Abstract
GmACP3 from Geobacter metallireducens is a specialized acyl carrier protein (ACP) whose gene, gmet_2339, is located near genes encoding many proteins involved in lipopolysaccharide (LPS) biosynthesis, indicating a likely function for GmACP3 in LPS production. By overexpression in Escherichia coli, about 50% holo-GmACP3 and 50% apo-GmACP3 were obtained. Apo-GmACP3 exhibited slow precipitation and non-monomeric behavior by 15N NMR relaxation measurements. Addition of 4′-phosphopantetheine (4′-PP) via enzymatic conversion by E. coli holo-ACP synthase, resulted in stable >95% holo-GmACP3 that was characterized as monomeric by 15N relaxation measurements and had no indication of conformational exchange. We have determined a high-resolution solution structure of holo-GmACP3 by standard NMR methods, including refinement with two sets of NH residual dipolar couplings, allowing for a detailed structural analysis of the interactions between 4′-PP and GmACP3. Whereas the overall four helix bundle topology is similar to previously solved ACP structures, this structure has unique characteristics, including an ordered 4′-PP conformation that places the thiol at the entrance to a central hydrophobic cavity near a conserved hydrogen-bonded Trp-His pair. These residues are part of a conserved WDSLxH/N motif found in GmACP3 and it’s orthologs. The helix locations and the large hydrophobic cavity are more similar to medium- and long-chain acyl-ACPs than to other apo- and holo-ACP structures. Taken together, structural characterization along with bioinformatic analysis of nearby genes suggest that GmACP3 is involved in lipid A acylation, possibly by atypical long-chain hydroxy fatty acids, and potentially involved in synthesis of secondary metabolites.
Bacterial acyl carrier proteins (ACPs) are conserved abundant proteins that transport acyl intermediates during fatty acid synthesis (FAS) as well as lipopolysaccharide (LPS) synthesis and polyketide synthesis (PKS). Geobacter metallireducens is an anaerobic Gram-negative metal-reducing proteobacterium (1, 2) that could have important applications to bioremediation of contaminated environments. In contrast to the well-studied single ACP from Escherichia coli K-12 (3), G. metallireducens possesses at least three genes that produce ACP-like proteins, GmACP1 (gmet_1602), GmACP2 (gmet_1689) and GmACP3 (gmet_2339). Additional G. metallireducens ACPs may be present that do not have the canonical DSL motif (such as gmet_2021). GmACP1 likely serves as the primary ACP for standard FAS, with 70% sequence identity to E. coli ACP and whose gene is similarly located in the fab gene cluster encoding enzymes for FAS. Some bacteria have secondary ACPs that carry specialized fatty acids or ketides that can be either released as secondary metabolites or transferred to other molecules during LPS or polyketide biosynthesis (reviewed in (4–8)).
Most bacteria and plants, as well as eukaryotic plastids and mitochondria, possess a type II (dissociative) FAS system. They utilize a small acidic ACP to shuttle the growing fatty acid chain between component enzymes during chain elongation and modification. To enable this, the active form, holo-ACP, has a long 4′-phosphopantetheine (4′-PP) arm of ~18 Å that carries the growing acyl intermediates as thioesters attached to the terminal thiol of the 4′-PP. Conversion to holo-ACP is carried out by holo-ACP synthase (AcpS), which covalently attaches a 4′-PP moiety of coenzyme-A to the hydroxyl group of a serine residue located within a highly conserved DSL sequence via a phosphodiester bond. Following initial transfer of the acyl group from acetyl- or malonyl-CoA to the 4′-PP thiol, the remainder of the fatty acid is synthesized through a cycle of condensation reactions involving several enzymes. Movement of the flexible 4′-PP alternately shelters the intermediate within the hydrophobic core of the ACP or exposes it to enzymes for further modification.
GmACP3 was selected for study by the Northeast Structural Genomics Consortium (NESG, www.nesg.org, ID GmR141) after its gene was identified in the G. metallireducens genome, which was sequenced (2) with the support of the Department of Energy’s Microbial Genome Program (http://microbialgenomics.energy.gov). In Swiss-Prot (9) GmACP3 is annotated as a “putative acyl carrier protein”. The gmet_2339 gene encoding GmACP3 is located near genes predicted to play a role in LPS biosynthesis. LPS is the major surface component (> 90%) in the outer membrane of Gram-negative bacteria and functions to modulate interactions with the external environment. LPS contains three discrete structural domains: lipid A (the endotoxin), a core oligosaccharide (OS), and a distal polysaccharide (or O-antigen). LPS toxicity depends primarily on lipid A composition and structure, but is also influenced by the core OS region (10). Diversity within lipid A molecules aides in survival of Gram-negative bacteria in various environments and is one of the major virulence factors in Gram-negative pathogens. In E. coli, holo-ACP thioesters play an essential role in hydroxy-fatty acid (OH-FA) and FA transfer at several steps in the multi-step enzymatic synthesis of lipid A. These include primary acylation of the sugar nucleotide UDP-GlcNAc and secondary acylation of the glucosamine disaccharide intermediate, resulting in the production of hexa-acylated lipid A (6, 11). In the “prototypical” lipid A synthesis, 3-hydroxymyristoyl- (3-OH-C14)-ACP is involved in the first two primary acylation reactions, and lauroyl- and myristoyl-ACPs provide the FA for secondary acylation reactions. Diversity in lipid A can come from incorporation of different acyl groups by homologous enzymes in different bacterial strains, including addition of secondary acyl chains (6, 10, 12). Following lipid A synthesis, core OSs are added, typically 3-deoxy-D-manno-oct-2-ulosonic-acid (Kdo) and heptose derivatives. The last step is ligation of a polysaccharide O-antigen to the lipid A core. The genomic context of GmACP3 suggests an involvement in LPS biosynthesis, possibly in lipid A acylation or core OS modification.
The acylation pattern of lipid A determines the shape and specific biological activity. For example, the primary and secondary acylation pattern of lipid A can vary with the primary fatty acids typically being 3-OH-FAs with 10, 12, 14, 16, or 18 carbon chains and the secondary fatty acid chains typically being deoxy-acyl chains with an even number of carbon atoms, although they can be 2-OH derivatives, odd numbered, branched, unsaturated, long chain, or even 1-OH-FAs (reviewed in (8)). In E. coli K-12, the single ACP is involved in both primary and secondary acylation of lipid A, but in other organisms, specialized ACPs can be involved in synthesis of lipid A variants. For example, the specialized ACP, AcpXL, in certain Rhizobium bacteria transfers an unusually long OH-FA, 27-hydroxyoctacosanoic acid (27-OH-C28), to the “piggy back” or secondary position (13, 14). Interestingly, LPS with distinct long-chain-length OH-FAs, 9-hydroxypalmitic (9-OH-C16) and 10-hydroxypalmitic (10-OH-C16) acids, comprise 10–15% of the total FAs have been characterized in G. metallireducens (1, 15). In addition, these OH-FAs are found at much lower levels (0.4 – 5%) in G. Sulfurreducens or G. Bemidjiensis (15), which do not contain homologs of GmACP3, suggesting a possible role of GmACP3 in primary or secondary acylation of the lipid A.
The three-dimensional (3D) structures of many type II FAS ACPs have been determined in apo-, holo-, and acylated-forms, including those of E. coli (Gram-negative), Streptomyces coelicolor, and Bacillus subtilis (Gram-positive) (16–25). In addition, several structures of ACPs that are not in a fab gene cluster have been determined, including the ACP1 isoform from spinach (plant) (26, 27), Plasmodium falciparum (protozoan) (28), and the bacteria Mycobacterium tuberculosis (Rv0033, PDB ID 2CGQ) and Thermatoga maritima (TM0175, PDB ID 1VKU). All of the ACPs have similar structures, featuring a bundle of four α helices with the 4′-PP attachment site at the N-terminus of helix α2. Most ACP-dependent enzymes interact with the conserved helix α2, which has been called the “recognition helix” (29). It has been suggested that structural adjustments of the ACP resulting from phosphopantetheinylation and subsequent acylation might result in allosteric regulation of interactions with other enzymes. For example, PKS actinorhodin apo-ACP had higher affinity than holo-ACP for AcpS, which could be due to conformational modulation of helices α2 and α3 upon phosphopantetheinylation (30). Since ACPs are involved in scores of biosynthetic pathways and are known to interact with dozens of proteins (31), competition and regulation is important. Structural characterization of specialized ACPs is key to understanding their specificity. Here we report the high-resolution NMR structure of the specialized ACP, holo-GmACP3, that appears to have an as yet uncharacterized role in LPS biosynthesis.
EXPERIMENTAL PROCEDURES
Expression, purification, and characterization of 50% holo-GmACP3
The GmACP3 protein (NESG ID GmR141), including eight non-native C-terminal residues (LEHHHHHH), was cloned, expressed and purified using standard protocols of the NESG consortium in order to prepare [U-13C, 15N] and U-15N, 5% biosynthetically directed 13C (NC5) samples (32, 33). The gmet_2339 gene was cloned into a pET21-23C expression vector (NESG ID GmR141-21.1), which has been deposited in the PSI Materials Repository (http://psimr.asu.edu). Expression and purification data are available in the NESG SPINE database (http://spine.nesg.org/target.cgi?id=GmR141). Briefly, [U-13C, 15N] and NC5 GmACP3 were expressed in E. coli strain BL21 (Gold DE3) grown in MJ9 medium (34) by induction with IPTG, and purified on a Ni-affinity column (HisTrap HP, 5 ml) followed by a gel filtration column (Superdex 75 26/60, GE Healthcare). The chromatography buffer was 50 mM Tris, 500 mM NaCl, 40 mM imidazole, 1 mM TCEP, pH 7.5, and the sample was eluted in the same buffer containing 500 mM imidazole. The protein was buffer-exchanged and concentrated by centrifugation to 0.8 mM in NMR buffer: 90% H2O/10% D2O solution containing 20 mM MES, 100 mM NaCl, 5 mM CaCl2, 10 mM dithiothreitol, 0.02% NaN3, at pH 6.5. Incubation with DTT converted any acyl-GmACP3 to holo- GmACP3. Analytical static light scattering measurements in-line with gel-filtration chromatography confirmed that the protein was monomeric in solution, although eluting sooner than expected (apparent MW 14.8 kDa). Sample purity was verified by SDS-PAGE with a single strong band (~10 kDa, > 95% purity). Biosynthetic incorporation of 4′-PP was determined to be ~25% for unlabeled GmACP3 grown in LB medium by electrospray mass spectrometry deconvoluted integrated peak intensities (The Ohio State University, Mass Spectrometry and Proteomics Facility). The masses 9876 and 10216 Da corresponded to apo- and holo–GmACP3 with the N-terminal methionine cleaved (as expected) and an additional 340 Da from 4′-PP on the holo-GmACP3 (Figure S1 of the Supporting Information). Biosynthetic incorporation of 4′-PP was determined to be ~30% in unlabeled GmACP3 and ~50% labeled in NC5 GmACP3 by 12 T Fourier transform ion cyclotron resonance (FTICR) mass spectrometry (Pacific Northwest National Laboratory).
Overexpression and Purification of E. coli AcpS
The p15Tv-L expression vector containing the gene encoding AcpS from E. coli K12 was transformed into competent cells of expression host E. coli BL21 (DE3). AcpS was expressed and purified based on the protocol of Lambalot and Walsh (35). A 1 L culture of transformed cells was grown at 37°C with vigorous shaking to an A600 nm of ~0.8 in LB medium supplemented with 30 μg/ml ampicillin. Protein overexpression was induced at 30°C with 100 μM IPTG, and cultures were grown for an additional 3 h before harvesting cells by centrifugation. Pelleted cells were resuspended in 25 ml of lysis buffer (50 mM Tris, 10 mM MgCl2, 5% glycerol, pH 8.0) and lysed by three passes through a French pressure cell (SLM Instruments). The resulting lysate was spun at 24,000 x g for 60 min. The supernatant was loaded onto a 10 ml Ni-NTA affinity column (Qiagen) and washed with 50 ml of lysis buffer containing 30 mM imidazole. The His6-tagged AcpS was eluted from the column with lysis buffer containing 300 mM imidazole, resulting in a final protein concentration of 21 μM.
Enzymatic conversion of 50% holo-GmACP3 to >95% holo-GmACP3
To prepare samples of >95% [U-13C, 15N] and NC5 holo-GmACP3, the previously described mixture of apo- and holo-GmACP3 was incubated with E. coli AcpS for Coenzyme A (CoA) and Mg+2 –dependent conversion. The reactions contained 10 mM MgCl2, 5% glycerol, 300 μM CoA, 357 μM GmACP3, 2.1 μM AcpS, and 50 mM Tris, pH 8.0, in a total volume of 10 ml. Reactions were incubated overnight at room temperature and conversion to holo-GmACP3 was confirmed by mass spectrometry (Bruker Autoflex III MALDI-TOF). Samples were then buffer-exchanged to NMR buffer and concentrated by centrifugation to 1 mM GmACP3. The >95% [U-13C, 15N] holo-GmACP3 contained 50% 13C- and 15N-labeled 4′-PP since isotopic enrichment was dependent on the amount of biosynthetic incorporation.
NMR Spectroscopy
NMR data were collected on both 50% and >95% holo-GmACP3 samples of about 250 μl in 5 mm Shigemi NMR tubes at 20°C on 600 MHz Varian Inova and 850 MHz Bruker Avance III NMR spectrometers. D2O-exchanged samples were prepared by freezing [U-13C, 15N] samples, followed by overnight lyophilization and resuspension in 99.9% D2O (Acros Organics). Spectra were processed with NMRPipe (36) and analyzed with Sparky 3.110 (Goddard, T. D. and Kneller, D. G.)
Average 15N relaxation times were determined from 15N-edited 1D experiments recorded at 600 MHz (37) by integration from 8.5 to 10.5 ppm and fit to an exponential decay. Longitudinal T1 relaxation delays were 100, 200, 300, 400, 700, 1000, 1500, and 2000 ms; transverse T1ρ relaxation delays were 10, 30, 50, 70, 110, 130, 150, and 170 ms; both experiments had 1.5 s recycle delays. The overall isotropic rotational correlation time, τc, was approximated using the equation (derived from (38)): , where υN is the 15N frequency in Hz. 15N{1H} heteronuclear NOE values were determined from the ratio of the intensities of the respective cross-peaks in 1H-15N correlation spectra obtained with and without 3 s of proton saturation and using a total 5 s recycle delay for both (37).
Holo-GmACP3 chemical shifts for 1H, 13C and 15N were determined from conventional triple-resonance spectra (39) including stereospecific Val and Leu assignments determined from the NC5 sample (40). Apo-GmACP3 chemical shifts for 20 amide crosspeaks observed as doubled resonances in the 50% holo-GmACP3 sample were also assigned by standard methods and by comparison with the >95% holo-GmACP3 1H-15N HSQC spectrum (Figure S2 of the Supporting Information).
Amide backbone residual dipolar coupling (RDC) data was collected for isotropic and two aligned samples of 1 mM >95% NC5 holo-GmACP3 in NMR buffer using NH J-modulation experiments to measure the one bond 1H-15N couplings (41). The first sample was aligned in a negatively charged compressed 7% polyacrylamide gel (42). Specifically, equal amounts of acrylamide and 2-acrylamido-2-methyl-1-propanesulfonic acid were polymerized in a 3.2 mm inner diameter plastic tube as described in (42). The gel was extensively washed with deionized water, NMR buffer to equilibrate the pH, and again with deionized water. The swelled gel (~7 mm diameter) was trimmed to a length of 35 mm and dried in open air for two days. The dry gel was swollen in a 5 mm Shigemi NMR tube with 240 μl protein solution in NMR buffer with 400 mM NaCl and compressed with the upper plunger to a height of 14 mm. A second sample was aligned in a neutral stretched 5% polyacrylamide gel prepared by polymerizing a mixture of acrylamide and N, N′-methylenebisacrylamide (19:1) utilizing a two-stage NMR tube (43). The gel was soaked in 400 μl of protein solution until it was absorbed. The gel was then sucked into the upper portion of the tube (4.2 mm inner diameter) for the isotropic measurements and then into the narrow portion (2.8 mm) for the aligned measurements in the stretched gel.
Structure determination and refinement of holo-GmACP3
Preliminary structures of holo-GmACP3 devoid of 4′-PP were calculated with CYANA 2.1 (44) using resonance assignments, NOESY peak lists from four NOESY spectra (τm = 70 ms), and dihedral restraints derived from TALOS+ (45) as input. NOE assignments were examined and manually evaluated. Intermediate structures were used to identify consistently or egregiously violated NOE restraints, which were then subjected to manual assessment, to end up with the final NOE restraint list. Subsequently, NOE restraints for 4′-PP were manually added and structures calculated with Xplor-NIH (version 2.25, (46)). Molecular parameter and topology files were created for the 4′-PP prosthetic group. Atom names were taken from the HIC-UP library with protons given the corresponding names to the heavy atoms. The Xplor patch feature was used to covalently attach 4′-PP to the Oγ of S36 in the protein structure file (in mkpsf.inp). For the final NMR ensemble, 30 structures were calculated with a standard simulated annealing protocol followed by refinement using the Xplor+ protocol (39), including refinement against the two sets of NH RDCs, and the 20 structures with the least constraint violations were selected.
Coordinates and NMR-derived constraints for the ensemble of 20 NMR structures of holo-GmACP3 were deposited in the Protein Data Bank with PDB ID 2KWM and the chemical shifts for the protein and 4′-PP, along with the NOESY FID and peak list data, were deposited in the BioMagResBank (University of Wisconsin-Madison) with BMRB code 16081.
RESULTS
Analysis of the GmACP3 sequence family
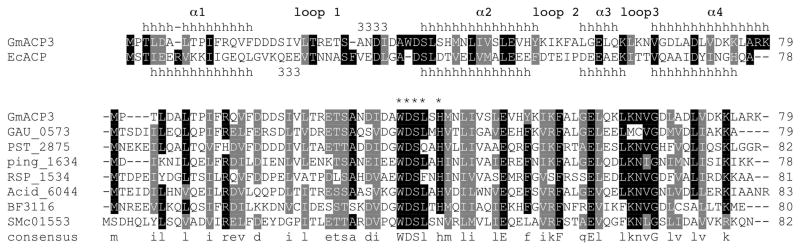
GmACP3 is a small acidic ACP (~10 kDa, pI 5.2) with no significant sequence similarity to the well-characterized E. coli ACP and whose gene is not found in the fab gene cluster. Dozens of Gram-negative bacteria contain a homolog of GmACP3 with greater than 40% sequence identity, identified with Blast (47). Multiple sequence alignment of GmACP3 with the nine closest homologs identified by a Blast search of the KEGG database (48) is shown in Figure 1. These include other soil bacteria such as Pseuodomonas stutzerii, which is also an antibiotic-resistant pathogen, and the pathogenic Bacteroides fragilis that is a frequent cause of clinical infections in hospitals. Several other pathogenic bacteria, including Vibrio cholerae, contain a homolog with less sequence similarity (Table S1 of the Supporting Information). GmACP3 homologs contain a conserved consensus sequence WDSLxH/N (Table S1 of the Supporting Information), which includes the DSL motif containing the serine that is the site of 4′-PP modification in all characterized ACPs. However, the conserved tryptophan and histidine (sometimes asparagine) are unique to this family. Interestingly, most bacteria with GmACP3 homologs also contain an ACP in a fab gene cluster that is more similar to the E. coli ACP. Therefore, the specialized function of GmACP3 orthologs is likely to be distinct from standard fatty acid synthesis.
FIGURE 1.
The GmACP3 sequence family. The aligned sequences and secondary structure from E. coli ACP (from PDB ID 2FAC-A) and G. metallireducens GmACP3 are shown. The nine most similar sequences to GmACP3 reported by a BLAST search of the KEGG database (48) are aligned. KEGG IDs are given for bacterial homologs from Gemmetimonas aurantiaca (GAU_0573), Pseudomonas stutzeri (PST_2875), Psychromonas ingrahamii (Ping_1634), Rhodobacter sphaeroides (RSP_1534), Solibacter usitatus (Acid_6044), Bacteroides fragilis (BF3116), and Sinorhizobium meliloti (SMc01553). Alignments were created with ClustalW2 (71). The consensus sequence “WDSLxH/N” is indicated by asterisks (*).
Analysis of possible Gmet_2339 function from genomic context
According to the DOOR web resource (Database for prOkaryotic OpeRons) (49), the gene encoding GmACP3, gmet_2339, is found in an operon with twenty-four open reading frames transcribed in the same direction (Operon ID 165016), from gmet_2337 to gmet_2360 (Table 2). This operon includes genes encoding E. coli homologs involved in lipid A and core OS biosynthesis (LpxA, LpxB, LpxD, LpxK, WaaA (formerly KdtA), WaaF, WaaQ). The E. coli LpxA ortholog, required for the first step of lipid A synthesis, is the most conserved with 50% identity. In addition, there are orthologs of E. coli proteins LpxB, LpxD, for lipid A synthesis, as well as MsbA (> 30%), the essential ABC transporter for lipid A export. LpxK phosphorylates the disaccharide backbone during lipid A synthesis and WaaA transfers Kdo residues to the nascent LPS. Other steps in inner core synthesis involve WaaF and WaaQ that are heptosyltransferases for addition of HepII, HepIII, respectively. Additional glycosyl transferases are involved in inner core modifications and outer core assembly. However, nothing is known about possible roles of ACP-mediated acylation in core oligosaccharide or distal polysaccharide assembly. In E. coli the two acyltransferases, LpxA and LpxD, transfer acyl groups from hydroxymyristoyl-ACP to each GlcNAc at two positions in a process referred to as primary acylation. After formation of the lipid A disaccharide, the resulting tetra-acylated lipid A can be further esterified at the 3-OH position on the primary fatty acid by secondary fatty acids, resulting in penta- or hexa-acylated lipid A. In E. coli, secondary acylation by so-called “late” acyltransferases (LpxL, LpxM and LpxP), which utilize acyl-ACP exclusively as donors, results in formation of the final lipid A. Since both primary and secondary acylation require acyl-ACPs, it seems likely that the specialized ACP, GmACP3, could be involved in synthesis of lipid A with unusual fatty acids. Given that the E. coli acyltransferases LpxL, LpxM and LpxP are not well conserved in G. metallireducens (all < 26% identity to the protein encoded by gmet_0091), alternate secondary acylation may involve other acyltransferases.
Table 2.
Neighboring genes of gmet_2339 from G. metallireducens.
| gmet gene no. | E. colia KEGG ID | %I | E. coli gene | Function of gene product b |
|---|---|---|---|---|
| 2337 | b3910 | 31% | yiiM, | GDSL-like lipase/acylhydrolase |
| 2338 | None | Membrane bound O-acyltransferase, MBOAT | ||
| 2339 | None | Acyl carrier protein | ||
| 2340 | b4069 | 34% | acsA | AMP-dependent acetyl-CoA synthetase and ligase |
| 2341 | b3632 | 31% | waaQ | glycosyl transferase (heptosyltransferase III) |
| 2342 | b3615 | 29% | yibD | glycosyl transferase |
| 2343 | b1994 | 22% | yeaP | (NodZ) diguanylate cyclase or fucosyl transferase |
| 2344 | b3615 | 28% | yibD | beta-1,4-glucosyltransferase |
| 2345 | b3620 | 32% | waaF | LPS heptosyltransferase II |
| 2346 | b0917 | 42% | ycaR | hypothetical |
| 2347 | b0915 | 38% | lpxK | lipid A disaccharide kinase |
| 2348 | b3633 | 37% | waaA | Kdo transferase |
| 2349 | b0914 | 38% | msbA | inner membrane ABC-transporter |
| 2350 | b0182 | 33% | lpxB | lipid A disaccharide synthase |
| 2351 | b2253 | 35% | arnB | L-AraN-modification of lipid A |
| 2352 | b1624 | 32% | ydgJ | oxidoreductase |
| 2353 | b0181 | 50% | lpxA | UDP-GlcNAc acyltransferase (AT) |
| 2354 | b0180 | 50% | fabZ | 3R-hydroxymyristoyl ACP dehydratase |
| 2355 | b0179 | 37% | lpxD | UDP-3-O-(3-hydroxymyristoyl)-GlcN-AT |
| 2356 | b0178 | 23% | skp | Outer membrane chaperone Skp (OmpH) |
| 2357 | b0177 | 28% | bamA | surface antigen (D15) |
| 2358 | b1117 | 52% | lolD | ABC transporter-related protein |
| 2359 | b1118 | 32% | lolE | lipoprotein releasing system transmembrane protein |
| 2360 | b2890 | 59% | lysS | lysyl-tRNA synthetase |
The E. coli homolog with the highest sequence similarity is listed along with the KEGG ID, the percent identity (%I), and the E. coli gene name.
The possible function is derived from KEGG annotation for G. metallireducens or E. coli genes.
In fact, the neighboring gmet_2338, is annotated as a membrane-bound O-acyl transferase (MBOAT) and is located in a gene cluster from gmet_2337 to gmet_2339, according to the KEGG database (48). Typically, MBOATs transfer fatty acyl chains to hydroxyl groups of membrane bound targets. The proteins encoded by gmet_2337 and gmet_2338 (Gmet_2337 and Gmet_2338) share sequence similarity with AlgJ and AlgI proteins identified in Pseudomonas aeruginosa involved in O acetylation of the polysaccharide alginate (26% and 38% identity, respectively). It seems probable that Gmet_2338 could transfer an acyl chain from GmACP3 to lipid A during synthesis of a specialized lipid A molecule. Alternatively, the role could involve acylation of sugars in the LPS backbone, the inner core, or even the polysaccharide outer core.
Although it is suggested that the MBOAT protein Gmet_2338 and possibly Gmet_2337 might transfer the acyl chain from GmACP3 to LPS, there is not a nearby enzyme for loading the initial acyl group onto the holo-GmACP3. In E. coli FAS, the malonyl-CoA:ACP transacylase (FabD) is responsible for synthesis of malonyl-ACP and the transacylase activity of FabH is responsible for synthesis of acetyl-ACP (reviewed in (7)). Presumably the same enzymes that load acyl chains onto holo-GmACP1 could also act on holo-GmACP3.
Several bacteria have a two-gene cluster similar to the one involving gmet_2338 and gmet_2339 and several have a cluster with genes homologous to gmet_2337 and gmet_2338. However, no other bacteria have been identified with a gmet_2339-like gene located near genes for LPS synthesis. Additional discussion of possible functions for gmet_2337 and gmet_2340 is available in Text S1 of the Supporting Information.
Holo-GmACP3 structure
NMR samples of >95% holo-GmACP3 were obtained by enzymatic conversion of mixed apo- and holo-GmACP3 samples following purification from E. coli and 4′-PP incorporation was confirmed by mass spectrometry. Monomeric behavior was confirmed by gel-filtration and correlation time estimates based on 15N relaxation measurements. The average τc of 4.8 ns and T1ρ of 113 ms are typical values expected for a monomer of about 10 kD. Chemical shift assignments were 97.8% and 96.2% complete for backbone and sidechain resonances of [U-13C, 15N]–holo-GmACP3, respectively. There was no evidence of conformational exchange in the manner of peak doubling or missing resonances for either the protein backbone or the 4′-PP moiety. The 15N{1H} heteronuclear NOE values are given in Figure S3 of the Supporting Information, including values for the NH36 and NH41 amide bonds of the 4′-PP. The values for the 4′-PP amides (0.67 and 0.74, respectively) were positive and of similar magnitude to those measured for amides in the holo-GmACP3 backbone (0.78 ± 0.08, average for residues 4–79), indicating that the 4′-PP has a rigid conformation and that NOEs could be used to determine a conformationally restricted structure. Due to the 50% isotopic enrichment of 4′-PP and the similarity of 4′-PP to a small peptide, multidimensional NMR experiments provided data that allowed assignment of the attached 4′-PP prosthetic group. Chemical shifts for 4′-PP (Figure 2) indicated interactions with the protein affecting the chemical environment of the 4′-PP nuclei. For example, the two methyl groups and two amide groups had unusually shielded chemical shifts. There were 23 unambiguous NOEs identified between GmACP3 and 4′-PP (Table S2 of the Supporting Information) as well as 21 intra-4′-PP NOEs.
FIGURE 2.
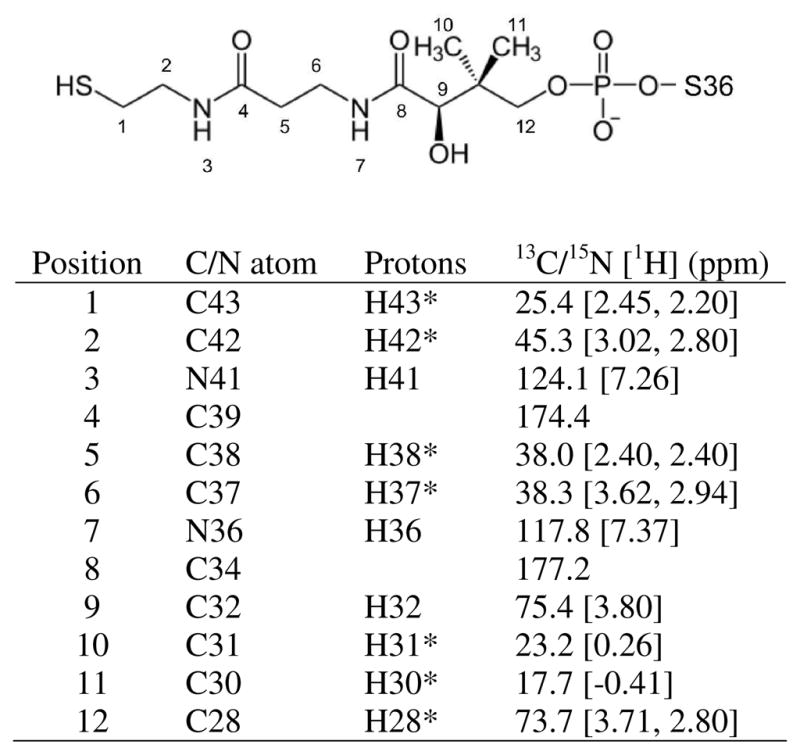
4′-PP chemical shifts. NMR assigned resonances in 4′-PP in holo-GmACP. Position of 13C and 15N atoms are shown as labeled on the structure of 4′-PP. The corresponding 13C and 15N atom names and the attached proton names are given in Xplor format. Methylene and methyl protons were not stereospecifically assigned and are named with an asterisk (*). Methyl groups with C30 and C31 were stereospecifically assigned based on NOEs.
The NMR solution structure of holo-GmACP3 features a right-twisted four-helix bundle with the helices in an up-down-up-down topology. The NMR ribbon and ensemble diagrams are shown in Figure 3A, B and the structural statistics are given in Table 1. Helix α2 (S36-Y50) is parallel and similar in length to helix α4 (V65-K79). On one side of this pair, the slightly shorter helix α1 (T3-F15) is antiparallel to helices α2 and α4. On the opposite side of helices α2 and α4 is helix α3 (L56-Q60), which is notably shorter in length and perpendicular to helix α1. All four helices are connected by loop regions of variable length, with the longest, loop 1 (20 residues, D16-D35), occurring between helices α1 and α2. The four-helix bundle is stabilized by a hydrophobic core created by interactions of interior aliphatic side chains. Additionally, the inter-helical loop regions are stabilized by electrostatic and hydrophobic interactions that cause them to be well defined in the NMR ensemble. This is the case for loop 1 that contains one turn of a 310 helix (A28-I31) and is stabilized by backbone hydrogen bonds to the N-terminus of helix α4 (V65 HN to T26 CO and G66 HN to R24 CO, Figure 3A).
FIGURE 3.
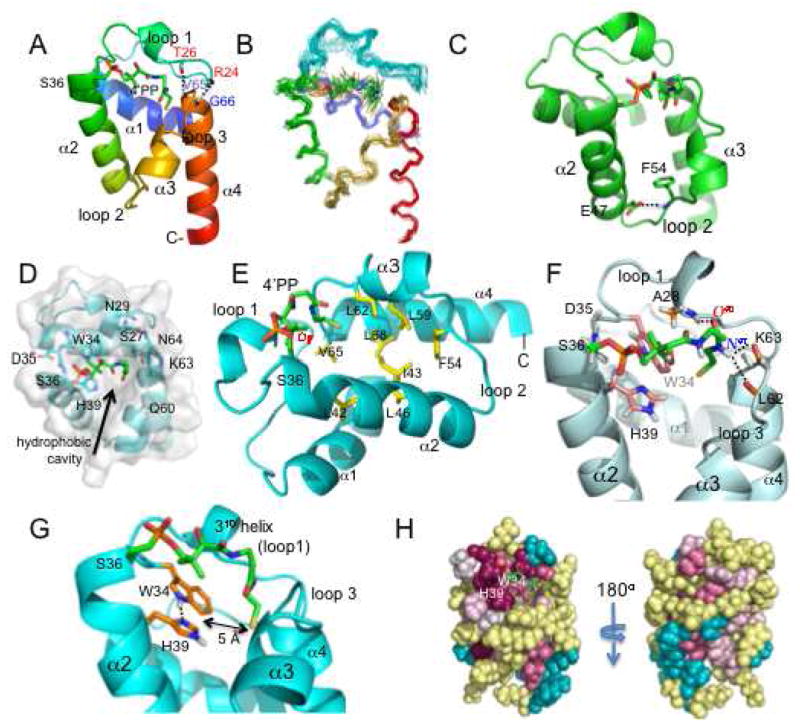
Structure of holo-GmAPC3. A. Ribbon diagram of representative structure is shown with backbone hydrogen bonds between loop 1 and helix 4 indicated. B. Backbone diagram of holo-GmACP3 ensemble, including the 4′-PP covalent modification. C. GmACP3 structure with the hydrogen bond between F54 HN and E47 side chain across loop 2 indicated. D. View into the hydrophobic cavity of holo-GmACP3 representative structure (top view), showing the side chains and backbone of residues at the opening of the cavity. E. Key residues in the hydrophobic cavity are shown. F. Detailed interactions between 4′-PP and GmACP3 (representative structure). Hydrogen bonds are indicated and all GmAPC3 protons with NOEs to 4′-PP atoms are shown. G. View of the hydrogen-bonded W34 and H39 and the distance to the 4′-PP sulfur atom. H. ConSurf (54) images, front view (same orientation as A) and back view. Magenta is highly conserved and cyan is variable. All structure figures in this paper were created with PyMol.
Table 1.
Summary of NMR and Structural Statistics for holo-GmACP3 a
| Completeness of resonance assignments b | ||
| Backbone | 97.8% | |
| Side chain | 96.2% | |
| Aromatic | 100% | |
| Stereospecific methyl (%) | 100% | |
| Conformationally restricting restraints c, d | ||
| NOE restraints | ||
| Intra-residue (i = j) | 315 | |
| Sequential (|i − j| = 1) | 250 | |
| Medium range (1 < |i − j| < 5) | 286 | |
| Long range (|i − j| ≥ 5) | 261 | |
| NOE restraints/residue d | 13.9 | |
| Restraints for 4′-PP | ||
| Intra - 4′-PP | 21 | |
| 4′-PP - protein | 23 | |
| Total | 1156 | |
| Dihedral angle restraints | 102 | |
| Hydrogen bond restraints | 52 | |
| Total restraints | 1310 | |
| Number of restraints/residue d | 15.8 | |
| Number of long range restraints/residue d | 3.3 | |
| Residual restraint violations c | ||
| Average distance restraint violations/structure | ||
| 0.1 – 0.2 Å | 5.5 | |
| 0.2 – 0.5 Å | 1.8 | |
| > 0.5 Å | 0 | |
| Average RMS of distance violation/restraint | 0.02 Å | |
| Maximum distance violation | 0.44 Å | |
| Average RMS dihedral angle violations/structure | ||
| > 1–10° | 0.6 | |
| > 10° | 0 | |
| Average RMS dihedral angle violation/restraint | 0.2° | |
| Maximum dihedral angle violation | 3.1° | |
| RMSD from the average coordinates c, e, f | ||
| Backbone atoms (N, Cα, C′) | 0.4 Å | |
| Heavy atoms | 1.0 Å | |
| RMSD from ideal geometry f | ||
| Bond length | 0.012 Å | |
| Bond angles | 1.5° | |
| Procheck Ramachandran plot statistics c, e | ||
| Most favored regions | 94.6% | |
| Additionally allowed regions | 4.8% | |
| Generously allowed regions | 0.5% | |
| Disallowed regions | 0.0% | |
| Global Quality Scores c | Raw | Z-score |
| Procheck G-factor (φ/ψ) d | 0.4 | 1.7 |
| Procheck G-factor (all dihedral angles) c | 0.3 | 1.8 |
| Verify3D | 0.3 | −3.2 |
| ProsaII | 0.1 | −2.3 |
| MolProbity clashscore | 15.1 | −1.1 |
| RPF Scores g | ||
| Recall | 0.96 | |
| Precision | 0.94 | |
| F measure | 0.95 | |
| DP score | 0.80 | |
Statistics were computed for the ensemble of 20 deposited structures (PDB ID 2KWM).
Computed for residues 2–82, using AVS software (68). Resonances that were not included were exchangeable protons (N-terminal NH3+, Lys NH3+, Arg NH2, Cys SH, Ser/Thr/Tyr OH) and Pro N, C-terminal carbonyl, side chain carbonyl and non-protonated aromatic carbons.
Calculated using PSVS 1.4 program (69). Average distance constraints were calculated using the sum of r−6.
For 80 residues with conformationally-restricting restraints, residues 2–81.
Ordered residues ranges: 2–16, 27–32, 39–60, 62–79, with the sum of φ and ψ order parameters > 1.8.
RMS, root mean square; RMSD, RMS deviation.
RPF scores (70) reflecting the goodness-of-fit of the final ensemble of structures (including disordered residues) to the NOESY data and resonance assignments.
Loop 2 (K51-A55) is stabilized by a hydrogen bond between F54 HN in loop 2 and the side-chain COO− of E47 in helix α2 (Figure 3C). The distance between these hydrogen bond donor and acceptor atoms is less than 3 Å in most structures. The hydrogen bond is supported by NOEs and the deshielded F54 HN resonance (9.61 ppm). Furthermore, the unusually deshielded resonance of one of E47 Hγs (3.09 ppm) can be explained by the influence of the nearby aromatic F54. It has been proposed that the comparable hydrogen bond observed in PfACP (E48 COO- to I55 HN) is responsible for the favorable movement of helix α3 in order to expand the hydrophobic cavity (50). Upadhyay, et al. (50) hypothesize that this hydrogen bond would be disrupted by insertion of an acyl chain into the cavity, allowing for expansion of the cavity via an outward movement of helix α3. E47 is conserved among the GmACP3 orthologs and F54 is conserved as Phe or Leu.
The hydrophobic pocket is a key feature of FAS ACPs, and has been shown to sequester the growing fatty-acid chain. The pocket in GmACP3 grows wide near the exterior of the protein and its opening is almost entirely surrounded by amino acids containing polar side-chains (S27, N29, W34, D35, S36, H39, K63, and N64) or by polar backbone atoms (S27, W34, D35, S36, Q60, K63, and N64) (Figure 3D). The arch-like structure of the 4′-PP results in the placement of its terminal thiol at the entrance of a deep hydrophobic pocket formed by conserved residues (L42, I43, L46, F54, L59, L62, V65, and L68) with F54 at the bottom of the cavity (Figure 3E). The 4′-PP moiety is covalently bound to the Oγ of S36, located in the conserved WDSLxH/N sequence. The 4′-PP is tethered by a phosphodiester bond to S36 at the N-terminus of helix α2 and extends across the “top” of the four-helix bundle making contacts with the 310 helix in loop 1 and with residues in loop 3 (connecting helices α3 and α4) (Figure 3F). Stabilization of the position of 4′-PP results from hydrophobic interactions with A28 and W34 and hydrogen bonding from 4′-PP O40 to A28 HN and from the 4′-PP H41 to the nearby L62 and K63 COs, observed in more than half the structures in the NMR ensemble.
Of special note is the alignment of two residues, W34 and H39, at the entrance to the hydrophobic cavity (Figure 3G). The WDSLxH/N motif is conserved among GmACP3 homologs and may be important for the function and/or local structural integrity in these ACPs. The side chains of W34 and H39 are well ordered and have a side-by-side planar orientation due to hydrogen bonding between the H39 Nδ1 and Hε1 of W34. This is supported by NOEs between the side-chain protons as well as the deshielded W34 Hε1 resonance (12.92 ppm). In addition, the deshielded Nδ1 resonance (254.0 ppm) and the pattern observed in the 3JNCH 1H-15N-HSQC centered on the histidine region (51) demonstrates that the Nε2-protonated neutral tautomer of H39 is favored at pH 6.5 and indicates a lower pKa compared with the His6 tag (Figure S4 of the Supporting Information). The H39 resonance of Hε2 was observed at 11.10 (typically too labile to be observed), which further supports this tautomer. Both W34 and H39 have NOEs to the methyl protons (H30* and H31*) in 4′-PP and are less than 5 Å from the sulfur atom of 4′-PP in most structures in the ensemble (Figure 3F). They are poised to interact with and stabilize the 4′-PP or acyl group in thioester intermediates of acyl-GmACP3 as the acyl chain is sequestered in the hydrophobic cavity. The Hε2 of H39 is available for hydrogen bonding to the carbonyl oxygen of the thioester and the side chain of W34 is available for hydrophobic interactions with the acyl chain or 4′-PP methyl groups. It has been similarly hypothesized that an active site histidine of FabB forms a hydrogen bond with thioester carbonyl based on the structure of FabB complexed with the malonyl-ACP mimic thiolactomycin (52, 54). Alternatively, the H39 Hε2 may hydrogen bond to OH or CO oxygen atoms in 4′-PP, in a similar manner to the T39 side chain in E. coli ACP (22), or possibly to hydroxy oxygens in OH-FAs. Analysis with ConSurf (54) revealed a large patch of conserved surface residues within and around the hydrophobic cavity (Figure 3H). The conserved residues W34 and H39 form a large portion of the surface at the entrance to the hydrophobic cavity. One side of the opening, formed by loop 1 and the N-terminus of helix α2, features the conserved surface patch, while the opposite side is comprised of variable residues.
Characterization of apo-GmACP3 in the 50% holo-GmACP3 sample
A mixed sample of apo- and holo-GmACP3 was obtained after standard E. coli expression and purification and characterized by mass spectrometry as 50% apo-protein. There were 20 doubled HN resonances and 3 sets of double NH2 resonances in the 1H-15N HSQC spectrum indicative of different chemical environments of the nuclei in the apo- versus holo-protein. Peaks belonging to the holo-protein were confirmed after assignment of the >95% holo-GmACP3. In the 15N-edited NOESY spectrum, apo-protein resonances had weaker NOEs than holo-protein resonances although there were no obvious NOE differences. For this reason, it was not possible to obtain a unique structure for apo-GmACP3. After conversion to > 95% holo-protein, there was just a single set of peaks.
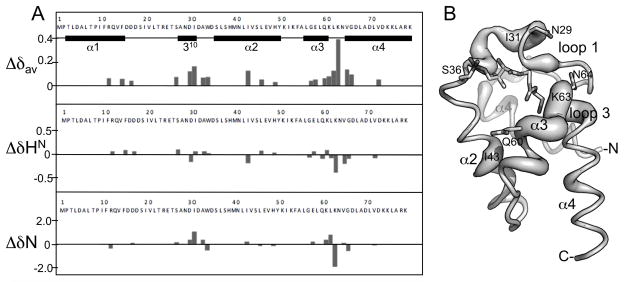
Chemical shifts differences between HN resonances for the apo- and holo-protein samples are plotted in Figure 4A. Doubled resonances included HNs of R12, F15, D17, S27, D30, I31, A33, W34, I43, L46, H49, G57, E58, Q60, K61, L62, K63, V65, G66, V72 and NH2s of N29, Q60, and N64. These residues are located in all four helices and in the two loops nearest to 4′-PP (loop 1 and loop 3) (Figure 4B). Resonances with the largest differences occurred in the loops nearest to 4′-PP (I31, K63, and nearby residues) (Figure 4B). Interestingly, we did not observe doubled resonances for D35, S36, or L37, the conserved residues that have often showed the largest chemical shifts difference between apo- and holo-conformations (19, 28, 55). For peaks that were not doubled, it was not possible to determine whether the apo-protein crosspeak was overlapped with the holo-protein crosspeak or if the apo-protein peak was invisible. However, since the resonances for D35, S36, and L37 in the apo-protein would be expected to be shifted significantly from those observed in the holo-protein, it is more likely that they were not observable in the apo-protein due to chemical exchange or conformational exchange broadening.
FIGURE 4.
Chemical shift changes in GmACP3 upon addition of 4′-PP. A. The chemical shift differences between 1HN and 15N resonances in holo- and apo-GmACP3 for doubled resonances are plotted as the weighted average (72), Δδav = {1/2[(ΔδHN)2 + (ΔδN/5)2]}1/2, ΔδHN = δHNapo − δHNholo and, ΔδN = δNapo − δNholo. B. Worm representation of GmACP3 scaled by the δΔav values. The side chains of N29, Q60, and N64, whose NH2 resonances also shifted, are shown.
There were two indications that apo-GmACP3 was unstable compared to holo-GmACP3. First, 15N nuclear relaxation measurements on the 50% apo-sample indicated an average τc of 11.4 ns and T1ρ of 50 ms, numbers that would be expected for a dimer of about 20 kDa. This means that apo-GmACP3 is either dimeric, or undergoing aggregation and/or chemical exchange. In addition to this non-monomeric behavior, the apo-protein peaks slowly disappeared from 1H-15N HSQC spectra over several weeks (data not shown) and white precipitation appeared. These results, taken together with observed HN peak doubling of resonances for residues nearest 4′-PP, indicated that 4′-PP covalent modification stabilized the holo-GmACP.
DISCUSSION
The structure of holo-GmACP3 is different compared to the structures of other holo-ACPs in that the 4′-PP is well ordered in the hydrophobic pocket as defined by many NOEs and hydrogen bonds. In contrast, in other ACP structures, few or no NOEs were observed between 4′-PP and the protein, including cases where holo-ACP was acylated (16, 19, 25, 28, 30, 50, 55–58). GmACP3 is also different from other characterized ACPs in that apo-GmACP3 was unstable and exhibited non-monomeric behavior (19, 30, 55, 58). Enzymatic addition of 4′-PP to GmACP3 resulted in a stable holo-protein that had an ordered 4′-PP conformation, allowing a detailed structural analysis of the interactions between 4′-PP and GmACP3. Chemical shifts for 4′-PP from other structures were typical of “unperturbed” values indicating only weak or transient interactions with the protein (28, 30, 57, 59, 60). The proton chemical shifts in these other cases are comparable to “free” 3,5-dioxo-hexyl-coenzyme A (see supplementary data for (60)). The holo-GmACP3 4′-PP chemical shifts are remarkable in that most are significantly shielded compared to the “free” values due to strong interactions between 4′-PP and GmACP3.
AcpS recognition
It was surprising to discover that GmACP3, which has < 25% sequence identity with E. coli ACP, was enzymatically converted to holo-GmACP3 by E. coli AcpS both in vivo and in vitro. It has been noted in previous studies that E. coli AcpS was capable of modifying ACP from spinach and M. tuberculosis (AcpM, Rv2244), but these are more similar to E. coli ACP (44% and 38% identical, respectively) (7, 55, 59). Residues identified in E. coli ACP for binding to AcpS (by comparison with B. subtilis ACP in complex with AcpS) are not well conserved in GmACP3 with the exception of the invariant DSL motif. The AcpS-binding residues of E. coli ACP are acidic residues D35, D38, E41, E48, D56, and E60 and the hydrophobic residue L37 and M44, all found in or near helices α2 and α3 (18). In GmACP3, the only acidic residues that are conserved in helices α2 and α3 are D35, E47 and E58.
Structural comparison of holo-GmACP3 and E. coli acyl-ACPs
A 3D structure similarity search using DALI (Distance matrix ALIgnment Server) (61), found many ACP structures that have a similar four-helix bundle geometry (Table S3 of the Supporting Information). Acylated E. coli X-ray crystal structures were among the best matches. The helix conformations in E. coli ACPs bound to long chain acyl intermediates were more similar to GmACP3 than the E. coli ACP apo-conformation, with the Cα RMSD decreasing from 1.8 Å for apo-structures (1LOH-A, 1T8K) to 1.6 Å for hexanoyl-, heptanoyl-, and decanoyl-structures (2FAC, 2FAD, 2FAE) (Figure 5A). RMSDs are reported in Figure 7. Interestingly, the RMSD with the E. coli acyl-ACPs bound to cytochrome P450 (3EJB, 3EJD, 3EJE) was only 1.4 Å. Cytochrome P450 is an enzyme from the biotin operon of B. subtilis that catalyzes synthesis of the 7-carbon pimelic acid, a precursor to biotin (24). Helices α1 and α3 are furthest from helix α2 in this bound conformation with a resulting expanded hydrophobic cavity (Figure 5A). In this conformation of the E. coli acyl-ACP, as in the holo-GmACP3 structure, the hydrophobic cavity is unoccupied by an acyl chain. This is because the 4′-PP-attached thioester is flipped out of the ACP and into the active site of P450.
FIGURE 5.
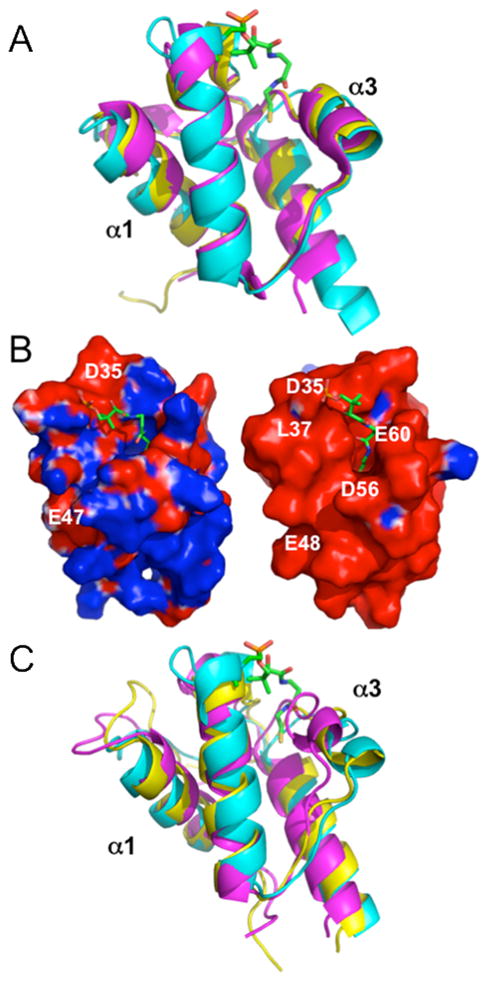
Comparison of GmACP3 with other ACPs. A. Structural comparison, highlighting the differences in helices α1 and α3 between holo-GmACP3 (cyan), E. coli heptanoyl-ACP (PDB ID 2FAD-A, magenta), and P450-bound E. coli tetradecanoyl-ACP (3EJB-C, yellow). B. Electrostatic surface potential diagrams (73) colored by electrostatic potential (−1 kT/e, red to +1 kT/e, blue) comparison between holoGmACP3 (left), E. coli heptanoyl-ACP (2FAD-A, right). C. Overlay, highlighting the differences in helices α1 and α3 between holoGmACP3 (cyan), S. coelicolor acetyl-actACP (2KG6, magenta), and S. coelicolor octanoyl-actACP (2KGC, yellow). Acyl chain and 4′-PP modifications are not shown for E. coli or S. coelicolor homologs.
In the X-ray crystal structure of E. coli heptanoyl-ACP, two alternate 4′-PP conformations were observed with molecule A having the sulfur atom 5 Å deeper into the hydrophobic cavity (22). The 4′-PP conformation in GmACP3 is more similar to the conformation of molecule B, although the sulfur atom is closer to the large loop 1 and loop 3 than in the E. coli heptanoyl-ACP molecule B. The X-ray structures of E. coli apo-, holo-, and butyryl-ACP, have no observable electron density for 4′-PP (22).
As previously noted, GmACP3 and E. coli ACP have similar helical geometries and hydrophobic cavities. Residues in the E. coli ACP hydrophobic cavity that interact with the acyl chain are conserved in GmACP3. These are the GmACP3/E. coli ACP residues A28/V29, L42/L42, I43/V43, L46/L46, F54/I54, L59/A59, L62/I62, L68/A68. Differences are found in helix α4, which is 4 residues longer in GmACP3, and in loop 1, which has a short 310 helix (Q19-E21) that is missing in GmACP3 (Figure 5A). Both structures have a corresponding short 310 helix in loop 1 next to 4′-PP (A28-I31/V29-L32) and both are stabilized by backbone hydrogen bonds to residues in the N-terminus of helix α4. It has been suggested that the longer helix α4 may allow ACPs to accommodate longer acyl chain lengths than the typical 10–14 carbon FAs used by E. coli ACP (55).
E. coli ACP is a flexible protein that can be stabilized by binding divalent cations at two low affinity sites at each end of helix α2 (62). This stabilization is attributed to partial neutralization of acidic residues (62). GmACP3 is less acidic and the Asp and Glu residues of these binding sites are not conserved. Addition of Mg+2 had no effect on the 1H-15N HSQC spectrum of GmACP3 (data not shown).
Helix α2 of E. coli ACP is of crucial importance in binding with AcpS as well as FAS enzymes and those involved in LPS synthesis (reviewed in (5, 29, 62)). However, unlike E. coli ACP, which has an electrostatic surface of mostly negative potential, GmACP3 has a smaller region of negative potential near the DSL motif, as illustrated in Figure 5B. The surface potential of GmACP3 resembles a dipole, with two distinct regions of opposing charge, situated at physically opposite regions of the protein. The area of negative potential is at the C-terminal end of helix α1, the residues of loop 1, and the N-terminal end of helix α2. These are near the WDSLxN/H motif and the entrance to the hydrophobic cavity. Residues creating the positive potential are located on the C-terminus of helix α2, loop 2, helix α3, loop 3, and helix α4, and are located at the opposite side of the protein. This dipole feature is not common to other ACPs, which typically have mostly negative electrostatic surface potential. The uncommon electrostatic surface properties of GmACP3 suggest a mechanism for proper orientation with enzymatic binding partners (ACP-dependent enzymes) that differs from that of E. coli ACP. However, the identity of the interacting proteins and whether GmACP3 will also interact with individual FAS proteins remains to be determined.
Structural comparison of holo-GmACP3 and other ACPs
The 3D structure of GmACP3 is also quite similar to the PKS ACP for actinorhodin synthesis (actACP) from Streptomyces coelicolor in the hexanoyl- and octanoyl-actACP complexes (60), with RMSDs of 1.5 Å. GmACP3 is less similar (> 2 Å RMSD) to the apo-, holo-, acetyl-, and malonyl-actACP structures as well as the 3-oxobutyl and 3,5-dioxohexyl actACPs designed to mimic polyketide intermediates (60). RMSDs are reported in Table S4 of the Supporting Information. Specifically, the distance between helix α3 and helices α1 and α2, and the resulting hydrophobic cavity, are smaller in the actACPs, with the exception of the hexanoyl- and octanoyl-actACPs. A comparison of holo-GmACP3 with octanoyl-actACP (1.5 Å RMSD) and acetyl-actACP (2.5 Å RMSD) is shown in Figure 5C. The conformation of 4′-PP determined in each of the actACP structures is very different than for holo-GmACP3. Although many NOEs to 4′-PP were observed for the acyl-actACPs, the resulting 4′-PP conformation is not oriented towards the inside of the ACP, but extends outwards and lies in a groove between helixes α2 and α3. The sulfur atom of 4′-PP is located outside the cavity and the covalently attached acyl chains are threaded towards the hydrophobic core through this groove. The placement of 4′-PP on the outside of the helices may explain the “free” chemical shifts observed for 4′-PP resonances (60). In fact, no currently deposited ACP NMR structures have the 4′-PP geometry observed in holo-GmACP3, and no deposited 4′-PP chemical shifts differ from the “free” values in contrast to those measured here for holo-GmACP3.
Other ACPs with structural similarity to holo-GmACP3, identified by DALI (61), included the secondary ACP from M. tuberculosis (RV033, PDB ID 2CGQ) and the decanoyl-ACP-I isoform from spinach (PDB ID 2FVF) with Z-scores 10.0 and 9.4. Neither of these proteins contains a WDSL sequence. However, M. tuberculosis has at least two secondary ACPs and plants, like spinach, have multiple isoforms resulting from mRNA splice variants as well as multiple copies of the ACP gene. The different ACPs are responsible for synthesizing various FAs that are preferentially supplied to different pathways (63). For example, ACP-I and ACP-II from spinach seeds provide FAs for different pathways by altered binding specificity with the pathway enzymes for triacylglycerol and membrane lipid synthesis (64).
There have been no previous structures of ACPs with the WDSLxH/N motif. However, there has been functional characterization of the protein SMc01553 (and the duplicate Smb22007), known as the sixth ACP from Sinorhizobium meliloti, with a WDSLxN sequence and a nearby MBOAT protein (65, 66). The mRNA for these proteins was detected in S. melitoti using RT-PCR, but no phenotype for DNA deletion mutants of the two genes was observed. Davila-Martinez, et al. noted that several bacteria encode a ACP homolog, a nearby MBOAT, and a FkbH homolog (66). FkbH-like proteins can catalyze the transfer of 1,3 -bisphosphoglycerate to a specialized ACP and dephosphorylate it, forming glyceryl-ACP in vitro (67). The resulting acyl-ACP provides a three-carbon intermediate as an extender unit for synthesis of natural product polyketides such as the antibiotics abyssomicin and tetronomycin, antitumor compound chlorothricin, and the immunosuppressant FK520. Although gmet_2339 does not have a nearby gene encoding a FkbH-like enzyme, Gmet_2338 and GmACP3 may be involved in synthesis and transportation of unusual “extender units” such as glyceryl for synthesis of polyketides as well as LPS acylation.
Possible role in LPS biosynthesis
There are many variations in lipid A composition and modifications that affect the structure and function of the resulting LPS (reviewed in (6, 8, 10)). The various lipid A modifications are often regulated by environmental conditions and aid bacterial survival under different environmental stresses, such as antibiotics. The unusual LPS OH-FAs observed in G. metallireducens have not been characterized in other bacteria, including G. Sulfurreducens and G. Bemidjiensis, and the biological significance of the LPS acylation by 9- and 10- hydroxyl palymitoyl-ACPs in G. metallireducens is unknown. We hypothesize that GmACP3, could provide this OH-FA for primary or secondary acylation of lipid A, and that it could hydrogen bond to the Hε2 of H39.
In summary, the structure of holo-GmACP3 has unique features that differentiate it from other bacterial ACP structures. Addition of 4′-PP resulted in stabilizing interactions with loop 1 and loop 3. These in turn may affect the position of the “recognition” helix α2 and helix α3, believed to affect the size of the hydrophobic cavity. This larger cavity may be adapted to bind bulkier acyl intermediates, such as longer, unsaturated, branched, or hydroxy fatty acids. In addition, the helical positions, which are similar to those in acyl-ACPs from E. coli and actACPs with 6 -10 carbon chains, are likely to be important for defining the specificity needed for protein recognition by enzymes in the synthetic pathway. GmACP3 may play a role in LPS biosynthesis as well as other roles in polyketide synthesis. It remains to be determined if these roles are conserved in bacteria with GmACP3 homologs containing the WDSLxH/N motif.
Supplementary Material
Acknowledgments
We thank Rosa Di Leo and Alexei Savchenko’s group at the Ontario Centre for Structural Proteomics (affiliated with the Midwest Center for Structural Genomics) for providing the E. coli AcpS clone. We thank Alex Eletski at SUNY Buffalo for providing the intav and τc macros for the Varian vnmr software. We also thank Rui Zhang and John Cort at Pacific Northwest National Laboratory for running the FTICR MS and for assistance with MS setup and interpretation.
Abbreviations
- ACP
acyl carrier protein
- NMR
nuclear magnetic resonance
- RDC
residual dipolar coupling
- NOE
nuclear Overhauser effect
- HSQC
heteronuclear single-quantum coherence spectroscopy
- RMSD
root mean squared deviation
- FAS
fatty acid synthesis
- PKS
polyketide synthesis
- LPS
lipopolysaccharide
- 4′-phosphopantetheine
4′-PP
- CoA
Coenzyme A
- LPS OH-FA
lipopolysaccharide hydroxy-fatty acids
- UDP
uridine diphosphate
- GlcNAc
N-acetylglucosamine
- Kdo
3-deoxy-D-manno-oct-2-ulosonic-acid
- acyl-ACPs
acetyl- (C2), malonyl- (C3), butyryl- (C4), hexanoyl- (C6), heptanoyl- (C7), octanoyl- (C8), decanoyl- (C10), lauroyl- (C12), myristoyl- (C14), palmitoyl- (C16), stearoyl- (C18), octacosanoyl- (C28) ACP
Footnotes
This work was supported by a grant from the Protein Structure Initiative of the National Institutes of Health (P50 GM 074958). FTICR mass spectrometry was performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the Department of Energy’s Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory in Richland, WA.
SUPPORTING INFORMATION AVAILABLE
Figures, tables and text containing mass spectrometry data, NMR 1H-15N HSQC spectra, GmACP3 homologs, possible functional importance of gmet_2337 and gmet_2340, 4′-PP NOEs, 1H-15N HSQC spectrum centered on histidine resonances, DALI hits, and RMSDs between holo-GMACP3 and ACPs structural homologs. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Lovley DR, Giovannoni SJ, White DC, Champine JE, Phillips EJ, Gorby YA, Goodwin S. Geobacter metallireducens gen. nov. sp. nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals. Arch Microbiol. 1993;159:336–344. doi: 10.1007/BF00290916. [DOI] [PubMed] [Google Scholar]
- 2.Aklujkar M, Krushkal J, DiBartolo G, Lapidus A, Land ML, Lovley DR. The genome sequence of Geobacter metallireducens: features of metabolism, physiology and regulation common and dissimilar to Geobacter sulfurreducens. BMC Microbiol. 2009;9:109. doi: 10.1186/1471-2180-9-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rawlings M, Cronan JE., Jr The gene encoding Escherichia coli acyl carrier protein lies within a cluster of fatty acid biosynthetic genes. J Biol Chem. 1992;267:5751–5754. [PubMed] [Google Scholar]
- 4.Lai JR, Koglin A, Walsh CT. Carrier protein structure and recognition in polyketide and nonribosomal peptide biosynthesis. Biochemistry. 2006;45:14869–14879. doi: 10.1021/bi061979p. [DOI] [PubMed] [Google Scholar]
- 5.Byers DM, Gong H. Acyl carrier protein: structure-function relationships in a conserved multifunctional protein family. Biochem Cell Biol. 2007;85:649–662. doi: 10.1139/o07-109. [DOI] [PubMed] [Google Scholar]
- 6.Raetz CR, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in gram-negative bacteria. Annu Rev Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cronan JE, Thomas J. Bacterial fatty acid synthesis and its relationships with polyketide synthetic pathways. Methods Enzymol. 2009;459:395–433. doi: 10.1016/S0076-6879(09)04617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holst O, Molinaro A. Core Region and Lipid A Components of Lipopolysaccharides. In: Moran Anthony P., editor. Microbial Glycobiology, Structures, Relevance, and Applications. Elsevier; 2009. pp. 29–56. [Google Scholar]
- 9.Boeckmann B, Blatter MC, Famiglietti L, Hinz U, Lane L, Roechert B, Bairoch A. Protein variety and functional diversity: Swiss-Prot annotation in its biological context. C R Biol. 2005;328:882–899. doi: 10.1016/j.crvi.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raetz CR, Guan Z, Ingram BO, Six DA, Song F, Wang X, Zhao J. Discovery of new biosynthetic pathways: the lipid A story. J Lipid Res. 2009;50(Suppl):S103–108. doi: 10.1194/jlr.R800060-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuszewski J, Gronenborn AM, Clore GM. A potential involving multiple proton chemical-shift restraints for nonstereospecifically assigned methyl and methylene protons. J Magn Reson B. 1996;112:79–81. doi: 10.1006/jmrb.1996.0113. [DOI] [PubMed] [Google Scholar]
- 13.Brozek KA, Carlson RW, Raetz CR. A special acyl carrier protein for transferring long hydroxylated fatty acids to lipid A in Rhizobium. J Biol Chem. 1996;271:32126–32136. doi: 10.1074/jbc.271.50.32126. [DOI] [PubMed] [Google Scholar]
- 14.Basu SS, Karbarz MJ, Raetz CR. Expression cloning and characterization of the C28 acyltransferase of lipid A biosynthesis in Rhizobium leguminosarum. J Biol Chem. 2002;277:28959–28971. doi: 10.1074/jbc.M204525200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hedrick DB, Peacock AD, Lovley DR, Woodard TL, Nevin KP, Long PE, White DC. Polar lipid fatty acids, LPS-hydroxy fatty acids, and respiratory quinones of three Geobacter strains, and variation with electron acceptor. J Ind Microbiol Biotechnol. 2009;36:205–209. doi: 10.1007/s10295-008-0486-7. [DOI] [PubMed] [Google Scholar]
- 16.Holak TA, Kearsley SK, Kim Y, Prestegard JH. Three-dimensional structure of acyl carrier protein determined by NMR pseudoenergy and distance geometry calculations. Biochemistry. 1988;27:6135–6142. doi: 10.1021/bi00416a046. [DOI] [PubMed] [Google Scholar]
- 17.Kim Y, Prestegard JH. Refinement of the NMR structures for acyl carrier protein with scalar coupling data. Proteins. 1990;8:377–385. doi: 10.1002/prot.340080411. [DOI] [PubMed] [Google Scholar]
- 18.Parris KD, Lin L, Tam A, Mathew R, Hixon J, Stahl M, Fritz CC, Seehra J, Somers WS. Crystal structures of substrate binding to Bacillus subtilis holo-(acyl carrier protein) synthase reveal a novel trimeric arrangement of molecules resulting in three active sites. Structure. 2000;8:883–895. doi: 10.1016/s0969-2126(00)00178-7. [DOI] [PubMed] [Google Scholar]
- 19.Xu GY, Tam A, Lin L, Hixon J, Fritz CC, Powers R. Solution structure of B. subtilis acyl carrier protein. Structure. 2001;9:277–287. doi: 10.1016/s0969-2126(01)00586-x. [DOI] [PubMed] [Google Scholar]
- 20.Roujeinikova A, Baldock C, Simon WJ, Gilroy J, Baker PJ, Stuitje AR, Rice DW, Slabas AR, Rafferty JB. X-ray crystallographic studies on butyryl-ACP reveal flexibility of the structure around a putative acyl chain binding site. Structure. 2002;10:825–835. doi: 10.1016/s0969-2126(02)00775-x. [DOI] [PubMed] [Google Scholar]
- 21.Qiu X, Janson CA. Structure of apo acyl carrier protein and a proposal to engineer protein crystallization through metal ions. Acta Crystallogr D Biol Crystallogr. 2004;60:1545–1554. doi: 10.1107/S0907444904015422. [DOI] [PubMed] [Google Scholar]
- 22.Roujeinikova A, Simon WJ, Gilroy J, Rice DW, Rafferty JB, Slabas AR. Structural studies of fatty acyl-(acyl carrier protein) thioesters reveal a hydrophobic binding cavity that can expand to fit longer substrates. J Mol Biol. 2007;365:135–145. doi: 10.1016/j.jmb.2006.09.049. [DOI] [PubMed] [Google Scholar]
- 23.Ploskon E, Arthur CJ, Kanari AL, Wattana-amorn P, Williams C, Crosby J, Simpson TJ, Willis CL, Crump MP. Recognition of intermediate functionality by acyl carrier protein over a complete cycle of fatty acid biosynthesis. Chem Biol. 2010;17:776–785. doi: 10.1016/j.chembiol.2010.05.024. [DOI] [PubMed] [Google Scholar]
- 24.Cryle MJ, Schlichting I. Structural insights from a P450 Carrier Protein complex reveal how specificity is achieved in the P450(BioI) ACP complex. Proc Natl Acad Sci U S A. 2008;105:15696–15701. doi: 10.1073/pnas.0805983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu BN, Zhang YM, Rock CO, Zheng JJ. Structural modification of acyl carrier protein by butyryl group. Protein Sci. 2009;18:240–246. doi: 10.1002/pro.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zornetzer GA, Fox BG, Markley JL. Solution structures of spinach acyl carrier protein with decanoate and stearate. Biochemistry. 2006;45:5217–522. doi: 10.1021/bi052062d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zornetzer GA, Tanem J, Fox BG, Markley JL. The length of the bound fatty acid influences the dynamics of the acyl carrier protein and the stability of the thioester bond. Biochemistry. 2010;49:470–477. doi: 10.1021/bi9014659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma AK, Sharma SK, Surolia A, Surolia N, Sarma SP. Solution structures of conformationally equilibrium forms of holo-acyl carrier protein (PfACP) from Plasmodium falciparum provides insight into the mechanism of activation of ACPs. Biochemistry. 2006;45:6904–6916. doi: 10.1021/bi060368u. [DOI] [PubMed] [Google Scholar]
- 29.Zhang YM, Marrakchi H, White SW, Rock CO. The application of computational methods to explore the diversity and structure of bacterial fatty acid synthase. J Lipid Res. 2003;44:1–10. doi: 10.1194/jlr.r200016-jlr200. [DOI] [PubMed] [Google Scholar]
- 30.Evans SE, Williams C, Arthur CJ, Burston SG, Simpson TJ, Crosby J, Crump MP. An ACP structural switch: conformational differences between the apo and holo forms of the actinorhodin polyketide synthase acyl carrier protein. Chembiochem. 2008;9:2424–2432. doi: 10.1002/cbic.200800180. [DOI] [PubMed] [Google Scholar]
- 31.Butland G, Peregrin-Alvarez JM, Li J, Yang W, Yang X, Canadien V, Starostine A, Richards D, Beattie B, Krogan N, Davey M, Parkinson J, Greenblatt J, Emili A. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 32.Acton TB, Gunsalus KC, Xiao R, Ma LC, Aramini J, Baran MC, Chiang YW, Climent T, Cooper B, Denissova NG, Douglas SM, Everett JK, Ho CK, Macapagal D, Rajan PK, Shastry R, Shih LY, Swapna GV, Wilson M, Wu M, Gerstein M, Inouye M, Hunt JF, Montelione GT. Robotic cloning and protein production platform of the Northeast Structural Genomics Consortium. Methods Enzymol. 2005;394:210–243. doi: 10.1016/S0076-6879(05)94008-1. [DOI] [PubMed] [Google Scholar]
- 33.Xiao R, Anderson S, Aramini J, Belote R, Buchwald WA, Ciccosanti C, Conover K, Everett JK, Hamilton K, Huang YJ, Janjua H, Jiang M, Kornhaber GJ, Lee DY, Locke JY, Ma LC, Maglaqui M, Mao L, Mitra S, Patel D, Rossi P, Sahdev S, Sharma S, Shastry R, Swapna GV, Tong SN, Wang D, Wang H, Zhao L, Montelione GT, Acton TB. The high-throughput protein sample production platform of the Northeast Structural Genomics Consortium. J Struct Biol. 2010;172:21–33. doi: 10.1016/j.jsb.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jansson M, Li YC, Jendeberg L, Anderson S, Montelione BT, Nilsson B. High-level production of uniformly 15N- and 13C-enriched fusion proteins in Escherichia coli. J Biomol NMR. 1996;7:131–141. doi: 10.1007/BF00203823. [DOI] [PubMed] [Google Scholar]
- 35.Lambalot RH, Walsh CT. Cloning, overproduction, and characterization of the Escherichia coli holo-acyl carrier protein synthase. J Biol Chem. 1995;270:24658–24661. doi: 10.1074/jbc.270.42.24658. [DOI] [PubMed] [Google Scholar]
- 36.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 37.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 38.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 39.Ramelot TA, Raman S, Kuzin AP, Xiao R, Ma LC, Acton TB, Hunt JF, Montelione GT, Baker D, Kennedy MA. Improving NMR protein structure quality by Rosetta refinement: a molecular replacement study. Proteins. 2009;75:147–167. doi: 10.1002/prot.22229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neri D, Szyperski T, Otting G, Senn H, Wuthrich K. Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry. 1989;28:7510–7516. doi: 10.1021/bi00445a003. [DOI] [PubMed] [Google Scholar]
- 41.Tjandra N, Grzesiek S, Bax A. Magnetic field dependence of nitrogen-proton J splittings in N-15-enriched human ubiquitin resulting from relaxation interference and residual dipolar coupling. J Am Chem Soc. 1996;118:6264–6272. [Google Scholar]
- 42.Cierpicki T, Bushweller JH. Charged gels as orienting media for measurement of residual dipolar couplings in soluble and integral membrane proteins. J Am Chem Soc. 2004;126:16259–16266. doi: 10.1021/ja046054g. [DOI] [PubMed] [Google Scholar]
- 43.Liu Y, Prestegard JH. A device for the measurement of residual chemical shift anisotropy and residual dipolar coupling in soluble and membrane-associated proteins. J Biomol NMR. 2010;47:249–258. doi: 10.1007/s10858-010-9427-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 45.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwieters CD, Kuszeski JJ, Clore GM. Using Xplor-NIH for NMR molecular structure determination. Prog Nucl Mag Res Sp. 2006;48:47–62. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 47.Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5–9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mao F, Dam P, Chou J, Olman V, Xu Y. DOOR: a database for prokaryotic operons. Nucleic Acids Res. 2009;37:D459–463. doi: 10.1093/nar/gkn757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Upadhyay SK, Misra A, Srivastava R, Surolia N, Surolia A, Sundd M. Structural insights into the acyl intermediates of the Plasmodium falciparum fatty acid synthesis pathway: the mechanism of expansion of the acyl carrier protein core. J Biol Chem. 2009;284:22390–22400. doi: 10.1074/jbc.M109.014829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pelton JG, Torchia DA, Meadow ND, Roseman S. Tautomeric states of the active-site histidines of phosphorylated and unphosphorylated IIIGlc, a signal-transducing protein from Escherichia coli, using two-dimensional heteronuclear NMR techniques. Protein Sci. 1993;2:543–558. doi: 10.1002/pro.5560020406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Price AC, Zhang YM, Rock CO, White SW. Structure of beta-ketoacyl-[acyl carrier protein] reductase from Escherichia coli: negative cooperativity and its structural basis. Biochemistry. 2001;40:12772–12781. doi: 10.1021/bi010737g. [DOI] [PubMed] [Google Scholar]
- 53.Price AC, Rock CO, White SW. The 1.3-Angstrom-resolution crystal structure of beta-ketoacyl-acyl carrier protein synthase II from Streptococcus pneumoniae. J Bacteriol. 2003;185:4136–4143. doi: 10.1128/JB.185.14.4136-4143.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wong HC, Liu G, Zhang YM, Rock CO, Zheng J. The solution structure of acyl carrier protein from Mycobacterium tuberculosis. J Biol Chem. 2002;277:15874–15880. doi: 10.1074/jbc.M112300200. [DOI] [PubMed] [Google Scholar]
- 56.Mayo KH, Prestegard JH. Acyl carrier protein from Escherichia coli. Structural characterization of short-chain acylated acyl carrier proteins by NMR. Biochemistry. 1985;24:7834–7838. doi: 10.1021/bi00347a049. [DOI] [PubMed] [Google Scholar]
- 57.Li Q, Khosla C, Puglisi JD, Liu CW. Solution structure and backbone dynamics of the holo form of the frenolicin acyl carrier protein. Biochemistry. 2003;42:4648–4657. doi: 10.1021/bi0274120. [DOI] [PubMed] [Google Scholar]
- 58.Kim Y, Kovrigin EL, Eletr Z. NMR studies of Escherichia coli acyl carrier protein: dynamic and structural differences of the apo- and holo-forms. Biochem Biophys Res Commun. 2006;341:776–783. doi: 10.1016/j.bbrc.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 59.Zornetzer GA, White RD, Markley JL, Fox BG. Preparation of isotopically labeled spinach acyl-acyl carrier protein for NMR structural studies. Protein Expr Purif. 2006;46:446–455. doi: 10.1016/j.pep.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 60.Evans SE, Williams C, Arthur CJ, Ploskon E, Wattana-amorn P, Cox RJ, Crosby J, Willis CL, Simpson TJ, Crump MP. Probing the Interactions of early polyketide intermediates with the Actinorhodin ACP from S. coelicolor A3 (2) J Mol Biol. 2009;389:511–528. doi: 10.1016/j.jmb.2009.03.072. [DOI] [PubMed] [Google Scholar]
- 61.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38(Suppl):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gong H, Murphy A, McMaster CR, Byers DM. Neutralization of acidic residues in helix II stabilizes the folded conformation of acyl carrier protein and variably alters its function with different enzymes. J Biol Chem. 2007;282:4494–4503. doi: 10.1074/jbc.M608234200. [DOI] [PubMed] [Google Scholar]
- 63.Mercer AC, Burkart MD. The ubiquitous carrier protein--a window to metabolite biosynthesis. Nat Prod Rep. 2007;24:750–773. doi: 10.1039/b603921a. [DOI] [PubMed] [Google Scholar]
- 64.Guerra DJ, Ohlrogge JB, Frentzen M. Activity of acyl carrier protein isoforms in reactions of plant fatty acid metabolism. Plant Physiol. 1986;82:448–453. doi: 10.1104/pp.82.2.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Geiger O, Lopez-Lara IM. Rhizobial acyl carrier proteins and their roles in the formation of bacterial cell-surface components that are required for the development of nitrogen-fixing root nodules on legume hosts. FEMS Microbiol Lett. 2002;208:153–162. doi: 10.1111/j.1574-6968.2002.tb11075.x. [DOI] [PubMed] [Google Scholar]
- 66.Davila-Martinez Y, Ramos-Vega AL, Contreras-Martinez S, Encarnacion S, Geiger O, Lopez-Lara IM. SMc01553 is the sixth acyl carrier protein in Sinorhizobium meliloti 1021. Microbiology. 2010;156:230–239. doi: 10.1099/mic.0.033480-0. [DOI] [PubMed] [Google Scholar]
- 67.Sun Y, Hong H, Gillies F, Spencer JB, Leadlay PF. Glyceryl-S-acyl carrier protein as an intermediate in the biosynthesis of tetronate antibiotics. Chembiochem. 2008;9:150–156. doi: 10.1002/cbic.200700492. [DOI] [PubMed] [Google Scholar]
- 68.Moseley HN, Sahota G, Montelione GT. Assignment validation software suite for the evaluation and presentation of protein resonance assignment data. J Biomol NMR. 2004;28:341–355. doi: 10.1023/B:JNMR.0000015420.44364.06. [DOI] [PubMed] [Google Scholar]
- 69.Bhattacharya A, Tejero R, Montelione GT. Evaluating protein structures determined by structural genomics consortia. Proteins. 2007;66:778–795. doi: 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- 70.Huang YJ, Powers R, Montelione GT. Protein NMR recall, precision, and F-measure scores (RPF scores): structure quality assessment measures based on information retrieval statistics. J Am Chem Soc. 2005;127:1665–1674. doi: 10.1021/ja047109h. [DOI] [PubMed] [Google Scholar]
- 71.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 72.Garrett DS, Seok YJ, Peterkofsky A, Clore GM, Gronenborn AM. Identification by NMR of the binding surface for the histidine-containing phosphocarrier protein HPr on the N-terminal domain of enzyme I of the Escherichia coli phosphotransferase system. Biochemistry. 1997;36:4393–4398. doi: 10.1021/bi970221q. [DOI] [PubMed] [Google Scholar]
- 73.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.