Abstract
Neurofibromatosis Type I (NF1) is a single-gene disorder characterized by a high incidence of complex cognitive symptoms, including learning disabilities, attention deficit disorder, executive function deficits, and motor coordination problems. Since the underlying genetic cause of this disorder is known, study of NF1 from a molecular, cellular, and systems perspective has provided mechanistic insights into the etiology of higher-order cognitive symptoms associated with the disease. In particular, studies of animal models of NF1 indicated that disruption of Ras regulation of inhibitory networks is critical to the etiology of cognitive deficits associated with NF1. Animal models of Nf1 identified mechanisms and pathways that are required for cognition, and represent an important complement to the complex neuropsychological literature on learning disabilities associated with this condition. Here, we review findings from NF1 animal models and human populations affected by NF1, highlighting areas of potential translation and discussing the implications and limitations of generalizing findings from this single-gene disease to idiopathic learning disabilities.
Keywords: Ras, GABA, LTP, animal model, neurodevelopmental disorder, ADHD
INTRODUCTION
Neurofibromatosis Type I (NF1) is one of a number of genetically determined neurocutaneous disorders that cause clusters of somatic and behavioral symptoms. Originally, these disorders were described according to their somatic symptoms, for example café au lait spots, Lisch nodules, and neurofibromas in NF1. Occurrence of these symptoms in a consistent pattern helped to identify a patient population affected by a single disease. Heading the human genetics revolution, linkage studies mapped the disease-causing gene in NF1 (NF1 (Neurofibromatosis Type I) to chromosome 17 (Barker et al. 1987, Seizinger et al. 1987). A small group of patients with NF1 were found to carry translocation breakpoints on chromosome 17, which greatly accelerated identification of the disease-causing gene (Fountain et al. 1989, Leach et al. 1989, Ledbetter et al. 1989). Further gene mapping within this region identified the Nf1 gene (Viskochil et al. 1990, Wallace et al. 1990). Although early on, these studies focused on the somatic symptoms of NF1, additional work within the NF1 patient population also revealed behavioral and learning problems (Hyman et al. 2005, North et al. 1997). These cognitive symptoms occur often, and have a significant impact on quality of life in individuals affected by NF1 (North et al. 1997).
NF1: Neurofibromatosis Type I
NF1 is a single gene--determined disorder characterized by multiple complex cognitive symptoms. Disease-causing mutations in the Nf1 gene (Viskochil et al. 1990, Wallace et al. 1990) result in loss of function of its protein product, Neurofibromin. Individuals affected by NF1 are heterozygous for the Nf1 gene mutation, as homozygous mutations appear to be lethal (Friedman 1999). Further study of these cognitive symptoms revealed that even in this single-gene disease, a complex picture of variable deficits and types of learning disabilities occurs. Here, we review human and animal studies that together have provided insight into the nature of cognitive symptoms associated with NF1 and the mechanisms underlying their expression and variability. Since NF1 and other neurocutaneous disorders are likely part of the broader cluster of learning disability disorders, we also discuss the current understanding of the evolution of cognitive symptoms in NF1 in the context of hypotheses regarding other learning disabilities with unknown etiology and poorly defined affected populations.
HUMAN GENETICS OF NF1
The Nf1 gene, located on chromosome 17q, is one of the largest in the genome, encompassing 60 exons (Figure 1) (Li et al. 1995, Marchuk et al. 1991). It encodes multiple biochemical domains, including a Ras-GAP domain, which requires exon 23a for its activity. The Nf1 gene has four splice variants, two of which are expressed in the CNS. The type I isoform of the Nf1 gene contains exon 23a, and as a result encodes a version of Neurofibromin with efficient Ras-GAP activity. This variant is predominantly expressed in neurons. The type II isoform of the gene is a splice variant without exon 23a, and encodes a protein with ten times less Ras-GAP activity than type I. This variant is highly expressed in glia. Therefore, differential splicing of the Nf1 gene confers different functions to its protein product across cell types.
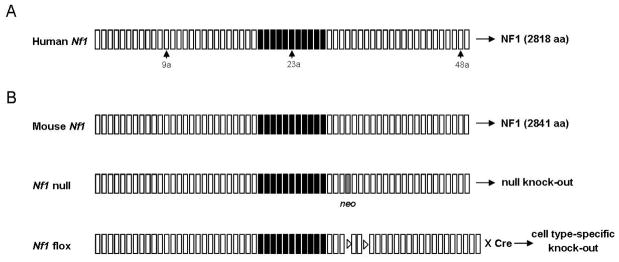
Figure 1.
Schematic representation of human and mouse Nf1. (a) Human Nf1 on chromosome 17 encodes a 2818--amino acid protein called neurofibromin. Small boxes represent exons. Black boxes indicate exons 21 to 27a that encode the GAP-related domain (GRD). Alternatively spliced exons are marked by arrows. Disease-causing mutations are widely distributed throughout the entire Nf1 gene and most of those result in loss of function of its protein product. (b) Top: the mouse Nf1 gene on chromosome 11 has a structure very similar to that of human Nf1 gene and encodes a protein with 98% identity to human NF1. Middle: Nf1 null mutant mice were generated by inserting a neomycin cassette in exon 31 (Jacks et al. 1994). Gray box indicates exon 31 including the neo gene. Bottom: conditional mutants were engineered by inserting loxP sites flanking exons 31 and 32 (Zhu et al. 2001). loxP sites are indicated by triangles. Delivery of Cre recombinase by crossing with Cre-expressing transgenic line or viral vector enables cell type-- specific deletion of Nf1.
Additionally, Nf1 expression occurs across multiple brain systems that participate in a broad variety of behaviors. Within the CNS, Nf1 transcription is seen in cortex, striatum, substantia nigra, brainstem, hippocampus, and cerebellum. In cortex and hippocampus, Nf1 is expressed in pyramidal neurons, interneurons, and glia. In cortex, Nf1 expression spans all cortical layers. In striatum, expression of Nf1 is sparse, occurring in a pattern suggestive of interneuronal expression (Gutmann et al. 1995). It is also highly expressed in the Purkinje neurons of the cerebellum. This broad range of brain systems in which Nf1 is expressed is also likely to contribute to the range of cognitive symptoms associated with its loss of function.
ROLE OF NEUROFIBROMIN IN CELL SIGNALING
The Nf1 protein product, Neurofibromin, has multiple biochemical roles (Figure 2). This protein acts as a Ras-GAP (GTPase activating protein) to negatively regulate Ras signaling (Weiss et al. 1999), and it can also serve as an activator of adenylate cyclase (Tong et al. 2002). Its role as a Ras- GAP has been most clearly implicated in regulating neuronal function in mammals.
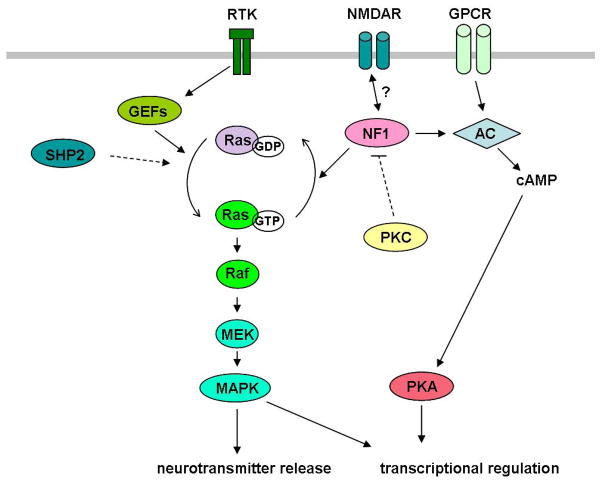
Figure 2.
Neurofibromin and cell signaling. Neurofibromin (NF1) is a GTPase activating protein (GAP) that functions as a negative regulator of Ras-MAPK signaling cascade. Guanine-nucleotide exchange factors (GEFs), such as SOS, may counteract the GAP function of NF1. On the other hand, NF1 acts as an activator of adenylate cyclase (AC). Note that the key pathways are grossly simplified in this diagram. For example, phosphatidylinositol 3-kinase (PI3K)-AKT-mTOR cascade is also modulated by NF1, but is omitted in the diagram. Arrows and barred lines indicate activation and suppression, respectively. MEK, mitogen-activated protein kinase or extracellular signal-regulated kinase kinase; NMDAR, N-methyl-D-aspartate receptor; GPCR, G proteincoupled receptor; RTK, receptor tyrosine kinase; PKA, protein kinase A; PKC, protein kinase C; SHP2, Src homology 2-containing tyrosine phosphatase.
The Ras protein is part of a large superfamily of GTPases that mediate signaling from membrane receptors to intracellular cascades of kinases. Ras cycles between the inactive GDP-bound state and the active GTP-bound state. The activity state of Ras is determined by a balance of activating proteins (GEFs, guanine nucleotide exchange factors, which allow bound GDP to be released so that GTP can bind) and inactivating proteins (GAPs, GTPase activating proteins, which increase the endogenous GTP hydrolyzing activity of Ras) (Bernards & Settleman 2004). Receptor tyrosine kinases, including the growth factor receptors (e.g., Trk B, the receptor for BDNF), are common upstream activators of Ras signaling (Bernards & Settleman 2005). This family of transmembrane receptors is characterized by phosphorylation of intracellular tyrosines upon ligand binding. Tyrosine phosphorylation then leads either to direct binding and inactivation of Ras-GAPs such as Neurofibromin, or to indirect activation of Ras-GEFs (Patapoutian & Reichardt 2001, Weiss et al. 1999). Since this complex of receptors, Ras-GAPs, and Ras-GEFs resides at the membrane, Ras must also be tethered to the cell membrane to be activated. At the membrane, Ras can interact with downstream effectors such as Raf kinase, which then activate signaling cascades such as the MEK/MAPK pathway (Boguski & McCormick 1993, Weiss et al. 1999). These kinases then induce a number of downstream processes, including transcription of immediate early genes, which lead to short- and long-term changes in neuronal function.
In neurons, loss of Neurofibromin results in constitutive increases in Ras intracellular signaling (Li et al. 2005). Further, Neurofibromin loss of function can allow Ras signaling to become decoupled from extracellular triggers such as growth factors. For example, Nf1−/− sensory neurons no longer require the extracellular growth factor BDNF to survive and mature. This is presumably due to constitutively increased levels of activity of the Ras pathway when it is no longer subject to negative regulation by Neurofibromin (Vogel et al. 1995). It is currently unclear exactly which receptors are upstream of Neurofibromin regulation of Ras signaling during behavior. However, Neurofibromin is known to be quickly regulated by PKC in response to growth factors that utilize the tyrosine kinase receptors (EGF), and by ubiquitin-dependent proteolysis in response to both Gprotein (LPA) and tyrosine kinase receptor ligands (EGF, PDGF).
The Ca2+-sensitive PKC-alpha isoform quickly (within 1 min) phosphorylates the N terminal region of Neurofibromin in neuronal cultures. This phosphorylation increases Neurofibromin association with actin and maximizes its activity. Thus, this phosphorylation could also make the Ras-GAP activity of Neurofibromin activity dependent (Leondaritis et al. 2009). In this context, Neurofibromin may normally act to downregulate the Ras response to growth factors in an activitydependent manner. The same growth factors can also quickly (by 5 min) but transiently induce ubiquitin-dependent proteolysis of Neurofibromin in an activity-independent manner. Thus, following growth factor release, Neurofibromin may play a key role in limiting and narrowing the time of Ras activation (Cichowski et al. 2003). As both forms of regulation of Neurofibromin were identified in embryonic culture systems, it remains to be seen whether similar regulation of Neurofibromin occurs in adult neuronal networks, whether it occurs downstream of growth factor receptors such as the BDNF receptors, and how either one or both mechanisms regulate neuronal networks during behavior.
Finally, although the receptors that utilize Neurofibromin to regulate Ras signaling in adult neurons have not been identified, one possible upstream receptor outside the growth factor receptors is the NMDA receptor. Neurofibromin is part of the NMDA receptor complex in the mouse forebrain (Husi et al. 2000). The NMDA receptor does interact with intracellular signaling cascades through its C terminus, in a manner that is independent of Ca2+ influx and critical to normal performance of delayed alternation, a behavior related to working memory (Bannerman et al. 2008). Although the functional relevance of Neurofibromin’s localization to the NMDA receptor is unknown, it suggests that this protein plays a role in NMDA receptor dependent regulation of Ras signaling.
Working memory: a flexible, continually updated system that maintains and manipulates information across short delay periods
In addition to its role as a Ras-GAP, Neurofibromin acts as an activator of adenylate cyclase (Tong et al. 2002). Learning defects in Nf1 null Drosophila melanogaster can be rescued by expression of a constitutively active form of PKA (Guo et al. 2000). These and other data suggested that the associative learning impairments in Nf1 null flies are due to decreased activation of adenylate cyclase. Neurofibromin can increase adenylate cyclase activity in a Ras-dependent manner. This form of signaling occurs in response to growth factors such as EGF. Neurofibromin can also directly increase adenylate cyclase activity in a Ras-independent manner, through its Cterminal domain. This function of Nf1 is required for stimulation of adenylate cyclase by neurotransmitters such as serotonin or histamine (Hannan et al. 2006). Nf1 loss in Drosophila causes specific phenotypes, such as small body size, through a direct decrease of adenylate cyclase activity (Tong et al. 2002). Other behavioral functions of Nf1, such as circadian rhythm modulation in Drosophila, are MAPK dependent and require the GAP domain (Williams et al. 2001). Still unclear is whether these biochemical effects of homozygous Nf1 deletion are relevant to the behavioral and cognitive deficits associated with NF1, which are caused by heterozygous mutations. In both mouse and Drosophila models, heterozygous deletion of Nf1 does not grossly affect regulation of the adenylate cyclase pathway (Tong et al. 2002). Nevertheless, it is possible that Nf1 plays a role in maintaining the relative balance between Ras- and cAMP-dependent signaling (Weeber & Sweatt 2002).
The Neurofibromin protein interacts with a number of upstream regulators of Ras signaling, and has the potential to play multiple roles within neurons as part of various intracellular pathways. This may contribute to the range of phenotypes that have been observed in Nf1+/−.
COGNITIVE PROFILE OF NF1 PATIENTS
NF1 is characterized by widespread symptoms affecting multiple organ systems (Lynch & Gutmann 2002, Williams et al. 2009). Among these symptoms are prominent cognitive impairments, which pose one of the most significant sources of lifetime morbidity for patients (North et al. 1997). NF1 specifically affects executive and other higher-order cognitive functions. In contrast, NF1 does not cause global cognitive impairments. In measures of global cognitive function, performance of affected individuals is comparable to unaffected siblings (Kayl & Moore 2000).
Executive function impairments are prevalent and can be severe in NF1. When specific cognitive domains are probed in neuropsychological batteries, 80%–90% of individuals affected by NF1 show impairments (Hyman et al. 2005, Krab et al. 2008a). In particular, NF1 affects planning, visuospatial function, reading/vocabulary, and motor coordination (Hofman et al. 1994, Hyman et al. 2005). Additional executive function deficits are prominently seen in working memory, cognitive flexibility, and inhibitory control (Rowbotham et al. 2009). There is also a high comorbidity between NF1 and attention deficit disorder (Hofman et al. 1994, Hyman et al. 2005, Mautner et al. 2002). In up to 40% of children with NF1 who are identified as academic underachievers, underachievement is likely due to deficits in attention, planning, and organizational skills (Dilts et al. 1996, Hyman et al. 2005, Kayl & Moore 2000, Koth et al. 2000, North 2000). Comorbid attention deficit disorder affects not only academic but also social development for children with NF1 (Barton & North 2004). Children with NF1 tend to have social problems and appear socially awkward and withdrawn, in comparison to their siblings and to children with other chronic, life-threatening illnesses (Kayl & Moore 2000). These social deficits could be related to poor interpersonal skills resulting from decreased attention to social cues.
Developmental learning disabilities are also highly associated with NF1. Overall, individuals with NF1 are fourfold more likely to require special education (Krab et al. 2008a), and learning disabilities are diagnosed in up to 65% of individuals affected by NF1(Rosser & Packer 2003). Learning disabilities are characterized by impairments restricted to specific domains of mental function, leading to a discrepancy between tests of intellectual capability and actual achievement(Kelly 2004, Kronenberger & Dunn 2003). Both intellectual capability and achievement are typically assessed using standardized, normalized tests. Learning disability is diagnosed by a discrepancy of one to two standard deviations between achievement and intelligence test scores.
DSM-IV places learning disabilities into five major categories: reading disorders, mathematical disorders, disorders of written expression, nonverbal learning disorders, and learning disorders not otherwise specified (Kelly 2004, Kronenberger & Dunn 2003, Palumbo & Lynch 2006). In addition, learning disabilities may be diagnosed according to specific symptoms, such as in dyslexia (reading disorder) or dyscalculia (math learning disorder). Thus, the criteria for diagnosis of learning disabilities are largely based on different systems of representing symptoms. However, a single learning disability category is often insufficient to describe all the symptoms of a given individual with idiopathic learning disability (Lagae 2008), or a group of individuals with an inherited learning disability such as NF1.
In fact, learning disabilities caused by NF1 show some characteristics of both nonverbal- and verbal-type learning disability. Components of nonverbal learning disability seen in NF1 include consistently poor performance in tests of visuospatial functioning and spatial learning (Kelly 2004), notable impairments in the ability to perceive social cues, poor organizational skills, and increased impulsiveness (Kayl & Moore 2000, North 2000). NF1 patients also show aspects of verbal learning disorder. Specifically, patients with NF1 have deficits in expressive and receptive language, vocabulary, visual naming, and phonologic awareness. In fact, reading and spelling are repeatedly found to be impaired more severely than predicted by IQ in NF1 patients (North 2000, North et al. 1997). Consistent with these impairments in language-based learning, NF1 patients show poorer academic achievement in reading and writing compared to their unaffected siblings. The pattern of learning disabilities seen in NF1 does not fall cleanly into one of the DSM-IV defined categories, but rather shares features with multiple learning disability categories. This shows that a single disorder, in this case NF1, can cause learning disabilities of varying phenotypes and presentations. Therefore, the various classifications and subdivisions of learning disabilities (for example, verbal versus nonverbal) may not actually represent different disease entities but rather different manifestations of a common disease cluster caused by a common set of underlying factors. In NF1, the drastic variability of symptom expression appears to be determined largely by independently inherited genetic modifiers.
GENETIC MODIFIERS OF BEHAVIORAL EXPRESSION OF NF1
Inheritance of NF1 shows an interesting pattern of complete genetic penetrance but variable expressivity (Ward & Gutmann 2005). Most people with an inactivating mutation in one allele of the Nf1 gene show some symptoms of NF1. However, the clinical presentation of NF1 ranges from minimal symptom load to extremely high severity across many types of symptoms. One important factor underlying variable expressivity of cognitive symptoms in the NF1 population is the contribution of modifying genes, some of which may significantly alter behavior in the presence of mutant Nf1, but not in a wild-type (WT) background. Studies in both mouse models and human patient populations suggest that inheritance of genetic modifiers accounts for the majority of variability in NF1 expression (Easton et al. 1993, Sabbagh et al. 2009).
Genetic penetrance: the proportion of individuals carrying a genetic variant that also show phenotypic/symptomatic manifestations associated with that variant
Variable genetic expression: indicates the range of phenotypic/symptomatic severity associated with the same genetic variant across phenotypes and different individuals; frequently seen in dominant genetic conditions
WT: wild type
In NF1 mouse models, the phenotypic effect of the Nf1 mutation depends on the background strain on which that mutation is expressed. This has been shown for both behavioral and somatic symptoms of NF1. Different strains of mice (for example, C57Bl/6J versus DBA/2J) carrying the Nf1+/− mutation have been shown to have different susceptibility to the formation of astrocytomas (Hawes et al. 2007). Similar studies have shown that the background strain also affects the behavioral phenotype of the Nf1 mutation (Costa 2002). Since these strains are engineered with the same Nf1 gene mutation, phenotypic differences across strains are attributed to differential expression of modifier genes across mouse strains that were inbred from genetically different founders. Such modifiers may interact with the gene directly, altering its level of expression, or, more likely, may interact on a more functional level to exacerbate or compensate for the signaling changes caused by loss of Nf1. The latter is likely to be most relevant as studies quantifying differences in levels of Nf1 expression across strains find that although expression levels can vary dramatically, they do not always correlate with phenotype expression (Hawes et al. 2007). On the other hand, introducing functional modifiers of learning pathways can exacerbate the learning phenotype of Nf1+/− mice. This was demonstrated using a heterozygous null mutation of a receptor (NMDA; NR1+/−) critical for learning and memory. Although the NR1+/− mutation alone does not have a spatial learning phenotype, it exacerbates the spatial learning phenotype of the Nf1+/− mutant mice (Silva et al. 1997). Therefore, mouse studies indicate that background genetic modifiers alter the expression of Nf1+/−-related phenotypes, likely through functional interactions with the cellular pathways altered by the Nf1 mutation.
In human patient studies, heritable genetic modifiers have also been found to affect expression of NF1, and current studies are in progress to identify specific loci encoding these modifiers. Analyses of large groups of extended families affected by NF1 suggest that the expression of NF1 symptoms is heritable. Variation in symptom expression between family members is consistent with a model of inheritance of modifier genes with little contribution from specific mutations to the Nf1 gene itself or from variations in the WT Nf1 allele (Easton et al. 1993, Sabbagh et al. 2009). Among multiple clinical symptoms of NF1, including learning disabilities and referral for remedial education, symptom expression shows high correlation among first-degree family members (who share 50% of their genes). Similarly, monozygotic twins affected by NF1 have a high correlation in symptom severity (Easton et al. 1993). As second- and third-degree family members are examined (who share 25% and 12.5% of their genes, respectively), the correlation in symptom expression decreases sharply. The strong genetic component in symptom expression in NF1 is consistent with modulation of disease symptoms by inherited genetic modifiers (Easton et al. 1993, Sabbagh et al. 2009). For example, proteins encoded by genetic modifiers may interact with the Neurofibromin signaling pathway to exacerbate effects of Nf1 mutations or confer protection by compensating for the loss of function.
These studies of NF1 highlight the regulation of clinical presentation and symptom severity by inheritable genetic modifiers, and suggest that these modifiers may have a more dramatic impact in the presence of the Nf1 mutation than in a WT background. Similar findings have been made in other genetically determined learning disabilities, such as Noonan syndrome (NS). NS is another common autosomal dominant genetic disorder with an incidence of 1 in ~2500 live births. It is characterized by facial abnormalities, short stature, webbed neck, motor delay, and cardiac disease (Noonan 1994, Tartaglia & Gelb 2005). Importantly, NS patients also show increased rates of learning disabilities and mental retardation (Lee et al. 2005, Money & Kalus 1979). Recently, Araki and colleagues generated NS mouse models by engineering NS-associated PTPN11 mutants. They demonstrated that the gain-of-function mutants show phenotypes similar to that found in NS patients (Araki et al. 2009, Araki et al. 2004). The penetrance of NS phenotypes is dependent on the specific Ptpn11 allele studied as well as the genetic background of the mutant mice (Araki et al. 2009). Ptpn11 mutations that modify tyrosine phosphatase SHP2 activity result in different heart defects. Half of the NS Ptpn11D61G/+ mutant mice are embryonic lethal on a mixed 129S4/SvJae X C57BL/6 genetic background (Araki et al. 2004). However, the Ptpn11D61G/+ mutation backcrossed to C57Bl\6 revealed high lethality. In contrast, when crossed into 129S6/SvEv genetic background, this mutation showed normal viability, suggesting that there are modifier allele(s) that affect the viability of the NS Ptpn11D61G/+ mutation (Araki et al. 2009). The examples described above illustrate the dramatic influence of modifier genes on the clinical presentation of a single-gene disorder.
NF1 AND PHENOTYPES OF ANIMAL MODELS
Mice with a heterozygous null mutation of the Nf1 gene (Nf1+/−) have been the dominant rodent model used to study NF1. The Nf1+/− mice show compelling genetic and behavioral parallels with human NF1, making them useful system in which to study the mechanisms of behavioral phenotypes associated with NF1. Nevertheless, a word of caution is in order: There is a very large evolutionary gulf between mice and humans, and therefore the behavioral phenotypic parallels between animal models and NF1 must not be overinterpreted. Much needs to be done to fully elucidate the apparent parallels between rodent and human behavior.
The sequence, transcriptional regulation, and downstream targets of Nf1 are conserved across species, including mouse and human (Bernards et al. 1993, Hajra et al. 1994). The majority of mutations (70%) of the Nf1 gene in humans affected by NF1 lead to synthesis of a truncated, nonfunctional version of the encoded Neurofibromin protein (Shen et al. 1996, Thomson et al. 2002). Accordingly, the Nf1+/−mouse model was made by inserting a neo gene in exon 31 of the Nf1 gene, which leads to an unstable, quickly degraded transcript (Figure 1) (Jacks et al. 1994). Like patients with NF1, the Nf1+/− mouse model is heterozygous for this loss-of-function mutation. Behavioral phenotypes of the Nf1+/− mouse show a pattern of specific impairments in certain domains and preserved function in others. This pattern of behavioral phenotypes is reminiscent of the pattern of behavioral and cognitive symptoms seen in humans affected by NF1. Based on the genetic, biochemical, and behavioral parallels between the Nf1+/− mouse model and human NF1, it is thought that this mouse offers a useful model of the behavioral and cognitive symptoms associated with the disorder. The Nf1+/− mouse has therefore been utilized to identify electrophysiological and molecular mechanisms that contribute critically to cognitive and behavioral changes associated with NF1.
Conditional mutants of the Nf1 gene have also been extensively studied to identify the effects of Nf1 deletion within specific neuronal types. Conditional mutations can be created in mice using the Cre-loxP system, a powerful tool widely used for restricting gene deletions to specific time frames, cell types, or areas. A mouse line was engineered with loxP sites flanking exons 31–32 of the Nf1 gene (Nf1flox/flox) (Figure 1). The floxed Nf1 gene acts like a WT allele prior to expression of Cre recombinase. Mice carrying one floxed Nf1 allele and one deleted Nf1 allele (Nf1flox/−) show the same phenotypes as Nf1+/− mice. For example, they have the same survival profile: Deletion of the floxed Nf1 allele following delivery of Cre recombinase caused embryonic lethality, similar to the Nf1−/− mutation (Zhu et al. 2001). Conditional Nf1 mutants have revealed cell-specific roles of Nf1 and interesting interactions between neighboring cells with different Nf1 gene doses. Such interactions are critical to the development of some of the somatic events associated with NF1 such as neurofibromas (Zhu et al. 2002).
Finally, mouse models targeting specific biochemical domains of the Nf1 gene have been examined to demonstrate the relative importance of the various regulatory functions of the Nf1 protein product, Neurofibromin. For example, exon 23a of the Nf1 gene is spliced out in peripheral tissue, but expressed in neuronal Nf1. Presence of exon 23a increases the Ras regulatory activity of Neurofibromin, suggesting that this represents a uniquely important aspect of Nf1 loss in the brain. Indeed, many of the behavior deficits identified in Nf1+/− mice were also found in Nf123a−/− mice, which carry a homozygous deletion restricted to exon 23 of the Nf1 gene (Costa et al. 2001). This deletion creates a version of Neurofibromin that is functional but has diminished Ras regulatory activity. Importantly, the Nf123a−/− mice show learning disability--related behavioral deficits, but none of the other somatic consequences of Nf1 mutation such as increased tumor predisposition (Costa et al. 2001). This not only demonstrates that Neurofibromin has a neuron-specific role as a Ras regulator, but also that altered behavioral performance in these mutant mice is a direct function of their Nf1 mutation, rather than a secondary effect of tumor formation. Detailed studies have characterized the behavioral consequences of Nf1 mutation.
TASKS USED TO ASSESS BEHAVIOR IN ANIMAL MODELS OF NF1
The behavioral tasks used to study disease mechanisms in Nf1+/− mice were chosen according to the following criteria: (a) face/construct validity to tasks performed less accurately by NF1 patients, (b) dependency on brain areas that contribute critically to cognitive functions affected by NF1, and (c) behaviors with well-established molecular and cellular underpinnings. To date, the behavioral paradigms used in the study of Nf1+/− mice were predominantly tests sensitive to hippocampal and parietal/prefrontal cortical function. This result reflects the prominence of symptoms thought to stem from dysfunction in these brain areas in NF1 patients. The Morris water maze was initially utilized in Nf1+/− mice as a test of spatial learning and memory that is highly sensitive to hippocampal function (Morris et al. 1982). Additionally, performance in the Morris water maze requires specific synaptic and cellular mechanisms, such as long-term potentiation (LTP) (Moser et al. 1998; Silva et al. 1992a,b). As such, this behavioral paradigm provided a bridge between functional/mechanistic changes in Nf1+/− mice and a mouse behavioral phenotype that could be related to the spatial learning deficits seen in NF1 patients. In the Morris water maze, mice are placed in a pool of opaque water in which there is an escape platform. The test subjects must use the spatial cues surrounding the pool to learn the location of the platform and navigate to it, a process that usually takes multiple trials over several days. Following training, memory of the spatial location of the platform is tested in probe trials where the platform is removed, and the experimenter measures the amount of time mice spend searching for the missing platform in the appropriate quadrant of the pool. In a control task (unperturbed by the Nf1+/− mutation), the position of the escape platform is directly marked by an object, and the mice just have to learn to go to it to escape from the water.
Long-term potentiation (LTP): a form of synaptic plasticity where high-frequency stimulation leads to increased strength of synaptic transmission. In hippocampus, LTP is thought to be required for learning and memory
Extensive studies with the water maze and other hippocampal-dependent tasks helped to define the behavioral deficits in Nf1 mice and unravel underlying mechanisms (Costa et al. 2001, 2002; Cui et al. 2008; Silva et al. 1997). Subsequent behavioral studies have begun to address some of the prominent cognitive symptoms of NF1. Specific behavioral paradigms were utilized that have been developed in rodents to model core cognitive functions commonly affected in human disease. For example, the lateralized reaction time task was used in Nf1+/− mice to test attention deficit/hyperactivity (Li et al. 2005), as attention deficits are highly associated with NF1 in humans. Although attention, like memory and other behaviors modeled in mice, has important differences across species, there are also conserved elements that the behavioral tasks used in rodent models utilize (see side bar: Animal models in human behavior). In this case, the lateralized reaction time task was designed to test attention processes in a manner that captures key features of tasks used to probe attention in humans (Robbins 2002). Further, performance of the rodent task requires the prefrontal cortex, an area thought to contribute to attention in humans. Performance of rats in the lateralized reaction time task is also improved by stimulants used to treat ADHD (attention deficit and hyperactivity disorder) in humans (Jentsch et al. 2009), supporting common mechanistic underpinnings. The lateralized reaction time task is implemented in an operant chamber and requires mice or rats to fixate on a central hole while attending to two potential cues, one on either side of the center. In multiple, serial, self-initiated trials, one of the cues is lit for varying lengths of time, and the rodent must maintain attention divided across both sides of space in order to notice and respond to the lit cues. Beyond attention, similar tasks have also been adapted to examine working memory in rodents (Aarde & Jentsch 2006), in a manner analogous to the Sternberg style working memory tests used in humans (Cannon et al. 2005). These tasks provide very useful methods that allow for parallel studies in rodents and humans of core symptoms of NF1, including attention deficit and the working memory impairments often found in learning disabilities. As more evidence accumulates from the use of these tasks in rodents and humans, we will be able to better understand which behavioral mechanisms have been conserved across species and which tasks are best able to probe conserved mechanisms in the context of disease. This offers exciting new prospects for understanding diseases, such as NF1, through an integration of cognitive, clinical, and basic neuroscience approaches (Chadman et al. 2009, Fossella & Casey 2006).
ADHD: attention deficit and hyperactivity disorder
Animal Models of Human Behaviors A key question often raised by animal models of human disease is whether the model captures all of the essential features of the disease. Despite the fact that some animal models only address a subset of the features of a disease, they can still be very useful for understanding the disease and developing treatments. To understand the complexity of the human condition a variety of different models is needed, not necessarily one with all of the features of the disorder. Another challenge is to extrapolate findings from animal models across species, despite differences between human and rodent control over cognition, timing, lifespan, and organization and development of brain structures such as prefrontal cortex. Currently, several approaches are being used in rodent models of disease. Behavioral batteries have been developed for animals to test for disease-relevant clusters of phenotypes.
Appropriate and high-throughput behavioral testing has been developed with demonstrated face, construct, and some predictive validity with human disease symptoms (Chadman et al. 2009). Also, specific behavioral tasks have been developed for animals to model cognitive functions commonly affected in human disease. For example, the 5-choice serial reaction time task (5CSRTT) is one example of a behavioral paradigm that is designed to test multiple attention processes in a manner highly analogous to the probing of those processes in humans (Robbins 2002). Mechanisms contributing to normal performance of this task have been extensively characterized, including the neuromodulatory systems and brain areas that mediate normal performance of the 5CSRTT in rodents (Robbins 2002). This paradigm has also been adapted to examine working memory in rodents (Aarde & Jentsch 2006), in a manner analogous to the Sternberg style working memory tests used in humans (Cannon et al. 2005). Tasks are also being developed for use in humans to parallel key features of commonly used rodent tasks (Demeter et al. 2008). The use of these tasks to obtain convergent evidence from multiple approaches offers exciting new prospects for understanding disease through the integration of the cognitive, clinical, and basic neurosciences (Chadman et al. 2009, Fossella & Casey 2006).
PHENOTYPES OF NF1 ANIMAL MODELS AND PARALLELS TO HUMAN COGNITIVE SYMPTOMS
The phenotypes identified in Nf1 animal models in the tasks described above demonstrate that mutation of the Nf1 gene in animal models causes impairments with striking analogy to that seen in the learning disabilities associated with NF1 in humans (Table 1). These phenotypes of the Nf1+/− mice can be improved with extended training, a feature also observed in human learning disabilities.
Table 1.
Cognitive phenotypes of NF1 mouse models
| Mouse Model | Behavioral phenotypes | Parallel human symptoms | References |
|---|---|---|---|
| Nf1+/− | Impaired in | ||
| Morris water maze (hidden platform | Learning deficits in specific domain (visual-spatial) | Silva et al. (1997) | |
| Contextual fear conditioning | Cui et al. (2008) | ||
| Lateralized reaction time task | Attention deficits | Li et al. (2005) | |
| Pre-pulse inhibition | |||
| Normal in | |||
| Open field and visible platform water maze | |||
| Cued fear conditioning | |||
| Nf123a−/− | Impaired in | Learning deficits in specific domain (visual-spatial) | Costa et al. (2001) |
| Morris water maze (hidden platform | |||
| Contextual discrimination | Motor deficits | ||
| Rota-rod | |||
| Normal in | |||
| Open field and visible platform water maze | |||
| Social transmission of food preference | |||
| Nf1flox/+;SynI-cre | Impaired in | Learning deficits in specific domain (visual-spatial) | Cui et al. (2008) |
| Nf1flox/+;Dlx5/6-cre | Morris water maze (hidden platform | ||
| Normal in | |||
| Visible platform water maze |
Nf1+/− mice show spatial learning deficits in the hidden version of the Morris water maze (Costa et al. 2001, 2002; Cui et al. 2008; Silva et al. 1997). Probe trials given early on during water maze training revealed that Nf1+/− mice require more training trials than controls to learn the position of the hidden platform. Thus, when searching for the missing platform in probe trials, the mutant mice spent less time in the appropriate quadrant compared to their WT littermates. Additional training led to continued improvement in spatial memory in Nf1+/− mice, such that probe trials given after additional days of training no longer revealed differences between genotypes. This sensitivity to overtraining is also a feature reported in NF1-associated learning disabilities in patients. Similarly, Nf1+/− mice also show deficits in contextual conditioning, a test where mice associated a novel chamber with a mild footshock (Cui et al. 2008). As with the water maze, contextual conditioning has a spatial learning component and requires hippocampal function.
In contrast to the significantly impaired spatial learning seen in Nf1+/− mice, neither visual learning nor simple associative learning is prominently disrupted in these mice (Costa et al. 2001, 2002; Cui et al. 2008; Silva et al. 1997). Performance of the Nf1+/− mice in the visual version of the Morris water maze is indistinguishable from WT. Additionally, Nf1+/− mice show no deficits in tone fear conditioning, where mice learn to associate a tone with a mild footshock. Unlike the Morris water maze, tone fear conditioning is a single-trial, simpler associative learning paradigm that requires amygdala function. Therefore, Nf1 mutation has very specific effects on spatial learning while sparing simple associative learning, possibly due to a greater effect on hippocampal function (Costa et al. 2002, Silva et al. 1997). This pattern of specific impairments is also seen in learning disabilities associated with NF1 in patients.
Finally, as NF1 subjects, Nf1+/− mice show attention deficits in the lateralized reaction time task (Li et al. 2005). ADHD is highly associated with NF1, and thought to contribute to academic and social changes in this patient population. In the lateralized reaction time task, Nf1+/− mice perform as accurately as WT mice when cues are lit for longer intervals (up to 1 s). However, as attention is taxed with shorter light presentations (0.5 s), a failure to maintain constant attention in the Nf1+/− mice is revealed. The Nf1+/− mice make significantly more omissions than do WT (Li et al. 2005). Humans with NF1 often show a type of ADHD characterized by increased omissions and lapses of attention (Hyman et al. 2005, Mautner et al. 2002).
These animal data demonstrate that mutation to the Nf1 gene is sufficient to directly cause a specific pattern of complex behavioral changes in mice. The behavioral phenotypes of the Nf1+/− mice in these tasks show analogy to the learning disabilities and other cognitive symptoms that characterize NF1. However, various differences clearly exist between mouse and human. For example, the anatomical organization and connectivity of prefrontal cortex and other regions likely to be critical for the NF1 phenotype are different between the two species. Additionally, the complexity of the human NF1 phenotype can only be partially captured in animal models. This and other factors make direct translation from mouse to human studies difficult and unpredictable. In the context of NF1, where a large literature of both animal and human studies exists, it will be interesting to follow future experiments evaluating which specific molecular mechanisms, cellular effects, and behavioral paradigms carry translational value.
MULTIPLE PHENOTYPES LINKED TO THE SAME MECHANISM
Cognitive effects of the Nf1+/− mutation in mouse have been seen in diverse processes, from hippocampal memory to quick, flexible, prefrontal/parietal cortex-dependent attention processes. Although distinct underlying circuits and brain mechanisms mediate these cognitive functions, in NF1 both memory and attention are impaired by the same genetic change. In Nf1+/− mice, both deficits in memory and in attention are caused by increased Ras signaling, resulting from the loss of regulation by Neurofibromin (Costa et al. 2002, Cui et al. 2008, Li et al. 2005). To demonstrate the Ras dependency of deficits in the Morris water maze, Nf1+/− mice were crossed to null Ras mutants (K-Ras+/− or N-Ras−/− mice). As a result of the decreased levels of Ras activity in the Ras mutants, the Nf1/Ras double mutants performed at the same level as did WT mice (Costa et al. 2002), even though each mutant individually (Nf1+/− and K-Ras+/−) showed deficits in this task. Similarly, performance of Nf1+/− mice in the Morris water maze was rescued with farnesyl transferase inhibitors under conditions that did not enhance the performance of controls. These inhibitors pharmacologically decrease levels of Ras signaling (Costa et al. 2002) by blocking the posttranslational modification that is necessary for Ras to associate with the cellular membrane and therefore be active. This mechanism of action can also be targeted with a class of drugs, the statins, which also decrease isoprenylation (Li et al. 2005). Statins are relatively safe and are widely used to control cholesterol levels. In both the Morris water maze and in the lateralized reaction time task, lovastatin, a drug in the statin class, normalized the performance of Nf1+/− mice to levels indistinguishable from those of WT (Li et al. 2005). Importantly, statins also normalized the signaling and the neurophysiological deficits of the Nf1+/− mice. Hence, regardless of method used, decreases in Ras signaling rescue both the learning impairments and the attention deficits of the Nf1+/− mice. These results also confirm the wide range of cognitive dimensions affected by alterations of Ras signaling. The observation that multiple symptoms are caused by disruption of a single biochemical process is not specific to NF1, but is also seen following mutations to other Rasregulatory genes. For example, SPRED1, another negative regulator of Ras-MEK/MAPK signaling, is responsible for a NF1-like syndrome (Legius syndrome) (Brems et al. 2007). Similar to Nf1, lossof- function mutations of SPRED1 also result in hyperactivity in MEK/MAPK signaling. The disorder associated with Spred1 mutations involves some of the same symptom dimensions (other than neurofibromas) seen in NF1(Pasmant et al. 2009). This includes NF1-like cognitive deficits in affected individuals (Brems et al. 2007, Pasmant et al. 2009, Spurlock et al. 2009). Spred1 knockout mice (Spred1−/−) have been generated (Brems et al. 2007, Denayer et al. 2008) and found to show deficits in hippocampus-dependent learning and memory tasks, which may model cognitive deficits in patients with Spred1 mutation (Denayer et al. 2008). Therefore, disruption of the Ras signaling pathway through SPRED1 can also lead to a complex, multifaceted disorder. These disorders demonstrate that multiple types of symptoms can all be caused by alterations in a single gene/biochemical component.
INCREASED GABA RELEASE FROM INTERNEURONS UNDERLIES Nf1 BEHAVIORAL PHENOTYPES
Nf1 deletion and increased Ras signaling has cell type--specific physiological effects that contribute to behavioral symptoms. In particular, Nf1expression in interneurons seems to be critical for behavior, whereas its role in pyramidal neurons appears to be nonessential for hippocampal learning in the strains of mice and behaviors tested (Cui et al. 2008). In the Morris water maze, heterozygous deletion of Nf1 from inhibitory interneurons is sufficient to cause behavioral impairments. Conversely, deletion of Nf1 from either pyramidal neurons or glia does not alter behavior in this task under the conditions tested. Thus, regulation of Ras signaling by Nf1 is particularly critical within interneurons, but perhaps less so in pyramidal neurons (Cui et al. 2008). Within the hippocampus of Nf1+/− mice, increased interneuronal Ras signaling causes an increase in activity-dependent GABA release (Figure 3). This increased activity-dependent release of GABA (Cui et al. 2008) leads to larger evoked inhibitory currents in CA1, shifting the balance between inhibitory and excitatory processes within hippocampal networks of the mutant mice. As a result, LTP is impaired in the Nf1+/− mice, perhaps because the increased inhibition prevents sufficient depolarization of NMDARs during learning (Cui et al. 2008). In the hippocampus, there is strong evidence that LTP is required for learning of spatial and contextual information (Lee & Silva 2009, Martin et al. 2000, Martin & Morris 2002, Richter-Levin et al. 1995). Additionally, the very manipulations that reverse the learning deficits of the Nf1+/− mice (decreases in ras signaling with statins or farnesyl transferase inhibitors), also reverse their LTP impairments. As additional training rescues the learning impairments of the mutants, additional synaptic stimulation (e.g., higher frequency) also rescues their LTP deficits. Therefore, the LTP deficits are thought to underlie the learning deficits seen in the Nf1+/− mice. Supporting this assertion, both the Nf1+/− LTP and water maze deficits can be improved using picrotoxin, a GABAA receptor antagonist (Cui et al. 2008), at concentrations that do not affect these phenomena in controls. These data demonstrate that Neurofibromin is an important regulator of inhibitory tone in hippocampal neuronal networks. This regulation of inhibition is functionally important as its disruption impairs behaviors, such as the acquisition of spatial and contextual information. Again, beyond manipulations of inhibition, Lovastatin, farnesyl transferase inhibitors, and the N-ras and K-ras mutations, all reverse both the LTP and the learning deficits of the Nf1+/− mutant mice, strengthening the link between the hippocampal LTP deficits of these mice and their hippocampal-dependent learning deficits (Costa et al. 2002, Li et al. 2005).
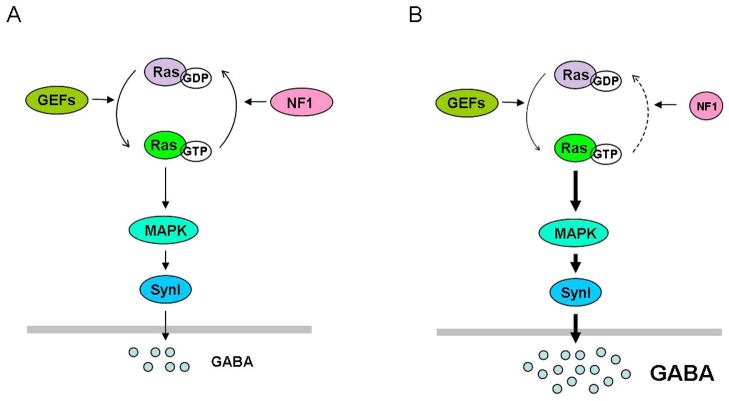
Figure 3.
Proposed cellular mechanism underlying learning deficits of Nf1 mutant mice. (a) Learning triggers interneuronal Ras signaling leading to increased GABA release. MAPKdependent phosphorylation of synapsin I (SynI) plays a critical role in GABA release. Wild-type NF1 restricts the increase in GABA release within an appropriate range that modulates learning. (b) In Nf1 mutants, reduced NF1 activity leads to abnormal hyperactivation of Ras signaling in inhibitory interneurons during learning, resulting in abnormally high GABA release. This increased activity-dependent GABA release shifts the balance between excitatory and inhibitory processes in neuronetworks of the mutant mice and impairs synaptic plasticity needed for learning and memory.
In other brain areas such as the prefrontal cortex and striatum, inhibition also plays an important role in regulating network phenomena that are critical for cognitive functions affected in NF1, including working memory and attention. In prefrontal cortex, working memory--related cellular activity involves a balance between a high degree of excitation (allowing inputs to induce persistent activity) and a coordinated set of inhibitory mechanisms (imposing specificity to the activity). This balance between excitation and inhibition allows prefrontal cortex to encode information across delays (Compte et al. 2000). Activity in prefrontal cortex is regulated by both feedforward and feedback inhibitory microcircuits (Constantinidis et al. 2002, Hasenstaub et al. 2005, Sanchez- Vives & McCormick 2000), which can affect the onset and duration of persistent activity (Compte et al. 2000, Fellous & Sejnowski 2003). In addition, inhibition acts to tune persistent activity of prefrontal neurons to specifically represent cue location, and to organize temporal interactions between prefrontal neurons (Constantinidis & Goldman-Rakic 2002, Constantinidis et al. 2002). Decreasing inhibition within prefrontal cortex leads to loss of spatial tuning, which results in inaccurate working memory performance. Decreasing inhibition also unmasks spatial tuning in previously silent neurons. During a working memory task, this spatial tuning results in inappropriate, pre-emptive behavioral responses (Rao et al. 2000). Therefore, inhibitory interneurons in the prefrontal cortex have multiple functions during behavior, an observation that could easily account for the attention and executive deficits in both Nf1 mice and NF1 subjects.
Similarly, striatal inhibitory interneurons modulate the activity of local medium spiny neurons. Inhibitory currents induced at the soma of medium spiny neurons by striatal interneurons can abolish or delay the onset of spiking (Koos & Tepper 1999, Tepper et al. 2008), making GABAergic interneurons a primary modulator of activity flow in the striatum. Importantly, striatal inteneurons are driven by glutamatergic inputs, including those descending from the cortex. Therefore, abnormally high levels of inhibition seen in Nf1+/− mice would be expected to significantly alter activity within the striatum, the prefrontal cortex, and the timing and flow of activity between striatum and cortex. Increased inhibition resulting from Nf1 mutation likely underlies not only learning and memory deficits but also other behavioral phenotypes of the Nf1+/− mice that are more closely related to dysfunction in frontal/cortical brain areas (e.g., attention, executive function).
Increased inhibition can lead to behavioral deficits by altering either the ongoing function of neuronal networks in the adult brain, or by affecting neuronal development. In the Nf1+/− mice, increased inhibition in the adult brain contributes critically to behavioral deficits, since these deficits can be rescued in adult Nf1+/− mice by decreasing inhibition (e.g., using picrotoxin), or by normalizing Ras signaling (e.g., with statins). This finding suggests that these behavioral deficits occur as a result of a reversible increase in inhibition. However, it is important to also consider potential effects of increased inhibition on development of neuronal networks, since most Nf1 gene mutations in patients and mice are present from birth. GABAergic inhibition plays an important role in the developmental patterning of neuronal networks, and so the Nf1 mutation may cause developmental defects that correlate with symptoms of NF1 such as learning disabilities. The distinction between developmental and adult causes of NF1 is relevant for deciding which features of NF1 could be targeted for treatment.
During development, experience-dependent plasticity mediates organization of neuronal networks using mechanisms shared with adult experience-dependent plasticity. Therefore developmental plasticity is likely to be disrupted by increased inhibition in NF1 by the same increase in inhibition that disrupts LTP in adult Nf1+/− mice. Further, GABAergic inhibitory networks play a role in regulating the developmental patterning of cortical networks by modulating the length of developmental sensitive periods. Sensitive periods are windows of time during development when experience has a greater effect on the organization of neuronal networks than in adulthood (Hensch 2004, Knudsen 2004). In primary sensory areas, the sensitive period is regulated by the maturation and activity of GABAergic interneurons. For example, tonic increases in GABAergic activity correlate with the closure of the critical period for ocular dominance in the visual cortex (Fagiolini & Hensch 2000, Hensch 2005). In NF1, increased inhibition may cause inappropriate, early closure of sensitive periods leading to altered patterning in cortical areas. Possibly related to increased inhibition, or to other effects of Nf1 deletion, cortical barrels fail to form in mice where Nf1 is homozygously deleted from neurons and astrocytes of the somtatosensory cortex (Lush et al. 2008). NF1 may be associated with changes in primary sensory processing that are caused by increased inhibition, but may not directly cause the higher-order cognitive symptoms such as learning disabilities. Consistent with this increased inhibition, learning disabilities are often found in association, but not correlated with, alterations in primary sensory processing (Ramus 2003). Because the severity of learning disabilities does not correlate well with severity of alterations in sensory processing, such sensory processing deficits are not likely to directly cause learning disabilities. In NF1, such associations may simply arise from a common underlying mechanism. Alternatively, it is possible that the increase in inhibition characterized in the Nf1+/− mutant mice is sufficient to cause learning deficits, but not irreversible developmental changes. Further studies are needed to better understand interactions between developmental effects of NF1, any associated sensory processing deficits, and behavioral effects of constitutively altered Ras signaling/interneuronal regulation.
LEARNING DISABILITIES AS A DISTINCT PATHOLOGICAL ENTITY: LESSONS FROM NF1
A key question concerning learning disabilities is whether they represent a distinct disease cluster or rather the low end of a spectrum of performance in the human population. This is a question that is common to discussions of many disorders of cognitive function. For example, similar discussions in schizophrenia, autism, and Alzheimer’s have concluded that these conditions are not part of a behavioral and cognitive phenotypic continuum in the human population, but that instead represent separate pathological clinical entities. Accordingly, individuals suffering from each of these conditions account for a very small percentage of the population (1%–2%). With respect to learning disabilities, there is little agreement in the literature on the prevalence of learning disabilities, but estimates on incidence range from 1% to 17% (Altarac & Saroha 2007, Lagae 2008). It is argued that an incidence of 10% is consistent with the idea that learning disability is not a single clinical entity, but reflects instead the wide distribution of cognitive traits in the population (normal spectrum hypothesis). Furthermore, the classification criteria used for these estimates are often broad, and these high manifestations of learning disability are also argued to be consistent with the normal spectrum hypothesis.
The studies of learning disabilities associated with single-gene disorders contradict these arguments. These studies demonstrate that the cognitive deficits defined in these individuals have unique pathological causes and can be classified into discrete clinical conditions with specific diagnoses. Despite their broad clinical manifestations, the studies with NF1, Fragile X, Tuberous Sclerosis, Noonan’s and Rett’s syndrome, to name only a few, indicate that a wide range of cognitive symptoms can be caused by single-gene mutations. Thus, broad behavioral and cognitive profiles do not necessarily reflect either the low end of the cognitive continuum of the general population, or the presence of an equally broad and diffuse pathological etiology. Rather, in these diseases, it is clear that a broad range of cognitive phenotypes are caused by specific genetic mutations leading to distinct clinical entities.
The results from single-gene disorders also demonstrate that a wide range of severities can be caused by the same mutation in a single gene. For example, learning disability and attention deficit symptoms occur in NF1 in a continuum of severity, and they are known to be caused by a single underlying genetic pathology. In fact, some individuals affected by NF1 show very few cognitive manifestations, whereas others reveal profound, albeit often specific, cognitive deficits. In NF1, genetic factors modify the severity and expression of symptoms, such that the variability in clinical presentation does not reflect the uniformity of the principle genetic cause (i.e., mutation in the NF1 gene). Among idiopathic learning disabilities, it is also likely that variability in expression between individuals occurs as a result of other unknown mutations in the genetic background of affected individuals (i.e., modifier genes), as well as environmental factors. Therefore, variability in expression of learning disabilities between individuals cannot be taken to imply variability in, or lack of, a distinct underlying clinical pathology. These considerations also lend support to the model of learning disabilities as a distinct syndrome with underlying pathological mechanisms that alter learning, rather than being a low end of the normal cognitive spectrum of the general population.
As in schizophrenia, autism, and Alzheimer’s, a small percentage of individuals with learning disabilities have known genetic mutations (e.g., mutations in the Nf1 gene), whereas the genetic etiology of most individuals is unknown. As in these other disorders, the severity and range of symptoms manifested in any one affected individual can vary, perhaps reflecting the interplay between genetic and environmental factors. Again, as in these well-described disorders, there is a cluster of phenotypes that represent the clinical diagnostic criteria, as well as other clinically important comorbidities that are not part of the disorder itself. For example, epilepsy and mental retardation are often associated with autism, but they are not an intrinsic part of the diagnosis of this condition, just as attention problems are often associated with learning disabilities. Thus, learning disabilities share these and other features with other complex mental health problems. This complexity and diversity is not the exception but the rule in current classifications of disorders of brain function.
Identifying learning disabilities as a disorder, rather than a variation of “normal” learning, has more than purely academic implications, as it bears on approaches to treatment. Learning disabilities have been identified as one of the primary factors affecting quality of life in the NF1 population, even given the high incidence of tumors and serious somatic symptoms that are also associated with this condition. Additionally, without intervention, individuals affected with NF1 more often than not have difficulties in effectively compensating for their deficits in learning and attention. From the perspective of affected individuals, family members, and professional caregivers, the cognitive and behavioral challenges associated with NF1 require professional intervention and are both unique and distinguishable from the normal cognitive spectrum in the general population. The difficulty in compensating for significant cognitive impairments also differentiates the learning problems associated with these disorders from those seen in “normal” populations. This need for specialized care, as well as the considerable negative repercussions of underlying pathologies on the life of affected individuals, further emphasizes the necessity to recognize learning disabilities as a distinct disorder that requires early detection, intervention, and treatment.
Importantly, early treatment in NF1 and other learning disabilities can be effective in improving cognition, as measured by neuropsychological testing as well as academic and social achievement. In the NF1 population, low doses of methylphenidate, which may address their attention problems, appear to improve performance in school as well as attention (Mautner et al. 2002). In idiopathic learning disabilities, current interventions include unified programs, involving phonology training, compensatory strategy training, as well as involvement of school teachers and parents. Such cognitive and behavioral therapy-based programs are also effective in improving academic outcomes, especially when initiated early (Lagae 2008). The benefits of therapeutic intervention for learning disabilities seem clearly to outweigh the risks of treatment. Therefore, it is critical to recognize learning disabilities as a disease entity that requires early diagnosis and intervention to improve quality of life in affected individuals.
ADULT TREATMENT OF NEURODEVELOPMENTAL DISORDERS: THE CASE FOR NF1
There is a reasonable and well-entrenched assumption that neurodevelopmental disorders such as NF1 disrupt developmental processes that cannot be fully reversed in adult individuals, and that hope for treatment rests in early (fetal stages) diagnosis and intervention (Ehninger et al. 2008). Indeed, there are compelling data that many of the genes that affect adult function also affect important developmental processes. For example, the Nf1 gene affects trophic function during rodent development (Luikart et al. 2008, Vogel et al. 1995). It is reasonable to propose that disruption of these developmental processes could lead to neurophysiological and neuroanatomical abnormalities irreversible in adult individuals. Additionally, there is compelling evidence that the development of the brain has critical periods, where key neurodevelopmental events must take place, outside of which these same processes no longer can be fully realized. For example, developmental studies of vision identified critical periods with narrow temporal windows, where high levels of sensory plasticity are important for the large-scale organization of specific areas of the visual cortex (Hensch 2005). The idea of developmental critical periods has further strengthened the bias that it may not be possible to reverse the pathologies associated with neurodevelopmental disorders in adult individuals.
Recent results with a number of animal models of neurodevelopmental disorders, including NF1, Fragile X, Tuberous Sclerosis, Rett syndrome, Lhermitte-Duclos disease/Cowden disease, Rubinstein-Taybi syndrome, Down syndrome, etc, demonstrate that it is possible to dramatically improve, if not fully reverse, the cognitive phenotypes associated with these neurodevelopmental disorders in adult animals (Ehninger et al. 2008). When the underlying biochemical and physiological pathologies are reversed in adult individuals, the cognitive phenotypes could be either dramatically improved or even fully reversed without necessarily intervening during development (Ehninger et al. 2008)! For example, studies reviewed above showed that brief treatments in adult mutants with either Lovastatin, a farnesyl transferase inhibitor, or picrotoxin could reverse the physiological and behavioral phenotypes of the Nf1+/− mutant mice. More importantly, recent clinical trials suggested that statins may be effective at reversing at least some of the cognitive phenotypes of NF1 patients (Krab et al. 2008b). Similarly, findings in Fragile X also suggest that treating a key physiological deficit in adult mice could reverse clinically relevant phenotypes in mouse models of this disease (Dolen et al. 2007). Additionally, results from pilot clinical trials are also starting to suggest that similar treatments may also be effective in Fragile X patients (Berry- Kravis et al. 2009, Hagerman et al. 2009).
It is too early to gauge the efficacy of adult treatments for neurodevelopmental disorders, since the interpretation of the few pilot clinical trials carried out so far is confounded by design flaws and limited numbers of subjects. Nevertheless, studies in animal models are starting to uncover the molecular, cellular, and systems mechanisms disrupted by neurodevelopmental disorders, as well as to use this information to develop targeted treatments. Importantly, these studies have led to recent results raising the exciting possibility of dramatically improving the life of individuals afflicted with neurodevelopmental disorders, even when treatments are started in adulthood.
Acknowledgments
This work is supported by the CTF Young Investigators Award, ARCS Foundation, and NIH MCNB Training Grant (2T32MH019384-11A2) to C.S. This work was supported by grants from the NIH (R01 NS38480), Neurofibromatosis Inc., the Children’s Tumor Foundation, and United States Army (W81XWH-06-1-0174) to A.J.S.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Aarde SM, Jentsch JD. Haploinsufficiency of the arginine-vasopressin gene is associated with poor spatial working memory performance in rats. Horm Behav. 2006;49:501–8. doi: 10.1016/j.yhbeh.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Altarac M, Saroha E. Lifetime prevalence of learning disability among US children. Pediatrics. 2007;119(Suppl 1):S77–83. doi: 10.1542/peds.2006-2089L. [DOI] [PubMed] [Google Scholar]
- Araki T, Chan G, Newbigging S, Morikawa L, Bronson RT, Neel BG. Noonan syndrome cardiac defects are caused by PTPN11 acting in endocardium to enhance endocardial-mesenchymal transformation. Proc Natl Acad Sci USA. 2009;106:4736–41. doi: 10.1073/pnas.0810053106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki T, Mohi MG, Ismat FA, Bronson RT, Williams IR, et al. Mouse model of Noonan syndrome reveals cell type- and gene dosage-dependent effects of Ptpn11 mutation. Nat Med. 2004;10:849–57. doi: 10.1038/nm1084. [DOI] [PubMed] [Google Scholar]
- Bannerman DM, Niewoehner B, Lyon L, Romberg C, Schmitt WB, et al. NMDA receptor subunit NR2A is required for rapidly acquired spatial working memory but not incremental spatial reference memory. J Neurosci. 2008;28:3623–30. doi: 10.1523/JNEUROSCI.3639-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker D, Wright E, Nguyen K, Cannon L, Fain P, et al. Gene for von Recklinghausen neurofibromatosis is in the pericentromeric region of chromosome 17. Science. 1987;236:1100–2. doi: 10.1126/science.3107130. [DOI] [PubMed] [Google Scholar]
- Barton B, North K. Social skills of children with neurofibromatosis type 1. Dev Med Child Neurol. 2004;46:553–63. doi: 10.1017/s0012162204000921. [DOI] [PubMed] [Google Scholar]
- Bernards A, Settleman J. GAP control: regulating the regulators of small GTPases. Trends Cell Biol. 2004;14:377–85. doi: 10.1016/j.tcb.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Bernards A, Settleman J. GAPs in growth factor signaling. Growth) Factors. 2005;23:143–49. doi: 10.1080/08977190500130480. [DOI] [PubMed] [Google Scholar]
- Bernards A, Snijders AJ, Hannigan GE, Murthy AE, Gusella JF. Mouse neurofibromatosis type 1 cDNA sequence reveals high degree of conservation of both coding and noncoding mRNA segments. Hum Mol Genet. 1993;2:645–50. doi: 10.1093/hmg/2.6.645. [DOI] [PubMed] [Google Scholar]
- Berry-Kravis E, Hessl D, Coffey S, Hervey C, Schneider A, et al. A pilot open label, single dose trial of fenobam in adults with Fragile X syndrome. J Med Genet. 2009;46:266–71. doi: 10.1136/jmg.2008.063701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguski MS, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–54. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- Brems H, Chmara M, Sahbatou M, Denayer E, Taniguchi K, et al. Germline loss-of-function mutations in SPRED1 cause a neurofibromatosis 1-like phenotype. Nat Genet. 2007;39:1120–26. doi: 10.1038/ng2113. [DOI] [PubMed] [Google Scholar]
- Cannon TD, Glahn DC, Kim J, Van Erp TG, Karlsgodt K, et al. Dorsolateral prefrontal cortex activity during maintenance and manipulation of information in working memory in patients with schizophrenia. Arch Gen Psychiatry. 2005;62:1071–80. doi: 10.1001/archpsyc.62.10.1071. [DOI] [PubMed] [Google Scholar]
- Chadman KK, Yang M, Crawley JN. Criteria for validating mouse models of psychiatric diseases. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:1–11. doi: 10.1002/ajmg.b.30777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichowski K, Santiago S, Jardim M, Johnson BW, Jacks T. Dynamic regulation of the Ras pathway via proteolysis of the NF1 tumor suppressor. Genes Dev. 2003;17:449–54. doi: 10.1101/gad.1054703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compte A, Brunel N, Goldman-Rakic PS, Wang XJ. Synaptic mechanisms and network dynamics underlying spatial working memory in a cortical network model. Cereb Cortex. 2000;10:910–23. doi: 10.1093/cercor/10.9.910. [DOI] [PubMed] [Google Scholar]
- Constantinidis C, Goldman-Rakic PS. Correlated discharges among putative pyramidal neurons and interneurons in the primate prefrontal cortex. J Neurophysiol. 2002;88:3487–97. doi: 10.1152/jn.00188.2002. [DOI] [PubMed] [Google Scholar]
- Constantinidis C, Williams GV, Goldman-Rakic PS. A role for inhibition in shaping the temporal flow of information in prefrontal cortex. Nat Neurosci. 2002;5:175–80. doi: 10.1038/nn799. [DOI] [PubMed] [Google Scholar]
- Costa R, Elgersma Y, Silva AJ. Modeling cognitive disorders: from genes to therapies. In: Fisch G, editor. Genetics and Genomics of Neurobehavioral Disorders. Totowa, NJ: Humana Press; 2003. [Google Scholar]
- Costa RM, Federov NB, Kogan JH, Murphy GG, Stern J, et al. Mechanism for the learning deficits in a mouse model of neurofibromatosis type 1. Nature. 2002;415:526–30. doi: 10.1038/nature711. [DOI] [PubMed] [Google Scholar]
- Costa RM, Yang T, Huynh DP, Pulst SM, Viskochil DH, et al. Learning deficits, but normal development and tumor predisposition, in mice lacking exon 23a of Nf1. Nat Genet. 2001;27:399–405. doi: 10.1038/86898. [DOI] [PubMed] [Google Scholar]
- Cui Y, Costa RM, Murphy GG, Elgersma Y, Zhu Y, et al. Neurofibromin regulation of ERK signaling modulates GABA release and learning. Cell. 2008;135:549–60. doi: 10.1016/j.cell.2008.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeter E, Sarter M, Lustig C. Rats and humans paying attention: cross-species task development for translational research. Neuropsychology. 2008;22:787–99. doi: 10.1037/a0013712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denayer E, Ahmed T, Brems H, Van Woerden G, Borgesius NZ, et al. Spred1 is required for synaptic plasticity and hippocampus-dependent learning. J Neurosci. 2008;28:14443–49. doi: 10.1523/JNEUROSCI.4698-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilts CV, Carey JC, Kircher JC, Hoffman RO, Creel D, et al. Children and adolescents with neurofibromatosis 1: a behavioral phenotype. J Dev Behav Pediatr. 1996;17:229–39. [PubMed] [Google Scholar]
- Dolen G, Osterweil E, Rao BS, Smith GB, Auerbach BD, et al. Correction of Fragile X syndrome in mice. Neuron. 2007;56:955–62. doi: 10.1016/j.neuron.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DF, Ponder MA, Huson SM, Ponder BA. An analysis of variation in expression of neurofibromatosis (NF) type 1 (NF1): evidence for modifying genes. Am J Hum Genet. 1993;53:305–13. [PMC free article] [PubMed] [Google Scholar]
- Ehninger D, Li W, Fox K, Stryker MP, Silva AJ. Reversing neurodevelopmental disorders in adults. Neuron. 2008;60:950–60. doi: 10.1016/j.neuron.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagiolini M, Hensch TK. Inhibitory threshold for critical-period activation in primary visual cortex. Nature. 2000;404:183–86. doi: 10.1038/35004582. [DOI] [PubMed] [Google Scholar]
- Fellous JM, Sejnowski TJ. Regulation of persistent activity by background inhibition in an in vitro model of a cortical microcircuit. Cereb Cortex. 2003;13:1232–41. doi: 10.1093/cercor/bhg098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossella JA, Casey BJ. Genes, brain, and behavior: bridging disciplines. Cogn Affect Behav Neurosci. 2006;6:1–8. doi: 10.3758/cabn.6.1.1. [DOI] [PubMed] [Google Scholar]
- Fountain JW, Wallace MR, Bruce MA, Seizinger BR, Menon AG, et al. Physical mapping of a translocation breakpoint in neurofibromatosis. Science. 1989;244:1085–7. doi: 10.1126/science.2543076. [DOI] [PubMed] [Google Scholar]
- Friedman JM. Epidemiology of neurofibromatosis type 1. Am J Med Genet. 1999;89:1–6. [PubMed] [Google Scholar]
- Guo HF, Tong J, Hannan F, Luo L, Zhong Y. A neurofibromatosis-1-regulated pathway is required for learning in Drosophila. Nature. 2000;403:895–98. doi: 10.1038/35002593. [DOI] [PubMed] [Google Scholar]
- Gutmann DH, Geist RT, Wright DE, Snider WD. Expression of the neurofibromatosis 1 (NF1) isoforms in developing and adult rat tissues. Cell Growth Differ. 1995;6:315–23. [PubMed] [Google Scholar]
- Hagerman RJ, Berry-Kravis E, Kaufmann WE, Ono MY, Tartaglia N, et al. Advances in the treatment of Fragile X syndrome. Pediatrics. 2009;123:378–90. doi: 10.1542/peds.2008-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajra A, Martin-Gallardo A, Tarle SA, Freedman M, Wilson-Gunn S, et al. DNA sequences in the promoter region of the NF1 gene are highly conserved between human and mouse. Genomics. 1994;21:649–52. doi: 10.1006/geno.1994.1328. [DOI] [PubMed] [Google Scholar]
- Hannan F, Ho I, Tong JJ, Zhu Y, Nurnberg P, Zhong Y. Effect of neurofibromatosis type I mutations on a novel pathway for adenylyl cyclase activation requiring neurofibromin and Ras. Hum Mol Genet. 2006;15:1087–98. doi: 10.1093/hmg/ddl023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasenstaub A, Shu Y, Haider B, Kraushaar U, Duque A, McCormick DA. Inhibitory postsynaptic potentials carry synchronized frequency information in active cortical networks. Neuron. 2005;47:423–35. doi: 10.1016/j.neuron.2005.06.016. [DOI] [PubMed] [Google Scholar]
- Hawes JJ, Tuskan RG, Reilly KM. Nf1 expression is dependent on strain background: implications for tumor suppressor haploinsufficiency studies. Neurogenetics. 2007;8:121–30. doi: 10.1007/s10048-006-0078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensch TK. Critical period regulation. Annu Rev Neurosci. 2004;27:549–79. doi: 10.1146/annurev.neuro.27.070203.144327. [DOI] [PubMed] [Google Scholar]
- Hensch TK. Critical period plasticity in local cortical circuits. Nat Rev Neurosci. 2005;6:877–88. doi: 10.1038/nrn1787. [DOI] [PubMed] [Google Scholar]
- Hofman KJ, Harris EL, Bryan RN, Denckla MB. Neurofibromatosis type 1: the cognitive phenotype. J Pediatr. 1994;124:S1–8. doi: 10.1016/s0022-3476(05)83163-4. [DOI] [PubMed] [Google Scholar]
- Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SG. Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat Neurosci. 2000;3:661–69. doi: 10.1038/76615. [DOI] [PubMed] [Google Scholar]
- Hyman SL, Shores A, North KN. The nature and frequency of cognitive deficits in children with neurofibromatosis type 1. Neurology. 2005;65:1037–44. doi: 10.1212/01.wnl.0000179303.72345.ce. [DOI] [PubMed] [Google Scholar]
- Jacks T, Shih TS, Schmitt EM, Bronson RT, Bernards A, Weinberg RA. Tumor predisposition in mice heterozygous for a targeted mutation in Nf1. Nat Genet. 1994;7:353–61. doi: 10.1038/ng0794-353. [DOI] [PubMed] [Google Scholar]
- Jentsch JD, Aarde SM, Seu E. Effects of atomoxetine and methylphenidate on performance of a lateralized reaction time task in rats. Psychopharmacology. 2009;202:497–504. doi: 10.1007/s00213-008-1181-0. [DOI] [PubMed] [Google Scholar]
- Kayl AE, Moore BD., 3rd Behavioral phenotype of neurofibromatosis, type 1. Ment Retard Dev Disabil Res Rev. 2000;6:117–24. doi: 10.1002/1098-2779(2000)6:2<117::AID-MRDD5>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Kelly DP. Neurodevelopmental Dysfunction in the School-Aged Child. In: Behrman RE, et al., editors. Nelson Textbook of Pediatrics. 17. chap 29. Philadelphia: Saunders; 2004. pp. 110–112. [Google Scholar]
- Knudsen EI. Sensitive periods in the development of the brain and behavior. J Cogn Neurosci. 2004;16:1412–25. doi: 10.1162/0898929042304796. [DOI] [PubMed] [Google Scholar]
- Koos T, Tepper JM. Inhibitory control of neostriatal projection neurons by GABAergic interneurons. Nat Neurosci. 1999;2:467–72. doi: 10.1038/8138. [DOI] [PubMed] [Google Scholar]
- Koth CW, Cutting LE, Denckla MB. The association of neurofibromatosis type 1 and attention deficit hyperactivity disorder. Child Neuropsychol. 2000;6:185–94. doi: 10.1076/chin.6.3.185.3155. [DOI] [PubMed] [Google Scholar]
- Krab LC, Aarsen FK, de Goede-Bolder A, Catsman-Berrevoets CE, Arts WF, et al. Impact of neurofibromatosis type 1 on school performance. J Child Neurol. 2008a;23:1002–10. doi: 10.1177/0883073808316366. [DOI] [PubMed] [Google Scholar]
- Krab LC, de Goede-Bolder A, Aarsen FK, Pluijm SM, Bouman MJ, et al. Effect of simvastatin on cognitive functioning in children with neurofibromatosis type 1: a randomized controlled trial. JAMA. 2008b;300:287–94. doi: 10.1001/jama.300.3.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberger WG, Dunn DW. Learning disorders. Neurol Clin. 2003;21:941–52. doi: 10.1016/s0733-8619(03)00010-0. [DOI] [PubMed] [Google Scholar]
- Lagae L. Learning disabilities: definitions, epidemiology, diagnosis, and intervention strategies. Pediatr Clin North Am. 2008;55:1259–68. vii. doi: 10.1016/j.pcl.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Leach RJ, Thayer MJ, Schafer AJ, Fournier RE. Physical mapping of human chromosome 17 using fragment-containing microcell hybrids. Genomics. 1989;5:167–76. doi: 10.1016/0888-7543(89)90043-8. [DOI] [PubMed] [Google Scholar]
- Ledbetter DH, Rich DC, O’Connell P, Leppert M, Carey JC. Precise localization of NF1 to 17q11.2 by balanced translocation. Am J Hum Genet. 1989;44:20–24. [PMC free article] [PubMed] [Google Scholar]
- Lee DA, Portnoy S, Hill P, Gillberg C, Patton MA. Psychological profile of children with Noonan syndrome. Dev Med Child Neurol. 2005;47:35–38. doi: 10.1017/s001216220500006x. [DOI] [PubMed] [Google Scholar]
- Lee YS, Silva AJ. The molecular and cellular biology of enhanced cognition. Nat Rev. 2009;10:126–40. doi: 10.1038/nrn2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leondaritis G, Petrikkos L, Mangoura D. Regulation of the Ras-GTPase activating protein neurofibromin by C-tail phosphorylation: implications for protein kinase C/Ras/extracellular signalregulated kinase 1/2 pathway signaling and neuronal differentiation. J Neurochem. 2009;109(2):573–83. doi: 10.1111/j.1471-4159.2009.05975.x. [DOI] [PubMed] [Google Scholar]
- Li W, Cui Y, Kushner SA, Brown RA, Jentsch JD, et al. The HMG-CoA reductase inhibitor lovastatin reverses the learning and attention deficits in a mouse model of neurofibromatosis type 1. Curr Biol. 2005;15:1961–67. doi: 10.1016/j.cub.2005.09.043. [DOI] [PubMed] [Google Scholar]
- Li Y, O’Connell P, Breidenbach HH, Cawthon R, Stevens J, et al. Genomic organization of the neurofibromatosis 1 gene (NF1) Genomics. 1995;25:9–18. doi: 10.1016/0888-7543(95)80104-t. [DOI] [PubMed] [Google Scholar]
- Luikart BW, Zhang W, Wayman GA, Kwon CH, Westbrook GL, Parada LF. Neurotrophindependent dendritic filopodial motility: a convergence on PI3K signaling. J Neurosci. 2008;28:7006–12. doi: 10.1523/JNEUROSCI.0195-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lush ME, Li Y, Kwon CH, Chen J, Parada LF. Neurofibromin is required for barrel formation in the mouse somatosensory cortex. J Neurosci. 2008;28:1580–87. doi: 10.1523/JNEUROSCI.5236-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch TM, Gutmann DH. Neurofibromatosis 1. Neurol Clin. 2002;20:841–65. doi: 10.1016/s0733-8619(01)00019-6. [DOI] [PubMed] [Google Scholar]
- Marchuk DA, Saulino AM, Tavakkol R, Swaroop M, Wallace MR, et al. cDNA cloning of the type 1 neurofibromatosis gene: complete sequence of the NF1 gene product. Genomics. 1991;11:931–40. doi: 10.1016/0888-7543(91)90017-9. [DOI] [PubMed] [Google Scholar]
- Martin SJ, Grimwood PD, Morris RG. Synaptic plasticity and memory: an evaluation of the hypothesis. Annu Rev Neurosci. 2000;23:649–711. doi: 10.1146/annurev.neuro.23.1.649. [DOI] [PubMed] [Google Scholar]
- Martin SJ, Morris RG. New life in an old idea: the synaptic plasticity and memory hypothesis revisited. Hippocampus. 2002;12:609–36. doi: 10.1002/hipo.10107. [DOI] [PubMed] [Google Scholar]
- Mautner VF, Kluwe L, Thakker SD, Leark RA. Treatment of ADHD in neurofibromatosis type 1. Dev Med Child Neurol. 2002;44:164–70. doi: 10.1017/s0012162201001876. [DOI] [PubMed] [Google Scholar]
- Money J, Kalus ME., Jr Noonan’s syndrome. IQ and specific disabilities. Am J Dis Child. 1979;133:846–50. [PubMed] [Google Scholar]
- Morris RG, Garrud P, Rawlins JN, O’Keefe J. Place navigation impaired in rats with hippocampal lesions. Nature. 1982;297:681–83. doi: 10.1038/297681a0. [DOI] [PubMed] [Google Scholar]
- Moser EI, Krobert KA, Moser MB, Morris RG. Impaired spatial learning after saturation of longterm potentiation. Science. 1998;281:2038–42. doi: 10.1126/science.281.5385.2038. [DOI] [PubMed] [Google Scholar]
- Noonan JA. Noonan syndrome. An update and review for the primary pediatrician. Clin Pediatr. 1994;33:548–55. doi: 10.1177/000992289403300907. [DOI] [PubMed] [Google Scholar]
- North K. Neurofibromatosis type 1. Am J Med Genet. 2000;97:119–27. doi: 10.1002/1096-8628(200022)97:2<119::aid-ajmg3>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- North KN, Riccardi V, Samango-Sprouse C, Ferner R, Moore B, et al. Cognitive function and academic performance in neurofibromatosis. 1: consensus statement from the NF1 Cognitive Disorders Task Force. Neurology. 1997;48:1121–27. doi: 10.1212/wnl.48.4.1121. [DOI] [PubMed] [Google Scholar]
- Palumbo D, Lynch PA. Psychological testing in adolescent medicine. Adolesc Med Clin. 2006;17:147–64. doi: 10.1016/j.admecli.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Pasmant E, Sabbagh A, Hanna N, Masliah-Planchon J, Jolly E, et al. SPRED1 germline mutations caused a neurofibromatosis type 1 overlapping phenotype. J Med Genet. 2009;46:425–30. doi: 10.1136/jmg.2008.065243. [DOI] [PubMed] [Google Scholar]
- Patapoutian A, Reichardt LF. Trk receptors: mediators of neurotrophin action. Curr Opin Neurobiol. 2001;11:272–80. doi: 10.1016/s0959-4388(00)00208-7. [DOI] [PubMed] [Google Scholar]
- Ramus F. Developmental dyslexia: specific phonological deficit or general sensorimotor dysfunction? Curr Opin Neurobiol. 2003;13:212–18. doi: 10.1016/s0959-4388(03)00035-7. [DOI] [PubMed] [Google Scholar]
- Rao SG, Williams GV, Goldman-Rakic PS. Destruction and creation of spatial tuning by disinhibition: GABAA blockade of prefrontal cortical neurons engaged by working memory. J Neurosci. 2000;20:485–94. doi: 10.1523/JNEUROSCI.20-01-00485.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter-Levin G, Canevari L, Bliss TV. Long-term potentiation and glutamate release in the dentate gyrus: links to spatial learning. Behav Brain Res. 1995;66:37–40. doi: 10.1016/0166-4328(94)00121-u. [DOI] [PubMed] [Google Scholar]
- Robbins TW. The 5-choice serial reaction time task: behavioural pharmacology and functional neurochemistry. Psychopharmacology. 2002;163:362–80. doi: 10.1007/s00213-002-1154-7. [DOI] [PubMed] [Google Scholar]
- Rosser TL, Packer RJ. Neurocognitive dysfunction in children with neurofibromatosis type 1. Curr Neurol Neurosci Rep. 2003;3:129–36. doi: 10.1007/s11910-003-0064-3. [DOI] [PubMed] [Google Scholar]
- Rowbotham I, Pitten Cate IM, Sonuga-Barke EJ, Huijbregts SC. Cognitive control in adolescents with neurofibromatosis type 1. Neuropsychology. 2009;23:50–60. doi: 10.1037/a0013927. [DOI] [PubMed] [Google Scholar]
- Sabbagh A, Pasmant E, Laurendeau I, Parfait B, Barbarot S, et al. Unraveling the genetic basis of variable clinical expression in neurofibromatosis 1. Hum Mol Genet. 2009;18:2779–90. doi: 10.1093/hmg/ddp212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Vives MV, McCormick DA. Cellular and network mechanisms of rhythmic recurrent activity in neocortex. Nat Neurosci. 2000;3:1027–34. doi: 10.1038/79848. [DOI] [PubMed] [Google Scholar]
- Seizinger BR, Rouleau GA, Ozelius LJ, Lane AH, Faryniarz AG, et al. Genetic linkage of von Recklinghausen neurofibromatosis to the nerve growth factor receptor gene. Cell. 1987;49:589–94. doi: 10.1016/0092-8674(87)90534-4. [DOI] [PubMed] [Google Scholar]
- Shen MH, Harper PS, Upadhyaya M. Molecular genetics of neurofibromatosis type 1 (NF1) J Med Genet. 1996;33:2–17. doi: 10.1136/jmg.33.1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AJ, Frankland PW, Marowitz Z, Friedman E, Laszlo GS, et al. A mouse model for the learning and memory deficits associated with neurofibromatosis type I. Nat Genet. 1997;15:281–84. doi: 10.1038/ng0397-281. [DOI] [PubMed] [Google Scholar]
- Silva AJ, Paylor R, Wehner JM, Tonegawa S. Impaired spatial learning in alpha-calciumcalmodulin kinase II mutant mice. Science. 1992a;257:206–11. doi: 10.1126/science.1321493. [DOI] [PubMed] [Google Scholar]
- Silva AJ, Stevens CF, Tonegawa S, Wang Y. Deficient hippocampal long-term potentiation in alpha-calcium-calmodulin kinase II mutant mice. Science. 1992b;257:201–6. doi: 10.1126/science.1378648. [DOI] [PubMed] [Google Scholar]
- Spurlock G, Bennett E, Chuzhanova N, Thomas N, Jim HP, et al. SPRED1 mutations (Legius syndrome): another clinically useful genotype for dissecting the neurofibromatosis type 1 phenotype. J Med Genet. 2009;46:431–37. doi: 10.1136/jmg.2008.065474. [DOI] [PubMed] [Google Scholar]
- Tartaglia M, Gelb BD. Noonan syndrome and related disorders: genetics and pathogenesis. Annu Rev Genomics Hum Genet. 2005;6:45–68. doi: 10.1146/annurev.genom.6.080604.162305. [DOI] [PubMed] [Google Scholar]
- Tepper JM, Wilson CJ, Koos T. Feedforward and feedback inhibition in neostriatal GABAergic spiny neurons. Brain Res Rev. 2008;58:272–81. doi: 10.1016/j.brainresrev.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson SA, Fishbein L, Wallace MR. NF1 mutations and molecular testing. J Child Neurol. 2002;17:555–61. doi: 10.1177/088307380201700803. discussion 71–72, 646–51. [DOI] [PubMed] [Google Scholar]
- Tong J, Hannan F, Zhu Y, Bernards A, Zhong Y. Neurofibromin regulates G protein-stimulated adenylyl cyclase activity. Nat Neurosci. 2002;5:95–96. doi: 10.1038/nn792. [DOI] [PubMed] [Google Scholar]
- Viskochil D, Buchberg AM, Xu G, Cawthon RM, Stevens J, et al. Deletions and a translocation interrupt a cloned gene at the neurofibromatosis type 1 locus. Cell. 1990;62:187–92. doi: 10.1016/0092-8674(90)90252-a. [DOI] [PubMed] [Google Scholar]
- Vogel KS, Brannan CI, Jenkins NA, Copeland NG, Parada LF. Loss of neurofibromin results in neurotrophin-independent survival of embryonic sensory and sympathetic neurons. Cell. 1995;82:733–42. doi: 10.1016/0092-8674(95)90470-0. [DOI] [PubMed] [Google Scholar]
- Wallace MR, Marchuk DA, Andersen LB, Letcher R, Odeh HM, et al. Type 1 neurofibromatosis gene: identification of a large transcript disrupted in three NF1 patients. Science. 1990;249:181–86. doi: 10.1126/science.2134734. [DOI] [PubMed] [Google Scholar]
- Ward BA, Gutmann DH. Neurofibromatosis 1: from lab bench to clinic. Pediatr Neurol. 2005;32:221–28. doi: 10.1016/j.pediatrneurol.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Weeber EJ, Sweatt JD. Molecular neurobiology of human cognition. Neuron. 2002;33:845–48. doi: 10.1016/s0896-6273(02)00634-7. [DOI] [PubMed] [Google Scholar]
- Weiss B, Bollag G, Shannon K. Hyperactive Ras as a therapeutic target in neurofibromatosis type 1. Am J Med Genet. 1999;89:14–22. [PubMed] [Google Scholar]
- Williams JA, Su HS, Bernards A, Field J, Sehgal A. A circadian output in Drosophila mediated by neurofibromatosis-1 and Ras/MAPK. Science. 2001;293:2251–56. doi: 10.1126/science.1063097. [DOI] [PubMed] [Google Scholar]
- Williams VC, Lucas J, Babcock MA, Gutmann DH, Korf B, Maria BL. Neurofibromatosis type 1 revisited. Pediatrics. 2009;123:124–33. doi: 10.1542/peds.2007-3204. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Ghosh P, Charnay P, Burns DK, Parada LF. Neurofibromas in NF1: Schwann cell origin and role of tumor environment. Science. 2002;296:920–22. doi: 10.1126/science.1068452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Romero MI, Ghosh P, Ye Z, Charnay P, et al. Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev. 2001;15:859–76. doi: 10.1101/gad.862101. [DOI] [PMC free article] [PubMed] [Google Scholar]