Abstract
A new generation of programmable diagnostic devices is needed to take advantage of information generated from the study of genomics, proteomics, metabolomics and glycomics. This report describes the ‘programmable nano-bio-chip’ with potential to bridge the significant scientific, technology and clinical gaps through the creation of a diagnostic platform to measure the molecules of life. This approach, with results at the point-of-care, possesses capabilities for measuring such diverse analyte classes as cells, proteins, DNA and small molecules in the same compact device. Applications such as disease diagnosis and prognosis for areas including cancer, heart disease and HIV are described. New diagnostic panels are inserted as ‘plug and play’ elements into the modular platform with universal assay operating systems and standard read out sequences. The nano-bio-chip ensemble exhibits excellent analytical performance and cost-effectiveness with extensive validation versus standard reference methods (R2 = 0.95–0.99). This report describes the construction and use of two major classes of nano-bio-chip designs that serve as cellular and chemical processing units, and provides perspective on future growth in this newly emerging field of programmable nano-bio-chip sensor systems.
Keywords: integrated sensor, lab-on-a-chip, Moore’s law, nano-bio-chip, point-of-care, programmable
Diagnostic tools are critical to the delivery of effective healthcare treatment, yet current in vitro diagnostic (IVD) devices are incapable of keeping pace with the rapidly increasing information content related to disease diagnosis and progression generated with advanced ‘omics’ methods such as genomics, proteomics, metabolomics and glycomics [1,2]. Here, a large number of biomarker ‘discovery’ papers (20,000 cancer and 6000 cardiac) have been reported, yet only approximately one biomarker per year received US FDA approval between 1995 and 2005 [3,4]. Unfortunately, most modern clinical analyzers are dedicated to single classes of analytes and are burdened by bulky, expensive, laboratory-confined instrumentation preventing broad access to these assays at the point-of-care (POC). Furthermore, large sample volume requirements and lack of standard instrumentation that is responsive to a broad range of analytes complicate clinical validation studies that need to follow the initial discoveries.
Traditional approaches to clinical analysis involve a well-appointed centralized laboratory, three degrees of separation from the patient. This hierarchy introduces a number of critical junctures in which errors may be introduced and delays incurred. To simplify and offer assay results immediately, research into devices that give results at the POC, specifically bedside, ambulance or remote location, currently flourishes - a situation advantageous to both patients and healthcare providers [5–9]. The ability to process large amounts of information at the point-of-need is common in the field of electronics, yet the ability to similarly process complex molecular disease signatures has not yet been fully demonstrated. The marriage of microelectronics and IVD provides huge opportunities to healthcare industries seeking affordable and accessible diagnostic infrastructures [10]. For example, although the approach to processor building typical of microelectronics via rapid prototyping and assembly translates well into IVD, the core substrate, a silicon wafer, does not owing to its high expense and the single-use nature of POC IVD.
While remarkable progress has been made toward POC clinical assay systems through microfluidic lab-on-a-chip (LOC) approaches and the micro total analysis system (μTAS) paradigm, completion and launch of workable systems based on these premises is largely incomplete [5,11–21]. While the scope of this manuscript does not allow for thorough treatment of the relevant background literature, the contributions of a few key efforts deserve mention with further analysis available in more complete reviews [5,12,22]. Whitesides’ work in the basic sciences defined the ideal materials, coatings and designs needed to create micro-channels and manipulate biological fluids [23]. Quake’s work has advanced the ‘large-scale integration’ of microfluidics, analogous to the electronics field [10]. This experimental design has been used to explore genetic and protein applications as well as the biophysical properties of single molecules. Others, such as Mirkin, Heath and Wang use nanowires, precious metal nanoparticles and magnetic techniques, respectively, to measure diverse sample types and create a variety of assembly types [9,24,25]. More integrated approaches by Sia via micro-electromechanical systems and Singh using chip-based separation and quantitation have continued to increase integration [26,27]. Both Singh and Ligler have extended their integrated approaches into the rapid, multiplexed detection of toxins and other biothreats [28]. Work by Madou and others has resulted in the LabCD, which eliminates traditional valves and pumps by using centrifugal and centripetal force to perform fluid movements [18]. Walt’s work with electronic noses uses arrays of optical fibers as the underlying infrastructure for biological sensing systems [29]. Finally, researchers in the Toner group have explored a number of novel methods for the isolation and enumeration of lymphocytes, erythrocytes and circulating tumor cells [30,31]. While these important activities serve as a basis to define an exciting new discipline, the area of integrated testing of real biological samples at the POC using medical microdevices remains in a state of infancy. Fundamental incongruousness with scalability, fouling of reactive surfaces and narrow, analyte-specific designs all prevent broad clinical acceptance of POC analysis systems based on these approaches [32,33]. Furthermore, some of these approaches require macroscopic laboratory-based infrastructure and, while their analysis core is substantially smaller than bench-top alternatives, the network of support structures required for sample processing, data collection and reagent handling imply that these platforms are best described as ‘chips in a lab,’ rather than true ‘labs-on-a-chip.’
In this report, we detail the ‘programmable nano-bio-chip’ (NBC), which has the capacity to serve as a highly flexible interface to gather health status information using small amounts of bodily fluids. This report summarizes the recent developments with this approach and provides projections for future growth in the area. The NBC methodology employs a 3D ‘nanonet’ that is part of a ‘microsponge’ composed of agarose webbing or polymer membrane, and nanoparticle-based signaling (nano) to selectively quantitate clinically relevant analytes (bio) from highly heterogeneous samples within a self-contained fluidic system (chip). A key feature differentiates the NBC from the analytical schema mentioned before. Unlike the approaches typical of microfluidics, biochip and LOC paradigms, in which one ‘chip’ is created specific to a type of cell/protein/oligo, the NBC has a broad portfolio of analytes that are measured with the same compact system. Assays for cells, proteins and nucleic acids can be completed within the universal compact, disposable reaction labcard that houses both ‘chemical processing units’ (CPU-1) for protein measurements and ‘cellular processing units’ (CPU-2) for cell differentiation and counting [34–37]. Targeted panels are created easily through the inclusion of modular reagent packages, bead capture elements and size-tuned cell collection ensembles. Another important distinction is the different mechanism of analyte capture. While most microfluidic approaches use planar arrays, the NBC employs high surface area 3D beads that serve to efficiently concentrate various analytes from biofluid. Results display in minutes and are available at the POC. Both systems reprogram quickly as new information related to disease signatures is obtained from research settings. Designed originally as an ‘electronic taste chip’, the chemical processing unit version of the NBC finds inspiration in nature as described below [38].
Background & goals of the system
Similar to the processes of smell and taste in which pattern recognition identifies the chemical signatures associated foods and beverages, the CPU-1-NBC contains artificial taste buds that capture various analyte classes present in complex fluid samples. Foodstuffs rarely contain a single dominant tastant (i.e., analyte resulting in taste perception), but rather express a combination of sweet, sour, salt, bitter and umami elements, which collaboratively engage the taste buds sensitive to these various stimuli to produce the resulting flavor sensation. Similarly, by measuring the levels of a collection of relatively nonspecific biomarkers via pattern recognition, the CPU-1-NBC yields disease status information from complex samples. Combinations of cells, proteins and small molecules are interpreted by advanced algorithms, which identify the biofingerprint corresponding to disease through a digital training process.
The NBC addresses such unmet clinical needs as accessibility and cost, yielding sensitive and selective information. Clinical biosensors, with a focus on POC measurement events, command quality results. We designate the acronym COMMAND QUALS to encapsulate those features of miniaturized diagnostic equipment most frequently required of POC analytical instrumentation. All compact systems seek to include the following characteristics:
Cheap – low cost increases access to diagnostic testing
Obvious – simple interfaces allow use by low-skill operators
Miniaturized – devices with a reduced footprint are amenable to POC use
Multiplexed – measuring many different analytes concurrently increases diagnostic efficacy
Automated – built-in sample preparation, reagent handling and data interpretation eases use
Nonperishable – rugged designs survive difficult conditions and eliminate the ‘cold-chain’
Dependable – extremely low rates of failure are critical
Quick – short turnaround times yield information quickly
Unobtrusive – noninvasive specimens and collection procedures reduce testing antipathy
Adaptable – devices should quantitate a variety of analytes (e.g., cells, proteins, nucleic acids, small molecules and ions)
Limited – small reagent and sample volumes decrease costs and minimize waste disposal
Self-contained – a closed system simplifies disposal and reduces exposure to biohazard waste
An accessible, scalable design
While the POC community has long envisioned a miniaturized test addressing all of the above COMMAND QUALS characteristics, a number of key concepts lacking in the areas of microfluidics, LOC, nanoscience and nanotechnology have prevented the development of such a universal, programmable minisensor [5,21]. Vastly different measurement procedures for molecular and cellular analytes, absence of high signal above background and noise, and the lack of budget-suitable construction materials have all hampered the straightforward creation of such a device. Although inroads have been made with LOC/μTAS, complete clinical acceptance of such approaches has yet to be realized [11,13,14]. Many systems based on these technologies are fundamentally incompatible with scalability – the ability to easily process increasing amounts of information. Another requirement of scalable systems, universality, is also absent and the current state of POC IVD is largely one of chips in a lab, usable only by highly trained research teams, specific to a single analyte.
The scientific and engineering communities have seen an analogous challenge previously. Among microelectronics researchers, much excitement followed the discovery of the three-point transistor, which had the potential to eliminate vacuum tubes, but the new elements were prone to failures, as connections remained dependent on manual soldering [10]. To overcome this hurdle, the new paradigm of photolithography allowed transistors, and other resistors and capacitors, to be fabricated as universal tools that scaled, earning Kilby the 2000 Nobel Prize in Physics [39]. While many researchers originally believed this paradigm could be neatly replicated for microfluidic structures, devices fabricated with silicon for clinical applications were neither necessary nor appropriate [11]. More importantly, the expense of silicon is difficult to reconcile with single-use diagnostic events where target costs of less than $1 per test are necessary.
The evolution of work leading to the NBC here described proceeded along three main avenues. The NBC uses the goals elucidated by μTAS and attributes of microfluidics such as small sample volumes and costs, and augments these features with new microsensor ensemble concepts that allow for efficient and rapid analyte capture, cell isolation, solid-state reagent dissolution and pressure-driven flow. By using miniaturized fluidic handling as an important tool for use in building POC equipment and not the cure-all to measurement challenges, the NBC has matured to fulfill an increasing number of the COMMAND QUALS descriptors for clinically relevant measurements [35,36,40–43]. Added to the microfluidics infrastructure are mass-produced agarose beads (i.e., 1 M beads per gram), chemically sensitized to a wide variety of analytes. Using the nanonet of agarose fibers within the micrometer-sized bead to sequester and concentrate analyte, a very high percentage of target molecules are isolated quickly from the sample in contrast to most planar arrays, even those employing next-generation capture agents [44,45]. The size of these fibers and distance between them are customized through synthetic techniques for tailored pore sizes and high surface area, resulting in maximized 3D coverage of reactive material both on and through the bead, producing rapid analysis events with quality analytical performance [46]. While previous NBC iterations used a silicon microchip to hold beads, the current design employs a stainless steel substrate. These lithographically processed metal layers offer an important change in thinking away from cost-prohibitive designs toward those that are broadly accessible. After immobilization on agarose strand by capture element, analyte presence is reported by nanoparticle quantum dots with signal amplification an order above conventional fluorophores.
Bridging size regimes: a universal system
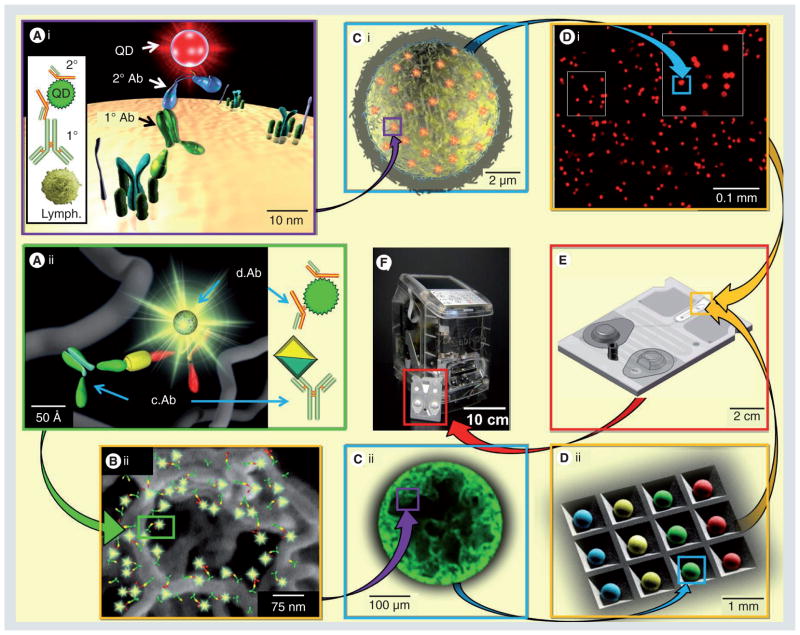
While there are a plethora of fascinating nano-sensors and other devices deploying nanotechnology for medical problems, very few integrate into practical, functional stand-alone systems. By contrast, the NBC employs two core functional units that bridge the nanometer, micrometer, millimeter and centimeter dimensionalities to create a highly functional, yet fully free-standing, reactive core. First (Figure 1Ai–Di), is a polycarbonate membrane filter with tunable pore sizes. Alternatively, 280 μm agarose beads (i.e., the microsponge) functionalized with IgG capture antibodies are used within the micro-fluidic chamber where reagents and sample flow both around and through the reactive material (Figure 1Aii–Dii). Tests for proteins, oligo-nucleotides and small molecules are done with CPU-1, while cells, spores and other particulates are measured with CPU-2 [43,47–49]. These CPUs are arranged either in parallel or in series [42], while initial optimization of the nano, bio and chip components was performed with a bench-top microscope. Recent work with translational partners has led to the compact scheme seen in Figure 1 [36,38].
Figure 1. Bridging gaps in healthcare: from nanometer to global.
The two CPU types explained in detail. The membrane-based, cellular processing unit (upper scheme (i)), is typified by cell counting applications, while the bead-based, chemical processing unit (lower scheme (ii)) has demonstrated utility in protein assays. Both utilize attributes of material across many different size scales. Nanometer-scale gaps between c.Ab. and d.Ab., and signaling probe (A & B) create bioconjugates for analyte fluorescence labeling (C) within the two CPU types (D). Expanded view in (B) illustrates the nanometer-scale network of capture agents. The modular labcard (E) houses the complete assay assembly for use in a portable, self-contained analyzer (F). Tracking of results of a global basis can become possible as these networked analyzers are distributed across the planet and secure diagnostic information that can be tied to geolocation. c.Ab: Capture antibody; d.Ab: Detecting antibody.
The fully integrated application employs the compact labcard (FIgure 1E), which condenses performance features across many size regimes, including coordination of nanometer scale antibody with micrometer-sized beads operating in tandem with the stainless steel support structure on the millimeter scale and the larger labcard, which also serves to hold solid-phase reagents, waste reservoir and a built-in metering capacity. Blister packs contain liquid-phase reagents and linear actuators control fluid flow, replacing the high-voltage power supplies typical of many μTAS systems [50]. Using inexpensive plastic materials for labcard construction along with similarly cost-effective stainless steel and agarose beads, the integrated structure has the potential to serve as an accessible and flexible bioassay system.
Each disposable labcard is designed for a single patient sample. After specimen collection and introduction into the compact NBC, the labcard is inserted into the analyzer, which serves to bridge the nanometer- and micrometer-sized components with the sizes typical of human experience. Through manipulations of the fluidic cartridges, reagents flow through the labcard and after data acquisition, is ejected with the standalone reader prepared for another analysis event. Critical to ease and cost, both use the same fluid, light and waste handling design, that is, the core of the NBC is universal to a broad class of analytes. As detailed below, four important features characterize the NBC: quality analytical behavior, a programmable (modular) design, the breadth of function and the inexpensive nature of the analysis ensemble.
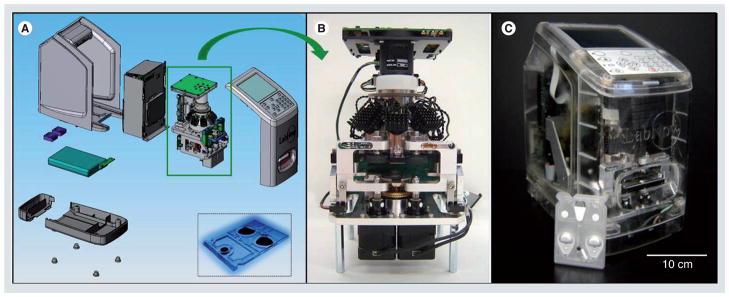
Specimen collection may be done by either venipuncture or finger-stick for blood samples or simple expectoration for saliva. In the case of the finger-stick, the sample is maintained briefly in capillary tube before introduction into the compact NBC via capillary action. The lab-card is then inserted into the analyzer where perturbations of the fluid-containing blister packs complete the assay. In Figure 2, the analyzer that interfaces with the disposable fluidic cards is illustrated. This universal instrumentation platform has a footprint about the size of a toaster with the cost approximately one-fifth of the current macroscopic instruments [101]. Assay output displays on a built-in screen shortly after completion. The device contains AC/DC power supplies with a battery life of several hours. Optical signal capture occurs via a 3.4x objective and downstream processing software, readout display, memory for up to 50,000 patient histories and USB/Ethernet/wireless communication features complete the device. Total weight is 13.5 lb making the system amenable to measurements at the POC (i.e., bedside, emergency room, ambulance and resource-poor settings).
Figure 2. Compact, integrated analyzer.
(A) Schematic of nano-bio-chip analyzer that is currently in development at LabNow. Assays completed with nano-bio-chip are performed with the compact, toaster-sized analyzer. The biochemical reactions are performed within the compact labcard (inset (A)). The standalone analzyer core (B) contains LED excitations source, actuators for fluid handling and a digital camera for image capture. The complete device and labcard (C) create an integrated, reaction approach.
The chemical processing unit
Like the sense of taste, the CPU-1-NBC has evolved to provide information about a broad range of analyte classes and is suitable for the interpretation of complex fluid samples. The CPU-1-NBC has been adapted to serve a variety of important health applications including cancer screening and monitoring, DNA quantitation, cardiac risk assessment and acute myocardial infarction diagnosis [35,38,51–55,101]. Similar to the software code in computing applications, the programmable NBC uses molecular-level code (i.e., the software) embedded in antibodies specific to analytes implicated in these diseases. With these antibodies bound to beads, automated placement of them in the NBC flow chamber (i.e., the hardware) allows for a huge variety of biomarker combinations. This positioning was performed with tweezers and a dissecting microscope in research-grade systems, but is currently performed via a piezo-controlled stage for the labcard approach. Importantly, bead exchange/regeneration is never necessary as all beads are single use. Figure 3A illustrates a typical photomicrograph containing data on the amount of biomarkers present in a particular sample. Both analyte-specific and isotype control beads are included in each experiment. The intensity of fluorescence emanating from the beads corresponds to the amount of fluorophore present and hence detecting antibody and analyte. For the approximately 40 currently validated protein assays, huge potential different arrangements are possible; however, a more focused approach through strategically created panels offers targeted diagnostic information.
Figure 3. Chemical processing unit.
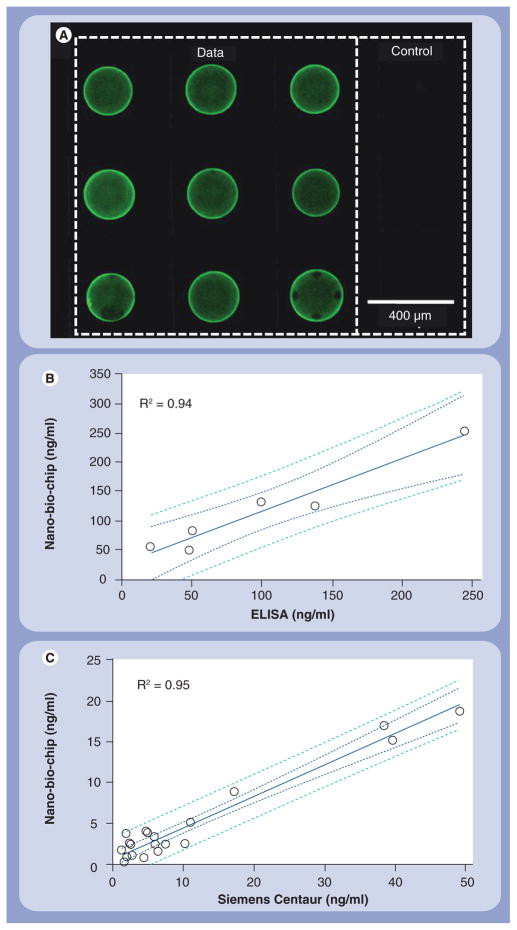
(A) The bead array is an image-based sensor that yields data in the form of a fluorescent photomicrograph. Various array sizes may be used including 4 × 5, 10 × 10 and the 3 × 4 shown here for a carcinoembryonic antigen assay (exposure time 1 s). Data collected correlates well (R2 > 0.94) for both (B) saliva and (C) serum samples [35].
Heart disease is the number one killer of humans worldwide and provides a case in point for the clinical utility of the CPU-1-NBC. Here, a cardiac theme chip employing seven cardiac-specific markers has been developed for analysis of both serum and saliva. Previous reports have demonstrated the use of both of these sample types and saliva in particular has advantages including ease of collection and storage [56,57]. The NBC is one of the few validated POC analytical designs validated for this emerging diagnostic fluid. Compared with control (n = 21) samples, logistic regression and AUC for receiver-operator characteristic analysis indicated that acute myocardial infarction-positive saliva specimens (n = 41) analyzed with the NBC gave AUC values of 0.85 (p < 0.0001) [40]. Upon the addition of EKG analysis, the AUC increased to 0.96 and with serum was 0.98. Thus, either sample type offers diagnostic relevance vastly superior to EKG alone whose AUC values are approximately 0.6.
Importantly, the cross-talk between the various beads in the microchip is low at less than 5% in multiplexed arrays [35,40]. For instance, when the cancer biomarkers carcinoembryonic antigen (CEA), Her-2 and CA125 were analyzed concurrently at concentrations one half the calibration curve maximum, the nonspecific signal was less than 5% of the specific signal [35]. The CEA assay has also been validated against conventional analysis techniques and indicates that the NBC correlates at R2 = 0.94 and 0.95 for saliva and serum samples, respectively, versus reference methods (Figure 3B & C). In addition to correlating to gold-standard methods, the analytical descriptors that characterize the NBC are highly competitive with benchtop systems. Table 1 illustrates the NBC assay for CEA versus other commercially available systems and shows the lower limit of detection, reduced turnaround time and narrow coefficient of variation that characterize this assay. Beyond proteins, the NBC has also been used for genomic analysis [43]. Hybridization times measured in minutes, with point mutation selectivity factors greater than 10,000, and limit of detection values of 10−13 M, are obtained readily with the NBC [43].
Table 1.
Nano-bio-chip performance.
| Instrument | Manufacturer | Method | LOL (ng/ml) | LOD (ng/ml) | Interassay CV | Intra-assay CV | Time (min) |
|---|---|---|---|---|---|---|---|
| NBC | McDevitt lab | Fluorescence immunoassay | 100 | 0.02 | <10% | 6.5% | 25 |
| ELISA | BioQuant | ELISA | 50 | 1.2 | 10% | <40% | 120 |
| Vitros Eci | Ortho | Electrochemiluminescence | 100 | 0.5 | 10% | 10% | 60 |
| IMMULITE | Siemens | Chemiluminescence | 550 | 0.2 | <10% | <10% | >60 |
| Elecsys | BMC/Roche | Electrochemiluminescence | 25,000 | 0.2 | 4.6% | 8.5% | 20 |
Here, the analytical performance of the NBC for the CEA assay is contrasted with other conventional, commercially available products. The NBC has a LOL, LOD, CV very competitive with similar technologies, as well as turnaround time superior to many of these current approaches to protein measurement.
CEA: Carcinoembryonic antigen; CV: Coefficients of variation; LOD: Limit of detection; LOL: Limit of linearity; NBC: Nano-bio-chip.
The cellular processing unit
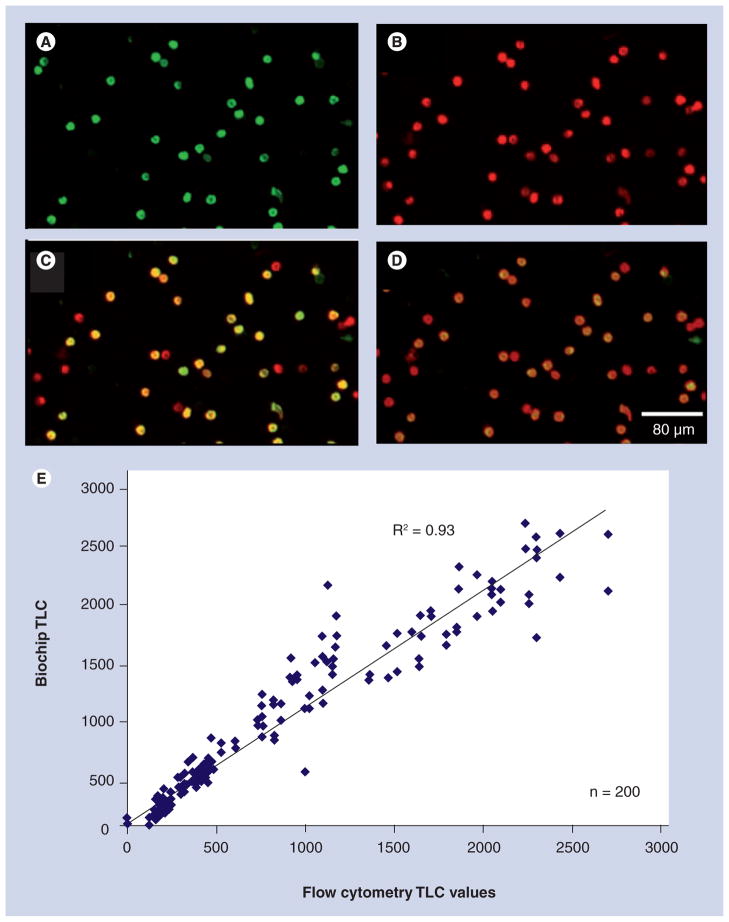
This quality clinical behavior extends beyond the CPU-1-NBC. By replacing the microchip and beads with a polycarbonate membrane filter, the NBC easily transforms into a cell enumeration system as previously demonstrated in pilot studies for HIV immune function testing using CD4 counts both in North America and Botswana (Africa) [34]. For these lymphocyte analysis, 3-μm pores within this membrane allow for passage of erythrocytes, platelets and plasma, while retaining the larger and more rigid leukocytes. To complete the CD4 cellular measurements, first a sample is collected either via venipuncture or finger-stick followed by introduction of a drop of the specimen into the labcard. Cell surface markers are then labeled with 2–3 μl of 1 mg/ml fluorescently tagged IgG antibodies allowing digital image capture and processing. Figure 4A–D illustrates this capability for counting CD4 T lymphocytes, cells infected with HIV and measured to determine a patient’s response to antiretroviral therapy. In resource-scarce environments, making such measurements is difficult or impossible due to the delicate and bulky nature of flow cytometry instrumentation. The development of affordable and accessible HIV immune function testing systems is one of the most important diagnostic challenges yet to be surmounted in global health area [58]. The NBC is a POC solution that allows T-lymphocyte counts below the commonly accepted 200 cell/μl discriminator between HIV and AIDS as well as across the region where increased testing is recommended [59]. The margin of error for the NBC is typically approximately 10%. Similar to the chemical processers, the CPU-2-NBC correlates well with existing technology, as typified in Figure 4E. In this mid-sized clinical study, the NBC correlates to flow cytometry at R2 = 0.93 for total lymphocyte count and R2 = 0.97 for T-lymphocyte counts [36,60].
Figure 4. Cellular processing unit.
Representative photomicrographs of whole blood labeled with a CD-specific antibody and quantum dot (QD) fluorescently tagged secondary antibody taken with a 10× objective and 3 s of exposure time. The QD 565 labels CD4+ cells including monocytes and T lymphocytes in the green channel (A) while QD 655 stains CD3+ lymphocytes red (B), as observed through separate filter cubes specific to each fluorophore. A digital overlap of the red and green images (C) shows monocytes (CD3+CD4+, green), T lymphocytes (CD3+CD4+, yellow) resulting from signal both in red and green channels, and remaining NK and CD8+CD3+ T-killer lymphocytes (CD3+CD4+, red). An alternative approach (D) utilizes a long-pass emission filter cube (520 nm and longer) allowing a single capture event to produce a similar image to that generated by separate photomicrographs. This membrane-based method was used to analyze a large sample set and found to correlate to flow cytometry for total lymphocyte counts at R2 = 0.93 and T lymphocyte counts at R2 = 0.97 (E) [36]. TLC: Total lymphocyte count.
In addition to lymphocyte enumeration in resource-poor settings, the same system can be tailored for cancer screens and other applications required in developed countries. Here, the CPU-2-NBC retasks to monitor oral muscosal cells for cancer, similar to a pathologist [41]. For this application, an oral brush biopsy collects cells for introduction into CPU-2-NBC to measure the nucleus, cytoplasm and EGF receptor (EGFR). By monitoring the size and shape of the cell as well as the EGFR levels, diagnostic information can be gleaned from the sample. When EGFR levels were monitored in three oral cancer cell types versus control cells, an increase in expression (p < 0.05) was observed above control cells. In correlation studies, NBC data matched flow cytometry (R2 = 0.98) and detected key differences (p < 0.01) between two of the oral cancer cell lines; a disparity of approximately 34,000 EGFR reporters per cell according to quantitative flow cytometry [41]. The cell surface limit of detection was determined to be 2500 copies of a given receptor. Another adaptation of the CPU-2-NBC approach is the detection of Bacillus globigii, a commonly used simulant of the bioterrorism threat, Bacillus anthracis. These experiments yielded limits of detection of approximately 500 spores, a value competitive with existing technology, but with turnaround times under 5 min [61].
Programmability: one core, many analytes
The creation of simple, programmable a la carte POC tool that provides near real-time results with integrated data outputs is needed both as a screening and prognostic tool. Of great significance would be the expansion of the strategic panels currently in use, with particular attention to current research aimed at identifying new biomarkers and incorporating those into use at the POC. Current POC devices such as the glucometer, pulse-oxygenation sensor and urine dipstick are limited in their inability to expand and address the dynamics of risk factors and biomarker discovery across all analyte classes. When fully developed, such tools would promise to make disease screening more user friendly for both the patient and physician.
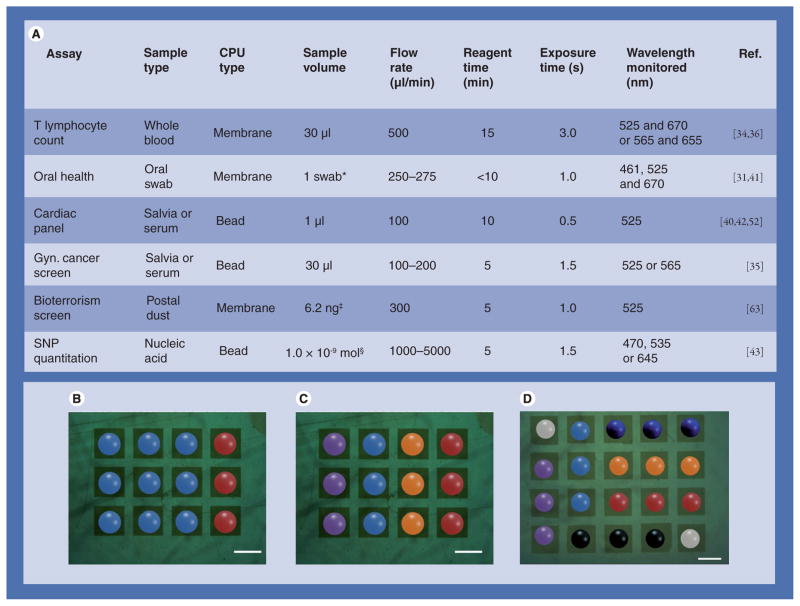
Figure 5A details how analytical programs for fluids, reagents, optics and samples are written in the NBC for various common laboratory procedures. Here, the same labcard and analyzer is used, but by programming the constituent experimental components and CPU type, markedly different analysis procedures result. The NBC’s documented capability in diverse medical areas including cardiac and cancer health, HIV monitoring, bioterrorism screens and nucleotide point mutation detection is significant and speaks to the system’s versatility. By maintaining the same fluid and light handling equipment across all of these assays, development times and costs are minimized and universality maintained. Assays designed in a strategic way from the same fundamental building blocks are integrated into the NBC, which now has a catalog of validated assays for easy expansion into customized applications and panels. Custom bead combinations, each with unique molecular-level code, change according to disease state. Figure 5B illustrates a one-analyte design, while Figure 5C indicates the easy expansion to three analytes. In cardiovascular medicine, for example, one could prepare different chip panels for the prevention visit (atherosclerosis or arteriosclerosis), the emergency department (chest pain etiology) and the congestive heart failure specialist. These profiles are created in a systematic fashion. By simply removing certain bead types and adding others, an entirely different panel is created, while retaining the core microchip and fluid flow of the chemical processer. Importantly, the chip is easily expandable as shown in Figure 5D. This 4 × 5 array allows more analytes as well as additional controls. Arrays sizes up to 10 × 10 have been prepared.
Figure 5. Programming the nano-bio-chip.
(A) Various hardware and software assay elements are programmed to result in the ideal combination suitable for analysis. The table in (A) highlights some of the experimental parameters that are optimized individually leading to ideal assay behavior in different nano-bio-chip programs. In addition to programming these parameters, the spatial code of beads placement and hence, molecular-level code of bioligands on the beads, is used to create programs specific to disease types. In (B) an assay for one analyte (blue) with control beads (red) is seen; (C) extends this to three different analytes (purple and orange) with the same negative control. (D) Larger arrays are also used and include positive control and calibrator beads. All scale bars are 300 μm. *Each oral swab contains thousands of epithelial cells suspended in buffer. Volumes injected into nano-bio-chip varied between 50 and 500 μl.
‡Validation studies used 1 ml Bacillus globigii at 6.2 μg/l.
§Sample volumes of 550 μl were used at a concentration of 3.2 μM 18-mer.
Conclusion & future perspective
With US per capita healthcare costs now above $7,400 annually, and rising 7–8% annually, it is clear that substantial changes are needed within the healthcare infrastructure [62]. A key factor in determining cost is clinical specimen testing and approaches that leverage the well-established production and performance reliability of microelectronics are clearly needed. The NBC and its use of mass produced, cost-effective materials is an important step in that direction. The use of stainless steel and agarose – two extremely common materials – serves as an essential step toward scalability. Scalable systems, easily adaptable to new analytical challenges, imply greater usage owing to universal access. Previously, the NBC was successfully deployed in resource-poor settings (Botswana) through the use of a cost-effective membrane CPU (type 2) [34]. With the motion from silicon to stainless steel type 1 CPUs, it is reasonable to project that similar broad uptake of this approach will occur in several other areas of significant clinical importance. When both developed and developing countries share a common diagnostic infrastructure as in electronics, significant healthcare progress can be made.
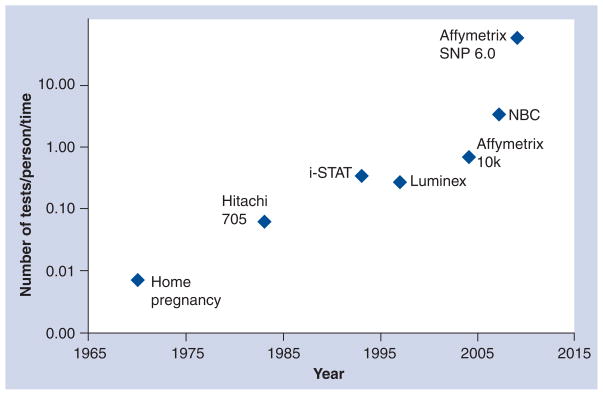
By retaining the core optical/fluid handling equipment and programming the NBC for panels specific to early detection and evaluation at the POC (or even home), substantial reductions are made versus existing approaches. Relatively nonspecific biomarkers yield selectivity and specificity as a collection, and future panels will incorporate even greater varieties of analytes into the same cohort. It seems reasonable to predict that genomic material will play an even greater role in the diagnosis and evaluation of disease. Finally, we project technological innovations including the NBC will lead to Moore’s Law type growth in POC diagnostic research [10]. We offer a new figure of merit that features the time needed to measure a collection of analytes for a given patient. Table 2 offers examples of both the time and assay components of various analysis systems and indicates that an increasing number of measurement events are occurring per unit time [63–65,102–105]. Figure 6 displays graphically the quotient, Q, of analysis events per unit time. As appropriate for POC diagnostics, time is defined here as assay time plus the transport (shipping) time plus the staging or preparation time. The total time value is used to compute a quotient describing the number of diagnostic measurements per patient that may made by each analysis approach. It is interesting to note that this figure continues to increase with each successive generation of equipment. While this analysis is not exhaustive and the time evolution of the data does not yield a perfect fit, the trend projected here is for a general tendency to provide more biomarker fingerprint data for each patient in a quicker time frame as history evolves similar to the Moore’s law trend for microelectronic devices.
Table 2.
An analysis of the amount of time required to complete a given number of tests by various techniques over time reveals an increasing trend in the quotient defined as number of tests that may be completed per patient in a unit of time.
| Manufacturer | Model | Use | Year | Tests | Transit time | Staging time | Analysis time | Total time | Q | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| Wampole | Home pregnancy | POC | 1970 | 1 | 0 | 20 | 120 | 140 | 0.01 | [102] |
| Hitachi | 705 | On-site | 1983 | 16 | 180 | 20 | 60 | 260 | 0.06 | [63] |
| I-STAT | PCA | POC | 1993 | 6 | 0 | 15 | 2 | 17 | 0.35 | [64] |
| Luminex | Flow metrix | On-site | 1997 | 64 | 180 | 20 | 30 | 230 | 0.28 | [65] |
| Affymetrix | 10K | Remote lab | 2004 | 10,240 | 4320 | 0 | 10,080 | 14,400 | 0.71 | [103] |
| NBC | NBC | POC | 2007 | 100 | 0 | 15 | 15 | 30 | 3.33 | [54] |
| Affymetrix | SNP 6.0 | Remote lab | 2009 | 1.8 × 106 | 4320 | 0 | 25,920 | 30,240 | 59.52 | [104] |
Transit time is defined as the shipping time for remote labs and or typical delivery time to the testing facility. For remote labs, a value of three days is used. For large dedicated instruments, once the sample is received it must wait its turn in the batch of samples. This is covered with the stage time. The final element is the sample processing and testing time. This is described as the analysis time.
NBC: Nano-bio-chip; POC: Point-of-care; Q: Quotient.
Figure 6. Moore’s Law-type growth.
The figure of merit detailed above describes tests per person per analytical system. Note the logarithmic increase in the past decades.
Just as computer microchip manufactures have allowed logarithmically increasing amounts of processing and storage to occur on smaller and cheaper chip footprint, so too have researchers in DNA sequencing and IVD. The electronics and software areas with proven scalability, target goals, standard operating systems and decrease in component costs serve as ideal models for future efforts in the micromedical device area.
Acknowledgments
The authors thank Jorge Wong for assistance in preparing Figure 1.
Footnotes
Financial & competing interests disclosure
The authors acknowledge funding provided by the NIH through the National Institute of Dental and Craniofacial Research (U01 DE017793). The content is solely the responsibility of the authors and does not necessarily represent or reflect views of the NIH or the Federal Government. John T McDevitt serves as the scientific founder for LabNow. The authors have applied for patents in areas related to nano-bio-chip sensor systems. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Contributor Information
Jesse V Jokerst, Email: jokerst@stanford.edu.
John T McDevitt, Email: mcdevitt@rice.edu.
Bibliography
- 1.Hanash SM, Pitteri SJ, Faca VM. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452(7187):571–579. doi: 10.1038/nature06916. [DOI] [PubMed] [Google Scholar]
- 2.Hunter DJ, Khoury MJ, Drazen JM. Perspective: letting the genome out of the bottle - will we get our wish? N Engl J Med. 2008;358(2):105–107. doi: 10.1056/NEJMp0708162. [DOI] [PubMed] [Google Scholar]
- 3.Anderson NL, Anderson NG. The human plasma proteome. History, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1(11):845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 4.Vitzthum F, Behrens F, Anderson NL, Shaw JH. Proteomics: from basic research to diagnostic application. A review of requirements and needs. J Proteome Res. 2005;4(4):1086–1097. doi: 10.1021/pr050080b. [DOI] [PubMed] [Google Scholar]
- 5.Yager P, Edwards T, Fu E, et al. Microfluidic diagnostic technologies for global public health. Nature. 2006;442(7101):412–418. doi: 10.1038/nature05064. [DOI] [PubMed] [Google Scholar]
- 6.Soper SA, Brown K, Ellington A, et al. Point-of-care biosensor systems for cancer diagnostics/prognostics. Biosens Bioelectron. 2006;21(10):1932–1942. doi: 10.1016/j.bios.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 7.Myers FB, Lee LP. Innovations in optical microfluidic technologies for point-of-care diagnostics. Lab Chip. 2008;8(12):2015–2031. doi: 10.1039/b812343h. [DOI] [PubMed] [Google Scholar]
- 8.Ouellette AL, Li JJ, Cooper DE, Ricco AJ, Kovacs GTA. Evolving point-of-care diagnostics using up-converting phosphor bioanalytical systems. Anal Chem. 2009;81(9):3216–3221. doi: 10.1021/ac900475u. [DOI] [PubMed] [Google Scholar]
- 9.Osterfeld SJ, Yu H, Gaster RS, et al. Multiplex protein assay based on real-time magnetic nanotag sensing. Proc Natl Acad Sci USA. 2008;105(52):20637–20640. doi: 10.1073/pnas.0810822105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thorsen T, Maerkl Sebastian J, Quake Stephen R. Microfluidic large-scale integration. Science. 2002;298(5593):580–584. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- 11.Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442(7101):368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 12.Vilkner T, Janasek D, Manz A. Micro total analysis systems. Recent developments. Anal Chem. 2004;76(12):3373–3385. doi: 10.1021/ac040063q. [DOI] [PubMed] [Google Scholar]
- 13.Mukhopadhyay R. Microfluidics: on the slope of enlightenment. Anal Chem. 2009;81(11):4169–4173. doi: 10.1021/ac900638w. [DOI] [PubMed] [Google Scholar]
- 14.Ligler FS. Perspective on optical biosensors and integrated sensor systems. Anal Chem. 2009;81(2):519–526. doi: 10.1021/ac8016289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toriello NM, Douglas ES, Thaitrong N, et al. Integrated microfluidic bioprocessor for single-cell gene expression analysis. Proc Natl Acad Sci USA. 2008;105(51):20173–20178. doi: 10.1073/pnas.0806355106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sia SK, Kricka LJ. Microfluidics and point-of-care testing. Lab Chip. 2008;8(12):1982–1983. doi: 10.1039/b817915h. [DOI] [PubMed] [Google Scholar]
- 17.McCarley RL, Vaidya B, Wei SY, et al. Resist-free patterning of surface architectures in polymer-based microanalytical devices. J Am Chem Soc. 2005;127(3):842–843. doi: 10.1021/ja0454135. [DOI] [PubMed] [Google Scholar]
- 18.Madou M, Zoval J, Jia G, et al. Lab on a CD. Annu Rev Biomed Eng. 2006;8:601–628. doi: 10.1146/annurev.bioeng.8.061505.095758. [DOI] [PubMed] [Google Scholar]
- 19.Waggoner PS, Craighead HG. Micro- and nanomechanical sensors for environmental, chemical, and biological detection. Lab Chip. 2007;7(10):1238–1255. doi: 10.1039/b707401h. [DOI] [PubMed] [Google Scholar]
- 20.de Mello AJ, Wootton RCR. Chemistry at the crossroads. Nat Chem. 2009;1(1):28–29. doi: 10.1038/nchem.156. [DOI] [PubMed] [Google Scholar]
- 21.Sanhai WR, Sakamoto JH, Canady R, Ferrari M. Seven challenges for nanomedicine. Nat Nanotech. 2008;3(5):242–244. doi: 10.1038/nnano.2008.114. [DOI] [PubMed] [Google Scholar]
- 22.Melin J, Quake SR. Microfluidic large-scale integration: the evolution of design rules for biological automation. Annu Rev Biophys Biomol Struc. 2007;36:213–231. doi: 10.1146/annurev.biophys.36.040306.132646. [DOI] [PubMed] [Google Scholar]
- 23.Holmesfarley SR, Bain CD, Whitesides GM. Wetting of functionalized polyethylene film having ionizable organic-acids and bases at the polymer water interface – relations between functional-group polarity, extent of ionization, and contact-angle with water. Langmuir. 1988;4(4):921–937. [Google Scholar]
- 24.Goluch ED, Nam J-M, Georganopoulou DG, et al. A bio-barcode assay for on-chip attomolar-sensitivity protein detection. Lab Chip. 2006;6(10):1293–1299. doi: 10.1039/b606294f. [DOI] [PubMed] [Google Scholar]
- 25.Qin L, Vermesh O, Shi Q, Heath JR. Self-powered microfluidic chips for multiplexed protein assays from whole blood. Lab Chip. 2009;9(14):2016–2020. doi: 10.1039/b821247c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Srivastava N, Brennan JS, Renzi RF, et al. Fully integrated microfluidic platform enabling automated phosphoprofiling of macrophage response. Anal Chem. 2009;81(9):3261–3269. doi: 10.1021/ac8024224. [DOI] [PubMed] [Google Scholar]
- 27.Chin CD, Linder V, Sia SK. Lab-on-a-chip devices for global health: Past studies and future opportunities. Lab Chip. 2007;7(1):41–57. doi: 10.1039/b611455e. [DOI] [PubMed] [Google Scholar]
- 28.Kim JS, Anderson GP, Erickson JS, et al. Multiplexed detection of bacteria and toxins using a microflow cytometer. Anal Chem. 2009;81(13):5426–5432. doi: 10.1021/ac9005827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walt DR. Chemistry: miniature analytical methods for medical diagnostics. Science. 2005;308(5719):217–219. doi: 10.1126/science.1108161. [DOI] [PubMed] [Google Scholar]
- 30.Cheng X, Gupta A, Chen C, et al. Enhancing the performance of a point-of-care CD4+ T-cell counting microchip through monocyte depletion for HIV/AIDS diagnostics. Lab Chip. 2009;9(10):1357–1364. doi: 10.1039/b818813k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer. N Engl J Med. 2008;359(4):366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mukhopadhyay R. When microfluidic devices go bad. Anal Chem. 2005;77(21):429A–432A. [PubMed] [Google Scholar]
- 33.Janasek D, Franzke J, Manz A. Scaling and the design of miniaturized chemical-analysis systems. Nature. 2006;442(7101):374–380. doi: 10.1038/nature05059. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez WR, Christodoulides N, Floriano PN, et al. A microchip CD4 counting method for HIV monitoring in resource-poor settings. PLoS Med. 2005;2(7):e182. doi: 10.1371/journal.pmed.0020182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jokerst JV, Raamanathan A, Christodoulides N, et al. Nano-bio-chips for high performance multiplexed protein detection: determinations of cancer biomarkers in serum and saliva using quantum dot bioconjugate labels. Biosens Bioelectron. 2009;24(12):3622–3629. doi: 10.1016/j.bios.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jokerst JV, Floriano PN, Christodoulides N, Simmons GW, McDevitt JT. Integration of semiconductor quantum dots into nano-bio-chip systems for enumeration of CD4+ T cell counts at the point-of-need. Lab Chip. 2008;8(12):2079–2090. doi: 10.1039/b817116e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goodey A, Lavigne JJ, Savoy SM, et al. Development of multianalyte sensor arrays composed of chemically derivitized polymeric microspheres localized in micromachined cavities. J Am Chem Soc. 2001;123:2559–2570. doi: 10.1021/ja003341l. [DOI] [PubMed] [Google Scholar]
- 38.Lavigne JJ, Savoy S, Clevenger MB, et al. Solution-based analysis of multiple analytes by a sensor array: toward the development of an ‘electronic tongue’. J Am Chem Soc. 1998;120(25):6429–6430. [Google Scholar]
- 39.Kilby JSC. Turning potential into realities: the invention of the integrated circuit. ChemPhysChem. 2001;2(8/9):482–489. doi: 10.1002/1439-7641(20010917)2:8/9<482::AID-CPHC482>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 40.Floriano P, Christodoulides N, Miller C, et al. Use of saliva-based nano-biochip tests for acute myocardial infarction at the point of care: a feasibility study. Clin Chem. 2009;55(8):1530–1538. doi: 10.1373/clinchem.2008.117713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weigum SE, Floriano PN, Christodoulides N, McDevitt JT. Cell-based sensor for analysis of EGFR biomarker expression in oral cancer. Lab Chip. 2007;7(8):995–1003. doi: 10.1039/b703918b. [DOI] [PubMed] [Google Scholar]
- 42.Christodoulides N, Floriano PN, Acosta SA, et al. Toward the development of a lab-on-a-chip dual-function leukocyte and C-reactive protein analysis method for the assessment of inflammation and cardiac risk. Clin Chem. 2005;51(12):2391–2395. doi: 10.1373/clinchem.2005.054882. [DOI] [PubMed] [Google Scholar]
- 43.Ali MF, Kirby R, Goodey AP, et al. DNA hybridization and discrimination of single-nucleotide mismatches using chip-based microbead arrays. Anal Chem. 2003;75:4732–4739. doi: 10.1021/ac034106z. [DOI] [PubMed] [Google Scholar]
- 44.Agnew HD, Rohde RD, Millward SW, et al. Iterative in situ click chemistry creates antibody-like protein-capture agents. Angew Chem Int Ed. 2009;48(27):4944–4948. doi: 10.1002/anie.200900488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Christodoulides N, Dharshan P, Wong J, et al. A microchip-based assay for interleukin-6. Meth Mol Bio. 2007;385:131–144. doi: 10.1007/978-1-59745-426-1_10. [DOI] [PubMed] [Google Scholar]
- 46.Gustavsson P-E, Axelsson A, Larsson P-O. Superporous agarose beads as a hydrophobic interaction chromatography support. J Chromatography A. 1999;830(2):275–284. doi: 10.1016/s0021-9673(98)00899-1. [DOI] [PubMed] [Google Scholar]
- 47.McCleskey SC, Griffin MJ, Schneider SE, McDevitt JT, Anslyn EV. Differential receptors create patterns diagnostic for ATP and GTP. J Am Chem Soc. 2003;125(5):1114–1115. doi: 10.1021/ja021230b. [DOI] [PubMed] [Google Scholar]
- 48.Wright AT, Anslyn EV, McDevitt JT. A differential array of metalated synthetic receptors for the analysis of tripeptide mixtures. J Am Chem Soc. 2005;127(49):17405–17411. doi: 10.1021/ja055696g. [DOI] [PubMed] [Google Scholar]
- 49.Christodoulides N, Mohanty S, Miller CS, et al. Application of microchip assay system for the measurement of C-reactive protein in human saliva. Lab Chip. 2005;5(3):261–269. doi: 10.1039/b414194f. [DOI] [PubMed] [Google Scholar]
- 50.Jackson DJ, Naber JF, Roussel TJ, et al. Portable high-voltage power supply and electrochemical detection circuits for microchip capillary electrophoresis. Anal Chem. 2003;75(14):3643–3649. doi: 10.1021/ac0206622. [DOI] [PubMed] [Google Scholar]
- 51.Christodoulides N, Floriano PN, Miller CS, et al. Lab-on-a-chip methods of point-of-care measurements of salivary biomarkers of periodontitis. Ann NY Acad Sci. 2007;1098:411–428. doi: 10.1196/annals.1384.035. [DOI] [PubMed] [Google Scholar]
- 52.Christodoulides N, Tran M, Floriano PN, et al. A microchip-based multianalyte assay system for the assessment of cardiac risk. Anal Chem. 2002;74(13):3030–3036. doi: 10.1021/ac011150a. [DOI] [PubMed] [Google Scholar]
- 53.Curey T, Goodey A, Tsao A, et al. Characterization of multicomponent monosaccharide solutions using an enzyme-based sensor array. Anal Biochem. 2001;293(2):178–184. doi: 10.1006/abio.2001.5114. [DOI] [PubMed] [Google Scholar]
- 54.Goodey A, Lavigne JJ, Savoy SM, et al. Development of multianalyte sensor arrays composed of chemically derivatized polymeric microspheres localized in micromachined cavities. J Am Chem Soc. 2001;123(11):2559–2570. doi: 10.1021/ja003341l. [DOI] [PubMed] [Google Scholar]
- 55.Goodey AP, McDevitt JT. Multishell microspheres with integrated chromatographic and detection layers for use in array sensors. J Am Chem Soc. 2003;125(10):2870–2871. doi: 10.1021/ja029696h. [DOI] [PubMed] [Google Scholar]
- 56.Kirby R, Cho EJ, Gehrke B, et al. Aptamer-based sensor arrays for the detection and quantitation of proteins. Anal Chem. 2004;76(14):4066–4075. doi: 10.1021/ac049858n. [DOI] [PubMed] [Google Scholar]
- 57.Wong DT. Salivary diagnostics powered by nanotechnologies, proteomics and genomics. JADA. 2006;137(3):313–321. doi: 10.14219/jada.archive.2006.0180. [DOI] [PubMed] [Google Scholar]
- 58.Herr AE, Hatch AV, Throckmorton DJ, et al. Microfluidic immunoassays as rapid saliva-based clinical diagnostics. Proc Natl Acad Sci USA. 2007;104(13):5268–5273. doi: 10.1073/pnas.0607254104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cohen J. Monitoring treatment: at what cost? Science. 2004;304(5679):1936–1936. doi: 10.1126/science.304.5679.1936. [DOI] [PubMed] [Google Scholar]
- 60.CDC. 1997 revised guidelines for performing CD4+ T-cell determinations in persons infected with human immunodeficiency virus (HIV) MMWR Morb Mortal Wkly Rep. 1997;46(RR-2):1–29. [PubMed] [Google Scholar]
- 61.Rodriguez WR, Christodoulides N, Floriano PN, et al. A microchip CD4 counting method for HIV monitoring in resource-poor settings. PloS Med. 2005;2(7):663–672. doi: 10.1371/journal.pmed.0020182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Floriano PN, Christodoulides N, Romanovicz DK, et al. Membrane-based on-line optical analysis system for rapid detection of bacteria and spores. Biosens Bioelectron. 2005;20(10):2079–2088. doi: 10.1016/j.bios.2004.08.046. [DOI] [PubMed] [Google Scholar]
- 63.Ginsburg PB, Strunk BC, Banker MI, Cookson JP. Tracking health care costs: continued stability but at high rates in 2005. Health Aff. 2006;25(6):w486–w495. doi: 10.1377/hlthaff.25.w486. [DOI] [PubMed] [Google Scholar]
- 64.Kineiko RW, Floering DA, Morrissey M. Laboratory evaluation of the Boehringer Mannheim ‘Hitachi 705’ automatic analyzer. Clin Chem. 1983;29(4):688–691. [PubMed] [Google Scholar]
- 65.Erickson KA, Wilding P. Evaluation of a novel point-of-care system, the i-STAT portable clinical analyzer. Clin Chem. 1993;39(2):283–287. [PubMed] [Google Scholar]
- 66.Fulton RJ, McDade RL, Smith PL, Kienker LJ, Kettman JR., Jr Advanced multiplexed analysis with the FlowMetrix system. Clin Chem. 1997;43(9):1749–1756. [PubMed] [Google Scholar]
Websites
- 101. [Accessed 07/30/2009];LabNow. www.labnow.com/
- 102.NIH. http://tinyurl.com/yfz8352.
- 103.Affymetrix. http://tinyurl.com/yh3yn43.
- 104.Affymetrix. http://tinyurl.com/ygxbks8.
- 105.McDevitt Research Laboratory. http://www.tastechip.com.