ER stress induces expression of miR-708, which suppresses the production of rhodopsin to prevent ER overloading in retinal epithelial cells.
Abstract
Using genome-wide microribonucleic acid (microRNA [miRNA]) expression profiling, bioinformatics, and biochemical analyses, we identified miR-708, an endoplasmic reticulum (ER) stress-inducible miRNA whose expression is regulated by the transcription factor CCAAT enhancer-binding protein homologous protein (CHOP) in vertebrates. miR-708 is encoded within an intron of the CHOP-regulated gene Odz4, a member of the highly conserved teneurin family of developmental regulators. Odz4 and mir-708 expression is coregulated by CHOP, and the two transcripts are coexpressed in the brain and eyes of mice, suggesting common physiological functions in these tissues. We validated rhodopsin as a target of miR-708 through loss- and gain-of-function experiments. Together, our data implicate miR-708 in the homeostatic regulation of ER function in mammalian rod photoreceptors, whereby miR-708 may help prevent an excessive rhodopsin load from entering the ER. Hence, miR-708 may function analogously to other unfolded protein response controls that throttle protein influx into the ER to avoid ER stress through mechanisms, such as general translational attenuation by protein kinase RNA–like ER kinase or membrane-bound messenger RNA decay by inositol-requiring enzyme 1.
Introduction
MicroRNAs (miRNAs) are small endogenous RNAs of ∼22 nucleotides in length that posttranscriptionally regulate gene expression in several biological processes, including cell differentiation, proliferation, and survival (Bartel, 2004). miRNAs direct the RNA-induced silencing complex (RISC) to their target mRNAs to repress translation, degrade the transcript, or both (Wightman et al., 1993; Olsen and Ambros, 1999; Lim et al., 2005; Guo et al., 2010). Sequence-specific interactions between the target mRNA and a complementary “seed” within the 5′ terminus of the miRNA determine target recognition (Lewis et al., 2003). Sequences encoding miRNAs are found in intergenic regions and within protein-coding genes (Griffiths-Jones et al., 2006). As a result, miRNA expression can be under the control of autonomous promoters or depend on the regulation of a neighboring or host gene (Baskerville and Bartel, 2005). Transcription initially produces a larger miRNA precursor subsequently processed into a mature RNA duplex (miRNA–miRNA*) that gets loaded onto the RISC. The “passenger strand” (miRNA*) is removed and degraded, freeing the “guide strand” (miRNA) for transcript targeting (Khvorova et al., 2003; Schwarz et al., 2003).
miRNAs have been linked to various cellular stresses, among them processes that impinge on the function of the ER (e.g., hypoxia, insulin secretion, and B cell differentiation; Poy et al., 2004; Vigorito et al., 2007; Huang et al., 2009a). ER stress stems from an imbalance of the ER’s protein-folding capacity, typically resulting from an increased protein load or expression of mutant proteins that cannot fold properly. Accumulation of mis- or unfolded proteins within the ER results in the activation of the unfolded protein response (UPR). In metazoans, three ER transmembrane sensors, inositol-requiring enzyme 1 (IRE1), activating transcription factor 6 (ATF6), and protein kinase RNA–like ER kinase (PERK), initiate the UPR by sensing protein-folding perturbations in the ER (Ron and Walter, 2007). Together, these sensors activate an adaptive transcriptional program that adjusts ER abundance and its protein-folding capacity according to need. The transcription factors XBP1, a downstream effector of IRE1, and ATF6 regulate the expression of genes, including chaperones and foldases (Okada et al., 2002; Lee et al., 2003). Likewise, the transcription factor ATF4, a downstream effector of PERK, activates genes involved in amino acid metabolism and redox homeostasis as well as the transcription factor CCAAT enhancer-binding protein homologous protein (CHOP; Harding et al., 2003).
Besides increasing the ER protein-folding capacity, the UPR sensors also minimize the protein load in the ER. IRE1, for example, engages in the degradation of ER-bound mRNAs through a process known as regulated IRE1-dependent decay (Hollien and Weissman, 2006; Han et al., 2009; Hollien et al., 2009), and PERK reduces protein synthesis by the phosphorylation of the translation initiation factor eIF2-α (Harding et al., 2000).
If ER stress remains unmitigated and homeostasis is not restored, the UPR switches from a cytoprotective to an apoptotic role (Lin et al., 2007). CHOP expression is linked to ER stress-induced apoptosis (Zinszner et al., 1998), yet its role in the UPR extends beyond this function. For example, CHOP regulates transcription of GADD34, a component of a phosphatase acting on eIF2-α to restore translation after PERK activation (Marciniak et al., 2004), as well as ODZ4, a plasma membrane protein essential in development (Wang et al., 1998).
With the discovery of a UPR-regulated miRNA, miR-708, we further our understanding of the mammalian UPR and expand the role of CHOP, the transcription factor controlling its expression. Our results suggest that miR-708 helps balance the ER protein-folding capacity with the load of newly synthesized rhodopsin molecules entering the ER in rod photoreceptor cells. This new level of control may tune the UPR to meet specific physiological demands of mammalian photoreceptors.
Results and discussion
CHOP controls miR-708 transcription during prolonged ER stress
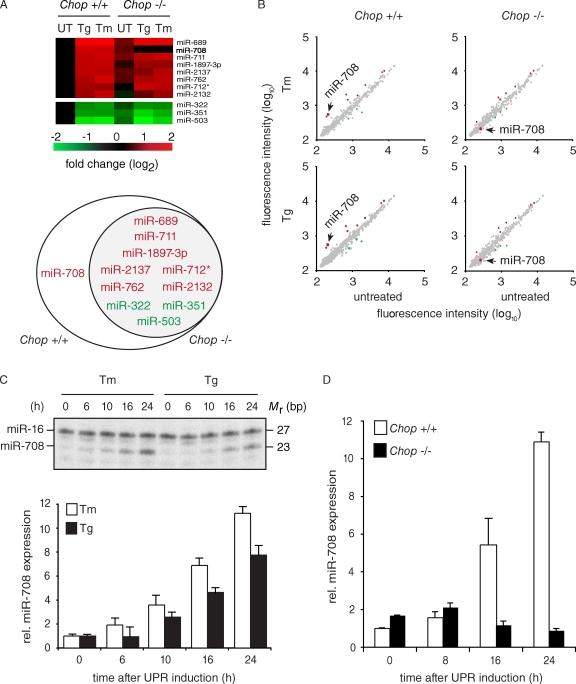
To determine whether miRNAs are regulated during ER stress, we profiled their expression levels in wild-type mouse embryonic fibroblasts (MEFs) treated with the ER stress inducers tunicamycin (Tm) or thapsigargin (Tg) for 24 h. We found 11 miRNAs differentially expressed greater than twofold during ER stress (Fig. 1 A, Chop+/+). Eight of these exhibited an increase in expression 24 h after ER stress induction (miR-689, miR-708, miR-711, miR-1897-3p, miR-2137, miR-762, miR-712*, and miR-2132), whereas three showed a decrease in expression (miR-503, miR-351, and miR-322). Interestingly, analogous experiments in CHOP-deficient MEFs showed that expression of only one of these, miR-708, was strictly dependent on CHOP (Fig. 1, A and B). miR-708 expression increased greater than threefold with the addition of either Tm or Tg in Chop+/+ MEFs, and this induction was not observed with either drug in Chop−/− MEFs. Notably, the increased expression of miR-708 during ER stress was restricted to a late time point. Indeed, after only 10 h, we observed no significant expression changes of miR-708 or any other miRNA in 3T3 mouse fibroblasts in which we induced ER stress under identical conditions (Fig. S1 A).
Figure 1.
CHOP regulates miR-708 expression during ER stress. (A) Heat maps and Venn diagrams of miRNAs differentially regulated during ER stress in Chop+/+ and Chop−/− MEFs. The applied criterium for differential expression was a more than twofold change in treated versus untreated conditions, represented as logarithmic values. Red, increase in differential expression during ER stress; Green, decrease in expression. miR-708 is indicated in bold. Cells were treated with 5 µg/ml tunicamycin (Tm) or 500 nM thapsigargin (Tg) for 24 h. UT, untreated. (B) Scatter plots illustrating the changes in expression of the miRNAs in A. (C) RNase protection assay in Chop+/+ MEFs treated with 5 µg/ml Tm or 500 nM Tg for 24 h. The loading control used was miR-16. (bottom) Quantification of the data (miR-708/miR-16). Error bars are SDs of two independent experiments. (D) TaqMan miRNA assay of miR-708 (normalized to snoRNA 202) in Chop+/+ and Chop−/− MEFs treated with 5 µg/ml Tm. Error bars are SDs of three independent experiments.
To validate our microarray data, we examined the expression of miR-708 using an RNase protection assay. The analysis confirmed the ER stress-mediated regulation of miR-708 (Fig. 1 C). miR-708 expression increased 11-fold in MEFs treated with Tm and eightfold in MEFs treated with Tg. Interestingly, the increased expression of miR-708 was delayed compared with canonical markers of UPR activation, such as the induction of the ER chaperone Grp78 or the splicing of Xbp1 (Fig. S1 B).
TaqMan miRNA assays detected the mature form of miR-708 in Chop+/+ and Chop−/− MEFs undergoing prolonged ER stress (Fig. 1 D). Corroborating the RNase protection data, we observed an 11-fold increase in miR-708 expression in Chop+/+ MEFs, whereas no such increase was observed in Chop−/− MEFs. Detection of the mature form of the miRNA in Chop−/− MEFs suggests that mechanisms independent of CHOP may be responsible for maintaining the basal levels of miR-708 observed in our experiments. Together, our data show that miR-708 is transcriptionally regulated by CHOP during the ER stress response.
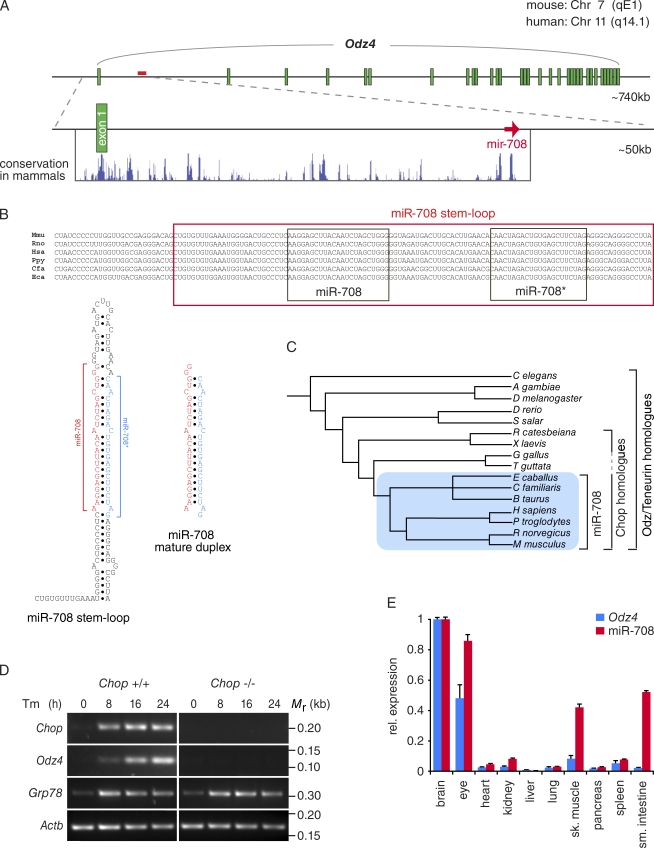
mir-708 is an intronic miRNA residing within the CHOP-inducible gene Odz4
The gene encoding miR-708 (mir-708) resides in intron 1 of Odz4, an evolutionarily conserved gene (Fig. 2 A). The encoded protein, ODZ4, is a vertebrate homologue of the teneurin family of developmental regulators essential for nervous system development (Ben-Zur et al., 2000). Odz4 was originally characterized as one of several genes regulated by CHOP (Wang et al., 1998). Consequently, mir-708 may therefore be a transcriptional target of CHOP carved out of the Odz4 transcript produced during ER stress.
Figure 2.
miR-708 is a conserved intronic miRNA highly expressed in neuroectodermal tissues. (A) Schematic of the locus encoding Odz4 indicating that mir-708 resides within its first intron. University of California, Santa Cruz Genome Browser conservation in mammals is shown. (B, top) Sequence alignment of the miR-708 stem loop in mammals (Mmu, Mus musculus; Rno, Rattus norvegicus; Hsa, Homo sapiens; Ppy, Pongo pygmaeus; Cfa, Canis familiaris; and Eca, Equus caballus). The guide (miR-708) and passenger strands (miR-708*) are outlined in black boxes. (bottom left) Stem loop structure and mature duplex of murine miR-708. (C) Phylogenetic tree of bilateral animals in which Odz/Teneurin homologues are found. Chop homologues are found only in amphibians and mammals, and miR-708 homologues are found only in mammals (blue box). Bioinformatics analyses were performed using the HomoloGene database (National Center for Biotechnology Information). (D) RT-PCR analyses in Chop+/+ and Chop−/− MEFs treated with 5 µg/ml Tm for 24 h. Grp78 mRNA indicates activation of the UPR. The loading control used was β-actin (Actb). (E) Gene expression analyses of miR-708 (TaqMan miRNA assay) and Odz4 (qRT-PCR) in adult mouse tissues normalized to snoRNA 202 and Rps26, respectively. Variations in their relative expressions (rel. expression) can be attributed to (a) undetected Odz4 isoforms, (b) differential regulation of the miRNA and host gene, and/or (c) experimental variation between TaqMan and SYBR green–based assays. Error bars are SDs of two independent experiments. sk., skeletal; sm., small.
Bioinformatics analyses indicate mir-708 is conserved in mammals within the Odz4 intron, suggesting that mir-708 has co-opted the ER stress-dependent regulation of Odz4 (Fig. 2 A). Alluding to its functional relevance, the precursor miR-708 stem loop is highly conserved, and more importantly, the guide strand (miR-708) is strictly conserved in mammals (Fig. 2 B). Although mir-708 appears to be more recently evolved, Odz4 and its homologues exhibit broader conservation in bilateral animals (Fig. 2 C). Therefore, we asked whether miR-708 is indeed coexpressed with its ancestral host gene, Odz4. RT-PCR analyses in Chop+/+ and Chop−/− MEFs subjected to ER stress indicated a CHOP-dependent expression of Odz4 (Fig. 2 D) that mirrored the delayed kinetics of accumulation of miR-708 (compare Figs. 1, C and D; and 2 D). As expected for CHOP-regulated transcripts, the UPR-induced expression of both Odz4 and miR-708 closely lagged behind Chop. Moreover, expression of miR-708 correlated well with the expression of Odz4 in adult mouse tissues (Fig. 2 E). Strikingly, we observed a significant accumulation of both transcripts in the brain and eyes, strongly suggesting a physiological role for miR-708 in tissues in which Odz4 is expressed.
miR-708 is loaded on the RISC
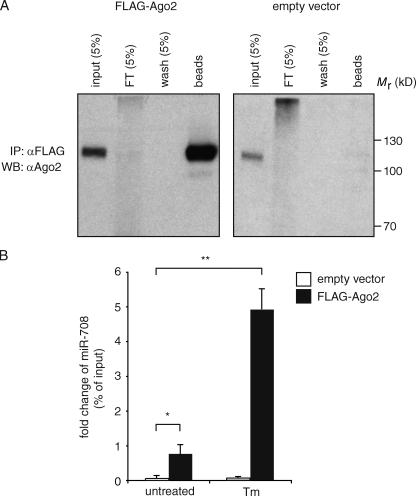
If miR-708 is indeed functional, as suggested by sequence conservation, it should be loaded onto the RISC. To test this, we performed immunoprecipitations in 3T3 cells stably expressing FLAG-tagged Argonaute 2 (Ago2; FLAG-Ago2), an essential component of the RISC, followed by TaqMan-based detection of miR-708 (Fig. 3 A). Analyses in untreated cells revealed a 75-fold enrichment of miR-708 loaded onto the RISC compared with cells expressing the empty vector, indicating that even the low steady-state level of miR-708 is efficiently loaded onto the complex (Fig. 3 B). In ER stress-induced cells, there was a 500-fold enrichment in RISC-associated miR-708 as compared with the empty vector, indicating that the strong transcriptional induction of mir-708 upon ER stress results in a concomitant increase of miR-708 loaded onto the RISC (Fig. 3 B). Together, these results show that miR-708 is engaged with the cellular components expected for a functional miRNA.
Figure 3.
Mature miR-708 is loaded on the RISC. (A) Immunoprecipitation (IP) of FLAG-tagged Ago2 (FLAG-Ago2) from 3T3 fibroblasts stably expressing it. (right) 3T3 cells transduced with an empty vector. FT, flow through. WB, Western blot. (B) TaqMan miRNA assay of miR-708 from FLAG-immunoprecipitated fractions obtained from lysates of the cells in A. Error bars are SDs of two independent experiments. *, P < 0.0005; **, P < 0.008. P-values were derived from a t test for independent samples.
Rhodopsin is a functional target of miR-708
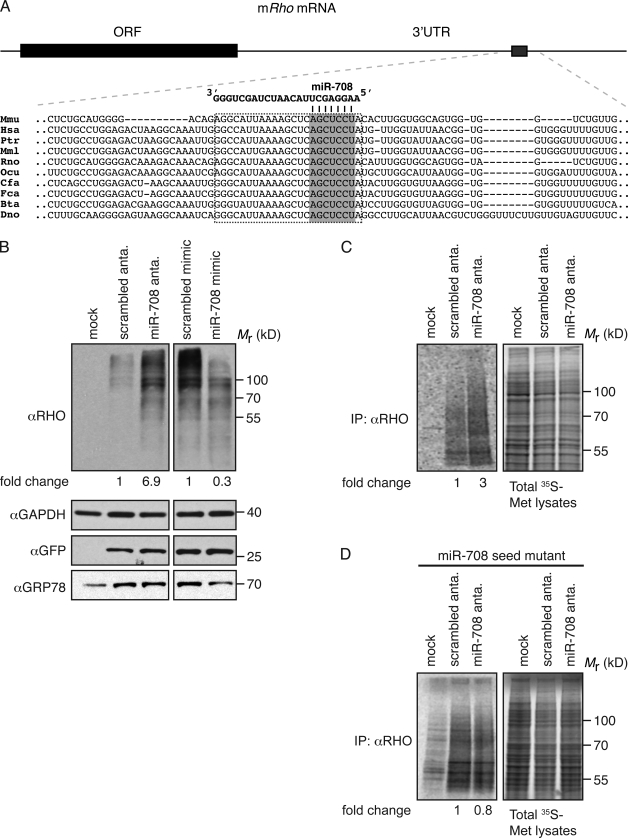
To address the biological role of miR-708, we used the miRNA target prediction program TargetScan (Lewis et al., 2005; Friedman et al., 2009) to generate a list of candidate transcripts with putative miR-708 binding sites (Table S1). Gene ontology analyses on the predicted targets revealed the highest enrichment of genes involved in vision (FOXJ3, RPGRIP1L, RHO, and RCVRN; P = 0.003) and phototransduction (RHO and RCVRN; P = 0.005). Because Odz4 and miR-708 show enhanced expression in the eyes (Fig. 2 E), we reasoned that genes involved in vision may be targeted by miR-708. We focused on rhodopsin because its synthesis relies on ER function and its expression is correlated with CHOP induction (see following paragraphs). Moreover, bioinformatics analyses revealed a highly conserved putative miR-708 binding site in the 3′ untranslated region (UTR) of rhodopsin, which, like miR-708 itself, is highly conserved among mammals (Fig. 4 A).
Figure 4.
miR-708 targets rhodopsin for posttranscriptional inhibition. (A, top) Schematic of full-length mouse rhodopsin mRNA. (bottom) Sequence alignment of the region containing the predicted conserved miR-708 site in mammals. The gray box represents the putative site complementary to the seed sequence; the black dotted line encircles the entire putative site. Mmu, M. musculus; Hsa, H. sapiens; Ptr, Pan troglodytes; Mml, Macaca mulatta; Rno, R. norvegicus; Ocu, Oryctolagus cuniculus; Cfa, C. familiaris; Fca, Felis catus; Bta, Bos taurus; and Dno, Dasypus novemcinctus. (B) Immunoblots of lysates from 293T cells transfected with plasmids encoding full-length mouse Rho or GFP along with an miR-708 antagomir (anta.) or mimic. Overexpression of rhodopsin results in the expected aggregates observed when resolved by SDS-PAGE. GRP78 was used to show activation of the UPR. GFP was used as a control for transfection efficiency and off-target effects of the antagomir/mimic. The loading control used was GAPDH. Numbers indicate the fold change in expression normalized to GAPDH. (C) Autoradiograms of 239T cells transfected with a plasmid encoding full-length mouse Rho along with an antagomir or scrambled control and pulse labeled with [35S]methionine (35S-Met) for 1 h. (left) Lysates immunoprecipitated (IP) with an anti-RHO antibody. (right) Total lysates. Numbers indicate relative amounts of radiolabeled rhodopsin normalized to total lysate. (D) Same experiment as in C except the 239T cells were transfected with a plasmid encoding full-length mouse Rho in which the miR-708 seed in the 3′UTR was replaced with a scrambled sequence (miR-708 seed mutant).
To test whether miR-708 regulates the expression of rhodopsin, we performed loss-of-function experiments by transiently transfecting 293T cells with a plasmid encoding full-length rhodopsin along with a single-stranded antisense inhibitor (antagomir) of miR-708 or a scrambled control. In complementary gain-of-function experiments, we cotransfected the aforementioned rhodopsin-encoding plasmid with a double-stranded RNA miR-708 mimic designed to imitate the miR-708–miR-708* duplex or a scrambled control. In both types of experiments, a plasmid encoding GFP was used as a transfection control. Because 293T cells exhibit higher basal levels of miR-708 than MEFs (Fig. S2), we expected 293T cells would allow us to examine the effects of miR-708 on rhodopsin expression even in the absence of ER stress. Indeed, reducing the levels of endogenous miR-708 with the antagomir resulted in a 6.9-fold increase of steady-state levels of rhodopsin protein (Fig. 4 B, left). Furthermore, expression of a miR-708 mimic resulted in a 2.5-fold decrease in rhodopsin expression (Fig. 4 B, right). Notably, GFP levels were unaffected by either the miR-708 antagomir or mimic, indicating that the effects observed were specific to rhodopsin (Fig. 4 B, bottom). To study miR-708’s effect on newly synthesized rhodopsin, we performed a pulse-labeling experiment in 293T cells transiently transfected with a plasmid encoding the full-length rhodopsin along with either an antagomir or scrambled control. Consistent with our immunoblotting data, we observed a net increase of rhodopsin (threefold) in cells transfected with the antagomir (Fig. 4 C). However, no such effect was observed in cells expressing rhodopsin encoded by a mutant mRNA in which the putative miR-708 seed binding site was replaced with a scrambled sequence (Fig. 4 D and not depicted). Together, our results suggest that miR-708 targets rhodopsin mRNA, resulting in its decreased expression in mammalian cells.
Here, we show that the intronic miRNA miR-708 is regulated by ER stress and provide evidence that one of its roles is to control expression of rhodopsin. mir-708 resides within the first intron of Odz4, a target of the UPR transcription factor CHOP (Wang et al., 1998). Odz4 and its paralogues (Odz1–3) are implicated in developmental processes, such as neurite growth, cell adhesion, and eye development (Zhou et al., 2003; Kinel-Tahan et al., 2007). Indeed, Odz4 is expressed in the developing eye (Ben-Zur et al., 2000) as well as in the adult brain and eyes (Fig. 2 E). Although little is known about the role of Odz4 during ER stress, our data show that miR-708 and Odz4 are coregulated by CHOP, thereby linking its regulation to the UPR.
It is attractive to speculate that mir-708 acquired an ER stress-regulated expression of its own by hitchhiking with the CHOP-regulated Odz4. The coupled expression of miR-708 and Odz4 in cells undergoing ER stress and their marked coexpression in the brain and eyes suggest a physiological function of miR-708 in these tissues (Lutter et al., 2010). As such, miR-708 joins other miRNAs that reside in introns of pre-mRNAs, such as miR-33, which regulates cholesterol biogenesis along with its host gene, SREBP, in macrophages and hepatocytes (Marquart et al., 2010; Najafi-Shoushtari et al., 2010; Rayner et al., 2010). Similarly, the identification of rhodopsin as a target of miR-708 links ER stress and the PERK pathway through CHOP to the regulation of rhodopsin biosynthesis.
Bioinformatics analyses suggest miR-708 targets several genes involved in vision. We focused on rhodopsin because this transmembrane protein must traverse the secretory pathway, relying on ER function for its delivery to the membranes of outer segments in rod photoreceptor cells (Mendes et al., 2005). Indeed, increases in Grp78 and Chop are observed in the developing retinas of rats (Lin et al., 2007), indicating heightened ER function and activation of the UPR. This suggests that an upsurge in Odz4 and mir-708 transcripts may also follow, although future experiments are required to test this prediction. Thus, it is plausible to assume that miR-708 may have evolved as an additional safeguard mechanism controlling the synthesis of rhodopsin, thereby balancing demand with the protein-folding capacity of the ER. In this way, miR-708 function would be conceptually similar to that of the regulated IRE1-dependent decay pathway, which reduces protein influx by degrading membrane-associated transcripts or eIF2-α phosphorylation by PERK, which achieves the same goal by reducing translation.
Moreover, miR-708 may also play an important role in retinal degenerative diseases that lead to blindness (e.g., retinitis pigmentosa). In some cases of retinitis pigmentosa, single missense mutations compromise the folding and trafficking of rhodopsin, leading to photoreceptor cell death (Anukanth and Khorana, 1994; Tam and Moritz, 2006). In such instances, UPR hyperactivation has been implicated in the apoptotic fate of the photoreceptor (Kosmaoglou et al., 2009). Indeed, genetic models of retinitis pigmentosa expressing constitutively misfolded rhodopsin show a late-phase burst of Chop expression (Lin et al., 2007), which may be coupled to increased miR-708 production. It will be interesting to explore whether less severe folding mutations are silent only because miR-708 keeps mutant rhodopsin expression levels low enough to prevent or delay cell death. Importantly, because miR-708 is also expressed outside the retina (Fig. 2 E), its role may extend beyond the control of rhodopsin to other gene expression programs involved in normal development or pathology (Tsang et al., 2010). Together, our data assign CHOP a cytoprotective function likely preceding its well-characterized apoptotic role, thus adding a new level of control for the UPR.
Materials and methods
Cells, cell culture, and drug treatments
Human embryonic kidney cells 293T (gift from M. Bassik and J. Weissman, University of California, San Francisco [UCSF], San Francisco, CA), wild-type MEFs (gift from L. Glimcher, Harvard University, Boston, MA), and MEFs derived from CHOP-deficient animals and their wild-type genetic counterparts (gift from D. Ron, New York University, New York, NY) were maintained in DME supplemented with 10% heat-inactivated FBS, 2 mM l-glutamine, and penicillin/streptomycin. For ER stress induction, cells were treated with Tg (Sigma-Aldrich) or Tm (EMD).
miRNA expression profiling
Total RNA was prepared using TRIZOL (Invitrogen) following the manufacturer’s recommendations. Sample preparation, labeling, and array hybridizations were performed according to standard protocols from the UCSF Shared Microarray Core Facilities and Agilent Technologies. Total RNA was labeled with Cy3-CTP using an miRNA power labeling kit (miRCURY LNA; Exiqon). Labeled RNA was hybridized for 16 h to custom UCSF miRNA v3.4 multispecies 8 × 15,000 feature inkjet arrays (Agilent Technologies). Arrays were scanned using a microarray scanner (Agilent Technologies), and raw signal intensities were extracted with Feature Extraction v10.1 software (Agilent Technologies). The median feature pixel intensity was used as the raw signal before quantile normalization (Bolstad et al., 2003). miRNA microarray expression analyses were performed at the Sandler Asthma Basic Research Center Functional Genomics Core Facility (R. Barbeau, A. Barczak, and D. Erle, UCSF, San Francisco, CA).
RNA protection assays, semiquantitative PCR (RT-PCR), real-time RT-PCR (quantitative RT-PCR [qRT-PCR]), and TaqMan miRNA assay
Total RNA for all procedures was prepared using TRIZOL. For the RNase protection assay, both an miRNA probe construction kit and an miRNA detection kit were used (mirVana; Invitrogen). The miR-708–specific probe was generated using the oligonucleotide 5′-AAGGAGCTTACAATCTAGCTGGGCCTGTCTC-3′ as a template for in vitro transcription. Gel-purified probes were then used for hybridization, digestion, and precipitation. The protected fragments were resolved on 15% polyacrylamide/8 M urea gels and visualized using the Typhoon 9400 Variable Mode Imager (GE Healthcare). Densitometric analyses of digital images were performed with ImageJ (National Institutes of Health).
For RT-PCR and qRT-PCR, 250 ng of total RNA was reverse transcribed with the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen), and 1% of the resulting cDNA was used for PCR reactions with gene-specific oligonucleotide primers. Forward and reverse primers used are as follows: 5′-GAGCCAGACCACTCGGCCCT-3′ and 5′-GCCGGGTCAGCGAGCGATAG-3′ (mouse Odz4), 5′-ACTGCCCATTGATCTCTATCG-3′ and 5′-TGCACTTCCTTCATCCTGCC-3′ (human Odz4), 5′-GTCAGTTATCTTGAGCCTAACACG-3′ and 5′-TGTGGTGGTGTATGAAGATGC-3′ (mouse Chop), 5′-TTAAGTCTAAGGCACTGAGCGTATC-3′ and 5′-TGCTTTCAGGTGTGGTGATG-3′ (human Chop), 5′-GAACCAGGAGTTAAGAACACG-3′ and 5′-AGGCAACAGTGTCAGAGTCC-3′ (total mouse Xbp1), 5′-ATAAACCCCGATGAGGCTGT-3′and 5′-AGCAGGAGGAATTCCAGTCA-3′ (mouse Grp78), 5′-GCCATCCATAGCAAGGTTGT-3′ and 5′-GCCTCTTTACATGGGCTTTG-3′ (mouse Rps26), 5′-CAGCTTCTTTGCAGCTCCTT-3′ and 5′-CACGATGGAGGGGAATACAG-3′ (mouse β-actin), 5′-TTCTACAATGAGCTGCGTGTG-3′ and 5′-AGGGCATACCCCTCGTAGAT-3′ (human β-actin), 5′-CACTTGGAGGTGAAATCGCCC-3′ and 5′-TCCAGGTGAAGACCACACCC-3′ (mouse Rho), and 5′-AGCCACACCGCTCAGACAC-3′ and 5′-TGGAAGATGGTGATGGGATT-3′ (human GAPDH). RT-PCR reactions were resolved on 2% agarose gels and quantified with ImageJ. qRT-PCR reactions were performed using iQ SYBR Green Supermix (Bio-Rad Laboratories). For TaqMan miRNA assays, 500 ng of total RNA was reverse transcribed with miRNA-specific primers (hsa-miR-708 and small nucleolar RNA 202 [snoRNA 202] or U6 RNA as controls) using the TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems). Both qRT-PCR and TaqMan reactions were run in a real-time PCR cycler (DNA Engine Opticon 2; Bio-Rad Laboratories) using the Opticon Monitor v3 software (Bio-Rad Laboratories).
Generation of stable cell lines, immunoprecipitations, transient transfections, and immunoblotting
N-terminally FLAG-tagged human Ago2 was excised from pIRES-neo FLAG/HA-Ago2 (gift from T. Tuschl, Rockefeller University, New York, NY) with EcoRI and HindIII and subcloned into the corresponding sites of the retroviral vector pLPCX (Takara Bio Inc.). High-titer retroviral supernatants produced by ecotropic cells (Phoenix; Orbigen) were used to transduce 3T3 cells. Stable expressors were selected by culturing the transduced cells in the presence of puromycin. To immunoprecipitate Ago2, cells expressing FLAG-Ago2 were lysed in 0.5% NP-40, 150 mM KCl, 25 mM Tris-HCl, pH 7.4, and 5 mM DTT supplemented with protease inhibitor tablets (Complete; Roche) and 100 U/ml RNase inhibitor (SUPERase-IN; Invitrogen) for 30 min on ice. After centrifugation at 14,000 rpm at 4°C for 30 min, lysate was incubated with anti-FLAG M2 agarose (Sigma-Aldrich) at 4°C for 4 h, and the immune complexes were washed four times with 0.5% NP-40, 150 mM KCl, and 25 mM Tris-HCl, pH 7.4. To assess the abundance of miRNAs in the immunoprecipitates, total RNA was TRIZOL extracted and used for TaqMan miRNA assays. Immunoprecipitation efficacy was determined by immunoblotting with anti-Ago2 antibody (ab57113; Abcam).
Transient transfections in 293T cells were performed with plasmids encoding full-length wild-type mouse rhodopsin (pSPORT6-mRHO, clone ID 4500760; Thermo Fisher Scientific) and a mutant variant of mouse rhodopsin or GFP (pcDNA3.1/NT-GFP; Invitrogen). The mutant rhodopsin includes a scrambled sequence replacing the putative miR-708 seed binding site in the 3′UTR (AGCTCCTA at positions 2,586–2,593 were replaced by CTAGAGCC). For gain- and loss-of-function experiments, the plasmids were cotransfected with an miR-708 antagomir or mimic with Lipofectamine 2000 (Invitrogen). miR-708 antagomir or mimic, including their respective scrambled controls, was purchased from Invitrogen: anti-miR inhibitor miR-708 (AM11161), Cy3-labeled anti-miR negative control (AM17011), pre-miR miRNA precursor miR-708 (PM11161), and Cy3-labeled pre-miR negative control (AM17120). Immunodetection of rhodopsin was performed 36 h after transfection. Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS) for 30 min at 4°C and clarified for 5 min. Lysates were separated on 4–12% SDS-PAGE gels, and immunoblots were probed with 1D4 anti-RHO (Abcam), anti-GFP (Roche), anti–glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Abcam), or anti-GRP78 (Cell Signaling Technology) antibodies. For pulse-labeling experiments, 293T cells transfected with rhodopsin plasmid along with either the antagomir or scrambled control were pulsed labeled with [35S]methionine for 1 h before lysis in RIPA buffer. 1D4 anti-RHO antibody was incubated with lysates for 2 h at 4°C followed by an additional 2-h incubation with protein A support (Affi-Prep; Bio-Rad Laboratories). Immune complexes were washed three times with RIPA buffer, boiled for 3 min, and resolved on a 4–12% SDS-PAGE gel.
Functional classification of predicted miR-708 targets
The National Institutes of Health Database for Annotation, Visualization, and Integrated Discovery program (Dennis et al., 2003; Huang et al., 2009b) was used to assign gene functional categories and to identify differentially enriched gene ontology terms among the miR-708 targets predicted by TargetScan.
Online supplemental material
Fig. S1 shows the inconspicuous changes in the expression of miRNAs in 3T3 fibroblasts exposed to ER stress for 10 h. Fig. S2 shows the expression of miR-708, Chop, and Odz4 in 293T cells compared with MEFs. Table S1 shows the top 30 candidate target genes of miR-708 defined by TargetScan. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201010055/DC1.
Acknowledgments
We thank Walter laboratory members for their critical review of the manuscript, David Ron for thoughtful insights and essential reagents, and Han Li for technical support. We also thank the members of the Sandler Asthma Basic Research Center Functional Genomics Core Facility, Rebecca Barbeau, Andrea Barczak, and David Erle, for miRNA expression arrays and analysis, which were funded in part by National Institutes of Health/National Center for Research Resources UCSF Clinical and Translational Science Institute. We thank Carolina Arias for assistance in the visualization of microarray data and Jiashun Zheng for assistance in bioinformatics analysis.
This work was supported by a National Science Foundation Predoctoral Fellowship (S. Behrman) and the Irvington Institute Postdoctoral Fellowship of the Cancer Research Institute (D. Acosta-Alvear). P. Walter is an investigator of the Howard Hughes Medical Institute.
Footnotes
Abbreviations used in this paper:
- Ago2
- Argonaute 2
- CHOP
- CCAAT enhancer-binding protein homologous protein
- GAPDH
- glyceraldehyde 3-phosphate dehydrogenase
- MEFs
- mouse embryonic fibroblasts
- miRNA
- microRNA
- PERK
- protein kinase RNA–like ER kinase
- qRT-PCR
- quantitative RT-PCR
- RIPA
- radioimmunoprecipitation assay
- RISC
- RNA-induced silencing complex
- snoRNA 202
- small nucleolar RNA 202
- Tg
- thapsigargin
- Tm
- tunicamycin
- UPR
- unfolded protein response
- UTR
- untranslated region
References
- Anukanth A., Khorana H.G. 1994. Structure and function in rhodopsin. Requirements of a specific structure for the intradiscal domain. J. Biol. Chem. 269:19738–19744 [PubMed] [Google Scholar]
- Bartel D.P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116:281–297 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Baskerville S., Bartel D.P. 2005. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 11:241–247 10.1261/rna.7240905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Zur T., Feige E., Motro B., Wides R. 2000. The mammalian Odz gene family: homologs of a Drosophila pair-rule gene with expression implying distinct yet overlapping developmental roles. Dev. Biol. 217:107–120 10.1006/dbio.1999.9532 [DOI] [PubMed] [Google Scholar]
- Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 19:185–193 10.1093/bioinformatics/19.2.185 [DOI] [PubMed] [Google Scholar]
- Dennis G., Jr, Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4:P3 10.1186/gb-2003-4-5-p3 [DOI] [PubMed] [Google Scholar]
- Friedman R.C., Farh K.K., Burge C.B., Bartel D.P. 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19:92–105 10.1101/gr.082701.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34(Suppl. 1):D140–D144 10.1093/nar/gkj112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Ingolia N.T., Weissman J.S., Bartel D.P. 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 466:835–840 10.1038/nature09267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D., Lerner A.G., Vande Walle L., Upton J.P., Xu W., Hagen A., Backes B.J., Oakes S.A., Papa F.R. 2009. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 138:562–575 10.1016/j.cell.2009.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding H.P., Zhang Y., Bertolotti A., Zeng H., Ron D. 2000. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell. 5:897–904 10.1016/S1097-2765(00)80330-5 [DOI] [PubMed] [Google Scholar]
- Harding H.P., Zhang Y., Zeng H., Novoa I., Lu P.D., Calfon M., Sadri N., Yun C., Popko B., Paules R., et al. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 11:619–633 10.1016/S1097-2765(03)00105-9 [DOI] [PubMed] [Google Scholar]
- Hollien J., Weissman J.S. 2006. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 313:104–107 10.1126/science.1129631 [DOI] [PubMed] [Google Scholar]
- Hollien J., Lin J.H., Li H., Stevens N., Walter P., Weissman J.S. 2009. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J. Cell Biol. 186:323–331 10.1083/jcb.200903014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Ding L., Bennewith K.L., Tong R.T., Welford S.M., Ang K.K., Story M., Le Q.T., Giaccia A.J. 2009a. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol. Cell. 35:856–867 10.1016/j.molcel.2009.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., Lempicki R.A. 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Khvorova A., Reynolds A., Jayasena S.D. 2003. Functional siRNAs and miRNAs exhibit strand bias. Cell. 115:209–216 10.1016/S0092-8674(03)00801-8 [DOI] [PubMed] [Google Scholar]
- Kinel-Tahan Y., Weiss H., Dgany O., Levine A., Wides R. 2007. Drosophila odz gene is required for multiple cell types in the compound retina. Dev. Dyn. 236:2541–2554 10.1002/dvdy.21284 [DOI] [PubMed] [Google Scholar]
- Kosmaoglou M., Kanuga N., Aguilà M., Garriga P., Cheetham M.E. 2009. A dual role for EDEM1 in the processing of rod opsin. J. Cell Sci. 122:4465–4472 10.1242/jcs.055228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.H., Iwakoshi N.N., Glimcher L.H. 2003. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 23:7448–7459 10.1128/MCB.23.21.7448-7459.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. 2003. Prediction of mammalian microRNA targets. Cell. 115:787–798 10.1016/S0092-8674(03)01018-3 [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 120:15–20 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 433:769–773 10.1038/nature03315 [DOI] [PubMed] [Google Scholar]
- Lin J.H., Li H., Yasumura D., Cohen H.R., Zhang C., Panning B., Shokat K.M., Lavail M.M., Walter P. 2007. IRE1 signaling affects cell fate during the unfolded protein response. Science. 318:944–949 10.1126/science.1146361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutter D., Marr C., Krumsiek J., Lang E.W., Theis F.J. 2010. Intronic microRNAs support their host genes by mediating synergistic and antagonistic regulatory effects. BMC Genomics. 11:224 10.1186/1471-2164-11-224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak S.J., Yun C.Y., Oyadomari S., Novoa I., Zhang Y., Jungreis R., Nagata K., Harding H.P., Ron D. 2004. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 18:3066–3077 10.1101/gad.1250704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquart T.J., Allen R.M., Ory D.S., Baldán A. 2010. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc. Natl. Acad. Sci. USA. 107:12228–12232 10.1073/pnas.1005191107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes H.F., van der Spuy J., Chapple J.P., Cheetham M.E. 2005. Mechanisms of cell death in rhodopsin retinitis pigmentosa: implications for therapy. Trends Mol. Med. 11:177–185 10.1016/j.molmed.2005.02.007 [DOI] [PubMed] [Google Scholar]
- Najafi-Shoushtari S.H., Kristo F., Li Y., Shioda T., Cohen D.E., Gerszten R.E., Näär A.M. 2010. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 328:1566–1569 10.1126/science.1189123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T., Yoshida H., Akazawa R., Negishi M., Mori K. 2002. Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem. J. 366:585–594 10.1042/BJ20020391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen P.H., Ambros V. 1999. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 216:671–680 10.1006/dbio.1999.9523 [DOI] [PubMed] [Google Scholar]
- Poy M.N., Eliasson L., Krutzfeldt J., Kuwajima S., Ma X., Macdonald P.E., Pfeffer S., Tuschl T., Rajewsky N., Rorsman P., Stoffel M. 2004. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 432:226–230 10.1038/nature03076 [DOI] [PubMed] [Google Scholar]
- Rayner K.J., Suárez Y., Dávalos A., Parathath S., Fitzgerald M.L., Tamehiro N., Fisher E.A., Moore K.J., Fernández-Hernando C. 2010. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 328:1570–1573 10.1126/science.1189862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D., Walter P. 2007. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8:519–529 10.1038/nrm2199 [DOI] [PubMed] [Google Scholar]
- Schwarz D.S., Hutvágner G., Du T., Xu Z., Aronin N., Zamore P.D. 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 115:199–208 10.1016/S0092-8674(03)00759-1 [DOI] [PubMed] [Google Scholar]
- Tam B.M., Moritz O.L. 2006. Characterization of rhodopsin P23H-induced retinal degeneration in a Xenopus laevis model of retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci. 47:3234–3241 10.1167/iovs.06-0213 [DOI] [PubMed] [Google Scholar]
- Tsang K.Y., Chan D., Bateman J.F., Cheah K.S. 2010. In vivo cellular adaptation to ER stress: survival strategies with double-edged consequences. J. Cell Sci. 123:2145–2154 10.1242/jcs.068833 [DOI] [PubMed] [Google Scholar]
- Vigorito E., Perks K.L., Abreu-Goodger C., Bunting S., Xiang Z., Kohlhaas S., Das P.P., Miska E.A., Rodriguez A., Bradley A., et al. 2007. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 27:847–859 10.1016/j.immuni.2007.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.Z., Kuroda M., Sok J., Batchvarova N., Kimmel R., Chung P., Zinszner H., Ron D. 1998. Identification of novel stress-induced genes downstream of chop. EMBO J. 17:3619–3630 10.1093/emboj/17.13.3619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B., Ha I., Ruvkun G. 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 75:855–862 10.1016/0092-8674(93)90530-4 [DOI] [PubMed] [Google Scholar]
- Zhou X.H., Brandau O., Feng K., Oohashi T., Ninomiya Y., Rauch U., Fässler R. 2003. The murine Ten-m/Odz genes show distinct but overlapping expression patterns during development and in adult brain. Gene Expr. Patterns. 3:397–405 10.1016/S1567-133X(03)00087-5 [DOI] [PubMed] [Google Scholar]
- Zinszner H., Kuroda M., Wang X., Batchvarova N., Lightfoot R.T., Remotti H., Stevens J.L., Ron D. 1998. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 12:982–995 10.1101/gad.12.7.982 [DOI] [PMC free article] [PubMed] [Google Scholar]