Abstract
Objective
Previous genome wide association study conducted in a population of European ancestry identified rs4963128, a KIAA1542 SNP 23kb telomeric to IRF7, in strong association with SLE. This study was undertaken to investigate whether genetic polymorphism within IRF7 is a risk factor for the development of SLE.
Methods
We genotyped one KIAA1542 SNP rs4963128 and one IRF7 SNP rs1131665 (Q412R) in an Asian population (cases vs. controls: 1302 vs.1479) to assess their association with SLE using custom-designed Beadstation Infinium II platform (Illumina). Subsequently, rs1131665 was further genotyped in independent panels of Chinese (528 vs.527), European American (EA) (446 vs.461) and African American (AA) (159 vs.115) by Taqman genotyping assay to seek confirmation of association in various ethnic groups. Luciferase reporter assay was used to assess the effect of Q412R polymorphism on the activation of IRF7.
Results
Consistent association of rs1131665 (Q412R) with SLE was identified in Asian, EA and AA populations (case vs. control: 2435 vs. 2582; Pmeta = 6.18×10−6, OR = 1.42[1.22–1.65]). Expression of IRF7 412Q risk allele resulted in a 2-fold increase in ISRE transcriptional activity compared with expression of IRF7 412R (P = 0.0003), suggesting IRF7 412Q confers elevated IRF7 activity and may therefore affect downstream IFN pathway.
Conclusion
We showed that the major allele of a nonsynonymous SNP rs1131665 (412Q) in IRF7 confers elevated IRF7 activation and predisposes to the development of SLE in multiple ethnic groups. This result provides direct genetic evidence supporting IRF7 may be a risk gene for human SLE.
Systemic lupus erythematosus (SLE) is a complex autoimmune disease with strong genetic components. Genetic studies, especially the recent genome wide association studies (GWAS), have identified and confirmed more than 25 gene variants (or gene complex) that are associated with SLE (1.2). Most of these genes are involved in immune complex processing, immune signal transduction in T, B lymphocytes, tissue damage, Toll-like receptor (TLR) function, or type I interferon (IFN) pathway.
Interferon regulatory factor 7 (IRF7) (MIM: 605047), a member of the interferon regulatory transcription factor family, is a key player in innate immunity. The complete loss of virus induced type I IFN expression in Irf7 knockout mice suggests that this is a master regulator of IFN inducible pathway (3). In a previous GWAS, rs4963128, a SNP 23kb telomeric to IRF7 in KIAA1542 (11p15.5) was found in strong association with SLE (P = 3.0X10−10, OR=0.78) in a population of European ancestry (4). Given the strong linkage disequilibrium (LD; r2=0.94) between SLE-associated SNP and rs702966, a 3’UTR KIAA1542 SNP within 0.6 kb of IRF7, the observed association was proposed to reflect the association signals within IRF7 (4,5). Supporting this notion, SLE patients carrying KIAA1542 risk genotypes (rs4963128 and/or rs702966) and expressing anti-dsDNA or anti-Sm showed higher serum IFN-α activity (6). However, there is no direct genetic evidence regarding the contribution of IRF7 polymorphisms to SLE.
Here, we report a nonsynonymous SNP (Q412R) in IRF7 regulates IFN response in vitro and is associated with SLE in multiple ethnic groups, supporting that IRF7 is a new susceptibility gene for SLE.
Subjects and Methods
Patients and controls
In the discovery panel of 1308 cases and 1480 controls of Asian ancestry, 1302 cases and 1479 controls (Koreans [cases vs. controls: 845 vs. 1022], Chinese [386 vs. 331], Japanese [26 vs. 47] and others including Philippines, Malaysians, Vietnamese and Cambodian [45 vs. 79]) were successfully genotyped for rs4963128 and rs1131665 (genotyping call rates>99%) . The participants were recruited from multiple centers in Eastern Asia and USA. Among them, 68 participants were enrolled into the Lupus Family Registry and Repository (http://lupus.omrf.org) at Oklahoma Medical Research Foundation (OMRF). Chinese case-control samples (528 vs. 527) genotyped in replication panel 1, were recruited from Renji Hospital, Shanghai Jiaotong University School of Medicine. EA (446 vs. 461) and AA (159 vs. 115) samples genotyped in replication panels 2 and 3 were enrolled from UCLA and University of Texas. The female/male ratio of SLE patients was 11.2 (discovery panel), 7.2 (Chinese), 9.6 (EA) and 8.9 (AA).
All SLE patients fulfilled the 1997 revised American College of Rheumatology (ACR) criteria for classification of SLE (7) and gave informed consent. The study was approved by the Human Subject Institutional Review Boards or the ethic committees at each participating location.
SNP selection and genotyping
In the discovery panel, rs4963128 was selected to replicate the previous GWAS finding. Within IRF7 gene, SNPs with allele frequency data are either located in intron or are synonymous polymorphisms except two nonsynonymous SNPs: rs1131665 and rs1061502. Given that rs1061502 failed to show significant association in a preliminary experiment conducted in Caucasians (417 vs. 306, P=0.510), rs1131665 was selected in this study. Genotyping was performed at the Lupus Genetics Studies Unit of OMRF using a custom-designed Infinium II platform on the BeadstationTm (Illumina, CA).
In replication panels, rs1131665 was genotyped using custom-designed TaqMan SNP Genotyping Assay (Applied Biosystems, CA). The end-point fluorescence readings were performed using an ABI Prism 7900 HT System.
Plasmid construction
Plasmid pISRE-Luc was purchased from Stratagene (La Jolla, CA). Plasmid pCMV-IRF7 wild type (412Q) was purchased from OriGene (Rockville, MD). 412Q to 412R mutated vector was made using the Quikchange mutagenesis kit (Stratagene, La Jolla, CA) and confirmed by sequencing.
Luciferase reporter assay
HEK293 cells seeded on 24-well plates (2×105 cells/well) were transiently transfected using lipofectamine 2000 (Invitrogen, Carlsbad, CA) with 1 µg of the luciferase reporter plasmid, 1 µg of IRF7 412Q or 412R vectors, and 100 ng Renilla luciferase reporter gene construct as an internal control. The luciferase activity in total cell lysates was measured after 24 h using dual-luciferase reporter assay system (Promega, Madison, WI). Luciferase data shown here represent three independent experiments.
Protein in silico analysis
The sequences of IRF7 protein from multiple species were obtained from UniProtKB database (http://www.expasy.uniprot.org/). The multiple sequence alignment was then created using the program MUSCLE (http://www.drive5.com/muscle/). 3D structure of IRF7 protein was predicted by using ESyPred3D (http://www.fundp.ac.be/sciences/biologie/urbm/bioinfo/esypred/) based on IRF3 [PDB:1J2F].
Statistical analysis
We used gPLINK 1.06 to evaluate the significance of differences in allelic frequencies of the single SNP in the case-control samples. Meta-analysis was used to calculate combined P value and OR. In luciferase reporter assay, the difference of relative luciferase activity among various groups was evaluated using ANOVA. A P value < 0.05 was considered to be statistically significant.
Results
Association of IRF7 SNP rs1131665 (Q412R) with SLE in multiple ethnic groups
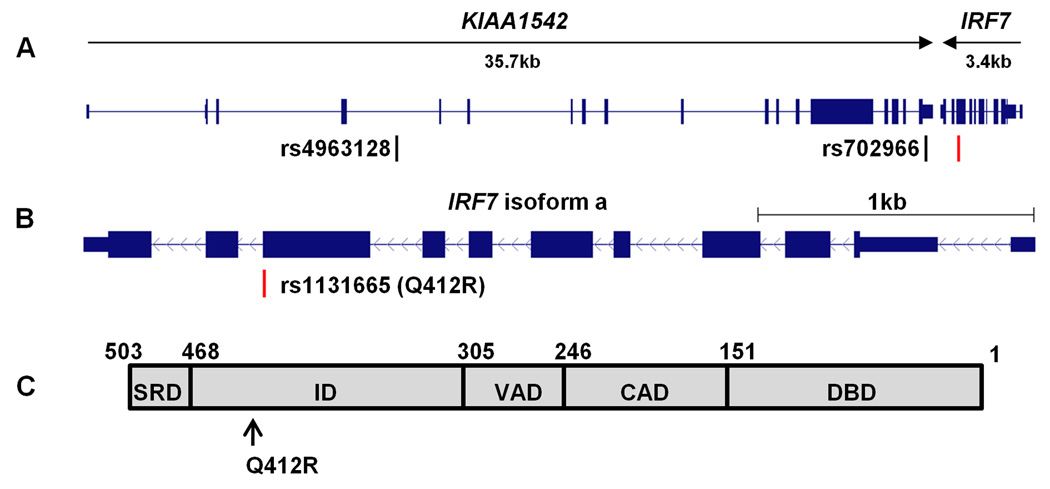
In the discovery panel, we first sought to replicate the previous GWAS finding in Asian population. While the previously reported KIAA1542 SNP rs4963128 showed no association with SLE (P=0.99, OR=1.00[0.83–1.21]) in Asians, significant association signal was observed in a nonsynonymous IRF7 SNP rs1131665 with SLE (risk allele frequency case vs. control: 98.4% vs. 97.1%, P=0.0016, OR=1.80[1.24–2.62]) (Table 1). This variation between the major allele A and minor allele G changes the amino acid residue at position 412 of IRF7 protein (NM_001572.3, NP_001563.2) from glutamine (Q) to arginine (R). The genomic structure of IRF7 and two SLE-associated SNPs in KIAA1542 were illustrated in Figure 1A and 1B. Since these two SNPs were genotyped together with other 300 SNPs in the custom-designed discovery panel, the association of rs1131665 was not statistically significant when multiple testing was considered. However, given that the KIAA1542-IRF7 locus was previously found to be a candidate susceptibility region in SLE, further replication and function studies were performed to verify the detected association signal.
Table 1.
Association of rs1131665 with SLE in multiple ethnic groups.
| Panels | Ethnic group | Sample size | A allele frequency | P | OR | ||
|---|---|---|---|---|---|---|---|
| Case | Control | Case | Control | ||||
| Discovery panel | Asian | 1302 | 1479 | 0.984 | 0.971 | 0.0017 | 1.80[1.24–2.62] |
| Replication panel 1 | Chinese | 528 | 527 | 0.989 | 0.976 | 0.018 | 2.31[1.25–4.72] |
| Replication panel 2 | EA | 446 | 461 | 0.715 | 0.658 | 0.009 | 1.30[1.07–1.59] |
| Replication panel 3 | AA | 159 | 115 | 0.497 | 0.430 | 0.124 | 1.31[0.93–1.83] |
| Meta-analysis | 2435 | 2582 | 6.18×10−6 | 1.42[1.22–1.65] | |||
Abbrevations: EA, European American; AA, African American; OR, odds ratio.
Figure 1.
Schematic structure of IRF7. A, Genomic structure of KIAA1542-IRF7 locus. Two KIAA1542 SNPs, rs4963128 and rs702966, are 23kb and 0.6kb telomeric to IRF7, respectively. In HapMap datasets (CEU, YRI, CHB and JPT), rs702966 is in strong LD (r2=1) with multiple SNPs in IRF7. B, Transcript structure of IRF7 isoform a. rs1131665 (Q412R) is highlighted. C, Conservative domain in IRF7 protein. DBD: DNA binding domain; CAD: constitutive activation domain; VAD: virus-activated domain; ID: inhibitory domain; SRD: signal response domain.
We conducted a replication study in 3 independent panels of individuals from various ethnic groups (Table 1). In Replication panel 1 of 528 Chinese cases and 527 controls, the major allele A (Q) showed a consistent significant association with SLE (98.9% vs. 97.6%, P=0.018, OR=2.31[1.25–4.72]). The association signal was also replicated in a population of EA (446 vs. 461), with a significant enrichment of the major allele A in cases (71.5%) compared with controls (65.8%) (P=0.009, OR=1.30[1.07–1.59]). Similarly, the A risk allele also showed higher frequency in SLE cases as compared to controls in AA population (50% vs. 43%), although the difference was not statistically significant (OR=1.31[0.93–1.83], P=0.12), which was probably due to the small sample size (159 vs. 115). The Hardy-Weinberg P value in each group was >0.01 except in the AA group, which was also probably due to the small sample size in the study. We then performed a meta-analysis to obtain an overall. Statistical analysis revealed no significant heterogeneity of ORs among various ethnic group (P=0.23), although the OR in Chinese panel appeared different from those in EA and AA. This was probably the fluctuation caused by the low MAF of rs1131665 and small number of minor allele carriers in Chinese panel. A larger sample size might reveal an OR closer to those in EA and AA as seen in our discovery Asian panel. The magnitude of the overall P value under the fixed effect model was Pmeta = 6.18×10−6 (OR=1.42[1.22–1.65]). Stratification analysis by clinical subsets was performed in a limit number of EA patients because of low minor allele frequency of rs1131665 in Asian population and inadequate clinical data available. Risk A allele showed a borderline association with anti-Ro antibody (55 vs. 165, P = 0.057, OR = 1.64[0.98–2.75]). There was no association between rs1131665 and presence of lupus nephritis, anti-dsDNA antibody or anti-Sm antibody. Taken together, the consistent association of rs1131665 with SLE across Asian, EA and AA populations provided evidence that this SNP or its linked variants confer risk of developing SLE, supporting that type I IFN pathway gene IRF7 is a novel susceptibility gene for SLE.
Protein in silico analysis
IRF7 contains five established domains as shown in Figure 1C (8). The functional polymorphism rs1131665 (Q412R) locates in exon 9 of the inhibitory domain (ID), which interferes with the binding and/or transactivation function of IRF7.
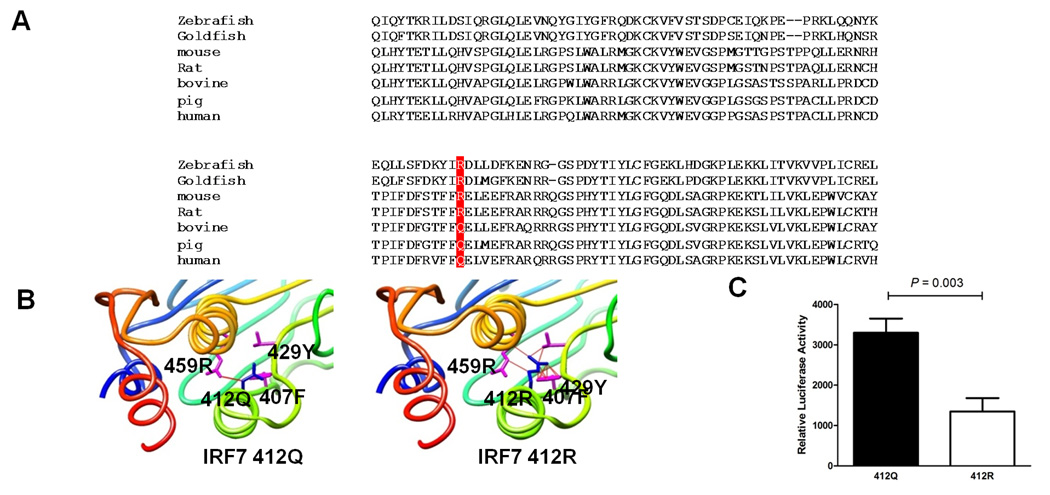
We next performed multiple sequence alignment of the IRF7 protein across species. IRF7 412Q is conserved in human, porcine, and bovine genomes, while R is found in zebrafish, goldfish, mouse and rat (Figure 2A). The fact that the 412Q allele rose to a higher frequency in relatively large mammals suggested that the IRF7 locus might be potentially functional in the evolution. Given that there is no IRF7 3D structure available in the current databases, we built a predicted 3D module of IRF7 protein by using ESyPred3D based on the structure of IRF3 [PDB:1J2F], another member from IRF family which shows significant homology with IRF7 and is closely related to IRF7 phylogenetically (9) (Figure 2B). The mutation from Q to R changes a polar but uncharged glutamine into a polar, positively charged arginine in a helix structure in the ID domain. While Q has only one contact with 459R in a nearby helix region adjacent to the signal response domain, R shows much more contacts and shorter distance with other amino acids including 407F and 429Y in a neighbouring coiled-coil region in the ID domain, which might lead to stronger inter-molecular interaction within ID domain. In unstimulated cells, ID helps suppress the constitutive activity of IRF7. When activated, the negative regulatory effect of ID on activity of IRF7 is relieved. One plausible consequence of Q/R change in the ID is influencing the structure of ID domain and its interaction with activity domains in IRF7, and therefore, affecting activation of IRF7 upon stimulation.
Figure 2.
Functional analysis of Q412R polymorphism. A, Protein sequence alignment of IRF7. The multiple sequence alignment of the IRF7 protein homologs was performed using MUSCLE. Q412R is highlighted. B, 3D structure prediction of IRF7 protein. Left panel: 412Q; right panel: 412R. The Q412R is depicted in blue and the nearby amino acids 459R, 407F and 429Y in plum. The contacts between Q412R and other amino acids are represented by red lines. R shows much more contacts with these amino acids as compared to Q. C, Q412R regulates IRF7 activity in vitro. ISRE reporter plasmid was co-transfected with either IRF7 412Q or 412R vector to HEK293 cells, and relative ISRE luciferase activity was measured after 24 hours. Expression of IRF7 412Q resulted in a 2-fold increase in ISRE transcriptional activity compared with that of IRF7 412R.
IRF7 Q412R polymorphism regulates IRF7 activation in vitro
To examine whether Q412R amino acid change is able to influence IRF7 mediated reaction, we tested the effects of IRF7 Q412R on the transcriptional activation of an IFN stimulated response element (ISRE)-dependent luciferase reporter. We co-transfected ISRE reporter plasmid together with IRF7 412Q or 412R vectors to HEK293 cells and measured the relative luciferase activity after 24h. As shown in Figure 2C, IRF7 over-expression in HEK293 cells enhanced ISRE luciferase activity. Expression of IRF7 412Q resulted in a 2-fold increase in ISRE transcriptional activity compared to that of 412R (P=0.0003), suggesting 412Q confers elevated IRF7 activity, affecting the downstream IFN pathway.
Discussion
IRF7 was first described as a protein that bound to and repressed the Qp promoter region of the Epstein-Barr virus encoded EBNA-1 gene, which contains an ISRE-like element in its promoter (9). Viral infection or DNA/RNA containing immune complexes, as in SLE, can trigger activation of Toll like receptor 7 (TLR7) or TLR9, leading to activation of transcriptional factors such as IRF3 and IRF7. Activated IRF7 translocates from cytoplasm to nucleus, where it induces the transcription of type I IFN genes (3).
Type I IFN pathway plays a crucial role in the pathogenesis of SLE. Significant elevation of various type I IFN-inducible genes, referred to as an IFN signature, was observed in peripheral blood cells from SLE patients and is closely associated with increased disease activity, specific autoantibody profiles and significant organ damage in SLE (10). Several genetic variants in IFN pathway such as risk hplotypes in IRF5 and STAT4 have been established in association with SLE susceptibility (reviewed in 1.2). Other identified SLE associated genes in type I IFN pathway include TREX1, TYK2, and SPP1 (reviewed in 1.2). The IRF7 SNP rs1131665 described here added a new member in this growing list and again, supported the pivotal role type I IFN system plays in the etiology of SLE.
The previously detected association of KIAA1542 SNP rs4963128 with SLE in European ancestry was not found in Asian populations. One possible explanation is that this SNP has different LD structure with IRF7 among various ethnic groups. Based on HapMap data (Release 27), rs4963128 has a much stronger LD with IRF7 SNP rs11246213 (located in intron 10, 241bp telomeric of rs1131665) (r2=0.89) in European (CEU) than that in Chinese (CHB) (r2 = 0.24) and African (YRI) ancestries (r2=0.18). Our current data does not exclude the possibility that KIAA1542 might be a causal gene itself, but does warrant a detailed study of this genomic region with saturating variant genotyping and locating the causal variants in multiple ethnic groups.
One of the limitations of the current study is that we have not investigated polymorphisms in IRF7 gene (3.4Kb) with sufficient density to be comprehensive. Given that multiple risk polymorphisms may exist within one gene, other IRF7 genetic variants might also contribute to the development of SLE. SLE patients carrying KIAA1542 SNP rs702966 risk genotype and expressing anti-dsDNA showed higher serum IFN-α activity. The LD structure and functional relationship between rs1131665 and rs702966 are not clear. Further fine-mapping studies are needed to elucidate this question.
In summary, we identified a nonsynonymous SNP (Q412R) in IRF7 that regulates IRF7 activation and predisposes to the development of SLE in multiple ethnic groups. At this point in time this polymorphism is thereby nominated as a candidate functional polymorphism. Whether IRF7 Q412R is causal awaits a complete assessment of the other possibly causal polymorphisms within the KIAA1542-IRF7 locus. This result provides direct genetic evidence that IRF7 may be a risk gene for human SLE, which again, supports the causal role of type I IFN pathway genes in the development of SLE.
Acknowledgement
We thank all subjects for participation in this study.
Grant support: This work was supported by US National Institute of Health grant AR043814, AR053483, RR015577, N01-AR062277, AI083194, RR020193, AR049084, AI24717, AR054459, 3AR42460 and Korea Healthcare Technology R&D Project, Ministry for Health and Welfare (A010252, A080588).
Footnotes
Note on the authors.
The members of TTSH Lupus Study group are: Bernard YH Thong, Kok Ooi Kong, Faith LA Chia, Hiok Hee Chng, Khai Pang Leong, Tsui Yee Lian, Weng Giap Law, Ee Tzun Koh, Grace YL Chan, Justina Tan and Wern Hui Yong.
Reference
- 1.Harley IT, Kaufman KM, Langefeld CD, Harley JB, Kelly JA. Genetic susceptibility to SLE: new insights from fine mapping and genome-wide association studies. Nat Rev Genet. 2009;10:285–290. doi: 10.1038/nrg2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moser KL, Kelly JA, Lessard CJ, Harley JB. Recent insights into the genetic basis of systemic lupus erythematosus. Genes Immun. 2009;10:373–379. doi: 10.1038/gene.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 4.Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crow MK. Collaboration, Genetic Associations, and Lupus Erythematosus. N Engl J Med. 2008;358:956–961. doi: 10.1056/NEJMe0800096. [DOI] [PubMed] [Google Scholar]
- 6.Salloum R, Franek BS, Kariuki SN, Rhee L, Mikolaitis RA, Jolly M, et al. Genetic variation at the IRF7/PHRF1 locus is associated with autoantibody profile and serum interferon-alpha activity in lupus patients. Arthritis Rheum. 2010;62:553–561. doi: 10.1002/art.27182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hochberg MC for the Diagnostic and Therapeutic Criteria Committee of the American College of Rheumatology. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 8.Zhang L, Pagano JS. Structure and function of IRF-7. J Interferon Cytokine Res. 2002;22(1):95–101. doi: 10.1089/107999002753452700. [DOI] [PubMed] [Google Scholar]
- 9.Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factor as regulators of host defense. Annu. Rev. Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 10.Feng X, Wu H, Grossman JM, Hanvivadhanakul P, FitzGerald JD, Park GS, et al. Association of increased interferon-inducible gene expression with disease activity and lupus nephritis in patients with systemic lupus erythematosus. Arthritis Rheum. 2006;54:2951–2962. doi: 10.1002/art.22044. [DOI] [PubMed] [Google Scholar]