Abstract
Blood pressure (BP), hypertension (HT) and cardiovascular disease (CVD) are common complex phenotypes, which are affected by multiple genetic and environmental factors. This article describes recent genome-wide association studies (GWAS) that have reported causative variants for BP/HT and CVD/heart traits and analyzes the overlapping associated gene polymorphisms. It also examines potential replication of findings from the HyperGEN data on African Americans and whites. Several genes involved in BP/HT regulation also appear to be involved in CVD. A better picture is emerging, with overlapping hot-spot regions and with interconnected pathways between BP/HT and CVD. A systemic approach to full understanding of BP/HT and CVD development and their progression to disease may lead to the identification of gene targets and pathways for the development of novel therapeutic interventions.
Keywords: Hypertension, Blood pressure, Cardiovascular disease, Single nucleotide polymorphisms, SNPs, Pathways, GWAS, Genome-wide association studies
Introduction
The World Health Organization (www.who.int/mediacentre/ factsheets/fs310/en/) reports that cardiovascular disease (CVD) is the number-1 cause of death in industrialized countries, accounting for 16% of deaths. In parallel, the prevalence of hypertension (HT), defined by systolic/ diastolic blood pressure (SBP/DBP) above 140/90 mm Hg, a well-established risk factor for CVD and mortality, increased in the United States to 29% between 1999 and 2008 [1]. Of hypertensive US adults, 63% were prescribed antihypertensive medication in 1999 to 2002, but a large proportion remain unaware, untreated, or both [2]. The rise of HT in the general population results from a multitude of risk factors such as obesity, excessive salt intake, aging, smoking, excessive alcohol intake, and stress. Prolonged HT can lead to adverse changes in the structure and function of the heart, including left ventricular hypertrophy (LVH), myocardial infarction (MI), atrial fibrillation (AF), and congestive heart failure; elevated blood pressure (BP) can lead to stroke and end-stage renal disease. A prolonged 5 mm Hg increase in DBP is associated with a 34% increase in the risk for stroke and a 21% increase in the risk for coronary events [3]. Thus, the study of genetic factors contributing to BP and HT is likely to show pathways interconnected with those predisposing to CVD.
Known Genetic Systems that Affect Blood Pressure
The discovery of several Mendelian forms of BP/HT has been critical in understanding pathways that regulate BP [4]. It is well known that the renin-angiotensin-aldosterone system (RAAS) regulates vascular volume homeostasis and vascular tone; the kallikrein-kinin system (KKS) influences vascular tone and renal salt handling; ion channels and receptors sense pressure changes in vessels; natriuretic proteins produced by brain and heart in response to increased blood pressure and adrenergic receptors influence cardiac rate, contraction, and relaxation; and nitric oxide causes vasodilatation, just as endothelin causes contraction. We briefly describe below several genes of these systems identified before the era of genome-wide association studies (GWAS).
The RAAS contributes to the regulation of blood volume and systemic vascular resistance, which together influence cardiac output and arterial pressure. As displayed in the Kyoto Encyclopedia of Genes and Genomes (KEGG) (KEGG 04614, http://kegg.jp/kegg-bin/show_pathway?map04614) [5•], the RAS pathway consists of a cascade of genes and gene products. Renin (REN, 1q32) is released by the kidneys and converts angiotensinogen (AGT, 1q42–q43) to the angiotensin I peptide. Haplotypes in the AGT gene are associated with differences in blood pressure and an increased prevalence of hypertension [6, 7]. In addition, reducing salt intake or weight lowers BP to a greater extent in those carrying the AGT risk alleles than in those with nonrisk alleles [8]. The AGT interactions are depicted in the diagram of the vascular smooth muscle contraction-relaxation pathway (KEGG 04270) at http://kegg.jp/kegg-bin/show_pathway?map04270. Angiotensin I is converted by angiotensin-converting enzyme (ACE, 17q23.3) into angiotensin II [9], which constricts resistance vessels by interacting with AGTR1 (3q24). Angiotensin II also stimulates the adrenal cortex to release aldosterone (CYP11B2, 8q21–q22), which in turn acts on the kidneys to increase sodium and fluid retention and also promotes cardiac and vascular hypertrophy.
Other systems regulating renal sodium and water handling, such as the KKS, are also important regulators of BP. Kallikrein 1 (KLK1, 19q13.3) releases a vasoactive peptide that can lower BP. The KKS is considered part of a larger picture of complement and coagulation cascades (KEGG 04610) depicted at http://kegg.jp/kegg-bin/show_pathway?map04610. Linking the KKS and RAAS, ACE serves also as a kinin/bradykinin–metabolizing enzyme. Kinins inhibit arginine vasopressin-stimulated water permeability and Na+ reabsorption in isolated collecting ducts [10]. It is interesting to note that a number of serpine genes also participate in the cascades of the coagulation and fibrinolytic systems. One of them, plasminogen activator 1 (PAI1, 7q22.1), is reported to be an important marker for metabolic syndrome [11].
Studies of renal tubular transporters and channels have provided important insights into the renal regulation of BP [12]. Ji et al. [13••] showed that rare, independent mutations in the renal salt-handling genes SLC12A1 (15q15–q21.1, Na-K-2Cl cotransporter gene), SLC12A3 (16q13, Na-Cl cotransporter gene), and KCNJ1 (11q24, inward K+ channel gene) contribute to monogenic syndromes characterized by HT or hypotension. Other ion transport genes reported to influence BP in humans are SLC12A4 (16q22.1), SLC12A5 (20q13.12), SLC12A6 (15q13), and SLC12A7 (5p15).
The epithelial sodium channel (ENaC), which is regulated by aldosterone, is another important regulator of sodium balance and thus of BP. The ENaC is encoded by four genes: SCNN1A (12p13), SCNN1B (16p12.2–p12.1), SCNN1D (1p36.3–p36.2), and SCNN1G (16p12). About 15 years ago, a rare monogenic form of salt-sensitive hypertension known as Liddle syndrome, associated with hypokalemia, low plasma aldosterone, and suppressed plasma renin, was found to be caused by deletions and/or mutations of the α and β subunits of ENaC [14]. In addition, SCNN1 is negatively regulated by NEDD4L (18q21), and loss of bindings sites in ENaC associates with the accumulation of active channels and more reabsorption of Na+ by the distal nephron, resulting in HT [14, 15]. Genetic studies in human and animal models have shown that ENaC in the kidney is under the control of aldosterone and vasopressin (AVP, 20p13). ENaC is depicted as part of the aldosterone-regulated sodium reabsorption pathway (KEGG 04960) at http://kegg.jp/ kegg-bin/show_pathway?map04960 and plays an important role in balancing the daily intake of salt and maintenance of BP [16•].
More recently, the discovery of the renal WNK kinases (lysine-deficient protein kinases) has explained the pathogenesis of a rare genetic form of hypertension known as familial hyperkalemic hypertension (FHHt, also known as pseudohypoaldosteronism type II [PHA2] or Gordon's syndrome). The WNKs are represented by four genes: WNK1 (12p13.3), WNK2 (9q22.3), WNK3 (Xp11.22), and WNK4 (17q21–q22). The mutations responsible for PHA2 are deletions within the first intron in WNK1 and WNK4 missense mutations downstream of the kinase domain [17, 18]. Kidney-specific WNK1 is predicted to be an important physiological regulator of renal K+ excretion, likely through its effects on the renal outer medullary potassium (ROMK) channel, KCNJ1, a potassium inwardly rectifying channel.
The group of interacting kinases with effects on BP is larger. SPAK kinase-inactive knockin mice, in which the phosphorylation site by WNK kinase is mutated, were reported to have lower blood pressure and a sodium-losing phenotype; a polymorphism of the STK39 gene (2q24.3), the human SPAK gene, is associated with human BP [19]. Another kinase, known as SGK1 (6q23- serum/glucocorticoid regulated kinase 1), activates certain potassium, sodium, and chloride channels, suggesting an involvement in the regulation of renal sodium excretion. High levels of expression of this gene may contribute to hypertension and diabetic nephropathy. Thus, it is possible that the WNK-OSR1/STK39-NCC and the SGK-Nedd4L-ENaC cascades may be novel effector systems of aldosterone action in the kidney and therefore important determinants of the BP level [20].
The RAAS, KKS, ion channels, and WNK kinases just described are important genetic systems with pleiotropic effects and complex interactions that influence BP. In the context of these complex interactions, Citterio et al. [21•] reviewed polymorphisms of adducin, with three known genes: ADD1, (4p16.3), ADD2 (2p13.3), and ADD3 (10q25.2). They provided evidence that adducin polymorphisms influence renal tubular sodium reabsorption and thus influence BP.
BP regulation is further influenced by regulators of vascular tone, which is ultimately dependent on ion channels present in the plasma membrane of vascular smooth muscle cells, such as several different types of K+ channels, voltage-gated Ca2+ channels, and Cl− channels [12].
Years of focused research on HT have produced more than 70 antihypertensive medications. They target products of important BP-influencing genes, although the mechanisms of action vary greatly. For example, the target of loop diuretics is SLC12A1; of thiazides, SLC12A3; of adrenergic beta-antagonists, ADRB1 (10q24–q26) and ADRB2 (5q31–q32); of adrenergic alpha-antagonists, ADRA1A (8p21.2), ADRA1B (5q33.3), and ADRA1D (20p13); of angiotensin-converting enzyme inhibitors, ACE; of calcium channel blockers, CACNA1S (1q32), CACNA1C (12p13.3), CACNA1D (3p14.3), and CACNA1F (Xp11.23); of angiotensin II receptor blockers, AGTR1 [22••, 23•]. Genes mentioned in this section represent only a few of the genes in the pathways related to BP. More genes are part of pathways cited above, and extra details of genes in Fig. 1 provide a larger pool of genes that regulate BP. The sections that follow will review evidence that recent GWAS contributions for BP/HT have further enriched our clinical knowledge aiming at the development of personalized anti-hypertensive treatment.
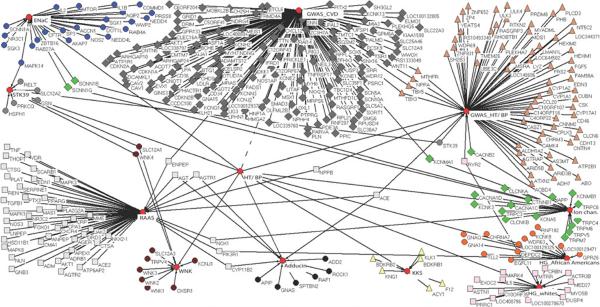
Fig. 1.
A schematic of interconnections matched by gene name reported by genome-wide association studies (GWAS) for BP/HT and GWAS for CVD/heart traits. Also included are gene names returned by a search using National Center for Biotechnology Information (NCBI) Entrez Gene, with keywords “renin angiotensin aldosterone”/ “enac”/“wnk”/“adducin”/“stk39”/“kallikrein-kinin” AND human, and “ion channels” AND hypertension AND human. Also included are selected results of the HyperGEN study for association tests with BP factors. (Kraja et al., unpublished data.)
Genetic Systems that Affect the Heart and Blood Pressure
CVD develops as a result of multiple contributing risks, including CVD family history, hypertension, hypercholesterolemia, obesity, smoking, metabolic syndrome, and diabetes. A pathologic increase of left ventricle mass (LVM) often results from HT and is considered a compensatory response to normalize increasing wall stress. Several genes are associated with LVM, such as ACE, PPARA (22q13.31), GNB3 (12p13), CYP11B2, IGF1 (12q23.2), and NPY (7p15.1) [24].
A few congenital cardiac defects that associate with abnormal BP level, or pathologic cardiac remodeling of the heart associated with HT, are not the only factors that influence and interact with BP. Molecules such as nitric oxide (NO), represented by the NOS1 (12q24.2–q24.31), NOS2 (17q11.2–q12), and NOS3 (7q36) genes, cause vasodilatation when produced by blood vessels. Thus, NO could be involved in cardiac changes associated with hemodynamic pressure overload. It has been shown that RGS2 (1q31), a regulator of G-protein signaling, serves as a mediator of NO action on BP and vasoconstrictor signaling [25]. NO also interacts with environmental factors such as diet. For example, Simão et al. [26] reported that NO increased and BP declined in clinical trials that included fish oil or soy protein in the diet of women with metabolic syndrome. Diet, especially increased salt intake in animal models, has been found experimentally to influence cardiovascular system functioning. For example, in a rat model of mild hypertension, treatment with angiotensin II and a high-salt diet increased expression of the sodium/ hydrogen exchanger isoform 1 (SLC9A1, 1p36.1–p35), a target of mineralocorticoid receptor activation. Wang et al. [27] concluded that mineralocorticoid receptor (MR) (NR3C2, 4q31.1) activation by oxidative stress caused left ventricular diastolic dysfunction. A schematic interaction between aldosterone and ENaC, by the way of NR3C2, is shown in http://kegg.jp/kegg-bin/show_pathway?map04960. Moreover, hypertension and aldosteronism cause cardiovascular tissue remodeling. Other potent vasoconstrictors are vasopressin, where its receptor AVPR1A (12q14–q15) mediates cell contraction and proliferation, platelet aggregation, and release of coagulation factor [28]; endothelin produced by vascular endothelial cells, by the genes EDN1 (6p24.1), EDN2 (1p34), and EDN3 (20q13.2–q13.3); and urotensin 2 (UTS2, 1p36), expressed only in brain tissues [29]. In addition, beta-adrenergic receptors, with genes ADRB1, ADRB2, ADRB3 (8p12), mediate the physiological effects of the hormone epinephrine and the neurotransmitter norepinephrine. They are common therapeutic targets for the treatment of hypertension, renal disease, and heart failure. Specific polymorphisms in beta-adrenergic receptor genes have been shown to affect the resting heart rate and can be involved in heart failure. All of these facts, from the era before GWAS, make a compelling case that the overlap of HT and CVD involves several genes.
A Case Study: Blood Pressure in the HyperGEN Study
The Hypertension Genetic Epidemiology Network (Hyper-GEN) is part of the Family Blood Pressure Program (FBPP), a multicenter study supported by the National Heart, Lung, and Blood Institute (NHLBI) [30] to study the genetics of hypertension. The HyperGEN study has special appeal for studying BP/HT because of its design of studying at least two siblings who were hypertensive before age 60. Also, parents and offspring of some of the hypertensive siblings were recruited, as well as a random population sample, serving as controls. Sample sizes used in this analysis of 908 African Americans and 1,025 whites constitute those study participants that had genotype data and no missing phenotypes for 11 traits. The BP-factor scores for each ethnicity were produced by using multivariate factor analysis on 11 traits, as described in the HyperGEN study on metabolic syndrome [31]. As a result, this analysis uses BP factor, a standardized variable where loadings (contributions) of SBP and DBP represent correlations among the factor scores and the original standardized SBP and DBP traits correspondingly (Supplemental Figure 1). The results of association tests of BP factors with a 500K Affymetrix chip 5.0 for whites and a 1M Affymetrix chip 6.0 for African Americans can be seen in Supplemental Figures 2 and 3 and Supplemental Tables 1 and 2.
Test results between BP-factor and genome-wide single nucleotide polymorphisms (SNPs) were produced under an additive genetic model. Statistical mixed regression analyses accounted for familial relationship. In addition, the model accounted for genetic heterogeneity for African Americans by using as covariates in the model the 10 first principal components produced on tag SNPs genotypes by EIGENSTRAT software ([32]; Kraja et al., unpublished data). Because BP factors had moderate heritabilities of 34% to 38% in African Americans and 33% to 37% in whites (similar to heritabilities of SBP/DBP), and because sample sizes were relatively small, the results in African Americans did not pass the genome-wide significance threshold. In comparison, one SNP in whites, rs17031343, in proximity to genes CRBN/IL5RA (3p26–p24), passed the 500K chip threshold (based on the number of tagSNPs counted as independent tests). But if the threshold is slightly liberalized by including findings with a P value <0.00001 (Supplemental Figures 2 and 3 and Supplemental Tables 1 and 2), 14 SNPs in African Americans and 18 in whites can be encompassed. This means at the same time that false positives can increase more than 5% genome-wide. Thus, these results will be used here only to see whether any HyperGEN findings are replicated in large GWAS studies.
Following is a brief partial listing of genes (and their function) that ranked at the top of associations with BP factor in the HyperGEN African Americans. (Parentheses around a gene name indicate that the SNP is near a gene region.) A nicotinic cholinergic receptor, (CHRNA7), is reported to stimulate a sympathetic cardiovascular response [33]; DEPDC2 is part of the phosphatidylinositol 3-kinase (PI3K) pathway [34]; GNA14 (9q21, rs6560608; minor allele frequency [MAF], 38.5%) is reported to be an HT-susceptible gene [35•]; (KCNK9, 8q24.3, rs2447409; MAF, 35.7%), a member of potassium channels that is highly expressed in the cerebellum, serves as a determinant of aldosterone [36]. In addition, SNP rs2468680 is near SNP rs2447409, associated with aortic root diameter [37]; and LOC1001129471, also known as FLJ36032, is a chloride channel gene; and PI3K (PIK3R1, 5q13.1, rs1490800; MAF, 27%) plays an important role in the metabolic actions of insulin, which also affects BP [38] (see Supplemental Table 1).
Two of the top-ranked genes in association with BP factor of HyperGEN whites are (CRBN/IL5RA, rs17031343; MAF, 3.4%), in which CRBN (3p26.2) product is found in the cytoplasm, localized with a calcium channel membrane protein; and RYR2 (1q43, rs604735; MAF, 48.9%), which is found in cardiac muscle sarcoplasmic reticulum. Mutations in RYR2 are associated with stress-induced polymorphic ventricular tachycardia and arrhythmogenic right ventricular dysplasia [39] (Supplemental Table 2).
In summary, the SNPs in association with BP factor in the HyperGEN study failed to reach genome-wide significance, with the exception of rs17031343. The relatively modest significance is likely the result of low heritability for BP factor and the relatively small sample size. Nonetheless, a few genes with suggestive findings were biologically interesting in that they probably contribute to BP regulation, and some of them have implications for heart function. Are any of these findings replicated in the recent larger GWAS studies for BP and CVD?
Findings from Large Studies on BP and CVD
GWAS for BP/HT started in 2007 with a few reports. Some of the early reports did not have BP/HT significant results [40–42]. The Wellcome Trust Case Control Consortium (WTCCC), with 2,000 HT cases and 3,000 controls, reasoned that, among other factors, HT may have few common risk alleles of detectably large effect sizes [40]. Supplemental Table 3 shows that without counting Hyper-GEN findings (used here only to see if they match with any previous reported findings), there are at least 39 cytogenetic reported locations from large GWAS with relatively small effect sizes significantly associated with SBP, DBP, HT, or pulse rate (the last one indicated as “beats per minute increase” in the column “Trait change” in the Supplemental Table 3). Actually, in the studies making up this table, there were 69 genes (and 1 SNP unassigned to genes) that were mentioned in the final results as possible unique causative genes or SNPs.
In the CVD studies (Supplemental Table 4), there were 142 CVD/heart traits with unique genes and 1 SNP unassigned to genes. It is quite possible that a number of these genes are redundant when a study reports one significant SNP and 3 to 5 gene names that surround that particular SNP as potential causative genes. Nevertheless, the number of genes contributing to BP/HT is not small. HyperGEN's r2 (variance explained by an additive model of a marker on BP factor) statistics were also relatively small (Supplemental Tables 1 and 2). A probable reason for the challenge in identifying BP/HT genes could be the number of SNPs studied, which, although large, may not have covered all regions of causative SNPs. In fact, genotyping platforms in 12 GWAS publications for BP/HT varied: four used Affymetrix platform of 100K SNPs, four used Affymetrix 500K platforms, and one used Illumina Infinium 550K; the remaining three publications (which include consortia collaborations) used larger arrays or imputed up to 2.5M SNPs based on HapMap. The HyperGEN study used the Affymetrix 5.0 platform with about 500K SNPs for whites and Affymetrix 6.0 with about 1M SNPs for African Americans. Two examples of integrating results from multiple studies with immense power for discovery are Levy et al. [22••] representing the CHARGE consortium with almost 30,000 participants, and Newton-Cheh et al. [43••] for the Global BPgen consortium, with more than 84,000 participants.
What were the new discoveries from these studies? Fig. 1 is a schematic of matching gene names of GWAS findings from BP/HT and CVD/heart traits. Additionally, a number of systems that affect BP were added by searching NCBI Entrez Gene, as explained in the figure legend. Also added were top-ranked findings of HyperGEN association tests on BP factors. One can see immediately that MTHFR, NPPA, TBX1, TBX5, KCNMA1, and ENPEP for BP/HT and CVD and STK39, CACNB2, ACE, NPPB, RYR2, PIK3R1, and several other known genes for HT/BP may be of special interest. We refer to these findings also via Supplemental Figure 4, which represents a coarse summary of overlaps among BP/HT and CVD/heart trait association results based on chromosomal cytogenetic band location. At first glance, this figure gives the impression that the BP/ HT recent studies have been able to co-localize (following chromosomal order) the SCNN1D (1p36.3–p36.2) member of the ENaC system, REN (1q32), AGT (1q42–q43), STK39 (2q24.3), NOS3 (7q36), ADRB1 (10q24–q26), NOS1 (12q24.2–q24.31), WNK4 (17q21–q22), ACE (17q23.3), and EDN3 (20q13.2–q13.3) genes. Actually, this is not the case for all of them. For example, Chung et al. [44] reported a significant association of rs4343, a coding-synonymous SNP of the ACE gene, with ACE activity in 400 individuals with young-onset hypertension, and confirmed it in an additional 623 such individuals and in a larger family study of 428 hypertensive families. These authors inferred a potential differential BP response to ACE activity. ACE has been extensively studied and it has been ascribed a role in HT, heart disease, and Alzheimer's disease [45, 46].
Another example is in the region 1q43. In this region, a good candidate for HT is AGT. WTCCC [40] reported on a case-control association test for HT with 2,000 cases and 3,000 controls, in which a top-ranked SNP, rs2820037 (P=7.7×10−7), was a possible proxy for RYR2 (ryanodine receptor 2). In HyperGEN whites, rs604735, an intron of RYR2, was associated with BP factor (P=4.0×10−6). Also, Smith et al. [47]reported a RYR2 SNP ranked in the top of associations with electrocardiographic (ECG) P-wave duration in an isolated founder population. Can we conclude from these modest significant associations that RYR2 is an important gene for BP/HT? RYR2 is 4.8 Mbps away from the AGT gene. Previous research evidence has recognized RYR2 as part of six different human pathways: calcium signaling (hsa04020), cardiac muscle contraction (hsa04260), pancreatic secretion (hsa04972), hypertrophic cardiomyopathy (hsa05410), arrhythmogenic right ventricular cardiomyopathy (hsa05412), and dilated cardiomyopathy (hsa05414). RYR2 protein is a component of a calcium channel that supplies calcium to cardiac muscle, and mutations in this gene are associated with stress-induced polymorphic ventricular tachycardia and arrhythmogenic right ventricular dysplasia. RYR2 is reported to interact with NOS3 [48] and also has been shown to interact with CACNA1D (3p14.3), a voltage-gated calcium channel [49], which is already a target for antihypertensive drugs that block neuronal voltage-gated L-type calcium channels. Recently, at 2.85 Mbps distant from RYR2, the rs17672135 (P=2×10−6), which is part of the FMN2 (formin 2, 1q43) gene, is reported to be associated with coronary heart disease [40]. Therefore the AGT-RYR2 chromosome 1q43 region is a “hot-spot” region for important candidates for both BP and heart functioning.
Findings of interest from large studies on BP/HT are located around the genes MTHFR (methylenetetrahydrofolate reductase [NAD(P)H], 1p36.3) and NPPA (natriuretic peptide precursor A, 1p36.21) but are about 10 Mbps distant from the SCNN1D (part of ENaC) candidate gene. MTHFR is reported to interact with PPAR-α, and MTHFR has been already been associated with occlusive vascular disease [50, 51]. Changes in the NPPA gene have been proposed as causal for atrial fibrillation (AF) (with negative results in another study [52]), and more recently MTHFR and NPPA again were reported to associate with AF [53]. Large studies have also identified RNF207 (~5.6 Mbps distant from NPPA) and nearby genes as putative candidates in association with QT-interval duration [54••, 55••].
In African Americans in the HyperGEN study, modest associations with BP factor were reported for rs6560608 (P=5.4×10−6 intron in GNA14, 9q21) and rs7025910 (P=3.86×10−6 located between GNA14 and GNAQ genes) (Kraja et al., unpublished data). The GNA14-BP association has also recently been reported by Kohara et al. [35•], who studied multiple candidate genes for hypertension in a case/control Japanese sample (N=9,700). However, if corrected for multiple comparisons in the Japanese sample, this finding would have not been deemed significant. A similar problem of top associations becoming nonsignificant when applying multiple comparison corrections was reported for DBP and HT by Adeyemo et al. [56], who studied an African American sample (N=1,017); in contrast, they reported several significant findings for associations with SBP. Among them, rs11160059 (P=1.54×10−8;MAF,17.82%), an intron in the SLC24A4 (sodium/potassium/calcium exchanger) gene on chromosome 14q32, was significantly associated with SBP. In HyperGEN African Americans, rs17783630 (P=0.00015; MAF, 49.48%) was ranked highest of SLC24A4 gene SNPs in association with the BP factor. Based on the NCBI build 36.3, these two SNPs of the same gene are 0.148 Mbps apart.
Other regions of interest were the REN - CD34 1q32 region (~4 Mpbs apart), where there are a number of genes/SNPs of interest, including REN for HT/BP [57]; CD34 rs2745967 (P=3.2×10−8, N=34,913) [58] and rs17706439 [59] for heart rate, and rs7512898 for ECG trait (PR segment) [47]; CD46 for pulse rate [60•]; the 3p22 region with ULK4 for DBP [22••]; SCN10A (3p22.2) for ECG traits (PR interval, QRS duration) [61–63]; SCN5A (3p21) for ECG traits (QT interval, PR interval) [47, 54••, 55••, 63]; and CMTM7 (93P22.3) for mortality among heart failure patients [64]; the 7q36 region with UBE3C in association with SBP [65•]; KCNH2 (7q36.1, a potassium channel) for ECG traits (QTc and QT interval) [54••, 55••, 61, 62]; NOS1 -12q24 region with SH2B3, recently associated with SBP and DBP [22••, 43••]; TBX3 and TBX5 with DBP [22••]; the CSK -15q24 region with CSK/ULK3 associated with DBP [22••, 43••]; and LOXL1 (15q22), associated with aortic root size [66] (Supplemental Tables 3 and 4).
This last region on chromosome 15 brings us back to an interesting theme of comparing findings between linkage and association results. Two papers published in 2005, one for HyperGEN [67] and one for the FBPP [68], showed associations for quantitative trait loci (QTLs) with four metabolic syndrome factors, and a third paper published in 2007 for HyperGEN showed QTLs for metabolic syndrome and echocardiographic phenotypes [69]. Among others, these three papers have reported a linkage peak for FBPP for Mexican Americans for BP factor that overlapped with the ACE location on chromosome 17 [68]. In addition, a QTL of HyperGEN for left ventricular (LV) wall thickness on chromosome 16q24.2–q24.3, with the local maximum LOD score at marker D16S402, which is positioned within the 5th intron of the CDH13 gene, is implicated in heart and vascular remodeling [69]. Recent GWAS have demonstrated that the CDH13 gene contributes to BP [70].
Finally, a QTL for BP factor was reported on chromosome 15 in HyperGEN whites, with a peak of 3.19 LOD score at GATA151F03, located at 65.340 Mbps [67]. Recent NCBI information shows that GATA151F03 marks a gene LOC748001 in Pan troglodytes. Searching the NCBI HomoloGene, LOC748001 matches with the OSTbeta gene in humans, which is located between 63.129–63.133 Mbps in NCBI build 36.3 but is not present in the HyperGEN GWAS data. The OSTbeta gene is an intestinal basolateral transporter that exports bile acid from enterocytes into portal blood. NR3C1 (glucocorticoid alpha) induces expression of OSTbeta, and OSTbeta interacts with NR1H4 (known also as FXR), which activates transcription of the bile-salt export pump ABCB11 [71]. It is not clear whether OSTbeta can contribute to BP regulation, because the linkage peak of interest extends from 55 cM to 70 cM. Two reports on 15q24.1 appear to be of interest for CSK and ULK3 [22••, 43••], but these genes are about 20 Mbps distant from OSTbeta. Thus, OSTbeta and nearby genes can be of interest to BP.
In summary, the large BP/HT GWAS included familial data, population cohorts, and case/control designs. These studies have mostly performed analyses using an additive genetic model and may have missed recessive/dominant variants. They have not addressed gene-gene or gene-environment interactions, and only a few made use of the gene expression information available in NCBI databases. They dealt with common variants, in contrast to studies from the pre-GWAS era, which relied mostly on rare phenotype variants or linkage results. The strongest findings of these studies were localized in introns or intragenic SNPs, with exception of 2 missense and 1 synonymous coding SNP (Supplemental Table 5), showing that summarized findings are most of the time not exactly in expressed variants [22••, 40, 43••, 44, 56, 60•, 65•, 70, 72–75]. To date, the additive variance explained by new findings of large GWAS is relatively small. For example, the CHARGE consortium [22••] and Global BPgen consortium [43••] reported 13 significant associations with common SNPs. Each association explained only a small proportion of the total variation in SBP or DBP (0.05%–0.10%), with a variation in SBP of about 1 mm Hg per allele, or 0.5 mm Hg per allele for DBP. All 13 SNPs together explained less than 2% of the BP variance.
A similar situation applies to CVD/heart traits. For example, 14 independent variants at 10 loci explained 5.4% to 6.5% of variation in the prolongation of electrocardiographic QT interval [54••], and most GWAS publications suggested that further investigations will be necessary to provide deeper functional understanding of the latest findings. Thus, cumulative knowledge explains a small fraction of the BP/HT variation and therefore is far from being translated into the benefit of personalized medicine [23•]. In addition, GWAS have not yet covered the full spectrum of cumulative genomic knowledge. For example, a search of the NCBI Entrez Gene for hypertension and human returned 438 gene IDs, versus 409 for mice and rats. New, large GWAS studies that will contain a larger number of SNPs especially in the exonic regions are likely to make additional progress. We envision new publications on BP/HT in the very near future, with imputations based on the 1000 Genomes Project (http://www.genome.gov/27528684), an international collaboration, with a much denser range of SNPs. Perhaps more likely, current efforts at identifying rare and low-frequency variants will contribute significantly to the identification of some of the missing heritability. In parallel, research in animal models is also likely to discover additional causative variants for BP/HT and CVD. These efforts, in combination with gene expression, functional, and proteomics studies, will further improve our understanding of BP/HT and CVD pathways.
Conclusions
Although discovering genetic loci for BP/HT is a challenging task, large GWAS consortia have made positive contributions. Several studies have embarked on candidate-gene approaches to improve power for their discoveries. These discoveries have contributed to a better understanding of BP regulation, where pathways of BP regulation and heart functioning have expanded with a more noticeable overlap between the two. Taken together, this review indicates that the identification of hypertension and CVD susceptible genes has a long road ahead.
Supplementary Material
Acknowledgment
This work was supported in part by the National Institutes of Health (NIH) HyperGEN Study grant U01 HL54471 and in part by the NIH Genetic Determinants of the LVH Phenotype grant RO1 HL07178205A.
Footnotes
Disclosure No potential conflicts of interest relevant to this article were reported.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Egan BM, Zhao Y, Axon RN. US trends in prevalence, awareness, treatment, and control of hypertension, 1988–2008. JAMA. 2010;303:2043–2050. doi: 10.1001/jama.2010.650. [DOI] [PubMed] [Google Scholar]
- 2.Gu Q, Paulose-Ram R, Dillon C, Burt V. Antihypertensive medication use among US adults with hypertension. Circulation. 2006;113:213–221. doi: 10.1161/CIRCULATIONAHA.105.542290. [DOI] [PubMed] [Google Scholar]
- 3.MacMahon S, Peto R, Cutler J, et al. Blood pressure, stroke, and coronary heart disease. Part 1, Prolonged differences in blood pressure: prospective observational studies corrected for the regression dilution bias. Lancet. 1990;335:765–774. doi: 10.1016/0140-6736(90)90878-9. [DOI] [PubMed] [Google Scholar]
- 4.Lifton RP, Gharavi AG, Geller DS. Molecular mechanisms of human hypertension. Cell. 2001;104:545–556. doi: 10.1016/s0092-8674(01)00241-0. [DOI] [PubMed] [Google Scholar]
- 5•.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38(Database issue):D355–360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article describes KEGG pathway databases.
- 6.Jeunemaitre X, Soubrier F, Kotelevtsev YV, et al. Molecular basis of human hypertension: role of angiotensinogen. Cell. 1992;71:169–180. doi: 10.1016/0092-8674(92)90275-h. [DOI] [PubMed] [Google Scholar]
- 7.Watkins WS, Hunt SC, Williams GH, et al. Genotype-phenotype analysis of angiotensinogen polymorphisms and essential hypertension: the importance of haplotypes. J Hypertens. 2010;28:65–75. doi: 10.1097/HJH.0b013e328332031a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hunt SC, Cook NR, Oberman A, et al. Angiotensinogen genotype, sodium reduction, weight loss, and prevention of hypertension: trials of hypertension prevention, phase II. Hyper-tension. 1998;32:393–401. doi: 10.1161/01.hyp.32.3.393. [DOI] [PubMed] [Google Scholar]
- 9.Easthope SE, Jarvis B. Candesartan cilexetil: an update of its use in essential hypertension. Drugs. 2002;62:1253–1287. doi: 10.2165/00003495-200262080-00016. [DOI] [PubMed] [Google Scholar]
- 10.Mukai H, Fitzgibbon WR, Bozeman G, et al. Bradykinin B2 receptor antagonist increases chloride and water absorption in rat medullary collecting duct. Am J Physiol. 1996;271:R352–360. doi: 10.1152/ajpregu.1996.271.2.R352. [DOI] [PubMed] [Google Scholar]
- 11.Kraja AT, Province MA, Arnett D, et al. Do inflammation and procoagulation biomarkers contribute to the metabolic syndrome cluster? Nutr Metab (Lond) 2007;4(1):28. doi: 10.1186/1743-7075-4-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adragna NC, Lauf PK. K-Cl cotransport function and its potential contribution to cardiovascular disease. Pathophysiology. 2007;14:135–146. doi: 10.1016/j.pathophys.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 13••.Ji W, Foo JN, O'Roak BJ, et al. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat Genet. 2008;40:592–599. doi: 10.1038/ng.118. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article emphasizes the importance of rare variants in BP variation.
- 14.Schild L, Lu Y, Gautschi I, et al. Identification of a PY motif in the epithelial Na channel subunits as a target sequence for mutations causing channel activation found in Liddle syndrome. EMBO J. 1996;15:2381–2387. [PMC free article] [PubMed] [Google Scholar]
- 15.Dunn DM, Ishigami T, Pankow J, et al. Common variant of human NEDD4L activates a cryptic splice site to form a frameshifted transcript. J Hum Genet. 2002;47:665–676. doi: 10.1007/s100380200102. [DOI] [PubMed] [Google Scholar]
- 16•.Schild L. The epithelial sodium channel and the control of sodium balance. Biochim Biophys Acta. 2010;1802:1159–65. doi: 10.1016/j.bbadis.2010.06.014. [DOI] [PubMed] [Google Scholar]; This is a good review on sodium channels.
- 17.Wilson FH, Disse-Nicodème S, Choate KA, et al. Human hypertension caused by mutations in WNK kinases. Science. 2001;293:1107–1112. doi: 10.1126/science.1062844. [DOI] [PubMed] [Google Scholar]
- 18.Uchida S. Pathophysiological roles of WNK kinases in the kidney. Pflugers Arch. 2010;460:695–702. doi: 10.1007/s00424-010-0848-7. [DOI] [PubMed] [Google Scholar]
- 19.Welling PA, Chang YP, Delpire E, Wade JB. Multigene kinase network, kidney transport, and salt in essential hypertension. Kidney Int. 2010;77:1063–1069. doi: 10.1038/ki.2010.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiga M, Rai T, Yang SS, et al. Dietary salt regulates the phosphorylation of OSR1/SPAK kinases and the sodium chloride cotransporter through aldosterone. Kidney Int. 2008;74:1403–1409. doi: 10.1038/ki.2008.451. [DOI] [PubMed] [Google Scholar]
- 21•.Citterio L, Lanzani C, Manunta P, Bianchi G. Genetics of primary hypertension: The clinical impact of adducin polymorphisms. Biochim Biophys Acta. 2010;1802:1285–98. doi: 10.1016/j.bbadis.2010.03.014. [DOI] [PubMed] [Google Scholar]; This article reviews the adducin role by expanding into the idea of system networks.
- 22••.Levy D, Ehret GB, Rice K, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is one of the largest GWAS for BP and HT.
- 23•.Arnett DK, Claas SA. Pharmacogenetics of antihypertensive treatment: detailing disciplinary dissonance. Pharmacogenomics. 2009;10:1295–1307. doi: 10.2217/pgs.09.61. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is a review of the lag of personalized antihypertensive treatment.
- 24.Vakili BA, Okin PM, Devereux RB. Prognostic implications of left ventricular hypertrophy. Am Heart J. 2001;141:334–341. doi: 10.1067/mhj.2001.113218. [DOI] [PubMed] [Google Scholar]
- 25.Osei-Owusu P, Sun X, Drenan RM, et al. Regulation of RGS2 and second messenger signaling in vascular smooth muscle cells by cGMP-dependent protein kinase. J Biol Chem. 2007;282:31656–31665. doi: 10.1074/jbc.M706360200. [DOI] [PubMed] [Google Scholar]
- 26.Simão AN, Lozovoy MA, Simão TN, et al. Nitric oxide enhancement and blood pressure decrease in patients with metabolic syndrome using soy protein or fish oil. Arq Bras Endocrinol Metabol. 2010;54:540–545. doi: 10.1590/s0004-27302010000600005. [DOI] [PubMed] [Google Scholar]
- 27.Wang H, Shimosawa T, Matsui H, et al. Paradoxical mineralocorticoid receptor activation and left ventricular diastolic dysfunction under high oxidative stress conditions. J Hypertens. 2008;26:1453–1462. doi: 10.1097/HJH.0b013e328300a232. [DOI] [PubMed] [Google Scholar]
- 28.Masuki S, Mori M, Tabara Y, et al. Shinshu University Genetic Research Consortium. Vasopressin V1a receptor polymorphism and interval walking training effects in middle-aged and older people. Hypertension. 2010;55:747–754. doi: 10.1161/HYPERTENSIONAHA.109.147728. [DOI] [PubMed] [Google Scholar]
- 29.Chai SB, Li XM, Pang YZ, et al. Increased plasma levels of endothelin-1 and urotensin-II in patients with coronary heart disease. Heart Vessels. 2010;25:138–143. doi: 10.1007/s00380-009-1178-6. [DOI] [PubMed] [Google Scholar]
- 30.Williams RR, Rao DC, Ellison RC, et al. NHLBI family blood pressure program: methodology and recruitment in the HyperGEN network. Hypertension genetic epidemiology network. Ann Epidemiol. 2000;10:389–400. doi: 10.1016/s1047-2797(00)00063-6. [DOI] [PubMed] [Google Scholar]
- 31.Kraja AT, Hunt SC, Pankow JS, et al. An evaluation of the metabolic syndrome in the HyperGEN study. Nutr Metab (Lond) 2005;2(1):2. doi: 10.1186/1743-7075-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 33.Li YF, LaCroix C, Freeling J. Specific subtypes of nicotinic cholinergic receptors involved in sympathetic and parasympathetic cardiovascular responses. Neurosci Lett. 2009;462:20–23. doi: 10.1016/j.neulet.2009.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fine B, Hodakoski C, Koujak S, et al. Activation of the PI3K pathway in cancer through inhibition of PTEN by exchange factor P-REX2a. Science. 2009;325:1261–1265. doi: 10.1126/science.1173569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35•.Kohara K, Tabara Y, Nakura J, et al. Identification of hypertension-susceptibility genes and pathways by a systemic multiple candidate gene approach: the millennium genome project for hypertension. Hypertens Res. 2008;31:203–212. doi: 10.1291/hypres.31.203. [DOI] [PubMed] [Google Scholar]; This article discusses an important initiative for detecting HT genes in Japan.
- 36.Brenner T, O'Shaughnessy KM. Both TASK-3 and TREK-1 two-pore loop K channels are expressed in H295R cells and modulate their membrane potential and aldosterone secretion. Am J Physiol Endocrinol Metab. 2008;295:E1480–1486. doi: 10.1152/ajpendo.90652.2008. [DOI] [PubMed] [Google Scholar]
- 37.Vasan RS, Larson MG, Aragam J, et al. Genome-wide association of echocardiographic dimensions, brachial artery endothelial function and treadmill exercise responses in the Framingham Heart Study. BMC Med Genet. 2007;8(Suppl 1):S2. doi: 10.1186/1471-2350-8-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barbour LA, Mizanoor Rahman S, et al. Increased P85alpha is a potent negative regulator of skeletal muscle insulin signaling and induces in vivo insulin resistance associated with growth hormone excess. J Biol Chem. 2005;280:37489–37494. doi: 10.1074/jbc.M506967200. [DOI] [PubMed] [Google Scholar]
- 39.Thomas NL, Maxwell C, Mukherjee S, Williams AJ. Ryanodine receptor mutations in arrhythmia: The continuing mystery of channel dysfunction. FEBS Lett. 2010;584:2153–2160. doi: 10.1016/j.febslet.2010.01.057. [DOI] [PubMed] [Google Scholar]
- 40.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research. Saxena R, Voight BF, Lyssenko V, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 42.Sabatti C, Service SK, Hartikainen AL, et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat Genet. 2009;41:35–46. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43••.Newton-Cheh C, Johnson T, Gateva V, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is one of the largest GWAS for BP.
- 44.Chung CM, Wang RY, Chen JW, et al. A genome-wide association study identifies new loci for ACE activity: potential implications for response to ACE inhibitor. Pharmacogenomics J. 2010:1–8. doi: 10.1038/tpj.2009.70. [DOI] [PubMed] [Google Scholar]
- 45.Johnson AD, Gong Y, Wang D, et al. Promoter polymorphisms in ACE (angiotensin I-converting enzyme) associated with clinical outcomes in hypertension. Clin Pharmacol Ther. 2009;85:36–44. doi: 10.1038/clpt.2008.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thameem F, Voruganti VS, He X, et al. Genetic variants in the reninangiotensin system genes are associated with cardiovascular-renal-related risk factors in Mexican Americans. Hum Genet. 2008;124:557–559. doi: 10.1007/s00439-008-0581-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith JG, Lowe JK, Kovvali S, et al. Genome-wide association study of electrocardiographic conduction measures in an isolated founder population: Kosrae. Heart Rhythm. 2009;6:634–641. doi: 10.1016/j.hrthm.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez-Moreno M, Alvarez-Barrientos A, Roncal F, et al. Direct interaction between the reductase domain of endothelial nitric oxide synthase and the ryanodine receptor. FEBS letters. 2005;579:3159–3163. doi: 10.1016/j.febslet.2005.04.078. [DOI] [PubMed] [Google Scholar]
- 49.Kim S, Yun HM, Baik JH, et al. Functional interaction of neuronal Cav1.3 L-type calcium channel with ryanodine receptor type 2 in the rat hippocampus. J Biol Chem. 2007;282:32877–32889. doi: 10.1074/jbc.M701418200. [DOI] [PubMed] [Google Scholar]
- 50.Mikael LG, Genest J, Jr, Rozen R. Elevated homocysteine reduces apolipoprotein A-I expression in hyperhomocysteinemic mice and in males with coronary artery disease. Circ Res. 2006;98:564–571. doi: 10.1161/01.RES.0000204825.66410.0b. [DOI] [PubMed] [Google Scholar]
- 51.Pizza V, Bisogno A, Lamaida E, et al. Migraine and coronary artery disease: an open study on the genetic polymorphism of the 5, 10 methylenetetrahydrofolate (MTHFR) and angiotensin I-converting enzyme (ACE) genes. Cent Nerv Syst Agents Med Chem. 2010;10:91–96. doi: 10.2174/187152410791196404. [DOI] [PubMed] [Google Scholar]
- 52.Roberts JD, Davies RW, Lubitz SA, et al. Evaluation of non-synonymous NPPA single nucleotide polymorphisms in atrial fibrillation. Europace. 2010;12:1078–1083. doi: 10.1093/europace/euq161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benjamin EJ, Rice KM, Arking DE, et al. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat Genet. 2009;41:879–881. doi: 10.1038/ng.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54••.Newton-Cheh C, Eijgelsheim M, Rice KM, et al. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nat Genet. 2009;41:399–406. doi: 10.1038/ng.364. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is one of the largest studies for cardiac traits.
- 55••.Pfeufer A, Sanna S, Arking DE, et al. Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nat Genet. 2009;41:407–414. doi: 10.1038/ng.362. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is one of the largest studies for cardiac traits.
- 56.Adeyemo A, Gerry N, Chen G, et al. A genome-wide association study of hypertension and blood pressure in African Americans. PLoS Genet. 2009;5(7):e1000564. doi: 10.1371/journal.pgen.1000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vangjeli C, Clarke N, Quinn U, et al. Confirmation that the renin gene distal enhancer polymorphism REN-5312C/T is associated with increased blood pressure. Circ Cardiovasc Genet. 2010;3:53–59. doi: 10.1161/CIRCGENETICS.109.899930. [DOI] [PubMed] [Google Scholar]
- 58.Eijgelsheim M, Newton-Cheh C, Sotoodehnia N, et al. Genome-wide association analysis identifies multiple loci related to resting heart rate. Hum Mol Genet. 2010;19:3885–3894. doi: 10.1093/hmg/ddq303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marroni F, Pfeufer A, Aulchenko YS, et al. EUROSPAN Consortium. A genome-wide association scan of RR and QT interval duration in 3 European genetically isolated populations: the EUROSPAN project. Circ Cardiovasc Genet. 2009;2:322–328. doi: 10.1161/CIRCGENETICS.108.833806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60•.Cho YS, Go MJ, Kim YJ, et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet. 2009;41:527–534. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]; This article reports a large initiative in South Korea for studying BP/HT genes, among others.
- 61.Chambers JC, Zhao J, Terracciano CM, et al. Genetic variation in SCN10A influences cardiac conduction. Nat Genet. 2010;42:149–152. doi: 10.1038/ng.516. [DOI] [PubMed] [Google Scholar]
- 62.Holm H, Gudbjartsson DF, Arnar DO, et al. Several common variants modulate heart rate, PR interval and QRS duration. Nat Genet. 2010;42:117–122. doi: 10.1038/ng.511. [DOI] [PubMed] [Google Scholar]
- 63.Pfeufer A, van Noord C, Marciante KD, et al. Genome-wide association study of PR interval. Nat Genet. 2010;42:153–159. doi: 10.1038/ng.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morrison AC, Felix JF, Cupples LA, et al. Genomic variation associated with mortality among adults of European and African ancestry with heart failure: the cohorts for heart and aging research in genomic epidemiology consortium. Circ Cardiovasc Genet. 2010;3:248–255. doi: 10.1161/CIRCGENETICS.109.895995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65•.Lowe JK, Maller JB, Pe'er I, et al. Genome-wide association studies in an isolated founder population from the Pacific Island of Kosrae. PLoS Genet. 2009;5(2):e1000365. doi: 10.1371/journal.pgen.1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article discusses BP/HT gene discovery in an isolated population.
- 66.Vasan RS, Glazer NL, Felix JF, et al. Genetic variants associated with cardiac structure and function: a meta-analysis and replication of genome-wide association data. JAMA. 2009;302:168–178. doi: 10.1001/jama.2009.978-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kraja AT, Rao DC, Weder AB, et al. Two major QTLs and several others relate to factors of metabolic syndrome in the Family Blood Pressure Program. Hypertension. 2005;46:751–757. doi: 10.1161/01.HYP.0000184249.20016.bb. [DOI] [PubMed] [Google Scholar]
- 68.Kraja AT, Hunt SC, Pankow JS, et al. Quantitative trait loci for metabolic syndrome in the Hypertension Genetic Epidemiology Network study. Obes Res. 2005;13:1885–1890. doi: 10.1038/oby.2005.231. [DOI] [PubMed] [Google Scholar]
- 69.Kraja AT, Huang P, Tang W, et al. QTLs of factors of the metabolic syndrome and echocardiographic phenotypes: the Hypertension Genetic Epidemiology Network study. BMC Med Genet. 2008;9:103. doi: 10.1186/1471-2350-9-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Org E, Eyheramendy S, Juhanson P, et al. Genome-wide scan identifies CDH13 as a novel susceptibility locus contributing to blood pressure determination in two European populations. Hum Mol Genet. 2009;18:2288–2296. doi: 10.1093/hmg/ddp135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Khan AA, Chow EC, Porte RJ, et al. Expression and regulation of the bile acid transporter, OSTalpha-OSTbeta in rat and human intestine and liver. Biopharm Drug Dispos. 2009;30:241–258. doi: 10.1002/bdd.663. [DOI] [PubMed] [Google Scholar]
- 72.Yang HC, Liang YJ, Wu YL, et al. Genome-wide association study of young-onset hypertension in the Han Chinese population of Taiwan. PLoS One. 2009;4(5):e5459. doi: 10.1371/journal.pone.0005459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Y, O'Connell JR, McArdle PF, et al. From the Cover: Whole-genome association study identifies STK39 as a hypertension susceptibility gene. Proc Natl Acad Sci U S A. 2009;106:226–231. doi: 10.1073/pnas.0808358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Turner ST, Bailey KR, Fridley BL, et al. Genomic association analysis suggests chromosome 12 locus influencing antihypertensive response to thiazide diuretic. Hypertension. 2008;52:359–365. doi: 10.1161/HYPERTENSIONAHA.107.104273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Levy D, Larson MG, Benjamin EJ, et al. Framingham Heart Study 100K Project: genome-wide associations for blood pressure and arterial stiffness. BMC Med Genet. 2007;8(Suppl 1):S3. doi: 10.1186/1471-2350-8-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.