Abstract
The phylum Actinobacteria hosts diverse high G + C, Gram-positive bacteria that have evolved a complex chemical language of natural product chemistry to help navigate their fascinatingly varied lifestyles. To date, 71 Actinobacteria genomes have been completed and annotated, with the vast majority representing the Actinomycetales, which are the source of numerous antibiotics and other drugs from genera such as Streptomyces, Saccharopolyspora and Salinispora. These genomic analyses have illuminated the secondary metabolic proficiency of these microbes – underappreciated for years based on conventional isolation programs – and have helped set the foundation for a new natural product discovery paradigm based on genome mining. Trends in the secondary metabolomes of natural product-rich actinomycetes are highlighted in this review article, which contains 199 references.
1 General features of Actinobacteria
The large bacterial phylum Actinobacteria is home to some of the most common soil microbial inhabitants that are responsible for degrading and recycling organic materials. This diverse group of high G + C, Gram-positive bacteria exhibits varied morphologies, physiologies, and metabolic properties that allow it to prosper in a wide range of environments. Actinobacteria are particularly noteworthy in human medicine where they encompass a number of devastating human pathogens as well as life-saving producers of antibiotics. The discovery of the antituberculosis agent streptomycin from the culture broth of Streptomyces griseus by Waksman in 1944 represented an important milestone in natural product chemotherapy1 and heralded the beginning of targeting the genus Streptomyces and related Actinomycetales microorganisms for antibiotics.
The taxonomy of the Actinobacteria is complex, and in 1997 was classified by Stackebrandt into five orders, 11 suborders, 42 families, 110 genera and more than 1000 species (Fig. 1).2 Microorganisms belonging to the order Actinomycetales are fascinatingly diverse and include human pathogens (e.g., Mycobacterium tuberculosis,3 Corynebacterium diphtheriae,4 Propionibacterium acnes,5 and Nocardia farcinica6), plant pathogens (Streptomyces scabies7 and Leifsonia xyli8), nitrogen-fixing symbionts (Frankia spp.9), terrestrial (Streptomyces spp.10-12) and marine (Salinispora spp.13) sediment inhabitants, and key members of the gut flora that reside in the colon (Bifidobacterium spp.14). A notable feature of the Actinomycetales is their metabolic versatility in the production of chemically diverse and biologically active secondary metabolites.15 Streptomyces in particular produces not only the majority of the clinical antibiotics of natural origin (e.g., vancomycin, erythromycin and tetracycline),16,17 but also gives rise to important antifungal (amphotericin B),18 anticancer (mitomycin C),19 antiparasitic (ivermectin),20 and immunosuppressive (rapamycin)21 agents. Other members of the Actinomycetales with robust metabolic features include Corynebacterium glutamicum, which is an important industrial producer of amino acids and vitamins,22 and Rhodococcus species, which have the ability to catabolize a wide range of organic compounds.23
Fig. 1.
Phylogenetic relatedness of Actinobacteria based on 16S rRNA sequence and the relative number of putative gene clusters for secondary metabolites. Strains with completed genomes are colored blue, those not yet published are noted with an asterisk, and those completed but with incomplete annotation are marked with double asterisks. Phylogenetic analysis of aligned sequences was done by a bootstrap method using bootstrap number 1000 and seed number 100.
2 Actinomycete genome projects
Since the completion by shotgun sequencing of the first genome in 1995, that of Haemophilus influenzae Rd,24 over 700 bacterial genomes have been deposited in the NCBI database. While genome-sequencing efforts were initially biased toward pathogens, more recent sequencing projects have focused on environmental and industrially relevant bacteria. To date, 53 Actinobacteria genomes have been completed and annotated, with the vast majority being in the order Actinomycetales (Table 1). Their genomes range from 1.93 Mb (Bifidobacterium animalis subsp. lactis) to 10.15 Mb (Streptomyces scabies) in size and contain 1605 (Mycobacterium leprae) to 8983 (S. scabies) protein-encoding genes. While the majority of their chromosomes are circular, most of the larger Actinomycetales chromosomes (e.g., Rhodococcus jostii RHA1,23 Rhodococcus opacus B4, and all Streptomyces strains25) form linear structures with distinct telomeres containing unique terminal-inverted repeats that bind terminal proteins.26 Although linear chromosomes, which are the predominant genetic elements in eukaryotes, have also been identified in other prokaryotes such as the Lyme disease spirochaete Borrelia burgdorferi27 and the α-proteobacterium Agrobacterium tumefaciens28,29 (the causal agent of Crown Gall disease in plants), terminal-inverted repeats and terminal proteins are absent in these chromosomes.
Table 1.
Features of Actinobacteria completed genomesa
| Microorganism | Subclass | Order | Size (bp) | GC (mol%) | Chr | Pls | Orf | %Coding | RNAs | rrn |
|---|---|---|---|---|---|---|---|---|---|---|
|
Acidimicrobium ferrooxidans DSM 10331 |
Acidimicrobidae | Acidimicrobiales | 2 145 416 | 68 | b | — | 2156 | 90 | 54 | 2 |
| Acidothermus cellulolyticus 11B | Actinobacteridae | Actinomycetales | 2 443 540 | 67 | 1C | — | 2157 | 89 | 56 | 1 |
| Actinosynnema mirum DSM 43827 | Actinobacteridae | Actinomycetales | 8 133 898 | 73 | b | — | 7258 | 86 | 75 | 5 |
| Arthrobacter aurescens TC1 | Actinobacteridae | Actinomycetales | 4 597 686 | 62 | 1C | 2C | 4041 | 88 | 73 | 6 |
| Arthrobacter chlorophenolicus A6 | Actinobacteridae | Actinomycetales | 4 395 537 | 66 | 1C | 2C | 3885 | 89 | 65 | 5 |
| Arthrobacter sp. FB24 | Actinobacteridae | Actinomycetales | 4 698 945 | 65 | 1C | 3C | 4146 | 89 | 69 | 5 |
| Beutenbergia cavernae DSM 12333 | Actinobacteridae | Actinomycetales | 4 669 183 | 73 | 1C | — | 4197 | 92 | 53 | 2 |
|
Bifidobacterium adolescentis ATCC 15703 |
Actinobacteridae | Bifidobacteriales | 2 089 645 | 59 | 1C | — | 1631 | 86 | 70 | 5 |
|
Bifidobacterium animalis subsp. lactis AD011 |
Actinobacteridae | Bifidobacteriales | 1 933 695 | 60 | 1C | — | 1528 | 84 | 59 | 2 |
|
Bifidobacterium animalis subsp. lactis ATCC SD5219 |
Actinobacteridae | Bifidobacteriales | 1 938 709 | 60 | 1C | — | 1567 | 85 | 64 | 4 |
|
Bifidobacterium animalis subsp. lactis DSM 10140 |
Actinobacteridae | Bifidobacteriales | 1 938 483 | 60 | 1C | — | 1566 | 85 | 63 | 4 |
| Bifidobacterium longum DJO10A | Actinobacteridae | Bifidobacteriales | 2 375 792 | 60 | 1C | 2C | 1990 | 86 | 73 | 4 |
| Bifidobacterium longum NCC2705 | Actinobacteridae | Bifidobacteriales | 2 256 640 | 60 | 1C | 1C | 1727 | 85 | 70 | 4 |
|
Bifidobacterium longum subsp. infantis ATCC 15697 |
Actinobacteridae | Bifidobacteriales | 2 832 748 | 59 | 1C | — | 2416 | 85 | 94 | 4 |
|
Brachybacterium faecium DSM 4810 |
Actinobacteridae | Actinomycetales | 3 608 624 | 72 | b | — | 3187 | 89 | 57 | b |
|
Clavibacter michiganensis subsp. michiganensis NCPPB 382 |
Actinobacteridae | Actinomycetales | 3 297 891 | 73 | 1C | 2C | 2984 | 89 | 60 | 2 |
|
Clavibacter michiganensis subsp. sepedonicus ATCC 33113 |
Actinobacteridae | Actinomycetales | 3 258 645 | 72 | 1C | 2C | 2941 | 86 | 51 | 2 |
|
Corynebacterium aurimucosum ATCC 700975 |
Actinobacteridae | Actinomycetales | 2 790 189 | 60 | 1C | 1C | 2531 | 88 | 67 | 4 |
|
Corynebacterium diphtheriae NCTC 13129 |
Actinobacteridae | Actinomycetales | 2 488 635 | 54 | 1C | — | 2272 | 87 | 69 | 5 |
| Corynebacterium efficiens YS-314 | Actinobacteridae | Actinomycetales | 3 147 090 | 63 | 1C | — | 2950 | 90 | 70 | 5 |
|
Corynebacterium glutamicum ATCC 13032 Bielefeld |
Actinobacteridae | Actinomycetales | 3 282 708 | 54 | 1C | — | 3002 | 87 | 86 | 6 |
|
Corynebacterium glutamicum ATCC 13032 Kitasato |
Actinobacteridae | Actinomycetales | 3 309 401 | 54 | 1C | — | 2993 | 86 | 80 | 6 |
| Corynebacterium glutamicum R | Actinobacteridae | Actinomycetales | 3 314 179 | 54 | 1C | 1C | 3052 | 86 | 76 | 6 |
| Corynebacterium jeikeium K411 | Actinobacteridae | Actinomycetales | 2 462 499 | 61 | 1C | 1C | 2104 | 89 | 61 | 3 |
|
Corynebacterium kroppenstedtii DSM 44385 |
Actinobacteridae | Actinomycetales | 2 446 804 | 57 | 1C | — | 2018 | 86 | 55 | 3 |
|
Corynebacterium urealyticum DSM 7109 |
Actinobacteridae | Actinomycetales | 2 369 219 | 64 | 1C | — | 2024 | 89 | 60 | 3 |
| Eggerthella lenta DSM 2243 | Coriobacteridae | Coriobacteriales | 3 610 731 | 64 | b | — | 3181 | 87 | 53 | 3 |
| Frankia alni ACN14a | Actinobacteridae | Actinomycetales | 7 497 934 | 73 | 1C | — | 6711 | 86 | 63 | 2 |
| Frankia sp. CcI3 | Actinobacteridae | Actinomycetales | 5 433 628 | 70 | 1C | — | 4499 | 84 | 70 | 2 |
| Frankia sp. EAN1pec | Actinobacteridae | Actinomycetales | 8 982 042 | 71 | 1C | — | 7191 | 83 | 59 | 3 |
|
Kineococcus radiotolerans SRS30216 |
Actinobacteridae | Actinomycetales | 4 761 183 | 74 | 1C | 2C | 4480 | 90 | 64 | 4 |
| Kocuria rhizophila DC2201 | Actinobacteridae | Actinomycetales | 2 697 540 | 71 | 1C | — | 2357 | 88 | 57 | 3 |
| Kytococcus sedentarius DSM 20547 | Actinobacteridae | Actinomycetales | 2 788 433 | 71 | b | — | 2703 | 90 | 51 | 2 |
|
Leifsonia xyli subsp. xyli str. CTCB07 |
Actinobacteridae | Actinomycetales | 2 584 158 | 68 | 1C | — | 2030 | 70 | 50 | 1 |
|
Micrococcus luteus Fleming NCTC 2665 |
Actinobacteridae | Actinomycetales | 2 501 097 | 72 | 1C | — | 2345 | 88 | 56 | 2 |
|
Mycobacterium abscessus CIP 104536 |
Actinobacteridae | Actinomycetales | 5 067 172 | 64 | 1C | 1C | 4920 | 92 | 50 | 1 |
| Mycobacterium avium 104 | Actinobacteridae | Actinomycetales | 5 475 491 | 69 | 1C | — | 5120 | 88 | 50 | 1 |
|
Mycobacterium avium subsp. paratuberculosis K-10 |
Actinobacteridae | Actinomycetales | 4 829 781 | 69 | 1C | — | 4350 | 91 | 49 | 1 |
| Mycobacterium bovis AF2122/97 | Actinobacteridae | Actinomycetales | 4 345 492 | 66 | 1C | — | 3920 | 90 | 50 | 1 |
|
Mycobacterium bovis BCG str. Pasteur 1173P2 |
Actinobacteridae | Actinomycetales | 4 374 522 | 66 | 1C | — | 3952 | 90 | 52 | 1 |
|
Mycobacterium bovis BCG str. Tokyo 172 |
Actinobacteridae | Actinomycetales | 4 371 711 | 65 | 1C | — | 3947 | 90 | 50 | 1 |
| Mycobacterium gilvum PYR-GCK | Actinobacteridae | Actinomycetales | 5 619 607 | 68 | 1C | 3C | 5241 | 92 | 55 | 2 |
| Mycobacterium leprae Br4923 | Actinobacteridae | Actinomycetales | 3 268 071 | 57 | 1C | — | 1604 | 49 | 47 | b |
| Mycobacterium leprae TN | Actinobacteridae | Actinomycetales | 3 268 203 | 58 | 1C | — | 1605 | 49 | 50 | 1 |
| Mycobacterium marinum M | Actinobacteridae | Actinomycetales | 6 636 827 | 66 | 1C | 1C | 5423 | 89 | 51 | 1 |
|
Mycobacterium smegmatis str. MC2 155 |
Actinobacteridae | Actinomycetales | 6 988 209 | 67 | 1C | — | 6716 | 90 | 54 | 2 |
| Mycobacterium sp. JLS | Actinobacteridae | Actinomycetales | 6 048 425 | 68 | 1C | — | 5739 | 92 | 57 | 2 |
| Mycobacterium sp. KMS | Actinobacteridae | Actinomycetales | 5 737 227 | 68 | 1C | 2C | 5460 | 92 | 57 | 2 |
| Mycobacterium sp. MCS | Actinobacteridae | Actinomycetales | 5 705 448 | 68 | 1C | 1C | 5391 | 92 | 61 | 2 |
|
Mycobacterium tuberculosis CDC1551 |
Actinobacteridae | Actinomycetales | 4 403 837 | 66 | 1C | — | 4189 | 90 | 48 | 1 |
| Mycobacterium tuberculosis F11 | Actinobacteridae | Actinomycetales | 4 424 435 | 66 | 1C | — | 3950 | 90 | 48 | 1 |
| Mycobacterium tuberculosis H37Ra | Actinobacteridae | Actinomycetales | 4 419 977 | 66 | 1C | — | 4034 | 90 | 50 | 1 |
| Mycobacterium tuberculosis H37Rv | Actinobacteridae | Actinomycetales | 4 411 532 | 66 | 1C | — | 4402 | 90 | 50 | 1 |
| Mycobacterium ulcerans Agy99 | Actinobacteridae | Actinomycetales | 5 631 606 | 66 | 1C | — | 4160 | 72 | 50 | 1 |
| Mycobacterium vanbaalenii PYR-1 | Actinobacteridae | Actinomycetales | 6 491 865 | 68 | 1C | — | 5979 | 91 | 58 | 2 |
| Nocardia farcinica IFM 10152 | Actinobacteridae | Actinomycetales | 6 021 225 | 71 | 1C | 2C | 5683 | 90 | 64 | 3 |
| Nocardioides sp. JS614 | Actinobacteridae | Actinomycetales | 4 985 871 | 71 | 1C | 1C | 4645 | 91 | 55 | 2 |
|
Propionibacterium acnes KPA171202 |
Actinobacteridae | Actinomycetales | 2 560 265 | 60 | 1C | — | 2297 | 89 | 54 | 3 |
|
Renibacterium salmoninarum ATCC 33209 |
Actinobacteridae | Actinomycetales | 3 155 250 | 56 | 1C | — | 3507 | 90 | 51 | 2 |
| Rhodococcus erythropolis PR4 | Actinobacteridae | Actinomycetales | 6 516 310 | 62 | 1C | 2C1L | 6030 | 91 | 74 | 5 |
| Rhodococcus jostii RHA1c | Actinobacteridae | Actinomycetales | 7 804 765 | 67 | 1L | 3L | 7211 | 91 | 63 | 4 |
| Rhodococcus opacus B4 | Actinobacteridae | Actinomycetales | 7 913 450 | 67 | 1L | — | 7246 | 91 | 62 | 4 |
|
Rubrobacter xylanophilus DSM 9941 |
Rubrobacteridae | Rubrobacterales | 3 225 748 | 71 | 1C | — | 3140 | 91 | 64 | 1 |
|
Saccharopolyspora erythraea NRRL 2338 |
Actinobacteridae | Actinomycetales | 8 212 805 | 71 | 1C | — | 7198 | 84 | 66 | 4 |
| Salinispora arenicola CNS-205 | Actinobacteridae | Actinomycetales | 5 786 361 | 70 | 1C | — | 4917 | 85 | 63 | 3 |
| Salinispora tropica CNB-440 | Actinobacteridae | Actinomycetales | 5 183 331 | 70 | 1C | — | 4536 | 88 | 61 | 3 |
| Streptomyces avermitilis MA-4680 | Actinobacteridae | Actinomycetales | 9 025 608 | 71 | 1L | 2L | 7577 | 86 | 89 | 6 |
| Streptomyces coelicolor A3(2) | Actinobacteridae | Actinomycetales | 8 667 507 | 72 | 1L | 1L1C | 7769 | 88 | 86 | 6 |
|
Streptomyces griseus subsp. griseus IFO13350 |
Actinobacteridae | Actinomycetales | 8 545 929 | 72 | 1L | — | 7136 | 87 | 86 | 6 |
| Streptomyces scabies 87.22d | Actinobacteridae | Actinomycetales | 10 148 695 | 72 | 1L | — | 8983 | 87 | 92 | 6 |
| Thermobifida fusca YX | Actinobacteridae | Actinomycetales | 3 642 249 | 68 | 1C | — | 3110 | 85 | 67 | 4 |
Abbreviations: Chr, chromosome; Pls, plasmid; C, circular replicon; L, linear replicon; Orf, open-reading frames; RNAs, functional RNA molecules (tRNA, rRNA, tmRNA, 4.5S RNA, RNaseP; rrn, ribosomal RNA operon (16S rRNA–23S rRNA–5S rRNA).
not defined.
The species name in the first report was not defined but the strain has since been classified as Rhodococcus jostii.
The detailed properties are not published but sequence and annotation data are available from the Sanger Centre (ftp://ftp.sanger.ac.uk/pub/pathogens/ssc/).
Homology searching of genes encoding known proteins involved in secondary metabolism reveals that Actinomycetales microbes contain a wide range of diverse secondary metabolic biosynthesis gene clusters from just a couple to well over 30 pathways (Fig. 1). Some families in particular, such as the Streptomycetaceae (genera Streptomyces and Kitasatospora), Micromonosporaceae (Salinispora and Micromonospora) and Pseudonocardiaceae (Saccharopolyspora), regularly possess more than twenty gene clusters for secondary metabolism and in doing so dedicate over 5% of their coding capacity to these processes. The natural product-rich Actinomycetales organisms generally have large genomes of >5 Mb and concentrate their secondary metabolic genes distal to the origin of replication gene dnaA (Fig. 2). Not all Actinomycetales, however, are prolific secondary metabolite producers, especially those with small genomes ranging from 2–4 Mb such as Kocuria rhizophila (2.7 Mb), which harbors just two secondary metabolic pathways.30
Fig. 2.
Circular representation of the actinomycete chromosomes S. coelicolor A3(2), S. erythraea NRRL2338 and S. tropica CNB-440 oriented to the dnaA gene (top). All genomes are scaled to their relative size. The inner rings show a normalized plot of GC skew, while the center rings show a normalized plot of GC content. The outer circles show the distribution of secondary metabolite gene clusters. Biosynthetic gene clusters associated with thiotemplate-based assembly (PKS, NRPS) are depicted in red, terpene clusters in blue, and loci encoding all other secondary metabolic pathways are marked in black. Clusters whose products have been isolated are labeled accordingly.
The Tuberculosis Database (TBDB) (http://www.tbdb.org/) was recently developed as a single repository to house genome sequence and annotation data together with gene expression data.31 As of September 2008, TBDB hosted 28 different M. tuberculosis strains as well as other mycobacteria and bacteria from related taxa such as Streptomyces coelicolor A3(2), Streptomyces avermitilis, Nocardia farcinica IFM 10152, and Rhodococcus jostii RHA1. Furthermore, TBDB houses nearly 1500 public tuberculosis and 260 Streptomyces microarrays. This database provides a suite of analytical tools for the comparative genomic analysis of M. tuberculosis and is intended to serve as a community hub for TB research but may also facilitate genomic research in the greater actinomycete scientific community. A number of other TB databases exist, including GenoMycDB (a database for comparative analysis of mycobacterial genes and genomes)32 and MGDD (Mycobacterium tuberculosis genome divergence database), yet they are at present more limited in scope.33 The rich secondary metabolism of mycobacteria, which is employed in the elaborate construction of its specialized lipid components, has been previously reviewed34 and is thus not covered in this review article. Rather, we focus our attention on reporting and discussing trends in select natural product-rich Actinobacteria with completed genome sequences.
2.1 Streptomyces
The Streptomyces genus is the largest within the Actinobacteria, with over 900 described species (http://www.ncbi.nlm.nih.gov/Taxonomy/). Their natural product biosynthetic prowess is arguably unsurpassed amongst the microbes, and hence they are of utmost importance in the pharmaceutical industry.15 To date, four Streptomyces genomes have been completed and are publicly available (Table 1), while numerous other Streptomyces genome projects have either been completed privately or are in draft assembly (such as the 16 draft Streptomyces genome sequences assembled at the Broad Institute – http://www.broadinstitute.org/annotation/genome/streptomyces_group). These studies revealed that Streptomyces have large (~8–10 Mb), linear chromosomes25 containing well over 20 secondary metabolic gene clusters, which encode the biosynthesis of polyketides by polyketide synthases (PKSs),35 peptides by nonribosomal peptide synthetases (NRPSs),36 bacteriocins,37 terpenoids, shikimate-derived metabolites, aminoglycosides, and other natural products. In this section we highlight the individual and comparative secondary metabolic features of Streptomyces coelicolor A3(2),10 Streptomyces avermitilis MA-4680,11,38 and Streptomyces griseus IFO 13350.12 The fourth species with a completed genome sequence, Streptomyces scabies strain 87.22, has not yet been published but is publicly available online at http://www.sanger.ac.uk/Projects/S_scabies/.
2.1.1 Streptomyces coelicolor A3(2)
Streptomyces coelicolor A3(2) is genetically the best known representative of the genus and has served as a model strain for the study of actinomycete chemistry and biology.39 Its single, linear chromosome from the derivative strain M145, which lacks the plasmids SCP1 and SCP2 of the parent strain A3(2), was sequenced and annotated in 2002 from an ordered cosmid library and contains 8 667 507 bp (72.1% GC content) coding for 7769 genes and 56 pseudogenes (Table 1).10 The linear SCP140 (356 023 bp; 69.1% GC content; 351 coding sequences) and the circular SCP241 (31 317 bp; 72.1% GC content; 34 coding sequences) plasmids were later sequenced separately.
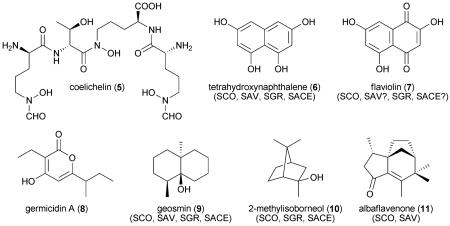
One of the main reasons why S. coelicolor A3(2) was developed as a model genetics system is its production of diffusible pigments that could serve as convenient genetic markers.42 At the time the genome sequence was released, the structures and biosynthetic gene clusters associated with the blue aromatic polyketide antibiotic actinorhodin (1),43 the red oligopyrrole prodiginine antibiotics (2),44,45 and the acidic lipopeptide calcium-dependent antibiotics (3)46 were known. Furthermore, the whiE type II PKS gene cluster encoding an unknown aromatic dodecaketide associated with the gray spore pigment was established47 as was the chemistry of the common streptomycete membrane lipid hopanoids hopene (4) and 29-(2′-hydroxyethyl)hopane.48 The initial genomic analysis of the S. coelicolor A3(2) chromosome predicted a further 17 secondary metabolic gene clusters.10 At present there are 29 predicted chromosomal gene clusters specifying secondary metabolites and another two from the linear plasmid SCP1 (Table 2). The majority of these gene clusters reside outside the central core of the chromosome in the subtelomeric regions.10 Their associated pathway products arise from a myriad of enzymatic processes and include polyketides, nonribosomal peptides, bacteriocins, terpenoids, and other natural products, which is a hallmark of streptomycete genomes. The most thoroughly investigated S. coelicolor A3(2) pathway involves the biosynthesis of the aromatic type II PKS product actinorhodin,49 which significantly was the target of the first genetic engineering experiment to yield an “unnatural” hybrid molecule50 and helped establish the scientific concept of smallmolecule rational engineering through combinatorial biosynthesis.51
Table 2.
Secondary metabolite gene clusters in Streptomyces coelicolor A3(2)
| No. | Cluster location | Actual or predicted producta |
|---|---|---|
| 1 | SCO0124-0129 | Eicosapentaenoic acid |
| 2 | SCO0185-0191 | Isorenieratene (12) |
| 3 | SCO0267-0270 | Lantibiotic |
| 4 | SCO0381-0401 | Deoxysugar |
| 5 | SCO0489-0499 | Coelichelin (5) |
| 6 | SCO0753-0756 | Bacteriocin |
| 7 | SCO1206-1208 | THN (6), flaviolin (7) |
| 8 | SCO1265-1273 | Aromatic polyketide |
| 9 | SCO1864-1867 | 5-Hydroxyectoine (14) |
| 10 | SCO2700-2701 | Melanin |
| 11 | SCO2782-2785 | Desferrioxamine (13) |
| 12 | SCO3210-3249 | Calcium-dep. antibiotic (3) |
| 13 | SCO5071-5092 | Actinorhodin (1) |
| 14 | SCO5222-5223 | Albaflavenone (11) |
| 15 | SCO5314-5320 | Gray spore pigment |
| 16 | SCO5799-5801 | Siderophore |
| 17 | SCO5877-5898 | Prodiginine (2) |
| 18 | SCO6073 | Geosmin (9) |
| 19 | SCO6266 | γ-Butyrolactone Scb1 (15) |
| 20 | SCO6273-6288 | Hexaketide |
| 21 | SCO6429-6438 | Dipeptide |
| 22 | SCO6681-6685 | Lanthionine-cont. peptide SapB |
| 23 | SCO6759-6771 | Hopene (4) |
| 24 | SCO6826-6827 | Polyketide |
| 25 | SCO6927-6932 | Lantibiotic |
| 26 | SCO7221 | Germicidin (8) |
| 27 | SCO7669-7671 | Aromatic polyketide |
| 28 | SCO7681-7691 | Coelibactin |
| 29 | SCO7700-7701 | 2-Methylisoborneol (10) |
| 30 | SCP1.228c-246 |
Methylenomycin (18), 2-alkyl-4- (hydroxymethyl)furan-3- carboxylic acids (17) |
Observed products are highlighted in bold.
The complete sequence and annotation of the S. coelicolor A3(2) genome allowed for its rational mining to identify many of the orphan pathway products.52 Structure prediction of the product of the trimodular NRPS associated with SCO0492 initially suggested a tripeptide siderophore53 that upon subsequent isolation yielded the new tetrapeptide iron chelator coelichelin (5).54 This seminal discovery of a novel natural product by genomic prediction helped usher in a new era of microbial “genome mining” for new chemical entities.52,55,56 In the case of 5, this discovery not only allowed the characterization of new biosynthetic reactions in NRPS biochemistry but also led the way to the characterization of further S. coelicolor A3(2) metabolites often with very intriguing structures, biosyntheses, and biological properties. Three type III PKS gene clusters are encoded in the genome, and the products of two of these pathways, 1,3,6,8-tetrahydroxynaphthalene (THN) (6) and its auto-oxidation product flaviolin (7) by THN synthase57,58 (SCO1206), and the germicidin (8) family of alkyl pyrones by germicidin synthase59 (SCO7221), have been characterized, while that of SCO7671 is unknown. Terpenoid biochemistry is also prevalent in S. coelicolor A3(2), with at least five gene clusters dedicated to this type of biosynthesis (Table 2). After the completion of the genome analysis, recombinant enzymes associated with three odorous-terpenoid compounds, namely geosmin60,61 (9), 2-methylisoborneol62 (10) and albaflavenone63-65 (11), were characterized and their pathway products confirmed. Geosmin and 2-methylisoborneol are widespread not only in Actinobacteria but also in Cyanobacteria and Myxobacteria as well as fungi.66 The remaining terpenoid pathway involves the carotenoids isorenieratene (12) and β-carotene, which were shown to be produced under blue light induction.67 Further genome mining efforts led to the tris-hydroxamate family of desferrioxiamine (13) siderophores,68 which are assembled by a new family of ATP-dependent oligomerization-macrocyclization biocatalysts.69 Production of ectoine and 5-hydroxyectoine (14), compatible solutes conferring dehydration protection in the genus Streptomyces, is encoded by a set of four genes (SCO1864–SCO1867) and can be triggered upon exposure to high salinity or elevated growth temperature.70 While no ribosomally-derived bacteriocins have been characterized from S. coelicolor A3(2), four putative gene clusters are likely (Table 2), with SCO6681–SCO6685 being very homologous to the SapB lantibiotic-like peptide from S. coelicolor J1501/pKN22.71
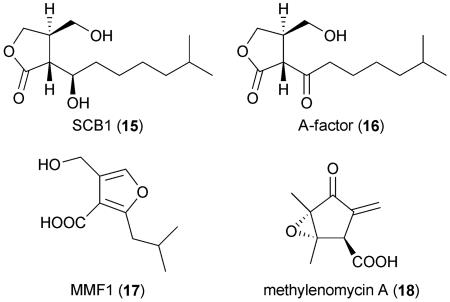
The biosynthesis of an unknown polyketide metabolite encoded by the cryptic modular PKS gene cluster SCO6273–SCO6288 is partially controlled by the signaling compound Scb1 (15),72-74 which is a γ-butyrolactone structurally related to the prototypical actinomycete hormone A-factor (16) that regulates antibiotic production and morphological differentiation in S. griseus.75 The Scb1 butenolide synthase gene scbA SCO6266 is located adjacent to the PKS gene set that it regulates. A homolog, mmfL, is located within the linear plasmid SCP1, where it facilitates the biosynthesis of 2-alkyl-4-(hydroxymethyl)furan-3-carboxylic acids (17).76,77 These molecules specifically induce the production of the antibiotic methylenomycin (18)78,79 and hence have been collectively termed the methylenomycin furans.76 Since glycosylated derivatives of 17 have been described from other streptomycetes80 and a related three-gene operon harboring mmfL is present in the S. avermitilis genome, these furans may constitute a new class of diffusible signaling molecules much like the better-known γ-butyrolactones.
2.1.2 Streptomyces avermitilis MA-4680
The draft genome sequence of the industrial microorganism S. avermitilis was published in 2001 and allowed the first genome-level glimpse into the extensive secondary metabolic dexterity of a streptomycete.38 Its completed nucleotide sequence was published two years later and showed that the linear genome contains 9 025 608 bp (70.7% GC content) and encodes 7582 protein coding sequences (Fig. 3).11 S. avermitilis also harbors two linear plasmids, SAP1 (94 287 bp; 69.2% GC content; 96 coding sequences) and SAP2 (ca. 200 kb).
Fig. 3.
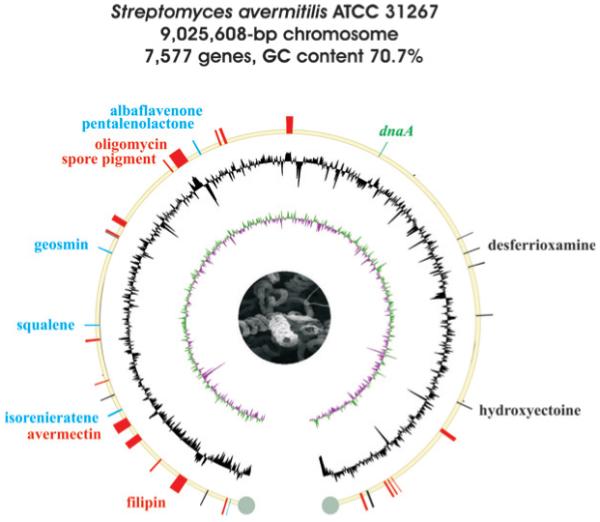
Schematic representation of the S. avermitilis MA-4680 chromosome. Drawing details are the same as for Fig. 2.
Streptomyces avermitilis was discovered in 1977 from a soil sample in Japan and produces the polyketide macrolide avermectin (19), which is a potent anthelmintic agent extremely effective against nematodes, insects and spiders. Its semisynthetic derivative ivermectin has been in use commercially since the 1980s in human and veterinary medicine.81,82 Prior to the availability of the genome sequence, the structure and biosynthesis of avermectin83 and that of another polyketide macrolide, oligomycin84 (20), had already been established. A further 11 PKS gene clusters (Table 3) were deduced from the genome sequence, leading to the subsequent isolation of a third group of macrolides,85 the filipins (21), which belong to the polyene family of antifungal agents previously isolated from Streptomyces filipinensis.86 As with S. coelicolor A3(2), S. avermitilis produces a brown spore pigment associated with the whiE-homologous type II PKS cluster spp, which upon disruption led to albino-colored spores (H. Ikeda, unpublished). Homologs of the whiE and spp clusters are prevalent in other Streptomyces microbes,47 yet not in S. griseus, which instead produces polyketide melanins via a type III PKS pathway.87
Table 3.
Secondary metabolite gene clusters in Streptomyces avermitilis MA-4680
| No. | Cluster location | Actual or predicted producta |
|---|---|---|
| 1 | SAV_76 | Terpene |
| 2 | SAV_100–101 | Polyketide |
| 3 | SAV_257–259 | Microcin |
| 4 | SAV_407–419 | Filipin (21) |
| 5 | SAV_603–609 | Peptidic siderophore |
| 6 | SAV_837–869 | Non-ribosomal peptide |
| 7 | SAV_935–953 | Avermectin (19) |
| 8 | SAV_1019–1025 | Isorenieratene (12) |
| 9 | SAV_1136–1137 | Melanin |
| 10 | SAV_1249–1251 | PK-NRP hybrid |
| 11 | SAV_1550–1552 | Polyketide |
| 12 | SAV_1650–1654 | Hopene (4) |
| 13 | SAV_2163 | Geosmin (9) |
| 14 | SAV_2267–2269 | 2-alkyl-4-hydroxymethylfuran-3- carboxylic acids |
| 15 | SAV_2277–2282 | Polyketide |
| 16 | SAV_2367–2369 | Polyketide |
| 17 | SAV_2372–2388 | Aromatic polyketide |
| 18 | SAV_2465–2467 | Siderophore |
| 19 | SAV_2835–2842 | Spore pigment |
| 20 | SAV_2890–2903 | Oligomycin (20) |
| 21 | SAV_2990–3002 | Neopentalenolactone (23) |
| 22 | SAV_3031–3032 | Albaflavenone (11) |
| 23 | SAV_3155–3164 | Non-ribosomal peptide |
| 24 | SAV_3193–3202 | Non-ribosomal peptide |
| 25 | SAV_3636–3651 | Non-ribosomal peptide |
| 26 | SAV_3653–3667 | Aromatic polyketide |
| 27 | SAV_5149 | Ochronotic pigment |
| 28 | SAV_5269–5272 | Desferrioxamine (13) |
| 29 | SAV_5361–5362 | Melanin |
| 30 | SAV_5686–5689 | Microcin |
| 31 | SAV_6395–6398 | 5-Hydroxyectoine (14) |
| 32 | SAV_6632–6633 | Non-ribosomal peptide |
| 33 | SAV_7130–7131 | THN, flaviolin |
| 34 | SAV_7161–7165 | Non-ribosomal peptide |
| 35 | SAV_7184 | Polyketide |
| 36 | SAV_7320–7323 | Vibrioferrin-like siderophore |
| 37 | SAV_7360–7362 | Polyketide |
Observed products are highlighted in bold.
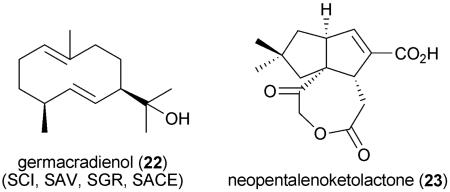
In addition to the enormous diversity of PKS pathways in S. avermitilis,88 the genome harbors secondary metabolic gene clusters for at least eight nonribosmal peptides, six terpenoids, and assorted pigments, osmolyte compounds, siderophores, and bacteriocins,89 for a total of 37 secondary metabolic gene clusters spanning 6.6% of the genome (Table 3). Most of the secondary metabolic genes are present in the chromosomal arms rather than in the core region of the chromosome. While no peptide products have yet been characterized, the majority of the terpenoids have either been characterized or their structures deduced based on biosynthetic precedent. The triterpene hopene (4) (H. Ikeda, unpublished), its abundant precursor squalene, the carotenoid isorenieratene67 (12), the odorous sesquiterpenoids geosmin90 (9), germacradienol90 (22), and albaflavenone (11) (via its precursor epi-isozizaene65), and derivatives of the sesquiterpene antibiotic neopentalenolactone91-93 (23) from Baeyer–Villiger monooxygenase chemistry, have all been characterized. While all the S. avermitilis terpenoid chemistry had been previously identified in other streptomycete strains, investigating the corresponding biosynthesis enzymes as recombinant proteins has been instrumental in shedding light onto the mechanisms by which these common streptomycete terpenoid molecules are assembled. In the case of 9 and 22, the gene product of SAV_2163 catalyzes their Mg-dependent formation from farnesyl diphosphate.90
Streptomyces avermitilis produces the desferrioxamine-related siderophore nocardamine (24) under iron-limiting conditions (H. Ikeda, unpublished). However, unlike S. coelicolor A3(2), it does not produce additional siderophores via NRPS logic such as coelichelin (5), although it possesses the genetic capacity. The NRPS SAV_603 exhibits 65% homology to a myxobacterial counterpart involved in the biosynthesis of the siderophore myxochelin94 and is associated with three putative iron(III)/siderophore transporter genes (SAV_600–SAV_602). The S. avermitilis genome also harbors another gene cluster (SAV_7320–SAV_7323) that may code for a novel streptomycete siderophore related to vibrioferrin from the Gram-negative bacterium Vibrio parahaemolyticus,95 yet the product of this pathway as well as the majority of the S. avermitilis secondary metabolome are presently unknown.
2.1.3 Streptomyces griseus IFO 13350
Streptomyces griseus IFO 13350 was discovered in S. A. Waksman’s laboratory more than 60 years ago and is famous for producing the aminoglycoside antibiotic streptomycin (25), which was instrumental in helping save people suffering from tuberculosis.1 Its complete genome sequence was published in 2008 and consists of a single linear chromosome of 8 546 929 bp with no plasmids (Fig. 4) and containing extremely long terminal inverted repeats of 132 910 bp each.12 In addition to its industrial importance in producing streptomycin, S. griseus is an important model bacterium for studying extracellular signaling by a diffusible, low-molecular weight compound, the γ-butyrolactone A factor (16).75,96
Fig. 4.
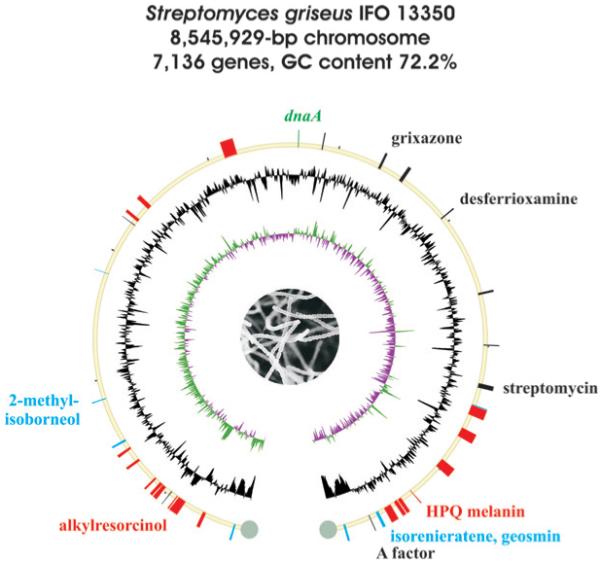
Schematic representation of the S. griseus IFO 13350 genome. Drawing details are the same as for Fig. 2.
Sequence analysis of the S. griseus genome and comparison against the other completed Streptomyces genomes suggested the existence of at least 36 genes or gene clusters associated with the biosynthesis of secondary metabolites (Table 4). As with S. coelicolor A3(2) and S. avermitilis, the majority reside outside the core region of the chromosome. In addition to streptomycin (25) whose biosynthesis had previously been characterized,97 biosynthetic genes and gene clusters were also known for A factor (16),98 the diffusible yellow pigmented grixazones (26),99 an uncharacterized carotenoid,100 and the 1,4,6,7,9,12-hexahydroxyperylene-3,10-quinone (HPQ) melanin (27)87,101 via the prototype bacterial type III PKS THN synthase (RppA).102 HPQ melanin is the dominant spore pigment of S. griseus and is distinct from aromatic polyketide spore pigments associated with the whiE gene cluster of S. coelicolor A3(2) and S. avermitilis which are absent in this strain. A second type III PKS pathway involving SGR470–SGR472 was later established to producealkylresorcinol (28) and alkylpyrone (29) phenolic lipids that confer penicillin resistance on the strain.103
Table 4.
Secondary metabolite gene clusters in Streptomyces griseus IFO13350
| No. | Cluster location | Actual or predicted producta |
|---|---|---|
| 1 | SGR_54–60 | Carotenoid |
| 2 | SGR_278–283 | PK-NRP hybrid |
| 3 | SGR_443–455 | Peptidic siderophore |
| 4 | SGR_470–472 | Alkylresorcinol (28) |
| 5 | SGR_527–528 | Melanin |
| 6 | SGR_574–593 | Non-ribosomal peptide |
| 7 | SGR_604-611 | 9-Membered enediyne |
| 8 | SGR_653–656 | Non-ribosomal peptide |
| 9 | SGR_810–815 | PK-NRP hybrid |
| 10 | SGR_895–901 | Non-ribosomal peptide |
| 11 | SGR_962–966 | Hopanoid |
| 12 | SGR_1268–1269 | 2-Methylisoborneol (10) |
| 13 | SGR_2079 | Terpene |
| 14 | SGR_2446–2447 | Melanin |
| 15 | SGR_2482–2489 | PK-NRP hybrid |
| 16 | SGR_2586–2598 | Non-ribosomal peptide |
| 17 | SGR_3239–3288 | PK-NRP hybrid |
| 18 | SGR_3845–3849 | Lantibiotic |
| 19 | SGR_4238–4250 | Grixazone (26) |
| 20 | SGR_4408–4421 | Thiazolylpeptide |
| 21 | SGR_4747–4751 | Desferrioxamine (13) |
| 22 | SGR_5285–5295 | Unknown |
| 23 | SGR_5632–5635 | Ectoine, 5-hydroxyectoine (14) |
| 24 | SGR_5914–5940 | Streptomycin (25) |
| 25 | SGR_6065 | Terpene |
| 26 | SGR_6071–6083 | Polyketide |
| 27 | SGR_6177–6183 | Polyketide |
| 28 | SGR_6360–6387 | Polyketide |
| 29 | SGR_6619–6620 | HPQ melanin (27) |
| 30 | SGR_6709–6717 | Peptidic siderophore |
| 31 | SGR_6730–6742 | Peptidic siderophore |
| 32 | SGR_6776–6786 | PK-NRP hybrid |
| 33 | SGR_6824–6830 | Isorenieratene (12) |
| 34 | SGR_6839 | Geosmin (9) |
| 35 | SGR_6889 | A factor (16) |
| 36 | SGR_7079–7085 | Carotenoid |
Observed products are highlighted in bold.
The majority of the S. griseus secondary metabolome is dominated by thiotemplate-based chemistry involving a total of at least nine PKS and nine NRPS gene clusters (Table 4). None of the natural product chemistry associated with the type I and type II PKS gene clusters, including the putative enediyne PKS (SGR604–SGR611) – which is similar to those common in Salinispora genome-sequenced strains104 – as well as from the NRPS pathways, has been characterized to date. Amongst the NRPS gene clusters, three (SGR443–SGR455, SGR6709–SGR6717 and SGR6730–SGR6742) are associated with siderophore-dependent iron transporter genes and thus are likely involved in the biosynthesis of peptidic siderophores as seen in S. coelicolor A3(2).
Terpenoid biosynthetic pathways also predominate in the S. griseus genome and include three copies of carotenoid gene clusters, four orthologs of terpene cyclases, and a cluster for hopanoid biosynthesis (Table 4). The carotenoid gene cluster of SGR54–SGR60 is identical to that of SGR7079–SGR7085 and together they reside in the long inverted-repeat telomere regions. Orthologs corresponding to the geosmin/germacradienol and 2-methylisoborneol terpene synthases have been identified, and the production of 2-methylisoborneol (10) was experimentally confirmed.66 The remaining terpene synthases, however, likely utilize farnesyl diphosphate for the synthesis of as-yet unknown sesquiterpenoid compounds. Further cryptic clusters associated with the biosynthesis of the siderophore nocardamine, melanins derived from l-dopamine, and bacteriocins are also predicted from the S. griseus genome.
2.1.4 Streptomyces comparative genomics
Sequence comparison of the three taxonomically distinct Streptomyces genome strains, S. coelicolor A3(2), S. avermitilis and S. griseus, suggests a conserved internal core region of approximately 6.4 Mb in which the majority of the genes are conserved and show global synteny (Fig. 5).12 Flanking the conserved core are variable subtelomeric regions of 0.5–2 Mb in size that carry mainly nonessential variable genes that include the majority of the secondary metabolic gene clusters. This variability was also reported in the S. ambofaciens chromosome,105 suggesting that this feature is common in Streptomyces linear chromosomes. Large DNA rearrangements involving gene duplication, elimination and acquisition are frequently observed in the chromosomal arms, suggesting that chromosomal instability and intermolecular recombination are important driving forces in Streptomyces evolution.106,107 Streptomyces typically carry large linear plasmids of >200 kb that can harbor a multitude of secondary metabolic gene clusters108 as in the case of pSLA2-L of S. rochei,109 and their chromosomal integration110 may provide the immediate opportunity for the production of a suite of bioactive products.
Fig. 5.

The comparative analysis of nucleotide sequences of S. griseus IFO 13350 (top), S. avermitilis MA-4680 (middle) and S. coelicolor A3(2) (bottom) using the MURASAKI program version 1.40 (weight; 45, length; 90). Predicted core regions (blue lines) account for 6.28 Mb of S. coelicolor A3(2), 6.50 Mb of S. avermitilis and 6.39 Mb of S. griseus and are centered about the replication of origin oriC (black arrow). The subtelomeric regions (bold lines) are less conserved with regard to sequence and ortholog distribution and contain more than half of the gene clusters for secondary metabolism (colored triangles).
The three Streptomyces genomes have seven conserved gene clusters, including those for hopanoid (4) and desferrioxamine (13) siderophore biosyntheses at the core region of their respective chromosomes (Fig. 6). Each gene cassette is arranged identically in terms of size and direction and is located within very similar genomic regions. On the other hand, other examples exist in which species-specific secondary metabolic gene clusters reside in genomic islands within conserved regions, as is the case with the oligomycin (20) cluster in S. avermitilis and the streptomycin (25) cluster in S. griseus (Fig. 7). The oligomycin biosynthesis gene cluster in S. avermitilis is inserted between the conserved F0–F1 ATPase operon and the teichoic acid biosynthetic gene tagO, while the S. griseus streptomycin gene cluster is inserted between the phenylalanyl-tRNA synthetase operon and the fatty acid catabolic gene fadC. Although the origins of these secondary metabolic gene clusters are not clear, they were most likely acquired through horizontal gene transfer. The genomic island concept linking secondary metabolism with functional adaptation and acquisition has recently been explored in the marine actinomycete genus Salinispora,104 and is further discussed in Section 2.3.
Fig. 6.

Conserved genomic regions amongst S. griseus IFO 13350, S. avermitilis MA-4680 and S. coelicolor A3(2), harboring (A) hopanoid and (B) desferrioxamine biosynthetic gene clusters.
Fig. 7.
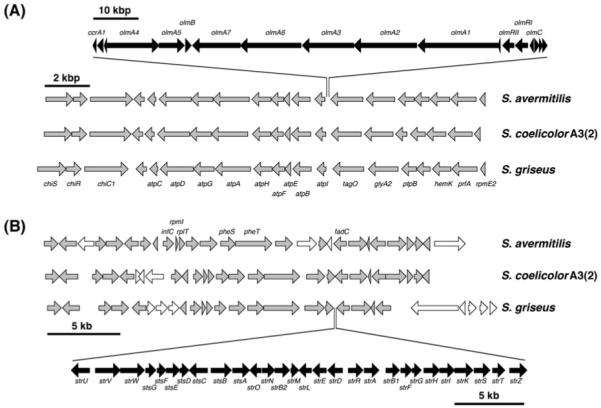
Examples of “island” secondary metabolic gene clusters in Streptomyces involving (A) oligomycin biosynthesis in S. avermitilis and (B) streptomycin in S. griseus.
2.2 Saccharopolyspora erythraea NRRL2338
The filamentous soil bacterium Saccharopolyspora erythraea is used for the industrial-scale production of the polyketide erythromycin A (30), which was the first macrolide antibiotic to be used clinically.111 This soil-dwelling actinomycete was originally identified as Streptomyces erythraeus but later assigned to the genus Saccharopolyspora based on chemotaxonomic observations.112 Despite this reclassification, the biology of S. erythraea had been considered similar to that of Streptomyces microbes, even though these distinct genera belong to different suborders. The sequence analysis of the 8 212 805 bp S. erythraea NRRL2338 genome established in 2007, however, revealed considerable divergence from the streptomycetes in both gene organization and function.113 Furthermore, while all Streptomyces genomes sequenced to date are linear, the S. erythraea genome has a circular topology like most other Actinobacteria (Table 1, Fig. 2).
The S. erythraea genome sequence suggests a very active secondary metabolism, with at least 27 biosynthesis genes or gene clusters associated with a diverse range of secondary metabolites of mostly unknown chemical compositions (Table 5). Only five of these gene sets, including that of erythromycin (ery), are distributed in the predicted core region of the genome that contains most essential genes.113 Expression profiling analyses using DNA microarrays revealed the differential expression of the core and non-core genes during the different growth stages of S. erythraea in which ery and six other secondary metabolism clusters were up-regulated.114 The erythromycin PKS encoding genes were first sequenced in the early 1990s,115,116 and established the paradigm for bacterial type I modular PKSs in the assembly of natural and engineered polyketides.117,118 Further S. erythraea polyketide potential includes modular and iterative type I PKSs, such as a second modular PKS (pke) of unknown function119 related to alkenyl furanone biosynthesis in streptomycetes120 and myxobacteria121 and a putative polyunsaturated fatty acid PKS pathway. While no type II PKS gene clusters typical of streptomycetes are evident in the S. erythraea genome,113 the common streptomycete type III PKS THN synthase (SACE1242–SACE1243) is associated with the flaviolin (7)-based reddish pigments particular to the S. erythraea red variant.122
Table 5.
Secondary metabolite gene clusters in Saccharopolyspora erythraea NRRL2338
| No. | Cluster location | Actual or predicted producta |
|---|---|---|
| 1 | SACE_0018–0028 | Polyunsaturated fatty acids |
| 2 | SACE_0483–0486 | Ectoine, 5-hydroxyectoine (14) |
| 3 | SACE_0712–0734 | Erythromycin (30) |
| 4 | SACE_1241–1244 | THN (6), flaviolin (7) |
| 5 | SACE_1304–1310 | Dipeptide |
| 6 | SACE_2342–2347 | Diketide |
| 7 | SACE_2618–2622 | PK-NRP hybrid |
| 8 | SACE_2628–2631 | Tetraketide |
| 9 | SACE_2692–2703 | Pentameric siderophore |
| 10 | SACE_2864–2879 | Diketide |
| 11 | SACE_3013–3017 | Non-ribosomal peptide |
| 12 | SACE_3029–3039 | Tetrapeptide siderophore |
| 13 | SACE_3187 | Terpene (geosmin?) |
| 14 | SACE_3223–3229 | Non-ribosomal peptide |
| 15 | SACE_3721–3722 | 2-methylisoborneol (10) |
| 16 | SACE_3976–3979 | Terpene (geosmin?) |
| 17 | SACE_4025–4029 | Lanthionine-containing peptide |
| 18 | SACE_4128–4145 | Nonaketide lactone |
| 19 | SACE_4230–4233 | Lanthionine-containing peptide |
| 20 | SACE_4275–4292 | Lipopeptide |
| 21 | SACE_4302–4307 | Polyketide |
| 22 | SACE_4327–4331 | Hopanoids |
| 23 | SACE_4471–4478 | Polyketide |
| 24 | SACE_4567–4578 | Aromatic polyketide |
| 25 | SACE_4906–4909 | Terpene (geosmin?) |
| 26 | SACE_5306–5309 | Methylsalicylic acid |
| 27 | SACE_5532 | Naphthoic acid |
Observed products are highlighted in bold.
Also absent is a homologous desferrioxamine siderophore biosynthetic pathway common to Streptomyces and Salinispora genera. Rather, S. erythraea produces a hydroxamate-type iron-siderophore (erythrobactin)123 of unknown structure and may produce two further peptidic siderophores associated with the NRPS gene clusters SACE2692–SACE2703 and SACE3029–SACE3039. Terpenoid biochemistry is also prevalent in S. erythraea, which harbors a putative hopanoid biosynthetic pathway and three copies of geosmin/germacradienol synthase genes (Table 5).66 While their functions have not yet been verified, S. erythraea produces geosmin (9) and germacradienol (22), and phylogenetic analyses suggest that SACE3187 may be involved in their biosynthesis. S. erythraea also produces 2-methylisoborneol (10) during sporulation on solid media, and SACE3721–SACE3722 have been identified as 2-methylisoborneol synthase and geranyl diphosphate methyltransferase genes, respectively.66
2.3 Salinispora tropica CNB-440 and Salinispora arenicola CNS-205
The genus Salinispora constitutes a discrete group of actinomycete bacteria that thrive in tropical and sub-tropical oceanic sediments.124,125 Phylogenetically associated with the family Micromonosporaceae, the Salinispora strains fall into three major phylotypes that exhibit different geographical distribution patterns. Salinispora arenicola has the widest distribution and appears to be ubiquitous in tropical and subtropical sediments, while Salinispora tropica and “Salinispora pacifica” have been retrieved from more restricted locations.13,126 A common feature of all Salinispora strains and what makes them of interest for biotechnological applications is the production of bioactive natural products for use in infectious disease and cancer treatments.127
The genomes of two representative strains, S. tropica CNB-440128 in 2007 (Fig. 2) and S. arenicola CNS-205104 in 2009 (Fig. 8), were assembled and functionally annotated. Ortholog plots of the fully sequenced chromosomes revealed a high degree of conservation and synteny across the entire Salinispora genomes, even though there are also several breakpoint regions that represent genomic islands or inversions. In contrast to the linear 7.7-Mb chromosome of their relative Micromonospora chalcea,129 the Salinispora genomes are significantly smaller with 5 183 331 bp and 5 786 361 bp, respectively, and exhibit a circular topology (Table 1). A large amount of coding capacity is devoted to the biosynthesis of secondary metabolites, at 8.8% in S. tropica and 10.9% in S. arenicola. Bioinformatic analyses revealed a total of 49 gene clusters in which the vast majority encode the biosynthesis of as-yet unknown chemical entities.104 Both genomes are particularly rich in polyketide pathways, including those for multimodular type I PKSs, iterative type I PKSs, hybrid type I PKS–NRPSs, heterodimeric type II PKSs, and homodimeric type III PKSs. While thiotemplate-based chemistry involving polyketide synthases and nonribosomal peptide synthetases appears to be the predominant theme in natural product assembly, the Salinispora chromosomes also harbor genes for the production of non-NRPS derived siderophores, bacteriocins, aminocyclitols, and terpenoids.
Fig. 8.
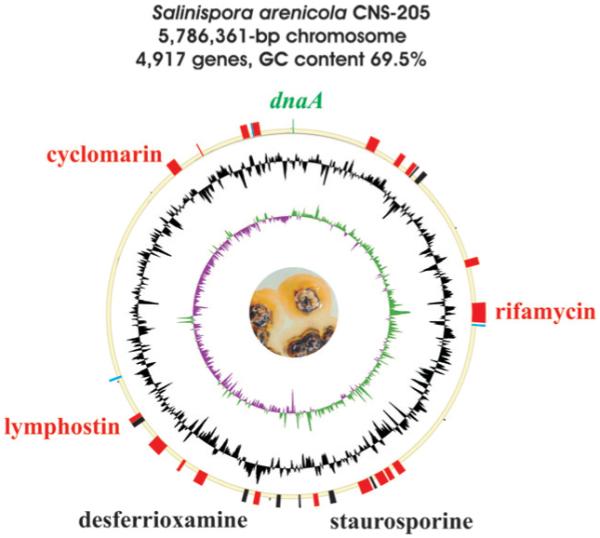
Schematic representation of the S. arenicola CNS-205 genome. Drawing details are the same as for Fig. 2.
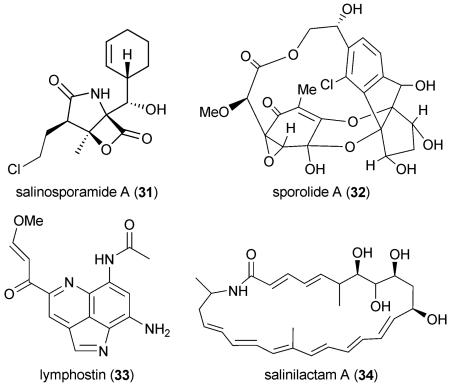
Altogether 19 biosynthetic gene clusters were annotated from the S. tropica genome (Table 6).128,130 Two are associated with novel natural products, namely the hybrid PKS–NRPS salinosporamide products131 (31) and the modified enediyne sporolide polyketides132 (32), and a third with the previously characterized streptomycete metabolite lymphostin133 (33). The analysis of the salinosporamide gene cluster gave rise to the elucidation of a new biological route of chlorination134 associated with the biosynthesis of these unusual γ-lactam-β-lactone proteasome inhibitors and further facilitated the engineered biosynthesis of unnatural salinosporamide analogs.135-137 Sequence analysis of the largest S. tropica secondary metabolic gene cluster, the 80 kb slm cluster, revealed that it was associated with the biosynthesis of the novel polyene macrolactam salinilactam A (34).128 Its structure elucidation was not only aided by the predictive nature of the genome sequence but also assisted in the sequence refinement of its highly repetitive modular PKS sequence. Twelve of the S. tropica gene clusters, including that putatively assigned to lymphostin biosynthesis, are also conserved in the phylogenetically related S. arenicola.
Table 6.
Secondary metabolite gene clusters in Salinispora tropica CNB-440
| No. | Cluster location | Actual or predicted producta |
|---|---|---|
| 1 | Stro0586–0610 | 10-Membered enediyne |
| 2 | Stro0667–0694 | Dipeptide |
| 3 | Stro1012–1043 | Salinosporamide (31) |
| 4 | Stro2174–2227 | Glycosylated decaketide |
| 5 | Stro2340–2346 | Aminocyclitol |
| 6 | Stro2428–2440 | Thiazolylpeptide |
| 7 | Stro2486–2510 | Aromatic polyketide |
| 8 | Stro2541–2555 | Desferrioxamine (13) |
| 9 | Stro2645–2659 | Yersiniabactin-like siderophore |
| 10 | Stro2691–2737 | Sporolide (32) |
| 11 | Stro2757–2781 | Salinilactam (34) |
| 12 | Stro2786–2813 | Dihydroaeruginoic acid |
| 13 | Stro2814–2842 | Coelibactin |
| 14 | Stro3042–3054 | Class I bacteriocin |
| 15 | Stro3055–3066 | Lymphostin (33) |
| 16 | Stro3244–3253 | Carotenoid pigment |
| 17 | Stro4264–4267 | Phenolic lipids |
| 18 | Stro4410–4429 | Tetrapeptide |
| 19 | Stro4437–4441 | Carotenoid pigment |
Observed products are highlighted in bold.
The majority of the isolated chemistry from S. arenicola CNS-205 is conversely associated with known actinomycete metabolites, including the polyketide antibiotic rifamycin (35), the cyclic heptapeptide cyclomarin A (36), and the indolocarbazole staurosporine (37),127 and their derivatives saliniketal138 (38) and cyclomarazine139 (39), which are unique to this strain. The functions of the remaining 27 biosynthetic gene clusters have not yet been reported, but represent a very diverse assemblage of secondary metabolic pathways that comprise a staggering 10.9% of the genes in the genome (Table 7).104 The S. arenicola genome encodes two enediyne type I PKS gene clusters reminiscent of the actinomycete anticancer agents calicheamicin and kedarcidin; however, in both cases, their associated gene clusters are fragmented across secondary metabolic-rich genomic islands that may signify that the respective pathways are no longer functional. Although associated enediyne-derived molecules have not yet been characterized from S. arenicola, this genus is known to produce enediyne-relevant chemistry in the form of the chlorinated metabolites sporolide A (32) from S. tropica and cyanosporaside A (40) from “S. pacifica”.132,140,141 The Salinispora genomes encode desferrioxamine and salicylate-primed NRPS siderophore biosynthetic pathways as in many terrestrial streptomycetes, yet are devoid of their characteristic terpenoid biosynthetic pathways that give rise to the “earthy” odiferous geosmin, 2-methylisoborneol and albaflavenone.
Table 7.
Secondary metabolite gene clusters in Salinispora arenicola CNS-205
| No. | Cluster location | Actual or predicted producta |
|---|---|---|
| 1 | Sare0345–0367 | Pentapeptide |
| 2 | Sare0478–0499 | PK-NRP hybrid |
| 3 | Sare0545–0560 | 9-Membered enediyne – A |
| 4 | Sare0570–0573 | Amino acid conjugate |
| 5 | Sare0602–0623 | Class I bacteriocin |
| 6 | Sare1041–1073 | Polyketide |
| 7 | Sare1240–1278 | Rifamycin (35) |
| 8 | Sare1286–1288 | Diterpene |
| 9 | Sare2017–2049 | 10-Membered enediyne – A |
| 10 | Sare2070–2081 | Yersiniabactin-like siderophore |
| 11 | Sare2088–2121 | 9-Membered enediyne – B |
| 12 | Sare2144–2151 | Amino acid conjugate |
| 13 | Sare2163–2206 | 10-Membered enediyne – B |
| 14 | Sare2326–2342 | Staurosporine (37) |
| 15 | Sare2400–2409 | PK-NRP hybrid |
| 16 | Sare2483–2491 | Aminocyclitol |
| 17 | Sare2583–2595 | Thiazolylpeptide |
| 18 | Sare2669–2694 | Aromatic polyketide |
| 19 | Sare2728–2744 | Desferrioxamine (13) |
| 20 | Sare2939–2968 | Tetrapeptide |
| 21 | Sare3051–3063 | Dipeptide |
| 22 | Sare3148–3163 | Macrolide |
| 23 | Sare3268–3280 | Class I bacteriocin |
| 24 | Sare3281–3293 | Lymphostin (33) |
| 25 | Sare3471–3480 | Carotenoid pigment |
| 26 | Sare4547–4569 | Cyclomarin (36) |
| 27 | Sare4694–4697 | Phenolic lipids |
| 28 | Sare4885–4904 | Tetrapeptide |
| 29 | Sare4927–4931 | Carotenoid pigment |
| 30 | Sare4932–4956 | 9-Membered enediyne – C |
Observed products are highlighted in bold.
The availability of two Salinispora genome sequences has allowed the genetic dissection of conserved and dissimilar pathways that are involved in secondary metabolism. Despite a close phylogenetic relationship between S. tropica and S. arenicola, comparative genomics indicated that the majority of their biosynthetic potential is species-specific.104 This result is consistent with previous fermentation studies showing that the Salinispora species produce distinct suites of secondary metabolites dependent on their geographic origin of isolation.142 Interestingly, most of the metabolic pathways that are unique to either one of the Salinispora genomes reside in genomic islands, whereas shared pathways conversely occur in the genus-specific core.104 A similar picture has also emerged in the Streptomyces, as discussed in Section 2.1.4. The Salinispora genomic islands are not contiguous regions of species-specific DNA. Indeed, gene acquisition, loss, duplication, inactivation, and rearrangement have shaped their genetic makeup.143 Surprisingly, the overall composition, evolutionary history, and function of the island genes are similar in both strains, with duplication and horizontal gene transfer accounting for the majority of genes, and secondary metabolism representing the largest functionally annotated category. It has been speculated recently that genomic islands house many of the adaptive traits that contribute to niche differentiation in bacteria.144 Applying this paradigm to Salinispora would support a functional role for secondary metabolites in environmental adaptation and ultimately speciation.
2.4 Frankia sp. strain CcI3, Frankia alni ACN14a, and Frankia sp. strain EAN1pec
Frankia is a nitrogen-fixing endophyte that forms symbiotic associations with actinorhizal plants, but can also survive as a free-living soil dweller. To clarify the genetic basis for the distinct host range of different phylotypes, the genomes of three Frankia strains, ACN14a, CcI3, and EAN1pec, were sequenced in 2007.145 All three chromosomes share a circular topology and do not contain any independently replicating plasmids. Correlating with host characteristics and biogeography, their sizes vary dramatically, ranging from 5.43 Mb for CcI3 to 9.04 Mb for EAN1pec (Table 1). Genes involved in secondary metabolism and regulation were found to be more common in the larger genomes of ACN14a and EAN1pec, respectively, suggesting that their acquisition has played a role in the process of host diversification and environmental adaptation.9 ACN14a and EAN1pec dedicate about 5% of their genomes to natural product assembly, a percentage only marginally smaller than the values that were previously reported for Streptomyces species. In contrast, the biosynthetic potential of CcI3 is significantly smaller (~3%). Analyses of codon adaptive indices have further indicated that CcI3 has fewer highly expressed biosynthesis genes than ACN14a and EAN1pec.146 As secondary metabolism is generally assumed to enhance the competitiveness of bacteria, these findings may provide the molecular basis for the limited biogeographical distribution of CcI3. Notwithstanding these differences, all sequenced Frankia strains were found to be quite versatile natural product producers, harboring a multitude of different pathways, including those for polyketide,128 non-ribosomal peptide, NRPS-independent siderophore, terpenoid, bacteriocin,89 and aminocyclitol147 biosynthesis. Homology-driven genome analysis has further revealed a unique pathway in ACN14a that potentially encodes the production of a novel phosphonate antibiotic.148 A conserved cassette of six genes, homologous to those in the phosphinothricin cluster from Streptomyces viridochromogenes, is involved in the biosynthesis of phosphonoformate (Fig. 9). Even though late-pathway enzymes required for phosphinothricin biosynthesis are not encoded on the ACN14a locus, it is likely that phosphonoformate is not the end product of the Frankia pathway, as evidenced by the presence of additional biosynthetic genes. Many phosphonate-containing natural products exhibit herbicidal properties,148 and the production of such a compound by a bacterial plant symbiont might be of ecological significance.
Fig. 9.

Gene organization and proposed biosynthesis of a phosphonoformate-like compound in Frankia alni ACN14a. (A) Phosphonate biosynthetic gene cluster in F. alni ACN14a. Genes that are responsible for the production of hydroxyethylphosphonate are depicted in grey (minimal cassette consisting of phosphoenolpyruvate mutase ppm, phosphonopyruvate decarboxylase ppd and phosphonoacetaldehyde dehydrogenase adh). (B) Proposed pathway to a hitherto unknown phosphonate antibiotic.
To date, the secondary metabolome of Frankia has not been explored beyond two reports concerning the production of anthraquinone- and calcimycin-type antibiotics.149,150 The genome sequences of Frankia now certainly provide a rationale to explore the chemistry of these intriguing microorganisms.
2.5 Nocardia farcinica IFM 10152
Members of the genus Nocardia are saprophytic soil bacteria, but also include opportunistic agents of nocardiosis. Together with the two pathogens Corynebacterium and Mycobacterium, Nocardia forms a monophyletic taxon within the order Actinomycetales.151 Organisms within this clade share an unusual cell wall composition, characterized by the presence of a waxy cell envelope containing mycolic acids, which distinguishes them from other bacteria. In 2004, the genome of the N. farcinica clinical isolate IFM 10152 was fully sequenced, revealing a single circular 6 021 225 bp chromosome and two smaller plasmids of 184 027 bp (pNF1) and 87 093 bp (pNF2) (Table 1).6 Consistent with previous phylogenetic analysis, ortholog plots confirmed the close relationship to C. glutamicum and M. tuberculosis. The synapomorphy between these taxa is genetically conserved in a pathway sustaining the synthesis of mycolic acids.152 Iron mobilization is crucial for many pathogenic bacteria, and therefore it is not surprising that the production of salicylate-derived siderophores is a prevailing feature of nocardial secondary metabolism.153-155 N. farcinica is no exception to this rule and possesses a gene set homologous to the mycobactin locus from M. tuberculosis,156 involving a hybrid PKS–NRPS assembly line (NFA7610–NFA7710), as well as mechanisms of lysine modification. In addition, the N. farcinica genome contains several orphan clusters that potentially encode the production of novel natural products. Except for an ortholog of the nrps5 region in S. avermitilis (SAV6631–SAV6633), these loci are unique to N. farcinica.152 Most striking is the discrete NRPS gene NFA50330, which encodes a 14,474-aa biosynthetic enzyme. This NRPS putatively catalyzes the assembly of at least twelve amino acid monomers, but lacks a terminating thioesterase to release the maturated peptidyl chain from covalent tethering on the final thiolation domain. Therefore it is not clear whether or not the associated cluster is functional.
Even though nocardial biosynthetic pathways are strongly biased towards thiotemplate-based assembly lines, there is also genomic evidence for the production of terpenoid structures. Ishikawa and coworkers identified a putative cluster encompassing genes for the mevalonate pathway associated with isoprenoid-related genes.6 While no secondary metabolites have yet been characterized from N. farcinica IFM 10152, its genetic potential clearly mirrors findings from previous chemical investigations in which nocardial strains yielded a large number of structurally diverse metabolites such as anthracyclines,157,158 angucyclines,159 macrolides,160 monobactams,161 indole alkaloids,162 and terpenoids.163,164
2.6 Rhodococcus jostii RHA1
Rhodococcus jostii RHA1 is a versatile polychlorinated biphenyl-degrading soil microbe that has emerged as a model actinomycete for the biodegradation of a wide range of organic compounds.165 Its complete nucleotide sequence was reported in 2006, revealing a linear 7 804 765 bp chromosome and three linear plasmids of 1 123 075 bp (pRH1), 442 536 bp (pRH2), and 332 361 bp (pRH3) (Table 1).23 Although its linear chromosomal topology is similar to that of Streptomyces microbes, comparative genome analyses with other actinomycetes revealed that R. jostii is more closely related to M. tuberculosis. The genome is exceptionally rich in oxygenases (203) and ligases (192) that are predicted to contribute to its catabolic versatility in the degradation of xenobiotics. For instance, R. jostii RHA1 produces an impressive array of Baeyer–Villiger monooxygenases that catalyze diverse regio- and enantioselective reactions.166 While rhodococci are known for their catabolic prowess, they are largely unknown for their secondary metabolic abilities. Although the closely related R. jostii K01-B0171 produces the anti-mycobacterial, lasso peptides lariatins A and B via a ribosomal process,167 there are no reports of natural products derived from R. jostii RHA1. Surprisingly, when the R. jostii RHA1 genome was completed, two PKS, a hybrid PKS–NRPS, and 14 NRPS gene clusters were identified, suggesting the possibility of a robust secondary metabolism (Table 8).23 Six of the NRPS genes are >25 kb in length, and one cluster (ro02318–ro02320) may be involved in the biosynthesis of an NRPS-based siderophore because of co-clustered siderophore-iron complex transporter genes. Terpene biosynthesis genes involved in common actinomycete metabolites such as carotenoid, hopanoid, and odoriferous sesquiterpenes are, however, noticeably absent.
Table 8.
Secondary metabolite gene clusters in Rhodococcus jostii RHA1
| No. | Cluster location | Predicted product |
|---|---|---|
| 1 | RHA1_ro00071–00073 | Non-ribosomal peptide |
| 2 | RHA1_ro00232–00235 | Non-ribosomal peptide |
| 3 | RHA1_ro00429–00435 | Non-ribosomal peptide |
| 4 | RHA1_ro01305–01307 | Ectoine |
| 5 | RHA1_ro02207–02310 | PK-NRP hybrid |
| 6 | RHA1_ro02318–02320 | Peptidic siderophore |
| 7 | RHA1_ro02391–02397 | Non-ribosomal peptide |
| 8 | RHA1_ro02492–02494 | Non-ribosomal peptide |
| 9 | RHA1_ro04063–04066 | Polyketide |
| 10 | RHA1_ro04230–04231 | Polyketide |
| 11 | RHA1_ro04379–04382 | Non-ribosomal peptide |
| 12 | RHA1_ro04713–04717 | Non-ribosomal peptide |
| 13 | RHA1_ro05093–05103 | Non-ribosomal peptide |
| 14 | RHA1_ro05430–05431 | Non-ribosomal peptide |
| 15 | RHA1_ro05452–05453 | Non-ribosomal peptide |
| 16 | RHA1_ro05466–05468 | Non-ribosomal peptide |
| 17 | RHA1_ro06098–06103 | Non-ribosomal peptide |
| 18 | RHA1_ro06663–06665 | Non-ribosomal peptide |
3 Actinomycete genome scanning for new natural product biosynthesis gene clusters
Since genes encoding biosynthetic enzymes, resistance proteins, and regulatory systems associated with microbial natural products are typically clustered, genome sequencing has emerged as a powerful tool to quickly identify biosynthetic gene clusters associated with known metabolites. Natural product biosynthetic gene clusters are typically identified by targeting proposed biosynthetic or resistance genes or by various in vivo strategies such as transposon mutagenesis or by complementing nonproducing mutants. Whole-genome scanning offers an alternative method that is direct and reliable, especially for those gene sets associated with natural products without specific biosynthetic reactions for which to conveniently probe. Several examples exist from the actinomycete literature, and some of these are highlighted below.
The distinctive C5N unit in the phosphoglycolipid moenomycin A (41) facilitated a PCR-guided discovery of a three-gene operon in Streptomyces ghanaensis associated with its biosynthesis from aminolevulinate.168 However, sequence analysis of flanking genes did not reveal the remainder of the moenomycin A biosynthetic gene cluster. After failing to identify the complete gene cluster by standard approaches, the genome of S. ghanaensis was shotgun sequenced and partially assembled to give 1018 contigs covering 7.4 Mb. Genome scanning for candidate genes quickly identified the main moenomycin A gene cluster of 20 open reading frames, which was subsequently sequenced in full on two overlapping cosmid clones and verified by gene disruption and heterologous expression. This split cluster differs from other carbohydrate-containing natural product gene clusters in that most of its biosynthetic building blocks are derived from primary metabolism and thereby represents an alternative paradigm for the biosynthesis of glycosylated natural products.
Sequence analysis of approximately 3.0 Mb of the two chromosomal arms of S. ambofaciens ATCC23877107 (http://www.weblgm.scbiol.uhp-nancy.fr/ambofaciens/) revealed at least nine secondary metabolic gene clusters, including those for the siderophore coelichelin (5),169 the structurally unknown angucycline antibiotic alpomycin,170,171 and the pyrrole-amide antibiotic congocidine172 (42), whose structure had been determined prior to genome sequencing. Congocidine is synthesized by an atypical iterative NRPS, and its gene cluster represents the first example responsible for the biosynthesis of a pyrrole-amide natural product.
The prenylated alkaloid diazepinomicin/ECO-4601 (43), which contains a very unusual dibenzodiazepinone core that was of unknown biosynthetic origin, was discovered from two Micromonospora strains including one obtained from a marine ascidian.173 Genome scanning of strain 046Eco-11 revealed five distinct natural product biosynthetic gene clusters, one of which spanning 50 kb and coding for 42 genes was shown to be involved in the synthesis and coupling of isoprenoid (via the mevalonate pathway), 3-hydroxyanthranilic acid, and 3-amino-5-hydroxybenzoic acid residues in the biosynthesis of 43.174
Thiopeptide antibiotics such as the prototypical thiostrepton (44), a topical veterinary antibiotic from Streptomyces azureus and Streptomyces laurentii, were long suspected to be of either ribosomal or nonribosomal origin.175 Attempts since the early 1990s to identify the biosynthetic gene cluster had been unsuccessful176 until two groups in 2009 independently established that thiostrepton is indeed a highly modified bacteriocin.177,178 Partial genome sequencing of S. laurentii was pursued in one study to yield 7.26 Mb on over 3600 fragments that led to the identification of the chromosomal gene tsrA encoding the thiostrepton prepeptide.177 Gene inactivation of tsrA resulted in the loss of thiostrepton biosynthesis and subsequently led to the cloning and sequencing of its gene cluster from two overlapping fosmids. In addition to thiostrepton, other thiopeptide bacteriocin biosynthetic gene clusters recently identified include those for siomycin A from Streptomyces sioyaensis,178 thiocillin from Bacillus cereus,178,179 and the thiomuracins from the rare actinomycete Nonomuraea,180 that collectively illuminate the extensive posttranslational modification reactions associated with these ribosomally-derived peptides.
4 Actinomycete genome mining for new natural product chemical entities
Genome sequencing has also proved to be a powerful method to quickly access the genome-wide natural product biosynthetic capacity of diverse bacteria in order to guide the discovery of new chemical entities.52,55,56,181-183 Various strategies have been employed to mine natural product-laden genomes, as exemplified in Section 2 of this article on completed actinomycete genomes. One of the first reports of genome mining from partial sequences of actinomycete genomes involved the characterization of enediyne polyketide chemistry in 2003.184 Since many polyketides, peptides and hybrids thereof are synthesized on large, modular synthases in an ordered, assembly-line process,185,186 their products or fragments can be predicted often with high accuracy to greatly aid in their isolation and structure elucidation.187 Numerous databases and in silico computational approaches have been developed to analyze orphan pathways and assist in the identification of their products.188-192 Examples abound from the actinomycete literature on mined polyketides, which include the 28-membered macrolide halstoctacosanolide A (45) from Streptomyces halstedii HC34,193,194 the linear polyketide ECO-02301 (46) from Streptomyces aizunensis NRRL B-11277,195 the antimicrobial glycosidic polyketide ECO-0501 (47) from the vancomycin-producer Amycolatopsis orientalis in which genomic analysis revealed at least ten genetic loci associated with secondary metabolism,196 and three new 5-alkenyl-3,3(2H)-furanones such as E-837 (48) from two Streptomyces species.120 E-837 from S. aculeolatus NRRL 18422 is identical to actinofuranone A, which was isolated by a standard approach from the marine-derived strain CNQ766,197 that like S. aculeolatus also produces the type III PKS-derived napyradiomycin family of meroterpenoid antibiotics.198 Analysis of the recently sequenced genome of the thermophilic actinomycete Thermobifida fusca further led to the discovery of the fuscachelin (49) family of novel NRPS-derived siderophores.199
5 Outlook
Actinomycetes have long been known for their biosynthetic prowess in the production of complex natural products. In recent years, genomics has transformed the ways in which we think about these chemically prolific microorganisms and has greatly influenced the manner in which we probe their chemistry and biology. We now know that conventional natural product isolation programs vastly underestimated their biosynthetic ability, in many cases by upwards of 80–90%. As a consequence, orphan secondary metabolic gene clusters deduced from genome sequencing efforts represent a large, untapped resource of new chemical entities that promise to illuminate new biology as well as provide a novel source of drug candidates. This latter point is especially relevant to actinomycetes such as Frankia that are not known for their natural product chemistry, but on the basis of genomic analyses are predicted to be nearly as rich in natural products as the streptomycetes. One of the greatest future challenges in this field will be in the development of general methods to awaken these silent gene clusters and characterize the chemical biology of their pathway products. What is certain is that microbial genomics has made an indelible mark in our understanding of the chemistry and biology of the actinomycetes, and that future sequencing efforts will continue to stimulate the broader scientific community.
6 Acknowledgements
B.S.M. gratefully acknowledges the National Institutes of Health (CA127622 and GM085770) for supporting research involving actinomycete genomics and genome-inspired drug discovery. H.I. acknowledges support from a Grant-In-Aid for Scientific Research on Priority Areas “Applied Genomics” from the Ministry of Education, Culture, Sports, Science and Technology, Japan (MEXT) and by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (20310122)
Biography

Markus Nett (born 1977) studied pharmacy, and obtained his Ph.D. on the subject of natural product chemistry under the supervision of Professor Gabriele M. König at Rheinische-Friedrich-Wilhelms-Universität Bonn. In 2007, he continued with postdoctoral research in the group of Professor Bradley S. Moore, now focussing on the engineering of biosynthetic pathways in marine actinomycete bacteria. In 2008, he gained a fellowship of the German Academic Exchange Service and became a DAAD fellow at the Scripps Institution of Oceanography, San Diego. Since January 2009, he has been head of a junior research group at the Leibniz Institute for Natural Product Research and Infection Biology (Hans-Knöll-Institute) in Jena. His research interests are in exploring the secondary metabolic capabilities of predatory bacteria, mutasynthesis and the role of natural products in microbial communication processes.

Haruo Ikeda was born in Tokyo, Japan (1954), receiving his B.S. (1977) and M.S. (1979) in pharmaceutical sciences at Kitasato University. He obtained his Ph.D. (1982) in pharmaceutical sciences from Kitasato University, where he studied biosynthesis of 16-membered macrolide antibiotics produced by Streptomyces spp. with Professor Satoshi Omura. He began studying Streptomyces genetics with Professors Sir David A. Hopwood and Keith F. Chater at the Department of Genetics, John Innes Institute, UK, as a postdoctoral fellow, and then took a faculty position in the School of Pharmaceutical Sciences, Kitasato University (1983–2002), becoming Associate Professor in 1992. Since 2002 he has been a full Professor at Kitasato Institute for Life Sciences and the Graduate School of Infection Control Sciences, Kitasato University. His research interests are in studying the biosynthesis of microbial secondary metabolites using bioinformatics and in engineering and designing the Streptomyces chromosome for the evaluation and optimization of the secondary metabolism.

Bradley S. Moore is currently Professor of Oceanography and Pharmaceutical Sciences at the Scripps Institution of Oceanography and the Skaggs School of Pharmacy and Pharmaceutical Sciences at University of California, San Diego. He was first introduced to natural product research as a chemistry undergraduate student at the University of Hawaii, where he explored the chemistry and biosynthesis of cyanobacterial natural products with the late Prof. R. E. Moore. Fascinated by the beauty and complexity of natural product structures, he went on to conduct graduate (Ph.D. 1994 in bioorganic chemistry with Prof. H. G. Floss at the University of Washington) and postdoctoral research (1994–1995 with Prof. J. A. Robinson at the University of Zürich) on the biosynthesis of bacterial natural products. Prior to moving to the University of California at San Diego in 2005, he held academic appointments at the University of Washington (1996–1999) and the University of Arizona (1999–2005). His research interests involve exploring and exploiting marine microbial genomes to discover new biosynthetic enzymes, secondary metabolic pathways, and natural products for drug discovery and development.
Footnotes
This article is part of a themed issue on genomics.
7 References
- 1.Waksman SA. Science. 1953;118:259–266. doi: 10.1126/science.118.3062.259. [DOI] [PubMed] [Google Scholar]
- 2.Stackebrandt E, Sproer C, Rainey FA, Burghardt J, Pauker O, Hippe H. Int. J. Syst. Bacteriol. 1997;47:1134–1139. doi: 10.1099/00207713-47-4-1134. [DOI] [PubMed] [Google Scholar]
- 3.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, III, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver S, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton S, Squares S, Sqares R, Sulston JE, Taylor K, Whitehead S, Barrell BG. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 4.Cerdeño-Tárraga AM, Efstratiou A, Dover LG, Holden MT, Pallen M, Bentley SD, Besra GS, Churcher C, James KD, De Zoysa A, Chillingworth T, Cronin A, Dowd L, Feltwell T, Hamlin N, Holroyd S, Jagels K, Moule S, Quail MA, Rabbinowitsch E, Rutherford KM, Thomson NR, Unwin L, Whitehead S, Barrell BG, Parkhill J. Nucleic Acids Res. 2003;31:6516–6523. doi: 10.1093/nar/gkg874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brüggemann H, Henne A, Hoster F, Liesegang H, Wiezer A, Strittmatter A, Hujer S, Dürre P, Gottschalk G. Science. 2004;305:671–673. doi: 10.1126/science.1100330. [DOI] [PubMed] [Google Scholar]
- 6.Ishikawa J, Yamashita A, Mikami Y, Hoshino Y, Kurita H, Hotta K, Shiba T, Hattori M. Proc. Natl. Acad. Sci. U. S. A. 2004;101:14925–14930. doi: 10.1073/pnas.0406410101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loria R, Kers J, Joshi M. Annu. Rev. Phytopathol. 2006;44:469–487. doi: 10.1146/annurev.phyto.44.032905.091147. [DOI] [PubMed] [Google Scholar]
- 8.Monteiro-Vitorello CB, Camargo LE, Van Sluys MA, Kitajima JP, Truffi D, do Amaral AM, Harakava R, de Oliveira JC, Wood D, de Oliveira MC, Miyaki C, Takita MA, da Silva AC, Furlan LR, Carraro DM, Camarotte G, Almeida NFJ, Carrer H, Coutinho LL, El-Dorry HA, Ferro MI, Gagliardi PR, Giglioti E, Goldman MH, Goldman GH, Kimura ET, Ferro ES, Kuramae EE, Lemos EG, Lemos MV, Mauro SM, Machado MA, Marino CL, Menck CF, Nunes LR, Oliveira RC, Pereira GG, Siqueira W, de Souza AA, Tsai SM, Zanca AS, Simpson AJ, Brumbley SM, Setúbal JC. Mol. Plant-Microbe Interact. 2004;17:827–836. doi: 10.1094/MPMI.2004.17.8.827. [DOI] [PubMed] [Google Scholar]
- 9.Normand P, Queiroux C, Tisa LS, Benson DR, Rouy Z, Cruveiller S, Medigue C. Physiol. Plant. 2007;130:331–343. [Google Scholar]
- 10.Bentley SD, Chater KF, Cerdeño-Tárraga A-M, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang C-H, Kieser T, Larke L, Murphy L, Oliver K, O’Niel S, Rabbinowitsch E, Rajandream M-A, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA. Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 11.Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sasaki Y, Hattori M, Omura S. Nat. Biotechnol. 2003;21:526–531. doi: 10.1038/nbt820. [DOI] [PubMed] [Google Scholar]
- 12.Ohnishi Y, Ishikawa J, Hara H, Suzuki H, Ikenoya M, Ikeda H, Yamashita A, Hattori M, Horinouchi S. J. Bacteriol. 2008;190:4050–4060. doi: 10.1128/JB.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jensen PR, Mafnas C. Environ. Microbiol. 2006;8:1881–1888. doi: 10.1111/j.1462-2920.2006.01093.x. [DOI] [PubMed] [Google Scholar]
- 14.Barrangou R, Briczinski EP, Traeger LL, Loquasto JR, Richards M, Horvath P, Coûté-Monvoisin AC, Leyer G, Rendulic S, Steele JL, Broadbent JR, Oberg T, Dudley EG, Schuster S, Romero DA, Roberts RF. J. Bacteriol. 2009 doi: 10.1128/JB.00155-09. DOI: 10.1128/JB.00155-00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berdy J. J. Antibiot. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 16.Watve MG, Tickoo R, Jog MM, Bhole BD. Arch. Microbiol. 2001;176:386–390. doi: 10.1007/s002030100345. [DOI] [PubMed] [Google Scholar]
- 17.Baltz RH. Curr. Opin. Pharmacol. 2008;8:557–563. doi: 10.1016/j.coph.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 18.Caffrey P, Aparicio JF, Malpartida F, Zotchev SB. Curr. Top. Med. Chem. 2008;8:639–653. doi: 10.2174/156802608784221479. [DOI] [PubMed] [Google Scholar]
- 19.Olano C, Mendez C, Salas JA. Nat. Prod. Rep. 2009;26:628–660. doi: 10.1039/b822528a. [DOI] [PubMed] [Google Scholar]
- 20.Shiomi K, Omura S. Proc. Jpn. Acad., Ser. B, Phys. Biol. Sci. 2004;80:245–258. [Google Scholar]
- 21.Graziani EI. Nat. Prod. Rep. 2009;26:602–609. doi: 10.1039/b804602f. [DOI] [PubMed] [Google Scholar]
- 22.Kalinowski J, Bathe B, Bartels D, Bischoff N, Bott M, Burkovski A, Dusch N, Eggeling L, Eikmanns BJ, Gaigalat L, Goesmann A, Hartmann M, Huthmacher K, Krämer R, Linke B, McHardy AC, Meyer F, Möckel B, Pfefferle W, Pühler A, Rey DA, Rückert C, Rupp O, Sahm H, Wendisch VF, Wiegräbe I, Tauch A. J. Biotechnol. 2003;104:5–25. doi: 10.1016/s0168-1656(03)00154-8. [DOI] [PubMed] [Google Scholar]
- 23.McLeod MP, Warren RL, Hsiao WWL, Araki N, Myhre M, Fernandes C, Miyazawa D, Wong W, Lillquist AL, Wang D, Dosanjh M, Hara H, Petrescu A, Morin RD, Yang G, Stott JM, Schein JE, Shin H, Smailus D, Siddiqui AS, Marra MA, Jones SJM, Holt R, Brinkman FSL, Miyauchi K, Fukuda M, Davies JE, Mohn WW, Eltis LD. Proc. Natl. Acad. Sci. U. S. A. 2006;103:15582–15587. doi: 10.1073/pnas.0607048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM, McKenney K, Sutton G, Fitzhugh W, Fields C, Gocayne JD, Scott J, Shirley R, Liu LI, Glodek A, Kelley JM, Weidman JF, Phillips CA, Spriggs T, Hedblom E, Cotton MD, Utterback TR, Hanna MC, Nguyen DT, Saudek DM, Brandon RC, Fine LD, Fritchman JL, Fuhrmann JL, Geoghagen NSM, Gnehm CL, McDonald LA, Small KV, Fraser CM, Smith HO, Venter JC. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 25.Hopwood DA. Annu. Rev. Genet. 2006;40:1–23. doi: 10.1146/annurev.genet.40.110405.090639. [DOI] [PubMed] [Google Scholar]
- 26.Bao K, Cohen SN. Genes Dev. 2001;15:1518–1527. doi: 10.1101/gad.896201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb JF, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fuji C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO, Venter JC. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 28.Wood DW, Setubal JC, Kaul R, Monks DE, Kitajima JP, Okura VK, Zhou Y, Chen L, Wood GE, Almeida NFJ, Woo L, Chen Y, Paulsen IT, Eisen JA, Karp PD, Bovee DS, Chapman P, Clendenning J, Deatherage G, Gillet W, Grant C, Kutyavin T, Levy R, Li MJ, McClelland E, Palmieri A, Raymond C, Rouse G, Saenphimmachak C, Wu Z, Romero P, Gordon D, Zhang S, Yoo H, Tao Y, Biddle P, Jung M, Krespan W, Perry M, Gordon-Kamm B, Liao L, Kim S, Hendrick C, Zhao ZY, Dolan M, Chumley F, Tingey SV, Tomb JF, Gordon MP, Olson MV, Nester EW. Science. 2001;294:2317–2323. doi: 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- 29.Goodner B, Hinkle G, Gattung S, Miller N, Blanchard M, Qurollo B, Goldman BS, Cao Y, Askenazi M, Halling C, Mullin L, Houmiel K, Gordon J, Vaudin M, Iartchouk O, Epp A, Liu F, Wollam C, Allinger M, Doughty D, Scott C, Lappas C, Markelz B, Flanagan C, Crowell C, Gurson J, Lomo C, Sear C, Strub G, Cielo C, Slater S. Science. 2001;294:2323–2328. doi: 10.1126/science.1066803. [DOI] [PubMed] [Google Scholar]
- 30.Takarada H, Sekine M, Kosugi H, Matsuo Y, Fujisawa T, Omata S, Kishi E, Shimizu A, Tsukatani N, Tanikawa S, Fujita N, Harayama S. J. Bacteriol. 2008;190:4139–4146. doi: 10.1128/JB.01853-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reddy TB, Riley R, Wymore F, Montgomery P, DeCaprio D, Engels R, Gellesch M, Hubble J, Jen D, Jin H, Koehrsen M, Larson L, Mao M, Nitzberg M, Sisk P, Stolte C, Weiner B, White J, Zachariah ZK, Sherlock G, Galagan JE, Ball C, Schoolnik GK. Nucleic Acids Res. 2009;37:D499–D508. doi: 10.1093/nar/gkn652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Catanho M, Mascarenhas D, Degrave W, Miranda AB. Genet. Mol. Res. 2006;5:115–126. [PubMed] [Google Scholar]
- 33.Vishnoi A, Srivastava A, Roy R, Bhattacharya A. BMC Genomics. 2008;9:373–376. doi: 10.1186/1471-2164-9-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gokhale RS, Saxena P, Chopra T, Mohanty D. Nat. Prod. Rep. 2007;24:267–277. doi: 10.1039/b616817p. [DOI] [PubMed] [Google Scholar]
- 35.Staunton J, Weissman KJ. Nat. Prod. Rep. 2001;18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- 36.Marahiel MA, Essen LO. Methods Enzymol. 2009;458:337–351. doi: 10.1016/S0076-6879(09)04813-7. [DOI] [PubMed] [Google Scholar]
- 37.Moore BS. Angew. Chem., Int. Ed. 2008;47:9386–9388. doi: 10.1002/anie.200803868. [DOI] [PubMed] [Google Scholar]
- 38.Omura S, Ikeda H, Ishikawa J, Hanamoto A, Takahashi C, Shinose M, Takahashi Y, Horikawa H, Nakazawa H, Osonoe T, Kikuchi H, Shiba T, Sakaki Y, Hattori M. Proc. Natl. Acad. Sci. U. S. A. 2001;98:12215–12220. doi: 10.1073/pnas.211433198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. The John Innes Foundation; Norwich: 2000. [Google Scholar]
- 40.Bentley SD, Brown S, Murphy LD, Harris DE, Quail MA, Parkhill J, Barrell BG, McCormick JR, Santamaria RI, Losick R, Yamasaki M, Kinashi H, Chen CW, Chandra G, Jakimowicz D, Kieser HM, Kieser T, Chater KF. Mol. Microbiol. 2004;51:1615–1628. doi: 10.1111/j.1365-2958.2003.03949.x. [DOI] [PubMed] [Google Scholar]
- 41.Haug I, Weissenborn A, Brolle D, Bentley SD, Kieser T, Altenbuchner J. Microbiology. 2003;149:505–513. doi: 10.1099/mic.0.25751-0. [DOI] [PubMed] [Google Scholar]
- 42.Hopwood DA. Microbiology. 1999;145:2183–2202. doi: 10.1099/00221287-145-9-2183. [DOI] [PubMed] [Google Scholar]
- 43.Malpartida F, Hopwood DA. Nature. 1984:309. doi: 10.1038/309462a0. [DOI] [PubMed] [Google Scholar]
- 44.Cerdeño AM, Bibb MJ, Challis GL. Chem. Biol. 2001;8:817–829. doi: 10.1016/s1074-5521(01)00054-0. [DOI] [PubMed] [Google Scholar]
- 45.Williamson NR, Fineran PC, Leeper FJ, Salmond GPC. Nat. Rev. Microbiol. 2006;4:887–899. doi: 10.1038/nrmicro1531. [DOI] [PubMed] [Google Scholar]
- 46.Hojati Z, Milne C, Harvey B, Gordon L, Borg M, Flett F, Wilkinson B, Sidebottom PJ, Rudd BAM, Hayes MA, Smith CP, Micklefield J. Chem. Biol. 2002;9:1175–1187. doi: 10.1016/s1074-5521(02)00252-1. [DOI] [PubMed] [Google Scholar]
- 47.Yu T-W, Shen Y, McDaniel R, Floss HG, Khosla C, Hopwood DA, Moore BS. J. Am. Chem. Soc. 1998;120:7749–7759. [Google Scholar]
- 48.Poralla K, Muth G, Härtner T. FEMS Microbiol. Lett. 2000;189:93–95. doi: 10.1111/j.1574-6968.2000.tb09212.x. [DOI] [PubMed] [Google Scholar]
- 49.Das A, Khosla C. Acc. Chem. Res. 2009;42:631–639. doi: 10.1021/ar8002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hopwood DA, Malpartida F, Kieser HM, Ikeda H, Duncan J, Fujii I, Rudd BAM, Floss HG, Omura S. Nature. 1985;314:642–644. doi: 10.1038/314642a0. [DOI] [PubMed] [Google Scholar]
- 51.McDaniel R, Ebert-Khhosla S, Hopwood DA, Khosla C. Science. 1993;262:1546–1550. doi: 10.1126/science.8248802. [DOI] [PubMed] [Google Scholar]
- 52.Challis GL. Microbiology. 2008;154:1555–1569. doi: 10.1099/mic.0.2008/018523-0. [DOI] [PubMed] [Google Scholar]
- 53.Challis GL, Ravel J. FEMS Microbiol. Lett. 2000;187:111–114. doi: 10.1111/j.1574-6968.2000.tb09145.x. [DOI] [PubMed] [Google Scholar]
- 54.Lautru S, Deeth RJ, Bailey LM, Challis GL. Nat. Chem. Biol. 2005;1:265–269. doi: 10.1038/nchembio731. [DOI] [PubMed] [Google Scholar]
- 55.Challis GL. J. Med. Chem. 2008;51:2618–2628. doi: 10.1021/jm700948z. [DOI] [PubMed] [Google Scholar]
- 56.Gross H. Curr. Opin. Drug Discov. Devel. 2009;12:207–219. [PubMed] [Google Scholar]
- 57.Izumikawa M, Shipley PR, Hopke J, O’Hare T, Xiang L, Noel JP, Moore BS. J. Ind. Microbiol. Biotechnol. 2003;30:510–515. doi: 10.1007/s10295-003-0075-8. [DOI] [PubMed] [Google Scholar]
- 58.Austin MB, Izumikawa M, Bowman ME, Udwary DW, Ferrer J-L, Moore BS, Noel JP. J. Biol. Chem. 2004;279:45162–45174. doi: 10.1074/jbc.M406567200. [DOI] [PubMed] [Google Scholar]
- 59.Song L, Barona-Gomez F, Corre C, Xiang L, Udwary DW, Austin MB, Noel JP, Moore BS, Challis GL. J. Am. Chem. Soc. 2006;128:14754–14755. doi: 10.1021/ja065247w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. Proc. Natl. Acad. Sci. U. S. A. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiang J, He X, Cane DE. Nat. Chem. Biol. 2007;3:711–715. doi: 10.1038/nchembio.2007.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang CM, Cane DE. J. Am. Chem. Soc. 2008;130:8908–8909. doi: 10.1021/ja803639g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin X, Hopson R, Cane DE. J. Am. Chem. Soc. 2006;128:6022–6023. doi: 10.1021/ja061292s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin X, Cane DE. J. Am. Chem. Soc. 2009;131:6332–6333. doi: 10.1021/ja901313v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao B, Lin X, Lei L, Lamb DC, Kelly SL, Waterman MR, Cane DE. J. Biol. Chem. 2008;283:8183–8189. doi: 10.1074/jbc.M710421200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Komatsu M, Tsuda M, Omura S, Oikawa H, Ikeda H. Proc. Natl. Acad. Sci. U. S. A. 2008;105:7422–7427. doi: 10.1073/pnas.0802312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takano H, Obitsu S, Beppu T, Ueda K. J. Bacteriol. 2005;187:1825–1832. doi: 10.1128/JB.187.5.1825-1832.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barona-Gomez F, Wong U, Giannakopulos AE, Derrick PJ, Challis GL. J. Am. Chem. Soc. 2004;126:16282–16283. doi: 10.1021/ja045774k. [DOI] [PubMed] [Google Scholar]
- 69.Kadi N, Oves-Costales D, Barona-Gomez F, Challis GL. Nat. Chem. Biol. 2007;3:652–656. doi: 10.1038/nchembio.2007.23. [DOI] [PubMed] [Google Scholar]
- 70.Bursy J, Kuhlmann AU, Pittelkow M, Hartmann H, Jebbar M, Pierik AJ, Bremer E. Appl. Environ. Microbiol. 2008;74:7286–7296. doi: 10.1128/AEM.00768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kodani S, Hudson ME, Durrant MC, Buttner MJ, Nodwell JR, Willey JM. Proc. Natl. Acad. Sci. U. S. A. 2004;101:11448–11453. doi: 10.1073/pnas.0404220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takano E, Nihira T, Hara Y, Jones JJ, Gershater CJL, Yamada Y, Bibb M. J. Biol. Chem. 2000;275:11010–11016. doi: 10.1074/jbc.275.15.11010. [DOI] [PubMed] [Google Scholar]
- 73.Takano E, Kinoshita H, Mersinias V, Bucca G, Hotchkiss G, Nihira T, Smith CP, Bibb M, Wohlleben W, Chater K. Mol. Microbiol. 2005;56:465–479. doi: 10.1111/j.1365-2958.2005.04543.x. [DOI] [PubMed] [Google Scholar]
- 74.Pawlik K, Kotowska M, Chater KF, Kuczek K, Takano E. Arch. Microbiol. 2007;187:87–99. doi: 10.1007/s00203-006-0176-7. [DOI] [PubMed] [Google Scholar]
- 75.Takano E. Curr. Opin. Microbiol. 2006;9:287–294. doi: 10.1016/j.mib.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 76.Corre C, Song L, O’Rourke S, Chater KF, Challis GL. Proc. Natl. Acad. Sci. U. S. A. 2008;105:17510–17515. doi: 10.1073/pnas.0805530105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.O’Rourke S, Wietzorrek A, Fowler K, Corre C, Challis GL, Chater KF. Mol. Microbiol. 2009;71:763–778. doi: 10.1111/j.1365-2958.2008.06560.x. [DOI] [PubMed] [Google Scholar]
- 78.Challis GL, Chater KF. Chem. Commun. 2001:935–936. [Google Scholar]
- 79.Corre C, Challis GL. ChemBioChem. 2005;6:2166–2170. doi: 10.1002/cbic.200500243. [DOI] [PubMed] [Google Scholar]
- 80.J. F. Hu, D. Wunderlich, I. Sattler, A. Härtl, I. Papastavrou, S. Grond, S. Grabley, X. Z. Feng and R. Thiericke, 2000.
- 81.Omura S, Crump A. Nat. Rev. Microbiol. 2004;2:984–989. doi: 10.1038/nrmicro1048. [DOI] [PubMed] [Google Scholar]
- 82.Omura S. Int. J. Antimicrob. Agents. 2008;31:91–98. doi: 10.1016/j.ijantimicag.2007.08.023. [DOI] [PubMed] [Google Scholar]
- 83.Ikeda H, Nonomiya T, Usami M, Ohta T, Omura S. Proc. Natl. Acad. Sci. U. S. A. 1999;96:9509–9514. doi: 10.1073/pnas.96.17.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ikeda H, Takada Y, Pang CH, Tanaka Y, Omura S. J. Bacteriol. 1993;175:2077–2082. doi: 10.1128/jb.175.7.2077-2082.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lamb DC, Ikeda H, Nelson DR, Ishikawa J, Skaug T, Jackson C, Omura S, Waterman MR, Kelly SL. Biochem. Biophys. Res. Commun. 2003;307:610–619. doi: 10.1016/s0006-291x(03)01231-2. [DOI] [PubMed] [Google Scholar]
- 86.Rochet P, Lancelin JM. Magn. Reson. Chem. 1997;35:538–542. [Google Scholar]
- 87.Funa N, Ohnishi Y, Fujii I, Shibuya M, Ebizuka Y, Horinouchi S. Nature. 1999;400:897–899. doi: 10.1038/23748. [DOI] [PubMed] [Google Scholar]
- 88.Jenke-Kodama H, Börner T, Dittmann E. PLoS Comput. Biol. 2006;2:e132. doi: 10.1371/journal.pcbi.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Severinov K, Semenova E, Kazakov A, Kazakov T, Gelfand MS. Mol. Microbiol. 2007;65:1380–1394. doi: 10.1111/j.1365-2958.2007.05874.x. [DOI] [PubMed] [Google Scholar]
- 90.Cane DE, He X, Kobayashi S, Omura S, Ikeda H. J. Antibiot. 2006;59:471–479. doi: 10.1038/ja.2006.66. [DOI] [PubMed] [Google Scholar]
- 91.Tetzlaff CN, You Z, Cane DE, Takamatsu S, Omura S, Ikeda H. Biochemistry. 2006;45:6179–6186. doi: 10.1021/bi060419n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.You Z, Omura S, Ikeda H, Cane DE. Arch. Biochem. Biophys. 2007;459:233–240. doi: 10.1016/j.abb.2006.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jiang J, Tetzlaff CN, Takamatsu S, Iwatsuki M, Komatsu M, Ikeda H, Cane DE. Biochemistry. 2009;48:6431–6440. doi: 10.1021/bi900766w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Silakowski B, Kunze B, Nordsiek G, Blöcker H, Höfle G, Müller R. Eur. J. Biochem. 2000;267:6476–6485. doi: 10.1046/j.1432-1327.2000.01740.x. [DOI] [PubMed] [Google Scholar]
- 95.Tanabe T, Funahashi T, Nakao H, Miyoshi SI, Shinoda S, Yamamoto S. J. Bacteriol. 2003;185:6938–6949. doi: 10.1128/JB.185.23.6938-6949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hara H, Ohnishi Y, Horinouchi S. Microbiology. 2009;155:2197–2210. doi: 10.1099/mic.0.027862-0. [DOI] [PubMed] [Google Scholar]
- 97.Distler JK, Mansouri K, Mayer G, Stockmann M, Piepersberg W. Gene. 1992;115:105–111. doi: 10.1016/0378-1119(92)90547-3. [DOI] [PubMed] [Google Scholar]
- 98.Kato J, Funa N, Watanabe H, Ohnishi Y, Horinouchi S. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2378–2383. doi: 10.1073/pnas.0607472104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Higashi T, Iwasaki Y, Ohnishi Y, Horinouchi S. J. Bacteriol. 2007;189:3515–3524. doi: 10.1128/JB.00055-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lee H-S, Ohnishi Y, Horinouchi S. J. Mol. Microbiol. Biotechnol. 2001;3:95–101. [PubMed] [Google Scholar]
- 101.Funa N, Funabashi M, Ohnishi Y, Horinouchi S. J. Bacteriol. 2005;187:8149–8155. doi: 10.1128/JB.187.23.8149-8155.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Moore BS, Hopke JN. ChemBioChem. 2001;2:35–38. doi: 10.1002/1439-7633(20010105)2:1<35::AID-CBIC35>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 103.Funabashi M, Funa N, Horinouchi S. J. Biol. Chem. 2008;283:13983–13991. doi: 10.1074/jbc.M710461200. [DOI] [PubMed] [Google Scholar]
- 104.Penn K, Jenkins C, Nett M, Udwary DW, Gontang EA, McGlinchey RP, Foster B, Lapidus A, Podell S, Allen E, Moore BS, Jensen PR. ISME J. 2009 doi: 10.1038/ismej.2009.58. DOI: 10.1038/ismej.2009.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Choulet F, Gallois A, Aigle B, Mangenot S, Gerbaud C, Truong C, Francou FX, Borges F, Fourrier C, Guérineau M, Decaris B, Barbe V, Pernodet JL, Leblond P. J. Bacteriol. 2006;188:6599–6610. doi: 10.1128/JB.00734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen CW, Huang CH, Lee HH, Tsai HH, Kirby R. Trends Genet. 2002;18:522–529. doi: 10.1016/s0168-9525(02)02752-x. [DOI] [PubMed] [Google Scholar]
- 107.Choulet F, Aigle B, Gallois A, Mangenot S, Gerbaud C, Truong C, Francou FX, Fourrier C, Guérineau M, Decaris B, Barbe V, Pernodet JL, Leblond P. Mol. Biol. Evol. 2006;23:2361–2369. doi: 10.1093/molbev/msl108. [DOI] [PubMed] [Google Scholar]
- 108.Kinashi H, Shimaji M, Sakai A. Nature. 1987;328:454–456. doi: 10.1038/328454a0. [DOI] [PubMed] [Google Scholar]
- 109.Mochizuki S, Hiratsu K, Suwa M, Ishii T, Sugino F, Yamada K, Kinashi H. Mol. Microbiol. 2003;48:1501–1510. doi: 10.1046/j.1365-2958.2003.03523.x. [DOI] [PubMed] [Google Scholar]
- 110.Yamasaki M, Kinashi H. J. Bacteriol. 2004;186:6553–6559. doi: 10.1128/JB.186.19.6553-6559.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhanel GG, Dueck M, Hoban DJ, Vercaigne LM, Embil JM, Gin AS, Karlowsky JA. Drugs. 2001;61:443–498. doi: 10.2165/00003495-200161040-00003. [DOI] [PubMed] [Google Scholar]
- 112.Labeda DP. Int. J. Syst. Bacteriol. 1987;37:19–22. [Google Scholar]
- 113.Oliynyk M, Samborskyy M, Lester JB, Mironenko T, Scott N, Dickens S, Haydock SF, Leadlay PF. Nat. Biotechnol. 2007;25:447–453. doi: 10.1038/nbt1297. [DOI] [PubMed] [Google Scholar]
- 114.Peano C, Bicciato S, Corti G, Ferrari F, Rizzi E, Bonnal RJP, Bordoni R, Albertini A, Bernardi LR, Donadio S, De Bellis G. Microb. Cell Factories. 2007;6:37. doi: 10.1186/1475-2859-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cortes J, Haydock SF, Roberts GA, Bevitt DJ, Leadlay PF. Nature. 1990;348:176–178. doi: 10.1038/348176a0. [DOI] [PubMed] [Google Scholar]
- 116.Donadio S, Staver MJ, McAlpine JB, Swanson SJ, Katz L. Science. 1991;252:675–679. doi: 10.1126/science.2024119. [DOI] [PubMed] [Google Scholar]
- 117.Staunton J, Wilkinson B. Chem. Rev. 1997;97:2611–2630. doi: 10.1021/cr9600316. [DOI] [PubMed] [Google Scholar]
- 118.Katz L. Methods Enzymol. 2009;459:113–142. doi: 10.1016/S0076-6879(09)04606-0. [DOI] [PubMed] [Google Scholar]
- 119.Boakes S, Oliynyk M, Cortés J, Bohm I, Rudd BAM, Revill WP, Staunton J, Leadlay PF. J. Mol. Microbiol. Biotechnol. 2004;8:73–80. doi: 10.1159/000084562. [DOI] [PubMed] [Google Scholar]
- 120.Banskota AH, McAlpine JB, Sorensen D, Aouidate M, Piraee M, Alarco AM, Omura S, Shiomi K, Farnet CM, Zazopoulos E. J. Antibiot. 2006;59:168–176. doi: 10.1038/ja.2006.24. [DOI] [PubMed] [Google Scholar]
- 121.Frank B, Wenzel SC, Bode HB, Scharfe M, Blöcker H, Müller R. J. Mol. Biol. 2007;374:24–38. doi: 10.1016/j.jmb.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 122.Cortés J, Velasco J, Foster G, Blackaby AP, Rudd BAM, Wilkinson B. Mol. Microbiol. 2002;44:1213–1224. doi: 10.1046/j.1365-2958.2002.02975.x. [DOI] [PubMed] [Google Scholar]
- 123.Oliveira PH, Batagov A, Ward J, Baganz F, Krabben P. Lett. Appl. Microbiol. 2006;42:375–380. doi: 10.1111/j.1472-765X.2006.01849.x. [DOI] [PubMed] [Google Scholar]
- 124.Jensen PR, Dwight R, Fenical W. Appl. Environ. Microbiol. 1991;57:1102–1108. doi: 10.1128/aem.57.4.1102-1108.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mincer TJ, Jensen PR, Kauffman CA, Fenical W. Appl. Environ. Microbiol. 2002;68:5005–5011. doi: 10.1128/AEM.68.10.5005-5011.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Maldonado L, Fenical W, Goodfellow M, Jensen PR, Ward AC. Int. J. System. Appl. Microbiol. 2005;55:1759–1766. doi: 10.1099/ijs.0.63625-0. [DOI] [PubMed] [Google Scholar]
- 127.Fenical W, Jensen PR. Nat. Chem. Biol. 2006;2:666–673. doi: 10.1038/nchembio841. [DOI] [PubMed] [Google Scholar]
- 128.Udwary DW, Zeigler L, Asolkar RN, Singan V, Lapidus A, Fenical W, Jensen PR, Moore BS. Proc. Natl. Acad. Sci. U. S. A. 2007;104:10376–10381. doi: 10.1073/pnas.0700962104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Redenbach M, Scheel J, Schmidt U. Antonie van Leeuwenhoek. 2000;78:227–235. doi: 10.1023/a:1010289326752. [DOI] [PubMed] [Google Scholar]
- 130.Nett M, Moore BS. Pure Appl. Chem. 2009;81:1075–1084. [Google Scholar]
- 131.Eustáquio AS, McGlinchey RP, Liu Y, Hazzard C, Beer LL, Florova G, Alhamadsheh MM, Lechner A, Kale AJ, Kobayashi Y, Reynolds KA, Moore BS. Proc. Natl. Acad. Sci. U. S. A. 2009;106:12295–12300. doi: 10.1073/pnas.0901237106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.McGlinchey RP, Nett M, Moore BS. J. Am. Chem. Soc. 2008;130:2406–2407. doi: 10.1021/ja710488m. [DOI] [PubMed] [Google Scholar]
- 133.Aotani Y, Nagata H, Yoshida M. J. Antibiot. 1997;50:543–545. doi: 10.7164/antibiotics.50.543. [DOI] [PubMed] [Google Scholar]
- 134.Eustáquio AS, Pojer F, Noel JP, Moore BS. Nat. Chem. Biol. 2008;4:69–74. doi: 10.1038/nchembio.2007.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.McGlinchey RP, Nett M, Eustáquio AS, Asolkar RN, Fenical W, Moore BS. J. Am. Chem. Soc. 2008;130:7822–7823. doi: 10.1021/ja8029398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Eustáquio AS, Moore BS. Angew. Chem., Int. Ed. 2008;47:3936–3938. doi: 10.1002/anie.200800177. [DOI] [PubMed] [Google Scholar]
- 137.Liu Y, Hazzard C, Eustáquio AS, Reynolds KA, Moore BS. J. Am. Chem. Soc. 2009;131:10376–10377. doi: 10.1021/ja9042824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Williams PG, Asolkar RN, Kondratyuk T, Pezzuto JM, Jensen PR, Fenical W. J. Nat. Prod. 2007;70:83–88. doi: 10.1021/np0604580. [DOI] [PubMed] [Google Scholar]
- 139.Schultz AW, Oh D-C, Carney JR, Williamson RT, Udwary DW, Jensen PR, Gould SJ, Fenical W, Moore BS. J. Am. Chem. Soc. 2008;130:4507–4516. doi: 10.1021/ja711188x. [DOI] [PubMed] [Google Scholar]
- 140.Oh D-C, Williams PG, Kauffman CA, Jensen PR, Fenical W. Org. Lett. 2006;8:1021–1024. doi: 10.1021/ol052686b. [DOI] [PubMed] [Google Scholar]
- 141.Perrin CL, Rodgers BL, O’Connor JM. J. Am. Chem. Soc. 2007;129:4795–4799. doi: 10.1021/ja070023e. [DOI] [PubMed] [Google Scholar]
- 142.Jensen PR, Williams PG, Oh D-C, Zeigler L, Fenical W. Appl. Environ. Microbiol. 2007;73:1146–1152. doi: 10.1128/AEM.01891-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fischbach MA, Walsh CT, Clardy J. Proc. Natl. Acad. Sci. U. S. A. 2008;105:4601–4608. doi: 10.1073/pnas.0709132105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Coleman ML, Sullivan MB, Martiny AC, Steglich C, Barry K, DeLong EF, Chisholm SW. Science. 2006;311:1768–1770. doi: 10.1126/science.1122050. [DOI] [PubMed] [Google Scholar]
- 145.Normand P, Lapierre P, Tisa LS, Gogarten JP, Alloisio N, Bagnarol E, Bassi CA, Berry AM, Bickhart DM, Choisne N, Couloux A, Cournoyer B, Cruveiller S, Daubin V, Demange N, Francino MP, Goltsman E, Huang Y, Kopp OR, Labarre L, Lapidus A, Lavire C, Marechal J, Martinez M, Mastronunzio JE, Mullin BC, Niemann J, Pujic P, Rawnsley T, Rouy Z, Schenowitz C, Sellstedt A, Tavares F, Tomkins JP, Vallenet D, Valverde C, Wall LG, Wang Y, Medigue C, Benson DR. Genome Res. 2007;17:7–15. doi: 10.1101/gr.5798407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sen A, Sur S, Bothra AK, Benson DR, Normand P, Tisa LS. Antonie van Leeuwenhoek. 2008;93:335–346. doi: 10.1007/s10482-007-9211-1. [DOI] [PubMed] [Google Scholar]
- 147.Wu X, Flatt PM, Xu H, Mahmud T. ChemBioChem. 2009;10:304–314. doi: 10.1002/cbic.200800527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Shao Z, Blodgett JAV, Circello BT, Eliot AC, Woodyer R, Li GL, van der Donk WA, Metcalf WW, Zhao H. J. Biol. Chem. 2008;283:23161–23168. doi: 10.1074/jbc.M801788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hauser FM, Caringal Y. J. Org. Chem. 1990;55:555–559. [Google Scholar]
- 150.Klika KD, Haansuu JP, Ovcharenko VV, Haahtela KK, Vuorela PM, Sillanpää R, Pihlaja K. Z. Naturforsch., B. 2003;58:1210–1215. [Google Scholar]
- 151.Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, van Sinderen D. Microbiol. Mol. Biol. Rev. 2007;71:495–548. doi: 10.1128/MMBR.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Donadio S, Monciardini P, Sosio M. Nat. Prod. Rep. 2007;24:1073–1109. doi: 10.1039/b514050c. [DOI] [PubMed] [Google Scholar]
- 153.Nemoto A, Katsukiyo Y, Yazawa K, Ando A, Mikami Y. J. Antibiot. 2002;55:593–597. doi: 10.7164/antibiotics.55.593. [DOI] [PubMed] [Google Scholar]
- 154.Tsuda M, Yamakawa M, Oka S, Tanaka Y, Hoshino Y, Mikami Y, Sato A, Fujiwara H, Ohizumi Y, Kobayashi J. J. Nat. Prod. 2005;68:462–464. doi: 10.1021/np0496385. [DOI] [PubMed] [Google Scholar]
- 155.Mukai A, Fukai T, Matsumoto Y, Ishikawa J, Hoshino Y, Yazawa K, Harada K, Mikami Y. J. Antibiot. 2006;59:366–369. doi: 10.1038/ja.2006.53. [DOI] [PubMed] [Google Scholar]
- 156.Quadri LEN, Sello J, Keating TA, Weinreb PH, Walsh CT. Chem. Biol. 1998;5:631–645. doi: 10.1016/s1074-5521(98)90291-5. [DOI] [PubMed] [Google Scholar]
- 157.Mikami Y, Yazawa K, Ohashi S, Maeda A, Akao M. J. Antibiot. 1992;45:995–997. doi: 10.7164/antibiotics.45.995. [DOI] [PubMed] [Google Scholar]
- 158.Tanaka Y, Gräfe U, Yazawa K, Mikami Y, Ritzau M. J. Antibiot. 1997;50:822–827. doi: 10.7164/antibiotics.50.822. [DOI] [PubMed] [Google Scholar]
- 159.Tsuda M, Sato H, Tanaka Y, Yazawa K, Mikami Y, Sasaki T, Kobayashi J. J. Chem. Soc., Perkin Trans. 1. 1996:1773–1775. [Google Scholar]
- 160.Shigemori H, Tanaka Y, Yazawa K, Mikami Y, Kobayashi J. Tetrahedron. 1996;52:9031–9034. [Google Scholar]
- 161.Hashimoto M, Komori T, Kamiya T. J. Am. Chem. Soc. 1976;98:3023–3025. doi: 10.1021/ja00426a063. [DOI] [PubMed] [Google Scholar]
- 162.Kobayashi J, Tsuda M, Nemoto A, Tanaka Y, Yazawa K, Mikami Y. J. Nat. Prod. 1997;60:719–720. doi: 10.1021/np970132e. [DOI] [PubMed] [Google Scholar]
- 163.Shigemori H, Komaki H, Yazawa K, Mikami Y, Nemoto A, Tanaka Y, Sasaki T, In Y, Ishida T, Kobayashi J. J. Org. Chem. 1998;63:6900–6904. doi: 10.1021/jo9807114. [DOI] [PubMed] [Google Scholar]
- 164.Tsuda M, Nemoto A, Komaki H, Tanaka Y, Yazawa K, Mikami Y, Kobayashi J. J. Nat. Prod. 1999;62:1640–1642. doi: 10.1021/np970132e. [DOI] [PubMed] [Google Scholar]
- 165.Martinkova L, Uhnakova B, P. M, Nesvera J, Kren V. Environ. Int. 2009;35:162–177. doi: 10.1016/j.envint.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 166.Szolkowy C, Eltis LD, Bruce NC, Grogan G. ChemBioChem. 2009;10:1208–1217. doi: 10.1002/cbic.200900011. [DOI] [PubMed] [Google Scholar]
- 167.Iwatsuki M, Tomoda H, Uchida R, Gouda H, Hirono S, Omura S. J. Am. Chem. Soc. 2006;128:7486–7491. doi: 10.1021/ja056780z. [DOI] [PubMed] [Google Scholar]
- 168.Ostash B, Saghatelian A, Walker S. Chem. Biol. 2007;14:257–267. doi: 10.1016/j.chembiol.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Barona-Gomez F, Lautru S, Francou FX, Leblond P, Pernodet JL, Challis GL. Microbiology. 2006;152:3355–3366. doi: 10.1099/mic.0.29161-0. [DOI] [PubMed] [Google Scholar]
- 170.Pang X, Aigle B, Girardet J-M, Mangenot S, Pernodet JL, Decaris B, Leblond P. Antimicrob. Agents Chemother. 2004;48:575–588. doi: 10.1128/AAC.48.2.575-588.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Bunet R, Mendes MV, Rouhier N, Pang X, Hotel L, Leblond P, Aigle B. J. Bacteriol. 2008;190:3293–3305. doi: 10.1128/JB.01989-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Juguet M, Lautru S, Francou FX, Nezbedová S, Leblond P, Gondry M, Pernodet JL. Chem. Biol. 2009;16:421–431. doi: 10.1016/j.chembiol.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 173.Charan RD, Schlingmann G, Janso J, Bernan V, Feng X, Carter GT. J. Nat. Prod. 2004;67:1431–1433. doi: 10.1021/np040042r. [DOI] [PubMed] [Google Scholar]
- 174.McAlpine JB, Banskota AH, Charan RD, Schlingmann G, Zazopoulos E, Piraee M, Janso J, Bernan VS, Aouidate M, Farnet CM, Feng X, Zhao Z, Carter GT. J. Nat. Prod. 2008;71:1585–1590. doi: 10.1021/np800376n. [DOI] [PubMed] [Google Scholar]
- 175.Bagley MC, Dale JW, Merritt EA, Xiong X. Chem. Rev. 2005;105:685–714. doi: 10.1021/cr0300441. [DOI] [PubMed] [Google Scholar]
- 176.Smith TM, Jiang Y-F, Shipley PR, Floss HG. Gene. 1995;164:137–142. doi: 10.1016/0378-1119(95)00442-9. [DOI] [PubMed] [Google Scholar]
- 177.Kelly WL, Pan L, Li C. J. Am. Chem. Soc. 2009;131:4327–4334. doi: 10.1021/ja807890a. [DOI] [PubMed] [Google Scholar]
- 178.Liao RJ, Duan L, Lei C, Pan H, Ding Y, Zhang Q, Chen D, Shen B, Yu Y, Liu W. Chem. Biol. 2009;16:141–147. doi: 10.1016/j.chembiol.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Brown LCW, Acker MG, Clardy J, Walsh CT, Fischbach MA. Proc. Natl. Acad. Sci. U. S. A. 2009;106:2549–2553. doi: 10.1073/pnas.0900008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Morris RP, Leeds JA, Naegeli HU, Oberer L, Memmert K, Weber E, LaMarche MJ, Parker CN, Burrer N, Esterow S, Hein A, Schmitt EK, Krastel P. J. Am. Chem. Soc. 2009;131:5946–5955. doi: 10.1021/ja900488a. [DOI] [PubMed] [Google Scholar]
- 181.Zerikly M, Challis GL. ChemBioChem. 2009;10:625–633. doi: 10.1002/cbic.200800389. [DOI] [PubMed] [Google Scholar]
- 182.Wilkinson B, Micklefield J. Nat. Chem. Biol. 2007;3:379–386. doi: 10.1038/nchembio.2007.7. [DOI] [PubMed] [Google Scholar]
- 183.Scherlach K, Hertweck C. Org. Biomol. Chem. 2009;7:1753–1760. doi: 10.1039/b821578b. [DOI] [PubMed] [Google Scholar]
- 184.Zazopoulos E, Huang KX, Staffa A, Liu W, Bachmann BO, Nonaka K, Ahlert J, Thorson JS, Shen B, Farnet CM. Nat. Biotechnol. 2003;21:187–190. doi: 10.1038/nbt784. [DOI] [PubMed] [Google Scholar]
- 185.Fischbach MA, Walsh CT. Chem. Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 186.Haynes SW, Challis GL. Curr. Opin. Drug Discov. Dev. 2007;10:203–218. [PubMed] [Google Scholar]
- 187.Van Lanen SG, Shen B. Curr. Opin. Microbiol. 2006;9:252–260. doi: 10.1016/j.mib.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 188.Jenke-Kodama H, Dittmann E. Nat. Prod. Rep. 2009;26:874–883. doi: 10.1039/b810283j. [DOI] [PubMed] [Google Scholar]
- 189.Yadav G, Gokhale RS, Mohanty D. PLoS Comput. Biol. 2009;5:e1000351. doi: 10.1371/journal.pcbi.1000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Li MH, Ung PM, Zajkowski J, Garneau-Tsodikova S, Sherman DH. BMC Bioinformatics. 2009;10:185. doi: 10.1186/1471-2105-10-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Starcevic A, Zucko J, Simunkovic J, Long PF, Cullum J, Hranueli D. Nucleic Acids Res. 2008;36:6882–6892. doi: 10.1093/nar/gkn685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Weber T, Rausch C, Lopez P, Hoof I, Gaykova V, Huson DH, Wohlleben W. J. Biotechnol. 2009;140:13–17. doi: 10.1016/j.jbiotec.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 193.Tohyama S, Eguchi T, Dhakal RP, Akashi T, Otsuka M, Kakinuma K. Tetrahedron. 2004;60:3999–4005. [Google Scholar]
- 194.Tohyama S, Kakinuma K, Eguchi T. J. Antibiot. 2006;59:44–52. doi: 10.1038/ja.2006.7. [DOI] [PubMed] [Google Scholar]
- 195.McAlpine JB, Bachmann BO, Piraee M, Tremblay S, Alarco AM, Zazopoulos E, Farnet CM. J. Nat. Prod. 2005;68:493–496. doi: 10.1021/np0401664. [DOI] [PubMed] [Google Scholar]
- 196.Banskota AH, McAlpine JB, Sorensen D, Ibrahim A, Aouidate M, Piraee M, Alarco AM, Farnet CM, Zazopoulos E. J. Antibiot. 2006;59:533–542. doi: 10.1038/ja.2006.74. [DOI] [PubMed] [Google Scholar]
- 197.Cho JY, Kwon HC, Williams PG, Kauffman CA, Jensen PR, Fenical W. J. Nat. Prod. 2006;69:425–428. doi: 10.1021/np050402q. [DOI] [PubMed] [Google Scholar]
- 198.Winter JM, Moffitt MC, Zazopoulos E, McAlpine JB, Dorrestein PC, Moore BS. J. Biol. Chem. 2007;282:16362–16368. doi: 10.1074/jbc.M611046200. [DOI] [PubMed] [Google Scholar]
- 199.Dimise EJ, Widboom PF, Bruner SD. Proc. Natl. Acad. Sci. U. S. A. 2008;105:15311–15316. doi: 10.1073/pnas.0805451105. [DOI] [PMC free article] [PubMed] [Google Scholar]