The use of new candidate markers for yeast quiescence reveals that quiescence entry and exit primarily rely on cellular metabolic status and can be uncoupled from the cell cycle.
Abstract
Quiescence is defined as a temporary arrest of proliferation, yet it likely encompasses various cellular situations. Our knowledge about this widespread cellular state remains limited. In particular, little is known about the molecular determinants that orchestrate quiescence establishment and exit. Here we show that upon carbon source exhaustion, budding yeast can enter quiescence from all cell cycle phases. Moreover, using cellular structures that are candidate markers for quiescence, we found that the first steps of quiescence exit can be triggered independently of cell growth and proliferation by the sole addition of glucose in both Saccharomyces cerevisiae and Schizosaccharomyces pombe. Importantly, glucose needs to be internalized and catabolized all the way down to glycolysis to mobilize quiescent cell specific structures, but, strikingly, ATP replenishment is apparently not the key signal. Altogether, these findings strongly suggest that quiescence entry and exit primarily rely on cellular metabolic status and can be uncoupled from the cell cycle.
Introduction
Most cells spend the majority of their life in quiescence, a state defined as a temporary absence of proliferation. Establishing quiescence and maintaining the capacity to reenter the proliferation cycle are critical for cell survival, and must be tightly orchestrated to avoid pathological proliferation. Remarkably little is known about the molecular bases of the transitions between proliferation and quiescence. For most organisms, it is acknowledged that quiescence entry takes place in the G1 phase, before or at the restriction point, a point after which cells are committed to the cell cycle (Hartwell et al., 1974; Pardee, 1974). The importance of such a point where quiescent cells synchronously arrest is still under debate (Cooper, 2003).
An essential issue is to elucidate whether quiescence results from a passive adaptation to adverse conditions or if it is actively driven by a “program” involving specific genes conserved among eukaryotes. The characterization of quiescent cell transcription profiles in a variety of organisms has demonstrated that specific genes are expressed upon entry into quiescence (Martinez et al., 2004; Wu et al., 2004; Radonjic et al., 2005; Coller et al., 2006; Shimanuki et al., 2007; Sambasivan et al., 2008), these genes being different from those expressed in G1. Importantly, when cells have entered quiescence by different routes, the transcriptional response is partly independent of the input signal (Wu et al., 2004; Coller et al., 2006; Sambasivan et al., 2008). Together, these findings suggest the existence of a cellular “program” that commits cells to quiescence. However, a unified view of the molecular determinants of the quiescent state in eukaryotes is clearly missing.
In unicellular eukaryotes like Saccharomyces cerevisiae, quiescence entry is triggered by nutrient limitation, and is controlled, at least in part, by a complex interplay between various nutrient-sensitive protein kinases such as PKA and Tor (Smets et al., 2010). Quiescence is associated with a variety of features, including a decrease of protein synthesis rate, acquisition of thermo-resistance, and accumulation of storage molecules (Gray et al., 2004). We have shown that yeast starved for carbon reorganize their actin cytoskeleton into F-actin reservoirs called actin bodies (Sagot et al., 2006). Additionally, we have found that upon carbon exhaustion, yeast relocalize their proteasome from the nucleus into a cytoplasmic structure, named the proteasome storage granule (PSG; Laporte et al., 2008). Recently, using a systematic approach, Narayanaswamy et al. (2009) have shown that many proteins that are localized diffusely in the cytoplasm of dividing yeast relocalize into cytoplasmic foci upon quiescence entry, most of these proteins being metabolic enzymes. Therefore, a broad range of cellular machineries seems to be drastically reorganized upon quiescence entry.
Here, using a combination of criteria, we show that upon carbon source exhaustion, quiescence entry can occur not only in G1, but also in all other cell cycle phases. This suggests that if a “quiescence program” does exist, it has to be independent of the cell cycle. Then, we show that quiescence exit can be triggered independently of cell growth and proliferation by the sole addition of glucose. Finally, we demonstrate that the glucose-induced signal triggering the disassembly of quiescent cells specific structures depends on glucose catabolism through glycolysis. Together, these findings strongly suggest that in yeast, the metabolic status is the critical signal that regulates quiescence establishment and exit.
Results and discussion
Stationary phase cultures are composed of quiescent cells arrested in all cell cycle stages
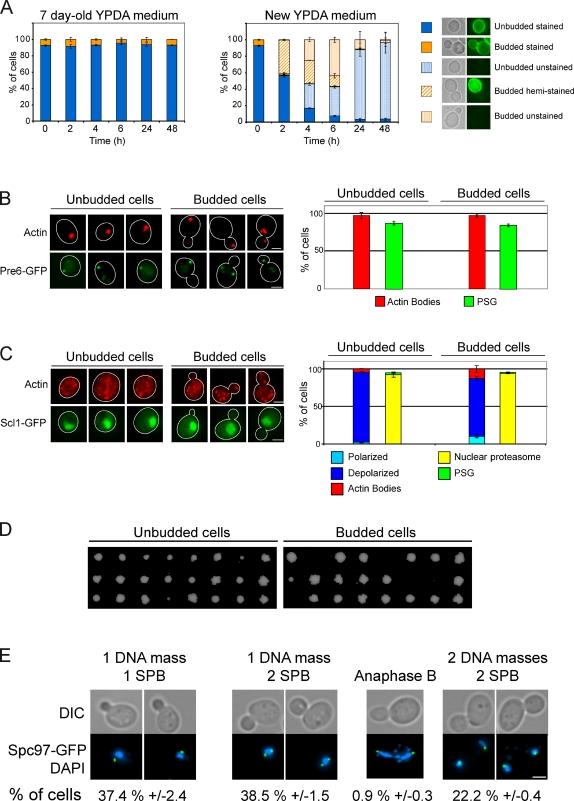
Stationary phase haploid yeast cultures have been shown to be heterogeneous based on physical properties (Allen et al., 2006; Aragon et al., 2008). To describe these cultures at the individual cell level, we characterized wild type (WT) yeast cells grown for 7 d in liquid glucose containing rich medium (YPDA), a condition for which carbon is the limiting nutrient (Gray et al., 2004; Smets et al., 2010). After 7 d, the population was composed of both unbudded (90 ± 5%) and budded (10% ± 5) cells. The presence of budded cells in a stationary phase population was previously described for different genetic backgrounds, different growth conditions, and various strain ploidy (Johnston et al., 1977; Sudbery et al., 1980; Allen et al., 2006; Sagot et al., 2006; Sahin et al., 2008). This observation suggested the possible existence of quiescent budded cells, challenging the commonly accepted idea that S. cerevisiae cells enter quiescence as unbudded cells in G1.
Given that quiescence is defined as a reversible nonproliferative state, we investigated whether these properties applied to the budded cell subpopulation. First, methylene blue staining revealed that after 7 d, 92 ± 2% of the budded cells and 95 ± 4% of unbudded cells were alive (n > 300 cells, three experiments), which indicates that budded cells have the same propensity to survive when starved for carbon as unbudded cells. Moreover, when cells were pulse-labeled with concanavalin-A–FITC (ConA-FITC; a conjugated lectin that stains the cell wall) and re-inoculated in their 7-d-old medium, no unstained or hemi-stained cells were observed even after an additional 48 h (Fig. 1 A). This experiment demonstrates that after 7 d, cells no longer proliferate. This was further confirmed by the fact that neither unbudded nor budded cells displayed features required for proliferation, namely a polarized actin cytoskeleton or nuclear proteasome. Instead, almost all cells had reorganized these machineries into actin bodies and PSG (Fig. 1 B).
Figure 1.
A yeast stationary phase population contains quiescent cells arrested in all cell cycle stages. (A) No cell growth or division could be observed after 7 d in YPDA. A 7-d-old culture of WT prototroph strain was stained with ConA-FITC, then washed and transferred back into either old or new YPDA medium. Stained, unstained, or hemi-stained cells were counted. (B and C) Actin cytoskeleton organization revealed by phalloidin staining and proteasome localization in WT cells grown for 7 d in YPDA (B) and 30 min after refeeding with YPDA (C). The proteasome localization was followed using Pre6p-GFP (B) or Scl1p-GFP (C). Cell outlines are drawn in white. (D) Colony formation after microseparation of unbudded and budded cells grown for 7 d in YPDA. (E) Budded quiescent cells can be arrested in all cell cycle stages. WT cells expressing Spc97p, a spindle pole body component, fused to GFP were grown for 7 d in YPDA and stained with DAPI (numbers are percentages of budded cells). For each time point, n > 200 cells, two experiments. Errors bars indicate SEM. Bars, 2 µm.
Second, we asked whether these subpopulations were able to reenter the proliferation cycle. Cells were grown for 7 d then re-fed with YPDA. As shown in Fig. 1 C, in both cell types, actin bodies disassembled and the cytoskeleton reorganized into depolarized actin cables and patches. In parallel, proteasome subunits relocalized from PSGs into the nucleus (Fig. 1, B and C). Of note, budded cells analyzed 30 min after refeeding were not newly budding cells because upon quiescence exit, new bud emergence required at least 60 min (see Fig. 3 A; Sagot et al., 2006; Sahin et al., 2008). Together, these observations revealed that the stationary phase budded cells subpopulation was resuming the early steps of cell proliferation. In fact, 2 h after refeeding, 30% of these budded cells had repolarized their actin cytoskeleton (Fig. S1, A and B).
Figure 3.
Quiescence exit can be triggered independently of reentry into the proliferation cycle. (A) WT cells were grown for 7 d in YPDA then transferred into the indicated medium. The budding index and the actin cytoskeleton organization are shown. (B) WT cells expressing Pre4p, a proteasome subunit, fused to GFP, were grown for 7 d in YPDA then transferred into the indicated medium. For each time point, the budding index and proteasome localization were monitored. (C) S. pombe expressing Pad1-GFP, a proteasome subunit, fused to GFP, were grown for 4 d and then transferred into the indicated medium. For each time point, proteasome localization was monitored. Cell outlines are drawn in white. n > 200 cells, two experiments. Errors bars indicate SEM. Bars, 2 µm.
Finally, to conclusively establish that the budded cells subpopulation found in stationary phase cultures upon carbon source exhaustion was quiescent, we tested their capacity to exit quiescence and give rise to progeny. Cells from a 7-d-old culture were isolated by micromanipulation, and their ability to form a colony was tested. This experiment showed that 95.1 ± 1.7% of unbudded cells (n = 135) and 65.7 ± 9.2% of budded cells (n = 162) could give rise to a colony (Fig. 1 D), which demonstrates that stationary phase budded cells, although apparently more fragile than the unbudded ones, are indeed bona fide quiescent cells. Importantly, DAPI staining and spindle pole body localization revealed that the quiescent budded cell subpopulation is composed of cells arrested in all cell cycle stages (Fig. 1 E). All together, these observations establish that upon carbon source exhaustion, quiescence entry can occur in all cell cycle phases, which strongly suggests that if a quiescent program exists in S. cerevisiae, it is independent of the cell cycle.
Quiescence establishment can occur independently of cell cycle signals
In a precursor study, Wei et al. (1993) had shown that upon carbon or nitrogen starvation, yeast cdc mutants were able to acquire heat shock resistance in whatever cell cycle stage they were arrested in, and that these arrested cells were capable of giving rise to a progeny. Although thermo-tolerance is one feature of yeast quiescence, resistance to heat can be induced in proliferating cells by several other routes (Parsell and Lindquist, 1993; Lindquist and Kim, 1996; Piper, 1998; Lu et al., 2009), and this property by itself cannot be used as a method to identify quiescent cells at the individual level. In contrast, actin bodies and PSGs are structures that are specifically assembled upon entry into quiescence after carbon source exhaustion (Fig. S1 C; Sagot et al., 2006; Laporte et al., 2008). In fact, although actin bodies do not form upon nitrogen starvation, probably because of the activation of the autophagy process (Fig. S1 D), both actin bodies and PSGs do form when proliferating cells are transferred to water, a condition where all nutrients are missing (Fig. S1, E and F), which strongly suggests that carbon limitation is a preeminent signal in yeast.
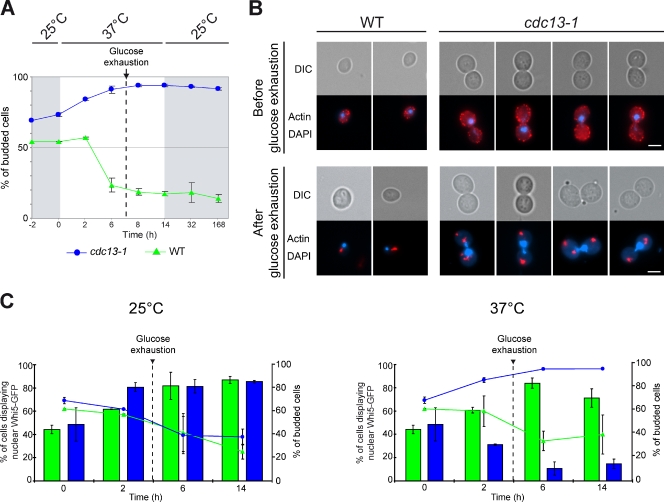
To definitively prove that cells arrested in cell cycle stages other than G1 can enter quiescence, we revisited the experiment of Wei et al. (1993) using actin body formation as a read-out. The cdc13-1 temperature-sensitive mutant is affected for telomere function and, when shifted to 37°C, arrests in G2/M (Hartwell et al., 1974; Burke and Church, 1991). WT and cdc13-1 rho0 cells were grown in YPDA at 25°C to OD600nm 2 and then shifted to 37°C for 14 h. As expected, after the shift, cdc13-1 cells arrested homogeneously as budded cells with a single nucleus (Fig. 2, A and B). Although cell cycle progression was blocked, cdc13-1 cells continued to increase in size and gave rise to large budded cells (Fig. 2 B; Johnston et al., 1977). Before glucose exhaustion, 95.3 ± 0.6% of cdc13-1 rho0 cells displayed a depolarized actin cytoskeleton (Fig. 2 B, top). However, after glucose exhaustion, the cdc13-1 rho0 population was composed of 95.2 ± 1.1% of budded cells, among which 99.3 ± 0.2% displayed actin bodies (Fig. 2 B, bottom). Importantly, these G2/M-arrested cdc13-1 rho0 cells displaying actin bodies were capable of giving rise to progeny (their ability to form colonies was 62.2 ± 2.7% of that of the WT cells). Thus the majority of the G2/M-arrested cdc13-1 cells could reenter the proliferation cycle and therefore meet the criteria of quiescence. To further prove that these cells were in G2/M and not in a pseudo-G1 state, we analyzed the localization of Whi5p, an inhibitor of the G1/S transition that mirrors Cdk activity. Indeed, Whi5p localizes in the nucleus in G1, exits the nucleus upon Cdk activation at the G1/S transition, and reenters the nucleus only in late mitosis (Costanzo et al., 2004). Consistently, in WT unbudded quiescent cells, Whi5p localizes in the nucleus (Fig. 2 C; Costanzo et al., 2004). In contrast, when glucose was exhausted 14 h after the shift to 37°C, the majority of the G2/M-arrested cdc13-1 rho0 cells did not display any detectable Whi5p-GFP in the nucleus (96.3 ± 0.2% of the cdc13-1 were large budded cells, among which 96.2 ± 1.2% did not show detectable nuclear Whi5p-GFP). Accordingly, Whi5p was not required for quiescence establishment, maintenance, or exit (Fig. S2, A to D). Together, these experiments demonstrate that carbon source exhaustion is sufficient to trigger entry into quiescence even if cells are arrested in G2/M.
Figure 2.
Cells can enter quiescence in G2/M upon glucose exhaustion. (A) Diagram representing the experimental protocol: WT and cdc13-1 rho0 cells were grown in YPDA at 25°C. At OD600nm 2, cells were transferred to 37°C for 14 h, then transferred back to 25°C. Budding indexes for WT and cdc13-1 rho0 strains are shown. (B) Actin cytoskeleton organization in WT and cdc13-1 rho0 before and after glucose exhaustion, respectively, 6 and 32 h after the shift to 37°C. Bars, 2 µm. (C) Whi5p-GFP localization in WT (green) and cdc13-1 rho0 cells (blue). WT and cdc13-1 rho0 Whi5p-GFP–expressing cells were grown in YPDA at 25°C. At OD600nm 2, cells were transferred to 37°C or to 25°C for 14 h. The budding index is indicated for each time point (WT, green triangles; cdc13-1 rho0, blue circles). Glucose exhaustion is indicated. n > 200 cells for each time point, two experiments. Errors bars indicate SEM.
Finally, using a cdc24-3 strain, we verified that a sole arrest in G1 is not sufficient to trigger actin body formation (Fig. S2, E and F). This is in good agreement with the fact that in both yeast and mammals, cell cycle arrest in G1 is not sufficient to induce a quiescence-specific gene expression profile (Coller et al., 2006; Aragon et al., 2008). Therefore, although quiescence entry and G1 arrest are generally concomitant, an arrest in G1 is neither necessary nor sufficient for quiescence establishment. Protein synthesis is crucial for reaching the critical cell size required for the passage through “start.” In fact, in S. cerevisiae, limited protein synthesis extends G1 length but has little effect on other cell cycle stages (Unger and Hartwell, 1976; Hartwell and Unger, 1977; Johnston et al., 1977; Slater et al., 1977; Popolo et al., 1982). Thus, the fact that quiescence entry occurs preferentially in G1 could simply be the passive result of a translation rate slowdown.
Glucose can trigger quiescence exit
Quiescence establishment is a slow phenomenon; therefore, the identification of signaling cascades involved in this process is difficult. In contrast, both actin body and PSG mobilization occur within seconds after nutrient addition (Sagot et al., 2006; Laporte et al., 2008; Sahin et al., 2008). In this context, we made the striking observation that the sole addition of glucose was leading to the mobilization of these structures. When cells grown for 7 d were transferred into a 2% glucose solution, both actin bodies and PSGs disassembled just as rapidly as upon transfer to YPDA (Fig. 3, A and B). This is reminiscent of a pioneer work by Granot and Snyder (1991, 1993) showing that pre-incubation of starved yeast cells in glucose accelerates bud emergence upon transfer to rich medium, which indicates that glucose-treated cells are somehow “primed” to reenter proliferation. However, a prolonged incubation in glucose alone resulted in massive cell death, and this loss of viability shows that these cells were no longer quiescent (Granot and Snyder, 1991, 1993).
Interestingly, we found that glucose is also sufficient to trigger quiescent cells’ specific structure mobilization in Schizosaccharomyces pombe. When S. pombe cells were starved for carbon, they arrested in G2 with a 2N DNA content (Costello et al., 1986) and assembled PSGs (Fig. 3 C; Laporte et al., 2008). As shown in Fig. 3 C, in S. pombe, just as in S. cerevisiae, glucose could promote PSGs mobilization. Importantly, in both yeast species, cell growth was only observed when quiescence exit was triggered by the addition of YPDA (Table I), and in S. cerevisiae, no new bud emergence was observed in the presence of glucose alone. This indicated that the sole addition of glucose was sufficient to induce the early steps of quiescence exit but did not suffice to trigger cell growth and proliferation. Consistently, we have previously demonstrated that both actin bodies and PSGs can be mobilized upon nutrient replenishment even in the absence of de novo protein synthesis (Sagot et al., 2006; Laporte et al., 2008), a condition preventing cell growth and synthesis of cell cycle regulators. Collectively, these results strongly suggest that the first steps of quiescence exit can be triggered independently of cell growth and proliferation by the addition of glucose.
Table I.
Glucose addition does not trigger cell growth.
| Species | Before transfer | H2O | YPDA | 2% glucose |
| S. cerevisiaea | 34.2 ± 1.9 µm3 | 30.5 ± 1.9 µm3 | 51.8 ± 2.0 µm3 | 32.9 ± 1.9 µm3 |
| S. pombeb | 8.76 ± 1.33 µm | 8.8 ± 1.2 µm | 13.2 ± 2.3 µm | 9.52 ± 1.49 µm |
Cells were grown for 7 d in YPDA and then transferred into the indicated medium. The cell volume was measured 2 h after transfer (mean volume, n = 50,000 cells ± SEM, two experiments).
Cells were grown for 4 d in YPDA then transferred into the indicated medium. The cell length was measured 3 h after transfer (mean length ± SEM, n > 150, two experiments).
Quiescence exit requires glucose catabolism but appears to be independent of ATP replenishment
We then asked whether glucose-induced mobilization of quiescent cell structures was caused by the activation of nutrient-sensing cascades or whether it was a consequence of glucose metabolism. First, we showed that glucose-induced actin body mobilization could occur in the absence of de novo protein synthesis (Fig. S3 A). Then, we demonstrated that actin bodies could be mobilized upon glucose addition even if the Ras or the Tor pathways were inactivated (Fig. S3, B–E). This is in good agreement with the fact that all known glucose sensors were dispensable for glucose-induced actin body mobilization (Fig. 4 B, Δsensors strain). In contrast, yeast strains impaired for glucose uptake (Fig. 4 B, Δtransporters strain; Wieczorke et al., 1999) or glucose phosphorylation (Fig. 4 B, Δhexokinases strain) were totally unable to disassemble actin bodies. Finally, various mutants that block individual glycolysis steps were tested, and none of them could remobilize actin bodies when fed with glucose (Fig. 4 B). This demonstrates that glucose has to be catabolized through the glycolytic pathway at least into pyruvate to trigger actin body mobilization. This was further supported by the fact that no actin body mobilization was observed with nonmetabolizable glucose analogues or when glucose was added together with iodo-acetate, a drug that inhibits glycolysis (Fig. S3 F).
Figure 4.
Actin body mobilization requires glucose catabolism. (A) Schematic representation of the glycolysis pathway. Genes encoding each activity in S. cerevisiae are indicated in green. G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; F1,6BP, fructose 1,6 bisphosphate; DHAP, dihydroxy acetone phosphate; G3P, glyceraldehyde 3 phosphate; 1,3BPG, 1,3 bis phosphoglycerate; 3-PG, 3 phosphoglycerate; 2-PG, 2 phosphoglycerate; PEP, phosphoenolpyruvate. (B) WT, snf3Δ, rgt2Δ, gpr1Δ (Δsensors) and hxk1Δ, hxk2Δ, and glk1Δ (Δhexokinases) strains were grown in galactose-rich medium. WT and a strain that cannot uptake glucose (Δtransporters strain; Wieczorke et al., 1999) were grown in maltose-rich medium. WT and pgi1Δ mutant were grown in fructose-rich medium. WT, pfk1Δ, pfk2Δ double mutant, and pgk1Δ mutants were grown in lactate-rich medium. After 7 d, cells were transferred into glucose (2%) and the actin cytoskeleton remodeling was monitored by phalloidin staining. (C) UV chromatograms from WT cells grown for 7 d in YPDA (black line), then transferred for 15 min into 2% glucose (red line) or into water (green line). Blue insets show enlarged views of the of AICAR and SAICAR peaks, the black inset shows amperometric detection of G6P and F1,6BP. (D and E) WT and pgi1 strains were grown for 7 d in rich medium containing 2% fructose, then transferred into either a 2% glucose or fructose solution. (D) ATP, G6P, and F1,6BP concentration measured before (7 d) and 15 min after transfer. (E) Actin cytoskeleton organization revealed by phalloidin staining. For each time point, n > 200 cells, at least two experiments. Error bars indicate SEM.
To get an overview of the metabolic consequences of glucose addition onto quiescent cells, we monitored the intracellular concentration of several metabolites. As shown in Fig. 4 C, glucose addition onto WT quiescent cells caused a significant increase in 5′-phosphoribosyl-5-amino-4-imidazole carboxamide (AICAR), SAICAR (succinyl-AICAR), inosine, GTP, CTP, and UTP, and a drop of AMP. Using mutants or drugs, we showed that none of these variations were required for glucose-induced actin body mobilization (Fig. S3, F and H). Further, as expected, glucose addition also caused a drastic ATP concentration increase in WT cells (Fig. 4, C and D). In contrast, in pgi1Δ cells, ATP concentration did not increase 15 min after transfer into either a glucose- or a fructose-containing solution (Fig. 4 D). Yet, upon fructose, but not glucose, addition, this mutant mobilized its actin bodies just as efficiently as the WT strain (Fig. 4 E). This surprising result clearly establishes that an ATP concentration increase is not required for actin body mobilization. In addition, we found that when quiescent cells were transferred into water, the ATP concentration transiently increased, whereas no actin body disassembly was observed (Fig. 3 A and Fig. 4 C). We therefore conclude that ATP replenishment is neither sufficient nor necessary for actin body mobilization. At the present time, we do not know which metabolites resulting from glucose catabolism are responsible for this critical signaling step; yet our study adds to the emerging idea that metabolism is not just for fueling cells, but that small molecules are directly involved in the regulation of a myriad of cellular processes (McKnight, 2010), including proliferation/quiescence transitions.
Materials and methods
Yeast strains and growth conditions
All the S. cerevisiae strains used in this study are isogenic to BY4741, BY4742, or BY4743, available from Euroscarf unless specified. Yeast strains carrying GFP fusions were obtained from Invitrogen. The S. pombe strain expressing Pad1-GFP from the endogenous locus was from J.P Javerzat (University de Bordeaux, Centre National de la Recherche Scientifique, Institut de Biochimie et Génétique Cellulaires, UMR 5095, Bordeaux, France; Wilkinson et al., 1998). Thermo-sensitive cdc13-1 rho0 and cdc24-3 strains were from C. Boone (The Donnelly Centre for Cellular and Biomolecular Research, University of Toronto, Toronto, Ontario, Canada). The strain referred as to Δtransporters is leu2-3,112 ura3-52 trp1-289 his3-Δ1 MAL2-8C hxt514Δ::loxP hxt2Δ::loxP hxt367Δ::loxP hxt8Δ::loxP hxt9Δ::loxP hxt10Δ::loxP hxt11Δ::loxP hxt12Δ::loxP hxt13Δ::loxP hxt14Δ::LoxP hxt15Δ::loxP hxt16Δ::loxP hxt17Δ gal2Δ stl1Δ::loxP agt1Δ::loxP mph2Δ::loxP mph3Δ::loxP snf3Δ::loxP rgt2Δ::loxP, and is from E. Boles (Institute of Molecular Biosciences, Goethe-University Frankfurt am Main, Frankfurt am Main, Germany; Wieczorke et al., 1999). The strain expressing Tpk1 (M164G), Tpk2 (M147G), Tpk3 (M165G), and Sch9 (T492G) was a gift from J. Broach (Princeton University, Princeton, NJ; Zaman et al., 2009). The YPDA medium contained 1% yeast extract, 2% peptone, 2% glucose, and 0.04 g/liter adenine. Unless specified, yeast cells were grown in liquid medium at 30°C in flasks with 220 rpm shaking. For refeeding experiments, cells were centrifuged and washed twice with water, then inoculated into the indicated medium at a final OD600nm of 0.6–1 and incubated at 30°C unless specified otherwise.
Cell staining
For regrowth experiments, a 7-d-old culture was centrifuged, and the remaining supernatant, i.e., the “old” YPDA medium, was filtered to remove nonpelleted cells. ConA-FITC (Sigma-Aldrich) was added to a final concentration of 0.2 g/liter. Cells were then incubated for 1 h at 25°C, then washed twice with old filtered YPDA, and finally transferred into the indicated medium. For actin phalloidin staining, cells were fixed with formaldehyde (3.7% final), washed, and stained overnight with Alexa Fluor 568–phalloidin (Invitrogen). Cells were then washed twice, resuspended in a mounting solution containing 70% glycerol and 5 mg/liter paraphenylenediamine, and imaged at room temperature (Sagot et al., 2002). For live cell imaging, 2 µl of the culture was directly spotted onto a glass slide and immediately imaged at room temperature.
Microscopy
Cells were observed in a fully automated inverted microscope (Zeiss 200M; Carl Zeiss, Inc.) equipped with an MS-2000 stage (Applied Scientific Instrumentation), a Lambda LS 300 W xenon light source (Sutter Instrument), a 100× 1.4 NA Plan-Apochromat objective lens, and a five-position filter cube turret with an FITC filter (excitation, HQ487/25; emission, HQ535/40; Beamsplitter, Q505lp), a Cy3 filter (excitation, HQ535/50; emission, HQ610/75; Beamsplitter, Q565lp), and a DAPI filter (excitation, 360/40; emission, 460/50; Beamsplitter, 400) from Chroma Technology Corp. Images were acquired using a CoolSnap HQ camera (Roper Scientific). The microscope, camera and shutters (Uniblitz) were controlled by SlideBook software 5.10. (Intelligent Imaging Innovations). Images are maximal projection of z-stacks performed using a 0.2 µm step using SlideBook.
Metabolomics
Cellular extracts were prepared by an ethanol extraction method adapted from Loret et al. (2007). In brief, cells were harvested by rapid filtration on nitrocellulose filter, immediately dropped into 5 ml ethanol/10 mM Hepes, pH 7.2 (4/1 vol/vol), and incubated at 80°C for 3 min. Samples were evaporated using a rotavapor apparatus. The residue was resuspended in 500 µl of water and insoluble particles were eliminated by centrifugation. Separation of metabolites was performed on a CarboPac PA1 column, linked to an ICS3000 chromatography workstation (Dionex), by using a NaOH/Na acetate gradient. Sugar phosphate and nucleotide derivatives were, respectively, detected by amperometry (IntAmp, carbohydrates standard quad potential ED50; Dionex) and UV diode array (Ultimate 3000; Dionex) detectors.
Miscellaneous
Yeast strains were micro-manipulated using a Singer MSM system (Singer Instrument Co. Ltd.). Glucose concentration was measured using the d-glucose/d-fructose UV test kit (Roche). Cell volume was measured using a Multi-sizer 4 (Beckman Coulter).
Online supplemental material
Fig. S1 shows actin cytoskeleton and proteasome reorganization upon various nutrient exhaustions and upon cell refeeding. Fig. S2 shows that quiescence establishment does not require Whi5 and is not triggered by a sole arrest in G1. Fig. S3 shows that quiescence exit requires glucose catabolism but appears to be independent of Ras or Tor pathways and of nucleotide replenishment. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201009028/DC1.
Acknowledgments
We are grateful to J.P. Javerzat, D. McCusker, and D. Pellman for helpful discussions, and A. Feytout for FACS experiments. We are thankful to E. Boles, C. Boone, J. Broach, and J.P. Javerzat, for providing reagents.
This work was supported by Université Bordeaux 2, Conseil Regional d’Aquitaine, a Young Investigator grant from the Agence Nationale pour la Recherche (JC08 310804), a grant form Association pour la Recherche sur le Cancer (ARC; SFI20101201558), and a PEPS grant from the Centre National de la Recherche Scientifique. D. Laporte was supported by an ARC fellowship. A. Lebaudy was supported by a fellowship from La Fondation pour la Recherche Medicale.
Footnotes
Abbreviations used in this paper:
- AICAR
- 5′-phosphoribosyl-5-amino-4-imidazole carboxamide
- ConA-FITC
- concanavalin-A–FITC
- PSG
- proteasome storage granule
- SAICAR
- succinyl-AICAR
- WT
- wild type
References
- Allen C., Büttner S., Aragon A.D., Thomas J.A., Meirelles O., Jaetao J.E., Benn D., Ruby S.W., Veenhuis M., Madeo F., Werner-Washburne M. 2006. Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures. J. Cell Biol. 174:89–100 10.1083/jcb.200604072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragon A.D., Rodriguez A.L., Meirelles O., Roy S., Davidson G.S., Tapia P.H., Allen C., Joe R., Benn D., Werner-Washburne M. 2008. Characterization of differentiated quiescent and nonquiescent cells in yeast stationary-phase cultures. Mol. Biol. Cell. 19:1271–1280 10.1091/mbc.E07-07-0666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D.J., Church D. 1991. Protein synthesis requirements for nuclear division, cytokinesis, and cell separation in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:3691–3698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller H.A., Sang L., Roberts J.M. 2006. A new description of cellular quiescence. PLoS Biol. 4:e83 10.1371/journal.pbio.0040083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper S. 2003. Reappraisal of serum starvation, the restriction point, G0, and G1 phase arrest points. FASEB J. 17:333–340 10.1096/fj.02-0352rev [DOI] [PubMed] [Google Scholar]
- Costanzo M., Nishikawa J.L., Tang X., Millman J.S., Schub O., Breitkreuz K., Dewar D., Rupes I., Andrews B., Tyers M. 2004. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell. 117:899–913 10.1016/j.cell.2004.05.024 [DOI] [PubMed] [Google Scholar]
- Costello G., Rodgers L., Beach D. 1986. Fission yeast enters the stationary phase G0 state from either mitotic G1 or G2. Curr. Genet. 11:119–125 10.1007/BF00378203 [DOI] [Google Scholar]
- Granot D., Snyder M. 1991. Glucose induces cAMP-independent growth-related changes in stationary-phase cells of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 88:5724–5728 10.1073/pnas.88.13.5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granot D., Snyder M. 1993. Carbon source induces growth of stationary phase yeast cells, independent of carbon source metabolism. Yeast. 9:465–479 10.1002/yea.320090503 [DOI] [PubMed] [Google Scholar]
- Gray J.V., Petsko G.A., Johnston G.C., Ringe D., Singer R.A., Werner-Washburne M. 2004. “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 68:187–206 10.1128/MMBR.68.2.187-206.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L.H., Unger M.W. 1977. Unequal division in Saccharomyces cerevisiae and its implications for the control of cell division. J. Cell Biol. 75:422–435 10.1083/jcb.75.2.422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L.H., Culotti J., Pringle J.R., Reid B.J. 1974. Genetic control of the cell division cycle in yeast. Science. 183:46–51 10.1126/science.183.4120.46 [DOI] [PubMed] [Google Scholar]
- Johnston G.C., Pringle J.R., Hartwell L.H. 1977. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp. Cell Res. 105:79–98 10.1016/0014-4827(77)90154-9 [DOI] [PubMed] [Google Scholar]
- Laporte D., Salin B., Daignan-Fornier B., Sagot I. 2008. Reversible cytoplasmic localization of the proteasome in quiescent yeast cells. J. Cell Biol. 181:737–745 10.1083/jcb.200711154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S., Kim G. 1996. Heat-shock protein 104 expression is sufficient for thermotolerance in yeast. Proc. Natl. Acad. Sci. USA. 93:5301–5306 10.1073/pnas.93.11.5301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loret M.O., Pedersen L., François J. 2007. Revised procedures for yeast metabolites extraction: application to a glucose pulse to carbon-limited yeast cultures, which reveals a transient activation of the purine salvage pathway. Yeast. 24:47–60 10.1002/yea.1435 [DOI] [PubMed] [Google Scholar]
- Lu C., Brauer M.J., Botstein D. 2009. Slow growth induces heat-shock resistance in normal and respiratory-deficient yeast. Mol. Biol. Cell. 20:891–903 10.1091/mbc.E08-08-0852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez M.J., Roy S., Archuletta A.B., Wentzell P.D., Anna-Arriola S.S., Rodriguez A.L., Aragon A.D., Quiñones G.A., Allen C., Werner-Washburne M. 2004. Genomic analysis of stationary-phase and exit in Saccharomyces cerevisiae: gene expression and identification of novel essential genes. Mol. Biol. Cell. 15:5295–5305 10.1091/mbc.E03-11-0856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight S.L. 2010. On getting there from here. Science. 330:1338–1339 10.1126/science.1199908 [DOI] [PubMed] [Google Scholar]
- Narayanaswamy R., Levy M., Tsechansky M., Stovall G.M., O’Connell J.D., Mirrielees J., Ellington A.D., Marcotte E.M. 2009. Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc. Natl. Acad. Sci. USA. 106:10147–10152 10.1073/pnas.0812771106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee A.B. 1974. A restriction point for control of normal animal cell proliferation. Proc. Natl. Acad. Sci. USA. 71:1286–1290 10.1073/pnas.71.4.1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsell D.A., Lindquist S. 1993. The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu. Rev. Genet. 27:437–496 10.1146/annurev.ge.27.120193.002253 [DOI] [PubMed] [Google Scholar]
- Piper P. 1998. Differential role of Hsps and trehalose in stress tolerance. Trends Microbiol. 6:43–44 10.1016/S0966-842X(97)01190-6 [DOI] [PubMed] [Google Scholar]
- Popolo L., Vanoni M., Alberghina L. 1982. Control of the yeast cell cycle by protein synthesis. Exp. Cell Res. 142:69–78 10.1016/0014-4827(82)90410-4 [DOI] [PubMed] [Google Scholar]
- Radonjic M., Andrau J.C., Lijnzaad P., Kemmeren P., Kockelkorn T.T., van Leenen D., van Berkum N.L., Holstege F.C. 2005. Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Mol. Cell. 18:171–183 10.1016/j.molcel.2005.03.010 [DOI] [PubMed] [Google Scholar]
- Sagot I., Klee S.K., Pellman D. 2002. Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat. Cell Biol. 4:42–50 [DOI] [PubMed] [Google Scholar]
- Sagot I., Pinson B., Salin B., Daignan-Fornier B. 2006. Actin bodies in yeast quiescent cells: an immediately available actin reserve? Mol. Biol. Cell. 17:4645–4655 10.1091/mbc.E06-04-0282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin A., Daignan-Fornier B., Sagot I. 2008. Polarized growth in the absence of F-actin in Saccharomyces cerevisiae exiting quiescence. PLoS ONE. 3:e2556 10.1371/journal.pone.0002556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambasivan R., Pavlath G.K., Dhawan J. 2008. A gene-trap strategy identifies quiescence-induced genes in synchronized myoblasts. J. Biosci. 33:27–44 10.1007/s12038-008-0019-6 [DOI] [PubMed] [Google Scholar]
- Shimanuki M., Chung S.Y., Chikashige Y., Kawasaki Y., Uehara L., Tsutsumi C., Hatanaka M., Hiraoka Y., Nagao K., Yanagida M. 2007. Two-step, extensive alterations in the transcriptome from G0 arrest to cell division in Schizosaccharomyces pombe. Genes Cells. 12:677–692 10.1111/j.1365-2443.2007.01079.x [DOI] [PubMed] [Google Scholar]
- Slater M.L., Sharrow S.O., Gart J.J. 1977. Cell cycle of Saccharomyces cerevisiae in populations growing at different rates. Proc. Natl. Acad. Sci. USA. 74:3850–3854 10.1073/pnas.74.9.3850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smets B., Ghillebert R., De Snijder P., Binda M., Swinnen E., De Virgilio C., Winderickx J. 2010. Life in the midst of scarcity: adaptations to nutrient availability in Saccharomyces cerevisiae. Curr. Genet. 56:1–32 10.1007/s00294-009-0287-1 [DOI] [PubMed] [Google Scholar]
- Sudbery P.E., Goodey A.R., Carter B.L. 1980. Genes which control cell proliferation in the yeast Saccharomyces cerevisiae. Nature. 288:401–404 10.1038/288401a0 [DOI] [PubMed] [Google Scholar]
- Unger M.W., Hartwell L.H. 1976. Control of cell division in Saccharomyces cerevisiae by methionyl-tRNA. Proc. Natl. Acad. Sci. USA. 73:1664–1668 10.1073/pnas.73.5.1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Nurse P., Broek D. 1993. Yeast cells can enter a quiescent state through G1, S, G2, or M phase of the cell cycle. Cancer Res. 53:1867–1870 [PubMed] [Google Scholar]
- Wieczorke R., Krampe S., Weierstall T., Freidel K., Hollenberg C.P., Boles E. 1999. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 464:123–128 10.1016/S0014-5793(99)01698-1 [DOI] [PubMed] [Google Scholar]
- Wilkinson C.R., Wallace M., Morphew M., Perry P., Allshire R., Javerzat J.P., McIntosh J.R., Gordon C. 1998. Localization of the 26S proteasome during mitosis and meiosis in fission yeast. EMBO J. 17:6465–6476 10.1093/emboj/17.22.6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Zhang N., Hayes A., Panoutsopoulou K., Oliver S.G. 2004. Global analysis of nutrient control of gene expression in Saccharomyces cerevisiae during growth and starvation. Proc. Natl. Acad. Sci. USA. 101:3148–3153 10.1073/pnas.0308321100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman S., Lippman S.I., Schneper L., Slonim N., Broach J.R. 2009. Glucose regulates transcription in yeast through a network of signaling pathways. Mol. Syst. Biol. 5:245. [DOI] [PMC free article] [PubMed] [Google Scholar]