Abstract
Endosomal Toll-like receptors (TLRs) 7 and 9 recognize viral pathogens and induce signals leading to the activation of nuclear factor κB (NF-κB)–dependent proinflammatory cytokines and interferon regulatory factor 7 (IRF7)–dependent type I interferons (IFNs). Recognition of viral nucleic acids by TLR9 requires its cleavage in the endolysosomal compartment. Here, we show that TLR9 signals leading to the activation of type I IFN, but not proinflammatory cytokine genes, require TLR9 trafficking from endosomes to a specialized lysosome-related organelle. Furthermore, we identify adapter protein-3 as the protein complex responsible for the trafficking of TLR9 to this subcellular compartment. Our results reveal an intracellular mechanism for bifurcation of TLR9 signals by selective receptor trafficking within the endosomal system.
Endosomal Toll-like receptors (TLRs) 7 and 9 are used by plasmacytoid dendritic cells (pDCs) for recognition of viral nucleic acids (1). These endosomal TLRs require trafficking by the endoplasmic reticulum (ER) membrane protein UNC93B (2, 3). UNC93B physically interacts with TLR3, TLR7, and TLR9 via the transmembrane domain (2) and transports these TLRs from the ER to the endosome. In order for TLR9 to signal, the ectodomain of TLR9 must be first cleaved in the endolysosomes by cathepsins (4, 5). Although both TLR7 and TLR9 signal through the adapter protein MyD88 (myeloid differentiation primary response gene 88), two bifurcating pathways emanate from these TLRs. The first pathway leads to the transcriptional activation of proinflammatory cytokines and requires nuclear factor κB (NF-κB) (6). The second pathway leads to the activation of type I interferon (IFN) genes through phosphorylation of interferon regulatory factor 7 (IRF7) (7). Although both pathways depend on MyD88 (8) and UNC93B (3), the latter pathway also requires additional molecules (9–15). Retention of multimeric A-type CpG DNA (CpG-A) in early endosome is associated with IFN production in pDCs, whereas rapid translocation of monomeric B-type CpG DNA (CpG-B) to the lysosome is associated with NF-κB activation (16, 17). These studies suggested that the intracellular location of TLR9 activation could dictate the nature of its downstream signaling. The mechanism by which these two types of signals are mediated through these TLRs, however, remains poorly defined.
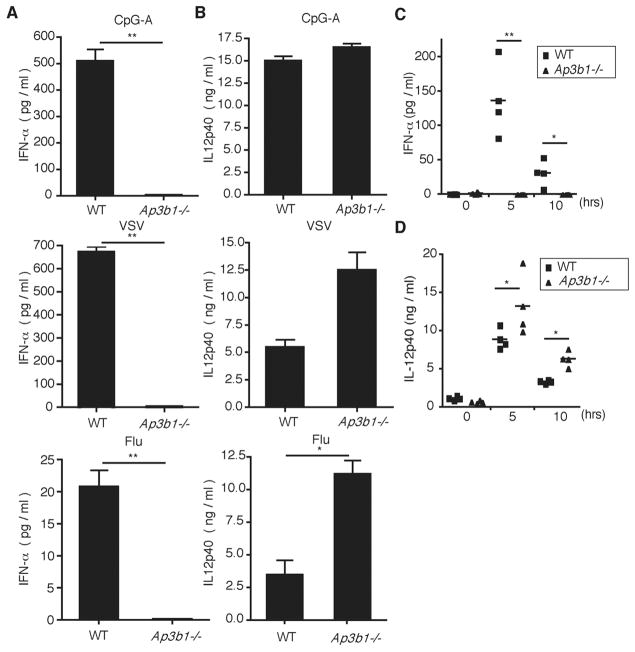
To gain an insight into the cell biological basis of TLR9 signaling, we examined adaptor proteins (APs) that could mediate trafficking of TLR9 (18). APs select cargo for inclusion into coated vesicles in the late secretory and endocytic pathways. Of the four adaptors identified for coated vesicles (AP-1, AP-2, AP-3, and AP-4), the AP-3 complex (consisting of four subunits: δ, β3A, μ3A, and σ3) is found on the endosomal membranes and has been shown to recruit its cargo in the endosomes and deliver them to lysosome-related organelles (LROs) (19). LROs share features of late endosome/lysosomes but are functionally, morphologically, and compositionally distinct. To better understand the nature of the endosome from which TLR7 and TLR9 signal, cultured bone marrow (BM) pDCs from wild-type (WT) or AP-3–deficient (Ap3b1−/−) mice were incubated with CpG-A (TLR9 agonist), vesicular stomatitis virus (VSV), or influenza virus (TLR7 agonists). AP-3–deficient pDCs failed to secrete IFN-α (Fig. 1A) despite normal to moderately enhanced production of interleukin (IL)–12p40, which is an NF-κB–dependent cytokine (Fig. 1B). Similar observations were made in pDCs from mice that have mutations in AP-3β1 (pearl mice) or AP-3δ (mocha mice), which render the AP-3 complex defective (fig. S1, A and B). This was not due to selective block in IFN protein secretion but was due to the failure of AP-3–deficient pDCs to activate transcription of IFN genes (fig. S1C). To examine whether this phenotype was restricted to pDCs, we took advantage of the fact that liposome-mediated delivery of CpG-A to conventional DCs and macrophages enables production of type I IFNs through IRF7 activation by TLR9 (17). Upon stimulation with CpG-A coupled to DOTAP (a liposomal transfection reagent) (Roche Diagnostics, Indianapolis, IN) or with TLR7 agonists poly uridine (poly U) or R848, Ap3b1−/− BM DCs were also found to have impaired IFN production while maintaining the production of IL-12p40 (fig. S2, A to C). Both cytokine and IFN responses after DOTAP–CpG-A stimulation were largely dependent on TLR9, albeit low levels were detected at higher CpG-A doses (fig. S2D), as reported previously (17). To determine whether the requirement for AP-3 also applies to other TLRs known to engage type I IFN stimulation from the endosome (20, 21), we stimulated BM macrophages (BMMs) from WT or Ap3b1−/− mice with lipopolysaccharide (LPS) (a TLR4 agonist) or polyinosinic:polycytidylic acid (poly I:C) (a TLR3 agonist). IFN-β expression was partially impaired in the absence of AP-3 (fig. S2, E and F), suggesting the involvement of this adaptor complex in signaling from other endosomal TLRs. Next, we examined the in vivo relevance of AP-3 deficiency in TLR9 and TLR7 signaling. AP-3–deficient mice did not have any obvious defects in pDCs or other DC subset development (fig. S3). Mice were injected systemically with CpG-A, which is known to trigger pDCs in the spleen to secrete high amounts of type I IFNs (22). Serum concentrations of IFN-α were undetectable in Ap3b1−/−mice, whereas they peaked around 5 hours after injection in WT mice (Fig. 1C). In contrast, high amounts of IL-12p40 were detected in both groups, with slightly elevated amounts found in the Ap3b1−/−group (Fig. 1D). These data demonstrated that the AP-3 complex is required for signaling through TLR7 and TLR9 to induce activation of IFN but not proinflammatory cytokine genes.
Fig. 1.
Type-I IFN production is impaired in AP-3–deficient pDCs after endosomal TLR stimulation. Flt3L-cultured BM pDCs from WT and Ap3b1−/− mice were stimulated with (top) CpG-A DNA (3 μM), (middle) VSV (MOI 10), or (bottom) influenza A virus infection (MOI 5) for 24 hours. Amounts of (A) IFN-α and (B) IL-12p40 were measured by means of enzyme-linked immunosorbent assay (ELISA). WT and Ap3b1−/− mice were injected intravenously with CpG-A DNA (25 μg), and at the indicated time points serum concentrations of (C) IFN-α and (D) IL-12p40 were measured with ELISA. *P < 0.05. **P < 0.01. Results are mean ± SEM (n = 4 mice) and are representative of [(A) and (B)] four and [(C) and (D)] three independent experiments.
Next, we examined whether the defective signaling of TLR9 in Ap3b1−/− cells could be explained by transcriptional regulation of signaling molecules downstream of TLR9. Quantitative reverse transcription polymerase chain reaction (RT-PCR) comparison of mRNA encoding MyD88, TRAF6, TRAF3, IRAK4, IRAK1, and IKK-α revealed no differences in either steady state or CpG-induced levels of these molecules (fig. S4, A and B). CpG-A–mediated transcriptional activation of IFN-responsive genes, however, was highly dependent on the presence of AP-3 in pDCs (fig. S4C).
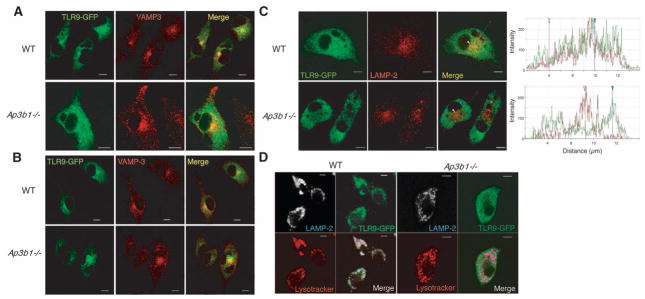
These results led us to focus on the cell biological basis for the requirement of AP-3 in TLR signaling. We hypothesized that TLR9 signals from two distinct endosomal compartments, one that engages NF-κB (NF-κB endosome) and the other that engages IRF7 (IRF7 endosome). Whereas both of these compartments must be sufficiently proteolytic to allow TLR9 cleavage to occur (4, 5), AP-3 is only required for the function of the IRF7 endosome. To test this hypothesis, we tracked TLR9 by transducing primary BMM with a TLR9–green fluorescent protein (GFP) fusion protein in which GFP was fused to the C terminus of TLR9. TLR9-GFP retained signaling activity (fig. S5). Under steady-state conditions, TLR9 was found in intracellular compartments, including the ER in both WT and AP-3–deficient cells (fig. S6), as shown previously (23–25). TLR9 entered VAMP3+ endosomes after 3 hours of DOTAP–CpG-A stimulation in both WT and Ap3b1−/− cells (Fig. 2A). VAMP3 marked early endosomes, which are distinct from the major histocompatibility complex (MHC) class II+ late endosomes and lysosomes (26) (fig. S7). In contrast to WT cells, however, TLR9 failed to migrate into the LAMP2+ endolysosomal compartment at 6 hours after DOTAP–CpG-A stimulation in Ap3b1−/− cells (Fig. 2, B and C). Trafficking of DOTAP-conjugated CpG-A and CpG-B was not impaired in the absence of AP-3 (fig. S8). These data indicated that AP-3 is required for trafficking of TLR9 to the LAMP2+, but not VAMP3+, compartment. A previous study showed that a small portion of transmembrane proteins including LAMP2 are mislocalized to the plasma membrane in AP-3–deficient cells (27). Therefore, to exclude the possibility that the lack of TLR9 and LAMP2 colocalization in Ap3b1−/−cells (Fig. 2C) is due to mislocalization of LAMP2 and not to defective TLR9 trafficking we labeled the late endolysosomal compartment using LysoTracker (Invitrogen, Carlsbad, California), which labels acidic compartments. TLR9 localized to LAMP2+ LysoTracker+ vesicles in WT macrophages after DOTAP–CpG-A stimulation. In contrast, even though LAMP2 localized within the acidic compartment TLR9 did not localize in LAMP2+ LysoTracker+ compartment in Ap3b1−/− cells (Fig. 2D). Collectively, these data indicated that the majority of LAMP2 still localized to intracellular acidified vesicles in Ap3b1−/− cells and that TLR9 trafficking to these vesicles requires AP-3.
Fig. 2.
TLR9 fails to enter the acidic LRO compartment in AP-3–deficient cells after CpG-A stimulation. BMM from WT and Ap3b1−/− mice were transduced with TLR9-GFP retrovirus. Cells were stimulated with DOTAP–CpG-A and analyzed for TLR9-GFP (green) and VAMP-3 (an early endosomal marker) (red) at (A) 3 hours or (B) 6 hours by means of confocal microscopy. (C and D) TLR9-GFP–transduced BMM were stimulated with DOTAP–CpG-A for 6 hours and analyzed by means of confocal microscopy to detect (C) TLR9-GFP (green) and LAMP-2 (late endosomal marker) (red). The histograms depict cross-line scans of the fluorescence intensities of the merged panel. (D) To visualize the acidic compartment, the cells were incubated with LysoTracker (red) followed by staining for TLR9-GFP (green) and LAMP-2 (white/blue). The data are representative of three independent experiments. Scale bars, 5 μm.
The ER transmembrane protein UNC93B associates with TLR3, TLR7, and TLR9 in the ER and facilitates their transport to the endolysosome (2, 3). We next examined whether the defect in TLR9 trafficking in Ap3b1−/− cells was due to mis-localization of UNC93B. Primary BMM from WT and Ap3b1−/− mice were cotransduced with retroviruses encoding TLR9-GFP and UNC93B-HA. Six hours after DOTAP–CpG-A stimulation, the localization of TLR9 (fig. S9, green), UNC93B (fig. S9, white), and LAMP2 (fig. S9, red) was assessed with confocal microscopy. Whereas UNC93B colocalized with TLR9 in the LAMP2+ endosome in WT cells, neither UNC93B nor TLR9 entered the LAMP2+ compartment in AP-3–deficient cells (fig. S9). These data indicated that AP-3 is required for trafficking of both UNC93B and TLR9 to the LAMP2+ compartment after DOTAP–CpG-A stimulation.
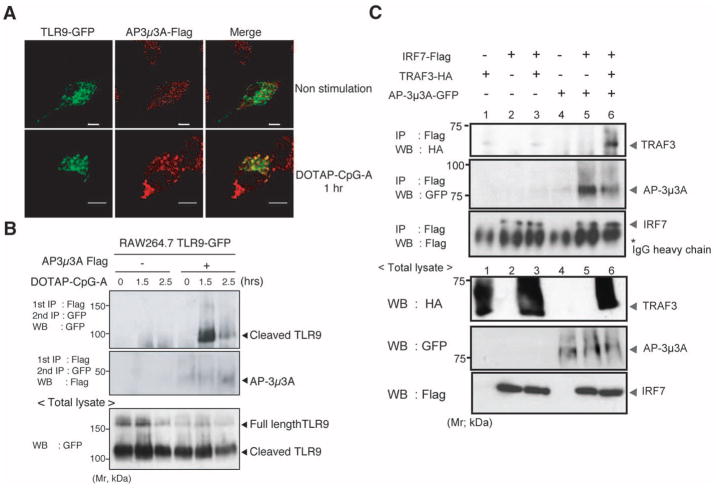
We next examined a possible interaction between TLR9 and AP-3. AP-3 is normally localized to the trans Golgi network and the tubular endosomal sorting compartment (28). Accordingly, whereas little colocalization of the AP-3 complex with TLR9 was observed at steady state, upon CpG-A stimulation substantial colocalization between AP-3 and TLR9 was seen (Fig. 3A). In contrast, we observed minimal colocalization of AP-3 and TLR9 at any time during the 120-min observation period when cells were stimulated with DOTAP alone, DOTAP-GpC, or DOTAP–CpG-B (fig. S10). To further test whether TLR9 and AP-3 interact, TLR9-GFP and a Flag-tagged AP-3μ3A subunit were coexpressed in RAW264.7 macrophages, and their association after DOTAP–CpG-A stimulation was determined by means of coimmunoprecipitation (Fig. 3B). These data indicated that AP-3μ3A preferentially associated with cleaved TLR9 in the presence of DOTAP–CpG-A (Fig. 3B) but not DOTAP alone, DOTAP-GpC, or DOTAP–CpG-B stimulation (fig. S11). We hypothesized that AP-3 might mediate this through the recruitment of downstream signaling proteins capable of activating IRF-7. In this regard, tumor necrosis factor (TNF) receptor–associated factor 3 (TRAF3) has been shown to specifically trigger activation of type I IFN genes upon TLR engagement (10, 11). AP-3 associated with IRF7 and was capable of promoting interaction between IRF7 and TRAF3 (Fig. 3C). These data indicated that besides associating with the cleaved form of TLR9 and facilitating its trafficking to the LAMP2+ compartment, AP-3 supports the association of TRAF3 and IRF7.
Fig. 3.
AP-3 forms a complex with TLR9 and promotes association of TRAF3 and IRF7. (A) RAW264.7 cells expressing TLR9-GFP and AP-3μ3A were left unstimulated or stimulated with DOTAP–CpG-A (3 μM) for 1 hour. The cells were stained and analyzed for TLR9-GFP (green) and AP-3μ3A (red). Scale bars, 5 μm. (B) RAW264.7 cells transduced with a combination of retroviruses encoding TLR9-GFP or AP-3μ3A-Flag or both were stimulated with DOTAP–CpG-A for the indicated time points, and lysates were prepared and analyzed either (bottom) directly or (top) after immunoprecipitation with an antibody to Flag and immunoblotted with an antibody to GFP. (C) Human embryonic kidney (HEK) 293 T cells transfected with the indicated plasmids for 24 hours were lysed and immunoprecipitated with antibody to Flag, separated by means of SDS–polyacrylamide gel electrophoresis (SDS-PAGE), and blotted with the indicated antibodies. The data are representative of (A) four, (B) five, and (C) three independent experiments.
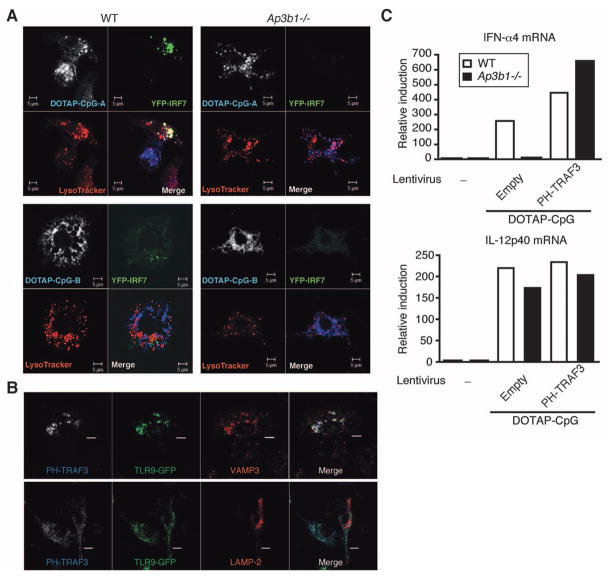
To directly examine from which intracellular compartment TLR9 signals to activate IRF7, we performed a confocal microscopy analysis of IRF7 engagement by TLR9 at various time points after DOTAP-CpG stimulation. BMM from WT and Ap3b1−/− mice were transduced with IRF7–yellow fluorescent protein (YFP) (fig. S12A). In unstimulated cells and cells stimulated with DOTAP–CpG-B, IRF7 signal remained diffuse throughout the cytosol. In contrast, IRF7 was recruited to CpG-A–containing LysoTracker+ vesicles in cells stimulated with DOTAP–CpG-A (Fig. 4A). IRF7 failed to form around CpG-A–containing vesicles in AP-3–deficient cells. Quantitative analysis revealed that 50% of WT cells and 5% of Ap3b1−/− cells had IRF7 recruited to CpG-A–containing vesicles (fig. S12B). These data further supported the notion that AP-3 is required to recruit IRF7 to TLR9.
Fig. 4.
Targeting of TRAF3 to PI(3,5)P2+ compartment bypasses the requirement for AP-3 in TLR9 signaling. (A) BMM from Ap3b1−/− mice transduced with lentivirus encoding YFP-IRF7 were stimulated with DOTAP–CpG-A-Cy5 for 6 hours and analyzed by means of confocal microscopy to detect YFP-IRF7 (green), LysoTracker (red), and CpG-A (white/blue). Scale bars, 5 μm. (B) BMM from Ap3b1−/− mice transduced with retrovirus encoding TLR9-GFP and PH-TRAF3 were stimulated with DOTAP–CpG-A for 4 hours (VAMP-3 staining) or 6 hours (LAMP-2 staining) and analyzed by means of confocal microscopy in order to detect TLR9-GFP (green), VAMP3 (red), or LAMP-2 (red), and PH-TRAF3 (blue). Scale bars, 5 μm. (C) BMMs from Ap3b1−/− mice were transduced with lentivirus encoding TRAF3 or PH-TRAF3. IFN-α and IL-12p40 mRNA expression was assessed by means of quantitative RT-PCR at 12 hours after DOTAP–CpG-A stimulation. Results are representative of four independent experiments.
Our data thus far suggested that the failure of TLR9 to induce IFN expression in AP-3–deficient cells is due to the inability of TLR9 to traffic to a specialized LAMP2+ LRO and engage IRF7. If this is the case, it should be possible to bypass the requirement for AP-3 by targeting TRAF3 to the VAMP3+ compartment from which TLR9 normally engages NF-κB activation. To accomplish this goal, we searched for a targeting motif that allowed molecules to be specifically recruited to such an endosomal compartment. Phosphoinositides (PIs) are a key component of cell membranes. Every organelle is equipped with a distinct array of PI kinases and PI phosphatases, which produce a specific set of PI species. Phosphatidylinositol 3,5-bisphosphates [PI(3,5)P2] are synthesized in the early endosomes (29) and are also found on the membrane of multivesicular bodies (30). Thus, we generated a chimeric molecule in which a PI(3,5)P2-binding plextrin homology (PH) domain of centaurin β2 (31) is fused to the N-terminal end of TRAF3 (PH-TRAF3). PH-TRAF3 associated with the VAMP3+LAMP2−TLR9+ endosomes in response to DOTAP–CpG-A stimulation in Ap3b1−/− cells (Fig. 4B). Combined with our finding that TLR9 localization in VAMP3+ compartment is sufficient to trigger IL-12p40 production (Figs. 1 and 2), these data suggested that PI(3,5)P2 decorate the VAMP3+LAMP2− endosome from which TLR9 signals to activate NF-κB (NF-κB endosome). To determine whether TRAF3 targeting to the NF-κB endosome could overcome the deficient IFN induction seen in Ap3b1−/− cells, Ap3b1−/− primary BMM were transduced with PH-TRAF3, and its ability to induce IFN expression after DOTAP–CpG-A stimulation was assessed. We found that targeting of TRAF3 to the NF-κB endosome was sufficient to reconstitute TLR9 signaling for IFN induction in Ap3b1−/−cells (Fig. 4C). Further, IL-12p40 gene expression was not substantially altered by PH-TRAF3 expression, indicating that the levels of ectopic PH-TRAF3 expression in primary macrophages were not sufficient to affect NF-κB signaling. Collectively, our data indicated that the defective TLR9 signaling in the absence of AP-3 is due to the inability of TLR9 to enter the specialized LRO capable of recruiting TRAF3 and IRF7 to the MyD88 signaling complex.
On the basis of our current study, we propose the following mechanism for TLR9 trafficking and signaling (fig. S13). TLR9 synthesized in the ER traffics through the Golgi to enter the early endosome—a process facilitated by UNC93B. This VAMP3+LAMP2− PI(3,5)P2+ endosome (NF-κB endosome) is proteolytically active, enabling TLR9 to be cleaved and signal through NF-κB to activate proinflammatory cytokine genes. AP-3 complex interacts with cleaved TLR9 in the NF-κB endosome and facilitates its trafficking to the LAMP2+ LRO compartment, where TLR9 can engage the TRAF3 and IRF7 signaling pathway and induce transcription of IFN genes (IRF7 endosome). This specialized IFN-inducing LRO compartment is probably further downstream of the NF-κB–inducing endosome because transcription of IL-12 p40 occurred as early as 2 hours after CpG-A stimulation in pDCs, whereas mRNA for IFN-α was not detected until 4 to 6 hours (fig. S14). Moreover, intracellular cytokine analysis of pDCs stimulated with CpG-A revealed that cells that secrete IFN-α always also produced TNF-α but not vice versa (fig. S15). Given that AP-3 is dispensable for conventional maturation of endosomes but is required for the formation of LRO in specialized cell types such as melanocytes (melanosomes) and platelets (platelet granules) (19), it is tempting to speculate that pDCs are naturally equipped with an LRO dedicated for IFN production and that DCs and macrophages are capable of generating such LROs when CpG-A is delivered by cationic liposomes. Our data are consistent with the idea that the temporal and spatial coordination of a single receptor could be used to send signals for transcriptional activation of a distinct set of genes and that an adaptor protein can regulate this process.
Supplementary Material
Acknowledgments
We thank R. Medzhitov for critical reading of the manuscript and W. Mothes for helpful discussion. This work is supported by NIH grants to A.I. (AI054359, AI081884, and AI064705). M.S. is a Japan Society for the Promotion of Science fellow and a recipient of Uehara Memorial Foundation fellowship. A.I. acknowledges support by Gilead Sciences. A.I. is a recipient of the Burroughs Wellcome Investigators in Pathogenesis of Infectious Disease.
Footnotes
References and Notes
- 1.Liu YJ. Annu Rev Immunol. 2005;23:275. doi: 10.1146/annurev.immunol.23.021704.115633. [DOI] [PubMed] [Google Scholar]
- 2.Brinkmann MM, et al. J Cell Biol. 2007;177:265. doi: 10.1083/jcb.200612056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tabeta K, et al. Nat Immunol. 2006;7:156. doi: 10.1038/ni1297. [DOI] [PubMed] [Google Scholar]
- 4.Ewald SE, et al. Nature. 2008;456:658. doi: 10.1038/nature07405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park B, et al. Nat Immunol. 2008;9:1407. doi: 10.1038/ni.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takaoka A, et al. Nature. 2005;434:243. doi: 10.1038/nature03308. [DOI] [PubMed] [Google Scholar]
- 7.Honda K, et al. Nature. 2005;434:772. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 8.Medzhitov R, et al. Mol Cell. 1998;2:253. doi: 10.1016/s1097-2765(00)80136-7. [DOI] [PubMed] [Google Scholar]
- 9.Uematsu S, et al. J Exp Med. 2005;201:915. doi: 10.1084/jem.20042372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Häcker H, et al. Nature. 2006;439:204. doi: 10.1038/nature04369. [DOI] [PubMed] [Google Scholar]
- 11.Oganesyan G, et al. Nature. 2006;439:208. doi: 10.1038/nature04374. [DOI] [PubMed] [Google Scholar]
- 12.Hoshino K, et al. Nature. 2006;440:949. doi: 10.1038/nature04641. [DOI] [PubMed] [Google Scholar]
- 13.Shinohara ML, et al. Nat Immunol. 2006;7:498. doi: 10.1038/ni1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watarai H, et al. Proc Natl Acad Sci USA. 2008;105:2993. doi: 10.1073/pnas.0710351105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gotoh K, et al. J Exp Med. 2010;207:721. doi: 10.1084/jem.20091776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guiducci C, et al. J Exp Med. 2006;203:1999. doi: 10.1084/jem.20060401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honda K, et al. Nature. 2005;434:1035. doi: 10.1038/nature03547. [DOI] [PubMed] [Google Scholar]
- 18.Materials and methods are available as supporting material on Science Online.
- 19.Bonifacino JS, Traub LM. Annu Rev Biochem. 2003;72:395. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 20.Matsumoto M, et al. J Immunol. 2003;171:3154. doi: 10.4049/jimmunol.171.6.3154. [DOI] [PubMed] [Google Scholar]
- 21.Kagan JC, et al. Nat Immunol. 2008;9:361. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asselin-Paturel C, et al. Nat Immunol. 2001;2:1144. doi: 10.1038/ni736. [DOI] [PubMed] [Google Scholar]
- 23.Latz E, et al. Nat Immunol. 2004;5:190. doi: 10.1038/ni1028. [DOI] [PubMed] [Google Scholar]
- 24.Leifer CA, et al. J Immunol. 2004;173:1179. doi: 10.4049/jimmunol.173.2.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim YM, Brinkmann MM, Paquet ME, Ploegh HL. Nature. 2008;452:234. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- 26.Trombetta ES, Mellman I. Annu Rev Immunol. 2005;23:975. doi: 10.1146/annurev.immunol.22.012703.104538. [DOI] [PubMed] [Google Scholar]
- 27.Dell’Angelica EC, Shotelersuk V, Aguilar RC, Gahl WA, Bonifacino JS. Mol Cell. 1999;3:11. doi: 10.1016/s1097-2765(00)80170-7. [DOI] [PubMed] [Google Scholar]
- 28.Peden AA, et al. J Cell Biol. 2004;164:1065. doi: 10.1083/jcb.200311064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ikonomov OC, Sbrissa D, Shisheva A. Am J Physiol Cell Physiol. 2006;291:C393. doi: 10.1152/ajpcell.00019.2006. [DOI] [PubMed] [Google Scholar]
- 30.De Matteis MA, Godi A. Nat Cell Biol. 2004;6:487. doi: 10.1038/ncb0604-487. [DOI] [PubMed] [Google Scholar]
- 31.Dowler S, et al. Biochem J. 2000;351:19. doi: 10.1042/0264-6021:3510019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.