Abstract
Chlamydiae are well known for their species specificity and tissue tropism, and yet the individual species and strains show remarkable genomic synteny and share an intracellular developmental cycle unique in the microbial world. Only a relatively few chlamydial genes have been linked to specific disease or tissue tropism. Here we show that chlamydial species associated with human infections, Chlamydia trachomatis and C. pneumoniae, exhibit unique requirements for Src-family kinases throughout their developmental cycle. Utilization of Src-family kinases by C. trachomatis includes tyrosine phosphorylation of the secreted effector Tarp during the entry process, a functional role in microtubule-dependent trafficking to the microtubule organizing center, and a requirement for Src-family kinases for successful initiation of development. Nonhuman chlamydial species C. caviae and C. muridarum show none of these requirements and, instead, appear to be growth restricted by the activities of Src-family kinases. Depletion of Src-family kinases triggers a more rapid development of C. caviae with up to an 800% increase in infectious progeny production. Collectively, the results suggest that human chlamydial species have evolved requirements for tyrosine phosphorylation by Src-family kinases that are not seen in other chlamydial species. The requirement for Src-family kinases thus represents a fundamental distinction between chlamydial species that would not be readily apparent in genomic comparisons and may provide insights into chlamydial disease association and species specificity.
IMPORTANCE
Chlamydiae are well known for their species specificity and tissue tropism as well as their association with unique diseases. A paradox in the field relates to the remarkable genomic synteny shown among chlamydiae and the very few chlamydial genes linked to specific diseases. We have found that different chlamydial species exhibit unique requirements for Src-family kinases. These differing requirements for Src-family kinases would not be apparent in genomic comparisons and appear to be a previously unrecognized distinction that may provide insights to guide research in chlamydial pathogenesis.
INTRODUCTION
Chlamydiae are Gram-negative obligate intracellular bacteria that are the causative agents of several significant human diseases. Chlamydia trachomatis includes over 15 serologically defined variants, or serovars, associated with different diseases and tissue tropisms. These include the etiologic agents of trachoma, the most common cause of infectious blindness, while other serovars cause sexually transmitted diseases (1). C. pneumoniae is a common cause of community-acquired pneumonia (2). C. muridarum and C. caviae were isolated from mice and guinea pigs, respectively (3, 4), and are not known to infect humans.
Chlamydiae are characterized by a biphasic life cycle, alternating between infectious elementary bodies (EBs) and replicative reticulate bodies (RBs) (5). Following endocytosis by a host cell, chlamydiae reside within a vacuole termed an inclusion, which is nonfusogenic with the endosomal/lysosomal pathway but acquires sphingomyelin and cholesterol from the Golgi apparatus (6–8). Once modified by de novo-synthesized chlamydial proteins, the nascent inclusion is trafficked to the perinuclear region, where it is typically observed in close apposition with the host cell centrosomes (9–14). This initial trafficking is dependent upon the host microtubule network and the minus-end microtubule motor, dynein (14, 15). Recently, we have shown that active forms of the protein tyrosine kinases Src and Fyn are recruited to the chlamydial inclusion membrane, where they colocalize in microdomains that are thought to serve as a platform for interactions between dynein and the chlamydial inclusion (16).
Src and Fyn belong to the Src family of nonreceptor membrane-associated tyrosine kinases. Src, Yes, and Fyn are ubiquitously expressed, with some functional redundancy, whereas the other family members are expressed in subsets of specialized cells. Src and Fyn are involved in a variety of signaling pathways that govern a broad range of cellular processes (17).
Here we show that Src-family kinases (SFKs) have multiple functions in C. trachomatis development, including trafficking of nascent inclusions to the microtubule organizing center (MTOC) and a necessary role in completion of the C. trachomatis intracellular developmental cycle. The functional implications of these phenotypes are supported by the observation that chlamydial species that do not recruit Src-family kinases to their inclusion membranes (16) do not traffic to the MTOC and show enhanced inclusion development in cells deficient in Src-family kinases.
RESULTS
SFK activation is upregulated in C. trachomatis-infected cells.
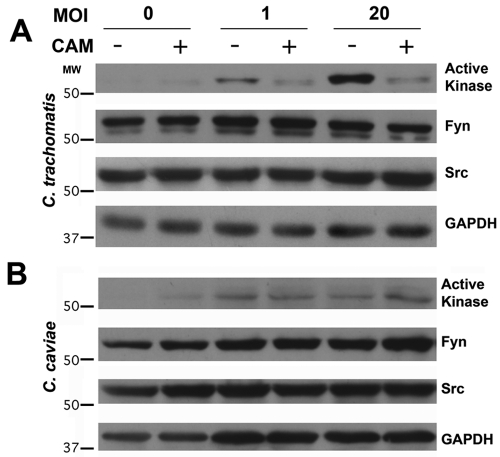
Src-family kinase activity is regulated by the phosphorylation state of tyrosine residues within the kinase. Enzymatic activity is upregulated in kinases that are phosphorylated at Tyr419 (human Src numbering) and downregulated when phosphorylated at Tyr530 (17). Active Src-family kinases are recruited to the C. trachomatis inclusion membrane, where they are observed in microdomains enriched in cholesterol and specific chlamydial inclusion membrane proteins (16). The intensity of the active Src kinase signal at these microdomains indicates a significant enrichment at the inclusion membrane (16). Src-family kinase activity is upregulated in C. trachomatis-infected cells (Fig. 1A). This increase does not occur in the presence of chloramphenicol and thus is not simply a response by the host cell to the challenge of chlamydial infection. C. caviae, a species that does not recruit active kinase to the inclusion membrane, does not induce activation of Src-family kinases (Fig. 1B).
FIG 1 .
Src-family kinase activation in C. trachomatis-infected cells. Immunoblot analysis of cell lysate 24 h after infection with C. trachomatis L2 (A) or C. caviae GPIC (B) in the absence or presence of chloramphenicol (CAM) shows that active Src-family kinase is upregulated after C. trachomatis, but not C. caviae, infection. Upregulation is dependent upon bacterial protein synthesis, as it is inhibited by chloramphenicol. The concentrations of total kinase (Fyn and Src) are unchanged. GAPDH is included as a loading control. MW, molecular weight (in thousands).
Src-family kinases are not required for C. trachomatis attachment or entry.
The role of Src-family kinases at various stages of the chlamydial development cycle was examined. SYF cells are mouse embryonic fibroblast cells that are null mutants for the three ubiquitous Src-family kinases (Src, Yes, and Fyn). Src was subsequently reintroduced to create the SYF + Src cell line (18). Mouse L929 fibroblasts were used as an additional control. Comparison of the numbers of attached and internalized C. trachomatis L2 EBs indicates that the SYF cells showed no significant defect in bacterial attachment or invasion compared to L929 and SYF + Src cells (see Fig. S1 in the supplemental material). This is in agreement with previous reports showing that inhibition of Src-family kinases does not reduce C. trachomatis L2 invasion (19, 20).
Nascent C. trachomatis inclusions require SFKs for proper trafficking to the MTOC.
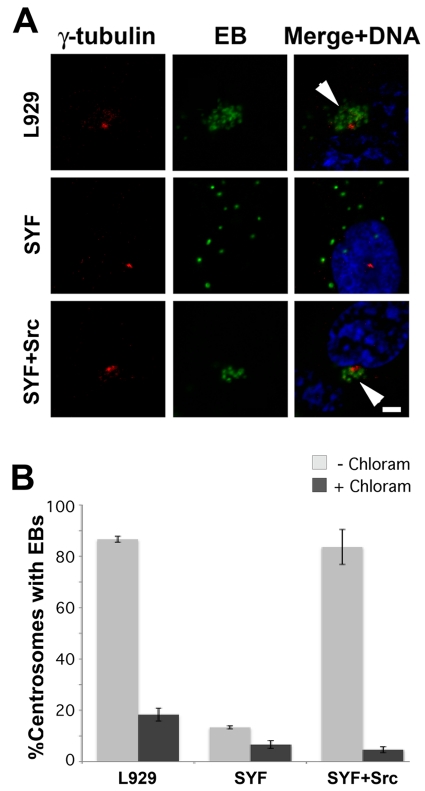
Trafficking of C. trachomatis L2 inclusions to the MTOC was examined in L929, SYF, and SYF + Src cells (Fig. 2). Nascent inclusions rapidly cluster at the MTOC in L929 cells and SYF + Src cells. This is in contrast to the dispersed inclusions observed in SYF cells or cultures in which bacterial translation was blocked with chloramphenicol (Fig. S2). Approximately the same percentages of cells with C. trachomatis EBs aggregated at the MTOC were observed in L929 and SYF + Src cells, whereas in SYF cells this value was reduced to the minimal levels seen in chloramphenicol-treated cultures. Src-family kinase activity is thus required for the dynein-dependent trafficking of the nascent C. trachomatis inclusion to the MTOC.
FIG 2 .
Trafficking of nascent C. trachomatis inclusions to the MTOC requires Src-family kinases. (A) L929, SYF, and SYF + Src cells were infected with C. trachomatis L2 in the absence or presence of chloramphenicol. Five hours postinfection, samples were fixed and stained for EBs (green) and γ-tubulin (red). Arrowheads indicate early inclusions converged at the MTOC. Bar, 10 µm. (B) The percentage of cells with inclusions clustered at the MTOC is significantly decreased in SYF cells compared to that in L929 and SYF + Src cells and approximates the percentage observed in chloramphenicol-treated cells. One hundred infected cells were counted per sample (n = 3); error bars show standard deviations.
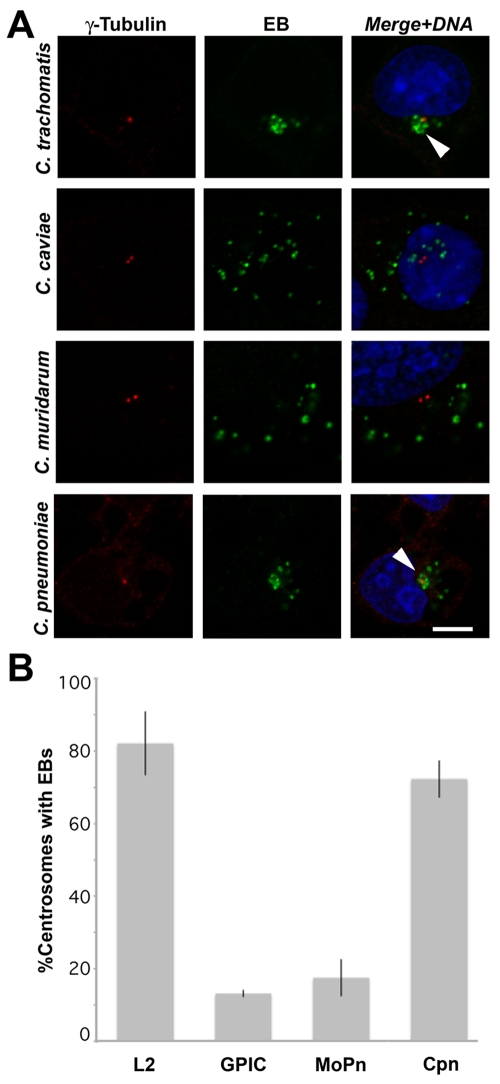
C. trachomatis serovars L2 and D and C. pneumoniae recruit active Src-family kinases to the inclusion membrane, whereas C. caviae and C. muridarum do not (16). We investigated whether Src-family kinase recruitment might correlate with trafficking to the MTOC in a species-specific manner. Examination of infected L929 cells confirms that C. trachomatis and C. pneumoniae inclusions consistently traffic to the MTOC, whereas C. caviae and C. muridarum inclusions do not (Fig. 3). These findings support the conclusion that Src-family kinases play an essential role in the trafficking of chlamydial inclusions to the MTOC. Examination of C. caviae and C. muridarum inclusions at later times confirms a lack of trafficking to the MTOC (Fig. S3).
FIG 3 .
Trafficking of nascent chlamydial inclusions to the MTOC is species specific. (A) L929 cells were infected with C. trachomatis L2, C. caviae GPIC, C. muridarum MoPn, or C. pneumoniae AR39. Five hours postinfection, the cells were fixed and labeled with anti-EB antisera (green) and anti-γ-tubulin antibodies (red) followed by DyLight 488 and 594 secondary antibodies as well as DRAQ5 DNA stain (blue). Arrowheads indicate early inclusions converged at the MTOC. Bar, 10 µm. (B) The percentage of infected cells with EBs clustered at the MTOC was determined using confocal fluorescent microscopy. C. trachomatis L2 and C. pneumoniae consistently traffic to the centrosome, whereas C. caviae and C. muridarum do not. One hundred cells were counted per sample (n = 3); error bars show standard deviations. Cpn, C. pneumoniae.
Differential effects of SFK deficiency on chlamydial development.
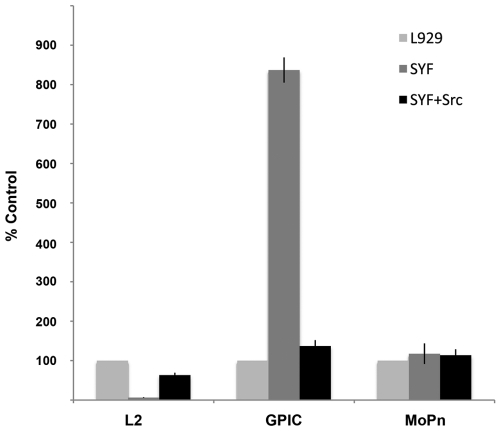
Although attachment and entry are unaffected, development of C. trachomatis inclusions is significantly reduced in SYF cells compared to development in L929 or SYF + Src cells (Fig. S4). Consistent with the observed inhibition of inclusion development, the number of infectious progeny is decreased in SYF cells to less than 10% of that obtained from L929 cells (Fig. 4). Reconstitution of Src largely restores infectious progeny production, although a modest decrease is seen from the SYF + Src cells relative to L929 cells, suggesting that Fyn or Yes may play specific roles during chlamydial development for which Src does not provide functional redundancy.
FIG 4 .
Chlamydial dependency on Src-family kinases for productive infection is species specific. Confluent monolayers of L929, SYF, and SYF + Src cells were infected with C. trachomatis L2, C. caviae GPIC, or C. muridarum MoPn. Cultures were lysed at 30 h postinfection, and the progeny EBs were replated onto confluent monolayers of HeLa cells. At 24 h postinfection, the number of inclusions per field was determined (10 fields per sample; n = 3) using fluorescent microscopy. Values reported are relative to those obtained from L929 cells (L2, 4.7 × 107 IFU/ml; GPIC, 5.3 × 107 IFU/ml; MoPn, 7.4 × 107 IFU/ml). Error bars show standard deviations.
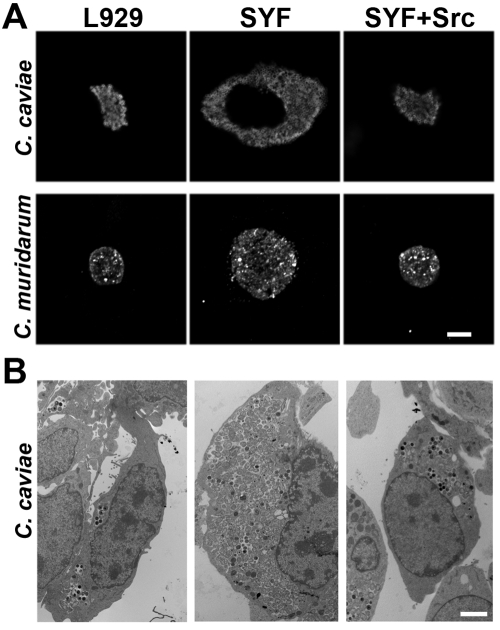
Neither C. caviae nor C. muridarum displays a decrease in the number of developed inclusions or in infectious progeny produced from SYF cells. Indeed, C. caviae and C. muridarum inclusions in SYF cells appear significantly larger than those in L929 or SYF + Src cells (Fig. 5). There was a corresponding increase in the number of infectious progeny produced in SYF cells, with approximately an 800% and 20% increase over those produced in L929 cells for C. caviae and C. muridarum, respectively (Fig. 4).
FIG 5 .
Enhanced development of non-C. trachomatis inclusions in SYF cells. L929, SYF, and SYF + Src cells were infected with C. caviae GPIC or C. muridarum MoPn. At 24 h postinfection, samples were visualized using fluorescent confocal microscopy (bar, 10 µm) (A) or transmission electron microscopy (bar, 5 µm) (B). Inclusions in SYF cells are much larger than corresponding inclusions in L929 or SYF + Src cells.
Because depletion of Src-family kinases blocks both trafficking of the nascent C. trachomatis inclusions to the MTOC and normal inclusion development, we asked whether inhibition of trafficking to the MTOC might be responsible for the lack of inclusion development in SYF cells. As has been shown previously (14, 21), nocodazole inhibits trafficking of C. trachomatis inclusions to the MTOC but does not impair subsequent inclusion development or production of infectious progeny (Fig. S5).
Src-family kinases are not required for Inc microdomain formation.
Although few C. trachomatis inclusions develop in SYF cells, those that do appear to develop normally (Fig. S6). In those inclusions, Inc microdomains are observed on the inclusion membrane. This is in agreement with previous observations that Src-family kinases are not required for Inc microdomain formation (16). Active kinase colocalizes with Inc microdomains in L929 cells, and the signal intensity is enhanced in cells overexpressing Src (SYF + Src), but the signal is not detected in cells lacking Src-family kinases. However, tyrosine-phosphorylated proteins are observed in microdomains at the inclusion membrane in all three cell lines, although they appear more abundant in the SYF + Src cells and less abundant in the SYF cells. Active Src and Fyn are themselves tyrosine phosphorylated, which may account for the reduced phosphotyrosine signal in SYF cells and the increased signal observed in SYF + Src cells. However, it is likely that there are tyrosine-phosphorylated proteins other than Src-family kinases present in the microdomains.
Src-family kinase inhibitors mimic the inhibitory effects observed in SYF cells.
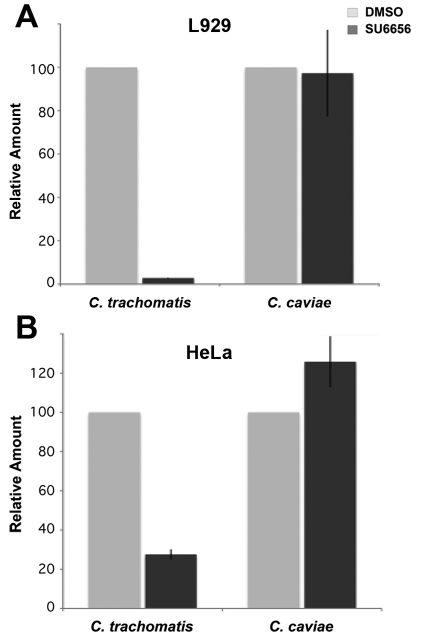
Given the phenotypes observed in SYF cells, we investigated the ability of Src-family kinase inhibitors to recapitulate these phenotypes. Dynein-dependent trafficking of C. trachomatis early inclusions to the MTOC was inhibited by the protein tyrosine kinase inhibitor SU6656 (Fig. S7). The effect of SU6656 on C. trachomatis L2 and C. caviae progeny production was also examined (Fig. 6A). Consistent with the effects observed in SYF cells, the production of C. trachomatis progeny EBs was reduced by over 95% by inhibition of Src-family kinases. Conversely, production of infectious progeny from C. caviae-infected cells was unaffected by Src-family kinase inhibitors. The decreasing viability of the host cells in the extended presence of inhibitor prevented accurate quantitation of inclusion-forming units (IFU) at later time points where the largest increase in IFU from SYF cells was observed.
FIG 6 .
Src-family kinase inhibitors inhibit production of C. trachomatis infectious progeny. Confluent monolayers of L929 (A) or HeLa (B) cells were infected with C. trachomatis L2 or C. caviae GPIC. One hour postinfection, cells were treated with DMSO (gray bars) or SU6656 (black bars). Cells were lysed, and progeny was replated onto HeLa cell monolayers. Cells were fixed and fluorescently labeled, and the number of inclusions per field was counted using fluorescent microscopy. Values reported are relative to those obtained from mock-treated (DMSO) controls (HeLa cells: L2, 1.1 × 107 IFU/ml; GPIC, 2.3 × 107 IFU/ml; L929 cells: L2, 1.0 × 108 IFU/ml; GPIC, 6.3 × 106 IFU/ml). Ten fields per sample (n = 3). Error bars show standard deviations.
To further delineate the requirements for Src-family kinases during C. trachomatis intracellular trafficking and development, we allowed nascent inclusions to traffic to the MTOC before inhibiting Src-family kinase activity with SU6656. SU6656 reduced progeny EB formation to 1.9% ± 0.6% and 3.1% ± 0.6% of untreated control values when added at 0 and 5 h postinfection, respectively. Complementary to the results above demonstrating that trafficking to the MTOC is not necessary for C. trachomatis development, these results indicate that trafficking to the MTOC is not sufficient to promote normal development in the absence of Src-family kinase activity. The growth inhibition of C. trachomatis by Src-family kinase inhibition thus appears unrelated to effects on microtubule trafficking.
Because C. trachomatis and C. caviae have different natural hosts, we examined the possibility that these species-specific chlamydial phenotypes might be related to the murine origin of the Src-family kinase-knockout cells. Similar inhibition of C. trachomatis by SU6566 was observed in the human cells, while C. caviae infectious progeny production was not disrupted (Fig. 6B). This result indicates that the differential effects of Src-family kinases on C. trachomatis and C. caviae replication are independent of the host cell species.
DISCUSSION
Tyrosine phosphorylation of proteins is an important posttranslational regulator of activity. Src-family kinases comprise a prominent family of protein tyrosine kinases that play a central role in multiple signaling pathways that regulate a diverse array of cellular activities, including adhesion, migration, differentiation, cell cycle progression, apoptosis, transcription, and intracellular trafficking (17). The intimate relationship of intracellular pathogens with eukaryotic host cells necessitates that they adapt to and manipulate host cellular processes. Accordingly, intracellular pathogens subvert cellular signaling pathways at several stages of infection to promote entry, replication, or release during their life cycles. Src-family kinases have been shown to play important roles in the pathogenesis of a wide range of intracellular pathogens, including viruses, parasites, and bacteria (22–31).
C. trachomatis interacts with Src-family kinases at multiple stages during its intracellular development. The C. trachomatis type III effector Tarp, which is secreted upon attachment and implicated in the entry process due to its actin-nucleating activity, is tyrosine phosphorylated by Src-family or other tyrosine kinases upon translocation into the host cytosol (19, 20, 32, 33). Although the role(s) of Tarp remains to be fully elucidated, the functions associated with it are independent of chlamydial transcription or translation (34) and thus can be readily distinguished from events that are dependent upon chlamydial protein synthesis, such as trafficking to the MTOC (14), fusion with Golgi apparatus-derived vesicles (13), differentiation of chlamydial developmental forms (35), and activation of Src-family kinases.
C. trachomatis and C. pneumoniae also appear to have evolved a dependency on Src-family kinases for intracellular trafficking and development. Recently, microdomains on the C. trachomatis inclusion membrane were identified and shown to be comprised of at least four inclusion membrane proteins as well as activated Src-family kinases. These microdomains are thought to participate in the interactions of the chlamydial inclusion with the microtubule motor complex and function in the trafficking of the nascent inclusion to the MTOC (16). In SYF cells or cells in which Src-family kinases are inhibited, nascent C. trachomatis L2 inclusions are not trafficked to the MTOC and show a reduction in inclusion development with a corresponding decrease in infectious progeny. The two defects induced by a Src-family kinase deficiency appear to be unrelated, since the microtubule inhibitor nocodazole effectively blocks trafficking of nascent C. trachomatis inclusions to the MTOC without negatively affecting inclusion development or production of infectious progeny. Nascent inclusion trafficking is dependent upon microtubules and dynein (14). Src-family kinases have been shown to phosphorylate tubulin and bind to dynein-associated proteins (36–38), suggesting that recruitment of Src-family kinases by chlamydiae may directly or indirectly mediate linkage of the inclusion to the microtubule network of the host cell. Those species examined that do not display active Src-family kinase-enriched microdomains on the inclusion membrane, C. muridarum and C. caviae, do not exhibit microtubule-dependent trafficking to or association with the MTOC. Whether Src-family kinase activity is required for processivity of the dynein motor complex or whether tyrosine phosphorylation contributes to the recruitment of necessary components remains to be determined. An improved understanding of C. trachomatis trafficking may help to delineate the role of Src-family kinases in dynein motor function and microtubule trafficking.
Src-family kinases also appear to serve a separate function later in infection that is essential for normal C. trachomatis inclusion development. The observation that those few EBs which differentiate and commit to development do so normally suggests that this critical initial step of differentiation may be a stochastic process that does not favor C. trachomatis development in the absence of Src-family kinases. This developmental defect is not observed in SYF cells infected with C. caviae or C. muridarum. Indeed, C. caviae and C. muridarum replicate more efficiently in SYF cells. Although the mechanism of growth enhancement of C. caviae or C. muridarum in cells deficient in Src-family kinases is unknown, one hypothesis might be that Src-family kinases play a role in a pathway that restricts growth of these pathogens and that the absence of this pathway in SYF cells permits unregulated, enhanced growth.
Chlamydial species are well known for their distinct tissue tropisms and species specificity, despite a high degree of synteny of their genomes (39). Few loci have been associated with disease or tissue tropism (40–44). Divergence occurs primarily within a hypervariable replication termination region or plasticity zone (45). A key distinction between human and murine chlamydial species involves their adaptations to gamma interferon (IFN-γ) (46–49). In murine cell lines and in vivo, C. muridarum is resistant to the effects of IFN-γ, whereas C. trachomatis is inhibited. IFN-γ-mediated inhibition of C. trachomatis in murine cells has been attributed to the presence of p47 or immunity-related GTPases (IRGs), which are present in mice but largely absent from human cells (47, 49). Resistance of C. muridarum to IFN-γ is thought to be related to the inactivation of specific IRGs by the cytotoxin that is absent or truncated in C. trachomatis (47). However, treatment of a human cell line with a Src-family kinase inhibitor led to a decrease in C. trachomatis replication similar to that observed in murine cells; thus, the mechanisms of the Src-family kinase appear to be acting though pathways other than mouse-specific IRGs. IFN-γ treatment was not investigated here; however, the plasticity zone remains a highly plausible locus for chlamydial genes responsible for the divergent responses to Src-family kinase depletion.
Early studies of tyrosine phosphorylation during chlamydial infection described two or more proteins that were tyrosine phosphorylated very early in infection and located in close apposition to the invading EBs (50, 51). Although the proteins were not identified, they were presumed to be of host origin. To date, ezrin has been the only host protein identified that is tyrosine phosphorylated during chlamydial invasion (52). A more complete accounting of host and chlamydial proteins that are tyrosine phosphorylated throughout the developmental cycle will undoubtedly refine future studies designed to understand the mechanisms of growth inhibition of C. trachomatis and growth enhancement of C. caviae and C. muridarum by depletion of Src-family kinases.
Protein tyrosine kinases are activated and functional at multiple stages of C. trachomatis intracellular development. By all parameters analyzed, C. caviae and C. muridarum showed no activation of, recruitment of, or requirement for Src-family kinases and thus have apparently evolved life-styles that circumvent the need for Src-family kinases. Src-family kinases therefore play an important, but species-specific, role in the chlamydial developmental cycle. It is apparent that different combinations of requirements occur between species. For example, C. pneumoniae Tarp is not phosphorylated (32), and yet its inclusions contain tyrosine-phosphorylated microdomains (16) and are trafficked to the MTOC. It is likely that additional functions regulated by Src-family kinases will be identified in chlamydia-infected cells. A more complete understanding of host and chlamydial tyrosine-phosphorylated proteins and the pathways associated with them should help elucidate significant questions in chlamydial biology and pathogenesis.
MATERIALS AND METHODS
Organisms and cell culture.
Chlamydia trachomatis serovar L2 (LGV 434) and serovar D (UW-3-Cx), Chlamydia muridarum Nigg, Chlamydophila caviae GPIC, and Chlamydia pneumoniae AR-39 were propagated in HeLa 229 cells and purified by Renografin density gradient centrifugation as previously described (53). L929, SYF, and SYF + Src cells (American Type Culture Collection [ATCC], Manassas, VA) were grown in Dulbecco’s modified Eagle’s medium (DMEM; ATCC) with 10% fetal bovine serum (FBS; ATCC) and gentamicin (Gibco, Carlsbad, CA). The SYF cells used were between passage numbers 22 and 30.
Antibodies.
The antichlamydia antibodies used were polyclonal rabbit anti-C. trachomatis L2 EB, anti-C. pneumoniae EB, and anti-C. caviae EB. Monoclonal antibody L2-i-45 against C. trachomatis L2 major outer membrane protein (MOMP) was kindly provided by H. D. Caldwell. Rabbit anti-Inc101 was previously described (16). Anti-phospho-Src-family Tyr416 clone 9A6 (Millipore, Billerica, MA) was used to detect active Src-family kinases. Monoclonal antibody 4G10 (Millipore) was used to detect phosphotyrosine. Centrosomes were detected using mouse anti-γ-tubulin, clone GTU-88 (Sigma, St. Louis, MO). Anti-mouse or anti-goat DyLight 594 and anti-rabbit DyLight 488 (Jackson ImmunoResearch Laboratories, West Grove, PA) were used as secondary antibodies for indirect immunofluorescence. Anti-Src (Cell Signaling, Danvers, MA), anti-Fyn (BD Biosciences, San Diego, CA), anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH; Sigma), and horseradish peroxidase (HRP)-conjugated donkey anti-rabbit IgG and HRP-conjugated donkey anti-mouse IgG (Jackson ImmunoResearch Laboratories) were used for immunoblotting.
Western blotting.
HeLa cells were mock infected or infected with C. trachomatis or C. caviae at a multiplicity of infection (MOI) of 1 or 20. At 24 h postinfection, the cells were incubated with 1 mM sodium vanadate for 30 minutes; rinsed with 50 mM NaPO4, 150 mM NaCl, pH 7.4 (phosphate-buffered saline [PBS]); and lysed in Laemmli buffer (54) with 5% beta-mercaptoethanol and 1 mM sodium vanadate. The lysates were separated by SDS-PAGE for immunoblotting as previously described (16).
Attachment and invasion assays.
Monolayers of L929, SYF, and SYF + Src cells on glass coverslips in 24-well plates were chilled to 4°C on ice for 30 minutes prior to addition of C. trachomatis L2 at an MOI of ~20 followed by a 30-minute incubation at 4°C. For attachment assays, cells were rinsed once in cold Hanks’ balanced salt solution (HBSS) and fixed with methanol before immunostaining. Images of attached chlamydiae were obtained using a Zeiss LSM 510 Meta laser confocal scanning microscope using a 20×, 0.8-numerical-aperture (NA) objective (Carl Zeiss MicroImaging, Inc., Maple Grove, MN), and the number of attached bacteria was determined for 10 fields per sample. For invasion assays, cells were inoculated as described above, prewarmed medium was added, and cultures were incubated at 37°C for 30 minutes prior to fixation with 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) in PBS. Intracellular and extracellular bacteria were differentially stained by indirect immunofluorescence, and the percentage of internalized bacteria was calculated by subtracting the number of extracellular bacteria from the total number of bacteria and dividing by the total number of bacteria as previously described (55). A minimum of 10 cells and 100 bacteria were counted for each sample.
Inclusion development and infectious progeny (IFU) assays.
Confluent monolayers of L929, SYF, and SYF + Src cells on glass coverslips or Costar Cellbind (Corning, Lowell, MA) 24-well plates were infected with C. trachomatis serovar L2 or D, C. muridarum, or C. caviae as previously described (16) at an MOI of ~0.5. At the indicated times postinfection, the cultures were fixed in methanol or rinsed once and lysed in water and disrupted by passage through a 26-gauge needle, before dilution in HBSS and replating on HeLa cell monolayers for determination of inclusion-forming units (IFU). Where indicated, nocodazole (Sigma) was added to 400 ng/ml at the time of infection. For Src inhibitor experiments, cells were treated with dimethyl sulfoxide (DMSO; Sigma) or 10 µM SU6656 (Sigma) 1 h after the initial infection and maintained in the continuous presence of the drug.
Centrosomal clustering assays.
Subconfluent monolayers of L929, SYF, or SYF + Src cells were infected with C. trachomatis serovar L2 or D, C. pneumoniae, C. muridarum, or C. caviae at an MOI of ~20. At 5 h postinfection, cells were rinsed and fixed with methanol prior to labeling with rabbit anti-EB and mouse anti-γ-tubulin (Sigma). The specimens were counterstained with goat anti-rabbit IgG-DyLight 488 and goat anti-mouse IgG-DyLight 594. The number of infected cells that had a minimum of two EBs adjacent to at least one host centrosome was determined by fluorescent microscopy using a Zeiss LSM 510 Meta laser confocal scanning microscope with a 63×, 1.4-NA oil objective. A minimum of 100 infected cells were counted for each sample, and experiments were performed in triplicate. Cells were untreated or treated with DMSO, nocodazole (400 ng/ml), or chloramphenicol (50 µg/ml) concurrently with infection. For the Src inhibitor experiments, L929 cells were pretreated with DMSO or 10 µM SU6656 for 1 h prior to infection and maintained in the presence of inhibitor.
Electron microscopy.
L929, SYF, and SYF + Src monolayers on Thermanox coverslips were infected with C. caviae. At 24 h postinfection, cells were fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer and processed for transmission electron microscopy as previously described (16).
SUPPLEMENTAL MATERIAL
ACKNOWLEDGMENTS
This work was supported by the Intramural Research Program of the NIAID/NIH.
We thank V. Nair for electron microscopy and Janet Sager for excellent technical assistance. We also thank members of the Hackstadt laboratory and R. Heinzen for helpful discussions and critical review of the manuscript.
Footnotes
Citation Mital J, Hackstadt T. 2011. Diverse requirements for Src-family tyrosine kinases distinguish chlamydial species. mBio 2(2):e00031-11. doi:10.1128/mBio.00031-11.
REFERENCES
- 1. Schachter J. 1999. Infection and disease epidemiology, p. 139–169 In Stephens RS, Chlamydia: intracellular biology, pathogenesis, and immunity. ASM Press, Washington, DC [Google Scholar]
- 2. Kuo CC, Jackson LA, Campbell LA, Grayston JT. 1995. Chlamydia pneumoniae (TWAR). Clin. Microbiol. Rev. 8:451–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Murray ES. 1964. Guinea pig inclusion conjunctivitis virus. I. Isolation and identification as a member of the psittacosis-lymphogranuloma-trachoma group. J. Infect. Dis. 114:1–12 [DOI] [PubMed] [Google Scholar]
- 4. Nigg C. 1942. An unidentified virus which produces pneumonia and systemic infection in mice. Science 95:49–50 [DOI] [PubMed] [Google Scholar]
- 5. Moulder JW. 1991. Interaction of chlamydiae and host cells in vitro. Microbiol. Rev. 55:143–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carabeo RA, Mead DJ, Hackstadt T. 2003. Golgi-dependent transport of cholesterol to the Chlamydia trachomatis inclusion. Proc. Natl. Acad. Sci. U. S. A. 100:6771–6776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hackstadt T, Scidmore MA, Rockey DD. 1995. Lipid metabolism in Chlamydia trachomatis-infected cells: directed trafficking of Golgi-derived sphingolipids to the chlamydial inclusion. Proc. Natl. Acad. Sci. U. S. A. 92:4877–4881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Heinzen RA, Scidmore MA, Rockey DD, Hackstadt T. 1996. Differential interaction with endocytic and exocytic pathways distinguish parasitophorous vacuoles of Coxiella burnetii and Chlamydia trachomatis. Infect. Immun. 64:796–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Campbell S, Richmond SJ, Yates PS. 1989. The effect of Chlamydia trachomatis infection on the host cell cytoskeleton and membrane compartments. J. Gen. Microbiol. 135:2379–2386 [DOI] [PubMed] [Google Scholar]
- 10. Hackstadt T, Rockey DD, Heinzen RA, Scidmore MA. 1996. Chlamydia trachomatis interrupts an exocytic pathway to acquire endogenously synthesized sphingomyelin in transit from the Golgi apparatus to the plasma membrane. EMBO J. 15:964–977 [PMC free article] [PubMed] [Google Scholar]
- 11. Higashi N. 1955. Electron microscopic studies on the mode of reproduction of trachoma virus and psittacosis virus in cell cultures. Exp. Mol. Pathol. 4:24–39 [DOI] [PubMed] [Google Scholar]
- 12. Majeed M, Kihlström E. 1991. Mobilization of F-actin and clathrin during redistribution of Chlamydia trachomatis to an intracellular site in eucaryotic cells. Infect. Immun. 59:4465–4472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Scidmore MA, Rockey DD, Fischer ER, Heinzen RA, Hackstadt T. 1996. Vesicular interactions of the Chlamydia trachomatis inclusion are determined by chlamydial early protein synthesis rather than route of entry. Infect. Immun. 64:5366–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grieshaber S, Grieshaber N, Hackstadt T. 2003. Chlamydia trachomatis uses host cell dynein to traffic to the microtubule organizing center in a p50 dynamitin independent process. J. Cell Sci. 116:3793–3802 [DOI] [PubMed] [Google Scholar]
- 15. Clausen JD, Christiansen G, Holst HU, Birkelund S. 1997. Chlamydia trachomatis utilizes the host cell microtubule network during early events of infection. Mol. Microbiol. 25:441–449 [DOI] [PubMed] [Google Scholar]
- 16. Mital J, Miller NJ, Fischer ER, Hackstadt T. 2010. Specific chlamydial inclusion membrane proteins associate with active Src family kinases in microdomains that interact with the host microtubule network. Cell. Microbiol. 12:1235–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thomas SM, Brugge JS. 1997. Cellular functions regulated by Src family kinases. Annu. Rev. Cell Dev. Biol. 13:513–609 [DOI] [PubMed] [Google Scholar]
- 18. Klinghoffer RA, Sachsenmaier C, Cooper JA, Soriano P. 1999. Src family kinases are required for integrin but not PDGFR signal transduction. EMBO J. 18:2459–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jewett TJ, Dooley CA, Mead DJ, Hackstadt T. 2008. Chlamydia trachomatis tarp is phosphorylated by src family tyrosine kinases. Biochem. Biophys. Res. Commun. 371:339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mehlitz A, Banhart S, Hess S, Selbach M, Meyer TF. 2008. Complex kinase requirements for Chlamydia trachomatis Tarp phosphorylation. FEMS Microbiol. Lett. 289:233–240 [DOI] [PubMed] [Google Scholar]
- 21. Schramm N, Wyrick PB. 1995. Cytoskeletal requirements in Chlamydia trachomatis infection of host cells. Infect. Immun. 63:324–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Backert S, Selbach M. 2005. Tyrosine-phosphorylated bacterial effector proteins: the enemies within. Trends Microbiol. 13:476–484 [DOI] [PubMed] [Google Scholar]
- 23. Baumgartner M, et al. 2003. Constitutive exclusion of Csk from Hck-positive membrane microdomains permits Src kinase-dependent proliferation of Theileria-transformed B lymphocytes. Blood 101:1874–1881 [DOI] [PubMed] [Google Scholar]
- 24. Bruce-Staskal PJ, Weidow CL, Gibson JJ, Bouton AH. 2002. Cas, Fak and Pyk2 function in diverse signaling cascades to promote Yersinia uptake. J. Cell Sci. 115:2689–2700 [DOI] [PubMed] [Google Scholar]
- 25. Gillrie MR, et al. 2007. Src-family kinase dependent disruption of endothelial barrier function by Plasmodium falciparum merozoite proteins. Blood 110:3426–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Greenway AL, et al. 2003. HIV-1 Nef control of cell signalling molecules: multiple strategies to promote virus replication. J. Biosci. 28:323–335 [DOI] [PubMed] [Google Scholar]
- 27. Liang Y, Roizman B. 2006. State and role of SRC family kinases in replication of herpes simplex virus 1. J. Virol. 80:3349–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mounier J, et al. 2009. The IpaC carboxyterminal effector domain mediates Src-dependent actin polymerization during Shigella invasion of epithelial cells. PLoS Pathog. 5:e1000271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Munter S, Way M, Frischknecht F. 2006. Signaling during pathogen infection. Sci. STKE 2006:re5 . [DOI] [PubMed] [Google Scholar]
- 30. Newsome TP, Scaplehorn N, Way M. 2004. SRC mediates a switch from microtubule- to actin-based motility of vaccinia virus. Science 306:124–129 [DOI] [PubMed] [Google Scholar]
- 31. Reeves PM, et al. 2011. Variola and monkeypox viruses utilize conserved mechanisms of virion motility and release that depend on Abl and Src family tyrosine kinases. J. Virol. 85:21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Clifton DR, et al. 2005. Tyrosine phosphorylation of chlamydial Tarp is species specific and not required for the recruitment of actin. Infect. Immun. 73:3860–3868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Elwell CA, Ceesay A, Kim JH, Kalman D, Engel JN. 2008. RNA interference screen identifies Abl kinase and PDGFR signaling in Chlamydia trachomatis entry. PLoS Pathog. 4:e1000021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Clifton DR, et al. 2004. A chlamydial type III translocated protein is tyrosine phosphorylated at the site of entry and associated with recruitment of actin. Proc. Natl. Acad. Sci. U. S. A. 101:10166–10171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Grieshaber NA, Fischer ER, Mead DJ, Dooley CA, Hackstadt T. 2004. Chlamydial histone-DNA interactions are disrupted by a metabolite in the methylerythritol phosphate pathway of isoprenoid biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 101:7451–7456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Levi M, Shalig R. 2010. The role of Fyn kinase in the release from metaphase in mammalian oocytes. Mol. Cell. Endocrinol. 2:228–233 [DOI] [PubMed] [Google Scholar]
- 37. Macurek L, et al. 2008. Regulation of microtubule nucleation from membranes by complexes of membrane-bound gamma-tubulin with Fyn kinase and phosphoinositide 3-kinase. Biochem. J. 416:421–430 [DOI] [PubMed] [Google Scholar]
- 38. Colello D, et al. 2010. Androgen and Src signaling regulate centrosome activity. J. Cell Sci. 123:2094–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stephens RS. 1999. Genomic autobiographies of chlamydiae, p. 9–27 In Stephens RS, Chlamydia: intracellular biology, pathogenesis, and immunity. ASM Press, Washington, DC [Google Scholar]
- 40. Belland RJ, et al. 2001. Chlamydia trachomatis cytotoxicity associated with complete and partial cytotoxin genes. Proc. Natl. Acad. Sci. U. S. A. 98:13984–13989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brunelle BW, Nicholson TL, Stephens RS. 2004. Microarray-based genomic surveying of gene polymorphisms in Chlamydia trachomatis. Genome Biol. 5:R42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fehlner-Gardiner C, et al. 2002. Molecular basis defining human Chlamydia trachomatis tissue tropism. A possible role for tryptophan synthase. J. Biol. Chem. 277:26893–26903 [DOI] [PubMed] [Google Scholar]
- 43. Gomes JP, et al. 2006. Polymorphisms in the nine polymorphic membrane proteins of Chlamydia trachomatis across all serovars: evidence for serovar Da recombination and correlation with tissue tropism. J. Bacteriol. 188:275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lutter EI, et al. 2010. Phylogenetic analysis of Chlamydia trachomatis Tarp and correlation with clinical phenotype. Infect. Immun. 78:3678–3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Read TD, et al. 2003. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res. 31:2134–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Morrison RP. 2000. Differential sensitivities of Chlamydia trachomatis strains to inhibitory effects of gamma interferon. Infect. Immun. 68:6038–6040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nelson DE, et al. 2005. Chlamydial interferon gamma immune evasion is linked to host infection tropism. Proc. Natl. Acad. Sci. U. S. A. 102:10658–10663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Roshick C, Wood H, Caldwell HD, McClarty G. 2006. Comparison of gamma interferon-mediated antichlamydial defense mechanisms in human and mouse cells. Infect. Immun. 74:225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Coers J, et al. 2008. Chlamydia muridarum evades growth restriction by the IFN-gamma-inducible host resistance factor Irgb10. J. Immunol. 180:6237–6245 [DOI] [PubMed] [Google Scholar]
- 50. Birkelund S, Johnsen H, Christiansen G. 1994. Chlamydia trachomatis serovar L2 induces protein tyrosine phosphorylation during uptake by HeLa cells. Infect. Immun. 62:4900–4908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fawaz FS, van Ooij C, Homola E, Mutka SC, Engel JN. 1997. Infection with Chlamydia trachomatis alters the tyrosine phosphorylation and/or localization of several host cell proteins including cortactin. Infect. Immun. 65:5301–5308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Swanson KA, Crane DD, Caldwell HD. 2007. Chlamydia trachomatis species-specific induction of ezrin tyrosine phosphorylation functions in pathogen entry. Infect. Immun. 75:5669–5677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Caldwell HD, Kromhout J, Schachter J. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 31:1161–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 55. Carabeo RA, Dooley CA, Grieshaber SS, Hackstadt T. 2007. Rac interacts with Abi-1 and WAVE2 to promote an Arp2/3-dependent actin recruitment during chlamydial invasion. Cell. Microbiol. 9:2278–2288 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.