Abstract
We have determined the three-dimensional structure of a non-selfcomplementary nonanucleotide duplex which contains an abasic (apyrimidinic) site in the centre, i.e. a deoxyribose residue opposite an adenosine. The majority of the base and sugar proton resonances were assigned by NOESY, COSY and 2DQF spectra in D2O and H2O. We have measured the initial slope of buildup of NOEs in NOESY spectra at very short mixing times (25 to 50 ms), and from these were able to establish interproton distances for the central part of the duplex. We propose a different strategy for proton-proton distance determinations which takes into account the observed variations in correlation times for particular proton-proton vectors. A set of 31 measured interproton distances was incorporated into the refinement of the oligonucleotide structure by molecular mechanics calculations. Two structures were obtained which retain all aspects of a classical B DNA in which the unpaired adenine and the abasic deoxyribose lie inside the helix. We observe that the non-hydrogen bonded adenine is held well in the helix, the Tm of this base being the same as that of the A.T base pairs in the same duplex.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Feigon J., Leupin W., Denny W. A., Kearns D. R. Two-dimensional proton nuclear magnetic resonance investigation of the synthetic deoxyribonucleic acid decamer d(ATATCGATAT)2. Biochemistry. 1983 Dec 6;22(25):5943–5951. doi: 10.1021/bi00294a038. [DOI] [PubMed] [Google Scholar]
- Frechet D., Cheng D. M., Kan L. S., Ts'o P. O. Nuclear Overhauser effect as a tool for the complete assignment of nonexchangeable proton resonances in short deoxyribonucleic acid helices. Biochemistry. 1983 Oct 25;22(22):5194–5200. doi: 10.1021/bi00291a020. [DOI] [PubMed] [Google Scholar]
- Gelin B. R., Karplus M. Mechanism of tertiary structural change in hemoglobin. Proc Natl Acad Sci U S A. 1977 Mar;74(3):801–805. doi: 10.1073/pnas.74.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
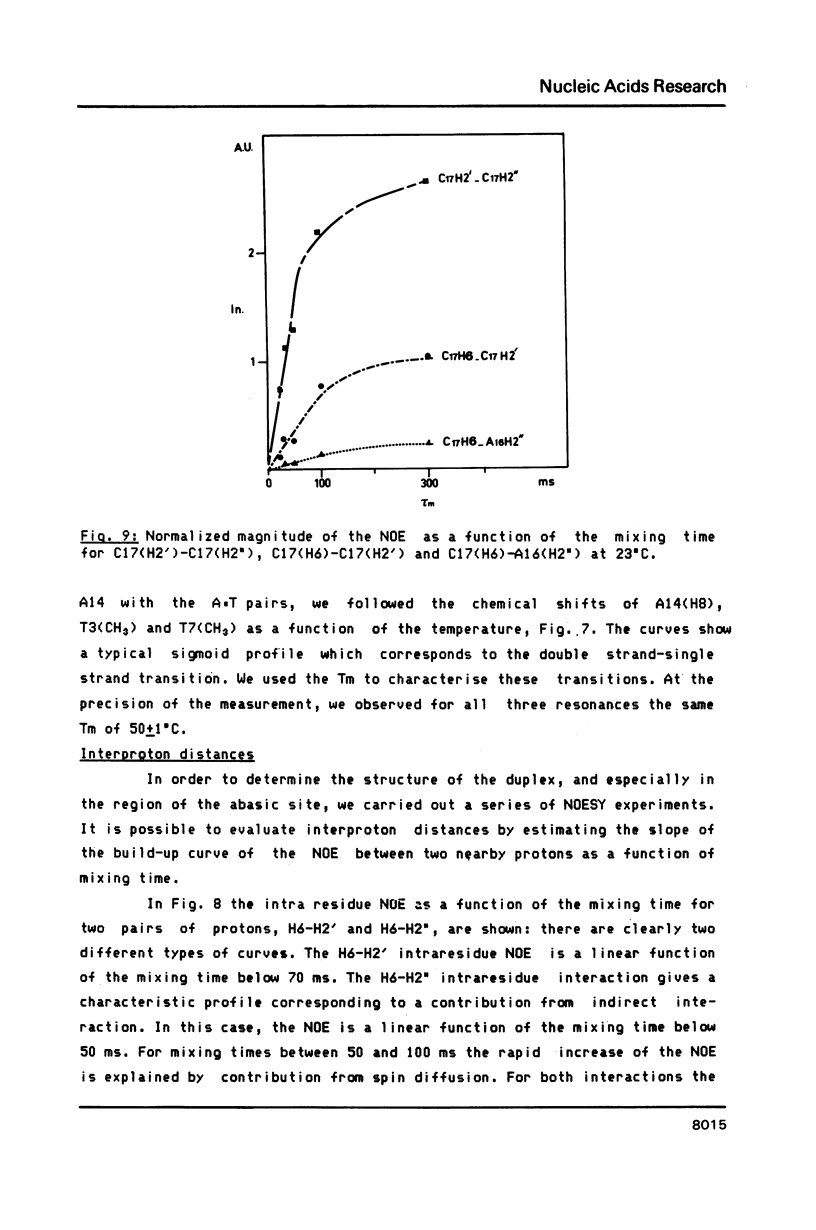
- Gronenborn A. M., Clore G. M., Kimber B. J. An investigation into the solution structures of two self-complementary DNA oligomers, 5'-d(C-G-T-A-C-G) and 5'-d(A-C-G-C-G-C-G-T), by means of nuclear-Overhauser-enhancement measurements. Biochem J. 1984 Aug 1;221(3):723–736. doi: 10.1042/bj2210723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare D. R., Reid B. R. Three-dimensional structure of a DNA hairpin in solution: two-dimensional NMR studies and distance geometry calculations on d(CGCGTTTTCGCG). Biochemistry. 1986 Sep 9;25(18):5341–5350. doi: 10.1021/bi00366a053. [DOI] [PubMed] [Google Scholar]
- Hare D. R., Wemmer D. E., Chou S. H., Drobny G., Reid B. R. Assignment of the non-exchangeable proton resonances of d(C-G-C-G-A-A-T-T-C-G-C-G) using two-dimensional nuclear magnetic resonance methods. J Mol Biol. 1983 Dec 15;171(3):319–336. doi: 10.1016/0022-2836(83)90096-7. [DOI] [PubMed] [Google Scholar]
- Hare D., Shapiro L., Patel D. J. Extrahelical adenosine stacks into right-handed DNA: solution conformation of the d(C-G-C-A-G-A-G-C-T-C-G-C-G) duplex deduced from distance geometry analysis of nuclear Overhauser effect spectra. Biochemistry. 1986 Nov 18;25(23):7456–7464. doi: 10.1021/bi00371a030. [DOI] [PubMed] [Google Scholar]
- Hogan M. E., Jardetzky O. Internal motions in deoxyribonucleic acid II. Biochemistry. 1980 Jul 22;19(15):3460–3468. doi: 10.1021/bi00556a009. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Mutational specificity of depurination. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1494–1498. doi: 10.1073/pnas.81.5.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt M. Computer simulation of DNA double-helix dynamics. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):251–262. doi: 10.1101/sqb.1983.047.01.030. [DOI] [PubMed] [Google Scholar]
- Lindahl T. DNA glycosylases, endonucleases for apurinic/apyrimidinic sites, and base excision-repair. Prog Nucleic Acid Res Mol Biol. 1979;22:135–192. doi: 10.1016/s0079-6603(08)60800-4. [DOI] [PubMed] [Google Scholar]
- Lindahl T. DNA repair enzymes. Annu Rev Biochem. 1982;51:61–87. doi: 10.1146/annurev.bi.51.070182.000425. [DOI] [PubMed] [Google Scholar]
- Lockhart M. L., Deutsch J. F., Yamaura I., Cavalieri L. F., Rosenberg B. H. Termination of DNA synthesis in vitro at apurinic sites but not at ethyl adducts on the template. Chem Biol Interact. 1982 Oct;42(1):85–95. doi: 10.1016/0009-2797(82)90144-2. [DOI] [PubMed] [Google Scholar]
- Millican T. A., Mock G. A., Chauncey M. A., Patel T. P., Eaton M. A., Gunning J., Cutbush S. D., Neidle S., Mann J. Synthesis and biophysical studies of short oligodeoxynucleotides with novel modifications: a possible approach to the problem of mixed base oligodeoxynucleotide synthesis. Nucleic Acids Res. 1984 Oct 11;12(19):7435–7453. doi: 10.1093/nar/12.19.7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall S. K., Eritja R., Kaplan B. E., Petruska J., Goodman M. F. Nucleotide insertion kinetics opposite abasic lesions in DNA. J Biol Chem. 1987 May 15;262(14):6864–6870. [PubMed] [Google Scholar]
- Richards R. G., Sowers L. C., Laszlo J., Sedwick W. D. The occurrence and consequences of deoxyuridine in DNA. Adv Enzyme Regul. 1984;22:157–185. doi: 10.1016/0065-2571(84)90013-x. [DOI] [PubMed] [Google Scholar]
- Sagher D., Strauss B. Insertion of nucleotides opposite apurinic/apyrimidinic sites in deoxyribonucleic acid during in vitro synthesis: uniqueness of adenine nucleotides. Biochemistry. 1983 Sep 13;22(19):4518–4526. doi: 10.1021/bi00288a026. [DOI] [PubMed] [Google Scholar]
- Schaaper R. M., Kunkel T. A., Loeb L. A. Infidelity of DNA synthesis associated with bypass of apurinic sites. Proc Natl Acad Sci U S A. 1983 Jan;80(2):487–491. doi: 10.1073/pnas.80.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheek R. M., Boelens R., Russo N., van Boom J. H., Kaptein R. Sequential resonance assignments in 1H NMR spectra of oligonucleotides by two-dimensional NMR spectroscopy. Biochemistry. 1984 Mar 27;23(7):1371–1376. doi: 10.1021/bi00302a006. [DOI] [PubMed] [Google Scholar]
- Tidor B., Irikura K. K., Brooks B. R., Karplus M. Dynamics of DNA oligomers. J Biomol Struct Dyn. 1983 Oct;1(1):231–252. doi: 10.1080/07391102.1983.10507437. [DOI] [PubMed] [Google Scholar]
- Weiner M. J. Case 22-1984: seminoma. N Engl J Med. 1984 Sep 20;311(12):800–801. doi: 10.1056/nejm198409203111219. [DOI] [PubMed] [Google Scholar]


