and mature adipocytes in vivo and in vitro [13,14,41], but no similar human data have been reported. In this study, adipose ATGL expression was increased by pioglitazone in human subjects, and this appeared to be a direct effect on adipocytes, rather than an indirect effect of improved insulin sensitivity, since the addition of pioglitazone to differentiated adipocytes induced ATGL mRNA level in cell culture. The magnitude of the increase in ATGL was higher in vitro, which could be due to compensatory changes induced in vivo that attenuate the effects of the thiazolidinedione, or to lower concentrations of drug achieved in vivo. Pioglitazone improves insulin sensitivity and results in an overall reduction in plasma FFA, along with an increase in expression of many adipocyte lipogenic enzymes [42]. The impact of the ATGL increase, with no increase in CGI-58, following pioglitazone treatment is not clear.
It is possible that pioglitazone positively regulates ATGL as part of an overall increase in adipocyte gene expression, yet the improved adipocyte insulin sensitivity may downregulate overall lipolytic activity. Pioglitazone had no effect on muscle ATGL expression. Although muscle expresses PPARγ, the level of expression is low compared to adipose tissue [43] and previous changes in IMCL from pioglitazone have not been accompanied by changes in muscle oxidative capacity [21]. Pioglitazone improves insulin sensitivity, but this change in insulin sensitivity is not sufficient to alter muscle ATGL expression. Future study is needed to define the role of ATGL in the balance of adipose and muscle triglyceride metabolism.
In summary, this study examined the regulation of ATGL and CGI-58 in human adipose and muscle, and in response to training and pioglitazone treatment. Adipose ATGL protein was decreased with obesity and insulin resistance, even though there was no change in ATGL mRNA levels, and pioglitazone increased adipose ATGL mRNA. Muscle ATGL and CGI-58 were associated with other features of muscle fatty acid oxidation, and ATGL increased in muscle with a training/weight loss program. These data indicate that both ATGL and CGI-58 may be important components of adipose tissue and muscle control of insulin resistance.
Acknowledgements
We wish to thank Regina Dennis for assistance with recruitment, and the nurses on the GCRC for assistance with the procedures. We wish to thank Dr. Susan Fried for providing adipose tissue for RNA extraction.
Funding.
A Merit Review Grant from the Veterans Administration (N.R., G.R.), grants from the National Institutes of Health M01RR14288, RO1 DK 39176 (PAK), RO1 DK71349 (PAK), and KO1 DK 64716-01 (RHC). We would also like to acknowledge support from the American Heart Association SDA 0335172N (RHC).
Abbreviations
- ATGL
Adipose triglyceride lipase
- CGI-58
Comparative Gene Identification-58
- BMI
body mass index
- SI
Insulin sensitivity index
- T2D
type 2 diabetes
- TAG
triacylglycerol
- FFA
free fatty acid
- OGTT
oral glucose tolerance test
- VAT
visceral adipose tissue
- SAT
subcutaneous adipose tissue
- FSIGT
frequently sampled IV glucose tolerance test
- CPT-1
carnitine palmitoyltransferase I
- AdipoR1
adiponectin receptor 1
- AdipoR2
adiponectin receptor 2
- IMCL
intramyocellular lipid
- SVF
stromal vascular fraction
- LPL
lipoprotein lipase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures:
Dr. Coker has received consultant fees from Pfizer, Inc.
Reference List
- 1.Kahn BB, Flier JS. Obesity and insulin resistance. J Clin Invest. 2000;106(4):473–481. doi: 10.1172/JCI10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore KJ, Rosen ED, Fitzgerald ML, et al. The role of PPAR-gamma in macrophage differentiation and cholesterol uptake. Nature Medicine. 2001;7(1):41–47. doi: 10.1038/83328. [DOI] [PubMed] [Google Scholar]
- 3.Zimmermann R, Strauss JG, Haemmerle G, et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306(5700):1383–1386. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
- 4.Jenkins CM, Mancuso DJ, Yan W, et al. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J Biol Chem. 2004;279(47):48968–48975. doi: 10.1074/jbc.M407841200. [DOI] [PubMed] [Google Scholar]
- 5.Haemmerle G, Lass A, Zimmermann R, et al. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science. 2006;312(5774):734–737. doi: 10.1126/science.1123965. [DOI] [PubMed] [Google Scholar]
- 6.Zimmermann R, Haemmerle G, Wagner EM, et al. Decreased fatty acid esterification compensates for the reduced lipolytic activity in hormone-sensitive lipase-deficient white adipose tissue. J Lipid Res. 2003;44(11):2089–2099. doi: 10.1194/jlr.M300190-JLR200. [DOI] [PubMed] [Google Scholar]
- 7.Kershaw EE, Hamm JK, Verhagen LA, et al. Adipose triglyceride lipase: function, regulation by insulin, and comparison with adiponutrin. Diabetes. 2006;55(1):148–157. [PMC free article] [PubMed] [Google Scholar]
- 8.Lass A, Zimmermann R, Haemmerle G, et al. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in ChanarinDorfman Syndrome. Cell Metab. 2006;3(5):309–319. doi: 10.1016/j.cmet.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Granneman JG, Moore HP, Granneman RL, et al. Analysis of Lipolytic Protein Trafficking and Interactions in Adipocytes. J Biol Chem. 2007;282(8):5726–5735. doi: 10.1074/jbc.M610580200. [DOI] [PubMed] [Google Scholar]
- 10.Miyoshi H, Perfield JW, Souza SC, et al. Control of adipose triglyceride lipase action by serine 517 of perilipin A globally regulates protein kinase A-stimulated lipolysis in adipocytes. J Biol Chem. 2007;282(2):996–1002. doi: 10.1074/jbc.M605770200. [DOI] [PubMed] [Google Scholar]
- 11.Radner FP, Streith IE, Schoiswohl G, et al. Growth retardation, impaired triacylglycerol catabolism, hepatic steatosis, and lethal skin barrier defect in mice lacking comparative gene identification-58 (CGI-58) J Biol Chem. 2010;285(10):7300–7311. doi: 10.1074/jbc.M109.081877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Festuccia WT, Laplante M, Berthiaume M, et al. PPARgamma agonism increases rat adipose tissue lipolysis, expression of glyceride lipases, and the response of lipolysis to hormonal control. Diabetologia. 2006;49(10):2427–2436. doi: 10.1007/s00125-006-0336-y. [DOI] [PubMed] [Google Scholar]
- 13.Shen WJ, Patel S, Yu Z, et al. Effects of rosiglitazone and high fat diet on lipase/esterase expression in adipose tissue. Biochim Biophys Acta. 2007;1771(2):177–184. doi: 10.1016/j.bbalip.2006.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kershaw EE, Schupp M, Guan HP, et al. PPARgamma regulates adipose triglyceride lipase in adipocytes in vitro and in vivo. Am J Physiol Endocrinol Metab. 2007;293(6):E1736–E1745. doi: 10.1152/ajpendo.00122.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watt MJ, van Denderen BJ, Castelli LA, et al. Adipose triglyceride lipase regulation of skeletal muscle lipid metabolism and insulin responsiveness. Mol Endocrinol. 2008;22(5):1200–1212. doi: 10.1210/me.2007-0485. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Ryden M, Jocken J, Van H V, et al. Comparative studies of the role of hormone-sensitive lipase and adipose triglyceride lipase in human fat cell lipolysis. Am J Physiol Endocrinol Metab. 2007;292(6):E1847–E1855. doi: 10.1152/ajpendo.00040.2007. [DOI] [PubMed] [Google Scholar]
- 17.Jocken JW, Langin D, Smit E, et al. Adipose triglyceride lipase and hormone-sensitive lipase protein expression is decreased in the obese insulin-resistant state. J Clin Endocrinol Metab. 2007;92(6):2292–2299. doi: 10.1210/jc.2006-1318. [DOI] [PubMed] [Google Scholar]
- 18.Steinberg GR, Kemp BE, Watt MJ. Adipocyte triglyceride lipase expression in human obesity. Am J Physiol Endocrinol Metab. 2007;293(4):E958–E964. doi: 10.1152/ajpendo.00235.2007. [DOI] [PubMed] [Google Scholar]
- 19.Berndt J, Kralisch S, Kloting N, et al. Adipose triglyceride lipase gene expression in human visceral obesity. Exp Clin Endocrinol Diabetes. 2008;116(4):203–210. doi: 10.1055/s-2007-993148. [DOI] [PubMed] [Google Scholar]
- 20.Jocken JW, Smit E, Goossens GH, et al. Adipose triglyceride lipase (ATGL) expression in human skeletal muscle is type I (oxidative) fiber specific. Histochem Cell Biol. 2008;129(4):535–538. doi: 10.1007/s00418-008-0386-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rasouli N, Raue U, Miles LM, et al. Pioglitazone improves insulin sensitivity through reduction in muscle lipid and redistribution of lipid into adipose tissue. Am J Physiol Endocrinol Metab. 2005;288(5):E930–E934. doi: 10.1152/ajpendo.00522.2004. [DOI] [PubMed] [Google Scholar]
- 22.Varma V, Yao-Borengasser A, Bodles AM, et al. Thrombospondin-1 is an adipokine associated with obesity, adipose inflammation, and insulin resistance. Diabetes. 2008;57(2):432–439. doi: 10.2337/db07-0840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coker RH, Williams RH, Yeo SE, et al. The impact of exercise training compared to caloric restriction on hepatic and peripheral insulin resistance in obesity. J Clin Endocrinol Metab. 2009;94(11):4258–4266. doi: 10.1210/jc.2008-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Gregorio GB, Yao-Borengasser A, Rasouli N, et al. Expression of CD68 and Macrophage Chemoattractant Protein-1 Genes in Human Adipose and Muscle Tissues: Association With Cytokine Expression, Insulin Resistance, and Reduction by Pioglitazone. Diabetes. 2005;54(8):2305–2313. doi: 10.2337/diabetes.54.8.2305. [DOI] [PubMed] [Google Scholar]
- 25.Bodles AM, Banga A, Rasouli N, et al. Pioglitazone increases secretion of high-molecular-weight adiponectin from adipocytes. Am J Physiol Endocrinol Metab. 2006;291(5):E1100–E1105. doi: 10.1152/ajpendo.00187.2006. [DOI] [PubMed] [Google Scholar]
- 26.Halvorsen YD, Bond A, Sen A, et al. Thiazolidinediones and glucocorticoids synergistically induce differentiation of human adipose tissue stromal cells: biochemical, cellular, and molecular analysis. Metab Clin Experimental. 2001;50(4):407–413. doi: 10.1053/meta.2001.21690. [DOI] [PubMed] [Google Scholar]
- 27.Bergman RN, Finegood DT, Ader M. Assessment of insulin sensitivity in vivo. Endocr Rev. 1985;6:45–86. doi: 10.1210/edrv-6-1-45. [DOI] [PubMed] [Google Scholar]
- 28.Boston RC, Stefanovski D, Moate PJ, et al. MINMOD Millennium: a computer program to calculate glucose effectiveness and insulin sensitivity from the frequently sampled intravenous glucose tolerance test. Diabetes Technol Ther. 2003;5(6):1003–1015. doi: 10.1089/152091503322641060. [DOI] [PubMed] [Google Scholar]
- 29.Zechner R, Kienesberger PC, Haemmerle G, et al. Adipose triglyceride lipase and the lipolytic catabolism of cellular fat stores. J Lipid Res. 2009;50(1):3–21. doi: 10.1194/jlr.R800031-JLR200. [DOI] [PubMed] [Google Scholar]
- 30.Schoiswohl G, Schweiger M, Schreiber R, et al. Adipose triglyceride lipase plays a key role in the supply of the working muscle with fatty acids. J Lipid Res. 2010;51(3):490–499. doi: 10.1194/jlr.M001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fischer J, Lefevre C, Morava E, et al. The gene encoding adipose triglyceride lipase (PNPLA2) is mutated in neutral lipid storage disease with myopathy. Nat Genet. 2007;39(1):28–30. doi: 10.1038/ng1951. [DOI] [PubMed] [Google Scholar]
- 32.Lefevre C, Jobard F, Caux F, et al. Mutations in CGI-58, the gene encoding a new protein of the esterase/lipase/thioesterase subfamily, in Chanarin-Dorfman syndrome. Am J Hum Genet. 2001;69(5):1002–1012. doi: 10.1086/324121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langin D, Dicker A, Tavernier G, et al. Adipocyte lipases and defect of lipolysis in human obesity. Diabetes. 2005;54(11):3190–3197. doi: 10.2337/diabetes.54.11.3190. [DOI] [PubMed] [Google Scholar]
- 34.Mairal A, Langin D, Arner P, et al. Human adipose triglyceride lipase (PNPLA2) is not regulated by obesity and exhibits low in vitro triglyceride hydrolase activity. Diabetologia. 2006;49(7):1629–1636. doi: 10.1007/s00125-006-0272-x. [DOI] [PubMed] [Google Scholar]
- 35.Kim JY, Tillison K, Lee JH, et al. The adipose tissue triglyceride lipase ATGL/PNPLA2 is downregulated by insulin and TNF-alpha in 3T3-L1 adipocytes and is a target for transactivation by PPARgamma. Am J Physiol Endocrinol Metab. 2006;291(1):E115–E127. doi: 10.1152/ajpendo.00317.2005. [DOI] [PubMed] [Google Scholar]
- 36.Kralisch S, Klein J, Lossner U, et al. Isoproterenol, TNFalpha, and insulin downregulate adipose triglyceride lipase in 3T3-L1 adipocytes. Mol Cell Endocrinol. 2005;240(1-2):43–49. doi: 10.1016/j.mce.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 37.Huijsman E, van de PC, Economou C, et al. Adipose triacylglycerol lipase deletion alters whole body energy metabolism and impairs exercise performance in mice. Am J Physiol Endocrinol Metab. 2009;297(2):E505–E513. doi: 10.1152/ajpendo.00190.2009. [DOI] [PubMed] [Google Scholar]
- 38.Alsted TJ, Nybo L, Schweiger M, et al. Adipose triglyceride lipase in human skeletal muscle is upregulated by exercise training. Am J Physiol Endocrinol Metab. 2009;296(3):E445–E453. doi: 10.1152/ajpendo.90912.2008. [DOI] [PubMed] [Google Scholar]
- 39.Yamaguchi T, Omatsu N, Matsushita S, et al. CGI-58 interacts with perilipin and is localized to lipid droplets. Possible involvement of CGI-58 mislocalization in Chanarin-Dorfman syndrome. J Biol Chem. 2004;279(29):30490–30497. doi: 10.1074/jbc.M403920200. [DOI] [PubMed] [Google Scholar]
- 40.Subramanian V, Rothenberg A, Gomez C, et al. Perilipin A mediates the reversible binding of CGI-58 to lipid droplets in 3T3-L1 adipocytes. J Biol Chem. 2004;279(40):42062–42071. doi: 10.1074/jbc.M407462200. [DOI] [PubMed] [Google Scholar]
- 41.Liu LF, Purushotham A, Wendel AA, et al. Regulation of adipose triglyceride lipase by rosiglitazone. Diabetes Obes Metab. 2009;11(2):131–142. doi: 10.1111/j.1463-1326.2008.00916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerhold DL, Liu F, Jiang G, et al. Gene expression profile of adipocyte differentiation and its regulation by peroxisome proliferator-activated receptor-gamma agonists. Endocrinology. 2002;143(6):2106–2118. doi: 10.1210/endo.143.6.8842. [DOI] [PubMed] [Google Scholar]
- 43.Vidalpuig AJ, Considine RV, Jimenezlinan M, et al. Peroxisome proliferator-activated receptor gene expression in human tissues - effects of obesity, weight loss, and regulation by insulin and glucocorticoids. J Clin Invest. 1997;99(10):2416–2422. doi: 10.1172/JCI119424. [DOI] [PMC free article] [PubMed] [Google Scholar]