Abstract
There is a substantial genetic component to birth weight variation. We tested eighteen single nucleotide polymorphisms (SNPs) in the IGF2, H19, and IGF2R genes for associations with birth weight variation in 342 mother-newborn pairs (birth weight 2.1 – 4.7 Kg at term) and 527 parent-newborn trios (birth weight 2.1 – 5.1 Kg) across three localities. SNPs in the IGF2R (rs8191754; maternal genotype), IGF2 (rs3741205; newborn genotype) and in the 5' region of the H19 (rs2067051, rs2251375, and rs4929984) genes were associated with birth weight. Detailed analyses to distinguish direct maternal, direct newborn, and parent of origin effects for the most strongly associated H19 SNP (rs4929984) determined that the association of maternal genotype with newborn birth weight was due to parent of origin effects, not direct maternal effects. That SNP is located near the CTCF binding sites that influence expression of the maternally-imprinted IGF2 and paternally-imprinted H19 locus, and there are statistically significant and independent opposite effects of the same rs4929984 allele, depending on the parent from which it was inherited.
Introduction
Term, low birth weight infants are second only to premature infants in their rates of morbidity and mortality (1). As adults, individuals born small for gestational age (SGA) are at elevated risk of multiple chronic illnesses (2). The link between reduced birth weight and adult illness might be explained by uteroplacental insufficiency that alters organ function and hormonal milieu to make the individual more susceptible to disease (3). Alternatively, genetic factors may exist that both reduce fetal growth and increase predisposition to disease (4).
Birth weight variation has a substantial genetic component. The likelihood of an SGA individual giving birth to an SGA child is 3.5x for men, 4.7x for women, or 16.3x if both parents were SGA, implying a multiplicative effect (5). Total heritability for birth weight is 42–45%, with a slightly stronger maternal versus paternal component (6–7). Additionally, SGA births both cluster in families and recur in successive generations (8–9).
Insulin-like growth factor 2 (IGF2), its receptor (IGF2R) and H19 are strong candidate genes for influencing birth weight variation. The maternal allele of IGF2 is imprinted in the fetus, meaning that the maternally-transmitted allele is mostly silenced. Mouse knockouts of the paternal Igf2 allele are growth retarded, while fetuses over-expressing Igf2 exhibit overgrowth (10–11). In humans, the fetal growth restriction of Silver-Russell and fetal overgrowth of Beckwith-Wiedemann Syndromes (BWS) are associated with depressed versus elevated levels of IGF-II (12–13). H19 is an untranslated gene about 100 Kb from IGF2 for which the RNA transcript appears to be the functional product. H19 is growth-inhibitory (14) and functionally antagonistic to IGF2. The paternal allele of H19 is imprinted, and the binding of the CTCF protein to an imprinting control region upstream of H19, regulated to a great extent by DNA methylation, reduces the IGF2 gene’s access to a key enhancer located 3’ of H19 (15–16). Finally, IGF2R exhibits inconsistent paternal imprinting in humans and functions mainly to remove IGF-II from the circulation (17–19). Correspondingly, mouse knockouts of IGF2R exhibit fetal overgrowth (11). Numerous studies have shown association between birth weight or adult body mass and variation in the IGF2, H19, and IGF2R genes, although replication has been problematic (20–28).
Using three independent populations, we tested the association between single nucleotide polymorphisms (SNPs) in the IGF2, H19 and IGF2R genes and birth weight. While some previously associated SNPs failed to replicate in our cohorts, we identify at least one SNP in each gene that associates with birth weight when present in the maternal or newborn genome. We also make the novel observation that maternal versus paternal transmission of the same H19 allele has an opposite effect on birth weight.
Methods
Subjects
From 2003–2005 maternal blood, umbilical cord blood and placental tissue were obtained from healthy pairs of mothers and newborns (Table 1) during labor and delivery at the University of Mississippi Medical Center (Jackson, MS; N = 104 pairs) and the Regional Medical Center (Memphis, TN; N = 123). From 2007–2008 healthy women (N = 115) were recruited during prenatal care into a longitudinal cohort study of cognitive development in Memphis, TN. Maternal blood was obtained from these women during prenatal care, and umbilical cord blood was obtained at delivery. Finally, from 2008–2009 maternal and umbilical cord blood was collected from mothers and newborns (N = 527) and mouthwash saliva (Listerine Cool Mint) from fathers (N = 502) at the Naval Medical Center Portsmouth (Virginia). All newborns were healthy at the time of delivery. Inclusion criteria were: singleton pregnancy, >36 weeks gestation, 18–40 years old. Exclusion criteria eliminated many additional factors influencing birth weight and included, among others: diabetes (type 1, type 2, gestational), hypertension/vascular disease, preeclampsia, autoimmune disease, infectious disease (i.e., hepatitis, HIV), sickle-cell disease, uterine infection, illicit drug use, smoking, and birth defects. The Institutional Review Boards of the University of Tennessee Health Science Center, University of Mississippi Medical School and Naval Medical Center at Portsmouth approved this study, and informed consent was obtained from all mothers and fathers.
Table 1.
Characteristics of the participants.
| Variable | Memphis TN & Jackson MS |
Portsmouth VA |
||
|---|---|---|---|---|
| Median | Range | Median | Range | |
| Race (Maternal) | ||||
| African-American (N) | 286 | 94 | ||
| Caucasian (N) | 56 | 406 | ||
| Other (N) | 0 | 27 | ||
| Mothers | ||||
| Age (years) | 22 | 17 – 40 | 26 | 18 – 44 |
| Parity | 2 | 1 – 11 | 2 | 1 – 6 |
| Weight gain (Kg) | 15.3 | −4.1 – 47.6 | 15.4 | 0 – 37.2 |
| BMI (Kg/m2) | 24.9 | 16.1 – 56.7 | 24.7 | 15.4 – 45.2 |
| Newborns | ||||
| Gestational age (weeks) | 39 | 36 – 42 | 39 | 36 – 42 |
| Female (%) | 44 | 49 | ||
| Weight (Kg) | 3.3 | 2.1 – 4.7 | 3.4 | 2.1 – 5.1 |
| Z | −0.1 | −2.24 – 3.09 | 0.0 | −2.74 – 3.48 |
Genotyping
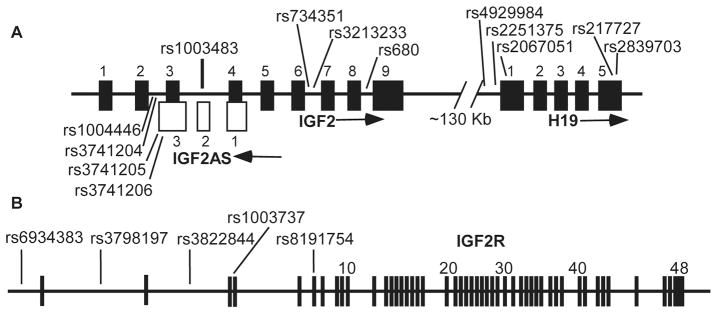
SNPs were selected based on previously published associations with birth weight or body mass-related phenotypes or for the purpose of tagging haplotypes (Table 2; Figure 1). Haplotype tagging SNPs were selected using SNPbrowser software v3.5 (Applied Biosystems, Inc., Carlsbad CA, USA) and data from the International HapMap Project with the criteria r2 ≥ 0.8 with untyped SNPs and minor allele frequency > 20% in the Yoruban population (or Caucasian, if Yoruban data unavailable). IGF2R is a large gene, and haplotypes were tagged for only the 5’ one-third of the gene. All genotyping used TaqMan reagents (Applied Biosystems Inc.). Genotyping was ambiguous for H19 SNP rs2251375, and this was verified by either direct DNA sequencing or allele-specific PCR. Only 203 mother-newborn pairs were genotyped for rs2251375, all African-American. Genotyping was attempted for IGF2 SNP rs3741211, but allelic discrimination was poor and this site is not reported.
Table 2.
Statistical analyses of SNPs in Memphis and Jackson populations (N = 342).
| Minor allele frequency* |
Association with birth weight (p value) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Maternal |
Fetal |
|||||||
| African-American | Caucasian | Additive | Dominant | Recessive | Additive | Dominant | Recessive | |
| IGF2R | ||||||||
| rs6934383 | 0.36 | 0.18 | 0.8452 | 0.8122 | 0.9152 | 0.9916 | 0.9243 | 0.9612 |
| rs3798197 | 0.41 | 0.26 | 0.2263 | 0.3740 | 0.2938 | 0.7921 | 0.5283 | 0.9276 |
| rs3822844 | 0.44 | 0.30 | 0.8446 | 0.6223 | 0.8994 | 0.8848 | 0.3191 | 0.5468 |
| rs1003737 | 0.48 | 0.54 | 0.6813 | 0.4419 | 0.9567 | 0.9426 | 0.8541 | 0.7725 |
| rs8191754 | 0.13 | 0.07 | 0.0462 | 0.9911 | 0.0283 | 0.7728 | 0.4536 | 0.9065 |
| IGF2-H19 | ||||||||
| rs2839703 | 0.12 | 0.42 | 0.2200 | 0.6911 | 0.2025 | 0.1672 | 0.8648 | 0.0800 |
| rs217727 | 0.08 | 0.13 | 0.1734 | 0.4075 | 0.2073 | 0.1856 | 0.2289 | 0.2670 |
| rs2067051 | 0.09 | 0.28 | 0.0318 | 0.5225 | 0.0305 | 0.9735 | 0.1093 | 0.5073 |
| rs2251375 | 0.38 | - | 0.0140 | 0.1689 | 0.0172 | 0.3672 | 0.7188 | 0.3177 |
| rs4929984 | 0.31 | 0.48 | 0.0013 | 0.0009 | 0.0276 | 0.2268 | 0.5223 | 0.2307 |
| rs680 | 0.07** | 0.21 | 0.3523 | 0.1610 | 0.5963 | 0.6281 | 0.3878 | 0.8071 |
| rs3213233 | 0.10 | 0 | 0.9765 | 0.7003 | 0.8837 | 0.8319 | 0.5404 | 0.9467 |
| rs734351 | 0.25 | 0.30 | 0.7531 | 0.8741 | 0.6393 | 0.6327 | 0.3547 | 0.9209 |
| rs1003483 | 0.16 | 0.50 | 0.6032 | 0.9680 | 0.5427 | 0.1579 | 0.2132 | 0.2805 |
| rs3741206 | 0.28 | 0.45 | 0.1436 | 0.9632 | 0.0621 | 0.7680 | 0.9763 | 0.6904 |
| rs3741205 | 0.30 | 0.26 | 0.1754 | 0.2712 | 0.2668 | 0.0383 | 0.2944 | 0.0385 |
| rs3741204 | 0.43 | 0.30 | 0.7660 | 0.8057 | 0.8123 | 0.1842 | 0.2582 | 0.2966 |
| rs1004446 | 0.44 | 0.31 | 0.9567 | 0.9054 | 0.8566 | 0.1475 | 0.4209 | 0.1332 |
Among newborns.
Test of Hardy-Weinberg genotypic expectations, p < 0.05 among newborns
Figure 1.
A) IGF2, IGF2AS, and H19 genes. B) IGF2R locus. The locations of genotyped SNPs are shown. Although not to scale, the relative spacing of exons is represented. Arrows indicate the direction of transcription.
Statistical Analysis
Potential confounders and effect modifiers
All statistical analyses were performed using Stata SE v.10.1 (StataCorp, College Station, TX, USA). Before inclusion of genetic data, a non-genetic model was constructed of potential confounders or effect modifiers, including gestational age (nearest week), maternal age, parity, newborn gender, maternal body mass index (MBMI), and pregnancy weight gain. Each variable was plotted against birth weight to determine how best to present it. The relationship between MBMI and birth weight is non-linear with birth weight increasing up to an MBMI of 27 and then decreasing beyond that. Therefore, this variable was represented as a linear spline of MBMI and MBMI27 (coded as 0 when MBMI ≤ 27 and MBMI-27 when MBMI > 27). The final model was identified by iteratively removing the least significant variable via “backward selection” until all remaining variables were significant (p ≤ 0.05). The final model included gestational age, newborn gender, MBMI, and MBMI27.
Association between birth weight and each SNP (0, 1, or 2 copies of the minor allele) was tested in the combined Memphis and Jackson populations by addition of the genetic data to the non-genetic multiple linear regression model. Differences in African and European allele frequencies (admixture) can produce false positive associations (29–30). To adjust for admixture, we included as a covariable an estimate of the proportion of African genomic ancestry in each newborn calculated by the program Admixmap (31). These estimates were based on European and African allele frequencies and used the candidate gene SNPs, 50 SNPs from 13 other protein-coding loci (32–34), and 12 unlinked ancestry-informative SNPs (minor allele frequency difference >80% between Yoruban and European).
Whether an additive, recessive, or dominant genetic model best fits the data is unknown a priori. Performing all three tests at each SNP introduces a multiple testing burden. While Bonferroni correction is too stringent due to non-independence among SNPs in a small genomic region, permutation can determine empirical significance levels without a loss of power (35). We tested all three genetic models and calculated P values by permuting the dependent variable (birth weight) 100,000 times and determining the rank of the original regression t statistic (36). For the most strongly associated SNP (rs4929984), we genotyped the family trios in our replication sample from Portsmouth VA and employed the polytomous logistic regression approach of Kistner et al. (37) to distinguish between direct maternal and parent of origin genetic effects. Because that method does not allow the inclusion of covariables, birth weight Z scores (standard deviations from the average birth weight for race, gender and gestational age using national statistics for 2003) were used as the dependent variable. Some possibility for confounding exists due to a negative correlation between African-American-specific birth weight Z scores and genetically estimated proportions of African genomic ancestry (34). Hardy-Weinberg genotypic expectations were tested via an exact test (command genhwi), and measures of linkage disequilibrium were calculated using the pwld command (authored by David Clayton, Cambridge Institute for Medical Research) in Stata.
Results
General Characteristics of the Population
Overall characteristics of the mothers and newborns (Table 1) were similar among recruitment sites. Birth weights covered a broad range, with 2–4% (N = 13 Memphis & Jackson, N = 13 Portsmouth) SGA (≤10th percentile for gestational age, gender, and ethnicity) and 4–9% (N = 12 Memphis & Jackson, N = 47 Portsmouth) large for gestational age (LGA; ≥90th percentile). Likely, the proportion of SGA and LGA newborns was cumulatively less than 20% due to our selection of only healthy mothers and newborns, a decision intended to enrich for the genetic component of normal birth weight variation.
Genetic Variants
IGF2 SNP rs680 deviated (p = 0.01) from Hardy-Weinberg genotypic expectations among African-American newborns, with a deficit of eight heterozygotes and nearly equal excesses of both homozygotes. Given that 34 statistical tests were performed, this could be a result of type I error.
In Memphis and Jackson, four SNPs (IGF2R rs8191754 and H19 rs2067051, rs2251375, and rs4929984) among mothers and one SNP (IGF2 rs3741205) among newborns showed significant association with birth weight (Table 2). Linkage disequilibrium is high among the H19 SNPs (D’ ≥ 0.97; R2 = 0.20 – 0.72) but low between the H19 and IGF2 SNPs (D’ ≤ 0.27; R2 = 0). Maternal genotypes of rs4929984 significantly (p = 0.04) associated with birth weight, but neither paternal nor newborn genotypes were associated (p > 0.27), among the Caucasian family trios from our replication population from Portsmouth VA, who should exhibit few effects of admixture. Using polytomous logistic regression, which is insensitive to population stratification, in all Portsmouth trios, parent of origin (p = 0.016), but not direct maternal genetic association (p = 0.396), was significant for rs4929984. In regression (Table 3) of birth weight on gestational age, gender, MBMI, MBMI27, ethnicity, and alleles of inferred parental origin in only the African-American and Caucasian participants from all locations, both the maternally-(p=0.007) and paternally-transmitted (p=0.012) alleles were significant, indicating independent associations with both alleles. Maternal transmission of the A allele was associated with an 89g decrease in birth weight, while paternal transmission was associated with a 78g increase. This implies opposite associations with the same allele, depending on whether it was paternally or maternally transmitted. We can assume that the paternal genotype does not directly affect fetal growth. However, direct maternal, direct fetal, and parent-of-origin effects (both maternal and paternal alleles) may be operating singly or in any combination, making it very difficult to isolate one effect from the others. One approach is to eliminate variation in some of these factors, while testing for the others. Therefore, to better understand the possibility of opposing parent-of-origin associations, we considered only homozygous mothers from all localities, which holds constant both the maternal genotype and the maternally-transmitted allele (Table 4). Newborns whose mothers were CC (maternally-transmitted C allele) versus AA had significantly higher birth weight Z scores (N=468, t=2.25, two-sided p=0.025), as would be expected (Table 2). Within each category of homozygous mother, newborns who received an A allele from their father had a higher average Z score. The significance of this difference was marginal, likely due to small sample sizes within these narrow subcategories.
Table 3.
Multiple linear regression of birth weight (g) on parental origin of alleles at rs4929984 in the Memphis, Jackson and Portsmouth African-American and Caucasian participants.
| Variable | All (N = 686) |
Primiparous (N=228) |
Multiparous (N = 458) |
|||
|---|---|---|---|---|---|---|
| β | P | β | P | β | P | |
| Gestational age (weeks) | 124 | <0.001 | 127 | <0.001 | 135 | <0.001 |
| Ethnicity (African-American/Caucasian) | 249 | <0.001 | 226 | <0.001 | 265 | <0.001 |
| Gender (Male/Female) | −119 | <0.001 | −76 | 0.135 | −147 | <0.001 |
| MBMI | 30 | <0.001 | 39 | <0.001 | 26 | <0.001 |
| MBMI27 | −33 | <0.001 | −50 | 0.004 | −29 | 0.005 |
| Paternally-transmitted allele (C/A) | 78 | 0.012 | 178 | 0.001 | 33 | 0.392 |
| Maternally-transmitted allele (C/A) | −89 | 0.007 | −86 | 0.136 | −84 | 0.039 |
Table 4.
Effect of the paternally-transmitted allele of rs4929984 when maternal genotype and her transmitted allele are held constant.
| Maternal Genotype (Z̄) | Paternal Allele | Z̄ (SD) | N* | P** |
|---|---|---|---|---|
| CC (0.02) | C | −0.05 (0.06) | 197 | 0.09 |
| A | 0.11 (0.08) | 127 | ||
| AA (−0.19) | C | −0.32 (0.09) | 79 | 0.05 |
| A | −0.02 (0.12) | 65 |
Memphis, Jackson and Portsmouth combined.
2-sided t-test
Discussion
IGF2R
The association of IGF2R with birth weight is inconsistent. Among 884 Japanese mothers and newborns, rs8191754 CC homozygotes had the highest birth weight (p = 0.02), with heterozygotes intermediate between homozygotes (23). In contrast, among 97 Greek newborns, neither neonatal rs8191754 genotypes nor parental origin of alleles were associated with birth weight (24). We found association between maternal, but not newborn, rs8191754 genotypes and birth weight. In our case, GG homozygotes were heaviest (3,425g), but this was based on only five individuals and is probably unreliable. A contrast (t-test, , p = 0.04) between the genotypes CC (3,301g; N = 242) and CG (3,186g; N = 79) should be more accurate and is consistent with the Japanese pattern. Although consistent between Japanese and our cohort, the association of rs8191754 with birth weight remains equivocal given the conflicting association with neonatal versus maternal genotypes and the modest effect size estimated.
IGF2
We surveyed eight SNPs in the IGF2/IGF2AS (IGF2 antisense transcript) genes, some of which have shown association with birth weight, fetal growth disorders, or mature body mass. Polymorphism rs680 is associated with birth weight among Japanese (23), but not Brazilians or Belgians (21, 38), and with adult male BMI among some British cohorts (20, 22, 27–28), but not others (38). In Memphis and Jackson, we found no association between birth weight and either maternal or newborn rs680 genotypes or in analyses of individual newborn genders (p > 0.08).
Polymorphism rs3741205 is the defining SNP of a CAGA haplotype (rs1004446, rs3741204, rs3741205, and rs3741206) in a differentially methylated region (DMR) termed DMR0 in intron 2 of IGF2 that is enriched in sporadic BWS patients (39). BWS is characterized by pre- and postnatal overgrowth and is commonly associated with altered methylation of DMRs near H19 and KCNQ1 (500 Kb from IGF2) and their loss of imprinting. Among Japanese, inferred paternal inheritance of IGF2 haplotype CTG at rs1003484, rs3741211 and rs3741206 was associated with a higher frequency of SGA births (25). We observed a significant association between birth weight and newborn genotypes at rs3741205 for both an additive and recessive (TT vs. GT and GG) model. The presence of a G allele is associated with an increase of 74g after adjustment for covariables. Unlike among the Japanese population, we find no significant association with inheritance of the inferred paternal allele (p > 0.2 in all subjects and within newborn genders).
The CAGA haplotype more frequent among BWS patients was associated with loss of methylation of an exon 10 DMR (KvDMR1) in the maternally imprinted KCNQ1 gene (39). Lack of maternal methylation of KvDMR1 permits expression of an antisense transcript (KCNQ1OT1) which reduces expression of the KCNQ1 and CDKN1C genes, and this is observed in about half of BWS cases. Genetic variation in the region of IGF2 DMR0 may be associated with changes in DNA methylation at KvDMR1. Alternatively, given its proximity to the gene, this variant may influence the expression of IGF2AS, whose function is presently unclear.
H19 region
Among British mothers and mother-newborn pairs, rs217727 in the paternally imprinted H19 gene exhibited independent associations of maternal and newborn rs217727 genotypes with birth weight and a further association of maternal rs217727 genotypes with umbilical cord IGF-II levels (26). There was no evidence of a parent of origin effect. We found no birth weight association with either maternal or newborn genotypes of rs217727. The minor allele of rs217727 is rare (8 – 13%; 5 newborn and 3 maternal TT homozygotes) among our subjects, reducing statistical power in regression analyses. Nevertheless, restricting attention to CC (268 mothers, 275 newborns) and CT (59 mothers, 51 newborns) individuals, the relationship to birth weight remains nonsignificant (p > 0.3), despite a 45g (newborn, adjusted for covariables) or 55g (maternal) increase in birth weight among CT heterozygotes.
In Memphis and Jackson, we found strong association between birth weight and maternal genotypes at rs4929984 and weaker association with rs2067051 and rs2251375. Linkage disequilibrium is high (D’ = 0.94 – 1) among those SNPs, meaning that the association with each site is not independent and indicating: 1) only one of the SNPs is functionally important, 2) they are functionally important as a haplotype, or 3) all three are in linkage disequilibrium with a functional genetic variant that was not surveyed.
The association of the H19 rs4929984 polymorphism was replicated in the Portsmouth VA family trios. Polytomous logistic regression (37) is capable of disentangling direct maternal, direct fetal, and parent of origin associations and reveals that the association is due to parent of origin, not to direct association with maternal genotypes. Analyses of the effect of the maternally- and paternally-transmitted alleles at rs4929984 indicate that maternal inheritance of the A allele results in an average 89g decrease (adjusted for covariables; Table 3), while paternal inheritance of the same allele results in an average 78g increase in birth weight. These associations remain if only the paternally- or maternally-transmitted alleles are included in the regression model, but with smaller effect sizes (maternal allele, 74g, p = 0.02; paternal allele, 60g, p = 0.05), consistent with the possibility of mutual confounding due to opposite associations with the same allele depending on parental origin. This may be why paternal genotypes are not directly associated with birth weight in the Virginia population. The maternally-transmitted allele may confound the ability to detect an association with paternal genotype due to its opposite and slightly larger effect. Analyses within maternal genotypic classes (Table 4) that eliminate variation due to the maternal genotype and maternally-transmitted allele illustrate that the paternal allele has a consistent association in the opposite direction of the maternal allele.
The association of rs217727 with birth weight (26) was strongest for the first born child, suggesting a contribution from in utero growth constraint in first pregnancies. To determine if a similar effect exists among our participants, we distinguished between first and subsequent pregnancies (Table 3). Among multiparous women, the maternally transmitted allele had a significant effect, but not among primiparous women. However, the estimated effect size of the maternal allele was similar within both groups, and the nonsignificant result may be due to lack of statistical power in the comparatively small subset (N = 228). Interestingly, the paternally-transmitted allele was significantly associated only among primiparous mothers, suggesting a pattern similar to that previously observed for maternal genotypes of rs217727.
A functional basis for the opposite direction of association of parentally-transmitted alleles of rs4929984 requires explanation. The imprinting of both IGF2 and H19 is under the control of an imprinting control region (ICR) with seven CTCF binding sites that resides 500 – 5,200 nucleotides upstream of H19. The significantly associated SNPs near H19 flank the ICR (rs4929984 upstream, rs2251375 and rs2067051 downstream). Therefore, it is unlikely that these SNPs directly disrupt the insulator function of CTCF binding. However, these SNPs may be in linkage disequilibrium with variants that alter CTCF binding or affect the methylation of the ICR and its ability to block IGF2 access to an enhancer 3’ of H19. Alternatively, these SNPs, or ones in linkage disequilibrium with them, may affect the level of H19 transcription or stability. Given the competitive use of the H19 ICR by IGF2 and H19 and the activity of those loci on different parental chromosomes, it is quite feasible that the same genetic variant could promote fetal growth when on the paternally-inherited chromosome and retard fetal growth when on the maternally-inherited chromosome. Interestingly, the pattern of birth weight variation we have observed with respect to rs4929984 closely resembles the “bipolar” (homozygotes similar, with one type of heterozygote smaller and the other larger) dominance pattern of imprinting for postnatal growth found at some genomic regions in an intercross of the large vs. small inbred mouse lines (40).
In summary, we identified polymorphisms in IGF2R, IGF2, and near H19 that are associated with birth weight. Even though the H19 association was replicated in a second cohort, the sample sizes were small, and these results should be validated in larger, independent populations. Recently, a large genome-wide association meta-analysis (41) was performed that identified two strong candidates for birth weight association, neither of which were in the loci studied here. However, that study focused entirely on the child's genotype and would not be expected to identify the associations we primarily observed with maternal genotypes.
Acknowledgments
Financial support: This study was funded by a grants from the US National Institute of Child Health and Human Development (HD055462), the Children’s Foundation Research Center of Memphis, University of Tennessee Health Science Center’s Clinical Translational Science Institute, and Accredo Foundation [to R.M.A.], from The Urban Child Institute (Memphis, TN) [to G.S.], and from the US National Center for Research Resources (M01-RR00211). The contents of this study are solely the responsibility of the authors and do not necessarily represent the official views of the NICHD.
We gratefully acknowledge the laboratory expertise of Jeanette Peeples, the participant recruitment and sample collection by CANDLE staff, and the mothers who consented to participate. We are particularly indebted to Dr. Emily Kistner-Griffin (Medical University of South Carolina) for her assistance in understanding the polytomous logistic regression method.
Abbreviations
- ICR
imprinting control region
- IGF2R
IGF2 receptor
- IGF2AS
IGF2 antisense
- SNP
single nucleotide polymorphism
- SGA
small for gestational age
- BWS
Beckwith-Wiedemann Syndrome
- DMR
differentially methylated region
- MBMI
maternal body mass index
- MBMI27
maternal body mass index in excess of 27
References
- 1.Hay WW, Jr, Catz CS, Grave GD, Yaffe SJ. Workshop summary: fetal growth: its regulation and disorders. Pediatrics. 1997;99:585–591. doi: 10.1542/peds.99.4.585. [DOI] [PubMed] [Google Scholar]
- 2.Gillman MW, Barker D, Bier D, Cagampang F, Challis J, Fall C, Godfrey K, Gluckman P, Hanson M, Kuh D, Nathanielsz P, Nestel P, Thornburg KL. Meeting report on the 3rd International Congress on Developmental Origins of Health and Disease (DOHaD) Pediatr Res. 2007;61:625–629. doi: 10.1203/pdr.0b013e3180459fcd. [DOI] [PubMed] [Google Scholar]
- 3.Barker DJ. Mother, babies, and health in later life. Churchill Livingstone; London: 1998. pp. 13–42. [Google Scholar]
- 4.Basso O, Wilcox AJ, Weinberg CR. Birth Weight and Mortality: Causality or Confounding? Am J Epidemiol. 2006;164:303–311. doi: 10.1093/aje/kwj237. [DOI] [PubMed] [Google Scholar]
- 5.Jaquet D, Swaminathan S, Alexander GR, Abajian C. Significant paternal contribution to the risk of small for gestational age. BJOG. 2005;112:153–159. doi: 10.1111/j.1471-0528.2004.00313.x. [DOI] [PubMed] [Google Scholar]
- 6.Clausson B, Lichtenstein P, Cnattingius S. Genetic influence on birthweight and gestational length determined by studies in offspring of twins. BJOG. 2000;107:375–381. doi: 10.1111/j.1471-0528.2000.tb13234.x. [DOI] [PubMed] [Google Scholar]
- 7.Svensson AC, Pawitan Y, Cnattingius S, Reilly M, Lichtenstein P. Familial aggregation of small-for-gestational-age births: the importance of fetal genetic effects. Am J Obstet Gynecol. 2006;194:475–479. doi: 10.1016/j.ajog.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 8.Ananth CV, Kaminsky L, Getahun D, Kirby RS, Vintzileos AM. Recurrence of fetal growth restriction in singleton and twin gestations. J Matern Fetal Neonatal Med. 2009;22:654–661. doi: 10.1080/14767050902740207. [DOI] [PubMed] [Google Scholar]
- 9.Selling KE, Carstensen J, Finnstrom O, Sydsjo G. Intergenerational effects of preterm birth and reduced intrauterine growth: a population-based study of Swedish mother-offspring pairs. BJOG. 2006;113:430–440. doi: 10.1111/j.1471-0528.2006.00872.x. [DOI] [PubMed] [Google Scholar]
- 10.DeChiara TM, Efstratiadis A, Robertson EJ. A growth-deficiency phenotype in heterozygous mice carrying an insulin-like growth factor II gene disrupted by targeting. Nature. 1990;345:78–80. doi: 10.1038/345078a0. [DOI] [PubMed] [Google Scholar]
- 11.Lau MM, Stewart CE, Liu Z, Bhatt H, Rotwein P, Stewart CL. Loss of the imprinted IGF2/cation-independent mannose 6-phosphate receptor results in fetal overgrowth and perinatal lethality. Genes Dev. 1994;8:2953–2963. doi: 10.1101/gad.8.24.2953. [DOI] [PubMed] [Google Scholar]
- 12.Gicquel C, Rossignol S, Cabrol S, Houang M, Steunou V, Barbu V, Danton F, Thibaud N, Le Merrer M, Burglen L, Bertrand AM, Netchine I, Le Bouc Y. Epimutation of the telomeric imprinting center region on chromosome 11p15 in Silver-Russell syndrome. Nat Genet. 2005;37:1003–1007. doi: 10.1038/ng1629. [DOI] [PubMed] [Google Scholar]
- 13.Weksberg R, Smith AC, Squire J, Sadowski P. Beckwith-Wiedemann syndrome demonstrates a role for epigenetic control of normal development. Hum Mol Genet. 2003;12:R61–R68. doi: 10.1093/hmg/ddg067. [DOI] [PubMed] [Google Scholar]
- 14.Gabory A, Ripoche MA, Le Digarcher A, Watrin F, Ziyyat A, Forne T, Jammes H, Ainscough JF, Surani MA, Journot L, Dandolo L. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development. 2009;136:3413–3421. doi: 10.1242/dev.036061. [DOI] [PubMed] [Google Scholar]
- 15.Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 16.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 17.Kalscheuer VM, Mariman EC, Schepens MT, Rehder H, Ropers HH. The insulin-like growth factor type-2 receptor gene is imprinted in the mouse but not in humans. Nat Genet. 1993;5:74–78. doi: 10.1038/ng0993-74. [DOI] [PubMed] [Google Scholar]
- 18.Ogawa O, McNoe LA, Eccles MR, Morison IM, Reeve AE. Human insulin-like growth factor type I and type II receptors are not imprinted. Hum Mol Genet. 1993;2:2163–2165. doi: 10.1093/hmg/2.12.2163. [DOI] [PubMed] [Google Scholar]
- 19.Xu Y, Goodyer CG, Deal C, Polychronakos C. Functional polymorphism in the parental imprinting of the human IGF2R gene. Biochem Biophys Res Commun. 1993;197:747–754. doi: 10.1006/bbrc.1993.2542. [DOI] [PubMed] [Google Scholar]
- 20.Gaunt TR, Cooper JA, Miller GJ, Day IN, O'Dell SD. Positive associations between single nucleotide polymorphisms in the IGF2 gene region and body mass index in adult males. Hum Mol Genet. 2001;10:1491–1501. doi: 10.1093/hmg/10.14.1491. [DOI] [PubMed] [Google Scholar]
- 21.Gomes MV, Soares MR, Pasqualim-Neto A, Marcondes CR, Lobo RB, Ramos ES. Association between birth weight, body mass index and IGF2/ApaI polymorphism. Growth Horm IGF Res. 2005;15:360–362. doi: 10.1016/j.ghir.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 22.Gu D, O'Dell SD, Chen XH, Miller GJ, Day IN. Evidence of multiple causal sites affecting weight in the IGF2-INS-TH region of human chromosome 11. Hum Genet. 2002;110:173–181. doi: 10.1007/s00439-001-0663-5. [DOI] [PubMed] [Google Scholar]
- 23.Kaku K, Osada H, Seki K, Sekiya S. Insulin-like growth factor 2 (IGF2) and IGF2 receptor gene variants are associated with fetal growth. Acta Paediatr. 2007;96:363–367. doi: 10.1111/j.1651-2227.2006.00120.x. [DOI] [PubMed] [Google Scholar]
- 24.Kukuvitis A, Georgiou I, Syrrou M, Andronikou S, Dickerman Z, Islam A, McCann J, Polychronakos C. Lack of association of birth size with polymorphisms of two imprinted genes, IGF2R and GRB10. J Pediatr Endocrinol Metab. 2004;17:1215–1220. doi: 10.1515/jpem.2004.17.9.1215. [DOI] [PubMed] [Google Scholar]
- 25.Nagaya K, Makita Y, Taketazu G, Okamoto T, Nakamura E, Hayashi T, Fujieda K. Paternal allele of IGF2 gene haplotype CTG is associated with fetal and placental growth in Japanese. Pediatr Res. 2009;66:135–139. doi: 10.1203/PDR.0b013e3181a9e818. [DOI] [PubMed] [Google Scholar]
- 26.Petry CJ, Ong KK, Barratt BJ, Wingate D, Cordell HJ, Ring SM, Pembrey ME, Reik W, Todd JA, Dunger DB ALSPAC Study Team. Common polymorphism in H19 associated with birthweight and cord blood IGF-II levels in humans. BMC Genet. 2005;6:22. doi: 10.1186/1471-2156-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodríguez S, Gaunt TR, Dennison E, Chen XH, Syddall HE, Phillips DI, Cooper C, Day IN. Replication of IGF2-INS-TH*5 haplotype effect on obesity in older men and study of related phenotypes. Eur J Hum Genet. 2006;14:109–116. doi: 10.1038/sj.ejhg.5201505. [DOI] [PubMed] [Google Scholar]
- 28.Zhang W, Maniatis N, Rodriguez S, Miller GJ, Day IN, Gaunt TR, Collins A, Morton NE. Refined association mapping for a quantitative trait: weight in the H19-IGF2-INS-TH region. Ann Hum Genet. 2006;70:848–856. doi: 10.1111/j.1469-1809.2006.00290.x. [DOI] [PubMed] [Google Scholar]
- 29.Ewens WJ, Spielman RS. The transmission/disequilibrium test: history, subdivision, and admixture. Am J Hum Genet. 1995;57:455–464. [PMC free article] [PubMed] [Google Scholar]
- 30.Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- 31.Hoggart CJ, Parra EJ, Shriver MD, Bonilla C, Kittles RA, Clayton DG, McKeigue PM. Control of confounding of genetic associations in stratified populations. Am J Hum Genet. 2003;72:1492–1504. doi: 10.1086/375613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adkins RM, Klauser CK, Magann EF, Krushkal J, Boyd TK, Fain JN, Morrison JC. Site -2548 of the leptin gene is associated with gender-specific trends in newborn size and cord leptin levels. Int J Pediatr Obes. 2007;2:130–137. doi: 10.1080/17477160701286734. [DOI] [PubMed] [Google Scholar]
- 33.Adkins RM, Krushkal J, Klauser CK, Magann EF, Morrison JC, Somes G. Association between small for gestational age and paternally inherited 5' insulin haplotypes. Int J Obes (Lond) 2008;32:372–380. doi: 10.1038/sj.ijo.0803700. [DOI] [PubMed] [Google Scholar]
- 34.Adkins RM, Krushkal J, Magann EF, Klauser CK, Morrison JC, Ramsey R, Somes G. Association of Maternally Inherited GNAS Alleles with African-American Male Birth Weight. Int J Pediatr Obes. 2010;5:177–184. doi: 10.3109/17477160903111714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lettre G, Lange C, Hirschhorn JN. Genetic model testing and statistical power in population-based association studies of quantitative traits. Genet Epidemiol. 2007;31:358–362. doi: 10.1002/gepi.20217. [DOI] [PubMed] [Google Scholar]
- 36.Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kistner EO, Infante-Rivard C, Weinberg CR. A method for using incomplete triads to test maternally mediated genetic effects and parent-of-origin effects in relation to a quantitative trait. Am J Epidemiol. 2006;163:255–261. doi: 10.1093/aje/kwj030. [DOI] [PubMed] [Google Scholar]
- 38.Heude B, Ong KK, Luben R, Wareham NJ, Sandhu MS. Study of association between common variation in the insulin-like growth factor 2 gene and indices of obesity and body size in middle-aged men and women. J Clin Endocrinol Metab. 2007;92:2734–2738. doi: 10.1210/jc.2006-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murrell A, Heeson S, Cooper WN, Douglas E, Apostolidou S, Moore GE, Maher ER, Reik W. An association between variants in the IGF2 gene and Beckwith-Wiedemann syndrome: interaction between genotype and epigenotype. Hum Mol Genet. 2004;13:247–255. doi: 10.1093/hmg/ddh013. [DOI] [PubMed] [Google Scholar]
- 40.Wolf JB, Cheverud JM, Roseman C, Hager R. Genome-wide analysis reveals a complex pattern of genomic imprinting in mice. PLoS Genet. 2008;4:e1000091. doi: 10.1371/journal.pgen.1000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Freathy RM, Mook-Kanamori DO, Sovio U, Prokopenko I, Timpson NJ, Berry DJ, Warrington NM, Widen E, Hottenga JJ, Kaakinen M, Lange LA, Bradfield JP, Kerkhof M, Marsh JA, Magi R, Chen CM, Lyon HN, Kirin M, Adair LS, Aulchenko YS, Bennett AJ, Borja JB, Bouatia-Naji N, Charoen P, Coin LJ, Cousminer DL, de Geus EJ, Deloukas P, Elliott P, Evans DM, Froguel P, Glaser B, Groves CJ, Hartikainen AL, Hassanali N, Hirschhorn JN, Hofman A, Holly JM, Hypponen E, Kanoni S, Knight BA, Laitinen J, Lindgren CM, McArdle WL, O'Reilly PF, Pennell CE, Postma DS, Pouta A, Ramasamy A, Rayner NW, Ring SM, Rivadeneira F, Shields BM, Strachan DP, Surakka I, Taanila A, Tiesler C, Uitterlinden AG, van Duijn CM, Wijga AH, Willemsen G, Zhang H, Zhao J, Wilson JF, Steegers EA, Hattersley AT, Eriksson JG, Peltonen L, Mohlke KL, Grant SF, Hakonarson H, Koppelman GH, Dedoussis GV, Heinrich J, Gillman MW, Palmer LJ, Frayling TM, Boomsma DI, Smith GD, Power C, Jaddoe VW, Jarvelin MR, McCarthy MI. Variants in ADCY5 and near CCNL1 are associated with fetal growth and birth weight. Nat Genet. 2010;42:430–435. doi: 10.1038/ng.567. [DOI] [PMC free article] [PubMed] [Google Scholar]