Abstract
Summary: Deinococcus radiodurans is a robust bacterium best known for its capacity to repair massive DNA damage efficiently and accurately. It is extremely resistant to many DNA-damaging agents, including ionizing radiation and UV radiation (100 to 295 nm), desiccation, and mitomycin C, which induce oxidative damage not only to DNA but also to all cellular macromolecules via the production of reactive oxygen species. The extreme resilience of D. radiodurans to oxidative stress is imparted synergistically by an efficient protection of proteins against oxidative stress and an efficient DNA repair mechanism, enhanced by functional redundancies in both systems. D. radiodurans assets for the prevention of and recovery from oxidative stress are extensively reviewed here. Radiation- and desiccation-resistant bacteria such as D. radiodurans have substantially lower protein oxidation levels than do sensitive bacteria but have similar yields of DNA double-strand breaks. These findings challenge the concept of DNA as the primary target of radiation toxicity while advancing protein damage, and the protection of proteins against oxidative damage, as a new paradigm of radiation toxicity and survival. The protection of DNA repair and other proteins against oxidative damage is imparted by enzymatic and nonenzymatic antioxidant defense systems dominated by divalent manganese complexes. Given that oxidative stress caused by the accumulation of reactive oxygen species is associated with aging and cancer, a comprehensive outlook on D. radiodurans strategies of combating oxidative stress may open new avenues for antiaging and anticancer treatments. The study of the antioxidation protection in D. radiodurans is therefore of considerable potential interest for medicine and public health.
INTRODUCTION
Deinococcus radiodurans is unparalleled among all known species in its capacity to overcome oxidative stress that affects all cellular macromolecules (62, 108, 120). In humans, the oxidative modification of cellular macromolecules underlies a variety of degenerative diseases, cancer, and aging. Oxidative stress is incurred by reactive oxygen species (ROS), which can be produced metabolically or can form upon exposure to physical and chemical agents such as desiccation (495), ionizing radiation (114), UV radiation (280), mitomycin C (MMC) (376), or hydrogen peroxide (268). D. radiodurans displays remarkable resistance to all ROS-generating agents (37). ROS generated by desiccation and ionizing radiation damage proteins, lipids, nucleic acids, and carbohydrates and induce potentially lethal double-strand DNA breaks (DSBs) in the bacterial genome. D. radiodurans can survive high doses of ionizing radiation, which break its genome into several hundred fragments (up to 2,000 DSBs per multigenomic cell) without causing considerable protein damage (79, 122, 135, 307). The robustness of this bacterium is due to strong oxidative stress resistance mechanisms that protect proteins from oxidative damage (122) and a DNA repair process that accomplishes an efficient and precise reassembly of DNA fragments (572, 676). The antioxidation protection of DNA repair and other proteins enables them to retain their catalytic activity and to provide a swift response under conditions of oxidative stress.
Traditionally, DNA has been considered the primary radiation target (264). Modern research in the D. radiodurans field, however, has shown that this bacterium is as susceptible to radiation-induced DSBs as all other species (206), whereas its proteome is better protected against ROS-induced oxidative damage than radiation-sensitive species (122). These findings suggest that the level of protein damage, and not DNA damage, together with the cellular ROS-scavenging capacity determine the radiation survival of bacteria (120, 122). In D. radiodurans, manganese complexes were recently identified as the most powerful ROS scavengers (121). Their ability to protect human cell lines from radiation-induced death opens several potential applications in the fields of medicine and biotechnology (121). Here we give a comprehensive outlook on D. radiodurans strategies of combating oxidative stress, and we discuss how antioxidation protection pathways in D. radiodurans could provide means for delaying aging and preventing cancer.
GENERAL PROPERTIES OF DEINOCOCCUS RADIODURANS
Common Features of a “Strange” Bacterium
Deinococcus radiodurans (from the Greek deinos, meaning strange or unusual, and coccus, meaning a grain or berry) is a Gram-positive, red-pigmented, nonsporulating, nonpathogenic bacterium occurring in diads and tetrads with an average cell diameter of 1 μm (range, 0.5 to 3.5 μm) (452). It is a mesophile with a thermal limitation above 39°C. D. radiodurans is conventionally grown at 32°C in rich TGY medium (0.5% tryptone, 0.1% glucose, 0.15% yeast extract) with aeration, where cell doubling takes approximately 100 min and colonies require 3 days for adequate development. A high cell density for large-scale production is accomplished at 37°C with HEPES-buffered (pH 7) TGY medium (1% tryptone, 1% glucose, 0.5% yeast extract) supplemented with magnesium and manganese (246). D. radiodurans is sensitive to antibiotics inhibiting RNA synthesis (e.g., actinomycin D), protein synthesis (e.g., chloramphenicol, streptomycin, neomycin, kanamycin, and erythromycin), and cell wall synthesis (e.g., penicillin, bacitracin, and vancomycin) (245).
The 3.28-megabase genome of D. radiodurans consists of two chromosomes of 2,648,638 and 412,348 bp and two plasmids of 177,466 and 45,704 bp (653). The genome contains 3,187 open reading frames (ORFs) (653) and has a high GC content of 66.6% (447, 653). Although D. radiodurans DNA has been considered methyl deficient, lacking methylated bases and DNA methyltransferase activity (Dam− Dcm−) (540), Prasad et al. (497) detected N6-methyladenine in the D. radiodurans genome as well as adenine methyltransferase activity in the cell extract. The average DNA content of D. radiodurans in exponentially grown cells is 3.4 × 107 bases, with each cell containing up to 10 copies of the genome (155). The genome copy number depends on the growth stage (234) and the culture medium (241). Whereas an older report claimed that cells grown in TGY medium have a lower genome copy number than cells grown in minimal medium (241), a more recent analysis showed that the DNA content is approximately 2-fold lower in cells grown in minimal medium (J. R. Battista, unpublished data). The minimum copy number of 2 is characteristic of stationary-phase cells (234). Although there is no correlation between the genome copy number and radiation resistance (241), 2 genome copies are always available as a substrate for recombinational repair.
Ecology of D. radiodurans
D. radiodurans was first isolated from gamma-irradiated canned meat in Oregon by Anderson et al. (14) and was named Micrococcus radiodurans on the basis of its morphological and physiological characteristics (502). A morphologically similar M. radiodurans strain, strain Sark, was later isolated as an air contaminant in a hospital in Ontario, Canada (R. G. E. Murray and C. F. Robinow, presented at the Seventh International Congress of Microbiology, 1958). However, 16S rRNA analysis indicated that M. radiodurans forms a unique phylogenetic group of bacteria (584); hence, it was included in a new family, the Deinococcaceae, and renamed Deinococcus radiodurans (76). Forty-three Deinococcus species have been isolated to date, often by radiation selection (Table 1), although desiccation can also serve as a selective feature for isolation (533). The Deinococcus lineage comprises mesophilic, thermophilic, and psychrophilic representatives, which can be found in a variety of habitats, such as animal gut, hot springs, deserts, alpine environments, and Antarctica (Table 1). Unlike bacterial (e.g., Bacillus subtilis) spores, dried D. radiodurans does not tolerate high humidity (150, 151). All Deinococcus species are distinguished by their extraordinary ability to tolerate the lethal effects of DNA-damaging agents, particularly those of ionizing radiation and UV radiation (37).
TABLE 1.
Deinococcus species isolated to datea
| Deinococcus species (reference[s]) | D10 (kGy) (reference[s]) | Site(s) of isolation |
|---|---|---|
| Deinococcus radiodurans (14, 97, 271, 325, 330) | 12.7 (561) | Gamma-irradiated canned meat |
| 16 (123) | Ground beef and pork, the hides and hair of live beef, creeks | |
| Sawdust | ||
| Air of clean rooms and laboratories of an institute | ||
| Clothes of people working in an institute | ||
| Deinococcus radiopugnans (133) | 5.3 (133) | Haddock tissue |
| Deinococcus radiophilus (1, 107, 355) | >16 (355) | Mumbai duck in India |
| Weathered granite in Antarctica | ||
| Hypersaline oil-polluted microbial mat in Saudi Arabia | ||
| Deinococcus proteolyticus (311) | 10.3 (561) | Feces of a llama |
| Deinococcus grandis (89, 478) | 11 (89, 478) | Feces of an elephant |
| 7 (561) | Tataouine desert | |
| Deinococcus geothermalis (187, 299, 389, 527) | 5.1 (187) | Hot spring in Italy and Portugal |
| 10 (37°C) (123) | Subterranean hot springs in Iceland | |
| 16 (50°C) (383) | Hydrothermal vents in Papua New Guinea | |
| Industrial paper machine water | ||
| Deinococcus murrayi (187) | 9.1 (187) | Hot springs in Portugal |
| Deinococcus indicus (595) | 4.2 (561) | Groundwater in India |
| Deinococcus frigens (254) | NA | Antarctic soil |
| Deinococcus saxicola (254) | NA | Antarctic sandstone |
| Deinococcus marmoris (254, 591) | NA | Antarctic marble |
| Surfaces of historic Scottish monuments | ||
| Deinococcus hohokamensis (501) | ND | Sonoran desert soil |
| Deinococcus navajonensis (501) | ND | |
| Deinococcus hopiensis (501) | ND | |
| Deinococcus apachensis (501) | ND | |
| Deinococcus maricopensis (501) | ND | |
| Deinococcus pimensis (501) | ND | |
| Deinococcus yavapaiensis (501) | ND | |
| Deinococcus papagonensis (501) | ND | |
| Deinococcus sonorensis (501) | ND | |
| Deinococcus deserti (139) | >7.5 (139) | Sahara desert sand |
| Deinococcus ficus (334) | 11 (561) | Rhizosphere of Ficus religiosa |
| Deinococcus mumbaiensis (559) | 17 (559) | Contaminated agar plate in India |
| Deinococcus peraridilitoris (500) | ND | Coastal desert in Chile |
| Deinococcus radiomollis (82) | 2.2 (82) | Alpine environments |
| Deinococcus claudionis (82) | 3.6 (82) | |
| Deinococcus altitudinis (82) | 3.8 (82) | |
| Deinococcus alpinitundrae (82) | 4 (82) | |
| Deinococcus aquaticus (265) | NA | Freshwater in South Korea |
| Deinococcus caeni (265) | NA | Activated sludge in South Korea |
| Deinococcus aquatilis (284) | NA | Water in Germany |
| Deinococcus aquiradiocola (27) | ND | Radioactive site in Japan |
| Deinococcus xinjiangensis (487) | ND | Desert soil in China |
| Deinococcus gobiensis (675) | 12.7 (675) | Gobi desert |
| Deinococcus aerius (669) | 4.9 (669) | High atmosphere in Japan |
| Deinococcus piscis (560) | 7.4 (560) | Marine fish |
| Deinococcus aetherius (670) | >8 (670) | Stratosphere in Japan |
| Deinococcus aerolatus (673) | NA | Air in South Korea |
| Deinococcus aerophilus (673) | NA | Air in South Korea |
| Deinococcus wulumuquiensis (647) | ND | Radiation-polluted soil in China |
| Deinococcus xibeiensis (647) | ND | Radiation-polluted soil in China |
| Deinococcus guangriensis (594) | 9.8 (594) | Radiation Centre of Guangxi University, Nanning, China |
| Deinococcus depolymerans (28) | ND | Radioactive freshwater site in Japan |
D10 is the gamma radiation dose that yields a 10% survival rate. ND denotes a radiation-resistant species for which the D10 has not been determined. NA refers to species that have not been characterized with regard to radiation resistance.
Radiation resistance is dispersed among the three kingdoms of life and is not in correlation with the prevalence of these organisms and the characteristics of their habitats (108). However, a generalization can be made for bacteria with respect to Gram staining: most of the radiation-resistant bacteria reported are Gram positive, with the exception of a radiation-resistant Gram-negative cyanobacterium, Chroococcidiopsis (gamma radiation dose that yields a 10% survival rate [D10] of 5 kGy) (57), while most of the radiation-sensitive bacteria are Gram negative, with one of the exceptions being the radiation-sensitive Gram positive Micrococcus luteus (Sarcina lutea) (409). Other radiation-resistant bacteria unrelated to members of the Deinococcaceae include Rubrobacter radiotolerans (674), Rubrobacter xylanophilus (186), Kocuria rosea (76), Methylobacterium radiotolerans (214), Lactobacillus plantarum (244), Acinetobacter radioresistens (461), Enterococcus faecium (634), Hymenobacter actinosclerus (103), and Kineococcus radiotolerans (492). Radiation resistance is widespread among hyperthermophilic archaea, for example, Pyrococcus furiosus (145), Desulfurococcus amylolyticus, Thermococcus stetteri (315), Thermococcus gammatolerans (276), and Halobacterium (321). Radioresistance is also shared by some eukaryotes: a green alga, Dunaliella bardawil (46); a slime mold, Dictyostelium discoideum (138); fungi such as Ustilago maydis (341), Cryptococcus neoformans (113), Curvularia geniculata, and Alternaria alternata (528); tardigrades such as Milnesium tardigradum (258); and bdelloid rotifers such as Adineta vaga and Philodina roseola (211).
D. radiodurans Physical Structure and Cell Division
Although D. radiodurans is Gram positive, the cell envelope is reminiscent of Gram-negative bacteria due to its multilayered structure (335, 615) and lipid composition (310, 661). Gram-positive cells usually have a cytoplasmic membrane and a mucopeptide- or peptidoglycan-containing layer, whereas Gram-negative bacteria also have an outer membrane that contains lipopolysaccharide. The D. radiodurans cell envelope consists of at least five layers with a total thickness of 150 nm: (i) the cytoplasmic membrane, (ii) the rigid peptidoglycan-containing holey layer, (iii) the compartmentalized layer, (iv) the interior layer, and (v) the fragile soft layer, containing hexagonally packed subunits (S layer) (335, 662). The holey layer is composed of a mucopeptide containing glucosamine, muramic acid, and four main amino acids (glutamic acid, alanine, glycine, and lornithine) (662). The diamino acid l-ornithine occurs very rarely in bacterial cell walls (661). The hexagonally packed layer contains carotenoids, lipids, proteins, and polysaccharides. The polysaccharide contains galactose and glucose, with traces of rhamnose and mannose, but not heptose. Forty-three percent of membrane lipids are composed of phosphoglycolipids that contain alkylamines as structural components, which are considered unique to D. radiodurans (17). Common bacterial phospholipids, such as phosphatidylethanolamine, phosphatidylserine, phosphatidylcholine, and phosphatidylinositol, are absent (613). Lipoproteins contain mainly even-numbered, straight-chain, saturated fatty acids, with palmitoleate being predominant (210), but also a significant amount of odd-numbered saturated fatty acids and monounsaturated fatty acids, which are not common components of bacteria (310). Unsaturated fatty acids, which increase the fluidity of cell membranes, may accommodate significant changes in volume experienced during desiccation. Polyunsaturated, cyclopropyl, and branched-chain fatty acids are not detectable.
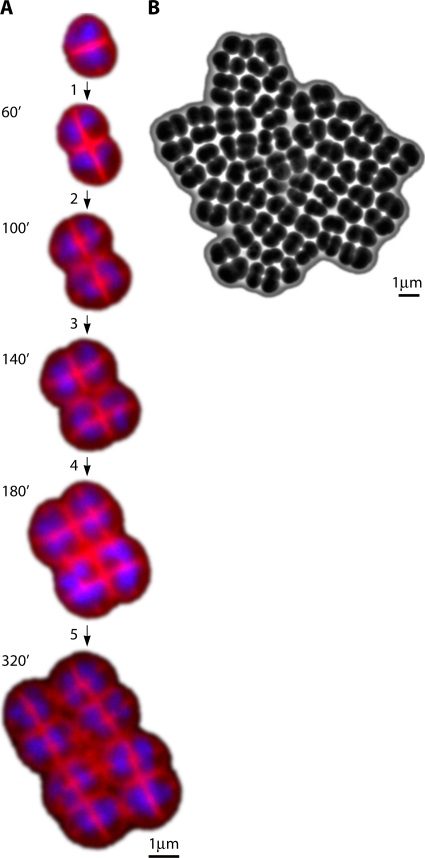
D. radiodurans cells divide alternately in two planes (453) (Fig. 1, and see Video S1 in the supplemental material). When the subsequent division begins, the next cross wall is formed perpendicular to the previous one (615; Murray and Robinow, presented at the Seventh International Congress of Microbiology, 1958). The cell division of D. radiodurans is unlike that of other cocci, because two septa sweep across the cell from opposite sides to form a slit closure (453). The new septation begins before the septal curtains meet such that rapidly dividing cells may have four communicating compartments (452). Figure 1A shows the development of a tetrad, with newly forming septa invaginating into two contiguous cells and with nuclear material in the process of separating into the daughter cells (662). Only the cytoplasmic membrane and the peptidoglycan layer are involved in septum formation during cell division. During division, when the cross wall is complete, it splits into two layers, and sheath material is laid between the layers (615). Microcolonies of D. radiodurans form a rectangular array during growth (156) (Fig. 1). This ordered growth pattern is maintained for at least five generations.
FIG. 1.
D. radiodurans cell division on TGY agar at 30°C. (A) Cell division monitored by time-lapse fluorescent microscopy starting from a single diad (see Video S1 in the supplemental material). Images were obtained with a fluorescent microscope (Zeiss Axiovert 200 M). Membranes were stained with FM 4-64 (red), and nucleoids were stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). Septa are formed perpendicular to the cross wall, separating the two cells in a diad (step 1). Septation creates two new cross walls, and two diads are formed as a result. Perpendicular to the previously formed cross walls, new septa invaginate into four cells with nuclear material in the process of separating into daughter cells (step 2). This stage of cell division represents two tetrads. Complete septation results in four new cross walls that give rise to four diads (step 3). New septa invaginate perpendicular to the previously formed cross walls, giving rise to four tetrads (step 4). The following round of cell division produces eight tetrads (step 5). (B) Phase contrast-image of a microcolony obtained from a single tetrad.
Metabolic Configuration of D. radiodurans
D. radiodurans is an organotrophic bacterium with a proteolytic life-style (383, 452). The productive growth of D. radiodurans is accomplished with the following minimal requirements: a carbon source, nicotinic acid, a sulfur source, a nitrogen source, and a source of manganese (Mn). Amino acids are a preferred primary carbon energy source (246), while carbohydrates are preferred in the following order: fructose > pyruvate > lactate > glucose > oxaloacetate > glycerol (637). Carbohydrates are presumably imported via a phosphoenolpyruvate phosphotransferase system encoded on the megaplasmid (653). D. radiodurans is dependent on exogenous nicotinic acid because it lacks key enzymes for NAD biosynthesis (637). When a morpholinepropanesulfonic acid (MOPS) defined medium is used, biotin is required in addition to nicotinic acid (256). Its methionine auxotrophy (557, 637) can be alleviated with vitamin B12, which is required as a cofactor for methionine synthase (256). In the presence of vitamin B12, sulfate can be used as the sole sulfur source (256). Growth on ammonium as the sole nitrogen source in the presence of vitamin B12 has been demonstrated, although it was not possible to achieve reproducibly good growth in the absence of at least one amino acid as a nitrogen source (256). Serine together with glutamate or glutamine supported the best growth (256). It appears that there is a defect in the assimilation of ammonia, although glutamine synthetase (glnA), which assimilates ammonia into glutamate, and glutamate synthase genes (gltB and gltD) are harbored in the genome (637, 653). Enzymes involved in the production of ammonia, such as xanthine permease and xanthine dehydrogenase, which produce urate, and ATP-binding cassette (ABC) transporters with specificity for urea also appear in the genome (653).
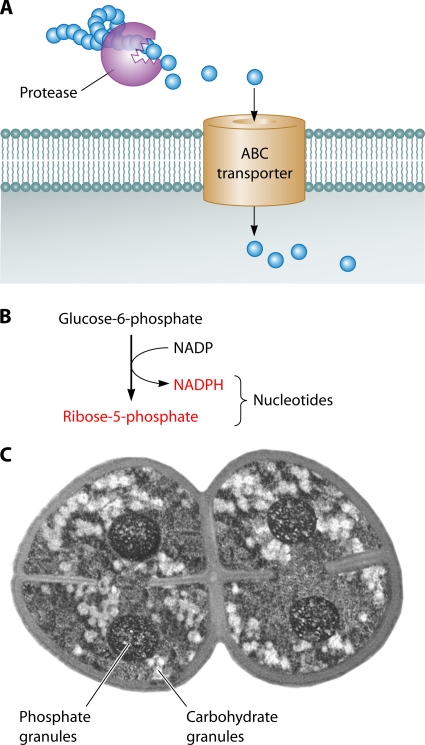
Several metabolic properties help D. radiodurans to surmount oxidative stress: (i) proteolysis and the import of exogenous peptides and amino acids, (ii) the conversion of glucose into precursors for deoxynucleoside triphosphates (dNTPs), (iii) the suppression of ROS production by the induction of the glyoxylate bypass of the tricarboxylic acid (TCA) cycle and a reduction in the number of respiratory chain enzymes and enzymes with iron-sulfur clusters, (iv) metabolic defects resulting in metabolite accumulation, and (v) carbohydrate and polyphosphate storage granules (Fig. 2) (discussed in detail below).
FIG. 2.
Sources of energy and building blocks in oxidatively stressed D. radiodurans. (A) Extracellular proteolysis. D. radiodurans contains many secreted proteases and ABC transporters, which provide exogenous amino acids as protein building blocks and peptides as components of manganese complexes. (B) The pentose phosphate pathway (PPP) for the conversion of glucose into dNTPs. Glucose is essential for recovery from oxidative stress; glucose-6-phosphate dehydrogenase (the marker enzyme of PPP) converts glucose into the precursors of dNTPs as DNA building blocks and, possibly, components of Mn complexes. (C) Carbohydrate and polyphosphate granules. Shown is a schematic representation of a D. radiodurans tetrad viewed by electron microscopy (based on data from reference 615 and unpublished data from J. R. Battista). The small light granules contain carbohydrates, while the large dark circles represent polyphosphate granules. Both can be used as sources of energy, while polyphosphates also serve as a phosphate source for the synthesis of nucleic acids and, presumably, for manganese-phosphate complexes.
D. radiodurans derives its energy mainly from proteolysis (i.e., amino acids and peptides). The biomass formation during growth in TGY medium is supported largely by tryptone and yeast extract (i.e., amino acids), with only a marginal contribution from glucose in the later stages of growth (246, 683). D. radiodurans displays a high level of proteolytic activity when grown on plates with skimmed milk, forming large halo-shaped areas of clearing around colonies (207). As a proteolytic bacterium, D. radiodurans encodes systems for protein degradation and amino acid catabolism, most of which were acquired by horizontal gene transfer (HGT) (475). Intracellular proteolytic activity is induced following ionizing radiation (121). The increase in proteolytic activity may expedite the recovery process by providing amino acids and peptides from the degraded proteins or proteins from the cells that did not survive the stress conditions. Protein recycling is expected to (i) minimize biosynthetic demands (207, 368) and (ii) contribute to the antioxidant complexes of amino acids and peptides with manganese (121) (see “Manganese complexes”). D. radiodurans encodes 3 hemolysin genes, 10 secreted subtilisin-like proteases, and 4 peptide and amino acid ABC transporters that are highly induced after irradiation (207, 368, 380, 653) (Fig. 2A). Energy import systems are particularly pronounced in Deinococcus deserti, which encodes 90 ABC transporters for amino acids and peptides and 54 ABC transporters for sugars (140). The expansion of ABC transporters in D. deserti is presumably a mechanism of adaptation to a nutrient-poor desert environment (140).
Although D. radiodurans is largely proteolytic, glucose metabolism is important for recovery from DNA-damaging agents (683). In D. radiodurans, carbohydrates are stored in granules (615), appearing as light areas in electron micrographs, which give a positive reaction with periodic acid-Schiff (PAS) stain (Murray and Robinow, presented at the Seventh International Congress of Microbiology, 1958) and with Thiery reagent (574) (Fig. 2C). D. radiodurans has different pathways for the utilization of glucose: glycolysis, gluconeogenesis, the pentose phosphate pathway (PPP), the tricarboxylic acid cycle, and glyoxylate bypass (381) (Table 2). The PPP is highly active in D. radiodurans (502), with the activity of glucose-6-phosphate dehydrogenase (G6PDH) (the marker enzyme for PPP) being 4-fold higher than that in Escherichia coli (683). G6PDH is upregulated in gamma-irradiated cells (677), and mutants defective in G6PDH are sensitive to UV light, hydrogen peroxide (H2O2), and MMC (367, 682). This pathway is employed for metabolizing glucose in TGY-grown cells into ribose-5-phosphate, glyceraldehyde-3-phosphate, and NADPH, which are all precursors for dNTPs (Fig. 2B). NADPH is additionally important as a cofactor for glutathione and thioredoxin reductases, which regenerate reduced glutathione and thioredoxin (230). Glutathione and thioredoxin are antioxidants, which reduce hydrogen peroxide and oxidized cysteines. By providing substrates for DNA repair and for protection against ROS toxicity, glucose enhances resistance to UV radiation (683), whereas glucose depletion results in an increased susceptibility to the lethal effects of UV and ionizing radiation, MMC, and N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) (96, 362). D. radiodurans normally incorporates 8% of the transported 14C from the glucose into its DNA, which increases to 18% after UV irradiation (683). When glucose is depleted from the growth medium by the addition of a high concentration of manganese (100 μM MnCl2), the incorporation of 14C from glucose into DNA is reduced to 4% and 6% before and after UV irradiation, respectively (683). The addition of Mn to stationary-phase cultures results in a rapid disappearance of glucose from the growth medium without incorporation into biomass due to the induction of the Embden-Meyerhof-Parnas (EMP) pathway (as measured by the activity of the marker enzyme d-fructose 1,6-biphosphate aldolase [ALD]), which oxidizes glucose into CO2 (683). Massive glucose oxidation in Mn-accumulating cells may create oxidative stress and explain the higher catalase and superoxide dismutase (SOD) activities after exposure to high levels of Mn as well as the higher sensitivity to DNA-damaging agents (96).
TABLE 2.
Metabolic pathways and genes important for the recovery of D. radiodurans from ionizing radiation, their transcriptional regulators, and their expression levelsa
| Pathway and enzyme | Locus tag | Regulation | PHXb | Phase(s) of constitutive expression |
Type(s) of stressc | Induced after IR stressd |
||
|---|---|---|---|---|---|---|---|---|
| DM | RM | Gene | Protein | |||||
| Glycolysis and gluconeogenesis | ||||||||
| Glucose kinase (Glk) | DR2296 | E | − | C | + | |||
| Glucose-6-phosphate isomerase (Pgi) | DR1742 | ES | ES | OHSAC | − | |||
| 6-Phosphofructokinase (PfkA) | DR0635 | S | ES | O | + | |||
| Fructose-bisphosphate aldolase (FbaA) | DR1589 | + | ES | ES | OHSAC | − | ||
| Triosephosphate isomerase (Tpi) | DR1339 | IrrE | ES | ES | OHSAC | − | ||
| Glyceraldehyde 3-phosphate dehydrogenase (GapA) | DR1343 | IrrE | + | ES | ES | OHSAC | − | + |
| Phosphoglycerate kinase (Pgk) | DR1342 | ES | ES | OHSAC | − | |||
| Phosphoglucomutase (Pgm) | DR0303 | |||||||
| Enolase (Eno) | DR2637 | + | ES | ES | OHSAC | − | ||
| Pyruvate kinase (PykA) | DR2635 | ES | ES | OHSAC | − | |||
| Phosphoenolpyruvate carboxykinase (PckA) | DR0977 | + | ES | ES | OHSAC | − | ||
| Phosphoenolpyruvate synthase (PpsA) | DRC0002 | S | − | C | − | |||
| Fructose 1,6-bisphosphatase (GlpX) | DR2013 | |||||||
| Pentose phosphate and Entner-Doudoroff | ||||||||
| Glucose-6-phosphate dehydrogenase (Zwf) | DR1596 | ES | ES | OSAC | − | + | ||
| 6-Phosphogluconate dehydrogenase (Gnd) | DR1595 | ES | ES | OHSAC | − | |||
| Transketolase (TktA) | DR2256 | DdrO? | ES | ES | OHSAC | + | ||
| Transaldolase (TalA) | DR1337 | ES | ES | OHSA | − | |||
| Ribulose-phosphate 3-epimerase (YhfD) | DR1401 | E | − | SAC | − | |||
| Ribose 5-phosphate isomerase (RpiA) | DR0845 | ES | ES | OHSAC | − | |||
| Deoxyribose-phosphate aldolase (DeoC) | DR1205 | ES | ES | OHSAC | − | |||
| Dihydroxy acid dehydratase (Edd) | DR1132 | ES | E | OHSAC | − | |||
| TCA cycle | ||||||||
| Citrate synthase (GltA) | DR0757 | IrrE | + | ES | ES | OHSAC | − | + |
| Aconitase (AcnA) | DR1720 | IrrE | + | ES | ES | OHSAC | + | + |
| Isocitrate dehydrogenase (Icd) | DR1540 | + | ES | ES | OHSAC | − | ||
| 2-Oxoglutarate dehydrogenase (SucA) | DR0287 | + | ES | ES | OHSAC | − | ||
| Dihydrolipoyltranssuccinylase (SucB) | DR0083 | + | ES | ES | OHSAC | − | ||
| Succinyl-CoA synthetase (SucC) | DR1247 | + | ES | ES | OHSAC | + | ||
| Succinyl-CoA ligase (SucD) | DR1248 | + | ES | ES | OHSAC | + | ||
| Succinate dehydrogenase | ||||||||
| SdhA | DR0952 | + | ES | ES | OHSAC | − | ||
| SdhB | DR0951 | + | ES | ES | OHSAC | − | ||
| SdhD | DR0953 | + | − | |||||
| Fumarase (FumC) | DR2627 | + | ES | ES | OHSAC | − | ||
| Malate dehydrogenase (Mdh) | DR0325 | + | ES | ES | OHSAC | + | + | |
| Glyoxylate bypass | ||||||||
| Malate synthase (GlcB) | DR1155 | − | E | H | + | |||
| DRA0277 | ES | ES | OHSAC | − | ||||
| Isocitrate lyase (AceA) | DR0828 | + | ES | ES | OHSAC | + | ||
| ATP hydrolysis | ||||||||
| V-type ATP synthase subunit | ||||||||
| A | DR0700 | IrrE | + | ES | ES | OHSAC | + | |
| B | DR0701 | + | ES | ES | OHSAC | + | ||
| C | DR0698 | ES | ES | OHSAC | ++ | + | ||
| D | DR0702 | + | ES | ES | OHSC | − | ||
| E | DR0697 | IrrE | + | ES | ES | OHSA | ++ | |
| F | DR0699 | S | ES | HC | + | |||
| I | DR0695 | + | ES | ES | OHSAC | + | ||
| K | DR0696 | + | S | − | OHS | ++ | ||
| Sulfur metabolism | ||||||||
| Ferredoxin-nitrite reductase (CysI) | DRA0013 | ES | ES | OHSAC | ++ | |||
| Adenylylsulfate kinase (Ask) | DRA0014 | ES | E | OHSAC | + | |||
| Phosphoadenosine phosphosulfate reductase (PAPS reductase) | DRA0015 | S | − | S | ++ | |||
| Sulfate adenylyltransferase (Sat) | DRA0016 | ES | ES | OHSC | ++ | |||
| Thioesterase | DRA0017 | OxyR | ES | ES | OHS | + | ||
Shown are data for the constitutive expression of proteins in a defined medium (DM) or a rich medium (RM) in exponential (E) or stationary (S) phase, stress-induced expression of proteins, and radiation-induced expression at the gene and protein levels. Blank spaces indicate no reported data. (Based on data from references 94, 285, 364, 368, 377, 381, 383, 602, 645, and 677.)
PHX, predicted to be highly expressed.
O, oxidative stress; H, heat shock; S, starvation; A, alkaline stress; C, cold stress.
IR, ionizing radiation; ++, high level of postirradiation induction.
Endogenous ROS production in D. radiodurans is constitutively reduced due to fewer respiratory chain enzymes (cytochromes and flavoproteins) and enzymes with iron-sulfur clusters. Compared to the radiation-sensitive bacterium Shewanella oneidensis, D. radiodurans encodes 65% fewer proteins with iron-sulfur clusters ([2Fe-2S] and [4Fe-4S]) and 56% fewer cytochromes and flavoproteins (207). Furthermore, D. radiodurans lacks most of the Fe-chelating and Fe transport systems found in radiation-sensitive bacteria (207, 383), while iron is confined to the area outside the cytosol in the septum between dividing cells (122, 382). Enzymes with iron-sulfur clusters can be inactivated by oxidative stress or iron deprivation, leading to the release of free iron from the enzyme, which exacerbates oxidative stress (189). Free iron engages in Fenton-type chemistry, producing hydroxyl radicals, the most reactive ROS (268). However, one of the enzymes with an iron-sulfur ([4Fe-4S]) cluster, aconitase, is important for the oxidative stress response. Aconitase is highly expressed under normal growth conditions (285, 286, 364) and after gamma irradiation (677). As a sensor of ROS (524), the TCA cycle enzyme aconitase mediates the response to oxidative stress via transcription regulation in both E. coli (604) and B. subtilis (7). The major source of endogenous ROS is the auto-oxidation of respiratory chain enzymes, where the transfer of electrons from NADH and reduced flavin adenine dinucleotide (FADH2) to oxygen yields superoxide radicals and/or hydrogen peroxide (268). Apart from having fewer respiratory chain enzymes, D. radiodurans limits the release of NADH and FADH2 by inducing the glyoxylate bypass of the TCA cycle when grown in a defined minimal medium and in response to radiation (207, 368). Glyoxylate bypass is induced via the upregulation of isocitrate lyase (aceA), which converts isocitrate into succinate and glyoxylate. Glyoxylate is subsequently converted into malate by two predicted malate synthases in D. radiodurans (207). The glyoxylate pathway bypasses the formation of 2-oxoglutarate and succinyl coenzyme A (CoA) and the release of NADH and FADH2. Glyoxylate bypass has also been described for E. coli and Mycobacterium tuberculosis (381).
D. radiodurans has unusual metabolic defects that are also acquired to some extent by radiation-resistant strains of Salmonella enterica serovar Typhimurium, which emerged after laboratory exposures to multiple cycles of ionizing radiation (132). The inability to synthesize NAD and to employ nucleosides (e.g., uridine and adenosine) and TCA cycle products (e.g., α-ketoglutarate, succinate, fumarate, or malate) as carbon sources may lead to the accumulation of nucleotides as NAD precursors and amino acids as derivatives of TCA cycle products (121, 207). By acting as ROS scavengers, nucleotides and amino acids were found to protect proteins against oxidative damage in complex with divalent manganese ions (Mn2+) (121). The D. radiodurans metabolic configuration thus not only suppresses endogenous ROS production but also protects against exogenously induced ROS by endowing high concentrations of common cellular metabolites with ROS-scavenging properties when combined with Mn2+. High concentrations of “small molecules” in the D. radiodurans cytosol are expected to create great osmotic pressures, which need to be contained by thick cell walls such as the one found in D. radiodurans (121) (see “D. radiodurans Physical Structure and Cell Division”). The D. radiodurans physical structure may therefore present another element contributing to its oxidative stress artillery.
D. radiodurans exponential-phase cells contain electron-dense granules for the likely storage of polyphosphates (polyPs) (177, 453, 615) (Fig. 2C). PolyP granules are spherical aggregates of high-molecular-weight linear phosphates that can be found in bacteria, fungi, protozoa, plants, and mammals (316, 660). PolyP can serve as a source of energy for ATP synthesis; as a phosphorylating agent for sugars, nucleosides, and proteins; as a means of activating the precursors of nucleic acids, phospholipids, polypeptides, and fatty acids; for the chelation of metals; as a buffer against alkali; for bacterial Ca2+-induced transformation; for the virulence of pathogens; for chromosome condensation; and for the regulation of growth, development, and the stress response (8, 316, 508, 660). In D. radiodurans, polyphosphates not only may function as a source of energy and building blocks but also may provide orthophosphate that acts synergistically with Mn2+ to scavenge superoxide radicals and thereby protect proteins against oxidative stress (121) (see “Manganese complexes”). Mn-phosphate complexes can compensate for the loss of superoxide dismutase in Lactobacillus plantarum (22) and in the yeast Saccharomyces cerevisiae, where the Mn-orthophosphate but not the Mn-polyphosphate complex exhibits antioxidant properties (402).
D. radiodurans possesses an enzymatic system encoded on chromosome II for the degradation of fatty acids into acetyl-CoA after carbon sources have been exhausted (653). It also encodes aerobic-type carbon monoxide dehydrogenase on the megaplasmid, which may provide energy through the oxidation of CO (475). It has a functional RelA activity, producing ppGpp in response to amino acid deprivation, which reduces protein synthesis by repressing RNA synthesis (637).
Phylogeny of D. radiodurans
Based on the analysis of 16S rRNA sequences (250, 650, 657), 5S rRNA sequences (33), and protein signature sequences (222) and phylogenetic analyses of several conserved proteins (475), D. radiodurans was found to be phylogenetically related to Thermus thermophilus. D. radiodurans and T. thermophilus have similar GC contents; are both naturally transformable, red pigmented, nonsporulating, and aerobic; and share an A3β murein-type peptidoglycan (l-ornithine as the diamino acid and glycylglycine as the interpeptide bridge) (249). There is some evidence that the species from the Deinococcus-Thermus phylum are related to the Cyanobacteria and Actinobacteria (41, 658). Bacteria of the genera Thermus and Deinococcus have mixed phenotypes. While the Thermus species stain Gram negative and have a cell wall which resembles that of Gram-negative bacteria, there are many features that appear Gram positive, such as the presence of ornithine in the peptidoglycan (483) and branched-chain fatty acids in the lipids (148). Similarly, although Deinococcus species stain Gram positive, their fatty acid profile is similar to those of Gram-negative bacteria, and they have an outer cell membrane as a unique and defining characteristic of Gram-negative species.
The Deinococcus-Thermus phylum was recently expanded to include another family, the Trueperaceae (6). Truepera radiovictrix is a radiation-resistant sphere-shaped thermophile found in hot spring runoffs in the Azores (6). This bacterium thus shares phenotypic traits with D. radiodurans, a radiation-resistant Gram-positive coccal mesophile, and T. thermophilus, a radiation-sensitive Gram-negative rod-shaped thermophile. The Deinococcus-Thermus phylum is a classic case of phenotypic features not always correlating with phylogenies in bacteria (657).
The common ancestor of the Deinococcus-Thermus phylum was either a mesophile or a moderate thermophile (381, 475). After divergence from their common ancestor, Thermus adapted to high temperatures through horizontal gene transfer from archaea and thermophilic bacteria, whereas Deinococcus acquired numerous stress response genes from various bacteria (475). Sixty-five proteins were found to be unique to the Deinococcus-Thermus phylum, 206 proteins are found only in members of the Deinococcaceae (e.g., PprA and DdrB), and 399 proteins are unique to D. radiodurans (216).
It was estimated that 10 to 15% of the D. radiodurans genome has been acquired through HGT (381). The natural competence of D. radiodurans is compatible with a significant amount of horizontal gene acquisition. Many horizontally acquired genes are on the megaplasmid (475). Among the genes acquired from archaea or eukaryotes are vacuolar-type A/V ATPases; prolyl-, glycyl-, isoleucyl-, arginyl-, and aspartyl-tRNA synthetases (473); topoisomerase IB, adjacent to a uracil-DNA glycosylase of eukaryotic origin; and late-embryogenesis-abundant (LEA) proteins from plants (see “Desiccation Resistance of D. radiodurans”) (381).
Genetics of D. radiodurans
D. radiodurans is amenable to genetic manipulation owing to its natural transformability. Its cells are competent throughout the exponential growth stage. The frequency of transformants falls only during stationary phase (448, 620), whereas other bacteria have maximal competence just before they enter stationary phase (509). CaCl2 (30 mM) increases the frequency of transformation 40 times, with marker-specific efficiencies ranging from 0.01 to 3% (620). A family of 13 proteins that are homologous to the putative nuclease inhibitor DinB in B. subtilis (50, 232, 380) may enhance the transformation proficiency of D. radiodurans. The efficiency of transformation is higher with double-stranded DNA (dsDNA) than with single-stranded DNA (ssDNA) (444) and with plasmid DNA than with linearized DNA (448). Transformation with nonmethylated donor DNA passed through a dam dcm E. coli strain significantly increases the transformation efficiency (404). UV-irradiated transforming DNA has a much higher transforming ability in D. radiodurans than in Haemophilus influenzae due to the efficiency of D. radiodurans at repairing irradiated transforming DNA in the same manner as its own irradiated DNA (448). Although the transformation frequency decreases in UV- or gamma-irradiated D. radiodurans, the repair of irradiated transforming DNA is not affected (444). D. radiodurans extract improves the transforming ability of H. influenzae by a factor of up to 20 (448). Transformation in D. radiodurans seems to involve the processing of donor DNA into ssDNA fragments, which are subsequently assimilated into homologous dsDNA recipients through the action of RecA.
The first described introduction of heterologous DNA sequences into the D. radiodurans genome was achieved by a process of duplication insertion of E. coli drug resistance genes (kanamycin and chloramphenicol) flanked by a direct repeat of host DNA sequences (578). Up to 50 amplifications of this structure gave rise to a 500-kb insertion (>10% of the genome). The remarkable genome plasticity of D. radiodurans, which extends to 3 Mb of foreign DNA introduced by duplication insertion, shows that D. radiodurans is able to maintain, replicate, and express extremely large segments of foreign DNA (74). Without selective pressure, duplication insertions are lost via intrachromosomal recombination (578).
The genetic manipulation of D. radiodurans was facilitated by the construction of shuttle plasmids. The first shuttle plasmids that can replicate in D. radiodurans and E. coli were constructed by inserting an E. coli plasmid carrying the kanamycin resistance gene into plasmids pUE10 (37 kb) and pUE11 (45 kb), plasmids which naturally occur in D. radiodurans strain Sark (577). D. radiodurans promoter sequences upstream of the E. coli kanamycin resistance gene are required for the expression of this heterologous gene in D. radiodurans (577). The absence of a deinococcal promoter can sometimes be circumvented by the amplification of the drug resistance gene by duplication insertion (578, 579). Subsequently developed shuttle plasmids contained a wider choice of restriction sites (pI3) but were hampered by their size (16 kb) (393). Meima and Lidstrom (403) constructed a smaller (6.3-kb) shuttle plasmid, pRAD1, containing the minimal replicon from pUE10 and a chloramphenicol resistance cassette driven by a pUE10-derived promoter, which is poorly maintained in the absence of selective pressure. Regulated gene expression in D. radiodurans was accomplished with the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible Spac system, containing a Pspac promoter and a lacI repressor gene, which can control the expression of a lacZ reporter gene over 2 orders of magnitude (342).
D. radiodurans mutants can be generated by targeted insertional mutagenesis (225), by duplication insertion mutagenesis (388), by gene disruption using an in vitro transposition system (171), and by gene replacement (64, 342). The multigenomic nature of D. radiodurans often requires several passages through selective pressure (antibiotic) to obtain a strain homozygous for the respective mutation.
Mobile Genetic Elements in D. radiodurans
The D. radiodurans genome contains various mobile genetic elements such as inteins (protein splicing elements typically inserted into genes involved in DNA metabolism), insertion sequences (ISs), small noncoding repeats (SNRs), and two prophages (381). D. radiodurans contains one of the largest numbers of insertion sequences found in bacterial genomes. It has 52 ISs (Deinococcus geothermalis has 84 ISs [383]) and 247 SNRs, the number and distribution of which are comparable to those found in the evolutionarily distant E. coli (384). ISs are more prevalent on the two plasmids, whereas SNRs reside on the chromosomes. SNRs do not seem to be involved in transcriptional/translational regulation (384) or in the recovery from radiation or desiccation due to the fact that they are absent in D. geothermalis and D. deserti, which are as resistant to ionizing radiation as D. radiodurans (382, 383).
Insertion sequences encode a transposase and have inverted terminal repeats and/or internal repeats. Their transposition can disrupt a gene or activate a downstream gene by its internal promoter. Furthermore, they are expected to cause genome instability and result in high levels of genome rearrangement (381). IS elements are transcriptionally active in D. radiodurans and were found to induce mutations in uvrA (455), pprI (260), and thyA (408). One of the IS elements, ISDra2 (IS200/IS605 family), is present at different copy numbers in different D. radiodurans strains (14, 270, 381). Whereas the level of ISDra2 transposition is very low in the absence of stress, it is highly induced in irradiated cells (408, 484). Single-stranded DNA intermediates formed during postirradiation genome reconstitution (see “Recombinational Processes in D. radiodurans DNA Repair”) favor the transposition of ISDra2, the excision of which proceeds via a single-stranded DNA circle (484). Another IS family, ISDra5, is responsible for the translocation of the small plasmid into the large chromosome, which occurs spontaneously in the recA mutant cells and after high doses of ionizing radiation in wild-type cells (514) (see “Fidelity of DNA Repair in Irradiated D. radiodurans”).
The propensity of D. radiodurans to amplify DNA sequences flanked by direct repeats (578) and the abundance of repeat elements in the genome provide the basis for the expansion and regulation of genomic regions in response to environmental changes (381).
DNA DAMAGE RESISTANCE AND DNA REPAIR MECHANISMS OF D. RADIODURANS
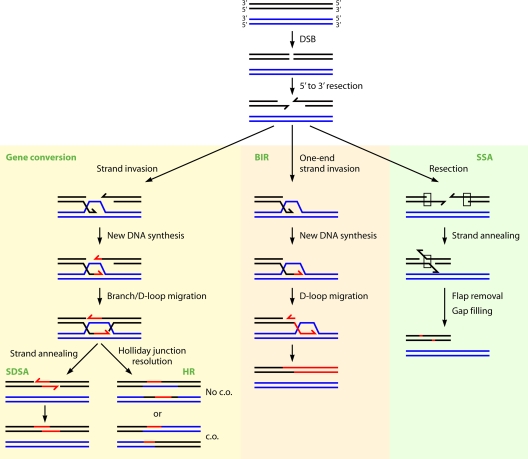
D. radiodurans is extremely resistant to various DNA-damaging agents inducing different forms of DNA damage. Ionizing radiation and desiccation generate double-strand breaks (DSBs), single-strand breaks (SSBs), and base damage; MMC generates DNA interstrand cross-links; UV radiation forms diverse pyrimidine dimers; and hydrogen peroxide, methyl methane sulfonate (MMS), N-methyl-N′-nitro-N-nitrosoguanidine (MNNG), nitrous acid, and hydroxylamine induce severe base and nucleotide damage. The diversity of DNA damage withstood by D. radiodurans is commensurate with the selection of DNA repair pathways at its disposal: direct damage reversal, base and nucleotide excision repair, mismatch repair, and recombinational repair (Table 3). Photoreactivation (441) and translesion synthesis (381) are absent. Ionizing radiation disintegrates the D. radiodurans genome by double-strand breakage into multiple fragments but also introduces at least 10-times more SSBs and many more sites of base damage. Extremely efficient excision repair systems enable the repair of base and nucleotide damage with a broad range of substrate specificities (381). DSBs, as the most severe form of DNA damage, are patched up by a sequential action of two mechanisms: “extended synthesis-dependent strand annealing” (ESDSA) and homologous recombination by crossovers (676).
TABLE 3.
DNA repair pathways in D. radiodurans, the genes involved, their transcriptional regulators, and their expression levelsa
| Pathway and enzyme | Locus tag | Regulation | PHXb | Constitutively expressed |
Induced after IR stressc |
||
|---|---|---|---|---|---|---|---|
| DM | RM | Gene | Protein | ||||
| DR | |||||||
| O6-Methylguanine DNA methyltransferase (Ogt) | DR0428 | + | − | − | |||
| Xanthosine triphosphate pyrophosphatase (YggV) | DR0179 | − | − | − | |||
| BER | |||||||
| 3-Methyladenine DNA glycosylase (AlkA) | DR2074 | − | |||||
| DR2584 | − | ||||||
| Uracil-DNA glycosylases (Ung) | DR0689 | − | + | ||||
| DR1751 | + | + | − | ||||
| Mug | DR0715 | − | − | ||||
| Endonuclease V (Nfi) | DR2162 | − | |||||
| 8-Oxoguanine DNA glycosylase (MutY) | DR2285 | − | + | ||||
| Formamidopyrimidine and 8-oxoguanine DNA glycosylase (MutM) | DR0493 | − | − | − | − | ||
| Endonuclease III (Nth) | DR0928 | − | − | ||||
| DR2438 | − | + | − | − | |||
| DR0289 | − | − | − | − | |||
| Exonuclease III (Xth) | DR0354 | − | + | − | − | ||
| NER | |||||||
| UvrA1 | DR1771 | DdrO? | − | + | + | +D | |
| UvrB | DR2275 | DrRRA | − | + | + | ++D | |
| DdrO? | |||||||
| UvrC | DR1354 | − | + | + | + | ||
| UvsE | DR1819 | − | − | ||||
| Mfd | DR1532 | − | − | ||||
| DNA or RNA helicase of superfamily II (Rad25) | DRA0131 | + | − | − | |||
| HR | |||||||
| RecA | DR2340 | IrrE | + | + | + | ++D | + |
| DrRRA | |||||||
| DdrO? | |||||||
| RecF | DR1089 | − | − | ||||
| RecO | DR0819 | − | + | − | |||
| RecR | DR0198 | − | + | + | − | ||
| RecJ | DR1126 | − | + | + | − | ||
| RecN | DR1477 | − | + | − | − | ||
| RecQ | DR1289 | DdrO? | − | − | + | − | |
| RecD | DR1902 | − | − | + | − | ||
| SbcC | DR1922 | − | + | + | − | ||
| SbcD | DR1921 | DdrO? | − | − | + | − | |
| RuvA | DR1274 | − | − | ||||
| RuvB | DR0596 | DdrO? | − | +D | |||
| RuvC | DR0440 | − | − | − | − | ||
| RecG | DR1916 | − | + | − | + | ||
| RadA | DR1105 | − | + | + | − | ||
| Rad54 DNA helicase | DR1259 | − | − | − | |||
| DdrA | DR0423 | DdrO? | − | − | ++D | ||
| PprA | DRA0346 | IrrE | − | − | +D | + | |
| DrRRA | |||||||
| DdrO? | |||||||
| MR | |||||||
| MutL | DR1696 | DdrO? | − | + | + | − | |
| MutS1 | DR1039 | DdrO? | − | + | − | − | |
| MP | |||||||
| DNA polymerase I (PolA) | DR1707 | − | + | + | − | ||
| DNA polymerase III | |||||||
| DnaE (alpha) | DR0507 | − | + | + | − | ||
| DnaQ (epsilon) | DR0856 | + | |||||
| DnaN (beta) | DR0001 | + | − | + | |||
| Tau/gamma | DR2410 | + | − | − | |||
| DNA ligase (LigA) | DR2069 | − | + | + | − | ||
| SSB | DR0099 | IrrE | − | + | − | + | + |
| DdrO? | |||||||
| DdrB | DR0070 | DdrO? | − | − | + | + | |
| UvrD | DR1775 | DdrO? | − | + | − | + | |
| DNA gyrase | |||||||
| GyrA | DR1913 | DdrO? | + | + | +D | ||
| GyrB | DR0906 | DrRRA | + | + | ++D | ||
| DdrO? | |||||||
Shown are data for constitutive expression in a defined medium (DM) or a rich medium (RM) and radiation-induced expression at the transcriptional and translational levels. DR, direct damage reversal; BER, base excision repair; NER, nucleotide excision repair; HR, homologous recombination; MR, mismatch repair; MP, multiple pathways. Blank spaces indicate no reported data. (Based on data from references 94, 285, 364, 368, 377, 383, 464, 602, 645, and 677.)
PHX, predicted to be highly expressed.
IR, ionizing radiation; ++, high level of postirradiation induction; +D or ++D, induction after ionizing radiation and desiccation.
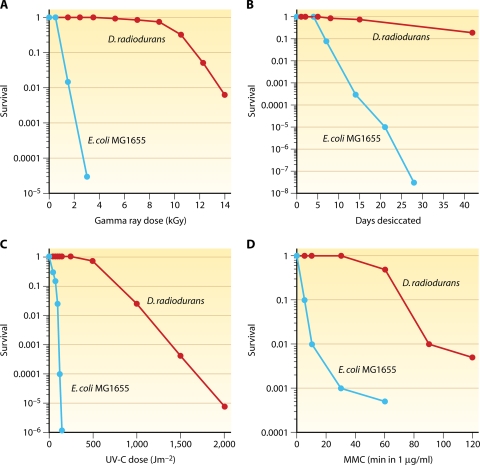
D. radiodurans is 30-fold and 1,000-fold more resistant to ionizing radiation than E. coli and humans, respectively, and can repair approximately 200 DSBs or 190 cross-links per genome copy without a loss of viability (37, 241, 303, 439). D. radiodurans displays sigmoidal survival curves against DNA-damaging agents, which are characterized by large shoulders followed by exponential slopes (Fig. 3). The extreme DNA damage resistance of this organism has been traditionally attributed to a remarkable DNA repair capacity and to exceptional DNA repair systems. However, according to the present genomic and genetic analysis, the enzymology of deinococcal DNA repair does not seem to be exceptional. DNA repair systems appear even less complex than those reported for E. coli or Shewanella oneidensis (120, 123, 381, 383), and the most radiation-sensitive DNA repair mutants in D. radiodurans can be fully (polA [224]) or partially (recA and recO [541, 665]) complemented with the respective E. coli genes. Moreover, the mechanism of DSB repair in D. radiodurans is remarkably similar to those of yeast cells (124, 126, 127, 572, 676). However, there remains a possibility that the evolutionary advantage of DNA repair in D. radiodurans resides in unrecognized Deinococcus-specific DNA repair enzymes, which are not found in other organisms (140, 216), and in a high degree of redundancy in DNA repair genes. D. radiodurans encodes 11 DNA glycosylases, 2 UV repair pathways (UvrABC and UV damage endonuclease [UVDE]), 2 divergent SSB proteins (464), and 23 genes belonging to the Nudix family of nucleoside triphosphate pyrophosphorylases (653). Nonetheless, recent developments in the Deinococcus field demonstrate that the DNA damage resistance of bacteria correlates with the level of protection of DNA repair and other enzymes against oxidative damage and not with the amount of DNA damage or the capacity to repair it (120-122) (see “Antioxidation Protection in D. radiodurans” and What Determines the Extreme Radiation Resistance of D. radiodurans?).
FIG. 3.
Extreme resistance of D. radiodurans to gamma rays (A), desiccation (B), UV-C radiation (100 to 295 nm) (C), and mitomycin C (D) in comparison with E. coli. (Panel B is based on data from references 123 and 398, panel C is based in part on E. coli data from reference 238, and panel D is based in part on D. radiodurans data from reference 239.)
Ionizing Radiation Resistance of D. radiodurans
There are two types of ionizing radiation, both produced by the decay of radioactive elements: electromagnetic (X and gamma radiation) and particulate (α and β particles) (108). Gamma rays are photons that generate ions, which react with other molecules to produce free radicals. Reaction with water molecules gives rise to hydroxyl radicals (OH·), the most reactive ROS (230, 268). Ionizing radiation generates multiple types of DNA damage: base damage, SSBs, DSBs, and interstrand cross-links. DNA bases are most affected, with more than 80 different types of structural modifications induced by ionizing radiation (59). Approximately 10% to 20% of the time, the sugar-phosphate moiety is affected, which can lead to a single-strand break (72). On average, for every 20 SSBs induced by gamma rays in DNA, there is 1 DSB (328). If not repaired, DSBs prevent the replication of genomes and lead to cell death. DSBs can arise when SSBs occur in close proximity on opposite strands, when a cluster of radicals introduces strand breaks in both strands at one location, or through the excision repair of damaged bases present in close proximity on both strands. In irradiated D. radiodurans, DSBs are a result of a single event, as opposed to a double event, where two close SSBs in opposite chains yield a DSB (79). In the case of “double-event” breaks, the number of breaks would be proportional to the square of the dose and would be dependent on ionic strength, which was not found (79). At 7 kGy, less than 3% of the DSBs are a result of double-event breaks (79). Radiation-resistant and radiation-sensitive species have remarkably similar numbers of DSBs per Gy per genome (0.002 to 0.006 DSBs/Gy/Mbp) (67, 79, 206, 211, 499, 516, 522) but differ in the amounts of oxidative DNA base damage (302). Furthermore, in D. radiodurans, aerobic conditions yield only 2.5-fold more DSBs/Gy than do anaerobic conditions (123). Consequently, it appears that the majority of DSBs are caused by a direct interaction between gamma photons and DNA and are not affected by the antioxidant status of the cells. Nevertheless, there can be significant differences in base damage between gamma-irradiated cells, as base damage is also affected by radiation-induced OH· and may thus reflect the cellular antioxidant status.
The rate of DSB formation in D. radiodurans is proportional to the gamma radiation dose (79). A dose of 6 kGy induces approximately 200 DSBs (79), over 3,000 SSBs (65, 79), and many more sites of base damage per D. radiodurans genome (236). D. radiodurans survives 5 kGy of ionizing radiation without lethality or mutagenesis (135, 161, 597) (Fig. 3A). Under nutrient-rich conditions, D. radiodurans can also grow in the presence of chronic irradiation of 60 Gy/h with no effect on its growth rate (336).
E. coli and most other bacterial cells are killed if there are less than a dozen radiation-induced DSBs per chromosome (327, 515). However, E. coli can endure dozens of DSBs produced by the transient expression of the restriction endonuclease EcoRI (248). Analogously, yeast and human cells can repair up to 200 and 400 DSBs during meiosis, respectively (78, 153), but succumb to 40 ionizing-radiation-induced DSBs (194, 522). The discrepancies in the tolerances of endogenous and radiation-induced DSBs across species indicate that DNA is not the prime target site of the lethal radiochemical lesion as traditionally believed (264, 442), as ionizing radiation also targets proteins and cellular membranes (45) (see What Determines the Extreme Radiation Resistance of D. radiodurans?). Oxidative protein damage is diverse, including the carbonylation of certain amino acids (proline, arginine, lysine, and threonine), the oxidation of sulfhydryl groups in methionine and cysteine, the introduction of hydroxyl groups into phenylalanine and tyrosine, tryptophan ring cleavage, peptide bond cleavage, and cross-linked species such as dityrosine (252).
In D. radiodurans, proteins are protected against radiation-induced oxidative damage via ROS scavengers dominated by manganese complexes (120-122) (see “Manganese complexes”). Cellular membranes, which are composed of lipids, proteins, and carbohydrates, are damaged by radiation. In D. radiodurans the outermost hexagonal cell wall layer appears to be a major site of cell wall radiation damage, releasing both an exonuclease (205, 419) and polysaccharides in response to ionizing radiation (420). D. radiodurans irradiated with 4 kGy loses up to 30% of its wet weight due to the loss of polysaccharides into the growth medium from a hydrophobic site in the hexagonal cell wall layer (420). This is likely due to a radiolytic cleavage of a covalently linked lipid, which normally serves to anchor these substances to the membrane (422). A large loss of water during irradiation suggests a shrinkage and/or permeability alteration in the cell envelope (420). The loss of water may reduce ROS levels by concentrating the cytosolic pool of small molecules needed to form ROS-scavenging Mn2+ complexes (121).
Factors influencing radiation resistance.
The extent of D. radiodurans resistance to ionizing radiation depends strongly on physiological conditions, such as the age of the culture, the cell concentration, the growth medium, the pH, the irradiation medium, the irradiation temperature, and the plating medium (435). The full recovery of irradiated D. radiodurans is highly dependent on a rich source of nutrients, which are required for growth and the generation of Mn2+ complexes (121, 207). Survival following chronic exposure is affected profoundly by the metabolic state of cells, with a total delivered dose of 3 kGy (50 h at 60 Gy/h) being lethal (637). Cells grown in minimal medium and chronically irradiated with 60 Gy/h rapidly lose viability coupled with severe DNA degradation, unless high concentrations of amino acids are provided (637).
Earlier studies established that stationary-phase cells are 3-fold more resistant than exponentially grown cells (287, 548). However, according to recent studies, late-stationary-phase cells are 4-fold more sensitive than exponential- and early-stationary-phase cells (592). The phasic response of cells is a result of the physiological changes occurring as a result of the passage of the organism from one metabolic state to another (548). The perturbation of exponential growth with a 1-h incubation in phosphate buffer increases radiation resistance (198, 548). Such a growth disruption may allow the cessation or completion of chromosome replication prior to irradiation and/or delay the onset of growth after irradiation, thereby increasing the opportunity for the repair of cell damage (198).
The multicellularity of D. radiodurans, which forms tetrads during exponential growth, does not contribute to its radiation resistance, judging by the high survival rate of single cells observed by microscopic examination, as opposed to the CFU scores on agar plates (161). However, cell clumping significantly increases survival as a result of the shielding of cells inside a cluster from oxygen effects (416). Analogously, a high concentration of bacteria protects the cells and increases the survival by lowering the oxygen content (165). Aeration of the culture maintains the sensitivity to radiation regardless of the cell concentration (220).
D. radiodurans is 3-fold more resistant when irradiated in a dry state than in aqueous suspensions, presumably due to lower water radiolysis generating hydroxyl radicals (292). With regard to the growth medium, survival increases in the following order: phosphate buffer < minimal medium < TGY broth (165, 241). The addition of a tryptic digest of casein to the growth medium lowers ionizing radiation resistance 2-fold (326). Changing the pH of the phosphate buffer to pH 5, 7, or 9 does not affect radiation sensitivity (166). However, irradiation in TGY broth at pH 10.5 substantially lowers the rate of survival relative to that at pH 7 (122). The reduction in survival by the pH increase is correlated with the increase in oxidative protein damage and has been linked to the disruption of manganese redox cycling (122) (see “Manganese complexes”).
Irradiation in the presence of oxidizing agents (e.g., ascorbate) increases radiation sensitivity, whereas reducing agents (e.g., cysteine) reduce it (166). The irradiation of D. radiodurans in the presence of iodoacetamide, an enzyme poison that alkylates sulfhydryl (thiol) groups in cysteine, producing carboxyamidomethylcysteine (580), causes the shoulder of the survival curve to disappear and the exponential part to become steeper (134). This sensitizer thus enhances the radiation-induced inactivation of proteins required for DNA repair and cell survival. Sensitization by iodoacetamide may include the repression of DNA degradation through the inhibition of an enzyme that excises DNA from a DNA-membrane complex (643) (see “DNA Degradation in Irradiated D. radiodurans”) and the inhibition of the DNA reassociation with the membranes (129). The incorporation of 5-bromouracil into DNA also sensitizes the cells to the effects of X-ray radiation (147) by inhibiting DNA degradation and removing the shoulder of the survival curve (350). Furthermore, the addition of 100 μM manganese to the growth medium during early stationary phase perturbs Mn-redox cycling and increases the susceptibility of the cells to the lethal and mutagenic effects of UV and ionizing radiation, MMC, and MNNG due to glucose oxidation and an increase in oxidative stress (96, 122, 362). Chloramphenicol blocks the repair of DSBs and prevents postirradiation recovery by blocking protein synthesis (307).
In contrast to data from previous reports showing that frozen conditions do not offer much protection against radiation damage in D. radiodurans (166), more recent data show that D. radiodurans cells irradiated on dry ice (−70°C) have a higher gamma ray survival rate than do cells irradiated on ice or at room temperature (128, 130, 519). The increase in radiation resistance under frozen irradiation conditions may be due to the diminished oxygen effects on proteins (120, 122). Conversely, preirradiation heat treatment and irradiation at higher temperatures (40°C) lower the survival rate (166).
According to data reported previously by Moseley and Laser (442), cells recovered from the initial irradiation damage are again as resistant to X-ray radiation as unirradiated cells, whereas Driedger et al. (161) reported a slight increase in the radiation sensitivity of the progeny of irradiated cells, possibly due to a higher dose of irradiation of the parental cells. In other bacteria, one or more doses of radiation can cause an increase in radiation resistance (132, 358, 481), an increase in antibiotic resistance (498), morphological changes (173, 358), biochemical changes (173), and decreased pathogenicity (180, 498). Irradiated or chemically treated D. radiodurans cells usually give rise to colonies identical to those of unirradiated bacteria, although some colonies are smaller, pigmentless, and rough with irregular edges (161, 434). This may result from a partial defect in recombination-type repair, which is presumably required for the regulation of cell division (438). Microscopic observation of dying cells revealed that only about 10% of the cells that fail to give rise to a colony after 7 to 10 kGy of radiation do not divide at all, while the majority undergo between one and six divisions, after which the entire microcolony lyses (161). The phenomenon of abortive colony formation suggests that the DNA repair is either incomplete, with an inaccurate partitioning of genetic material to the progeny upon division, or inaccurate, with the production of mutations (161).
Puzzlingly, when held in liquid media, irradiated D. radiodurans cells show a loss in viability, which is not shown when cells are plated immediately (436). Cells treated with MMC and incubated in a liquid medium also show lower viability than cells plated immediately after the treatment (304).
Evolution of radiation resistance in D. radiodurans.
As noted above, the Deinococcaceae can be found in organic-rich environments, such as soil, animals and their feces, sewage, and the plant rhizosphere, but also under extreme environmental conditions, such as deserts, rocks, hot springs, hydrothermal vents, and permafrost. Since dry environments are more prevalent on Earth than those that generate a high flux of ionizing radiation, D. radiodurans ionizing radiation resistance is, according to the “desiccation adaptation hypothesis,” only an incidental consequence of the adaptation to dehydration, which is a common physiological stress (398). Namely, all 41 examined ionizing-radiation-sensitive strains of D. radiodurans were also found to be proportionally sensitive to desiccation, indicating that radiation resistance and desiccation resistance are functionally related phenomena (398). The recovery of large numbers of extremely radiation-resistant bacteria from an arid soil sample found in the Sonoran desert, compared to a nonarid soil sample found in a Louisiana forest, corroborates the hypothesis that the radiation resistance phenotype is a consequence of the evolution of cellular systems that protect cells from desiccation (501). A cyanobacterium, Chroococcidiopsis (57), and E. faecium (123) are also both desiccation and ionizing radiation tolerant. Further support for the coevolution of desiccation and radiation resistance is provided by Shukla et al. (569), who showed that ionizing-radiation-resistant bacteria from various habitats are also highly resistant to desiccation, MMC, and H2O2. Although all known naturally occurring radiation-resistant bacteria are also desiccation resistant, radiation-resistant strains of E. coli evolved under laboratory conditions are not desiccation resistant (238; J. R. Battista, personal communication). Similarly, desiccation tolerance mechanisms do not always result in radiation resistance, as desiccation-tolerant bacteria from the Actinobacteria, Arthrobacter, and Rhodococcus families are not radiation resistant (569).
Alternatively, the extreme radiation resistance may have evolved as an adaptation to permafrost or semifrozen conditions where background radiation-induced DNA damage is accumulated (519) or to high natural ionizing radiation levels in deeply buried manganese-rich marine sediments (“radiation adaptation hypothesis”) (555). D'Hondt et al. (140b) found microbial activities in these deeply buried sediments with high ionizing radiation levels. A Martian origin of D. radiodurans and other radiation-resistant bacteria has been proposed to explain the development of radiation resistance in these bacteria under highly radiating Martian conditions, followed by the transfer of the “trained” bacteria to Earth via meteorites (485). Diaz and Schulze-Makuch (141) showed that D. radiodurans could survive Martian-like conditions of low temperatures (−35°C for 10 days), low pressure (83.3 kPa for 10 days), and high UV radiation (37 W/m2 for 24 h), which occur near the surface of Mars. The “training” of radiation resistance was demonstrated experimentally on different bacteria (E. coli, Bacillus pumilus, and Salmonella enterica serovar Typhimurium) exposed to multiple cycles of high radiation doses that yielded a permanently acquired increase in radiation resistance (132, 238, 358, 481).
Kinetics of DNA Repair in Gamma-Irradiated D. radiodurans
Exposure to ionizing radiation induces a dose-dependent growth arrest in D. radiodurans cells (440). During growth arrest, oxidative stress is alleviated, cells are cleansed of the damaged biomolecules, damaged cellular components are recovered, proteins are resynthesized, and damaged DNA is accurately repaired. Cellular recovery is inhibited under conditions forestalling protein synthesis, such as a nonnutrient buffer (160), chloramphenicol (157), or nalidixic acid at high concentrations (158), and under conditions that impair DNA repair, such as a recombination deficiency (439) or DNA synthesis deficiency (446, 572). Growth-promoting conditions are essential for the removal of lesions from DNA (128, 413).
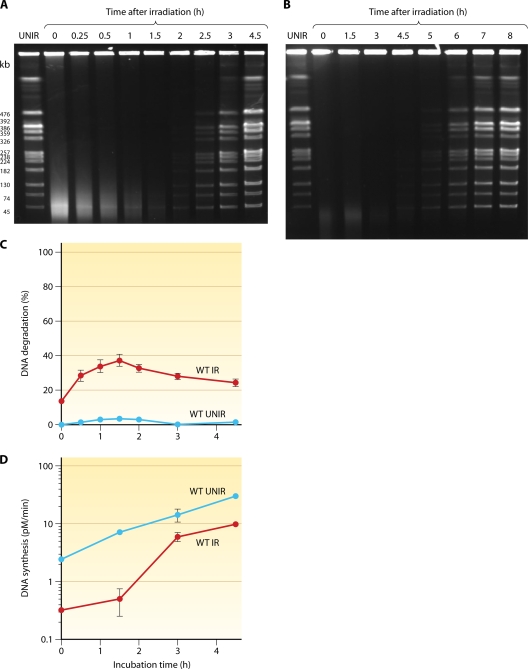
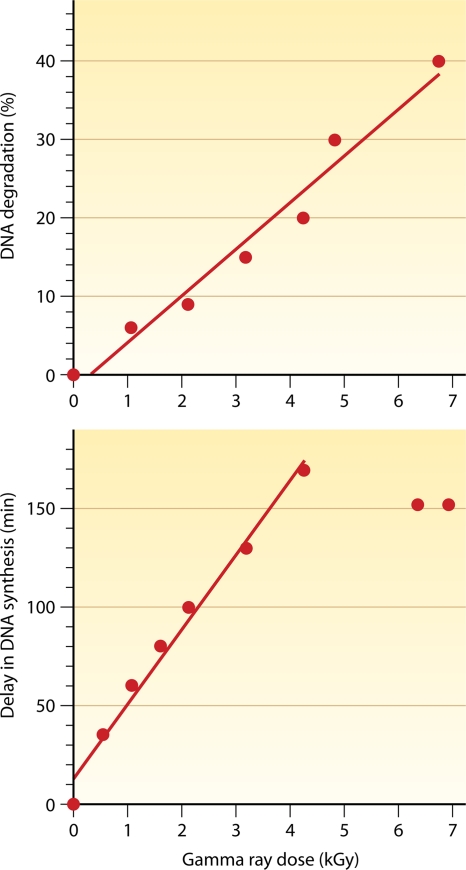
Radiation incurs massive DNA damage, breaking D. radiodurans chromosomal DNA into many fragments (represented by a broad smear of lower-molecular-weight material in Fig. 4 A and B). The repair of DSBs can be monitored by neutral sucrose gradients (79, 307) and by pulsed-field gel electrophoresis (PFGE) (217). Digestion of the D. radiodurans genome with NotI yields 12 fragments that can be resolved by PFGE (Fig. 4A and B). Following doses of 7 and 14 kGy of ionizing radiation, D. radiodurans DNA is shattered by double-strand breakage into 30- to 40-kb and 10- to 20-kb fragments, respectively (Fig. 4A and B). An extremely efficient DNA repair process rapidly reassembles all the fragments into functional chromosomes (413, 435). Postirradiation recovery comprises three distinct phases: (i) a DNA degradation period with little repair (Fig. 4C), (ii) the rejoining of DNA fragments in parallel with extensive DNA synthesis (Fig. 4D), and (iii) the resumption of growth upon the completion of DNA repair. Kinetic parameters associated with the recovery of D. radiodurans cells from ionizing radiation-induced DNA damage are all linearly correlated with the dose of ionizing radiation. These parameters include the rate and the extent of DNA degradation (161, 350, 351) (Fig. 5, top), a delay in DNA synthesis (135, 350, 351, 440) (Fig. 5, bottom), and the rate of DNA synthesis (350).
FIG. 4.
Kinetics of DNA fragment joining in D. radiodurans after 7 kGy (A) and 14 kGy (B) of gamma rays monitored by PFGE. Samples of unirradiated and irradiated wild-type cells were taken to prepare DNA plugs, which were digested with NotI, generating 12 visible fragments. Lane “UNIR” shows the NotI restriction pattern of DNA from unirradiated cells, lane “0” shows the NotI restriction pattern of DNA from irradiated cells immediately after irradiation, and subsequent lanes show the NotI restriction patterns of DNA from cells at different time points after irradiation, expressed in hours. (C) DNA degradation measured in 3H-prelabeled unirradiated (blue) and 7-kGy-irradiated (red) wild-type (WT) cells. (D) Rate of DNA synthesis in unirradiated (blue) and 7-kGy-irradiated (red) wild-type cells. The rate of DNA synthesis is expressed as the amount of [3H]thymidine (pM) incorporated into DNA per minute. (Panels C and D are modified from reference 572 with permission from Elsevier.)
FIG. 5.
Linear relationship between the dose of ionizing radiation and, from top to bottom, the extent of DNA degradation and the delay in DNA synthesis. The delay in DNA synthesis is linearly correlated with a dose of up to 4 kGy, above which it reaches a plateau. (Modified from reference 351 with permission of the publisher and based in part on data from references 572 and 643.)
Oxidized bases are rapidly repaired by base excision repair (BER). In the 2.5-kGy-irradiated archaeon Halobacterium salinarum, oxidized bases are repaired within 2 h, whereas DSBs are repaired within 8 h (302). Similarly, the rejoining of strand breaks in D. radiodurans follows biphasic kinetics, with a rapid initial step of rejoining the SSBs and a slow subsequent step where the DSBs are repaired (65, 136, 306). Following a dose of 2 kGy in anoxia, about 50% of the SSBs are restituted during irradiation at 0°C (136). Of the remaining breaks, half are rapidly restituted within 5 min of postirradiation incubation, whereas the broken strands are rejoined more slowly (136). SSBs are directly rejoined without DNA synthesis and without a requirement for postirradiation protein synthesis, possibly via polynucleotide kinase and DNA ligase (157). The direct repair of SSBs is further corroborated by the partial repair upon incubation in a nonnutrient buffer (160). The repair of SSBs in D. radiodurans can be monitored by using alkaline sucrose gradients (79, 154, 157, 350). The repair of DSBs is more difficult than the damage that affects a single strand (SSB or base damage) and relies upon interchromosomal recombination (125, 445, 572, 676).
DNA Degradation in Irradiated D. radiodurans
DNA degradation is considered an integral part of the DNA repair process and not just a consequence of cell death (643). The decrease in DNA degradation in recombination-deficient mutants (recA and recFOR) that fail to reassemble their broken genomes (49, 572) substantiates the importance of DNA degradation in DSB repair. Ionizing radiation induces DNA degradation not only in bacteria (178, 213, 351, 400, 588) but also in yeast (172), plant protoplasts (229), mouse cells (589), rat cells (630, 684), and human cells (101, 571).
Irradiated D. radiodurans cells recovered under optimal growth conditions show no net increase in DNA during the early period of incubation (Fig. 4C and D). This arrest in DNA synthesis coincides with DNA degradation, which reaches a maximum when DNA synthesis resumes (135). In the nonlethal-dose range the delay in DNA synthesis increases linearly with the dose and is longer than the time necessary for maximum DNA degradation to occur (351). In the lethal-dose range the delay in DNA synthesis rapidly reaches its maximum, while the time required for maximum DNA degradation continues to increase linearly (350) (Fig. 5). The extensive DNA breakdown causes a loss of viability during the postirradiation incubation of irradiated cells (136). The contribution of DNA degradation products from dying cells may be delayed until many hours after irradiation and does not account for the DNA degradation observed immediately after irradiation (161).
DNA degradation results from exonucleolytic activity at the broken DNA ends and from the excision repair of the damaged bases. Base damage is the predominant type of lesion caused by ionizing radiation in D. radiodurans (236). The release of damaged thymine (5-hydroperoxy-6-hydroxy-5,6-dihydrothymine and 5,6-dihydroxy-5,6-dihydrothymine) is biphasic: a rapid release during the first 30 min after 1.5 kGy of radiation and a slow release beginning after 60 min (235, 236). The first-phase release may be due to exonucleolytic DNA degradation at the broken DNA ends, while the excision process (BER) may account for the second-phase release (235). DNA degradation is severalfold enhanced in the polA mutant, which attests to the importance of polymerase I (Pol I) in sealing excision repair-generated gaps and preventing further DNA degradation (65). Conversely, DNA degradation is diminished in the recA and the recFOR mutants (49, 572) and when cells are irradiated in a dry state (29). It is completely inhibited when irradiated cells are incubated in a phosphate buffer (65, 136), in 5-bromouracil (350), in hydroxyurea (572), and when irradiated stationary-phase cells are incubated in the “exhausted” TGY medium in which they were grown prior to irradiation (K. Zahradka, unpublished data).
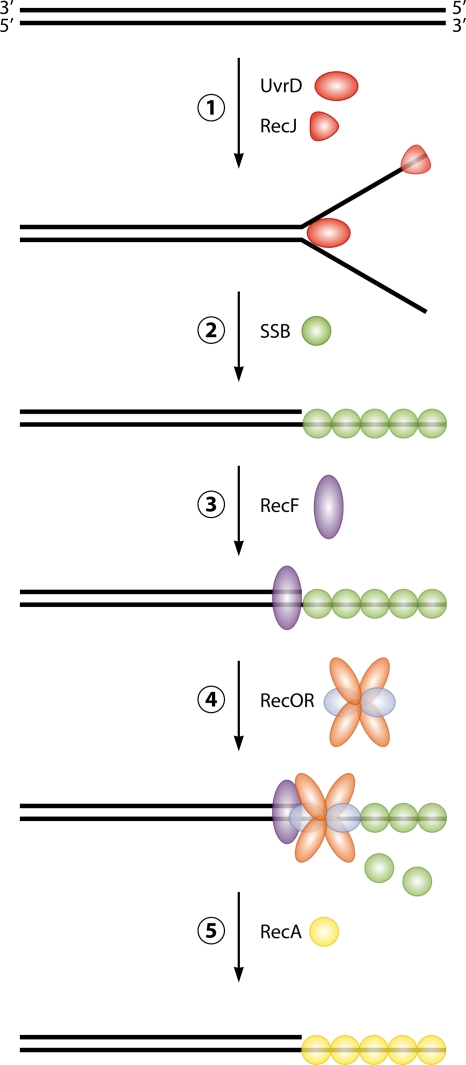
Exonucleases play important roles in DNA repair by the end recession of damaged DNA to generate 3′ overhangs, which are able to engage in recombinational repair (Fig. 6, step 2). The 5′-to-3′ polarity of end recession in D. radiodurans was indirectly demonstrated by expression in trans of the SbcB 3′-5′ exonuclease from E. coli, which rendered D. radiodurans radiation sensitive and inhibited DNA double-strand-break repair (418). Among possible candidates for 5′-3′ exonuclease activity in D. radiodurans are RecJ and the 5′-3′ exonuclease activity of Pol I. In E. coli, RecJ is required for the repair of DSBs (535) to prepare ssDNA for the subsequent loading of the RecA protein when RecA loading and nuclease functions of RecBCD are inactivated (272). RecJ serves a more important function in D. radiodurans, where RecJ is the only 5′-3′ exonuclease, the depletion of which results in lethality (49). The 5′-3′ exonuclease activity of E. coli Pol I is required for the removal of RNA primers from Okazaki fragments (253, 314, 471), for excision repair (106), for nick translation (520), for the repair of DNA cross-links (573), and for the repair of DSBs (558).
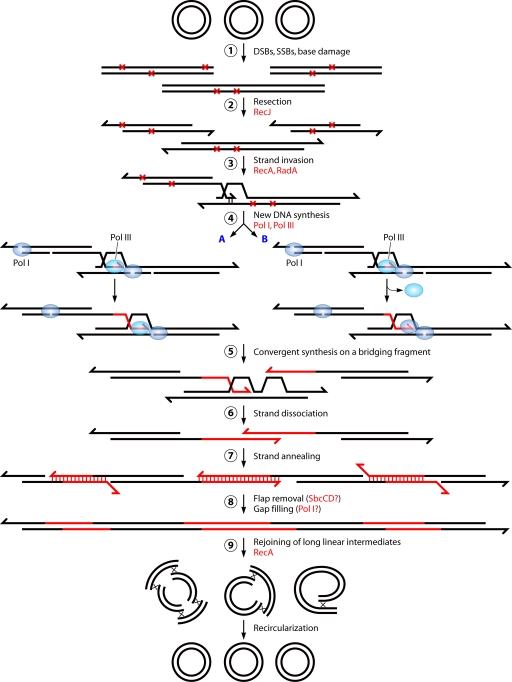
FIG. 6.
Two-step mechanism of DNA repair in D. radiodurans shattered by ionizing radiation. Several genomic copies of D. radiodurans undergo random DNA double-strand breakage, producing numerous fragments (step 1). The fragmented DNA is recessed in a 5′-to-3′ direction, presumably by RecJ, liberating single-stranded 3′ overhangs (step 2), which, through RecA- and RadA-mediated strand invasion, prime synthesis on overlapping fragments through a migrating D loop (step 3). DNA synthesis is initiated by Pol III (step 4) and elongated by Pol III, with Pol I filling up gaps arising from the excision repair of damaged bases (A), or by Pol I alone (B).Two noncontiguous fragments are linked by convergent elongations on a third “bridging” fragment (step 5). Newly synthesized single strands dissociate from the template (step 6) and anneal to complementary single-stranded extensions, forming dsDNA intermediates (step 7). The flaps are removed (by SbcCD?), and the gaps are filled (by Pol I?) (step 8). Long linear intermediates are joined into circular chromosomes by RecA-dependent crossovers (step 9). (Modified from reference 572 with permission from Elsevier and based in part on data from reference 676.)
The degraded DNA components are exported from the cells. The removal of the damaged nucleotides from the cells could represent a survival strategy to protect the organism from excessive mutagenesis by preventing the reincorporation of damaged bases into the genome (37). After 5 kGy of radiation, large oligonucleotides (∼1 kb) are immediately released into the surrounding medium and are rapidly degraded to nucleotides by an extracellular exonuclease (643). Nondamaged degraded products can be reincorporated into the DNA upon the DNA synthesis restart (159, 572). The export of the damaged nucleotides may be mediated by UvrA homologs, which are closely related to ABC transporter proteins (149, 363) and which serve as a site of attachment of nucleotide excision repair (NER) to the cell membrane in E. coli (361). D. radiodurans UvrA2 does not contribute to UV radiation resistance (603) but may be involved in the export process (653). Iodoacetamide inhibits the release of oligonucleotides (643), presumably by inhibiting the export protein. The exported oligonucleotides may be degraded extracellularly into nucleotides by a cell surface exonuclease (419, 423, 424). This 117-kDa extracellular nuclease (DRB0067) is encoded on the megaplasmid (653) and is most closely related to exonucleases from the Cyanobacteria. It degrades both DNA and RNA to 5′-mononucleotides, has a distinct preference for single-stranded DNA, is active at alkaline pHs, has tightly bound Ca2+, and is bound to the carotenoid-containing hexagonal layer (205, 423, 424). Hydroxyl radicals generated by ionizing radiation attack the cell wall (419) and initiate the release of this enzyme into the medium (205, 421, 601). The expression level of this exonuclease is increased following 1 kGy of ionizing radiation (677).
Recombinational Processes in D. radiodurans DNA Repair
Whereas radiation-induced SSBs are rapidly rejoined, the repair of DSBs is a slow recombinational process, dependent on DNA and protein synthesis (136, 157). In homologous recombination, DNA lesions are repaired by using an intact homologous donor molecule as a template. Because strand breaks are generated randomly, the probability of DNA damage occurring at the same site on every chromosome is very low. The multigenomic nature of D. radiodurans with a minimum copy number of 2 (241) thus provides a prerequisite for the repair of DSBs via recombinational processes (234). Increasing the chromosomal copy number in D. radiodurans does not enhance radiation resistance (241). In fact, many radiation-sensitive bacterial species have multiple genome copies, such as M. luteus, Micrococcus sodonensis (435), Azotobacter vinelandii (379), and T. thermophilus (469). Chromosome multiplicity enhances radiation resistance in E. coli and S. cerevisiae, with the former being more radiation resistant when grown in rich medium, which gives rise to multigenomic cells (327), and the latter being more radiation resistant in the diploid than in the haploid form (433). However, as ploidy increases beyond the diploid form in S. cerevisiae, the radiation resistance is progressively decreased (433).
The recombinational repair of DSBs in D. radiodurans proceeds via two homologous recombination processes, ESDSA and homologous recombination by crossovers, both of which rely on the RecA recombinase (128, 572, 676) (Fig. 6). DSB repair is initiated by the RecFOR pathway, as D. radiodurans lacks RecBC (49). The ends of DSBs are presumably processed by UvrD and RecJ into 3′ single-stranded DNA substrates onto which the RecFOR complex loads RecA (49). RecA and its homolog, RadA, prime DNA repair synthesis on partially overlapping fragments as templates (572, 676) (Fig. 6, step 3). RecA is essential, as RadA cannot replace RecA-mediated DNA synthesis priming. Following RecA-RadA-catalyzed priming, DNA Pol III initiates DNA repair synthesis, whereas the elongation step is performed by either (i) Pol III alone, with Pol I protecting the DNA fragments' integrity following BER, or (ii) Pol I after a Pol III dissociation caused, for instance, by unrepaired lesions in the template (572) (Fig. 6, step 4). The unwinding of the dsDNA template during D-loop migration may be mediated by UvrD, RecD, RecQ, RuvAB, and/or other helicases, such as Rad54 (DR1258/DR1259), a member of the eukaryotic Swi2/Snf2 family of SF2 helicases, which promotes heteroduplex DNA extension during strand exchange and displaces Rad51 from duplex DNA in yeast (582, 583). DNA repair synthesis generates long newly synthesized single strands, which processively dissociate from the migrating D loops, aided by DNA helicases, and can readily anneal with complementary strands (Fig. 6, steps 6 and 7). Whether single-strand annealing is a spontaneous process promoted by the proximity of the annealing strands or requires a specialized protein, such as Rad52 in eukaryotes, has yet to be examined. The 3′ flaps generated after the annealing of single strands could be incised by SbcCD (259) (Fig. 6, step 8). The long linear products of ESDSA require RecA-mediated crossovers within overlapping homologies to mature into circular chromosomes (323, 676) (Fig. 6, step 9).
Physical scaffolds for DNA repair in D. radiodurans.
The recombinational repair of DSBs entails a search for homologous DNA sites by RecA-DNA filaments. The prevailing puzzle in recombinational repair is how RecA searches for homologies among numerous dsDNA fragments generated by ionizing radiation. The homology search by the RecA-DNA filament is encumbered by the small cellular concentration of the target compared to the vast excess of the nontarget DNA along with the low diffusion coefficients of DNA (201). A homologous search conducted under such circumstances would entail a repetitive and futile reinspection of multiple randomly dispersed DNA fragments, making the process of the reconstruction of chromosomes seem like an insurmountable task (415). Several models explain how structural aspects may contribute to the observed rapidity and efficiency of the RecA-mediated homology search in D. radiodurans: (i) genome condensation, (ii) ring-like nucleoid morphology, (iii) DNA-membrane association, and (iv) chromosome alignment. Although DNA repair proteins in D. radiodurans are enzymatically very similar to those in other bacteria, their remarkable efficiency in assembling DNA fragments may be partially imparted by structural facilities.
(i) Genome condensation in D. radiodurans.
The general observation that nucleoids in stationary-phase cells are compact, whereas exponentially grown cells have diffuse nucleoids, also applies to D. radiodurans (177). Genomic DNA from exponential-phase cells of radiation-resistant species is still more condensed than that of radiation-sensitive species (688). Genome condensation is thought to preserve DNA linear continuity even in the presence of numerous breaks by (i) restricting the diffusion of DNA fragments, (ii) protecting the fragments from free radicals generated in the cytoplasm by water radiolysis, and (iii) limiting accessibility to degradation enzymes (354, 688). Apart from restricting the diffusion of broken fragments, genome condensation may also prevent the occurrence of DSBs by stabilizing proximal SSBs and preventing the separation of DNA ends (108).
In E. coli, the histone-like protein HU (162, 525) and the Dps family of proteins (200, 294) serve an important function in the compaction of the nucleoid. In vivo studies showed that the D. radiodurans HU ortholog (DRA0065) plays a major role in nucleoid organization and DNA compaction in D. radiodurans (459). It is essential for cell viability, as its progressive depletion leads to the decondensation of DNA, the fractionation of the nucleoid, and cell lysis (459). However, the nucleoid condensation role of HU is not supported by data from in vitro studies, which showed that HU cannot bend DNA or bind to DNA with nicks or gaps (209). HU binds to four-way DNA junctions and may be additionally optimized for DNA recombination events by stabilizing recombination intermediates (208, 209). Contrary to HU, one of the D. radiodurans Dps (“DNA protection during starvation”) proteins, Dps1 (DR2263), was shown to promote DNA compaction in vitro (54) but not in vivo, as its depletion has no effect on genome condensation and cell viability (459). The D. radiodurans SMC (“structural maintenance of chromosomes”) protein (DR1471), which is important for genome stabilization and DNA repair in eukaryotes (100, 365) and bacteria (640), has no role in genome condensation in D. radiodurans either (140a). Although manganese ions bind D. radiodurans chromosomes (346) and neutralize the repulsion between phosphate groups, they are not essential for chromosomal condensation, as Mn-depleted radiosensitive D. radiodurans nucleoids display normal levels of condensation (123).
(ii) Ring-like nucleoids in D. radiodurans.
Cross sections through highly condensed stationary-phase D. radiodurans cells reveal circular-shaped structures (123, 453) in which DNA is folded around a proteinaceous core possibly consisting of SMC proteins (688). This unusual toroidal (ring-like) morphology of the D. radiodurans genome was also revealed by transmission electron microscopy (354) as well as epifluorescence and deconvolution microscopy (688). It was proposed to restrict the diffusion of DNA fragments and provide a scaffold for DNA repair through high-fidelity DNA end-joining processes (179, 354). However, nonhomologous end joining (NHEJ) has never been observed for D. radiodurans (125, 676). The toroidal DNA organization is more pronounced in stationary-phase D. radiodurans cells, which is consistent with a 3-fold superior resistance to ionizing radiation compared to that of exponentially grown cells (179, 354). Nevertheless, several observations refuted the contribution of toroidal DNA organization to radiation resistance (38): (i) the resistance of D. radiodurans cultures grown in TGY broth that contain cells without ringlike nucleoids is superior to the resistance of cultures grown in a defined minimal medium that contains cells with DNA toroids (123); (ii) apart from D. radiodurans, D. radiophilus, D. proteolyticus, D. grandis, and D. murrayi, other radiation-resistant Deinococcaceae, such as D. radiopugnans and D. geothermalis, lack a specific nucleoid organization (688); and (iii) cryoelectron microscopy of vitreous sections (CEMOVIS) failed to reveal ring-like nucleoids and showed that DNA fragments are diffusible (177). The mobility of chromosomal fragments is also supported by the high efficiency of recombination between homologous DNA fragments, whether located adjacently on the same chromosome (125), separated on different chromosomes (126), or present on a chromosome and a plasmid (127).
(iii) DNA-membrane association in D. radiodurans.
Another model proposes that DNA is attached to the membrane, which acts as a support structure to ensure the correct sequential restitution of the genome (79). According to this model, DNA fragments attached to the fragments of the plasma membrane are rejoined by attachment to the piece of DNA that remains bound to a larger membrane structure (79). DNA from unirradiated D. radiodurans cells was indeed found to be associated with the membrane complex (79, 154), while DNA shattered by X-ray radiation dissociates from the membranes and reassociates upon the rejoining of the broken DNA fragments (79). This reassociation is decreased by chloramphenicol and inhibited by iodoacetamide, thereby increasing sensitivity to radiation (129). The DNA-membrane association may be promoted by CinA, which binds RecA and locates it to the cell membrane in Streptococcus pneumoniae (395). The DNA-membrane association may also contribute to the high transformation competence of D. radiodurans. It is worth noting that DNA-membrane complexes were also found in E. coli (131) and murine lymphoma (mammalian) cells, which show the same effect of radiation on the release of DNA from the membrane (477).
(iv) Chromosome alignment in D. radiodurans.
Murray proposed in 1986 that the error-free repair in D. radiodurans is mediated by some aspect of the nuclear structure that allows the juxtaposition of homologous regions of multiple genome copies so that recombinational exchange can take place (452). Minton and Daly elaborated a model where chromosomes are linked to each other by thousands of Holliday junctions (415). Such a prealignment would ensure immediate access to the substrate during recombinational repair and would thus facilitate the “search for homology.” However, molecular studies have shown high levels of recombination between homologous DSB fragments irrespective of their genomic origin, which argues against the existence of structures linking chromosomes (124-127). Apart from the Holliday junctions, chromosome alignment could also be achieved by sequence-specific DNA binding proteins. Although there is no evidence for a preexisting alignment of D. radiodurans chromosomes, we have yet to inspect the possibility that the alignment of homologous fragments occurs after exposure to ionizing radiation (“postalignment”). Deinococcal RecA, which preferentially binds double-stranded DNA (296), may bring together DNA fragments with overlapping homology. Such a RecA-mediated postirradiation fragment alignment would facilitate the accurate priming of strand extension during ESDSA.
Enzymatic tools of recombinational repair in D. radiodurans. (i) The RecFOR pathway.
In bacterial cells the processing of double-stranded DNA fragments in order to obtain single-stranded 3′ DNA ends onto which RecA can be loaded is carried out by the RecBCD or the RecFOR pathway. The RecBCD heterotrimer acts at the same time as helicase, ATP-dependent dsDNA exonuclease, ssDNA exonuclease, and ssDNA endonuclease (143, 323). D. radiodurans does not encode RecB and RecC homologs, or SbcB (a 3′-5′ ssDNA nuclease), and the levels of nuclease activity in deinococcal cell extracts are consequently much lower than that in E. coli extracts (288). The overexpression of E. coli RecBC (288) and SbcB (418) in D. radiodurans sensitizes the cells to ionizing radiation and delays or inhibits DSB repair in gamma-irradiated cells, respectively. RecBC-expressing D. radiodurans cells suffer from an extensive degradation of DNA ends, while the inhibition of DSB repair in SbcB-expressing cells suggests that 3′ ssDNA ends are indispensable for the recombinational repair of DSBs (418). Moreover, the ATP-sensitive 3′-5′ exonuclease activity of a dual-function esterase/nuclease, DR0505, is attenuated during the initial stages of postirradiation recovery by higher ATP levels (282, 318). The RecBC activities that are missing in D. radiodurans are replaced with the RecFOR pathway (49). D. radiodurans contains all components of the RecFOR pathway, RecF (DR1089), RecO (DR0819), RecR (DR0198), and RecJ (DR1126), and is devoid of SbcB, which inhibits this pathway. All four proteins, in particular RecJ, are required for normal cellular growth and recombinational repair (49). Recombinational repair is facilitated by RecN (DR1477), an SMC family member which tethers DNA molecules in a cohesin-like fashion and prevents the separation of DNA fragments (518). In B. subtilis, RecN is one of the first proteins recruited to DSBs (531). In vitro, D. radiodurans RecN also stimulates the intermolecular ligation of linear DNA molecules in the presence of DNA ligase (518).
In E. coli the RecFOR pathway governs the initiation of recombination. The RecQ helicase unwinds DNA in the 3′-to-5′ direction, and the RecJ 5′-3′ exonuclease digests the 5′ end, producing single-stranded 3′ ends (374). In D. radiodurans, the UvrD helicase (DR1775) rather than RecQ (DR1289) appears to be involved in the initial steps of DSB repair (49) (Fig. 7, step 1). Whereas recQ-deficient cells exhibit wild-type properties, uvrD-deficient cells are moderately sensitive to gamma rays and display a significant delay in DNA synthesis and DNA fragment reassembly (49) (Fig. 8). The requirement for UvrD in DSB repair in D. radiodurans is surprising given its antirecombination properties in E. coli (191, 636). Although the recQ-deficient strain is only slightly sensitive to ionizing radiation, it is highly sensitive to MMC, H2O2, and UV radiation (261) (Fig. 8). The D. radiodurans RecQ helicase contains three C-terminal HRD (helicase-RNase D) domains also found in Neisseria meningitidis and Neisseria gonorrhoeae, which are critical for high-affinity DNA binding and DNA unwinding (261, 293). Its involvement in DNA repair in D. radiodurans remains to be elucidated. For D. radiodurans, RecJ (DR1226) is an essential protein, and a heterozygous recJ mutant (with 60% of the genome copies retaining the recJ gene) is modestly sensitive to gamma rays and only slightly sensitive to UV and H2O2 (49, 85). As its E. coli homolog, D. radiodurans RecJ exonuclease has 5′-3′ single-strand-specific exonuclease activity (85). Whereas RecJ is the only 5′-3′ exonuclease in D. radiodurans, E. coli has two other 5′-3′ exonucleases (RecBCD and XseAB), which may explain the lethality of the recJ mutation in D. radiodurans (49). The 3′-tailed ssDNA produced by UvrD and RecJ is coated with SSB proteins (Fig. 7, step 2), which are highly induced in response to ionizing radiation and which are thought to protect ssDNA during long periods of desiccation (52) [see “Enzymatic tools of recombinational repair in D. radiodurans. (iv) The SSB proteins”]. Apart from acting upstream of RecJ and UvrD, RecN may also stabilize 3′ ssDNA tails as in E. coli and B. subtilis (369, 530). A RecN deficiency in D. radiodurans leads to a slightly increased sensitivity to gamma rays, UV radiation, and MMC (202) (Fig. 8).
FIG. 7.
Model for the processing of DSB ends and the initiation of recombination in D. radiodurans by the RecFOR pathway. UvrD unwinds the ends of DSBs in the 3′-to-5′ direction, and RecJ digests the 5′ end (step 1). 3′ single-stranded tails are coated by SSB proteins (step 2). RecF binds to the ssDNA-dsDNA junction (step 3) and promotes the assembly of the RecOR complex (RecO-to-RecR ratio, 4:2) onto the junctions (step 4). RecOR displaces SSB proteins and loads RecA onto 3′ ssDNA (step 5). (Based on data from references 49 and 619.)
FIG. 8.
Effect of mutations in DNA repair-related genes on the resistance of D. radiodurans to ionizing radiation (A) (based on data from references 48, 49, 170, 202, 225, 261, 305, 308, 343, 407, 551, 570, 602, 645, 663, and 665), UV-C radiation resistance (B) (based on data from references 43, 91, 170, 202, 225, 305, 308, 343, 551, 603, 645, and 665 and unpublished data from D. Slade), and MMC resistance (C) (based on data from references 4, 48, 202, 223, 225, 239, 261, 277, 305, 308, 343, 407, and 551 and unpublished data from D. Slade). Genes are classified into three groups according to the sensitivity of their mutants: extremely sensitive genes are depicted in black, highly sensitive genes are depicted in red, and moderately/slightly sensitive genes are depicted in blue. Designations for radiation sensitivity are relative to wild-type D. radiodurans, as even the most sensitive repair-deficient D. radiodurans mutants are more resistant than many bacteria which encode a full complement of DNA repair genes. The compiled data were not obtained under the same experimental conditions.
In E. coli the RecFOR complex loads RecA onto the 3′-tailed DNA coated with SSB (430) (Fig. 7, steps 3 to 5). The D. radiodurans recF, recO, and recR mutants are extremely sensitive to gamma rays, as much as the recA mutant, and share the same kinetics of DSB repair characterized by an incomplete genome reconstitution, a reduced level of DNA breakdown, and an absence of DNA synthesis (49, 665) (Fig. 8). E. coli recO can only partially compensate for the gamma radiation sensitivity of the deinococcal recO mutant (665). Although weaker than the E. coli protein, deinococcal RecO has ssDNA and dsDNA binding and strand-annealing properties and forms a stable complex with RecR (347, 385). The selective binding to 3′-overhanging DNA requires the assembly of the RecOR complex, as RecO and RecR alone do not display this binding preference (619) (Fig. 7, step 4). Deinococcal RecR is a homotetramer with a ring-shaped architecture containing a central hole, which is large enough to accommodate dsDNA and may act as a DNA-holding clamp that is capable of opening and closing (344). Although the RecOR complex alone displays a high binding affinity for the ssDNA-dsDNA junctions, RecF may stabilize the RecOR assembly on ssDNA-dsDNA junctions through binding dsDNA and interacting with RecR (619) (Fig. 7, steps 3 and 4). Overall, the loading of RecA at the processed DSB site is mediated by RecR interactions with RecO, which binds SSB-coated ssDNA, and with RecF, which in turn recognizes the ssDNA-dsDNA junctions (386).
(ii) RecA.
In E. coli the RecA protein binds to DNA ends processed by the RecBCD or the RecFOR pathway and forms a nucleoprotein filament that searches for homology in dsDNA (16). The nucleoprotein filament aligns the bound single strand with a homologous duplex and promotes a strand exchange between them. The RecA-mediated invasion of a homologous duplex by ssDNA forms a D loop (109).
RecA-mediated homologous recombination is critical to DSB repair and cellular survival in D. radiodurans (125, 128). A reduced frequency of recombination in a recA mutant is commensurate with the reduction in radiation resistance (445), while the shoulder of the survival curve is completely eliminated for ionizing radiation, UV radiation, and MMC (223, 439) (Fig. 8). The recA mutant has a doubling time in TGY broth of 200 min, an irregular cell morphology, and a transformation deficiency (439). D. radiodurans recA mutants were obtained by MNNG treatment (rec30; G224S) (439), targeted insertion (223), or a replacement of the recA operon with an antibiotic resistance cassette (64, 342). The three recA mutants share similar degrees of sensitivity to ionizing radiation but are not phenotypically equivalent with respect to UV and MMC sensitivity, with the rec30 mutant being less sensitive than either the insertional or the replacement mutants (88, 223, 439, 541, 665). Surprisingly, the insertional D. radiodurans recA mutant is slightly more radiation resistant than wild-type E. coli (414), which may be explained by the high level of antioxidation protection of the DNA repair enzymes in D. radiodurans that can somewhat compensate for the absence of RecA (see “Antioxidation Protection in D. radiodurans” and “The RecA-Independent pathway of DSB Repair in D. radiodurans”). In the rec30 mutant there is no radiation-induced DNA degradation at 500 Gy (439), while in the replacement recA mutant DNA degradation is reduced compared to the wild type (49, 572). Whereas D. radiodurans uvrA (4) and polA (225) can be fully complemented by the respective E. coli genes, this is not the case with rec30 (88), while a deletion mutant of recA can be partially complemented with E. coli recA (541). Conversely, deinococcal RecA can fully restore recombinational repair in an E. coli recA mutant (457). However, the gamma ray resistance of E. coli recA complemented with deinococcal recA is not enhanced over that of a recAEC+ strain, which suggests that deinococcal RecA is insufficient for imparting the resistance phenotype (457).
The recA locus (DR2340) in D. radiodurans forms a polycistronic operon with the upstream competence/damage-inducible protein CinA along with a 2′-5′ RNA ligase (LigT) (457). In S. pneumoniae, CinA binds RecA and locates it to the cell membrane (395), which may be relevant for the membrane association of the D. radiodurans genome that is seemingly involved in DSB repair (79, 154). The inactivation of deinococcal CinA has only a marginal (2-fold) effect on transformation efficiency (64). The 2′-5′ RNA ligase is homologous to the E. coli enzyme (26) and is highly induced following radiation (368). It is likely involved in the repair of damaged RNA (26).
RecA is expressed constitutively at low levels (364) and is transiently expressed at high levels following extreme DNA damage (64, 88, 364, 458). D. deserti has two different RecA proteins encoded by three different genes (168). The chromosomal RecA protein is present in unirradiated cells, whereas both the chromosomal and the plasmidic RecA proteins are induced after gamma radiation (168). The basal level of RecA in D. radiodurans has been estimated to be 11,000 monomers per cell (64). RecA gene expression is induced 8 times following 15 kGy of ionizing radiation (368), whereas protein expression is induced 2 to 2.5 times (458) and 4 times (64) following 2 kGy and 7 kGy of ionizing radiation, respectively. The induction of RecA begins immediately after exposure to ionizing radiation and continues at a constant rate until it reaches a plateau (64). The recA mutant cells that produce RecA at a low concentration from an IPTG-inducible promoter (2,500 molecules per cell) are as radiation resistant as wild-type bacteria but sensitive to MMC (277). However, when the postirradiation recovery is monitored in a liquid medium, a limited concentration of RecA delays DSB repair and leads to cell death (277).
Neither CinA and LigT nor the two LexA proteins are involved in the regulation of expression or function of RecA (64, 458). LexA1 (DRA0344) undergoes RecA-mediated cleavage but does not repress the expression of RecA, as the level of RecA in unirradiated cells is the same in wild-type and lexA mutant cells (64, 458, 537). Furthermore, the deletion of lexA does not affect the repair of DSBs (277). The N-terminal domain of deinococcal LexA is distinct from the corresponding E. coli domain that is involved in DNA binding (263), suggesting that the deinococcal LexA DNA binding motif is different from the E. coli SOS box (458). D. radiodurans encodes another LexA homolog, LexA2 (DRA0074), which is also cleavable by RecA and not involved in the regulation of RecA expression (565). The lexA2 disruptant strain exhibits a higher level of resistance to ionizing radiation than the wild type (537). The transcription of the recA gene following exposure to ionizing radiation is stimulated by a novel regulatory protein, IrrE (DR0167) (170), also referred to as PprI (260). Although recA is strongly induced after irradiation, pprI is constitutively expressed, with no postirradiation induction (368). Under nonstress conditions, the transcriptional regulator DrRRA positively controls the expression of RecA, with no effect under radiation stress (645). The transcriptional regulator DdrO may be responsible for the induction of recA in gamma-irradiated cells (383). RecX is a negative regulator of recA expression and also reduces RecA recombination activities in vivo (564). RecX directly inhibits RecA activities in vitro, including ATPase activity, LexA cleavage, and strand exchange (564).
Deinococcal RecA is exceptional in that it binds dsDNA with a higher affinity than that for ssDNA and promotes an inverse strand exchange relative to other known RecA proteins (296). Like E. coli RecA (RecAEC), D. radiodurans RecA (RecADR) (i) forms filaments on ssDNA (297), (ii) possesses a coprotease activity (536), (iii) has a binding stoichiometry of 1 RecADR monomer per 3 nucleotides of ssDNA (297), and (iv) can use dATP as a cofactor, which permits an easier displacement of SSBs from ssDNA, while with ATP as a cofactor, SSB inhibits the binding of RecA to ssDNA (297). However, RecADR differs from RecAEC in many aspects. RecADR promotes an inverse DNA strand exchange reaction, binding the dsDNA first and the homologous ssDNA substrate second (296). Although the binding of RecADR to ssDNA is kinetically rapid, with no lag time, the complexes are not thermodynamically favorable, whereas the binding to dsDNA is slow, with a lag time, but the complexes are favorable (295). Consistently, with ssDNA as a cofactor, ATP hydrolysis begins immediately; with dsDNA as a cofactor, a lag is observed before the steady state is established, while with both ssDNA and dsDNA, the lag is absent (297). Inversely to the strand exchange reaction with RecAEC, which is maximized when sufficient RecA is provided to saturate the binding sites on ssDNA, the maximum reaction with RecADR is achieved when it saturates dsDNA (296). The mechanistic features of deinococcal RecA regarding the preference for an inverse strand exchange reaction and for binding dsDNA may have a structural basis, with three outstanding structural differences compared to the E. coli enzyme: (i) a large reorientation of the C-terminal domain, which may bind ssDNA instead of dsDNA; (ii) an increased positive electrostatic potential along the central axis of the filament, which may dictate the binding of dsDNA instead of ssDNA; and (iii) unique amino acids around a flexible β-hairpin that is implicated in DNA binding (503). Although RecADR shares some of its distinct conserved residues with Thermus RecA proteins (383), Thermus aquaticus RecA exhibits the same DNA strand exchange properties as those of RecAEC (18). Overall, RecADR seems to have evolved to efficiently promote the repair of DSBs by finding overlapping dsDNA fragments and splicing them together (296).
(iii) RadA, RuvABC, and RecG.
RadA is a highly conserved eubacterial protein absent from eukaryotic genomes (44). The RadA protein sequence contains three characteristic regions, an N-terminal zinc finger, a middle region related to the RecA strand exchange protein, and the DnaB helicase with Walker A and B boxes characteristic of ATPases, while the C-terminal region is related to the Lon protease (44). Both E. coli (44, 146, 535) and D. radiodurans (572, 687) radA mutants are moderately sensitive to ionizing radiation and have a delay in repairing DSBs induced by ionizing radiation (Fig. 8). Unlike E. coli radA, the D. radiodurans radA mutant is also highly sensitive to MMC (D. Slade, unpublished data). While the biochemical activity of RadA awaits analysis, in vivo studies of E. coli point to its involvement in the processing of branched DNA molecules or stalled replication forks (44, 372). In D. radiodurans, RadA was found to assist RecA in priming DNA repair synthesis during ESDSA, although it cannot replace RecA (572) (Fig. 6, step 3). In this sense, deinococcal RadA resembles the family of Rad51 paralogs that function in concert with the Rad51 strand exchange protein (392) and is most similar to Dmc1 (44). RadA functions in D. radiodurans seem to diverge depending on the availability of RecA, as in the absence of RecA, RadA functions in a different pathway, which is responsible for a partial reconstitution of the NotI fragments in the recA mutant (572) (see “The RecA-Independent Pathway of DSB Repair in D. radiodurans”).
The DNA heteroduplex formed by RecA-mediated strand exchange is extended by branch migration. In E. coli, RuvAB (273, 623) and RecG (370) are specialized DNA helicases that possess branch migration activities. Whereas the RuvAB complex stimulates the branch migration of Holliday junctions in the 5′-to-3′ direction (624), the migration promoted by RecG is in the opposite 3′-to-5′ direction, with the latter leading to the destruction of the heteroduplex (652). D. radiodurans possesses the corresponding homologs: RuvA (DR1274), RuvB (DR0596), and RecG (DR1916). The ruvB mutant is modestly sensitive to UV radiation, gamma rays, and MMC (305), while the recG mutant is highly sensitive to gamma rays and H2O2 (663) (Fig. 8).
(iv) The SSB proteins.
In DNA strand exchange reactions, the SSB (ssDNA binding) protein is required (i) to protect ssDNA from nucleolytic degradation and remove the secondary structure in the ssDNA to allow complete RecA filament formation (324, 399) and (ii) to block the reversal of the strand exchange reaction by binding to the displaced strand (338) (Fig. 7). The E. coli SSB (SSBEC) and D. radiodurans SSB (SSBDR) proteins have a positive effect on the RecAEC ATPase activity, thereby promoting filament formation, while in the case of RecADR, the ATPase activity is slowly reduced after an initial increase as RecADR is replaced by either SSB protein (464). The D. radiodurans SSB protein (DR0099) is different from standard bacterial homologs. It is double the size of the E. coli SSB protein, forms dimers instead of tetramers, and contains two oligonucleotide/oligosaccharide-binding (OB) folds for binding ssDNA per monomer instead of one (174). The deinococcal SSB arrangement of two DNA binding domains per polypeptide allows an independent evolution of the two domains, which may have specialized differently in protecting ssDNA (52). The structure of the deinococcal SSB protein shows that adjacent dimers bind each other by using an extensive surface area that is formed by their N-terminal OB domains and β-hairpin connectors between the domains (52). This interface may promote the assembly of multiple SSB dimers on long ssDNA molecules, protecting ssDNA during long periods of desiccation until nutrients become available (52). The deinococcal SSB protein binds 47 to 54 nucleotides (656) and has the capacity to denature the DNA helix at the ssDNA-dsDNA junctions of partially duplex DNAs, preferentially with 3′ overhangs (175). It stimulates RecADR- and RecAEC-mediated DNA strand exchange reactions with the same efficiency as that of the E. coli SSB protein (174). E. coli maintains 200 to 3,000 SSB tetramers per cell, whereas in D. radiodurans the 19,500 dimers per cell increase to 56,000 dimers in response to ionizing radiation (52). The expression of ssb is also induced following MMC treatment but not after UV treatment, H2O2 treatment, or desiccation (629). IrrE upregulates the SSB protein in irradiated cells (377), while RecX represses it under normal conditions (563). While D. radiodurans and D. geothermalis have one SSB protein homolog, D. deserti has four (140). The D. murrayi SSB protein can complement the E. coli SSB protein and is the most thermostable SSB-like protein identified to date (188).
The Deinococcaceae are unique in having an alternative SSB protein from a new family, referred to as DdrB (DR0070) (464). It forms a pentamer with a novel ssDNA binding fold that is distinct from the OB fold (590) and binds 42 nucleotides per pentamer (464). DdrB has the same qualitative effect as SSB proteins on RecA ATPase activity during filament formation and elongation but appears to have a stronger affinity for ssDNA, making it harder for RecA to replace it on ssDNA, while DdrB can readily displace RecA from ssDNA (464). Unlike the SSB protein, DdrB displays single-stranded DNA-annealing activity, even in the presence of SSB (664). Following 3 kGy (602) and 15 kGy (368) of gamma radiation, ddrB gene expression is highly upregulated, and Western blot analyses detected the protein in 4-kGy-irradiated cells but not in unirradiated cells (464). The inactivation of ddrB modestly sensitizes the cells to ionizing radiation (602) (Fig. 8), without any effect on the kinetics of DNA fragment reassembly (664). However, a ddrB deletion increases the sensitivity of the recA mutant (602) and completely abolishes fragment rejoining (664). DdrB is therefore proposed to act in the RecA-independent DNA repair pathway, where fragments are rejoined by single-strand annealing (664) (see “The RecA-Independent Pathway of DSB Repair in D. radiodurans”).
(v) RecD.
E. coli RecD is a DNA-dependent ATPase as well as a 5′-3′ helicase (144, 609). Although D. radiodurans does not encode RecB and RecC homologs, it does encode RecD (DR1902). RecD is also present in the absence of RecBC in firmicutes, mollicutes, Streptomyces, and Desulfovibrio (429, 521). RecD2 (RecD present in genomes lacking RecBC) differs from RecD1 by an N-terminal extension (521) and represents an ancestral form of RecD (429). Deinococcal RecD has a helicase activity with 5′-to-3′ polarity and low processivity (556, 644). Its deletion enhances the efficiency of transformation by exogenous homologous DNA, which suggests that RecD has antirecombinogenic properties in D. radiodurans (551). According to data described previously by Zhou et al. (686), a RecD-deficient strain was sensitive to H2O2 but resistant to UV and ionizing radiation, whereas Servinsky and Julin (551) reported moderate sensitivity to ionizing and UV radiation as well as H2O2 (Fig. 8). RecD is not essential for DSB repair, as the kinetics of DSB repair remain unaltered in gamma-irradiated or H2O2-stressed recD mutant cells (49, 429). RecD was also proposed to take part in antioxidant processes by stimulating catalase activity and ROS scavenging in D. radiodurans (686).
(vi) SbcCD.
Homologs of Rad50/SbcC (DR1922) and Mre11/SbcD (DR1921) do not seem crucial for the recombinational repair of DSBs in D. radiodurans. The sbcCD mutant cells are resistant to doses as high as 10 kGy and exhibit only a slight delay in the reconstitution of the radiation-fragmented genome (48) (Fig. 8). As in E. coli (104), the SbcCD complex has single-stranded endonuclease and 3′-5′ double-stranded exonuclease activities and can cleave hairpin DNA (259). Deinococcal SbcCD was also shown to cleave 3′-flap ssDNA in vitro (259). A similar hairpin-modulated 3′-5′ exonuclease activity was observed for Pol X, an X family DNA polymerase (61). Another nuclease, DR0505, can act on hairpin structures as an ATP-dependent ssDNA-dsDNA junction endonuclease (318). While individual deletions of SbcCD, Pol X, and DR0505 have no effects on DNA damage sensitivity, a coupled SbcCD-Pol X deficiency in D. radiodurans yields an additive radiation-sensitive phenotype and an additive delay in genome reconstitution (48, 318, 343). As all three have hairpin-modulated nuclease activities, SbcCD, Pol X, and DR0505 were proposed to have redundant roles in the processing of damaged DNA ends in cells containing DNA lesions that are excessively numerous or difficult to repair (48). Additionally, SbcCD is likely involved in DNA folding together with the SMC protein, as a deficiency of both proteins sensitizes the cells to gyrase inhibitors (140a).
(vii) Novel deinococcal DNA repair proteins.
PprA (“pleiotropic protein promoting DNA repair”) (DRA0346) is a DNA repair protein (456) unique to the Deinococcaceae (216). PprA is highly expressed after ionizing radiation and desiccation (368, 602) and repressed by LexA2 (537), PprM (468), and RecX (563). Whereas a lexA2 deletion increases radiation resistance (537), a pprM deletion reduces it significantly, which suggests that PprM regulates other proteins as well (468). In vitro, PprA stimulates DNA end-joining reactions catalyzed by ATP- and NAD-dependent DNA ligases while inhibiting E. coli exonuclease III activity and may thus protect DNA ends from extensive degradation (456). It binds DNA ends with a greater affinity than internal DNA regions and may even promote DNA looping (451). Its ability to bind dsDNA carrying strand breaks was harnessed to visualize radiation-induced DNA strand breaks in mammalian cell cultures by immunofluorescence, where permeabilized irradiated cells were treated with PprA and fluorescently labeled with an anti-PprA antibody (538). The pprA mutant strain grows slower than the wild type, with a doubling time of 2.4 h (602). It is highly sensitive to ionizing radiation, MMC (456), and UV-A radiation (43) (Fig. 8). A pprA recA double mutant is as sensitive to ionizing radiation as the recA mutant, which suggests that PprA is epistatic to RecA (i.e., they function in the same pathway) (602). A transgenic pprA-expressing E. coli strain is 2- to 3-fold more tolerant to H2O2 due to the PprA-mediated stimulation of the E. coli catalase (KatE) (320). PprA also stimulates E. coli catalase activity in vitro (320). By acting as a Ku-like protein, PprA was proposed to stimulate NHEJ in D. radiodurans by the ATP-dependent ligase DRB0100 (456). The ATP-dependent ligase activity of DRB0100 was demonstrated at 60°C (340) but not at 30°C (60).
DdrA (“DNA damage response”) (DR0423) is another highly radiation- and desiccation-induced protein also found in T. thermophilus (475). DdrA protects 3′ ssDNA overhangs from degradation by E. coli exonuclease I (239), presumably ensuring long-lived recombination substrates and the recycling of RecA (277). Its absence results in excessive DNA degradation following radiation exposure (239). Hence, DdrA is proposed to preserve genome integrity by protecting DNA fragments generated by ionizing radiation from nuclease degradation, particularly when nutrients are scarce and when DNA repair processes are hindered as a result (239). The disruption of this gene results in only modest sensitivity to ionizing radiation and to MMC but causes a substantial increase in DNA degradation (239) (Fig. 8). High concentrations of RecA can partially suppress the radiation sensitivity of ddrA mutant cells, which suggests that RecA can protect ssDNA tails from degradation by polymerization on ssDNA or that a more rapid recombinational repair can compensate for the defects in alternative DSB repair mechanisms (277). Due to its distant relation to Rad52, DdrA could be a component of the single-strand annealing (SSA) system that functions in conjunction with RecA-dependent homologous recombination or independently of RecA (239) (see “The RecA-Independent Pathway of DSB Repair in D. radiodurans”). Although DdrA does not display any Rad52-like single-strand annealing activity (239), the crystal structure of D. deserti DdrA shows similarity with Rad52 (227).
Replication Processes in D. radiodurans DNA Repair
DNA repair in irradiated D. radiodurans is stringently dependent on DNA synthesis, which coincides with the fragment reassembly observed with PFGE gels (676) (Fig. 4A and D). On this basis, several described double-strand-break repair mechanisms that involve little DNA synthesis, such as nonhomologous end joining (NHEJ) of DNA fragments, homologous recombination via crossovers, and SSA, were discounted as major DNA repair pathways for the reconstitution of shattered chromosomes in this bacterium (676) (Fig. 9). The SSA model was further refuted by UV photolysis experiments with cellular DNA repaired in bromodeoxyuridine (BrdU), a photosensitive thymidine analog which yields SSBs if present in only one strand for a given region of DNA or DSBs when present in both strands. UV photolysis degrades D. radiodurans DNA repaired in BrdU by double-strand breakage, which is in agreement with synthesis-dependent strand annealing (SDSA) and the conservative version of break-induced replication (BIR) but in disagreement with SSA (676). UV breakage generates fragment sizes similar to those seen after the initial radiation-induced DNA breakage, which suggests that reassembled fragments are linked together via double-stranded blocks of newly synthesized DNA and that reassembled chromosomes are patchworks of old (synthesized before radiation) and new (synthesized after radiation) double-stranded DNA blocks (Fig. 6, steps 7 and 8). SDSA and BIR, the two synthesis-dependent DSB repair mechanisms, can be differentiated by a dose gradient of UV light in the photolysis experiment, where a saturating dose of UV is reached with the fragment size corresponding to the initial radiation-induced breakage (676). Thus, BIR, which would leave little, if any, original DNA duplex within the repaired chromosome, was largely dismissed, although some fragment reassembly events may still proceed by a conservative version of BIR. In a template-switching variant of BIR, strand invasion creates a D loop that migrates down the template, and the extended 3′ end undergoes several rounds of strand invasion until captured by annealing with a complementary strand. This variant of BIR is highly similar to SDSA (576). The SDSA repair process in D. radiodurans was termed “extensive SDSA,” or ESDSA, due to the extensive DNA synthesis between dispersed fragments that belong to different chromosomal copies and have overlapping sequence homology (676). Overall, DSB repair in D. radiodurans is remarkably similar to that of yeast, where (i) DSBs are repaired by gene conversion (SDSA or homologous recombination by crossovers) 98% of the time (480) and (ii) SDSA is the main mechanism of mitotic DSB repair (269), while (iii) most of the meiotic DSBs are repaired by crossovers (480). Similarities with yeast meiotic repair were first demonstrated by Daly and Minton, who adapted a reporter system used in yeast to monitor interplasmidic, interchromosomal, and plasmid-chromosome recombination between loci with physical and genetic polymorphisms. They found that recombination occurs by gene conversion with and without crossovers and that the kinetics of the recombinant products are similar to those of yeast cells undergoing meiosis (124-127, 359).
FIG. 9.
Mechanisms of DSB repair involving homologous sequences. DSB repair commences with the exonucleolytic resection of the ends of DSBs in the 5′-to-3′ direction to produce 3′ single-stranded tails. In gene conversion and break-induced replication (BIR), 3′ tails invade a homologous template. Strand invasion creates a D loop, where 3′ ends prime new DNA synthesis. Gene conversion further branches into two different mechanisms. In synthesis-dependent strand annealing (SDSA), newly synthesized DNA strands formed by D-loop migration are displaced from the template and anneal to each other to restore a contiguous chromosome in a noncrossover configuration. In the homologous recombination (HR) model, two Holliday junctions are formed as the D loop created by strand invasion pairs with the other side of the DSB (second-end capture) and the 3′ end of the noninvading strand is extended by DNA synthesis. Two Holliday junctions can branch migrate to enlarge the heteroduplex region. Holliday junctions can be cleaved by a resolvase by cutting either the two noncrossed strands or the two crossed strands. If both Holliday junctions are cleaved in the same way, gene conversion is not associated with crossover (No c.o.), whereas differential cleavage results in crossover (c.o.). BIR occurs when only one end of a DSB is available for recombination. Such one-ended strand invasion results in extensive DNA synthesis, which may extend hundreds of kilobases. Intrachromosomal single-strand annealing (SSA) enables the repair of a DSB that occurs between two flanking homologous regions (direct repeats). Resection produces single-stranded tails in which complementary strands of the duplicated sequence are exposed and can reanneal, resulting in a deletion of the intervening sequence. Interchromosomal SSA occurs between two chromosomal copies or between homologous sequences on different genomic elements (Fig. 10C) and may not result in a deletion. (Based on data from reference 480.)
Three DNA polymerases have been identified in D. radiodurans: Pol I, Pol III, and Pol X. D. radiodurans Pol I and Pol III share 41% and 44% sequence identity with the E. coli enzymes, respectively. As in E. coli, Pol I is required for SSB repair (65, 225) and for excision repair (436). The DNA polymerase function of deinococcal Pol I is modulated by Mn2+, which enables a bypass of DNA lesions that is accompanied by some loss of DNA polymerase selectivity (247). D. radiodurans polA mutants are extremely sensitive to the lethal effects of MMC and ionizing and UV radiation (65, 224, 225) (Fig. 8). D. radiodurans polA mutants obtained by MNNG mutagenesis (65, 434, 437) are typically less sensitive to DNA-damaging agents than is the mutant obtained by insertional mutagenesis (225). Deinococcal Pol I does not have special properties that are essential for the extreme resistance to DNA damage, as the expression of E. coli polA by duplication insertion can restore wild-type resistance in the D. radiodurans polA mutant (224). The contribution of Pol I to global DNA synthesis is negligible, as similar amounts of radioactive [3H]thymidine are incorporated into both wild-type and polA cell lysates (65) as well as intact cells (572). The predominant contributor to global DNA synthesis in D. radiodurans is the Pol III holoenzyme (572), an essential multiprotein complex that carries out high-fidelity processive DNA replication (275). The importance of Pol III for DNA repair in D. radiodurans was analyzed with temperature-sensitive Pol III mutants that are unable to grow at 37°C due to their inability to synthesize DNA. Temperature-sensitive Pol III mutants were isolated after MNNG treatment (446) and were constructed by gene engineering of the catalytic subunit of Pol III (dnaE) to introduce the same amino acid change as those in the respective E. coli mutants (572). In these mutants, DNA synthesis ceases immediately upon the temperature upshift and resumes immediately after the temperature downshift (446, 572). When kept at the restrictive temperature prior to irradiation, temperature-sensitive Pol III mutants are sensitized to ionizing and UV radiation and lose their recombination function (445).
Both Pol I and Pol III are central to DNA repair synthesis in D. radiodurans recovering from radiation-induced DNA damage. Fragment reassembly is delayed in the absence of Pol I (65, 436, 572) and abrogated in the absence of Pol III (572). DNA repair synthesis is primed at the 3′ end of the strand, invading the homologous region of another DNA fragment and forming a structure similar to a D loop (Fig. 6, step 3). Pol III initiates single-strand elongation, whereas Pol I participates either directly in strand extension synthesis (e.g., resumes DNA synthesis when Pol III stumbles upon base or nucleotide damage) or indirectly by contributing only to the maintenance of fragments (572) (Fig. 6, step 4). Pol I, which is required for excision repair (436) and single-strand-break repair (65, 225) in D. radiodurans, is expected to protect the integrity of the DNA fragments produced by ionizing radiation by repairing all nicks, including those created by the BER enzymes. If unrepaired, DNA base and sugar damage generated by radiation-induced ROS can result in SSBs through BER and in DSBs through proximal BER events.
In E. coli, Pol I appears to be the central polymerase in recombination-dependent replication, a mechanism for the repair of DSBs and for the restoration of collapsed replication forks (410). Pol I has all the properties necessary for performing DNA synthesis in recombination-dependent replication: (i) Pol I is essential for replication in some dnaE(Ts) strains, which suggests that Pol I is involved in an alternative pathway of DNA replication (462) by recruitment into a replisome-like holoenzyme (83); (ii) Pol I may be able to perform processive DNA replication in vivo, just like it does in vitro (397); and (iii) the Pol I polymerase domain can act on D-loop structures and use the 3′ terminus of an invading strand to initiate strand synthesis (primer extension) (366). Conversely, in D. radiodurans, Pol III is of equal, if not greater, importance for DSB repair, as it is required for the initiation of DNA repair synthesis (572). In this regard the deinococcal repair of DSBs resembles yeast SDSA repair, which also requires yeast Pol III equivalents, Polɛ and Polδ (257), and involves only leading-strand synthesis (648). When DNA damage is limited at low doses of ionizing radiation, Pol III can efficiently replace Pol I, while Pol I can compensate for the Pol III deficiency only weakly (572). However, at higher doses Pol I cannot replace Pol III in deinococcal ESDSA, whereas a Pol I deficiency causes a long delay in the Pol III-dependent reconstitution of the broken genome (572). The absolute requirement for Pol III in the initiation of DNA repair synthesis at high radiation doses presupposes (i) a higher copy number of Pol III molecules in D. radiodurans than the 10 to 20 molecules of Pol III present per E. coli cells (317), (ii) the protection of Pol III against radiation-induced damage (122), or (iii) rapid resynthesis of radiation-damaged Pol III through the preservation of transcription and translation machinery (278). The catalytic alpha subunit of Pol III is not induced following ionizing radiation, while the beta subunit (beta clamp required for processivity) and the 3′-5′ exonuclease subunit are induced immediately after irradiation (368).
Apart from Pol I and Pol III, D. radiodurans also possesses Pol X (Pol XDR) (DR0467), a DNA polymerase of the X family, endowed with the Mn2+-dependent polymerase, 3′-5′ exonuclease activity modulated by a stem-loop structure (61, 343), and 5′-deoxyribose phosphate lyase activity required for BER (289). There are 2,000 to 4,000 molecules of Pol X per exponentially growing cell (343). Pol X-deficient cells are sensitized to gamma rays only at doses exceeding 10 kGy and are capable of reconstituting the intact genome with a 30-min delay after exposure to 6.8 kGy of gamma rays (343) (Fig. 8). According to data reported previously by Lecointe et al. (343), a Pol X-deficient strain has wild-type resistance to H2O2, UV radiation, and MMC, whereas Khairnar and Misra (289) reported a sensitivity of the Pol X mutant to these DNA-damaging agents and a higher sensitivity to gamma rays. Unlike other DNA polymerases, which are characterized by a closed-right-hand conformation, Pol XDR adopts a novel extended conformation (352). The involvement of Pol X in DSB repair in D. radiodurans seems restricted to its structure-specific 3′-5′ exonuclease activity at higher doses of ionizing radiation when secondary structures and clustered lesions arise in DNA (61). Akin to its homologs the eukaryotic Polβ (331) and B. subtilis Pol X (34), the physiological role of Pol X seems to reside in short-patch BER (289). Pol X shows 5′-deoxyribose phosphate lyase activity and short-patch DNA synthesis activity, which function in BER in conjunction with a glycosylase and an apurinic/apyrimidinic (AP) endonuclease (289). Pol X was also shown to improve the UV radiation and MMC survival of E. coli mutants defective in BER (alkA) and NER (uvrA) functions (289).
DNA Repair Checkpoints and Cell Division Control in D. radiodurans
The balance between DNA degradation and DNA synthesis is essential for the survival of D. radiodurans. It necessitates a tight regulation of the extent of DNA degradation and the time of the onset of DNA synthesis and repair. DNA degradation is an essential part of the repair process (643), which needs to be terminated by a protein(s) synthesized de novo after irradiation (136). Uncontrolled exonucleolytic activity would otherwise completely degrade damaged DNA and thereby preclude any possibility for repair. With this point in mind, DNA synthesis was suggested to be an important factor in the regulation of DNA degradation in D. radiodurans (136). In one of the scenarios for the termination of DNA degradation, exonuclease(s) proceeds along the DNA template and is presumably halted by factors responsible for the reestablishment of DNA synthesis before excessive amounts of DNA are digested (351). DNA Pol III and Pol I are indeed involved in the cessation of DNA degradation. At the end of the DNA degradation period, when DNA synthesis is expected to recommence, the absence of Pol III and the combined absence of Pol III and Pol I augment DNA degradation by 20% and 50%, respectively (572). DNA is also extensively degraded in UV-irradiated E. coli cells lacking Pol I and Pol III (599). The absence of Pol I enhances DNA degradation through an accumulation of single-strand breaks and gaps generated by the excision of defective bases and nucleotides, which present new sites for exonucleolytic attack (65). Pol III may attenuate DNA degradation through direct competition with exonucleases for free DNA ends generated by ionizing radiation (572). Whereas DNA polymerases counterbalance DNA degradation by DNA synthesis, the termination of DNA degradation may be accomplished by an inhibitor protein that actively restricts the erosion of DNA. Among such proteins in D. radiodurans are IrrI, which is activated shortly after DNA damage and limits the extent of DNA degradation (628); DdrA (239) and PprA (456), which protect DNA ends from exonucleases; and RecN, which tethers DNA ends in a cohesin-like manner (518). Alternatively, DNA degradation may be terminated by the reassociation of DNA with the cytoplasmic membrane (137). Small nuclear repeats (SNRs), which are scattered throughout the genome, may also be important for limiting the extent of DNA degradation (653). Significantly, an electrophoretic mobility shift assay identified a DNA binding activity from soluble cell extracts specific for the D. radiodurans genomic repeat (653). The binding of this factor to the repeats may prevent exhaustive chromosomal degradation after irradiation and may also prevent unfavorable recombination events between the repeats (see “Fidelity of DNA Repair in Irradiated D. radiodurans”) (653). For example, DNA binding proteins in E. coli recognize certain SNRs (181). However, SNRs are absent from the genomes of D. geothermalis and D. deserti (382), which excludes them as a general strategy for controlling DNA degradation among the Deinococcaceae.
DNA degradation in D. radiodurans seems to be under both negative and positive regulation. Whereas the above-mentioned proteins and mechanisms restrict DNA degradation, the deinococcal RecA protein was found to promote DNA degradation (572). Moseley and Copland (439) also found that there is no radiation-induced DNA degradation at 500 Gy in the rec30 mutant. The effect of deinococcal RecA on DNA degradation is controversial with respect to E. coli RecA, which restricts DNA degradation. After UV radiation, total chromosomal DNA in excision repair-proficient E. coli cells becomes susceptible to limited degradation, whereas in the absence of RecA, chromosomal degradation after UV irradiation is uncontrollable (99). Deinococcal RecA may promote DNA degradation through a helicase activity (55), thereby allowing access to exonucleases (572). By allowing access to exonucleases that liberate the 3′ overhangs required for RecA-mediated strand invasion and the priming of DNA synthesis, deinococcal RecA may embody a coupled regulatory mechanism controlling two opposing processes: DNA degradation and DNA synthesis. The importance of RecA in ensuring rapid and timely DNA repair is corroborated by the observation that a limited concentration of RecA delays DNA repair and induces cell death, even though the complete genome is eventually reconstituted (277).
Apart from the meticulous temporal coordination between DNA degradation and DNA synthesis, the dose-dependent duration of the lag period before the onset of cellular replication suggests the existence of a checkpoint that induces growth arrest and delays the initiation of semiconservative DNA synthesis to prevent the replication of a damaged genome (37, 39). This checkpoint control mechanism presumably detects and responds to DSBs in a manner similar to that of the RecA-initiated SOS response in E. coli (73) or the ATM (ataxia telangiectasia, mutated)- and ATR (ATM and Rad3 related)-initiated DNA damage checkpoint in eukaryotes (240). In D. radiodurans, as in E. coli and eukaryotes (87), ssDNA may serve as a common checkpoint signal, as it is formed at stalled replication forks, during nucleotide and base excision repair, and during end recession in double-strand-break repair. One of the checkpoint players is the ClpPX protease, which was posited to control chromosome segregation and cell division in D. radiodurans cells recovering from DNA damage (550). Without ClpPX, cells showed decondensed nucleoids and abnormal septa, with some cells being devoid of DNA altogether (550). The inactivation of the ClpPX protease reduces the ionizing radiation resistance at doses above 10 kGy, delays DSB repair, and extends the lag phase before the resumption of cell division (550). ClpPX-mediated cell division control may involve the degradation of proteins that need to be removed to restore proper cell division or the degradation of a damage-responsive checkpoint akin to SulA in E. coli (550). SulA is an SOS-inducible inhibitor of cell division (262), which must be expressed only transiently during the recovery process to avoid a lethal inhibition of septation and which is removed by the Lon protease (426). A SulA homolog is, however, absent from the D. radiodurans genome.
The RecA-Independent Pathway of DSB Repair in D. radiodurans
In the absence of RecA, approximately one-third of the DSBs generated by ionizing radiation can be rejoined by a RecA-independent pathway (125, 413, 572, 676). The RecA-independent effects are also manifest at early postirradiation times in wild-type cells, yielding a small increase in the chromosomal fragment size (128). The mechanism of RecA-independent repair is kinetically separate from that of RecA-dependent repair, with the absence of significant DNA synthesis and a lesser extent of DNA degradation (572). The present data suggest that the RecA-independent capacity to mend DSBs in the recA mutant and in the wild type at early postirradiation times consists of single-strand annealing reactions (125, 572). In wild-type cells, early annealing reactions would increase the physical length of many chromosomal DNA fragments, thereby reducing the damage caused by exonucleases and facilitating RecA-mediated DNA repair. NHEJ as an alternative to SSA has never been observed for D. radiodurans (125, 676).
The RecA-independent SSA pathway may involve proteins such as DdrA, which protects 3′ ssDNA ends from degradation (239); DdrB, an SSB-like protein with strand-annealing properties (464, 664); and RadA, a distant RecA homolog (572). The importance of DdrA and DdrB in the absence of RecA is reflected in (i) an increased sensitivity of the recA mutant when ddrA or ddrB is deleted (602), (ii) an increased sensitivity of ddrA mutant cells when the concentration of RecA is limited (277), and (iii) a complete failure in DNA fragment rejoining in the ddrB recA mutant (664). The lesser extent of DNA degradation observed for the recA mutant (572) is congruent with the importance of protecting the DNA fragments' ends in the absence of RecA before annealing with overlapping fragments can occur. In the absence of RecA, RadA also seems to contribute to the RecA-independent pathway of DSB repair, although its role remains unclear (572).
Fidelity of DNA Repair in Irradiated D. radiodurans
DNA repair in D. radiodurans is not only very efficient but also very faithful. Although point mutations and the transposition of mobile genetic elements are induced by ionizing radiation (408), the repaired DNA is free from detectable gross chromosomal aberrations below extremely high radiation doses (514). Ionizing radiation of 2 kGy increases point mutations to rifampin resistance 16-fold and the frequency of spontaneously arising trimethoprim-resistant mutants 10-fold (408). Resistance to trimethoprim arises more frequently by an insertion sequence (IS) insertion into the thyA gene than by point mutations (408). Point mutations are induced predominantly at low irradiation doses, while transposition is induced at high doses (408). Whereas point mutations and transpositions occur at various genomic locations, chromosomal rearrangements arise at the site of DSBs. Rearrangements lead to genomic instability, which in mammalian cells may instigate neoplastic transformation and tumor growth. Regions containing repetitive sequences are particularly prone to rearrangements. In humans, recombination between repeats is at the origin of disease-causing deletions, such as α-thalassemias, Duchenne muscular dystrophy, and familial hypercholesterolemia (242, 345, 593). D. radiodurans contains many repetitive sequences, including 52 ISs and 247 small noncoding repeats (SNRs) (384), and is therefore susceptible to rearrangements between repetitive sequences. However, DNA fragment reassembly in irradiated D. radiodurans cells appears to be accurate, as inferred by the faithful reconstitution of all NotI fragments in PFGE gels. The fidelity of DNA fragment assembly in ESDSA requires that all fragments' overhangs be extended by copying a fragment that is contiguous in the intact chromosome, as each mispriming of strand elongation within a repeated sequence of a wrong fragment followed by SSA results in chromosomal rearrangements. RecA was found to be crucial for ensuring the fidelity of DNA fragment assembly in D. radiodurans by preventing chromosomal rearrangements, as cells devoid of the recA gene are riddled with rearrangements (514). In recA mutant cells rearrangements arise spontaneously and in response to ionizing (but not UV) radiation, whereas in wild-type cells rearrangements are induced only with extremely high doses of radiation (e.g., 25 kGy) (514). DNA and cellular damage at high radiation doses may saturate RecA-dependent repair, impair the resynthesis of proteins required for faithful repair, and produce malfunctioning proteins contributing to erroneous DNA repair. The patterns of rearrangements in recA and wild-type cells are highly similar, which indicates the existence of hotspots as regions of genomic instability prone to DSBs (514). One of the identified rearrangements involves an ISDra5-mediated insertion of the small plasmid into the large chromosome (514). Rearrangements arising in the absence of RecA may be generated by an SSA mechanism (125) (see “The RecA-Independent Pathway of DSB Repair in D. radiodurans”), which was found to be responsible for RecA-independent sequence rearrangements in E. coli, specifically for deletions associated with palindromic sequences (80) (Fig. 10 C). In mammalian cells rearrangements are mediated by SSA and NHEJ (649), but there is no evidence for NHEJ in D. radiodurans (125). SSA in E. coli depends on the SbcCD nuclease to introduce a DSB at the hairpin structure formed by the inverted repeats (80). D. radiodurans also encodes SbcCD, which, analogously to E. coli SbcCD, may be responsible for SSA-mediated chromosomal rearrangements in regions of genomic instability, such as palindromes.
FIG. 10.
Fidelity of DSB repair in D. radiodurans. (A and B) The fidelity of DSB repair can be compromised by insertion sequences (ISs) at the level of the priming of new DNA synthesis (A) and the annealing of newly synthesized single strands (B). (A) Mispriming could occur between identical ISs present on noncontiguous DNA fragments. RecA may ensure the fidelity of priming by aligning homologous DNA fragments. (B) Misannealing could occur when newly synthesized single strands from noncontiguous fragments contain identical ISs. The fidelity of annealing is ensured via the synthesis of long single-stranded overhangs, which are much longer than the longest IS. (C) Rearrangements observed in the absence of a functional RecA protein may result from the single-strand annealing (SSA) of noncontiguous fragments that share identical ISs.
RecA may ensure the fidelity of ESDSA at two levels: the priming of single-strand synthesis and strand annealing (Fig. 10A and B). The unique property of deinococcal RecA to bind preferentially to double-stranded DNA (296) may be physiologically relevant for bracing homologous dsDNA fragments before the initiation of D loops to form a platform for rapid and accurate fragment assembly (“postalignment”) [see “Physical scaffolds for DNA repair in D. radiodurans. (iv) Chromosome alignment in D. radiodurans”] (Fig. 10A). This RecA-promoted chromosome alignment model is supported by the observed rapid annealing of newly synthesized single strands in ESDSA (572, 676). The rapid annealing of newly synthesized single strands suggests that the synthesis of complementary single strands is coincident in space and time and that two noncontiguous fragments are reassembled by copying a third “bridging” fragment (Fig. 6, step 5). The fidelity of annealing is additionally ensured via the synthesis of long single-stranded overhangs, which are much longer than the longest D. radiodurans repetitive sequences (1,322 bp) (384) and which ensure the correct assembly of a large number of DNA fragments (Fig. 10B). The avoidance of lethal erroneous DNA fragment assembly may have provided sufficient selective pressure for the evolution of the ESDSA mechanism (with long single-stranded overhangs) rather than a mechanistically more simple interchromosomal SSA (with shorter overhangs). D. radiodurans may additionally possess proteins that bind to repetitive sequences, preventing them from becoming single stranded or from annealing. A binding activity of soluble cell extracts with specificity for the genomic repeat sequences was identified experimentally (653). Editing of the pairing process by mismatch repair proteins is also expected to forestall recombination between nonhomologous fragments (373, 490, 510). Finally, tethering the ends of a given DSB to prevent them from dissociating via the cohesin-like RecN protein (518) or via genome condensation-promoting proteins, such as HU (459) and Dps1 (54) [see “Physical scaffolds for DNA repair in D. radiodurans. (i) Genome condensation in D. radiodurans”], may prohibit chromosomal rearrangements and thus contribute to the maintenance of genomic stability in D. radiodurans.
Desiccation Resistance of D. radiodurans
D. radiodurans is also highly resistant to desiccation (dehydration) (Fig. 3B). Desiccation-tolerant organisms include bacteria, yeast, rotifers, brine shrimp, the shrimp Artemia salina, tardigrades, plant seeds, mosses, nematodes, some plants (the resurrection plants Craterostigma plantagineum and Selaginella lepidophylla and the bryophyte Tortula ruralis), and algae. Desiccation is defined as a water content below 0.1 g H2O g−1 dry mass. The removal of water from a cell is a severe, often lethal stress (496) due to protein denaturation and the formation of ROS, which cause lipid peroxidation, protein oxidation, and oxidative DNA damage (152, 233). Desiccation-tolerant organisms have different means of protecting their cellular macromolecules from the deleterious effects of desiccation. Trehalose and sucrose replace water molecules that are lost following desiccation and form glasses (“a supercooled liquid with extreme viscosity”) in the dry state, which stabilize a dried cytoplasm (110, 111) and reduce the release of free radicals (193). Small acid-soluble proteins (SASPs) coat DNA molecules in B. subtilis spores and promote a tight packaging of the nucleoprotein helices into a ring-like assembly that is excluded from the water (215). Bacillus spores are also radiation resistant (460) and accumulate high levels of manganese ions (176). Hydrophilins, intrinsically unstructured proteins of high glycine content and a high hydrophilicity index which include late-embryogenesis-abundant (LEA) proteins, accumulate in cells following desiccation or hyperosmotic stress (204). Several functions have been proposed for these proteins: molecular chaperones, hydration buffers, membrane stabilizers, and an ion sink (12, 111).
D. radiodurans survives 6 weeks in a desiccator (relative humidity, <5%) with 85% viability (398). These conditions give rise to approximately 60 DSBs per genome (398). After water supply, D. radiodurans can reconstitute its genome and revitalize all metabolic activities. The degree of resistance to desiccation is governed by the level of oxidative protein damage caused during desiccation (122, 196). Both radiation- and desiccation-resistant bacteria are marked with a high Mn/Fe ratio, and their proteins are less susceptible to protein oxidation than are those of sensitive bacteria (196). Recently identified manganese complexes acting as the most efficient D. radiodurans ROS scavengers are expected to protect proteins against desiccation-induced damage (121). Protection from desiccation may also be mediated by trehalose, an osmoprotective disaccharide that plays a major role in the desiccation resistance of E. coli (651). In D. radiodurans, trehalose is probably synthesized via trehalose synthase (DR2036) and/or via maltooligosyl trehalose synthase (DR0463) and degraded by trehalohydrolase (DR0464) (651). D. radiodurans is unique among bacteria in possessing as many as four homologs of plant desiccation resistance-associated proteins (381). The DR1372 protein belongs to the LEA-14 family, with members among plants and bacteria, while DR1172 and DR0105 belong to the LEA-76 family, with orthologs in plants, nematodes, Drosophila melanogaster, protozoa, yeast, fungi, and bacteria (381). The DRB0118 protein is a homolog of a desiccation-related protein from the resurrection plant C. plantagineum, an extremely desiccation-resistant plant (381). Homologs of this protein can also be found in other plants and bacteria (40). Of these proteins, DR1172 was detected under various culture conditions, while DR0105 was detected only in a defined medium; DR1372 and DRB0118 were not detected (364). However, in D. deserti three of the four homologs were detected after standard cultivation, which suggests their importance for adaptation to desert conditions (140). The inactivation of DR1172 and DRB0118 results in a 75% reduction in the viability of desiccated cultures, with no effect on ionizing radiation resistance (40). However, these proteins are not induced in response to desiccation (40). Furthermore, the inactivation of DR0105 has no significant effect on ionizing radiation resistance (383). Several other proteins with intrinsically disordered hydrophilic segments in D. radiodurans were recently identified and include DNA polymerase III; a Nudix hydrolase, DR0550; and an ABC transporter, DR2145 (30). Hydrophilic tails presumably increase the probability of these proteins to stay solvated, while other proteins denature due to dehydration (30).
Resistance of D. radiodurans to UV-C Radiation
D. radiodurans is extremely resistant to UV-C radiation (100 to 295 nm) and can efficiently repair UV-induced bipyrimidine photoproducts (BPPs): cyclobutane pyrimidine dimers (CPDs) and pyrimidine-(6-4)pyrimidone photoproducts (6-4 PPs) (554). The major BPP in UV-irradiated D. radiodurans is CPD TC (47.2%), while 6-4 PPs are least represented (428). UV-induced DNA damage is repaired by two nucleotide excision repair mechanisms, UvrABC and UVDE (63, 413, 441), and a genetic recombination mechanism (439, 445). Accordingly, the most UV-sensitive DNA repair mutants in D. radiodurans are recA, recO, polA, and uvrA uvsE mutants (171, 223-225, 434, 439, 441, 603, 665) (Fig. 8B). Whereas UV-irradiated E. coli exhibits an exponential decline in viability, D. radiodurans has a shoulder of resistance that extends to 500 J/m2 (37, 597) (Fig. 3C). The 20-fold-superior resistance of D. radiodurans is not due to the protection of its DNA against BPPs but is based on the protection of proteins against UV-induced oxidative damage (329). As in the case of ionizing radiation-induced DSBs, the yields of UV-induced BPPs are similar for D. radiodurans and E. coli: 500 J/m2 yields 14,000 and 19,500 BPPs/107 bases, respectively (428). This slight difference can be explained by a higher genomic pyrimidine dinucleotide frequency in E. coli, the 2-fold-higher TT frequency of which is linked to its lower GC content (428, 554). It therefore appears that, similar to ionizing-radiation-induced DSBs, UV-induced BPPs are caused mainly by direct effects and are not determined by the cellular antioxidant status.
Kinetics and enzymology of the repair of UV-induced DNA damage in D. radiodurans.
UV radiation induces a dose-dependent delay in DNA synthesis due to the collapse or stalling of replication forks (333). If a replication fork encounters a UV-induced nick or a single-stranded DNA gap resulting from the excision of a BPP, the continuation of DNA replication creates a DSB separating one branch of the fork from the parental DNA, thereby causing a replication fork collapse (332). Alternatively, replication forks stall upon encountering BPPs while fork regression enables the excision repair of BPPs and promotes a fork restart (411). Collapsed or stalled replication forks are eventually restored by recombination (322, 333, 411). Before DNA replication can resume, BPPs must be removed by excision repair (440, 553, 554). Following 500 J/m2 of radiation, more than 80% of thymine-derived photoproducts are removed from D. radiodurans cells within 90 min (635) and appear in the form of di- and trinucleotides in the medium outside the cells (63). In addition to the elimination of dimers, more DNA is degraded and found outside the cells (63). Exposure to 500 J/m2 of radiation is accompanied by the degradation of about 9% of cellular DNA, which is equal to the loss of 50 bases per thymine-containing photoproduct (635). This value exceeds the excision patch size, which is known in E. coli to be only 12 to 13 bases per bulky lesion (545). UV-irradiated cells suffer not only from DNA degradation but also from DNA fragmentation. The DSBs that can be observed with neutral sucrose gradients or PFGE in the course of post-UV repair may result from the stalling or collapse of replication forks and from the excision of closely spaced BPPs on opposite strands (66, 333, 411, 514, 614). Although the rate of DNA synthesis is markedly reduced in UV-irradiated cells (291, 440, 446, 554), the undamaged origins of replication continue to fire (56, 526) and generate DSBs. Recombination is required for the repair of DSBs and the reestablishment of DNA replication.
As in E. coli, the repair of UV-induced DNA damage in D. radiodurans therefore relies on excision (63) and recombination (439, 445) mechanisms. The completion of excision repair precedes the completion of recombinational repair (440). D. radiodurans possesses two nucleotide excision repair pathways for the removal of pyrimidine dimers: the classical nucleotide excision repair pathway (UvrABC) and the UV damage endonuclease (UVDE) pathway (413, 441). A tandem of UvrABC and UVDE pathways also exists in B. subtilis (653), D. geothermalis (383), and D. deserti (140). The UvrABC pathway involves a protein complex (UvrABC excinuclease) that recognizes the structural changes in DNA caused by UV damage and creates the dual incisions 5′ and 3′ to the damaged site. This pathway is also involved in the removal of many other bulky lesions such as mitomycin C adducts (489). D. radiodurans encodes homologs of UvrA (UvrA1 [DR1771] and UvrA2 [DRA0188]), UvrB (DR2275), and UvrC (DR1354). UvrA2 does not contribute to UV radiation resistance (603). E. coli UvrA can restore MMC resistance in UvrA-deficient D. radiodurans cells, which suggests that the two bacteria have similar UvrABC systems (4). The UVDE pathway is mediated by endonuclease β (uvsE [DR1819]), which has a novel requirement for manganese ions and an endonucleolytic mode of action that is different from that of UvrABC (182-184, 441). Endonuclease β shares a 30% sequence identity with Schizosaccharomyces pombe Uve1p, which introduces a nick immediately 5′ to the lesion, subsequent to which other repair proteins digest the damaged strand and fill in the gap (9). In S. pombe, the activity of Uve1p also overlaps with the UvrABC pathway (672). Whereas the nucleolytic activity associated with UvrA is terminated by a UV-induced protein, the nucleolytic activity associated with endonuclease β is slower and does not require the inducible terminator (184). UVDE efficiently removes both CPDs and 6-4 PPs, whereas UvrABC is more specific for 6-4 PPs (603). The two pathways have overlapping functions, as both need to be inactivated to produce a UV-sensitive phenotype (184, 441). A uvrA uvsE double mutant is 100-fold more sensitive to 250 J/m2 UV than the wild type and loses the shoulder of UV resistance (171) (Fig. 8B). The slightly higher UV sensitivity of the uvrA mutant than the uvsE mutant suggests that, as in S. pombe (672), UvrABC is more important for UV resistance than is UVDE (603). In addition, UvrABC is constitutively expressed, unlike endonuclease β, which indicates that UvrABC is important for the continuous removal of damaged nucleotides from the cells (364) and which explains why excision repair can act in the absence of protein synthesis (63, 184). Both the UvrABC and UVDE pathways require Pol I, as the polA mutant is extremely sensitive to UV radiation (224, 225) (Fig. 8B). While the UV sensitivity of polA equals the sensitivity of the uvrA uvsE double mutant (224-226), the recombination-deficient recA mutant is more sensitive to UV radiation than is the uvrA uvsE mutant, which suggests that recombinational repair is more significant than the two excision repair pathways for UV radiation resistance in D. radiodurans (444, 603) (Fig. 8B). Furthermore, both CPDs and 6-4 PPs are eliminated even in the absence of UvrABC and UVDE (603). Mutations in other recombination genes, recO and recF, also result in UV-sensitive phenotypes (91, 665). In E. coli, recombinational repair is essential for replication fork restart in UV-damaged cells (322, 333). The recombination-dependent replication fork restart likely underlies the primary significance of recombinational repair for the UV survival of D. radiodurans as well. The ability of the cells to repair UV damage decreases proportionally when cells are exposed to increasing preirradiation doses of X-rays, and vice versa (443), which confirms that the enzymatic repair of both ionizing radiation and UV damage includes overlapping components.
Unlike ionizing radiation, UV does not induce point mutations in D. radiodurans, even at doses as high as 1,485 J/m2 (596, 603, 612). The absence of translesion synthesis (TLS) DNA polymerases in D. radiodurans (381) contributes to the high fidelity of the repair of UV lesions. Conversely, D. deserti possesses two TLS polymerases, which are responsible for the UV-induced point mutations to Rifr observed for this deinococcal species (168). UV-induced mutagenesis in D. deserti is also dependent on the chromosomal RecA protein, which induces the expression of TLS polymerases by cleaving LexA, as in E. coli (168). Although UV radiation does not induce point mutations in D. radiodurans, it can induce IS transposition (408). However, unlike ionizing radiation, UV does not induce rearrangements in RecA-deficient cells (514).
Sensitivity of D. radiodurans to UV-A Radiation and Singlet Oxygen
Although resistant to UV-C, D. radiodurans is more sensitive to UV-A (320 to 400 nm), which comprises about 95% of terrestrial UV radiation, than is E. coli (43, 69, 81, 494). For the same inactivation level the amount of BPPs is higher in UV-A- than in UV-C-irradiated D. radiodurans cells (494). Surprisingly, the amount of BPPs and UV-A survival are the same for the wild type and the excision and recombination repair mutants (uvrA uvsE and recA) (494). While UV-C causes mainly direct DNA damage due to a strong absorption at wavelengths below 320 nm, UV-A causes only indirect damage to DNA through ROS (185). For example, photons from UV-A radiation can generate singlet oxygen through type II photosensitization reactions with endogenous photosensitizers (e.g., flavin) or metals (627). Although very efficient in removing other ROS, D. radiodurans appears highly sensitive to singlet oxygen generated in the presence of photosensitizers (539). D. radiodurans is 100-fold more sensitive than E. coli to photodynamic treatment with the sensitizer Rose Bengal, which generates singlet oxygen species (1O2) and superoxide radicals (O2·−) (539). The accumulation of DNA lesions in UV-A-irradiated D. radiodurans and the inefficiency of DNA repair enzymes at removing them may indicate (i) a saturation of the DNA repair machinery with numerous BPPs and/or (ii) excessive damage of DNA repair and other cellular proteins. In fact, a recent study of E. coli showed that proteins are the primary cellular targets implicated in the lethal effects of UV-A (69, 70). UV-A generates oxidative stress that irreversibly damages proteins required for vital cellular functions such as transcription, translation, glycolysis, the TCA cycle, and respiration (70). The proteins with iron-sulfur clusters, most of which belong to the respiratory chain enzymes involved in energy production, are highly susceptible to UV-A (69). Oxidative damage to respiratory chain enzymes can amplify oxidative stress through uncontrolled production and the release of ROS during electron transfer as well as through the release of iron from iron-sulfur clusters (69, 268). Although D. radiodurans has a reduced number of respiratory chain enzymes and proteins with iron-sulfur clusters (see “Metabolic Configuration of D. radiodurans”) along with extremely efficient ROS-scavenging mechanisms (see “Antioxidation Protection in D. radiodurans”), its sensitivity to singlet oxygen generated in the presence of photosensitizers may overcome its antioxidant mechanisms and thereby account for its high level of sensitivity to UV-A. Additionally, UV-A-induced singlet oxygen may target lipid components of the D. radiodurans cell wall, the unsaturated fatty acids in particular, the double bonds of which are susceptible to oxidation (68, 631). Lipid peroxidation disrupts membrane integrity, which may lead to the loss of chromosome anchoring to the cell membrane (79, 154) [see “Physical scaffolds for DNA repair in D. radiodurans. (iii) DNA-membrane association in D. radiodurans”] and to cell death.
Resistance of D. radiodurans to Mitomycin C
MMC is a cross-linking agent that reacts with guanine to form interstrand cross-links (506). The cross-links formed with MMC are bis-guanine adducts, arising through the consecutive alkylation of two guanines at their N2 positions (621). Cross-links act as absolute blocks to DNA replication and transcription. D. radiodurans can tolerate interstrand cross-links in DNA induced by MMC (303) without mutations and with a higher survival rate than that of E. coli (597) (Fig. 3D). Incubation with 20 μg/ml MMC for 10 min generates 100 to 200 cross-links per genome without a loss of viability (303). When cells are pretreated with a lower concentration of MMC, the disappearance of cross-links is almost normal, even when chloramphenicol is present during postincubation, which suggests that the repair of MMC-induced damage is inducible (303). After the induction of cross-links, DSBs are observed in a neutral sucrose gradient (304). UvrA (4, 441, 611) and RecA (223, 439) are essential for the repair of MMC-induced damage (Fig. 8C), which suggests that, as in E. coli, interstrand cross-links are repaired through the concerted action of NER and homologous recombination (102, 163, 463). The high level of sensitivity of the uvrA mutant to MMC is not increased in the uvrA polA double mutant, as Pol I acts in the same pathway as UvrA for the repair of MMC lesions (437). MMC-sensitive mutants in D. radiodurans also include irrE (170), uvrD (407), radA (Slade, unpublished), recQ (261), and recR (308) mutants (Fig. 8C). D. radiodurans is also highly resistant to trans-platinum-induced DNA-protein cross-links (81).
Resistance of D. radiodurans to Base-Damaging Chemicals
D. radiodurans is extremely resistant to various types of base damage: alkylation (caused by MMS and MNNG), deamination (caused by hydroxylamine and nitrous acid), and oxidation (caused by ROS and ionizing radiation). Compared to E. coli, D. radiodurans is 15 times more resistant to the lethal effects of MNNG, 62 times more resistant to nitrous acid, and 7 times more resistant to hydroxylamine but sensitive to ethyl methane sulfonate (EMS) (597). EMS and MMS are alkylating agents that add ethyl and methyl radicals, respectively, at N and O atoms in DNA. MNNG methylates guanine on N7 and O6 and adenine on N3, and O6-methylated guanine mispairs with thymine. Hydroxylamine and nitrous acid cause the deamination of adenine and cytosine. D. radiodurans is also very resistant to 4-nitroquinoline-N-oxide (4NQO), which has lethal and mutagenic effects on E. coli similar to those of UV radiation (445). The exposure of D. radiodurans to 50 μg/ml 4NQO for 4 h does not reduce viability or induce mutations, whereas an equivalent exposure of E. coli reduces viability to 0.1% (445).
Base excision repair in D. radiodurans.
The BER pathway protects cells from the deleterious effects of endogenous and exogenous DNA damage induced by hydrolysis, ROS, ionizing radiation, and strong alkylating agents (544). The existence of BER in D. radiodurans was first documented by Masters et al. (394), who showed AP endonuclease and uracil glycosylase activities in D. radiodurans. Alkylated bases are presumably removed by two AlkA glycosylases (DR2074 and DR2584), whereas deaminated and oxidized bases are removed by an ensemble of nine DNA glycosylases (381) (Table 3). Abasic sites generated by glycosylases are presumably removed by the AP endonuclease Xth (DR0354) (381).
Uracil, which is generated by the deamination of cytosine, is removed by uracil-DNA glycosylases. The major uracil-DNA glycosylase in D. radiodurans is a family 1 uracil-DNA glycosylase (ung [DR0689]), which removes uracil from U·G and U·A base pairs in dsDNA and ssDNA, whereas a family 2 enzyme (DR1751) is more active on ssDNA (534). The mismatch-specific uracil-DNA glycosylase DR0715 (mug) poorly removes uracil from a U/G pair (534) and has a broader substrate specificity than does the E. coli Mug homolog (427). The fourth putative homolog (DR0022), which is highly induced after ionizing radiation, does not show any uracil-DNA glycosylase activity (534). Xanthine (deaminated guanine) and hypoxanthine (deaminated adenine) are removed by Nfi (endonuclease V [DR2162]).
BER also repairs oxidative base damage caused by ROS. BER removes oxidized bases generating SSBs, which can be repaired by de novo synthesis using the undamaged strand as a template. 7,8-Dihydro-8-oxoguanine (also called 8-oxoguanine [GO]) is the most stable product known to be caused by oxidative damage to DNA and one of the most abundant oxidative lesions in the genome which is often used as a marker of the extent of oxidative stress (309). If not repaired, GO lesions in DNA can produce A/GO mismatches during DNA replication (566) and can result in transversions from G·C to T·A (431). MutM, also called formamidopyrimidine-DNA glycosylase (FaPy-DNA glycosylase), excises GO and other oxidized purines from the GO/C base pair (610). However, if this does not occur and replication takes place, the DNA glycosylase MutY intercepts the resultant GO/A mismatches and removes the inappropriate adenine, leaving AP/GO, which is a substrate for MutM (622). D. radiodurans MutY (DR2285) is a monofunctional 8-oxoguanine glycosylase active on A/G, A/C, and A/GO mismatches, with a binding preference for A/GO mismatches and with kinetic parameters similar to those of E. coli MutY (356). D. radiodurans MutY complements the E. coli mutY mutant (356). In contrast to E. coli MutM, deinococcal MutM (DR0493) is able to excise GO from A/GO mismatches (546). The excision rates for formamidopyrimidines (2,6-diamino-4-hydroxyl-5-formamidopyrimidine [FapyGua] and 4,6-diamino-5-formamidopyrimidine [FapyAde]) are significantly greater than those for GO when these lesions are paired with C (42, 546). The calculated yield of 8-oxoguanine, FapyGua, and FapyAde in irradiated D. radiodurans is ∼0.6/kGy/Mbp (302). D. radiodurans also encodes three endonucleases III (nth, archaeon-type DR0928 and DR2438, and yeast-type DR0289) for the removal of thymine glycol, one of the major stable oxidative modifications of thymine. The excision of thymine glycols by thymine glycol glycosylase following UV or ionizing radiation has been demonstrated (450, 607).
Chemical mutagenesis in D. radiodurans.
D. radiodurans is nonmutable by MMC, hydroxylamine, 2-aminopurine, and zebularine; only slightly sensitive to mutation by nitrous acid and EMS; and highly mutable by MNNG and 5-azacytidine (298, 597). MNNG has therefore been widely used for the generation of D. radiodurans mutants (88, 202, 225, 304, 434, 437, 445, 446, 628). Compared to E. coli, D. radiodurans is equally sensitive to the lethal effects of EMS but not its mutagenic effects. Conversely, it is sensitive to the mutagenic effects of MNNG but not its lethal effects (597). D. radiodurans may be particularly susceptible to MNNG-induced mutations due to a high content of sulfhydryl groups (199), which catalyze the cellular activation of MNNG (339). Alternatively, MNNG-induced O6-methylguanine, which causes G·C-to-A·T transition mutations (639), may not be well recognized by D. radiodurans (413). The system for the repair of MNNG-induced damage appears to be constitutive, as an adaptive response for lowering the mutation rate after preexposure to MNNG is absent (511).
Mismatch Repair in D. radiodurans
The mismatch repair system enhances the fidelity of both replication and recombination processes by recognizing replication-generated mismatches and mismatches in recombination intermediates. D. radiodurans has a functional mismatch repair system, which involves MutS1 (DR1039), MutL (DR1696), and UvrD (DR1775) (407). The absence of MutH homologs suggests that strand discrimination is different from that in E. coli. MutH homologs are also absent from the eukaryotic mismatch repair system (313). Cells devoid of MutS1 or MutL display a mutator phenotype and homeologous recombination, with a 7-fold increase in the frequency of spontaneous Rifr mutagenesis and a 10-fold increase in the efficiency of the integration of a donor point mutation marker during transformation (407). The inactivation of UvrD increases the level of spontaneous mutagenesis but has no effect on marker integration (407). Another MutS homolog, MutS2, has no effect on mutagenesis or recombination (407). Cells devoid of MutS1 or MutL are as resistant to gamma rays, UV, and MMC as wild-type cells, while UvrD-deficient cells are moderately sensitive to gamma rays and extremely sensitive to MMC (407) (Fig. 8). Given that the spontaneous mutation rate for D. radiodurans of 3.78 × 10−8 per cell per generation is 15-fold higher than that reported for E. coli and that the inactivation of mismatch repair in E. coli results in a 15-fold-higher increase in spontaneous Rifr mutagenesis (212, 407), mismatch repair in D. radiodurans would appear to be inefficient.
Nonhomologous End Joining in D. radiodurans
Nonhomologous end joining (NHEJ) indicates recombination between sequences with little or no sequence homology, which is often error prone (633). NHEJ is the dominant mechanism for DSB repair in the G1 phase of the eukaryotic cell cycle (480). NHEJ was also identified in several bacteria (e.g., M. tuberculosis, B. subtilis, and Pseudomonas aeruginosa) but not in D. radiodurans (21). The existence of NHEJ in D. radiodurans was investigated by using a direct insertion of a plasmid carrying an antibiotic resistance cassette into the chromosome (125). The failure to circularize such a plasmid after irradiation, which can occur only by NHEJ, demonstrates the absence of this error-prone DNA repair pathway. Furthermore, the massive DNA repair synthesis observed during DSB repair in irradiated D. radiodurans cells is incompatible with NHEJ (572, 676).
STRESS RESPONSE IN D. RADIODURANS
Along with an efficient DNA repair mechanism, a prompt and efficient DNA damage response to upregulate the expression of genes needed to remedy DNA and other cellular damage constitutes another important factor in DNA damage resistance. DNA repair in D. radiodurans is damage inducible and requires de novo protein synthesis following ionizing radiation (307). Pretreatment with sublethal levels of ionizing and UV radiation (600), H2O2 (646), and MMC (303) induces resistance to their lethal effects. The genes required for oxidative, general, and desiccation stress responses are listed in Table 4.
TABLE 4.
Stress response-related genes in D. radiodurans, their transcriptional regulators, and their expression levelsa
| Stress and enzyme | Locus tag | Regulation | PHXb | Constitutively expressed |
Type(s) of stressc | Induced after IR stressd |
||
|---|---|---|---|---|---|---|---|---|
| DM | RM | Gene | Protein | |||||
| Oxidative | ||||||||
| Catalases | ||||||||
| KatE | DR1998 | DrRRA | + | ES | ES | OHSAC | + | + |
| OxyR | ||||||||
| KatA | DRA0259 | DrRRA | + | ES | ES | OHSAC | − | |
| DRA0146 | ES | ES | OHSAC | − | ||||
| Peroxidase | DRA0145 | DrRRA | ES | ES | OHSAC | − | ||
| Cytochrome c peroxidase | DRA0301 | ES | − | − | + | |||
| Superoxide dismutases | ||||||||
| SodA | DR1279 | DrRRA | + | ES | ES | OHSAC | − | |
| SodC | DR1546 | DrRRA | + | ES | E | OHSA | + | |
| DRA0202 | + | ES | ES | OHSAC | + | |||
| DR0644 | ES | ES | OHSAC | + | ||||
| Chloride peroxidase | DR0791 | + | − | ES | SC | ++ | ||
| Organic hydroperoxidase resistance protein | DR1857 | + | ES | ES | HAC | − | ||
| Peptide methionine sulfoxide reductases | ||||||||
| MsrA | DR1849 | DrRRA | + | ES | ES | OS | + | |
| MsrB | DR1378 | − | ||||||
| Peroxiredoxins | DR2242 | E | − | OA | − | |||
| DR1208 | + | |||||||
| DR1209 | − | |||||||
| DR0846 | ES | ES | OHSAC | − | ||||
| Thioredoxin | DR0944 | DrRRA | + | ES | ES | OHSAC | − | |
| DRA0164 | ES | E | SA | − | ||||
| DR1832 | ES | ES | OHSAC | − | ||||
| Thioredoxin reductases | DR1982 | ES | ES | OHSAC | − | |||
| DR2623 | − | + | ||||||
| DR0412 | S | ES | HSC | + | ||||
| Glutaredoxin | DR2085 | DrRRA | − | − | A | + | ||
| DRA0072 | − | |||||||
| Osmotically induced protein involved in alkylperoxide | DR1538 | ES | ES | HSAC | + | |||
| and oxidative stress response (OsmC, YhfA) | DR1857 | + | ES | ES | HAC | − | ||
| DR1177 | − | |||||||
| DNA protection during starvation proteins (Dps) | ||||||||
| Dps1 | DR2263 | DrRRA | + | OHSAC | ++ | |||
| Dps2 | DRB0092 | + | ||||||
| Dessication | ||||||||
| LEA-14 family | DR1372 | − | ||||||
| LEA-76 family | DR0105 | DrRRA | ES | S | − | + | ||
| Desiccation-related protein from C. plantagineum | DR1172 | DrRRA | ES | ES | OHSAC | − | ||
| DRB0118 | DrRRA | − | ||||||
| General/heat | ||||||||
| Chaperone | ||||||||
| GroES | DR0606 | + | ES | ES | OHSAC | − | ||
| GroEL | DR0607 | + | ES | ES | OHSAC | − | ||
| GrpE | DR0128 | DrRRA | + | ES | ES | OHSC | − | |
| DnaK | DR0129 | DrRRA | + | ES | ES | OHSAC | − | |
| DnaJ | DR0126 | DrRRA | ES | E | HC | |||
| DR1424 | E | − | H | − | ||||
| Peptidyl-prolyl cis-trans isomerase (cyclophilin) | ||||||||
| Cyclophilin type | DR0237 | IrrE | + | ES | ES | OHSAC | + | |
| DR2542 | + | ES | ES | OHSAC | − | |||
| FKBP type | DR2464 | + | ES | E | HS | − | ||
| C type | DR1063 | IrrE | + | ES | ES | OHSC | + | + |
| Small heat shock protein (IbpA) | DR1114 | IrrE | + | +D | ||||
| DR1691 | − | |||||||
| Related to heat shock protein HSLJ | DR2056 | − | ||||||
| DR1940 | E | E | O | + | ||||
| Clp protease | ||||||||
| ClpA | DR0588 | − | S | HS | − | |||
| ClpB | DR1046 | ES | ES | OHSAC | − | |||
| ClpC | DR1117 | ES | E | OHSAC | − | |||
| ClpP | DR1972 | ES | ES | OHSAC | − | |||
| ClpX | DR1973 | + | ES | ES | OHSA | − | ||
| Lon protease | ||||||||
| Lon1 | DR1974 | DrRRA | + | ES | E | S | − | |
| Lon2 | DR0349 | + | ES | ES | OHSAC | +D | ||
| Periplasmic serine protease with regulatory | DR0327 | ES | − | H | − | |||
| PDZ domain (HtrA) | DR0745 | S | E | S | − | |||
| DR1756 | ES | E | OHSC | − | ||||
| DR0984 | ||||||||
| DR0300 | ||||||||
| General/heat | ||||||||
| Tail-specific periplasmic serine protease | DR1308 | − | ||||||
| DR1491 | + | |||||||
| DR1551 | + | ES | ES | OHSC | − | |||
| Subtilisin-like proteases | DR0812 | + | E | − | A | + | ||
| DR1459 | ES | ES | OHSAC | + | ||||
| DR1536 | E | − | O | − | ||||
| DR1937 | ES | ES | OHSAC | − | ||||
| DR2322 | + | ++ | ||||||
| DR2325 | E | − | OHAC | ++ | ||||
| DRA0064 | OxyR | E | E | OS | − | |||
| DRA0283 | IrrE | + | ES | ES | OHSAC | − | ||
| DRA0341 | + | |||||||
| DRB0069 | S | − | OHA | + | ||||
| Membrane-associated Zn-dependent protease I (YaeL) | DR1507 | E | S | OSAC | − | |||
| ATP-dependent Zn protease (cell division protein FtsH) | DR0583 | − | E | − | − | |||
| DR1020 | − | |||||||
| DRA0290 | DrRRA | + | ES | ES | OHSAC | − | ||
| Zn-dependent proteases (HtpX) | DR0190 | − | ||||||
| DR0194 | DrRRA | E | E | − | +D | |||
| Protease I related to general stress protein 18 (ThiJ) | DR1199 | + | ES | ES | OHSAC | − | + | |
| DR0491 | DrRRA | ES | − | OHS | + | |||
| Membrane chaperone (SugE) | DR1004 | − | ||||||
| DR1005 | − | |||||||
| Diadenosine tetraphosphate hydrolase (Hit) | DR1621 | + | + | OS | − | |||
| ABC transporters (ATP binding periplasmic peptides, | DR0095 | OxyR | ES | ES | OHSAC | + | ||
| branched-chain amino acids, maltose) | DR0096 | DrRRA | S | − | HAC | + | ||
| OxyR | ||||||||
| DR0205 | ES | S | OHSC | ++ | ||||
| DR1356 | − | − | − | ++ | ||||
| DR1357 | ++ | |||||||
| DR1358 | − | E | C | ++ | ||||
| DR1359 | ES | ES | OHSC | ++ | ||||
| DR0363 | IrrE | ES | ES | OHSAC | + | |||
| DR0365 | S | − | − | − | ||||
| DR2118 | − | S | — | +D | ||||
| DR0280 | + | ES | − | OHSAC | − | |||
| DR0561 | + | ES | ES | OHSAC | + | |||
| Zn binding protein of the ABC-type Zn transport system (YebL) | DR2523 | DrRRA | ES | ES | OHSA | |||
| GTPase (HflX) | DR0139 | S | E | OC | − | |||
| DR0646 | S | − | − | − | ||||
| Nucleotide binding universal stress protein (UspA) | DR2363 | DrRRA | ES | − | H | − | ||
| DR2132 | ES | ES | OHSAC | − | ||||
Shown are data for the constitutive expression of proteins in a defined medium (DM) or a rich medium (RM) in exponential (E) or stationary (S) phase, stress-induced expression of proteins, and radiation-induced expression at the gene and protein levels. Blank spaces indicate no reported data. (Based on data from references 94, 285, 364, 368, 377, 383, 601, 602, 645, and 677.)
PHX, predicted to be highly expressed.
O, oxidative stress; H, heat shock; S, starvation; A, alkaline stress; C, cold stress.
IR, ionizing radiation; ++, high level of postirradiation induction; +D or ++D, induction after ionizing radiation and desiccation.
Stress Response Transcriptional Regulators
Although the error-prone SOS response was not observed for D. radiodurans, a specific damage response regulon is likely to exist in this bacterium. Indeed, a set of genes associated with DNA damage resistance that are upregulated after irradiation contains a common palindromic DNA motif, named the radiation/desiccation response motif (RDRM), also found in D. geothermalis (383) and D. deserti (140). The RDR regulon is predicted to comprise a minimum of 29 genes in D. radiodurans, among which are recA, recQ, ssb, ruvB, sbcD, ddrA, ddrB, pprA, uvrB, uvrC, uvrD, mutS, mutL, gyrA, and gyrB (383). The RDR promoter was shown to govern the induction of the ssb gene following ionizing radiation and MMC treatment (629). DdrO is proposed to be the global regulator of the RDR regulon in D. radiodurans, as it is the only gene for a predicted transcriptional regulator that is preceded by an RDRM site (383). Its effect on the expression of genes within the RDR regulon awaits future analysis.
Transcription and translation elongation factors, along with RNA polymerase, are induced in response to ionizing radiation to enable the rapid synthesis of damaged proteins, particularly those essential for the recovery process (364, 368, 677). Several transcriptional regulators are also induced after irradiation to selectively upregulate the recovery genes. D. radiodurans has an extended array of genes involved in transcriptional regulation and signal transduction (380). Among the radiation-induced transcriptional regulators, the most highly induced are DdrO (DR2574), belonging to the Xre family of transcriptional regulators (602); IrrI (DR0171), belonging to a specific deinococcal family of transcriptional regulators (368); and DrRRA (DR2418) (677). IrrE (377) and DrRRA (645) control the expression of many genes following ionizing radiation.
IrrE (170), also referred to as PprI (260), is a novel regulatory protein considered to be a general switch that efficiently enhances the DNA repair capability and the extreme radiation resistance of D. radiodurans via the regulation of a series of pathways (377). Its inactivation provokes a dramatic increase in sensitivity to ionizing radiation, UV radiation, and MMC (170, 260) (Fig. 8) and a considerable delay in genome reassembly (377). A constitutively high concentration of RecA (100,000 monomers) can fully restore the resistance of IrrE-deficient bacteria to MMC without any effect on ionizing radiation resistance (277). IrrE upregulates 31 proteins and downregulates 4 proteins in response to 1 kGy ionizing radiation (377). Among the upregulated proteins are those involved in DNA repair (RecA, PprA, and the SSB protein), transcription and translation, metabolism, signal transduction, cell cycle control, stress response, proteases, and chaperones (377). However, IrrE does not seem to bind to the promoter region of recA or other induced genes (203) and may regulate the transcription of genes through a more classic transcription factor (642). The expression of irrE in E. coli slightly enhances the expression of RecA (1.5-fold), radiation resistance (1.6-fold), the scavenging of O2·− and H2O2, and both the constitutive (2.4-fold) and radiation-induced (4.3-fold) activities of KatG (203). Furthermore, the heterologous expression of irrE increases stress tolerance and ethanol production in E. coli; in a crop plant, Brassica napus; and in the ethanologenic bacterium Zymomonas mobilis (479, 681).
Another novel stress response transcriptional regulator, DrRRA, regulates the expression of numerous genes under normal and ionizing radiation conditions, such as DNA damage-related genes (e.g., recA, pprA, ligT, cinA, gyrB, and uvrB), dps1, genes homologous to plant desiccation resistance proteins, and genes for antioxidant proteins, chaperones, and proteases (645). DrRRA was shown to bind the promoter region of ddrI (DR0997), a DNA damage response gene, the expression of which is significantly reduced in the DrRRA gene mutant under normal and radiation conditions (645). The mutant strain is highly sensitive to ionizing radiation, with a delayed genome reassembly, and moderately sensitive to UV, H2O2, and desiccation (645) (Fig. 8). Superoxide dismutase and catalase activities are decreased and RecA and PprA levels are reduced in the DrRRA gene mutant (645).
The transcriptional regulator OxyR (DR0615 and DRA0336), found in many bacteria, is important for mounting a stress response to H2O2 in D. radiodurans as well (94, 671). The two OxyR homologs regulate catalase activities and ROS scavenging in D. radiodurans exposed to H2O2 (94, 671). Their deletion results in an increased sensitivity to H2O2 and an accumulation of ROS (671). Surprisingly, DR0615 acts as both a positive regulator and a negative regulator of gene expression; microarray data revealed that in this oxyR mutant, 130 genes are induced (e.g., oxidoreductases such as thioredoxin) and 150 genes are repressed (e.g., the catalase DR1998, N-acetyltransferases, and the electron transport genes, ligT, recA, cinA, and the ddr genes) under normal conditions (94).
Induction of Gene Expression and Protein Synthesis in Response to Ionizing Radiation
Protein synthesis in the period subsequent to irradiation is essential for the survival of bacterial cells (136). The reestablishment of protein synthesis after irradiation is vital for the termination of DNA degradation (see “DNA Degradation in Irradiated D. radiodurans”), for the complete restitution of genomic integrity, and for cell survival. Many genes involved in DNA repair, oxidative stress resistance, and metabolism are induced at the gene expression and protein synthesis levels in D. radiodurans cells recovering from ionizing radiation (368, 602, 677). Transcriptome analysis of exponentially grown cells recovering from 3 kGy of ionizing radiation (602) and of stationary-phase cells exposed to a dose of 15 kGy of ionizing radiation (368) revealed 72 and 832 upregulated genes, respectively. Mass spectrometry coupled with two-dimensional (2D) electrophoresis showed both quantitative and qualitative changes in the proteome; in response to 1 kGy of ionizing radiation, the expression level of 21 proteins increased, and the expression level of five new proteins was detected (677). The expression of genes after irradiation is inversely related to the size of genomic elements, where the 45-kb plasmid as the smallest genomic element has the largest number of genes with the highest expression levels (368). However, the genes most critical to recovery are not carried on the smaller genomic elements, while the disruption of many of the most highly induced genes has little effect on radiation resistance (602).
The most highly induced DNA repair genes are ddrA (DR0423), ddrB (DR0070), pprA (DRA0346), recA (DR2340), uvrA (DR1771), uvrB (DR2275), gyrA (DR1913), gyrB (DR0906) (368, 602), a DNA repair operon (DRB0100 to DRB0098) with end-healing and end-processing functions (60), a VSR (very short patch repair)-like nuclease possibly involved in very-short-patch repair (DR2566), mutT (DR0261 and DR1776), ruvB (DR0596), and ssb (DR0099) (368). The maximum response occurs around the time when DNA repair commences (368). The McrA-like nuclease DR2483 and the ComA protein (DR0207), involved in DNA transformation competence, also show a high level of induction (368). Among the five most highly induced genes in 3-kGy-irradiated cells (ddrA to ddrD and pprA), ddrC and ddrD encode proteins of unknown function, and their deletion strains are as resistant to ionizing radiation as the wild type (602).
Among the highly induced genes implicated in oxidative stress resistance are the catalase gene katE (DR1998), terB (DR2220), terZ (DR2224), msrA (DR1849), dps2 (DRB0092) (602), and five genes of the Nudix hydrolase family, including the MutT ortholog (368). The tellurium resistance proteins TerB and TerZ maintain the intracellular reducing environment in E. coli, possibly by directly reversing disulfide bonds (626), and confer resistance to various damaging agents, such as heavy metal ions, MMS, MMC, and UV radiation (31). MsrA is a methionine sulfoxide reductase, while Dps2 is a DNA binding protein, which presumably protects DNA from oxidative damage (11, 390, 668).
An overlap between the postirradiation increases in gene and protein expression levels was found for RecA (364, 368, 602), the SSB protein (368, 677), DdrB (368, 464, 602), PprA (368, 602, 677), DNA-dependent RNA polymerase (364, 368), KatE (601, 602), TerB (368, 602, 677), aconitase, malate dehydrogenase, and V-type ATP synthase (368, 677) (Tables 2, 3, and 4).
Protein Regulation in Response to Radiation Damage
The DNA damage response involves not only the transcriptional regulation of gene expression but also the modulation of protein function via posttranslational modifications. Among those, protein phosphorylation is known to be important for intracellular signaling in DNA damage repair both in eukaryotes (529) and in prokaryotes, where tyrosine phosphorylation of the SSB protein induces its activity nearly 200-fold (412). In D. radiodurans, DNA repair proteins appear to interact with protein kinases and phosphoproteins in a multiprotein complex (319). Global phosphorylation levels, ATP levels, and protein kinase activities are increased while phosphatase and phosphodiesterase activities are decreased following ionizing radiation (282). Specifically, an elevated level of protein phosphorylation was found to attenuate nucleolytic activity in radiation-damaged D. radiodurans cells (282). According to our limited present knowledge, protein phosphorylation in response to radiation in D. radiodurans is mediated by a serine/threonine protein kinase (DR2518) (504, 505) and by IrrE (PprI) (377). Although the target proteins and the regulatory pathways of the DR2518 protein kinase are yet to be identified, its physiological significance is reflected in the considerable radiation sensitivity of kinase-deficient D. radiodurans cells due to delayed DSB repair (504). Its expression is induced in irradiated cells, while in vitro activity is stimulated by the coenzyme pyrroloquinoline-quinone (PQQ) and by linear (i.e., fragmented) but not circular DNA, which is again suggestive of its involvement in the DNA damage response (504, 505). Although global phosphorylation levels are increased (282), the level of serine/threonine phosphorylation decreases immediately after radiation damage in D. radiodurans (505), which was also observed for the tyrosine phosphorylation of the E. coli SSB protein (412). This indicates differential protein regulation in response to DNA damage depending on the type of phosphorylation modification.
PREVENTION AND TOLERANCE OF DNA AND PROTEIN DAMAGE IN D. RADIODURANS
Although DNA has been considered the primary target of radiation damage and the repair of damaged DNA has been considered the primary determinant of the survival capacity (264), DNA-damaging agents induce oxidative damage not only to DNA but also to all cellular macromolecules via the production of ROS (114, 280, 376, 495). Radiation-resistant and radiation-sensitive bacteria are equally susceptible to DSBs (though not base damage) induced by ionizing radiation (123, 206, 302) but differ in the amount of protein damage, which is significantly more pronounced in radiation-sensitive bacteria (122). This observation has given rise to a new outlook on radiation toxicity, whereby protein oxidation is considered the main cause of radiation-induced cell death and the capacity to prevent and tolerate protein damage is a major determinant of radiation and desiccation resistance (115, 120-123, 164). D. radiodurans is endowed with strong oxidative stress prevention and tolerance mechanisms, which protect proteins from oxidative damage and sanitize the cells from toxic oxidized products.
Alongside DNA repair, D. radiodurans employs several other strategies to prevent and cope with oxidative stress: (i) cell cleaning through the elimination of damaged (oxidized) macromolecules, (ii) the selective protection of some proteins against oxidative damage, (iii) the suppression of endogenous ROS production, and (iv) antioxidant defense systems (Fig. 11). Oxidized nucleotides are detoxified and recycled by Nudix hydrolases (666) and nucleotidases (318), and oxidized oligonucleotides are excreted (643), while oxidized proteins are degraded by proteases (550). Endogenous ROS production is suppressed by the reduction in the number of respiratory chain enzymes and enzymes with iron-sulfur clusters as the major sources of endogenous ROS (207). Antioxidant defense is mediated by nonenzymatic (e.g., divalent manganese complexes and carotenoids) and enzymatic (e.g., catalases, superoxide dismutases, and peroxidases) scavengers. D. radiodurans has an expanded repertoire of genes for cellular cleansing and antioxidation protection, which serve to reinforce the capacities of D. radiodurans for the prevention and repair of damage to DNA, RNA, and proteins (380). Many of these protective and salvage genes are expressed at high constitutive levels (364) and are induced after irradiation (368, 602, 677).
FIG. 11.
Factors contributing to ionizing radiation resistance in D. radiodurans: cellular cleansing, antioxidant defenses, and DNA repair. Cellular cleansing involves the degradation of oxidized nucleotides by Nudix hydrolases, the export of damaged oligonucleotides, and the proteolytic degradation of damaged proteins. The antioxidant defense system consists of nonenzymatic scavengers, such as manganese complexes and carotenoids, and enzymatic scavengers, such as catalases and superoxide dismutases. Within the DNA repair machinery, base excision repair (BER) acts on damaged DNA bases, and nucleotide excision repair (NER) removes damaged nucleotides, while extended synthesis-dependent strand annealing (ESDSA) and homologous recombination (HR) mend DNA double-strand breaks.
Cell-Cleaning Proteins in D. radiodurans
Oxidative stress damages DNA and generates potentially toxic and mutagenic oxidized derivatives within the nucleotide pool. The export and degradation of damaged (oligo)nucleotides are highly pronounced components of an antimutagenic system in D. radiodurans that is responsible for sanitizing the cells from oxidative DNA damage. Damaged oligonucleotides are exported from the cells after ionizing radiation (643) and UV radiation (63) (see “DNA Degradation in Irradiated D. radiodurans” and “Kinetics and enzymology of the repair of UV-induced DNA damage in D. radiodurans”). UvrA2, which is closely related to the ABC transporters, may be in charge of oligonucleotide export (653). Damaged nucleotides and potentially toxic nucleoside diphosphate derivatives are detoxified and recycled by Nudix (“nucleoside diphosphate linked to some other moiety x”) hydrolases (53, 666) and by nucleotidases (nucleoside monophosphate phosphohydrolases) (318). The Nudix family is markedly expanded in D. radiodurans, with 23 members (653), 5 of which are induced following exposure to ionizing radiation (368). As the prototype of the Nudix family of housecleaning and gatekeeping enzymes, MutT hydrolyzes 8-oxo-dGTP and 8-oxo-GTP into 8-oxo-dGMP and 8-oxo-GMP, thereby preventing their misincorporation into DNA and RNA (387, 598). Nucleotidases can subsequently dephosphorylate 8-oxo-dGMP and 8-oxo-GMP into 8-oxo-dG and 8-oxo-G that can be excreted (105). The first characterized 5′ nucleotidase in D. radiodurans, DR0505, is an ATP-sensitive phosphomonoesterase and phospodiesterase that exhibits differential activity on normal, oxidized, and cyclic nucleotides (318). In addition to their importance for the cellular recycling of nucleotides, periplasmically located bacterial nucleotidases facilitate the uptake of extracellular nucleosides that can be used as carbon sources (667).
Cellular sanitization entails not only the degradation and export of damaged DNA but also the degradation of damaged proteins. Oxidatively damaged proteins are dysfunctional and need to be proteolytically removed and rapidly resynthesized. D. radiodurans adheres to the pattern of selective sensibility of certain proteins to degradation found in distantly related organisms such as E. coli, yeast, and Arabidopsis thaliana (116). Degradation affects the chaperones GroEL and DnaK, along with the TCA cycle enzymes aconitase and citrate synthase, which are also the first to be resynthesized (278, 677). Proteolytic functions are highly diverse and redundant in D. radiodurans, enabling efficient cellular sanitization from the oxidized proteins (285, 550). The level of intracellular proteolytic activity is increased following radiation exposure (121). Proteases may be triggered by aconitase, which acts as an oxidative stress sensor (524). The majority of bacterial proteolysis is carried out by ATP-dependent proteases from the Lon and Clp families, which are two-component enzymes comprising a proteolytic subunit and an ATPase subunit. D. radiodurans encodes two Lon protease homologs, Lon1 (DR1974) and Lon2 (DR0349); one putative proteolytic subunit, ClpP (DR1972); and four putative ATPase subunits, ClpA, ClpB, ClpC, and ClpX (DR0588, DR1046, DR1117, and DR1973, respectively) (550). The inactivation of the Lon1 and Lon2 proteases has no effect on radiation resistance but reduces resistance to puromycin (550). Puromycin is a tRNA analog that induces the premature release of polypeptide chains from ribosomes, resulting in the production of misfolded peptides. This suggests that Lon1 and Lon2 are important for the removal of misfolded proteins in D. radiodurans but not those damaged by radiation (550). Whereas Lon proteases are important for removing dysfunctional proteins, the ClpPX protease is important for the regulation of cell division (550) and quite possibly for providing amino acids and peptides, important constituents of manganese complexes (121) (see “Manganese complexes”).
Whereas some proteins are degraded (and rapidly resynthesized) following ionizing radiation, others are protected against degradation (278). Among the most prominent proteins to escape degradation following 6 kGy of ionizing radiation are a serine protease, EF-Tu, and, quite possibly, DNA-directed RNA polymerase subunits β (RpoB) and β′ (RpoC) (278). The selective protection of RNA polymerase and elongation factors would enable swift protein resynthesis and thus ensure the rapid postirradiation recovery of all cellular functions. Some proteins may be impervious to oxidative damage due to their structural design, where amino acids that are susceptible to oxidative damage are buried in the interior and inaccessible to ROS.
Antioxidation Protection in D. radiodurans
In general, oxidative protein damage judged from the carbonylation levels is lower in unirradiated or irradiated D. radiodurans than in radiation-sensitive organisms (122). For both the radiation-resistant organism D. radiodurans and the radiation-sensitive organism E. coli, the amount of carbonylated proteins increases sigmoidally with the ionizing and UV radiation doses and is negatively correlated with survival (329). Carbonylation is the most common oxidative modification of proteins, often used as a biomarker of oxidative stress. The accumulation of oxidative damage to proteins alters their catalytic activities and interactions, which leads to the disruption of cellular functions and culminates in cell death (Fig. 12). The oxidation of DNA repair proteins causes error-prone activities, which result in DNA mutations (122). Highly diverse and redundant antioxidant defense systems prevent protein oxidation and alleviate oxidative stress in D. radiodurans by neutralizing reactive oxygen species. The D. radiodurans antioxidant defense machinery is active against all three primary reactive oxygen species: hydroxyl radicals (OH·), superoxide radicals (O2·−), and hydrogen peroxide (H2O2). Apart from water radiolysis, OH· is also produced by the decomposition of H2O2 in the Fenton or Haber-Weiss reaction started by traces of transition metal ions, principally iron (169, 406). While OH· is highly reactive with all cellular macromolecules, O2·− and H2O2 are not particularly reactive with DNA but affect proteins with exposed iron-sulfur or heme groups and with cation-binding sites where iron-catalyzed oxidation can occur (121, 266). O2·− can liberate Fe2+ from iron-sulfur clusters, which can react with H2O2 in the Fenton reaction to produce OH· (267). D. radiodurans protein extracts have 30-fold-, >17-fold-, and 6-fold-higher scavenging effects on H2O2, OH·, and O2·−, respectively, than do those of E. coli (617). Enzymatic ROS scavenging is mediated by three catalases, four superoxide dismutases, two peroxidases, and two Dps proteins, whereas nonenzymatic scavengers include divalent manganese complexes and carotenoids (Fig. 13). The D. radiodurans metabolic configuration and metabolic control suppress endogenous ROS production via a reduction in the number of respiratory chain enzymes and enzymes with iron-sulfur clusters along with the induction of the glyoxylate bypass step of the TCA cycle (123, 207) (see “Metabolic Configuration of D. radiodurans”). Of all the ROS scavengers identified in D. radiodurans, manganese complexes are the most powerful antioxidants (121).
FIG. 12.
Schematic representation of a protein network (I) and the consequences of increasing carbonylation of proteins (II and III) on its activity. Red dots represent single carbonyl groups.
FIG. 13.
Enzymatic (A) and nonenzymatic (B and C) antioxidant defenses in D. radiodurans. (A) D. radiodurans has higher levels of catalase and superoxide dismutase (SOD) activities than does E. coli. SOD converts superoxide ions (O2·−) into hydrogen peroxide (H2O2), which is converted by catalase into water and oxygen. (Based on data from reference 617.) (B) The D. radiodurans major carotenoid, deinoxanthin, has a higher level of H2O2-scavenging activity than does β-carotene. Carotenoids scavenge peroxyl radicals by the radical adduct formation mechanism. (Modified from reference 618 with permission from Elsevier.) (C) A high intracellular Mn/Fe ratio is correlated with a high radiation resistance level and a low protein oxidation (carbonylation) level among bacteria. Divalent manganese ions (Mn2+) can scavenge O2·− in complex with phosphates and H2O2 in complex with amino acids and bicarbonate. (Modified from reference 120 with permission of the publisher and based on data from references 36, 51, 120, 122, and 123.)
ROS scavenging in D. radiodurans is under both positive and negative control. RecD (686) and RecQ (92) stimulate ROS scavenging, whereas RecX (DR1310) (562) and the Mn-dependent transcriptional regulator TroR/DtxR (93) repress it. In recD mutant cells, levels of catalase activity and ROS scavenging are lower than those in wild-type cells (686). Similarly, recQ mutant cells are more susceptible to oxidative stress than are wild-type cells, with higher ROS levels under both normal and H2O2-stressed conditions and with a more vigorous stress response (92). Conversely, RecX was shown to repress the expression of the catalase DRA0146, the superoxide dismutase DR1279 (562), and ferredoxin (DR2075) (563). ROS scavenging activity is induced in irradiated wild-type cells but not in recX mutant cells, in which the scavenging activity is already enhanced under nonirradiated conditions (562). DtxR is another negative regulator of catalase (DR1998) activity; the mutant strain has a higher catalase activity and is more resistant to H2O2 than the wild type as a result (93). Finally, OxyR is both a positive and a negative regulator of ROS scavenging in D. radiodurans cells exposed to H2O2 (94).
Catalases, superoxide dismutases, and peroxidases.
D. radiodurans is highly resistant to H2O2 (646) and O2·− (3) and has high levels of constitutive catalase activity (14, 96, 364) and superoxide dismutase activity (364) (Fig. 13A). Catalases and peroxidases remove H2O2, whereas superoxide dismutases eliminate superoxide radicals from the cells. D. radiodurans encodes three catalases (katE catalases DR1998 and DRA0259 and eukaryotic-type DRA0146), four superoxide dismutases (Mn-dependent DR1279 and Cu/Zn-dependent DR1546, DRA0202, and DR0644), a cytochrome c peroxidase (DRA0301), and an iron-dependent peroxidase (DRA0145) (381) (Table 4).
D. radiodurans is much more resitant to H2O2 than is E. coli, with a large shoulder in the survival curve (646). According to data reported previously by Wang and Schellhorn (646), the catalase activities during exponential and stationary phases are 127 and 32 times higher those in E. coli, respectively, whereas Tian et al. (617) reported a 15-fold-higher catalase activity in D. radiodurans than in E. coli (Fig. 13A). Catalase activity is affected by H2O2 (98), ionizing radiation (601), the addition of manganese (96), and the growth phase (243), with a higher level of catalase activity in stationary-phase cells than in exponential-phase cells (646). Catalase activity is positively controlled by the transcriptional regulator DrRRA (645) and negatively controlled by OxyR (94). DR1998 is induced in response to ionizing radiation (601, 602) and is more stable in the presence of H2O2 than commercially available Aspergillus niger or bovine liver catalases (312). Among the SOD proteins, DR1279 is constitutively expressed (364). DR1279 efficiently eliminates higher O2·− concentrations than Mn-SODs in E. coli and humans due to the more rapid protonation and release of H2O2 (3). D. radiodurans catalase (DR1998) and superoxide dismutase (DR1279) mutants are sensitive to H2O2 and paraquat, respectively, but not to ionizing radiation at doses lower than 16 kGy (388). While D. proteolyticus, D. indicus, and D. grandis also have high levels of catalase and SOD activities, D. ficus and D. mumbaiensis have a 15-fold-lower level of catalase activity but high levels of SOD activity (561). The absence of a strong positive correlation between catalase activity and (i) the MIC of H2O2 or (ii) ionizing radiation resistance across Deinococcus species suggests that other (nonenzymatic) antioxidants (such as manganese complexes) contribute to the scavenging of H2O2 (561).
D. radiodurans also encodes other oxidative defense proteins, such as glutaredoxin, thioredoxin, thioredoxin reductase, and alkyl hydroperoxide reductase, while glutathione, glutathione reductase, and glutathione peroxidase are absent (653) (Table 4). In E. coli, the alkyl hydroperoxide reductase is the primary scavenger of endogenous H2O2 (543). Thioredoxin (DRA0164) reduces oxidized cysteines in proteins and is reverted from its oxidized form by thioredoxin reductase (DR1982) in an NADPH-dependent reaction (466, 547). D. radiodurans also possesses two peptide methionine sulfoxide reductases, MsrA (DR1849) and MsrB (DR1378) (475), which are important for the reduction of oxidized methionine in proteins. MsrA is transcriptionally induced following ionizing radiation (602).
Dps proteins.
Apart from promoting nucleoid compaction, E. coli Dps proteins also protect DNA from oxidative damage by binding DNA, chelating ferrous ions (Fe2+), and reducing H2O2 to H2O (11, 390). The elimination of Fe2+ and H2O2 precludes Fenton chemistry, whereby H2O2 is reduced in the presence of Fe2+ into the highly damaging OH·. D. radiodurans encodes two Dps homologs, Dps1 (DR2263) and Dps2 (DRB0092), which share only 14% sequence identity. A dimeric form of Dps1 protects DNA from hydroxyl radical cleavage (218), which may also be true for Dps2, as the dps2 mutant is sensitive to H2O2 (668). Both Dps1 and Dps2 are induced in response to ionizing radiation (368, 602).
Carotenoids.
Carotenoids are efficient scavengers of ROS, especially of singlet oxygen (1O2) and peroxyl radicals (ROO·) (587, 608). 1O2 transfers its energy to the carotenoid to generate the ground state of oxygen (3O2) and the triplet state of the carotenoid, which returns to its ground state by releasing its energy to environmental substances (192). Peroxyl radicals are scavenged by radical adduct formation (360, 432). Carotenoids protect DNA from oxidative damage, proteins from carbonylation (680), and membranes from lipid peroxidation (586). In vitro, D. radiodurans carotenoids can scavenge all types of ROS (OH·, O2·−, H2O2, and 1O2) (618, 679) as well as RNS (reactive nitrogen species) such as 2,2-diphenyl-1-picrylhydrazyl (DPPH·) (616). D. radiodurans harbors 13 genes involved in carotenoid biosynthesis (381). Deinoxanthin, a major product in the carotenoid synthesis pathway in D. radiodurans (348), has a stronger scavenging ability on H2O2 and 1O2 than two carotenes (lycopene and β-carotene) and two xanthophylls (zeaxanthin and lutein) (618) (Fig. 13B) and inhibits protein oxidation in vitro at lower concentrations than other carotenoids (616). Deinoxanthin also exerts some protective effect (20%) on plasmid DNA exposed to OH· by recovering the supercoiled plasmid form, which is otherwise completely shattered (618). However, carotenoids seem to have little effect on radiation resistance, as pigmentless D. radiodurans and D. radiopugnans mutants are as resistant to ionizing and UV radiation as the wild type (396, 447, 472). Pigmentless colonies occasionally appear among D. radiodurans survivors after extreme radiation exposures. Moreover, a targeted mutation of the phytoene synthase gene (crtB [DR0862]), which blocks the carotenoid synthesis pathway, only slightly increased sensitivity to ionizing radiation, desiccation, and UV radiation (618, 679). This suggests that (i) other ROS scavengers can successfully replace carotenoids, (ii) carotenoids are not as efficient in vivo as they are in vitro, or (iii) membranes are not the primary targets of ionizing or UV radiation-induced ROS.
Although D. radiodurans carotenoids have a strong scavenging ability on singlet oxygen (1O2) and superoxide radicals (O2·−) in vitro (618, 679), D. radiodurans is 100-fold more sensitive than E. coli to photodynamic treatment with the sensitizer Rose Bengal, which generates both 1O2 and O2·− (539). This may additionally indicate that, although efficient in vitro, carotenoids are ineffective as singlet oxygen scavengers in vivo. The differential effect of Rose Bengal on the two species may also be attributed to the higher sensitivity of the Deinococcus cell membrane to singlet oxygen (539) due to the presence of unsaturated fatty acids, which are prone to peroxidation (68, 631).
Manganese complexes.
In vitro, divalent manganese ions (Mn2+) can scavenge O2·− in complex with phosphate (25, 36) and H2O2 in complex with bicarbonate and amino acids or peptides (51) (Fig. 13C). Mn-orthophosphate acts as a true catalytic superoxide scavenger; Mn-pyrophosphate is a stoichiometric scavenger, whereas Mn-polyphosphate is inefficient as a superoxide scavenger (25, 36, 402). Mn may not only act as a chemical scavenger but may also replace Fe in Fe-loaded enzymes, thereby precluding protein oxidation resulting from the iron-driven Fenton reaction (19). The removal of O2·− in vitro is dependent on a threshold concentration of Mn (25, 36, 121, 122). D. radiodurans has an exceptionally high intracellular manganese content (0.2 to 4 mM) (122, 123, 346) and a high intracellular manganese-to-iron (Mn/Fe) ratio of 0.24 (123). A high Mn/Fe ratio correlates with extreme levels of ionizing radiation and desiccation resistance among bacteria (123, 196) as well as a low level of oxidative protein damage (122) (Fig. 13C). However, late-stationary-phase D. radiodurans cells are 4-fold more radiation sensitive than are early-stationary-phase cells but have the same Mn/Fe ratio (592). The growth of D. radiodurans in a defined minimal medium is dependent on Mn (123, 654). An optimal concentration of manganese in the growth medium (e.g., 5 μM) is necessary for ensuring the antioxidant properties of manganese complexes, as both manganese depletion (e.g., in potassium phosphate-buffered TGY medium) (246) and manganese overaccumulation (e.g., 100 μM MnCl2) (96, 683) can cause oxidative stress. D. radiodurans cells grown in defined rich media without Mn have a 6-fold-lower Mn/Fe ratio and are consequently 6-fold more sensitive to ionizing radiation with a high level of oxidative protein damage (122, 123). Furthermore, Mn is required for postirradiation recovery, as 10-kGy-irradiated D. radiodurans cells incubated without Mn display a 1,000-fold reduction in cell survival compared to cells recovered on TGY medium (123).
The majority of manganese in D. radiodurans is present as small complexes with orthophosphate and peptides, which, by scavenging O2·− and H2O2, specifically protect proteins against oxidative damage (121, 122) (Fig. 14). Nucleosides in D. radiodurans, such as uridine, also form complexes with Mn2+ and orthophosphate, which scavenge ROS more efficiently than Mn2+-orthophosphate alone but less efficiently than Mn2+-orthophosphate-peptide complexes (121) (Fig. 14). Individually, nucleosides, amino acids, peptides, and other small organic metabolites scavenge OH· efficiently but do not remove O2·− or H2O2 (121, 641). Compared to radiation-sensitive bacteria, protein-free cell extracts (ultrafiltrates) of D. radiodurans are enriched in all these radioprotective metabolites (Mn2+, orthophosphate, peptides, and nucleosides), which enhance the survival of irradiated E. coli and human cells when applied ex vivo (121). When reconstituted in vitro at concentrations approximating those in D. radiodurans, the phosphate buffer (13 mM), MnCl2 (1 mM), and decapeptide (3 mM) synergistically preserve the activity of irradiated enzymes in vitro (121). A similar mixture of phosphate buffer (13 mM), MnCl2 (1 μM), uridine (3 mM), and 3% dimethyl sulfoxide (DMSO) (an OH· scavenger) increases the survival of 3-kGy-irradiated E. coli 10,000 times and promotes its growth under conditions of high-level chronic irradiation of 42 Gy/h (121). While manganese complexes have emerged as the most powerful protein antioxidants in D. radiodurans (121), a comparative analysis of Deinococcus species failed to show a strong correlation between the Mn/Fe ratio, catalase activity, and carotenoid content on one side and H2O2 tolerance and ionizing radiation resistance on the other. This suggests that oxidative stress resistance of the Deinococcaceae is a cumulative contribution of both nonenzymatic and enzymatic antioxidants (561).
FIG. 14.
Manganese-based chemical antioxidants in D. radiodurans. Divalent manganese (Mn2+) complexes scavenge long-lived (O2·− and H2O2) and short-lived (OH·) ROS, thereby preventing their interconversion and proliferation in cells. Mn2+ catalytically scavenges superoxide radicals (O2·−) in complex with orthophosphate (red). Free amino acids or peptides in complex with Mn2+ and orthophosphate (or bicarbonate) catalytically decompose hydrogen peroxide (H2O2) and scavenge O2·− (green). Nucleosides (and their analogs) containing two carbonyl groups separated by an amino group (e.g., uridine) complex with Mn2+-orthophosphate and scavenge O2·− (blue). Nucleosides, free amino acids, peptides, and other small organic metabolites stoichiometrically scavenge hydroxyl radicals (OH·). Note that a single Mn2+ cannot bind all three ligands at the same time. It has been proposed that the DNA repair proteins of D. radiodurans work so efficiently because they are protected from ROS by Mn2+ complexes. (Based on data from reference 121.)
Complexes between manganese and cellular metabolites are present at high concentrations throughout the D. radiodurans cytosol and provide rapid and immediate scavenging of ROS during and after irradiation (121-123). Mn2+ complexes are therefore more efficient in protecting DNA repair enzymes and other recovery proteins than enzymatic ROS-scavenging systems, which are damaged and need to be resynthesized after irradiation (122, 123). Depending on growth conditions, D. radiodurans has 100-fold-higher Mn levels (121, 346), 5-fold-higher phosphate levels, 35-fold-higher levels of nucleosides and bases, and up to 100-fold-higher concentrations of amino acids than E. coli (121). The highest cellular Mn concentration coincides with electron-dense granules located at the center of D. radiodurans nucleoids (122). Electron-dense granules presumably contain polyphosphates (615), which may provide orthophosphate for Mn2+-phosphate complexes (120, 121). The accumulation of uridine and adenosine may stem from the inability of D. radiodurans to use these nucleosides as carbon sources (121). Similarly, the inability to use TCA cycle products (e.g., α-ketoglutarate) as carbon sources may lead to the accumulation of amino acids as derivatives of the TCA cycle products (121). It is significant that nucleosides, amino acids, and peptides all accumulate after irradiation due to high levels of nucleolytic (159, 235, 236, 572) and proteolytic (121) activity.
The antioxidation protection strategy based on manganese complexes may not be limited to the Deinococcaceae. In yeast cells and in yeast mutants with 7-fold-higher Mn levels, 47% and 60% of the intracellular Mn, respectively, forms a complex with orthophosphate, and 25% of the intracellular Mn forms a complex with polyphosphate (402). Whereas the Mn-polyphosphate complex is ineffective as an antioxidant, the Mn-orthophosphate complex is a strong ROS scavenger, the concentration of which correlates with oxidative stress resistance (402). Yeast mutants with higher concentrations of Mn-orthophosphate can compensate for the loss of a superoxide dismutase, whereas mutants with lower concentrations of Mn or Mn-orthophosphate are inviable in the absence of SOD (402). Even exogenously supplied Mn can compensate for the loss of SOD in yeast cells (90, 532), in N. gonorrhoeae (625), and in E. coli (10). In the radiation-resistant bacterium L. plantarum, which is unusual in lacking a superoxide dismutase, Mn (30 mM) presumably forms a complex with phosphates (65 mM) and proteins that can compensate for the absence of SOD by scavenging O2·− (22, 23). Lactobacillus bulgaricus and Lactobacillus acidophilus, which have neither SOD nor Mn, are highly oxygen intolerant (24). Desiccation- and radiation-resistant spores of Bacillus species accumulate Mn, dipicolinic acid, and small proteins (176, 552), while radiation-resistant Cyanobacteria accumulate Mn and trehalose (568). Mitochondria (221) and chloroplasts (659) also accumulate high levels of manganese. It therefore appears that Mn complexes represent a widespread nonenzymatic strategy for combating oxidative stress by providing levels of protection from ROS that equal or exceed the ROS-scavenging capacities of enzymes (121).
Even though Fe levels in D. radiodurans are four times higher than Mn levels (123), D. radiodurans chromosomes preferentially bind Mn (346). Nonetheless, while efficiently protecting proteins against oxidative damage, neither manganese nor any other agent seems to protect DNA from double-strand breakage during irradiation, as the numbers of DSBs per Gy per genome are similar for radiation-resistant and radiation-sensitive species (0.002 to 0.006 DSBs/Gy/Mbp) (67, 79, 206, 211, 499, 516, 522). DNA bases may still be protected against oxidation, as the amounts of oxidative DNA base damage per radiation dose per genome differ among organisms. For example, the more radiation-resistant D. radiodurans has a higher level of oxidized bases/Gy/Mbp than does H. salinarum (302). The high levels of intracellular halides of H. salinarum protect DNA bases (302), while in the desiccation-resistant cyanobacterium Nostoc commune a high concentration of trehalose enhances DNA protection against ROS (568). As Mn inhibits the formation of thymine-containing dimers and therefore contributes to the high level of UV resistance of D. radiodurans (346), it remains possible that Mn or another protective agent shields DNA bases in D. radiodurans against oxidative base damage.
Apart from its antioxidant properties, manganese is required for the activity of enzymes such as the UVDE endonuclease (183), the superoxide dismutase (279), DNA polymerase X (61), the NAD-dependent DNA ligase (60), and the dual-function esterase/nuclease DR0505 (318). Mn homeostasis in D. radiodurans is regulated by an ABC-type Mn transporter (DR2283), an NRAMP family Mn transporter (DR1709), and an Mn-dependent transcriptional regulator, TroR/DtxR (DR2539) (383, 475). DR1709 is an essential gene (383) and is induced as D. radiodurans recovers from both ionizing radiation and desiccation (602). DR2539 is a negative regulator of DR2283 and a positive regulator of Fe-dependent transporter genes (DR1219 and DRB0125) (93). D. deserti has several manganese ABC transporters and three Mn-dependent transcriptional regulators, TroR/DtxR (140). As in yeast, there may be a division of roles between Mn transporters, where some provide Mn for Mn-requiring enzymes, while others are critical under conditions of oxidative stress (512, 513).
WHAT DETERMINES THE EXTREME RADIATION RESISTANCE OF D. RADIODURANS?
As long as the genome can be reconstituted, cellular life can be restored. This is the basis of a long-held DNA-centric dogma postulating that survival following exposure to DNA-damaging agents, such as ionizing radiation and desiccation, depends principally on the capacity to repair damaged DNA. This position would seem to be at variance with the observation that the rates of DSB production are markedly similar among radiation-resistant and radiation-sensitive organisms (67, 79, 206, 211, 499, 516). Moreover, while yeast and human cells can survive hundreds of endogenous DSBs (78, 153), they succumb to ionizing radiation doses that generate the same number of DSBs but which also cause protein damage (121). Together, these observations suggested that cellular macromolecules other than DNA determine survival following exposure to ionizing radiation, and this in turn called for a reassessment of the molecular basis of oxidative stress resistance.
In recent years a new paradigm of radiation resistance has emerged based on the finding that in irradiated bacteria the level of protein oxidation is negatively correlated with survival and that protein oxidation is the cause, and not the consequence, of cell death (122, 329). This suggests that proteins are the primary target of radiation toxicity and that the ability to protect proteins against oxidation distinguishes radiation-resistant from radiation-sensitive species (120-122, 383). In D. radiodurans the major contribution to protein protection comes from complexes between Mn2+, orthophosphate, peptides, and possibly nucleosides that synergistically scavenge all ROS (121) (Fig. 14). The efficient protection of DNA repair and other proteins against oxidative damage enables conventional DNA repair enzymes to function with greater efficiency and to provide a timely response under conditions of oxidative stress, such as exposure to ionizing radiation or long periods of desiccation. In radiation-sensitive bacteria, the inactivation of DNA repair proteins by ROS is presumably the cause of their limited efficiency (123, 207). While Mn complexes, particularly those consisting of Mn2+, orthophosphate, and peptides, were shown to confer in vitro radioprotection to enzymes such as BamHI and glutamine synthetase (121), it would be interesting to compare their protective effects on DNA repair enzymes such as RecA and PolA from radiation-sensitive and radiation-resistant organisms. This would rule out the possibility that enzymes from radiation-resistant species have special structural properties that shield them from oxidative damage.
Although it is now clear that proteins are the primary target of radiation damage and that antioxidative protein protection is the primary determinant of bacterial radiation resistance (69, 70, 122, 329), whether DNA repair systems in radiation-resistant organisms are endowed with advantageous properties compared to the radiation-sensitive organisms remains open to debate. Similarities with conventional DNA repair systems are supported by the following observations: (i) D. radiodurans seems to possess a relatively standard set of DNA repair proteins also found in radiation-sensitive bacteria such as E. coli (381, 653), and orthologs from E. coli can complement several highly radiation-sensitive D. radiodurans DNA repair mutants, such as polA (224) and uvrA (4) mutants; (ii) a comparative analysis of the genomes of the radiation-resistant bacteria D. radiodurans, D. geothermalis, L. plantarum, and R. xylanophilus did not reveal a shared group of uncharacterized genes that might be responsible for radiation resistance (123); and (iii) the mechanism of DSB repair in irradiated D. radiodurans is remarkably similar to mitotic and meiotic DSB repair in yeast (124, 126, 127, 572, 676). However, it is difficult to conceive that the reassembly of numerous DNA fragments generated at high radiation doses can be accomplished by standard DNA repair machinery. Although the previously identified DNA repair enzymes of D. radiodurans are seemingly indistinct from those of radiation-sensitive bacteria, there are still many uncharacterized genes which may encode functions that enhance the efficiency of DNA repair and render the D. radiodurans DNA repair machinery distinct from those of standard organisms (108, 216). The Deinococcaceae have 206 (216) or 230 (140) unique proteins; only 5 are identified DNA repair-related proteins, DdrB, DdrC, DdrD, PprA, and DdrO (140, 216), of which DdrB (464) and PprA (456) have been characterized. Many of the Deinococcus-specific proteins are highly expressed, suggesting their importance in general metabolism, cell viability, and stress resistance (140). Furthermore, D. radiodurans shows a high degree of redundancy in DNA repair-related genes, which is expected to potentiate the effectiveness of DNA repair processes. D. radiodurans has at its disposal 11 DNA glycosylases (381), 2 pathways (UvrABC and UVDE) for the repair of UV-induced DNA damage (171, 413, 441, 603), 2 distinct SSB proteins (the classic SSB and DdrB) (464), and 23 Nudix hydrolases (653, 666). Redundancy is a striking feature of D. deserti, which has three recA genes coding for two different proteins (140, 168). Another sequenced radioresistant bacterium, K. radiotolerans, is also abundant in BER and NER enzymes (32). A concerted action of enzymes with overlapping functions is expected to enhance the efficacy of the DNA repair processes. The earlier finding that UV-irradiated transforming DNA has a much greater transforming ability in D. radiodurans than in H. influenzae due to the efficiency of D. radiodurans at repairing irradiated transforming DNA in the same manner as that of its own irradiated DNA (448) also suggests that D. radiodurans possesses a more efficient DNA repair machinery. Finally, the evolutionary advantage of DSB repair in D. radiodurans may rely on structural elements that enable the rapid and accurate assembly of broken DNA fragments (see “Physical scaffolds for DNA repair in D. radiodurans”).
The induction of DNA damage without causing any protein damage should reveal whether D. radiodurans is as efficient at DNA repair as radiation-sensitive species. If D. radiodurans is endowed with special DNA repair properties, it is expected to repair the same amount of DNA damage more efficiently than a radiation-sensitive bacterium such as E. coli. The difficulty of such an approach is that all known DNA-damaging agents concomitantly induce oxidative damage to other cellular macromolecules via ROS production, including ionizing radiation (114), UV radiation (280), desiccation (495), and the quinone anticancer drugs mitomycin C, bleomycin, doxorubicin, and daunorubicin (228, 281, 376). An alternative approach is to compare the efficiencies of double-strand-break repair in vitro by using a cellular extract from D. radiodurans and E. coli. However, Craig Venter and colleagues were unsuccessful when trying to reproduce in vitro the fragment assembly process with a deinococcal extract, with the failure possibly being due to the extract being made from non-stress-induced cells. The concept of introducing damaged DNA into an intact cell remains the best approach to evaluating the source of the extreme radiation resistance of D. radiodurans. A newly developed technique that combines atomic force microscopy with nanofluidics has been employed to deliver dyes into live neuroblastoma cells (405). A potential application of this technique is to deliver overlapping DNA fragments into D. radiodurans and E. coli and assay their assembly into functional plasmids.
Until an assay that distinguishes unequivocally between the contribution of the DNA repair process and protection against oxidative damage at the protein and global macromolecular levels is available, we have to concede that the extreme radiation resistance of D. radiodurans is imparted synergistically by efficient protection against oxidative stress and by an efficient DNA repair mechanism, enhanced by functional redundancies in both systems (572). Indeed, an analysis of the evolved radiation-resistant E. coli strains failed to pinpoint a unique mechanism responsible for the acquisition of radiation resistance and showed that multiple pathways impart the radioresistant phenotype (238).
SOME RAMIFICATIONS OF D. RADIODURANS OXIDATIVE STRESS RESISTANCE
Can D. radiodurans Provide a Means for Circumventing Aging and Cancer?
The properties of the extreme resistance of D. radiodurans to ionizing radiation and other oxidative agents due to extremely efficient protection against oxidative damage and extremely efficient DNA repair are expected to provide means for avoiding or alleviating DNA, RNA, and protein damage and the oxidative stress that is tightly linked with aging and cancer. Aging and cancer are associated with increased DNA and protein oxidation due to ROS generation, a decline in the robustness of antioxidant defenses and DNA repair, and an accumulation of the end products of oxidative damage (45). Aging and cancer could perhaps be delayed or prevented by interventions designed to prevent DNA damage accumulation and the production of oxidized proteins. A focal point of aging and cancer research is to identify factors that antagonize the aging process and carcinogenesis and to design adequate therapeutic strategies; the D. radiodurans strategies of combating oxidative stress may open new avenues.
Oxidative stress, aging, and cancer.
The free radical theory of aging and cancer postulates that the damage caused by the production of ROS is the underlying cause of aging (237) and cancer (309). A more rapid aging process (progeria) and diseases such as Alzheimer's disease (251, 493, 575) and Parkinson's disease (401) are also clearly associated with oxidative stress (655). While there is some evidence that protein oxidation is a cause of cell death in oxidatively stressed bacteria (329), for human cells it is still unclear whether oxidative stress is a cause or a consequence of the primary disease process (117, 337, 465). Oxidative stress occurs when ROS production is accelerated or when antioxidant defense enzymes are impaired, with the former being more frequent than the latter. Oxidative stress affects both DNA and proteins. As mentioned above, oxidative DNA damage leads to damaged bases, single-strand breaks, and double-strand breaks, while the most common oxidative modification of proteins is carbonylation. Whereas oxidative DNA damage can be repaired by a DNA repair apparatus, oxidized (carbonylated) proteins are destined for proteolytic degradation. Although accumulating evidence suggests that the recovery of D. radiodurans from oxidative stress is determined by protection against protein oxidation, researchers still argue whether aging and cancer are a consequence of the accumulation of DNA damage (371, 542) or protein damage (117, 353), as both genomic instability and the accumulation of protein damage are hallmarks of cancer and aging alike. The “death by protein damage” hypothesis of oxidative stress in bacteria (120, 121) remains to be tested with human cells by determining whether there exists a similar quantitative relationship between protein oxidation and cell survival following radiation-induced oxidative damage.
DNA damage, aging, and cancer.
Empirical evidence from diverse lines of research suggests that aging is a process of a gradual increase in somatic mutations leading eventually to frailty and an increased risk of a spectrum of age-associated diseases (300, 301). For example, 8-oxoguanine, a biomarker of oxidative DNA damage, accumulates with age (231). Premature-aging diseases and progeroid syndromes are associated with defects in genes involved in DNA repair and genome maintenance (542). Cancer is a genetic disease, and individuals with defects in DNA repair pathways are often predisposed to various cancers (685). Carcinogenesis can be driven by mutational or chromosomal genomic instability (349). The mutational-instability phenotype is characterized by point mutations or small deletions, whereas the chromosomal-instability phenotype is characterized by the gross rearrangement of chromosomes, likely initiated by DSBs (632). Individuals with germ line mutations in DNA repair genes involved in base excision repair, nucleotide excision repair, mismatch repair, double-strand-break repair, and interstrand cross-links repair have a high predisposition for cancer (255, 486).
Efficient DNA repair mechanisms therefore comprise a critical component in the protection against aging and cancer. Evidence from mice and humans suggests a division of tasks among DNA repair pathways: transcription-coupled repair and interstrand cross-link repair of cytotoxic lesions are predominantly responsible for longevity, whereas excision repair of mutagenic lesions provides protection against cancer (425). Biallelic germ line mutations in the MUTYH gene, which encodes a DNA glycosylase (E. coli MutY homolog) that is involved in the repair of the mutagenic consequence of oxidative DNA damage, strongly predispose humans to a rare hereditary form of colorectal cancer called familial adenomatous polyposis (FAP) (13). Congenital defects in NER give rise to rare photosensitive, recessively inherited genetic disorders that are either cancer prone, progeroid, or both, including xeroderma pigmentosum, trichothiodystrophy, and Cockayne syndrome (425). Hereditary nonpolyposis colorectal cancer (HNPCC) is associated with germ line mutations in the mismatch repair genes MLH1 and MSH2 (482). DNA interstrand cross-link repair defects underlie a cancer-progeroid condition called Fanconi anemia (274, 605). The improper response to cytotoxic DSBs plays a role in cancer-progeroid conditions, including ataxia telangiectasia (AT), characterized by mutations in the ATM kinase, and the Nijmegen breakage syndrome (NBS), which is associated with defects in NBS1 (491, 523, 567). Defects in the family of RecQ DNA helicases, which are required for DNA repair, replication bypass of DNA damage, and recombination, underlie the progeroid-cancer conditions Werner syndrome (WS), Bloom syndrome (BLS), and Rothmund-Thomson syndrome (RTS) (507). Inherited defects in BRCA1 and BRCA2 strongly predispose individuals to breast cancer (255), while defects in BRCA1 also cause progeroid phenotypes in mice (84).
Protein damage, aging, and cancer.
Aging and the premature aging diseases Werner syndrome and Hutchinson-Gilford progeria syndrome as well as cancer and other disorders such as Alzheimer's and Parkinson's diseases, diabetes, atherosclerosis, caractogenesis, sepsis, cystic fibrosis, psoriasis, rheumatoid arthritis, chronic lung disease, and chronic renal failure are also characterized by increased carbonylation levels (117, 474, 585). Carbonylation is an irreversible oxidative process (117). Protein carbonylation can be a consequence of oxidative stress but can also be a targeted event to eliminate the protein when it is no longer needed or when it is misfolded and dysfunctional (465). Carbonylated proteins are marked for proteolysis by the eukaryotic 20S proteasome and the bacterial Lon protease but can escape degradation and form high-molecular-weight aggregates. The accumulation of carbonyls does not occur linearly with age but instead increases dramatically in the last one-third of the life span (353, 474). The carbonylation curve in the reproductive stage of the life cycle is phenomenologically equivalent to the shoulder of the survival curve in D. radiodurans in that they both reflect little oxidative damage (Fig. 15). The accumulation of oxidative damage, at high radiation doses in D. radiodurans or during the postreproductive age in humans, causes the shoulder of the survival curve to fall exponentially and carbonylation levels to rise exponentially. The shoulder of the survival curve, which distinguishes D. radiodurans from damage-sensitive species, suggests that the DNA repair systems and other proteins necessary for normal cellular function are protected from the oxidative effects of DNA-damaging agents (120, 121, 211). The high level of oxidative stress causing mortality at high doses of oxidative agents in D. radiodurans and the age-dependent dramatic increase in the carbonyl content in animals and humans can be attributed to (i) an increased production of ROS (5, 581), (ii) a decline in antioxidant cell defenses (95, 391, 581), (iii) a diminished capacity for the removal of oxidized proteins by the proteasome (86, 142, 219, 570), or (iv) an increased susceptibility of aberrant (misfolded) proteins to oxidative damage (167).
FIG. 15.
The shoulder of the survival curve (red) of D. radiodurans to various DNA-damaging agents such as gamma rays (A) is phenomenologically equivalent to the shoulder of the protein oxidation curve (red) in the reproductive stage of the life cycle in humans and animals (B), in that both reflect little oxidative damage. The accumulation of oxidative damage, at high radiation doses in D. radiodurans or during the last one-third of the life span of humans, causes the shoulder of the survival curve to fall and carbonylation levels to rise exponentially. (Modified from reference 353 with permission from Elsevier.)
Harnessing of D. radiodurans for the development of antioxidants.
Given that oxidative stress plays a significant role in the aging and cancer processes, strategies aimed at either reducing the oxidative burden or boosting defense mechanisms against oxidative damage would have significant antiaging and anticancer effects. The environmental robustness of D. radiodurans and the processes underlying the most efficient biological protection mechanisms against oxidation may be harnessed for these crucial purposes: to delay aging and prevent cancer and other age-related diseases. The challenge for future research is to employ deinococcal antioxidants to prevent or reduce age- and cancer-related protein modifications and DNA mutations.
A significant radioprotective effect of D. radiodurans protein-free extracts on E. coli cells and human Jurkat T cells was recently demonstrated (121). The radioprotective effect pertains to small molecules (<3 kDa) and is lost upon their removal (121). Daly and colleagues identified manganese, orthophosphate, peptides, and nucleosides as the radioprotective components of D. radiodurans ultrafiltrates, which are synergistically responsible for the antioxidant protection of proteins but not DNA (121). In vitro, Mn2+-based metabolite complexes protect irradiated enzymes against inactivation by radiation-induced ROS and irradiated human and E. coli cells against radiation-induced death (121). D. radiodurans Mn2+ appears to consist of inorganic and organic ligands, which scavenge a variety of ROS. Mn2+-orthophosphate complexes, which may include certain nucleosides, catalytically remove O2·− (36, 122), while Mn2+-amino acid and Mn2+-peptide complexes decompose H2O2 (51) (Fig. 14). As the organic components of the complexes scavenge OH· efficiently, the presence of Mn2+ complexes prevents the interconversion and proliferation of ROS during irradiation and recovery (120, 121). The finding that antioxidant protection is imparted by high concentrations of common cellular metabolites raises the prospect of the metabolic manipulation of oxidative-stress-sensitive organisms to render them oxidative stress resistant (121). Indeed, radiation-resistant variants evolved from naturally sensitive species after multiple cycles of high radiation doses exhibit metabolic deficiencies similar to those of D. radiodurans (132, 207, 481). Metabolic interventions at the cellular level to induce the accumulation of manganese and metabolites should show whether such an approach may enhance oxidative stress resistance in animals as well. Although technical and ethical restrictions preclude such considerations for humans, the pharmacological application of manganese complexes remains a worthy challenge in fields of cancer medicine, aging, and radiation protection. However, appropriate preparations will need to be formulated to circumvent the toxicity of manganese overexposure, which can lead to manganism, a Parkinson's disease-like condition that causes severe neurological damage (35, 47). While orthophosphate complexes of Mn2+ act as antioxidants (25, 36), purine complexes may induce neurodegeneration (190).
Among other, less efficient deinococcal antioxidants are deinoxanthin, a deinococcal carotenoid which is more effective in scavenging ROS than other known carotenoids (618), and a pyrroloquinoline-quinone (PQQ) coenzyme, which detoxifies ROS more efficiently than other natural antioxidants (417). PQQ was found to prevent cognitive deficits caused by oxidative stress in rats (470) and to prevent oxidative injury in rat cardiomyocytes (606). Deinococcal PQQ safeguards E. coli against ROS, directly or via the stimulation or induction of the antioxidant enzymes catalase and superoxide dismutase (290), and is potentially valuable in antioxidant therapy. Lipoic acid and folates may also partake in the D. radiodurans arsenal of antioxidants (678). In addition, sulfhydryl-based antioxidants were proposed to contribute to D. radiodurans radioresistance (77). Iodoacetamide, which alkylates cysteine sulfhydryl groups, sensitizes D. radiodurans to ionizing radiation (134). Sulfhydryl-containing compounds that confer protection against gamma rays to E. coli were found in D. radiodurans extracts (77). However, extracts from stationary-phase cultures, while highly radioprotective, are devoid of sulfur compounds (549).
To date, various attempts to decelerate aging, cancer, and other conditions with oxidative stress phenomenology by administering antioxidant therapy (58, 476, 638), or even genetically overexpressing antioxidant activities (337, 449, 488), have failed to show any benefit with respect to disease outcome and have occasionally shown adverse effects. As the remarkable antioxidant properties of manganese-metabolite mixtures derived from D. radiodurans have already been proven effective in protecting human cell lines against oxidative damage (121), their medical potential for fighting aging and cancer is eminent.
Do D. radiodurans and Cancer Cells Share the Same Mechanism of Radioresistance?
In addition to providing new routes for antioxidant therapy for cancer, the extreme resistance of D. radiodurans to ionizing radiation may provide insights into resistance to cytotoxic and radiotherapy treatments (517). The radiation resistance of human cancer cells evolves in the course of radiotherapy. In the case of the human osteosarcoma cell line HS-Os-1, the origin of radiation resistance has been attributed to strong ROS scavenging following irradiation (467). Based on the deinococcal model of efficient ROS scavenging, the enhancement of ROS scavenging in cancer cells may stem from a change in manganese-driven redox cycling, the accumulation of secondary metabolites, and/or an upregulation of catalase and superoxide dismutase activities. The development of radiation resistance is not likely to involve the acquisition of new genes and protein functions but may operate on a transcription level to enhance the selective expression of protective or reparatory proteins. Mitochondria share genes with manganese-accumulating bacteria and accumulate as much manganese as radiation-resistant bacteria (119). In contrast with Mn-rich radiation-resistant bacteria, Mn-driven redox cycling in mitochondria leads to the release of H2O2 within cells, which is consistent with the high radiation sensitivities of most eukaryotic cells (119). The increase in the radiation resistance of cancer cells may therefore result from the modulation of manganese redox cycling in mitochondria combined with the accumulation of metabolites in the cytoplasm to buttress ROS scavenging.
Biotechnological Application of D. radiodurans in Bioremediation
The radiation resistance of D. radiodurans also makes it an ideal candidate for the bioremediation of sites contaminated with radionuclides such as uranium 235U, toxic organic solvents such as toluene, heavy metal ions such as mercury Hg2+ and Cr6+, and endocrine-disrupting compounds such as di-n-butyl phthalate (DBP) (20, 74, 75, 118, 336, 357). D. radiodurans has already been isolated from highly radioactive sediments beneath a waste tank that had leaked high-level radioactive waste (197).
Large areas of soils, sediments, and groundwaters are contaminated with radionuclides (uranium, strontium, and cesium), heavy metals (chromium, lead, and mercury), and toxic solvents (benzene, toluene, xylenes, and chlorinated hydrocarbons). The decontamination of vast waste sites with physicochemical cleanup technologies is expensive and dangerous. An alternative technology for the treatment of these waste sites is in situ bioremediation using specialized microorganisms that can detoxify both metallic and organic contaminants and thus prevent or minimize the dissemination of contaminants before they become widely dispersed in the environment (75, 118).
Genetically engineered D. radiodurans harboring phoN, which encodes a nonspecific acid phosphatase from Salmonella enterica, efficiently precipitates uranium from dilute nuclear waste (20). PhoN hydrolyzes organic phosphates to release inorganic phosphate, which interacts with the metal and precipitates it on the cell surface as insoluble metal phosphate (378). D. radiodurans tolerates up to 800 mg/liter of the organic solvent toluene (336). A recombinant strain of D. radiodurans expressing toluene dioxygenase (tod) from Pseudomonas putida can degrade toluene, indole, chlorobenzene, and 3,4-dichloro-1-butene in a highly irradiating environment of 60 Gy/h (336). Another recombinant strain expressing a mercuric ion (Hg2+) reductase (merA) can detoxify highly toxic Hg2+ to a much less toxic and nearly inert and volatile Hg in the presence of 60 Gy/h (74). D. radiodurans also has the potential for metal reduction under anaerobic conditions (195). Generally, the solubility of metals is reduced at lower oxidation states (375). D. radiodurans can reduce iron Fe3+ coupled to the oxidation of lactate, pyruvate, or succinate and can also reduce Fe3+, uranium U6+, and technetium Tc7+ in combination with humic acid or synthetic electron shuttle agents (195). Cr6+, a known human carcinogen, can be reduced in the absence of humic acids to the less toxic Cr3+ (195). As toluene frequently occurs together with Cr6+ in radionuclide-contaminated sites, D. radiodurans engineered for complete toluene degradation by expressing the P. putida tod operon is capable of using energy derived from toluene metabolism to efficiently reduce metals such as Cr6+ (75). Finally, D. radiodurans is naturally efficient in degrading di-n-butyl phthalate (DBP), which is used in plastics, coatings, cosmetics, and other industrial activities and which consistently accumulates in wastewater (357). While bioengineered D. radiodurans is already established as an effective decontaminator of radioactive waste sites, the finding that radioprotective components of D. radiodurans extracts can impart radiation resistance to species such as E. coli gives rise to an alternative approach of shielding naturally bioremediation-competent species such as P. putida with radioprotective compounds (121).
CONCLUSIONS AND PERSPECTIVES
D. radiodurans is a robust organism that survives conditions of extreme desiccation and irradiation, which damage all cellular macromolecules by generating oxidative stress. D. radiodurans is not a conventional extremophile such as thermophiles and psychrophiles, which thrive under extreme temperature conditions. D. radiodurans thrives in moderate temperatures and organic-rich environments but is capable of withstanding and recovering from excessive damage inflicted by the desiccation that frequently occurs in its natural environment. As desiccation, radiation, and toxic chemicals have common effects on cellular macromolecules, they therefore require common defense mechanisms. While conventional genetic studies of the phenotypic limitations caused by specific gene mutations inform us which genes and proteins are required for the normal functioning of the “wild type,” D. radiodurans biology should teach us how to improve its performances. Indeed, the recent discovery that small-molecule antioxidants composed of manganese, orthophosphate, peptides, and nucleosides are primarily responsible for the resistance of D. radiodurans to oxidative stress elicits the distinct possibility that these antioxidant compounds could be applied to and adopted by other species, including humans (121).
The radiation biology of D. radiodurans has been instructive in many aspects. The observation that proteins in D. radiodurans and other radiation-resistant bacteria are shielded against oxidative damage, while DNA is as susceptible to breakage as in the radiation-sensitive species (120, 121, 206), challenged the traditional concept of DNA being the primary target of lethal radiation damage (264). Based on the lessons from D. radiodurans, a new concept of radiation toxicity dubbed “death by protein damage” has emerged, whereby protein damage and protection against protein damage determine the level of radiation resistance (120-122). The extreme repair efficiency of radiation-induced DNA breaks, which seems to rely on enzymes that are also present in radiation-sensitive species (49, 572, 676), has therefore been attributed to the antioxidation protection of DNA repair enzymes rather than to the special properties of the DNA repair mechanism (120, 122, 383). Antioxidant defense systems comprise an arsenal of ROS-scavenging enzymes, such as catalases and superoxide dismutases (14, 96, 364), and nonenzymatic scavengers, such as carotenoids (618) and manganese complexes (120, 123). Although oxidative stress resistance appears to be a collective contribution from both enzymatic and nonenzymatic antioxidants (561), mutation studies have shown that enzymatic scavengers (388) and carotenoids (396, 447, 472, 618, 679) are dispensable in D. radiodurans, whereas manganese depletion greatly diminishes postirradiation cell survival due to extensive protein damage (122, 123). By efficiently scavenging all reactive oxygen species, manganese complexes stand out as the most powerful protein-protective antioxidants that preserve protein activity under conditions of extreme oxidative stress (121). Another important aspect of the unequaled effectiveness of D. radiodurans in preventing protein damage and repairing DNA damage is the high degree of redundancy in DNA repair-related enzymes (e.g., glycosylases, UV excinucleases, SSB proteins, and Nudix hydrolases) and antioxidant enzymes (e.g., catalases and superoxide dismutases). Its genomic redundancy with 2 to 10 genome copies in each cell enables accurate DNA repair based on sequence homology. Finally, in times of stress, D. radiodurans limits its biosynthetic demands by importing amino acids derived from extracellular proteins (207, 368, 380, 653) and converting glucose into the precursors of nucleotides by the pentose phosphate pathway (683). The energy required for the recovery process can be retrieved from carbohydrate storage granules (574, 615) and polymetaphosphate granules (615).
To date, the practical application of the exceptional radiation resistance of D. radiodurans has been explored solely for the decontamination of radioactive sites from radionuclides, heavy metals, and organic solvents (118). Many other applications of the biology of its high level of resistance to radiation and desiccation are conceivable in areas of medicine, health, and biotechnology. The isolation of small manganese-based complexes from D. radiodurans extracts, which are highly protective against oxidation damage when administered ex vivo to human cell lines, opens numerous possibilities for their application (121). Deinococcal antioxidants could solve many problems in the following areas: (i) the preparation of safe but potent vaccines from radiation-inactivated bacteria or viruses with preserved antigens; (ii) transplantation and cellular therapies by improved organ, tissue, and cell conservation; (iii) food conservation; (iv) anti-inflammatory treatments, therapeutic radiation treatments, and the treatments of burn victims; and (v) the robustness of both natural and engineered strains in biotechnology and bioremediation. The demonstration that manganese complexes consist of common cellular metabolites, which accumulate in D. radiodurans as a result of its inherent metabolic deficiencies, instigates an alternative approach in protecting cells against oxidative damage by endogenous rather than exogenous manipulations. The genetic tuning of the metabolic functions toward an accumulation of manganese, phosphate, peptides, and nucleosides is expected to create cells that are continuously protected against oxidative stress. As such an approach is inapplicable to humans, the focus of pharmacological research will be to devise the appropriate combinations of antioxidants that are both bioavailable and nontoxic. Given that oxidative stress and the accumulation of reactive oxygen species lead to aging and cancer, by preventing or reducing oxidative stress deinococcal antioxidants are expected to antagonize and thus retard the aging process and carcinogenesis. The continued study of D. radiodurans as a model organism for oxidative stress resistance is therefore of considerable potential interest and sustainable global significance for the future of medicine and public health.
Supplementary Material
Acknowledgments
D.S. thanks Rupert Thorough for the editing of English. We acknowledge Michael Collins's help in generating time-lapse movies demonstrating cell division in D. radiodurans. We thank John R. Battista and Ksenija Zahradka for sharing unpublished results and Kira S. Makarova for editing the section on phylogeny. D.S. thanks Željko Glavor for help in assembling Fig. 2C, and M.R. thanks Mikula Radman for help with Fig. 12.
D.S. was a Boehringer Ingelheim Fonds Ph.D. fellow and holds an AXA Research Fund postdoctoral fellowship.
Footnotes
Supplemental material for this article may be found at http://mmbr.asm.org/.
REFERENCES
- 1.Abed, R. M., B. Zein, A. Al-Thukair, and D. de Beer. 2007. Phylogenetic diversity and activity of aerobic heterotrophic bacteria from a hypersaline oil-polluted microbial mat. Syst. Appl. Microbiol. 30:319-330. [DOI] [PubMed] [Google Scholar]
- 2.Reference deleted.
- 3.Abreu, I. A., et al. 2008. The kinetic mechanism of manganese-containing superoxide dismutase from Deinococcus radiodurans: a specialized enzyme for the elimination of high superoxide concentrations. Biochemistry 47:2350-2356. [DOI] [PubMed] [Google Scholar]
- 4.Agostini, H. J., J. D. Carroll, and K. W. Minton. 1996. Identification and characterization of uvrA, a DNA repair gene of Deinococcus radiodurans. J. Bacteriol. 178:6759-6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aguilaniu, H., L. Gustafsson, M. Rigoulet, and T. Nystrom. 2001. Protein oxidation in G0 cells of Saccharomyces cerevisiae depends on the state rather than rate of respiration and is enhanced in pos9 but not yap1 mutants. J. Biol. Chem. 276:35396-35404. [DOI] [PubMed] [Google Scholar]
- 6.Albuquerque, L., et al. 2005. Truepera radiovictrix gen. nov., sp. nov., a new radiation resistant species and the proposal of Trueperaceae fam. nov. FEMS Microbiol. Lett. 247:161-169. [DOI] [PubMed] [Google Scholar]
- 7.Alen, C., and A. L. Sonenshein. 1999. Bacillus subtilis aconitase is an RNA-binding protein. Proc. Natl. Acad. Sci. U. S. A. 96:10412-10417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allen, M. M. 1984. Cyanobacterial cell inclusions. Annu. Rev. Microbiol. 38:1-25. [DOI] [PubMed] [Google Scholar]
- 9.Alleva, J. L., S. Zuo, J. Hurwitz, and P. W. Doetsch. 2000. In vitro reconstitution of the Schizosaccharomyces pombe alternative excision repair pathway. Biochemistry 39:2659-2666. [DOI] [PubMed] [Google Scholar]
- 10.Al-Maghrebi, M., I. Fridovich, and L. Benov. 2002. Manganese supplementation relieves the phenotypic deficits seen in superoxide-dismutase-null Escherichia coli. Arch. Biochem. Biophys. 402:104-109. [DOI] [PubMed] [Google Scholar]
- 11.Almiron, M., A. J. Link, D. Furlong, and R. Kolter. 1992. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 6:2646-2654. [DOI] [PubMed] [Google Scholar]
- 12.Alsheikh, M. K., B. J. Heyen, and S. K. Randall. 2003. Ion binding properties of the dehydrin ERD14 are dependent upon phosphorylation. J. Biol. Chem. 278:40882-40889. [DOI] [PubMed] [Google Scholar]
- 13.Al-Tassan, N., et al. 2002. Inherited variants of MYH associated with somatic G:C→T:A mutations in colorectal tumors. Nat. Genet. 30:227-232. [DOI] [PubMed] [Google Scholar]
- 14.Anderson, A. W., H. C. Nordan, R. F. Cain, G. Parrish, and D. Duggan. 1956. Studies on a radio-resistant microccous. I. Isolation, morphology, cultural characteristics, and resistance to gamma radiation. Food Technol. 10:575-577. [Google Scholar]
- 15.Reference deleted.
- 16.Anderson, D. G., and S. C. Kowalczykowski. 1997. The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a chi-regulated manner. Cell 90:77-86. [DOI] [PubMed] [Google Scholar]
- 17.Anderson, R., and K. Hansen. 1985. Structure of a novel phosphoglycolipid from Deinococcus radiodurans. J. Biol. Chem. 260:12219-12223. [PubMed] [Google Scholar]
- 18.Angov, E., and R. D. Camerini-Otero. 1994. The recA gene from the thermophile Thermus aquaticus YT-1: cloning, expression, and characterization. J. Bacteriol. 176:1405-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anjem, A., S. Varghese, and J. A. Imlay. 2009. Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli. Mol. Microbiol. 72:844-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Appukuttan, D., A. S. Rao, and S. K. Apte. 2006. Engineering of Deinococcus radiodurans R1 for bioprecipitation of uranium from dilute nuclear waste. Appl. Environ. Microbiol. 72:7873-7878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aravind, L., and E. V. Koonin. 2001. Prokaryotic homologs of the eukaryotic DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system. Genome Res. 11:1365-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Archibald, F. S., and I. Fridovich. 1982. Investigations of the state of the manganese in Lactobacillus plantarum. Arch. Biochem. Biophys. 215:589-596. [DOI] [PubMed] [Google Scholar]
- 23.Archibald, F. S., and I. Fridovich. 1981. Manganese and defenses against oxygen toxicity in Lactobacillus plantarum. J. Bacteriol. 145:442-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Archibald, F. S., and I. Fridovich. 1981. Manganese, superoxide dismutase, and oxygen tolerance in some lactic acid bacteria. J. Bacteriol. 146:928-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Archibald, F. S., and I. Fridovich. 1982. The scavenging of superoxide radical by manganous complexes: in vitro. Arch. Biochem. Biophys. 214:452-463. [DOI] [PubMed] [Google Scholar]
- 26.Arn, E. A., and J. N. Abelson. 1996. The 2′-5′ RNA ligase of Escherichia coli. Purification, cloning, and genomic disruption. J. Biol. Chem. 271:31145-31153. [DOI] [PubMed] [Google Scholar]
- 27.Asker, D., T. S. Awad, T. Beppu, and K. Ueda. 2009. Deinococcus aquiradiocola sp. nov., isolated from a radioactive site in Japan. Int. J. Syst. Evol. Microbiol. 59:144-149. [DOI] [PubMed] [Google Scholar]
- 28.Asker, D., T. S. Awad, L. McLandsborough, T. Beppu, and K. Ueda. 2010. Deinococcus depolymerans sp. nov., a gamma- and UV-radiation-resistant bacterium, isolated from a radioactive site in Japan. Int. J. Syst. Evol. Microbiol., in press. [DOI] [PubMed]
- 29.Auda, H., and C. Emborg. 1973. Studies on post-irradiation DNA degradation in Micrococcus radiodurans, strain R 11 5. Radiat. Res. 53:273-280. [PubMed] [Google Scholar]
- 30.Awile, O., A. Krisko, I. F. Sbalzarini, and B. Zagrovic. 2010. Intrinsically disordered regions may lower the hydration free energy in proteins: a case study of nudix hydrolase in the bacterium Deinococcus radiodurans. PLoS Comput. Biol. 6:e1000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azeddoug, H., and G. Reysset. 1994. Cloning and sequencing of a chromosomal fragment from Clostridium acetobutylicum strain ABKn8 conferring chemical-damaging agents and UV resistance to E. coli recA strains. Curr. Microbiol. 29:229-235. [DOI] [PubMed] [Google Scholar]
- 32.Bagwell, C. E., et al. 2008. Survival in nuclear waste, extreme resistance, and potential applications gleaned from the genome sequence of Kineococcus radiotolerans SRS30216. PLoS One 3:e3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bakeeva, L. E., K. M. Chumakov, A. L. Drachev, A. L. Metlina, and V. P. Skulachev. 1986. The sodium cycle. III. Vibrio alginolyticus resembles Vibrio cholerae and some other vibriones by flagellar motor and ribosomal 5S-RNA structures. Biochim. Biophys. Acta 850:466-472. [DOI] [PubMed] [Google Scholar]
- 34.Banos, B., J. M. Lazaro, L. Villar, M. Salas, and M. de Vega. 2008. Characterization of a Bacillus subtilis 64-kDa DNA polymerase X potentially involved in DNA repair. J. Mol. Biol. 384:1019-1028. [DOI] [PubMed] [Google Scholar]
- 35.Barceloux, D. G. 1999. Manganese. J. Toxicol. Clin. Toxicol. 37:293-307. [DOI] [PubMed] [Google Scholar]
- 36.Barnese, K., E. B. Gralla, D. E. Cabelli, and J. S. Valentine. 2008. Manganous phosphate acts as a superoxide dismutase. J. Am. Chem. Soc. 130:4604-4606. [DOI] [PubMed] [Google Scholar]
- 37.Battista, J. R. 1997. Against all odds: the survival strategies of Deinococcus radiodurans. Annu. Rev. Microbiol. 51:203-224. [DOI] [PubMed] [Google Scholar]
- 38.Battista, J. R., et al. 2003. The structure of D. radiodurans. Science 302:567-568. [DOI] [PubMed] [Google Scholar]
- 39.Battista, J. R., A. M. Earl, and M. J. Park. 1999. Why is Deinococcus radiodurans so resistant to ionizing radiation? Trends Microbiol. 7:362-365. [DOI] [PubMed] [Google Scholar]
- 40.Battista, J. R., M. J. Park, and A. E. McLemore. 2001. Inactivation of two homologues of proteins presumed to be involved in the desiccation tolerance of plants sensitizes Deinococcus radiodurans R1 to desiccation. Cryptobiology 43:133-139. [DOI] [PubMed] [Google Scholar]
- 41.Battistuzzi, F. U., A. Feijao, and S. B. Hedges. 2004. A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol. Biol. 4:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bauche, C., and J. Laval. 1999. Repair of oxidized bases in the extremely radiation-resistant bacterium Deinococcus radiodurans. J. Bacteriol. 181:262-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bauermeister, A., E. Bentchikou, R. Moeller, and P. Rettberg. 2009. Roles of PprA, IrrE, and RecA in the resistance of Deinococcus radiodurans to germicidal and environmentally relevant UV radiation. Arch. Microbiol. 191:913-918. [DOI] [PubMed] [Google Scholar]
- 44.Beam, C. E., C. J. Saveson, and S. T. Lovett. 2002. Role for radA/sms in recombination intermediate processing in Escherichia coli. J. Bacteriol. 184:6836-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beckman, K. B., and B. N. Ames. 1998. The free radical theory of aging matures. Physiol. Rev. 78:547-581. [DOI] [PubMed] [Google Scholar]
- 46.Ben-Amotz, A., and M. Avron. 1990. Dunaliella bardawil can survive especially high irradiance levels by the accumulation of beta-carotene. Trends Biotechnol. 8:121-126. [Google Scholar]
- 47.Benedetto, A., C. Au, and M. Aschner. 2009. Manganese-induced dopaminergic neurodegeneration: insights into mechanisms and genetics shared with Parkinson's disease. Chem. Rev. 109:4862-4884. [DOI] [PubMed] [Google Scholar]
- 48.Bentchikou, E., P. Servant, G. Coste, and S. Sommer. 2007. Additive effects of SbcCD and PolX deficiencies in the in vivo repair of DNA double-strand breaks in Deinococcus radiodurans. J. Bacteriol. 189:4784-4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bentchikou, E., P. Servant, G. Coste, and S. Sommer. 2010. A major role of the RecFOR pathway in DNA double-strand-break repair through ESDSA in Deinococcus radiodurans. PLoS Genet. 6:e1000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berka, R. M., et al. 2002. Microarray analysis of the Bacillus subtilis K-state: genome-wide expression changes dependent on ComK. Mol. Microbiol. 43:1331-1345. [DOI] [PubMed] [Google Scholar]
- 51.Berlett, B. S., P. B. Chock, M. B. Yim, and E. R. Stadtman. 1990. Manganese(II) catalyzes the bicarbonate-dependent oxidation of amino acids by hydrogen peroxide and the amino acid-facilitated dismutation of hydrogen peroxide. Proc. Natl. Acad. Sci. U. S. A. 87:389-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bernstein, D. A., et al. 2004. Crystal structure of the Deinococcus radiodurans single-stranded DNA-binding protein suggests a mechanism for coping with DNA damage. Proc. Natl. Acad. Sci. U. S. A. 101:8575-8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bessman, M. J., D. N. Frick, and S. F. O'Handley. 1996. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J. Biol. Chem. 271:25059-25062. [DOI] [PubMed] [Google Scholar]
- 54.Bhattacharyya, G., and A. Grove. 2007. The N-terminal extensions of Deinococcus radiodurans Dps-1 mediate DNA major groove interactions as well as assembly of the dodecamer. J. Biol. Chem. 282:11921-11930. [DOI] [PubMed] [Google Scholar]
- 55.Bianchi, M. E., and C. M. Radding. 1983. Insertions, deletions and mismatches in heteroduplex DNA made by recA protein. Cell 35:511-520. [DOI] [PubMed] [Google Scholar]
- 56.Billen, D., R. R. Hewitt, T. Lapthisophon, and P. M. Achey. 1967. Deoxyribonucleic acid repair replication after ultraviolet light or X-ray exposure of bacteria. J. Bacteriol. 94:1538-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Billi, D., E. I. Friedmann, K. G. Hofer, M. G. Caiola, and R. Ocampo-Friedmann. 2000. Ionizing-radiation resistance in the desiccation-tolerant cyanobacterium Chroococcidiopsis. Appl. Environ. Microbiol. 66:1489-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bjelakovic, G., D. Nikolova, L. L. Gluud, R. G. Simonetti, and C. Gluud. 2007. Mortality in randomized trials of antioxidant supplements for primary and secondary prevention: systematic review and meta-analysis. JAMA 297:842-857. [DOI] [PubMed] [Google Scholar]
- 59.Bjelland, S., and E. Seeberg. 2003. Mutagenicity, toxicity and repair of DNA base damage induced by oxidation. Mutat. Res. 531:37-80. [DOI] [PubMed] [Google Scholar]
- 60.Blasius, M., R. Buob, I. V. Shevelev, and U. Hubscher. 2007. Enzymes involved in DNA ligation and end-healing in the radioresistant bacterium Deinococcus radiodurans. BMC Mol. Biol. 8:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blasius, M., I. Shevelev, E. Jolivet, S. Sommer, and U. Hubscher. 2006. DNA polymerase X from Deinococcus radiodurans possesses a structure-modulated 3′→5′ exonuclease activity involved in radioresistance. Mol. Microbiol. 60:165-176. [DOI] [PubMed] [Google Scholar]
- 62.Blasius, M., S. Sommer, and U. Hubscher. 2008. Deinococcus radiodurans: what belongs to the survival kit? Crit. Rev. Biochem. Mol. Biol. 43:221-238. [DOI] [PubMed] [Google Scholar]
- 63.Boling, M. E., and J. K. Setlow. 1966. The resistance of Micrococcus radiodurans to ultraviolet radiation. 3. A repair mechanism. Biochim. Biophys. Acta 123:26-33. [DOI] [PubMed] [Google Scholar]
- 64.Bonacossa de Almeida, C., G. Coste, S. Sommer, and A. Bailone. 2002. Quantification of RecA protein in Deinococcus radiodurans reveals involvement of RecA, but not LexA, in its regulation. Mol. Genet. Genomics 268:28-41. [DOI] [PubMed] [Google Scholar]
- 65.Bonura, T., and A. K. Bruce. 1974. The repair of single-strand breaks in a radiosensitive mutant of Micrococcus radiodurans. Radiat. Res. 57:260-275. [PubMed] [Google Scholar]
- 66.Bonura, T., and K. C. Smith. 1975. Enzymatic production of deoxyribonucleic acid double-strand breaks after ultraviolet irradiation of Escherichia coli K-12. J. Bacteriol. 121:511-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bonura, T., and K. C. Smith. 1976. The involvement of indirect effects in cell-killing and DNA double-strand breakage in gamma-irradiated Escherichia coli K-12. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 29:293-296. [DOI] [PubMed] [Google Scholar]
- 68.Bose, B., S. Agarwal, and S. N. Chatterjee. 1990. Membrane lipid peroxidation by UV-A: mechanism and implications. Biotechnol. Appl. Biochem. 12:557-561. [PubMed] [Google Scholar]
- 69.Bosshard, F., M. Bucheli, Y. Meur, and T. Egli. 2010. The respiratory chain is the cell's Achilles' heel during UVA inactivation in Escherichia coli. Microbiology 156:2006-2015. [DOI] [PubMed] [Google Scholar]
- 70.Bosshard, F., et al. 2010. Protein oxidation and aggregation in UVA-irradiated Escherichia coli cells as signs of accelerated cellular senescence. Environ. Microbiol. 12:2931-2945. [DOI] [PubMed] [Google Scholar]
- 71.Reference deleted.
- 72.Breen, A. P., and J. A. Murphy. 1995. Reactions of oxyl radicals with DNA. Free Radic. Biol. Med. 18:1033-1077. [DOI] [PubMed] [Google Scholar]
- 73.Bridges, B. A. 2005. Error-prone DNA repair and translesion DNA synthesis. II. The inducible SOS hypothesis. DNA Repair (Amst.) 4:725-726, 739. [DOI] [PubMed] [Google Scholar]
- 74.Brim, H., et al. 2000. Engineering Deinococcus radiodurans for metal remediation in radioactive mixed waste environments. Nat. Biotechnol. 18:85-90. [DOI] [PubMed] [Google Scholar]
- 75.Brim, H., et al. 2006. Deinococcus radiodurans engineered for complete toluene degradation facilitates Cr(VI) reduction. Microbiology 152:2469-2477. [DOI] [PubMed] [Google Scholar]
- 76.Brooks, B. W., and R. G. E. Murray. 1981. Nomenclature for “Micrococcus radiodurans” and other radiation-resistant cocci: Deinococcaceae fam. nov. and Deinococcus gen. nov., including five species. Int. J. Syst. Bacteriol. 31:353-360. [Google Scholar]
- 77.Bruce, A. K. 1964. Extraction of the radioresistant factor of Micrococcus radiodurans. Radiat. Res. 22:155-164. [PubMed] [Google Scholar]
- 78.Burgoyne, P. S., S. K. Mahadevaiah, and J. M. Turner. 2007. The management of DNA double-strand breaks in mitotic G2, and in mammalian meiosis viewed from a mitotic G2 perspective. Bioessays 29:974-986. [DOI] [PubMed] [Google Scholar]
- 79.Burrell, A. D., P. Feldschreiber, and C. J. Dean. 1971. DNA-membrane association and the repair of double breaks in X-irradiated Micrococcus radiodurans. Biochim. Biophys. Acta 247:38-53. [DOI] [PubMed] [Google Scholar]
- 80.Bzymek, M., and S. T. Lovett. 2001. Instability of repetitive DNA sequences: the role of replication in multiple mechanisms. Proc. Natl. Acad. Sci. U. S. A. 98:8319-8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Caimi, P., and A. Eisenstark. 1986. Sensitivity of Deinococcus radiodurans to near-ultraviolet radiation. Mutat. Res. 162:145-151. [DOI] [PubMed] [Google Scholar]
- 82.Callegan, R. P., et al. 2008. Description of four novel psychrophilic, ionizing radiation-sensitive Deinococcus species from alpine environments. Int. J. Syst. Evol. Microbiol. 58:1252-1258. [DOI] [PubMed] [Google Scholar]
- 83.Camps, M., and L. A. Loeb. 2004. When pol I goes into high gear: processive DNA synthesis by pol I in the cell. Cell Cycle 3:116-118. [PubMed] [Google Scholar]
- 84.Cao, L., W. Li, S. Kim, S. G. Brodie, and C. X. Deng. 2003. Senescence, aging, and malignant transformation mediated by p53 in mice lacking the Brca1 full-length isoform. Genes Dev. 17:201-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cao, Z., C. W. Mueller, and D. A. Julin. 2010. Analysis of the recJ gene and protein from Deinococcus radiodurans. DNA Repair (Amst.) 9:66-75. [DOI] [PubMed] [Google Scholar]
- 86.Carney, J. M., et al. 1991. Reversal of age-related increase in brain protein oxidation, decrease in enzyme activity, and loss in temporal and spatial memory by chronic administration of the spin-trapping compound N-tert-butyl-alpha-phenylnitrone. Proc. Natl. Acad. Sci. U. S. A. 88:3633-3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Carr, A. M. 2002. DNA structure dependent checkpoints as regulators of DNA repair. DNA Repair (Amst.) 1:983-994. [DOI] [PubMed] [Google Scholar]
- 88.Carroll, J. D., M. J. Daly, and K. W. Minton. 1996. Expression of recA in Deinococcus radiodurans. J. Bacteriol. 178:130-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chanal, A., et al. 2006. The desert of Tataouine: an extreme environment that hosts a wide diversity of microorganisms and radiotolerant bacteria. Environ. Microbiol. 8:514-525. [DOI] [PubMed] [Google Scholar]
- 90.Chang, E. C., and D. J. Kosman. 1989. Intracellular Mn(II)-associated superoxide scavenging activity protects Cu,Zn superoxide dismutase-deficient Saccharomyces cerevisiae against dioxygen stress. J. Biol. Chem. 264:12172-12178. [PubMed] [Google Scholar]
- 91.Chang, X., et al. 2010. Involvement of recF in 254 nm ultraviolet radiation resistance in Deinococcus radiodurans and Escherichia coli. Curr. Microbiol. 61:458-464. [DOI] [PubMed] [Google Scholar]
- 92.Chen, H., et al. 2009. Pleiotropic effects of RecQ in Deinococcus radiodurans. Genomics 94:333-340. [DOI] [PubMed] [Google Scholar]
- 93.Chen, H., et al. 2010. DR2539 is a novel DtxR-like regulator of Mn/Fe ion homeostasis and antioxidant enzyme in Deinococcus radiodurans. Biochem. Biophys. Res. Commun. 396:413-418. [DOI] [PubMed] [Google Scholar]
- 94.Chen, H., et al. 2008. A novel OxyR sensor and regulator of hydrogen peroxide stress with one cysteine residue in Deinococcus radiodurans. PLoS One 3:e1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen, T. S., J. P. Richie, H. T. Nagasawa, and C. A. Lang. 2000. Glutathione monoethyl ester protects against glutathione deficiencies due to aging and acetaminophen in mice. Mech. Ageing Dev. 120:127-139. [DOI] [PubMed] [Google Scholar]
- 96.Chou, F. I., and S. T. Tan. 1990. Manganese(II) induces cell division and increases in superoxide dismutase and catalase activities in an aging deinococcal culture. J. Bacteriol. 172:2029-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Christensen, E. A., and H. Kristensen. 1981. Radiation-resistance of micro-organisms from air in clean premises. Acta Pathol. Microbiol. Scand. B 89:293-301. [DOI] [PubMed] [Google Scholar]
- 98.Christman, M. F., R. W. Morgan, F. S. Jacobson, and B. N. Ames. 1985. Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell 41:753-762. [DOI] [PubMed] [Google Scholar]
- 99.Clark, A. J., and M. Chamberlin. 1966. Abnormal metabolic response to ultraviolet light of a recombination deficient mutant of Escherichia coli K12. J. Mol. Biol. 19:442-454. [DOI] [PubMed] [Google Scholar]
- 100.Cobbe, N., and M. M. Heck. 2000. Review: SMCs in the world of chromosome biology—from prokaryotes to higher eukaryotes. J. Struct. Biol. 129:123-143. [DOI] [PubMed] [Google Scholar]
- 101.Cole, L. J., and M. E. Ellis. 1957. Radiation-induced changes in tissue nucleic acids; release of soluble deoxypolynucleotides in the spleen. Radiat. Res. 7:508-517. [PubMed] [Google Scholar]
- 102.Cole, R. S. 1973. Repair of DNA containing interstrand crosslinks in Escherichia coli: sequential excision and recombination. Proc. Natl. Acad. Sci. U. S. A. 70:1064-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Collins, M. D., R. A. Hutson, I. R. Grant, and M. F. Patterson. 2000. Phylogenetic characterization of a novel radiation-resistant bacterium from irradiated pork: description of Hymenobacter actinosclerus sp. nov. Int. J. Syst. Evol. Microbiol. 50:731-734. [DOI] [PubMed] [Google Scholar]
- 104.Connelly, J. C., E. S. de Leau, and D. R. Leach. 1999. DNA cleavage and degradation by the SbcCD protein complex from Escherichia coli. Nucleic Acids Res. 27:1039-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cooke, M. S., and M. D. Evans. 2007. 8-Oxo-deoxyguanosine: reduce, reuse, recycle? Proc. Natl. Acad. Sci. U. S. A. 104:13535-13536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cooper, P. K., and P. C. Hanawalt. 1972. Role of DNA polymerase I and the rec system in excision-repair in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 69:1156-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Counsell, T. J., and R. G. Murray. 1986. Polar lipid profiles of the genus Deinococcus. Int. J. Syst. Bacteriol. 36:202-206. [Google Scholar]
- 108.Cox, M. M., and J. R. Battista. 2005. Deinococcus radiodurans—the consummate survivor. Nat. Rev. Microbiol. 3:882-892. [DOI] [PubMed] [Google Scholar]
- 109.Cox, M. M., et al. 2000. The importance of repairing stalled replication forks. Nature 404:37-41. [DOI] [PubMed] [Google Scholar]
- 110.Crowe, J. H., J. F. Carpenter, and L. M. Crowe. 1998. The role of vitrification in anhydrobiosis. Annu. Rev. Physiol. 60:73-103. [DOI] [PubMed] [Google Scholar]
- 111.Crowe, J. H., F. A. Hoekstra, and L. M. Crowe. 1992. Anhydrobiosis. Annu. Rev. Physiol. 54:579-599. [DOI] [PubMed] [Google Scholar]
- 112.Reference deleted.
- 113.Dadachova, E., et al. 2004. Susceptibility of the human pathogenic fungi Cryptococcus neoformans and Histoplasma capsulatum to gamma-radiation versus radioimmunotherapy with alpha- and beta-emitting radioisotopes. J. Nucl. Med. 45:313-320. [PubMed] [Google Scholar]
- 114.Dainton, F. S. 1948. On the existence of free atoms and radicals in water and aqueous solutions subjected to ionizing radiation. J. Phys. Colloid. Chem. 52:490-517. [DOI] [PubMed] [Google Scholar]
- 115.Dale, W. M. 1940. The effect of X-rays on enzymes. Biochem. J. 34:1367-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dalle-Donne, I., et al. 2006. Protein carbonylation, cellular dysfunction, and disease progression. J. Cell. Mol. Med. 10:389-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dalle-Donne, I., D. Giustarini, R. Colombo, R. Rossi, and A. Milzani. 2003. Protein carbonylation in human diseases. Trends Mol. Med. 9:169-176. [DOI] [PubMed] [Google Scholar]
- 118.Daly, M. J. 2000. Engineering radiation-resistant bacteria for environmental biotechnology. Curr. Opin. Biotechnol. 11:280-285. [DOI] [PubMed] [Google Scholar]
- 119.Daly, M. J. 2006. Modulating radiation resistance: insights based on defenses against reactive oxygen species in the radioresistant bacterium Deinococcus radiodurans. Clin. Lab. Med. 26:491-504. [DOI] [PubMed] [Google Scholar]
- 120.Daly, M. J. 2009. A new perspective on radiation resistance based on Deinococcus radiodurans. Nat. Rev. Microbiol. 7:237-245. [DOI] [PubMed] [Google Scholar]
- 121.Daly, M. J., et al. 2010. Small-molecule antioxidant proteome-shields in Deinococcus radiodurans. PLoS One 5:e12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Daly, M. J., et al. 2007. Protein oxidation implicated as the primary determinant of bacterial radioresistance. PLoS Biol. 5:e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Daly, M. J., et al. 2004. Accumulation of Mn(II) in Deinococcus radiodurans facilitates gamma-radiation resistance. Science 306:1025-1028. [DOI] [PubMed] [Google Scholar]
- 124.Daly, M. J., O. Ling, and K. W. Minton. 1994. Interplasmidic recombination following irradiation of the radioresistant bacterium Deinococcus radiodurans. J. Bacteriol. 176:7506-7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Daly, M. J., and K. W. Minton. 1996. An alternative pathway of recombination of chromosomal fragments precedes recA-dependent recombination in the radioresistant bacterium Deinococcus radiodurans. J. Bacteriol. 178:4461-4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Daly, M. J., and K. W. Minton. 1995. Interchromosomal recombination in the extremely radioresistant bacterium Deinococcus radiodurans. J. Bacteriol. 177:5495-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Daly, M. J., and K. W. Minton. 1997. Recombination between a resident plasmid and the chromosome following irradiation of the radioresistant bacterium Deinococcus radiodurans. Gene 187:225-229. [DOI] [PubMed] [Google Scholar]
- 128.Daly, M. J., L. Ouyang, P. Fuchs, and K. W. Minton. 1994. In vivo damage and recA-dependent repair of plasmid and chromosomal DNA in the radiation-resistant bacterium Deinococcus radiodurans. J. Bacteriol. 176:3508-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dardalhon-Samsonoff, M., and N. Rebeyrotte. 1975. Role of DNA-membrane attachment in repair of radiation damage in Micrococcus radiodurans. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 27:157-169. [DOI] [PubMed] [Google Scholar]
- 130.Dartnell, L. R., S. J. Hunter, K. V. Lovell, A. J. Coates, and J. M. Ward. 2010. Low-temperature ionizing radiation resistance of Deinococcus radiodurans and Antarctic Dry Valley bacteria. Astrobiology 10:717-732. [DOI] [PubMed] [Google Scholar]
- 131.Davern, C. I. 1966. Isolation of the DNA of the E. coli chromosome in one piece. Proc. Natl. Acad. Sci. U. S. A. 55:792-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Davies, R., and A. J. Sinskey. 1973. Radiation-resistant mutants of Salmonella typhimurium LT2: development and characterization. J. Bacteriol. 113:133-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Davis, N. S., G. J. Silverman, and E. B. Masurovsky. 1963. Radiation-resistant, pigmented coccus isolated from haddock tissue. J. Bacteriol. 86:294-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dean, C. J., and P. Alexander. 1962. Sensitization of radio-resistant bacteria to X-rays by iodoacetamide. Nature 196:1324-1326. [Google Scholar]
- 135.Dean, C. J., P. Feldschreiber, and J. T. Lett. 1966. Repair of X-ray damage to the deoxyribonucleic acid in Micrococcus radiodurans. Nature 209:49-52. [DOI] [PubMed] [Google Scholar]
- 136.Dean, C. J., J. G. Little, and R. W. Serianni. 1970. The control of post irradiation DNA breakdown in Micrococcus radiodurans. Biochem. Biophys. Res. Commun. 39:126-134. [DOI] [PubMed] [Google Scholar]
- 137.Dean, C. J., M. G. Ormerod, R. W. Serianni, and P. Alexander. 1969. DNA strand breakage in cells irradiated with X-rays. Nature 222:1042-1044. [DOI] [PubMed] [Google Scholar]
- 138.Deering, R. A. 1968. Dictyostelium discoideum: a gamma-ray resistant organism. Science 162:1289-1290. [DOI] [PubMed] [Google Scholar]
- 139.de Groot, A., et al. 2005. Deinococcus deserti sp. nov., a gamma-radiation-tolerant bacterium isolated from the Sahara Desert. Int. J. Syst. Evol. Microbiol. 55:2441-2446. [DOI] [PubMed] [Google Scholar]
- 140.de Groot, A., et al. 2009. Alliance of proteomics and genomics to unravel the specificities of Sahara bacterium Deinococcus deserti. PLoS Genet. 5:e1000434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140a.de la Tour, C. B., et al. 2009. The Deinococcus radiodurans SMC protein is dispensable for cell viability yet plays a role in DNA folding. Extremophiles 13:827-837. [DOI] [PubMed] [Google Scholar]
- 140b.D'Hondt, S., et al. 2004. Distributions of microbial activities in deep subseafloor sediments. Science 306:2216-2221. [DOI] [PubMed] [Google Scholar]
- 141.Diaz, B., and D. Schulze-Makuch. 2006. Microbial survival rates of Escherichia coli and Deinococcus radiodurans under low temperature, low pressure, and UV-irradiation conditions, and their relevance to possible Martian life. Astrobiology 6:332-347. [DOI] [PubMed] [Google Scholar]
- 142.Dice, J. F. 1982. Altered degradation of proteins microinjected into senescent human fibroblasts. J. Biol. Chem. 257:14624-14627. [PubMed] [Google Scholar]
- 143.Dillingham, M. S., and S. C. Kowalczykowski. 2008. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 72:642-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dillingham, M. S., M. Spies, and S. C. Kowalczykowski. 2003. RecBCD enzyme is a bipolar DNA helicase. Nature 423:893-897. [DOI] [PubMed] [Google Scholar]
- 145.DiRuggiero, J., N. Santangelo, Z. Nackerdien, J. Ravel, and F. T. Robb. 1997. Repair of extensive ionizing-radiation DNA damage at 95°C in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 179:4643-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Diver, W. P., N. J. Sargentini, and K. C. Smith. 1982. A mutation (radA100) in Escherichia coli that selectively sensitizes cells grown in rich medium to X- or u.v.-radiation, or methyl methanesulphonate. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 42:339-346. [DOI] [PubMed] [Google Scholar]
- 147.Djordjevic, B., and W. Szybalski. 1960. Genetics of human cell lines. III. Incorporation of 5-bromo- and 5-iododeoxyuridine into the deoxyribonucleic acid of human cells and its effect on radiation sensitivity. J. Exp. Med. 112:509-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Donado, M. M., E. A. Seleiro, and M. S. Costa. 1990. Polar lipid and fatty acid composition of strains of the genus Thermus. Syst. Appl. Microbiol. 13:234-239. [Google Scholar]
- 149.Doolittle, R. F., et al. 1986. Domainal evolution of a prokaryotic DNA repair protein and its relationship to active-transport proteins. Nature 323:451-453. [DOI] [PubMed] [Google Scholar]
- 150.Dose, K., et al. 1995. ERA—experiment “Space Biochemistry.” Adv. Space Res. 16:119-129. [PubMed] [Google Scholar]
- 151.Dose, K., et al. 2001. Survival of microorganisms under the extreme conditions of the Atacama Desert. Orig. Life Evol. Biosph. 31:287-303. [DOI] [PubMed] [Google Scholar]
- 152.Dose, K., A. Bieger-Dose, M. Labusch, and M. Gill. 1992. Survival in extreme dryness and DNA-single-strand breaks. Adv. Space Res. 12:221-229. [DOI] [PubMed] [Google Scholar]
- 153.Dresser, M. E., et al. 1997. DMC1 functions in a Saccharomyces cerevisiae meiotic pathway that is largely independent of the RAD51 pathway. Genetics 147:533-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Driedger, A. A. 1970. Are there multiple attachments between bacterial DNA and the cell membrane? Can. J. Microbiol. 16:881-882. [DOI] [PubMed] [Google Scholar]
- 155.Driedger, A. A. 1970. The DNA content of single cells of Micrococcus radiodurans. Can. J. Microbiol. 16:1136-1137. [DOI] [PubMed] [Google Scholar]
- 156.Driedger, A. A. 1970. The ordered growth pattern of microcolonies of Micrococcus radiodurans: first generation sectoring of induced lethal mutations. Can. J. Microbiol. 16:1133-1135. [DOI] [PubMed] [Google Scholar]
- 157.Driedger, A. A., and M. J. Grayston. 1971. Demonstration of two types of DNA repair in X-irradiated Micrococcus radiodurans. Can. J. Microbiol. 17:495-499. [DOI] [PubMed] [Google Scholar]
- 158.Driedger, A. A., and M. J. Grayston. 1971. The effects of nalidixic acid on X-ray-induced DNA degradation and repair in Micrococcus radiodurans. Can. J. Microbiol. 17:501-505. [DOI] [PubMed] [Google Scholar]
- 159.Driedger, A. A., and M. J. Grayston. 1971. The enhancement of X-ray-induced DNA degradation in Micrococcus radiodurans by phenethyl alcohol. Can. J. Microbiol. 17:487-493. [DOI] [PubMed] [Google Scholar]
- 160.Driedger, A. A., and M. J. Grayston. 1970. Rapid lysis of cell walls of Micrococcus radiodurans with lysozyme: effects of butanol pretreatment on DNA. Can. J. Microbiol. 16:889-893. [DOI] [PubMed] [Google Scholar]
- 161.Driedger, A. A., A. P. James, and M. J. Grayston. 1970. Cell survival and X-ray-induced DNA degradation in Micrococcus radiodurans. Radiat. Res. 44:835-845. [PubMed] [Google Scholar]
- 162.Drlica, K., and J. Rouviere-Yaniv. 1987. Histonelike proteins of bacteria. Microbiol. Rev. 51:301-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Dronkert, M. L., and R. Kanaar. 2001. Repair of DNA interstrand cross-links. Mutat. Res. 486:217-247. [DOI] [PubMed] [Google Scholar]
- 164.Du, J., and J. M. Gebicki. 2004. Proteins are major initial cell targets of hydroxyl free radicals. Int. J. Biochem. Cell Biol. 36:2334-2343. [DOI] [PubMed] [Google Scholar]
- 165.Duggan, D. E., A. W. Anderson, and P. R. Elliker. 1963. Inactivation of the radiation-resistant spoilage bacterium Micrococcus radiodurans. I. Radiation inactivation rates in three meat substrates and in buffer. Appl. Microbiol. 11:398-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Duggan, D. E., A. W. Anderson, and P. R. Elliker. 1963. Inactivation of the radiation-resistant spoilage bacterium Micrococcus radiodurans. II. Radiation inactivation rates as influenced by menstruum temperature, preirradiation heat treatment, and certain reducing agents. Appl. Microbiol. 11:413-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Dukan, S., et al. 2000. Protein oxidation in response to increased transcriptional or translational errors. Proc. Natl. Acad. Sci. U. S. A. 97:5746-5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Dulermo, R., S. Fochesato, L. Blanchard, and A. de Groot. 2009. Mutagenic lesion bypass and two functionally different RecA proteins in Deinococcus deserti. Mol. Microbiol. 74:194-208. [DOI] [PubMed] [Google Scholar]
- 169.Dunford, H. B. 1987. Free radicals in iron-containing systems. Free Radic. Biol. Med. 3:405-421. [DOI] [PubMed] [Google Scholar]
- 170.Earl, A. M., M. M. Mohundro, I. S. Mian, and J. R. Battista. 2002. The IrrE protein of Deinococcus radiodurans R1 is a novel regulator of recA expression. J. Bacteriol. 184:6216-6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Earl, A. M., S. K. Rankin, K. P. Kim, O. N. Lamendola, and J. R. Battista. 2002. Genetic evidence that the uvsE gene product of Deinococcus radiodurans R1 is a UV damage endonuclease. J. Bacteriol. 184:1003-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Eckstein, H., S. Ahnefeld, and K. Albietz-Loges. 1974. Acid-soluble deoxynucleotides and DNA synthesis in growing yeast after X-irradiation. II. Synthesis of deoxynucleoside tri- and monophosphates in synchronized and asynchronously growing cells. Z. Naturforsch. C 29:272-282. [DOI] [PubMed] [Google Scholar]
- 173.Edman, I. E., F. S. Thatcher, and K. F. McQueen. 1961. Studies on the irradiation of microorganisms in relation to food preservation. II. Irradiation resistant mutants. Can. J. Microbiol. 7:207-215. [DOI] [PubMed] [Google Scholar]
- 174.Eggington, J. M., N. Haruta, E. A. Wood, and M. M. Cox. 2004. The single-stranded DNA-binding protein of Deinococcus radiodurans. BMC Microbiol. 4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Eggington, J. M., A. G. Kozlov, M. M. Cox, and T. M. Lohman. 2006. Polar destabilization of DNA duplexes with single-stranded overhangs by the Deinococcus radiodurans SSB protein. Biochemistry 45:14490-14502. [DOI] [PubMed] [Google Scholar]
- 176.Eisenstadt, E., S. Fisher, C. L. Der, and S. Silver. 1973. Manganese transport in Bacillus subtilis W23 during growth and sporulation. J. Bacteriol. 113:1363-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Eltsov, M., and J. Dubochet. 2005. Fine structure of the Deinococcus radiodurans nucleoid revealed by cryoelectron microscopy of vitreous sections. J. Bacteriol. 187:8047-8054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Emmerson, P. T., and P. Howard-Flanders. 1965. Post-irradiation degradation of DNA following exposure of UV-sensitive and resistant bacteria to X-rays. Biochem. Biophys. Res. Commun. 18:24-29. [DOI] [PubMed] [Google Scholar]
- 179.Englander, J., et al. 2004. DNA toroids: framework for DNA repair in Deinococcus radiodurans and in germinating bacterial spores. J. Bacteriol. 186:5973-5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Epps, N. A., and E. S. Idziak. 1970. Radiation treatment of foods. II. Public health significance of irradiation-recycled Salmonella. Appl. Microbiol. 19:338-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Espeli, O., and F. Boccard. 1997. In vivo cleavage of Escherichia coli BIME-2 repeats by DNA gyrase: genetic characterization of the target and identification of the cut site. Mol. Microbiol. 26:767-777. [DOI] [PubMed] [Google Scholar]
- 182.Evans, D. M., and B. E. Moseley. 1988. Deinococcus radiodurans UV endonuclease beta DNA incisions do not generate photoreversible thymine residues. Mutat. Res. 207:117-119. [DOI] [PubMed] [Google Scholar]
- 183.Evans, D. M., and B. E. Moseley. 1985. Identification and initial characterisation of a pyrimidine dimer UV endonuclease (UV endonuclease beta) from Deinococcus radiodurans; a DNA-repair enzyme that requires manganese ions. Mutat. Res. 145:119-128. [DOI] [PubMed] [Google Scholar]
- 184.Evans, D. M., and B. E. Moseley. 1983. Roles of the uvsC, uvsD, uvsE, and mtcA genes in the two pyrimidine dimer excision repair pathways of Deinococcus radiodurans. J. Bacteriol. 156:576-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Fernandez Zenoff, V., F. Sineriz, and M. E. Farias. 2006. Diverse responses to UV-B radiation and repair mechanisms of bacteria isolated from high-altitude aquatic environments. Appl. Environ. Microbiol. 72:7857-7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ferreira, A. C., et al. 1999. Characterization and radiation resistance of new isolates of Rubrobacter radiotolerans and Rubrobacter xylanophilus. Extremophiles 3:235-238. [DOI] [PubMed] [Google Scholar]
- 187.Ferreira, A. C., et al. 1997. Deinococcus geothermalis sp. nov. and Deinococcus murrayi sp. nov., two extremely radiation-resistant and slightly thermophilic species from hot springs. Int. J. Syst. Bacteriol. 47:939-947. [DOI] [PubMed] [Google Scholar]
- 188.Filipkowski, P., A. Duraj-Thatte, and J. Kur. 2007. Identification, cloning, expression, and characterization of a highly thermostable single-stranded-DNA-binding protein (SSB) from Deinococcus murrayi. Protein Expr. Purif. 53:201-208. [DOI] [PubMed] [Google Scholar]
- 189.Flint, D. H., J. F. Tuminello, and M. H. Emptage. 1993. The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J. Biol. Chem. 268:22369-22376. [PubMed] [Google Scholar]
- 190.Florence, T. M., and J. L. Stauber. 1989. Manganese catalysis of dopamine oxidation. Sci. Total Environ. 78:233-240. [DOI] [PubMed] [Google Scholar]
- 191.Flores, M. J., N. Sanchez, and B. Michel. 2005. A fork-clearing role for UvrD. Mol. Microbiol. 57:1664-1675. [DOI] [PubMed] [Google Scholar]
- 192.Foote, C. S. 1968. Mechanisms of photosensitized oxidation. There are several different types of photosensitized oxidation which may be important in biological systems. Science 162:963-970. [DOI] [PubMed] [Google Scholar]
- 193.Franca, M. B., A. D. Panek, and E. C. Eleutherio. 2007. Oxidative stress and its effects during dehydration. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 146:621-631. [DOI] [PubMed] [Google Scholar]
- 194.Frankenberg-Schwager, M., and D. Frankenberg. 1990. DNA double-strand breaks: their repair and relationship to cell killing in yeast. Int. J. Radiat. Biol. 58:569-575. [DOI] [PubMed] [Google Scholar]
- 195.Fredrickson, J. K., H. M. Kostandarithes, S. W. Li, A. E. Plymale, and M. J. Daly. 2000. Reduction of Fe(III), Cr(VI), U(VI), and Tc(VII) by Deinococcus radiodurans R1. Appl. Environ. Microbiol. 66:2006-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Fredrickson, J. K., et al. 2008. Protein oxidation: key to bacterial desiccation resistance? ISME J. 2:393-403. [DOI] [PubMed] [Google Scholar]
- 197.Fredrickson, J. K., et al. 2004. Geomicrobiology of high-level nuclear waste-contaminated vadose sediments at the Hanford site, Washington State. Appl. Environ. Microbiol. 70:4230-4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Freedman, M. L., and A. K. Bruce. 1972. Deoxyribonucleic acid-protein ratio and radioresistance in Micrococcus radiodurans. J. Bacteriol. 109:1310-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Freedman, M. L., P. L. Freedman, and A. K. Bruce. 1969. Distinctive sulphydryl responses in X-irradiated and in heat-treated Micrococcus radiodurans. Nature 224:183-184. [DOI] [PubMed] [Google Scholar]
- 200.Frenkiel-Krispin, D., et al. 2004. Nucleoid restructuring in stationary-state bacteria. Mol. Microbiol. 51:395-405. [DOI] [PubMed] [Google Scholar]
- 201.Frenkiel-Krispin, D., and A. Minsky. 2006. Nucleoid organization and the maintenance of DNA integrity in E. coli, B. subtilis and D. radiodurans. J. Struct. Biol. 156:311-319. [DOI] [PubMed] [Google Scholar]
- 202.Funayama, T., et al. 1999. Identification and disruption analysis of the recN gene in the extremely radioresistant bacterium Deinococcus radiodurans. Mutat. Res. 435:151-161. [DOI] [PubMed] [Google Scholar]
- 203.Gao, G., et al. 2003. Expression of Deinococcus radiodurans PprI enhances the radioresistance of Escherichia coli. DNA Repair (Amst.) 2:1419-1427. [DOI] [PubMed] [Google Scholar]
- 204.Garay-Arroyo, A., J. M. Colmenero-Flores, A. Garciarrubio, and A. A. Covarrubias. 2000. Highly hydrophilic proteins in prokaryotes and eukaryotes are common during conditions of water deficit. J. Biol. Chem. 275:5668-5674. [DOI] [PubMed] [Google Scholar]
- 205.Gentner, N. E., and R. E. Mitchel. 1975. Ionizing radiation-induced release of a cell surface nuclease from Micrococcus radiodurans. Radiat. Res. 61:204-215. [PubMed] [Google Scholar]
- 206.Gerard, E., E. Jolivet, D. Prieur, and P. Forterre. 2001. DNA protection mechanisms are not involved in the radioresistance of the hyperthermophilic archaea Pyrococcus abyssi and P. furiosus. Mol. Genet. Genomics 266:72-78. [DOI] [PubMed] [Google Scholar]
- 207.Ghosal, D., et al. 2005. How radiation kills cells: survival of Deinococcus radiodurans and Shewanella oneidensis under oxidative stress. FEMS Microbiol. Rev. 29:361-375. [DOI] [PubMed] [Google Scholar]
- 208.Ghosh, S., and A. Grove. 2006. The Deinococcus radiodurans-encoded HU protein has two DNA-binding domains. Biochemistry 45:1723-1733. [DOI] [PubMed] [Google Scholar]
- 209.Ghosh, S., and A. Grove. 2004. Histone-like protein HU from Deinococcus radiodurans binds preferentially to four-way DNA junctions. J. Mol. Biol. 337:561-571. [DOI] [PubMed] [Google Scholar]
- 210.Girard, A. E. 1971. A comparative study of the fatty acids of some micrococci. Can. J. Microbiol. 17:1503-1508. [DOI] [PubMed] [Google Scholar]
- 211.Gladyshev, E., and M. Meselson. 2008. Extreme resistance of bdelloid rotifers to ionizing radiation. Proc. Natl. Acad. Sci. U. S. A. 105:5139-5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Glickman, B. W., and M. Radman. 1980. Escherichia coli mutator mutants deficient in methylation-instructed DNA mismatch correction. Proc. Natl. Acad. Sci. U. S. A. 77:1063-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Grady, L. J., and E. C. Pollard. 1968. Ionizing radiation-initiated degradation of deoxyribonucleic acid in bacteria. A possible role for defective prophage. Radiat. Res. 36:68-86. [PubMed] [Google Scholar]
- 214.Green, P. N., and I. J. Bousfield. 1983. Emendation of Methylobacterium Patt, Cole, and Hanson 1976; Methylobacterium rhodinum (Heumann 1962) comb. nov. corrig.; Methylobacterium radiotolerans (Ito and Iizuka 1971) comb. nov. corrig.; and Methylobacterium mesophilicum (Austin and Goodfellow 1979) comb. nov. Int. J. Syst. Bacteriol. 33:875-877. [Google Scholar]
- 215.Griffith, J., A. Makhov, L. Santiago-Lara, and P. Setlow. 1994. Electron microscopic studies of the interaction between a Bacillus subtilis alpha/beta-type small, acid-soluble spore protein with DNA: protein binding is cooperative, stiffens the DNA, and induces negative supercoiling. Proc. Natl. Acad. Sci. U. S. A. 91:8224-8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Griffiths, E., and R. S. Gupta. 2007. Identification of signature proteins that are distinctive of the Deinococcus-Thermus phylum. Int. Microbiol. 10:201-208. [PubMed] [Google Scholar]
- 217.Grimsley, J. K., C. I. Masters, E. P. Clark, and K. W. Minton. 1991. Analysis by pulsed-field gel electrophoresis of DNA double-strand breakage and repair in Deinococcus radiodurans and a radiosensitive mutant. Int. J. Radiat. Biol. 60:613-626. [DOI] [PubMed] [Google Scholar]
- 218.Grove, A., and S. P. Wilkinson. 2005. Differential DNA binding and protection by dimeric and dodecameric forms of the ferritin homolog Dps from Deinococcus radiodurans. J. Mol. Biol. 347:495-508. [DOI] [PubMed] [Google Scholar]
- 219.Grune, T., T. Jung, K. Merker, and K. J. Davies. 2004. Decreased proteolysis caused by protein aggregates, inclusion bodies, plaques, lipofuscin, ceroid, and ‘aggresomes’ during oxidative stress, aging, and disease. Int. J. Biochem. Cell Biol. 36:2519-2530. [DOI] [PubMed] [Google Scholar]
- 220.Gunter, S. E., and H. I. Kohn. 1956. The effect of X-rays on the survival of bacteria and yeast. I. A comparative study of the dose-survival curves of Azotobacter agile, Escherichia coli, Pseudomonas fluorescens, Rhodopseudomonas spheroides, and Saccharomyces cerevisiae irradiated in the resting state. J. Bacteriol. 71:571-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Gunter, T. E., C. E. Gavin, and K. K. Gunter. 2009. The case for manganese interaction with mitochondria. Neurotoxicology 30:727-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Gupta, R. S. 1998. Protein phylogenies and signature sequences: a reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol. Mol. Biol. Rev. 62:1435-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Gutman, P. D., J. D. Carroll, C. I. Masters, and K. W. Minton. 1994. Sequencing, targeted mutagenesis and expression of a recA gene required for the extreme radioresistance of Deinococcus radiodurans. Gene 141:31-37. [DOI] [PubMed] [Google Scholar]
- 224.Gutman, P. D., P. Fuchs, and K. W. Minton. 1994. Restoration of the DNA damage resistance of Deinococcus radiodurans DNA polymerase mutants by Escherichia coli DNA polymerase I and Klenow fragment. Mutat. Res. 314:87-97. [DOI] [PubMed] [Google Scholar]
- 225.Gutman, P. D., P. Fuchs, L. Ouyang, and K. W. Minton. 1993. Identification, sequencing, and targeted mutagenesis of a DNA polymerase gene required for the extreme radioresistance of Deinococcus radiodurans. J. Bacteriol. 175:3581-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Gutman, P. D., H. L. Yao, and K. W. Minton. 1991. Partial complementation of the UV sensitivity of Deinococcus radiodurans excision repair mutants by the cloned denV gene of bacteriophage T4. Mutat. Res. 254:207-215. [DOI] [PubMed] [Google Scholar]
- 227.Gutsche, I., et al. 2008. Complex oligomeric structure of a truncated form of DdrA: a protein required for the extreme radiotolerance of Deinococcus. Biochim. Biophys. Acta 1784:1050-1058. [DOI] [PubMed] [Google Scholar]
- 228.Gutteridge, J. M., and D. J. Shute. 1981. Iron-dioxygen-dependent changes to the biological activities of bleomycin. J. Inorg. Biochem. 15:349-357. [DOI] [PubMed] [Google Scholar]
- 229.Hall, R. D., G. J. Rouwendal, and F. A. Krens. 1992. Asymmetric somatic cell hybridization in plants. II. Electrophoretic analysis of radiation-induced DNA damage and repair following the exposure of sugarbeet (Beta vulgaris L.) protoplasts to UV and gamma rays. Mol. Gen. Genet. 234:315-324. [PubMed] [Google Scholar]
- 230.Halliwell, B., and J. M. Gutteridge. 1999. Free radicals in biology and medicine, 3rd ed. Oxford University Press, Oxford, United Kingdom.
- 231.Hamilton, M. L., et al. 2001. Does oxidative damage to DNA increase with age? Proc. Natl. Acad. Sci. U. S. A. 98:10469-10474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Hamoen, L. W., W. K. Smits, A. de Jong, S. Holsappel, and O. P. Kuipers. 2002. Improving the predictive value of the competence transcription factor (ComK) binding site in Bacillus subtilis using a genomic approach. Nucleic Acids Res. 30:5517-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Hansen, J. M., Y. M. Go, and D. P. Jones. 2006. Nuclear and mitochondrial compartmentation of oxidative stress and redox signaling. Annu. Rev. Pharmacol. Toxicol. 46:215-234. [DOI] [PubMed] [Google Scholar]
- 234.Hansen, M. T. 1978. Multiplicity of genome equivalents in the radiation-resistant bacterium Micrococcus radiodurans. J. Bacteriol. 134:71-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Hariharan, P. P., and P. A. Cerutti. 1971. Repair of gamma-ray-induced thymine damage in Micrococcus radiodurans. Nat. New Biol. 229:247-249. [DOI] [PubMed] [Google Scholar]
- 236.Hariharan, P. V., and P. A. Cerutti. 1972. Formation and repair of gamma-ray induced thymine damage in Micrococcus radiodurans. J. Mol. Biol. 66:65-81. [DOI] [PubMed] [Google Scholar]
- 237.Harman, D. 1956. Aging: a theory based on free radical and radiation chemistry. J. Gerontol. 2:298-300. [DOI] [PubMed] [Google Scholar]
- 238.Harris, D. R., et al. 2009. Directed evolution of ionizing radiation resistance in Escherichia coli. J. Bacteriol. 191:5240-5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Harris, D. R., et al. 2004. Preserving genome integrity: the DdrA protein of Deinococcus radiodurans R1. PLoS Biol. 2:e304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Harrison, J. C., and J. E. Haber. 2006. Surviving the breakup: the DNA damage checkpoint. Annu. Rev. Genet. 40:209-235. [DOI] [PubMed] [Google Scholar]
- 241.Harsojo, S. Kitayama, and A. Matsuyama. 1981. Genome multiplicity and radiation resistance in Micrococcus radiodurans. J. Biochem. 90:877-880. [DOI] [PubMed] [Google Scholar]
- 242.Harteveld, K. L., M. Losekoot, R. Fodde, P. C. Giordano, and L. F. Bernini. 1997. The involvement of Alu repeats in recombination events at the alpha-globin gene cluster: characterization of two alphazero-thalassaemia deletion breakpoints. Hum. Genet. 99:528-534. [DOI] [PubMed] [Google Scholar]
- 243.Hassan, H. M., and I. Fridovich. 1978. Regulation of the synthesis of catalase and peroxidase in Escherichia coli. J. Biol. Chem. 253:6445-6450. [PubMed] [Google Scholar]
- 244.Hastings, J. W., W. H. Holzapfel, and J. G. Niemand. 1986. Radiation resistance of lactobacilli isolated from radurized meat relative to growth and environment. Appl. Environ. Microbiol. 52:898-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Hawiger, J., and J. Jeljaszewicz. 1967. Antibiotic sensitivity of Micrococcus radiodurans. Appl. Microbiol. 15:304-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.He, Y. 2009. High cell density production of Deinococcus radiodurans under optimized conditions. J. Ind. Microbiol. Biotechnol. 36:539-546. [DOI] [PubMed] [Google Scholar]
- 247.Heinz, K., and A. Marx. 2007. Lesion bypass activity of DNA polymerase A from the extremely radioresistant organism Deinococcus radiodurans. J. Biol. Chem. 282:10908-10914. [DOI] [PubMed] [Google Scholar]
- 248.Heitman, J., N. D. Zinder, and P. Model. 1989. Repair of the Escherichia coli chromosome after in vivo scission by the EcoRI endonuclease. Proc. Natl. Acad. Sci. U. S. A. 86:2281-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Henne, A., et al. 2004. The genome sequence of the extreme thermophile Thermus thermophilus. Nat. Biotechnol. 22:547-553. [DOI] [PubMed] [Google Scholar]
- 250.Hensel, R., W. Demharter, O. Kandler, R. M. Kroppendstedt, and E. Stackebrandt. 1986. Chemotaxonomic and molecular-genetic studies of the genus Thermus: evidence for a phylogenetic relationship of Thermus aquaticus and Thermus ruber to the genus Deinococcus. Int. J. Syst. Bacteriol. 36:444-453. [Google Scholar]
- 251.Hensley, K., et al. 1995. Brain regional correspondence between Alzheimer's disease histopathology and biomarkers of protein oxidation. J. Neurochem. 65:2146-2156. [DOI] [PubMed] [Google Scholar]
- 252.Hipkiss, A. R. 2006. Accumulation of altered proteins and ageing: causes and effects. Exp. Gerontol. 41:464-473. [DOI] [PubMed] [Google Scholar]
- 253.Hirose, S., R. Okazaki, and F. Tamanoi. 1973. Mechanism of DNA chain growth. XI. Structure of RNA-linked DNA fragments of Escherichia coli. J. Mol. Biol. 77:501-517. [DOI] [PubMed] [Google Scholar]
- 254.Hirsch, P., et al. 2004. Deinococcus frigens sp. nov., Deinococcus saxicola sp. nov., and Deinococcus marmoris sp. nov., low temperature and draught-tolerating, UV-resistant bacteria from continental Antarctica. Syst. Appl. Microbiol. 27:636-645. [DOI] [PubMed] [Google Scholar]
- 255.Hoeijmakers, J. H. 2001. Genome maintenance mechanisms for preventing cancer. Nature 411:366-374. [DOI] [PubMed] [Google Scholar]
- 256.Holland, A. D., H. M. Rothfuss, and M. E. Lidstrom. 2006. Development of a defined medium supporting rapid growth for Deinococcus radiodurans and analysis of metabolic capacities. Appl. Microbiol. Biotechnol. 72:1074-1082. [DOI] [PubMed] [Google Scholar]
- 257.Holmes, A. M., and J. E. Haber. 1999. Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell 96:415-424. [DOI] [PubMed] [Google Scholar]
- 258.Horikawa, D. D., et al. 2006. Radiation tolerance in the tardigrade Milnesium tardigradum. Int. J. Radiat. Biol. 82:843-848. [DOI] [PubMed] [Google Scholar]
- 259.Hu, Y., et al. 2009. Characteristics of nuclease activity of the SbcCD complex from Deinococcus radiodurans. J. Biochem. 6:6. [DOI] [PubMed] [Google Scholar]
- 260.Hua, Y., et al. 2003. PprI: a general switch responsible for extreme radioresistance of Deinococcus radiodurans. Biochem. Biophys. Res. Commun. 306:354-360. [DOI] [PubMed] [Google Scholar]
- 261.Huang, L., et al. 2007. Three tandem HRDC domains have synergistic effect on the RecQ functions in Deinococcus radiodurans. DNA Repair (Amst.) 6:167-176. [DOI] [PubMed] [Google Scholar]
- 262.Huisman, O., and R. D'Ari. 1981. An inducible DNA replication-cell division coupling mechanism in E. coli. Nature 290:797-799. [DOI] [PubMed] [Google Scholar]
- 263.Hurstel, S., M. Granger-Schnarr, M. Daune, and M. Schnarr. 1986. In vitro binding of LexA repressor to DNA: evidence for the involvement of the amino-terminal domain. EMBO J. 5:793-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Hutchinson, F. 1966. The molecular basis for radiation effects on cells. Cancer Res. 26:2045-2052. [PubMed] [Google Scholar]
- 265.Im, W. T., et al. 2008. Deinococcus aquaticus sp. nov., isolated from fresh water, and Deinococcus caeni sp. nov., isolated from activated sludge. Int. J. Syst. Evol. Microbiol. 58:2348-2353. [DOI] [PubMed] [Google Scholar]
- 266.Imlay, J. A. 2008. Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 77:755-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Imlay, J. A. 2006. Iron-sulphur clusters and the problem with oxygen. Mol. Microbiol. 59:1073-1082. [DOI] [PubMed] [Google Scholar]
- 268.Imlay, J. A. 2003. Pathways of oxidative damage. Annu. Rev. Microbiol. 57:395-418. [DOI] [PubMed] [Google Scholar]
- 269.Ira, G., D. Satory, and J. E. Haber. 2006. Conservative inheritance of newly synthesized DNA in double-strand break-induced gene conversion. Mol. Cell. Biol. 26:9424-9429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Islam, S. M., et al. 2003. Characterization and distribution of IS8301 in the radioresistant bacterium Deinococcus radiodurans. Genes Genet. Syst. 78:319-327. [DOI] [PubMed] [Google Scholar]
- 271.Ito, H. 1977. Isolation of Micrococcus radiodurans occurring in radurized sawdust culture media of mushroom. Agric. Biol. Chem. 41:35-41. [Google Scholar]
- 272.Ivancic-Bace, I., et al. 2003. RecFOR function is required for DNA repair and recombination in a RecA loading-deficient recB mutant of Escherichia coli. Genetics 163:485-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Iwasaki, H., M. Takahagi, A. Nakata, and H. Shinagawa. 1992. Escherichia coli RuvA and RuvB proteins specifically interact with Holliday junctions and promote branch migration. Genes Dev. 6:2214-2220. [DOI] [PubMed] [Google Scholar]
- 274.Joenje, H., and K. J. Patel. 2001. The emerging genetic and molecular basis of Fanconi anaemia. Nat. Rev. Genet. 2:446-457. [DOI] [PubMed] [Google Scholar]
- 275.Johnson, A., and M. O'Donnell. 2005. Cellular DNA replicases: components and dynamics at the replication fork. Annu. Rev. Biochem. 74:283-315. [DOI] [PubMed] [Google Scholar]
- 276.Jolivet, E., S. L'Haridon, E. Corre, P. Forterre, and D. Prieur. 2003. Thermococcus gammatolerans sp. nov., a hyperthermophilic archaeon from a deep-sea hydrothermal vent that resists ionizing radiation. Int. J. Syst. Evol. Microbiol. 53:847-851. [DOI] [PubMed] [Google Scholar]
- 277.Jolivet, E., et al. 2006. Limited concentration of RecA delays DNA double-strand break repair in Deinococcus radiodurans R1. Mol. Microbiol. 59:338-349. [DOI] [PubMed] [Google Scholar]
- 278.Joshi, B., R. Schmid, K. Altendorf, and S. K. Apte. 2004. Protein recycling is a major component of post-irradiation recovery in Deinococcus radiodurans strain R1. Biochem. Biophys. Res. Commun. 320:1112-1117. [DOI] [PubMed] [Google Scholar]
- 279.Juan, J. Y., S. N. Keeney, and E. M. Gregory. 1991. Reconstitution of the Deinococcus radiodurans aposuperoxide dismutase. Arch. Biochem. Biophys. 286:257-263. [DOI] [PubMed] [Google Scholar]
- 280.Jurkiewicz, B. A., and G. R. Buettner. 1994. Ultraviolet light-induced free radical formation in skin: an electron paramagnetic resonance study. Photochem. Photobiol. 59:1-4. [DOI] [PubMed] [Google Scholar]
- 281.Kalyanaraman, B., E. Perez-Reyes, and R. P. Mason. 1980. Spin-trapping and direct electron spin resonance investigations of the redox metabolism of quinone anticancer drugs. Biochim. Biophys. Acta 630:119-130. [DOI] [PubMed] [Google Scholar]
- 282.Kamble, V. A., Y. S. Rajpurohit, A. K. Srivastava, and H. S. Misra. 2009. Increased synthesis of signaling molecules coincides with reversible inhibition of nucleolytic activity during postirradiation recovery of Deinococcus radiodurans. FEMS Microbiol. Lett. 303:18-25. [DOI] [PubMed] [Google Scholar]
- 283.Reference deleted.
- 284.Kampfer, P., N. Lodders, B. Huber, E. Falsen, and H. J. Busse. 2008. Deinococcus aquatilis sp. nov., isolated from water. Int. J. Syst. Evol. Microbiol. 58:2803-2806. [DOI] [PubMed] [Google Scholar]
- 285.Karlin, S., and J. Mrazek. 2001. Predicted highly expressed and putative alien genes of Deinococcus radiodurans and implications for resistance to ionizing radiation damage. Proc. Natl. Acad. Sci. U. S. A. 98:5240-5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Karlin, S., and J. Mrazek. 2000. Predicted highly expressed genes of diverse prokaryotic genomes. J. Bacteriol. 182:5238-5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Keller, L. C., and R. B. Maxcy. 1984. Effect of physiological age on radiation resistance of some bacteria that are highly radiation resistant. Appl. Environ. Microbiol. 47:915-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Khairnar, N. P., V. A. Kamble, and H. S. Misra. 2008. RecBC enzyme overproduction affects UV and gamma radiation survival of Deinococcus radiodurans. DNA Repair (Amst.) 7:40-47. [DOI] [PubMed] [Google Scholar]
- 289.Khairnar, N. P., and H. S. Misra. 2009. DNA polymerase X from Deinococcus radiodurans implicated in bacterial tolerance to DNA damage is characterized as a short patch base excision repair polymerase. Microbiology 155:3005-3014. [DOI] [PubMed] [Google Scholar]
- 290.Khairnar, N. P., H. S. Misra, and S. K. Apte. 2003. Pyrroloquinoline-quinone synthesized in Escherichia coli by pyrroloquinoline-quinone synthase of Deinococcus radiodurans plays a role beyond mineral phosphate solubilization. Biochem. Biophys. Res. Commun. 312:303-308. [DOI] [PubMed] [Google Scholar]
- 291.Khidhir, M. A., S. Casaregola, and I. B. Holland. 1985. Mechanism of transient inhibition of DNA synthesis in ultraviolet-irradiated E. coli: inhibition is independent of recA whilst recovery requires RecA protein itself and an additional, inducible SOS function. Mol. Gen. Genet. 199:133-140. [DOI] [PubMed] [Google Scholar]
- 292.Kilburn, R. E., W. D. Bellamy, and S. A. Terni. 1958. Studies on a radiation-resistant pigmented Sarcina sp. Radiat. Res. 9:207-215. [PubMed] [Google Scholar]
- 293.Killoran, M. P., and J. L. Keck. 2006. Three HRDC domains differentially modulate Deinococcus radiodurans RecQ DNA helicase biochemical activity. J. Biol. Chem. 281:12849-12857. [DOI] [PubMed] [Google Scholar]
- 294.Kim, J., et al. 2004. Fundamental structural units of the Escherichia coli nucleoid revealed by atomic force microscopy. Nucleic Acids Res. 32:1982-1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Kim, J. I. 2006. Analysis of double stranded DNA-dependent activities of Deinococcus radiodurans RecA protein. J. Microbiol. 44:508-514. [PubMed] [Google Scholar]
- 296.Kim, J. I., and M. M. Cox. 2002. The RecA proteins of Deinococcus radiodurans and Escherichia coli promote DNA strand exchange via inverse pathways. Proc. Natl. Acad. Sci. U. S. A. 99:7917-7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Kim, J. I., et al. 2002. RecA protein from the extremely radioresistant bacterium Deinococcus radiodurans: expression, purification, and characterization. J. Bacteriol. 184:1649-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Kim, M., et al. 2004. Developing a genetic system in Deinococcus radiodurans for analyzing mutations. Genetics 166:661-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Kimura, H., R. Asada, A. Masta, and T. Naganuma. 2003. Distribution of microorganisms in the subsurface of the Manus Basin hydrothermal vent field in Papua New Guinea. Appl. Environ. Microbiol. 69:644-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Kirkwood, T. B. 2002. Evolution of ageing. Mech. Ageing Dev. 123:737-745. [DOI] [PubMed] [Google Scholar]
- 301.Kirkwood, T. B. 2005. Understanding the odd science of aging. Cell 120:437-447. [DOI] [PubMed] [Google Scholar]
- 302.Kish, A., et al. 2009. Salt shield: intracellular salts provide cellular protection against ionizing radiation in the halophilic archaeon, Halobacterium salinarum NRC-1. Environ. Microbiol. 11:1066-1078. [DOI] [PubMed] [Google Scholar]
- 303.Kitayama, S. 1982. Adaptive repair of cross-links in DNA of Micrococcus radiodurans. Biochim. Biophys. Acta 697:381-384. [DOI] [PubMed] [Google Scholar]
- 304.Kitayama, S., S. Asaka, and K. Totsuka. 1983. DNA double-strand breakage and removal of cross-links in Deinococcus radiodurans. J. Bacteriol. 155:1200-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Kitayama, S., M. Kohoroku, A. Takagi, and H. Itoh. 1997. Mutation of D. radiodurans in a gene homologous to ruvB of E. coli. Mutat. Res. 385:151-157. [DOI] [PubMed] [Google Scholar]
- 306.Kitayama, S., and A. Matsuyama. 1976. DNA synthesis and repair in permeable cells of Micrococcus radiodurans. Biochim. Biophys. Acta 418:321-329. [DOI] [PubMed] [Google Scholar]
- 307.Kitayama, S., and A. Matsuyama. 1968. Possibility of the repair of double-strand scissions in Micrococcus radiodurans DNA caused by gamma-rays. Biochem. Biophys. Res. Commun. 33:418-422. [DOI] [PubMed] [Google Scholar]
- 308.Kitayama, S., I. Narumi, M. Kikuchi, and H. Watanabe. 2000. Mutation in recR gene of Deinococcus radiodurans and possible involvement of its product in the repair of DNA interstrand cross-links. Mutat. Res. 461:179-187. [DOI] [PubMed] [Google Scholar]
- 309.Klaunig, J. E., and L. M. Kamendulis. 2004. The role of oxidative stress in carcinogenesis. Annu. Rev. Pharmacol. Toxicol. 44:239-267. [DOI] [PubMed] [Google Scholar]
- 310.Knivett, V. A., J. Cullen, and M. J. Jackson. 1965. Odd-numbered fatty acids in Micrococcus radiodurans. Biochem. J. 96:2C-3C. [DOI] [PubMed] [Google Scholar]
- 311.Kobatake, M., S. Tanabe, and S. Hasegawa. 1973. New Micrococcus radioresistant red pigment, isolated from Lama glama feces, and its use as microbiological indicator of radiosterilization. C. R. Seances Soc. Biol. Fil. 167:1506-1510. (In French.) [PubMed] [Google Scholar]
- 312.Kobayashi, I., et al. 2006. Characterization of monofunctional catalase KatA from radioresistant bacterium Deinococcus radiodurans. J. Biosci. Bioeng. 101:315-321. [DOI] [PubMed] [Google Scholar]
- 313.Kolodner, R. D., and G. T. Marsischky. 1999. Eukaryotic DNA mismatch repair. Curr. Opin. Genet. Dev. 9:89-96. [DOI] [PubMed] [Google Scholar]
- 314.Konrad, E. B., and I. R. Lehman. 1974. A conditional lethal mutant of Escherichia coli K12 defective in the 5′ leads to 3′ exonuclease associated with DNA polymerase I. Proc. Natl. Acad. Sci. U. S. A. 71:2048-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Kopylov, V. M., E. A. Bonch-Osmolovskaya, V. A. Svetlichnyi, M. L. Miroshnichenko, and V. S. Skobkin. 1993. Gamma irradiation resistance and UV-sensitivity of extremely thermophilic archaebacteria and eubacteria. Mikrobiologiia 62:90-95. (In Russian.) [Google Scholar]
- 316.Kornberg, A. 1995. Inorganic polyphosphate: toward making a forgotten polymer unforgettable. J. Bacteriol. 177:491-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Kornberg, A., and T. Baker. 1992. DNA replication, 2nd ed. W. H. Freeman and Co., New York, NY.
- 318.Kota, S., C. V. Kumar, and H. S. Misra. 2010. Characterization of an ATP regulated DNA processing enzyme and thermotolerant phosphoesterase in a radioresistant bacterium Deinococcus radiodurans. Biochem. J. 431:149-157. [DOI] [PubMed] [Google Scholar]
- 319.Kota, S., and H. S. Misra. 2008. Identification of a DNA processing complex from Deinococcus radiodurans. Biochem. Cell Biol. 86:448-458. [DOI] [PubMed] [Google Scholar]
- 320.Kota, S., and H. S. Misra. 2006. PprA: a protein implicated in radioresistance of Deinococcus radiodurans stimulates catalase activity in Escherichia coli. Appl. Microbiol. Biotechnol. 72:790-796. [DOI] [PubMed] [Google Scholar]
- 321.Kottemann, M., A. Kish, C. Iloanusi, S. Bjork, and J. DiRuggiero. 2005. Physiological responses of the halophilic archaeon Halobacterium sp. strain NRC1 to desiccation and gamma irradiation. Extremophiles 9:219-227. [DOI] [PubMed] [Google Scholar]
- 322.Kowalczykowski, S. C. 2000. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 25:156-165. [DOI] [PubMed] [Google Scholar]
- 323.Kowalczykowski, S. C., D. A. Dixon, A. K. Eggleston, S. D. Lauder, and W. M. Rehrauer. 1994. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 58:401-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Kowalczykowski, S. C., and R. A. Krupp. 1987. Effects of Escherichia coli SSB protein on the single-stranded DNA-dependent ATPase activity of Escherichia coli RecA protein. Evidence that SSB protein facilitates the binding of RecA protein to regions of secondary structure within single-stranded DNA. J. Mol. Biol. 193:97-113. [DOI] [PubMed] [Google Scholar]
- 325.Krabbenhoft, K. L., A. W. Anderson, and P. R. Elliker. 1965. Ecology of Micrococcus radiodurans. Appl. Microbiol. 13:1030-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Krabbenhoft, K. L., A. W. Anderson, and P. R. Elliker. 1967. Influence of culture media on the radiation resistance of Micrococcus radiodurans. Appl. Microbiol. 15:178-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Krasin, F., and F. Hutchinson. 1977. Repair of DNA double-strand breaks in Escherichia coli, which requires recA function and the presence of a duplicate genome. J. Mol. Biol. 116:81-98. [DOI] [PubMed] [Google Scholar]
- 328.Krisch, R. E., M. B. Flick, and C. N. Trumbore. 1991. Radiation chemical mechanisms of single- and double-strand break formation in irradiated SV40 DNA. Radiat. Res. 126:251-259. [PubMed] [Google Scholar]
- 329.Krisko, A., and M. Radman. 2010. Protein damage and death by radiation in Escherichia coli and Deinococcus radiodurans. Proc. Natl. Acad. Sci. U. S. A. 107:14373-14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Kristensen, H., and E. A. Christensen. 1981. Radiation-resistant micro-organisms isolated from textiles. Acta Pathol. Microbiol. Scand. B 89:303-309. [DOI] [PubMed] [Google Scholar]
- 331.Kubota, Y., et al. 1996. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. EMBO J. 15:6662-6670. [PMC free article] [PubMed] [Google Scholar]
- 332.Kuzminov, A. 1995. Collapse and repair of replication forks in Escherichia coli. Mol. Microbiol. 16:373-384. [DOI] [PubMed] [Google Scholar]
- 333.Kuzminov, A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev. 63:751-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Lai, W. A., et al. 2006. Deinococcus ficus sp. nov., isolated from the rhizosphere of Ficus religiosa L. Int. J. Syst. Evol. Microbiol. 56:787-791. [DOI] [PubMed] [Google Scholar]
- 335.Lancy, P., Jr., and R. G. Murray. 1978. The envelope of Micrococcus radiodurans: isolation, purification, and preliminary analysis of the wall layers. Can. J. Microbiol. 24:162-176. [DOI] [PubMed] [Google Scholar]
- 336.Lange, C. C., L. P. Wackett, K. W. Minton, and M. J. Daly. 1998. Engineering a recombinant Deinococcus radiodurans for organopollutant degradation in radioactive mixed waste environments. Nat. Biotechnol. 16:929-933. [DOI] [PubMed] [Google Scholar]
- 337.Lapointe, J., and S. Hekimi. 2009. When a theory of aging ages badly. Cell. Mol. Life Sci. 67:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Lavery, P. E., and S. C. Kowalczykowski. 1992. A postsynaptic role for single-stranded DNA-binding protein in recA protein-promoted DNA strand exchange. J. Biol. Chem. 267:9315-9320. [PubMed] [Google Scholar]
- 339.Lawley, P. D., and C. J. Thatcher. 1970. Methylation of deoxyribonucleic acid in cultured mammalian cells by N-methyl-N′-nitro-N-nitrosoguanidine. The influence of cellular thiol concentrations on the extent of methylation and the 6-oxygen atom of guanine as a site of methylation. Biochem. J. 116:693-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Le, D., et al. 2008. Biochemical characterization of two DNA ligases from Deinococcus radiodurans. Protein Pept. Lett. 15:600-605. [DOI] [PubMed] [Google Scholar]
- 341.Leaper, S., M. A. Resnick, and R. Holliday. 1980. Repair of double-strand breaks and lethal damage in DNA of Ustilago maydis. Genet. Res. 35:291-307. [DOI] [PubMed] [Google Scholar]
- 342.Lecointe, F., G. Coste, S. Sommer, and A. Bailone. 2004. Vectors for regulated gene expression in the radioresistant bacterium Deinococcus radiodurans. Gene 336:25-35. [DOI] [PubMed] [Google Scholar]
- 343.Lecointe, F., I. V. Shevelev, A. Bailone, S. Sommer, and U. Hubscher. 2004. Involvement of an X family DNA polymerase in double-stranded break repair in the radioresistant organism Deinococcus radiodurans. Mol. Microbiol. 53:1721-1730. [DOI] [PubMed] [Google Scholar]
- 344.Lee, B. I., et al. 2004. Ring-shaped architecture of RecR: implications for its role in homologous recombinational DNA repair. EMBO J. 23:2029-2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Lehrman, M. A., et al. 1985. Mutation in LDL receptor: Alu-Alu recombination deletes exons encoding transmembrane and cytoplasmic domains. Science 227:140-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Leibowitz, P. J., L. S. Schwartzberg, and A. K. Bruce. 1976. The in vivo association of manganese with the chromosome of Micrococcus radiodurans. Photochem. Photobiol. 23:45-50. [DOI] [PubMed] [Google Scholar]
- 347.Leiros, I., J. Timmins, D. R. Hall, and S. McSweeney. 2005. Crystal structure and DNA-binding analysis of RecO from Deinococcus radiodurans. EMBO J. 24:906-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Lemee, L., E. Peuchant, M. Clerc, M. Brunner, and H. Pfander. 1997. Deinoxanthin: a new carotenoid isolated from Deinococcus radiodurans. Tetrahedron 53:919-926. [Google Scholar]
- 349.Lengauer, C., K. W. Kinzler, and B. Vogelstein. 1998. Genetic instabilities in human cancers. Nature 396:643-649. [DOI] [PubMed] [Google Scholar]
- 350.Lett, J. T., I. Caldwell, and J. G. Little. 1970. Repair of X-ray damage to the DNA in Micrococcus radiodurans: the effect of 5-bromodeoxyuridine. J. Mol. Biol. 48:395-408. [DOI] [PubMed] [Google Scholar]
- 351.Lett, J. T., P. Feldschreiber, J. G. Little, K. Steele, and C. J. Dean. 1967. The repair of X-ray damage to the deoxyribonucleic acid in Micrococcus radiodurans: a study of the excision process. Proc. R. Soc. Lond. B Biol. Sci. 167:184-201. [DOI] [PubMed] [Google Scholar]
- 352.Leulliot, N., et al. 2009. The family X DNA polymerase from Deinococcus radiodurans adopts a non-standard extended conformation. J. Biol. Chem. 284:11992-11999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Levine, R. L. 2002. Carbonyl modified proteins in cellular regulation, aging, and disease. Free Radic. Biol. Med. 32:790-796. [DOI] [PubMed] [Google Scholar]
- 354.Levin-Zaidman, S., et al. 2003. Ringlike structure of the Deinococcus radiodurans genome: a key to radioresistance? Science 299:254-256. [DOI] [PubMed] [Google Scholar]
- 355.Lewis, N. F. 1971. Studies on a radio-resistant coccus isolated from Bombay duck (Harpodon nehereus). J. Gen. Microbiol. 66:29-35. [DOI] [PubMed] [Google Scholar]
- 356.Li, X., and A. L. Lu. 2001. Molecular cloning and functional analysis of the MutY homolog of Deinococcus radiodurans. J. Bacteriol. 183:6151-6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Liao, C. S., L. C. Chen, B. S. Chen, and S. H. Lin. 2010. Bioremediation of endocrine disruptor di-n-butyl phthalate ester by Deinococcus radiodurans and Pseudomonas stutzeri. Chemosphere 78:342-346. [DOI] [PubMed] [Google Scholar]
- 358.Licciardello, J. J., J. T. Nickerson, S. A. Goldblith, C. A. Shannon, and W. W. Bishop. 1969. Development of radiation resistance in Salmonella cultures. Appl. Microbiol. 18:24-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Lichten, M., R. H. Borts, and J. E. Haber. 1987. Meiotic gene conversion and crossing over between dispersed homologous sequences occurs frequently in Saccharomyces cerevisiae. Genetics 115:233-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Liebler, D. C., and T. D. McClure. 1996. Antioxidant reactions of beta-carotene: identification of carotenoid-radical adducts. Chem. Res. Toxicol. 9:8-11. [DOI] [PubMed] [Google Scholar]
- 361.Lin, C. G., O. Kovalsky, and L. Grossman. 1997. DNA damage-dependent recruitment of nucleotide excision repair and transcription proteins to Escherichia coli inner membranes. Nucleic Acids Res. 25:3151-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Lin, C. L., and S. T. Tan. 1996. Isolation and characterization of a novel Deinococcus radiodurans mutant abnormally susceptible to mutation induction by UV, gamma-ray, and mitomycin C. Int. J. Radiat. Biol. 69:493-502. [DOI] [PubMed] [Google Scholar]
- 363.Linton, K. J., and C. F. Higgins. 1998. The Escherichia coli ATP-binding cassette (ABC) proteins. Mol. Microbiol. 28:5-13. [DOI] [PubMed] [Google Scholar]
- 364.Lipton, M. S., et al. 2002. Global analysis of the Deinococcus radiodurans proteome by using accurate mass tags. Proc. Natl. Acad. Sci. U. S. A. 99:11049-11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Lisby, M., and R. Rothstein. 2004. DNA repair: keeping it together. Curr. Biol. 14:R994-R996. [DOI] [PubMed] [Google Scholar]
- 366.Liu, L. F., and J. C. Wang. 1975. In vitro DNA synthesis on primed covalently closed double-stranded templates. I. Studies with Escherichia coli DNA polymerase I. ICN-UCLA Symp. Mol. Cell. Biol. 3:38-63. [Google Scholar]
- 367.Liu, X., et al. 2008. Resistance of Deinococcus radiodurans to mutagenesis is facilitated by pentose phosphate pathway in the mutS1 mutant background. Curr. Microbiol. 57:66-71. [DOI] [PubMed] [Google Scholar]
- 368.Liu, Y., et al. 2003. Transcriptome dynamics of Deinococcus radiodurans recovering from ionizing radiation. Proc. Natl. Acad. Sci. U. S. A. 100:4191-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Lloyd, R. G., C. Buckman, and F. E. Benson. 1987. Genetic analysis of conjugational recombination in Escherichia coli K12 strains deficient in RecBCD enzyme. J. Gen. Microbiol. 133:2531-2538. [DOI] [PubMed] [Google Scholar]
- 370.Lloyd, R. G., and G. J. Sharples. 1993. Dissociation of synthetic Holliday junctions by E. coli RecG protein. EMBO J. 12:17-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Lombard, D. B., et al. 2005. DNA repair, genome stability, and aging. Cell 120:497-512. [DOI] [PubMed] [Google Scholar]
- 372.Lovett, S. T. 2006. Replication arrest-stimulated recombination: dependence on the RecA paralog, RadA/Sms and translesion polymerase, DinB. DNA Repair (Amst.) 5:1421-1427. [DOI] [PubMed] [Google Scholar]
- 373.Lovett, S. T., and V. V. Feschenko. 1996. Stabilization of diverged tandem repeats by mismatch repair: evidence for deletion formation via a misaligned replication intermediate. Proc. Natl. Acad. Sci. U. S. A. 93:7120-7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Lovett, S. T., and R. D. Kolodner. 1989. Identification and purification of a single-stranded-DNA-specific exonuclease encoded by the recJ gene of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 86:2627-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Lovley, D. R., and J. D. Coates. 1997. Bioremediation of metal contamination. Curr. Opin. Biotechnol. 8:285-289. [DOI] [PubMed] [Google Scholar]
- 376.Lown, J. W., S. K. Sim, and H. H. Chen. 1978. Hydroxyl radical production by free and DNA-bound aminoquinone antibiotics and its role in DNA degradation. Electron spin resonance detection of hydroxyl radicals by spin trapping. Can. J. Biochem. 56:1042-1047. [DOI] [PubMed] [Google Scholar]
- 377.Lu, H., et al. 2009. Deinococcus radiodurans PprI switches on DNA damage-response and cellular survival networks after radiation damage. Mol. Cell. Proteomics 8:481-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Macaskie, L. E., K. M. Bonthrone, and D. A. Rouch. 1994. Phosphatase-mediated heavy metal accumulation by a Citrobacter sp. and related enterobacteria. FEMS Microbiol. Lett. 121:141-146. [DOI] [PubMed] [Google Scholar]
- 379.Majumdar, S., and A. K. Chandra. 1985. UV-repair and mutagenesis in Azotobacter vinelandii. II. Repair and mutagenesis. Zentralbl. Mikrobiol. 140:255-258. [PubMed] [Google Scholar]
- 380.Makarova, K. S., L. Aravind, M. J. Daly, and E. V. Koonin. 2000. Specific expansion of protein families in the radioresistant bacterium Deinococcus radiodurans. Genetica 108:25-34. [DOI] [PubMed] [Google Scholar]
- 381.Makarova, K. S., et al. 2001. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol. Mol. Biol. Rev. 65:44-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Makarova, K. S., and M. J. Daly. 2010. Comparative genomics of stress response systems in Deinococcus bacteria, 2nd ed. ASM Press, Washington, DC.
- 383.Makarova, K. S., et al. 2007. Deinococcus geothermalis: the pool of extreme radiation resistance genes shrinks. PLoS One 2:e955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Makarova, K. S., Y. I. Wolf, O. White, K. Minton, and M. J. Daly. 1999. Short repeats and IS elements in the extremely radiation-resistant bacterium Deinococcus radiodurans and comparison to other bacterial species. Res. Microbiol. 150:711-724. [DOI] [PubMed] [Google Scholar]
- 385.Makharashvili, N., O. Koroleva, S. Bera, D. P. Grandgenett, and S. Korolev. 2004. A novel structure of DNA repair protein RecO from Deinococcus radiodurans. Structure 12:1881-1889. [DOI] [PubMed] [Google Scholar]
- 386.Makharashvili, N., T. Mi, O. Koroleva, and S. Korolev. 2008. RecR-mediated modulation of RecF dimer specificity for single- and double-stranded DNA. J. Biol. Chem. 284:1425-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Maki, H., and M. Sekiguchi. 1992. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature 355:273-275. [DOI] [PubMed] [Google Scholar]
- 388.Markillie, L. M., S. M. Varnum, P. Hradecky, and K. K. Wong. 1999. Targeted mutagenesis by duplication insertion in the radioresistant bacterium Deinococcus radiodurans: radiation sensitivities of catalase (katA) and superoxide dismutase (sodA) mutants. J. Bacteriol. 181:666-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Marteinsson, V. T., et al. 2001. Phylogenetic diversity analysis of subterranean hot springs in Iceland. Appl. Environ. Microbiol. 67:4242-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Martinez, A., and R. Kolter. 1997. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J. Bacteriol. 179:5188-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Martinez, M., M. L. Ferrandiz, A. Diez, and J. Miquel. 1995. Depletion of cytosolic GSH decreases the ATP levels and viability of synaptosomes from aged mice but not from young mice. Mech. Ageing Dev. 84:77-81. [DOI] [PubMed] [Google Scholar]
- 392.Masson, J. Y., and S. C. West. 2001. The Rad51 and Dmc1 recombinases: a non-identical twin relationship. Trends Biochem. Sci. 26:131-136. [DOI] [PubMed] [Google Scholar]
- 393.Masters, C. I., and K. W. Minton. 1992. Promoter probe and shuttle plasmids for Deinococcus radiodurans. Plasmid 28:258-261. [DOI] [PubMed] [Google Scholar]
- 394.Masters, C. I., B. E. Moseley, and K. W. Minton. 1991. AP endonuclease and uracil DNA glycosylase activities in Deinococcus radiodurans. Mutat. Res. 254:263-272. [DOI] [PubMed] [Google Scholar]
- 395.Masure, H. R., B. J. Pearce, H. Shio, and B. Spellerberg. 1998. Membrane targeting of RecA during genetic transformation. Mol. Microbiol. 27:845-852. [DOI] [PubMed] [Google Scholar]
- 396.Mathews, M. M., and N. I. Krinsky. 1965. The relationship between carotenoid pigments and resistance to radiation in non-photosynthetic bacteria. Photochem. Photobiol. 4:813-817. [DOI] [PubMed] [Google Scholar]
- 397.Matson, S. W., and R. A. Bambara. 1981. Short deoxyribonucleic acid repair patch length in Escherichia coli is determined by the processive mechanism of deoxyribonucleic acid polymerase I. J. Bacteriol. 146:275-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Mattimore, V., and J. R. Battista. 1996. Radioresistance of Deinococcus radiodurans: functions necessary to survive ionizing radiation are also necessary to survive prolonged desiccation. J. Bacteriol. 178:633-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.McEntee, K., G. M. Weinstock, and I. R. Lehman. 1980. recA protein-catalyzed strand assimilation: stimulation by Escherichia coli single-stranded DNA-binding protein. Proc. Natl. Acad. Sci. U. S. A. 77:857-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.McGrath, R. A., R. W. Williams, and D. C. Swartzendruber. 1966. Breakdown of DNA in X-irradiated Escherichia coli. Biophys. J. 6:113-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.McNaught, K. S., C. W. Olanow, B. Halliwell, O. Isacson, and P. Jenner. 2001. Failure of the ubiquitin-proteasome system in Parkinson's disease. Nat. Rev. Neurosci. 2:589-594. [DOI] [PubMed] [Google Scholar]
- 402.McNaughton, R. L., et al. 2010. Probing in vivo Mn2+ speciation and oxidative stress resistance in yeast cells with electron-nuclear double resonance spectroscopy. Proc. Natl. Acad. Sci. U. S. A. 107:15335-15339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Meima, R., and M. E. Lidstrom. 2000. Characterization of the minimal replicon of a cryptic Deinococcus radiodurans SARK plasmid and development of versatile Escherichia coli-D. radiodurans shuttle vectors. Appl. Environ. Microbiol. 66:3856-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Meima, R., H. M. Rothfuss, L. Gewin, and M. E. Lidstrom. 2001. Promoter cloning in the radioresistant bacterium Deinococcus radiodurans. J. Bacteriol. 183:3169-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Meister, A., et al. 2009. FluidFM: combining atomic force microscopy and nanofluidics in a universal liquid delivery system for single cell applications and beyond. Nano Lett. 9:2501-2507. [DOI] [PubMed] [Google Scholar]
- 406.Mello Filho, A. C., and R. Meneghini. 1984. In vivo formation of single-strand breaks in DNA by hydrogen peroxide is mediated by the Haber-Weiss reaction. Biochim. Biophys. Acta 781:56-63. [DOI] [PubMed] [Google Scholar]
- 407.Mennecier, S., G. Coste, P. Servant, A. Bailone, and S. Sommer. 2004. Mismatch repair ensures fidelity of replication and recombination in the radioresistant organism Deinococcus radiodurans. Mol. Genet. Genomics 272:460-469. [DOI] [PubMed] [Google Scholar]
- 408.Mennecier, S., P. Servant, G. Coste, A. Bailone, and S. Sommer. 2006. Mutagenesis via IS transposition in Deinococcus radiodurans. Mol. Microbiol. 59:317-325. [DOI] [PubMed] [Google Scholar]
- 409.Merrick, T. P., and A. K. Bruce. 1965. Radiation response of potassium efflux in Micrococcus radiodurans and Sarcina lutea. Radiat. Res. 24:612-618. [PubMed] [Google Scholar]
- 410.Michel, B., et al. 2001. Rescue of arrested replication forks by homologous recombination. Proc. Natl. Acad. Sci. U. S. A. 98:8181-8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Michel, B., G. Grompone, M. J. Flores, and V. Bidnenko. 2004. Multiple pathways process stalled replication forks. Proc. Natl. Acad. Sci. U. S. A. 101:12783-12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Mijakovic, I., et al. 2006. Bacterial single-stranded DNA-binding proteins are phosphorylated on tyrosine. Nucleic Acids Res. 34:1588-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Minton, K. W. 1994. DNA repair in the extremely radioresistant bacterium Deinococcus radiodurans. Mol. Microbiol. 13:9-15. [DOI] [PubMed] [Google Scholar]
- 414.Minton, K. W. 1996. Repair of ionizing-radiation damage in the radiation resistant bacterium Deinococcus radiodurans. Mutat. Res. 363:1-7. [DOI] [PubMed] [Google Scholar]
- 415.Minton, K. W., and M. J. Daly. 1995. A model for repair of radiation-induced DNA double-strand breaks in the extreme radiophile Deinococcus radiodurans. Bioessays 17:457-464. [DOI] [PubMed] [Google Scholar]
- 416.Misra, H., and I. Fridovich. 1976. Superoxide dismutase and the oxygen enhancement of radiation lethality. Arch. Biochem. Biophys. 176:577-581. [DOI] [PubMed] [Google Scholar]
- 417.Misra, H. S., et al. 2004. Pyrroloquinoline-quinone: a reactive oxygen species scavenger in bacteria. FEBS Lett. 578:26-30. [DOI] [PubMed] [Google Scholar]
- 418.Misra, H. S., et al. 2006. An exonuclease I-sensitive DNA repair pathway in Deinococcus radiodurans: a major determinant of radiation resistance. Mol. Microbiol. 59:1308-1316. [DOI] [PubMed] [Google Scholar]
- 419.Mitchel, R. E. 1975. Involvement of hydroxyl radicals in the release by ionizing radiation of a cell surface nuclease from Micrococcus radiodurans. Radiat. Res. 64:321-330. [PubMed] [Google Scholar]
- 420.Mitchel, R. E. 1976. Ionizing radiation damage in Micrococcus radiodurans cell wall: release of polysaccharide. Radiat. Res. 66:158-169. [PubMed] [Google Scholar]
- 421.Mitchel, R. E. 1975. Origin of cell surface proteins released from Micrococcus radiodurans by ionizing radiation. Radiat. Res. 64:380-387. [PubMed] [Google Scholar]
- 422.Mitchel, R. E. 1981. Loss of covalently linked lipid as the mechanism for radiation-induced release of membrane-bound polysaccharides and exonuclease from Micrococcus radiodurans. Radiat. Res. 87:341-349. [PubMed] [Google Scholar]
- 423.Mitchel, R. E. 1980. Micrococcus radiodurans surface exonuclease. Dimer to monomer conversion by ionizing radiation-generated aqueous free radicals. Biochim. Biophys. Acta 621:138-146. [DOI] [PubMed] [Google Scholar]
- 424.Mitchel, R. E. 1978. The radiation-releasable cell wall nuclease of Micrococcus radiodurans. Purification and properties of the native enzyme. Biochim. Biophys. Acta 524:362-372. [DOI] [PubMed] [Google Scholar]
- 425.Mitchell, J. R., J. H. Hoeijmakers, and L. J. Niedernhofer. 2003. Divide and conquer: nucleotide excision repair battles cancer and ageing. Curr. Opin. Cell Biol. 15:232-240. [DOI] [PubMed] [Google Scholar]
- 426.Mizusawa, S., and S. Gottesman. 1983. Protein degradation in Escherichia coli: the lon gene controls the stability of sulA protein. Proc. Natl. Acad. Sci. U. S. A. 80:358-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Moe, E., I. Leiros, A. O. Smalas, and S. McSweeney. 2006. The crystal structure of mismatch-specific uracil-DNA glycosylase (MUG) from Deinococcus radiodurans reveals a novel catalytic residue and broad substrate specificity. J. Biol. Chem. 281:569-577. [DOI] [PubMed] [Google Scholar]
- 428.Moeller, R., et al. 2010. Genomic bipyrimidine nucleotide frequency and microbial reactions to germicidal UV radiation. Arch. Microbiol. 192:521-529. [DOI] [PubMed] [Google Scholar]
- 429.Montague, M., C. Barnes, H. O. Smith, R. Y. Chuang, and S. Vashee. 2009. The evolution of RecD outside of the RecBCD complex. J. Mol. Evol. 69:360-371. [DOI] [PubMed] [Google Scholar]
- 430.Morimatsu, K., and S. C. Kowalczykowski. 2003. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol. Cell 11:1337-1347. [DOI] [PubMed] [Google Scholar]
- 431.Moriya, M. 1993. Single-stranded shuttle phagemid for mutagenesis studies in mammalian cells: 8-oxoguanine in DNA induces targeted G.C→T.A transversions in simian kidney cells. Proc. Natl. Acad. Sci. U. S. A. 90:1122-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Mortensen, A. 2002. Scavenging of benzylperoxyl radicals by carotenoids. Free Radic. Res. 36:211-216. [DOI] [PubMed] [Google Scholar]
- 433.Mortimer, R. K. 1958. Radiobiological and genetic studies on a polyploid series (haploid to hexaploid) of Saccharomyces cerevisiae. Radiat. Res. 9:312-326. [PubMed] [Google Scholar]
- 434.Moseley, B. E. 1967. The isolation and some properties of radiation-sensitive mutants of Micrococcus radiodurans. J. Gen. Microbiol. 49:293-300. [DOI] [PubMed] [Google Scholar]
- 435.Moseley, B. E. 1983. Photobiology and radiobiology of Micrococcus (Deinococcus) radiodurans. Photochem. Photobiol. Rev. 7:223-274. [Google Scholar]
- 436.Moseley, B. E. 1969. Repair of ultraviolet radiation damage in sensitive mutants of Micrococcus radiodurans. J. Bacteriol. 97:647-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Moseley, B. E., and H. F. Copland. 1978. Four mutants of Micrococcus radiodurans defective in the ability to repair DNA damaged by mitomycin-C, two of which have wild-type resistance to ultraviolet radiation. Mol. Gen. Genet. 160:331-337. [DOI] [PubMed] [Google Scholar]
- 438.Moseley, B. E., and H. J. Copland. 1975. Involvement of a recombination repair function in disciplined cell division of Micrococcus radiodurans. J. Gen. Microbiol. 86:343-357. [DOI] [PubMed] [Google Scholar]
- 439.Moseley, B. E., and H. J. Copland. 1975. Isolation and properties of a recombination-deficient mutant of Micrococcus radiodurans. J. Bacteriol. 121:422-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Moseley, B. E., and H. J. Copland. 1976. The rate of recombination repair and its relationship to the radiation-induced delay in DNA synthesis in Micrococcus radiodurans. J. Gen. Microbiol. 93:251-258. [DOI] [PubMed] [Google Scholar]
- 441.Moseley, B. E., and D. M. Evans. 1983. Isolation and properties of strains of Micrococcus (Deinococcus) radiodurans unable to excise ultraviolet light-induced pyrimidine dimers from DNA: evidence for two excision pathways. J. Gen. Microbiol. 129:2437-2445. [DOI] [PubMed] [Google Scholar]
- 442.Moseley, B. E., and H. Laser. 1965. Repair of X-ray in Micrococcus radiodurans. Proc. R. Soc. Lond. B Biol. Sci. 162:210-222. [DOI] [PubMed] [Google Scholar]
- 443.Moseley, B. E., and H. Laser. 1965. Similarity of repair of ionizing and ultra-violet radiation damage in Micrococcus radiodurans. Nature 206:373-375. [DOI] [PubMed] [Google Scholar]
- 444.Moseley, B. E., and A. Mattingly. 1971. Repair of irradiation transforming deoxyribonucleic acid in wild type and a radiation-sensitive mutant of Micrococcus radiodurans. J. Bacteriol. 105:976-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Moseley, B. E., A. Mattingly, and H. J. Copland. 1972. Sensitization to radiation by loss of recombination ability in a temperature-sensitive DNA mutant of Micrococcus radiodurans held at its restrictive temperature. J. Gen. Microbiol. 72:329-338. [DOI] [PubMed] [Google Scholar]
- 446.Moseley, B. E., A. Mattingly, and M. Shimmin. 1972. Isolation and some properties of temperature-sensitive mutants of Micrococcus radiodurans defective in DNA synthesis. J. Gen. Microbiol. 70:399-409. [DOI] [PubMed] [Google Scholar]
- 447.Moseley, B. E., and A. H. Schein. 1964. Radiation resistance and deoxyribonucleic acid base composition of Micrococcus radiodurans. Nature 203:1298-1299. [DOI] [PubMed] [Google Scholar]
- 448.Moseley, B. E., and J. K. Setlow. 1968. Transformation in Micrococcus radiodurans and the ultraviolet sensitivity of its transforming DNA. Proc. Natl. Acad. Sci. U. S. A. 61:176-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Muller, F. L., M. S. Lustgarten, Y. Jang, A. Richardson, and H. Van Remmen. 2007. Trends in oxidative aging theories. Free Radic. Biol. Med. 43:477-503. [DOI] [PubMed] [Google Scholar]
- 450.Mun, C., J. Del Rowe, M. Sandigursky, K. W. Minton, and W. A. Franklin. 1994. DNA deoxyribophosphodiesterase and an activity that cleaves DNA containing thymine glycol adducts in Deinococcus radiodurans. Radiat. Res. 138:282-285. [PubMed] [Google Scholar]
- 451.Murakami, M., I. Narumi, K. Satoh, A. Furukawa, and I. Hayata. 2006. Analysis of interaction between DNA and Deinococcus radiodurans PprA protein by atomic force microscopy. Biochim. Biophys. Acta 1764:20-23. [DOI] [PubMed] [Google Scholar]
- 452.Murray, R. G. 1986. Family II. Deinococcaceae, p. 1035-1043. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams & Wilkins Co., Baltimore, MD. [Google Scholar]
- 453.Murray, R. G., M. Hall, and B. G. Thompson. 1983. Cell division in Deinococcus radiodurans and a method for displaying septa. Can. J. Microbiol. 29:1412-1423. [DOI] [PubMed] [Google Scholar]
- 454.Reference deleted.
- 455.Narumi, I., K. Cherdchu, S. Kitayama, and H. Watanabe. 1997. The Deinococcus radiodurans uvrA gene: identification of mutation sites in two mitomycin-sensitive strains and the first discovery of insertion sequence element from deinobacteria. Gene 198:115-126. [DOI] [PubMed] [Google Scholar]
- 456.Narumi, I., et al. 2004. PprA: a novel protein from Deinococcus radiodurans that stimulates DNA ligation. Mol. Microbiol. 54:278-285. [DOI] [PubMed] [Google Scholar]
- 457.Narumi, I., et al. 1999. Molecular analysis of the Deinococcus radiodurans recA locus and identification of a mutation site in a DNA repair-deficient mutant, rec30. Mutat. Res. 435:233-243. [DOI] [PubMed] [Google Scholar]
- 458.Narumi, I., et al. 2001. The LexA protein from Deinococcus radiodurans is not involved in RecA induction following gamma irradiation. J. Bacteriol. 183:6951-6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Nguyen, H. H., et al. 2009. The essential histone-like protein HU plays a major role in Deinococcus radiodurans nucleoid compaction. Mol. Microbiol. 73:240-252. [DOI] [PubMed] [Google Scholar]
- 460.Nicholson, W. L., N. Munakata, G. Horneck, H. J. Melosh, and P. Setlow. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64:548-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Nishimura, Y., T. Ino, and H. Iizuka. 1988. Acinetobacter radioresistens sp. nov. isolated from cotton and soil. Int. J. Syst. Bacteriol. 38:209-211. [Google Scholar]
- 462.Niwa, O., S. K. Bryan, and R. E. Moses. 1981. Alternate pathways of DNA replication: DNA polymerase I-dependent replication. Proc. Natl. Acad. Sci. U. S. A. 78:7024-7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Noll, D. M., T. M. Mason, and P. S. Miller. 2006. Formation and repair of interstrand cross-links in DNA. Chem. Rev. 106:277-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Norais, C. A., S. Chitteni-Pattu, E. A. Wood, R. B. Inman, and M. M. Cox. 2009. DdrB Protein, an alternative Deinococcus radiodurans SSB induced by ionizing radiation. J. Biol. Chem. 284:21402-21411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Nystrom, T. 2005. Role of oxidative carbonylation in protein quality control and senescence. EMBO J. 24:1311-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Obiero, J., V. Pittet, S. A. Bonderoff, and D. A. Sanders. 2010. The thioredoxin system from Deinococcus radiodurans. J. Bacteriol. 192:494-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Ogawa, Y., et al. 2004. Immunocytochemical characteristics of human osteosarcoma cell line HS-Os-1: possible implication in apoptotic resistance against irradiation. Int. J. Mol. Med. 14:397-403. [PubMed] [Google Scholar]
- 468.Ohba, H., K. Satoh, H. Sghaier, T. Yanagisawa, and I. Narumi. 2009. Identification of PprM: a modulator of the PprI-dependent DNA damage response in Deinococcus radiodurans. Extremophiles 13:471-479. [DOI] [PubMed] [Google Scholar]
- 469.Ohtani, N., M. Tomita, and M. Itaya. 2010. An extreme thermophile, Thermus thermophilus, is a polyploid bacterium. J. Bacteriol. 192:5499-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Ohwada, K., et al. 2008. Pyrroloquinoline quinone (PQQ) prevents cognitive deficit caused by oxidative stress in rats. J. Clin. Biochem. Nutr. 42:29-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Okazaki, R., M. Arisawa, and A. Sugino. 1971. Slow joining of newly replicated DNA chains in DNA polymerase I-deficient Escherichia coli mutants. Proc. Natl. Acad. Sci. U. S. A. 68:2954-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Okazawa, Y., and A. Matsuyama. 1967. A note on radiation resistance of Micrococcus radiodurans. Agric. Biol. Chem. 31:1505-1508. [Google Scholar]
- 473.Olendzenski, L., et al. 2000. Horizontal transfer of archaeal genes into the Deinococcaceae: detection by molecular and computer-based approaches. J. Mol. Evol. 51:587-599. [DOI] [PubMed] [Google Scholar]
- 474.Oliver, C. N., B. W. Ahn, E. J. Moerman, S. Goldstein, and E. R. Stadtman. 1987. Age-related changes in oxidized proteins. J. Biol. Chem. 262:5488-5491. [PubMed] [Google Scholar]
- 475.Omelchenko, M. V., et al. 2005. Comparative genomics of Thermus thermophilus and Deinococcus radiodurans: divergent routes of adaptation to thermophily and radiation resistance. BMC Evol. Biol. 5:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Omenn, G. S., et al. 1996. Effects of a combination of beta carotene and vitamin A on lung cancer and cardiovascular disease. N. Engl. J. Med. 334:1150-1155. [DOI] [PubMed] [Google Scholar]
- 477.Ormerod, M. G., and A. R. Lehmann. 1971. The release of high molecular weight DNA from a mammalian cell (L-5178Y). Attachment of the DNA to the nuclear membrane. Biochim. Biophys. Acta 228:331-343. [PubMed] [Google Scholar]
- 478.Oyaizu, H. E., et al. 1987. A radiation-resistant rod-shaped bacterium, Deinobacter grandis gen. nov., sp. nov., with peptidoglycan containing ornithine. Int. J. Syst. Bacteriol. 37:62-67. [Google Scholar]
- 479.Pan, J., et al. 2009. IrrE, a global regulator of extreme radiation resistance in Deinococcus radiodurans, enhances salt tolerance in Escherichia coli and Brassica napus. PLoS One 4:e4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Paques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Parisi, A., and A. D. Antoine. 1974. Increased radiation resistance of vegetative Bacillus pumilus. Appl. Microbiol. 28:41-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Parsons, R., et al. 1993. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell 75:1227-1236. [DOI] [PubMed] [Google Scholar]
- 483.Pask-Hughes, R., and R. A. Williams. 1978. Cell envelope components of strains belonging to genus Thermus. J. Gen. Microbiol. 107:65-72. [Google Scholar]
- 484.Pasternak, C., et al. 2010. Irradiation-induced Deinococcus radiodurans genome fragmentation triggers transposition of a single resident insertion sequence. PLoS Genet. 6:e1000799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Pavlov, A. K., V. L. Kalinin, A. N. Konstantinov, V. N. Shelegedin, and A. A. Pavlov. 2006. Was Earth ever infected by martian biota? Clues from radioresistant bacteria. Astrobiology 6:911-918. [DOI] [PubMed] [Google Scholar]
- 486.Paz-Elizur, T., et al. 2008. DNA repair of oxidative DNA damage in human carcinogenesis: potential application for cancer risk assessment and prevention. Cancer Lett. 266:60-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Peng, F., et al. 2009. Deinococcus xinjiangensis sp. nov., isolated from desert soil. Int. J. Syst. Evol. Microbiol. 59:709-713. [DOI] [PubMed] [Google Scholar]
- 488.Perez, V. I., et al. 2009. The overexpression of major antioxidant enzymes does not extend the lifespan of mice. Aging Cell 8:73-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Petit, C., and A. Sancar. 1999. Nucleotide excision repair: from E. coli to man. Biochimie 81:15-25. [DOI] [PubMed] [Google Scholar]
- 490.Petit, M. A., J. Dimpfl, M. Radman, and H. Echols. 1991. Control of large chromosomal duplications in Escherichia coli by the mismatch repair system. Genetics 129:327-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Petrini, J. H. 2000. The Mre11 complex and ATM: collaborating to navigate S phase. Curr. Opin. Cell Biol. 12:293-296. [DOI] [PubMed] [Google Scholar]
- 492.Phillips, R. W., et al. 2002. Kineococcus radiotolerans sp. nov., a radiation-resistant, Gram-positive bacterium. Int. J. Syst. Evol. Microbiol. 52:933-938. [DOI] [PubMed] [Google Scholar]
- 493.Picklo, M. J., T. J. Montine, V. Amarnath, and M. D. Neely. 2002. Carbonyl toxicology and Alzheimer's disease. Toxicol. Appl. Pharmacol. 184:187-197. [DOI] [PubMed] [Google Scholar]
- 494.Pogoda de la Vega, U., P. Rettberg, T. Douki, J. Cadet, and G. Horneck. 2005. Sensitivity to polychromatic UV-radiation of strains of Deinococcus radiodurans differing in their DNA repair capacity. Int. J. Radiat. Biol. 81:601-611. [DOI] [PubMed] [Google Scholar]
- 495.Potts, M. 1994. Desiccation tolerance of prokaryotes. Microbiol. Rev. 58:755-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Potts, M. 2001. Desiccation tolerance: a simple process? Trends Microbiol. 9:553-559. [DOI] [PubMed] [Google Scholar]
- 497.Prasad, B. J., K. Sabnis, D. D. Deobagkar, and D. N. Deobagkar. 2005. Deinococcus radiodurans strain R1 contains N6-methyladenine in its genome. Biochem. Biophys. Res. Commun. 335:412-416. [DOI] [PubMed] [Google Scholar]
- 498.Previte, J. J., Y. Chang, and H. M. el-Bisi. 1971. Effects of radiation pasteurization on Samonella. 3. Radiation lethality and the frequency of mutation to antibiotic resistance. Can. J. Microbiol. 17:385-389. [DOI] [PubMed] [Google Scholar]
- 499.Prise, K. M., et al. 1998. A review of dsb induction data for varying quality radiations. Int. J. Radiat. Biol. 74:173-184. [DOI] [PubMed] [Google Scholar]
- 500.Rainey, F. A., et al. 2007. Deinococcus peraridilitoris sp. nov., isolated from a coastal desert. Int. J. Syst. Evol. Microbiol. 57:1408-1412. [DOI] [PubMed] [Google Scholar]
- 501.Rainey, F. A., et al. 2005. Extensive diversity of ionizing-radiation-resistant bacteria recovered from Sonoran Desert soil and description of nine new species of the genus Deinococcus obtained from a single soil sample. Appl. Environ. Microbiol. 71:5225-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Raj, H. D., et al. 1960. Utilization of carbohydrates and amino acids by Micrococcus radiodurans. Can. J. Microbiol. 6:289-298. [DOI] [PubMed] [Google Scholar]
- 503.Rajan, R., and C. E. Bell. 2004. Crystal structure of RecA from Deinococcus radiodurans: insights into the structural basis of extreme radioresistance. J. Mol. Biol. 344:951-963. [DOI] [PubMed] [Google Scholar]
- 504.Rajpurohit, Y. S., R. Gopalakrishnan, and H. S. Misra. 2008. Involvement of a protein kinase activity inducer in DNA double strand break repair and radioresistance of Deinococcus radiodurans. J. Bacteriol. 190:3948-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Rajpurohit, Y. S., and H. S. Misra. 2010. Characterization of a DNA damage inducible membrane protein kinase from Deinococcus radiodurans and its role in bacterial radioresistance and DNA strand break repair. Mol. Microbiol. 77:1470-1482. [DOI] [PubMed] [Google Scholar]
- 506.Rajski, S. R., and R. M. Williams. 1998. DNA cross-linking agents as antitumor drugs. Chem. Rev. 98:2723-2796. [DOI] [PubMed] [Google Scholar]
- 507.Ramirez, C. L., J. Cadinanos, I. Varela, J. M. Freije, and C. Lopez-Otin. 2007. Human progeroid syndromes, aging and cancer: new genetic and epigenetic insights into old questions. Cell. Mol. Life Sci. 64:155-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.Rao, N. N., M. R. Gomez-Garcia, and A. Kornberg. 2009. Inorganic polyphosphate: essential for growth and survival. Annu. Rev. Biochem. 78:605-647. [DOI] [PubMed] [Google Scholar]
- 509.Ravin, A. W. 1961. The genetics of transformation. Adv. Genet. 10:61-163. [DOI] [PubMed] [Google Scholar]
- 510.Rayssiguier, C., D. S. Thaler, and M. Radman. 1989. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature 342:396-401. [DOI] [PubMed] [Google Scholar]
- 511.Rebeyrotte, N. 1983. Induction of mutation in Micrococcus radiodurans by N-methyl-N′-nitro-N-nitrosoguanidine. Mutat. Res. 108:57-66. [DOI] [PubMed] [Google Scholar]
- 512.Reddi, A. R., L. T. Jensen, and V. C. Culotta. 2009. Manganese homeostasis in Saccharomyces cerevisiae. Chem. Rev. 109:4722-4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513.Reddi, A. R., et al. 2009. The overlapping roles of manganese and Cu/Zn SOD in oxidative stress protection. Free Radic. Biol. Med. 46:154-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Repar, J., et al. 2010. RecA protein assures fidelity of DNA repair and genome stability in Deinococcus radiodurans. DNA Repair (Amst.) 9:1151-1161. [DOI] [PubMed] [Google Scholar]
- 515.Resnick, M. A. 1978. Similar responses to ionizing radiation of fungal and vertebrate cells and the importance of DNA doublestrand breaks. J. Theor. Biol. 71:339-346. [DOI] [PubMed] [Google Scholar]
- 516.Resnick, M. A., and P. Martin. 1976. The repair of double-strand breaks in the nuclear DNA of Saccharomyces cerevisiae and its genetic control. Mol. Gen. Genet. 143:119-129. [DOI] [PubMed] [Google Scholar]
- 517.Rew, D. A. 2003. Deinococcus radiodurans. Eur. J. Surg. Oncol. 29:557-558. [DOI] [PubMed] [Google Scholar]
- 518.Reyes, E. D., P. L. Patidar, L. A. Uranga, A. S. Bortoletto, and S. L. Lusetti. 2010. RecN is a cohesin-like protein that stimulates intermolecular DNA interactions in vitro. J. Biol. Chem. 285:16521-16529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519.Richmond, R. C., R. Sridhar, and M. J. Daly. 1999. Physicochemical survival pattern for the radiophile Deinococcus radiodurans: a polyextremophile model for life on Mars. SPIE 3755:210-222. [Google Scholar]
- 520.Rigby, P. W., M. Dieckmann, C. Rhodes, and P. Berg. 1977. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J. Mol. Biol. 113:237-251. [DOI] [PubMed] [Google Scholar]
- 521.Rocha, E. P., E. Cornet, and B. Michel. 2005. Comparative and evolutionary analysis of the bacterial homologous recombination systems. PLoS Genet. 1:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Rothkamm, K., and M. Lobrich. 2003. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low X-ray doses. Proc. Natl. Acad. Sci. U. S. A. 100:5057-5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Rotman, G., and Y. Shiloh. 1998. ATM: from gene to function. Hum. Mol. Genet. 7:1555-1563. [DOI] [PubMed] [Google Scholar]
- 524.Rouault, T. A., and R. D. Klausner. 1996. Iron-sulfur clusters as biosensors of oxidants and iron. Trends Biochem. Sci. 21:174-177. [PubMed] [Google Scholar]
- 525.Rouviere-Yaniv, J., M. Yaniv, and J. E. Germond. 1979. E. coli DNA binding protein HU forms nucleosomelike structure with circular double-stranded DNA. Cell 17:265-274. [DOI] [PubMed] [Google Scholar]
- 526.Rudolph, C. J., A. L. Upton, and R. G. Lloyd. 2007. Replication fork stalling and cell cycle arrest in UV-irradiated Escherichia coli. Genes Dev. 21:668-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 527.Saarimaa, C., et al. 2006. Characterization of adhesion threads of Deinococcus geothermalis as type IV pili. J. Bacteriol. 188:7016-7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 528.Saleh, Y. G., M. S. Mayo, and D. G. Ahearn. 1988. Resistance of some common fungi to gamma irradiation. Appl. Environ. Microbiol. 54:2134-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.Sancar, A., L. A. Lindsey-Boltz, K. Unsal-Kacmaz, and S. Linn. 2004. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73:39-85. [DOI] [PubMed] [Google Scholar]
- 530.Sanchez, H., and J. C. Alonso. 2005. Bacillus subtilis RecN binds and protects 3′-single-stranded DNA extensions in the presence of ATP. Nucleic Acids Res. 33:2343-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531.Sanchez, H., D. Kidane, M. Castillo Cozar, P. L. Graumann, and J. C. Alonso. 2006. Recruitment of Bacillus subtilis RecN to DNA double-strand breaks in the absence of DNA end processing. J. Bacteriol. 188:353-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 532.Sanchez, R. J., et al. 2005. Exogenous manganous ion at millimolar levels rescues all known dioxygen-sensitive phenotypes of yeast lacking CuZnSOD. J. Biol. Inorg. Chem. 10:913-923. [DOI] [PubMed] [Google Scholar]
- 533.Sanders, S. W., and R. B. Maxcy. 1979. Isolation of radiation-resistant bacteria without exposure to irradiation. Appl. Environ. Microbiol. 38:436-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.Sandigursky, M., S. Sandigursky, P. Sonati, M. J. Daly, and W. A. Franklin. 2004. Multiple uracil-DNA glycosylase activities in Deinococcus radiodurans. DNA Repair (Amst.) 3:163-169. [DOI] [PubMed] [Google Scholar]
- 535.Sargentini, N. J., and K. C. Smith. 1986. Quantitation of the involvement of the recA, recB, recC, recF, recJ, recN, lexA, radA, radB, uvrD, and umuC genes in the repair of X-ray-induced DNA double-strand breaks in Escherichia coli. Radiat. Res. 107:58-72. [PubMed] [Google Scholar]
- 536.Satoh, K., et al. 2002. Characterization of RecA424 and RecA670 proteins from Deinococcus radiodurans. J. Biochem. 131:121-129. [DOI] [PubMed] [Google Scholar]
- 537.Satoh, K., H. Ohba, H. Sghaier, and I. Narumi. 2006. Down-regulation of radioresistance by LexA2 in Deinococcus radiodurans. Microbiology 152:3217-3226. [DOI] [PubMed] [Google Scholar]
- 538.Satoh, K., et al. 2006. Method for detecting DNA strand breaks in mammalian cells using the Deinococcus radiodurans PprA protein. Mutat. Res. 596:36-42. [DOI] [PubMed] [Google Scholar]
- 539.Schafer, M., C. Schmitz, and G. Horneck. 1998. High sensitivity of Deinococcus radiodurans to photodynamically-produced singlet oxygen. Int. J. Radiat. Biol. 74:249-253. [DOI] [PubMed] [Google Scholar]
- 540.Schein, A., B. J. Berdahl, M. Low, and E. Borek. 1972. Deficiency of the DNA of Micrococcus radiodurans in methyladenine and methylcytosine. Biochim. Biophys. Acta 272:481-485. [DOI] [PubMed] [Google Scholar]
- 541.Schlesinger, D. J. 2007. Role of RecA in DNA damage repair in Deinococcus radiodurans. FEMS Microbiol. Lett. 274:342-347. [DOI] [PubMed] [Google Scholar]
- 542.Schumacher, B., G. A. Garinis, and J. H. Hoeijmakers. 2008. Age to survive: DNA damage and aging. Trends Genet. 24:77-85. [DOI] [PubMed] [Google Scholar]
- 543.Seaver, L. C., and J. A. Imlay. 2001. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 183:7173-7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Seeberg, E., L. Eide, and M. Bjoras. 1995. The base excision repair pathway. Trends Biochem. Sci. 20:391-397. [DOI] [PubMed] [Google Scholar]
- 545.Selby, C. P., and A. Sancar. 1990. Structure and function of the (A)BC excinuclease of Escherichia coli. Mutat. Res. 236:203-211. [DOI] [PubMed] [Google Scholar]
- 546.Senturker, S., C. Bauche, J. Laval, and M. Dizdaroglu. 1999. Substrate specificity of Deinococcus radiodurans Fpg protein. Biochemistry 38:9435-9439. [DOI] [PubMed] [Google Scholar]
- 547.Seo, H. J., and Y. N. Lee. 2006. Occurrence of thioredoxin reductase in Deinococcus species, the UV resistant bacteria. J. Microbiol. 44:461-465. [PubMed] [Google Scholar]
- 548.Serianni, R. W., and A. K. Bruce. 1968. Radioresistance of Micrococcus radiodurans during the growth cycle. Radiat. Res. 36:193-207. [PubMed] [Google Scholar]
- 549.Serianni, R. W., and A. K. Bruce. 1968. Role of sulphur in radioprotective extracts of Micrococcus radiodurans. Nature 218:485-487. [DOI] [PubMed] [Google Scholar]
- 550.Servant, P., et al. 2007. The ClpPX protease is required for radioresistance and regulates cell division after gamma-irradiation in Deinococcus radiodurans. Mol. Microbiol. 66:1231-1239. [DOI] [PubMed] [Google Scholar]
- 551.Servinsky, M. D., and D. A. Julin. 2007. Effect of a recD mutation on DNA damage resistance and transformation in Deinococcus radiodurans. J. Bacteriol. 189:5101-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 552.Setlow, B., S. Atluri, R. Kitchel, K. Koziol-Dube, and P. Setlow. 2006. Role of dipicolinic acid in resistance and stability of spores of Bacillus subtilis with or without DNA-protective alpha/beta-type small acid-soluble proteins. J. Bacteriol. 188:3740-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553.Setlow, J. K., and M. E. Boling. 1965. The resistance of Micrococcus radiodurans to ultraviolet radiation. II. Action spectra for killing, delay in DNA synthesis, and thymine dimerization. Biochim. Biophys. Acta 108:259-265. [DOI] [PubMed] [Google Scholar]
- 554.Setlow, J. K., and D. E. Duggan. 1964. The resistance of Micrococcus radiodurans to ultraviolet radiation. I. Ultraviolet-induced lesions in the cell's DNA. Biochim. Biophys. Acta 87:664-668. [DOI] [PubMed] [Google Scholar]
- 555.Sghaier, H., I. Narumi, K. Satoh, H. Ohba, and H. Mitomo. 2007. Problems with the current deinococcal hypothesis: an alternative theory. Theory Biosci. 126:43-45. [DOI] [PubMed] [Google Scholar]
- 556.Shadrick, W. R., and D. A. Julin. 2010. Kinetics of DNA unwinding by the RecD2 helicase from Deinococcus radiodurans. J. Biol. Chem. 285:17292-17300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 557.Shapiro, A., D. DiLello, M. C. Loudis, D. E. Keller, and S. H. Hutner. 1977. Minimal requirements in defined media for improved growth of some radio-resistant pink tetracocci. Appl. Environ. Microbiol. 33:1129-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 558.Sharma, R. C., and K. C. Smith. 1987. Role of DNA polymerase I in postreplication repair: a reexamination with Escherichia coli delta polA. J. Bacteriol. 169:4559-4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 559.Shashidhar, R., and J. R. Bandekar. 2006. Deinococcus mumbaiensis sp. nov., a radiation-resistant pleomorphic bacterium isolated from Mumbai, India. FEMS Microbiol. Lett. 254:275-280. [DOI] [PubMed] [Google Scholar]
- 560.Shashidhar, R., and J. R. Bandekar. 2009. Deinococcus piscis sp. nov., a radiation-resistant bacterium isolated from marine fish. Int. J. Syst. Evol. Microbiol. 59:2714-2717. [DOI] [PubMed] [Google Scholar]
- 561.Shashidhar, R., S. A. Kumar, H. S. Misra, and J. R. Bandekar. 2010. Evaluation of the role of enzymatic and nonenzymatic antioxidant systems in the radiation resistance of Deinococcus. Can. J. Microbiol. 56:195-201. [DOI] [PubMed] [Google Scholar]
- 562.Sheng, D., et al. 2005. RecX is involved in antioxidant mechanisms of the radioresistant bacterium Deinococcus radiodurans. FEMS Microbiol. Lett. 244:251-257. [DOI] [PubMed] [Google Scholar]
- 563.Sheng, D., J. Jao, M. Li, P. Xu, and J. Zhang. 2009. RecX is involved in the switch between DNA damage response and normal metabolism in D. radiodurans. J. Biochem. 146:337-342. [DOI] [PubMed] [Google Scholar]
- 564.Sheng, D., et al. 2005. Dual negative regulatory mechanisms of RecX on RecA functions in radiation resistance, DNA recombination and consequent genome instability in Deinococcus radiodurans. DNA Repair (Amst.) 4:671-678. [DOI] [PubMed] [Google Scholar]
- 565.Sheng, D., Z. Zheng, B. Tian, B. Shen, and Y. Hua. 2004. LexA analog (dra0074) is a regulatory protein that is irrelevant to recA induction. J. Biochem. 136:787-793. [DOI] [PubMed] [Google Scholar]
- 566.Shibutani, S., M. Takeshita, and A. P. Grollman. 1991. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature 349:431-434. [DOI] [PubMed] [Google Scholar]
- 567.Shiloh, Y. 1997. Ataxia-telangiectasia and the Nijmegen breakage syndrome: related disorders but genes apart. Annu. Rev. Genet. 31:635-662. [DOI] [PubMed] [Google Scholar]
- 568.Shirkey, B., et al. 2003. Genomic DNA of Nostoc commune (Cyanobacteria) becomes covalently modified during long-term (decades) desiccation but is protected from oxidative damage and degradation. Nucleic Acids Res. 31:2995-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 569.Shukla, M., et al. 2007. Multiple-stress tolerance of ionizing radiation-resistant bacterial isolates obtained from various habitats: correlation between stresses. Curr. Microbiol. 54:142-148. [DOI] [PubMed] [Google Scholar]
- 570.Sitte, N., K. Merker, T. Von Zglinicki, T. Grune, and K. J. Davies. 2000. Protein oxidation and degradation during cellular senescence of human BJ fibroblasts. Part I. Effects of proliferative senescence. FASEB J. 14:2495-2502. [DOI] [PubMed] [Google Scholar]
- 571.Skalka, M., J. Matyasova, and M. Cejkova. 1976. DNA in chromatin of irradiated lymphoid tissues degrades in vivo into regular fragments. FEBS Lett. 72:271-274. [DOI] [PubMed] [Google Scholar]
- 572.Slade, D., A. B. Lindner, G. Paul, and M. Radman. 2009. Recombination and replication in DNA repair of heavily irradiated Deinococcus radiodurans. Cell 136:1044-1055. [DOI] [PubMed] [Google Scholar]
- 573.Sladek, F. M., M. M. Munn, W. D. Rupp, and P. Howard-Flanders. 1989. In vitro repair of psoralen-DNA cross-links by RecA, UvrABC, and the 5′-exonuclease of DNA polymerase I. J. Biol. Chem. 264:6755-6765. [PubMed] [Google Scholar]
- 574.Sleytr, U. B., M. T. Silva, M. Kocur, and N. F. Lewis. 1976. The fine structure of Micrococcus radiophilus and Micrococcus radioproteolyticus. Arch. Microbiol. 107:313-320. [DOI] [PubMed] [Google Scholar]
- 575.Smith, C. D., et al. 1991. Excess brain protein oxidation and enzyme dysfunction in normal aging and in Alzheimer disease. Proc. Natl. Acad. Sci. U. S. A. 88:10540-10543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 576.Smith, C. E., B. Llorente, and L. S. Symington. 2007. Template switching during break-induced replication. Nature 447:102-105. [DOI] [PubMed] [Google Scholar]
- 577.Smith, M. D., R. Abrahamson, and K. W. Minton. 1989. Shuttle plasmids constructed by the transformation of an Escherichia coli cloning vector into two Deinococcus radiodurans plasmids. Plasmid 22:132-142. [DOI] [PubMed] [Google Scholar]
- 578.Smith, M. D., E. Lennon, L. B. McNeil, and K. W. Minton. 1988. Duplication insertion of drug resistance determinants in the radioresistant bacterium Deinococcus radiodurans. J. Bacteriol. 170:2126-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 579.Smith, M. D., C. I. Masters, E. Lennon, L. B. McNeil, and K. W. Minton. 1991. Gene expression in Deinococcus radiodurans. Gene 98:45-52. [DOI] [PubMed] [Google Scholar]
- 580.Smythe, C. V. 1936. The reactions of iodoacetate and of iodoacetamide with various sulfhydryl groups, with urease, and with yeast preparations. J. Biol. Chem. 114:601-612. [Google Scholar]
- 581.Sohal, R. S., S. Agarwal, and B. H. Sohal. 1995. Oxidative stress and aging in the Mongolian gerbil (Meriones unguiculatus). Mech. Ageing Dev. 81:15-25. [DOI] [PubMed] [Google Scholar]
- 582.Solinger, J. A., K. Kiianitsa, and W. D. Heyer. 2002. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol. Cell 10:1175-1188. [DOI] [PubMed] [Google Scholar]
- 583.Solinger, J. A., G. Lutz, T. Sugiyama, S. C. Kowalczykowski, and W. D. Heyer. 2001. Rad54 protein stimulates heteroduplex DNA formation in the synaptic phase of DNA strand exchange via specific interactions with the presynaptic Rad51 nucleoprotein filament. J. Mol. Biol. 307:1207-1221. [DOI] [PubMed] [Google Scholar]
- 584.Stackebrandt, E., and C. R. Woese. 1981. Towards a phylogeny of the actinomycetes and related organisms Curr. Microbiol. 5:197-202. [Google Scholar]
- 585.Stadtman, E. R. 1992. Protein oxidation and aging. Science 257:1220-1224. [DOI] [PubMed] [Google Scholar]
- 586.Stahl, W., et al. 1998. Carotenoid mixtures protect multilamellar liposomes against oxidative damage: synergistic effects of lycopene and lutein. FEBS Lett. 427:305-308. [DOI] [PubMed] [Google Scholar]
- 587.Stahl, W., and H. Sies. 2003. Antioxidant activity of carotenoids. Mol. Aspects Med. 24:345-351. [DOI] [PubMed] [Google Scholar]
- 588.Stuy, J. H. 1960. Studies on the radiation inactivation of microorganisms. VI. X-ray-induced breakdown of deoxyribonucleic acid in Haemophilus influenzae and in other bacteria. J. Bacteriol. 79:707-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 589.Suciu, D., and O. Bojan. 1981. Autodigestion of chromatin in some radiosensitive and radioresistant mouse cells. Role of proteolysis and endonucleolysis. Int. J. Radiat. Biol. Relat. Stud. Phys Chem. Med. 39:273-279. [DOI] [PubMed] [Google Scholar]
- 590.Sugiman-Marangos, S., and M. S. Junop. 2010. The structure of DdrB from Deinococcus: a new fold for single-stranded DNA binding proteins. Nucleic Acids Res. 38:3432-3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 591.Suihko, M. L., et al. 2007. Characterization of aerobic bacterial and fungal microbiota on surfaces of historic Scottish monuments. Syst. Appl. Microbiol. 30:494-508. [DOI] [PubMed] [Google Scholar]
- 592.Sukhi, S. S., R. Shashidhar, S. A. Kumar, and J. R. Bandekar. 2009. Radiation resistance of Deinococcus radiodurans R1 with respect to growth phase. FEMS Microbiol. Lett. 297:49-53. [DOI] [PubMed] [Google Scholar]
- 593.Suminaga, R., et al. 2000. Non-homologous recombination between Alu and LINE-1 repeats caused a 430-kb deletion in the dystrophin gene: a novel source of genomic instability. J. Hum. Genet. 45:331-336. [DOI] [PubMed] [Google Scholar]
- 594.Sun, J., P. Shen, H. Chao, and B. Wu. 2009. Isolation and identification of a new radiation-resistant bacterium Deinococcus guangriensis sp. nov. and analysis of its radioresistant character. Wei Sheng Wu Xue Bao 49:918-924. (In Chinese.) [PubMed] [Google Scholar]
- 595.Suresh, K., G. S. Reddy, S. Sengupta, and S. Shivaji. 2004. Deinococcus indicus sp. nov., an arsenic-resistant bacterium from an aquifer in West Bengal, India. Int. J. Syst. Evol. Microbiol. 54:457-461. [DOI] [PubMed] [Google Scholar]
- 596.Sweet, D. M., and B. E. Moseley. 1974. Accurate repair of ultraviolet-induced damage in Micrococcus radiodurans. Mutat. Res. 23:311-318. [DOI] [PubMed] [Google Scholar]
- 597.Sweet, D. M., and B. E. Moseley. 1976. The resistance of Micrococcus radiodurans to killing and mutation by agents which damage DNA. Mutat. Res. 34:175-186. [DOI] [PubMed] [Google Scholar]
- 598.Taddei, F., et al. 1997. Counteraction by MutT protein of transcriptional errors caused by oxidative damage. Science 278:128-130. [DOI] [PubMed] [Google Scholar]
- 599.Tait, R. C., A. L. Harris, and D. W. Smith. 1974. DNA repair in Escherichia coli mutants deficient in DNA polymerases I, II and-or 3. Proc. Natl. Acad. Sci. U. S. A. 71:675-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 600.Tan, S. T., and R. B. Maxcy. 1986. Simple method to demonstrate radiation-inducible radiation resistance in microbial cells. Appl. Environ. Microbiol. 51:88-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 601.Tanaka, A., H. Hirano, M. Kikuchi, S. Kitayama, and H. Watanabe. 1996. Changes in cellular proteins of Deinococcus radiodurans following gamma-irradiation. Radiat. Environ. Biophys. 35:95-99. [DOI] [PubMed] [Google Scholar]
- 602.Tanaka, M., et al. 2004. Analysis of Deinococcus radiodurans's transcriptional response to ionizing radiation and desiccation reveals novel proteins that contribute to extreme radioresistance. Genetics 168:21-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 603.Tanaka, M., et al. 2005. Characterization of pathways dependent on the uvsE, uvrA1, or uvrA2 gene product for UV resistance in Deinococcus radiodurans. J. Bacteriol. 187:3693-3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 604.Tang, Y., and J. R. Guest. 1999. Direct evidence for mRNA binding and post-transcriptional regulation by Escherichia coli aconitases. Microbiology 145:3069-3079. [DOI] [PubMed] [Google Scholar]
- 605.Taniguchi, T., and A. D. D'Andrea. 2006. Molecular pathogenesis of Fanconi anemia: recent progress. Blood 107:4223-4233. [DOI] [PubMed] [Google Scholar]
- 606.Tao, R., et al. 2007. Pyrroloquinoline quinone preserves mitochondrial function and prevents oxidative injury in adult rat cardiac myocytes. Biochem. Biophys. Res. Commun. 363:257-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 607.Targovnik, H. S., and P. V. Hariharan. 1980. Excision repair of 5,6-dihydroxydihydrothymine from the DNA of Micrococcus radiodurans. Radiat. Res. 83:360-363. [PubMed] [Google Scholar]
- 608.Tatsuzawa, H., T. Maruyama, N. Misawa, K. Fujimori, and M. Nakano. 2000. Quenching of singlet oxygen by carotenoids produced in Escherichia coli—attenuation of singlet oxygen-mediated bacterial killing by carotenoids. FEBS Lett. 484:280-284. [DOI] [PubMed] [Google Scholar]
- 609.Taylor, A. F., and G. R. Smith. 2003. RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature 423:889-893. [DOI] [PubMed] [Google Scholar]
- 610.Tchou, J., et al. 1991. 8-Oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc. Natl. Acad. Sci. U. S. A. 88:4690-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 611.Tempest, P. R., and B. E. Moseley. 1980. Defective excision repair in a mutant of Micrococcus radiodurans hypermutable by some monofunctional alkylating agents. Mol. Gen. Genet. 179:191-199. [DOI] [PubMed] [Google Scholar]
- 612.Tempest, P. R., and B. E. Moseley. 1982. Lack of ultraviolet mutagenesis in radiation-resistant bacteria. Mutat. Res. 104:275-280. [DOI] [PubMed] [Google Scholar]
- 613.Thompson, B. G., R. Anderson, and R. G. Murray. 1980. Unusual polar lipids of Micrococcus radiodurans strain Sark. Can. J. Microbiol. 26:1408-1411. [DOI] [PubMed] [Google Scholar]
- 614.Thoms, B., and W. Wackernagel. 1998. Interaction of RecBCD enzyme with DNA at double-strand breaks produced in UV-irradiated Escherichia coli: requirement for DNA end processing. J. Bacteriol. 180:5639-5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 615.Thornley, M. J., R. W. Horne, and A. M. Glauert. 1965. The fine structure of Micrococcus radiodurans. Arch. Mikrobiol. 51:267-289. [DOI] [PubMed] [Google Scholar]
- 616.Tian, B., et al. 2009. Effects of carotenoids from Deinococcus radiodurans on protein oxidation. Lett. Appl. Microbiol. 49:689-694. [DOI] [PubMed] [Google Scholar]
- 617.Tian, B., et al. 2004. Chemiluminescence assay for reactive oxygen species scavenging activities and inhibition on oxidative damage of DNA in Deinococcus radiodurans. Luminescence 19:78-84. [DOI] [PubMed] [Google Scholar]
- 618.Tian, B., Z. Xu, Z. Sun, J. Lin, and Y. Hua. 2007. Evaluation of the antioxidant effects of carotenoids from Deinococcus radiodurans through targeted mutagenesis, chemiluminescence, and DNA damage analyses. Biochim. Biophys. Acta 1770:902-911. [DOI] [PubMed] [Google Scholar]
- 619.Timmins, J., I. Leiros, and S. McSweeney. 2007. Crystal structure and mutational study of RecOR provide insight into its mode of DNA binding. EMBO J. 26:3260-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 620.Tirgari, S., and B. E. Moseley. 1980. Transformation in Micrococcus radiodurans: measurement of various parameters and evidence for multiple, independently segregating genomes per cell. J. Gen. Microbiol. 119:287-296. [Google Scholar]
- 621.Tomasz, M., et al. 1987. Isolation and structure of a covalent cross-link adduct between mitomycin C and DNA. Science 235:1204-1208. [DOI] [PubMed] [Google Scholar]
- 622.Tsai-Wu, J. J., H. F. Liu, and A. L. Lu. 1992. Escherichia coli MutY protein has both N-glycosylase and apurinic/apyrimidinic endonuclease activities on A.C and A.G mispairs. Proc. Natl. Acad. Sci. U. S. A. 89:8779-8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 623.Tsaneva, I. R., B. Muller, and S. C. West. 1992. ATP-dependent branch migration of Holliday junctions promoted by the RuvA and RuvB proteins of E. coli. Cell 69:1171-1180. [DOI] [PubMed] [Google Scholar]
- 624.Tsaneva, I. R., B. Muller, and S. C. West. 1993. RuvA and RuvB proteins of Escherichia coli exhibit DNA helicase activity in vitro. Proc. Natl. Acad. Sci. U. S. A. 90:1315-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 625.Tseng, H. J., Y. Srikhanta, A. G. McEwan, and M. P. Jennings. 2001. Accumulation of manganese in Neisseria gonorrhoeae correlates with resistance to oxidative killing by superoxide anion and is independent of superoxide dismutase activity. Mol. Microbiol. 40:1175-1186. [DOI] [PubMed] [Google Scholar]
- 626.Turner, R. J., J. H. Weiner, and D. E. Taylor. 1999. Tellurite-mediated thiol oxidation in Escherichia coli. Microbiology 145:2549-2557. [DOI] [PubMed] [Google Scholar]
- 627.Tyrrell, R. M., and S. M. Keyse. 1990. New trends in photobiology. The interaction of UVA radiation with cultured cells. J. Photochem. Photobiol. B 4:349-361. [DOI] [PubMed] [Google Scholar]
- 628.Udupa, K. S., P. A. O'Cain, V. Mattimore, and J. R. Battista. 1994. Novel ionizing radiation-sensitive mutants of Deinococcus radiodurans. J. Bacteriol. 176:7439-7446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 629.Ujaoney, A. K., A. A. Potnis, P. Kane, R. Mukhopadhyaya, and S. K. Apte. 2010. Radiation desiccation response motif-like sequences are involved in transcriptional activation of the deinococcal ssb gene by ionizing radiation but not by desiccation. J. Bacteriol. 192:5637-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 630.Umansky, S. R., B. A. Korol, and P. A. Nelipovich. 1981. In vivo DNA degradation in thymocytes of gamma-irradiated or hydrocortisone-treated rats. Biochim. Biophys. Acta 655:9-17. [DOI] [PubMed] [Google Scholar]
- 631.Valenzeno, D. P. 1987. Photomodification of biological membranes with emphasis on singlet oxygen mechanisms. Photochem. Photobiol. 46:147-160. [DOI] [PubMed] [Google Scholar]
- 632.van Gent, D. C., J. H. Hoeijmakers, and R. Kanaar. 2001. Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. Genet. 2:196-206. [DOI] [PubMed] [Google Scholar]
- 633.van Gent, D. C., and M. van der Burg. 2007. Non-homologous end-joining, a sticky affair. Oncogene 26:7731-7740. [DOI] [PubMed] [Google Scholar]
- 634.van Gerwen, S. J., F. M. Rombouts, K. van't Riet, and M. H. Zwietering. 1999. A data analysis of the irradiation parameter D10 for bacteria and spores under various conditions. J. Food Prot. 62:1024-1032. [DOI] [PubMed] [Google Scholar]
- 635.Varghese, A. J., and R. S. Day III. 1970. Excision of cytosine-thymine adduct from the DNA of ultraviolet-irradiated Micrococcus radiodurans. Photochem. Photobiol. 11:511-517. [DOI] [PubMed] [Google Scholar]
- 636.Veaute, X., et al. 2005. UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J. 24:180-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 637.Venkateswaran, A., et al. 2000. Physiologic determinants of radiation resistance in Deinococcus radiodurans. Appl. Environ. Microbiol. 66:2620-2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 638.Verrax, J., and P. B. Calderon. 2008. The controversial place of vitamin C in cancer treatment. Biochem. Pharmacol. 76:1644-1652. [DOI] [PubMed] [Google Scholar]
- 639.Vidal, A., N. Abril, and C. Pueyo. 1998. DNA sequence analysis of spontaneous lacI-d mutations in O6-alkylguanine-DNA alkyltransferase-proficient and -deficient Escherichia coli. Mutagenesis 13:367-373. [DOI] [PubMed] [Google Scholar]
- 640.Volkov, A., J. Mascarenhas, C. Andrei-Selmer, H. D. Ulrich, and P. L. Graumann. 2003. A prokaryotic condensin/cohesin-like complex can actively compact chromosomes from a single position on the nucleoid and binds to DNA as a ring-like structure. Mol. Cell. Biol. 23:5638-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 641.von Frijtag, J. K., D. Kunzel, J. van der Zee, and A. P. Ijzerman. 1996. Radical scavenging properties of adenosine and derivatives in vitro. Drug Develop. Res. 37:48-54. [Google Scholar]
- 642.Vujicic-Zagar, A., et al. 2009. Crystal structure of the IrrE protein, a central regulator of DNA damage repair in Deinococcaceae. J. Mol. Biol. 386:704-716. [DOI] [PubMed] [Google Scholar]
- 643.Vukovic-Nagy, B., B. W. Fox, and M. Fox. 1974. The release of a deoxyribonucleic acid fragment after X-irradiation of Micrococcus radiodurans. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 25:329-337. [DOI] [PubMed] [Google Scholar]
- 644.Wang, J., and D. A. Julin. 2004. DNA helicase activity of the RecD protein from Deinococcus radiodurans. J. Biol. Chem. 279:52024-52032. [DOI] [PubMed] [Google Scholar]
- 645.Wang, L., et al. 2008. DrRRA: a novel response regulator essential for the extreme radioresistance of Deinococcus radiodurans. Mol. Microbiol. 67:1211-1222. [DOI] [PubMed] [Google Scholar]
- 646.Wang, P., and H. E. Schellhorn. 1995. Induction of resistance to hydrogen peroxide and radiation in Deinococcus radiodurans. Can. J. Microbiol. 41:170-176. [DOI] [PubMed] [Google Scholar]
- 647.Wang, W., et al. 2009. Deinococcus wulumuqiensis sp. nov., and Deinococcus xibeiensis sp. nov., isolated from radiation-polluted soil. Int. J. Syst. Evol. Microbiol. 60:2006-2010. [DOI] [PubMed] [Google Scholar]
- 648.Wang, X., et al. 2004. Role of DNA replication proteins in double-strand break-induced recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 24:6891-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 649.Weinstock, D. M., C. A. Richardson, B. Elliott, and M. Jasin. 2006. Modeling oncogenic translocations: distinct roles for double-strand break repair pathways in translocation formation in mammalian cells. DNA Repair (Amst.) 5:1065-1074. [DOI] [PubMed] [Google Scholar]
- 650.Weisburg, W. G., S. J. Giovannoni, and C. R. Woese. 1989. The Deinococcus-Thermus phylum and the effect of rRNA composition on phylogenetic tree construction. Syst. Appl. Microbiol. 11:128-134. [DOI] [PubMed] [Google Scholar]
- 651.Welsh, D. T., and R. A. Herbert. 1999. Osmotically induced intracellular trehalose, but not glycine betaine accumulation promotes desiccation tolerance in Escherichia coli. FEMS Microbiol. Lett. 174:57-63. [DOI] [PubMed] [Google Scholar]
- 652.Whitby, M. C., L. Ryder, and R. G. Lloyd. 1993. Reverse branch migration of Holliday junctions by RecG protein: a new mechanism for resolution of intermediates in recombination and DNA repair. Cell 75:341-350. [DOI] [PubMed] [Google Scholar]
- 653.White, O., et al. 1999. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286:1571-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 654.Wierowski, J. V., and A. K. Bruce. 1980. Modification of radiation resistance by manganese in Micrococcus radiodurans. Radiat. Res. 83:384. [Google Scholar]
- 655.Willcox, J. K., S. L. Ash, and G. L. Catignani. 2004. Antioxidants and prevention of chronic disease. Crit. Rev. Food Sci. Nutr. 44:275-295. [DOI] [PubMed] [Google Scholar]
- 656.Witte, G., C. Urbanke, and U. Curth. 2005. Single-stranded DNA-binding protein of Deinococcus radiodurans: a biophysical characterization. Nucleic Acids Res. 33:1662-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 657.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 658.Wolf, Y. I., I. B. Rogozin, N. V. Grishin, R. L. Tatusov, and E. V. Koonin. 2001. Genome trees constructed using five different approaches suggest new major bacterial clades. BMC Evol. Biol. 1:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 659.Wolfe-Simon, F., V. Starovoytov, J. R. Reinfelder, O. Schofield, and P. G. Falkowski. 2006. Localization and role of manganese superoxide dismutase in a marine diatom. Plant Physiol. 142:1701-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 660.Wood, H. G., and J. E. Clark. 1988. Biological aspects of inorganic polyphosphates. Annu. Rev. Biochem. 57:235-260. [DOI] [PubMed] [Google Scholar]
- 661.Work, E. 1964. Amino acids of walls of Micrococcus radiodurans. Nature 201:1107-1109. [DOI] [PubMed] [Google Scholar]
- 662.Work, E., and H. Griffiths. 1968. Morphology and chemistry of cell walls of Micrococcus radiodurans. J. Bacteriol. 95:641-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 663.Wu, Y., W. Chen, Y. Zhao, H. Xu, and Y. Hua. 2009. Involvement of RecG in H2O2-induced damage repair in Deinococcus radiodurans. Can. J. Microbiol. 55:841-848. [DOI] [PubMed] [Google Scholar]
- 664.Xu, G., et al. 2010. DdrB stimulates single-stranded DNA annealing and facilitates RecA-independent DNA repair in Deinococcus radiodurans. DNA Repair (Amst.) 9:805-812. [DOI] [PubMed] [Google Scholar]
- 665.Xu, G., et al. 2008. RecO is essential for DNA damage repair in Deinococcus radiodurans. J. Bacteriol. 190:2624-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 666.Xu, W., J. Shen, C. A. Dunn, S. Desai, and M. J. Bessman. 2001. The Nudix hydrolases of Deinococcus radiodurans. Mol. Microbiol. 39:286-290. [DOI] [PubMed] [Google Scholar]
- 667.Yagil, E., and I. R. Beacham. 1975. Uptake of adenosine 5′-monophosphate by Escherichia coli. J. Bacteriol. 121:401-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 668.Yan, Z. Y., Z. J. Xu, G. Z. Xu, B. Tian, and Y. J. Hua. 2007. Construction of a dps mutant and its functional analysis in Deinococcus radiodurans. Wei Sheng Wu Xue Bao 47:610-615. (In Chinese.) [PubMed] [Google Scholar]
- 669.Yang, Y., et al. 2009. Deinococcus aerius sp. nov., isolated from the high atmosphere. Int. J. Syst. Evol. Microbiol. 59:1862-1866. [DOI] [PubMed] [Google Scholar]
- 670.Yang, Y., et al. 2010. Deinococcus aetherius sp. nov., isolated from the stratosphere. Int. J. Syst. Evol. Microbiol. 60:776-779. [DOI] [PubMed] [Google Scholar]
- 671.Yin, L., et al. 2010. DRA0336, another OxyR homolog, involved in the antioxidation mechanisms in Deinococcus radiodurans. J. Microbiol. 48:473-479. [DOI] [PubMed] [Google Scholar]
- 672.Yonemasu, R., et al. 1997. Characterization of the alternative excision repair pathway of UV-damaged DNA in Schizosaccharomyces pombe. Nucleic Acids Res. 25:1553-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 673.Yoo, S. H., et al. 2009. Deinococcus aerolatus sp. nov. and Deinococcus aerophilus sp. nov., isolated from air samples. Int. J. Syst. Evol. Microbiol. 60:1191-1195. [DOI] [PubMed] [Google Scholar]
- 674.Yoshinaka, T., K. Yano, and H. Yamaguchi. 1973. Isolation of highly radio-resistant bacterium, Arthrobacter radiotolerans nov. sp. Agric. Biol. Chem. 37:2269-2275. [Google Scholar]
- 675.Yuan, M., et al. 2009. Deinococcus gobiensis sp. nov., an extremely radiation-resistant bacterium. Int. J. Syst. Evol. Microbiol. 59:1513-1517. [DOI] [PubMed] [Google Scholar]
- 676.Zahradka, K., et al. 2006. Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature 443:569-573. [DOI] [PubMed] [Google Scholar]
- 677.Zhang, C., et al. 2005. Proteomic analysis of Deinococcus radiodurans recovering from gamma-irradiation. Proteomics 5:138-143. [DOI] [PubMed] [Google Scholar]
- 678.Zhang, H. Y., X. J. Li, N. Gao, and L. L. Chen. 2009. Antioxidants used by Deinococcus radiodurans and implications for antioxidant drug discovery. Nat. Rev. Microbiol. 7:476. [DOI] [PubMed] [Google Scholar]
- 679.Zhang, L., et al. 2007. Knockout of crtB or crtI gene blocks the carotenoid biosynthetic pathway in Deinococcus radiodurans R1 and influences its resistance to oxidative DNA-damaging agents due to change of free radicals scavenging ability. Arch. Microbiol. 188:411-419. [DOI] [PubMed] [Google Scholar]
- 680.Zhang, P., and S. T. Omaye. 2000. Beta-carotene and protein oxidation: effects of ascorbic acid and alpha-tocopherol. Toxicology 146:37-47. [DOI] [PubMed] [Google Scholar]
- 681.Zhang, Y., et al. 2010. irrE, an exogenous gene from Deinococcus radiodurans, improves the growth of and ethanol production by a Zymomonas mobilis strain under ethanol and acid stress. J. Microbiol. Biotechnol. 20:1156-1162. [DOI] [PubMed] [Google Scholar]
- 682.Zhang, Y. M., J. K. Liu, and T. Y. Wong. 2003. The DNA excision repair system of the highly radioresistant bacterium Deinococcus radiodurans is facilitated by the pentose phosphate pathway. Mol. Microbiol. 48:1317-1323. [DOI] [PubMed] [Google Scholar]
- 683.Zhang, Y. M., T. Y. Wong, L. Y. Chen, C. S. Lin, and J. K. Liu. 2000. Induction of a futile Embden-Meyerhof-Parnas pathway in Deinococcus radiodurans by Mn: possible role of the pentose phosphate pathway in cell survival. Appl. Environ. Microbiol. 66:105-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 684.Zhivotovsky, B. D., N. B. Zvonareva, and K. P. Hanson. 1981. Characteristics of rat thymus chromatin degradation products after whole-body X-irradiation. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 39:437-440. [DOI] [PubMed] [Google Scholar]
- 685.Zhou, B. B., and S. J. Elledge. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408:433-439. [DOI] [PubMed] [Google Scholar]
- 686.Zhou, Q., X. Zhang, H. Xu, B. Xu, and Y. Hua. 2007. A new role of Deinococcus radiodurans RecD in antioxidant pathway. FEMS Microbiol. Lett. 271:118-125. [DOI] [PubMed] [Google Scholar]
- 687.Zhou, Q., X. Zhang, H. Xu, B. Xu, and Y. Hua. 2006. RadA: a protein involved in DNA damage repair processes of Deinococcus radiodurans R1. Chin. Sci. Bull. 51:2993-2999. [Google Scholar]
- 688.Zimmerman, J. M., and J. R. Battista. 2005. A ring-like nucleoid is not necessary for radioresistance in the Deinococcaceae. BMC Microbiol. 5:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.