C/EBPβ mediates GH stimulation of multiple early response genes. A subset of these genes requires phosphorylation by ERKs and p300 recruitment in response to GH.
Abstract
Regulation of c-Fos transcription by GH is mediated by CCAAT/enhancer binding protein β (C/EBPβ). This study examines the role of C/EBPβ in mediating GH activation of other early response genes, including Cyr61, Btg2, Socs3, Zfp36, and Socs1. C/EBPβ depletion using short hairpin RNA impaired responsiveness of these genes to GH, as seen for c-Fos. Rescue with wild-type C/EBPβ led to GH-dependent recruitment of the coactivator p300 to the c-Fos promoter. In contrast, rescue with C/EBPβ mutated at the ERK phosphorylation site at T188 failed to induce GH-dependent recruitment of p300, indicating that ERK-mediated phosphorylation of C/EBPβ at T188 is required for GH-induced recruitment of p300 to c-Fos. GH also induced the occupancy of phosphorylated C/EBPβ and p300 on Cyr61, Btg2, and Socs3 at predicted C/EBP-cAMP response element-binding protein motifs in their promoters. Consistent with a role for ERKs in GH-induced expression of these genes, treatment with U0126 to block ERK phosphorylation inhibited their GH-induced expression. In contrast, GH-dependent expression of Zfp36 and Socs1 was not inhibited by U0126. Thus, induction of multiple early response genes by GH in 3T3-F442A cells is mediated by C/EBPβ. A subset of these genes is regulated similarly to c-Fos, through a mechanism involving GH-stimulated ERK 1/2 activation, phosphorylation of C/EBPβ, and recruitment of p300. Overall, these studies suggest that C/EBPβ, like the signal transducer and activator of transcription proteins, regulates multiple genes in response to GH.
Analysis of transcription factors regulated by GH has improved our understanding of the mechanistic basis of growth and metabolism in health and disease. Most studies of GH-regulated gene expression have focused on signal transducer and activator of transcription (Stat) family factors, particularly Stat5a and Stat5b, which mediate expression of multiple genes in response to GH (1–4). However, GH signaling to the nucleus uses other transcription factors as well (5). Among these, CCAAT/enhancer binding protein β (C/EBPβ) is a critical factor for GH-induced transcription of the protooncogene c-Fos. C/EBPβ is a B-Zip transcription factor that has been implicated in regulation of genes that control cellular differentiation, proliferation, metabolism, and inflammation (6, 7). Among its diverse functions, C/EBPβ is a key component of a transcription factor cascade that contributes to early adipogenesis (8, 9) and has been implicated in breast, myeloid, and other cancers (10–13). Mice with targeted deficiency of C/EBPβ are smaller than wild-type (WT) littermates and exhibit deficiencies in lactation and immune function (7, 14).
The C/EBPβ dependence of GH-induced c-Fos gene expression is evident in the dramatic impairment of c-Fos expression in GH-responsive cells made deficient in C/EBPβ by RNA interference (15). C/EBPβ dimerizes with other B-Zip family factors and binds to a C/EBP site on c-Fos (16); C/EBPβ is also reported to associate with a c-Fos cAMP response element (CRE) (17, 18). Chromatin immunoprecipitation (ChIP), EMSA, and genome-wide approaches show that endogenous C/EBPβ occupies the c-Fos promoter constitutively (15, 19–22). To modulate its function, the C/EBPβ associated with c-Fos DNA is regulated through posttranslational modifications, such as phosphorylation and acetylation, which are critical for C/EBPβ to activate transcription (20, 23–25). Such modifications can be initiated by a variety of hormones and growth factors, including GH (20, 26–28). The stimulation of c-Fos by GH depends on phosphorylation of murine C/EBPβ at T188 (P-C/EBPβ), a substrate site for the MAPKs ERK1 and ERK2; T188 in murine C/EBPβ corresponds to T235 in human C/EBPβ, which is also phosphorylated by ERKs 1 and 2 (20, 26, 28). C/EBPβ-dependent gene activation is often associated with recruitment of coactivators such as p300 or CRE-binding protein (CREB) binding protein (CBP) to the promoters of its target genes (15, 29, 30) and coincides with their coactivation of gene transcription (15, 18, 31). CREB and the c-Fos CRE have also been found to participate in GH-induced c-Fos transcription; activation by both CREB and C/EBPβ are mediated by stimulation of ERKs 1 and 2 (ERK 1/2) (18).
To identify other GH-regulated genes that are dependent on C/EBPβ and examine transcriptional mechanisms involved, the present study uses cells deficient in endogenous C/EBPβ. In addition, the mechanisms by which C/EBPβ mediates induction of these genes in the context of GH regulation, including phosphorylation of C/EBPβ and recruitment of the coactivator p300, are investigated. The findings implicate C/EBPβ in the activation of multiple GH-induced early response genes. A subset of the GH-regulated early response genes that utilize C/EBPβ show occupancy of phosphorylated C/EBPβ and recruitment of p300 in response to GH. Overall, these studies suggest that C/EBPβ, as well as Stat5, is a GH-regulated transcription factor that can mediate the transcription of multiple GH target genes.
Results
Multiple early response genes are induced by GH
To identify GH-dependent genes that are regulated by C/EBPβ, a gene expression profile was examined which contained over 500 genes induced or repressed by GH in time-dependent waves in 3T3-F442A adipocytes (19). The present investigation focuses on a cluster of early response genes that includes the C/EBPβ-dependent gene c-Fos, with the view that genes showing similar patterns of response to GH might be transcriptionally coregulated, i.e. regulated by similar factors. In the microarray profile, the genes in this cluster exhibited a transient increase in mRNA expression detected 30 min after GH treatment but not 4 or 48 h later. Fifteen of the genes in the cluster showed responses to GH that were at least 150% of control expression (Table 1); these include some known GH target genes, such as those encoding the signaling molecules suppressor of cytokine signaling (Socs)3 and Socs1 (32), IL-6 (33), and the transcription factors JunB and early growth response (Egr)1 (34–37).
Table 1.
GH-induced early response genes
| Gene symbol | GH/Con | C/EBP-CREB |
|---|---|---|
| Socs3 | 17.47 | Yes |
| Fos | 4.39 | Yes |
| Egr2 | 3.66 | Yes |
| Cyr61 | 2.28 | Yes |
| Egr1 | 2.19 | Yes |
| Junb | 2.14 | Yes |
| Btg2 | 2.13 | Yes |
| Cited2 | 1.89 | Yes |
| Frat2 | 1.83 | Yes |
| Il6 | 1.66 | Yes |
| Klf4 | 1.50 | Yes |
| Phlda1 | 5.32 | No |
| Socs1 | 2.82 | No |
| Zfp36 | 2.68 | No |
| Ier2 | 2.41 | No |
Based on microarray analysis (19), the expression of the genes listed was stimulated by GH at least 150% above control (GH/Con) in 3T3-F442A adipocytes. They were present in a cluster of genes stimulated by GH at 30 min but not at 4 or 48 h. Sequence analysis was performed as described in Materials and Methods to identify predicted C/EBP-CREB motifs that were conserved in both mouse and human gene promoters (−1500 to +200).
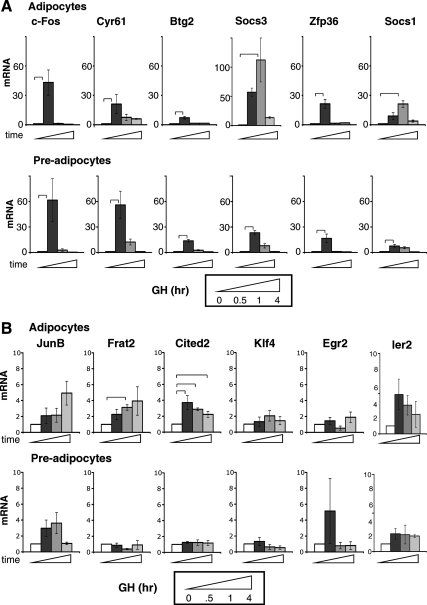
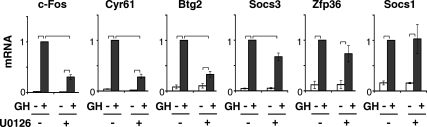
To verify that the genes in this cluster were transiently stimulated by GH, as suggested by the microarray data, mRNA expression was measured by quantitative real-time PCR (qPCR) in 3T3-F442A adipocytes and preadipocytes treated with GH for 0, 0.5, 1, or 4 h (Fig. 1). Typical of early response genes, their expression was induced 30–60 min after GH and subsided by 4 h. The six most responsive (10- to 100-fold increase) genes encode c-Fos, cysteine-rich 61/Cyr61,CTGF, NOV family of genes 1 (Cyr61/CCN1), B-cell translocation gene 2 (Btg2), Socs3, zinc finger protein 36 (Zfp36), and Socs1 (Fig. 1A). Among these, genes not previously recognized as GH targets encode the secreted extracellular matrix-associated protein Cyr61/CCN1 (38, 39), the p53-activated antiproliferative factor Btg2 (40), and the zinc finger transcription factor Zfp36 (41, 42). The responses to GH were temporally consistent with the microarray. Other genes, including JunB, Frat2, Cited2, Klf4, Egr2, and Ier2 (Fig. 1B) showed lower responses to GH in preadipocytes or adipocytes, and few were statistically significant. GH dependence of Il6 and Egr1 expression was reported previously (35–37, 43). Of the verified GH-dependent early response genes, sequence analysis predicted C/EBP or CREB motifs (referred to here as C/EBP-CREB motifs), which are conserved in mouse and human promoters for c-Fos, Cyr61, Btg2, and Socs3 but not for Zfp36 and Socs1 (Table 1), suggesting potential differences in the regulation of these two sets of genes. These six early response genes with mRNA expression most responsive to GH were analyzed further to evaluate whether the genes were coregulated by similar transcriptional mechanisms.
Fig. 1.
GH rapidly and transiently induces expression of multiple genes. A, Genes highly responsive to GH. B, Genes with lower responsiveness to GH. 3T3-F442A adipocytes (top) or preadipocytes (bottom) were treated with GH for various times as indicated. RNA was analyzed by qPCR. For each gene tested, mRNA induced by GH is shown after 0, 0.5, 1, or 4 h (open, dark, medium, and light gray bars, respectively). Each bar shows mean ± se for three independent experiments. mRNA expression at time 0 is set equal to 1. Brackets show responses that are significantly (P < 0.05) different between bracketed bars, in this and subsequent figures.
C/EBPβ mediates GH-induced expression of multiple early response genes
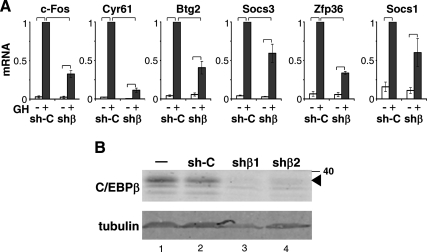
For insight into whether C/EBPβ contributes to their GH-induced expression, this subset of early response genes was studied in cells deficient in endogenous C/EBPβ. C/EBPβ was stably knocked down in GH-responsive 3T3-F442A preadipocytes by retroviral infection of a short hairpin RNA (shRNA) targeting C/EBPβ (shβ); control cells were infected with a nontargeting sequence [sh-control (sh-C)]. When the mRNA expression of the GH-responsive genes was compared in the shβ and sh-C cells to determine their dependence on C/EBPβ, the deficiency of C/EBPβ significantly reduced GH-induced expression of c-Fos, Cyr61, Btg2, and Socs3 (Fig. 2), which are predicted to contain conserved C/EBP-CREB sites (Table 2). The induction of Zfp36 and Socs1 by GH was also impaired by C/EBPβ deficiency. The endogenous C/EBPβ protein was markedly reduced, with only residual levels barely detectable in immunoblots of the shβ cells (Fig. 2B, lanes 3 and 4), whereas the endogenous C/EBPβ was evident in sh-C cells at a level comparable with that in parental noninfected 3T3-F442A cells (Fig. 2B, lanes 2 and 1, respectively). Because C/EBPβ is a critical factor during adipogenesis, the failure of the shβ cells to differentiate into adipocytes when incubated with adipogenic medium (data not shown) while the sh-C cells differentiated readily under these conditions provides evidence of functional C/EBPβ deficiency. In contrast, GH-regulated expression of the apoptotic early response gene Phlda1, which does not contain a predicted C/EBP-CREB site, is not impaired by C/EBPβ deficiency (Supplemental Fig. 1, published on The Endocrine Society's Journals Online web site at http://mend.endojournals.org). These findings indicate that the impaired transcription observed for this panel of GH-sensitive early response genes in shβ cells does not reflect generalized transcription impairment. Overexpression of C/EBPβ (Supplemental Fig. 2) led to increased expression of mRNA for c-Fos, Cyr61, Btg2, and Socs3 in 293T cells; however, Socs1 and Zfp36 were not induced by C/EBPβ overexpression. Overall, these findings are consistent with multiple GH-regulated early response genes being dependent on C/EBPβ, likely through multiple mechanisms.
Fig. 2.
GH-induced expression of early response genes is impaired by C/EBPβ deficiency. A, Pooled C/EBPβ-deficient 3T3-F442A preadipocytes expressing shβ, or control sh-C cells, were treated without (open bar) or with GH (black bar) for 30 min. RNA was analyzed by qPCR as for Fig. 1. Each bar shows the mean ± se for three independent experiments. mRNA expression in sh-C cells with GH treatment is set equal to 1. The response to GH in shβ cells is significantly (P < 0.01) less than the response in sh-C cells as indicated by brackets. B, Endogenous C/EBPβ protein, shown by immunoblot, is almost completely deficient in 3T3-F442A preadipocytes stably expressing shβ, clones 1 and 2 (lanes 3 and 4), compared with control cells expressing nontargeting shRNA (sh-C, lane 2) or noninfected parental 3T3-F442A cells (−, lane 1). Tubulin (lower panel) indicates sample loading. The shβ1 cells were used in A and in subsequent experiments.
Table 2.
Predicted C/EBP-CREB motifs in promoters of GH-regulated early response genes
| 1) c-fos promoter |
| M -374 T G G C T G C A G C C G G C G A G C T G T T C C C G T C A A T C C C T C C C t c c t T T A C A C A G |
| H -367 T G G C T G C A G C C C G C G A G C A G T T C C C G T C A A T C C C T C C C c c c - T T A C A C A G |
| M -324 G A T G T C C A T A T T A G G A C A T C T G C G T C A G C A G G T T T C C A C G G C C G G T C C C |
| H -318 G A T G T C C A T A T T A G G A C A T C T G C G T C A G C A G G T T T C C A C G G C C T T T C C C |
| 2) Cyr61 promoter |
| Cyr61-1 |
| M -606 T G C C A C T C C G G G T A T T A A T T T G C A A T A C A C T t c t C T T G G C T A A T A A A C A T |
| H -752 T G C C A C T G T G G G T A T T A A T T T G C A A T T C A C T g a a C T T T G C T A A T A A A C A T |
| Cyr61-2 |
| M -89 A G A A T T C T a G A A C G C G C C G A C A G A G C t - A C G T C A C T G C A A C A C G C G G C G C C T |
| H -109 A G A A T T C T g G A A C G C G C A G A C A G A G C c gA C G T C A C T G C A A C A C G C G G C G C C T |
| 3) Btg2 promoter |
| M -199 c a c a g c a c g g g a g t c c g g t g c t t G T T C C C A A T A A T G A C G TC A G T G A G C G A T G A C C T C A G C G C C t g G G A G G C G C G G |
| H -254 c a g c t c a g c c g g g c g c a g G T T C C T A G C A C T G A C G A C A G C G A G C G A T G A C C T C A G C G C C g c c a a g g c t g c g a g g g c g |
| 4) SOCS3 promoter |
| M -632 C T G T C T A C A G G T A A A T G T C G C G C A T C C C C C T C C T C c a c t t c c t a g g t c c c |
| H -622 C T G C C C G C A G G T G A C T G T C G C A C G T C T C C A A C C T C c g g c t c c c g g g t c t g |
Sequences in GH-induced early response genes that were identified as C/EBP-CREB motifs in promoters (−1500 to +200 bp relative to transcription start site), and that were conserved in mouse (M) and human (H) genes, were obtained using Genomatix software. The conserved sequences are in capital letters, and predicted C/EBP-CREB motifs are underlined. Location of each sequence from transcription start site is indicated on left.
Phosphorylation of C/EBPβ and recruitment of p300 mediate c-Fos stimulation by GH
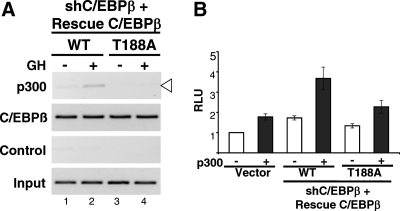
For insight into the mechanism(s) by which C/EBPβ mediates induction of GH-stimulated genes, c-Fos provides an informative model. As demonstrated by ChIP using phospho-specific antibodies (15, 18), GH induces the occupancy of murine C/EBPβ phosphorylated at the ERK substrate site at T188 on c-Fos DNA. Further, occupancy of P-C/EBPβ on c-Fos is impaired in the presence of the MAPK kinase (MEK) inhibitor U0126 (18), consistent with ERK pathways mediating the induction of P-C/EBPβ occupancy by GH. The activation of the c-Fos promoter in the presence of C/EBPβ is enhanced by recruitment of the coactivator p300 to the c-Fos C/EBP site (15, 31). If the GH-induced P-C/EBPβ is required for recruitment of p300 to c-Fos in response to GH, then induced recruitment of p300 would not be expected on c-Fos if T188 of C/EBPβ is mutated. Accordingly, the importance of P-C/EBPβ for the GH-induced recruitment of p300 to c-Fos was examined. When endogenous C/EBPβ was silenced in GH-responsive 293T cells and rescued with WT C/EBPβ (Supplemental Fig. 3), GH induced the recruitment of p300 to c-Fos (Fig. 3A, top panel, lane 2 vs. lane 1). In contrast, T188A C/EBPβ did not rescue GH-induced recruitment of p300 to c-Fos (Fig. 3A, lane 4 vs. lane 2). The observed differences in occupancy of p300 coincide with the detection of P-C/EBPβ in cells in which WT C/EBPβ is rescued, but absence of P-C/EBPβ in T188A C/EBPβ rescued cells (data not shown) (20). These experiments demonstrate that the GH-induced recruitment of p300 to c-Fos depends on phosphorylation of C/EBPβ at T188. The p300 and C/EBPβ are likely in a complex, because they are induced by GH to occupy the same c-Fos DNA (15). The functional importance of such a complex for c-Fos promoter activation is demonstrated by the ability of p300 to coactivate C/EBPβ in inducing c-Fos transcription: c-Fos promoter activity is enhanced when p300 is coexpressed in shβ cells with WT rescue C/EBPβ but not when p300 is coexpressed with T188A rescue C/EBPβ (Fig. 3B). In a complementary approach, p300 is recruited in response to GH to a greater extent in cells overexpressing WT C/EBPβ than in cells overexpressing T188A C/EBPβ (Supplemental Fig. 4A). As predicted, coexpression of p300 with WT C/EBPβ coactivates the c-Fos promoter, whereas T188A C/EBPβ does not (Supplemental Fig. 4B). Taken together, these data indicate that activation of c-Fos by GH involves activation of ERKs 1/2, P-C/EBPβ, and recruitment of p300.
Fig. 3.
GH-induced recruitment of p300 to c-Fos depends on an intact phosphorylation site at T188 of C/EBPβ. A, Plasmids for shβ, and for rescue constructs expressing WT or T188A C/EBPβ, as well as for GHR, were expressed in 293T cells; 48 h later, cells were treated with GH for 30 min. Nuclear extracts were analyzed by ChIP using antibodies against p300 or C/EBPβ and probed for the c-Fos C/EBP site. Control without antibody and 1% input are shown. B, Plasmids for shβ and rescue constructs for WT or T188A C/EBPβ, or vector, were transiently transfected into 293T cells, with (black bar) or without (open bar) plasmid for CMV-p300. Plasmids for Fos-luc and RSV-β-gal were also transfected; 48 h later, cells were lysed and used for luciferase assay. The c-Fos promoter activity in cells expressing control pcDNA3 without p300 is set equal to 1. Each bar shows mean ± se for three independent experiments. In C/EBPβ-deficient cells transiently expressing shβ, significant coactivation of c-Fos-luc by p300 occurs with rescue of WT but not T188A C/EBPβ.
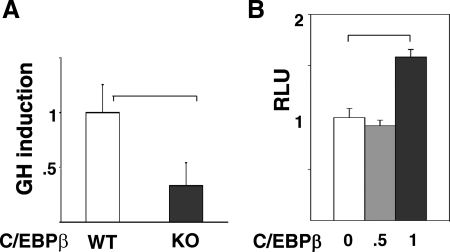
In support of the GH-induced expression of c-Fos being dependent on C/EBPβ, c-Fos activation by GH is impaired by 60% in murine embryonic fibroblasts (MEFs) from mice with a targeted deletion of C/EBPβ [knockout (KO)] (Fig. 4A and Supplemental Fig. 5). Reintroduction of C/EBPβ in the KO MEFs increased c-Fos promoter activity (Fig. 4B). In C/EBPβ KO mice, well-recognized GH-regulated signaling events (5), including phosphorylation of Stats 5 and 3, and ERKs 1/2, were generally intact in liver and/or MEFs after GH treatment, when compared with signaling in GH-treated C/EBPβ heterozygous mice (Supplemental Fig. 6).
Fig. 4.
Deficiency of endogenous C/EBPβ in vivo impairs GH-stimulated c-Fos expression. A, Primary MEF from WT or C/EBPβ deficient (KO) mice were transfected with plasmids for c-Fos-luc, rat GHR, and β-gal; 48 h later, cells were treated with GH for 4 h, and luciferase was measured. Bars represent GH-induced increment in c-Fos promoter activation (mean ± se, n = 3 independent experiments). B, C/EBPβ activates c-Fos promoter in C/EBPβ KO MEF. MEFs from C/EBPβ KO mice were cotransfected with plasmids for c-Fos-luc and the indicated amounts of plasmid for C/EBPβ (μg/well). After 48 h, cells were lysed and luciferase [expressed as relative luciferase units (RLU)] measured. Bars show mean ± se for three independent experiments.
Regulation mediated by ERK 1/2 activation distinguishes a subset of GH-stimulated early response genes
To determine whether stimulation by GH of early response genes in addition to c-Fos is dependent on ERK activation, 3T3-F442A preadipocytes were incubated before GH treatment with or without U0126, a MEK inhibitor that prevents GH activation of ERKs 1 and 2. In the panel of C/EBPβ-regulated genes tested, U0126 significantly impaired the GH-stimulated expression of a subset that included c-Fos, Cyr61, Btg2, and Socs3 (Fig. 5). GH-induced expression of Zfp36 and Socs1 genes was not inhibited by U0126 (Fig. 5). These observations distinguish Zfp36 and Socs1 from the other GH-stimulated genes dependent on C/EBPβ and suggest that induction by GH of Zpf36 and Socs1, which do not contain predicted C/EBP-CREB motifs in their promoters, is not dependent on the ERK pathway. Zfp36 and Socs1 are also distinguished from the other four genes in their lack of stimulation by C/EBPβ overexpression (Supplemental Fig. 2). Together, these observations suggest that a subset of the GH-regulated genes identified in shβ cells as dependent on C/EBPβ are distinguished as two subsets, one of which is also regulated by ERKs. In this regard, it is of note that the transient GH-activated phosphorylation of ERKs 1/2 was intact in the shβ cells (Supplemental Fig. 7); GH-stimulated phosphorylation of Stat5 and Stat3, as well as of ERKs, was also observed in the shβ cells (Supplemental Fig. 8).
Fig. 5.
GH-induced expression of multiple early response genes is dependent on ERK 1/2. 3T3-F442A preadipocytes were treated with the MEK inhibitor U0126 or with dimethylsulfoxide vehicle for 30 min as indicated and then were treated without (open bars) or with GH (black bars) for 30 min. RNA was analyzed by qPCR for the genes indicated. Bars show mean ± se for three independent experiments. mRNA expression with GH treatment is set equal to 1 for each gene tested.
Phosphorylation of C/EBPβ and recruitment of p300 contribute to stimulation by GH of a subset of early response genes
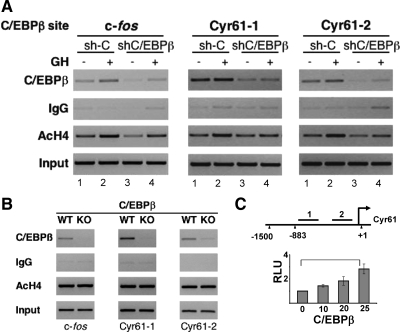
To determine the extent to which regulation of other early response genes by GH involves mechanisms similar to those for c-Fos, Cyr61 was examined in more detail. Expression of Cyr61, a novel GH target gene, showed substantial impairment in basal and GH-induced expression with C/EBPβ deficiency (Fig. 2A); the response of Cyr61 to GH was inhibited almost 90% in the shβ cells. In addition, Cyr61 stimulation by GH was impaired by ERK inhibition to a similar extent as c-Fos stimulation (Fig. 5). The gene product Cyr61/CCN1 is a secreted protein that associates with the extracellular matrix and can activate integrin signaling to regulate events such as angiogenesis, cell adhesion, and cell proliferation (38, 44, 45). Cyr61/CCN1 protein expression was detected in sh-C but not shβ cells (Supplemental Fig. 9). In the Cyr61 promoter, two motifs predicted by Genomatix software to recognize C/EBP or CREB family proteins are conserved in the mouse and human gene sequences (Table 2). Endogenous C/EBPβ was found to occupy cellular Cyr61 DNA in 3T3-F442A cells on both of the Cyr61 sites (Cyr61-1 and Cyr61-2) predicted to recognize C/EBP-CREB family proteins (Fig. 6A). The occupancy of C/EBPβ at these sites was constitutive in intact sh-C 3T3-F442A preadipocytes (Fig. 6A, lane 1). In contrast, C/EBPβ occupancy at both sites on Cyr61 was greatly reduced in cells with reduced levels of C/EBPβ (shβ) (Fig. 6A, lane 3 vs. lane 1). C/EBPβ also occupies Cyr61 DNA under physiological conditions in vivo in mouse liver (Fig. 6B, WT), where endogenous C/EBPβ occupancy was observed on both of the Cyr61 sites as on the c-Fos promoter. Further, C/EBPβ occupancy on Cyr61 was completely absent in liver from C/EBPβ KO mice (Fig. 6B, KO). A Cyr61 promoter sequence (−883 to +1) that contains the predicted C/EBP-CREB sites upstream of luciferase was stimulated when C/EBPβ was expressed (Fig. 6C), supporting functional activation of the Cyr61 promoter by C/EBPβ. Together, these findings indicate that C/EBPβ specifically occupies the Cyr61 promoter under physiological conditions in vitro and in vivo and that the Cyr61 promoter is functionally stimulated by C/EBPβ.
Fig. 6.
C/EBPβ occupies predicted C/EBP sites on the Cyr61 promoter. A, 3T3-F442A preadipocytes stably expressing shβ or control sh-C cells were treated without (lanes 1 and 3) or with GH (lanes 2 and 4) for 30 min. Nuclear extracts were analyzed by ChIP using antibodies against C/EBPβ or acetylated H4 (AcH4), or IgG, with probes for the C/EBP site of c-Fos or the two predicted sites on Cyr61; 1% input is also shown. B, Liver from C/EBPβ deficient (KO) or control (WT) mice was used to prepare nuclear extracts for ChIP. Samples were analyzed as in A. C, Diagram of Cyr61 promoter shows locations (bars) of predicted C/EBP-CREB motifs that are conserved in mouse and human sequences (see Table 2). Diagram is not to scale. To test Cyr61 promoter activation, plasmid for Cyr61-luc (0.5 μg/well) containing the predicted C/EBP sites, and the indicated amounts of plasmid for C/EBPβ (ng/well) and RSV-β-gal (20 ng/well), were transfected into 293T cells in 24-well plates; 48 h later, lysates were analyzed for luciferase activity. Each bar shows the mean of three to five independent experiments. RLU, Relative luciferase units.
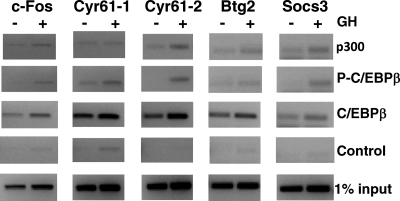
Among the panel of GH-regulated early response genes studied, Btg2 and Socs3, as well as c-Fos and Cyr61, contain predicted C/EBP-CREB motifs in their promoters that are conserved in the mouse and human DNA sequences (Table 2). For this subset of genes, endogenous C/EBPβ was found to occupy the predicted C/EBP-CREB motifs in Btg2 and Socs3, as in c-Fos and Cyr61, in 3T3-F442A preadipocytes (C/EBPβ in Fig. 7, third panel). This was demonstrated by ChIP using probes encompassing the predicted C/EBP-CREB motif sequences in their promoters. Constitutive occupancy of C/EBPβ was detected in the absence of GH and increased after 30 min GH treatment on these four early response genes. For c-Fos, GH-stimulated phosphorylation of the C/EBPβ occupying the promoter is critical for activation. It is thus notable that GH induced the occupancy of phosphorylated C/EBPβ on Cyr61 and Socs3 as well as on c-Fos (P-C/EBPβ in Fig. 7, second panel), detected by ChIP using antibodies specific for P-C/EBPβ. These findings suggest that P-C/EBPβ contributes to the mechanism by which GH stimulates expression these early response genes. An additional mechanistic feature that appears to be shared among these genes is GH-induced recruitment of p300 to their promoters: ChIP using antibodies against p300 showed that p300 occupancy increased in response to GH on Cyr61-2 and Socs3 as well as on c-Fos (p300 in Fig. 7, top panel). Occupancy of p300 and P-C/EBPβ on Btg2 is modestly increased by GH. These observations suggest that for the subset of GH-stimulated early response genes comprised of c-Fos, Cyr61, Btg2, and Socs3, whose GH-induced expression is reduced by inhibitors of ERKs 1/2, transcriptional activation is mediated by C/EBPβ, involves ERK-stimulated phosphorylation of C/EBPβ at T188, and recruitment of p300 to C/EBP-CREB motifs on their promoters.
Fig. 7.
GH induces occupancy of phosphorylated C/EBPβ and p300 on promoters of multiple early response genes. 3T3-F442A preadipocytes were treated without (−) or with (+) GH for 30 min. Nuclear extracts were analyzed by ChIP using antibodies against p300, P-C/EBPβ, or C/EBPβ. Controls contain no antibody; 1% input is also shown. ChIP samples were analyzed with probes specific to the known or predicted C/EBP-CREB sites in the c-Fos, Cyr61, Btg2, and Socs3 promoters.
Discussion
C/EBPβ mediates the activation of multiple GH-stimulated early response genes
This work demonstrates that C/EBPβ mediates GH-induced expression of multiple genes. Dependence on C/EBPβ was initially demonstrated for GH stimulation of the early response protooncogene c-Fos (15, 46). Many of the genes that clustered with c-Fos in a gene profile showing rapid and transient induction by GH in adipocytes were predicted to contain C/EBP-CREB motifs in their upstream regulatory sequences. Some of the most responsive of these early response genes include known GH target genes that encode transcription factors such as c-Fos, JunB, Egr1, Egr2 (34–37), and signaling molecules (Socs1, Socs3, and IL-6) (32, 33, 47). In addition, other genes were unrecognized GH targets or had appeared in GH-regulated gene profiles (48–51) but were uncharacterized, such as the gene for the antiproliferative factor Btg2 (40) and for Cyr61/CCN1, which interacts with extracellular matrix (38). The shRNA knockdown of C/EBPβ in GH-responsive 3T3-F442A cells showed that mRNA expression of the six genes tested that were most responsive to GH (c-Fos, Cyr61, Btg2, Socs3, Zfp36, and Socs1) was dependent on C/EBPβ, suggesting that early response genes stimulated by GH are coregulated by a mechanism involving C/EBPβ. The gene encoding activating transcription factor 3 also shows an early response to GH in preadipocytes, which is mediated by C/EBPβ (19). Other genes that generally show slower responses to GH, such as Igf1 and Igfbp1, have also been reported to be regulated by C/EBPβ (52, 53), although their C/EBPβ dependence for GH regulation has not been reported. The degree of dependence on C/EBPβ varied somewhat among the genes studied, because their GH stimulation was impaired to different extents in cells expressing shβ. Different degrees of inhibition may reflect overlapping functions of other B-Zip family transcription factors; for some of the genes, such as Cyr61 and Btg2, CREB also occupied C/EBP-CREB sites and/or stimulated promoter activation (data not shown) (18, 54, 55). CREB can be rapidly phosphorylated and activated by GH and other signals in an ERK 1/2-dependent manner; further, CREB can cooperate with C/EBPβ in stimulating c-Fos (18, 56). The variability in impairment in shβ cells may also reflect contributions of other transcription regulatory factors with C/EBPβ and/or that their combined functions are perturbed differentially by reduced levels of C/EBPβ in shβ cells.
Phosphorylation of C/EBPβ at an ERK substrate site and recruitment of p300 mediate GH-stimulated expression of a subset of early response genes
C/EBPβ can mediate transcriptional activation in a complex with the coactivator p300 (29, 31), which on c-Fos can be recruited in response to GH and can enhance C/EBPβ-mediated transcriptional activation (15). This study shows that GH-induced recruitment of p300 is dependent on the integrity of the phosphorylation site at T188 of C/EBPβ, which is a substrate site for ERKs 1 and 2. Mutation of T188 of murine C/EBPβ impaired the recruitment of p300 to c-Fos through rescue and overexpression approaches. This study also defines a subset of three other GH-stimulated early response genes, Cyr61, Btg2, and Socs3, which are coregulated by similar transcriptional mechanisms. Their expression was significantly impaired by C/EBPβ deficiency and by inhibition of ERKs 1 and 2. Moreover, GH induced the occupancy of P-C/EBPβ and of p300 on their promoters. The increased occupancy of C/EBPβ may reflect recruitment, in response to GH, of newly phosphorylated P-C/EBPβ to the c-Fos DNA, although the phosphorylation may also occur on the C/EBPβ that constitutively occupies c-Fos.
Because transcription is regulated by complexes of nuclear proteins, C/EBPβ is likely to coordinate with other GH-regulated transcription factors. Stats, particularly Stat5, are implicated in GH-regulated gene transcription. Sequence analysis that includes the upstream promoters of the early response genes in the present study predicted not only C/EBP-CREB sites but in some cases Stat sites. Two genes most impaired by C/EBPβ deficiency, Cyr61 and Btg2, were predicted to contain C/EBP sites but no Stat sites. The gene for Socs3 was predicted to contain both C/EBP-CREB and Stat sites; its dependence on Stat5 has been demonstrated (32), and other reports indicate that C/EBPβ can also mediate Socs3 expression (32, 57, 58). Here, the roles of phosphorylation of C/EBPβ at an ERK substrate site and recruitment of p300 in the regulation of Socs3 by C/EBPβ in response to GH are demonstrated. On the other hand, expression of two genes whose sequences were predicted to contain Stat but not C/EBP sites, Zfp36 and Socs1, was also impaired in GH-treated shβ cells. These two genes appear to form a different subset relative to the others studied here. They were not stimulated by C/EBPβ overexpression, and their stimulation by GH was not impaired by ERK inhibition. These genes may contain C/EBPβ responsive sites not detected in the locations searched under conditions of the present study. The contribution of C/EBPβ to regulating these genes may be indirect, e.g. by participating in transcription regulatory complexes without binding to DNA directly, or because other factors are also required. Additional C/EBP sites and Stat sites in any of the genes studied may be present but not predicted by the present computational approaches. Further, such sites may be functional in sequences other than the region that was analyzed in silico in the genes in this study, e.g. multiple GH-responsive Stat5 sites have been identified throughout the gene for IGF-I (59–61). The relatively smaller responses to GH of genes such as Cited2 and Frat2 do not necessarily signify that their contributions to GH physiology are not substantive but remain to be studied further. Differences in mechanisms for regulation among these genes may be important for conferring specificity in their responses to GH.
Taken together, these studies demonstrate that multiple early response genes stimulated by GH are coregulated by mechanisms involving C/EBPβ and related B-Zip family transcription factors. A subset of these genes further share a mechanism dependent on rapid activation of ERK signaling, induced occupancy of P-C/EBPβ, and recruitment of the coactivator p300 to their promoters in response to GH. Expression of other GH-stimulated early response genes are responsive to C/EBPβ deficiency but appear to utilize distinct transcription mechanisms. Given that gene transcription involves combined contributions of multiple regulatory factors, the present observations are consistent with C/EBPβ and B-Zip-dependent events joining Stat5 as transcriptional mechanisms mediating gene expression in response to GH.
Materials and Methods
Materials
Murine 3T3-F442A preadipocytes were provided by H. Green (Harvard University, Cambridge, MA) and M. Sonenberg (Memorial Sloan-Kettering Cancer Center, New York, NY). The 293T human kidney cell line was provided by M. Lazar (University of Pennsylvania, Philadelphia, PA) and O. MacDougald (University of Michigan). Chinese hamster ovary (CHO) cells stably expressing a truncated GH receptor (GHR) (GHR1-454) were provided by G. Norstedt (Karolinska Institutet, Stockholm, Sweden) and N. Billestrup (Novo Nordisk, Bagsværd, Denmark) (62); they are designated CHO-GHR cells (15, 36, 63). Deoxyribonuclease, trypsin, and reagents for preparation and genotyping MEF and mice were from Invitrogen (Carlsbad, CA). [32P]dATP was purchased from PerkinElmer Life Sciences (Waltham, MA). Human GH was a generous gift from Eli Lilly, Inc. (Indianapolis, IN) culture media; L-glutamine and antibiotic-antimycotic were purchased from Invitrogen. Fetal bovine serum was from Invitrogen or Irvine Scientific (Santa Ana, CA) and calf serum from Invitrogen or Atlanta Biologicals (Lawrenceville, GA). BSA (CRG7) was purchased from Serologicals Corp. (Norcross, GA). Lipofectamine and TRIzol Reagent were purchased from Invitrogen and TaqMan Reverse Transcription kit from Applied Biosystems (Foster City, CA). MEK inhibitor U0126 and Luciferin were purchased from Promega (Madison, WI) and β-galactosidase (β-gal) chemiluminescence reagents from Tropix (Bedford, MA). Protease inhibitor cocktail tablets (mini-EDTA free) and the protease inhibitors leupeptin and aprotinin were purchased from Roche (Indianapolis, IN), phenylmethylsulfonylfluoride was purchased from Mallinckrodt (Hazelwood, MO). Puromycin, sodium orthovanadate, formaldehyde, and SYBR green I were purchased from Sigma (St. Louis, MO). Immobilized protein A was purchased from RepliGen (Waltham, MA), protein G beads from GE Healthcare (Princeton, NJ), sonicated salmon sperm DNA from Stratagene (La Jolla, CA), and Qiaquick PCR Purification kits from QIAGEN (Valencia, CA). Polyvinylidene fluoride membrane was purchased from Millipore (Bedford, MA). Kits for enhanced chemiluminescence were obtained from Amersham (Piscataway, NJ). Protein molecular weight standards were purchased from Invitrogen and Bio-Rad Laboratories (Hercules, CA).
Plasmids and antibodies
The reporter plasmid fos-luc containing the mouse c-fos sequence from −379 to +1 (referred to as “promoter” throughout) upstream of the luciferase gene was generously provided by B. Cochran (Tufts University, Medford, MA). The expression plasmid for full-length rat C/EBPβ, also known as liver-enriched activating protein (64), was provided by U. Schibler (University of Geneva, Geneva, Switzerland) and L. Sealy (Vanderbilt University, Nashville, Tennessee). The plasmid for murine C/EBPβ (residues 22-296) tagged with human influenza hemagglutinin (HA) at the N terminus (designated C/EBPβ or HA-C/EBPβ) was a gift from J. R. Cardinaux (University of Lausanne, Lausanne, Switzerland) (31). The phosphorylation site mutant T188A-C/EBPβ (T188A) was created by introducing a Thr to Ala mutation at T188 in HA-C/EBPβ (23). The plasmid for human C/EBPβ was a gift from S. Akira (Osaka, Japan) and L. Sealy (Vanderbilt University). Plasmids for shβ and mU6pro vector, provided by D. Turner (University of Michigan), used in transient transfections were described previously (15). The rescue plasmids murine HA-C/EBPβ WT (rescue WT) and HA-C/EBPβ T188A (rescue T188A) were generated using the QuikChange mutagenesis kit (Stratagene) to introduce silent mutations (underlined) in the shβ recognition region of C/EBPβ [WT, G AGC GAC GAG TAC AAG ATG (residues 223-226: D-E-Y-K); rescue mutant, G AGC GAT GAA TAT AAA ATG], which allows production of C/EBPβ protein in the presence of shβ. The human cyr61 promoter (−883 to +1) plasmids in pGL3 (cyr61-luc) were kindly provided by H. S. Kim (Seoul National University College of Medicine, Seoul, Korea) (65) and by G. Fisher (University of Michigan) (55). The plasmid for murine socs3-luciferase was generously provided by S. Melmed (Cedars-Sinai Medical Center, Los Angeles, CA) (66). Cytomegalovirus (CMV)-driven expression plasmid for rat GHR has been described (67); the plasmid for human p300-FLAG was provided by R. Kwok (University of Michigan). The vector pcDNA3 (Invitrogen) was used as a control. The plasmids for β-gal driven by rous sarcoma virus (RSV) or CMV were provided by M. Uhler (University of Michigan).
Antibodies against C/EBPβ (C-19), against C/EBPδ (amino acids 115-130), Stat5 (C-17), Cyr61 hinge (H-78), Cyr61 N terminus (N-16), p300 (N-15), α-tubulin (TU-02), and normal rabbit IgG were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Antibodies targeting the C-terminal tyrosine phosphorylation site (Tyr694/699) of murine Stat5 (P-Stat5), phospho-tyrosine 705 of Stat3 (P-Stat3), and total Stat3 were purchased from Upstate/Millipore (Lake Placid, NY). The anti-P-C/EBPβ antibody against murine C/EBPβ phosphorylated at T188 (equivalent to T235 of human C/EBPβ) as well as antibody against phospho-Thr202/Tyr204 of human p44/42 MAPK (P-ERK 1/2) and the antibody against total p44/42 MAPKs (ERK 1/2) were from Cell Signaling (Beverly, MA). Antibodies against acetylated histone 4 (Ac-H4) and phospho-RNA polymerase II were from Millipore. Anti-HA antibody was from Covance (Richmond, CA); secondary antibody labeled with horseradish peroxidase was from Santa Cruz Biotechnology, Inc. The secondary antibodies conjugated with IRDye800 and IRDye700, which were used in immunoblots scanned by Licor Odyssey, were obtained from Rockland, Inc. (Gilbertsville, PA) and Fisher Scientific (Auburn, AL).
Cell culture and treatment
Native 3T3-F442A preadipocytes, 293T cells and CHO-GHR cells were grown and maintained as described previously (15, 20). Where indicated, 3T3-F442A preadipocytes were differentiated into adipocytes (19). Before all experiments with in vitro GH treatment, cells were deprived of serum for 16–18 h in medium containing 1% BSA instead of serum. Cells were then treated without or with human GH (500 ng/ml = 22 nm) for the times indicated; adipocytes were treated with GH on d 7 after initiation of adipogenesis. In some experiments, 3T3-F442A cells were pretreated with U0126 (20 μm) for 30 min before GH treatment; controls were treated with equivalent amounts of dimethylsulfoxide vehicle (0.1% vol/vol) for equivalent times.
C/EBPβ-deficient cells
Stable deficiency of endogenous C/EBPβ in 3T3-F442A cells was achieved retrovirally using shRNA. The targeting sequence for murine C/EBPβ (shβ) was 5′-GAGCGACGAGTACAAGATG-3′ (15). The shRNA control 3T3-F442A preadipocytes (designated sh-C) stably expressing a nontargeting shRNA (5′-UUCUCCGAACGUGUCACGU-3′) (QIAGEN-Xeragon, Germantown, MD) have been described (68). The appropriate oligonucleotides were annealed and subcloned into the vector pSuperior.retro.puro (Oligoengine, Seattle, WA) at BamHI and HindIII sites. The sequences were confirmed by the University of Michigan DNA Sequencing Core. The retroviral constructs were transfected into 293T cells with packaging vectors SV-E-MLV-env and SVψ-E-MLV. Medium was collected 24 and 48 h later, mixed with polybrene (1 μg/ml), and applied to 3T3-F442A preadipocytes. Virus-infected cells were cultured in selection medium containing puromycin (4 μg/ml). Deficiency of the C/EBPβ in the 3T3-F442A cells stably expressing shRNA was verified by immunoblotting and by qPCR to compare the protein and mRNA expression levels of C/EBPβ with those in the control sh-C cells and the parental 3T3-F442A cells. A small amount of residual C/EBPβ protein was barely detectable (Fig. 2B). The 3T3-F442A cells expressing shβ or control sh-C were maintained in DMEM containing 2 μg/ml of puromycin.
In experiments analyzing transient C/EBPβ deficiency, 293T cells in 10-cm dishes were transfected by calcium phosphate coprecipitation (15, 63) with plasmids for shβ (5 μg) or vector (mU6pro), along with rescue plasmids for WT or T188A C/EBPβ (2 μg each) and rat GHR (2 μg). In complementary experiments, 293T cells were transfected with plasmid for WT or T188A C/EBPβ (2 μg each) along with plasmid for rat GHR (2 μg); 24 h later, cells were cultured with 1% BSA-DMEM for an additional 18 h, then were treated with GH for 15 min before lysis for ChIP or protein analysis.
Quantitative real-time PCR
Quantitative real-time PCR was performed on total RNA isolated from cells and analyzed using iCycler iQ real-time detection system software (Bio-Rad Laboratories) as described (15, 19). The sequences of primers are based on published studies or were designed using National Center for Biotechnology Information (NCBI) Primer Tool (Supplemental Table 2). Gene expression was normalized to glyceraldehyde-3-phosphate dehydrogenase or TATA-box binding protein as control. In each experiment, the reference control is set equal to 1 for relative expression of other conditions. Results of at least three independent experiments were averaged for statistical evaluation unless indicated otherwise. Statistical analysis was performed using one-way ANOVA with Dunnet or Bonferroni correction for multiple testing (Prism version 3) as described (18, 69). Statistical significance (P < 0.05) is indicated on figures by brackets between bars and in figure legends.
Immunoblotting analysis
Livers were homogenized (4 C) and sonicated in ice-cold lysis buffer [10 mm Tris-HCl (pH 7.5), 5 mm EDTA, 150 mm NaCl, 30 mm sodium pyrophosphate, 50 mm NaF, 1 mm Na3VO4, 10% glycerol, and 0.5% Nonidet P-40] containing protease inhibitors as described (20). Lysates were centrifuged at 15,000 × g at 4C for 15 min to pellet insoluble material. To detect cellular protein in primary MEFs, as well as 3T3-F442A, 293T, and other cells lines, cells were lysed with sodium dodecyl sulfate lysis buffer [60 mm Tris-HCl (pH 6.8) and 1% sodium dodecyl sulfate] and boiled as described (70). Samples were separated by SDS-PAGE and transferred to polyvinylidene fluoride membrane. Immunoblotting for endogenous or expressed protein was performed at 4 C overnight as described (70) with the following antibodies: anti-C/EBPβ (1:1000), anti-P-Stat5 (1:1000), anti-Stat5 (1:1000), anti-P-Stat3, anti-Stat3, anti-P-Akt (1:1000), anti-Akt (1:1000), anti-Cyr61/CCN1 (1:1000), anti-P-ERK 1/2 (1:1000), anti-ERK 1/2 (1:1000), or anti-α-tubulin (1:1000). The proteins were visualized using enhanced chemiluminescence or the Odyssey scanning system (LI-COR Biosciences, Lincoln, NE) (15). Molecular weight was estimated using Cruz Marker or MagicMark Western Standard.
Computational prediction of C/EBP sites in GH-regulated genes
To analyze GH-regulated genes whose stimulation might be mediated by C/EBPβ, the promoter sequences of a set of GH-regulated genes in 3T3-F442A adipocytes (19) were subjected to computational analysis using Genomatix (Munich, Germany) MatInspector tools. The analysis was applied to a cluster of genes defined by hierarchical clustering, which included c-fos and 14 other early response genes that were stimulated by GH within 30 min but not changed at 4 or 48 h after GH treatment. The mouse and human promoter sequences (−1500 to +200 bp) for the 15 genes were downloaded from the NCBI website (http://www.ncbi.nlm.nih.gov/) using the transcription start site defined by RefSeq. Using the Genomatix Dialign tool, the conserved C/EBP motifs in the mouse and human promoter sequence pairs were analyzed and predicted (Table 1), using the default stringency of 0.75. Because B-Zip transcription factors such as C/EBPβ and CREB can dimerize and/or bind to similar sequences, the motifs for C/EBP and CREB were categorized interchangeably with the software used and are referred to as C/EBP or C/EBP-CREB target motifs.
Chromatin immunoprecipitation
ChIP was carried out as described (15) on 3T3-F442A preadipocytes stably expressing retroviral shβ or sh-C, or 293T cells transiently expressing shβ in mU6Pro or vector. Frozen mouse liver was minced in 1% formaldehyde for 15 min at room temperature and was centrifuged (1500 rpm for 2 min) to collect liver cells. In each immunoprecipitation, 100 μg of nuclear protein from cell lines or 300 μg protein from mouse liver lysate was incubated overnight at 4 C with 4 μg of one of the following antibodies: anti-C/EBPβ, anti-p300, anti-HA, anti-AcH4, or anti-P-Pol II. Samples incubated without antibody or with normal rabbit IgG served as a negative control; 1% input was used to indicate the relative amount of each sample used for individual ChIP analysis. The mouse c-Fos promoter containing a C/EBP motif was amplified by PCR (15). The human c-Fos C/EBP region was amplified using primers 5′-CCC GAC CTC GGG AAC AAG GG-3′ and 5′-ATG AGG GGT TTC GGG GAT GG-3′. For mouse Cyr61, Genomatix analysis described above predicted two C/EBP sites referred to as Cyr61-1 and Cyr61-2, each containing one predicted C/EBP motif. The Cyr61 C/EBP sites were amplified by PCR with the primers 5′-TGGGCTGGAACTAAAACTGG-3′ and 5′-GCCAGGGATCTATTTGTGGA-3′ for Cyr61-1 and primers 5′-ATCAAAATCATCACCCTCGC-3′ and 5′-GCCCTTTATAATGCCTGCCT-3′ for Cyr61-2. Mouse Btg2 C/EBP-CREB site was amplified by PCR with the primers 5′-GGCGAGGGCATCCTGGAGGA-3′ and 5′- AGAGGGCCTTGGACCTGGGC-3′. The Socs3 C/EBP-CREB site predicted by Genomatix was amplified by PCR with the primers 5′-ATGTCGCGCATCCCCCTCCT-3′ and 5′-GACGCTCCCCTCCCTCTGCA-3′. The ChIP PCR primers were designed using NCBI Primer Tool. In each experiment, all of the immunoprecipitated samples were analyzed using the same PCR conditions to evaluate relative amounts of each protein associated with the promoter. PCR products were separated on 2% agarose gels and stained with ethidium bromide. Input served as the internal control in each experiment. In some experiments, ChIP eluates were analyzed by immunoblotting as described (15).
Transcriptional activation
The 293T or CHO-GHR cells were used to measure transcription activation. For Cyr61 promoter activation, 293T cells in 24-well plates were transfected with plasmid for Cyr61-luc (0.5 μg/well) using calcium phosphate coprecipitation as described (15, 63). For c-Fos coactivation, 293T, or CHO-GHR cells were transfected with plasmids for Fos-luc (0.5 μg), along with shβ (1 μg) and WT or T188A C/EBPβ expression constructs (10 ng each) or with WT or T188A C/EBPβ expression constructs, with or without p300-FLAG (0.5 μg) in six-well plates. pcDNA3 was used to normalize the amount of DNA among the conditions, and CMV- or RSV-β-gal was coexpressed as an indicator of transfection efficiency. In some experiments, primary MEFs (25,000 cells/35-mm well) were transfected by calcium phosphate coprecipitation (71) or with Lipofectamine according to the manufacturer's protocol, using c-Fos-luc and rat GHR plasmid (each 3 μg/well). In some experiments, plasmid encoding C/EBPβ was also cotransfected in amounts indicated.
Where indicated, cells were treated with GH as described above. After 4 h, cells were lysed and analyzed for luciferase activity using an Opticomp Luminometer as described previously (15, 46, 63). Luciferase activity is expressed as relative luciferase units. Each condition was tested in triplicate in each experiment. The reporter activity in the presence of the pcDNA3 control was set equal to 1 to calculate relative expression among experimental conditions. Data from at least three independent experiments are presented as mean ± se. Statistical analysis was performed as described above for qPCR. Statistical significance (P < 0.05) is indicated on figures by brackets between bars and in figure legends.
C/EBPβ-deficient mice and MEF
C/EBPβ −/− mice were obtained by crossing heterozygous C/EBPβ +/− males (generously provided by V. Poli; Dundee, Scotland, and courtesy of F. Costantini; Columbia University, New York, NY) on a 129sv/ev background. Females were purchased from Taconic Laboratories (Hudson, NY). Animals were maintained on 12-h light, 12-h dark cycle and were fed rodent laboratory chow 5001 (Ralston Purina Co., St. Louis, MO) ad libitum. All procedures were approved by the University of Michigan Committee on Use and Care of Animals.
To generate MEFs, C/EBPβ +/− females were crossed with +/− males. Embryos were collected at embryonic d13.5–17.5, carcasses were individually minced and trypsinized in the presence of deoxyribonuclease for 1–3 h. Resulting cells from each embryo were plated separately in DMEM (4.5 g/liter glucose) with 10% fetal bovine serum, 1% glutamine, antibiotic, and antimycotic, and incubated at 37 C in an atmosphere of 5% CO2-95% air. After 16 h, cells were washed with PBS, and fresh medium was added. Cells were subsequently grown to 70–80% confluence and then split at a density of 250,000 cells/100-mm plate and used for experiments or stored in liquid nitrogen. The genotypes of mice, embryos and MEFs were verified by PCR using the primers: P1, AAG ACG GTG GAC AAG CTG AG; P2, GGC AGC TGC TTG AAC AAG TTC; P3, CAT CAG AGC AGC CGA TTG TC; to amplify the C/EBPβ region with (P1 and P3) or without (P1 and P2) the inserted neomycin cassette (Poli, V., personal communication).
C/EBPβ −/− mice and cells were compared with C/EBPβ-intact samples from WT (+/+) or heterozygous (+/−) littermate donors. For the analysis of GH responses in vivo, mice were fasted for 16–18 h and injected ip with human GH (1.5 mg/kg body weight) or with vehicle. Tissues were obtained 15 min after injection and flash frozen until analysis. For ChIP, liver tissue from C/EBPβ KO mice bred from C/EBPβ +/− mice from The Jackson Laboratory (Bar Harbor, ME), and from WT control mice, was kindly provided by J. D. Lin and D. Ma (University of Michigan).
Acknowledgments
We thank the excellent contributions of Michelle Richardson, Stacy Richardson, and Christina Fulton to these studies. We also thank Dr. Jeffrey Huo for microarray data and Dr. Yili Chen for advice on promoter computational analysis; Dr. Jiandie Lin and Di Ma for tissues from C/EBPβ-deficient mice; Dr. Ormond MacDougald and Dr. Isabel Gerin for advice on retroviral infection; and Dr. T. Saunders and E. Hughes of University of Michigan Transgenic Core for advice on preparation of MEFs.
Present address for T.X.C.: Department of Surgery, University of Michigan, Ann Arbor, Michigan 48109.
Present address for G.P-.P.: Instituto de Biología y Medicina Experimental, Buenos Aires 1428, Argentina.
Present address for N.L.: Van Andel Institute, Grand Rapids, Michigan 49503.
Present address for H.J.: Department of Pharmacology, Case-Western Reserve University, Cleveland, Ohio 44106.
Present address for Z.S.Q.: Department Statistics, Emory University, Atlanta, Georgia 30303.
J.S. was supported by the National Science Foundation Grant IBN 00-80193, the National Institutes of Health (NIH) Grant DK46072, the American Diabetes Association Grant 7-09-BS-168), the University of Michigan Center for Computational Biology and Medicine, and the Biomedical Research Council; Z.S.Q. by the NIH Grant HG05119; C.C.-S. by the NIH Grant DK34171; G.L. by the NIH T32 Grant GM07315, the University of Michigan Cancer Biology Training Program, and a Rackham Predoctoral Fellowship (University of Michigan); C.R.L. by the University of Michigan Center for Organogenesis Grant T32 HD007505; M.R. by a fellowship from the University of Michigan Undergraduate Research Opportunity Program; G.P.-P. by the University of Michigan Center for Organogenesis Fellowship T32 HD007505; N.L. by the NIH Grant T32 GM07315, a Rackham Regents fellowship, and the University of Michigan Center for Organogenesis Grant T32 HD007505; and H.J. by the NIH Grant K01 DK07791.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- C/EBPβ
- CCAAT/enhancer binding protein β
- ChIP
- chromatin immunoprecipitation
- CHO
- Chinese hamster ovary
- CMV
- cytomegalovirus
- CRE
- cAMP response element
- CREB
- CRE-binding protein
- Cyr61/CCN1
- cysteine-rich 61/Cyr61, CTGF, NOV family of genes 1
- Egr
- early growth response
- β-gal
- β-galactosidase
- GHR
- GH receptor
- KO
- knockout
- MEF
- murine embryonic fibroblast
- MEK
- MAPK kinase
- NCBI
- National Center for Biotechnology Information
- P-C/EBPβ
- phosphorylation of C/EBPβ at T188
- qPCR
- quantitative real-time PCR
- RSV
- rous sarcoma virus
- shβ
- shRNA targeting C/EBPβ
- sh-C
- sh-control
- shRNA
- short hairpin RNA
- Socs
- suppressor of cytokine signaling
- Stat
- signal transducer and activator of transcription
- WT
- wild type
- Zfp
- zinc finger protein.
References
- 1. Herrington J, Smit LS, Schwartz J, Carter-Su C. 2000. The role of STAT proteins in growth hormone signaling. Oncogene 19:2585–2597 [DOI] [PubMed] [Google Scholar]
- 2. Hennighausen L, Robinson GW. 2008. Interpretation of cytokine signaling through the transcription factors STAT5A and STAT5B. Genes Dev 22:711–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rotwein P, Chia DJ. 2010. Gene regulation by growth hormone. Pediatr Nephrol 25:651–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Waxman DJ, O'Connor C. 2006. Growth hormone regulation of sex-dependent liver gene expression. Mol Endocrinol 20:2613–2629 [DOI] [PubMed] [Google Scholar]
- 5. Ceseña TI, Cui TX, Piwien-Pilipuk G, Kaplani J, Calinescu AA, Huo JS, Iñiguez-Lluhí JA, Kwok R, Schwartz J. 2007. Multiple mechanisms of growth hormone-regulated gene transcription. Mol Genet Metab 90:126–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Darlington GJ, Ross SE, MacDougald OA. 1998. The role of C/EBP genes in adipocyte differentiation. J Biol Chem 273:30057–30060 [DOI] [PubMed] [Google Scholar]
- 7. Poli V. 1998. The role of C/EBP isoforms in the control of inflammatory and native immunity functions. J Biol Chem 273:29279–29282 [DOI] [PubMed] [Google Scholar]
- 8. Farmer SR. 2006. Transcriptional control of adipocyte formation. Cell Metab 4:263–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rosen ED, MacDougald OA. 2006. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol 7:885–896 [DOI] [PubMed] [Google Scholar]
- 10. Zahnow CA. 2009. CCAAT/enhancer-binding protein β: its role in breast cancer and associations with receptor tyrosine kinases. Expert Rev Mol Med 11:e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sebastian T, Johnson PF. 2006. Stop and go: anti-proliferative and mitogenic functions of the transcription factor C/EBPβ. Cell Cycle 5:953–957 [DOI] [PubMed] [Google Scholar]
- 12. Grimm SL, Contreras A, Barcellos-Hoff MH, Rosen JM. 2005. Cell cycle defects contribute to a block in hormone-induced mammary gland proliferation in CCAAT/enhancer-binding protein (C/EBPβ)-null mice. J Biol Chem 280:36301–36309 [DOI] [PubMed] [Google Scholar]
- 13. Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, Lasorella A, Aldape K, Califano A, Iavarone A. 2010. The transcriptional network for mesenchymal transformation of brain tumours. Nature 463:318–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Seagroves TN, Krnacik S, Raught B, Gay J, Burgess-Beusse B, Darlington GJ, Rosen JM. 1998. C/EBPβ, but not C/EBPα, is essential for ductal morphogenesis, lobuloalveolar proliferation, and functional differentiation in the mouse mammary gland. Genes Dev 12:1917–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cui TX, Piwien-Pilipuk G, Huo JS, Kaplani J, Kwok R, Schwartz J. 2005. Endogenous CCAAT/enhancer binding protein β and p300 are both regulated by growth hormone to mediate transcriptional activation. Mol Endocrinol 19:2175–2186 [DOI] [PubMed] [Google Scholar]
- 16. Metz R, Ziff E. 1991. cAMP stimulates the C/EBP-related transcription factor rNFIL-6 to trans-locate to the nucleus and induce c-fos transcription. Genes Dev 5:1754–1766 [DOI] [PubMed] [Google Scholar]
- 17. Wilson HL, Roesler WJ. 2002. CCAAT/enhancer binding proteins: do they possess intrinsic cAMP-inducible activity? Mol Cell Endocrinol 188:15–20 [DOI] [PubMed] [Google Scholar]
- 18. Cui TX, Kwok R, Schwartz J. 2008. Cooperative regulation of endogenous cAMP-response element binding protein and CCAAT/enhancer-binding protein β in GH-stimulated c-fos expression. J Endocrinol 196:89–100 [DOI] [PubMed] [Google Scholar]
- 19. Huo JS, McEachin RC, Cui TX, Duggal NK, Hai T, States DJ, Schwartz J. 2006. Profiles of growth hormone (GH)-regulated genes reveal time-dependent responses and identify a mechanism for regulation of activating transcription factor 3 by GH. J Biol Chem 281:4132–4141 [DOI] [PubMed] [Google Scholar]
- 20. Piwien-Pilipuk G, MacDougald O, Schwartz J. 2002. Dual regulation of phosphorylation and dephosphorylation of C/EBPβ modulate its transcriptional activation and DNA binding in response to growth hormone. J Biol Chem 277:44557–44565 [DOI] [PubMed] [Google Scholar]
- 21. Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, Denissov S, Børgesen M, Francoijs KJ, Mandrup S, Stunnenberg HG. 2008. Genome-wide profiling of PPARγ:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev 22:2953–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lefterova MI, Zhang Y, Steger DJ, Schupp M, Schug J, Cristancho A, Feng D, Zhuo D, Stoeckert CJ, Jr, Liu XS, Lazar MA. 2008. PPARγ and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev 22:2941–2952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ceseña TI, Cardinaux JR, Kwok R, Schwartz J. 2007. CCAAT/enhancer-binding protein (C/EBP)β is acetylated at multiple lysines: acetylation of C/EBPβ at lysine 39 modulates its ability to activate transcription. J Biol Chem 282:956–967 [DOI] [PubMed] [Google Scholar]
- 24. Xu M, Nie L, Kim SH, Sun XH. 2003. STAT5-induced Id-1 transcription involves recruitment of HDAC1 and deacetylation of C/EBPβ. EMBO J 22:893–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schwartz C, Beck K, Mink S, Schmolke M, Budde B, Wenning D, Klempnauer KH. 2003. Recruitment of p300 by C/EBPβ triggers phosphorylation of p300 and modulates coactivator activity. EMBO J 22:882–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakajima T, Kinoshita S, Sasagawa T, Sasaki K, Naruto M, Kishimoto T, Akira S. 1993. Phosphorylation at threonine-235 by a ras-dependent mitogen-activated protein kinase cascade is essential for transcription factor NF-IL6. Proc Natl Acad Sci USA 90:2207–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Buck M, Poli V, van der Geer P, Chojkier M, Hunter T. 1999. Phosphorylation of rat serine 105 or mouse threonine 217 in C/EBPβ is required for hepatocyte proliferation induced by TGFα. Mol Cell 4:1087–1092 [DOI] [PubMed] [Google Scholar]
- 28. Park BH, Qiang L, Farmer SR. 2004. Phosphorylation of C/EBPβ at a consensus extracellular signal-regulated kinase/glycogen synthase kinase 3 site is required for the induction of adiponectin gene expression during the differentiation of mouse fibroblasts into adipocytes. Mol Cell Biol 24:8671–8680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mink S, Haenig B, Klempnauer KH. 1997. Interaction and functional collaboration of p300 and C/EBPβ. Mol Cell Biol 17:6609–6617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ohoka N, Kato S, Takahashi Y, Hayashi H, Sato R. 2009. The orphan nuclear receptor RORα restrains adipocyte differentiation through a reduction of C/EBPβ activity and perilipin gene expression. Mol Endocrinol 23:759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kovács KA, Steinmann M, Magistretti PJ, Halfon O, Cardinaux JR. 2003. CCAAT/enhancer-binding protein family members recruit the coactivator CREB-binding protein and trigger its phosphorylation. J Biol Chem 278:36959–36965 [DOI] [PubMed] [Google Scholar]
- 32. Barclay JL, Anderson ST, Waters MJ, Curlewis JD. 2007. Regulation of suppressor of cytokine signaling 3 (SOC3) by growth hormone in pro-B cells. Mol Endocrinol 21:2503–2515 [DOI] [PubMed] [Google Scholar]
- 33. Fasshauer M, Klein J, Lossner U, Paschke R. 2003. Interleukin (IL)-6 mRNA expression is stimulated by insulin, isoproterenol, tumour necrosis factor α, growth hormone, and IL-6 in 3T3-L1 adipocytes. Horm Metab Res 35:147–152 [DOI] [PubMed] [Google Scholar]
- 34. Slootweg MC, de Groot RP, Herrmann-Erlee MP, Koornneef I, Kruijer W, Kramer YM. 1991. Growth hormone induces expression of c-jun and junB oncogenes and employs a protein kinase C signal transduction pathway for the induction of c-fos gene expression. J Mol Endocrinol 6:179–188 [DOI] [PubMed] [Google Scholar]
- 35. Hodge C, Liao J, Stofega M, Guan K, Carter-Su C, Schwartz J. 1998. Growth hormone stimulates phosphorylation and activation of Elk-1 and expression of c-fos, egr-1, and junB through activation of extracellular signal-regulated kinases 1 and 2. J Biol Chem 273:31327–31336 [DOI] [PubMed] [Google Scholar]
- 36. Gong TW, Meyer DJ, Liao J, Hodge CL, Campbell GS, Wang X, Billestrup N, Carter-Su C, Schwartz J. 1998. Regulation of glucose transport and c-fos and egr-1 expression in cells with mutated or endogenous growth hormone receptors. Endocrinology 139:1863–1871 [DOI] [PubMed] [Google Scholar]
- 37. Clarkson RW, Shang CA, Levitt LK, Howard T, Waters MJ. 1999. Ternary complex factors Elk-1 and Sap-1a mediate growth hormone-induced transcription of egr-1 (early growth response factor-1) in 3T3-F442A preadipocytes. Mol Endocrinol 13:619–631 [DOI] [PubMed] [Google Scholar]
- 38. Chen CC, Lau LF. 2009. Functions and mechanisms of action of CCN matricellular proteins. Int J Biochem Cell Biol 41:771–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Leask A, Abraham DJ. 2006. All in the CCN family: essential matricellular signaling modulators emerge from the bunker. J Cell Sci 119:4803–4810 [DOI] [PubMed] [Google Scholar]
- 40. Winkler GS. 2010. The mammalian anti-proliferative BTG/Tob protein family. J Cell Physiol 222:66–72 [DOI] [PubMed] [Google Scholar]
- 41. Bouchard L, Tchernof A, Deshaies Y, Marceau S, Lescelleur O, Biron S, Vohl MC. 2007. ZFP36: a promising candidate gene for obesity-related metabolic complications identified by converging genomics. Obes Surg 17:372–382 [DOI] [PubMed] [Google Scholar]
- 42. Cao H, Urban JF, Jr, Anderson RA. 2008. Insulin increases tristetraprolin and decreases VEGF gene expression in mouse 3T3-L1 adipocytes. Obesity 16:1208–1218 [DOI] [PubMed] [Google Scholar]
- 43. Chen Y, Lin G, Huo JS, Barney D, Wang Z, Livshiz T, States DJ, Qin ZS, Schwartz J. 2009. Computational and functional analysis of growth hormone (GH)-regulated genes identifies the transcriptional repressor B-cell lymphoma 6 (Bc16) as a participant in GH-regulated transcription. Endocrinology 150:3645–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang GP, Lau LF. 1991. Cyr61, product of a growth factor-inducible immediate early gene, is associated with the extracellular matrix and the cell surface. Cell Growth Differ 2:351–357 [PubMed] [Google Scholar]
- 45. Kireeva ML, Lam SC, Lau LF. 1998. Adhesion of human umbilical vein endothelial cells to the immediate-early gene product Cyr61 is mediated through integrin αvβ3. J Biol Chem 273:3090–3096 [DOI] [PubMed] [Google Scholar]
- 46. Liao J, Piwien-Pilipuk G, Ross SE, Hodge CL, Sealy L, MacDougald OA, Schwartz J. 1999. CCAAT/enhancer-binding protein β (C/EBPβ) and C/EBPδ contribute to growth homone-regulated transcription of c-fos. J Biol Chem 274:31597–31604 [DOI] [PubMed] [Google Scholar]
- 47. Adams TE, Hansen JA, Starr R, Nicola NA, Hilton DJ, Billestrup N. 1998. Growth hormone preferentially induces the rapid, transient expression of SOCS-3, a novel inhibitor of cytokine receptor signaling. J Biol Chem 273:1285–1287 [DOI] [PubMed] [Google Scholar]
- 48. Sjögren K, Leung KC, Kaplan W, Gardiner-Garden M, Gibney J, Ho KK. 2007. Growth hormone regulation of metabolic gene expression in muscle: a microarray study in hypopituitary men. Am J Physiol Endocrinol Metab 293:E364–E371 [DOI] [PubMed] [Google Scholar]
- 49. Qin Y, Tian YP. 2010. A microarray gene analysis of peripheral whole blood in normal adult male rats after long-term GH gene therapy. Cell Mol Biol Lett 15:177–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Govoni KE, Lee SK, Chadwick RB, Yu H, Kasukawa Y, Baylink DJ, Mohan S. 2006. Whole genome microarray analysis of growth hormone-induced gene expression in bone: T-box3, a novel transcription factor, regulates osteoblast proliferation. Am J Physiol Endocrinol Metab 291:E128–E136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kawai M, Namba N, Mushiake S, Etani Y, Nishimura R, Makishima M, Ozono K. 2007. Growth hormone stimulates adipogenesis of 3T3-L1 cells through activation of the Stat5A/5B-PPARγ pathway. J Mol Endocrinol 38:19–34 [DOI] [PubMed] [Google Scholar]
- 52. Nolten LA, van Schaik FM, Steenbergh PH, Sussenbach JS. 1994. Expression of the insulin-like growth factor I gene is stimulated by the liver-enriched transcription factors C/EBPα and LAP. Mol Endocrinol 8:1636–1645 [DOI] [PubMed] [Google Scholar]
- 53. Guo S, Cichy SB, He X, Yang Q, Ragland M, Ghosh AK, Johnson PF, Unterman TG. 2001. Insulin suppresses transactivation by C/EBPβ: signaling to p300/CBP by protein kinase B disrupts interaction with the major activation domain of C/EBPβ. J Biol Chem 276:8516–8523 [DOI] [PubMed] [Google Scholar]
- 54. Dobroff AS, Wang H, Melnikova VO, Villares GJ, Zigler M, Huang L, Bar-Eli M. 2009. Silencing cAMP-response element-binding protein (CREB) identifies CYR61 as a tumor suppressor gene in melanoma. J Biol Chem 284:26194–26206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Quan T, Qin Z, Xu Y, He T, Kang S, Voorhees JJ, Fisher GJ. 2010. Ultraviolet irradiation induces CYR61/CCN1, a mediator of collagen homeostasis, through activation of transcription factor AP-1 in human skin fibroblasts. J Invest Dermatol 130:1697–1706 [DOI] [PubMed] [Google Scholar]
- 56. Chen Y, Zhuang S, Cassenaer S, Casteel DE, Gudi T, Boss GR, Pilz RB. 2003. Synergism between calcium and cyclic GMP in cyclic AMP response element-dependent transcriptional regulation requires cooperation between CREB and C/EBPβ. Mol Cell Biol 23:4066–4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yarwood SJ, Borland G, Sands WA, Palmer TM. 2008. Identification of CCAAT/enhancer-binding proteins as exchange protein activated by cAMP-activated transcription factors that mediate the induction of the SOCS-3 gene. J Biol Chem 283:6843–6853 [DOI] [PubMed] [Google Scholar]
- 58. Borland G, Bird RJ, Palmer TM, Yarwood SJ. 2009. Activation of protein kinase Cα by EPAC1 is required for the ERK- and CCAAT/enhancer-binding protein β-dependent induction of the SOCS-3 gene by cyclic AMP in COS1 cells. J Biol Chem 284:17391–17403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Woelfle J, Chia DJ, Rotwein P. 2003. Mechanisms of growth hormone (GH) action. Identification of conserved Stat5 binding sites that mediate GH-induced insulin-like growth factor-I gene activation. J Biol Chem 278:51261–51266 [DOI] [PubMed] [Google Scholar]
- 60. Chia DJ, Ono M, Woelfle J, Schlesinger-Massart M, Jiang H, Rotwein P. 2006. Characterization of distinct Stat5b binding sites that mediate growth hormone-stimulated IGF-I gene transcription. J Biol Chem 281:3190–3197 [DOI] [PubMed] [Google Scholar]
- 61. Eleswarapu S, Gu Z, Jiang H. 2008. Growth hormone regulation of insulin-like growth factor-I gene expression may be mediated by multiple distal signal transducer and activator of transcription 5 binding sites. Endocrinology 149:2230–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Möller C, Hansson A, Enberg B, Lobie PE, Norstedt G. 1992. Growth hormone (GH) induction of tyrosine phosphorylation and activation of mitogen-activated protein kinase in cells transfected with rat GH receptor cDNA. J Biol Chem 267:23403–23408 [PubMed] [Google Scholar]
- 63. Piwien-Pilipuk G, Van Mater D, Ross SE, MacDougald OA, Schwartz J. 2001. Growth hormone regulates phosphorylation and function of C/EBPβ by modulating Akt and glycogen synthase kinase-3. J Biol Chem 276:19664–19671 [DOI] [PubMed] [Google Scholar]
- 64. Descombes P, Chojkier M, Lichtsteiner S, Falvey E, Schibler U. 1990. LAP, a novel member of the C/EBP gene family, encodes a liver-enriched transcriptional activator protein. Gene Dev 4:1541–1551 [DOI] [PubMed] [Google Scholar]
- 65. Lee HY, Chung JW, Youn SW, Kim JY, Park KW, Koo BK, Oh BH, Park YB, Chaqour B, Walsh K, Kim HS. 2007. Forkhead transcription factor FOXO3a is a negative regulator of angiogenic immediate early gene CYR61, leading to inhibition of vascular smooth muscle cell proliferation and neointimal hyperplasia. Circ Res 100:372–380 [DOI] [PubMed] [Google Scholar]
- 66. Auernhammer CJ, Bousquet C, Melmed S. 1999. Autoregulation of pituitary corticotroph SOCS-3 expression: characterization of the murine SOCS-3 promoter. Proc Nat Acad Sci USA 96:6964–6969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang X, Uhler MD, Billestrup N, Norstedt G, Talamantes F, Nielsen JH, Carter-Su C. 1992. Evidence for association of the cloned liver growth hormone receptor with a tyrosine kinase. J Biol Chem 267:17390–17396 [PubMed] [Google Scholar]
- 68. Jin H, Lanning NJ, Carter-Su C. 2008. JAK2, but not Src family kinases, is required for STAT, ERK and Akt signaling in response to growth hormone in preadipocytes and hepatoma cells. Mol Endocrinol 22:1825–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ceseña TI, Cui TX, Subramanian L, Fulton CT, Iñiguez-Lluhí JA, Kwok RP, Schwartz J. 2008. Acetylation and deacetylation regulate CCAAT/enhancer binding protein β at K39 in mediating gene transcription. Mol Cell Endocrinol 289:94–101 [DOI] [PubMed] [Google Scholar]
- 70. Liao J, Hodge C, Meyer D, Ho PS, Rosenspire K, Schwartz J. 1997. Growth hormone regulates ternary complex factors and serum response factor associated with the c-fos serum response element. J Biol Chem 272:25951–25958 [DOI] [PubMed] [Google Scholar]
- 71. Chen C, Okayama H. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol 7:2745–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]