Abstract
The understanding of integral membrane protein (IMP) structure and function is hampered by the difficulty of handling these proteins. Aqueous solubilization, necessary for many types of biophysical analysis, generally requires a detergent to shield the large lipophilic surfaces displayed by native IMPs. Many proteins remain difficult to study owing to a lack of suitable detergents. We introduce a class of amphiphiles, each of which is built around a central quaternary carbon atom derived from neopentyl glycol, with hydrophilic groups derived from maltose. Representatives of this maltose-neopentyl glycol (MNG) amphiphile family display favorable behavior relative to conventional detergents, as tested on multiple membrane protein systems, leading to enhanced structural stability and successful crystallization. MNG amphiphiles are promising tools for membrane protein science because of the ease with which they may be prepared and the facility with which their structures may be varied.
Integral membrane proteins (IMPs) play crucial roles in many aspects of biology by mediating the transfer of material and signals between cells and their environment. It is estimated that 20-30% of all open reading frames in the human genome encode membrane proteins, and that more than 50% of current pharmaceutical agents target IMPs1. Our understanding of IMP structure and function, however, is hampered by difficulties associated with handling these proteins2. Most IMPs are not soluble in aqueous buffer because they display large hydrophobic surfaces when properly folded; therefore, detergents are required to extract IMPs from the lipid bilayer and to maintain the native state in solution. Mild detergents are widely used for IMP manipulation, but many membrane proteins solubilized with these agents tend to denature and/or aggregate3, making it difficult to conduct functional studies, spectroscopic analysis or crystallization trials.
Prior efforts to develop amphiphiles tailored for IMP applications have involved diverse strategies and achieved varying levels of success. Several peptide-based designs have been explored (peptitergents4, lipopeptide detergents5, short peptide surfactants6), but so far have not gained broad acceptance. Amphiphilic polymers (“amphipols”7,8) and discoidal lipid bilayers stabilized by an amphiphilic protein scaffold (“nanodiscs”9,10) have proven to be versatile tools for studying IMPs in native-like states in aqueous solution. It is not clear, however, whether either of these approaches can yield high-quality crystals for diffraction analysis, a prominent objective of IMP studies. Furthermore, neither amphipols nor nanodiscs are designed to extract IMPs from biological membranes. Recently reported agents of low molecular weight, such as hemifluorinated surfactants (HFS)8,11 and cholic acid-based amphiphiles12, have displayed promising properties, but the scope of their utility remains to be explored. Thus there has been a need for amphiphiles that can extract, stabilize, and promote crystallization of IMPs more effectively than do current detergents. Amphiphiles with this combination of capabilities would have to be easily prepared on a large scale, which would be extremely challenging for peptide- or protein-based agents.
Here we report a class of amphiphiles that display favorable behavior with a diverse set of membrane proteins. The design of these amphiphiles features a central quaternary carbon, which is intended to place subtle restraints on conformational flexibility13-15. Since the quaternary carbon was derived from neopentyl glycol and since the hydrophilic groups in the examples discussed here are derived from maltose, we designate these compounds Maltose-Neopentyl Glycol (MNG) amphiphiles. The quaternary carbon distinguishes MNG architecture from conventional detergent structures and enables the incorporation of two hydrophilic and two lipophilic subunits. We hypothesized that the modulation of flexibility and distinctive orientations of hydrophilic and lipophilic surfaces would cause MNG amphiphiles to display properties distinct from those of analogous conventional detergents. These amphiphiles are also readily synthesized. We have evaluated their performance with multiple membrane proteins in diverse applications, including maintenance of native IMP folding, association and function, extraction from a native membrane, growth of high-quality crystals, and support of cell-free translation.
Results
MNG amphiphile architecture
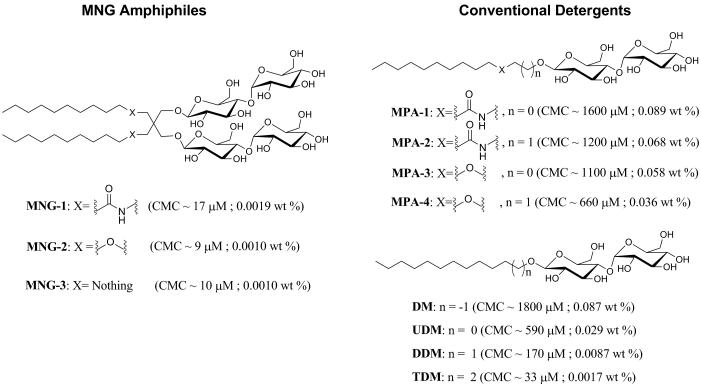
We performed extensive preliminary studies that identified MNG-1, MNG-2 and MNG-3 (Fig. 1) as displaying particularly promising behavior. Each of these amphiphiles features two maltose units in the hydrophilic portion and two n-decyl chains in the lipophilic portion. The lipophilic unit attachment varies, with amide linkages in MNG-1, ether linkages in MNG-2 and direct connection to the quaternary center in MNG-3. Synthesis was straightforward and efficient (Supplementary Note). We prepared analogues with conventional detergent architecture, MPA-1 to MPA-4 (for monopodal amphiphile), for comparison with MNG-1 and MNG-2 (Fig. 1). The comparison compounds for MNG-3 are commercially available: n-undecyl-β-D-maltoside (UDM) and n-dodecyl-β-D-maltoside (DDM); we examined lower homologue decyl-β-D-maltoside (DM) and higher homologue tridecyl-β-D-maltoside (TDM) as well. DDM is currently one of the most widely used detergents in membrane protein research.
Figure 1.
Chemical structures of MNG amphiphiles (MNG-1, MNG-2, and MNG-3) and their linear counterparts (MPA-1, MPA-2, MPA-3, MPA-4, DM, UDM, DDM, and TDM). The critical micelle concentration (CMC) value for each agent, measured via hydrophobic dye solubilization, is indicated in parentheses.
MNG amphiphiles stabilize diverse membrane proteins
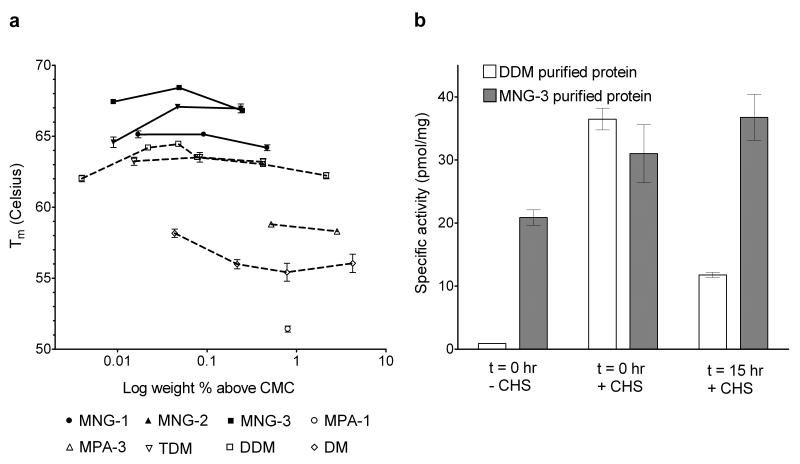
We first examined the thermal stability of a human β2 adrenergic receptor-T4-lysozyme fusion protein (β2AR-T4L)16 solubilized with an MNG amphiphile or conventional detergent. Stability was assessed via optical absorption measurements of β2AR-T4L bound to the inverse agonist carazolol (fluorescence emission maximum at 341 nm in the receptor-bound state, and at 356 nm after carazolol release from the receptor). The receptor was initially solubilized and purified with DDM, which was then exchanged for the amphiphile or detergent to be evaluated. The 341:356 nm peak intensity ratio was used to monitor the relative amounts of intact and denatured β2AR-T4L (Supplementary Fig. 1). We evaluated the effect of amphiphile concentration on the melting temperature (Tm) of β2AR-T4L (Supplementary Fig. 2 and Supplementary Table 1). All three MNG amphiphiles were superior to DDM and other conventional detergents (including DM and TDM) in terms of β2AR-T4L thermal stability (Fig. 2a). The concentration ranges that confer optimal stabilization in each case were similar in terms of wt % (between 0.05 and 0.1 wt %), but differ somewhat on a CMC-based scale (Supplementary Fig. 2b). The most successful amphiphile was MNG-3. MNG-3 was superior to DDM also in terms of maintaining a solubilized form of the muscarinic M3 acetylcholine receptor (M3AchR) in an active state (Fig. 2b). Together, the observations with β2AR-T4L and M3AchR raise the possibility that MNG amphiphiles should be generally useful for GPCR stabilization.
Figure 2.
GPCR stability in MNG amphiphiles or conventional detergents. (a) Tm values of the β2AR-T4L plotted in terms of wt % of the MNG amphiphiles (MNG-1, MNG-2, and MNG-3) or conventional detergents (MPA-1, MPA-3, DM, DDM, and TDM). β2AR-T4L with bound carazolol (an inverse agonist) was incubated with various agents at the various concentrations at indicated temperatures for 5 min prior to fluorescence emission measurements. Normalized results are expressed as mean ± s.e.m. (n = 3, 4, or 5). (b) Specific activities (pmol/mg) of M3AchR in DDM and MNG-3. The activity of the protein was evaluated after being washed and eluted with buffer including DDM or MNG-3, but without CHS, via a binding assay involving the antagonist [3H] N-methyl scopolamine, in the absence (t = 0 hr, − CHS; the first bar) or presence of CHS (t = 0 hr, + CHS; the second bar). The DDM- and MNG-3-purified M3AchR samples were stored at 4 °C for 15 hours, and then activities were measured again in the presence of CHS (t = 15 hr, + CHS; the third bar). Results are expressed as mean ± s.d. (n = 3).
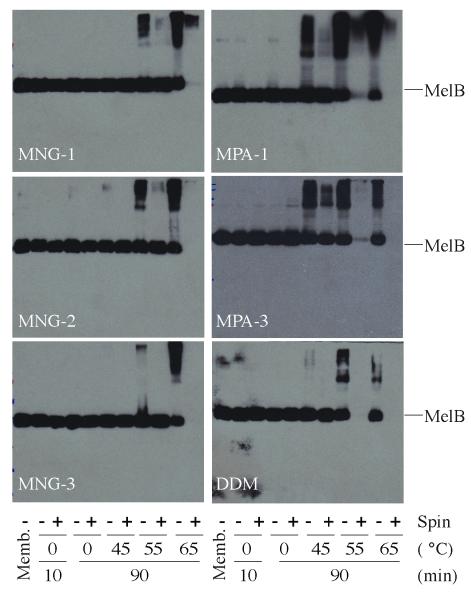
We turned next to melibiose permease (MelB), which catalyzes the accumulation of α-D-galactopyranosides by a cation-solute symport mechanism17. Treatment of membrane preparations containing overexpressed MelB (from E. coli DW2 cells) with solutions containing 1.5 wt % amphiphile or detergent at 0 °C for 10 min quantitatively extracted MelB (Fig. 3). We incubated the solubilized samples on ice or at elevated temperatures for 90 min prior to ultracentrifugation, in order to assess protein thermostability. For the conventional detergents (MPA-1, MPA-3, and DDM), we observed MelB aggregation when the protein was incubated at 45 °C for 90 min, and the protein disappeared from solution when treated at 55 °C or 65 °C followed by ultracentrifugation (Fig. 3). In contrast, all three MNG amphiphiles provided large amounts of soluble protein even after treatment at 55 °C. In particular, MNG-3 was unique in preventing aggregation at 55 °C.
Figure 3.
SDS-12% PAGE analysis and Western blot detection of MelB. MelB samples were subjected to SDS-PAGE analysis, and MelB was detected by Western blotting using anti-His tag antibody. 10 μg membrane proteins were applied for the untreated membrane (memb.) or detergent extracts prior to ultracentrifugation (−), and equal volume of solutions were loaded for samples that the ultracentrifugation were applied (+).
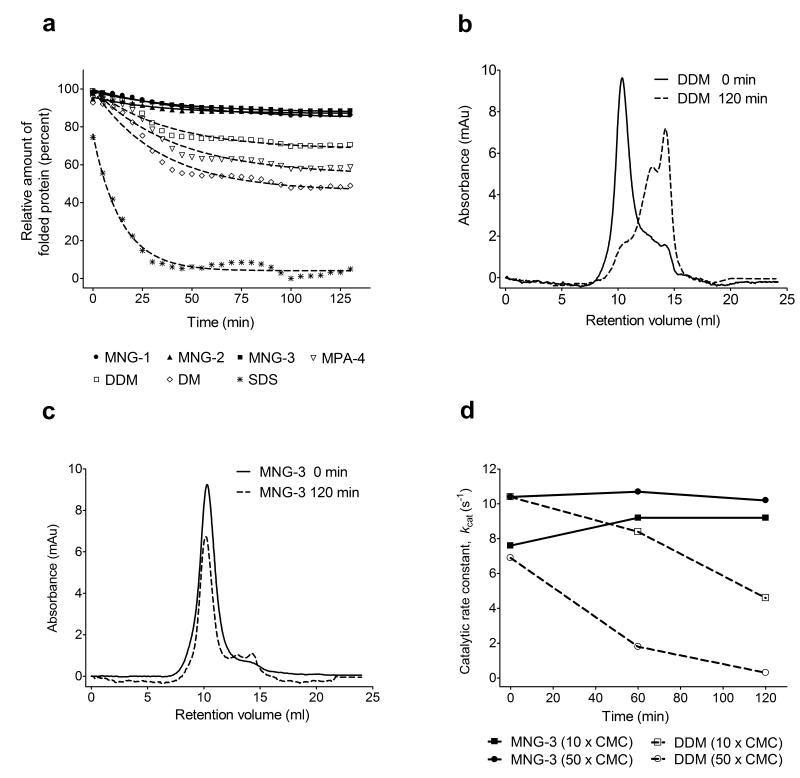
We assessed the thermostabilities of additional membrane protein systems via a fluorescence assay. N-[4-(7-diethylamino-4-methyl-3-coumarinyl)phenyl]maleimide (CPM)18 can be selectively and covalently attached to side chain thiol groups of solvent-accessible cysteine residues. The maleimide unit of CPM quenches coumarin fluorescence; however, the coumarin unit becomes fluorescent after the maleimide reacts with a thiol. This assay provides insight on unfolding for membrane proteins that have buried cysteine residues in the native conformation, because cysteine side chains that become exposed as a result of unfolding are reactive. We applied the CPM assay to two prokaryotic respiratory complexes, succinate:quinone oxidoreductase (SQR)19 and cytochrome bo3 ubiquinol oxidase (cytochrome bo3)20 from E. coli, and to the murine cytidine-5′-monophosphate-sialic acid transporter (CMP-Sia)21. SQR was purified with the conventional detergent C12E9, and cytochrome bo3 and CMP-Sia were purified with DDM. The purified membrane protein-detergent preparations were individually diluted into solutions containing each amphiphile at 10 × CMC, and the unfolding in each protein sample was monitored over time at 40 °C. In addition to DDM, we evaluated MPA-4, DM and sodium dodecyl sulfate (SDS). SDS is widely recognized to be highly disruptive toward native protein conformations, and this detergent caused the most rapid and extensive unfolding of each protein among the agents we examined (Fig. 4a and Supplementary Fig. 3). The level of fluorescence observed with SDS after 130 min at 40 °C was taken to indicate a limiting denatured state, and a lack of fluorescence was taken to indicate a native state for each protein.
Figure 4.
Stability of SQR solubilized with MNG amphiphiles or conventional detergents. (a) CPM assays for SQR solubilized with MNG amphiphiles (MNG-1, MNG-2, and MNG-3) or conventional detergents (MPA-4, DDM, DM and SDS) at 10 × CMC. The unfolding of the each protein was monitored at 40 °C for 130 min using a microplate spectrofluorometer. Gel filtration analysis of SQR in (b) DDM or (c) MNG-3 at 10 × CMC. SQR in DDM or MNG-3 was incubated for 120 min at 40 °C. (d) Time course of SQR activity in MNG-3 or DDM. Each agent was used at 10 × CMC (0.01 wt % for MNG-3, 0.087 wt % for DDM) and 50 × CMC (0.05 wt % for MNG-3, 0.44 wt % for DDM). Note that 50 × CMC MNG-3 is comparable to DDM at 10 × CMC in terms of wt %. The catalytic rate constant (kcat) is plotted as a function of incubation time. Data at t = 0 correspond to the activity of SQR following thermal activation performed at 30 °C for 20 min. Protein solubilized with each agent was incubated at 40 °C for a further 120 min, and activity of the protein was measured at the designated times. The kcat values at each time point were calculated by analyzing reaction data according to Michaelis-Menten kinetics.
All three MNG amphiphiles appeared to be superior to conventional detergents at maintaining native protein structure, as indicated by CPM assay results for SQR (Fig. 4a), and comparable results for cytochrome bo3 and CMP-Sia (Supplementary Fig. 3a,b). DDM and MPA-4 were nearly as effective as the MNG amphiphiles, but DM was noticeably inferior. We next used gel filtration analysis to determine whether MNG-3 or DDM could maintain quaternary interactions among the four SQR subunits. After 120 min at 40 °C in the presence of 10 × CMC DDM, the native quaternary structure of SQR was almost completely destroyed (Fig. 4b). In contrast, the quaternary structure remained largely intact after 130 min at 40 °C in the presence of 10 × CMC MNG-3 (Fig. 4c). We also compared MNG-3 and DDM in an SQR functional assay. SQR must be thermally activated (incubation at 30 °C for 20 min) to remove bound oxaloacetate from the active site prior to assay. We incubated activated SQR with either MNG-3 or DDM at 40 °C, and measured the catalytic efficacy of the protein (kcat) immediately after activation (0 min) and then at 60 min intervals. Both MNG-3 and DDM gave fairly high initial kcat values, but SQR activity steadily declined with DDM, while activity was maintained or even slightly improved with MNG-3 (Fig. 4d). Overall, these results show that MNG-3 maintains the SQR quaternary assembly in a fully native state, and that the MNG amphiphile is superior to DDM in stabilizing catalytically competent SQR.
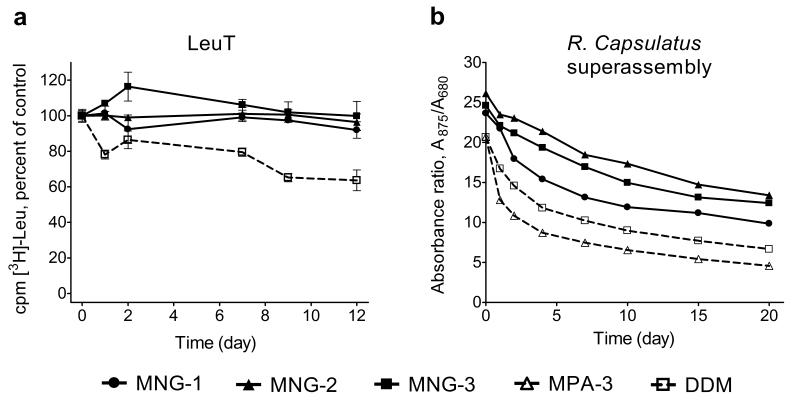
We turned to bacterial leucine transporter (LeuT)22 to evaluate protein stability as a function of time at room temperature (rather than as an ability to resist thermal denaturation). LeuT, a bacterial member of the neurotransmitter:sodium symporter family (NSS family) proteins23, was solubilized and purified with DDM and then transferred into individual amphiphile solutions. We assessed activity in terms of its ability to bind [3H]leucine via scintillation proximity assay24. Preliminary studies, conducted at 10 × CMC, indicated that LeuT solubilized with several conventional detergents displayed a rapid decline in activity, while LeuT solubilized with DDM displayed a more gradual loss of activity; all three MNG amphiphiles were superior to DDM in terms of maintaining LeuT activity over time (Supplementary Fig. 4a,b). We conducted further studies involving the MNG amphiphiles and DDM with each agent at 0.026 wt % above its CMC (Fig. 5a and Supplementary Fig. 4c). Under these conditions, each MNG amphiphile kept LeuT fully soluble and fully active over the 12-day study period. In contrast, LeuT activity declined to ~65% after 12 days in the presence of DDM (Fig. 5a).
Figure 5.
Long-term stability of LeuT and R. capsulatus superassembly in MNG amphiphiles or conventional detergents. (a) Time course of activity ([3H]-Leu binding) assay for LeuT solubilized with MNG amphiphiles (MNG-1, MNG-2, and MNG-3) and DDM at 0.026 wt % above its critical micelle concentration (CMC) (total concentrations: 0.035 wt % DDM, 0.028 wt % MNG-1, 0.027 wt % MNG-2 and 0.027 wt % MNG-3). LeuT activity was monitored at indicated time points, using a scintillation proximity assay (SPA), for protein stored at the room temperature. Results are expressed as % activity relative to the appropriate day 0 measurement. Normalized results are expressed as mean ± s.e.m. (n = 2). (b) Time course of stability of R. capsulatus superassembly purified with MNG amphiphiles (MNG-1, MNG-2, and MNG-3) and conventional detergents (MPA-3 and DDM) at 1 × CMC. The absorption ratios (A875/A680) of the detergent or amphiphile samples were followed as a function of time.
MNG amphiphiles extract IMPs from native membranes
In order to assess the ability of MNG amphiphiles to extract intrinsic proteins from their native membranes, we examined the photosynthetic superassembly of Rhodobacter capsulatus25. These studies employed membranes isolated from an R. capsulatus strain that lacks light harvesting complex II26; in this case the superassembly comprises the very labile light harvesting complex I (LHI) and the more resilient reaction center complex (RC). This system is well-suited for assessing extraction and stabilization properties of detergents and amphiphiles because the superassembly can be detected and its composition can be qualitatively monitored via optical measurements15: intact LHI-RC superassembly has a strong absorbance at 875 nm and 875/760 absorption ratio > 7. (Absorbance at 760 nm arises from bacteriochlorophyll units that have dissociated from LHI.) We treated intracytoplasmic R. capsulatus membranes enriched in LHI-RC complex with solutions containing 1 wt % detergent or amphiphile for 30 min at 32 °C. MNG-2 and MNG-3 were effective at extracting the intact superassembly (strong absorption at 875 nm; 875/760 ratio ~9-10; Supplementary Fig. 5). Comparable efficacy was observed with MPA-3 and DDM (875/760 ratio ~8.5), but other conventional detergents were less successful. After purification of solubilized samples (Supplementary Fig. 6), we monitored the superassembly stability over time at room temperature based on the 875/680 absorbance (absorbance at 680 nm arises from oxidation of bacteriochlorophyll that has been released from LHI). We compared the three MNG amphiphiles to conventional detergents (DDM and MPA-3), with each agent at its CMC (Fig. 5b; comparisons involving other concentrations may be found in Supplementary Fig. 7). For all samples, the 875/680 absorbance ratio declined over 20 days; however, at every time point, the ratio was higher for the samples solubilized with an MNG amphiphile than for the samples solubilized with a conventional detergent. Overall, the results with R. capsulatus photosynthetic proteins show that MNG amphiphiles can extract a protein quaternary structure intact from its native membrane and then provide superior structural stability over time relative to DDM or other conventional detergents.
We performed additional studies to evaluate the use of MNG-3 for extraction of other proteins from membranes. MNG-3 was comparable to conventional detergents DDM and TDM, at 1 wt % or 2 wt %, for the extraction of wild type β2AR from Sf9 insect cell membranes. We evaluated the activity of β2AR receptor via a binding assay with antagonist [3H]-dihydroalprenolol (Supplementary Fig. 8a). At 1 wt %, MNG-3 was comparable to DDM and slightly inferior to TDM in terms of β2AR activity, but at 2 wt %, MNG-3 was superior to both conventional detergents. MNG-3 at 2 wt % yielded a receptor activity comparable to that of 1 wt % TDM. For extraction of LeuT from the bacterial membrane, MNG-3 proved to be somewhat inferior to DDM, providing only ~60% of the yield obtained with DDM. However, MNG-3-purified protein displayed identical substrate affinity to that of DDM-purified protein (Supplementary Fig. 8b). For extraction of a CMP-Sia fusion protein bearing green fluorescent protein (GFP) at the C terminus, expressed in Saccharomyces cerevisiae, MNG-3 and DDM provided comparable protein yields (70-80%); in each case, the protein was intact and homogenous (Supplementary Fig. 8c). Overall, based on results from several different systems, MNG-3 appears to be comparable to DDM for extraction of membrane proteins from biological membranes.
We observed that MNG amphiphiles enabled the expression and concomitant solubilization of a membrane protein, bacterio-opsin, (BO) from a cell-free wheat germ-based translation system (Supplementary Fig. 9). DDM and other conventional detergents could solubilize only limited amounts of translated BO at 0.1 wt %, and these detergents inhibited cell-free translation at 0.2 wt %. In contrast, 0.2 wt % MNG-2 or MNG-3 was compatible with translation and solubilized most of the BO.
MNG amphiphiles aid in membrane protein crystallization
Growth of high-quality crystals that allow structure determination is one of the most important and challenging goals of membrane protein research. We examined crystallization of the cytochrome b6f complex from Chlamydomonas reinhardtii with MNG-3. This protein assembly tends to denature in the presence of most conventional detergents, but DDM can maintain native structure and has previously enabled crystallization and structure determination via x-ray diffraction27. Cytochrome b6f crystallization requires 0.2 mM DDM (CMC = 0.17 mM); lower detergent concentrations promote protein aggregation while higher detergent concentration lead to dissociation of subunits. We found that cytochrome b6f in the presence of 0.5 mM MNG-3 exhibits stability comparable to that observed in the presence of 0.2 mM DDM. The tolerance for higher concentrations of MNG-3 relative to DDM provided a larger concentration window in which to attempt crystallization of solubilized cytochrome b6f. Protein-containing crystals appeared within 24 hours of setting up drops. After a few days, crystals reached a maximum size of 70 × 400 μm (data not shown).
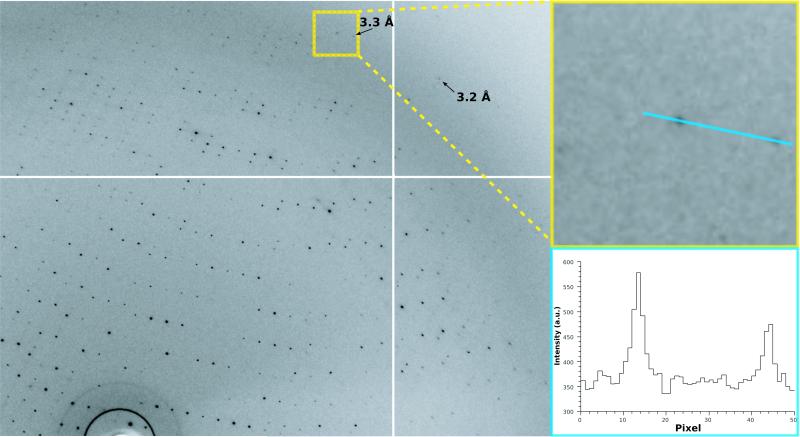
The diffraction data from cytochrome b6f crystals grown with MNG-3 extended up to ~3.4 Å resolution (Fig. 6 and Supplementary Table 2), a value similar to that commonly obtained for crystals grown with DDM. With DDM, extensive screening yielded crystals diffracting to 2.8 Å27. Fourier difference maps (crystals with DDM27 vs. crystals with MNG-3) exhibited no notable features in the protein region (data not shown). However, substantial differences (up to 6.8 σ) are observed in regions where detergent molecules have been localized in the crystals grown with DDM. These differences indicate that during the exchange of detergents, MNG-3 was able to displace the most strongly bound DDM molecules. Further analysis by refinement of the protein structure starting from the DDM structure confirmed that the electron density of two DDM molecules had vanished and that a new molecule with a slightly different maltoside headgroup position, presumably MNG-3, occupied this position. This electron density, however, was not sufficiently well defined to allow model building of MNG-3, despite otherwise good refinement statistics for the protein (Supplementary Table 2).
Figure 6.
Image and x-ray diffraction pattern from crystals of cytochrome b6f /MNG-3 complexes. X-ray diffraction by cytochrome b6f crystal obtained in the presense of MNG-3. The left panel represents a portion of the pattern (0.5 degree oscillation range). Resolution limits are marked with arrows (the white cross is due to the tiling of the detector). Top right: enlargement of the red square with two strong spots near the resolution limit. A section through the two strong spots is shown in the lower right corner.
Membrane protein crystallization from a lipidic cubic phase (LCP) is an increasingly popular strategy28 which has notably led to the recent structure solution of β2AR-T4L 16,29,30. We evaluated the ability of MNG-3 to promote LCP-based crystallization of two new forms of this GPCR, a fusion protein with a covalently attached agonist (unpublished data), and an agonist-bound β2AR-T4L stabilized by an antibody in an active state (unpublished data). Although efforts to crystallize DDM-solubilized agonist-bound receptor from a monoolein-water LCP yielded crystals (data not shown), it was impossible to grow them large enough to obtain high-resolution diffraction. In contrast, detergent exchange into MNG-3 in both cases facilitated incorporation into the LCP, from which larger crystals suitable for x-ray diffraction analysis were obtained (data not shown). In the latter case, the crystals were ~40 × 5 × 5 μm, and x-ray diffraction data allowed solution of the structure to 3.5 Å resolution. We speculate the enhanced stability of β2AR-T4L solubilized by MNG-3 relative to the DDM-solubilized form may be crucial for successful transfer of the protein into the LCP.
Discussion
Our results suggest that MNG amphiphiles will be generally useful for membrane protein biochemistry research. MNG amphiphiles can be readily prepared in multi-gram quantities, and this synthetic accessibility should enable their evaluation with many systems, including efforts directed toward structural analysis (e.g., - nuclear magnetic resonance spectroscopy31, mass spectrometry32) and the incorporation of MNG amphiphiles into new techniques for membrane protein purification and manipulation33. Given the diversity of shapes, sequences and properties of membrane proteins and their assemblies, it seems very unlikely that any single amphiphile will be ideal for all or even a large subset of membrane proteins. The ease with which MNG amphiphile structure may be varied should facilitate the development of a suite of agents that, collectively, display broad utility.
Many important questions remain to be addressed in subsequent studies. For example, it will be valuable to assess the sizes of micelles formed by MNG amphiphiles, and how micelle size is affected by changes in amphiphile structure. In addition, it will be useful to determine average numbers of amphiphile molecules per micelle. But even before these properties are elucidated, we anticipate, based on this first characterization study, that the MNG amphiphiles will become useful tools for membrane protein manipulation.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health (NIH) grant P01 GM75913 (S.H.G.), NS28471 (B.K.), the Lundbeck Foundation (S.G.F.R., C.J.L., U.G.), the Danish National Research Council (C.J.L., U.G.), the European Community’s Seventh Framework Programme FP7/2007-2013 under grant agreement n° HEALTH-F4-2007-201924, EDICT Consortium (K.G., U.G., B. B.) and by NIH grants GM083118 and NS28471 and NIH Protein Structure Initiative Grant U54 GM074901-01 (J.L. Markley, G.N. Phillips, and B.G.F.) and NIGMS GM-50853 (B.G.F.). This work was supported also by R21HL087895 from the National Heart, Lung, and Blood Institute and Texas Norman Hackerman Advanced Research Program under grant No. 010674-0034-2009 (to L.G.) and the Center for Membrane Protein Research, the Texas Tech University Health Sciences Center. R.R. was funded by the Defense, Science and Technology Laboratories. We thank P. Laible (Argonne National Laboratory, Chicago) for supplying membrane preparations from R. capsulatus. We acknowledge SOLEIL for provision of synchrotron radiation facilities and we would like to thank B. Guimaraes for assistance in using the beamline Proxima 1. We also thank R. Kaback (University of California, Los Angeles) and G. Leblanc (Institut de Biologie et technologies-Saclay) for the MelB expression system. M.A.G. acknowledges support from the National Science Foundation East Asia and Pacific Summer Institutes Fellowship program. We thank G. Cecchini (University of California, San Francisco) and J. Ruprecht (MRC Mitochondrial Biology Unit, Cambridge) for the purified SQR and the details of the SQR functional assay, and we acknowledge the assistance of P. Nixon in the analysis of the SQR functional data.
Footnotes
COMPETING INTERESTS STATEMENT P.S.C, S.G.F.R., B.K. and S.H.G. are co-inventors on a patent application that covers the MNG amphiphiles.
References
- 1.Liu J, Rost B. Comparing function and structure between entire proteomes. Protein Sci. 2001;10:1970–1979. doi: 10.1110/ps.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lacapere JJ, Pebay-Peyroula E, Neumann JM, Etchebest C. Determining membrane protein structures: still a challenge! Trends Biochem. Sci. 2007;32:259–270. doi: 10.1016/j.tibs.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Privé GG. Detergents for the stabilization and crystallization of membrane proteins. Methods. 2007;41:388–397. doi: 10.1016/j.ymeth.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Schafmeister CE, Meircke LJW, Stroud RM. Structure at 2.5 Å of a designed peptide that maintains solubility of membrane proteins. Science. 1993;262:734–738. doi: 10.1126/science.8235592. [DOI] [PubMed] [Google Scholar]
- 5.McGregor C-L, et al. Lipopeptide detergents designed for the structural study of membrane protein. Nat. Biotech. 2003;21:171–176. doi: 10.1038/nbt776. [DOI] [PubMed] [Google Scholar]
- 6.Zhao X, et al. Designer short peptide surfactants stabilize G protein-coupled receptor bovine rhodopsin. Proc. Natl. Acad. Sci. USA. 2006;103:17707–17712. doi: 10.1073/pnas.0607167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tribet C, Audebert R, Popot J-L. Amphipols: polymers that keep membrane proteins soluble in aqueous solutions. Proc. Natl. Acad. Sci. USA. 1996;93:15047–15050. doi: 10.1073/pnas.93.26.15047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Popot J-L. Amphipols, nanodiscs, and fluorinated surfactants: three non-conventional approaches to studying membrane proteins in aqueous solutions. Annu. Rev. Biochem. 2010;79:13.1–13.39. doi: 10.1146/annurev.biochem.052208.114057. [DOI] [PubMed] [Google Scholar]
- 9.Nath A, Atkins WM, Sligar SG. Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry. 2007;46:2059–2069. doi: 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]
- 10.Borch J, Hamann T. The nanodisc: a novel tool for membrane protein studies. Biol. Chem. 2009;390:805–814. doi: 10.1515/BC.2009.091. [DOI] [PubMed] [Google Scholar]
- 11.Breyton C, et al. Micellar and biochemical properties of (hemi)fluorinated surfactants are controlled by the size of the polar head. Biophys. J. 2009;97:1077–1086. doi: 10.1016/j.bpj.2009.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Q, et al. Designing facial amphiphiles for the stabilization of integral membrane protein. Angew. Chem. Int. Ed. 2007;119:7153–7155. doi: 10.1002/anie.200701556. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann RW. Flexible molecules with defined shape-conformational design. Angew. Chem. Int. Ed. Engl. 1992;31:1124–1134. [Google Scholar]
- 14.McQuade DT, et al. Rigid amphiphiles for membrane protein manipulation. Angew. Chem. Int. Ed. 2000;39:758–761. [PubMed] [Google Scholar]
- 15.Chae PS, Wander MJ, Bowling AP, Laible PD, Gellman SH. Glycotripod amphiphiles for solubilization and stabilization of a membrane protein superassembly: importance of branching in the hydrophilic portion. ChemBioChem. 2008;9:1706–1709. doi: 10.1002/cbic.200800169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenbaum DM, et al. GPCR engineering yields high-resolution structural insights into β2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 17.Bassilana M, Pourcher T, Lablanc G. Melibiose Permease of Escherichia coli. J. Biol. Chem. 1988;263:9663–9667. [PubMed] [Google Scholar]
- 18.Alexandrov A, Mileni M, Chien EY, Hanson MA, Stevens RC. Microscale fluorescent thermal stability assay for membrane proteins. Structure. 2008;16:351–359. doi: 10.1016/j.str.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 19.Horsefield R, Iwata S, Byrne B. Complex II from a structural perspective. Curr. Protein Pept. Sci. 2004;5:107–118. doi: 10.2174/1389203043486847. [DOI] [PubMed] [Google Scholar]
- 20.Puustinen A, Finel M, Haltia T, Gennis RB, Wikström M. Properties of the two terminal oxidases of Escherichia coli. Biochemistry. 1991;30:3936–3942. doi: 10.1021/bi00230a019. [DOI] [PubMed] [Google Scholar]
- 21.Newstead S, Kim H, von Heijne G, Iwata S, Drew D. High-throughput fluorescent-based optimization of eukaryotic membrane protein overexpression and purification in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2007;104:13936–13941. doi: 10.1073/pnas.0704546104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deckert G, et al. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 23.Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature. 2005;437:215–223. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- 24.Quick M, Javitch JA. Monitoring the function of membrane transport proteins in detergent-solubilized form. Proc. Natl. Acad. Sci. USA. 2007;104:3603–3608. doi: 10.1073/pnas.0609573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu XC, Ritz T, Damjanovic A, Authenrieth F, Schulten K. Photosynthetic apparatus of purple bacteria. Quart. Rev. Biophys. 2002;35:1–62. doi: 10.1017/s0033583501003754. [DOI] [PubMed] [Google Scholar]
- 26.Youvan DC, Ismail S, Bylina EJ. Chromosomal deletion and plasmid complementation of the photosynthetic reaction center and light-harvesting genes from Rhodopseudomonas capuslata. Gene. 1985;38:19–30. doi: 10.1016/0378-1119(85)90199-4. [DOI] [PubMed] [Google Scholar]
- 27.Stroebel D, Choquet Y, Popot J-L, Picot D. An atypical haem in the cytochrome b6f complex. Nature. 2003;426:413–418. doi: 10.1038/nature02155. [DOI] [PubMed] [Google Scholar]
- 28.Rosenbaum DM, Rasmussen SGF, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cherezov V, et al. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanson MA, et al. A specific cholesterol binding site is established by the 2.8 Å structure of the human β2-adrenergic receptor. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanders CR, Sonnichsen F. Solution NMR of membrane proteins: practice and challenges. Magn. Reson. Chem. 2006;44:S24–S40. doi: 10.1002/mrc.1816. [DOI] [PubMed] [Google Scholar]
- 32.Barrera NP, Di Bartolo N, Booth PJ, Robinson CV. Micelles protect membrane complexes from solution to vacuum. Science. 2008;321:243–246. doi: 10.1126/science.1159292. [DOI] [PubMed] [Google Scholar]
- 33.Li L, et al. Simple host-guest chemistry to modulate the process of concentration and crystallization of membrane proteins by detergent capture in a microfluidic device. J. Am. Chem. Soc. 2008;130:14324–14328. doi: 10.1021/ja805361j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pourcher T, Leclercq S, Brandolin G, Leblanc G. Melibiose permease of Escherichia coli: large scale purification and evidence that H+, Na+, and Li+ sugar symport is catalyzed by a single polypeptide. Biochemistry. 1995;34:4412–4420. doi: 10.1021/bi00013a033. [DOI] [PubMed] [Google Scholar]
- 35.Drew D, Newstead S, Sonoda Y, Kim H, von Heijne G, Iwata S. GFP-based optimization scheme for the overexpression and purification of eukaryotic membrane proteins in Saccharomyces cerevisiae. Nat. Protoc. 2008;3:784–798. doi: 10.1038/nprot.2008.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14:673–681. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 37.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Apppl. Crys. 1993;26:795–800. [Google Scholar]
- 38.Collaborative Computational Project, Number 4 The CC4 Suite: Programs for Protein Crystallography. Acta Cryst. 1994;D50:760–763. [Google Scholar]
- 39.Adams PD, et al. PHENIX: bulding new software for automated crystallographic structure determination. Acta. Cryst. 2002;D58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 40.Bricogne G, et al. BUSTER, version 2.8.0. Global Phasing Ltd; Cambridge, United Kindom: 2009. [Google Scholar]
- 41.Pierre Y, Breyton C, Kramers D, Popot J-L. Purification and characterization of the cytochrome b6f complex from Chlamydomonas reinhardtii. J. Biol. Chem. 1995;270:29342–29349. doi: 10.1074/jbc.270.49.29342. [DOI] [PubMed] [Google Scholar]
- 42.Goren MA, Fox BG. Wheat germ cell-free translation, purification, and assembly of a functional human stearoyl-CoA desaturase complex. Protein Expr. Purif. 2008;62:171–178. doi: 10.1016/j.pep.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.