Abstract
Polycystin-1 (PC1) is a large, membrane-bound protein that localizes to the cilia and is implicated in the common ciliopathy autosomal-dominant polycystic kidney disease. The physiological function of PC1 is dependent upon its subcellular localization as well as specific cleavages that release soluble fragments of its C-terminal tail. The techniques described here allow visualization and quantification of these aspects of the biology of the PC1 protein. To visualize PC1 at the plasma membrane, a live-cell surface labeling immunofluorescence protocol paired with the labeling of an internal antigen motif allows a robust detection of the surface population of this protein. This technique is modified to generate a surface enzyme-linked immunosorbent assay (ELISA), which quantitatively measures the amount of surface protein as a fraction of the total amount of the protein expressed in that cell population. These assays are powerful tools in the assessment of the small but biologically important pool of PC1 that reaches the cell surface. The C-terminal tail cleavage of PC1 constitutes an interesting modification that allows PC1 to extend its functional role into the nucleus. A reporter assay based on Gal4/VP16 luciferase can be used to quantitate the amount of PC1 C-terminal tail that reaches the nucleus. This assay can be paired with quantitative measurement of the protein expression in the cell, allowing a more complete understanding of the pattern of PC1 cleavage and the nuclear localization of the resultant.
I. Introduction
Mounting evidence illuminates the crucial role that cilia play in mechanosensation and signal transduction, linking extracellular conditions to changes in intracellular signaling pathways. Ciliary proteins such as polyductin and the polycystins localize to the cilia and plasma membrane where they can alter intracellular conditions directly, through changes in ion concentration, or indirectly, by releasing soluble cytoplasmic fragments that can partner with intracellular signaling molecules to affect processes such as gene transcription. Regulation of the correct physiological functioning of these ciliary proteins therefore involves their localization and cleavage; the immunofluorescence and reporter-driven assays described here provide ways to analyze these crucial aspects of ciliary protein biology.
Polycystin-1 (PC1) is the product of the polycystic kidney disease 1 (PKD1) gene that, along with PKD2 (encoding PC2), harbors the mutations that cause autosomal-dominant polycystic kidney disease (ADPKD). This common genetic disease affects approximately 1 in 1000 individuals. A significant manifestation of the disease is the progressive appearance and growth of renal cysts. These cysts displace and destroy adjacent renal parenchyma, leading to end-stage renal disease in approximately 50% of cases. There are also cardiovascular, musculoskeletal, and gastrointestinal abnormalities associated with ADPKD (Gabow, 1993). The connection between the PKD1 and PKD2 genes and ADPKD was first shown by genetic linkage studies and later verified in animal models. Cysts form when both somatic copies of either polycystin gene are mutated or knocked out (Lu et al., 1997; Qian et al., 1996). Cysts can also arise when the level of PKD1 expression is significantly up- or downregulated (Lantinga-van Leeuwen et al., 2004; Pritchard et al., 2000).
The complex subcellular localization of PC1 reflects the broad range of this protein’s cellular functions. There is an extensive literature documenting the localization of PC1 and PC2 to primary cilia. This localization is thought to permit the PC1/PC2 complex to play a role in sensing fluid flow (Chauvet et al., 2004; Nauli et al., 2003). PC1 may also play a role in establishing cell–cell connections; it is found at desmosomes and in the basolateral membrane of Madin–Darby canine kidney (MDCK) cells (Bukanov et al., 2002) and can stimulate junction formation by binding E-cadherin (Streets et al., 2009). It is also thought to contribute to ion channel activity when it localizes with PC2 to the plasma membrane (Hanaoka et al., 2000). These studies have primarily used costaining with specific antibodies to identify PC1’s presence in specific membrane domains through colocalization, but until now there has been no technique that has allowed a quantitative assessment of the overall delivery of PC1 to the plasma membrane.
PC1 participates in a variety of signaling pathways in the cell, and the cleavage of the PC1 protein’s C-terminal tail may allow PC1 to affect a variety of diverse intracellular processes in response to stimuli such as extracellular fluid flow. One cleavage occurs within the cytoplasmic tail and releases a protein fragment that translocates to the nucleus and interacts with STAT6 and p100 (Low et al., 2006). Another cleavage releases a larger soluble portion of the tail that activates the activator protein 1 pathway (Chauvet et al., 2004) and that inhibits canonical Wnt signaling (Lal et al., 2008). Rates of cleavage at both cleavage sites increase with the cessation of fluid flow, suggesting a link between PC1’s roles in mechanosensation and the modulation of signaling pathways. To date, the principal approaches to studying the C-terminal PC1 cleavage and nuclear translocation have involved Western blotting and immunofluorescence, which can report on the occurrence of cleavage and translocation under some conditions but do not allow a very nuanced understanding of how environmental cues affect the extent of cleavage and the subcellular location of the soluble PC1 tail fragment. The Gal4/VP16 assay as described below permits the measurement of PC1 cleavage and nuclear translocation, allowing a more detailed analysis of these processes under varying physiological conditions.
II. Assay Rationale and History
A. Surface Immunofluorescence and Enzyme-Linked Immunosorbent Assay
PC1 has two distinct subcellular distributions when it is exogenously expressed by transfection in cell culture. A significant portion of the protein is found in the endoplasmic reticulum when heterologously expressed in cell culture, but it has also been shown to localize to the plasma membrane and the primary cilium and this localization is likely critical for the function of the PC1 protein as a channel or flow sensor. Since the location of PC1 may have an effect on its function, it is useful to know under what conditions the protein reaches the plasma membrane and whether this can be altered by coexpression of other proteins or the application of drugs to change the intra- or extracellular environment. While cell surface biotinylation is often used as a standard method for quantifying the amount of a given protein that reaches the surface (Hurley et al., 1985), PC1’s large size and relatively low level of detectable surface delivery led us to develop another method for identifying and quantifying surface PC1.
Visualizing the surface pool of PC1 protein is most effectively accomplished using an immunofluorescence protocol that yields a view of protein distribution in which the surface protein is tagged with one fluorescent marker and the internal pool is labeled with another color (Fig. 1). This provides a clear and relatively simple way to image the distribution of PC1 and provides an assay system that can then be perturbed with drugs or coexpressed proteins to reveal the effects of these manipulations on PC1’s surface localization. The Alexa class of fluorescent dyes produce a bright signal that renders it easy to detect even small populations of surface-localized PC1, thus ensuring a high degree of sensitivity of the assay for surface localization.
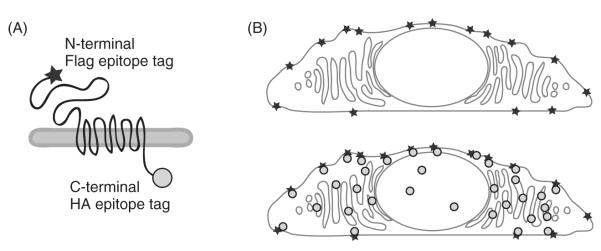
Fig. 1.
Surface immunofluorescence detects the pool of polycystin-1 (PC1) at the plasma membrane. The N-terminal Flag epitope tag on PC1 is marked with one antibody (represented by a star), while the intracellular C-terminal HA epitope tag is detected with a second antibody (represented by a circle) (A). The protocol for surface immunofluorescence is optimized to mark only the PC1 that has reached the plasma membrane (B, upper panel), while adding the HA antibody after permeabilizing the cells allows a visualization of the total amount of PC1 expressed in the same cell (B, lower panel).
Although immunofluorescence is an ideal method for qualitatively looking at protein distribution at the level of individual cells, effort is required to ensure that it is quantifiable. Quantitative immunofluorescence requires both a lengthy labeling protocol and a substantial investment of time to examine each coverslip on a microscope. We therefore developed a whole-cell enzyme-linked immunosorbent assay (ELISA) to provide a high-throughput assay system that quantifies the amount of surface PC1 relative to the total amount expressed in the cells. This assay can easily be performed in 96-well plates, allowing us to screen up to 16 conditions. Since the assay is sensitive to changes in cell number, it is best suited to assessing the consequences of manipulations that do not cause cell death.
B. Gal/VP Luciferase Assay
Regulated intramembrane proteolysis is a mechanism by which a membrane-bound protein, such as Notch, amyloid β-protein precursor, or EpCAM (Ebinu and Yankner, 2002; Maetzel et al., 2009), is cleaved within a transmembrane domain to release a soluble cytoplasmic peptide that translocates to the nucleus and modifies gene expression or alters intracellular signaling pathways. The study of these types of cleavage and translocation events was facilitated through the generation of a complementary DNA (cDNA) reporter construct that could be transcriptionally activated by the nuclear translocation of a tagged protein. The DNA-binding domain of the yeast Gal4 transcription factor, combined with the transcriptional activation domain of the viral VP16 protein, drives strong expression of genes that are downstream of the Gal4-binding upstream activating sequence (UAS) (Sadowski et al., 1988). This allows a membrane protein of interest, fused to both Gal4 and VP16, to induce the expression of a UAS-driven reporter protein upon the cleavage, release, and nuclear translocation of its cytoplasmic domain. Since the development of this system, various UAS-driven reporters have been used in a variety of assay systems to illuminate processes such as the in vivo location of Notch cleavage in the developing Drosophila embryo (Struhl and Greenwald, 1999) and to assay for small molecules that affect the cleavage and translocation of the amyloid precursor protein (Bakshi et al., 2005).
The cleavage and trafficking of PC1 have many parallels with the life cycles of other proteins cleaved by regulated intramembrane proteolysis, in that the soluble C-terminal tail is cleaved and translocates to the nucleus (see review by Guay-Woodford, 2004). Studying PC1 cleavage has been complicated by the fact that there are at least two different C-terminal fragments that can be released by cleavage (Chauvet et al., 2004; Low et al., 2006). Expressing a soluble form of the C-terminal fragment could yield information about the effects of these peptides on intracellular signaling pathways, but this gives no insight into the processes that generate the soluble peptides and may actually produce results that are difficult to interpret in a physiological context (Basavanna et al., 2007). The endogenous fragments produced by these cleavage events within the full-length protein are often short-lived and found in low abundance. Their relatively small size makes it difficult to use sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gels to quantify their production in relation to the amount of the full-length protein or to determine the relative percent of the cleaved fragments that translocate to the nucleus under various physiological conditions. These limitations made PC1 an ideal candidate for a Gal4/VP16 reporter assay to quantify its cleavage and nuclear translocation.
To create PC1-Gal4/VP16, we inserted the binding and activation domains of Gal4/VP16 at the C-terminus of a cDNA encoding full-length PC1 in the mammalian expression vector pcDNA3.1, using a unique restriction site in a 3 × hemagglutinin (HA) tag already present at the end of the PC1 sequence. The placement of the Gal4/VP16 sequence means that any fragment produced by a C-terminal cleavage contains the binding and activating domains required to drive the transcription of the chosen reporter construct. To study changes in PC1 cleavage over relatively short time courses in cell culture-based assays, we chose to use a UAS-luciferase from Promega containing a sequence of amino acids rich in proline, glutamic acid, serine and threonine (a PEST sequence) that promotes protein degradation. It is important to note that this reporter system requires robust cotransfection and expression of both the PC1-Gal4/VP16 and reporter proteins, so experimental conditions or treatments that alter transfection efficiency or protein synthesis could give false-positive results in assays evaluating manipulations that might perturb PC1 cleavage. To compensate for this, we cotransfected the Renilla luciferase gene under the constitutively active herpes simplex virus (HSV) promoter. We can then compensate for variations in transfection efficiency and protein synthesis by normalizing the PC1-induced firefly luciferase signal to the amount of Renilla luciferase activity (Fig. 2).
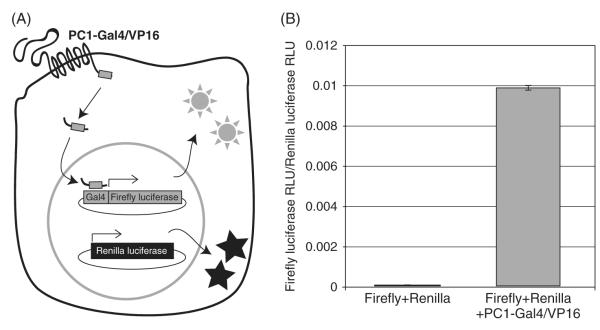
Fig. 2.
The PC1-Gal4/VP16 reporter assay. A diagram illustrating the cleavage of PC1-Gal4/VP16 releasing a soluble C-terminal fragment that, upon nuclear translocation, binds to the Gal4 UAS DNA-binding domain and activates the transcription of the downstream firefly luciferase gene (A). Constitutive expression Renilla luciferase from a cotransfected plasmid allows normalization to account for variations in transfection efficiency. The assay has a very low background, as measured by the relative light units (RLUs) produced by cells expressing only the firefly and Renilla plasmids (B). Expression of PC1-Gal4/VP16 provides a specific increase in the firefly luciferase RLU, as normalized to the Renilla RLU count.
III. Materials
A. Enzyme-Linked Immunosorbent Assay
Blocking buffer: 5% FBS, 0.5% BSA in PBS++ (phosphate buffered saline with 100 μM CaCl2 and 1 mM MgCl2)
Permeabilization buffer: 5% FBS, 0.5% BSA, and 0.5% TritonX-100
Antibody against an epitope that is extracellular when the protein of interest reaches the plasma membrane (for illustrative purposes these instructions will refer to polyclonal anti-Flag, Sigma-Aldrich, St. Louis, MO)
Secondary antibody, conjugated to horseradish peroxidase (HRP), directed against the primary antibody’s isotype
Ultra tetramethylbenzidine (TMB)-ELISA (Thermo Scientific, Waltham, MA)
Sulfuric acid, 1 N
Plate reader that can read absorbance at 450 nm
B. Surface Immunofluorescence
Primary antibody directed against an epitope that is exposed at the extracellular surface when the protein of interest reaches the plasma membrane (we use the same antibody as for the ELISA, which is polyclonal anti-Flag from Sigma-Aldrich)
Primary antibody against an intracellular epitope of the protein of interest. This antibody should not cross-react with the primary antibody against the extracellular epitope and should be produced in a different species (we use a monoclonal anti-HA from Covance, Princeton, NJ)
PBS++ with 100 μM CaCl2 and 1 mM MgCl2
Blocking Buffer: 0.1% BSA in PBS++
Permeabilization buffer: 0.3% TritonX-100, 0.1% BSA in PBS++
Goat serum dilution buffer (GSDB): 16% goat serum, 120 mM sodium phosphate, 0.3% Triton X-100, and 450 mM NaCl
Secondary antibodies, conjugated to fluorophores of choice, against immunoglobulin G (IgG) from the appropriate species used for the primary antibodies (we use the Alexa Fluor 594 anti-rabbit IgG and 488 anti-mouse IgG for the polyclonal anti-Flag and monoclonal anti-HA, respectively, both supplied by Invitrogen, Carlsbad, CA)
C. Gal/VP Luciferase Assay
UAS-promoted firefly luciferase cDNA plasmid [pGL4.31(luc2p/GAL4UAS/Hygro) from Promega, Madison, WI].
Constitutively expressed Renilla luciferase cDNA plasmid (pRL-TK from Promega)
PC1-Gal/VP: PC1 with the Gal4 DNA-binding domain and VP16-activation domains added at the C-terminus of the protein
Kit for reading the luciferase signals (Dual Luciferase kit from Promega).
Luminometer, either single-read or configured as a plate-reader. If using a plate reader, whether or not it has injection capabilities will influence your choice of assay kit, since some require addition of a buffer immediately before reading the light output
IV. Methods
A. Surface Immunofluorescence
This protocol is optimized for use on adherent cell lines grown on either glass coverslips or filters. The broad outline of the protocol requires that the surface antibody be applied in the cold prior to fixation, and then the internal antibody is added after fixation and permeabilization. As long as the surface and internal antibodies are from different source species, the secondary antibodies can be added together in the final labeling step. For PC1, immunofluorescence against the N-terminal Flag tag illuminates the pool of PC1 at the plasma membrane, while postpermeabilization immunofluorescence against the C-terminal HA tag detects the total population of PC1 (Fig. 3). This protocol is also applicable to polarized porcine kidney cells (LLC-PK) in which the ciliary localization of PC1 is demonstrated by the ciliary staining of both externally applied anti-Flag and internal anti-HA antibodies (Fig. 3).
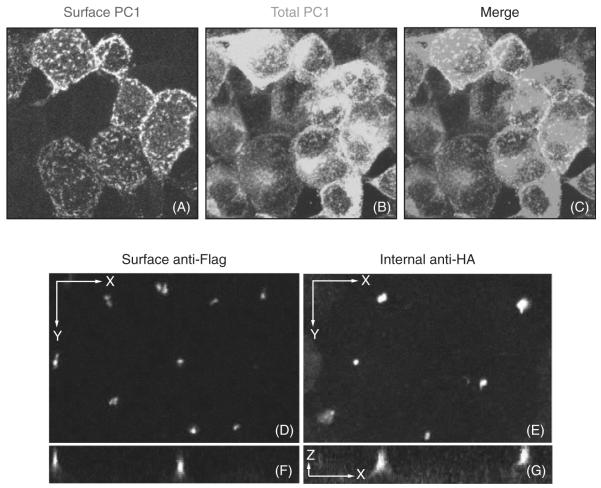
Fig. 3.
Examples of surface immunofluorescence under conditions that promote PC1 delivery to the plasma membrane and cilia. In nonpolarized HEK293 stably expressing PC1, the anti-Flag antibody shows the surface pool of PC1 (A), while the internal anti-HA antibody shows the overall PC1 expression (B). The merged image reveals the colocalization between internal and external markers (C). Images (A–C) were generated by flattening a z-stack of images obtained using confocal microscopy. Polarized LLCPK cells stably expressing PC1 have a ciliary localization of PC1, as revealed with both the surface anti-Flag antibody (D, F) and the internal anti-HA antibody (E, G); images shown in x–y and x–y projections. A slice along the z-axis shows that the HA antibody labels both ciliary and intracellular protein pools (G), while the surface anti-Flag antibody labels only cilia (F). The scale bar for (D–G) is 10 μm. (See Plate no. 15 in the Color Plate Section.)
Cells for surface immunofluorescence should be grown on either filters or glass coverslips, although cells that are weakly adherent should be plated on coverslips coated with poly-L-lysine to minimize cell loss. Cells should be transfected according to the method of choice for the particular cell type when the cells have reached an appropriate level of confluency. The cells should then be allowed to express the protein of interest for 24 h before beginning the immunofluorescence protocol. When the cells are ready to be assayed, they may be first incubated in the blocking buffer at 4°C for 30 min to prevent nonspecific antibody binding. We have found that signal-to-noise ratio of the polyclonal anti-Flag antibody we use does not improve dramatically with this surface-blocking step, but it has been a useful step for antibodies that have higher amounts of nonspecific binding. Cells not being blocked are rinsed once with blocking buffer, and then all coverslips are inverted onto a small volume of blocking buffer containing primary antibody against the extracellular epitope for 1 h at 4°C in a humidified chamber. If the cells are grown on filters, then the surface antibody may be selectively applied to the plasma membrane domain (apical or basolateral) in which the protein of interest resides. The primary antibody is applied at 4°C to prevent trafficking and redistribution of the surface population of the antigen during the course of the primary antibody incubation. After this incubation, the coverslips are returned to their wells and gently washed a minimum of one time in PBS++ and fixed in 4% paraformaldehyde for 20 min. All steps including and following the fixation are performed at room temperature. The protocol may be paused after the paraformaldehyde is washed out and the coverslips may be stored overnight at 4°C in PBS.
After fixation, the coverslips are washed three times in PBS++ and then incubated at room temperature with permeabilization buffer for 15 min. They are then blocked with GSDB for 30 min before an hour-long incubation with the antibody against the intracellular epitope diluted in GSDB. After this incubation, the cells are washed three times with permeabilization buffer and then incubated in a darkened, humidified chamber on a drop of the secondary antibodies diluted in GSDB for 45 min. We have found that the choice of secondary antibody makes a difference when detecting proteins, such as PC1, that have low levels of surface localization under some conditions. The high signal-to-noise obtained with bright fluorophores such as the Alexa dyes makes it much easier to detect the surface signal than does the use of rhodamine- or fluorescein isothiocyanate (FITC)-conjugated secondary antibodies.
Following incubation in secondary antibody, the coverslips are washed again in PBS++ and then the nuclei are stained. We routinely use Hoechst stain to mark the nuclei, although the nuclear stain should be chosen based on which secondary antibodies are being used and on the detection capabilities of the microscope. The stained cells are then mounted on a drop of anti-fade mounting media on glass slides, the edges are sealed with nail polish, and the slides are then ready to be viewed on the microscope of choice. We use an upright Zeiss Axiophot microscope equipped with a charge-coupled device (CCD) camera for viewing the slides and taking pictures for routine quantification, but we use a Zeiss LSM 510 Meta confocal microscope when we wish to generate images of higher quality.
In addition to providing a means to visualize the presence or absence of protein at the surface, this technique can also be adapted to quantify how the pool of surface protein changes in response to biological or biochemical treatments. To quantify the intensity of surface immunofluorescence signal, we take images of each experimental condition with identical settings for zoom and exposure. While taking images with confocal microscopy, we image the entire cell in the vertical dimension, generating a z-stack of images. The thickness of each slice and the number of slices should be optimized for the cell type being used in the experiment. We then flatten the resulting stack by exporting an image from the extended focus view in Volocity (Improvision, PerkinElmer). Once flattened, the confocal image file can be treated like that of an image obtained using an epifluorescence microscope.
To quantify the average pixel intensity per cell, an image’s total pixel intensity is calculated and divided by the total number of cells in each image that are positive for surface immunofluorescence signal. We perform this calculation by exporting the image’s pixel intensity histogram from Image J (National Institutes of Health), then multiplying each intensity value (0–255) by the number of pixels that have that intensity. Summing all values above background then gives the total pixel intensity for the image. The cutoff for the background should be calculated by evaluating the intensity histogram for an image taken of a microscopic field without surface signal and determining the pixel intensity value below which most of this background signal is found. The number of surface-positive cells can be counted from the raw images or, more accurately, it can be counted after subtracting the background cutoff value from each pixel using Image J, thereby showing exactly how many of the cells are being included in the final pixel intensity sum. Having calculated the average pixel intensity per cell, independent experiments can then be compared to determine the effects that specific treatments have on the surface pool of protein.
B. Enzyme-Linked Immunosorbent Assay
The ELISA, to compare amounts of surface and total protein, uses a spectrophotometric assay to measure the amount of protein detected using conditions in which labeling is performed with or without permeabilization. Given that this assay is amenable to being used in relatively high-throughput applications, we plate cells into 96-well plates and use six wells per experimental condition, subdivided so that three wells provide triplicate measurements of the surface protein and the other three measure the total amount of expressed protein. After correcting for background, the data can provide an accurate measurement of the amount of surface protein relative to the total amount present in the cells.
To prepare for a surface ELISA, cells are grown in a 96-well plate and subjected to transfection for an appropriate length of time prior to beginning the assay. We routinelyuse Lipofectamine 2000 (Invitrogen) and transfect cells approximately 24 h prior to performing the assay. Each assay condition should be repeated over six wells, with an additional six wells receiving no treatment (or transfection with a blank plasmid, if appropriate). These wells will be used to measure the background signal.
An important consideration for the entire ELISA procedure is that variations in the number of cells per well can produce significant effects on the assay values. When plating the cells, care should be taken to utilize a uniform cell suspension so that the plated volume yields equivalent numbers of cells per well. In addition, cell lines such as human embryonic kidney cells (HEK293) that do not adhere strongly to plastic should be grown on PLL-coated wells in order to minimize cell loss during prefixation washes. Additionally, when washing the wells, the experimenter should be as consistent as possible about whether or where the suction pipette touches the bottom of the well during aspiration of media and wash liquids, since touching the cell layer will remove cells and affect the reading.
Once cells have been transfected and treated, all cells are carefully washed with cold PBS++, after which the protocol for each set of wells (surface, total, and control) diverges for the labeling (Fig. 4). In general, 25 μl of diluted antibody is sufficient to cover the bottom surface in a 96-well plate.
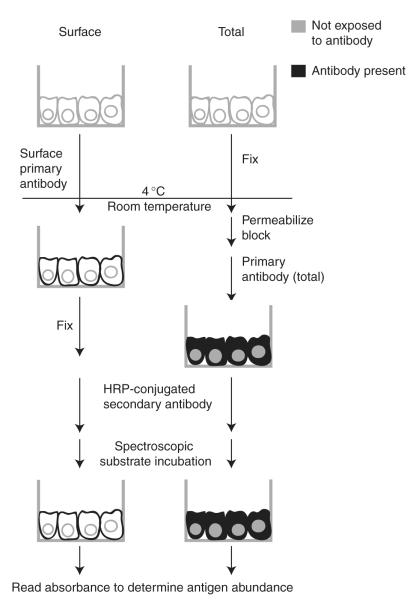
Fig. 4.
Diagram of the ELISA protocol. A representation of cells grown in small-welled tissue culture plates illustrates the treatments for wells measuring the amount of PC1 protein on the surface and the total amount present in the cell. The requisite wash steps are not depicted here, and the protocol for control wells (without primary antibody) is also not shown.
Wells used to measure surface expression are first blocked at 4°C in blocking buffer for 30 min. Antibody against the extracellular epitope, diluted in blocking buffer, is then applied at 4°C for 1 h. An antibody dilution similar to that used for immunofluorescence experiments is generally suitable for this application. After incubation with the primary antibody, the cells are washed once in cold PBS++ and fixed for 20 min in 4% paraformaldeyhde. After fixation, the cells are washed three times with PBS and are then ready for the secondary antibody.
Wells used for the measurement of total PC1 protein are fixed in 4% paraformaldehyde after the initial wash with PBS++. If the assay is being done in a single plate, these “total” wells will need to be allowed to fix while the “surface” wells are in the primary antibody at 4°C, so the fixation should last for 1 h. After three washes with PBS++, the cells are permeabilized and blocked using a 30-min incubation in permeabilization buffer. The same primary antibody that is used for surface labeling is then diluted in the blocking buffer and applied for 1 h at room temperature. After this step, the cells are washed three times with permeabilization buffer and are then ready for the secondary antibody.
Control wells do not receive any primary antibody, but are fixed in 4% paraformaldehyde and then washed along with the other wells. The control wells measuring the surface background are left to sit in the blocking buffer until receiving the secondary antibody, but the control wells measuring the total background are permeabilized for 30 min after fixation and washing.
The secondary antibody is applied to all wells. The secondary antibody is HRP-conjugated antibody against the appropriate animal source of the primary antibody; in our case a goat anti-rabbit HRP is used to detect the polyclonal anti-Flag antibody. The dilution for the secondary antibody is 1:3000 for this rabbit HRP, but can be up to 1:10,000 for some mouse HRP conjugates. The secondary antibody is diluted in the blocking buffer and applied for 45 min at room temperature. After this incubation, the wells are washed three times with PBS and then 80 μl Ultra-TMB substrate, which has been equilibrated to room temperature, is added for up to 15 min. The wells that have been permeabilized will react with this HRP substrate relatively quickly and care should be taken to stop the reaction before it proceeds far enough that the reaction product becomes saturating, after which the linearity of the assay is compromised. This time course is best determined empirically. The reaction is stopped after 10–15 min using 80 μl of 1 N sulfuric acid and the absorbance at 450 nm is measured using a spectrophotometric plate reader.
Due to endogenous peroxidase activity that is present to varying degrees in most cell types, there will be a small but measurable amount of background absorbance generated even in the untransfected control wells. Some published ELISA protocols use a mild hydrogen peroxide treatment before the secondary antibody incubation step to reduce this background, but we did not find this to be significantly effective in our system. To compensate for the background, we use the controls wells that do not receive the primary antibody. Utilizing a set of control wells for each treatment condition is especially useful if the experimental treatments change the total number of cells in any way, since that will alter background signal. If the experimental treatments have no effect on cell number, then it is feasible to do just one set of surface and total control wells for the entire plate. To analyze the data, we subtract the average of the appropriate control wells from each experimental reading, then average the readings across the experimental conditions, and take the ratio of surface:total absorbance signal.
C. Gal/VP Luciferase Assay
This PC1-Gal/VP-driven luciferase assay provides a tool to assay the nuclear translocation of the cleaved PC1 C-terminal tail and has the advantage that it permits simultaneous assessment of the total expressed protein in the cells by Western blot. Given the range of possible assays, the experiments can be set up in wells of almost any size, depending on the goals of a particular experiment. A 96-well plate can be used to effectively screen several conditions with biological replicates, while carrying out the experiment in 12-well or even 6-well plates allows one to both obtain a reading of luciferase while leaving enough lysate to be for Western blots or immunoprecipitations. If the assay results are to be read with a plate reader, then the cells should be plated in clear-bottomed, black-sided wells to minimize leakage of extraneous luciferase signal from adjoining wells.
The prerequisites for this assay include the cotransfection of cDNAs encoding UAS-promoted firefly luciferase, constituitively expressed Renilla luciferase, and PC1-Gal/VP. Once cells have reached an appropriate density in the plate of choice, they are transfected with the plasmids using Lipofectamine 2000 or a similar transfection agent. Since the plasmids are of different sizes and express at different levels, we routinely use a 1:20:60 ratio of Renilla:firefly:PC1-Gal4/VP16 in order to keep the luciferase signals within the linear detection range of our luminometer and maximize the signal from PC1-Gal4/VP16. The control condition for all assays is the transfection of only Renilla and firefly luciferase plasmids, replacing the PC1-Gal4/VP16 with an “empty” plasmid to keep the total micrograms of transfected DNA the same across all experimental conditions.
After transfection, the cells should be left for a minimum of 24 h, including the time required for any experimental treatment, before assaying. We routinely use the Dual-Luciferase kit from Promega, but alter the protocol for lysis depending on the goals of the assay. If the same lysate is to be used both for luciferase readings and for SDS-PAGE gel and Western blotting, then it is best to add protease inhibitors to the 1X passive lysis buffer provided with the luciferase kit. To detect the entire pool of protein, including any cleavage fragments that have translocated to the nucleus, we then sonicate the lysate and spin for 15 min at 18,000 g at 4°C before preparing an aliquot of the lysate for SDS-PAGE. The luciferase signal can be read either before or after sonication. Once the Renilla and luciferase readings are taken, the data are analyzed by dividing each sample’s firefly luciferase signal by its corresponding Renilla luciferase value. This ratio per well is then normalized to the averaged ratio of all control wells, yielding a quantified measurement for each well that is effectively the fold-increase of the signal above the backgound. The ratioed values for each well can then be averaged for each experimental condition and statistically evaluated for significance.
V. Discussion
A reliable method to detect and quantify expression of proteins on the cell surface is of value in a wide range of applications. Measuring the size of the surface pool of a target protein can reveal cell biological processes that would not be seen with an immunofluorescence protocol that only recognizes the total pool of cell-associated protein. This is especially true in experiments that utilize cell-culture-based protein overexpression systems, since in these settings there may be dramatic and biologically interesting experimental effects on protein localization that would otherwise be missed. The visual signal produced by the population of protein trapped in the endoplasmic reticulum can mask the shift of a fraction of that protein to the cell surface, making a surface immunofluorescence protocol more illuminating than a technique that only sees the total pool. We have successfully used this surface immunofluorescence technique to demonstrate that PC1 is brought to the cell surface with different efficiencies as a function of the proteins with which it is coexpressed. A clean signal derived exclusively from the surface pool of protein is also useful in determining the protein’s residence in specialized membrane subdomains. For example, pairing the surface antibody with an internal antibody against ciliary or junctional proteins may allow a more precise measurement of colocalization for proteins of interest that have both plasma membrane and intracellular distributions.
A key advantage of the surface immunofluorescence approach is that it is temporally specific. Since the surface labeling protocol is performed in the cold, and thus in the absence of ongoing membrane traffic, the technique effectively gives a snapshot of protein localization at a specific point in time, allowing the surface delivery to be analyzed over a time course. One hypothetical application of this capability involves examining several time points over the course of increasing cell confluency to assess the delivery of a protein of interest to junctions as they form. We have also used similarly designed time course experiments to assess the change in surface localization of PC1 protein following treatment with environmental stimuli and chemical compounds.
The surface labeling protocol described here is similar to that outlined by Bengtsson et al. (2008), with the added advantage that both secondary antibodies are employed in a single, simultaneous, postfixation incubation step. This cuts down on the time required for the overall protocol, and if greater speed is necessary, the times used here could be further optimized for a faster throughput. It should be noted that our protocol also allows for a blocking step before the surface antibody incubation to minimize the background signal that may improve the specificity of some primary antibodies.
While the immunofluorescence protocol is useful for detecting the changes in surface localization that occur at the level of individual cells, the ELISA protocol described here allows for a much more quantitative assessment of the amount of protein reaching the cell surface in a population of cells. While a whole-cell ELISA technique was developed quite some time ago to quantify surface proteins in bacteria (Elder et al. 1982), that uses a room temperature incubation to label the antigen and measures only the surface protein pool. Our protocol assays the ratio of the surface protein to the total amount expressed, thereby taking into account that the amount of surface protein may be affected by experimental treatments that alter the general stability or expression of the protein of interest. Additionally, by pairing each experimental condition with the appropriate controls, this protocol permits quantification of changes effected by a range of experimental conditions, including protein coexpression or drugs applied in the media. The miniaturization of the protocol allows for easy completion of a number of biological replicates, increasing confidence in the significance of experimental observations.
The biological importance of PC1’s C-terminal cleavage has been recognized for several years, and the signaling effects of the soluble fragment have been suggested to be initiated by abnormal fluid flow past ciliated cells. Until the application of the Gal4/VP16 assay, though, there had been no way to quantify the nuclear translocation of the PC1 cleavage fragments. This Gal4/VP16 assay has now been utilized to demonstrate that the C-terminal cleavage of PC1 is enhanced by coexpression with PC2, a finding that had been suggested by Western blot and immunofluorescence, but had not been quantified using either technique. Furthermore, the relative ease and speed of assay completion allowed screening of conditions that altered intracellular calcium, revealing that the PC2 enhancement of PC1 cleavage is independent of intracellular calcium concentrations (Bertuccio et al., 2009). These results show an interesting contrast between PC1 and fibrocystin, another ciliary protein that causes autosomal-recessive PKD when mutated. A luciferase assay based on a fibrocystin–Gal4/VP16 fusion protein revealed that fibrocystin undergoes a C-terminal cleavage, but that this cleavage requires activation of protein kinase C as well as increased cytoplasmic calcium concentrations (Hiesberger et al., 2006). Similarly to PC1, the released C-terminal fragment of fibrocystin contains a nuclear localization sequence that mediates the nuclear translocation of the cleaved fragment. In combination, these results suggest a common mechanism of membrane-bound, ciliary proteins influencing intracellular signaling events through the cleavage and release of soluble, cytoplasmic fragments.
VI. Summary
The biology of ciliary proteins is fascinating and complex, and it is becoming clear that the localization and cleavage of these proteins play a key role in regulating their function. The techniques outlined here provide methods to visualize and quantify the localization and cleavage of membrane-bound proteins, allowing a more detailed understanding of how proteins such as PC1 are delivered to the cilia and plasma membrane, and what influences the cleavage that releases their cytoplasmic fragments. The PC1-Gal4/VP16 assay has been successfully used to show the calcium-independence of PC1 cleavage (Bertuccio et al., 2009), laying the groundwork for subsequent exploration of the regulation of PC1 cleavage. In sum, these techniques contribute to the panoply of techniques available for the study of the dynamic cell biology of ciliary proteins.
References
- Bakshi P, Liao YF, Gao J, Ni J, Stein R, Yeh LA, Wolfe MS. A high-throughput screen to identify inhibitors of amyloid beta-protein precursor processing. J. Biomol. Screen. 2005;10:1–12. doi: 10.1177/1087057104270068. [DOI] [PubMed] [Google Scholar]
- Basavanna U, Weber KM, Hu Q, Ziegelstein RC, Germino GG, Sutters M. The isolated polycystin-1 COOH-terminal can activate or block polycystin-1 signaling. Biochem. Biophys. Res. Commun. 2007;359:367–372. doi: 10.1016/j.bbrc.2007.05.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson DC, Sowa KM, Arnot DE. Dual fluorescence labeling of surface-exposed and internal proteins in erythrocytes infected with the malaria parasite Plasmodium falciparum. Nat. Protoc. 2008;3:1990–6. doi: 10.1038/nprot.2008.196. [DOI] [PubMed] [Google Scholar]
- Bertuccio CA, Chapin HC, Cai Y, Mistry K, Chauvet V, Somlo S, Caplan MJ. Polycystin-1 C-terminal cleavage is modulated by polycystin-2 expression. J. Biol. Chem. 2009;284:21011–26. doi: 10.1074/jbc.M109.017756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukanov N, Husson H, Dackowski WR, Lawrence BD, Clow PA, Roberts BL, Klinger KW, Ibraghimov-Beskrovnaya O. Functional polycystin-1 expression is developmentally regulated during epithelial morphogenesis in vitro: Downregulation and loss of membrane localization during cystogenesis. Hum. Mol. Genet. 2002;11:923–936. doi: 10.1093/hmg/11.8.923. [DOI] [PubMed] [Google Scholar]
- Chauvet V, Tian X, Husson H, Grimm DH, Wang T, Hiesberger T, Igarashi P, Bennett AM, Ibraghimov-Beskrovnaya O, Somlo S, Caplan MJ. Mechanical stimuli induce cleavage and nuclear translocation of the polycystin-1 C terminus. J. Clin. Invest. 2004;114:1433–1443. doi: 10.1172/JCI21753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebinu JO, Yankner BA. A RIP tide in neuronal signal transduction. Neuron. 2002;34:499–502. doi: 10.1016/s0896-6273(02)00704-3. [DOI] [PubMed] [Google Scholar]
- Elder BL, Boraker DK, Fives-Taylor PM. Whole-bacterial cell enzyme-linked immunosorbent assay for Streptococcus sanguis fimbrial antigens. J. Clin. Microbiol. 1982;16:141–4. doi: 10.1128/jcm.16.1.141-144.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabow PA. Autosomal dominant polycystic kidney disease. N. Engl. J. Med. 1993;329:332–342. doi: 10.1056/NEJM199307293290508. [DOI] [PubMed] [Google Scholar]
- Guay-Woodford LM. RIP-ed and ready to dance: New mechanisms for polycystin-1 signaling. J. Clin. Invest. 2004;114:1404–1406. doi: 10.1172/JCI23544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka K, Qian F, Boletta A, Bhunia AK, Piontek K, Tsiokas L, Sukhatme VP, Guggino WB, Germino GG. Co-assembly of polycystin-1 and -2 produces unique cation-permeable currents. Nature. 2000;408:990–994. doi: 10.1038/35050128. [DOI] [PubMed] [Google Scholar]
- Hiesberger T, Gourley E, Erickson A, Koulen P, Ward CJ, Masyuk TV, Larusso NF, Harris PC, Igarashi P. Proteolytic cleavage and nuclear translocation of fibrocystin is regulated by intracellular Ca2+ and activation of protein kinase C. J. Biol. Chem. 2006;281:34357–34364. doi: 10.1074/jbc.M606740200. [DOI] [PubMed] [Google Scholar]
- Hurley WL, Finkelstein E, Holst BD. Identification of surface proteins on bovine leukocytes by a biotin-avidin protein blotting technique. J. Immunol. Methods. 1985;85:195–202. doi: 10.1016/0022-1759(85)90287-x. [DOI] [PubMed] [Google Scholar]
- Lal M, Song X, Pluznick JL, Di Giovanni V, Merrick DM, Rosenblum ND, Chauvet V, Gottardi CJ, Pei Y, Caplan MJ. Polycystin-1 C-terminal tail associates with beta-catenin and inhibits canonical Wnt signaling. Hum. Mol. Genet. 2008;17:3105–3117. doi: 10.1093/hmg/ddn208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantinga-van Leeuwen IS, Dauwerse JG, Baelde HJ, Leonhard WN, van de Wal A, Ward CJ, Verbeek S, Deruiter MC, Breuning MH, de Heer E, Peters DJ. Lowering of Pkd1 expression is sufficient to cause polycystic kidney disease. Hum. Mol. Genet. 2004;13:3069–3077. doi: 10.1093/hmg/ddh336. [DOI] [PubMed] [Google Scholar]
- Low SH, Vasanth S, Larson CH, Mukherjee S, Sharma N, Kinter MT, Kane ME, Obara T, Weimbs T. Polycystin-1, STAT6, and P100 function in a pathway that transduces ciliary mechanosensation and is activated in polycystic kidney disease. Dev. Cell. 2006;10:57–69. doi: 10.1016/j.devcel.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Lu W, Peissel B, Babakhanlou H, Pavlova A, Geng L, Fan X, Larson C, Brent G, Zhou J. Perinatal lethality with kidney and pancreas defects in mice with a targetted Pkd1 mutation. Nat. Genet. 1997;17:179–181. doi: 10.1038/ng1097-179. [DOI] [PubMed] [Google Scholar]
- Maetzel D, Denzel S, Mack B, Canis M, Went P, Benk M, Kieu C, Papior P, Baeuerle PA, Munz M, Gires O. Nuclear signalling by tumour-associated antigen EpCAM. Nat. Cell Biol. 2009;11:162–171. doi: 10.1038/ncb1824. [DOI] [PubMed] [Google Scholar]
- Nauli SM, Alenghat FJ, Luo Y, Williams E, Vassilev P, Li X, Elia AE, Lu W, Brown EM, Quinn SJ, Ingber DE, Zhou J. Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat. Genet. 2003;33:129–137. doi: 10.1038/ng1076. [DOI] [PubMed] [Google Scholar]
- Pritchard L, Sloane-Stanley JA, Sharpe JA, Aspinwall R, Lu W, Buckle V, Strmecki L, Walker D, Ward CJ, Alpers CE, Zhou J, Wood WG, et al. A human PKD1 transgene generates functional polycystin-1 in mice and is associated with a cystic phenotype. Hum. Mol. Genet. 2000;9:2617–2627. doi: 10.1093/hmg/9.18.2617. [DOI] [PubMed] [Google Scholar]
- Qian F, Watnick TJ, Onuchic LF, Germino GG. The molecular basis of focal cyst formation in human autosomal dominant polycystic kidney disease type I. Cell. 1996;87:979–987. doi: 10.1016/s0092-8674(00)81793-6. [DOI] [PubMed] [Google Scholar]
- Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- Streets AJ, Wagner BE, Harris PC, Ward CJ, Ong AC. Homophilic and heterophilic polycystin 1 interactions regulate E-cadherin recruitment and junction assembly in MDCK cells. J. Cell Sci. 2009;122:1410–1417. doi: 10.1242/jcs.045021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G, Greenwald I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature. 1999;398:522–525. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]