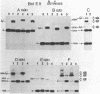
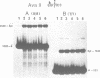
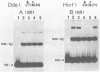
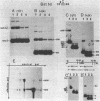
Abstract
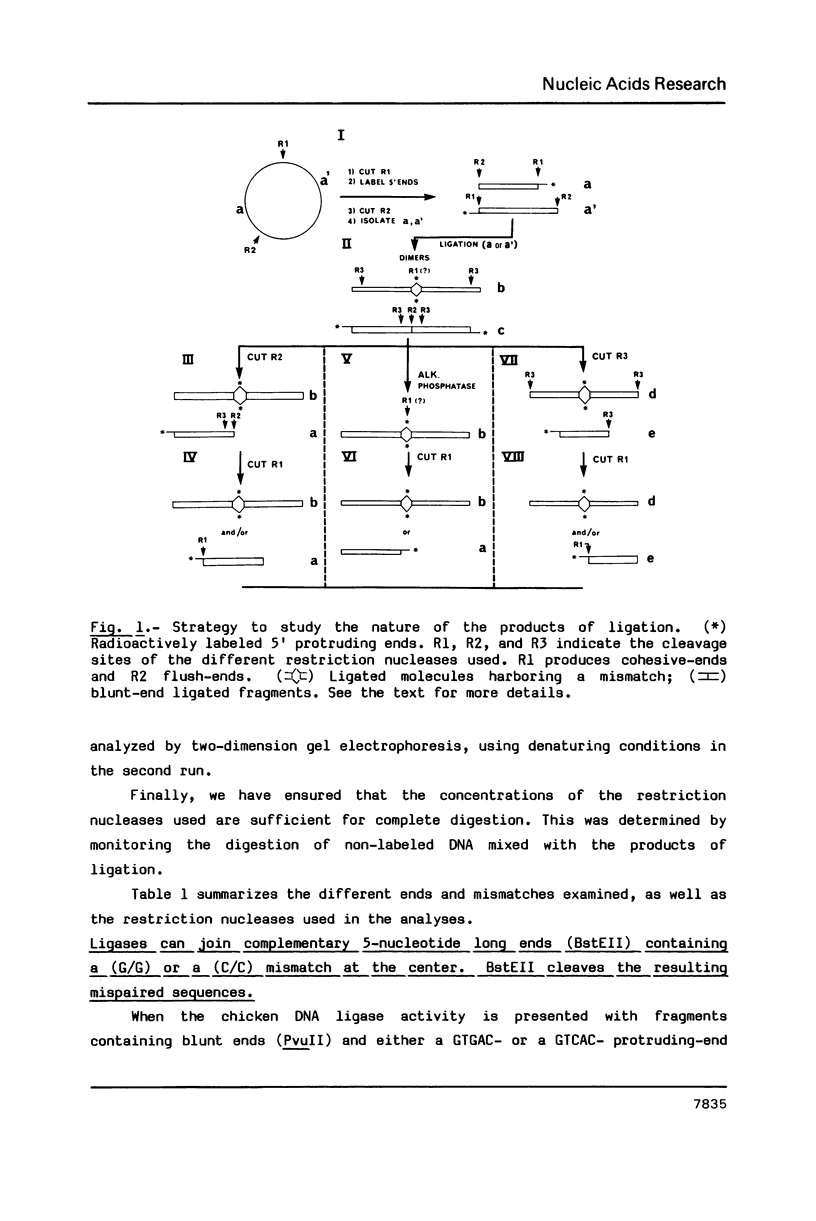
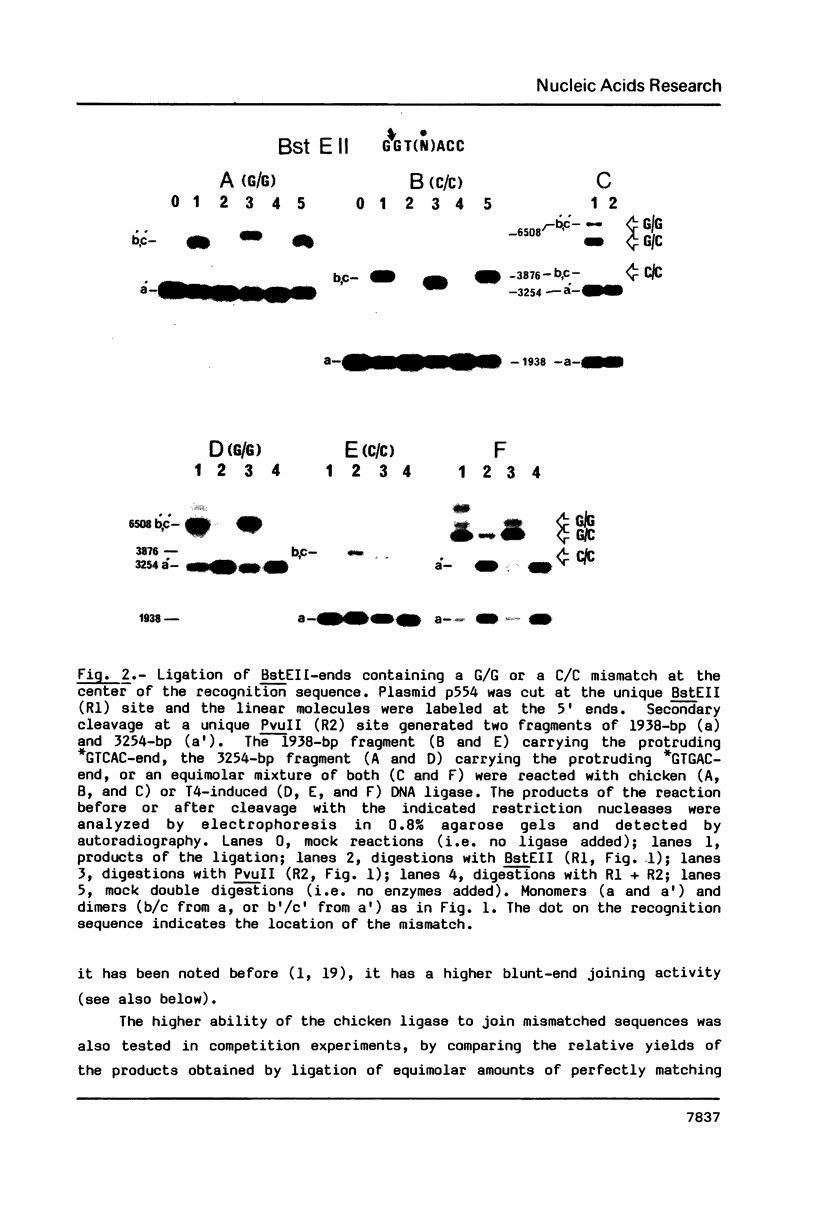
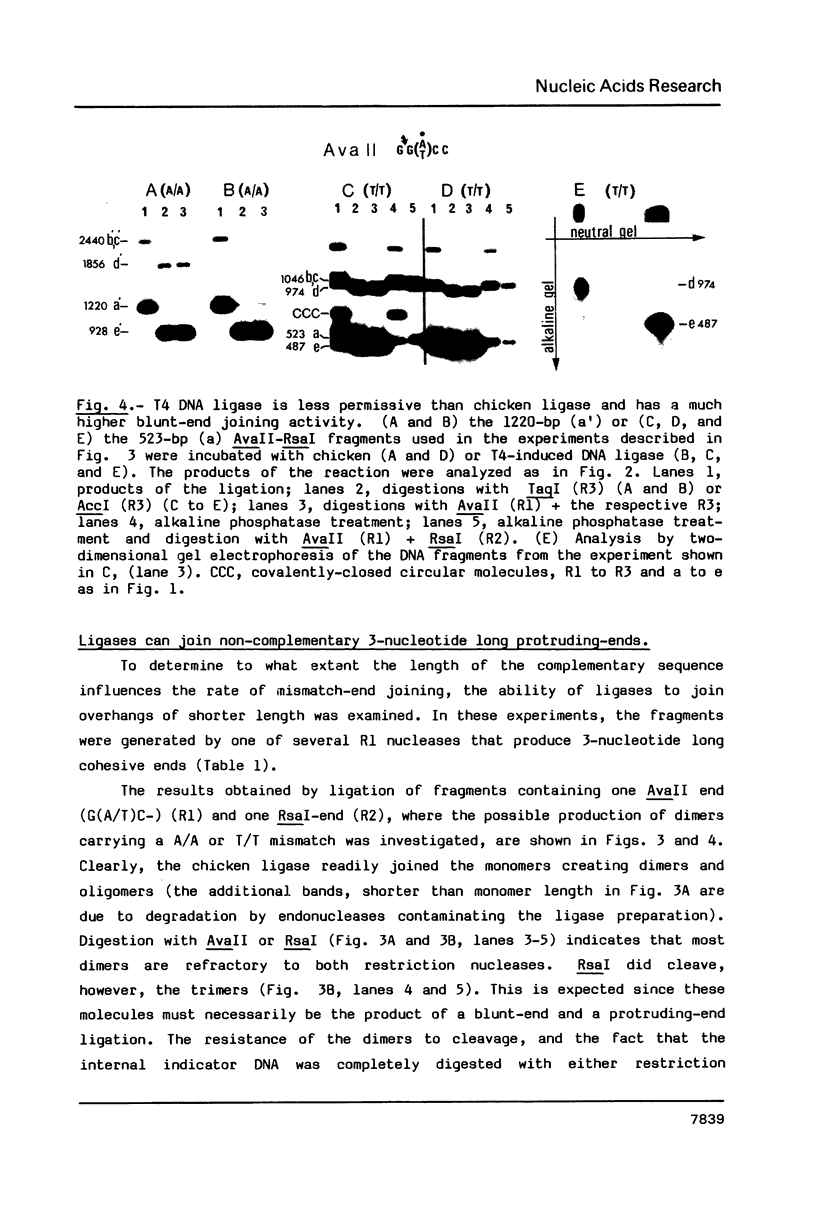
A nuclear DNA ligase activity from immature chicken erythrocytes, and to a lesser extent T4-induced DNA ligase, can join cohesive-ends (3 and 5-nucleotides long) having one of the mismatches, A/A, T/T, C/C, G/G, at the middle position. The rate of ligation depends on the length and stability of the mispaired intermediate (G/G, T/T greater than A/A, C/C). When the non-complementary overhanging-ends are short (i.e. 1-nucleotide) both ligases catalyze the joining of the single-stranded protruding-end with a blunt-end. This reaction occurs at low but significant rates compared to blunt-end ligation. The chicken ligase has lower flush-end joining activity than T4 DNA ligase, but it is more permissive since it joins C/C or A/A mismatched-ends, whereas the prokaryotic ligase does not. Possible biological implications of the reactions are discussed. We have also found that BstEII easily cleaves at sites harboring a C/C or a G/G mismatch at the center of its recognition sequence, whereas AvaII (T/T or A/A), HinfI (G/G) and DdeI (G/G) do not.
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aboul-ela F., Koh D., Tinoco I., Jr, Martin F. H. Base-base mismatches. Thermodynamics of double helix formation for dCA3XA3G + dCT3YT3G (X, Y = A,C,G,T). Nucleic Acids Res. 1985 Jul 11;13(13):4811–4824. doi: 10.1093/nar/13.13.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrand J. E., Willis A. E., Goldsmith I., Lindahl T. Different substrate specificities of the two DNA ligases of mammalian cells. J Biol Chem. 1986 Jul 15;261(20):9079–9082. [PubMed] [Google Scholar]
- Bodnar J. W., Zempsky W., Warder D., Bergson C., Ward D. C. Effect of nucleotide analogs on the cleavage of DNA by the restriction enzymes AluI, DdeI, HinfI, RsaI, and TaqI. J Biol Chem. 1983 Dec 25;258(24):15206–15213. [PubMed] [Google Scholar]
- Claverys J. P., Méjean V., Gasc A. M., Sicard A. M. Mismatch repair in Streptococcus pneumoniae: relationship between base mismatches and transformation efficiencies. Proc Natl Acad Sci U S A. 1983 Oct;80(19):5956–5960. doi: 10.1073/pnas.80.19.5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohet C., Wagner R., Radman M. Repair of defined single base-pair mismatches in Escherichia coli. Proc Natl Acad Sci U S A. 1985 Jan;82(2):503–505. doi: 10.1073/pnas.82.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins N. P., Cozzarelli N. R. DNA-joining enzymes: a review. Methods Enzymol. 1979;68:50–71. doi: 10.1016/0076-6879(79)68006-0. [DOI] [PubMed] [Google Scholar]
- Jiricny J., Martin D. Restriction endonucleases HindII and TaqI cleave DNA with mismatched nucleotides within their recognition sequences. Nucleic Acids Res. 1986 Mar 11;14(5):1943–1949. doi: 10.1093/nar/14.5.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapp L. N., Brown S. L., Klevecz R. R. Detecting small quantities of DNA on CsCl gradients. Biochim Biophys Acta. 1974 Aug 29;361(2):140–143. doi: 10.1016/0005-2787(74)90341-4. [DOI] [PubMed] [Google Scholar]
- Kramer B., Kramer W., Fritz H. J. Different base/base mismatches are corrected with different efficiencies by the methyl-directed DNA mismatch-repair system of E. coli. Cell. 1984 Oct;38(3):879–887. doi: 10.1016/0092-8674(84)90283-6. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miller C. K., Temin H. M. High-efficiency ligation and recombination of DNA fragments by vertebrate cells. Science. 1983 May 6;220(4597):606–609. doi: 10.1126/science.6301012. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. A. Genetic and enzymatic characterization of conditional lethal mutants of the yeast Schizosaccharomyces pombe with a temperature-sensitive DNA ligase. J Mol Biol. 1979 May 25;130(3):273–284. doi: 10.1016/0022-2836(79)90541-2. [DOI] [PubMed] [Google Scholar]
- Nilsson S. V., Magnusson G. Sealing of gaps in duplex DNA by T4 DNA ligase. Nucleic Acids Res. 1982 Mar 11;10(5):1425–1437. doi: 10.1093/nar/10.5.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Carrillo A., Affolter M., Renaud J. Genomic organization of the genes coding for the six main histones of the chicken: complete sequence of the H5 gene. J Mol Biol. 1983 Nov 15;170(4):843–859. doi: 10.1016/s0022-2836(83)80191-0. [DOI] [PubMed] [Google Scholar]
- Ruiz-Carrillo A., Renaud J. Endonuclease G: a (dG)n X (dC)n-specific DNase from higher eukaryotes. EMBO J. 1987 Feb;6(2):401–407. doi: 10.1002/j.1460-2075.1987.tb04769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Vazquez R., Ruiz-Carillo A. Construction of chimeric plasmids containing histone H5 cDNA from hen erythrocyte. DNA sequence of a fragment derived from the 5' region of H5 mRNA. Nucleic Acids Res. 1982 Mar 25;10(6):2093–2108. doi: 10.1093/nar/10.6.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgaramella V., Khorana H. G. CXII. Total synthesis of the structural gene for an alanine transfer RNA from yeast. Enzymic joining of the chemically synthesized polydeoxynucleotides to form the DNA duplex representing nucleotide sequence 1 to 20. J Mol Biol. 1972 Dec 28;72(2):427–444. doi: 10.1016/0022-2836(72)90155-6. [DOI] [PubMed] [Google Scholar]
- Sgaramella V., Van de Sande J. H., Khorana H. G. Studies on polynucleotides, C. A novel joining reaction catalyzed by the T4-polynucleotide ligase. Proc Natl Acad Sci U S A. 1970 Nov;67(3):1468–1475. doi: 10.1073/pnas.67.3.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy S., Daigle K., Ehrlich K. C., Gehrke C. W., Ehrlich M. Hydrolysis by restriction endonucleases at their DNA recognition sequences substituted with mismatched base pairs. Nucleic Acids Res. 1986 Jun 11;14(11):4407–4420. [PMC free article] [PubMed] [Google Scholar]
- Söderhäll S., Lindahl T. DNA ligases of eukaryotes. FEBS Lett. 1976 Aug 1;67(1):1–8. doi: 10.1016/0014-5793(76)80858-7. [DOI] [PubMed] [Google Scholar]
- Söderhäll S., Lindahl T. Mammalian DNA ligases. Serological evidence for two separate enzymes. J Biol Chem. 1975 Nov 10;250(21):8438–8444. [PubMed] [Google Scholar]
- Söderhäll S., Lindahl T. Mammalian deoxyribonucleic acid ligase. Isolation of an active enzyme-adenylate complex. J Biol Chem. 1973 Jan 25;248(2):672–675. [PubMed] [Google Scholar]
- Söderhäll S., Lindahl T. Two DNA ligase activities from calf thymus. Biochem Biophys Res Commun. 1973 Aug 6;53(3):910–916. doi: 10.1016/0006-291x(73)90178-2. [DOI] [PubMed] [Google Scholar]
- Teraoka H., Sumikawa T., Tsukada K. Purification of DNA ligase II from calf thymus and preparation of rabbit antibody against calf thymus DNA ligase II. J Biol Chem. 1986 May 25;261(15):6888–6892. [PubMed] [Google Scholar]
- Tsiapalis C. M., Narang S. A. On the fidelity of phage T4-induced polynucleotide ligase in the joining of chemically synthesized deoxyribooligonucleotides. Biochem Biophys Res Commun. 1970 May 22;39(4):631–636. doi: 10.1016/0006-291x(70)90251-2. [DOI] [PubMed] [Google Scholar]
- Werntges H., Steger G., Riesner D., Fritz H. J. Mismatches in DNA double strands: thermodynamic parameters and their correlation to repair efficiencies. Nucleic Acids Res. 1986 May 12;14(9):3773–3790. doi: 10.1093/nar/14.9.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J. H., Berget P. B., Pipas J. M. Somatic cells efficiently join unrelated DNA segments end-to-end. Mol Cell Biol. 1982 Oct;2(10):1258–1269. doi: 10.1128/mcb.2.10.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman S. B., Pheiffer B. H. Macromolecular crowding allows blunt-end ligation by DNA ligases from rat liver or Escherichia coli. Proc Natl Acad Sci U S A. 1983 Oct;80(19):5852–5856. doi: 10.1073/pnas.80.19.5852. [DOI] [PMC free article] [PubMed] [Google Scholar]