SUMMARY
Cdc48/p97 is an essential ATPase whose role in targeting substrates to the ubiquitin-proteasome system (UPS) remains unclear. Existing models posit that Cdc48 acts upstream of UPS receptors. To address this hypothesis, we examined the association of ubiquitin (Ub) conjugates with 26S proteasomes. Unexpectedly, proteasomes isolated from cdc48 mutants contain high levels of Ub conjugates and mass spectrometry identified numerous non-proteasomal proteins, including Rpb1, the largest subunit of RNA Pol II. UV-induced turnover of Rpb1 depends upon Cdc48–Ufd1–Npl4, Ubx4 and the uncharacterized adaptor, Ubx5. Ubiquitinated Rpb1, proteasomes, and Cdc48 accumulate on chromatin in UV-treated wildtype cells and the former two accumulate to higher levels in mutant cells, suggesting that degradation of Rpb1 is facilitated by Cdc48 at sites of stalled transcription. These data reveal an intimate coupling of function between proteasomes and Cdc48 that we suggest is necessary to sustain processive degradation of unstable subunits of some macromolecular protein complexes.
INTRODUCTION
Budding yeast Cdc48 is a member of the AAA (ATPases associated with various cellular activities) protein family. Cdc48 has been implicated in a plethora of functions that include cell cycle regulation, membrane fusion, the stress response, and ER-associated degradation (ERAD) (Hirsch et al., 2009; Vembar and Brodsky, 2008). Its highly conserved mammalian counterpart, p97, has been additionally implicated in reformation of the nucleus (Ramadan et al., 2007), organelle biogenesis (Halawani and Latterich, 2006), myofibril organization (Janiesch et al., 2007) and the degradation of proteins such as Hif1-α (Alexandru et al., 2008) and HMG-CoA reductase (DeBose-Boyd, 2008).
Two underlying properties of Cdc48/p97 contribute to its myriad functions: its ATPase activity and the ability to bind Ub (Ye, 2006). Together, these activities are thought to underpin a ‘segregase’ function that separates ubiquitinated polypeptides from tightly-bound partner proteins, enabling selective degradation of the former (Braun et al., 2002; Johnson et al., 1990). The Ub-binding activity of Cdc48 is synergistically enhanced by the binding of adaptors belonging to the UFD (Ub-fusion degradation) pathway (Johnson et al., 1995) and/or to the UBX (Ub regulatory X) family, members of which possess Ub-binding domains (Schuberth and Buchberger, 2008). In its best-understood function – ERAD – Cdc48, in conjunction with its cofactors Ufd1 and Npl4, extracts misfolded proteins from the ER membrane by virtue of its ATPase activity. The ubiquitinated substrates subsequently engage the UBA (Ub-associated) domain-containing receptors Rad23 and Dsk2, which in turn bind to the proteasome via their UbL (Ub-like) domains, thereby delivering substrates for degradation (Raasi and Wolf, 2007).
The breadth of Cdc48’s involvement in the UPS remains poorly understood. Whereas Cdc48 function clearly plays a prominent role in turnover of ER proteins (Jarosch et al., 2002; Rabinovich et al., 2002; Ravid et al., 2006; Ye et al., 2001), the connection between Cdc48 and the UPS was first discovered based on the requirement of Cdc48 and UFD proteins for turnover of non-ER, soluble reporter substrates (Ghislain et al., 1996). Fractionation studies and live imaging of Cdc48-GFP indicate that only a portion of yeast Cdc48 is peripherally bound to the ER/nuclear envelope, with most being partitioned between the cytosol and nucleus (Madeo et al., 1998)(Huh et al., 2003). Cdc48 has recently been shown to be required for the degradation of the cytosolic protein fructose-1,6-bisphosphatase [FBPase] (Barbin et al., 2010). Given the relative dearth of soluble (and physiological) UPS substrates, we wished to identify substrates whose degradation depends on Cdc48 and determine at what stage in their degradation Cdc48 is required.
Although there is general agreement that Cdc48 functions between Ub ligases and the proteasome, the only data that address the specific targeting step on which Cdc48 acts to promote turnover of soluble proteins are those underlying the ‘escort’ model (Richly et al., 2005). This model posits that Cdc48 takes over from Ub ligases by coordinating the elongation of a size-restricted yet degradation-competent chain upon substrate, whereupon the substrate is handed off to an Ub chain receptor such as Rad23 for delivery to the proteasome. Sculpting of the Ub chain is achieved by a combination of trimming by deubiquitinating enzymes (DUBs) such as Otu1 and chain extension by the ‘E4’ enzyme Ufd2 (Rumpf and Jentsch, 2006). A central prediction of the escort model is that proteasome-bound Ub conjugates should become depleted in Cdc48 mutants, much as is seen in rad23Δdsk2Δ double mutants (Elsasser et al., 2004), but this has not been evaluated.
Here, we characterize 26S proteasomes isolated from wildtype and temperature-sensitive conditional cdc48-3 mutant cells. Contrary to expectation, Ub conjugates and many non-proteasomal proteins accumulated to high levels on proteasomes from cdc48-3 mutants. In-depth analysis of one particular Cdc48-dependent turnover substrate, Rpb1, reveals that Cdc48 and its adaptor Ubx5 function downstream of Cul3 Ub ligase to facilitate degradation of chromatin-bound Rpb1.
RESULTS
Ub conjugates and numerous proteins accumulate on proteasomes isolated from cdc48-3 mutants
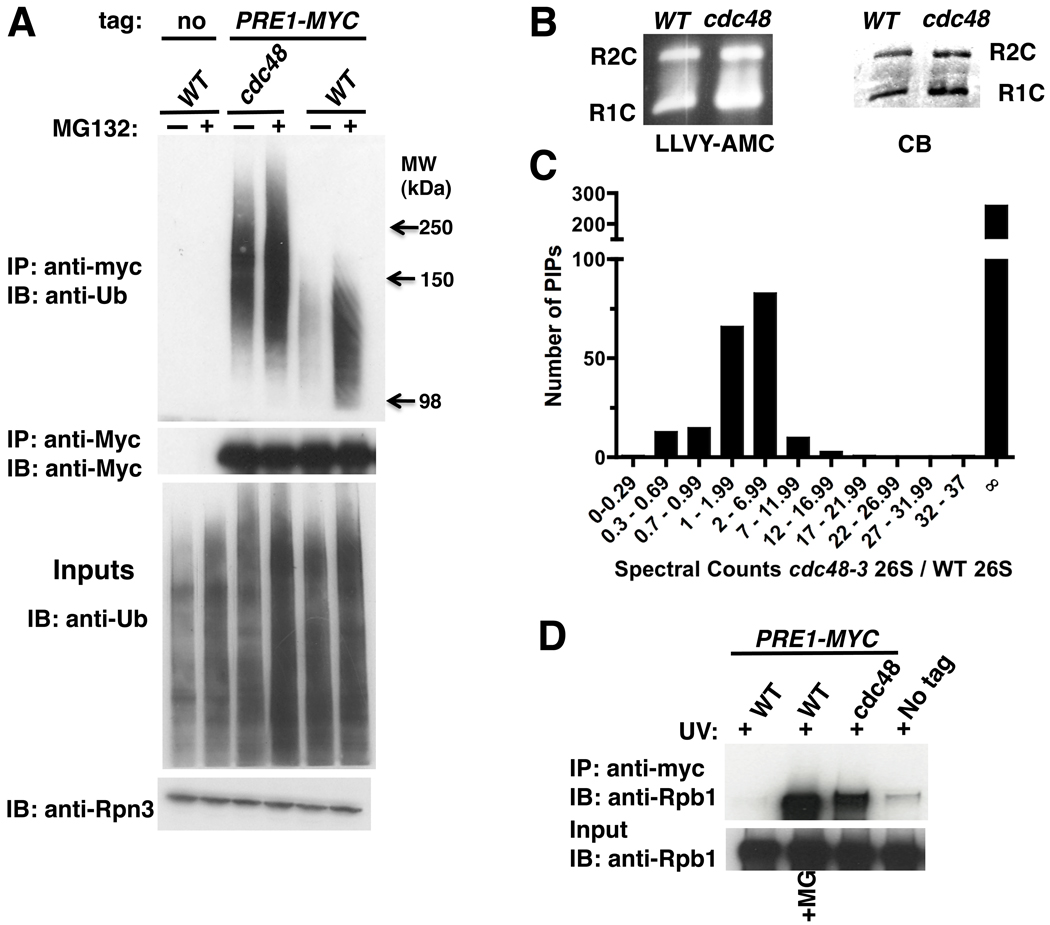
To determine if Ub conjugates are targeted to proteasomes in the absence of Cdc48 function, we isolated 26S proteasomes from wildtype and cdc48-3 cells. Cdc48 was not required for proteasome assembly (Figure S1A). We next evaluated Ub by immunoblotting and surprisingly observed that proteasomes purified from cdc48-3 cells contained an increased level of Ub conjugates compared to wildtype (Figure 1A). Indeed, the level of conjugates associated with proteasomes isolated from mutant cells was comparable to that observed with proteasomes purified from wildtype cells treated with the proteasome inhibitor MG132. Additionally, the conjugates were of unusually high molecular weight (HMW; 110 kDa and greater).
Figure 1. Ubiquitinated proteins are targeted to 26S proteasomes in cdc48-3 mutant cells.
(A) Ub conjugates accumulated on proteasomes in cdc48-3. Wildtype (WT; RJD3437) and mutant (RJD3454) cells expressing myc-tagged Pre1 (or not, RJD4090) were shifted to 37°C for 90 min and treated with 40 µM MG132 for 30 min. Proteasomes were isolated by anti-myc immunoprecipitation (IP) and immunoblotted (IB) for Ub and Pre1-myc (upper panels). Input extracts were blotted for Ub and Rpn3 (lower panels).
(B) Accumulation of conjugates on proteasome was not due to a defect in chymotryptic activity in cdc48-3 mutants. Isolated 26S proteasomes fractionated by native PAGE were either stained with Coomassie Blue (CB) or processed for in-gel peptidase activity using the fluorescent reporter substrate LLVY-AMC. R1C and R2C refer to 20S complexes capped by 1 or 2 19S regulatory particles, respectively.
(C) Numerous proteins accumulated on proteasomes in cdc48-3 mutants. Multidimensional mass spectrometry (MudPIT) was performed on an LTQ mass spectrometer using 26S proteasomes isolated from WT and mutant (RJD2902) cells grown at 37°C for 2 hours. The actual ratio of total spectral counts of proteasomal subunits from mutant to WT was 0.8 and was normalized to one. The same normalization factor was applied for spectral counts obtained for the proteasome-interacting proteins (PIPs). ∞ refers to proteins found in cdc48-3 but not WT proteasomes.
(D) Rpb1 accumulated on proteasome in cdc48-3 mutant treated with UV. WT and mutant cells were shifted to 37°C for 90 minutes and UV irradiated. Cells were recovered at 37°C for 30 minutes in the absence or presence of MG132. Input cell extracts and proteasomes immunoprecipitated (IP) with anti-myc were blotted (IB) for Rpb1 using 4H8 antibody.
See also Figure S1.
One explanation for this result is that Cdc48 was required for proteasomal peptidase activity, but this was ruled out by an in-gel peptidase assay on 26S proteasomes fractionated by native PAGE (Figure 1B).
The temperature-sensitive cdc48-3 allele bears two point mutations, P257L and R387K (Jeffrey Laney, personal communication). The molecular effects of these mutations are not known, but the mutant protein is stable at the restrictive temperature (not shown). To assess more directly if the ATPase activity of Cdc48 contributed to degradation of proteasome-bound Ub conjugates, mutant cells were transformed with plasmids that expressed either wild type or ATPase-dead (pcdc48Q2) Cdc48 (Ye et al., 2003). Ub conjugate accumulation on 26S proteasomes isolated from cdc48-3 was suppressed by co-expression of wild type Cdc48 but was further enhanced in cells expressing the ATPase-deficient Cdc48 (Figure S1B).
To compare the protein composition of 26S proteasome complexes affinity-purified from wild type and cdc48-3 cells, we subjected the preparations to multidimensional mass spectrometry (MudPIT) (Graumann et al., 2004; Mayor et al., 2005) using an LTQ (Figures 1C and S1D) or LCQ (Figure S1C) mass spectrometer and measured spectrum counts, which correlate with protein abundance (Weiss et al., 2010). The ratio of total proteasomal subunit spectral counts from mutant to wildtype was close to 1, indicating consistent recovery of equivalent amounts of proteasome from wildtype and cdc48-3 cells. Moreover, the spectrum counts observed for individual subunits (Figure S1D) were similar for both preparations. We next determined the spectral counts for proteasome-interacting proteins (PIPs) in each preparation and calculated the ratio as above. Notably, and in striking contrast to proteasome subunits, spectrum counts for most PIPs were higher in cdc48-3 proteasomes and a large number of PIPs were found only in the mutant preparation (Figures 1C and S1C).
We reasoned that proteins found at elevated levels in proteasomes isolated from cdc48-3 cells might be substrates that could not be degraded due to incomplete unfolding or extraction from binding partners. To identify candidate substrates, we mined data from four datasets: (i) all proteins in yeast with a half-life of less than 55 min (Belle et al., 2006); (ii) proteins reported to be ubiquitinated in a large-scale proteomic study (Peng et al., 2003); (iii) proteins that accumulate as Ub conjugates upon UPS inhibition (Mayor et al., 2007) and (iv) proteins reported in the Saccharomyces Genome Database (SGD, www.yeastgenome.org) to have physical or genetic links with Ub ligases. Several PIPs were found to overlap between two datasets, an example being the septin-localized checkpoint kinase Hsl1 that is degraded via APC/C in G1 (Burton and Solomon, 2000). However, one candidate – the largest subunit of RNA polymerase II, Rpb1 – was identified in all four datasets. Although we chose to focus the remaining study on Rpb1, we did confirm that Hsl1 degradation was dependent on Cdc48 (Figure S2).
Rpb1 is ubiquitinated in the absence of any inducing signal (Daulny et al., 2008; Peng et al., 2003) and also upon stalling of transcription by drugs such as 6-azauracil (6-AU) that deplete intracellular nucleotide pools (Somesh et al., 2007). These data suggest that there may be basal turnover of Rpb1 fueled by stalling of transcription throughout the genome (Sigurdsson et al., 2010). However, upon induction of DNA damage by ultraviolet radiation (UV) or by the UV-mimetic 4-nitroquinoline-1-oxide (4-NQO), there is a large induction of Rpb1 turnover (Beaudenon et al., 1999; Chen et al., 2007; Ribar et al., 2006, 2007; Somesh et al., 2005). To evaluate whether Rpb1 accumulates on proteasomes in UV-treated cdc48-3 cells, we irradiated wild type (+/−MG132) and cdc48-3 cells with UV, isolated proteasomes, and immunoblotted for Rpb1. As shown in Figure 1D, MG132 induced accumulation of Rpb1 on proteasomes from UV-irradiated wildtype cells. Strikingly, there was also a strong accumulation of Rpb1 on proteasomes isolated from UV-irradiated cdc48-3 cells.
UV-induced degradation of Rpb1 is dependent on Cdc48
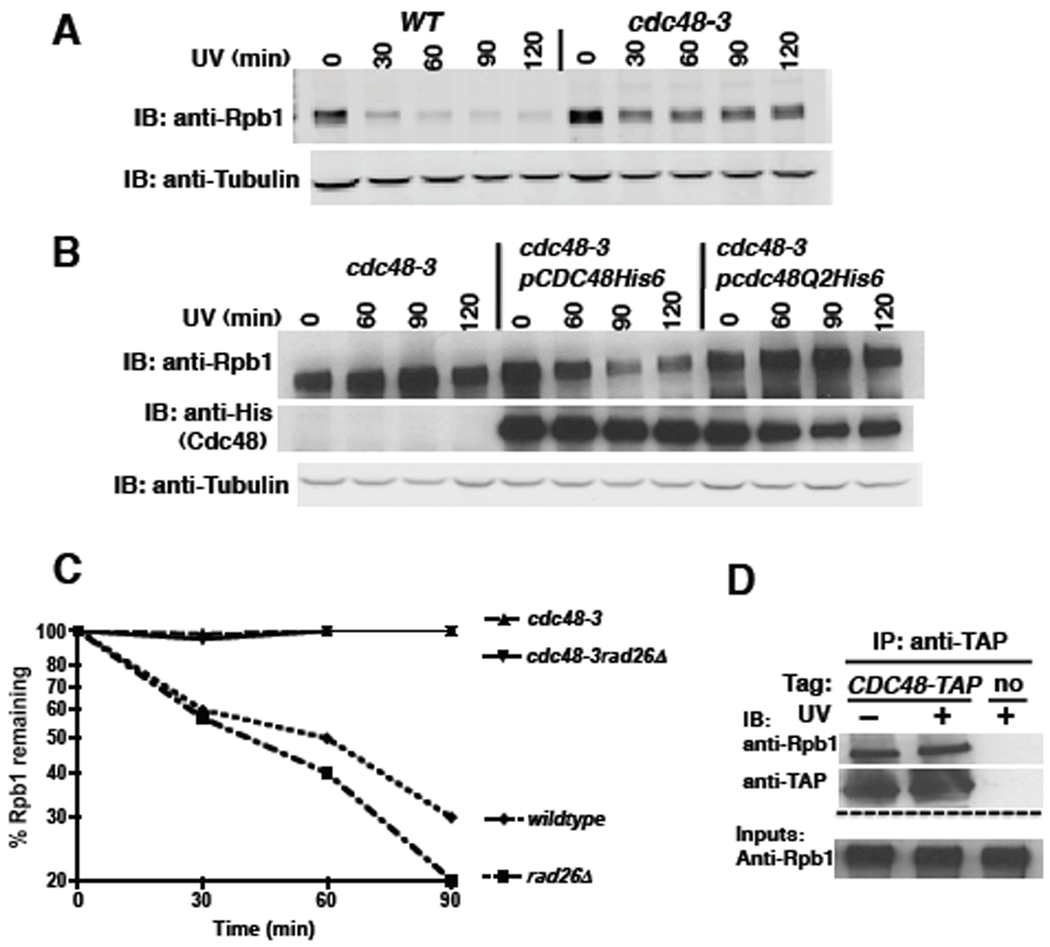
To address whether Cdc48 is involved in the degradation of Rpb1, we treated wildtype and cdc48-3 cells with UV and allowed them to recover in the presence of cycloheximide to block protein synthesis. Aliquots were collected at various time intervals and Rpb1 levels monitored by immunoblotting. Rpb1 was degraded upon UV irradiation of wildtype but not cdc48-3 mutant cells (Figure 2A). Quantification of the immunoblot and normalization with tubulin yielded data similar to Figure 2C (not shown). A similar stabilization was observed when Rpb1 turnover was induced by 4-NQO, and Rpb1 accumulated on 26S proteasomes isolated from 4-NQO-treated cdc48-3 cells to levels comparable to wildtype cells treated simultaneously with 4NQO and MG132 (Figures S3C, D). Stabilization of Rpb1 in UV-treated cdc48-3 was not due to arrest of this mutant at the mitosis checkpoint (Cheng and Chen, 2010), because we also observed stabilization in checkpoint-deficient cdc48-3mad2Δ mutants (Figure S3A) and in cdc48-3 at a temperature (30°C) that is permissive for mitosis (Figure S3B). To determine if the ATPase activity of Cdc48 was required for Rpb1 degradation, cdc48-3 mutants transformed with the same plasmids used for the experiment in Figure S1B were analyzed as described above. Whereas wildtype CDC48 rescued the degradation defect of the cdc48-3 allele (only allele tested), the ATPase mutant did not (Figure 2B).
Figure 2. The ATPase activity of Cdc48 is required for UV-induced turnover of the largest subunit of RNA Polymerase II, Rpb1.
(A) Rpb1 was stabilized in cdc48-3 cells irradiated with UV. WT (RJD360) and mutant (RJD3411) cells were shifted to 37°C for one hour, irradiated with UV, and transferred to medium containing 100 µg/ml cycloheximide. At the indicated time-points, cells were harvested and lysed for immunoblot (IB) analysis of Rpb1 using 8WG antibody. Tubulin served as the loading control.
(B) WT, but not ATPase-deficient Cdc48, restored Rpb1 turnover in UV-irradiated cdc48-3 cells. Mutant cdc48-3 (RJD3411) cells containing plasmid-borne WT GAL-CDC48His6 (RJD4996) or the Q2 mutant (RJD4997) were grown in galactose for two hours to induce expression of ectopic Cdc48. Induced cells were shifted to 35°C (the minimum restrictive temperatue in this medium) for one hour, UV irradiated, and then sampled at the indicated time-points.
(C) Stabilization of Rpb1 in cdc48-3 was not suppressed by rad26Δ. Single (RJD3411 and 4523) and double (RJD4570) mutants were shifted to 37°C for one hour, UV irradiated, and processed as above. Quantification was performed on LI-COR Odyssey with normalization to tubulin. Following quantification, all data were plotted using Prism software on a logarithmic scale on the y-axis.
(D) Cdc48 associated with Rpb1. Cells were UV treated or not and lysates prepared for immunoprecipitation (IP). Aliquots of inputs and immunoprecipitates (IP) were analyzed for their content of Cdc48 and Rpb1 by blotting (IB) with TAP and 4H8 antibodies, respectively.
See also Figure S3.
Prior work has shown that Rpb1 ubiquitination and degradation is compromised in UV-irradiated def1Δ cells. Surprisingly, Def1’s function in Rpb1 degradation can be fully bypassed if RAD26 is deleted, suggesting that Rad26, which is a DNA-dependent ATPase homologous to mammalian Cockayne Syndrome B (CSB), prevents access to the ubiquitination machinery (Woudstra et al., 2002).We wished to determine if Cdc48 might function indirectly to overcome a RAD26-dependent barrier to Rpb1 degradation. Whereas Rpb1 degradation was accelerated in rad26Δ cells, the cdc48-3 rad26Δ double mutant was as compromised for degradation as cdc48-3 alone (Figure 2C). Given that Cdc48 remained essential for Rpb1 turnover in rad26Δ cells, we reasoned that its role might be direct. To assess this, we performed a coimmunoprecipitation experiment (Figure 2D). Rpb1 was specifically co-precipitated with Cdc48, but their interaction was UV-independent. This observation suggests an additional, yet-to-be-discovered role for Cdc48 in Rpb1 transactions independent of UV damage (see below).
Dependency of Rpb1 degradation on Cdc48 adaptor proteins
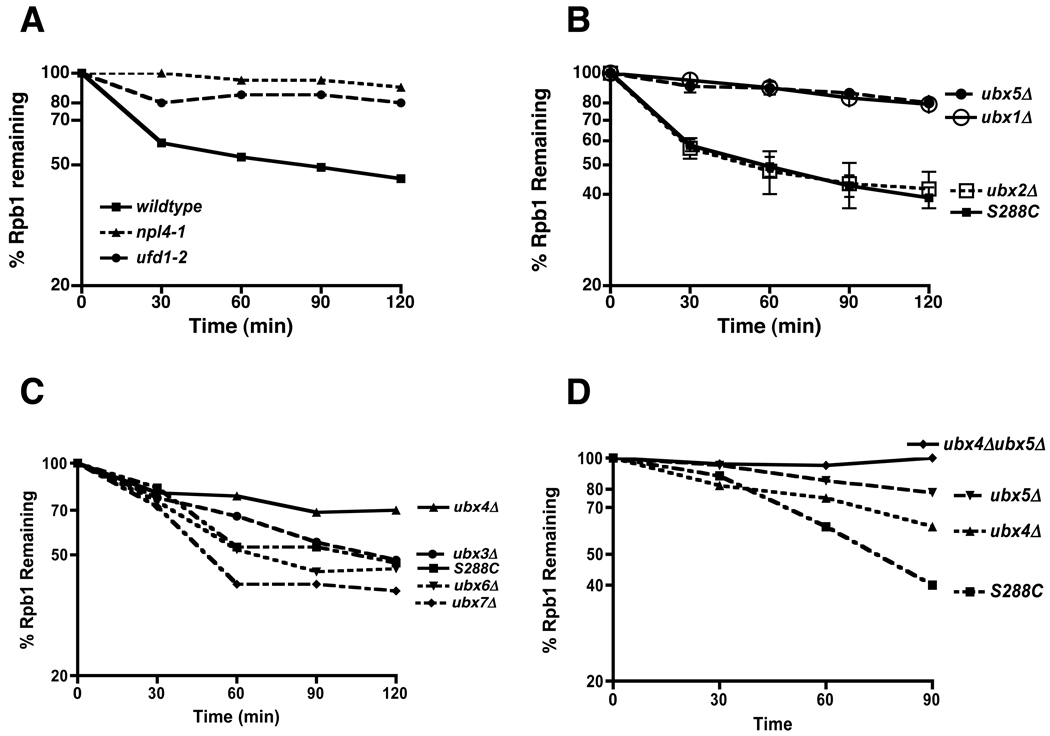
Cdc48 is a mechanochemical transducer that is coupled to its ubiquitinated substrates by adaptor proteins. As shown in Figure 3A, UV-dependent turnover of Rpb1 was also dependent on the Ufd1–Npl4 adaptor complex. Cdc48/p97 also interacts with a second set of putative substrate receptors, the UBX domain proteins. Shp1/Ubx1 and its mammalian homolog p47 are believed to be substrate recruitment factors for Cdc48/p97 that are mutually exclusive with Ufd1–Npl4 (Table S2), and Ubx1 is required for the degradation of UFD pathway reporter substrates in budding yeast (Schuberth et al., 2004). We evaluated UV-induced Rpb1 turnover in mutants individually lacking each one of the UBX proteins and found that Rpb1 was stabilized in ubx1Δ, ubx5Δ, and ubx4Δ, whereas degradation was unimpeded in ubx2Δ, ubx3Δ, ubx6Δ, and ubx7Δ mutants (Figures 3B and C). As will be shown below, the defect in ubx1Δ was due to an indirect effect on UV signaling and thus we focused our effort on Ubx4 and Ubx5. To determine if these proteins function in the same or parallel pathways, we quantified UV-induced Rpb1 turnover in single and double mutants. Rpb1 was more stable in ubx4Δ ubx5Δ than in either single mutant, suggesting that these proteins act in parallel to promote Rpb1 degradation (Figure 3D).
Figure 3. Multiple Cdc48 adaptors are involved in Rbp1 degradation.
(A) Npl4 and Ufd1 contributed to Rpb1 turnover. Wildtype and mutant cells (npl4-1/RJD2589 maintained at 30°C, and ufd1-2/RJD3264 shifted to 37°C for one hour) were UV irradiated at 0 min. Cultures were processed as described in Figure 2C.
(B) Ubx1 and Ubx5 contributed to Rpb1 turnover. WT (S288C) and mutant cells (RJD4614, 2551, 3176, 3177) maintained at 30°C were irradiated with UV at 0 min. Cultures were processed as described in Figure 2C. Results are presented as means ± SEM of three independent experiments.
(C) Ubx4 contributed to Rpb1 turnover. WT and mutant cells (RJD5249, RJD5246, RJD5247, RJD5248) maintained at 30°C were irradiated with UV at 0 min. Cultures were processed as described in Figure 2C.
(D) Increased impairment of Rpb1 turnover in ubx4Δubx5Δ double mutants compared to single mutants. WT and mutant cells (RJD5259) maintained at 30°C were irradiated with UV at 0 min. Cultures were processed as described in Figure 2C.
Ongoing transcription is a prerequisite for UV-induced RNA Pol II degradation (Anindya et al., 2007). To evaluate the possibility that the UBX proteins were involved in global transcription, we determined the sensitivity of the adaptor null mutants to the nucleotide-depleting drug 6-AU, which affects both elongation rate and processivity of RNA Pol II (Mason and Struhl, 2005). Whereas a null mutant lacking Dst1/TFIIS, a general transcription elongation factor, was 6-AU-sensitive, the ubxΔ mutants were 6-AU-insensitive (Figure S4A). Additionally, because Rpb1 degradation was significantly impaired in cdc48-3 mutants even at the semi-restrictive temperature 30°C (Figure S3B), we evaluated 6-AU-, and UV-sensitivity of cdc48-3 at 30°C. The sensitivities of the mutant were akin to wildtype (Figures S4A, B). Thus, transcription elongation and UV signaling were intact in cdc48-3 cells under conditions where UV-induced degradation of Rpb1 was severely compromised.
Ubiquitinated Rpb1 accumulates in cdc48-3, ubx4Δ, and ubx5Δ cells
We have shown above that Cdc48 and three of its UBX domain adaptors were required for Rpb1 turnover. The dependency on Cdc48–Ubx could reflect a requirement for segregase (Braun et al., 2002) activity such that the Rpb1 degron(s) remains masked through interaction with its holoenzyme partners in cdc48-3 or ubxΔ cells, blocking access to Ub ligases, or the dependency could reflect a requirement for segregase/unfolding activity post-ubiquitination. To assess the modification status of Rpb1 in mutants, we immunoprecipitated Rpb1 from control and UV-irradiated wildtype cells and then blotted the sample with an Rpb1 antibody. A discrete, UV-stimulated modification of a small fraction of Rpb1 was reproducibly observed (Figure S5A). This modification was unaffected by cdc48-3, ubx4Δ, and ubx5Δ mutations but was compromised in ubx1Δ mutants (see Inputs/asterisk, Figure 4). Prior work established that this modification is due to sumoylation (Chen et al., 2009). We confirmed that Rpb1 was sumoylated in cdc48-3 but not ubx1Δ cells following UV irradiation (Figure S5B), and that sumoylation did not affect Rpb1 degradation (Figure S5C). These data establish that the UV damage response remains intact in cdc48-3, ubx4Δ, and ubx5Δ but not ubx1Δ mutants.
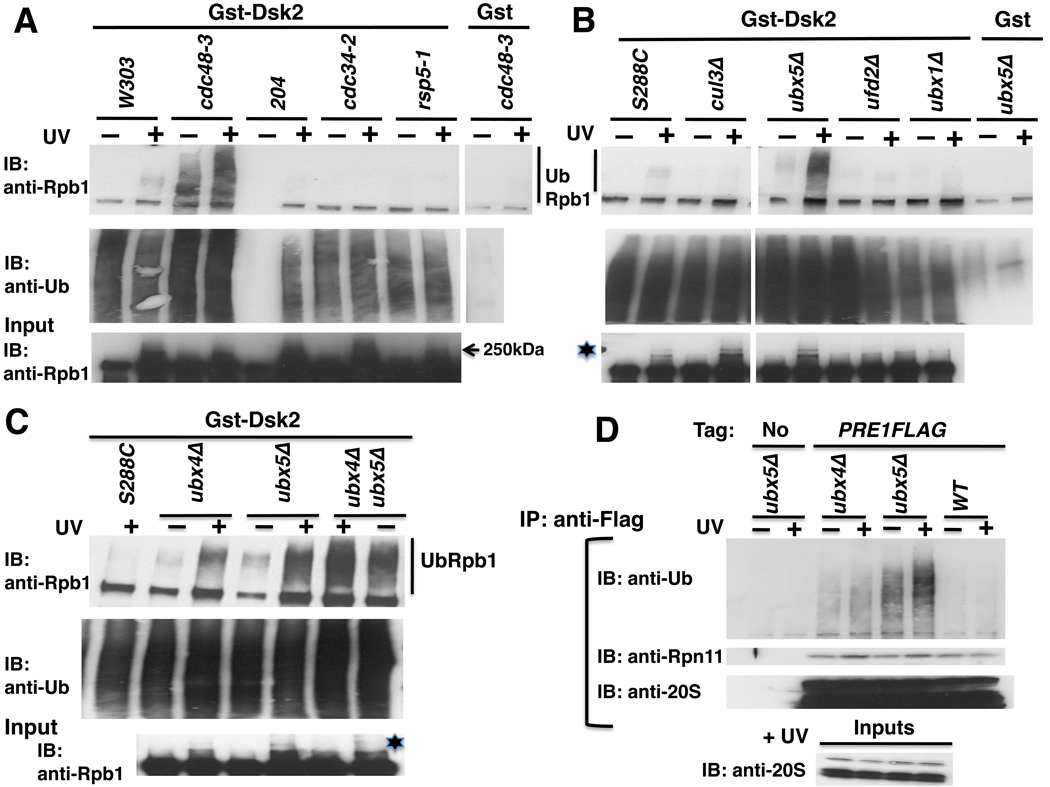
Figure 4. Rpb1 accumulates in the ubiquitinated state in cdc48-3, ubx4Δ, ubx5Δ and ubx4Δubx5Δ mutants.
(A) WT (W303 and 204) and mutant cells were shifted to 37°C for 90 minutes and UV irradiated. Lysates were fractionated on a GstDsk2 resin. The input extract and bound fractions were immunoblotted (IB) for Rpb1 (4H8) and Ub, as indicated. 204 is RJD487, a wild-type congenic to cdc43-2, and untreated sample and beads were partially lost during IP.
(B) Rpb1 Ub conjugates accumulated in ubx5Δ but not ubx1Δ, and were diminished in cul3Δ mutant cells. Methods same as (A) except that cells were maintained at 30°C. Asterisk indicates sumoylated Rpb1.
(C) Rpb1 Ub conjugates hyperaccumulated in a ubx4Δubx5Δ double mutan. Methods same as (A) except that cells were maintained at 30°C.
(D) Ub conjugates accumulated on proteasomes isolated from ubx4Δ and ubx5Δ mutants. Lysates from wildtype and mutant cells expressing Flag-tagged Pre1 (or not) were immunoprecipitated (IP) with anti-Flag and immunoblotted (IB) for Ub, Rpn11 and 20S. Input extracts were blotted for 20S.
See also Figure S5.
Typically, when degradation of a protein is blocked downstream of ubiquitination (e.g. with MG132), the fraction that accumulates as Ub conjugates is very small (Liu et al., 2007). To enhance our ability to detect the HMW polyubiquitinated pool of Rpb1 (UbRpb1), we exploited the ability of the UBA domain of Dsk2 to bind polyUb conjugates (Mayor et al., 2005). Lysates from wildtype and mutant cells were bound to GstDsk2. As expected, GstDsk2 retrieved Ub conjugates but Gst did not (middle panels, Figures 4A, B, C). Immunoblotting for Rpb1 revealed the accumulation of HMW conjugates in cdc48-3 cells. No accumulation of Rpb1 Ub conjugates was observed in either the Ub-conjugating enzyme mutant cdc34-2 or in the Ub ligase-deficient mutants rsp5-1 and cul3Δ. Cul3 generates the K48-linked Ub conjugates on Rpb1 that signal its degradation. It has been suggested that Cul3 functions either alone (Ribar et al., 2007) or downstream of Rsp5 (Harreman et al., 2009).
Amongst the ubxΔ mutants, UbRpb1 was recovered from UV-treated ubx4Δ and ubx5Δ cells but not ubx1Δ cells. Moreover, accumulation of constitutively ubiquitinated Rpb1 was also detected in cdc48-3, ubx4Δ, and ubx5Δ (Figures 4A, B and C), suggesting that Cdc48 and its adaptors act upon Rpb1 stalled at naturally occurring pause sites such as at the 5’ end of ORFs (Sigurdsson et al., 2010; Wade and Struhl, 2008) or at non-canonical DNA structures (Hanawalt and Spivak, 2008). Interestingly, accumulation of UbRpb1 was enhanced in ubx4Δubx5Δ double mutants compared to either single mutant (Figure 4C), providing further evidence that Ubx4 and Ubx5 function in parallel pathways. To determine if ubx4Δ or ubx5Δ elicits a more global defect in proteolysis, proteasomes were isolated from wildtype and mutant cells and immunoblotted for Ub conjugates. Strikingly, loss of either adaptor resulted in accumulation of HMW conjugates (Figure 4D), as was observed upon thermal inactivation of Cdc48 function (Figure 1A).
Ubx5 binds Rpb1 and Cul3 and functions downstream of Cul3
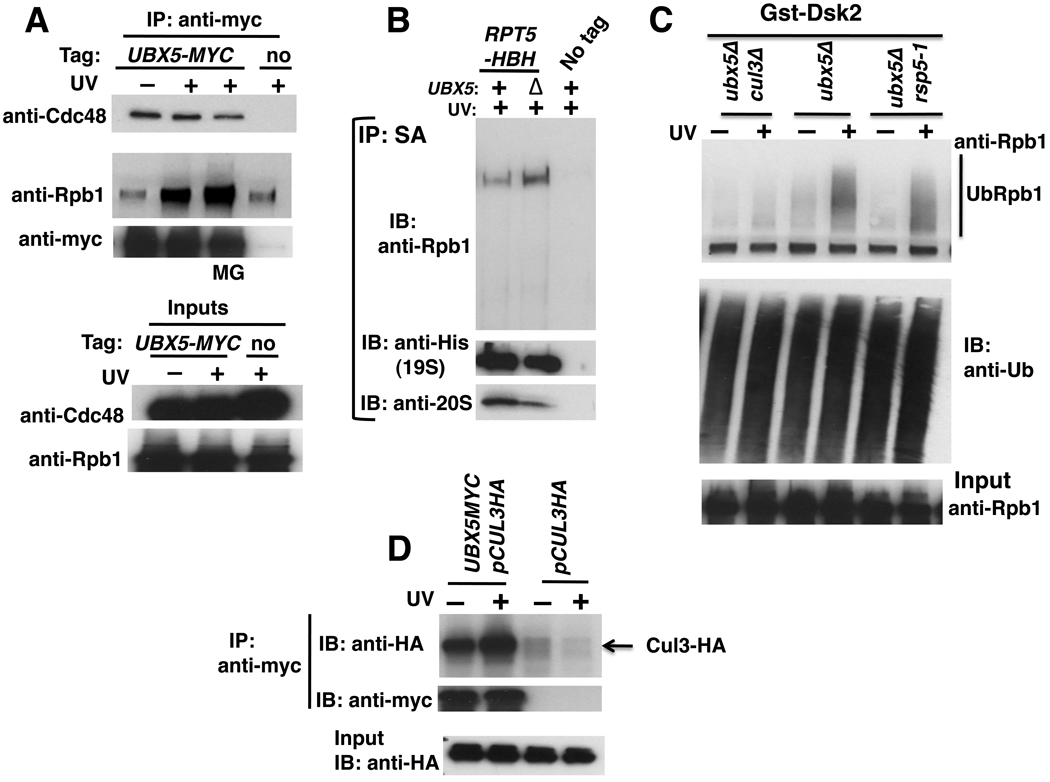
To investigate in greater detail how UBX protein function contributes to Rpb1 degradation, we focused our attention on Ubx5 because ubx5Δ exhibits a stronger phenotype than ubx4Δ in a) Rpb1 stabilization, b) UbRpb1 conjugate accumulation and c) Ub conjugate accumulation at the proteasome. Since Cdc48 bound Rpb1 (Figure 2D) we tested whether Ubx5 behaves likewise. Ubx5 bound Cdc48 constitutively, as expected, but its association with Rpb1 was strongly enhanced by UV irradiation (Figure 5A). This is an interesting counterpoint to Cdc48, which bound Rpb1 independently of DNA damage (Figure 2D). These observations suggest that Cdc48 engages in multiple transactions with Rpb1, with different transactions mediated by different adaptors. Additionally, as observed for cdc48-3 mutants, Rpb1 accumulation was enhanced on 26S proteasomes isolated under denaturing conditions from formaldehyde-crosslinked ubx5Δ cells (Figure 5B).
Figure 5. Ubx5–Cdc48 acts downstream of Cul3 in UV-dependent Rpb1 degradation.
(A) UV irradiation stimulated interaction of Ubx5 with Rpb1. Untagged (RJD4614), and UBX5-MYC (RJD4214) strains were UV irradiated or not, and lysates were immunoprecipitated (IP) with anti-myc antibody. Aliquots were blotted (IB) for Cdc48 and Rpb1 (using 4H8). MG: +MG132.
(B) Rpb1 accumulated on 26S proteasomes isolated from ubx5Δ cells. Cells expressing HBH-tagged Rpt5 (RJD4741 and RJD4742) were irradiated with UV and crosslinked with formaldehyde. Lysates were prepared in 6M guanidine-HCl and 26S proteasomes isolated by consecutive Ni2+-NTA and streptavidin (SA) affinity chromatography under denaturing conditions. Purified samples were blotted (IB) for Rpb1 (4H8), Rpt5-HBH (anti-His), and 20S (anti-a7).
(C) Ub conjugates that accumulated in ubx5Δ cells were substantially dependent on Cul3 but not Rsp5. Methods as in Figures 4A, B.
(D) Ubx5 and Cul3 interacted. Cells expressing HA-tagged Cul3 from a plasmid and Myc-tagged Ubx5 were irradiated with UV, or not. Input extracts and anti-myc immunoprecipitates (IP) were analyzed for their content of Cul3 by immunoblotting (IB) with anti-HA.
As noted above, both Rsp5 and Cul3 have been implicated in Rpb1 degradation. To test whether Ubx5 functioned downstream of one or both of these Ub ligases, we evaluated UbRpb1 levels in ubx5Δ mutants lacking one or the other enzyme. Interestingly, cul3Δ but not rsp5-1 severely attenuated recovery of UbRpb1 from ubx5Δ cells (Figure 5C). Given that Cul3 and Ubx5 could be coimmunoprecipitated (Figure 5D), we suggest that these proteins normally act sequentially, such that Rpb1 that has been ubiquitinated by Cul3 is ‘handed off’ to Ubx5. These data extend observations made in human cells, where UBXD7 is linked to the CUL2–VHL complex and its substrate HIF1α (Alexandru et al., 2008). Bioinformatic analysis suggests that Ubx5 is the ortholog of UBXD7 (Schuberth and Buchberger, 2008).
Accumulation of ubiquitinated Rpb1, Cdc48, and proteasome on chromatin
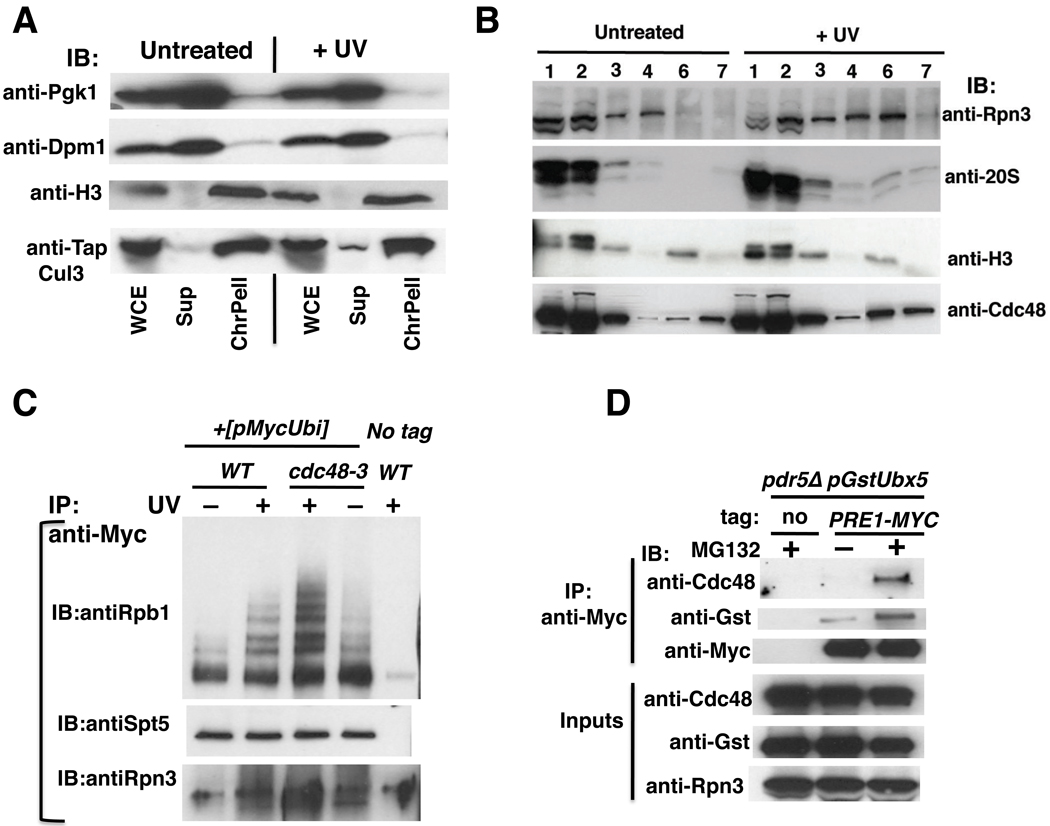
The results presented thus far suggest that Ubx5–Cdc48 acts on Rpb1, downstream of Cul3, to mediate degradation of ubiquitinated Rpb1 formed upon UV irradiation. The question that arises next is whether this sequence of events occurs on chromatin. If so, we would expect to detect the core components of the system – including Cul3, Cdc48, and the proteasome – on chromatin in UV-treated cells. To address this question, we employed a standard chromatin fractionation method (Liang and Stillman, 1997). Interestingly, the bulk of Cul3 fractionated with chromatin (Figure 6A). Cul3 was constitutively associated with chromatin, which is not surprising given that this enzyme is likely to have multiple substrates. To stringently probe the association of the abundant Cdc48 and proteasome complexes with chromatin, we subjected a crude chromatin pellet to limited micrococcal nuclease (MNase) digestion followed by centrifugation to yield a highly purified chromatin pellet consisting of polynucleosomes (Frc6), and the supernatant (Frc7). Frc6, which provides a stringent assessment of chromatin association (Liang and Stillman, 1997), was enriched for Cdc48 as well as both 19S and 20S proteasome components following UV irradiation, whereas Histone H3 was constitutively present and served as a loading control (Figure 6B). Given that both Cdc48 and the proteasome are likely to have many substrates, our data suggest that UV damage triggers a generalized response that may result in the removal or degradation of many chromatin proteins.
Figure 6. Ubiquitinated Rpb1 accumulates on chromatin in UV-treated cdc48-3 mutant.
(A) CUL3-TAP (RJD4680) cells were treated with UV or not, and harvested after the addition of 0.1% azide. Spheroplasts were prepared using 25 units of Zymolyase 100T for 2 × 109 cells. The three fractions: Whole cell extract (WCE), supernatant (Sup), and chromatin pellet (ChrPell) derived from spheroplasts were immunoblotted (IB) with the indicated antibodies.
(B) Cdc48 and 26S proteasome recruitment to chromatin was enhanced following UV. Crude chromatin pellets (Frc3) obtained by centrifuging WCE (Frc1) through a sucrose cushion were fractionated further following limited digestion with micrococcal nuclease (MNase) to generate high-speed pellet (Frc6) and supernatant (Frc7) fractions. Frc6 is enriched in released polynucleosomes. All fractions were immunoblotted (IB) for histone H3, 19S (Rpn3), 20S proteasome subunits, and Cdc48. Fraction numbers correspond to lane numbers.
(C) Ubiquitinated Rpb1 accumulated on chromatin in response to UV and inactivation of Cdc48. WT and mutant cells expressing Myc-tagged Ub were shifted to 37°C, and Cu2+ was added to induce expression of Ub. After two hours, cultures were irradiated with UV (or not) and harvested. Crude chromatin pellets (FrC) were isolated as in (B) above and treated with Benzonase. Solubilized material was immunoprecipitated (IP) with anti-myc, and aliquots were blotted (IB) for Rpb1 (4H8), Spt5, and Rpn3.
(D) Cells of the indicated genotype were galactose-induced for two hours, then treated (or not) with MG132 for 30 min, lysed, and subjected to immunoprecipitation with anti-myc. The resulting samples were immunoblotted with antibodies to detect the indicated antigens. pGST-UBX5 is a plasmid that expresses GST-tagged Ubx5 under the GAL promoter.
See also Figure S6
Based on the observation that Cul3, Cdc48, and the proteasome can all be found on chromatin in UV-stressed cells, we hypothesized that ubiquitination and degradation of Rpb1 occurs on chromatin. To test this idea, we examined chromatin-bound Rpb1. Crude chromatin (see Figures S6A, S6B for fractionation and loading controls) isolated from cells that expressed Myc-tagged Ub was solubilized with Benzonase and ubiquitinated chromatin proteins released into the supernatant were isolated by virtue of the Myc tag. Immunoblotting for Rpb1 revealed the presence of Ub ladders on chromatin-associated Rpb1 (Figure 6C). Importantly, the abundance of chromatin-bound Rpb1 Ub conjugates was greatly increased upon UV irradiation of wild type cells, and increased to even higher levels in UV irradiated cdc48-3 cells. Thus, it appears that Rpb1 is ubiquitinated on chromatin, consistent with the idea that the degradation cascade initiates with ubiquitination of Rpb1 arrested at a thymidine dimer.
Since the recruitment of both Cdc48 and the 26S proteasome to chromatin was enhanced following UV, we wished to determine if both complexes could be coimmunoprecipitated. Earlier work had relied on in vivo crosslinking to detect binding of Cdc48 to the proteasome (Guerrero et al., 2008). Immunoprecipitation of 26S proteasome under native conditions in the presence of ATP and 0.15M salt resulted in barely-detectable recovery of Cdc48 (Figure 6D), although Ubx5 could be readily detected after mild overproduction. Treating cells with the proteasome inhibitor MG132 enhanced interaction of both Cdc48 and Ubx5 with the proteasome, suggesting that binding was mediated in part by ubiquitinated substrates. This is consistent with the data in Figure 1A and supports the idea that Cdc48 can act upon substrates bound to the proteasome to enable their degradation.
DISCUSSION
Implicit in the models for Cdc48/p97 function based on its well-documented role in retrotranslocation of ubiquitinated proteins from the ER into the cytosol (Hirsch et al., 2009; Raasi and Wolf, 2007) and in the degradation of Ub fusion proteins (Richly et al., 2005) is the notion that it functions upstream of the proteasome shuttle receptors Rad23 and Dsk2. These models imply that Cdc48 is required for delivery of a subset of UPS substrates (i.e. Cdc48/p97-dependent substrates) to the proteasome. Our data in the current study demonstrating accumulation of Ub conjugates and PIPs on proteasomes isolated from cdc48-3 mutants challenge whether these models serve as a general paradigm for the temporal staging of Cdc48 function. Here, we validated Rpb1 as a bona-fide substrate of Cdc48 and show that in cells in which Rpb1 degradation was induced by UV radiation, Rpb1 was delivered to the proteasome in the absence of Cdc48 function.
The pathway for degradation of Rpb1 in UV-treated cells
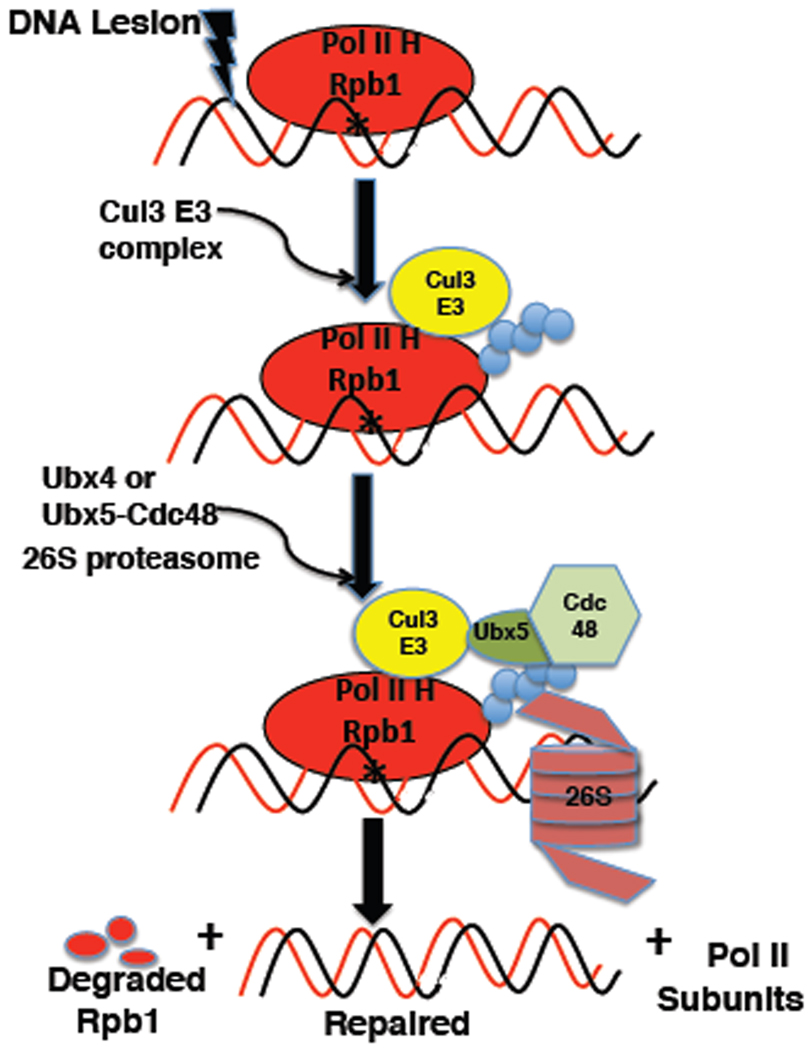
Our model for Rbp1 degradation (Figure 7) is that Rpb1 stalled at lesion sites is most often dislocated from the damage to enable repair followed by resumption of transcription. However, Rpb1 that is persistently blocked or refractory to dislocation is eventually modified by the Ub ligases Rsp5 and Cul3, of which the latter is believed to decorate Rpb1 with degradation-competent Ub chains. These chains enable recruitment of both Cdc48 and 26S proteasomes to the stalled complex, resulting in degradation of Rpb1 and recycling of the other holoenzyme subunits, which are not degraded (Malik et al., 2008). We envision that Cdc48 mediates extraction of UbRpb1 from chromatin-bound Pol II holoenzyme and that this extraction and subsequent Rpb1 degradation are tightly coupled, possibly even contemporaneous. Cdc48 carries out its function in conjunction with Ufd1–Npl4, Ubx4, and Ubx5. We do not know how the activities of Ufd1–Npl4 relate to those of the Ubx proteins. On the other hand, Ubx4 and Ubx5 appear to act in parallel pathways to promote Rpb1 degradation.
Figure 7. Hypothetical model for damage-dependent turnover of Rpb1.
The Rpb1 subunit of RNA Pol II holoenzyme (H) irreversibly stalled at sites of DNA lesions is ubiquitinated by the Cul3–RING ligase complex. UbRpb1 can independently recruit proteasome and Ubx5-Cdc48 complexes. UbRpb1 is extracted from its binding partners in an unfolding reaction dependent on Ubx4 or Ubx5-Cdc48 and is threaded into the 26S proteasome.
It has been proposed that Rsp5 attaches an initiator Ub to Rpb1, which primes polymerization of an Ub chain by Cul3 (Harreman et al., 2009). Alternatively, it has been suggested that Cul3 carries out the entire reaction and the role of Rsp5 is indirect (Ribar et al., 2007). Ubiquitinated Rpb1 that accumulated in ubx5Δ cells was greatly diminished by deletion of CUL3 but barely affected by rsp5-1. Either Rsp5 is not required to generate the UbRpb1 that accumulates in ubx5Δ or the rsp5-1 mutation was leaky under our conditions. Regardless of whether or not Cul3 extends Ub chains initiated by Rsp5, the epistasis data and the protein-protein interactions point to Ubx5 acting directly upon ubiquitinated Rpb1 formed by Cul3.
Components involved in the Rpb1 turnover pathway have been linked with chromatin regulation in other contexts. Cul3 functions as part of an Ub ligase that ubiquitinates the UV-damage sensor Rad4 (XPC)(Gillette et al., 2006; Ramsey et al., 2004). Cdc48 promotes dissociation of a repressor from its target promoter (Wilcox and Laney, 2009) and p97 extracts protein kinase Aurora B from chromatin during nuclear envelope reassembly (Ramadan et al., 2007). Similarly, 19S and 20S subunits of the proteasome have been shown to bind promoters, ORFs, and termination regions, both constitutively and in response to signals such as HO endonuclease-induced double strand breaks and UV (Auld et al., 2006; Collins and Tansey, 2006; Gillette et al., 2004; Krogan et al., 2004). These data suggest that extraction of proteins from chromatin by Cdc48/p97 – either coupled to proteasomal degradation or not – is likely to be a recurrent theme in chromatin regulation.
Proteasomal ATPases are insufficient for segregation and unfolding of unstable subunits of some macromolecular complexes: Implications for dependency on Cdc48
An unexpected insight from our work is that in cells deprived of Ubx5-Cdc48 function, substrates such as Rpb1 can gain access to the proteasome. This raises the question of why substrates accumulate on the proteasome in these cells? We suggest that extraction of UbRpb1 from chromatin-bound holoenzyme and targeting of UbRpb1 to the proteasome are normally coupled processes that do not occur obligately in a fixed order. Thus, depending upon the relative rates of disassembly versus targeting, in some instances the action of Cdc48 may precede substrate association with the proteasome (but this may be undesirable as explained below), whereas in other cases the substrate may associate with proteasome before Cdc48 can act. We suggest that the latter occurs far more commonly because disassembly/unfolding is likely to be much slower than targeting. For some substrates, like Rpb1 and the checkpoint kinase Hsl1, the disassembling and/or unfolding activity contributed by the resident proteasomal ATPases (Rpt1-6,(Finley, 2009)) may be insufficient to extract them from the macromolecular complexes in which they reside and thus they must wait for Cdc48 to complete its job before their degradation can commence. Coupling Cdc48 function to substrate degradation at the proteasome surface would reduce the probability that substrate that has been disassembled/unfolded by Cdc48 has an opportunity to reassemble, refold, or aggregate.
What is the molecular basis for determining whether or not a particular substrate requires Cdc48 for degradation? Prior work has established that stable domains within proteins are unfolded sequentially starting from the attachment point of the degradation signal (Lee et al., 2001; Prakash et al., 2004; Schrader et al., 2009). Rpb1 bound to multiple interacting proteins in the Pol II holoenzyme complex may lack an unstructured initiation site and thus may additionally require Cdc48 for preprocessing, as recently reported for stably folded UPS reporter substrates (Beskow et al., 2009). In stark contrast, the unfolding power of the 26S proteasome is sufficient to degrade to completion ubiquitinated Sic1 (UbSic1), even when it is tightly bound to S phase cyclin-dependent kinase (Verma et al., 2001). A potentially distinguishing feature of Sic1 is that it contains an intrinsically disordered N-terminal domain that mediates interaction with Ub ligase SCF (Mittag et al., 2008) and contains the primary sites of ubiquitination (Petroski and Deshaies, 2003). We speculate that because unraveling of UbSic1 initiates from the disordered region proximal to the Ub chain attachment site, the proteasome can complete the task without assistance. By contrast, we predict that Cdc48/p97 and its adaptors are required to sustain disassembly, unfolding, and processive degradation of soluble 26S proteasome substrates whose structural complexity overwhelms the proteasomal ATPases.
EXPERIMENTAL PROCEDURES
Turnover Analysis of UPS substrates
Aliquots of cultures treated as described in the respective figure legends (final A600 between 1–2) were collected and drop-frozen in liquid nitrogen. Frozen cell pellets were thawed and washed with ice-cold Buffer A (50 mM Tris, pH 7.5, 10 mM sodium azide, 10 mM EDTA, 10 mM EGTA, 1X protease inhibitor tablet (Roche), 10 mM NEM, 50 mM NaF, 60 mM β-glycerophosphate, 10 mM sodium pyrophoshate). The cell pellets were then immersed in boiling water for three minutes after which they were suspended in 1X SDS buffer (37.5 µl /O.D. unit). An equal volume of glass beads (Sigma, 425–600 microns, acid washed) was added, and cells were lysed by vortexing in Fast Prep-24 (MP) for 45 seconds at a setting of 6.5, and boiled again for 4 mins. Boiled lysates were centrifuged at 16,000 X g for 1 minute. Aliquots were resolved by SDS-PAGE, transferred to nitrocellulose, and stained with Ponceau S to determine equivalent loading of protein extracts. The nitrocellulose filters were immunoblotted with desired antibody and developed by ECL, or quantified by LI-COR Odyssey using IR dye-linked secondary antibodies (Invitrogen). Anti-PSTAIRE (Santa Cruz), and anti-tubulin (Sigma) served as loading controls. Following quantification, all data were plotted using Prism software on a logarithmic scale on the y-axis. Anti-Rpb1 (clones 8WG, 4H8), anti-Myc, and anti-HA antibodies were from Covance.
UV and 4-NQO treatment of yeast cells
Overnight cultures were diluted to an optical density (O.D.) of around 0.2. When cells reached an O.D. around 1, they were centrifuged and resuspended in 80% of the original culture volume in 2% Dextrose (or the sugar being used for the experiment). All the following steps were carried out in the dark, under red safe light. Cultures were exposed to 400 J/m2 of UV irradiation in either petri dishes, or glass trays, depending on volume. Irradiation was performed by using calibrated germicidal lamps (254 nm UV Lamp, UVP, Model # XX-405) or a pre-warmed and calibrated Stratalinker unit (Stratagene, Model 2400). The UV meter used for calibration was purchased from UVP, Inc, CA (Model # J225).
Cells were harvested immediately after UV, or when recovery was monitored, the 2% dextrose suspension was diluted into 20 % volume of 5X prewarmed media containing 100 ug /ml cycloheximde, and outgrowth continued in the dark. For 4-NQO treatment, exponential cultures were treated with the desired concentration from a 10 mg /ml stock stored in the dark.
Isolation of Chromatin Fraction
Essentially, the method of Liang and Stillman (Liang and Stillman, 1997) was followed with some modifications. In the original protocol, chromatin was isolated from yeast spheroplasts and this method was followed in Figure 6A. Because Rpb1 was considerably degraded during the process of spheroplasting (particularly in UV-treated cultures), the method was modified to isolate chromatin fractions from yeast cells ground with a mortar pestle chilled in liquid nitrogen as described earlier (Verma and Deshaies, 2005) for Figures 6B and 6C. The minimum culture volume that could be ground with easy recovery of cell powder was 350 ml. Ground powder was weighed, and 2X (wt /volume) Extraction Buffer (EB) containing 50 mM Hepes, pH 7.5, 100 mM KCl, 0.25 % Triton, 2.5 mM MgCl2, 25 mM NEM, 50 mM β-glycerophosphate, 5 mM sodium pyrophosphate, 1X protease inhibitor tablet +EDTA, 0.5 mM AEBSF was added. Unlysed cells were pelleted at 5000 r.p.m (all centrifugations in refrigerated Eppendorf model 5417R) for 1 min. Ubiquitin-aldehyde (1 µM) was added to the supernatant and an aliquot (25 % vol/vol) was saved as WCE (Frc1). Another 25 % of lysate was underlayered with 50 % volume of 30 % sucrose and centrifuged at 16,4000 rpm for 15 minutes. The pellet was saved as low speed pellet (Frc3) and the supernatant was designated “low-speed supernatant” (Frc2). The remaining 50 % of lysate was underlayered with sucrose and centrifuged to generate a second set of Fractions 2 and 3. Crude Fraction 3 was washed with EB at 12,000 rpm for 8 mins and the pellet was resuspended in EB. Resuspended chromatin pellet (375 µl) was pre-warmed to 37°C for 3 min after supplementing with CaCl2 (2 mM final) and digested with 2 µl 1:10 MNase (50 % glycerol stock solution; from Sigma, 204.3 Units / mg protein) for another 3 min. Digestion was stopped by the addition of EGTA to 5 mM. A low-speed micrococcal nuclease-treated pellet (Frc4) was generated by centrifuging at 10,000 rpm for 2 min at 4°C. The supernatant was centrifuged at 50,000 r.p.m. (Sorvall RC M120EX, Rotor # RP100AT4) for one hour to generate the high-speed pellet (Frc6), and supernatant (Frc7). Limited nuclease digestion retains chromatin proteins in the high-speed pellet fraction.
Solubilization of Chromatin
Washed crude chromatin pellet (FrC) was re-suspended in EB (750 µl) and NaCl was added to yield a final salt concentration of 0.6M. Ubiquitin aldehyde (1 µM) and benzonase (1 µl of 250 U/µl, Novagen) were added and the sample was incubated for 30 min on ice, sonicated for 15 sec (Amplitude at 20%), and then centrifuged at 10,000 rpm for 2 min. The supernatant was used for immunoprecipitations.
Supplementary Material
Acknowledgements
We thank D. G. Drubin, G. Hartzog, M. Hochstrasser, J. Huibregtse, E. Johnson, D. Kellogg, T. Miyakawa, Y. Saeki, W. Seufert, P. Silver, T. Sommer, A. Toh-E, A. Varshavsky, F. Winston and Y.Ye for yeast strains, expression plasmids, and antibodies. We thank the members of the Deshaies lab for helpful discussions. R.J.D. is an Investigator of the Howard Hughes Medical Institute, which supported this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alberts SM, Sonntag C, Schafer A, Wolf DH. Ubx4 modulates cdc48 activity and influences degradation of misfolded proteins of the endoplasmic reticulum. J Biol Chem. 2009;284:16082–16089. doi: 10.1074/jbc.M809282200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandru G, Graumann J, Smith GT, Kolawa NJ, Fang R, Deshaies RJ. UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1alpha turnover. Cell. 2008;134:804–816. doi: 10.1016/j.cell.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anindya R, Aygun O, Svejstrup JQ. Damage-induced ubiquitylation of human RNA polymerase II by the ubiquitin ligase Nedd4, but not Cockayne syndrome proteins or BRCA1. Mol Cell. 2007;28:386–397. doi: 10.1016/j.molcel.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Auld KL, Brown CR, Casolari JM, Komili S, Silver PA. Genomic association of the proteasome demonstrates overlapping gene regulatory activity with transcription factor substrates. Mol Cell. 2006;21:861–871. doi: 10.1016/j.molcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Barbin L, Eisele F, Santt O, Wolf DH. The Cdc48-Ufd1-Npl4 complex is central in ubiquitin-proteasome triggered catabolite degradation of fructose-1,6-bisphosphatase. Biochem Biophys Res Commun. 2010;394:335–341. doi: 10.1016/j.bbrc.2010.03.005. [DOI] [PubMed] [Google Scholar]
- Beaudenon SL, Huacani MR, Wang G, McDonnell DP, Huibregtse JM. Rsp5 ubiquitin-protein ligase mediates DNA damage-induced degradation of the large subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:6972–6979. doi: 10.1128/mcb.19.10.6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belle A, Tanay A, Bitincka L, Shamir R, O'Shea EK. Quantification of protein half-lives in the budding yeast proteome. Proc Natl Acad Sci U S A. 2006;103:13004–13009. doi: 10.1073/pnas.0605420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beskow A, Grimberg KB, Bott LC, Salomons FA, Dantuma NP, Young P. A conserved unfoldase activity for the p97 AAA-ATPase in proteasomal degradation. J Mol Biol. 2009;394:732–746. doi: 10.1016/j.jmb.2009.09.050. [DOI] [PubMed] [Google Scholar]
- Braun S, Matuschewski K, Rape M, Thoms S, Jentsch S. Role of the ubiquitin-selective CDC48(UFD1/NPL4)chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J. 2002;21:615–621. doi: 10.1093/emboj/21.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton JL, Solomon MJ. Hsl1p, a Swe1p inhibitor, is degraded via the anaphase-promoting complex. Mol Cell Biol. 2000;20:4614–4625. doi: 10.1128/mcb.20.13.4614-4625.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Ding B, LeJeune D, Ruggiero C, Li S. Rpb1 sumoylation in response to UV radiation or transcriptional impairment in yeast. PLoS One. 2009;4:e5267. doi: 10.1371/journal.pone.0005267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Ruggiero C, Li S. Yeast Rpb9 plays an important role in ubiquitylation and degradation of Rpb1 in response to UV-induced DNA damage. Mol Cell Biol. 2007;27:4617–4625. doi: 10.1128/MCB.00404-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YL, Chen RH. The AAA-ATPase Cdc48 and cofactor Shp1 promote chromosome bi-orientation by balancing Aurora B activity. J Cell Sci. 2010;123:2025–2034. doi: 10.1242/jcs.066043. [DOI] [PubMed] [Google Scholar]
- Collins GA, Tansey WP. The proteasome: a utility tool for transcription? Curr Opin Genet Dev. 2006;16:197–202. doi: 10.1016/j.gde.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Daulny A, Geng F, Muratani M, Geisinger JM, Salghetti SE, Tansey WP. Modulation of RNA polymerase II subunit composition by ubiquitylation. Proc Natl Acad Sci U S A. 2008;105:19649–19654. doi: 10.1073/pnas.0809372105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daulny A, Tansey WP. Damage control: DNA repair, transcription, and the ubiquitin-proteasome system. DNA Repair (Amst) 2009;8:444–448. doi: 10.1016/j.dnarep.2009.01.017. [DOI] [PubMed] [Google Scholar]
- DeBose-Boyd RA. Feedback regulation of cholesterol synthesis: sterol-accelerated ubiquitination and degradation of HMG CoA reductase. Cell Res. 2008;18:609–621. doi: 10.1038/cr.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decottignies A, Evain A, Ghislain M. Binding of Cdc48p to a ubiquitin-related UBX domain from novel yeast proteins involved in intracellular proteolysis and sporulation. Yeast. 2004;21:127–139. doi: 10.1002/yea.1071. [DOI] [PubMed] [Google Scholar]
- Elsasser S, Chandler-Militello D, Muller B, Hanna J, Finley D. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J Biol Chem. 2004;279:26817–26822. doi: 10.1074/jbc.M404020200. [DOI] [PubMed] [Google Scholar]
- Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislain M, Dohmen RJ, Levy F, Varshavsky A. Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cerevisiae. EMBO J. 1996;15:4884–4899. [PMC free article] [PubMed] [Google Scholar]
- Gillette TG, Gonzalez F, Delahodde A, Johnston SA, Kodadek T. Physical and functional association of RNA polymerase II and the proteasome. Proc Natl Acad Sci U S A. 2004;101:5904–5909. doi: 10.1073/pnas.0305411101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillette TG, Yu S, Zhou Z, Waters R, Johnston SA, Reed SH. Distinct functions of the ubiquitin-proteasome pathway influence nucleotide excision repair. EMBO J. 2006;25:2529–2538. doi: 10.1038/sj.emboj.7601120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann J, Dunipace LA, Seol JH, McDonald WH, Yates JR, 3rd, Wold BJ, Deshaies RJ. Applicability of tandem affinity purification MudPIT to pathway proteomics in yeast. Mol Cell Proteomics. 2004;3:226–237. doi: 10.1074/mcp.M300099-MCP200. [DOI] [PubMed] [Google Scholar]
- Guerrero C, Milenkovic T, Przulj N, Kaiser P, Huang L. Characterization of the proteasome interaction network using a QTAX-based tag-team strategy and protein interaction network analysis. Proc Natl Acad Sci U S A. 2008;105:13333–13338. doi: 10.1073/pnas.0801870105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halawani D, Latterich M. p97: The cell's molecular purgatory? Mol Cell. 2006;22:713–717. doi: 10.1016/j.molcel.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- Harreman M, Taschner M, Sigurdsson S, Anindya R, Reid J, Somesh B, Kong SE, Banks CA, Conaway RC, Conaway JW, et al. Distinct ubiquitin ligases act sequentially for RNA polymerase II polyubiquitylation. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0907052106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch C, Gauss R, Horn SC, Neuber O, Sommer T. The ubiquitylation machinery of the endoplasmic reticulum. Nature. 2009;458:453–460. doi: 10.1038/nature07962. [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Janiesch PC, Kim J, Mouysset J, Barikbin R, Lochmuller H, Cassata G, Krause S, Hoppe T. The ubiquitin-selective chaperone CDC-48/p97 links myosin assembly to human myopathy. Nat Cell Biol. 2007;9:379–390. doi: 10.1038/ncb1554. [DOI] [PubMed] [Google Scholar]
- Jarosch E, Taxis C, Volkwein C, Bordallo J, Finley D, Wolf DH, Sommer T. Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat Cell Biol. 2002;4:134–139. doi: 10.1038/ncb746. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Gonda DK, Varshavsky A. cis-trans recognition and subunit-specific degradation of short-lived proteins. Nature. 1990;346:287–291. doi: 10.1038/346287a0. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Ma PC, Ota IM, Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem. 1995;270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Lam MH, Fillingham J, Keogh MC, Gebbia M, Li J, Datta N, Cagney G, Buratowski S, Emili A, et al. Proteasome involvement in the repair of DNA double-strand breaks. Mol Cell. 2004;16:1027–1034. doi: 10.1016/j.molcel.2004.11.033. [DOI] [PubMed] [Google Scholar]
- Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- Liang C, Stillman B. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Apodaca J, Davis LE, Rao H. Proteasome inhibition in wild-type yeast Saccharomyces cerevisiae cells. Biotechniques. 2007;42:158, 160, 162. doi: 10.2144/000112389. [DOI] [PubMed] [Google Scholar]
- Madeo F, Schlauer J, Zischka H, Mecke D, Frohlich KU. Tyrosine phosphorylation regulates cell cycle-dependent nuclear localization of Cdc48p. Mol Biol Cell. 1998;9:131–141. doi: 10.1091/mbc.9.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Bagla S, Chaurasia P, Duan Z, Bhaumik SR. Elongating RNA polymerase II is disassembled through specific degradation of its largest but not other subunits in response to DNA damage in vivo. J Biol Chem. 2008;283:6897–6905. doi: 10.1074/jbc.M707649200. [DOI] [PubMed] [Google Scholar]
- Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Mayor T, Graumann J, Bryan J, MacCoss MJ, Deshaies RJ. Quantitative profiling of ubiquitylated proteins reveals proteasome substrates and the substrate repertoire influenced by the Rpn10 receptor pathway. Mol Cell Proteomics. 2007;6:1885–1895. doi: 10.1074/mcp.M700264-MCP200. [DOI] [PubMed] [Google Scholar]
- Mayor T, Lipford JR, Graumann J, Smith GT, Deshaies RJ. Analysis of polyubiquitin conjugates reveals that the Rpn10 substrate receptor contributes to the turnover of multiple proteasome targets. Mol Cell Proteomics. 2005;4:741–751. doi: 10.1074/mcp.M400220-MCP200. [DOI] [PubMed] [Google Scholar]
- Mittag T, Orlicky S, Choy WY, Tang X, Lin H, Sicheri F, Kay LE, Tyers M, Forman-Kay JD. Dynamic equilibrium engagement of a polyvalent ligand with a single-site receptor. Proc Natl Acad Sci U S A. 2008;105:17772–17777. doi: 10.1073/pnas.0809222105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuber O, Jarosch E, Volkwein C, Walter J, Sommer T. Ubx2 links the Cdc48 complex to ER-associated protein degradation. Nat Cell Biol. 2005;7:993–998. doi: 10.1038/ncb1298. [DOI] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Context of multiubiquitin chain attachment influences the rate of Sic1 degradation. Mol Cell. 2003;11:1435–1444. doi: 10.1016/s1097-2765(03)00221-1. [DOI] [PubMed] [Google Scholar]
- Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat Struct Mol Biol. 2004;11:830–837. doi: 10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- Raasi S, Wolf DH. Ubiquitin receptors and ERAD: a network of pathways to the proteasome. Semin Cell Dev Biol. 2007;18:780–791. doi: 10.1016/j.semcdb.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Rabinovich E, Kerem A, Frohlich KU, Diamant N, Bar-Nun S. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol Cell Biol. 2002;22:626–634. doi: 10.1128/MCB.22.2.626-634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadan K, Bruderer R, Spiga FM, Popp O, Baur T, Gotta M, Meyer HH. Cdc48/p97 promotes reformation of the nucleus by extracting the kinase Aurora B from chromatin. Nature. 2007;450:1258–1262. doi: 10.1038/nature06388. [DOI] [PubMed] [Google Scholar]
- Ramsey KL, Smith JJ, Dasgupta A, Maqani N, Grant P, Auble DT. The NEF4 complex regulates Rad4 levels and utilizes Snf2/Swi2-related ATPase activity for nucleotide excision repair. Mol Cell Biol. 2004;24:6362–6378. doi: 10.1128/MCB.24.14.6362-6378.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid T, Kreft SG, Hochstrasser M. Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J. 2006;25:533–543. doi: 10.1038/sj.emboj.7600946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid J, Svejstrup JQ. DNA damage-induced Def1-RNA polymerase II interaction and Def1 requirement for polymerase ubiquitylation in vitro. J Biol Chem. 2004;279:29875–29878. doi: 10.1074/jbc.C400185200. [DOI] [PubMed] [Google Scholar]
- Ribar B, Prakash L, Prakash S. Requirement of ELC1 for RNA polymerase II polyubiquitylation and degradation in response to DNA damage in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:3999–4005. doi: 10.1128/MCB.00293-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribar B, Prakash L, Prakash S. ELA1 and CUL3 are required along with ELC1 for RNA polymerase II polyubiquitylation and degradation in DNA-damaged yeast cells. Mol Cell Biol. 2007;27:3211–3216. doi: 10.1128/MCB.00091-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly H, Rape M, Braun S, Rumpf S, Hoege C, Jentsch S. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Rumpf S, Jentsch S. Functional division of substrate processing cofactors of the ubiquitin-selective Cdc48 chaperone. Mol Cell. 2006;21:261–269. doi: 10.1016/j.molcel.2005.12.014. [DOI] [PubMed] [Google Scholar]
- Schrader EK, Harstad KG, Matouschek A. Targeting proteins for degradation. Nat Chem Biol. 2009;5:815–822. doi: 10.1038/nchembio.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuberth C, Buchberger A. Membrane-bound Ubx2 recruits Cdc48 to ubiquitin ligases and their substrates to ensure efficient ER-associated protein degradation. Nat Cell Biol. 2005;7:999–1006. doi: 10.1038/ncb1299. [DOI] [PubMed] [Google Scholar]
- Schuberth C, Buchberger A. UBX domain proteins: major regulators of the AAA ATPase Cdc48/p97. Cell Mol Life Sci. 2008;65:2360–2371. doi: 10.1007/s00018-008-8072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuberth C, Richly H, Rumpf S, Buchberger A. Shp1 and Ubx2 are adaptors of Cdc48 involved in ubiquitin-dependent protein degradation. EMBO Rep. 2004;5:818–824. doi: 10.1038/sj.embor.7400203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson S, Dirac-Svejstrup AB, Svejstrup JQ. Evidence that transcript cleavage is essential for RNA polymerase II transcription and cell viability. Mol Cell. 2010;38:202–210. doi: 10.1016/j.molcel.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somesh BP, Reid J, Liu WF, Sogaard TM, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Multiple mechanisms confining RNA polymerase II ubiquitylation to polymerases undergoing transcriptional arrest. Cell. 2005;121:913–923. doi: 10.1016/j.cell.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Somesh BP, Sigurdsson S, Saeki H, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Communication between distant sites in RNA polymerase II through ubiquitylation factors and the polymerase CTD. Cell. 2007;129:57–68. doi: 10.1016/j.cell.2007.01.046. [DOI] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Deshaies RJ. Assaying degradation and deubiquitination of a ubiquitinated substrate by purified 26S proteasomes. Methods Enzymol. 2005;398:391–399. doi: 10.1016/S0076-6879(05)98032-4. [DOI] [PubMed] [Google Scholar]
- Verma R, McDonald H, Yates JR, 3rd, Deshaies RJ. Selective degradation of ubiquitinated Sic1 by purified 26S proteasome yields active S phase cyclin-Cdk. Mol Cell. 2001;8:439–448. doi: 10.1016/s1097-2765(01)00308-2. [DOI] [PubMed] [Google Scholar]
- Wade JT, Struhl K. The transition from transcriptional initiation to elongation. Curr Opin Genet Dev. 2008;18:130–136. doi: 10.1016/j.gde.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss M, Schrimpf S, Hengartner MO, Lercher MJ, von Mering C. Shotgun proteomics data from multiple organisms reveals remarkable quantitative conservation of the eukaryotic core proteome. Proteomics. 2010;10:1297–1306. doi: 10.1002/pmic.200900414. [DOI] [PubMed] [Google Scholar]
- Wilcox AJ, Laney JD. A ubiquitin-selective AAA-ATPase mediates transcriptional switching by remodelling a repressor-promoter DNA complex. Nat Cell Biol. 2009;11:1481–1486. doi: 10.1038/ncb1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woudstra EC, Gilbert C, Fellows J, Jansen L, Brouwer J, Erdjument-Bromage H, Tempst P, Svejstrup JQ. A Rad26-Def1 complex coordinates repair and RNA pol II proteolysis in response to DNA damage. Nature. 2002;415:929–933. doi: 10.1038/415929a. [DOI] [PubMed] [Google Scholar]
- Ye Y. Diverse functions with a common regulator: ubiquitin takes command of an AAA ATPase. J Struct Biol. 2006;156:29–40. doi: 10.1016/j.jsb.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J Cell Biol. 2003;162:71–84. doi: 10.1083/jcb.200302169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.