Abstract
Information processing algorithms require significant amounts of annotated data for training and testing. The availability of such data is often hindered by the complexity and high cost of production. In this paper, we investigate the benefits of a state-of-the-art tool to help with the semantic annotation of a large set of biomedical information queries.
Seven annotators were recruited to annotate a set of 10,000 PubMed® queries with 16 biomedical and bibliographic categories. About half of the queries were annotated from scratch, while the other half were automatically pre-annotated and manually corrected. The impact of the automatic pre-annotations was assessed on several aspects of the task: time, number of actions, annotator satisfaction, inter-annotator agreement, quality and number of the resulting annotations.
The analysis of annotation results showed that the number of required hand annotations is 28.9% less when using pre-annotated results from automatic tools. As a result, the overall annotation time was substantially lower when pre-annotations were used, while inter-annotator agreement was significantly higher. In addition, there was no statistically significant difference in the semantic distribution or number of annotations produced when pre-annotations were used. The annotated query corpus is freely available to the research community.
This study shows that automatic pre-annotations are found helpful by most annotators. Our experience suggests using an automatic tool to assist large-scale manual annotation projects. This helps speed-up the annotation time and improve annotation consistency while maintaining high quality of the final annotations.
Keywords: PubMed queries, biomedical entities, annotation standards, annotation methods
1. Background
1.1 The challenges of annotation tasks
Producing annotated data is a necessary step for many Natural Language Processing (NLP) or information processing tasks. Annotated data is useful for researchers in at least two respects: first, as a means to fully understand the task at hand, and second as an input to train and evaluate computational approaches developed to automatically address the task. While a limited amount of data could be sufficient for the former, the latter requires a much larger amount. In the biomedical domain, data curation and annotation not only provides a basis for computational analysis, but also directly supports experimental research scientists [1,2]. Current annotation projects cover many aspects of the life sciences ranging from annotation of genes and proteins [3,4], to more complex tasks like gene/protein interaction [5,6], event extraction [7] and others [8].
Manual annotation is expensive and highly time-consuming. It requires qualified annotators. In addition, expert knowledge is needed for domain-specific annotation. As a result, variable inter-annotator agreement (or consistency) is a well-know issue of manual annotations for virtually any type of task, with a negative impact on the reproducibility of the task by automated approaches.
A variety of research efforts have attempted to address these issues. Several efforts sought to improve the consistency of annotations through clear and specific definition of annotation guidelines [9,10]. Authors diverge on the benefit of letting annotators communicate on the material during the annotation process: Lu et al. [9] found it increases consistency, but Alex et al. [5] expressed concern that annotators might influence one another and bias the overall reported consistency. Other studies showed that the use of automatic tools could improve overall consistency [11,12]. However, one has to keep in mind that high consistency does not necessarily equal high annotation quality [13 – cited by 14]. In fact, in a study of part-of-speech (POS) tagging, Marcus et al. [12] found that while automatic pre-annotations increased inter-annotator consistency, a slight decrease in tagging quality was also observed. Other work takes the inevitable inter-annotator variation into account and shows that annotator disagreement can be modelled statistically [15].
Time saving is another benefit obtained by using automatic pre-annotations. In previous projects, the time saving was found to be 22% for the annotation of protein-protein interactions in biomedical texts [6] and 50% for part-of-speech tagging [12]. Finally, the use of automatic pre-annotations has been shown to increase the level of general annotator satisfaction for some annotators [5,16], and to generally support annotators and curators in a complex task. It was shown that the support of reliable automatic annotation tools could allow annotators to increase the scope of the resulting annotations by quickly revising automatic annotations and focusing on annotations beyond the coverage of the automatic tool [6,11]. Some annotation tools also allow curators to explore the literature related to the entities they are annotating [17].
1.2 Annotation of PubMed Queries
Characterizing the semantic content of user queries is fundamentally important to understanding PubMed users’ behavior and information needs, and to address them adequately [18]. In addition to developing a new corpus of PubMed queries annotated with 16 semantic categories, this study aims to determine how to best address this task using state-of-the-art NLP and annotation tools. To assess the benefits and caveats of using automatic pre-annotations for an annotation task in the biomedical domain, we seek answers to the following questions:
Do pre-annotations help accomplish an annotation task? Specifically, what are the implications on the time spent performing the task, the annotator satisfaction and the consistency of the annotations?
Do pre-annotations influence annotators in a negative way? Specifically, would pre-annotations induce a significant variation in the distribution of categories? Would the quality or number of annotations vary if pre-annotations were used?
Compared to previous work in the biomedical domain, our study covers a large spectrum of biomedical concepts including types of entities that were not part of any previous large-scale annotation task (e.g. “diseases” and “medical procedures”). This work is based on a large corpus of 10,000 PubMed information queries, a lesser studied biomedical text genre. Previous work was based on smaller corpora, e.g. 4 to 10 papers in [6]. Our study involves seven annotators, which is a larger number than in many previous studies where up to four annotators participated [6,12,19]. The remainder of this paper is organized as follows: in the Methods section, we describe the corpus, annotation scheme, and annotation tools that were used in the study. We also define evaluation measures such as inter-annotator agreement, annotation divergence and number of actions. The Results section presents and comments on the annotations obtained in the study. Finally, in the Discussion section, we highlight the main implications and limitations of our work.
2. Methods
In this section, we describe the corpus and tools used to perform the annotation task, including details on the development of the annotation scheme.
2.1 PubMed Queries
A set of 10,000 PubMed queries was randomly extracted from the PubMed logs for October 17, 2005. The queries were processed to convert any non-ASCII characters (e.g. accented letters were replaced by non-accented ASCII equivalents), in order to facilitate processing with some of the automatic tools described below. The ten sample PubMed queries below illustrate the content of the corpus.
qid00008|“Mammon Z”[Author]
qid00029|http://copernic.com
qid00042|suzuki
qid00386|Herbal preparations for obesity: are they useful?
qid02632|j.med.chem
qid03752|treatment obesity dog
qid07505|prenatal insult
qid07647|“Glyburide”[MeSH] AND “Metformin”[MeSH]
qid08130|autism
qid09048|“Cancer”[Jour] AND 2004[pdat]
Queries 8, 7647 and 9048 include PubMed tags for author, journal, MeSH term and publication date searches. Length varies from one token (e.g. queries 29, 42, 8130) to many (e.g., title query 386), and topics also vary from bibliographic queries (e.g., queries 8, 42, 2632, 9048) to biomedical topics (e.g., queries 3752, 7647, 8130), sometimes unlikely (e.g., 7505).
2.2 Annotation tool
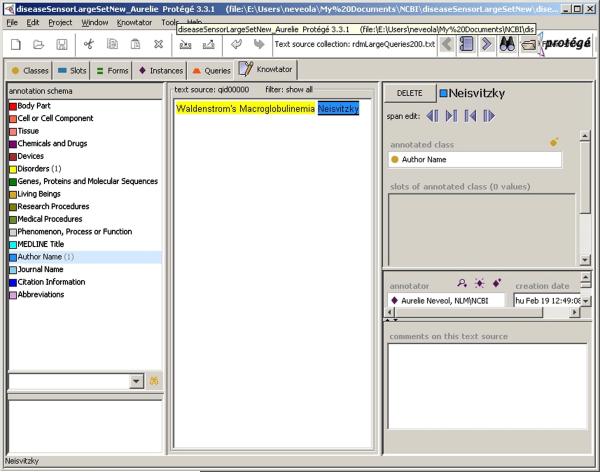
The Protégé plug-in Knowtator [20] was chosen as it provides a user-friendly interface to support the annotation. As shown in Figure 1, Knowtator divides the screen into three main windows. The annotation scheme can be seen on the left window, the queries can be viewed in the middle window, and annotations can be edited in the right window. Moreover, Knowtator allows us to compute inter-annotator agreement (IAA) for data analysis purposes.
Figure 1.
The sixteen category annotation scheme used to annotate PubMed queries is displayed on the left side of the Knowtator screen shot. A sample annotated query from the corpus is shown in the middle. Note that categories Devices and Living Beings correspond to the eponym UMLS Semantic Groups while categories Body Part, Cell or Cell Component and Tissue correspond to UMLS Semantic Types. Specific definitions and examples for all categories are given in Table 1 and in the supplementary data.
2.3 Annotation scheme
The rationale for annotating the query corpus is twofold. First, it can help understanding PubMed users’ information needs [18,21]. Second, it can be used in the development and evaluation of computational approaches for automatically analyzing PubMed queries. For instance, we adapted and evaluated two state-of-the-art tools for recognizing disease names in PubMed queries based on the corpus built in this study [22].
Given the aforementioned rationale, we developed a 16-category annotation scheme as follows: First, we manually identified different information needs in PubMed queries based on manual inspection of 200 sample queries. We reviewed the categories used in previous work [21], viz. Medical Subject Headings® (MeSH®) tree structures. We also considered semantic categories based on other existing terminological resources such as the UMLS® (Unified Medical Language System®) Semantic Groups. In order to ease the annotation task and keep it straightforward for human annotators, we avoided hierarchical relationships between categories. Our final annotation scheme had 16 categories, which we list in Table 1. Table 1 also gives brief definitions and examples of PubMed query terms that fall under each category. The final scheme as it appeared to annotators in Knowtator is shown in Figure 1. The supplementary data contains the specific definitions of each category in the annotation scheme provided to annotators. Shortly after the beginning of the annotation task, it was confirmed by the annotators that the scheme was suitable when applied outside of the initial 200 sample queries.
Table 1.
Brief definitions and examples for each of the 16 categories in the annotation scheme. More specific definitions and examples for all categories are given in the supplementary data.
| Annotation Category | Brief Definition | Examples |
|---|---|---|
| Body Part | A part/organ/limb of the human body |
finger, lung, heart |
| Cell or Cell Component | Type of cell or part of cell | Stem cell, membrane, nucleus |
| Tissue | Group of specialized cells | Abdominal muscle, subcutaneous tissue |
| Chemicals and Drugs | Antibiotic, drug or any chemical substance |
Aspirin, Metformin, lithium, calcium |
| Devices | Object used in research, diagnosis or treatment |
Adhesive bandage, insulin syringe |
| Disorders | Disease, syndrome, dysfunction, etc |
obesity, Heart Attack, autism, ankle fracture |
| Genes, Proteins and Molecular Sequences |
Name of any molecular sequence |
P450, lck, pex5, c-Myb transcription factor |
| Living Beings | Animal, human, organism | Male, alfalfa, mushroom, dog, marine bacteria |
| Research Procedures | Activity involving research, or experiment |
Real time PCR, bibliometric analysis |
| Medical Procedures | Activity involving diagnosis, or treatment |
Admission test, appendectomy, treatment |
| Phenomenon, Process or Function |
Biologic function, organism, cell or molecular function |
Mutation, apoptosis, protein interaction |
| MEDLINE Title | Title of a paper in MEDLINE | Herbal preparations for obesity: are they useful? |
| Author name | Name of authors | suzuki, Mammon Z, Jiang George |
| Journal name | Name of a journal referenced in MEDLINE |
Cancer, j.med.chem, Science |
| Citation information | Publication year, date, page number, etc |
2008, 2009 Feb;25(2) |
| Abbreviations | A shortened form of a word or phrase |
EEG, TB, DNA, NEJM, CRF, UGT2B1 |
2.4 Annotation guidelines
Along with the annotation scheme, annotation guidelines were developed. They contained definitions of the categories and examples of concepts that could be annotated with each category. Annotators were also encouraged to verify their annotations against reliable sources such as MEDLINE® or the UMLS Knowledge Source Server [23]. They were equally encouraged to perform these verifications when they worked with batches with or without pre-annotations.
One important guideline was that in general no annotations should be made for embedded entities. For example, three annotations could be derived from the query lung cancer: lung as a Body Part, and cancer and lung cancer as Disorders. According to our guidelines, only the annotation of the most specific Disorders, namely lung cancer, should be made. As embedded entities are not annotated, no Body Part annotation should be made in spite of the mention of lung in the query. This realizes our goal to identify the most specific entities in the queries. In addition, from an information retrieval perspective, we also expected the annotation of queries to provide a spectrum of what topics were searched by PubMed users. It seemed that embedded entities could hardly be defined as search topics. For example, a user querying for lung cancer is not likely to be searching documents related to lung. The only exception is for the Abbreviation category, where the guidelines advocate the annotation of the abbreviated form regardless. For instance, two annotations are expected for the query BRCA1: one for Genes and Proteins and the other for Abbreviations. As the annotation work progressed, the guidelines were enriched with examples of specific cases discussed in regular annotator meetings organized to ensure consistent interpretation of the guidelines. The supplementary data contains the guidelines and examples provided to annotators.
2.5 Conduct of the annotation task
Seven annotators with a variety of backgrounds were recruited to participate in the study. All annotators had previous experience with annotating biomedical documents. Three annotators have a computer science/information science background (annotators 1, 4 and 5), three a biology/medicine background (annotators 2, 3, 6), and one a bioinformatics background (annotator 7). In order to make the annotation task manageable and to facilitate the data analysis, the 10,000 queries were split into 50 batches of 200 queries. As previously mentioned, the first batch of 200 queries was used to define the annotation scheme, and was annotated by all seven annotators. Another 26 batches were distributed among the annotators to be annotated from scratch. The remaining 23 batches were automatically pre-annotated as follows:
MetaMap “mapping” results were converted to corresponding categories based on the semantic types of the concepts used in the definition of categories. For example, a concept of semantic type “diagnostic procedure” identified by MetaMap was automatically pre-annotated with the category Medical Procedures. For some recurring ambiguous concept that MetaMap associates with more than one semantic types, a consistent category assignment was chosen (e.g., MetaMap associates “cancer” to both the concepts “cancer genus” with the semantic type Invertebrate, which corresponds to the category Living Being and “neoplasm” with the semantic type Neoplastic Process, which corresponds to the category Disorder. In this case, we chose to have “cancer” pre-annotated as a Disorder category concept). A set of such equivalence rules (e.g. rule 1: if concept has semantic type “diagnostic procedure” annotate as Medical Procedures; rule 2: if concept has semantic type “Invertebrate”, unless concept is “cancer genus”, annotate as Living Being) was manually developed based on knowledge of the UMLS, and analysis of a sample query batch for fine-tuning. The full set of equivalence used is shown in supplementary Table 1. The rules were then implemented in a Perl script that automatically converted MetaMap results into xml files containing annotations according to our schema, which could be directly loaded into Knowtator. An example of automatic rule application on a sample query is shown in supplementary Table 2.
For the Author name, Journal name and Citation information categories, pre-annotations were created based on the MEDLINE search fields and tags in the queries [26], i.e. pre-annotations for these categories are only created if the corresponding PubMed search tags are present in the query.
For the Abbreviation category, pre-annotations were created for query words up to five characters long; if they contained all upper case letters and digits (e.g. BRCA1 and AIDS would be pre-annotated, but YCL069W would not, because it contains more than 5 characters. Similarly, p53 would not be recognized, because it contains a lower case letter, and neither would 123, because it contains only digits).
To assess the pre-annotation process, one of the pre-annotated batches was revised by all seven annotators. This batch was used to make sure that annotators would feel comfortable with the task of revising existing annotations vs. annotating from scratch. It was also used to measure inter-annotator agreement.
For each batch that they worked with, annotators were instructed to record the time they spent on producing or revising the annotations. The annotation task was distributed evenly among annotators, such that each annotator contributed annotations to the two shared batches and each annotator produced 4 or 5 batches of queries that included or did not include pre-annotations. As shown in Table 2, six annotators worked with a total of nine batches, two of which were also annotated by the others. One annotator worked with eight batches, including the two shared batches.
Table 2.
Batches assigned to each annotator participating in the study
 |
1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
|
Batches without pre-
annotations |
5 | 5 | 5 | 4 | 5 | 4 | 5 |
|
Batches with pre-
annotations |
4 | 4 | 4 | 5 | 4 | 4 | 4 |
| Total | 9 | 9 | 9 | 9 | 9 | 8 | 9 |
2.6 Creation of “Gold Standard” Annotations
To further assess the quality of the final annotations, two annotators (specifically, annotator 4 and 7) were involved in the creation of gold standard annotations for the two batches that were commonly annotated by all seven annotators. To obtain the “Gold Standard” annotations on each set, the two annotators were shown a pool of all the annotations made to queries in the set, without any indication of which annotator created the annotation, or how many annotators had agreed on a given annotation. The two annotators independently revised the pool of annotations. Then, the queries where their annotations still diverged (59 queries for the first batch without pre-annotations and 52 queries for the second batch with pre-annotations) were identified and they discussed the specific cases to arrive at a consensus, which could be different from any of the annotations initially produced by the seven annotators. In fact, prior annotations were used to compare and contrast in order to arrive at the best solution. For example, for one query “escaping the nuclear” some annotators had suggested the category “Phenomenon, Process or Function” while others had not produced any annotations. In the gold standard set, this query was finally annotated as a “MEDLINE Title” as it was considered to be a partial title query for the article “Escaping the nuclear confines: signal-dependent pre-mRNA splicing in anucleate platelets.
2.7 Data Analysis
Most of the data analysis was performed at the batch level (i.e. using one batch as a data point), using the Mann Whitney and Wilcoxon non-parametric tests (Prism5 [27]). We did not make the assumption that the observed data was normally distributed.
For the two batches of queries that were annotated by seven annotators, we used the Knowtator’s inter-annotator agreement (IAA). In Knowtator, IAA is computed as: IAA= number of matches/( number of matches + number of non-matches), where matches are computed at the category level, allowing string overlap. This means that span differences were counted as a match whereas category differences were not, as per the definitions below.
To assess the difference between pre-annotations and the final annotators’ annotations, we considered the following types of divergence between two annotation sets:
Category difference: a string was annotated with different categories (e.g. salmonella annotated as Living Beings vs. Disorders)
Span difference: overlapping strings were annotated with the same category (e.g. castleman disease multicentric vs. castleman disease annotated as Disorders)
Addition: an annotation was made by the annotator, but not by the automatic system
Removal: an annotation was made by the automatic system, but not by the annotator
For cases where both a span and a category differ (e.g. salmonella infection annotated as Disorders as a whole vs. salmonella as Living Beings by itself), we counted as one “removal” and one “addition” in our computation. Subsequently, the number of total actions performed by the annotators is computed as follows:
In batches without pre-annotations, the number of actions is the same as the number of total annotations
In batches with pre-annotations, the number of actions is computed as: 2*Category difference + Span difference + Addition + Removal The number of category differences was doubled because performing a category change in Knowtator requires the removal of the unwanted annotation and the subsequent addition of a new annotation, i.e. two actions in total. For other types of changes such as span difference, addition or removal of an existing annotation, only one Knowtator action was required.
Finally, each annotator reported the time spent annotating each of the batches of 200 queries, and their general satisfaction with the pre-annotation experiment.
3. Results
The corpus of 10,000 PubMed queries with 17,103 annotations is freely available for research purposes at http://www.ncbi.nlm.nih.gov/CBBresearch/Fellows/Neveol/Queries10K.zip. Overall, only 510 queries received no annotations (e.g. http://copernic.com or coreralation between the intracellular). The mean and standard deviation of queries without annotations was 10.2 +/−6.4 per batch, with at least one query without annotations in each of the 50 batches.
The impact of using automatic pre-annotations to assist manual annotations was studied from three aspects: annotation time, annotation quality, and annotator satisfaction.
3.1 Annotation time
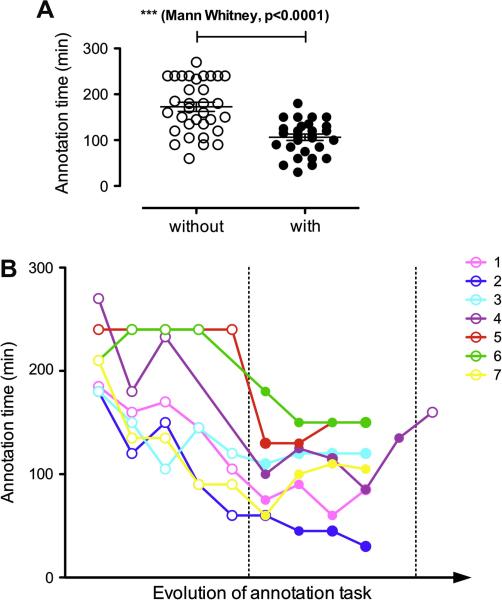
Figure 2 shows the time comparison from the perspective of batches and annotators. Figure 2A shows that the average annotation time in batches with pre-annotation was significantly lower compared to batches without pre-annotations (106 minutes vs. 173 minutes, a 38.7% decrease in time). Figure 2B shows the progression of annotation time for each individual annotator. As can be seen, there is a general trend of diminishing annotation time from one batch to the next for the first stage of our task where annotators created the annotations from scratch, suggesting that training (with enriched guidelines and examples) and experience play a role in shortening the annotation time. Furthermore, the average annotation time continued to decrease as can be seen in the latter stage of the process, where pre-annotations were used. Despite the general trend of decreasing annotation time, individual variations can be observed, but do not seem to be linked to the annotators’ background. For annotators 4, 5 (computer science/information science background), and 6 (biology/medical background), the pre-annotations seemed to have a greater impact on annotation time. The last two batches assigned to annotator 4 appear further to the right on Figure 2B, reflecting the fact that these batches were worked on some time after the main annotation period. In these two batches, the time spent on the batches with and without pre-annotations consistently in the same range as other batches for this annotator. However, the time difference between the two batches is smaller than between the average of this annotator’s other batches with and without pre-annotations. This might indicate that the impact of training is significant.
Figure 2.
Comparison of annotation time in minutes (a) overall and (b) for the seven annotators as the annotation task progresses. Full circles represent batches with pre-annotations while hollow circles represent batches without pre-annotations. Note that all the batches with pre-annotation appear between the vertical dashed lines.
Based on the observations, we hypothesized that the time saving shown in Figure 2 is due to two major factors: a) annotators became more efficient as they gained experience b) annotators also became more efficient as assistance became available in the form of pre-annotations (see discussion in Section 4.3). More specifically, we hypothesized that a lower number of actions needed to be performed for the pre-annotated batches.
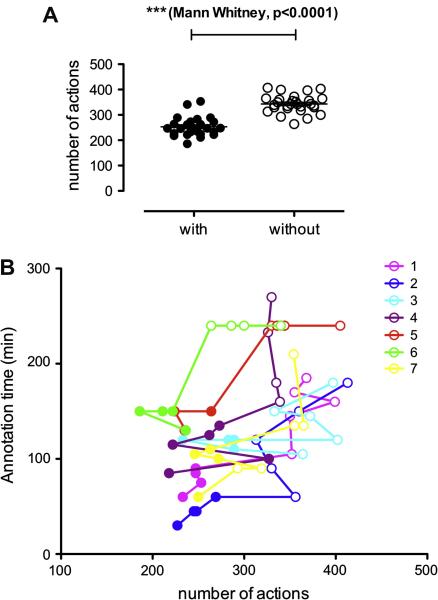
Figure 3A shows that overall, the number of actions to be performed is statistically lower for the pre-annotated batches (244 actions vs. 343, a 28.9% decrease in number of actions). On average, we can estimate that the average time per action is lower in batches with pre-annotations (~26 seconds) compared to batches without pre-annotations (~30 seconds). This could be due to training, but it could also be an indication that overall, annotators spent less time performing verifications on batches with pre-annotations if they felt confident that a pre-annotation was correct. However, the number of verifications performed, or the time spent on the verification was not recorded in our study.
Figure 3.
Comparison of the number of actions (a) overall and (b) for the seven annotators as a function of annotation time. Full circles represent batches with pre-annotations while hollow circles represent batches without pre-annotations.
Table 3 shows the distribution of the different divergence types (described previously in the Method Section) observed between the automatic annotations and the final annotations in pre-annotated batches. A large amount of divergence resulted from the “Addition” of annotations because many Author Name, Journal Name and Citation Information were missed by the rule-based algorithm we developed (automatic annotations for these categories relied entirely on the presence of tags in the queries). In Figure 3B, we can see that some batches requiring a high number of actions were still annotated relatively fast. In addition to individual variation between annotators, this could be due to the amount of verification that annotators performed, which was not recorded in our study.
Table 3.
Distribution of divergence types. (A) Between automatic pre-annotations and the final annotator’s annotations – result are shown as an average per batch over 23 batches where pre-annotations where shown to the annotators (B) between two annotators – results are shown as an average per batch and per annotator pair over two batches annotated by 7 annotators.
| Divergence type | Category difference |
Span difference |
Addition | Removal |
|---|---|---|---|---|
|
Between pre-
annotations and annotators |
13 | 12 | 156 | 40 |
|
Between two
annotators |
20 | 21 | 43 | 431 |
The number of addition and removal is similar because the comparison between two annotators was non-directional. Unlike the comparison between pre-annotations and annotators’ final annotations where the annotators’ work was considered as the “reference” for addition and removals, the comparison between two annotators was averaged by alternatively considering each annotator as a “reference”.
3.2 Annotation quality
To investigate the difference in quality between batches with and without pre-annotations, we observed the number of annotations, the inter-annotator agreement, and the category distribution in the two groups of batches.
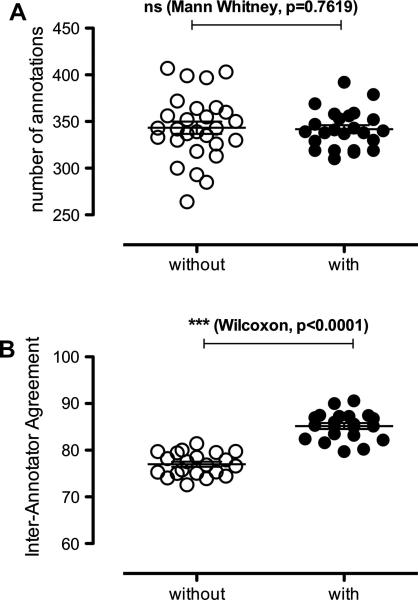
As shown on Figure 4A, there is no statistical difference in the number of annotations between batches with and without pre-annotations. Also, the smaller deviation between batches with pre-annotation suggests annotators are more consistent in producing annotations under such conditions. This is further illustrated in Figure 4B where the Inter-Annotator Agreements are significantly higher for batches with pre-annotations (on average, IAA=85.18 vs. 77.00).
Figure 4.
Comparison of (a) final number of annotations and (b) inter-annotator agreement for batches with and without pre-annotations. In (a) each circle represents a batch of queries; in (b) each circle represents an annotator pair.
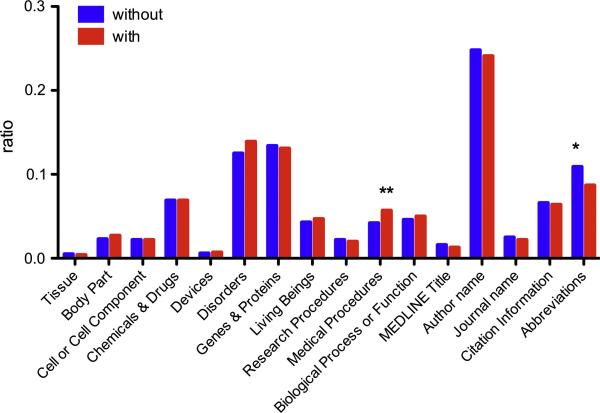
Figure 5 shows the distribution of annotations among the 16-category scheme in batches with and without pre-annotations. Overall, there is no statistical difference in the distribution (Wilcoxon, p=0.6233). However, at the category level, the number of Abbreviations annotations is statistically lower in batches with pre-annotations (*p=0.0107) whereas the number of Medical Procedures annotations is higher (**p=0.0040). We believe the decrease in Abreviations is due to the fact that this was the only category where overlaps with other annotations were permitted. As a result, the pre-annotations could have hidden the fact that an Abreviations annotation was missing.
Figure 5.
Comparison of the distribution of annotations over categories for batches with and without pre-annotations (stars indicate statistical differences in the distribution at the category level).
To further assess the quality of annotations, the agreement between the annotators and the “Gold Standard” annotations obtained for the two commonly annotated batches (cf. previous section) was computed. As shown in Table 4, the average agreement with the gold standard is high. It is higher in the batch with pre-annotations than in the batch without pre-annotations. In addition, in both batches, the average agreement with the gold standard is higher than the average inter-annotator agreement for the batch. These results indicate that the overall quality of annotations in the corpus is high. They also indicate that the higher inter-annotator agreement observed on the batch with pre-annotations is likely due to agreement on correct annotations, rather than a negative bias induced by the pre-annotations.
Table 4.
Overall agreement between annotators and gold standard annotations on sample 200-query sets with and without pre-annotations. The last column shows the average agreement for the batch.
| Annotator ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
|---|---|---|---|---|---|---|---|---|
|
Batch with pre-
annotations |
84.70 | 78.46 | 85.50 | 83.60 | 82.47 | 79.04 | 86.02 | 82.83 |
|
Batch without
pre-annotations |
89.46 | 85.11 | 83.02 | 84.71 | 86.67 | 79.07 | 87.41 | 85.89 |
3.3 Annotator satisfaction
Annotators were asked to assess their satisfaction with the use of pre-annotations on a five-point scale: very satisfied, satisfied, neither satisfied nor dissatisfied, dissatisfied, very dissatisfied. Three annotators were very satisfied with the pre-annotations (annotators 1, 4 and 5: those with computer science/information science background), two were satisfied (annotators 6 and 7), and two were neither satisfied nor dissatisfied (annotators 2 and 3). Overall, “satisfied” annotators liked that all explicit bibliographic annotations were already present, which saved time. They also reported that less manual looking up was required with pre-annotations while less satisfied annotators reported that the inconvenience of having to remove annotations (in particular for “title” type queries were several entities were usually pre-annotated) evened out the positive vs. negative aspects of pre-annotations. Satisfaction may be linked to annotator satisfaction with the pre-annotations as the annotators with a computer science/information science background were the most satisfied with pre-annotations. This may indicate that annotators with this type of background are more likely to trust and adopt automatic tools.
3.4 Annotation divergence
Table 3 shows the distribution of divergence types between pre-annotations and final annotations (in 23 batches with pre-annotations) as well as between 42 annotator pairs for the two batches that were annotated by all seven annotators. The main difference between annotator vs. annotator and annotator vs. pre-annotations is the number of additions – this is positive for the tool, as it tends to show that annotators mainly need to add annotations vs. remove or alter pre-annotations. The number of category and span divergence is smaller for annotator vs. pre-annotations, which reflects increased consistency when pre-annotations are used. An analysis of specific cases where annotations diverged shows that most cases fall into the following categories:
Entities belonging to multiple categories. Although categories were defined as specifically as possible, in some cases entities could still be reasonably assigned to more than one category. For example, in query cataract wild ginseng, wild ginseng can be annotated as Living beings (because it is a plant) as well as Chemical and Drug (because it is an alleged ingredient of medications for treating cataract). Such conflicts were not uncommon between category pairs such as Disorders vs. Living Being (e.g. Salmonella), Research Procedure vs. Medical Procedure (e.g. MRI), Gene or Protein vs. Chemical and Drugs (e.g. wortmannin).
Span divergence: the example above illustrates how span divergence could arise in the annotations. Span divergence can also occur in queries like alpha-adrenegic receptor subtypes, where two different annotations can be derived depending on whether the annotator regards the word subtypes as part of the gene name.
Complex entities: in some cases, annotators had different interpretations regarding the parsing of complex entities. For example, in query nfkb,IkB alpha degradation some annotators annotated IkB alpha degradation as Biological Process or Function whereas others separated IkB alpha as Gene and Protein and degradation as Biological Process or Function.
MEDLINE titles: if an annotator failed to identify the title of a MEDLINE publication, annotations for the biomedical entities in title query were usually created. Note that the identification of a “MEDLINE title” was not necessarily trivial, as some queries seem to contain truncated or slightly modified titles of MEDLINE citations, which can leave room for interpretation.
The number of “addition” and “removal” shown in Table 3 cover the differences of types 3 and 4 (complex entities and MEDLINE titles), the former being more frequent than the latter.
In order to reduce the different interpretations, regular annotator meetings were organized throughout the project. In addition to resolving differences for the two-shared sets, annotators were invited to present query examples that they found difficult to annotate in their own sets so that the group could agree on a consensus interpretation of the guidelines to apply to similar cases.
4. Discussion
The results above demonstrate the benefits of using pre-annotations produced by automatic tools to assist manual annotation.
4.1 Do pre-annotations induce negative bias in resulting annotations?
In typical conflict cases (e.g. Salmonella), the automatic pre-annotations always presented the same default choice to the annotators (e.g. a Disorders annotation). This likely resulted in higher regularity and consequently boosted the overall inter-annotator agreements in batches with pre-annotations as shown in Figure 4B. Although high inter-annotator agreement or a uniform distribution over the two types of batches are not a guarantee of the quality of annotations, the hypothesis of minimal negative influence from the pre-annotations is supported by the high agreement between the annotators and the gold standard. In addition, the agreement among annotators is almost twice as high as the agreement between annotators and the automatic system. This is consistent with the hypothesis that the annotators diverged with the pre-annotations that were erroneous or missing.
Specific attention can also be given to the diverse background of the annotators. In a recent effort involving annotations of pathology reports by linguists and pathologists [19], annotations produced by the linguists were found to be in higher agreement than annotations produced by the pathologists. Similarly, we observe a higher agreement between annotators with computer science/information science background compared to annotators with a biology/medicine background.
4.2 Implications of our annotation results
Being able to produce such a large-scale annotated corpus is not only important for our own mission at the National Library of Medicine (e.g. using such data for developing and testing PubMed sensors [28]), it would also be beneficial to the related research community and beyond. As detailed in previous work [18], this corpus served as the basis for understanding the information needs of PubMed users, including an analysis of query context which investigated which semantic classes are frequently queried together. The corpus could similarly benefit any research that is trying to improve the widely used search engine for biomedical literature. It would also benefit research that relies on the identification of biomedical entities [29] or author name disambiguation [30]. As mentioned before, we have used this annotated dataset for developing and evaluating two automatic approaches for recognizing disease names in biomedical text [22]. Similarly, this data can be used for other biomedical entity recognition problems ranging from widely studied entities like Gene & Proteins to rarely explored but important types like Medical/Research procedures.
4.3 Limitations of our study
The fact that batches with pre-annotations were given to the annotators after they had finished working on batches without pre-annotations makes it difficult to precisely characterize the two causes of the time saving that we observe (i.e. presence of pre-annotations vs. training and experience). As mentioned earlier in Section 3.1, by comparing sets with and without using pre-annotations, we found that there was an overall save of 38.7% in annotation time whereas the number of required annotation actions only dropped 28.9%, which led to a shorter time for each action (of 4 different types in Table 3) when using pre-annotations. Although it is likely that in this case, removal and changes with respects to category and spans should generally take less time to accomplish in Knowtator comparing to the action of addition, we cannot fully attribute the difference (38.7% vs. 28.9%) to this effect since they were not quantified directly. In order to precisely estimate the time saving due to each cause (i.e. presence of pre-annotations vs. training and experience), a future study should alternate work on batches with and without pre-annotations. Although annotators were encouraged to perform verifications to ensure annotation quality on all annotations in batches with and without annotations, recording the time allocated to verifications could also help determining whether pre-annotations also saved verification time.
Our analysis of pros and cons of using pre-annotations could be further enhanced by using additional sets of pre-annotations of known quality, such as random annotations (bad quality) and manual annotations (good quality). For example, Alex et al. [6] compared gold standard and the output of an automatic system as pre-annotations and found that gold standard annotations allowed a higher time saving than automatic system output.
5. Conclusions
In conclusion, this study shows that pre-annotations are found helpful by most annotators: it accelerates the annotation speed and improves annotation consistency. With such help, we were able to successfully annotate a large set of 10,000 PubMed queries, which is made freely available to the research community.
Supplementary Material
Acknowledgements
This research was supported by the Intramural Research Program of the NIH, National Library of Medicine. The authors would like to thank G. Jiang, S. Shooshan, T. Tao and WJ. Wilbur for their contribution to the annotation of the query corpus, colleagues in the NCBI engineering branch for their valuable feedback on the categorization scheme, D. Comeau for his editorial assistance, and P. Chappert at NIAID for his help using PRISM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary data
S 1 – Annotation guidelines
NeveolJBI2010_supplement1.doc contains the definitions each of the categories in the annotations scheme (with examples) as well as annotation guidelines provided to the annotators.
S 2 – Equivalence between UMLS Semantic Types and annotation scheme
NeveolJBI2010_supplement2.doc contains a table showing the list of equivalence rules used to automatically convert MetaMap results into pre-annotations. It also contains a table illustrating the use of these rules through application to MetaMap results obtained for a sample query.
S 3 – Annotated Corpus of 10,000 PubMed queries
Queries10K.zip is an archive containing the corpus of 10,000 PubMed queries described in this paper and the annotations in text format and Knowtator format. This data is available to the research community at the following location: http://www.ncbi.nlm.nih.gov/CBBresearch/Fellows/Neveol/Queries10K.zip
References
- [1].Peters B, Dirscherl S, Dantzer J, Nowacki J, Cross S, Li X, Cornetta K, Dinauer MC, Mooney MD. Automated analysis of viral integration sites in gene therapy research using the SeqMap web resource. Gene Ther. 2008 Sep;15(18):1294–8. doi: 10.1038/gt.2008.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Srinivasan P, Libbus B. Mining MEDLINE for implicit links between dietary substances and diseases. Bioinformatics. 2004 Aug 4;20(Suppl 1):i290–6. doi: 10.1093/bioinformatics/bth914. [DOI] [PubMed] [Google Scholar]
- [3].Morgan AA, Lu Z, Wang X, Cohen AM, Fluck J, Ruch P, Divoli A, Fundel K, Leaman R, Hakenberg J, Sun C, Liu HH, Torres R, Krauthammer M, Lau WW, Liu H, Hsu CN, Schuemie M, Cohen KB, Hirschman L. Overview of BioCreative II gene normalization. Genome Biol. 2008;9(Suppl 2):S3. doi: 10.1186/gb-2008-9-s2-s3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Smith L, Tanabe LK, Ando RJ, Kuo CJ, Chung IF, Hsu CN, Lin YS, Klinger R, Friedrich CM, Ganchev K, Torii M, Liu H, Haddow B, Struble CA, Povinelli RJ, Vlachos A, Baumgartner WA, Jr, Hunter L, Carpenter B, Tsai RT, Dai HJ, Liu F, Chen Y, Sun C, Katrenko S, Adriaans P, Blaschke C, Torres R, Neves M, Nakov P, Divoli A, Maña-López M, Mata J, Wilbur WJ. Overview of BioCreative II gene mention recognition. Genome Biol. 2008;9(Suppl 2):S2. doi: 10.1186/gb-2008-9-s2-s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Alex B, Grover C, Haddow B, Kabadjov M, Klein E, Matthews M, Roebuck S, Tobin R, Wang X. The ITI TXM Corpus: Tissue Expression and Protein-protein interactions. Proceedings of the LREC Workshop on Building and Evaluating Resources for Biomedical Text Mining; [Retrieved 08/18/09]. 2008b. from [ http://www.ltg.ed.ac.uk/np/publications/ltg/papers/Alex2008Corpora.pdf] [Google Scholar]
- [6].Alex B, Grover C, Haddow B, Kabadjor M, Klein E, Matthews M, Roebuck S, Tobin R, Wang X. Assisted Curation: Does Text Mining Really Help?. Proceedings of the Pacific Symposium on Biocomputing; 2008a. pp. 556–567. [PubMed] [Google Scholar]
- [7].Kim JD, Ohta T, Pyysalo S, Kano Y, Tsujii J. Overview of BioNLP’09 Shared Task on Event Extraction. Proceedings of NAACL BioNLP Workshop.2009. pp. 1–10. [Google Scholar]
- [8].Winnenburg R, Wächter T, Plake C, Doms A, Schroeder M. Facts from text: can text mining help to scale-up high-quality manual curation of gene products with ontologies? Brief Bioinform. 2008 Nov;9(6):466–78. doi: 10.1093/bib/bbn043. [DOI] [PubMed] [Google Scholar]
- [9].Lu Z, Bada M, Ogren P, Cohen KB, Hunter L. Improving Biomedical Corpus Annotation Guidelines. Proceedings of the Joint BioLink and 9th Bio-Ontologies Meeting; 5 August 2006. [Google Scholar]
- [10].Wilbur WJ, Rzhetsky A, Shatkay H. New directions in biomedical text annotation: definitions, guidelines and corpus construction. BMC Bioinformatics. 2006 Jul 25;7:356. doi: 10.1186/1471-2105-7-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].McShane M, Nirenburg S, Beale S, O’Hara T. Semantically Rich Human-Aided Machine Annotation. Proceedings of NAACL Workshop, Frontiers in Corpus Annotation II: Pie in the Sky; 2005. [Google Scholar]
- [12].Marcus MP, Santorini B, Marcinkiewicz MA. Building a large annotated corpus of English: the Penn Treebank. Computational Linguistics. 1993:19. [Google Scholar]
- [13].Cooper WS. Is Inter-indexer consistency a hobgoblin? American Documentation. 1969;20:268–278. [Google Scholar]
- [14].Funk ME, Reid CA. Indexing consistency in MEDLINE. Bull Med Libr Assoc. 1983 Apr;71(2):176–83. [PMC free article] [PubMed] [Google Scholar]
- [15].Rzhetsky A, Shatkay H, Wilbur WJ. How to get the most out of your curation effort. PLoS Comput Biol. 2009 May;5(5):e1000391. doi: 10.1371/journal.pcbi.1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ruiz ME, Aronson AR. Technical report. U.S. National Library of Medicine; [Retrieved 07/31/09]. 2007. User-centered Evaluation of the Medical Text Indexing (MTI) system. from [ http://ii.nlm.nih.gov/resources/MTIEvaluation-Final.pdf] [Google Scholar]
- [17].Couto FM, Silva MJ, Lee V, Dimmer E, Camon E, Apweiler R, Kirsch H, Rebholz-Schuhmann D. GOAnnotator: linking protein GO annotations to evidence text. J Biomed Discov Collab. 2006 Dec 20;1:19. doi: 10.1186/1747-5333-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Islamaj-Doğan R, Murray GC, Névéol A, Lu Z. Understanding PubMed user search behavior through log analysis. Database. 2009. bap018; doi:10.1093/database/bap018. [DOI] [PMC free article] [PubMed]
- [19].Patrick J, Scolyer R. Technical report. Health Information Technologies Research Laboratory; [Retrieved 08/11/09]. 2008. Information Extraction from Narrative Pathology Report on Melanoma. from [ http://www.it.usyd.edu.au/~hitru/] under the “Essays” section. [Google Scholar]
- [20].Ogren PV. Knowtator: A plug-in for creating training and evaluation data sets for Biomedical Natural Language systems. Proceedings of the 9th Intl. Protégé Conference; 2006. [Google Scholar]
- [21].Herskovic JR, Tanaka LY, Hersh W, Bernstam EV. A day in the life of PubMed: analysis of a typical day’s query log. J Am Med Inform Assoc. 2007;14(2):212–20. doi: 10.1197/jamia.M2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Névéol A, Kim W, Wilbur WJ, Lu Z. Exploring Two Biomedical Text Genres for Disease Recognition. Proceedings NAACL 2009, Workshop BioNLP. [Google Scholar]
- [23]. The UMLS Knowledge Source server [ http://umlsks.nlm.nih.gov/] [PubMed]
- [24].Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proceedings of AMIA Symp.; 2001; pp. 17–21. [PMC free article] [PubMed] [Google Scholar]
- [25].Aronson AR, Lang FM. The Evolution of MetaMap, a Concept Search Program for Biomedical Text. Proceedings of Amia Symp.; 2009. [Google Scholar]
- [26]. PubMed searchfields and tags [ http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=helppubmed&part=pubmedhel p#pubmedhelp.Search_Field_Descrip]
- [27]. GraphPad Prism [ http://www.graphpad.com/prism/Prism.htm]
- [28].National Library of Medicine Drug Sensor Added to PubMed® Results Page. NLM Technical Bulletin. 2008 Jun 18; http://www.nlm.nih.gov/pubs/techbull/ja08/ja08_drug_sensor.html.
- [29].Pafilis E, O’Donoghue SI, Jensen LJ, Horn H, Kuhn M, Brown NP, Schneider R. Reflect: augmented browsing for the life scientist. Nat Biotechnol. 2009 Jun;27(6):508–10. doi: 10.1038/nbt0609-508. [DOI] [PubMed] [Google Scholar]
- [30].Torvik VI, Smalheiser NR. Author Name Disambiguation in MEDLINE. ACM Trans Knowl Discov Data. 2009 Jul 1;3(3):pii, 11. doi: 10.1145/1552303.1552304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.