Abstract
Summary: Freshwater bacteria are at the hub of biogeochemical cycles and control water quality in lakes. Despite this, little is known about the identity and ecology of functionally significant lake bacteria. Molecular studies have identified many abundant lake bacteria, but there is a large variation in the taxonomic or phylogenetic breadths among the methods used for this exploration. Because of this, an inconsistent and overlapping naming structure has developed for freshwater bacteria, creating a significant obstacle to identifying coherent ecological traits among these groups. A discourse that unites the field is sorely needed. Here we present a new freshwater lake phylogeny constructed from all published 16S rRNA gene sequences from lake epilimnia and propose a unifying vocabulary to discuss freshwater taxa. With this new vocabulary in place, we review the current information on the ecology, ecophysiology, and distribution of lake bacteria and highlight newly identified phylotypes. In the second part of our review, we conduct meta-analyses on the compiled data, identifying distribution patterns for bacterial phylotypes among biomes and across environmental gradients in lakes. We conclude by emphasizing the role that this review can play in providing a coherent framework for future studies.
INTRODUCTION
The biogeochemical importance of bacteria in freshwater ecosystems was first recognized in the 1940s, when Lindeman put the “microbial ooze” at the center of a diagram depicting the trophic dynamics in a northern temperate lake, Cedar Creek Bog (121). Since this early recognition of the critical role of bacteria in regenerating and mobilizing nutrients in freshwater food webs, it has become clear that aquatic bacteria drive transformations and the cycling of most biologically active elements in these ecosystems. This key position held in essentially all biogeochemical cycles does not merely stem from the role of bacteria as the principal degraders and mineralizers of organic compounds to their inorganic constituents (30, 36) but also results from their biomass production and trophic coupling to eukaryote predators, which, by fueling the food web, has a profound impact on elemental fluxes and water quality in the ecosystem (154). Given the facts that lakes and other inland waters play a more critical role in the global carbon budget than previously recognized (31) and that lakes have been described as early indicators (i.e., sentinels) of both regional and global environmental change (133, 228), the role of microbes in these processes is of renewed interest.
Despite the recognition that bacteria occupy a prominent role in lake ecosystem processes and greatly impact lake water quality, the bacterial taxa participating in these activities remain largely undescribed. Molecular biology tools now provide us with unprecedented access to the diversity and composition of freshwater lake bacterial communities and have for the first time enabled our field to identify the numerically dominant organisms in these ecosystems and learn much about their distributions in time and space. Although every multilake study has shown that differences in bacterial community composition between lakes can be quite large (e.g., see references 108, 122, 234, and 235), there is a growing body of evidence that many bacterial groups are freshwater specific and have a global distribution (48, 63, 82, 123, 129, 178, 195, 223, 238, 242). These two viewpoints seem to stand in opposition and may be restated as, “each lake has a different bacterial community” and “many lakes have the same bacterial taxa.” This apparent dichotomy arises in part because different research groups inventory bacterial taxa using various levels of phylogenetic or taxonomic resolution, ranging from phylum-level fluorescent in situ hybridization (FISH) probes (23, 154, 176) to more narrowly identified phylotypes defined by 16S rRNA gene sequences or rRNA intergenic spacer regions (84, 140, 190, 232, 234). Thus, in order to engage in a meaningful discussion of freshwater lake taxa, we need a consistent and robust vocabulary with which to inventory taxonomic units.
To date, most studies employing molecular methods to examine freshwater lake bacterial taxa have been highly descriptive in nature, offering little more than “species” lists for a scattered set of freshwater ecosystems or the proverbial “stamp collection.” This descriptive approach is arguably a necessary early step in the progression of any natural science, but it cannot provide us with the predictive capabilities needed to create a synthetic understanding of freshwater microbial ecology. In order to continue our maturation as a discipline, we must develop a framework within which to explain taxon or phylotype distribution patterns based on processes and mechanisms. This review, in the form of a natural history guide (or “field guide”), as often compiled for macroscale organisms such as plants and animals, attempts to provide the starting point from which this framework can be developed. We note that previous freshwater bacterial compilations are either now outdated (238), heavily focused on methods and marine water and freshwater comparisons (154), or geared toward highlighting needed avenues of research in modern inland water microbiology (75). While acknowledging the importance of molecular methods for advancing microbial ecology, we refer readers to the many excellent reviews and books dedicated to their description (for example, see references 117, 145, and 154).
In light of the mismatched and overlapping naming structure for freshwater lake bacterial taxa, our approach to this natural history guide was to gather all published freshwater lake 16S rRNA gene sequences, construct high-quality phylogenies, and propose a new vocabulary that unifies taxon names and provides a platform with which to discuss these important freshwater lake taxa (akin to the approach taken by Zwart and colleagues [238]). We use a classification system that is hierarchical like Linnaean taxonomy but based on phylogenetics. Our classification system is also designed to maintain the phylogenetic context by which freshwater bacterial gene sequences historically have been identified, clustered, and named. With this natural history guide, we are not endorsing “phylogenetic” over “Linnaean” systematization (debate reviewed in reference 57). Instead, we choose a phylogenetic approach, because there are too few defined freshwater lake bacterial genera or species to have meaningful discussions using the Linnaean approach. We expect the number of described freshwater genera and species to grow rapidly, and indeed, this appears to be the trend, as a number of isolates have been described recently (76-79, 81, 113). As the database of described freshwater organisms matures, we envision that the phylogenetic groups described within this guide will be replaced by an organismal naming structure according to the conventions set forth in the bacteriological code. In the interim, this document provides the bridge between former naming conventions and newly described species. Furthermore, the phylogenetic approach allows us to synthesize the available published information on freshwater lake bacterial phylotypes, develop new cross-study comparisons, and discuss these phylotypes in an ecological context.
A natural history guide is a dynamic document, and this is a critical time for a new edition. As larger and larger molecular data sets are gathered through next-generation sequencing (both 16S rRNA gene tag sequencing and metagenomics), this natural history guide will enable cross-study comparisons and syntheses that are currently not feasible. In the following guide, we lay out our new naming structure with the controlled nomenclature of phylum/lineage/clade/tribe. We then review the current information on the ecology, metabolism, and distribution of the defined groups within each phylum. We also highlight previously unidentified bacterial distribution patterns and propose hypotheses to test in future studies. Finally, we review the current knowledge about ecological traits of different groups of freshwater lake bacteria.
Historical Perspectives
As recently as 1980, culture-based techniques led researchers to believe that the organisms inhabiting soil and aquatic environments were quite similar (170). Researchers now appreciate a clear distinction between those bacteria found in the soil and those found in aquatic habitats (130). Molecular methodology has since revealed distinct bacterial communities among aquatic environments that are chemically and physically unique (167, 242). The greatest community composition differences occur along salinity gradients, as manifested in the very distinct bacterial community compositions of the ocean and freshwater lakes (130, 167). Indeed, for other microorganisms (microeukaryotes, archaea, and viruses), evolutionary transitions across the salinity barrier also seem to have been quite rare (128, 129).
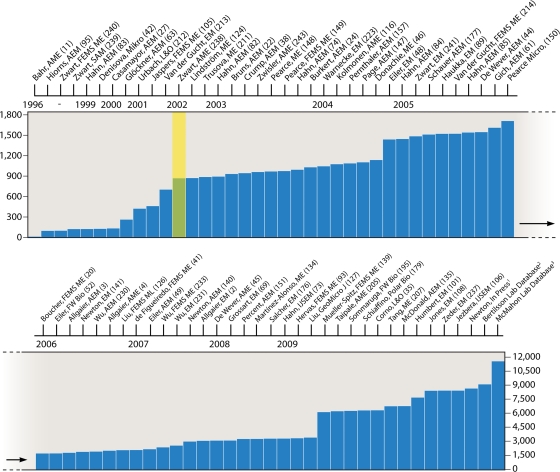
In 2002, Zwart and colleagues gathered the reported 16S rRNA gene sequences from all freshwater environments (238), which at the time consisted of 11 lakes and 2 rivers as part of 10 independent studies. This fruitful approach resulted in the collection of 689 bacterial 16S rRNA gene sequences and enabled the identification of 10 freshwater phyla and 34 putative bacterial freshwater clusters, defined as a monophyletic branch of a phylogenetic tree that contained at least two sequences with ≥95% gene identity from more than one freshwater environment. Many of the identified clusters were proposed to be common in freshwater and appeared to represent unique bacterial taxa found only in this biome. Over the past 8 years, many more researchers have retrieved and sequenced 16S rRNA genes from freshwater lake systems (Fig. 1), and a great number of new putative freshwater lake clusters have been proposed. However, the lack of a cohesive collection of known sequences and defined clusters has resulted in a mixture of naming conventions and many overlapping and incongruent clusters.
FIG. 1.
Timeline of 16S rRNA genes represented in the database. Lead author and journal abbreviation from papers contributing sequences are noted along the timeline. The yellow bar indicates the previous freshwater sequence collection review by Zwart and colleagues (238). Journal abbreviations are as follows: AEM, Applied and Environmental Microbiology; AME, Aquatic Microbial Ecology; EM, Environmental Microbiology; FEMS ME, FEMS Microbiology Ecology; FW Bio, Freshwater Biology; GeoMicro J, Geomicrobiology Journal; Mikro, Mikrobiologiia; L&O, Limnology and Oceanography; ME, Microbial Ecology; Micro, Microbiology; FEMS ML, FEMS Microbiology Letters; IJSEM, International Journal of Systematics and Evolutionary Microbiology; Polar Bio, Polar Biology; SAM, Systematic and Applied Microbiology. 1, manuscript in press at Environmental Microbiology (141a) (includes GenBank accession numbers FJ827781 to FJ828505); 2, GenBank accession numbers for the sequenced clones are HQ386253 to HQ386631; 3, GenBank accession numbers for the sequenced clones are FJ916807 to FJ916903 and HQ530565 to HQ532908, except HQ530583, HQ531638, and HQ532521. (Based on data from references 2, 3, 4, 11, 20, 22, 24, 27, 35, 38, 41, 42, 44, 45, 46, 48, 49, 52, 61, 63, 69, 73, 74, 82, 83, 84, 85, 89, 93, 95, 101, 105, 106, 108, 116, 124, 126, 127, 134, 135, 139, 140, 141, 147, 148, 149, 150, 151, 157, 176, 177, 179, 195, 205, 207, 211, 212, 213, 214, 223, 230, 231, 233, 237, 238, 239, 240, 241, and 243.)
CREATING A MODERN VIEW OF FRESHWATER LAKE BACTERIA
In this study, we present an overview and meta-analysis of published freshwater lake 16S rRNA gene sequences. To constrain our data set to a more consistent range of habitats, we limited our analysis to sequences collected from the epilimnia of lakes. We acknowledge that bacterial communities may be very different in other freshwater habitats such as wetlands or suboxic hypolimnetic water of stratified lakes. Microbial populations and communities in such environments are likely strongly influenced by the overall availability of electron acceptors, steep redox clines, and the release of organics from plant debris or sediments. An in-depth account of the phylogenetic composition and ecology of bacterial communities in these rather poorly characterized microbiomes is needed but lies beyond the scope of this review. We included 68 published papers that represent >8,400 16S rRNA gene sequences and have additionally added >2,600 unpublished sequences from the McMahon laboratory in Wisconsin and >400 unpublished sequences from the Bertilsson laboratory in Sweden (Fig. 1). The majority of these sequences are from projects reported within the last 5 years (Fig. 1). The increasing pace of lake studies containing larger numbers of 16S rRNA gene sequences follows the universal trend of an increasing rate of sequences deposited into the public databases (e.g., the Ribosomal Database [RDB] Project [222]), and with the explosion of next-generation sequencing technologies, the total number of sequences in this database is likely to be surpassed by each new project. We propose that our manually curated data set based largely on nearly full-length sequences generated by traditional Sanger technology will serve as a much-needed backbone for interpretations of the burgeoning collection of such comparatively short sequence reads.
Phylogenetic Tree Reconstructions and Cluster Classification
For each of the 69 published papers included in our synoptic analysis, 16S rRNA gene sequences from lake epilimnia were retrieved and downloaded from GenBank via their accession numbers. Taxonomic assignments were obtained for these published sequences and the unpublished McMahon laboratory and Bertilsson laboratory sequences using the Classifier program at the Ribosomal Database RDP II website (222). Based on the RDP taxonomic assignments, sequences were grouped by phylum and imported into the ARB software package (131), containing a 16S rRNA gene database from Greengenes (January 2007) (43). Sequences were initially automatically aligned by using the FAST_ALIGNER ARB tool before the alignment was heuristically adjusted using primary and secondary rRNA structures as a guide. Sequences that were thought to be chimeras (checked with the software program Mallard [10]) or those that contained many (>10) ambiguous bases were removed from the database. In total, ∼200 sequences were removed for either one of these reasons, leaving >11,500 sequences for further analyses.
Phylogenetic inferences were obtained by using only nearly full-length (≥1,300-nucleotide [nt]) sequences. A 50% base frequency filter, which excludes highly variable positions, was calculated for the sequence alignment in each phylogenetic analysis. Phylogenetic reconstructions were performed at CIPRES (www.phylo.org) using RAxML (197) maximum likelihood analysis with 100 bootstrap runs and with Bayesian inference in MrBayes v. 3.1.2 (172), using a general time-reversible gamma-distributed rate variation model with three independent Markov chain Monte Carlo analyses, each starting from random trees for each of four simultaneous chains until chain stabilization. Trees recovered before chain stabilization were discarded by using appropriate burn-in values, and a 50% majority-rule tree was calculated. Consensus trees obtained from the maximum likelihood and Bayesian inference methods were compared manually for the presence of nonstable branching patterns between methods. The final trees illustrated in all figures represent the reconstruction from Bayesian phylogenetic inference but are depicted with both the Bayesian clade credibility values and the maximum likelihood bootstrap values at nodes obtained by both methods. All partial sequences (<1,300 nt) were then added to the consensus trees in ARB using the maximum parsimony criteria with an appropriate 50% base frequency filter and without allowing changes in the tree topology.
Within the 68 published papers used to construct our database, >100 freshwater clusters were previously proposed. We integrated these clusters and many others resulting from our analyses into a single phylogenetic framework using a hierarchical naming structure (phylum/lineage/clade/tribe) illustrating the genetic identity relationships between groups. The phylum is synonymous to the long-established phylum defined by bacterial systematics. The lineage, the first of our phylogenetically defined groups, is the most broadly defined and consists of sequences that cluster as a monophyletic branch of a phylogenetic tree within a phylum (e.g., acI, Luna1, and acIII). Our defined lineages generally share maximum sequence identity differences of between 10 and 15% (calculated including all nucleotide positions over the shared length of the 16S rRNA gene). The clade definition is based upon the cluster definition used previously by Zwart and colleagues (238), thereby providing continuity between the last compiled database and this new rendition. As such, a clade represents a group of sequences clustering as a monophyletic branch of a phylogenetic tree that have ≥95% sequence identity to at least one other sequence of that branch (i.e., nearest neighbor). Our clade definition also requires that at least two sequences be obtained from different lakes (e.g., clades acI-A, acI-B, and acI-C). The most refined taxonomic group of our clustering system is the tribe. A tribe also consists of a group of sequences clustering as a monophyletic branch of a phylogenetic tree but requires each sequence to have ≥97% sequence identity to another sequence of that branch. As with the clade definition, the tribe definition also requires that at least two sequences originated from different lakes (e.g., tribes acI-B1, acI-B2, and acI-B3). This definition was chosen to avoid the clustering issues created from the large number of sequencing and curation errors contained in sequences deposited in the public databases while also closely representing the most common division (97% 16S rRNA gene identity) used to describe bacterial ecologically relevant groups in nature. When possible, tribes were more narrowly defined than with the minimum allowed 97% sequence identity. The program DOTUR (180) was used to identify suspected clade and tribe groupings observed in the phylogenetic reconstructions. Phyla/lineages/clades were excluded from detailed phylogenetic analyses when they did not contain at least one tribe.
Common Lineages of Freshwater Lake Bacteria
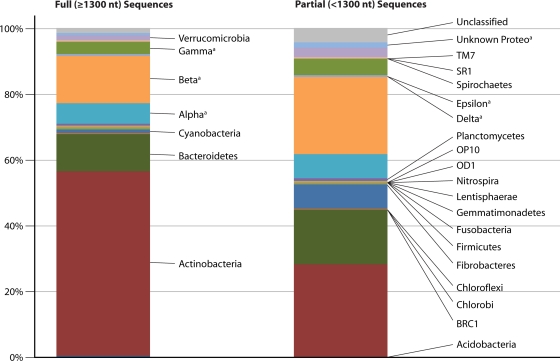
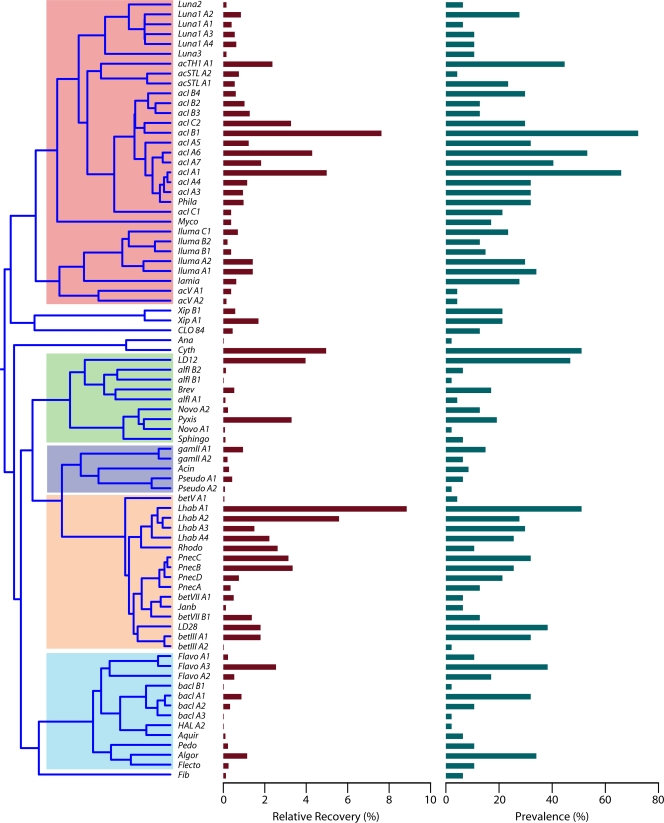
Examination of the database revealed that 21 phyla (based on RDP confidence assignments of ≥75%) have been recovered from lake epilimnia, with 5 phyla being recovered commonly (Proteobacteria, especially Betaproteobacteria, with >4,300 and >2,600 sequences, respectively; Actinobacteria, with >3,000 sequences; Bacteroidetes, with >1,900 sequences; Cyanobacteria, with >800 sequences; and Verrucomicrobia, with >300 sequences) (Fig. 2 and see Table S1 in the supplemental material). These five phyla were also the most numerous in the database compiled previously by Zwart and colleagues (238) and are in general agreement with the majority of FISH-based studies, which reported the Actinobacteria (4, 23, 63, 224) or the Betaproteobacteria (62, 93, 139, 153, 230) as being the most abundant bacterial phylum in lake epilimnia. The remaining 16 recovered phyla make up only ∼2.6% of the total sequences collected in our database and include Acidobacteria, BRC1, Chlorobi, Chloroflexi, Fibrobacteres, Firmicutes, Fusobacteria, Gemmatimonadetes, Lentisphaerae, Nitrospira, OD1, OP10, Planctomycetes, Spirochaetes, SR1, and TM7 (Fig. 2).
FIG. 2.
Stacked bar plot representing the distribution of sequences by phylum in the freshwater lake sequence database. Sequences were taxonomically classified by the RDP classifier (February 2010) (222) and were required to have 75% classifier confidence to be included as a representative of a phylum. Sequences with <75% confidence were termed unclassified. Phyla listed are present in both bar plots but are listed on only one side to aid in the visualization of the plots. a, the phylum Proteobacteria was split into its representative classes for sequences with a ≥75% confidence score at the class taxonomic assignment and unknown Proteobacteria for those sequences with confidence scores of <75%.
A large group of sequences in the data set was termed “unclassified” (∼500 sequences) (Fig. 2). An unclassified label signifies that a sequence had <75% confidence at the phylum taxonomic level with the RDP Classifier assignment. The majority of these unknown sequences were at least in part the result of the sequence read length being very short (<400 nt) (Fig. 2). It is not clear whether the 15 unclassified nearly full-length sequences represent bacterial 16S rRNA genes from undefined or poorly defined phyla, undetected chimeras, poor sequence reads, or nonspecific amplification. None of these 15 sequences met our criteria to be included in a clade or tribe.
In the following sections we review and summarize the currently available information regarding the ecology and diversity of each major bacterial phylum recovered in freshwater lakes. Along with basic information and the historical context, we present an updated phylogenetic framework that links back to prior findings and use this new framework as an outline for summarizing published studies of the ecology of each phylum and its more finely resolved groups. The length of each section reflects the currently available information and is also a fair proxy for the number of available sequences for each phylum. We conclude each section with a brief summarizing statement of unifying traits for freshwater lake bacteria in each phylum, even if it is clear that there is considerable ecological divergence at this higher taxonomic rank.
Phylum Actinobacteria
The phylum Actinobacteria (formerly part of the group of high-G+C Gram-positive bacteria) is made up of Gram-positive bacteria with a high mol% G+C DNA composition (generally ranging from 51 to 70% [218]). Historically, soils were considered the primary environment of residence and optimal activity for the Actinobacteria (66). The advent of molecular-based studies of aquatic systems has changed this perception. Initial 16S rRNA gene- and FISH-based studies revealed that members of the Actinobacteria are common and often numerically important component in a variety of freshwater habitats (42, 63, 95, 238, 239). Also clear from these studies was that the 16S rRNA genes recovered from the epilimnia of lakes were distinct from the “typical” Actinobacteria in soils or the more recently discovered Actinobacteria in marine systems (166). Further molecular surveys have since shown that these “freshwater” Actinobacteria can be found in a variety of limnic systems, such as rivers (37, 238), brackish seas (171), bays (186), and glacial ice (29). The emergent phylogenetic clustering by biome along with evidence that the Actinobacteria in freshwater lakes are actively synthesizing DNA (152, 224, 237) and proteins (23, 152) have provided ample evidence that this phylum is an indigenous resident of freshwaters.
Actinobacteria 16S rRNA gene-based phylogeny.
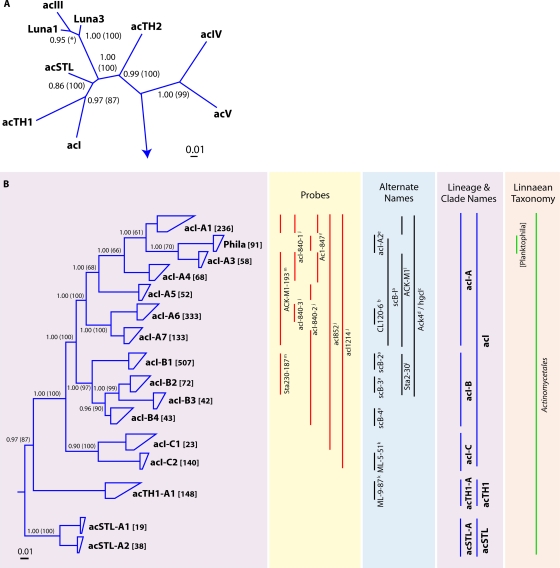
Since the first “typical” freshwater Actinobacteria 16S rRNA gene sequences were obtained in 1997 (95), >40 phylogenetic clusters have been postulated for this phylum (Fig. 3). In 2004, Warnecke and coauthors combined several previous 16S rRNA gene cluster-naming schemes (95) into a hierarchical classification system that has generally become fixed in the literature (223). Those authors named the most prominent lineages acI, acII, acIII, and acIV and subdivided these lineages into the clades acI-A to -C, acII-A to -D, and acIV-A and -B. The acI to acIII lineages are affiliated with the order Actinomycetales, and the acIV lineage is associated with the order Acidimicrobiales. Since this naming structure was implemented, numerous studies have contributed additional lineages (82, 233) and additional subdivisions of the acI to acIV lineages (3, 140, 233), many of which are overlapping (Fig. 3B to D).
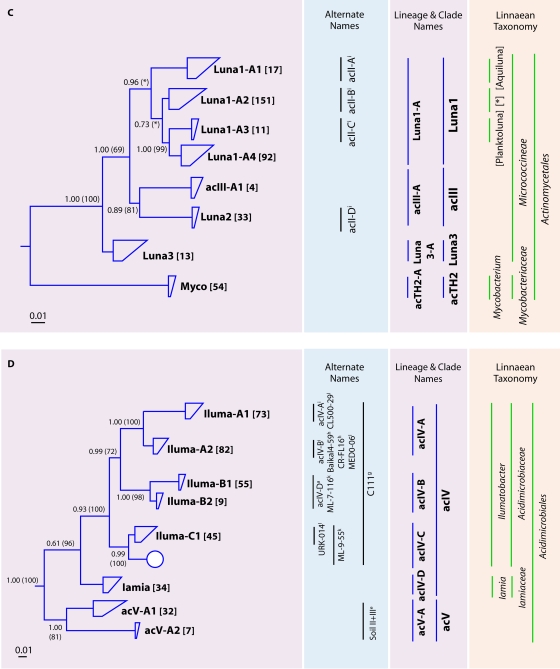
FIG. 3.
(A) Radial consensus phylogram of the freshwater lake lineages of the Actinobacteria. (B) Consensus phylogram of the tribes of lineages acI, acTH1, and acSTL. (C) Consensus phylogram of the tribes of lineages Luna1, acIII, Luna3, and acTH2. (D) Consensus phylogram of the tribes of lineages acIV and acV. All phylogenetic inference calculations were conducted with nearly full-length sequences (>1,300 nt) representing the lineages/clades/tribes. Frequency base filters were created to mask out highly variable positions, and representative sequences from members of the Archaea were used as the outgroup for all tree reconstructions. Bayesian clade credibility values and maximum likelihood bootstrap values (in parentheses) of >0.5 (50) are listed at each node. Tribe names are listed at the end of tree tips (trapezoids), with the number of sequences associated with each tribe listed in square brackets. Tree tips ending in ovals represent phylogenetic clusters that were named previously but did not meet the criteria to be called a tribe (≥2 sequences with ≥97% identity of ≥1,300 nt isolated from >1 lake). Current lineage and clade names are listed to the right of the phylograms and are highlighted in purple. The Linnaean taxonomy column is based on the RDP classification (222), where all sequences within the classified group must have ≥75% assigned confidence for the taxonomic group. Square brackets around a genus name indicate a candidate genus, and an asterisk indicates that the genus is described but not yet covered in the RDP classification. Alternate names from previous studies also covering the defined lineages/clades/tribes are listed. Probe coverage is based upon a perfect match to ≥75% of the sequences in a defined lineage/clade/tribe. References to past clades and probes are as follows: a, reference 3; b, reference 48; c, reference 63; d, reference 95; e, reference 140; f, reference 156; g, reference 212; h, reference 214; i, reference 223; j, reference 224; k, reference 233; l, reference 238; m, reference 242. *, tribe Luna1-A2 is associated with three “Candidatus” genera: “Candidatus Limnoluna,” “Candidatus Flaviluna,” and “Candidatus Rhodoluna” (73).
Our phylogenetic reconstruction yielded nine broadly defined lineages (acI, acTH1, acSTL, Luna1, acIII, Luna3, acTH2, acIV, and acV) (Fig. 3A), all of which were described previously. Six of the nine lineages were divided further into clades and/or tribes. Thirty-two tribes resulted from the reconstructions (Fig. 3B to D). The acI and acIV lineages are particularly abundant among the freshwater lake Actinobacteria, with the acI lineage being more abundant in the free-living fraction (2, 207), often comprising ≥90% of the identified actinobacterial taxa (224), and with the acIV lineage exhibiting more localized dominance (231). As the most-studied group, the acI lineage was also the most resolved, containing 13 narrowly defined tribes (some with >99% identity). Only one of the 13 acI tribes (Phila) contained a named organism, and this organism, “Candidatus Planktophila limnetica,” has been maintained solely in enrichment cultures (106). The acI-A and acI-B clades are nearly encompassed by the popular molecular probes acK-M1 and sta2-30, which were named after two of the first 16S rRNA gene clones recovered for this lineage (95, 238).
The acIV and Luna1 lineages were the next most abundant lineages in our database (Fig. 3C and D). The acIV lineage partitioned into four monophyletic clades (acIV-A, acIV-B, acIV-C, and acIV-D) and six tribes, many of which have received multiple names in the literature (Fig. 3D). The Luna1 lineage partitioned into only a single clade containing four tribes, one of which (Luna1-A2) was highly abundant in the sequence database. The acIV lineage does not contain cultured representatives, while the Luna1 lineage is well represented by previously described organisms (73). Using the same isolation process as that for the Luna2 tribe, Hahn and coworkers isolated or enriched for several bacteria from the Luna1 lineage. Seven species were obtained in these enrichment cultures and were named “Candidatus Planktoluna difficilis,” “Candidatus Aquiluna rubra,” “Candidatus Flaviluna lacus,” “Candidatus Rhodoluna limnophila,” “Candidatus Rhodoluna planktonica,” “Candidatus Rhodoluna lacicola,” and “Candidatus Limnoluna rubra” (73).
The acIII, acV, and acSTL lineages were the remaining three lineages containing clade and tribe divisions. All three of these lineages had a single clade harboring two tribes. The acIII lineage contains the Luna2 tribe, named after Lake Mondsee (meaning moon), the ecosystem from which the first isolate was obtained. This tribe is one of the few tribes to contain multiple isolates (82). The acV lineage is related to bacteria isolated from soils clustering with the defined soil lineage soil II+III but is only distantly related to cultured organisms (data not shown). The acSTL lineage name originates from clones retrieved from Lake Stechlin in Germany and, like many of the other lineages, does not contain cultured representatives.
The final three lineages of the Actinobacteria, acTH1, acTH2, and Luna3, are made up of sequences represented by only a single tribe. The acTH1 and acTH2 lineages are named after clone representatives obtained from Lake Taihu in China (231). All sequences obtained for acTH2 were closely affiliated with the genus Mycobacterium; therefore, we named the tribe representing this sequence cluster Myco. Unlike lineage acTH2, acTH1 does not contain cultured representatives. Luna3 is closely related to the other Luna lineages but does not have a cultured representative as of yet.
Ecology of the freshwater lake Actinobacteria.
Many studies employing a variety of methods, including fluorescent labeling of cells (3, 23, 224), PCR-based methods (2, 89, 123, 140, 141, 151, 212, 233, 242), and metagenomic profiling (39), have confirmed and expanded upon the initial observation that the Actinobacteria are ubiquitous and abundant in freshwater lakes. In fact, the Actinobacteria are often the numerically dominant phylum in lakes, where they can contribute >50% of the bacteria in the surface waters (epilimnion) (63). The freshwater Actinobacteria are also present in the bottom waters (hypolimnion) of lakes (20), but their abundance often decreases with decreasing oxygen concentrations (3, 205). This high level of abundance in the epilimnion has proven to be consistent across lake types, as the Actinobacteria are common among oligotrophic (101), mesotrophic (39, 101, 237), eutrophic (233), and dystrophic (141) lakes. Likewise, the dominance of the Actinobacteria is spread across the globe. Studies have shown high actinobacterial abundances in lakes in North America (140), Europe (63), Africa (44, 45, 101), Asia (82, 142, 230), Australia (84), South America (35), and Antarctica (149, 150).
Despite the ease with which 16S rRNA gene sequences from members of the Actinobacteria are recovered from the freshwater environment, the isolation of organisms representative of these sequences has proven difficult. The Actinobacteria were one of the first isolated and described groups from the pelagic lake environment (53, 194), but these first isolates were of the chromogenic actinomycete type, often Micromonospora-like, a group now recognized as not being particularly abundant in these environments (63). The first time a representative freshwater Actinobacteria isolate was available for study was in 2003, when Hahn and coworkers isolated several organisms affiliated with what were termed the Luna clusters (Luna clusters 1 and 2) (82). Nine Luna strains were isolated from lakes of contrasting trophic states (ranging from oligotrophic to hypereutrophic) located on two continents (Europe and Asia) (82). All strains were very small (<0.1 μm3) with a selenoid cell morphology and thin cell walls, the so-called “ultramicrobacteria” (82). This finding matched previously reported FISH observations that the freshwater lake Actinobacteria were generally free-living single cells of a very small size (63, 224). All isolates were also pigmented, either yellow or red, suggesting that like other soil-dwelling members of the Actinobacteria, these taxa might be capable of producing secondary metabolites. Since this initial isolation, Hahn and coworkers have isolated or enriched several other strains affiliated with the Luna clusters (84), giving several of these enrichments “Candidatus” names (73). During the course of isolating organisms from Luna1, those authors noted that they were unable to obtain pure cultures because a small percentage (often <1%) of another bacterial taxon was seemingly required to maintain the Actinobacteria in culture, although the identity of the cotaxon was highly variable and thus likely part of a nonspecific relationship (73). The enrichment of members of the freshwater Actinobacteria following the addition of signaling compounds (22) could represent one avenue for the apparent coculture requirement observed by Hahn and colleagues (73).
Although the isolation of the Luna clusters has already revealed many insights into the life-style of the freshwater lake Actinobacteria, no isolates have been obtained from the two lineages considered most abundant (lineages acI and acIV). Recently, Jezbera and coworkers established stable cocultures, including a member of the acI lineage, which was named “Candidatus Planktophila limnetica” (106). Unfortunately, this organism represented ≤5.6% of the cocultures; hence, little physiological insight could be attained.
Due to the dearth of freshwater Actinobacteria cultures, researchers have turned to molecular methods to gain insight into the ecophysiological roles that these organisms play in their natural habitat, with the majority of studies focusing on the distribution of lineages and clades in time and space. The first comparative study of the distribution of the Actinobacteria noted very similar 16S rRNA gene sequences from Europe and North America, suggesting that narrowly defined clades could have a worldwide distribution (239). Numerous studies based on 16S rRNA gene identification since then have confirmed a striking lack of geographic separation for taxa within the dominant Actinobacteria clades (84). However, as observed by Hahn and colleagues, strain distribution could be limited by environmental factors (84). In that study, Luna strains isolated from several continents but containing nearly identical rRNA operons exhibited very different adaptations to prevailing local temperatures, suggesting barriers to widespread colonization capabilities (84). It remains to be seen if this apparent lack of a biogeographic signal holds true for strain comparisons, protein-encoding genes, or whole-genome comparisons within some of the narrowly defined Actinobacteria clusters.
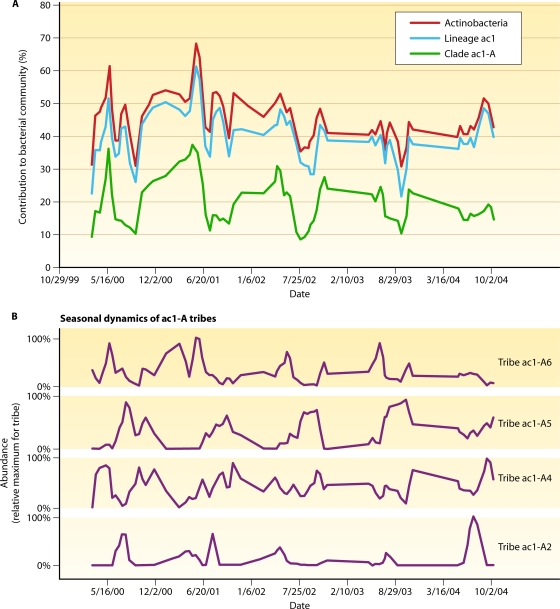
The cosmopolitan distribution and high level of abundance of actinobacterial 16S rRNA genes in freshwater lakes have prompted several investigations into the environmental factors and cell characteristics contributing to the success of these organisms. Through time, the phylum Actinobacteria, especially the acI lineage, appears to have comparatively smaller changes in biomass or prevalence across lake seasons (63), making it the most persistent lineage of freshwater lake bacteria. Those authors suggested that the comparatively minor fluctuations in the abundance of acI taxa point to a consistent source of energy generation for these organisms in spite of the marked seasonal changes observed for bacteria over time in lakes (183). Recently, metagenomic studies of freshwater lakes revealed an abundance of rhodopsin genes (a potential source of light-driven energy generation) in these systems, many of which are now termed actinorhodopsins due to their presence in clones containing freshwater Actinobacteria gene signatures (including the acI and Luna lineages [185]). These actinorhodopsin genes are also known to occur in several freshwater isolates (184). This potentially supplemental mode of energy generation may be partly responsible for the success that generates the cosmopolitan distribution and ubiquity of these organisms.
Another major mechanism for the ubiquity of the freshwater Actinobacteria seems to be related to their small cell size and cell wall composition. In a study by Pernthaler and coworkers the addition of the size-selective grazer Ochromonas (Poterioochromonas) sp. initiated acI blooms comprising up to 60% of the total bacterial community (156). In 2003, Hahn and coworkers confirmed this observation for the Luna lineages as well (82) but also observed that it was not simply size that reduced Ochromonas grazing on their isolates. Those authors posited that the cell wall structure, which included a visually identified S layer, might be an additional factor making these organisms less “edible.” In a subsequent study, attempts to disrupt the S layer resulted in increased grazing (4 to 5 times) upon members of the Luna2 tribe (209). The lack of members of the Actinobacteria in the food vacuoles of freshwater bacterivorous flagellates has also provided further evidence for a general grazing protection mechanism among these bacteria (106). The freshwater Actinobacteria are not impervious to grazing, however, as the addition of Cyclidium glaucoma, a bacteriovorous ciliate that efficiently feeds on small prey (163), resulted in equal grazing upon the acI lineage and other freshwater lake lineages (156). Others have also noted correlative decreases in the abundance of the Actinobacteria during periods of enhanced grazing pressure (141).
UV stress resistance has also been postulated to be one of the reasons for the success of the Actinobacteria in the upper waters of lakes, which often have high UV transparency. Warnecke and colleagues showed a significant positive relationship between Actinobacteria abundance and UV transparency in a series of mountain lakes (224). Those authors noted that both pigmentation and a high G+C content have been shown to increase protection from UV damage in bacteria and accordingly suggested that the freshwater Actinobacteria, which have a high G+C content and may be pigmented (82), might harbor similar protective mechanisms (224). However, no correlation between the freshwater Actinobacteria and UV transparency was observed in a second study involving high mountain lakes, casting doubt on this apparent relationship (230). In addition to UV protection, many members of the Actinobacteria are capable of producing spores (reviewed in reference 218), allowing them to survive long periods of desiccation. Although still not observed for the freshwater Actinobacteria, strong UV protection and desiccation resistance via spore formation, together with the known small cell size of these organisms, would make the freshwater Actinobacteria particularly suitable for aerial dispersal. This could explain their ubiquitous representation in globally dispersed lakes (93). However, to date, these taxa have not been identified in air samples (109).
The abundance and distribution of the freshwater Actinobacteria have been studied in relation to the chemical and physical properties of lakes and their resident microbiota. Actinobacteria taxa have not been found in physical association with members of the Cyanobacteria or other phytoplankton (116) but may become a greater part of the bacterioplankton community during phytoplankton blooms (3, 174, 237). Generally, increased nutrient concentrations select against the freshwater lake Actinobacteria (90). This disadvantage does not seemingly stem from an inability to assimilate common substrates, as the Actinobacteria have been shown to be capable of incorporating arginine, leucine, and thymidine at rates similar to those of other common freshwater lake taxa (23) or as the most active bacterial fraction for amino acid uptake (174). However, the growth rates for the freshwater Actinobacteria often do not match their capability for substrate uptake; generally, this group exhibits growth rates that are average to below average compared to those of other phyla or the bulk freshwater lake bacterioplankton community (188). The various growth and substrate incorporation rates observed for the freshwater Actinobacteria may be due to the preferences of individual tribes for different substrate sources. Substrate partitioning among common marine bacteria has been shown (102, 161). Likewise, members of the acI-A taxa of Lake Grosse Fuchskuhle were unable to assimilate acetate, while members of taxa associated with the acI-B clade readily assimilated this carbon source (23). The distribution of members of the Actinobacteria tribes also seems to partition by broad substrate source categories, such as the ratio of allochthonous to autochthonous carbon produced in a lake (108). This resource partitioning between clades and tribes has been postulated as a reason for shifts in the growth rates and biomass changes observed pre- to post-phytoplankton bloom in Lake Zurich (237).
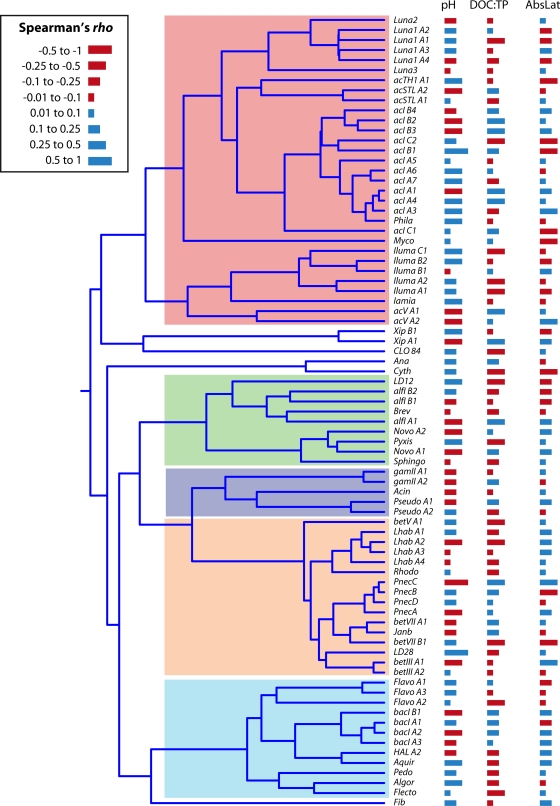
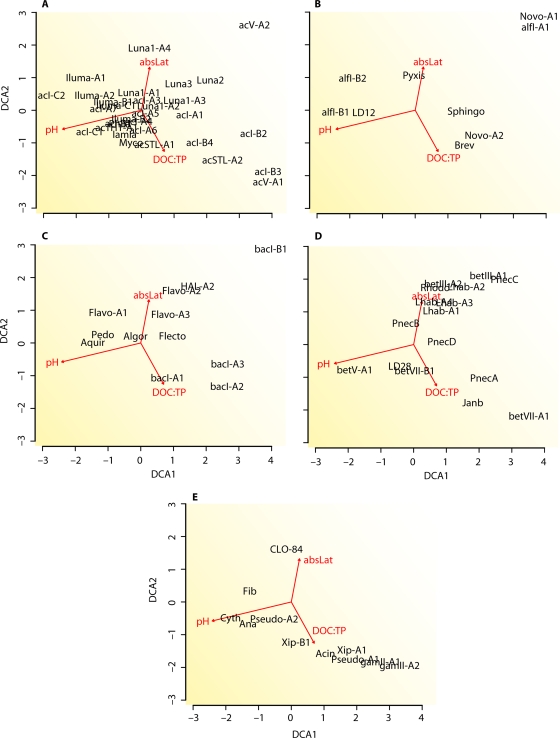
The pH of an ecosystem is often a master driver of bacterial community composition (55) and accordingly has been identified as being one of the major drivers of Actinobacteria clade and tribe distribution. In 2005, Lindström and colleagues recognized that members of clade ACK-M1 (similar to clade acI-A) (Fig. 3B) was more prevalent in high-pH lakes, while members of clade Sta2-30 (similar to clade acI-B) was more prevalent in low-pH lakes (123). Similarly, studies by Newton et al. and Taipale et al. found a majority of clade acI-B-related sequences in their humic (low-pH) lakes (141, 205). In an effort to identify more coherent response patterns, Newton and colleagues narrowly defined 11 tribes within clades acI-A and -B and found that individual tribes within the clades showed significant distribution differences based on lake pH (140). It is as yet unknown whether the other major lineages of the freshwater lake Actinobacteria show similar distribution differences based on pH.
Actinobacteria summary.
The phylum Actinobacteria contains several monophyletic lineages that are specialized to limnetic systems. These so-called “freshwater” lineages are highly abundant and ubiquitous in the epilimnia of lakes. The acI lineage appears particularly abundant in these systems and consequently has received the bulk of research attention, directed mainly at its distribution within and among lakes and its relationship to phytoplankton and bacteriovores. The isolation of representative taxa has proven elusive but has been achieved for the Luna lineages. Analysis of these isolates and FISH-based studies has shown that the freshwater Actinobacteria are small (<0.1 μm3) with a rod, coccus, or selenoid shape and are pigmented. Biogeographic signals have not been observed for taxa defined by any 16S rRNA gene groupings, but pH differences among lakes and particle attachment and carbon substrate preferences within lakes have been shown to differentiate the clades and tribes. A preliminary examination of metagenomic data has revealed that several members of the freshwater Actinobacteria have an overrepresentation of pathways for nucleic and amino acid metabolism (159) and harbor rhodopsins (actinorhodopsins), a potential mechanism for supplemental energy generation by light harvesting. Generally speaking, the freshwater lake Actinobacteria are free-living, open-water defense specialists, with possible photo- and heterotrophic energy generation life-styles.
Phylum Bacteroidetes
The phylum Bacteroidetes, formerly known as the Cytophaga-Flavobacterium-Bacteroides (CFB) phylum, exhibits enormous phenotypic and metabolic diversity. The members of this phylum occur in soil, in aquatic environments, or as symbionts of plants, animals, and humans. Most described isolates of the Bacteroidetes are chemoorganotrophs, but phototrophic capabilities have been described for two marine isolates (Polaribacter sp. strain MED152 and Dokdonia sp. strain MED134) that contain a photopigment related to the proteorhodopsin of the Proteobacteria (64, 65). Many representatives of the Bacteroidetes are known to have close relationships with animal and human hosts, where they can be either synergists or antagonists. Comparative phylogenomic analyses have provided strong support for the phylum Bacteroidetes and suggest that it shares many phenotypic characteristics and a common ancestor with the phyla Fibrobacteres and Chlorobi (72). Within the Bacteroidetes there are three distinct classes: Bacteroidales, Flavobacteriales, and Sphingobacteriales. Even if members of the Bacteroidetes sometimes dominate freshwater lake bacterial communities (157) and comprise the third most recovered phylum in this lake 16S rRNA gene data set (Fig. 2), few studies have examined the ecology of these organisms and/or defined monophyletic clusters.
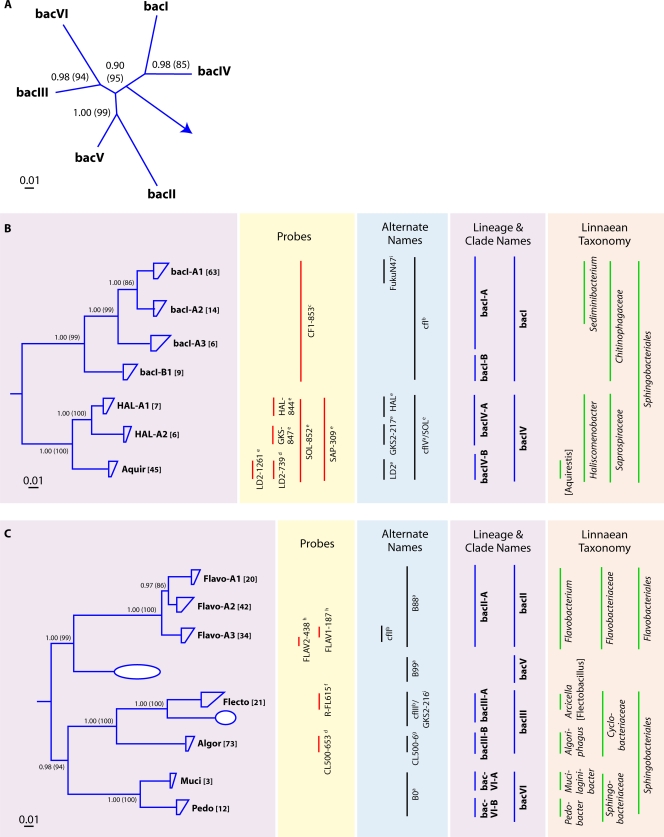
Bacteroidetes 16S rRNA gene-based phylogeny.
Altogether, five lineages and 23 clades and/or tribes could be identified within the Bacteroidetes (Fig. 4). The bacI, bacII (B88), and bacIII lineages of the Bacteroidetes contained the majority of 16S rRNA gene sequences in our data set (Fig. 4B and C). The bacII lineage was divided into three tribes made up entirely of the class Flavobacteria. The majority of the recovered freshwater lake Flavobacteria sequences were not monophyletically related to the isolates from freshwaters (e.g., Flavobacterium columnare and F. limicola) (data not shown). Likely, there are many more clades and tribes within the bacII lineage, as 24 operational taxonomic units (OTUs) were identified by terminal restriction fragment length polymorphism (tRFLP) patterns in a recent study of their seasonal dynamics in temperate lakes (49).
FIG. 4.
(A) Radial consensus phylogram of the freshwater lake lineages of the Bacteroidetes. (B) Consensus phylogram of the tribes of lineages bacI and bacIV. (C) Consensus phylogram of the tribes of lineages bacII, bacIII, bacV, and bacVI. All phylogenetic inference calculations were conducted with nearly full-length sequences (>1,300 nt) representing the lineages/clades/tribes. Frequency base filters were created to mask out highly variable positions, and representative sequences from members of the Archaea were used as the outgroup for all tree reconstructions. Bayesian clade credibility values and maximum likelihood bootstrap values (in parentheses) of >0.5 (50) are listed at each node. Tribe names are listed at the end of tree tips (trapezoids), with the number of sequences associated with each tribe listed in square brackets. Tree tips ending in ovals represent phylogenetic clusters that were named previously but did not meet the criteria to be called a tribe (≥2 sequences with ≥97% identity of ≥1,300 nt isolated from >1 lake). Current lineage and clade names are listed to the right of the phylograms and are highlighted in purple. The Linnaean taxonomy column is based on the RDP classification (222), where all sequences within the classified group must have ≥75% assigned confidence to the taxonomic group. Square brackets around a genus name indicate a candidate genus, and an asterisk indicates that the genus is described but not yet covered in the RDP classification. Alternate names from previous studies also covering the defined lineages/clades/tribes are listed. Probe coverage is based upon a perfect match to ≥75% of the sequences in a defined lineage/clade/tribe. References to past clades and probes are as follows: a, reference 49; b, reference 63; c, reference 156; d, reference 157; e, reference 177; f, reference 192; g, reference 233; h, reference 237; i, reference 238.
Another dominant sequence group within the Bacteroidetes was the bacI lineage, which is most closely related to the bacIV lineage (Fig. 4B). Both of these lineages are members of the order Sphingobacteriales. No studies have focused on the four tribes comprising the better-represented bacI lineage, but several studies have examined the bacIV lineage (157, 177, 178) and previously divided it into 3 clusters (HAL, GKS2-217, and LD2) (Fig. 4B). Two isolates from this lineage have been described, “Candidatus Aquirestis calciphila” and “Candidatus Haliscomenobacter calcifugiens” (86).
The bacIII and bacVI lineages are also members of the order Sphingobacteriales. As with many of the tribes within the Bacteroidetes, each of the tribes in these lineages (two each in bacIII and bacVI) contained previously described species. The Algor tribe (Algoriphagus-like) of the bacIII lineage was represented by the most sequences among all Bacteroidetes tribes. The Flecto (Flectobacillus-like) tribe is known to be filamentous and is targeted by the frequently used probe R-FL615 (188, 192). The two bacVI tribes (Muci for Mucilaginibacter-like and Pedo for Pedobacter-like) were not recovered frequently in our database.
Ecology of the freshwater lake Bacteroidetes.
In lake epilimnia, the Bacteroidetes may comprise a large proportion of particle-associated bacteria (120, 143) and seem to play a particularly important role in the degradation of complex biopolymers (115). In agreement with this inferred niche, several studies using PCR-based clone libraries and/or quantitative PCR (qPCR) have documented an increase in the propensity of the Bacteroidetes to occur during periods or at sites characterized by high external dissolved organic carbon (DOC) loading or alga-derived DOC inputs (48, 49, 116, 237). Using batch culture experiments, a relationship between bacterial community composition patterns and humic substance loading revealed that a DOC concentration above 0.54 mM favored tribes bacI-A1 and bacI-A2 of the phylum Bacteroidetes (51). Following a humic matter enrichment of lake water in another experiment, members of the Bacteroidetes related to Flexibacter (Chitinophaga) became the most abundant bacterial group, suggesting that they may play a significant role in humic matter degradation (104).
The freshwater lake Bacteroidetes are often found in high abundance during periods following cyanobacterial blooms. In one case, a majority of the bacterial community measured using 16S rRNA gene-based tRFLP and qPCR was made up of the Bacteroidetes Flavobacterium-like lineages B88 (bacII) and B99 (bacV) following the senescence and decline of an intense cyanobacterial bloom (49). It was concluded that many Flavobacterium-like populations are favored during periods of high heterotrophic activity and enhanced growth, where resource availability is the main force structuring the distribution of these populations. Furthermore, evidence for this copiotroph life-style (adaptation to high-nutrient conditions) among certain groups of the Flavobacteria was provided recently by Zeder and colleagues (237). In dilution cultures, group FLAV2 (within Flavo-A3 and targeted by probe FLAV2-438) (Fig. 4B) exhibited rapid growth. This rapid growth was enhanced even further when samples were obtained and diluted during and after a spring phytoplankton bloom. However, the net increase in FLAV2 cell numbers was much lower in the environment than predicted from the dilution bioassays conducted in the laboratory, suggesting a strong top-down control of the rather large FLAV2 cells by mixotrophic grazers (237).
The influences of microeukaryote predation on the freshwater Bacteroidetes have been studied in some detail. Environmental surveys from various freshwater environments have shown an increase in the abundance of members of the Bacteroidetes during periods of enhanced grazing (32, 191). However, conflicting results have been reported. In those studies, enrichments of microeukaryote predators resulted in a decrease in the abundance of members of the Bacteroidetes (111, 175, 192). In planktonic environments, predator-prey interactions are particularly influenced by the size-structure of microbial prey and predators (71). Size-selective grazing by bacteriovorous nanoflagellates allows the smallest bacterial cells to largely avoid or altogether escape predation. Elongated and filamentous morphologies can also provide resistance against grazing, as cells are too large to be ingested by nanoflagellates (70). In fact, a number of field studies and experiments on the occurrence of elongated bacteria during periods of strong protozoan grazing pressure provide strong support for filamentous morphologies as an efficient bacterial defense strategy against predation (70, 83, 112, 118).
In a well-designed study by Pernthaler and coauthors (157), the authors noted a significant increase in numbers of filamentous bacterial cells during the late spring and early summer, which is generally a period of increased grazing by phagotrophic protists, known as the “clear-water” phase (32, 157). During this period, those authors observed a bloom of a phylogenetically narrow cluster (≥99% 16S rRNA gene identity) of members of the Bacteroidetes. These filamentous bacteria, identified as LD2 (tribe Aquir; Fig. 4B), made up more than 40% of the total bacterial biomass in the lake water, as measured by using FISH probes. A very interesting hypothesis is that filaments are formed by bacteria capable of a high level of morphological plasticity under high grazing pressure. Strong support for this hypothesis has been provided by independent experimental studies that observed an increase in filamentous morphotypes and a decrease in rod-shaped morphotypes belonging to Flectobacillus-like organisms when under strong grazing pressure (33, 34, 83). A phylogenetically broader group of the Bacteroidetes, which includes LD2 and has filamentous morphotypes, was described by Schauer and Hahn (177) and was named the SOL cluster (lineage bacIV; Fig. 4B). Although the genus Flectobacillus and SOL bacteria represent facultative and obligate filamentous bacteria, they possess highly contrasting growth characteristics, with the latter group having higher growth rates (193). In members of the class Bacteroidetes and the family Saprospiraceae, three filamentous cell groups within the SOL cluster have been identified: the HAL-A1 tribe (HAL), the HAL-A2 tribe (GKS2-217), and the Aquir tribe (LD2). A population study based on FISH revealed that 84 out of 115 lakes in Europe and Asia contained members of the SOL cluster (177). On average, this lineage made up only 1% of the cells in the epilimnion but contributed a much larger biomass, since the cells were filaments (178). The tribes within the SOL cluster seem to exhibit habitat preferences. The HAL-A2 tribe (GKS2-217) was found only in soft-water lakes (<50 μS cm−1) at a pH range of 6.5 to 7.3, the Aquir tribe (LD2) was found only in hard-water lakes at a pH range of 7.7 to 9.5, and the HAL-A1 tribe (HAL) was found in a variety of lakes with a pH range of 6.7 to 8.6 (177). A pure culture is available for the HAL-A1 tribe (215), and enrichments have been obtained recently for the Aquir and HAL-A2 tribes (86), revealing low growth rates under natural conditions, an obligate aerobic life-style, and often very large (up to 150 μm) filament lengths.
Bacteroidetes summary.
Unlike other common freshwater lake groups, the tribes of the Bacteroidetes have not exhibited any perceivable seasonal or lake-specific occurrence patterns (49). This finding might be related to their strong dependency on organic matter load or phytoplankton blooms, which are more likely to occur during sporadic and somewhat stochastic disturbance events rather than as part of a predictable seasonal trajectory. Also, the connection between Bacteroidetes taxa and grazing pressure is striking (178). Future studies targeting the phylogenetically more narrowly defined tribes could reveal intriguing and possibly diversified roles for members of this phylum in specialized organic matter degradation and in freshwater food webs.
Phylum Cyanobacteria
The conspicuous nature and long-recognized ecological importance of the Cyanobacteria have led to a much longer and more complex history of classification than that for the majority of the other bacterial phyla. Common freshwater lake genera include Microcystis, Anabaena, Aphanizomenon, Oscillatoria, Planktothrix, Synechococcus, and Cyanothece. Similar to eukaryotic phytoplankton, the freshwater Cyanobacteria perform oxygenic photosynthesis but rely only on chlorophyll a and an assortment of phycobilins for photosynthesis (132, 198). Some aquatic Cyanobacteria contain heterocysts, which are cells dedicated solely to nitrogen fixation (198). These Cyanobacteria along with many others that do not contain heterocysts, but fix nitrogen, play a key part in nutrient cycling in lakes. Cyanobacteria species are often considered nuisance organisms, as some species form large floating mats and may release toxins into lake waters (100). Numerous studies have been dedicated to an understanding of the environmental factors controlling cyanobacterial blooms and the induction of toxin production in freshwaters (reviewed in references 100, 144, and 146). A more detailed discussion of the large body of knowledge concerning these phototrophic freshwater organisms can be found elsewhere (25, 67, 100, 168, 198, 200).
Cyanobacteria 16S rRNA gene-based phylogeny.
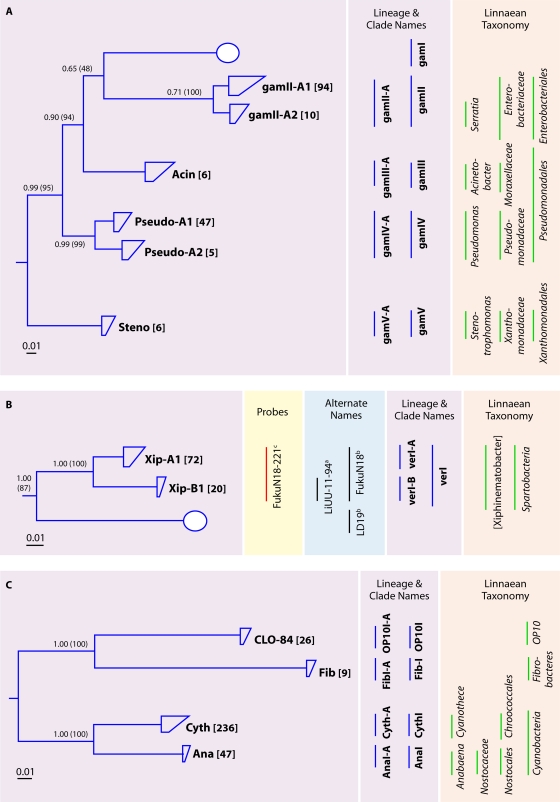
The lineages of the freshwater Cyanobacteria are much more thoroughly defined and include a vastly greater number of described species than for other freshwater lake bacterial phyla (182). For this reason, we did not wish to recreate the already defined phylogeny but instead point to the many excellent reviews on this subject (see references 182 and 203 and citations therein). More than 800 16S rRNA genes (the majority were <1,300 nt) were obtained from studies included in our data set (Fig. 2). Most of these sequences were closely related to the genera Cyanothece and Microcystis (data not shown). Two tribes, CyanI-A1 (Cyanothece related) and AnaI-A1 (Anabaena related) were identified in the data set (see Fig. 7C).
Phylum Proteobacteria
The phylum Proteobacteria is a group of Gram-negative bacteria encompassing the majority of recognized agriculturally, industrially, and medically relevant organisms and therefore is the most studied of the bacterial phyla. Six classes of Proteobacteria are currently recognized: the Alphaproteobacteria, Betaproteobacteria, Deltaproteobacteria, Epsilonproteobacteria, Gammaproteobacteria, and Zetaproteobacteria. The long history of study and wealth of knowledge pertaining to these organisms have led to the independent examination of the proteobacterial classes in most freshwater lake studies. We will follow this convention and discuss them separately below.
Class Alphaproteobacteria
The class Alphaproteobacteria has a special role in evolution and microbiology, as eukaryote mitochondria are believed to have originated from an alphaproteobacterial symbiont (8). The Alphaproteobacteria are found in all imaginable habitats and display enormous plasticity in their genomes and in their life-styles. The large variation in genome organization and size among members of this class (1 to >9 Mbp) is likely related to their common roles as endosymbionts and intracellular parasites (13). The Alphaproteobacteria are also at the hub of the global nitrogen cycle, because symbiotic members of this phylum (e.g., the Rhizobiales) often facilitate atmospheric nitrogen fixation by plants. The Alphaproteobacteria are numerically dominant in many marine ecosystems (137). In freshwater lakes, the Alphaproteobacteria are also ubiquitous although less numerous.
Alphaproteobacteria 16S rRNA gene-based phylogeny.
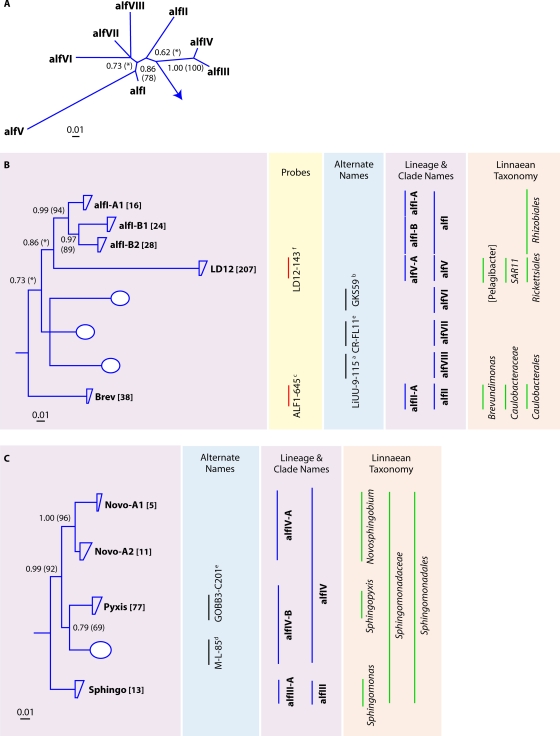
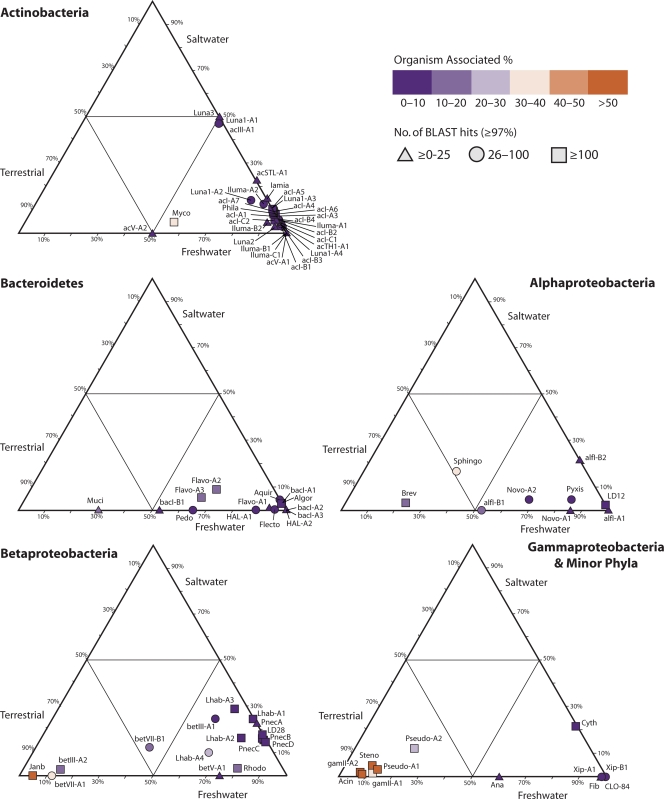
In an early study by Glöckner and colleagues, the phylogeny of the freshwater Alphaproteobacteria was broken down into six clusters (alphaI to alphaVI) (63). Since then, eight additional clusters have been described. Our phylogenetic analyses yielded nine tribes from five lineages (Fig. 5B and C). The most widely distributed freshwater lake tribe of the Alphaproteobacteria is LD12 (alfV lineage), the freshwater sister group to marine SAR11 (128), and this tribe also contained the most Alphaproteobacteria sequences in our database (Fig. 5B). The alfIII lineage, overlapping with the genus Sphingomonas sensu stricto (206), contained few sequences, whereas the alfIV lineage was more abundant in our combined freshwater lake data set. This lineage features tribes Novo-A1 and -A2, corresponding largely to the genus Novosphingobium, and tribe Pyxis, which overlaps with the genus Sphingopyxis (206). The alfI (related to the Rhizobiales) and alfII (related to Caulobacter and Brevundimonas) lineages were also fairly common in freshwater lake clone libraries.
FIG. 5.
(A) Radial consensus phylogram of the freshwater lake lineages of the Alphaproteobacteria. (B) Consensus phylogram of the tribes of lineages alfI, alfII, and alfV. (C) Consensus phylogram of the tribes of lineages alfIII and alfIV. All phylogenic inference calculations were conducted with nearly full-length sequences (>1,300 nt) representing the lineages/clades/tribes. Frequency base filters were created to mask out highly variable positions, and representative sequences from members of the Archaea were used as the outgroup for all tree reconstructions. Bayesian clade credibility values and maximum likelihood bootstrap values (in parentheses) of >0.5 (50) are listed at each node. Tribe names are listed at the end of tree tips (trapezoids), with the number of sequences associated with each tribe listed in square brackets. Tree tips ending in ovals represent phylogenetic clusters that were named previously but did not meet the criteria to be called a tribe (≥2 sequences with ≥97% identity of ≥1,300 nt isolated from >1 lake). Current lineage and clade names are listed to the right of the phylograms and are highlighted in purple. The Linnaean taxonomy column is based on the RDP classification (222), where all sequences within the classified group must have ≥75% assigned confidence to the taxonomic group. Square brackets around a genus name indicate a candidate genus, and an asterisk indicates that the genus is described but not yet covered in the RDP classification. Alternate names from previous studies also covering the defined lineages/clades/tribes are listed. Probe coverage is based upon a perfect match to ≥75% of the sequences in a defined lineage/clade/tribe. References to past clades and probes are as follows: a, reference 48; b, reference 155; c, reference 156; d, reference 233; e, reference 238; f, reference 242.
Ecology of the freshwater lake Alphaproteobacteria.
Quantitative assays based on FISH have consistently shown that the Alphaproteobacteria, at least at the class level, are resistant to predation: their relative abundance increases in response to enhanced microeukaryote grazing (32, 110, 119, 175, 191). The increase in abundance has often been accompanied by a tendency of members of the Alphaproteobacteria to form filaments, aggregates, or Caulobacter-like stalked cells that sometimes can make up a majority of the Alphaproteobacteria population (83, 175, 191). It is currently not known whether these changes are due to the phenotypic plasticity of persistent populations or shifts in the Alphaproteobacteria community composition. Even if top-down control via predation favors the Alphaproteobacteria, members of this class are generally not very abundant, suggesting that unknown mechanisms are keeping the abundance of the Alphaproteobacteria low in freshwater lakes. One potential mechanism may relate to the competitive ability of the Alphaproteobacteria for organic and inorganic substrates. Indeed, some experiments with lake bacterioplankton suggest that the low availability of organic nutrients, comparable to the nutrient availability characteristic of oligotrophic oceans, favors certain members of the Alphaproteobacteria (51, 160).
A second strategy for substrate acquisition among members of the Alphaproteobacteria may involve the capacity to degrade recalcitrant organic compounds such as humic substances. This strategy has been suggested for freshwater Novosphingobium (clade alfIV-A) and Sphingopyxis (clade alfIV-B) based on the isolation of bacteria in phenol-enriched cultures from humic lakes (104). The physical interaction with primary producers or other organisms that release organic and inorganic nutrients is another strategy used by some bacteria to satisfy their carbon and nutrient demands. This has not been investigated in any detail for the freshwater lake Alphaproteobacteria but may be expected in light of the frequent parasitic and symbiotic life-styles within the class in general (13). Accordingly, some members of the Alphaproteobacteria have been identified in freshwater clone libraries from cyanobacterial phycospheres (52). Furthermore, freshwater Alphaproteobacteria isolates affiliated with Brevundimonas (clade alfII) have been shown to either promote or inhibit the growth of the coexisting blooming Cyanobacteria, implying strong functional interactions (15).
Little is known about the ecology and functional role of LD12 (clade alfV-A), which is the freshwater sister group to SAR11 (11). This tribe was first discovered in Toolik Lake (11) but often appears as the most abundant member of the Alphaproteobacteria in freshwater lake 16S rRNA libraries (238). It is widely distributed in lakes all over the world (38, 48, 123, 230, 242), and a recent phylogenetic study revealed surprisingly low global diversity within the tribe, indicating either slow diversification or global dispersal between freshwater ecosystems (129). This study also points to an ancient diversification of this tribe from their marine sister group and few historical freshwater-marine water transitions.
Alphaproteobacteria summary.
In general, the freshwater Alphaproteobacteria are poorly studied, but the available data suggest that their dominant freshwater lake members are (i) resistant to grazing, (ii) competitive under conditions of low nutrient/substrate availability but also capable of degrading complex organic compounds, and (iii) widely distributed in lakes all over the globe.
Class Betaproteobacteria
The class Betaproteobacteria, like other members of the phylum, is broadly recognized for its morphological and physiological diversity. In contrast to the related Alphaproteobacteria, the Betaproteobacteria are often the numerically dominant group in freshwater lakes (assessed with FISH [23, 63, 95, 243]) but are in relatively low abundance in the ocean (173). The high abundance and amenability to culturing have contributed to the Betaproteobacteria being the best-studied group in freshwater lakes.
Betaproteobacteria 16S rRNA gene-based phylogeny.
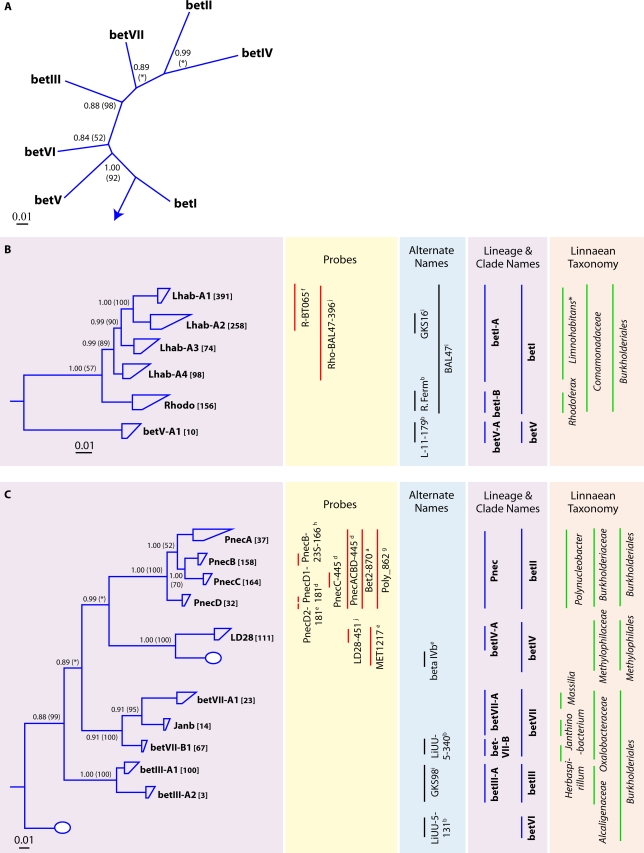
A number of freshwater lake Betaproteobacteria-associated monophyletic clusters have been identified, and many of these clusters are widely distributed among lakes. Following previous efforts to organize the freshwater lake Betaproteobacteria into coherent clusters (63, 242), we have divided the subclass into seven lineages, betI, betII, betIII, betIV, betV, betVI, and betVII, six of which are comprised of monophyletic clades containing a total of 16 tribes (Fig. 6B and C). The betVI lineage was reported previously (48); thus, it was included in the tree inference process but did not contain any tribes, so we excluded it from further analyses.
FIG. 6.
(A) Radial consensus phylogram of the freshwater lake lineages of the Betaproteobacteria. (B) Consensus phylogram of the tribes of lineages betI and betV. (C) Consensus phylogram of the tribes of lineages betII, betIII, betIV, and betVII. All phylogenic inference calculations were conducted with nearly full-length sequences (>1,300 nt) representing lineages/clades/tribes. Frequency base filters were created to mask out highly variable positions, and representative sequences from members of the Archaea were used as the outgroup for all tree reconstructions. Bayesian clade credibility values and maximum likelihood bootstrap values (in parentheses) of >0.5 (50) are listed at each node. Tribe names are listed at the end of tree tips (trapezoids), with the number of sequences associated with each tribe listed in square brackets. Tree tips ending in ovals represent phylogenetic clusters that were named previously but did not meet the criteria to be called a tribe (≥2 sequences with ≥97% identity of ≥1,300 nt isolated from >1 lake). Current lineage and clade names are listed to the right of the phylograms and are highlighted in purple. The Linnaean taxonomy column is based on the RDP classification (222), where all sequences within the classified group must have ≥75% assigned confidence to the taxonomic group. Square brackets around a genus name indicate a candidate genus, and an asterisk indicates that the genus is described but not yet covered in the RDP classification. Alternate names from previous studies also covering the defined lineages/clades/tribes are listed. Probe coverage is based upon a perfect match to ≥75% of the sequences in a defined lineage/clade/tribe. References to past clades and probes are as follows: a, reference 24; b, reference 48; c, reference 80; d, reference 85; e, reference 176; f, reference 192; g, reference 216; h, reference 229; i, reference 238; j, reference 242.
The betI and betII lineages contained the most sequences (Fig. 6B and C). The betI lineage, which is sometimes known as the BAL47 cluster for its affiliation with an initial isolate from the Baltic Sea (Rhodoferax sp. strain BAL47), encompasses two clades (clades betI-A and betI-B), of which clade betI-A has been well studied and further partitioned into tribes (Fig. 6B). Tribe Lhab-A2 is known as the GKS16 cluster based on an original clone retrieved from Lake Gossenkollesee in Austria (63). Both Lhab-A1 and Lhab-A2 are circumscribed by the popular FISH probe R-BT065 (192). Representatives of the betI lineage were recently obtained in pure cultures and systematically described and are now known as members of the genus Limnohabitans within the family Comamonadaceae (76, 77). The first described species, Limnohabitans curvus strain MWH-C5T, shares 96.6% and 95.7% 16S rRNA gene sequence identities with the previously described species Curvibacter delicates and Rhodoferax fermentans, respectively (76), and is a member of the Lhab-A3 tribe. Interestingly, its 16S rRNA gene sequence does not match with the popular R-BT065 FISH probe. Limnohabitans australis strain MWH-BRAZ-DAM2DT was described as a very similar species with different substrate utilization characteristics (77). Limnohabitans planktonicus strain II-D5T and Limnohabitans parvus strain II-B4T, both of tribe Lhab-A1, are the first cultured and described members of the well-studied R-BT065 cluster and are targeted by the R-BT065 FISH probe (113). They share only 97.0 to 97.2% 16S rRNA gene sequence identity with L. curvus.
The betII lineage includes perhaps the best-studied freshwater lake clade within the Betaproteobacteria, Pnec. The name originates from its association with the genus Polynucleobacter, which was originally proposed for an obligate symbiont of ciliates (Polynucleobacter necessarius) (91). The genus was recently emended to also encompass four strains of a free-living subspecies (P. necessarius subsp. asymbioticus) (80). The Pnec clade has been subdivided into several phylogenetically refined tribes (tribes PnecA to PnecD). The PnecC tribe is arguably the best studied and includes P. necessarius (80). Several clade members were recently obtained in pure cultures from both temperate and subtropical freshwater habitats and systematically identified (80). The first described species of the PnecA tribe, Polynucleobacter rarus MT-CBb6A5T, shares 96.0%, 95.6%, and 96.0% 16S rRNA gene sequence identities with P. necessarius subsp. asymbioticus, a sequence representing the endosymbiotic P. necessarius subsp. necessarius E24, and Polynucleobacter cosmopolitanus, respectively (81). This species appears to be rare compared to other members of the genus. The first cultured isolate of the PnecB tribe, Polynucleobacter acidiphobus MWH-PoolGreenA3T, shares 97.8%, 97.5%, 97.3%, and 96.7% 16S rRNA gene sequence identities with P. necessarius subsp. asymbioticus, P. necessarius subsp. necessarius E24, the type strain of P. cosmopolitanus, and P. rarus MT-CBb6A5T, respectively (79).
P. cosmopolitanus is a member of PnecD and shares many phenotypic, chemotaxonomic, and ecological traits with P. necessarius (78). Representatives have been cultured from freshwater habitats in Austria, France, Uganda, China, and New Zealand. The only available genome sequence from common freshwater lake bacteria described in this data set (other than Cyanobacteria) is derived from a PnecC member (M. W. Hahn, unpublished data).
The betIII lineage encompasses one clade subdivided into two tribes and was originally named the GKS98 cluster based on sequences retrieved from Lake Gossenkollesee (63). Isolates have been obtained from lineage betIII (225, 230). The betIV and betV lineages are each defined by only one clade and tribe. Lineage betIV contains the LD28 tribe, based on a clone recovered from Lake Loosdrecht in the Netherlands (238). No cultured representatives of the tribes in lineages betIV or betV have been reported to date. In contrast, each of the three tribes in lineage betVII are represented by isolates: the betVII-A1 tribe is represented by the genus Massilia, Janb is represented by the genus Janthinobacterium, and betVII-B1 is represented by the genus Herbaspirillum.
Ecology of the freshwater lake Betaproteobacteria.
The Betaproteobacteria are abundant in many different freshwater lake habitats, sometimes constituting up to 60 to 70% of the total number of 4′,6-diamidino-2-phenylindole (DAPI)-stained cells, as measured by using FISH probes (63, 176, 187). The relative abundances of various lineages and clades within the Betaproteobacteria vary among lakes, within lakes (with depth and horizontally), and with time. Members of the Betaproteobacteria are often cocultured with algae such as Cryptomonas sp. (156), are associated with the Cyanobacteria (52), and are often particle associated (120, 191, 227).
We know the most about the ecology of the betI-A clade and the Pnec clade, based on their distribution patterns in time and space, interaction with other members of the aquatic food web, mesocosm experiments, and available pure cultures. Both clades are considered to be cosmopolitan freshwater lake clades because they are found in nearly all lakes that have been studied. The betI-A clade is most frequently studied by using a 16S rRNA-targeted FISH probe that specifically hybridizes to the Lhab-A1 and Lhab-A2 tribes (together also known as R-BT065; Fig. 6B). This probe-defined cluster was found to be present in 98 of 102 diverse freshwater habitats in Europe that were surveyed for their presence, in abundances of up to 29% of the total number of DAPI-stained bacteria (190). This clade has also been detected by its 16S rRNA gene sequences in nearly every lake studied by using this technique, including Lake Gossenkollesee in Austria (63), Lake Grosse Fuchskuhle in Germany (63), Lake Baikal in Russia (63), deep Patagonian lakes (35), Tibetan lakes (127), eutrophic Swedish lakes (48), shallow eutrophic lakes in Belgium (214), shallow eutrophic Lake Taihu in China (233), oligotrophic Crater Lake in Oregon (212), oligotrophic Toolik Lake in Alaska (38), and a variety of lakes in Wisconsin (this study). The cluster seems to be more abundant under circumneutral to alkaline pH conditions than in acidic or humic habitats and most abundant in small shallow ponds (190). The R-BT065 cluster has also been found distributed throughout stratified water columns (23, 176) but is most abundant at shallower depths (176). A few studies have focused on the substrate assimilation preferences of R-BT065 bacteria using microautoradiography-FISH. In the experimentally partitioned Lake Grosse Fuchskuhle, that group assimilated glucose and leucine but very little acetate (23). R-BT065 also readily used leucine in a reservoir under both P-limiting and P-replete conditions (96). Substantial evidence for both top-down and bottom-up controls on their abundances is available based on both field observations and experiments. The R-BT065 bacteria are known for their relatively short population turnover times and ability to respond rapidly to changing environmental conditions (6, 187, 188, 192). They grow quickly when exposed to nutrient pulses in mesocosms (187, 188), and their abundance was positively associated with watercolor metrics that are proxies for low-molecular-weight alga-derived substrates (190). The latter suggests that they are particularly competitive in photosynthetically active planktonic systems regardless of trophic status (189). Members of the cluster were also highly susceptible to grazing (97, 187, 192) and had higher growth rates in the presence of flagellates (226). Some researchers have proposed that these bacteria are particularly competitive in freshwaters because of their opportunistic life-style, in which they balance their vulnerability to grazing with fast growth rates (96, 176). We note that very little is known about ecophysiological differences between tribes Lhab-A1 and Lhab-A2 because most studies of the betI-A clade have been conducted by using the R-BT065 FISH probe, which targets both tribes.
The four described Limnohabitans species within the betI lineage can be discriminated based on their substrate utilization characteristics as well as their 16S rRNA gene sequences and extent of genomic DNA-DNA hybridization (76, 77, 113). The first described species, L. curvus, is a chemoorganotrophic, aerobic, and facultative anaerobe capable of assimilating glucose and a variety of small organic acids such as acetate, pyruvate, and fumarate but not amino acids (76). Notably, L. parvus cannot assimilate acetate, while the other three species are capable of doing so (113).
The Pnec clade has also been the subject of a large number of studies directed toward an understanding of their ecophysiology, evolutionary ecology, and role in aquatic food webs. The four tribes, named PnecA to PnecD (P. rarus, P. acidiphobus, P. necessarius, and P. cosmopolitanus, respectively) are well delineated phylogenetically (229) and seem to be similarly distinct in their distributions within and among lakes and over time. In two large lakes, Lake Taihu in China and Lake Mondsee in Austria, the FISH-identified PnecB cluster was the most abundant of the four tribes, although the tribe exhibited peak abundances in opposite seasons in the two lakes (232). PnecB exhibited remarkably consistent seasonal dynamics, which mirrored the change in water temperature over 3 years (232). A study of the effect of depth on PnecB abundance showed that its abundance consistently decreased as the depth below the surface increased (232). PnecB and PnecC seem to ecologically partition themselves along a pH gradient, with PnecB being most successful under neutral to alkaline conditions and PnecC being more abundant in more acidic habitats. PnecB was identified with FISH in 51 of 65 lakes that were circumneutral or alkaline but in 0 of 16 lakes that were acidic (230). Likewise, the abundance of PnecB increased with increasing pH, as did the prevalence of PnecA (123). On the other hand, the abundance of PnecC decreased with increasing pH (230). Although PnecC exhibits a preference for low-pH lakes, it is ubiquitous in lake systems (80, 107). A large survey of 137 freshwater lakes and ponds over a 2,000-km2 area in the Czech Republic, Austria, and Sweden showed that PnecC could be recovered from lakes with pHs as low as 3.8 and as high as 8 (107).
There is strong evidence that PnecC is particularly competitive in environments rich in humic acids, since they are frequently the dominant betaproteobacterial tribe in dystrophic lakes (24, 69, 85, 107, 141, 205) and are at times known to comprise >50% of the DAPI-stained cells in these lakes (85, 107). PnecC is readily enriched upon exposure to elevated levels of allochthonous DOC (103, 104, 107, 225) and can assimilate small organic acids such as acetate, which are expected to be the breakdown products of humic acids (23). Their substrate incorporation patterns did vary over time and across basins of the partitioned Lake Grosse Fuchskuhle (23). PnecB and PnecC habitats also seem to partition with depth in stratified lakes, with both being present in the epilimnion but only PnecC existing in the hypolimnion of the prealpine oligomesotrophic Piburger See in Austria (176).
Hahn and colleagues have isolated and characterized a large number of isolates associated with the Pnec clade (genus Polynucleobacter), and this has provided valuable insight into substrate utilization preferences, temperature and pH ranges, and the ability to grow in the absence of oxygen (78, 80). The isolation of Polynucleobacter often requires a careful filtration-acclimatization method (87) or a dilution-acclimatization method (74), and these cells generally grow poorly on a single carbon source (80). However, a recent study indicated that PnecC could be isolated by using agar plates (225). It remains to be seen if this method is capable of retrieving the diversity of the Pnec clade or if it selects for certain members. Pnec isolates are described as being chemoorganotrophic and aerobic, with some strains being capable of facultative anaerobic growth. The grazing of flagellate bacteriovores upon the Pnec tribes appears to be governed by the size of the Pnec cell (18, 209), as low grazing rates were observed for ultramicrobacterial (cell volume, <0.1 μm3) PnecD organisms, but they increased with increasing cell size (18, 209). Isolates have been described for all Pnec tribes: P. rarus for PnecA (81), P. acidiphobus for PnecB (79), P. necessarius for PnecC (74, 80), and P. cosmopolitans for PnecD (78).
Less is known about the ecology of the betIII, betIV, betV, betVI, and betVII lineages. The betIII lineage (also known previously as the GKS98 cluster) and the LD28 tribe (lineage betIV) were each detected in 12 of 15 lakes in Sweden using reverse line blot hybridization (123). In another survey, the LD28 tribe was found in 59 of 81 lakes across Europe (242) and was present in high mountain Tibetan lakes (230), while the GKS98 cluster was found in both freshwater and hypersaline lakes (230). The betIV lineage in the Piburger See, Austria, was studied by using 16S rRNA gene sequencing and targeted FISH probes (176). This lineage was significantly more abundant in the anoxic hypolimnion than were other members of the Betaproteobacteria (i.e., betI and betII).
Betaproteobacteria summary.
The Betaproteobacteria are by far the most studied and often the most abundant bacteria inhabiting the upper waters of lakes. Two lineages, betI and betII, which are both abundant in lakes and exhibit a cosmopolitan distribution, have received the bulk of the research attention for the freshwater Betaproteobacteria. Depth, pH, carbon substrate preferences, and seasonal factors are all known to differentiate closely related organisms within these lineages. Generally speaking, the freshwater lake Betaproteobacteria are fast growing and nutrient loving and have a size-dependent vulnerability to grazing pressure. The recent isolation of numerous members of the genera Limnohabitans and Polynucleobacter has paved the way for a greater understanding of the roles that freshwater bacteria play in these ecosystems.
Class Gammaproteobacteria
The class Gammaproteobacteria contains the most studied of all bacterial organisms, the Enterobacteriales (commonly known as the enterics). Many enteric organisms, such as Escherichia coli, can be found in freshwater lakes; however, they are widely considered transient members dispensed from anthropogenic or zoonotic sources and are generally not seen in 16S rRNA gene libraries made from these environments (238). The Gammaproteobacteria are generally more abundant in saltwater environments such as the ocean (17, 173) or saline lakes (230) than in freshwater.
Gammaproteobacteria 16S rRNA gene-based phylogeny.
The majority of freshwater lake sequence-based studies do not contain data for members of the Gammaproteobacteria. Since relatively few of such gene sequences have been recovered from freshwater lakes, only two named clusters had been identified, the gamma I (Methylobacter psychrophilus) cluster (238) and the LiUU-3-334 cluster (48) (Fig. 7A). In our phylogenetic analyses we identified six tribes (Fig. 7A). Unlike the majority of freshwater lake clusters in our data set, many of these Gammaproteobacteria tribes fit into the described taxonomic framework for this group. For example, the majority of sequences fit into the families Enterobacteriaceae and Xanthomonadaceae or the orders Pseudomonadales and Legionellales (Fig. 7A). Many of the bacteria in these Gammaproteobacteria groupings were closely related to bacteria commonly cultured in the laboratory or those found associated with soil or animals.
FIG. 7.
(A) Consensus phylogram of the freshwater lake lineages of the Gammaproteobacteria. (B) Consensus phylogram of the freshwater lake lineages of the Verrucomicrobia. (C) Consensus phylogram of the freshwater lake lineages of OP10, the Fibrobacteres, and the Cyanobacteria. All phylogenic inference calculations were conducted with nearly full-length sequences (>1,300 nt) representing the lineages/clades/tribes. Frequency base filters were created to mask out highly variable positions, and representative sequences from members of the Archaea were used as the outgroup for all tree reconstructions. Bayesian clade credibility values and maximum likelihood bootstrap values (in parentheses) of >0.5 (50) are listed at each node. Tribe names are listed at the end of tree tips (trapezoids), with the number of sequences associated with each tribe listed in square brackets. Tree tips ending in ovals represent phylogenetic clusters that were named previously but did not meet the criteria to be called a tribe (≥2 sequences with ≥97% identity of ≥1,300 nt isolated from >1 lake). Current lineage and clade names are listed to the right of the phylograms and are highlighted in purple. The Linnaean taxonomy column is based on the RDP classification (222), where all sequences within the classified group must have ≥75% assigned confidence to the taxonomic group. Square brackets around a genus name indicate a candidate genus, and an asterisk indicates that the genus is described but not yet covered in the RDP classification. Alternate names from previous studies also covering the defined lineages/clades/tribes are listed. Probe coverage is based upon a perfect match to ≥75% of the sequences in a defined lineage/clade/tribe. References to past clades and probes are as follows: a, reference 48; b, reference 238; c, reference 242.
Ecology of the freshwater lake Gammaproteobacteria.
The Gammaproteobacteria, like the Alphaproteobacteria, are abundant in the ocean (17) but are not particularly abundant in freshwater lakes (238). Although the total numbers of sequences recovered in our data set are relatively equal for the Alphaproteobacteria and Gammaproteobacteria, the diversity among the Gammaproteobacteria sequences (i.e., the number of sequences that are not closely [>97%] related to another sequence) is greater (data not shown). This patchy recovery of a wide range of members of the Gammaproteobacteria suggests that they may be transient members of lake communities brought in from the surrounding environment (e.g., enterics) (124), or they could be common lake members existing at low abundances, making regular retrieval via clone libraries unlikely. Many Gammaproteobacteria strains are regularly used as microbial genetic tools in the laboratories of those researchers carrying out the isolation of DNA from freshwaters, implying that the recovery of their 16S rRNA gene sequences in clone libraries derived from lakes could be a product of methodological contamination.
Zavarzin et al. suggested that the Gammaproteobacteria are copiotrophs, which could explain why they are disproportionately isolated in culture-based studies (236). Upholding that hypothesis, members of the Gammaproteobacteria grew significantly faster than the average lake bacterioplankton and exhibited even faster growth rates when nitrogen and phosphorus were added to enclosures (59, 188). To our knowledge, few studies have further examined the freshwater lake Gammaproteobacteria tribes described here or other members of the Gammaproteobacteria as resident members of the upper waters in lakes (for notable exceptions, see references 164 and 221). There is, however, vast literature on the use of members of the Gammaproteobacteria, specifically members of the Enterobacteriaceae, in the source tracking of pollutants in surface waters (reviewed in reference 201).
Phylum Verrucomicrobia
Members of the Verrucomicrobia were identified and cultured as early as the 1970s (genus Prosthecobacter) (196), but the phylum, which took its name from its first aerobic freshwater isolate (Verrucomicrobia spinosum), was not created until 1997 (92). Verrucomicrobia species have been identified in lakes, soil, oceans, and human feces and even as ecto- and endosymbionts of eukaryotes (220). Recently, methane-oxidizing representatives of the Verrucomicrobia have been isolated from extremely low-pH, thermophilic environments (181). The Verrucomicrobia have an elevated degree of phylogenetic relationship to members of the Planctomycetes and Chlamydiae. Some representatives of these three groups also have compartmentalized intracellular structures (220).
Verrucomicrobia 16S rRNA gene-based phylogeny.
In our phylogenetic reconstructions, one lineage, two clades, and two tribes were defined (Fig. 7B). Despite being ubiquitous, little is known about freshwater lake Verrucomicrobia taxa. The three previously described clusters of the freshwater Verrucomicrobia are FukuN18, subdivision 3, and CL120-10 (Opitutaceae). These three clusters each harbor many sequences, suggesting that a finer resolution of clusters could reveal more phylogenetic structures within each lineage. In our database, only lineage verI (FukuN18) could be further divided (Fig. 7B) into clades (clades verI-A and verI-B) and tribes (tribes Xip-A1 and Xip-B1).
Ecology of the freshwater lake Verrucomicrobia.
Members of the Verrucomicrobia do not seem particularly abundant in lakes, ranging between <1% and 6% in abundance based on clone recovery. However, this may reflect poor primer coverage or PCR bias, since a recent study based on newly developed FISH probes revealed abundances of up to 20% of the total bacterial community in a humic lake (9). Still, this phylum does have a cosmopolitan distribution in lakes. In a study by Zwart and colleagues (242), the freshwater cluster CL0-14 was found in 73 of 81 studied lakes in Europe. The prevalence of this cluster was negatively correlated with pH and positively correlated with hydraulic retention time and temperature (123). Members of the Verrucomicrobia have also been observed in both surface and hypolimnetic waters, suggesting a variety of metabolic strategies within the group (45, 125). Some members of the phylum seem to be associated with high-nutrient environments or algal blooms (48, 90, 116).
Minor Freshwater Lake Phyla
Sixteen phyla, the Acidobacteria, BRC1, Chlorobi, Chloroflexi, Fibrobacteres, Firmicutes, Fusobacteria, Gemmatimonadetes, Lentisphaerae, Nitrospira, OD1, OP10, Planctomycetes, Spirochaetes, SR1, and TM7, were recovered infrequently by the studies included in our database (Fig. 2). Of these phyla, members of the Planctomycetes and the Firmicutes were recovered most often. Few studies examining the presence, distribution, or activity of any of the 16 minor phyla have been conducted for the epilimnion of freshwater lakes, although a few studies have identified putative monophyletic freshwater clusters (48, 212, 233). Notably, members of the Acidobacteria and Firmicutes are commonly found in freshwater sediments, and members of the Chloroflexi (green nonsulfur bacteria) and Chlorobi (green sulfur bacteria) are commonly present in the metalimnia or hypolimnia of deeper lakes.
16S rRNA gene-based phylogenies of minor phyla.
Two of the minor phyla in our data set, OP10 and the Fibrobacteres, were represented by tribes (Fig. 7C). The OP10 tribe CL0-84 was described previously (238), while the Fibrobacteres tribe was not. Certainly, all 16 minor phyla would show greater phylogenetic structures with further sequencing efforts.
Ecology of minor freshwater lake phyla.
In this section we attempt to summarize the little that is known about the more prevalent and/or more studied freshwater lake representatives from the minor phyla listed above. In particular, we will focus on the Fibrobacteres, Chloroflexi, and Planctomycetes and the candidate division OP10. Studies of bacterial 16S rRNA gene clone libraries generated from geographically and geochemically distinct freshwater lake samples have reported sequences related to each of these phyla (48). Even though isolates have been obtained for three of these phyla and several are known to exhibit unusual functional traits and/or suspicious intracellular structures, they have not received much attention from freshwater studies.
Using a Fibrobacteres-specific quantitative PCR assay, McDonald and colleagues (135) showed a depth-dependent distribution of the freshwater Fibrobacteres never exceeding 1% of the total bacterioplankton. Interestingly, those authors were able to enrich members of the Fibrobacteres in a biofilm that developed on cotton strings incubated in a eutrophic lake. Since all cultured strains assigned to the Fibrobacteres are capable of cellulose hydrolysis, those authors suggested that they could play a role in cellulose hydrolysis in freshwater environments. However, the freshwater lake Fibrobacteres sequences fall into the narrow clade FibI-A, which is not closely related to any of the described isolates (data not shown). Members of the Fibrobacteres are more common in sediments, which is consistent with the obligate anaerobic physiology of all known cultured members of the phylum.
The majority of Chloroflexi 16S rRNA gene sequences belong to the cluster called SAR202, which are commonly recovered from soils, sediments, and deep-subsurface terrestrial environments. Chloroflexi SAR202-related bacterioplankton also seems to be common in the meso- and bathypelagic areas of the open ocean (138) and has been identified in the hypolimnia and epilimnia of freshwater systems (212). However, of the 39 Chloroflexi sequences recovered in our database, only the two sequences recovered from Crater Lake by Urbach and colleagues (212) fell into the SAR202 clade. Since there are currently no cultured representatives of the Chloroflexi that are closely related to the obtained environmental sequences and since very little is known about their distribution in aquatic environments, our understanding of their ecology and physiology is purely speculative.
The Planctomycetes are understudied despite the fact that they are common in freshwater lake environments and are able to provide important ecosystem functions. Part of the reason for the lack of studies examining this phylum stems from the fact that the Planctomycetes are underrepresented in 16S rRNA gene clone libraries, as many members have mismatches to commonly used universal primers (219). In fact, members of the Planctomycetes may be very abundant in freshwater lakes. In one study of an acidic bog lake using FISH probes to quantify the major phyla, the Planctomycetes were the most abundant phylum (40). In the field of biotechnology, research on the Planctomycetes has recently gained increased notoriety because of the group's general ability to carry out the anaerobic ammonium oxidation (anammox) reaction (202). Furthermore, genomic and proteomic analyses of Planctomycetes isolates suggest that they can degrade phytoplankton-derived carbohydrates (165). For more details on the ecology of members of the Planctomycetes, we refer the readers to two excellent reviews (58, 220).
“Candidatus” phylum OP10 lacks cultured representatives, and its existence is generally verified only by 16S rRNA gene sequences. Sequences representing the OP10 phylum have been reported for a wide diversity of habitats (162) but very rarely make up ≥5% of the sequences from 16S rRNA gene libraries. Since representative isolates are lacking and their dynamics have not been a focus of any freshwater lake studies, the abundance and role of OP10 in freshwater lakes are uncertain. A first hint at their ecological role was provided by observations that some taxa follow or are associated with phytoplankton blooms (58). OP10 members have also been implicated in metal cycling, as a narrow but abundant phylogenetic cluster was retrieved from iron- and manganese-rich particles in the hypolimnion of a reservoir during late-summer stratification (199).
Summary of minor phyla.
Studies involving the less abundant phyla of freshwater lakes represent a particular avenue of research that is understudied and should be explored. Very little is known about the diversity of these phyla or the roles that members of these phyla play in lakes. Some phyla, such as the Planctomycetes, may even be very abundant in particular freshwater lake systems (40). Members of this so-called “rare biosphere” could prove to be important contributors to crucial biochemical reactions and ecosystem functions or may harbor unique properties amenable to biotechnology applications (75). The advent of cheaper sequencing technologies may for the first time provide a way to access the potentially incredible bacterial diversity sustained by freshwater lakes.
META-ANALYSIS: LIFE HISTORY PATTERNS OF FRESHWATER LAKE BACTERIA
Natives, Vagabonds, or Tourists?
Freshwater lakes are intimately linked to the terrestrial and marine biomes via the hydrological cycle and other modes of dispersal. Hence, it is relevant to address the question of whether the putative freshwater lake tribes that we have defined with phylogenetic tools represent true indigenous populations characteristic of the freshwater biome (natives), if they have a wider distribution and/or niche in the biosphere (vagabonds), or if they are just there by chance (tourists). To do this, we used blastn to map the distribution of representative sequences from each tribe to GenBank sequences from terrestrial, freshwater, and marine habitats (Fig. 8). This synoptic analysis clearly shows that the majority of putative freshwater lake tribes identified within the Actinobacteria, Bacteroidetes, Alphaproteobacteria, and Betaproteobacteria appear to be restricted (natives) to lake epilimnia. However, there are some notable exceptions within these phyla and classes that will be discussed below. In particular, the Gammaproteobacteria tribes found in freshwater lake ecosystems appear to be abundant in terrestrial ecosystems with an emphasis on organism-associated habitats (Fig. 8), pointing to their potential role as “tourists” in freshwater lakes.
FIG. 8.
Habitat specificity of putative freshwater lake bacterial tribes. Two nearly full-length (>1,300 nt) 16S rRNA gene sequences from each tribe were chosen as representatives for a subsequent blastn analysis against the NCBI nonredundant (nr) database. Each tribe's sequence representatives were chosen based on obtaining a maximum or near-maximum within-tribe phylogenetic distance between the two representatives. The top 5,000 blastn hits were returned for each sequence representative, and the database sequence records with ≥97% identity were retained. The “isolation source” field was parsed from each sequence record, allowing categorization by the biome from which the sequence was obtained (freshwater, saltwater, or terrestrial) and by organism association (whether or not the sequence was isolated from a bacterium living on or within another organism). Two environments required special biome characterization: a brackish isolation source was considered half freshwater and half saltwater, and a sediment isolation source was considered half terrestrial and half aquatic (either freshwater or saltwater). Following categorization, the means of the biome and organism association results were calculated for each tribe. The percentage of sequences associated with each biome category for each tribe is visualized in ternary plots. Triangle vertices indicate a 100% relative abundance of the indicated biome for the plotted tribe. The symbol color for each tribe indicates the percentage of GenBank hits for the tribe that were noted as having been associated with another organism when the sequences were obtained (e.g., human skin or algal phycosphere). The symbol shape for each tribe reflects the mean number of GenBank hits with ≥97% sequence identity to the tribe reference sequences.
Although the identified Actinobacteria tribes are found almost exclusively in freshwaters, two tribes feature a partly terrestrial signal, the Myco (Mycobacterium) and acV-A2 tribes. The former group contains many parasitic and pathogenic representatives, which is possibly reflected in the higher incidence of organism-associated sources for this tribe. Tribe acV-A2 is also a member of the “soil II+III cluster,” and hence, it is not surprising that it is well represented in terrestrial habitats. It was surprising, however, that tribe acV-A1, the sister group to tribe acV-A2, which also groups with the “soil II+III cluster,” has not been found in terrestrial habitats. Three actinobacterial tribes (tribes Luna3, Luna1-A1, and acIII-A1) have a strong saltwater/marine signal (approximately 50% of BLAST hits). Among these, Luna1-A1 was postulated to be an indigenous freshwater lake bacterial group with a planktonic life-style (73), but our analysis suggests that members of this group are found in estuaries, which may suggest that this tribe can tolerate more saline environments (Fig. 8). Tribes Luna-A1 and acIII-A1 have similar freshwater-marine signals, but the closely related tribes Luna-A2 and Luna2 appear to be more exclusive freshwater groups, pointing to substantial diversification with regard to habitat preferences among related phylotypes.
None of the Bacteroidetes tribes described for freshwater lakes match a large proportion of sequences originating from marine habitats (<10% occurrence [previously noted in reference 5]). Instead, 5 of the 14 tribes have a terrestrial signal, pointing to a broader distribution across the terrestrial-aquatic continuum (Fig. 8). In particular, the closely related tribes Muci (Mucilaginibacter) and Pedo (Pedobacter) frequently match sequences from terrestrial systems. Both of these taxa are often detected and isolated in soils and wetlands and were also quite rare in the freshwater lake 16S rRNA gene data set.
Unlike the other major phyla present in freshwater lakes, the Proteobacteria have tribes that are seemingly less restricted to the freshwater biome. A strong freshwater-terrestrial distribution pattern is evident for several Alphaproteobacteria tribes: the Brev (Brevundimonas), Sphingo (Sphingomonas), and alfI-BI (within the Rhizobiales) tribes (Fig. 5B and C). Notably, the freshwater sister group of the marine SAR11 (LD12) clade appears to be an exclusively freshwater tribe. Few tribes within the Betaproteobacteria appear to be restricted to a single biome. Many of the groups commonly identified as “typical freshwater bacteria” (Polynucleobacter tribes, LD28, members of the genes Limnohabitans, and tribe betIII-A1 within the GKS98 cluster) frequently match sequences retrieved from marine ecosystems, particularly estuaries, pointing to a wider distribution than previously assumed. Some tribes also appear to be distributed across the aquatic-terrestrial interface; e.g., tribe betIII-A2 and all three tribes within the betVII lineage, known as the Oxalobacteraceae, almost exclusively match sequences from the terrestrial environment (Fig. 8). Hence, none of the putative freshwater lake tribes within the Burkholderiales appear to be indigenous freshwater bacteria but rather seem to have broad and divergent habitat preferences (vagabonds). This habitat divergence is also apparent for the Janthinobacteria (tribe JanB), where >50% of the GenBank matches are from organism-associated habitats coupled mainly to the terrestrial biome, although this genus has been implicated as a common contaminant in clone libraries (208). This result points to a physical association between this tribe and other organisms, where notably >60% of the closest matches in GenBank are from the human microbiome and another 38% are linked to miscellaneous terrestrial animals.
An even more pronounced association with other organisms is evident for the Gammaproteobacteria, where all but one (tribe Pseudo-A2) of the putative freshwater lake tribes have matching sequences associated mainly with other organisms from terrestrial habitats. Most of these tribes (tribe gamII-A2/Serratia, Stenotrophomonas, and Acinetobacter) contain mainly sequences from the human biome, whereas the more abundant freshwater lake tribe gamII-A1 is associated with sequences from other terrestrial animals (50% of the nearest GenBank matches). Hence, these tribes also seem to represent “tourists” rather than indigenous freshwater bacteria. This finding contrasts with the two Verrucomicrobia tribes, the OP10 tribe (CLO-84), and the Fibrobacteres (all minor components of freshwater lake bacterial communities), which match only freshwater sequences and hence likely represent true freshwater groups.
The emergent picture is that most putative freshwater lake tribes identified in our phylogenetic study are found exclusively in this biome. However, our results also point to considerable variation across phyla and families, where some groups are dominated by tribes with alternative native habitats. This alternative habitat is most often terrestrial, which is consistent with the dominant flow of water (and bacteria) from soils via freshwater and then further into the oceans. The stronger connection between the freshwater and terrestrial environments also makes sense in light of the salinity difference between lakes and marine habitats, which acts as an apparent habitat and evolutionary barrier for microorganisms (128). The absence of such a salinity barrier between the terrestrial and freshwater biomes may allow a more extensive exchange of microflora between these environments.
Biogeography of the Freshwater Lake Tribes
The extent to which “the environment selects” is a critical question in microbial ecology. We know that many biological, physical, and chemical features of lakes can influence the distribution of ecologically coherent bacterial populations. Previous bacterial 16S rRNA gene sequence database surveys making freshwater-marine comparisons (12) and comparisons across a diverse set of microbial systems (28) have observed strong compositional contrasts across ecosystems and have related these contrasts to environmental conditions. We undertake an approach similar to those of the studies described above to identify factors that correlate with tribe compositional differences among freshwater ecosystems. Biotic interactions that are likely to structure freshwater bacterial communities include predation (114), viral lysis (210), and competition for resources (93a). Specifically, the carbon source appears to be an important biological regulator of community composition (51). Physical features like landscape position (234) and lake retention time (123) have also been shown to strongly influence community structure. Finally, pH stands out as the strongest chemical determinant of freshwater lake bacterial community composition (123).
To date, the majority of freshwater microbial community ecology studies have focused on single groups (e.g., see reference 84), a limited number of systems (238), or a confined geographic range (234). In the phylogenetic framework presented in this review, we identified many narrow monophyletic freshwater groups, such as the described tribes (97% to 99.5% sequence similarity), which allowed us to explore their biogeographic patterns. To synthesize the patterns of the most common freshwater lake tribes, we gathered as much environmental information as possible about the lakes used to generate our freshwater lake sequence database. We used our environmental database along with tribe occurrence patterns and multivariate statistics to conduct a broad, cross-system comparison and reveal a more general picture of patterns of distribution along environmental gradients. By correlating the occurrence patterns of taxa to gradients under environmental conditions, we can approximate the niches of our tribes. The gradient analysis approach has been used with plant and animal systems for decades (e.g., see reference 60), but only recently have they been applied to microorganisms (123). We used three environmental state variables, pH and total phosphorus (TP) and dissolved organic carbon (DOC) concentrations, as well as the latitudinal position of the lakes. DOC and TP were most significant in explaining tribe distribution when considered as a ratio (DOC/TP ratio) that summarizes the relative importance of external (terrestrial) and internal (algal/cyanobacterial) carbon substrate supplies. In addition, we considered absolute latitude (AbsLat) as an indicator of climate. We initially compared the relative strengths of geographic and local environmental conditions using a partial Mantel test approach (19). Although both geographic (r = 0.24; P = 0.001 [partial Mantel test]) and environmental (r = 0.39; P = 0.001 [partial Mantel test]) variables were significantly correlated with bacterial community composition, the environmental effect was relatively stronger, especially when considering the limited number of environmental parameters examined in this analysis. In subsequent analyses we focused on local environmental characteristics in an attempt to characterize the environmental preferences of various bacterial tribes. However, we do remind the reader that geographic patterns could still play a weak but significant role in the structuring of global freshwater lake bacterial compositional patterns.
The tribe occurrence matrix that we used to characterize freshwater lake bacterial biogeographic patterns was populated by 79 freshwater lake bacterial tribes represented by 4,012 sequences collected from 47 lakes. Sites in the data set included lakes from all around the globe, such as Lake Taihu (China), Lake Mendota (Wisconsin), and Lake Grosse Fuchskuhle (Germany). Only one lake (Heywood Lake, Antarctica) is located in the southern hemisphere, and only seven lakes (six reservoirs in Burkina Faso and a Chinese lake) are located in the tropics. The majority of lakes were located in North America and Europe. Data sets were selected based on the amount of available sequence data and associated environmental-contextual data, which, with few exceptions, included pH, TP, DOC, global positioning system (GPS) coordinates, and trophic status. A study was included only when at least 40 clones were randomly sequenced, with the exception of a few cases where we used studies that provided clone counts based on ≥97% sequence similarity clusters. The choice of environmental variables was rather arbitrary, as they represent the most commonly measured environmental state variables. However, these variables are often thought to be the most significant drivers of bacterioplankton community composition. We used TP, which is a good indicator of nutrient availability and system productivity; lake pH, which has been repeatedly related to bacterial community composition and diversity patterns (140); and DOC, which provides information on the trophic status of the system, as high-DOC systems usually represent net heterotrophic humic lakes.
We used both correspondence analysis (CA) (94) and detrended correspondence analysis (DCA) (94) to visualize patterns of tribe distribution and community composition. CA and DCA are multivariate statistical techniques widely used by community ecologists to identify the main factors and/or gradients in species-rich but sparsely populated data matrices. They are both similar to principal-component analysis (PCA) (98) but assume Gaussian, rather than linear, responses along environmental gradients. DCA also incorporates a rescaling step to overcome distortions (“the arch effect”) commonly encountered with correspondence analyses (94). In general, DCA ordinations perform better with simulated data than do CA ordinations. Prior to conducting the CA and DCA, the library composition from each lake was scaled to a sum of 1 to control for sampling differences across studies. In addition, environmental variables were standardized to a mean of zero and scaled by their standard deviations (Z score).
To display the general abundance and occurrence patterns of all tribes, we plotted the relative recoveries of each tribe (number of tribe sequences/number of total sequences) from all lakes and their prevalence (number of lakes from which sequences from a tribe were recovered/number of lakes in the meta-analysis) in the 47 lakes (Fig. 9). Lineage acI appears to be the most common bacterioplankton lineage, with small changes in prevalence across lakes. Our findings suggest that the acI lineage is also the most persistent freshwater lake bacterial lineage, and this is in agreement with previous documentation of its low temporal dynamics over seasons (e.g., see reference 63). Other very abundant and widespread freshwater bacterial groups are the betaproteobacterial lineages Polynucleobacter (Pnec), where PnecC represents the most common tribe in this lineage, and Lhab, belonging to the Comamonadaceae. Tribe LD12, a freshwater lineage of the SAR11 group, showed the highest prevalence and relative recovery of all alphaproteobacterial lineages. However, tribe LD12 does not seem to be as ubiquitous and abundant in freshwater systems as its sister group SAR11 is in marine environments, where it was previously shown to dominate most bacterioplankton communities (50, 137).
FIG. 9.
Bar plots indicating the relative recovery (number of tribe sequences/number of total sequences) of each tribe from all lakes and their prevalence (number of lakes from which sequences from a tribe were recovered/total number of lakes) in the 47 lakes. For reference, bars are aligned with an ultrametric tree of all freshwater lake tribes observed for the 47 lakes.
In addition, Spearman rank correlation coefficients between the relative recoveries of each tribe from each lake and environmental state variables (pH, DOC/TP ratio, and AbsLat) were determined. In Fig. 10, positive (blue) and negative (red) correlations between each tribe and each of the three variables are indicated. The size of the symbol indicates the strength of the correlation. In general, a conservation of environmental responses at higher taxonomic levels (i.e., phylum) was rare, as suggested previously by others (108). However, many examples of conservation within more refined taxonomic groups can be observed (e.g., Iluma lineage response to pH, acI-B lineage response to pH and DOC/TP, and Flavo lineage response to DOC/TP). Coherent patterns in the environmental response appear to be more common in the phylum Actinobacteria, where tribes could be narrowly defined as a result of previous phylogenetic work and a large number of available sequences (140).
FIG. 10.
Sign and magnitude of Spearman rank correlation rho between the relative recovery of each tribe from each lake and environmental conditions (pH, the ratio between dissolved organic carbon and total phosphorus concentrations [DOC/TP], and absolute latitude [AbsLat]). Positive (blue) and negative (red) correlations between each tribe and each of the three variables are indicated. The size of the symbol indicates the magnitude of Spearman's rho. Environmental correlations are aligned with an ultrametric tree of all freshwater lake tribes observed for the 47-lake data set used for this analysis.
DCA and CA resulted in highly similar plots (for DCA, see Fig. 11; for CA, see the supplemental material); thus, we will discuss only DCA. The axis that explained the most variation in freshwater lake bacterial tribe distribution in our DCA was highly correlated with pH. Numerous studies have shown that pH is a major environmental covariate of taxon distribution patterns of zooplankton and phytoplankton (21) and was shown more recently to correlate strongly with biogeographic patterns of freshwater lake bacterioplankton (123). In a more detailed analysis of tribes within the phylum Actinobacteria, distribution patterns along pH gradients corresponded very well with our meta-analysis (140). Tribes belonging to the acI-B lineage seem to be more common in acidic lakes with high DOC concentrations, except for tribe acI-B1. This tribe and the tribes belonging to the acI-A lineage (except tribe acI-A1) seem to be more common in lakes with high pHs and low DOC concentrations. Tribes within the ubiquitous Polynucleobacter lineage revealed some clear distinctions. The abundance of PnecB, which has been suggested to be circumneutral or alkaline (230), likewise increased with increasing pH. On the other hand, the abundance of cluster PnecC was negatively correlated with pH and could be associated with acidic lakes. Such pH-dependent biogeography has also been described for members of the Bacteroidetes, where the HAL-A2 tribe (GKS2-217) was found only in soft-water lakes at a pH range of 6.5 to 7.3, the Aquir tribe (LD2) was found only in hard-water lakes at a pH range of 7.7 to 9.5, and the HAL-A1 tribe (HAL) was found in a variety of lakes at a pH range of 6.7 to 8.6 (178). Our DCA corroborates that HAL-A2 is more common in more acidic lakes than is Aquir. Other tribes that seem to be associated with acidic lakes are the bacI-A2, bacI-A3, bacI-B1, Flavo-A2, Flecto (R-FL615), and HAL-A2 (HAL) tribes, whereas the Flavo-A1, Algor, Aquir (LD2), and Pedo tribes are typical representatives in alkaline lakes (Fig. 11).
FIG. 11.
Detrended correspondence analysis (DCA) (94) to visualize patterns of tribe distribution along environmental gradients (pH, ratio between dissolved organic carbon and total phosphorus concentrations [DOC/TP], and absolute latitude [AbsLat]). The first axis explained ∼12% of the variance, and the second axis explained an additional 6%. All tribes were included in a single DCA, but each panel presents a limited number of tribes for clarity. (A) Actinobacteria; (B) Alphaproteobacteria; (C) Bacteroidetes; (D) Betaproteobacteria; (E) minor phyla.
In previous studies, resource availability has been linked to the distribution of freshwater lake bacterial tribes. The ratio of water color to chlorophyll a was recently suggested to be a useful way to summarize the relative contributions of within-lake (autochthonous) primary production versus terrestrial (allochthonous) organic matter (26, 108) supporting heterotrophic growth. In this meta-analysis the DOC-to-TP ratio appears to serve a similar purpose, where low ratios are indicative of autochthonous-dominated systems and high ratios are indicative of allochthonous systems. The Flavobacteriales especially seem to be favored in autochthonous systems, where phytoplankton blooms often occur (49). A metabolic capability to degrade recalcitrant organic compounds such as humic substances was suggested for Bacteroidetes tribes bacI-A1 and bacI-A2 (51) as well as for freshwater Novosphingobium (clade alfIV-A) and Sphingopyxis (clade alfIV-B) (104). These and numerous other lineages were associated with high DOC/TP ratios in the DCA plots, which suggests that these lineages are competitive under conditions where humic organic matter is abundant and possibly points to their role in the degradation of this large pool of recalcitrant (mostly allochthonous) organic carbon. Tribes within the betaproteobacterial lineage Lhab had strong negative correlations with the DOC/TP ratio, supporting their previously documented responses to nutrient enrichment (187). The two major tribes within the Verrucomicrobia appeared to contrast in the majority of their environmental responses, including their distribution along the gradient in DOC/TP.
Our meta-analysis provides a number of suggestive descriptions of the responses of tribes to environmental gradients. However, these relationships are correlative and may be confounded by correlation among other environmental variables. Hence, there is clearly a need for more comprehensive environmental sampling, including a higher resolution of the quality and quantity of resources, like nitrogen, phosphorus, trace elements, and organic compounds, but also biotic interactions, such as predation by grazers and viruses. Experimental approaches should be employed to make more mechanistic links between environmental drivers and tribe responses. Even though the defined tribes represent phylogenetic groups with broad ecological properties that seem to be congruent among the environmental variables used in our meta-analysis, we need to consider that in some cases, organisms with (almost) identical 16S rRNA genes have rather divergent genomes, allowing them to inhabit different environmental niches (e.g., see reference 88). There is clearly a need to verify the level of resolution defining ecologically coherent units among the freshwater lake bacteria. Having said that, we observe many promising and suggestive biogeographic patterns within the freshwater lake bacterial groups examined in this review and encourage others to test the hypotheses presented here. We have a long way ahead of us before we reach a robust understanding of the traits of freshwater lake bacterial groups and incorporate this understanding into a theoretical framework allowing predictive microbial ecology.
OUTLOOK: HOW TO USE THIS GUIDE
Lakes are foci for ecological processes and human activities in global landscapes (47). The resident bacterial communities of these ecosystems play a central role in many hydrological and biogeochemical cycles, thereby providing critical ecosystem services to humankind (136). Recently, lakes have been described as early indicators (i.e., sentinels) of both regional and global environmental change (228). Much of this sensitivity and responsiveness to environmental change can be attributed to the resident microorganisms, but with respect to bacteria, this has not been explored to its full potential (1). Given the importance of bacteria to lake ecosystems, few studies have been able to link bacterial identity to function in this environment. Most would agree that this research gap results largely from a lack of an understanding of the characteristics of the major taxa present in lake systems, which stems from the tendency of abundant freshwater lake bacteria to escape cultivation. Hence, a continued strategy to gather enrichments and pure cultures of the abundant groups of freshwater lake bacteria for further ecological and metabolic characterization is necessary, especially when in complement with the cultivation-independent identification of ecophysiological traits of these organisms in nature. The framework developed in this review provides a useful guide for this ongoing effort.
To date, much of the work carried out on connecting lake bacterial taxa to biogeochemical processes and other ecological traits has been done at the phylum level (reviewed in reference 154). Phylum-based analyses have certainly proven useful as an initial assessment of the types and abundances of bacteria that may be present in an environment and as a way to broadly partition the drivers and functions of bacterial taxa in their particular environments. However, combining a multitude of taxa into a single group, as is done when studying phyla in lakes, creates the unrealistic expectation that all taxa within a phylum behave similarly. Thus, the phylum level of taxonomic identity is a rather blunt tool with which to decipher the ecology of freshwater bacterioplankton and, as such, is more likely to generate conflicting data among studies. For freshwater lake bacteria, both the reviewed literature and synoptic phylogenetic analyses suggest substantial ecological divergence within phyla (for a visual representation of the ecological divergence of tribes within phyla, see Fig. 11 and 12). We do not altogether discard the possibility that there is also some level of basic ecological coherence at higher taxonomic ranks of bacteria (158). However, given the data at hand, this seems unlikely for the majority of bacterial phyla that dominate in freshwater lake ecosystems. The spatial and temporal separation of phyla and other higher taxonomic ranks has been observed repeatedly for freshwater lakes and many other environments. One interpretation of such patterns is that this reflects ecological coherence at this higher taxonomic level (158). We believe that it is much more likely that these apparent distribution patterns for phyla reflect the niches of particularly competitive lineages, clades, or tribes within a phylum that occasionally dominate the community, as previously observed for brackish bacterial communities in the Baltic Sea (7) and Actinobacteria in freshwater lakes (Fig. 12).
FIG. 12.
(A) Time course of the relative contributions by the acI lineage and acI-A clade (A) and the acI-A2, acI-A4, acI-A5, and acI-A6 tribes (B) of the phylum Actinobacteria to the bacterial community of Lake Mendota (by percentage of total fluorescence), inferred from automated ribosomal intergenic spacer analysis (ARISA) according to methods described previously (141). Sampling was conducted biweekly for 4 years during the ice-off period of the lake. The curves are constructed as a moving average (n = 2) of the relative contribution data. For comparison, the tribe plots are scaled to the maximum fluorescence observed for each tribe.
A few studies have addressed the question of whether or not community composition and, therefore, diversity are related to the functional traits of a community (e.g., see references 14, 16, 54, 56, 68, and 204). Those studies have provided conflicting results, thereby fueling the ecosystem diversity debate with arguments about extensive functional and ecological redundancy in bacterial communities but without fully acknowledging the diverse ecological and physiological properties of the individual populations within the studied communities. In fact, even the understanding and appreciation of mechanisms causing temporal and spatial biogeography patterns, like seasonal succession in freshwater lake bacterioplankton, are hampered by our currently very limited knowledge about the ecology and physiology of even the most abundant bacterial taxa in this ecosystem. During yearly cycles, lake bacterial communities undergo dramatic shifts in community composition (e.g., see reference 183), at least partly in response to disturbance events. It has been argued that such disturbances may alter community composition, thereby preventing competitive exclusion and the maintenance of community diversity (169). Diversity may also be maintained and community composition may be structured by internal community processes such as predation and concurrent competition between bacterial (species) tribes for multiple resources (99). However, without detailed genomic and experimental studies of the abundant bacterioplankton tribes, our understanding of these processes will remain uncertain. Again, the phylogenetic framework presented here provides an outline for gathering and synthesizing such information.
Now that many of the abundant pelagic lake bacteria have been identified and organized into phylogenetically narrow groups, we can focus on studying them in more detail using genomic and experimental approaches. We propose an emphasis on these defined freshwater groups instead of the previous emphasis on phyla, with the idea that these groups may be more similar at the level of biological organization to that of “species.” This will lead to more coherent responses and the development of more precise and more robust predictive models for their distribution patterns over time and space as well as for ecosystem-level functioning. Studies employing this more narrowly defined level of focus have already provided many insights into the ecology and physiology of common freshwater bacteria (as reviewed here). We do not suggest that all of the narrowly defined phylogenetic groups (i.e., tribes) represent congruent ecological units, as ecological differentiation within tribes has been shown previously. For example, there are known differences in habitat types among members of the PnecC tribe (217) and in salinity tolerance among members of the beIII-A1 tribe (230). This guide does, however, provide the framework within which questions about the identification of bacteria to the species level and ecological differentiation in freshwater bacterioplankton can be asked and explored while allowing for the continued refinement of the lineages, clades, and tribes until genera and species are properly described. We also note that our database has a major overrepresentation of lakes from northern latitudes, especially Europe and North America (see the supplemental material). In order to fully characterize the bacteria of freshwater lakes and further our understanding of their roles in this ecosystem, we emphasize the need to obtain more information on bacteria from tropical lakes and lakes of the southern hemisphere.
The results presented here illustrate the powerful nature of comparative cross-study analysis. Many new putative freshwater lake clusters were identified, but probably more importantly, the large numbers of described bacterial 16S rRNA clusters spread across the literature were brought together into a single phylogenetic framework. Phylogenies are always evolving as more sequences are obtained and added; thus, the phylogenies presented here are not intended to be static or constraining but provide a temporary structure for an ever-expanding collection of freshwater sequences. The results here also further support the notion that bacteria commonly found in freshwater lakes are not there accidentally but are the result of a suite of environmental forces that define freshwater lakes as a unique habitat that acts upon the global bacterial community, selecting for the taxa found living there.
The natural history guide to freshwater lake bacteria provides a sorely needed knowledge base and controlled vocabulary for future freshwater studies to build upon and explore. Our data set should provide a good starting point for further comparative studies among different chemically and physically defined epilimnia or between different freshwater environments, since many freshwater habitats (e.g., lake hypolimnia and littoral zones, rivers, and groundwater) were not included here. In addition, comparisons between more distantly related habitats such as the ocean or various terrestrial environments should now be more feasible. Those interested in learning more about lake microflora could benefit greatly from a synoptic table of shared ecological and metabolic traits for tribes, clades, lineages, and phyla. Our understanding of lake bacteria is still too superficial to enable the preparation of such a table, but the use of our phylogenetic framework to structure sequences from the growing number of surveys and experiments that employ next-generation sequencing might take us to that point in the near future. It is our hope that this effort spurs many new studies aimed at these potentially more ecologically relevant clusters so that the freshwater research community can learn more about the relationships between freshwater bacterial phylotypes and taxa, their habitat preferences, and their functions in these ecosystems.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences generated for this study have been deposited in GenBank under accession numbers FJ916807 to FJ916903, HQ386253 to HQ386631, and HQ530565 to HQ532908, except HQ530583, HQ531638, and HQ532521. The 16S rRNA gene ARB database used in this study is available upon request from R.J.N. (rjnewton@gmail.com) or the corresponding author.
Supplementary Material
Acknowledgments
We thank Tom Battin, Martin Polz, and Ashley Shade for their comments and suggestions on early versions of the manuscript. We also acknowledge a large number of researchers and groups for contributing unpublished contextual data for the lakes included in our synoptic study. We especially thank the anonymous reviewers for offering many wonderful suggestions that greatly improved the manuscript.
Funding was obtained from the Swedish Research Council (grants to S.B.), a U.S. National Science Foundation (NSF) doctoral dissertation improvement grant (grant DEB-0710059 to R.J.N.), the NSF Microbial Observatories Program (grant MCB-0702653 to K.D.M.), and the NSF Long Term Ecological Research Program (grants DEB-0217533 and DEB-0822700 to K.D.M.).
Biography
 Ryan J. Newton is an aquatic microbial ecologist having studied the distribution and genetic content of bacteria in both freshwater and marine systems. He received his B.S. in the Biological Sciences from the University of Nebraska—Lincoln in 2002 and his Ph.D. in Microbiology from the University of Wisconsin—Madison in 2008. After a postdoctoral fellowship at the University of Georgia Marine Sciences department, he is now currently employed as a postdoctoral research associate at the University of Wisconsin—Milwaukee Great Lakes WATER Institute.
Ryan J. Newton is an aquatic microbial ecologist having studied the distribution and genetic content of bacteria in both freshwater and marine systems. He received his B.S. in the Biological Sciences from the University of Nebraska—Lincoln in 2002 and his Ph.D. in Microbiology from the University of Wisconsin—Madison in 2008. After a postdoctoral fellowship at the University of Georgia Marine Sciences department, he is now currently employed as a postdoctoral research associate at the University of Wisconsin—Milwaukee Great Lakes WATER Institute.
 Stuart E. Jones received his Ph.D. in Limnology and Marine Sciences from the University of Wisconsin—Madison and conducted his postdoctoral work at Michigan State University's W. K. Kellogg Biological Station. He is currently an Assistant Professor in the Department of Biological Sciences at the University of Notre Dame. As an aquatic ecologist, Dr. Stuart investigates the role of microorganisms in the freshwater carbon cycle under current and future environmental scenarios.
Stuart E. Jones received his Ph.D. in Limnology and Marine Sciences from the University of Wisconsin—Madison and conducted his postdoctoral work at Michigan State University's W. K. Kellogg Biological Station. He is currently an Assistant Professor in the Department of Biological Sciences at the University of Notre Dame. As an aquatic ecologist, Dr. Stuart investigates the role of microorganisms in the freshwater carbon cycle under current and future environmental scenarios.
 Alexander Eiler is a microbial ecologist with a broad interest in the ecology and genetics of aquatic microbes. He has an M.S. in ecology from Vienna University (Austria), and a Ph.D. in Limnology from Uppsala University (Sweden). He is presently an Assistant Professor in Limnology at the Department of Ecology and Genetics, Uppsala University, Uppsala, Sweden.
Alexander Eiler is a microbial ecologist with a broad interest in the ecology and genetics of aquatic microbes. He has an M.S. in ecology from Vienna University (Austria), and a Ph.D. in Limnology from Uppsala University (Sweden). He is presently an Assistant Professor in Limnology at the Department of Ecology and Genetics, Uppsala University, Uppsala, Sweden.
 Katherine D. McMahon received her B.S. and M.S. at the University of Illinois at Urbana-Champaign and her Ph.D. at the University of California at Berkeley. She currently is an Associate Professor at the University of Wisconsin—Madison, where she has been since 2003. Dr. McMahon has been studying freshwater microbial ecology since she arrived at the University of Wisconsin—Madison and discovered both the beautiful lakes of Wisconsin and the rich local history of studying the microbes in them.
Katherine D. McMahon received her B.S. and M.S. at the University of Illinois at Urbana-Champaign and her Ph.D. at the University of California at Berkeley. She currently is an Associate Professor at the University of Wisconsin—Madison, where she has been since 2003. Dr. McMahon has been studying freshwater microbial ecology since she arrived at the University of Wisconsin—Madison and discovered both the beautiful lakes of Wisconsin and the rich local history of studying the microbes in them.
 Stefan Bertilsson received his B.Sc. and M.Sc. from Uppsala University, Uppsala, Sweden, and his Ph.D. from Linköping University, Linköping, Sweden. After a postdoctoral fellowship at the Massachusetts Institute of Technology he moved back to Uppsala University in 2002 and currently holds an Associate Professor position at the Department of Ecology and Genetics. Convinced that small things are very important, Dr. Bertilsson has studied bacteria in lakes and oceans since the beginning of his scientific career, first by looking at their biogeochemical roles in carbon transformation processes and during the last decade also by learning more about their diversity and molecular ecology.
Stefan Bertilsson received his B.Sc. and M.Sc. from Uppsala University, Uppsala, Sweden, and his Ph.D. from Linköping University, Linköping, Sweden. After a postdoctoral fellowship at the Massachusetts Institute of Technology he moved back to Uppsala University in 2002 and currently holds an Associate Professor position at the Department of Ecology and Genetics. Convinced that small things are very important, Dr. Bertilsson has studied bacteria in lakes and oceans since the beginning of his scientific career, first by looking at their biogeochemical roles in carbon transformation processes and during the last decade also by learning more about their diversity and molecular ecology.
Footnotes
Supplemental material for this article may be found at http://mmbr.asm.org/.
REFERENCES
- 1.Adrian, R., et al. 2009. Lakes as sentinels of climate change. Limnol. Oceanogr. 54:2283-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allgaier, M., S. Brückner, E. Jaspers, and H. P. Grossart. 2007. Intra- and inter-lake variability of free-living and particle-associated Actinobacteria communities. Environ. Microbiol. 9:2728-2741. [DOI] [PubMed] [Google Scholar]
- 3.Allgaier, M., and H. P. Grossart. 2006. Diversity and seasonal dynamics of Actinobacteria populations in four lakes in northeastern Germany. Appl. Environ. Microbiol. 72:3489-3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allgaier, M., and H. P. Grossart. 2006. Seasonal dynamics and phylogenetic diversity of free-living and particle-associated bacterial communities in four lakes in northeastern Germany. Aquat. Microb. Ecol. 45:115-128. [Google Scholar]
- 5.Alonso, C., F. Warnecke, R. Amann, and J. Pernthaler. 2007. High local and global diversity of Flavobacteria in marine plankton. Environ. Microbiol. 9:1253-1266. [DOI] [PubMed] [Google Scholar]
- 6.Alonso, C., M. Zeder, C. Piccini, D. Conde, and J. Pernthaler. 2009. Ecophysiological differences of betaproteobacterial populations in two hydrochemically distinct compartments of a subtropical lagoon. Environ. Microbiol. 11:867-876. [DOI] [PubMed] [Google Scholar]
- 7.Andersson, A., L. Riemann, and S. Bertilsson. 2010. Pyrosequencing reveals contrasting dynamics of taxa within Baltic Sea bacterioplankton. ISME J. 4:171-181. [DOI] [PubMed] [Google Scholar]
- 8.Andersson, S. G. E., et al. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133-140. [DOI] [PubMed] [Google Scholar]
- 9.Arnds, J., K. Knittel, U. Buck, M. Winkel, and R. Amann. 2010. Development of a 16S rRNA-targeted probe set for Verrucomicrobia and its application for fluorescence in situ hybridization in a humic lake. Syst. Appl. Microbiol. 33:139-148. [DOI] [PubMed] [Google Scholar]
- 10.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2006. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 72:5734-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bahr, M., J. E. Hobbie, and M. L. Sogin. 1996. Bacterial diversity in an arctic lake: a freshwater SAR11 cluster. Aquat. Microb. Ecol. 11:271-277. [Google Scholar]
- 12.Barberan, A., and E. O. Casamayor. 2010. Global phylogenetic community structure and beta-diversity patterns in surface bacterioplankton metacommunities. Aquat. Microb. Ecol. 59:1-10. [Google Scholar]
- 13.Batut, J., S. G. E. Andersson, and D. O'Callaghan. 2004. The evolution of chronic infection strategies in the alpha-proteobacteria. Nat. Rev. Microbiol. 2:933-945. [DOI] [PubMed] [Google Scholar]
- 14.Bell, T., J. A. Newman, B. W. Silverman, S. L. Turner, and A. K. Lilley. 2005. The contribution of species richness and composition to bacterial services. Nature 436:1157-1160. [DOI] [PubMed] [Google Scholar]
- 15.Berg, K. A., et al. 2009. High diversity of cultivable heterotrophic bacteria in association with cyanobacterial water blooms. ISME J. 3:314-325. [DOI] [PubMed] [Google Scholar]
- 16.Bertilsson, S., A. Eiler, A. Nordqvist, and N. O. G. Jørgensen. 2007. Links between bacterial production, amino-acid utilization and community composition in productive lakes. ISME J. 1:532-544. [DOI] [PubMed] [Google Scholar]
- 17.Biers, E. J., S. L. Sun, and E. C. Howard. 2009. Prokaryotic genomes and diversity in surface ocean waters: interrogating the global ocean sampling metagenome. Appl. Environ. Microbiol. 75:2221-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boenigk, J., P. Stadler, A. Wiedlroither, and M. W. Hahn. 2004. Strain-specific differences in the grazing sensitivities of closely related ultramicrobacteria affiliated with the Polynucleobacter cluster. Appl. Environ. Microbiol. 70:5787-5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borcard, D., P. Legendre, and P. Drapeau. 1992. Partialling out the spatial component of ecological variation. Ecology 73:1045-1055. [Google Scholar]
- 20.Boucher, D., L. Jardillier, and D. Debroas. 2006. Succession of bacterial community composition over two consecutive years in two aquatic systems: a natural lake and a lake-reservoir. FEMS Microbiol. Ecol. 55:79-97. [DOI] [PubMed] [Google Scholar]
- 21.Brezonik, P. L., et al. 1993. Experimental acidification of Little Rock Lake, Wisconsin: chemical and biological changes over the pH range 6.1 to 4.7. Can. J. Fish. Aquat. Sci. 50:1101-1121. [Google Scholar]
- 22.Bruns, A., U. Nübel, H. Cypionka, and J. Overmann. 2003. Effect of signal compounds and incubation conditions on the culturability of freshwater bacterioplankton. Appl. Environ. Microbiol. 69:1980-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buck, U., H. P. Grossart, R. Amann, and J. Pernthaler. 2009. Substrate incorporation patterns of bacterioplankton populations in stratified and mixed waters of a humic lake. Environ. Microbiol. 11:1854-1865. [DOI] [PubMed] [Google Scholar]
- 24.Burkert, U., F. Warnecke, D. Babenzien, E. Zwirnmann, and J. Pernthaler. 2003. Members of a readily enriched beta-proteobacterial clade are common in surface waters of a humic lake. Appl. Environ. Microbiol. 69:6550-6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Callieri, C. 2008. Picophytoplankton in freshwater ecosystems: the importance of small-sized phototrophs. Freshw. Rev. 1:1-28. [Google Scholar]
- 26.Carpenter, S. R., et al. 2005. Ecosystem subsidies: terrestrial support of aquatic food webs from 13C addition to contrasting lakes. Ecology 86:2737-2750. [Google Scholar]
- 27.Casamayor, E. O., H. Schafer, L. Baneras, C. Pedrós-Alió, and G. Muyzer. 2000. Identification of and spatio-temporal differences between microbial assemblages from two neighboring sulfurous lakes: comparison by microscopy and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 66:499-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaffron, S., H. Rehrauer, J. Pernthaler, and C. von Mering. 2010. A global network of coexisting microbes from environmental and whole-genome sequence data. Genome Res. 20:947-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng, S. M., and J. M. Foght. 2007. Cultivation-independent and -dependent characterization of bacteria resident beneath John Evans Glacier. FEMS Microbiol. Ecol. 59:318-330. [DOI] [PubMed] [Google Scholar]
- 30.Cole, J. J., S. Findlay, and M. L. Pace. 1988. Bacterial production in fresh and saltwater ecosystems—a cross-system overview. Mar. Ecol. Prog. Ser. 43:1-10. [Google Scholar]
- 31.Cole, J. J., et al. 2007. Plumbing the global carbon cycle: integrating inland waters into the terrestrial carbon budget. Ecosystems 10:171-184. [Google Scholar]
- 32.Comté, J., et al. 2006. Microbial community structure and dynamics in the largest natural French lake (Lake Bourget). Microb. Ecol. 52:72-89. [DOI] [PubMed] [Google Scholar]
- 33.Corno, G. 2006. Effects of nutrient availability and Ochromonas sp predation on size and composition of a simplified aquatic bacterial community. FEMS Microbiol. Ecol. 58:354-363. [DOI] [PubMed] [Google Scholar]
- 34.Corno, G., and K. Jürgens. 2006. Direct and indirect effects of protist predation on population size structure of a bacterial strain with high phenotypic plasticity. Appl. Environ. Microbiol. 72:78-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corno, G., et al. 2009. Bacterial diversity and morphology in deep ultraoligotrophic Andean lakes: role of UVR on vertical distribution. Limnol. Oceanogr. 54:1098-1112. [Google Scholar]
- 36.Cotner, J. B., and B. A. Biddanda. 2002. Small players, large role: microbial influence on biogeochemical processes in pelagic aquatic ecosystems. Ecosystems 5:105-121. [Google Scholar]
- 37.Crump, B. C., and J. E. Hobbie. 2005. Synchrony and seasonality in bacterioplankton communities of two temperate rivers. Limnol. Oceanogr. 50:1718-1729. [Google Scholar]
- 38.Crump, B. C., G. W. Kling, M. Bahr, and J. E. Hobbie. 2003. Bacterioplankton community shifts in an arctic lake correlate with seasonal changes in organic matter source. Appl. Environ. Microbiol. 69:2253-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Debroas, D., et al. 2009. Metagenomic approach studying the taxonomic and functional diversity of the bacterial community in a mesotrophic lake (Lac du Bourget-France). Environ. Microbiol. 11:2412-2424. [DOI] [PubMed] [Google Scholar]
- 40.Dedysh, S. N., T. A. Pankratov, S. E. Belova, I. S. Kulichevskaya, and W. Liesack. 2006. Phylogenetic analysis and in situ identification of Bacteria community composition in an acidic Sphagnum peat bog. Appl. Environ. Microbiol. 72:2110-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Figueiredo, D. R., et al. 2007. Bacterial community composition over a dry winter in meso- and eutrophic Portuguese water bodies. FEMS Microbiol. Ecol. 59:638-650. [DOI] [PubMed] [Google Scholar]
- 42.Denisova, L. Y., N. L. Bel'kova, I. I. Tulokhonov, and E. F. Zaichikov. 1999. Bacterial diversity at various depths in the southern part of Lake Baikal as revealed by 16S rDNA sequencing. Mikrobiologiia 68:475-483. [PubMed] [Google Scholar]
- 43.DeSantis, T. Z., et al. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Wever, A., et al. 2005. Bacterial community composition in Lake Tanganyika: vertical and horizontal heterogeneity. Appl. Environ. Microbiol. 71:5029-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Wever, A., K. Van der Gucht, K. Muylaert, S. Cousin, and W. Vyverman. 2008. Clone library analysis reveals an unusual composition and strong habitat partitioning of pelagic bacterial communities in Lake Tanganyika. Aquat. Microb. Ecol. 50:113-122. [Google Scholar]
- 46.Donachie, S. P., et al. 2004. The Hawaiian Archipelago: a microbial diversity hotspot. Microb. Ecol. 48:509-520. [DOI] [PubMed] [Google Scholar]
- 47.Downing, J. A., et al. 2006. The global abundance and size distribution of lakes, ponds, and impoundments. Limnol. Oceanogr. 51:2388-2397. [Google Scholar]
- 48.Eiler, A., and S. Bertilsson. 2004. Composition of freshwater bacterial communities associated with cyanobacterial blooms in four Swedish lakes. Environ. Microbiol. 6:1228-1243. [DOI] [PubMed] [Google Scholar]
- 49.Eiler, A., and S. Bertilsson. 2007. Flavobacteria blooms in four eutrophic lakes: linking population dynamics of freshwater bacterioplankton to resource availability. Appl. Environ. Microbiol. 73:3511-3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eiler, A., D. H. Hayakawa, M. J. Church, D. M. Karl, and M. S. Rappé. 2009. Dynamics of the SAR11 bacterioplankton lineage in relation to environmental conditions in the oligotrophic North Pacific subtropical gyre. Environ. Microbiol. 11:2291-2300. [DOI] [PubMed] [Google Scholar]
- 51.Eiler, A., S. Langenheder, S. Bertilsson, and L. J. Tranvik. 2003. Heterotrophic bacterial growth efficiency and community structure at different natural organic carbon concentrations. Appl. Environ. Microbiol. 69:3701-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eiler, A., J. A. Olsson, and S. Bertilsson. 2006. Diurnal variations in the auto- and heterotrophic activity of cyanobacterial phycospheres (Gloeotrichia echinulata) and the identity of attached bacteria. Freshw. Biol. 51:298-311. [Google Scholar]
- 53.Erikson, D. 1940. Studies on some lake-mud strains of Micromonospora. J. Bacteriol. 41:277-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fernandez, A., et al. 1999. How stable is stable? Function versus community composition. Appl. Environ. Microbiol. 65:3697-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fierer, N., and R. B. Jackson. 2006. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U. S. A. 103:626-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Findlay, S. E. G., R. L. Sinsabaugh, W. V. Sobczak, and M. Hoostal. 2003. Metabolic and structural response of hyporheic microbial communities to variations in supply of dissolved organic matter. Limnol. Oceanogr. 48:1608-1617. [Google Scholar]
- 57.Fitzhugh, K. 2008. Abductive inference: implications for ‘Linnean’ and ‘phylogenetic’ approaches for representing biological systematization. Evol. Biol. 35:52-82. [Google Scholar]
- 58.Fuerst, J. A. 1995. The Planctomycetes—emerging models for microbial ecology, evolution and cell biology. Microbiology 141:1493-1506. [DOI] [PubMed] [Google Scholar]
- 59.Gasol, J. M., et al. 2002. A transplant experiment to identify the factors controlling bacterial abundance, activity, production, and community composition in a eutrophic canyon-shaped reservoir. Limnol. Oceanogr. 47:62-77. [Google Scholar]
- 60.Gauch, H. G. J., G. B. Chase, and R. H. Whittaker. 1974. Ordination of vegetation samples by Gaussian species distributions. Ecology 55:1382-1390. [Google Scholar]
- 61.Gich, F., K. Schubert, A. Bruns, H. Hoffelner, and J. Overmann. 2005. Specific detection, isolation, and characterization of selected, previously uncultured members of the freshwater bacterioplankton community. Appl. Environ. Microbiol. 71:5908-5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Glöckner, F. O., B. M. Fuchs, and R. Amann. 1999. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65:3721-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Glöckner, F. O., et al. 2000. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of actinobacteria. Appl. Environ. Microbiol. 66:5053-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gómez-Consarnau, L., et al. 2007. Light stimulates growth of proteorhodopsin-containing marine Flavobacteria. Nature 445:210-213. [DOI] [PubMed] [Google Scholar]
- 65.González, J. M., et al. 2008. Genome analysis of the proteorhodopsin-containing marine bacterium Polaribacter sp. MED152 (Flavobacteria). Proc. Natl. Acad. Sci. U. S. A. 105:8724-8729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goodfellow, M., and S. T. Williams. 1983. Ecology of Actinomycetes. Annu. Rev. Microbiol. 37:189-216. [DOI] [PubMed] [Google Scholar]
- 67.Graham, J. E., L. W. Wilcox, and L. E. Graham. 2008. Algae, 2nd ed. Benjamin Cummings, San Francisco, CA.
- 68.Groffman, P. M., and P. J. Bohlen. 1999. Soil and sediment biodiversity—cross-system comparisons and large-scale effects. Bioscience 49:139-148. [Google Scholar]
- 69.Grossart, H. P., et al. 2008. Top-down and bottom-up induced shifts in bacterial abundance, production and community composition in an experimentally divided humic lake. Environ. Microbiol. 10:635-652. [DOI] [PubMed] [Google Scholar]
- 70.Gude, H. 1979. Grazing by protozoa as selection factor for activated sludge bacteria. Microb. Ecol. 5:225-237. [DOI] [PubMed] [Google Scholar]
- 71.Gude, H. 1989. The role of grazing on bacteria in plankton succession, p. 337-364. In U. Sommer (ed.), Plankton ecology. Springer-Verlag, Berlin, Germany.
- 72.Gupta, R. S. 2004. The phylogeny and signature sequences characteristics of Fibrobacteres, Chlorobi, and Bacteroidetes. Crit. Rev. Microbiol. 30:123-143. [DOI] [PubMed] [Google Scholar]
- 73.Hahn, M. W. 2009. Description of seven candidate species affiliated with the phylum Actinobacteria, representing planktonic freshwater bacteria. Int. J. Syst. Evol. Microbiol. 59:112-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hahn, M. W. 2003. Isolation of strains belonging to the cosmopolitan Polynucleobacter necessarius cluster from freshwater habitats located in three climatic zones. Appl. Environ. Microbiol. 69:5248-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hahn, M. W. 2006. The microbial diversity of inland waters. Curr. Opin. Biotechnol. 17:256-261. [DOI] [PubMed] [Google Scholar]
- 76.Hahn, M. W., et al. 2010. Limnohabitans curvus gen. nov., sp. nov., a planktonic bacterium isolated from freshwater lakes. Int. J. Syst. Evol. Microbiol. 60:1358-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hahn, M. W., V. Kasalicky, J. Jezbera, U. Brandt, and K. Šimek. 2010. Limnohabitans australis sp. nov., isolated from a freshwater pond, and emended description of the genus Limnohabitans. Int. J. Syst. Evol. Microbiol. 60:2946-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hahn, M. W., et al. 2010. Polynucleobacter cosmopolitanus sp. nov., free-living planktonic bacteria inhabiting freshwater lakes and rivers. Int. J. Syst. Evol. Microbiol. 60:166-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hahn, M. W., E. Lang, U. Brandt, and C. Sproer. 2010. Polynucleobacter acidiphobus sp. nov., a representative of an abundant group of planktonic freshwater bacteria. Int. J. Syst. Evol. Microbiol. [Epub ahead of print.] doi: 10.1099/ijs.0.023929-0. [DOI] [PMC free article] [PubMed]
- 80.Hahn, M. W., E. Lang, U. Brandt, Q. L. Wu, and T. Scheuerl. 2009. Emended description of the genus Polynucleobacter and the species Polynucleobacter necessarius and proposal of two subspecies, P. necessarius subsp. necessarius subsp. nov. and P. necessarius subsp. asymbioticus subsp. nov. Int. J. Syst. Evol. Microbiol. 59:2002-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hahn, M. W., E. Lang, M. Tarao, and U. Brandt. 2010. Polynucleobacter rarus sp. nov., a free-living planktonic bacterium isolated from an acidic lake. Int. J. Syst. Evol. Microbiol. [Epub ahead of print.] doi: 10.1099/ijs.0.017350-0. [DOI] [PMC free article] [PubMed]
- 82.Hahn, M. W., et al. 2003. Isolation of novel ultramicrobacteria classified as Actinobacteria from five freshwater habitats in Europe and Asia. Appl. Environ. Microbiol. 69:1442-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hahn, M. W., E. R. B. Moore, and M. G. Höfle. 1999. Bacterial filament formation, a defense mechanism against flagellate grazing, is growth rate controlled in bacteria of different phyla. Appl. Environ. Microbiol. 65:25-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hahn, M. W., and M. Pöckl. 2005. Ecotypes of planktonic Actinobacteria with identical 16S rRNA genes adapted to thermal niches in temperate, subtropical, and tropical freshwater habitats. Appl. Environ. Microbiol. 71:766-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hahn, M. W., M. Pöckl, and Q. L. L. Wu. 2005. Low intraspecific diversity in a Polynucleobacter subcluster population numerically dominating bacterioplankton of a freshwater pond. Appl. Environ. Microbiol. 71:4539-4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hahn, M. W., and M. Schauer. 2007. ‘Candidatus Aquirestis calciphila’ and ‘Candidatus Haliscomenobacter calcifugiens,’ filamentous, planktonic bacteria inhabiting natural lakes. Int. J. Syst. Evol. Microbiol. 57:936-940. [DOI] [PubMed] [Google Scholar]
- 87.Hahn, M. W., P. Stadler, Q. L. Wu, and M. Pöckl. 2004. The filtration-acclimatization method for isolation of an important fraction of the not readily cultivable bacteria. J. Microbiol. Methods 57:379-390. [DOI] [PubMed] [Google Scholar]
- 88.Handelsman, J. 2004. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68:669-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Haukka, K., E. Heikkinen, T. Kairesalo, H. Karjalainen, and K. Sivonen. 2005. Effect of humic material on the bacterioplankton community composition in boreal lakes and mesocosms. Environ. Microbiol. 7:620-630. [DOI] [PubMed] [Google Scholar]
- 90.Haukka, K., et al. 2006. Effect of nutrient loading on bacterioplankton community composition in lake mesocosms. Microb. Ecol. 51:137-146. [DOI] [PubMed] [Google Scholar]
- 91.Heckmann, K., and H. J. Schmidt. 1987. Polynucleobacter necessarius gen. nov., sp. nov., an obligately endosymbiotic bacterium living in the cytoplasm of Euplotes aediculatus. Int. J. Syst. Bacteriol. 37:456-457. [Google Scholar]
- 92.Hedlund, B. P., J. J. Gosink, and J. T. Staley. 1997. Verrucomicrobia div. nov., a new division of the bacteria containing three new species of Prosthecobacter. Antonie Van Leeuwenhoek 72:29-38. [DOI] [PubMed] [Google Scholar]
- 93.Hervas, A., and E. O. Casamayor. 2009. High similarity between bacterioneuston and airborne bacterial community compositions in a high mountain lake area. FEMS Microbiol. Ecol. 67:219-228. [DOI] [PubMed] [Google Scholar]
- 93a.Hibbing, M. E., C. Fuqua, M. R. Parsek, and S. B. Peterson. 2010. Bacterial competition: surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 8:15-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hill, M. O., and H. G. Gauch. 1980. Detrended correspondence analysis: an improved ordination technique. Vegetatio 42:47-58. [Google Scholar]
- 95.Hiorns, W. D., B. A. Methé, S. A. Nierzwicki-Bauer, and J. P. Zehr. 1997. Bacterial diversity in Adirondack Mountain lakes as revealed by 16S rRNA gene sequences. Appl. Environ. Microbiol. 63:2957-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Horňák, K., J. Jezbera, J. Nedoma, J. M. Gasol, and K. Šimek. 2006. Effects of resource availability and bacterivory on leucine incorporation in different groups of freshwater bacterioplankton, assessed using microautoradiography. Aquat. Microb. Ecol. 45:277-289. [Google Scholar]
- 97.Horňák, K., et al. 2005. Effects of decreased resource availability, protozoan grazing and viral impact on a structure of bacterioplankton assemblage in a canyon-shaped reservoir. FEMS Microbiol. Ecol. 52:315-327. [DOI] [PubMed] [Google Scholar]
- 98.Hotelling, H. 1933. Analysis of a complex of statistical variables into principal components. J. Educ. Psychol. 24:417-441. [Google Scholar]
- 99.Huisman, J., A. M. Johansson, E. O. Folmer, and F. J. Weissing. 2001. Towards a solution of the plankton paradox: the importance of physiology and life history. Ecol. Lett. 4:408-411. [Google Scholar]
- 100.Huisman, J., H. C. P. Matthijs, and P. M. Visser. 2005. Harmful cyanobacteria. Springer, Dordrecht, Netherlands.
- 101.Humbert, J. F., et al. 2009. Comparison of the structure and composition of bacterial communities from temperate and tropical freshwater ecosystems. Environ. Microbiol. 11:2339-2350. [DOI] [PubMed] [Google Scholar]
- 102.Hunt, D. E., et al. 2008. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science 320:1081-1085. [DOI] [PubMed] [Google Scholar]
- 103.Hutalle-Schmelzer, K. M. L., and H. P. Grossart. 2009. Changes in the bacterioplankton community of oligotrophic Lake Stechlin (northeastern Germany) after humic matter addition. Aquat. Microb. Ecol. 55:155-167. [Google Scholar]
- 104.Hutalle-Schmelzer, K. M. L., E. Zwirnmann, A. Krüger, and H. P. Grossart. 2010. Enrichment and cultivation of pelagic bacteria from a humic lake using phenol and humic matter additions. FEMS Microbiol. Ecol. 72:58-73. [DOI] [PubMed] [Google Scholar]
- 105.Jaspers, E., K. Nauhaus, H. Cypionka, and J. Overmann. 2001. Multitude and temporal variability of ecological niches as indicated by the diversity of cultivated bacterioplankton. FEMS Microbiol. Ecol. 36:153-164. [DOI] [PubMed] [Google Scholar]
- 106.Jezbera, J., A. K. Sharma, U. Brandt, W. F. Doolittle, and M. W. Hahn. 2009. ‘Candidatus Planktophila limnetica,’ an actinobacterium representing one of the most numerically important taxa in freshwater bacterioplankton. Int. J. Syst. Evol. Microbiol. 59:2864-2869. [DOI] [PubMed] [Google Scholar]
- 107.Jezberová, J., et al. 2010. Ubiquity of Polynucleobacter necessarius ssp asymbioticus in lentic freshwater habitats of a heterogenous 2000 km2 area. Environ. Microbiol. 12:658-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jones, S. E., R. J. Newton, and K. D. McMahon. 2009. Evidence for structuring of bacterial community composition by organic carbon source in temperate lakes. Environ. Microbiol. 11:2463-2472. [DOI] [PubMed] [Google Scholar]
- 109.Jones, S. E., R. J. Newton, and K. D. McMahon. 2008. Potential for atmospheric deposition of bacteria to influence bacterioplankton communities. FEMS Microbiol. Ecol. 64:388-394. [DOI] [PubMed] [Google Scholar]
- 110.Jürgens, K., and E. Jeppesen. 2000. The impact of metazooplankton on the structure of the microbial food web in a shallow, hypertrophic lake. J. Plankton Res. 22:1047-1070. [Google Scholar]
- 111.Jürgens, K., J. Pernthaler, S. Schalla, and R. Amann. 1999. Morphological and compositional changes in a planktonic bacterial community in response to enhanced protozoan grazing. Appl. Environ. Microbiol. 65:1241-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jürgens, K., and G. Stolpe. 1995. Seasonal dynamics of crustacean zooplankton, heterotrophic nanoflagellates and bacteria in a shallow, eutrophic lake. Freshw. Biol. 33:27-38. [Google Scholar]
- 113.Kasalicky, V., J. Jezbera, K. Šimek, and M. W. Hahn. 2010. Limnohabitans planktonicus sp. nov., and Limnohabitans parvus sp. nov., novel planktonic betaproteobacteria isolated from a freshwater reservoir, and emended description of the genus Limnohabitans. Int. J. Syst. Evol. Microbiol. 60:2710-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kent, A. D., et al. 2004. Annual patterns in bacterioplankton community variability in a humic lake. Microb. Ecol. 48:550-560. [DOI] [PubMed] [Google Scholar]
- 115.Kirchman, D. L. 2002. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 39:91-100. [DOI] [PubMed] [Google Scholar]
- 116.Kolmonen, E., K. Sivonen, J. Rapala, and K. Haukka. 2004. Diversity of cyanobacteria and heterotrophic bacteria in cyanobacterial blooms in Lake Joutikas, Finland. Aquat. Microb. Ecol. 36:201-211. [Google Scholar]
- 117.Kowalchuk, G. A., I. M. Head, A. D. L. Akkermans, and J. D. van Elsas. 2004. Molecular microbial ecology manual, 2nd ed. Kluwer Academic Publishers, Dordrecht, Netherlands.
- 118.Lampert, W. 1987. Predictability in lake ecosystems: the role of biotic interactions, vol. 61. Springer-Verlag, Berlin, Germany.
- 119.Langenheder, S., and K. Jürgens. 2001. Regulation of bacterial biomass and community structure by metazoan and protozoan predation. Limnol. Oceanogr. 46:121-134. [Google Scholar]
- 120.Lemarchand, C., et al. 2006. Community composition and activity of prokaryotes associated to detrital particles in two contrasting lake ecosystems. FEMS Microbiol. Ecol. 57:442-451. [DOI] [PubMed] [Google Scholar]
- 121.Lindeman, R. L. 1942. The trophic-dynamic aspect of ecology. Ecology 23:399-418. [Google Scholar]
- 122.Lindström, E. S. 2000. Bacterioplankton community composition in five lakes differing in trophic status and humic content. Microb. Ecol. 40:104-113. [DOI] [PubMed] [Google Scholar]
- 123.Lindström, E. S., M. P. Kamst-Van Agterveld, and G. Zwart. 2005. Distribution of typical freshwater bacterial groups is associated with pH, temperature, and lake water retention time. Appl. Environ. Microbiol. 71:8201-8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lindström, E. S., and E. Leskinen. 2002. Do neighboring lakes share common taxa of bacterioplankton? Comparison of 16S rDNA fingerprints and sequences from three geographic regions. Microb. Ecol. 44:1-9. [DOI] [PubMed] [Google Scholar]
- 125.Lindström, E. S., K. Vrede, and E. Leskinen. 2004. Response of a member of the Verrucomicrobia, among the dominating bacteria in a hypolimnion, to increased phosphorus availability. J. Plankton Res. 26:241-246. [Google Scholar]
- 126.Liu, Y. Q., et al. 2006. Microbial community structure in moraine lakes and glacial meltwaters, Mount Everest. FEMS Microbiol. Lett. 265:98-105. [DOI] [PubMed] [Google Scholar]
- 127.Liu, Y. Q., et al. 2009. Bacterial diversity of freshwater alpine Lake Puma Yumco on the Tibetan Plateau. Geomicrobiol. J. 26:131-145. [Google Scholar]
- 128.Logares, R., et al. 2009. Infrequent marine-freshwater transitions in the microbial world. Trends Microbiol. 17:414-422. [DOI] [PubMed] [Google Scholar]
- 129.Logares, R., J. Brate, F. Heinrich, K. Shalchian-Tabrizi, and S. Bertilsson. 2010. Infrequent transitions between saline and fresh waters in one of the most abundant microbial lineages (SAR11). Mol. Biol. Evol. 27:347-357. [DOI] [PubMed] [Google Scholar]
- 130.Lozupone, C. A., and R. Knight. 2007. Global patterns in bacterial diversity. Proc. Natl. Acad. Sci. U. S. A. 104:11436-11440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ludwig, W., et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Madigan, M. T., J. M. Martinko, P. V. Dunlap, and D. P. Clark. 2008. Brock biology of microorganisms, 12th ed. Prentice-Hall, Upper Saddle River, NJ.
- 133.Magnuson, J. J., B. J. Benson, and T. K. Kratz. 1990. Temporal coherence in the limnology of a suite of lakes in Wisconsin, USA. Freshw. Biol. 23:145-159. [Google Scholar]
- 134.Martínez-Alonso, M., et al. 2008. Spatial heterogeneity of bacterial populations in monomictic Lake Estanya (Huesca, Spain). Microb. Ecol. 55:737-750. [DOI] [PubMed] [Google Scholar]
- 135.McDonald, J. E., A. B. de Menezes, H. E. Allison, and A. J. McCarthy. 2009. Molecular biological detection and quantification of novel Fibrobacter populations in freshwater lakes. Appl. Environ. Microbiol. 75:5148-5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Millennium Ecosystems Assessment. 2005. Ecosystems and human well-being: current state and trends assessment. Island Press, Washington, DC.
- 137.Morris, R. M., et al. 2002. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420:806-810. [DOI] [PubMed] [Google Scholar]
- 138.Morris, R. M., M. S. Rappé, E. Urbach, S. A. Connon, and S. J. Giovannoni. 2004. Prevalence of the Chloroflexi-related SAR202 bacterioplankton cluster throughout the mesopelagic zone and deep ocean. Appl. Environ. Microbiol. 70:2836-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mueller-Spitz, S. R., G. W. Goetz, and S. L. McLellan. 2009. Temporal and spatial variability in nearshore bacterioplankton communities of Lake Michigan. FEMS Microbiol. Ecol. 67:511-522. [DOI] [PubMed] [Google Scholar]
- 140.Newton, R. J., S. E. Jones, M. R. Helmus, and K. D. McMahon. 2007. Phylogenetic ecology of the freshwater Actinobacteria acI lineage. Appl. Environ. Microbiol. 73:7169-7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Newton, R. J., A. D. Kent, E. W. Triplett, and K. D. McMahon. 2006. Microbial community dynamics in a humic lake: differential persistence of common freshwater phylotypes. Environ. Microbiol. 8:956-970. [DOI] [PubMed] [Google Scholar]
- 141a.Newton, R. J., and K. D. McMahon. 2010. Seasonal differences in bacterial community composition following nutrient additions in a eutrophic lake. Environ. Microbiol. [Epub ahead of print.] doi: 10.1111/j.1462-2920.2009.01977.x. [DOI] [PubMed]
- 142.Nishimura, Y., and T. Nagata. 2007. Alphaproteobacterial dominance in a large mesotrophic lake (Lake Biwa, Japan). Aquat. Microb. Ecol. 48:231-240. [Google Scholar]
- 143.Nold, S. C., and G. Zwart. 1998. Patterns and governing forces in aquatic microbial communities. Aquat. Ecol. 32:17-35. [Google Scholar]
- 144.Oliver, R. L., and G. G. Ganf. 2000. Freshwater blooms. Kluwer Academic Publishers, Dordrecht, Netherlands.
- 145.Osborn, A. M., and C. J. Smith. 2005. Molecular microbial ecology. Taylor & Francis Group, New York, NY.
- 146.Paerl, H. W., and J. Huisman. 2008. Blooms like it hot. Science 320:57-58. [DOI] [PubMed] [Google Scholar]
- 147.Page, K. A., S. A. Connon, and S. J. Giovannoni. 2004. Representative freshwater bacterioplankton isolated from Crater Lake, Oregon. Appl. Environ. Microbiol. 70:6542-6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pearce, D. A. 2003. Bacterioplankton community structure in a maritime Antarctic oligotrophic lake during a period of holomixis, as determined by denaturing gradient gel electrophoresis (DGGE) and fluorescence in situ hybridization (FISH). Microb. Ecol. 46:92-105. [DOI] [PubMed] [Google Scholar]
- 149.Pearce, D. A., C. J. van der Gast, B. Lawley, and J. C. Ellis-Evans. 2003. Bacterioplankton community diversity in a maritime Antarctic lake, determined by culture-dependent and culture-independent techniques. FEMS Microbiol. Ecol. 45:59-70. [DOI] [PubMed] [Google Scholar]
- 150.Pearce, D. A., C. J. van der Gast, K. Woodward, and K. K. Newsham. 2005. Significant changes in the bacterioplankton community structure of a maritime Antarctic freshwater lake following nutrient enrichment. Microbiology 151:3237-3248. [DOI] [PubMed] [Google Scholar]
- 151.Percent, S. F., et al. 2008. Bacterial community structure of acid-impacted lakes: what controls diversity? Appl. Environ. Microbiol. 74:1856-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Perez, M. T., P. Hörtnagl, and R. Sommaruga. 2010. Contrasting ability to take up leucine and thymidine among freshwater bacterial groups: implications for bacterial production measurements. Environ. Microbiol. 12:74-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Perez, M. T., and R. Sommaruga. 2006. Differential effect of algal- and soil-derived dissolved organic matter on alpine lake bacterial community composition and activity. Limnol. Oceanogr. 51:2527-2537. [Google Scholar]
- 154.Pernthaler, J., and R. Amann. 2005. Fate of heterotrophic microbes in pelagic habitats: focus on populations. Microbiol. Mol. Biol. Rev. 69:440-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pernthaler, J., et al. 1998. Seasonal community and population dynamics of pelagic bacteria and archaea in a high mountain lake. Appl. Environ. Microbiol. 64:4299-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Pernthaler, J., et al. 2001. Predator-specific enrichment of actinobacteria from a cosmopolitan freshwater clade in mixed continuous culture. Appl. Environ. Microbiol. 67:2145-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Pernthaler, J., E. Zollner, F. Warnecke, and K. Jürgens. 2004. Bloom of filamentous bacteria in a mesotrophic lake: identity and potential controlling mechanism. Appl. Environ. Microbiol. 70:6272-6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Phillippot, L., et al. 2010. The ecological coherence of high bacterial taxonomic ranks. Nat. Rev. Microbiol. 8:523-529. [DOI] [PubMed] [Google Scholar]
- 159.Philosof, A., G. Sabehi, and O. Béjà. 2009. Comparative analyses of actinobacterial genomic fragments from Lake Kinneret. Environ. Microbiol. 11:3189-3200. [DOI] [PubMed] [Google Scholar]
- 160.Pinhassi, J., and T. Berman. 2003. Differential growth response of colony-forming alpha- and gamma-proteobacteria in dilution culture and nutrient addition experiments from Lake Kinneret (Israel), the eastern Mediterranean Sea, and the Gulf of Eilat. Appl. Environ. Microbiol. 69:199-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Poretsky, R. S., et al. 2009. Comparative day/night metatranscriptomic analysis of microbial communities in the North Pacific subtropical gyre. Environ. Microbiol. 11:1358-1375. [DOI] [PubMed] [Google Scholar]
- 162.Portillo, M. C., and J. M. Gonzalez. 2009. Members of the candidate division OP10 are spread in a variety of environments. World J. Microbiol. Biotechnol. 25:347-353. [Google Scholar]
- 163.Posch, T., et al. 1999. Predator-induced changes of bacterial size-structure and productivity studied on an experimental microbial community. Aquat. Microb. Ecol. 18:235-246. [Google Scholar]
- 164.Power, M. L., J. Littlefield-Wyer, D. M. Gordon, D. A. Veal, and M. B. Slade. 2005. Phenotypic and genotypic characterization of encapsulated Escherichia coli isolated from blooms in two Australian lakes. Environ. Microbiol. 7:631-640. [DOI] [PubMed] [Google Scholar]
- 165.Rabus, R., et al. 2002. Analysis of N-acetylglucosamine metabolism in the marine bacterium Pirellula sp strain 1 by a proteomic approach. Proteomics 2:649-655. [DOI] [PubMed] [Google Scholar]
- 166.Rappé, M. S., D. A. Gordon, K. L. Vergin, and S. J. Giovannoni. 1999. Phylogeny of actinobacteria small subunit (SSU) rRNA gene clones recovered from marine bacterioplankton. Syst. Appl. Microbiol. 22:106-112. [Google Scholar]
- 167.Rappé, M. S., K. Vergin, and S. J. Giovannoni. 2000. Phylogenetic comparisons of a coastal bacterioplankton community with its counterparts in open ocean and freshwater systems. FEMS Microbiol. Ecol. 33:219-232. [DOI] [PubMed] [Google Scholar]
- 168.Reynolds, C. S. 2006. Ecology of phytoplankton. Cambridge University Press, Cambridge, United Kingdom.
- 169.Reynolds, C. S. 1998. What factors influence the species composition of phytoplankton in lakes of different trophic status? Hydrobiologia 370:11-26. [Google Scholar]
- 170.Rheinheimer, G. 1980. Aquatic microbiology. Wiley, New York, NY.
- 171.Riemann, L., et al. 2008. The native bacterioplankton community in the central Baltic sea is influenced by freshwater bacterial species. Appl. Environ. Microbiol. 74:503-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 173.Rusch, D. B., et al. 2007. The Sorcerer II global ocean sampling expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol. 5:398-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Salcher, M. M., J. Pernthaler, and T. Posch. 2010. Spatiotemporal distribution and activity patterns of bacteria from three phylogenetic groups in an oligomesotrophic lake. Limnol. Oceanogr. 55:846-856. [Google Scholar]
- 175.Salcher, M. M., J. Pernthaler, R. Psenner, and T. Posch. 2005. Succession of bacterial grazing defense mechanisms against protistan predators in an experimental microbial community. Aquat. Microb. Ecol. 38:215-229. [Google Scholar]
- 176.Salcher, M. M., J. Pernthaler, M. Zeder, R. Psenner, and T. Posch. 2008. Spatio-temporal niche separation of planktonic Betaproteobacteria in an oligo-mesotrophic lake. Environ. Microbiol. 10:2074-2086. [DOI] [PubMed] [Google Scholar]
- 177.Schauer, M., and M. W. Hahn. 2005. Diversity and phylogenetic affiliations of morphologically conspicuous large filamentous bacteria occurring in the pelagic zones of a broad spectrum of freshwater habitats. Appl. Environ. Microbiol. 71:1931-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Schauer, M., C. Kamenik, and M. W. Hahn. 2005. Ecological differentiation within a cosmopolitan group of planktonic freshwater bacteria (SOL cluster, Saprospiraceae, Bacteroidetes). Appl. Environ. Microbiol. 71:5900-5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Schiaffino, M. R., et al. 2009. Comparative analysis of bacterioplankton assemblages from maritime Antarctic freshwater lakes with contrasting trophic status. Polar Biol. 32:923-936. [Google Scholar]
- 180.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Semrau, J. D., A. A. DiSpirito, and J. C. Murrell. 2008. Life in the extreme: thermoacidophilic methanotrophy. Trends Microbiol. 16:190-193. [DOI] [PubMed] [Google Scholar]
- 182.Seo, P. S., and A. Yokota. 2003. The phylogenetic relationships of cyanobacteria inferred from 16S rRNA, gyrB, rpoC1 and rpoD1 gene sequences. J. Gen. Appl. Microbiol. 49:191-203. [DOI] [PubMed] [Google Scholar]
- 183.Shade, A. L., et al. 2007. Inter-annual dynamics and phenology of bacterial communities in a eutrophic lake. Limnol. Oceanogr. 52:487-494. [Google Scholar]
- 184.Sharma, A. K., et al. 2009. Actinorhodopsin genes discovered in diverse freshwater habitats and among cultivated freshwater Actinobacteria. ISME J. 3:726-737. [DOI] [PubMed] [Google Scholar]
- 185.Sharma, A. K., O. Zhaxybayeva, R. T. Papke, and W. F. Doolittle. 2008. Actinorhodopsins: proteorhodopsin-like gene sequences found predominantly in non-marine environments. Environ. Microbiol. 10:1039-1056. [DOI] [PubMed] [Google Scholar]
- 186.Shaw, A. K., et al. 2008. It's all relative: ranking the diversity of aquatic bacterial communities. Environ. Microbiol. 10:2200-2210. [DOI] [PubMed] [Google Scholar]
- 187.Šimek, K., et al. 2005. Influence of top-down and bottom-up manipulations on the R-BT065 subcluster of beta-proteobacteria, an abundant group in bacterioplankton of a freshwater reservoir. Appl. Environ. Microbiol. 71:2381-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Šimek, K., et al. 2006. Maximum growth rates and possible life strategies of different bacterioplankton groups in relation to phosphorus availability in a freshwater reservoir. Environ. Microbiol. 8:1613-1624. [DOI] [PubMed] [Google Scholar]
- 189.Šimek, K., et al. 2008. Spatio-temporal patterns of bacterioplankton production and community composition related to phytoplankton composition and protistan bacterivory in a dam reservoir. Aquat. Microb. Ecol. 51:249-262. [Google Scholar]
- 190.Šimek, K., et al. 2010. Broad habitat range of the phylogenetically narrow R-BT065 cluster, representing a core group of the betaproteobacterial genus Limnohabitans. Appl. Environ. Microbiol. 76:631-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Šimek, K., et al. 1999. Shifts in bacterial community composition associated with different microzooplankton size fractions in a eutrophic reservoir. Limnol. Oceanogr. 44:1634-1644. [Google Scholar]
- 192.Šimek, K., et al. 2001. Changes in bacterial community composition and dynamics and viral mortality rates associated with enhanced flagellate grazing in a mesoeutrophic reservoir. Appl. Environ. Microbiol. 67:2723-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Šimek, K., et al. 2007. Grazer and virus-induced mortality of bacterioplankton accelerates development of Flectobacillus populations in a freshwater community. Environ. Microbiol. 9:789-800. [DOI] [PubMed] [Google Scholar]
- 194.Snow, L. M., and E. B. Fred. 1926. Some characteristics of the bacteria of Lake Mendota. T. Wisc. Acad. Sci. 22:143-154. [Google Scholar]
- 195.Sommaruga, R., and E. O. Casamayor. 2009. Bacterial ‘cosmopolitanism’ and importance of local environmental factors for community composition in remote high-altitude lakes. Freshw. Biol. 54:994-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Staley, J. T., J. A. M. DeBont, and K. DeJonge. 1976. Prosthecobacter fusiformis nov. gen. et sp., the fusiform caulobacter. Anton. Leeuw. Int. J. G. 42:333-342. [DOI] [PubMed] [Google Scholar]
- 197.Stamatakis, A., P. Hoover, and J. Rougemont. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 57:758-771. [DOI] [PubMed] [Google Scholar]
- 198.Stanier, R. Y., and G. Cohen-Bazire. 1977. Phototrophic prokaryotes: the cyanobacteria. Annu. Rev. Microbiol. 31:225-274. [DOI] [PubMed] [Google Scholar]
- 199.Stein, L. Y., et al. 2002. Intriguing microbial diversity associated with metal-rich particles from a freshwater reservoir. FEMS Microbiol. Ecol. 42:431-440. [DOI] [PubMed] [Google Scholar]
- 200.Stockner, J. G., C. Callieri, and G. Cronberg. 2000. Picoplankton and non-bloom forming cyanobacteria in lakes. Kluwer Academic Publishers, Dordrecht, Netherlands.
- 201.Stoeckel, D. M., and V. J. Harwood. 2007. Performance, design, and analysis in microbial source tracking studies. Appl. Environ. Microbiol. 73:2405-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Strous, M., et al. 1999. Missing lithotroph identified as new planctomycete. Nature 400:446-449. [DOI] [PubMed] [Google Scholar]
- 203.Swingley, W. D., R. E. Blankenship, and J. Raymond. 2008. Integrating Markov clustering and molecular phylogenetics to reconstruct the cyanobacterial species tree from conserved protein families. Mol. Biol. Evol. 25:643-654. [DOI] [PubMed] [Google Scholar]
- 204.Szabo, K. E., P. O. B. Itor, S. Bertilsson, L. Tranvik, and A. Eiler. 2007. Importance of rare and abundant populations for the structure and functional potential of freshwater bacterial communities. Aquat. Microb. Ecol. 47:1-10. [Google Scholar]
- 205.Taipale, S., R. I. Jones, and M. Tiirola. 2009. Vertical diversity of bacteria in an oxygen-stratified humic lake, evaluated using DNA and phospholipid analyses. Aquat. Microb. Ecol. 55:1-16. [Google Scholar]
- 206.Takeuchi, M., K. Hamana, and A. Hiraishi. 2001. Proposal of the genus Sphingomonas sensu stricto and three new genera, Sphingobium, Novosphingobium and Sphingopyxis, on the basis of phylogenetic and chemotaxonomic analyses. Int. J. Syst. Evol. Microbiol. 51:1405-1417. [DOI] [PubMed] [Google Scholar]
- 207.Tang, X. M., et al. 2009. Characterization of bacterial communities associated with organic aggregates in a large, shallow, eutrophic freshwater lake (Lake Taihu, China). Microb. Ecol. 58:307-322. [DOI] [PubMed] [Google Scholar]
- 208.Tanner, M. A., B. M. Goebel, M. A. Dojka, and N. R. Pace. 1998. Specific ribosomal DNA sequences from diverse environmental settings correlate with experimental contaminants. Appl. Environ. Microbiol. 64:3110-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Tarao, M., J. Jezbera, and M. W. Hahn. 2009. Involvement of cell surface structures in size-independent grazing resistance of freshwater Actinobacteria. Appl. Environ. Microbiol. 75:4720-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Thingstad, T. F. 2000. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol. Oceanogr. 45:1320-1328. [Google Scholar]
- 211.Trusova, M. Y., and M. I. Gladyshev. 2002. Phylogenetic diversity of winter bacterioplankton of eutrophic Siberian reservoirs as revealed by 16S rRNA gene sequences. Microb. Ecol. 44:252-259. [DOI] [PubMed] [Google Scholar]
- 212.Urbach, E., et al. 2001. Unusual bacterioplankton community structure in ultra-oligotrophic Crater Lake. Limnol. Oceanogr. 46:557-572. [Google Scholar]
- 213.Van der Gucht, K., et al. 2001. Contrasting bacterioplankton community composition and seasonal dynamics in two neighbouring hypertrophic freshwater lakes. Environ. Microbiol. 3:680-690. [DOI] [PubMed] [Google Scholar]
- 214.Van der Gucht, K., et al. 2005. Characterization of bacterial communities in four freshwater lakes differing in nutrient load and food web structure. FEMS Microbiol. Ecol. 53:205-220. [DOI] [PubMed] [Google Scholar]
- 215.Vannini, C., G. Petroni, F. Verni, and G. Rosati. 2005. Polynucleobacter bacteria in the brackish-water species Euplotes harpa (Ciliata hypotrichia). J. Eukaryot. Microbiol. 52:116-122. [DOI] [PubMed] [Google Scholar]
- 216.Vannini, C., et al. 2007. Endosymbiosis in statu nascendi: close phylogenetic relationship between obligately endosymbiotic and obligately free-living Polynucleobacter strains (Betaproteobacteria). Environ. Microbiol. 9:347-359. [DOI] [PubMed] [Google Scholar]
- 217.van Veen, W. L., D. van der Kooij, E. C. W. A. Geuze, and A. W. van der Vlies. 1973. Investigations on the sheathed bacterium Haliscomenobacter hydrossis gen. n., sp. n., isolated from activated sludge. Antonie Van Leeuwenhoek 39:207-216. [DOI] [PubMed] [Google Scholar]
- 218.Ventura, M., et al. 2007. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylura. Microbiol. Mol. Biol. Rev. 71:495-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Vergin, K. L., et al. 1998. Screening of a fosmid library of marine environmental genomic DNA fragments reveals four clones related to members of the order Planctomycetales. Appl. Environ. Microbiol. 64:3075-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Wagner, M., and M. Horn. 2006. The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr. Opin. Biotechnol. 17:241-249. [DOI] [PubMed] [Google Scholar]
- 221.Walk, S. T., et al. 2009. Cryptic lineages of the genus Escherichia. Appl. Environ. Microbiol. 75:6534-6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Wang, Q., G. M. Garrity, J. M. Tiedje, and J. R. Cole. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Warnecke, F., R. Amann, and J. Pernthaler. 2004. Actinobacterial 16S rRNA genes from freshwater habitats cluster in four distinct lineages. Environ. Microbiol. 6:242-253. [DOI] [PubMed] [Google Scholar]
- 224.Warnecke, F., R. Sommaruga, R. Sekar, J. S. Hofer, and J. Pernthaler. 2005. Abundances, identity, and growth state of actinobacteria in mountain lakes of different UV transparency. Appl. Environ. Microbiol. 71:5551-5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Watanabe, K., N. Komatsu, Y. Ishii, and M. Negishi. 2009. Effective isolation of bacterioplankton genus Polynucleobacter from freshwater environments grown on photochemically degraded dissolved organic matter. FEMS Microbiol. Ecol. 67:57-68. [DOI] [PubMed] [Google Scholar]
- 226.Weinbauer, M. G., et al. 2007. Synergistic and antagonistic effects of viral lysis and protistan grazing on bacterial biomass, production and diversity. Environ. Microbiol. 9:777-788. [DOI] [PubMed] [Google Scholar]
- 227.Weiss, P., B. Schweitzer, R. Amann, and M. Simon. 1996. Identification in situ and dynamics of bacteria on limnetic organic aggregates (lake snow). Appl. Environ. Microbiol. 62:1998-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Williamson, C. E., W. Dodds, T. K. Kratz, and M. A. Palmer. 2008. Lakes and streams as sentinels of environmental change in terrestrial and atmospheric processes. Front. Ecol. Environ. 6:247-254. [Google Scholar]
- 229.Wu, Q. L., and M. W. Hahn. 2006. Differences in structure and dynamics of Polynucleobacter communities in a temperate and a subtropical lake, revealed at three phylogenetic levels. FEMS Microbiol. Ecol. 57:67-79. [DOI] [PubMed] [Google Scholar]
- 230.Wu, Q. L., G. Zwart, M. Schauer, M. P. Kamst-van Agterveld, and M. W. Hahn. 2006. Bacterioplankton community composition along a salinity gradient of sixteen high-mountain lakes located on the Tibetan Plateau, China. Appl. Environ. Microbiol. 72:5478-5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Wu, Q. L., et al. 2007. Submersed macrophytes play a key role in structuring bacterioplankton community composition in the large, shallow, subtropical Taihu Lake, China. Environ. Microbiol. 9:2765-2774. [DOI] [PubMed] [Google Scholar]
- 232.Wu, Q. L. L., and M. W. Hahn. 2006. High predictability of the seasonal dynamics of a species-like Polynucleobacter population in a freshwater lake. Environ. Microbiol. 8:1660-1666. [DOI] [PubMed] [Google Scholar]
- 233.Wu, X., W. Y. Xi, W. J. Ye, and H. Yang. 2007. Bacterial community composition of a shallow hypertrophic freshwater lake in China, revealed by 16S rRNA gene sequences. FEMS Microbiol. Ecol. 61:85-96. [DOI] [PubMed] [Google Scholar]
- 234.Yannarell, A. C., and E. W. Triplett. 2005. Geographic and environmental sources of variation in lake bacterial community composition. Appl. Environ. Microbiol. 71:227-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Yannarell, A. C., and E. W. Triplett. 2004. Within- and between-lake variability in the composition of bacterioplankton communities: investigations using multiple spatial scales. Appl. Environ. Microbiol. 70:214-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Zavarzin, G. A., E. Stackebrandt, and R. G. E. Murray. 1991. A correlation of phylogenetic diversity in the Proteobacteria with the influences of ecological forces. Can. J. Microbiol. 37:1-6. [DOI] [PubMed] [Google Scholar]
- 237.Zeder, M., S. Peter, T. Shabarova, and J. Pernthaler. 2009. A small population of planktonic Flavobacteria with disproportionally high growth during the spring phytoplankton bloom in a prealpine lake. Environ. Microbiol. 11:2676-2686. [DOI] [PubMed] [Google Scholar]
- 238.Zwart, G., B. C. Crump, M. P. K. V. Agterveld, F. Hagen, and S. K. Han. 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 28:141-155. [Google Scholar]
- 239.Zwart, G., et al. 1998. Nearly identical 16S rRNA sequences recovered from lakes in North America and Europe indicate the existence of clades of globally distributed freshwater bacteria. Syst. Appl. Microbiol. 21:546-556. [DOI] [PubMed] [Google Scholar]
- 240.Zwart, G., et al. 1998. Divergent members of the bacterial division Verrucomicrobiales in a temperate freshwater lake. FEMS Microbiol. Ecol. 25:159-169. [Google Scholar]
- 241.Zwart, G., et al. 2005. Molecular characterization of cyanobacterial diversity in a shallow eutrophic lake. Environ. Microbiol. 7:365-377. [DOI] [PubMed] [Google Scholar]
- 242.Zwart, G., et al. 2003. Rapid screening for freshwater bacterial groups by using reverse line blot hybridization. Appl. Environ. Microbiol. 69:5875-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Zwisler, W., N. Selje, and M. Simon. 2003. Seasonal patterns of the bacterioplankton community composition in a large mesotrophic lake. Aquat. Microb. Ecol. 31:211-225. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.