Abstract
The amyloid-β (Aβ) aggregation pathway is a key target in efforts to discover therapeutics that prevent or delay the onset of Alzheimer’s disease. Efforts at rational drug design, however, are hampered by uncertainties about the precise nature of the toxic aggregate. In contrast, high-throughput screening of compound libraries does not require a detailed understanding of the structure of the toxic species, and can provide an unbiased method for the discovery of small molecules that may lead to effective therapeutics. Here, we show that small molecule microarrays (SMMs) represent a particularly promising tool for identifying compounds that bind the Aβ peptide. Microarray slides with thousands of compounds immobilized on their surface were screened for binding to fluorescently labeled Aβ. 79 compounds were identified by the SMM screen, and then assayed for their ability to inhibit the Aβ-induced killing of PC12 cells. Further experiments focused on exploring the mechanism of rescue for one of these compounds: Electron microscopy and Congo red binding showed that the compound enhances fibril formation, and suggest that it may rescue cells by accelerating Aβ aggregation past an early toxic oligomer. These findings demonstrate that the SMM screen for binding to Aβ is effective at identifying compounds that reduce Aβ toxicity, and can reveal potential therapeutic leads without the biases inherent in methods that focus on inhibitors of aggregation.
INTRODUCTION
Considerable genetic and biochemical evidence indicates that aggregation of the amyloid-β peptide (Aβ) plays a causative role in the neurodegeneration, memory loss, and dementia associated with Alzheimer’s disease (AD).1–5 Although the precise structure of the toxic aggregate is still under investigation, the major features of the “amyloid cascade” pathway are understood: Proteolytic processing of the amyloid precursor protein (APP) results in the extracellular release of the 4 kDa Aβ peptide. Differential cleavage by secretase enzymes leads to the formation of Aβ variants ranging in size from 39 to 43 amino acids, with the 40-residue Aβ40 and 42-residue Aβ42 peptides being the most prevalent.6–8 Compared to Aβ40, Aβ42 is more prone to aggregation,9 more neurotoxic,10 and more closely correlated with symptomatic disease.2,11
Aβ42 is also the predominant component of the extracellular plaque that has long been viewed as the pathological hallmark of AD.12,13 While this insoluble plaque provides posthumous evidence of the disease, numerous findings over the past decade implicate earlier soluble intermediates as the neurotoxic agents. Aβ-derived diffusible ligands (ADDLs) have been observed to induce neuronal death in cell culture,14 and soluble Aβ oligomeric species are more closely associated with cognitive decline in AD patients and synapse loss in transgenic mice than fibril or plaque load.15–17 Most recently, a murine model in which ADDLs were endogenously expressed with sequence mutations that prevented subsequent plaque formation showed that cognitive deficits are dependent on soluble Aβ oligomers rather than end-stage aggregates.18
With the prevalence of Alzheimer’s disease projected to rise dramatically in the coming decades,19 there is increasing urgency to develop novel therapeutics to prevent and/or treat this debilitating disease. The pharmaceuticals currently used to treat AD include cholinesterase inhibitors and memantine, a modulator of glutamate receptors.20,21 These drugs provide symptomatic relief and may slow cognitive decline; however, they do not target the underlying molecular cause of AD. Therefore, currently approved drugs neither halt, nor reverse progression of the disease. In contrast to these existing drugs, novel compounds that interfere with Aβ aggregation, and thereby block the molecular events that cause AD, represent a promising approach to the prevention and/or cure of Alzheimer’s disease.
In principle, compounds that interfere with Aβ aggregation could be discovered by either of two approaches: Structure-based rational drug design or high-throughput screening. Since a range of intermediates along the Aβ aggregation pathway have been implicated as potential toxic species,22–25 and none of their precise structures are known, structure-based drug design is not yet possible. For the time being, high-throughput screening remains a more promising approach.
A number of high-throughput methods have been developed to screen for compounds that interfere with Aβ aggregation. Most screens use synthetic Aβ peptide and monitor aggregation by following the fluorescence of thioflavin-T or another dye that binds to fibrillar aggregates.26–28 Although relatively convenient to perform, these screens have limitations: First, they are hampered by the need for substantial quantities of synthetic peptide, which is expensive and hard to produce in a form that is free of pre-aggregated seeds. A second and more significant drawback of dye-binding assays is their reliance on a reporter that detects amyloid fibrils or protofibrils, rather than the early intermediates that are now thought to be the neurotoxic agent. The limitations of assays that screen for inhibitors of fibrillization are highlighted by recent reports that accelerating fibril formation may, in fact, be advantageous as a way to decrease the presence of toxic soluble Aβ oligomers.29,30
As an alternative to traditional assays using synthetic Aβ peptide, our lab previously reported the development of a high-throughput screen using an Aβ42-GFP fusion protein expressed in E. coli.31,32 In that system, aggregation of the Aβ42 sequence drags the entire fusion protein into misfolded insoluble aggregates, thereby preventing the folding and fluorescence of GFP. Compounds that inhibit the aggregation of Aβ42 enable the GFP part of the fusion to fold into its native structure, and thereby give rise to green fluorescent colonies (or liquid cultures) of E. coli. The Aβ42-GFP screen overcame two of the limitations of the dye binding assays: (i) Synthetic Aβ peptide was not required, and (ii) the assay reported Aβ misfolding and aggregation without requiring the formation of amyloid fibrils.
Application of the GFP-based screen enabled the identification of several aggregation inhibitors from a library of triazine derivatives,32 and from other libraries screened subsequently. Some of these compounds look promising and are being evaluated in a Drosophila model of AD. Nonetheless, the GFP-based screen also has several limitations: (i) Because the Aβ42-GFP fusion protein was expressed in E. coli, compounds that fail to cross the cell wall and membrane of E. coli, or which were toxic to E. coli, may have been missed. (ii) Because the folding and fluorescence of the GFP reporter is based on the solubility of the upstream sequence,33 the screen may not distinguish between compounds that block all aggregation (by favoring monomeric Aβ) and those that block fibrillization by favoring the toxic soluble oligomers. Thus, compounds that allow the Aβ42-GFP fusion protein to fold and fluoresce by enhancing the stability of soluble oligomers would be mistakenly identified as inhibitors of aggregation (hits) rather than as enhancers of the toxic species. (iii) Most importantly, because the Aβ42-GFP screen was designed to identify compounds that inhibit aggregation, it would miss potential therapeutics that remove toxic oligomers by enhancing their aggregation into insoluble fibrils.
These considerations motivated us to develop a novel method to screen compounds based solely on their ability to bind to monomeric Aβ. Ultimately, whether a small molecule reduces the toxicity of Aβ by inhibiting aggregation, accelerating it past the toxic species, or redirecting it to an alternate pathway, the compound must bind the Aβ peptide.
High-throughput identification of compounds that bind to specific proteins has been accomplished previously through the use of small molecule microarrays (SMMs). SMMs are glass slides on which libraries of small molecules are covalently immobilized in an array of microscopic spots.34 Compounds can be attached to the surface by any of several mild coupling reactions. For the current study, SMMs were prepared by installing an isocyanate on the surface of the slides and then attaching the small molecules to this group.35–37 Because isocyanate reacts with amines (primary and secondary), thiols, alcohols (primary and secondary), and several other groups, a compound possessing several different functional groups can attach to the surface in a range of orientations with various functional groups available to interact with the probe. To ensure that the small molecules are presented at a distance from the surface sufficient to enable interactions with a soluble probe (Aβ peptide in the current study), the isocyanate is separated from the surface by a polyethylene glycol spacer.
A single SMM slide typically displays 10,800 compounds, allowing for rapid and efficient screening of diverse libraries of small molecules. SMM slides are probed with a fluorophore- or epitope-tagged protein (in this case, fluorescently-labeled Aβ peptide), and compounds that bind the protein are detected by automated fluorescence read-out (a schematic is shown in figure 2a).35–37 SMMs have been used successfully to identify a wide range of protein-ligand interactions, including calmodulin and human immunoglobin G ligands,38,39 transcriptional regulators,40,41 and inhibitors of histone deacetylases.42
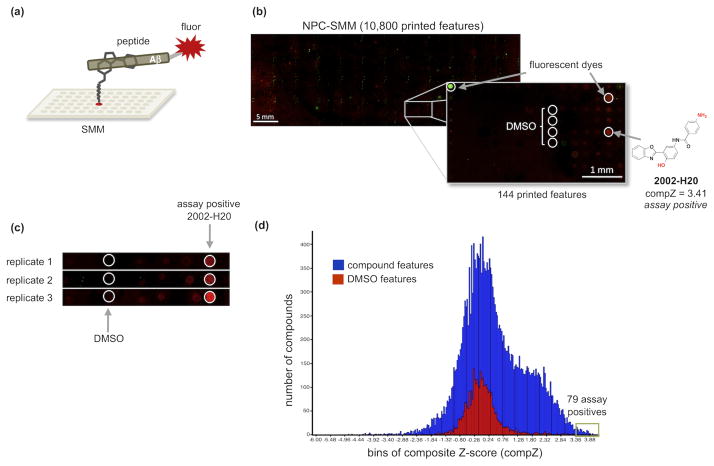
Figure 2.
(a) The SMM binding screen. Compounds are covalently attached in an array of spots on the surface of a slide, and probed with fluorescently-tagged Aβ peptide. Those compounds that bind Aβ and withstand several washes are revealed as fluorescent spots. (b) Fluorescent read-out of the NPC-SMM slide following incubation with fluorescent Aβ40. Enlargement of a grid section shows compound 2002-H20 binding the peptide (false-colored red) as well as fluorescent dyes used in grid alignment (false-colored green and red) and non-fluorescing DMSO control spots. The structure of 2002-H20 is shown with isocyanate-reactive functional groups colored red to indicate the positions available for attachment to the slide. Because two functional groups (an amine and a phenol) are available for cross-linking, the population displayed on the surface is assumed to include molecules displayed in more than one orientation, with some exposing the amine and others exposing the phenol for interaction with Aβ. (c) Three replicate SMM screens of the NPC compound set show that compound 2002-H20 binds fluorescently labeled Aβ40 reproducibly and consistently. (d) Histogram of the composite Z-scores of SMM fluorescence results from 3 replicates of the DIV and NPC slides. Results are divided into 254 bins with compounds shown in blue and DMSO controls in red. The green box surrounds bins for 79 assay positive compounds with composite Z-scores = 3.4.
The SMM technology has several features that make it ideal as a high-throughput screen for compounds that bind Aβ. First, it does not require knowledge of the protein structure or binding pocket. This unbiased approach is crucial since the structure of the toxic Aβ aggregate is not known, and a traditional active site pocket is not likely to exist. Second, the high sensitivity of the SMM assay allows the Aβ peptide to be used at very low concentrations. This is important because Aβ aggregation occurs via a nucleation-dependent mechanism,43 and keeping the peptide below the critical concentration favors the monomeric form, thereby facilitating the identification of compounds that interact with Aβ at or before the earliest steps of aggregation. An added benefit is that low concentrations of peptide make the assay relatively inexpensive.
Here we describe the development of SMM technology to screen libraries of small molecules for compounds that bind the Alzheimer’s Aβ peptide. As an initial step toward SMM assay development, we identified conditions that favor the majority of the Aβ probe being present as monomer. Next, two SMM slide sets – one with natural products and synthetic commercial compounds (NPC), and the other with diversity-oriented synthesis compounds (DIV) – were screened with fluorescently labeled Aβ40. Compounds in printed features that bound the peptide were identified as hits, as described previously.37,44 These compounds were subsequently assayed for their ability to rescue PC12 cells from Aβ42-induced toxicity. Further examination of a commercially available compound revealed dose-dependent rescue of PC12 cells. Finally, we demonstrated that this compound actually promotes, rather than inhibits, fibrillogenesis. These results validate the ability of the SMM screen to reveal compounds that interact with the Aβ peptide, and may provide leads for therapeutics that prevent or cure Alzheimer’s disease.
RESULTS AND DISCUSSION
Development of an SMM Assay for Binding to Aβ
Identifying compounds that interact with Aβ at or before the earliest stages of aggregation required that we develop conditions for the SMM assay wherein the peptide would occur predominantly in its monomeric form. Aβ, particularly Aβ42, is known to aggregate quickly, and aggregation is considerably faster when the sample is agitated,45–47 as required for the SMM assays. We assessed the aggregation of Aβ40 and Aβ42 with N-terminal fluorescent tags under conditions typically used for SMM screening, which include 30 minutes of gentle agitation (by orbital mixing) at room temperature. Monomeric and oligomeric states were estimated using Tris-Tricine SDS-PAGE. Such gels are used routinely to assess the oligomeric state of Aβ,48,49 and previous reports indicate that both cell-derived oligomers and those formed in vitro resist denaturation by SDS, particularly when gel analysis is performed at room temperature.50–52 Nonetheless, the possibility that SDS alters the distribution of oligomeric states cannot be ruled out. With this caveat in mind, we probed the oligomeric states of our samples using the standard Tris-Tricine SDS-PAGE assay.
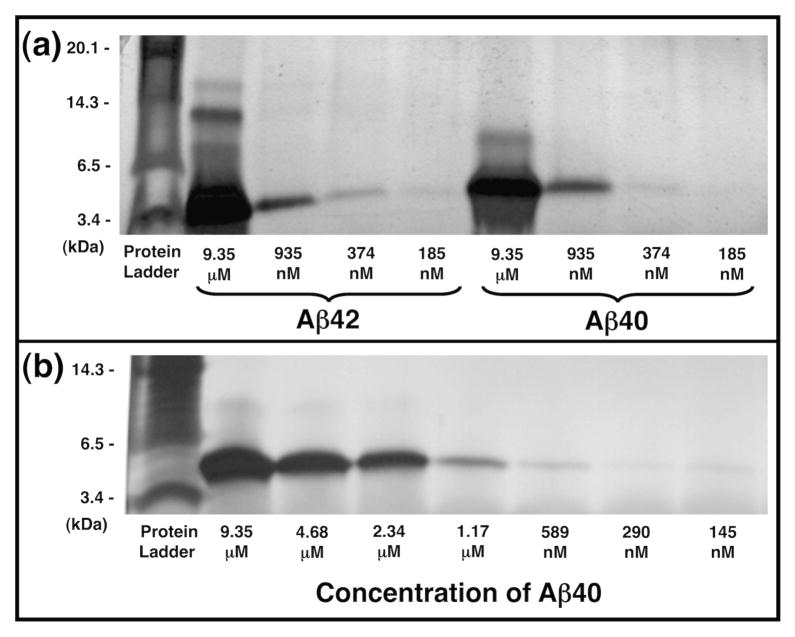
As shown in figure 1a, fluorescently labeled Aβ42 formed more oligomers and larger oligomers than Aβ40. Distinct monomer (4 kDa), dimer (8 kDa), trimer (12 kDa), and tetramer (16 kDa) bands are apparent for Aβ42 at 9.35 μM, with the trimer being the most intense oligomeric band, consistent with previous reports.48 Trimer and tetramer bands were also very faintly apparent for Aβ42 at 935 nM. In contrast to Aβ42, Aβ40 produced only monomer and dimer bands at 9.35 μM, and at concentrations below 1 μM, Aβ40 yielded only monomers.
Figure 1.
10–20% Tris-Tricine SDS-PAGE gels showing the oligomeric state of Aβ under SMM agitation conditions. (a) HiLyte Fluor 488-labeled Aβ42 (lanes 2–5) and HiLyte Fluor 647-labeled Aβ40 (lanes 6–9) after agitation for 30 minutes. (b) Fluorescently labeled Aβ40 after 60 minutes agitation. Monomeric Aβ appears at ~ 4 kDa. Gels were visualized by silver staining.
Due to its reduced propensity to aggregate, we opted to use Aβ40 for our SMM screens. Because Aβ40 lacks only two residues at its C-terminus and has the same core residues as Aβ42, we assumed that most compounds capable of binding Aβ40 would also bind Aβ42. To assess whether fluorescently labeled Aβ40 would remain monomeric under the conditions of the SMM assay, we incubated the peptide with gentle agitation for 60 minutes—twice as long as required for typical SMM binding experiments—and probed its oligomeric state by SDS-PAGE. As shown in figure 1b, at micromolar concentrations Aβ40 forms monomers and dimers, but at concentrations 1.17 μM or lower, only monomeric bands are seen. This is consistent with previous reports that the critical concentration of amyloid aggregation occurs in the μM or high nM range.53,54
These results suggest that fluorescently tagged Aβ40 is largely monomeric in solution at the concentration (185 nM) and incubation conditions used for SMM screening. Therefore, initial encounters between the immobilized small molecule and the peptide are likely to occur with the latter in its monomeric form. After the first peptide binds an immobilized small molecule and becomes attached to the surface, further copies may be recruited to aggregate onto this immobilized complex.
The SMM Screen Identifies Compounds that Bind Aβ
The SMM assay was used to screen two collections of compounds. The first collection contained natural product and commercial synthetic compounds (NPC), while the second contained a library of compounds from a diversity-oriented synthesis (DIV). A total of 17,905 compounds were screened, and all assays were performed in triplicate. Compounds that bound fluorescently labeled Aβ40 yielded spots exhibiting fluorescence at 632 nm (false-colored red). Rhodamine features exhibiting fluorescence at 532 nm (false-colored green) were used for grid alignment. Fluorescence read-out results for the NPC slide screened with the Aβ40 probe are shown in Figure 2b. Zooming in on a section of the slide shows the rhodamine grid alignment spots, nonfluorescing DMSO control spots, and the distinct, uniform red fluorescence of a representative compound that bound Aβ. Figure 2c shows that this compound, hereafter referred to as 2002-H20, bound Aβ40 in all three replicates.
Primary SMM assay positives were annotated using fluorescence signal as described previously by Seiler et al.44 The fluorescence intensity values from each replica slide were background subtracted and converted to a Z-score, or standard score representing the number of standard deviations that an observation—here, the fluorescence of a grid spot—is above or below the mean value.36,37,44 Z-scores from the three replicas performed for each slide set were combined to produce a composite Z-score for each compound. These values, and the composite Z-scores for DMSO control spots, were plotted in a histogram (Figure 2d) with the results divided into 254 bins. The resulting shape of the compound histogram, shown in blue, is normal with a center around zero and a small shoulder on the right side. The DMSO control histogram (red) also exhibits a normal distribution centered on zero. Compounds binding Aβ with a composite Z-score of 3.4 or greater were identified as “hits” and selected for follow-up testing. From the 17,905 compounds assayed from the DIV and NPC libraries, 79 compounds with a composite Z-score above the threshold value were chosen for additional studies.
Inhibition of Aβ42-Induced Cytotoxicity
The 79 hits identified in the SMM screens were assayed for their ability to inhibit Aβ42-induced killing of PC12 cells. We chose inhibition of cytotoxicity—rather than biophysical studies of Aβ aggregation—as our initial follow-up assay because it makes no assumptions about the precise mechanism of Aβ toxicity, and thus allows a less biased search for compounds that may ultimately lead to therapeutics. Because Aβ42 is known to be a more important contributor to AD-associated neurotoxicity than Aβ40, we used the longer peptide to assay for inhibition of Aβ-induced cell death. These assays were performed using pure peptide – not linked to the fluorescent probe used in the SMM screen. Therefore any compounds that were isolated in the SMM screen because of binding to the fluorophore tag would be weeded out at this stage.
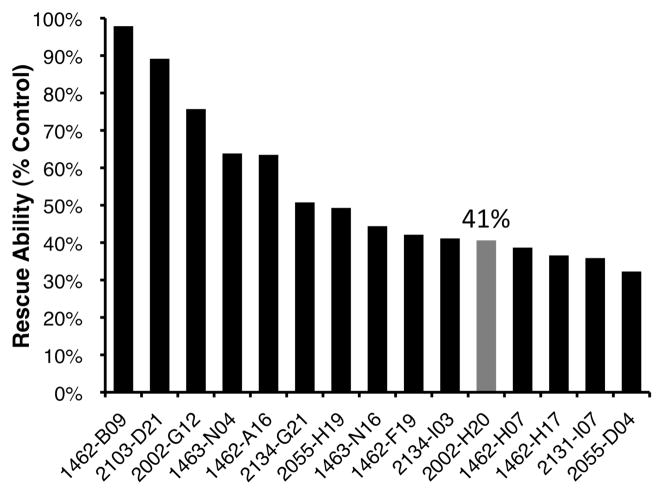
PC12 cells were incubated with Aβ42 and the SMM assay positive compounds, and cell viability was assessed using the MTT assay, which is based on the reduction of the yellow tetrazolium salt MTT (3-(4,5-dimethylthiazol-2yl)-2,5-diphenyltetrazolium bromide) to purple formazan crystals in metabolically active cells. Briefly, PC12 cells were grown to confluence and incubated for 24 hours in the presence of 20 μM Aβ42 and 100 μM of the hit compounds solubilized in DMSO. MTT was added and after incubation and solubilization, the difference between absorbance at the formazan wavelength (570 nm) and the MTT absorbance (670 nm) was taken as a measure of relative cell viability. At concentrations of 100 μM, 44 of the 79 SMM hit compounds were found to reduce the toxicity of Aβ42 in PC12 cells, with 15 of these compounds increasing cell viability by >30%. These results for the top 15 rescuers are shown in Figure 3 (toxicity rescue data for all 79 hit compounds can be found in Supporting Information, Table S1). As shown, one compound, 1462-B09, rescued cell viability to nearly 100%, almost eliminating Aβ-induced toxicity entirely. Another compound, 2002-H20, increases cell survival by 41%. Compound 2002-H20 was chosen for further study as it reproducibly binds Aβ40 in the SMM assays (Figure 2c), is readily soluble in DMSO, and is commercially available.
Figure 3.
Relative rescue scores for the top 15 compounds, which (at 100 μM) reduced the toxicity of Aβ42 to PC12 cells. Viability of cells in the absence of Aβ or exogenous compounds was scaled (A570-670 = 0.716) to 100% and viability of cells exposed to Aβ42 alone (A570-670 = 0.496) was scaled as 0%. As shown in gray, compound 2002-H20 increased viability by 41%. Cell viability was assayed using the MTT assay. Compound identities and toxicity data for all 79 compounds provided in Supporting Information.
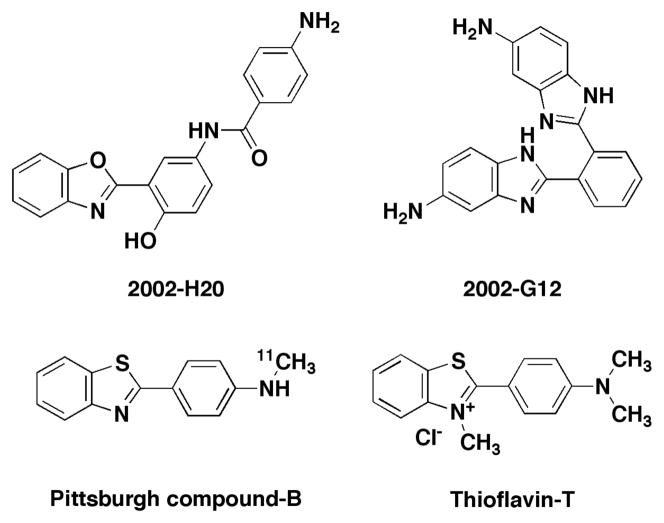
Structural Features of Compound 2002-H20
The structure of compound 2002-H20 is shown in Figure 4. With multiple aromatic groups and several sites for hydrogen bonding, it shares features with compounds identified previously as amyloid aggregation inhibitors.55–57 Its structure also bears some resemblance to thioflavin-T (ThT), a fluorescent dye used to bind and detect amyloid fibrils,58 and Pittsburgh Compound-B, which is used to image amyloid in the brains of AD patients.59 These compounds all contain a central phenyl ring and an attached benzoxazole or benzothiazole. These similarities to compounds previously known to bind Aβ support the efficacy of the SMM method for isolating Aβ binders. Similar structural features also occur in another SMM hit, compound 2002-G12, which contains a central phenyl ring with two benzimidizole substituents and reduced Aβ-induced toxicity by 76% (Figure 3). Though this compound was no longer available for purchase from the original commercial supplier, analogs with benzimidizole groups were tested and found to produce statistically significant improvements in the viability of PC12 cells exposed to Aβ42 (data not shown).
Figure 4.
The benzoxazole motif in compound 2002-H20 is similar to the benzothiazole motif in Pittsburgh Compound-B and thioflavin-T, two compounds that bind Aβ and are used routinely to detect amyloid. Another SMM hit compound, 2002-G12, contains similar benzimidozole motifs and reduced Aβ42 toxicity by 76% (Fig. 3).
Compound 2002-H20 Exhibits Dose Dependent Inhibition of Aβ42-Induced Cytotoxicity
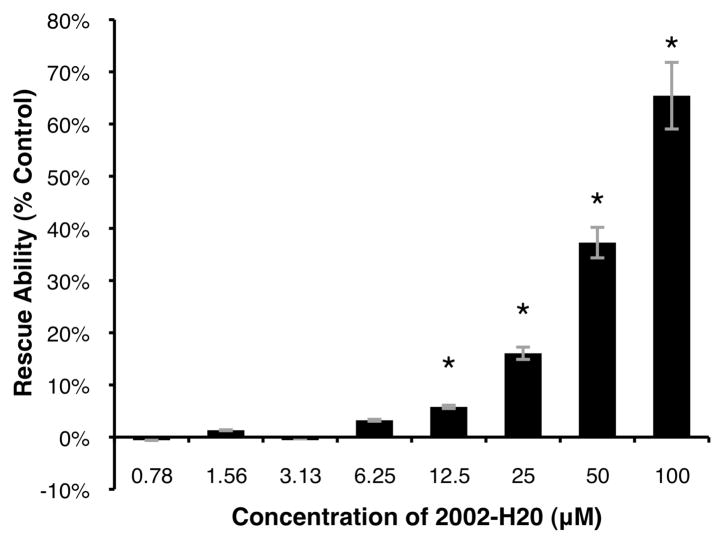
The MTT assay was used to assess the dose dependence of 2002-H20-mediated rescue of PC12 cells from Aβ42-induced toxicity. As shown in Figure 5, concentrations ≤ 3.13 μM were ineffective. However, beginning at 6.25 μM, a dose-dependent increase in cell viability was observed with concentrations ≤ 12.5 μM producing statistically significant rescue of PC12 cells exposed to Aβ42. The greatest improvement was at the high dose of 100 μM 2002-H20, with cell viability increased to 65% of the level of cells that were not incubated with Aβ42.
Figure 5.
Dose-dependence of compound 2002-H20’s ability to rescue PC12 cells from Aβ42-induced toxicity. 100% rescue scaled as the absorbance difference of cells incubated in buffer without peptide (A570–670 = 0.976) and 0% as absorbance difference of cells with 20 μM Aβ42 and no 2002-H20 (A570–670 = 0.389). An asterisk (*) indicates statistical significance as determined by the two-tailed Student’s T-test (p < 0.05) and error bars reflect standard error from the average of three measurements.
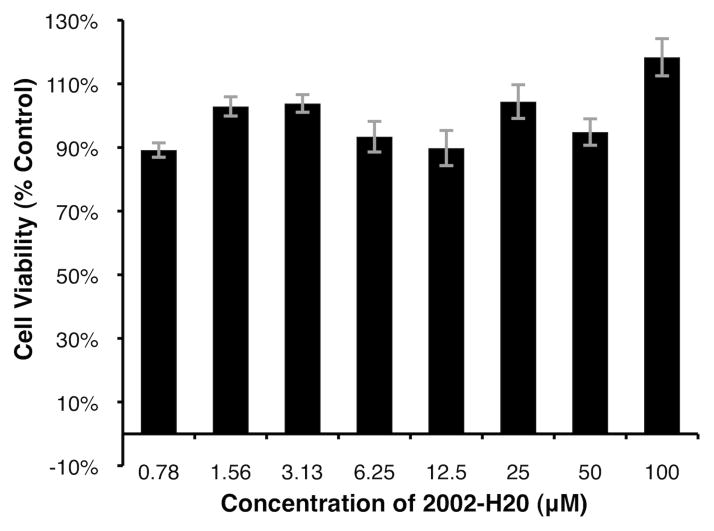
Compound 2002-H20 does not affect cell viability
To assess the effect of compound 2002-H20 on cell viability, it was added to PC12 cells in the absence of Aβ42. As shown in Figure 6, the compound does not appear to affect viability and there are no concentration-dependent trends. Variations in viability above and below 100% are likely due to the inherent noise of the MTT assay.
Figure 6.
Viability of PC12 cells exposed to compound 2002-H20 at varying concentrations. 100% is scaled as the measured viability of cells not exposed to exogenous compound or peptide, but incubated in buffer with 1% (v/v) DMSO (A570–670 = 0.976). Average of three measurements with standard error reported.
Analogs of Compound 2002-H20
To assess whether analogs of compound 2002-H20 inhibit Aβ42-induced cytotoxicity, we evaluated 21 related molecules. Compounds were tested at a concentration of 50 μM for their ability to reduce the toxicity of 20 μM Aβ42 in PC12 cells. 13 of these compounds showed statistically significant rescue, but no significant trends in functional groups were revealed. Only one analog was more effective than compound 2002-H20 itself (Supporting Information, Table S2 & Figure S1).
Mode of Action for 2002-H20-Mediated Rescue of Aβ42-induced Cytotoxicity
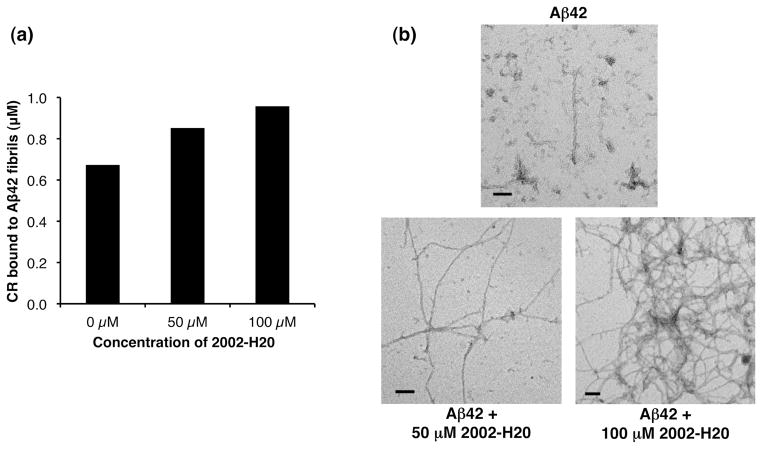
Compound 2002-H20 was identified by the SMM assay for its ability to bind Aβ42. Further characterization showed that it enhances the viability of PC12 cells exposed to Aβ42. To explore the mechanism by which 2002-H20 binding reduces Aβ42 toxicity, we investigated its impact on peptide aggregation in vitro. Compound 2002-H20 was incubated at concentrations of either 50 μM or 100 μM with synthetic Aβ42 at 20 μM. Amyloid formation was monitored by the Congo red (CR) spectral shift assay, and by transmission electron microscopy (TEM). When CR binds amyloid fibrils, it produces a characteristic spectral shift that enables quantization of amyloid.60 As shown in Figure 7a, compound 2002-H20 produces a dose-dependent increase in the CR signal. TEM images (Figure 7b) confirm that fibril formation is enhanced by compound 2002-H20. In the absence of 2002-H20, the predominant aggregates are short fibrils, protofibrils, and oligomeric aggregates. However, in the presence of 50 μM 2002-H20, Aβ42 forms long fibrils. Increasing the 2002-H20 concentration to 100 μM leads to dense fibrillar networks.
Figure 7.
Compound 2002-H20 increases amyloid fibril formation of synthetic Aβ42. (a) Congo red spectral shift. (b) Transmission electron microscopy. For both experiments, 20 μM of synthetic Aβ42 was incubated with the indicated concentration of compound 2002-H20. Incubations were at 37°C with gentle agitation for 24 hours.
The CR data and TEM images shown in Figure 7 confirm that compound 2002-H20 affects the aggregation pathway of Aβ42. However, rather than inhibiting aggregation, 2002-H20 seems to accelerate fibril formation. These results suggest that 2002-H20 inhibits cytotoxicity by driving Aβ42 aggregation towards fibrils, thereby reducing the presence of toxic oligomers.
A number of previous studies described compounds that increase Aβ aggregation. For example, Williams et al. screened a collection of 640 small molecules and isolated a compound that stabilizes protofibrils.61 Necula et al. examined compounds previously identified as aggregation inhibitors and found that several of these compounds reduced oligomer formation, but promoted fibrillization.62 In a related study of the phenothiazine compound, methylene blue, they proposed a causative link between the promotion of fibril formation and reduced oligomerization.30 The potential for the enhancement of fibril formation as a novel therapeutic strategy was suggested by Cheng et al., who found that accelerating fibril formation in transgenic AD mice led to reduced oligomerization and improved behavioral outcomes.29 Our finding that a compound isolated for its ability to bind Aβ42 can both increase fibril formation and decrease Aβ42-induced toxicity support the potential efficacy of this strategy.
Compound 2002-H20 Might Have Been Missed by Other High-Throughput Screens
The advantages of the SMM screen are highlighted by the realization that compound 2002-H20 would not have been found by previously reported high-throughput screens for inhibitors of Aβ aggregation.
We found that the most commonly used assay for Aβ aggregation—ThT fluorescence—was ineffective in the presence of 2002-H20 because the compound and ThT fluoresce at similar wavelengths (data not shown). Indeed, recent studies suggest that ThT assays are often prone to errors in the presence of compounds that absorb or fluoresce in the same region of the spectrum.63 However, even if the fluorescence of 2002-H20 and ThT did not overlap and the compound did not affect ThT fluorescnece, a ThT-based assay that screens for inhibition of fibrillization would have missed 2002-H20, which actually promotes fibril formation.
Most of the screens described in the literature aim to isolate compounds that alter the aggregation of Aβ.31,32,64 Screens that isolate inhibitors of aggregation would miss compounds like 2002-H20, which favor fibrils. Conversely, screens for enhancers of aggregation would miss compounds that stabilize monomeric Aβ. In both cases, novel molecules that may ultimately provide leads for Alzheimer’s therapeutics would have been missed by screens biased to favor either inhibitors or enhancers of aggregation. In contrast, the SMM screen described here makes no assumptions about whether aggregation to a particular state is advantageous or detrimental and isolates compounds based solely on their ability to interact with Aβ.
CONCLUSIONS
Our primary aim in this study was to develop the SMM assay as a screen for compounds that may ultimately lead to therapeutics for Alzheimer’s disease, and as described above, the assay effectively identified molecules that reduce the cytotoxicity of Aβ42. Further examination of one of those compounds revealed dose-dependent rescue of Aβ-challenged PC12 cells, with a concomitant increase in fibril formation. These findings highlight the benefits of a screen optimized to reveal binding to Aβ without bias towards a specific type of interaction, and further suggest the potential for the SMM method as a tool for exploring the Aβ aggregation pathway. For example, future examination of small molecules that bind Aβ in the SMM assay, but fail to rescue cells from Aβ-induced toxicity may reveal compounds that stabilize toxic aggregates. With further development, such compounds may prove useful as probes for the intermediates that cause Alzheimer’s disease. Additional applications can be imagined with the SMM assay having potential as an efficient and versatile screen for isolating compounds that interact with intermediates at various stages of the Aβ aggregation pathway.
MATERIALS AND METHODS
Materials
Fluorescently tagged amyloid-β peptide was purchased from Anaspec (San Jose, CA), diluted to a stock concentration of 9.35 μM in 8 mM NaOH, and used without additional purification. Aβ40 labeled at the N-terminus with HiLyte-Fluor 647 was reported by Anaspec to have a purity of greater than 90%, and Aβ42 labeled at the N-terminus with HiLyte-Fluor 488 had a reported purity of greater than 95%. Non-fluorescently labeled crude amyloid-β peptides, synthesized via FMOC synthesis, were purchased from the Keck Institute at Yale University (New Haven, CT) and purified as described below. SMM slides were prepared as described.65
Peptide Purification
Purification of crude peptides was accomplished via reverse phase-HPLC with a C4 column (Grace Vydac 214TP152022; Deerfield, IL). A two-solvent system was run at 60°C. Solvent A was 95% ultrapure H2O, 5% acetonitrile, and 0.1% trifluoroacetic acid (TFA); and solvent B was 50% ultrapure H2O, 50% acetonitrile, and 0.1% TFA. Purified peptides were disaggregated with TFA and 1,1,1,3,3,3-hexafluoro-2-isopropanol, which were applied successively and removed under a gentle argon stream.66 Disaggregated peptides were stored as a lyophilized film at −20°C.
SDS-PAGE Electrophoresis
9.35 μM stocks of fluorescently labeled peptides from Anaspec were diluted in 1× PBS to the indicated concentrations, and the pH of all dilutions was checked with pH paper and adjusted to 7–8 with 20% formic acid. The peptides were incubated for 30 or 60 minutes with gentle orbital shaking, combined with sample buffer, and loaded without boiling onto 10–20% Tris-Tricine gels (precast Ready Gels, Biorad; Hercules, CA). Electrophoresis was performed at 100 V and gels were developed with silver staining.
Small Molecule Microarray Assay
SMM slides were incubated with a slow circular orbit for 30 minutes at room temperature in 5 mL of 185 nM HiLyte 647-labeled Aβ40 (diluted from the 9.35 μM stock in 1× PBS-T, 0.1% Tween-20). Following incubation, slides were rinsed successively in PBS-T, PBS, and DI H2O for two minutes each, and then spun dry for 30 seconds with a slide spinner (Labnet International; Woodbridge, NJ). A GenePix 4200A microarray scanner (Molecular Devices; Sunnyvale, CA) was used to scan the slides with an excitation wavelength of 532 nm (green emission filter, 557–592 nm) and an excitation wavelength of 635 nm (red emission filter, 650–690 nm). All SMM experiments were performed in triplicate, and analyzed using a standard pipeline output for high-throughput screens at the Broad Institute.44
Cell Viability
Rat pheochromocytoma (PC12) cells, purchased from ATCC (Rockville, MD), were grown on collagen-coated Petri dishes (Falcon) in complete media (82.5% F12K, 15% horse serum, 2.5% fetal bovine serum; ATCC) in a humidified incubator at 37°C and 5% CO2 (HERAcell, Thermo Scientific). Once they reached confluence, cells were harvested by spraying through an 18.5 gauge needle, resuspended in fresh complete media and plated onto tissue culture-treated flat bottom 96-well plates (Costar) at a density of 10,000 cells per well (100 μL/well). The plates were incubated for an additional 24 hours to allow cell attachment. 1 mg of synthetic Aβ42, purified as described above, was dissolved in 100 μL of cell culture grade DMSO (ATCC), sonicated briefly to ensure complete solubilization, and then diluted to 200 μM with 1.0 mL of sterile 1× PBS (Invitrogen). 10 μL of this Aβ42 stock solution was then added to each 100 μL well to give a final peptide concentration of 20 μM.
In assaying for their ability to reduce Aβ42-induced toxicity, compounds solubilized in DMSO were added at 1% (v/v) to the wells to reach the final concentrations indicated. 1% (v/v) DMSO without compound was added to wells containing 20 μM Aβ42 for a toxicity control and to wells containing only buffer without peptide for a cell viability control. Plates were then incubated with Aβ42 and compounds for 24 more hours and cell viability was assessed with the MTT Cell Profileration kit from Roche, as directed.67,68 Following overnight incubation, absorbance was measured at 570 and 670 nm using a microplate reader (Thermo Varioskan), and A570nm – A670nm was calculated to evaluate relative formazan dye formation, which is directly correlated to cell viability. The absorbance difference without Aβ42 or exogenous compound was scaled as 100% cell viability in all experiments. In trials assessing the relative rescue ability of compounds, the absorbance difference calculated with Aβ42 only and no compound was scaled as 0% rescue ability. Where indicated, experiments were performed as above without peptide in order to assess the potential cytotoxicity of the compounds alone.
Congo Red Spectral Shift Assay
0.5 mg samples of purified Aβ42 were solubilized in 300 μL DMSO with sonication. 5 mL of 8 mM NaOH and 300 μL concentrated PBS were added (final concentration: 50 mM NaH2PO4/100 mM NaCl, pH 7.2–7.4) bringing the final peptide concentration to 20 μM. To remove contaminants and pre-existing aggregates, the samples were filtered with a 0.22 μm low-protein binding syringe filter (Millex®-GV; Millipore, Bedford, MA). Compound 2002-H20 (ChemDiv, San Diego, CA) was solubilized in DMSO to 10 mM and 5 mM concentrations, and added to samples to bring the final compound concentration to 100 μM and 50 μM. Samples were incubated in 15 mL horizontal polypropylene tubes with shaking (~200 rpm) on a Barnstead/Lab-Line titer plate shaker (Dubuque, IA). Samples containing only 2002-H20 without peptide were prepared and incubated in the same fashion in order to evaluate for any effect of the compound alone on the Congo red spectral shift. After 24 hours incubation, 400 μL aliquots were removed and mixed with 400 μL of a 34 μM Congo red (CR) solution (0.01M PBS/10% EtOH, solution syringe filtered and CR concentration measured as described by Klunk et al.69). Comparable samples with 0.01M PBS/10% EtOH lacking Congo red were also prepared to correct for light scattering due to Aβ and 2002-H20. After 30 min room temperature incubation, samples were mixed by pipetting and absorbance was measured at 403 nm and 541 nm. The concentration of CR bound to Aβ fibrils (in M) was calculated with background subtraction for light scattering from the absorbance of samples without CR69:
Electron Microscopy
The same samples prepared for the Congo red assay were also used for transmission electron microscopy (TEM). Formvar carbon-coated Cu grids were floated on a drop of each sample for 2 min, washed twice for 2 min with DI water, and stained for 2 min with 1% uranyl acetate. Grids were imaged with a Zeiss 912ab electron microscope (Thornwood, NY).
Supplementary Material
Acknowledgments
We thank Angela Fortner-McKoy for help with the cell viability assay and Margaret Bisher for assistance with electron microscopy. This research was funded by Grant 5R21AG028462 from the NIH and Award IIRG-08-89944 from the Alzheimer’s Association and Contract N01-CO-12400 from the NCI Initiative for Chemical Genetics.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Complete version of references 11 & 59; table of all 79 SMM hit compounds with composite Z and toxicity rescue scores; detailed results from study of structural analogs of compound 2002-H20. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hardy J, Allsop D. Trends Pharmacol Sci. 1991;12:383–388. doi: 10.1016/0165-6147(91)90609-v. [DOI] [PubMed] [Google Scholar]
- 2.Hsiao K, Chapman P, Nilsen S, Eckman C, Harigaya Y, Younkin S, Yang F, Cole G. Science. 1996;274:99–103. doi: 10.1126/science.274.5284.99. [DOI] [PubMed] [Google Scholar]
- 3.Oddo S, Caccamo A, Kitazawa M, Tseng BP, LaFerla FM. Neurobiol Aging. 2003;24:1063–1070. doi: 10.1016/j.neurobiolaging.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 4.Tanzi RE, Bertram L. Cell. 2005;120:545–555. doi: 10.1016/j.cell.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Lesne S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, Gallagher M, Ashe KH. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- 6.Shoji M, Golde T, Ghiso J, Cheung T, Estus S, Shaffer L, Cai X, McKay D, Tintner R, Frangione B, Younkin SG. Science. 1992;258:126–129. doi: 10.1126/science.1439760. [DOI] [PubMed] [Google Scholar]
- 7.Naslund J, Schierhorn A, Hellman U, Lannfelt L, Roses AD, Tjernberg LO, Silberring J, Gandy SE, Winblad B, Greengard P, Nordstedt C, Terenius L. Proc Natl Acad Sci USA. 1994;91:8378–8382. doi: 10.1073/pnas.91.18.8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller DL, Papayannopoulos IA, Styles J, Bobin SA, Lin YY, Biemann K, Iqbal K. Arch Biochem Biophy. 1993;301:41–52. doi: 10.1006/abbi.1993.1112. [DOI] [PubMed] [Google Scholar]
- 9.Jarrett JT, Berger EP, Lansbury PT. Biochemistry. 1993;32:4693–4697. doi: 10.1021/bi00069a001. [DOI] [PubMed] [Google Scholar]
- 10.El-Agnaf OMA, Mahil DS, Patel BP, Austen BM. Biochem Biophys Res Commun. 2000;273:1003–1007. doi: 10.1006/bbrc.2000.3051. [DOI] [PubMed] [Google Scholar]
- 11.Borchelt DR, et al. Neuron. 1996;17:1005–1013. doi: 10.1016/s0896-6273(00)80230-5. [DOI] [PubMed] [Google Scholar]
- 12.Roher AE, Lowenson JD, Clarke S, Woods AS, Cotter RJ, Gowing E, Ball MJ. Proc Natl Acad Sci USA. 1993;90:10836–10840. doi: 10.1073/pnas.90.22.10836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwatsubo T, Odaka A, Suzuki N, Mizusawa H, Nukina N, Ihara Y. Neuron. 1994;13:45–53. doi: 10.1016/0896-6273(94)90458-8. [DOI] [PubMed] [Google Scholar]
- 14.Lambert MP, Barlow AK, Chromy BA, Edwards C, Freed R, Liosatos M, Morgan TE, Rozovsky I, Trommer B, Viola KL, Wals P, Zhang C, Finch CE, Krafft GA, Klein WL. Proc Natl Acad Sci USA. 1998;95:6448–6453. doi: 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lue L-F, Kuo Y-M, Roher AE, Brachova L, Shen Y, Sue L, Beach T, Kurth JH, Rydel RE, Rogers J. Am J Pathol. 1999;155:853–862. doi: 10.1016/s0002-9440(10)65184-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mucke L, Masliah E, Yu GQ, Mallory M, Rockenstein EM, Tatsuno G, Hu K, Kholodenko D, Johnson-Wood K, McConlogue L. J Neurosci. 2000;20:4050–4058. doi: 10.1523/JNEUROSCI.20-11-04050.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLean CA, Cherny RA, Fraser FW, Fuller SJ, Smith MJ, Vbeyreuther K, Bush AI, Masters CL. Ann Neurol. 1999;46:860–866. doi: 10.1002/1531-8249(199912)46:6<860::aid-ana8>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 18.Gandy S, Simon AJ, Steele JW, Lublin AL, Lah JJ, Walker LC, Levey AI, Krafft GA, Levy E, Checler F, Glabe C, Bilker WB, Abel T, Schmeidler J, Ehrlich ME. Ann Neurol. 2010;68:220–230. doi: 10.1002/ana.22052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hebert LE, Beckett LA, Scherr PA, Evans DA. Alzheimer Dis Assoc Disord. 2001;15:169–173. doi: 10.1097/00002093-200110000-00002. [DOI] [PubMed] [Google Scholar]
- 20.van Marum RJ. Fundam Clin Pharmacol. 2008;22:265–274. doi: 10.1111/j.1472-8206.2008.00578.x. [DOI] [PubMed] [Google Scholar]
- 21.Omerovic M, Hampel H, Teipel SJ, Buerger K. World J Biol Psychiatry. 2008;9:69–75. doi: 10.1080/15622970701568412. [DOI] [PubMed] [Google Scholar]
- 22.Roher A, Chaney M, Kuo Y, Webster S, Stiner W, Haverkamp L, Woods A, Cotter R, Tuohy J, Krafft G, Bonnell B, Emmerling M. J Biol Chem. 1996;271:20631–20635. doi: 10.1074/jbc.271.34.20631. [DOI] [PubMed] [Google Scholar]
- 23.Hartley DM, Walsh DM, Ye CP, Diehl T, Vasquez S, Vassilev PM, Teplow DB, Selkoe DJ. J Neurosci. 1999;19:8876–8884. doi: 10.1523/JNEUROSCI.19-20-08876.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoshi M, Sato M, Matsumoto S, Noguchi A, Yasutake K, Yoshida N, Sato K. Proc Natl Acad Sci USA. 2003;100:6370–6375. doi: 10.1073/pnas.1237107100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shankar GM, Shaomin L, Mehta TH, Garcia-Munoz A, Shepardson NE, Smith I, Brett FM, Farrell MA, Rowan MJ, Lemere CA, Regan CM, Walsh DM, Sabatini BL, Selkoe DJ. Nat Med. 2008;14:837–842. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wood SJ, MacKenzie L, Maleeff B, Hurle M, Wetzel R. J Biol Chem. 1996;271:4086–4092. doi: 10.1074/jbc.271.8.4086. [DOI] [PubMed] [Google Scholar]
- 27.Nakagami Y, Nishimura S, Murasugi T, Kaneko I, Meguro M, Marumoto S, Kogen H, Koyama K, Oda T. Br J Pharmacol. 2002;137:676–682. doi: 10.1038/sj.bjp.0704911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dolphin GT, Ouberai M, Dumy P, Garcia J. ChemMedChem. 2007;2:1613–1623. doi: 10.1002/cmdc.200700103. [DOI] [PubMed] [Google Scholar]
- 29.Cheng IH, Scearce-Levie K, Legleiter J, Palop JJ, Gerstein H, Bien-Ly N, Puolivali J, Lesne S, Ashe KH, Muchowski PJ, Mucke L. J Biol Chem. 2007;282:23818–23828. doi: 10.1074/jbc.M701078200. [DOI] [PubMed] [Google Scholar]
- 30.Necula M, Breydo L, Milton S, Kayed R, van der Veer WE, Tone P, Glabe CG. Biochemistry. 2007;46:8850–8860. doi: 10.1021/bi700411k. [DOI] [PubMed] [Google Scholar]
- 31.Wurth C, Guimard NK, Hecht MH. J Mol Biol. 2002;319:1279–1290. doi: 10.1016/S0022-2836(02)00399-6. [DOI] [PubMed] [Google Scholar]
- 32.Kim W, Kim Y, Min J, Kim DJ, Chang YT, Hecht MH. ACS Chem Biol. 2006;1:461–469. doi: 10.1021/cb600135w. [DOI] [PubMed] [Google Scholar]
- 33.Waldo GS, Standish BM, Berendzen J, Terwilliger TC. Nat Biotechnol. 1999;17:691–695. doi: 10.1038/10904. [DOI] [PubMed] [Google Scholar]
- 34.Nicholson RL, Welch M, Ladlow M, Spring DR. ACS Chem Biol. 2007;2:24–30. doi: 10.1021/cb600321j. [DOI] [PubMed] [Google Scholar]
- 35.MacBeath G, Koehler AN, Schreiber SL. J Am Chem Soc. 1999;121:7967–7968. [Google Scholar]
- 36.Duffner J, Clemons P, Koehler AN. Curr Opin Chem Biol. 2007;11:74–82. doi: 10.1016/j.cbpa.2006.11.031. [DOI] [PubMed] [Google Scholar]
- 37.Vegas A, Fuller J, Koehler AN. Chem Soc Rev. 2008;37:1385–1394. doi: 10.1039/b703568n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barnes-Seeman D, Park SB, Koehler AN, Schreiber SL. Angew Chem, Int Ed Engl. 2003;42:2376–2379. doi: 10.1002/anie.200351043. [DOI] [PubMed] [Google Scholar]
- 39.Uttamchandani M, Walsh DP, Khersonsky SM, Huang X, Yao SQ, Chang YT. J Comb Chem. 2004;6:862–868. doi: 10.1021/cc049900s. [DOI] [PubMed] [Google Scholar]
- 40.Koehler AN, Shamji AF, Schreiber SL. J Am Chem Soc. 2003;125:8420–8421. doi: 10.1021/ja0352698. [DOI] [PubMed] [Google Scholar]
- 41.Kuruvilla FG, Shamji AF, Sternson SM, Hergenrother PJ, Schreiber SL. Nature. 2002;416:653–657. doi: 10.1038/416653a. [DOI] [PubMed] [Google Scholar]
- 42.Vegas AJ, Bradner JE, Tang W, McPherson OM, Greenberg EF, Koehler AN, Schreiber SL. Angew Chem, Int Ed Engl. 2007;46:7960–7964. doi: 10.1002/anie.200703198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lomakin A, Chung DS, Benedek GB, Kirschner DA, Teplow DB. Proc Natl Acad Sci USA. 1996;93:1125–1129. doi: 10.1073/pnas.93.3.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seiler KP, George GA, Happ MP, Bodycombe NE, Carrinski HA, Norton S, Brudz S, Sullivan JP, Muhlich J, Serrano M, Ferraiolo P, Tolliday NJ, Schreiber SL, Clemons PA. Nucleic Acids Res. 2008;36:D351–359. doi: 10.1093/nar/gkm843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morinaga A, Hasegawa K, Nomura R, Ookoshi T, Ozawa D, Goto Y, Yamada M, Naiki H. Biochim Biophys Acta Protein Proteomics. 2010;1804:986–995. doi: 10.1016/j.bbapap.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 46.Lee S, Fernandez EJ, Good TA. Protein Sci. 2007;16:723–732. doi: 10.1110/ps.062514807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paravastu AK, Petkova AT, Tycko R. Biophys J. 2006;90:4618–4629. doi: 10.1529/biophysj.105.076927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen Y, Glabe CG. J Biol Chem. 2006;281:24414–24422. doi: 10.1074/jbc.M602363200. [DOI] [PubMed] [Google Scholar]
- 49.Dahlgren KN, Manelli AM, Stine WB, Baker LK, Krafft GA, LaDu MJ. J Biol Chem. 2002;277:32046–32053. doi: 10.1074/jbc.M201750200. [DOI] [PubMed] [Google Scholar]
- 50.Podlisny MB, Ostaszewski BL, Squazzo SL, Koo EH, Rydell RE, Teplow DB, Selkoe DJ. J Biol Chem. 1995;270:9564–9570. doi: 10.1074/jbc.270.16.9564. [DOI] [PubMed] [Google Scholar]
- 51.Walsh DM, Klyubin I, Fadeeva JV, Cullen WK, Anwyl R, Wolfe MS, Rowan MJ, Selkoe DJ. Nature. 2002;416:535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 52.Bagriantsev SN, Kushnirov VV, Liebman SW. In: Methods in Enzymology. Kheterpal I, Wetzel R, editors. Vol. 412. Academic Press; San Diego: 2006. pp. 33–48. [DOI] [PubMed] [Google Scholar]
- 53.Harper JD, Lansbury PT. Annu Rev Biochem. 1997;66:385–407. doi: 10.1146/annurev.biochem.66.1.385. [DOI] [PubMed] [Google Scholar]
- 54.O’Nuallain B, Shivaprasad S, Kheterpal I, Wetzel R. Biochemistry. 2005;44:12709–12718. doi: 10.1021/bi050927h. [DOI] [PubMed] [Google Scholar]
- 55.Porat Y, Mazor Y, Efrat S, Gazit E. Biochemistry. 2004;43:14454–14462. doi: 10.1021/bi048582a. [DOI] [PubMed] [Google Scholar]
- 56.Porat Y, Abramowitz A, Gazit E. Chem Biol Drug Des. 2006;67:27–37. doi: 10.1111/j.1747-0285.2005.00318.x. [DOI] [PubMed] [Google Scholar]
- 57.Berhanu WM, Masunov AE. Biophys Chem. 2010;149:12–21. doi: 10.1016/j.bpc.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 58.Naiki H, Higuchi K, Hosokawa M, Takeda T. Anal Biochem. 1989;177:244–249. doi: 10.1016/0003-2697(89)90046-8. [DOI] [PubMed] [Google Scholar]
- 59.Klunk WE, et al. Ann Neurol. 2004;55:306–319. doi: 10.1002/ana.20009. [DOI] [PubMed] [Google Scholar]
- 60.Klunk W, Pettegrew J, Abraham D. J Histochem Cytochem. 1989;37:1273–1281. doi: 10.1177/37.8.2666510. [DOI] [PubMed] [Google Scholar]
- 61.Williams AD, Sega M, Chen M, Kheterpal I, Geva M, Berthelier V, Kaleta DT, Cook KD, Wetzel R. Proc Natl Acad Sci USA. 2005;102:7115–7120. doi: 10.1073/pnas.0408582102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Necula M, Kayed R, Milton S, Glabe CG. J Biol Chem. 2007;282:10311–10324. doi: 10.1074/jbc.M608207200. [DOI] [PubMed] [Google Scholar]
- 63.Hudson SA, Ecroyd H, Kee TW, Carver JA. FEBS J. 2009;276:5960–5972. doi: 10.1111/j.1742-4658.2009.07307.x. [DOI] [PubMed] [Google Scholar]
- 64.Esler WP, Stimson ER, Ghilardi JR, Felix AM, Lu YA, Vinters HV, Mantyh PW, Maggio JE. Nat Biotechnol. 1997;15:258–263. doi: 10.1038/nbt0397-258. [DOI] [PubMed] [Google Scholar]
- 65.Bradner JE, McPherson OM, Koehler AN. Nat Protoc. 2006;1:2344–2352. doi: 10.1038/nprot.2006.282. [DOI] [PubMed] [Google Scholar]
- 66.Zagorski MG, Yang J, Shao H, Ma K, Zeng H, Hong A. In: Methods in Enzymology. Wetzel R, editor. Vol. 309. Academic Press; San Diego: 1999. pp. 189–204. [DOI] [PubMed] [Google Scholar]
- 67.Maehara Y, Anai H, Tamada R, Sugimachi K. Eur J Cancer Clin Oncol. 1987;23:273–276. doi: 10.1016/0277-5379(87)90070-8. [DOI] [PubMed] [Google Scholar]
- 68.Vistica DT, Skehan P, Scudiero D, Monks A, Pittman A, Boyd MR. Cancer Res. 1991;51:2515–2520. [PubMed] [Google Scholar]
- 69.Klunk WE, Jacob RF, Mason RP. Anal Biochem. 1999;266:66–76. doi: 10.1006/abio.1998.2933. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.