Abstract
Differential compartmentalization of signaling molecules in cells and tissues is being recognized as an important mechanism for regulating the specificity of signal transduction pathways. A kinase anchoring proteins (AKAPs) direct the subcellular localization of protein kinase A (PKA) by binding to its regulatory (R) subunits. Dual specific AKAPs (D-AKAPs) interact with both RI and RII. A 372-residue fragment of mouse D-AKAP2 with a 40-residue C-terminal PKA binding region and a putative regulator of G protein signaling (RGS) domain was previously identified by means of a yeast two-hybrid screen. Here, we report the cloning of full-length human D-AKAP2 (662 residues) with an additional putative RGS domain, and the corresponding mouse protein less the first two exons (617 residues). Expression of D-AKAP2 was characterized by using mouse tissue extracts. Full-length D-AKAP2 from various tissues shows different molecular weights, possibly because of alternative splicing or posttranslational modifications. The cloned human gene product has a molecular weight similar to one of the prominent mouse proteins. In vivo association of D-AKAP2 with PKA in mouse brain was demonstrated by using cAMP agarose pull-down assay. Subcellular localization for endogenous mouse, rat, and human D-AKAP2 was determined by immunocytochemistry, immunohistochemistry, and tissue fractionation. D-AKAP2 from all three species is highly enriched in mitochondria. The mitochondrial localization and the presence of RGS domains in D-AKAP2 may have important implications for its function in PKA and G protein signal transduction.
Cyclic AMP-dependent protein kinase (PKA) plays a central role in one of the best understood signal transduction pathways in which the second-messenger, cAMP, is generated upon G protein coupled receptor activation by many different hormones and neurotransmitters. cAMP binds to the regulatory (R) subunits of PKA in the inactive, heterotetrameric holoenzyme, leading to the release of active catalytic subunits. PKA can phosphorylate a broad spectrum of substrates, including enzymes, membrane receptors, ion channels, and transcription factors, and regulate their function in cellular processes such as energy metabolism (1), steroidogenesis (2, 3), cell proliferation, differentiation, and cell death (4–6). With such diversified upstream stimulating signals and downstream substrate effectors, regulating the specificity of PKA signaling is critical. The discovery of a family of proteins known as A kinase anchoring proteins (AKAPs) (7–9) elucidated a mechanism by which PKA is compartmentalized, possibly with its substrates, so that individual signals can preferentially activate specific PKA pools. These types of regulatory assemblies have emerged as a common mechanism for governing the specificity of various signal transduction pathways (10).
AKAPs are multifunctional proteins that target to particular sites in the cell and recruit PKA by binding to a specific docking domain at the N terminus of the R subunit (11). So far, over two dozen AKAPs and their isoforms/homologs have been identified that localize to subcellular sites ranging from plasma membrane and actin cytoskeleton to nucleus, mitochondria, and endoplasmic reticulum (12, 13). The two types of R subunits of PKA, RI and RII, have distinct tissue and subcellular distributions. Although RIα and RIβ are predominantly cytoplasmic, RIIα and RIIβ are more often associated with cellular structures and organelles. Indeed, most of the AKAPs were isolated as RII-binding proteins until recently, when two dual-specific AKAPs (D-AKAP1 and -2) were cloned by using RIα as the bait in a yeast two-hybrid screen (14, 15). Other RI-specific AKAPs were subsequently identified (16, 17). The contrasted distribution pattern between RI and RII may be the result of their different affinities for AKAPs; however, regulation of the association between Rs and AKAPs is not well understood.
The original mouse D-AKAP2 cDNA clone encodes a 372-residue protein with a C-terminal PKA binding region that binds RIα, RIIα, RIIβ, but not RIβ in vitro (15), and a putative regulator of G protein signaling (RGS) domain. RGS domains have the potential to interact with Gα subunits and often act as GTPase-activating proteins (GAPs) (18–21); however, the role of the RGS domain in D-AKAP2 remains to be defined. When epitope-tagged mouse D-AKAP2 (1) was expressed in mouse fibroblasts, it displayed a diffuse pattern (L.W. and S.B., unpublished observations), which is somewhat unexpected for an AKAP. We later obtained antibodies against D-AKAP2 that detect bands of much larger size than a 372-residue protein in Western blots of mouse tissue extracts (see Results). Therefore, we suspected that the original mouse clone does not represent full-length D-AKAP2, possibly because of a defect in the cDNA library or an artifact in sequencing. Here, we report the cloning of full-length human D-AKAP2 as well as the mouse homolog less two exons at the N terminus, its association with PKA in vivo, and its subcellular localization determined by immunocytochemistry, immunohistochemistry, and tissue fractionation.
Experimental Procedures
Cloning of Full-Length Human D-AKAP2 and the Mouse Homolog.
Primers based on human D-AKAP2 sequence submitted to National Center for Biotechnology Information by Fischer and Chattergee, were used to amplify fragments of the coding region of D-AKAP2 from human brain cDNA (CLONTECH). A primer covering an EcoRI site, nine nucleotides 5′ to the initiating Met and the first four amino acid codons, was initially used together with a primer encoding Val512 to Lys518. In addition, an IMAGE consortium clone no. 838564 encoding the C terminus of human D-AKAP2 was obtained from Research Genetics (Huntsville, AL). The PCR-generated 1.6-kb fragment was digested with EcoRI and TaqI and ligated into pSP73 (Promega) and/or pFB-HTA (GIBCO/BRL) together with a 1-kb fragment of clone no. 838564 (TaqI, partial NheI digested). The 2.5-kb insert was sequenced in both directions. Most of the missing N-terminal region of mouse D-AKAP2 was PCR amplified from mouse brain cDNA (CLONTECH) by using primers based on known mouse and human sequence, and random mouse cDNA sequences homologous to the human gene from GenBank. All primer sequences are available on request.
Western Blot.
Mouse tissue extracts, mouse or rat tissue fractions, or in vitro-translated proteins were analyzed on SDS-PAGE, transferred to poly(vinylidene difluoride) membranes, and probed with polyclonal antibodies against D-AKAP2. Three different antigens were used individually to generate these antibodies: the 27-residue PKA binding motif as indicated in Fig. 1B, the N-terminal 260 residues of human D-AKAP2, and the C-terminal 372 residues of the mouse homologue. Adult mouse tissues were homogenized in cold homogenization buffer [20 mM Hepes, pH 7.4, 20 mM NaCl, 5 mM EDTA, 5 mM EGTA, 0.5% Triton X-100, 1 mM DTT, and protease inhibitors mixture (Calbiochem)] with a motor driven glass–Teflon homogenizer. The homogenates were centrifuged at 10,000 × g for 30 min at 4°C to obtain the supernatant. In vitro transcription–translation was done with the TnT quick-coupled kit from Promega.
Figure 1.
(A) cDNA and deduced protein sequence of human D-AKAP2. Amino acid numbers are shown on the right. The two putative RGS domains are underlined. The star indicates a stop codon. Vertical arrows indicate the beginning of each numbered exon. Exons 1 and 15 extend beyond the cDNA sequence we have. (B) Comparison of the sequence of D-AKAP2 from four species: HU, human; MS, mouse; FL, fly, drosophila; CE, C. elegans. The PKA-binding region is indicated by stars (*). Horizontal lines above the sequence indicate the two putative RGS domains. (C) Alignment of the RGS domains of the N- and C-terminal portions of D-AKAP2. Comparison of the RGS domains of various RGS molecules in a multiple sequence alignment were generated by CLUSTAL W. The sequences include the N-terminal domain of D-AKAP2 (amino acids 125–242), C-terminal domain of D-AKAP2 (amino acids 379–502), rat RGS4 (amino acids 62–174, GenBank no. AF117211), human axin (amino acids 88–207, GenBank no. AF009674), P115 (amino acids 48–166, GenBank no. U64105), Drosophila Rho GEF2 (amino acids 930-1052, GenBank no. AF032870), and human GRK2 (amino acids 54–171, GenBank no. P25098). Also illustrated is the secondary structure of the largely α-helical RGS4, based on the x-ray crystal structure described by Tesmer et al. (24). α-Helical domains are represented by horizontal lines.
Isolation of in Vivo Complexes Between D-AKAP2 and the R Subunits of PKA.
Mouse brain extract was prepared as described above. The supernatant was incubated with cAMP agarose (Sigma) in the presence or absence of 50 mM cAMP at 4°C overnight. The resin was then washed twice with high salt buffer (10 mM Hepes, pH 7.4, 1.5 mM MgCl2, 10 mM KCl, 0.5 M NaCl, 0.1% Nonidet P-40, 1 mM DTT, and protease inhibitors), four times with low salt buffer (high salt buffer without NaCl), and eluted twice with 25 mM cAMP by rotating 1 h at room temperature. The eluted protein was precipitated with 10% trichloroacetic acid, and analyzed by Western blot using anti-D-AKAP2 and commercial antibodies against RIα, RIIα (Transduction Laboratories, Lexington, KY), and RIIβ (Biomol, Plymouth Meeting, PA). After elution, the resin was boiled in SDS sample buffer for remaining bound proteins.
Isolation of Mitochondria from Mouse and Rat Tissues.
Mouse and rat brown or white adipose tissue mitochondria were isolated as described (22, 23). Tissues homogenized in 250 mM sucrose were spun at 8500 × g to obtain the cytosolic supernatant. The pellet was resuspended and spun at 800 × g to remove the nuclear pellet. Mitochondria were pelleted at 8500 × g and washed in 100 mM KCl with 1% BSA. The fractions were checked by electron microscopy to assure the purity and quality of mitochondria.
Electron Microscopy of Isolated Mitochondria.
Pelleted brown adipose tissue (BAT) mitochondria were fixed on ice with 2% paraformaldehyde/2.5% glutaraldehyde and postfixed with 1% osmium tetroxide. Ethanol was used for graded dehydration, and Durcupan ACM was used for embedment. Sections were cut with a thickness ≈50 nm and stained for 10 min in 2% aqueous uranyl acetate, followed by 2 min in lead salts. Images of nominally untilted sections were collected with a JEOL 100CX electron microscope operated at 80 kV.
Immunocytochemistry.
Cells were fixed with 4% paraformaldehyde for 30 min on ice, rinsed in PBS with 0.4 mg/ml glycine for 2 min, and permeabilized with blocking buffer (0.1% Triton X-100, 1% normal donkey serum, 1% fish gelatin, and 1% BSA in PBS). Cells were then stained with a mixture of primary antibodies (rabbit anti-D-AKAP2 or preimmune serum and antigen-blocked control, and mouse anti-cytochrome c from PharMingen) overnight at 4°C, followed by a mixture of secondary antibodies (fluorescein-labeled donkey anti-rabbit IgG and rhodamine red X-labeled donkey anti-mouse IgG from Jackson ImmunoResearch) for 1 h at 4°C in the dark. Antibodies were diluted in working buffer (blocking buffer diluted ten times with PBS and 0.1% Triton X-100), and cells were washed extensively with the same buffer after incubation with each antibody. Preimmune serum or antigen-blocked D-AKAP2 antibodies were used to control for nonspecific signals. Laser-scanning confocal microscopy was performed as described previously (13).
Immunohistochemistry.
Rats were whole-body perfused (intracardial) under anesthesia first with Ringer's solution and then with 4% paraformaldehyde and 0.1% glutaraldehyde in PBS. Cerebellar tissue was extracted and immersion fixed with the same aldehyde fixatives on ice for 2 h. Approximately 80-μm-thick vibratome sections were cut. These sections were extensively rinsed in PBS-glycine and labeled as described in the previous paragraph on immunocytochemistry.
Results
Cloning of Full-Length D-AKAP2.
D-AKAP2 was cloned from human brain cDNA. The cDNA encodes for a 662-aa residue protein with a predicted molecular weight of 74 kDa (Fig. 1A). Through a search in the BLAST human genome database, the gene for human D-AKAP2 was located to chromosome 17q, with 15 exons as indicated in Fig. 1A. The C-terminal 372 residues of human D-AKAP2 share 94% identity with the previously cloned mouse homologue (Fig. 1B) (15). However, endogenous D-AKAP2 detected in mouse tissue extracts migrated in SDS-PAGE with a much higher apparent molecular weight than predicted from the 372 residues (Fig. 2). We conclude that the previously cloned mouse gene is truncated at the N terminus resulting from a cDNA library artifact or a compression in the sequencing analysis, leading to a frame shift that caused a false stop codon in the readout. Exon 3–15 of the mouse protein was subsequently cloned based on homology with human D-AKAP2 and random mouse cDNA in the GenBank. Two additional D-AKAP2 homologs were identified in the Drosophila and Caenorhabditis elegans genomes, and are compared with those from human and mouse in Fig. 1B.
Figure 2.
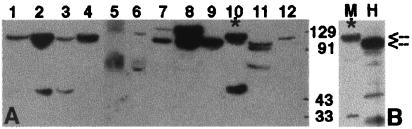
Expression of the D-AKAP2 protein in various mouse tissues. (A) Mouse tissue extracts were prepared as described in Experimental Procedures; 100 μg of total protein was loaded and probed with a polyclonal antibody against D-AKAP2 (anti-D-AKAP2). Antiserum, protein A, or antigen affinity-purified antibodies gave the same result, and none of the bands were detected by a number of unrelated control antibodies. The blots shown here used protein A purified antibody against the C terminus of mouse D-AKAP2. Lanes correspond to extracts from: white adipose tissue (1), BAT (2), skeletal muscle (3), tongue (4), small intestine (5), kidney (6), lung (7), brain (8), pancreas (9), heart (10), spleen (11), and liver (12). (B) Comparison of mouse and human full-length D-AKAP2. Human D-AKAP2 was in vitro translated by using a expression vector containing the cDNA as described in Experimental Procedures. It was then loaded onto SDS-PAGE along with a sample of mouse heart extract and probed with anti-D-AKAP2. M, mouse; H, human. The two arrows indicate the mobilities of the two full-length proteins. Lane 10 and M correspond to the same extract (*).
The C-terminal 40 residues of D-AKAP2 were shown to be responsible for the interaction with the R subunits of PKA (15). This motif is well conserved between mouse and human D-AKAP2: only three residues are different, one of which is a “positive” change between Ile and Val (Fig. 1B). Within this region, a 27-residue peptide, as indicated in Fig. 1B, was found to be sufficient for PKA binding (L. Burns, Y. Ma, D. Barraclough, and S.S.T., unpublished data). In addition, two putative RGS domains were identified in the full-length human D-AKAP2 (Fig. 1A). An alignment with other known RGS domains is shown in Fig. 1C. Also illustrated is the secondary structure of RGS4 based on the x-ray crystal structure (24). Overall, the RGS domains and the PKA binding motif represent the most conserved regions among all four species (Fig. 1B).
Tissue-Specific Expression Pattern of D-AKAP2.
To define the expression pattern of D-AKAP2, mouse tissue extracts were probed with polyclonal antibodies against mouse and human D-AKAP2 as described in Experimental Procedures. Fig. 2A shows a representative Western blot indicating that D-AKAP2 from different mouse tissues displays different molecular weights. D-AKAP2 in heart muscle and adipose tissues migrates more slowly than D-AKAP2 in other tissues such as spleen and pancreas, whereas brain shows at least two prominent bands. The bands below the 91-kDa marker are most likely degradation products because their relative abundance varies between different experiments. However, the differences in the full-length D-AKAP2 from different tissues are observed consistently. Such differences could result from alternative mRNA splicing or posttranslational modification. The protein is most abundant in brain, BAT, heart, and pancreas. The human protein generated by in vitro transcription–translation using the cloned cDNA has slightly different gel mobility as compared with the mouse heart isoform (Fig. 2B) but is similar to the faster migrating predominant isoform in other tissues. Like some other AKAPs, full-length D-AKAP2 has a higher apparent molecular weight on many gels than is predicted from the sequence. This is true for both endogenous proteins and for the cell-free translation products.
To assess the gene expression pattern of the D-AKAP2 gene in human tissues, we used a semiquantitative PCR-based analysis of a multitissue cDNA panel (CLONTECH). We detected a single cDNA species for D-AKAP2 that is expressed ubiquitously at slightly different levels (data not shown).
Association of D-AKAP2 with the R Subunits of PKA in Vivo.
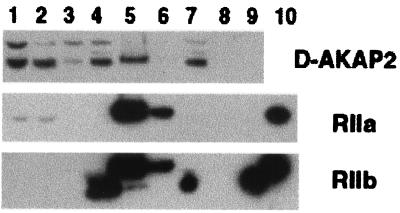
D-AKAP2 was originally discovered by yeast two-hybrid screen (15). We confirmed the in vivo binding of PKA to D-AKAP2 by using a cAMP agarose pull-down assay with mouse brain extract. As shown in Fig. 3, D-AKAP2 was pulled-down along with both RIIα and RIIβ, indicating complex formation under physiological conditions. Because D-AKAP2 is only one of the many AKAPs in brain, the amount that was pulled down is less than the total R subunits. The RIIβ antibody picked up a band that binds to the resin, even in the presence of cAMP and is therefore nonspecific. No detectable amount of RIα was found in any of the samples, indicating that RIα is not abundant in brain. This result represents evidence that D-AKAPs can bind to PKA in vivo.
Figure 3.
Isolation of complexes between D-AKAP2 and RII from mouse brain extract. cAMP pull-down assays were performed as described in Experimental Procedures. Lanes 1–3 are: total homogenate, supernatant, and resuspended pellet (a small fraction of the whole sample loaded). Lanes 4–6 and 7–9 are: flow through, cAMP elute and sample buffer elute after binding to the resin in the absence (lanes 4–6) or presence (lanes 7–9) of cAMP (entire amount of sample loaded). Lane 10 has purified RII standards.
Subcellular Localization of Endogenous D-AKAP2.
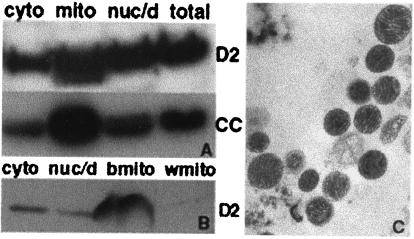
Because a primary function of AKAPs is to target PKA to specific subcellular locations, we investigated the subcellular distribution of D-AKAP2 by using multiple approaches. Subcellular fractionation of BAT from mouse (Fig. 4A) or rat (Fig. 4B) suggests that D-AKAP2 is enriched in the mitochondrial fraction along with cytochrome c, a mitochondria marker. Quality of the isolated mitochondria was routinely monitored by electron microscopy (Fig. 4C).
Figure 4.
Distribution of D-AKAP2 in mouse and rat adipose tissue fractions. Mitochondria were isolated from mouse BAT (A) or rat BAT (b) and WAT (w) (B) as described in Experimental Procedures to get the cytosolic supernatant, the nuclear and cell debris pellet and the mitochondria fraction. Then, 50 μg of total protein was loaded and probed with anti-D-AKAP2 (D2) or anti-cytochrome c (CC). Electron microscopy images were collected to indicate the quality of the isolated mitochondria and a representative image of BAT mitochondria is shown in C.
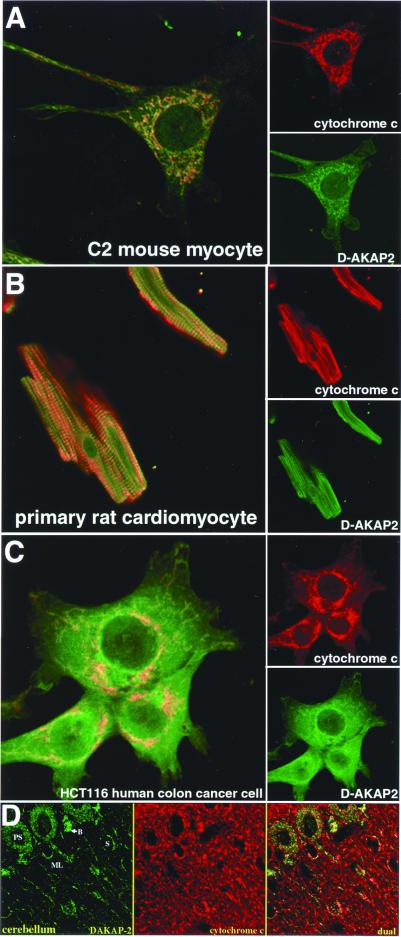
Immunocytochemistry coupled with confocal microscopy on a number of cell types from different species supports mitochondrial localization. In the C2 mouse skeletal muscle cell line (Fig. 5A) and HCT116 human colon cancer cell line (Fig. 5C), the majority of D-AKAP2 is colocalized with cytochrome c, an archetype mitochondria marker. Similar results were obtained from primary culture of rat cardiomyocytes, a more physiologically relevant system (Fig. 5B). Preimmune serum or specific antigen-blocked anti-D-AKAP2 gave no significant signal in control experiments (data not shown).
Figure 5.
Immunocytochemistry and immunohistochemistry of D-AKAP2. (A) Mouse skeletal muscle cell line; (B) rat cardiomyocyte primary culture; (C) human colon cancer cell line; (D) cerebellum tissue. D-AKAP2 codistributes with cytochrome c in a subset of mitochondria. Strong codistribution is found in the Purkinje cell soma (PS), basket cells (B), and stellate cells (S) in the molecular layer (ML). Much weaker D-AKAP2 labeling is present in the dendrites and fibers of the molecular layer. Cells and tissue slices were stained with anti-D-AKAP2 (green) and anti-cytochrome c (red). Yellow areas indicate colocalization to the mitochondria.
To confirm DAKAP-2 association with mitochondria in cultured cells, we performed confocal immunofluorescence microscopy on slices of rat brain tissue (cerebellum; Fig. 5D). Interestingly, it appears that DAKAP-2 associates with mitochondria in certain cellular subregions, but not others. By comparing the DAKAP-2 pattern with that of cytochrome c, we found that labeling of mitochondria occurred predominantly in the Purkinje cell soma, basket cells, and stellate cells. Labeling was either weak or nonexistent in the many mitochondria of the Purkinje cell dendrites, parallel fibers, and climbing fibers of the molecular layer. Little labeling was seen in the granule cell layer (not shown).
Discussion
Compartmentalization of signaling molecules through association with anchoring proteins ensures specificity in signal transduction by placing enzymes close to their appropriate effectors and substrates. In the case of PKA, subcellular targeting is achieved in part through the association between the R subunits and AKAPs. We have previously identified a cDNA from mouse that encodes a D-AKAP capable of binding both RI and RII (15). We report here the cloning of human D-AKAP2 that shares strong sequence homology to the previously cloned mouse D-AKAP2, but contains a substantially longer N terminus. We propose that the original mouse orthologue represents a truncated fragment of full-length D-AKAP2, of which the majority of the missing protein sequence is also reported here. By using multiple approaches, we demonstrated that D-AKAP2 is primarily localized to the mitochondria.
The observation that PKA protein or enzymatic activity is associated with mitochondria has been documented for decades. The enzyme was present in mitochondria purified from guinea pig and rat liver, as well as rat and porcine ovaries (25–27). In bovine heart, PKA activity was detected in the soluble matrix, or the matrix side of the inner membrane of mitochondria (28, 29). When probing for RII with immunogold in mammalian sperm, Lieberman et al. (30) found that almost all RII-specific labeling in the midpiece of the demembraned sperm tail can be assigned to the mitochondria. PKA was also observed in purified invertebrate and yeast mitochondria (31, 32). Consistent with these observations, it has been shown that cAMP can be transported into mitochondria. When incubated with rat liver mitochondria, labeled cAMP can apparently penetrate not only through the outer membranes, but also into mitoplasts, where it is accumulated mainly in the matrix (33).
It is likely that localization of PKA to mitochondria is mediated in part by specific AKAPs. We previously identified another dual-specific AKAP, D-AKAP1, a homolog of S-AKAP84 and AKAP121, that appears to target to the outer mitochondrial membrane (14, 34–36). Furthermore, only one of the two alternative N termini of D-AKAP1 targets to mitochondria (13). RIα was shown subsequently to bind to cytochrome c oxidase subunit Vb in a cAMP-dependent manner (37). In the present study, we provide evidence that D-AKAP2 is predominantly, but not exclusively, localized to mitochondria.
It is striking that DAKAP-2 associates with a defined subset of cerebellum mitochondria. These mitochondria are in the somas of Purkinje, basket, and stellate cells. Little labeling is found in the dendrites and fibers of the molecular layer. Because the basket and stellate cells function as inhibitory modulators of the action potential, it is possible that D-AKAP2 plays a role in this process.
The mechanism by which D-AKAP2 is targeted to the mitochondria is not clear. In most cases, the precursors of nuclear-encoded mitochondrial proteins have N-terminal presequences. Instead of sharing distinct consensus sequences, these presequences have common features in overall amino acid composition (38, 39). They are generally rich in positively charged residues, particularly arginine, and hydroxylated amino acids, and lack acidic residues. These presequences also have the potential to form amphipathic helical secondary structure. In the case of D-AKAP2, 7 of the first 30 residues are positively charged, 6 are Arg. There are also four Ser and Thr residues. In contrast, only one acidic residue (Asp) is found in this segment. Therefore, the N terminus of D-AKAP2 may serve as a mitochondrial targeting motif. The presence of four Pro residues, however, suggests that it will not be helical and the N terminus alone does not appear to target independently to mitochondria (unpublished results).
Subcellular localization of D-AKAP2 by using microscopy in all systems examined reveals a low level diffuse background staining and low levels of nuclear staining, in addition to the mitochondrial staining. This may indicate the existence of multiple pools of D-AKAP2 and perhaps a dynamic regulatory aspect of D-AKAP2 function. In addition, the human cell line we tested seems to have more background staining than the mouse and rat cells. One major mouse D-AKAP2 protein has a molecular weight similar to the protein expressed by the cloned human gene (Fig. 3). However, because endogenous mouse D-AKAP2 from other tissues including heart, and at least one isoform from brain, run higher than the cloned human full-length protein when visualized with an antibody against a C-terminal peptide (Fig. 3B), the N-terminal sequence (exons 1 and 2) of some isoforms may be different.
Our work demonstrated that the two D-AKAPs identified through two-hybrid screen can indeed bind to the R subunits of PKA in vivo (the data for D-AKAP1 is beyond the scope of this paper and not shown). Functional implications of such complexes remain to be elucidated.
The physiological relevance of interactions between PKA and AKAPs with mitochondria is not fully understood. Several PKA substrates involved in mitochondrial respiration have been identified recently (40, 41). In addition to its central role in aerobic energy production, mitochondria are also involved in triggering the apoptotic pathway (42, 43). PKA exhibits either pro-apoptotic or anti-apoptotic roles in different cell lines, or the same cell line under different conditions (44). Harada et al. (45) provide the most supportive evidence, whereby they demonstrate that BAD, a pro-apoptotic member, is phosphorylated and inactivated by mitochondria-anchored PKA. Furthermore, this process is inhibited if the association of PKA and AKAP is disrupted by the universal anchoring motif peptide mimetic, Ht31 (45). Future experiments will elucidate whether D-AKAP2 plays a direct role in apoptosis by mediating the localization of PKA to the mitochondria.
The full-length D-AKAP2 has two putative RGS domains. A multitude of RGS proteins have been recently identified, many of which possess the ability to interact with heterotrimeric G protein α subunits and function primarily as GAPs (18–21). A conserved core composed of approximately 120 amino acids has been identified as the RGS domain, a structural motif of largely α-helical composition. Although G protein and cAMP signaling pathways crosstalk directly through the regulation of adenylyl cyclase activity by various Gα subunits, D-AKAP2 may provide an additional link between the two pathways. Unfortunately, we have been unable to detect physical interactions between D-AKAP2 and any G proteins, nor any GAP activity in D-AKAP2 or its fragments encompassing one or both of the RGS domains (R.K.S. and A.K., unpublished observations). This may be explained by the unique features of D-AKAP2 compared with other RGS family members in terms of sequence homology. In fact, D-AKAP2 does not belong to any of the six subfamilies of RGS proteins categorized based on their conserved residues, domain structure organization, or phylogenetic relations (46). D-AKAP2 may therefore possess some unconventional characteristics related to or beyond Gα interaction and regulation. There are other exceptions in the RGS family that do not exhibit known G protein binding or GAP activity. One such example is axin, a negative regulator of the wnt signaling pathway that directly interacts with adenomatous polyposis coli (APC) through its RGS domain, and regulates the stabilization of β−catenin (47). A crystal structure of axin complexed with an axin-binding sequence from APC reveals that the APC binding surface is distinct from the surface that binds to classic G proteins (48). Thus, it is still unclear whether D-AKAP2 has a nonclassical G protein partner and/or whether it binds to another unrelated protein. The recent identification of a soluble adenylyl cyclase that is insensitive to G protein regulation (49) raised the possibility that additional cAMP and/or G protein signaling pathways may exist. It is also interesting to note that immunofluorescent studies on the adenylyl cyclase inhibitory G protein, Giα, revealed distinct mitochondrial and plasma membrane staining in multiple cell lines (50). Although the functional significance of this observation awaits further evaluation, it may eventually integrate into a mechanism by which D-AKAP2 facilitates PKA as well as G protein signal transduction.
Acknowledgments
We thank Dr. Palmer Taylor for providing the C2 cells, Dr. Larry Brunton and colleagues for the primary cardiomyocytes, and Dr. Degeng Wang for the HCT116 cells. This project was supported by National Institutes of Health Grant P01 DK54441.
Abbreviations
- AKAP
A kinase anchoring protein
- D-AKAP
dual-specific AKAP
- PKA
protein kinase A
- RI/RII
type I/II regulatory subunit of PKA
- RGS
regulator of G protein signaling
- GAP
GTPase-activating protein
- BAT
brown adipose tissue
Footnotes
References
- 1.Cummings D E, Brandon E P, Planas J V, Motamed K, Idzerda R L, McKnight G S. Nature (London) 1996;382:622–626. doi: 10.1038/382622a0. [DOI] [PubMed] [Google Scholar]
- 2.Jones P M, Sayed S B, Persaud S J, Burns C J, Gyles S, Whitehouse B J. J Mol Endocrinol. 2000;24:233–239. doi: 10.1677/jme.0.0240233. [DOI] [PubMed] [Google Scholar]
- 3.Whitehouse B J, Abayasekara D R. J Mol Endocrinol. 1994;12:195–202. doi: 10.1677/jme.0.0120195. [DOI] [PubMed] [Google Scholar]
- 4.Chen T C, Hinton D R, Zidovetzki R, Hofman F M. Lab Invest. 1998;78:165–174. [PubMed] [Google Scholar]
- 5.Tasken K, Skalhegg B S, Tasken K A, Solberg R, Knutsen H K, Levy F O, Sandberg M, Orstavik S, Larsen T, Johansen A K, et al. Adv Second Messenger Phosphoprotein Res. 1997;31:191–204. doi: 10.1016/s1040-7952(97)80019-5. [DOI] [PubMed] [Google Scholar]
- 6.Cross T G, Scheel-Toellner D, Henriquez N V, Deacon E, Salmon M, Lord J M. Exp Cell Res. 2000;256:34–41. doi: 10.1006/excr.2000.4836. [DOI] [PubMed] [Google Scholar]
- 7.Rubin C S. Biochim Biophys Acta. 1994;1224:467–479. [PubMed] [Google Scholar]
- 8.Coghlan V M, Bergeson S E, Langeberg L, Nilaver G, Scott J D. Mol Cell Biochem. 1993;127–128:309–319. doi: 10.1007/978-1-4615-2600-1_28. [DOI] [PubMed] [Google Scholar]
- 9.Scott J D, McCartney S. Mol Endocrinol. 1994;8:5–11. doi: 10.1210/mend.8.1.8152430. [DOI] [PubMed] [Google Scholar]
- 10.Pawson T, Scott J D. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 11.Newlon M G, Roy M, Hausken Z E, Scott J D, Jennings P A. J Biol Chem. 1997;272:23637–23644. doi: 10.1074/jbc.272.38.23637. [DOI] [PubMed] [Google Scholar]
- 12.Colledge M, Scott J D. Trends Cell Biol. 1999;9:216–221. doi: 10.1016/s0962-8924(99)01558-5. [DOI] [PubMed] [Google Scholar]
- 13.Huang L J, Wang L, Ma Y, Durick K, Perkins G, Deerinck T J, Ellisman M H, Taylor S S. J Cell Biol. 1999;145:951–959. doi: 10.1083/jcb.145.5.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang L J, Durick K, Weiner J A, Chun J, Taylor S S. J Biol Chem. 1997;272:8057–8064. doi: 10.1074/jbc.272.12.8057. [DOI] [PubMed] [Google Scholar]
- 15.Huang L J, Durick K, Weiner J A, Chun J, Taylor S S. Proc Natl Acad Sci USA. 1997;94:11184–11189. doi: 10.1073/pnas.94.21.11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Angelo R, Rubin C S. J Biol Chem. 1998;273:14633–14643. doi: 10.1074/jbc.273.23.14633. [DOI] [PubMed] [Google Scholar]
- 17.Miki K, Eddy E M. J Biol Chem. 1998;273:34384–34390. doi: 10.1074/jbc.273.51.34384. [DOI] [PubMed] [Google Scholar]
- 18.Berman D M, Kozasa T, Gilman A G. J Biol Chem. 1996;271:27209–27212. doi: 10.1074/jbc.271.44.27209. [DOI] [PubMed] [Google Scholar]
- 19.Berman D M, Wilkie T M, Gilman A G. Cell. 1996;86:445–452. doi: 10.1016/s0092-8674(00)80117-8. [DOI] [PubMed] [Google Scholar]
- 20.Watson N, Linder M E, Druey K M, Kehrl J H, Blumer K J. Nature (London) 1996;383:172–175. doi: 10.1038/383172a0. [DOI] [PubMed] [Google Scholar]
- 21.De Vries L, Gist Farquhar M. Trends Cell Biol. 1999;9:138–144. doi: 10.1016/s0962-8924(99)01515-9. [DOI] [PubMed] [Google Scholar]
- 22.Cannon B, Lindberg O. Methods Enzymol. 1979;55:65–78. doi: 10.1016/0076-6879(79)55010-1. [DOI] [PubMed] [Google Scholar]
- 23.Hovius R, Lambrechts H, Nicolay K, de Kruijff B. Biochim Biophys Acta. 1990;1021:217–226. doi: 10.1016/0005-2736(90)90036-n. [DOI] [PubMed] [Google Scholar]
- 24.Tesmer J J, Berman D M, Gilman A G, Sprang S R. Cell. 1997;89:251–261. doi: 10.1016/s0092-8674(00)80204-4. [DOI] [PubMed] [Google Scholar]
- 25.Kleitke B, Sydow H, Wollenberger A. Acta Biol Med Ger. 1976;35:K9–K17. [PubMed] [Google Scholar]
- 26.Dimino M J, Bieszczad R R, Rowe M J. J Biol Chem. 1981;256:10876–10882. [PubMed] [Google Scholar]
- 27.Inaba T, Wiest W G. Endocrinology. 1985;117:315–322. doi: 10.1210/endo-117-1-315. [DOI] [PubMed] [Google Scholar]
- 28.Burgess J W, Yamada E W. Biochem Cell Biol. 1987;65:137–143. doi: 10.1139/o87-019. [DOI] [PubMed] [Google Scholar]
- 29.Sardanelli A M, Technikova-Dobrova Z, Speranza F, Mazzocca A, Scacco S, Papa S. FEBS Lett. 1996;396:276–278. doi: 10.1016/0014-5793(96)01112-x. [DOI] [PubMed] [Google Scholar]
- 30.Lieberman S J, Wasco W, MacLeod J, Satir P, Orr G A. J Cell Biol. 1988;107:1809–1816. doi: 10.1083/jcb.107.5.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vallejo C G, Seguido A M, Fernandez-Renart M. Arch Biochem Biophys. 1997;339:9–16. doi: 10.1006/abbi.1996.9842. [DOI] [PubMed] [Google Scholar]
- 32.Muller G, Bandlow W. Z Naturforsch, C. 1987;42:1291–1302. doi: 10.1515/znc-1987-11-1224. [DOI] [PubMed] [Google Scholar]
- 33.Kulinskii V I, Zobova N V. Biokhimiya. 1985;50:1546–1552. [PubMed] [Google Scholar]
- 34.Lin R Y, Moss S B, Rubin C S. J Biol Chem. 1995;270:27804. doi: 10.1074/jbc.270.46.27804. [DOI] [PubMed] [Google Scholar]
- 35.Chen Q, Lin R Y, Rubin C S. J Biol Chem. 1997;272:15247–15257. doi: 10.1074/jbc.272.24.15247. [DOI] [PubMed] [Google Scholar]
- 36.Feliciello A, Rubin C S, Avvedimento E V, Gottesman M E. J Biol Chem. 1998;273:23361–23366. doi: 10.1074/jbc.273.36.23361. [DOI] [PubMed] [Google Scholar]
- 37.Yang W L, Iacono L, Tang W M, Chin K V. Biochemistry. 1998;37:14175–14180. doi: 10.1021/bi981402a. [DOI] [PubMed] [Google Scholar]
- 38.Hartl F U, Neupert W. Science. 1990;247:930–938. doi: 10.1126/science.2406905. [DOI] [PubMed] [Google Scholar]
- 39.Gavel Y, von Heijne G. Protein Eng. 1990;4:33–37. doi: 10.1093/protein/4.1.33. [DOI] [PubMed] [Google Scholar]
- 40.Qu W, Graves L M, Thurman R G. Am J Physiol. 1999;277:G1048–G1054. doi: 10.1152/ajpgi.1999.277.5.G1048. [DOI] [PubMed] [Google Scholar]
- 41.Papa S, Sardanelli A M, Scacco S, Technikova-Dobrova Z. FEBS Lett. 1999;444:245–249. doi: 10.1016/s0014-5793(99)00070-8. [DOI] [PubMed] [Google Scholar]
- 42.Waterhouse N J, Green D R. J Clin Immunol. 1999;19:378–387. doi: 10.1023/a:1020550716138. [DOI] [PubMed] [Google Scholar]
- 43.Kowaltowski A J. Braz J Med Biol Res. 2000;33:241–250. doi: 10.1590/s0100-879x2000000200014. [DOI] [PubMed] [Google Scholar]
- 44.Gu C, Ma Y C, Benjamin J, Littman D, Chao M V, Huang X Y. J Biol Chem. 2000;275:20726–20733. doi: 10.1074/jbc.M000152200. [DOI] [PubMed] [Google Scholar]
- 45.Harada H, Becknell B, Wilm M, Mann M, Huang L J, Taylor S S, Scott J D, Korsmeyer S J. Mol Cell. 1999;3:413–422. doi: 10.1016/s1097-2765(00)80469-4. [DOI] [PubMed] [Google Scholar]
- 46.Zheng B, De Vries L, Gist Farquhar M. Trends Biochem Sci. 1999;24:411–414. doi: 10.1016/s0968-0004(99)01474-7. [DOI] [PubMed] [Google Scholar]
- 47.Kishida S, Yamamoto H, Ikeda S, Kishida M, Sakamoto I, Koyama S, Kikuchi A. J Biol Chem. 1998;273:10823–10826. doi: 10.1074/jbc.273.18.10823. [DOI] [PubMed] [Google Scholar]
- 48.Spink K E, Polakis P, Weis W I. EMBO J. 2000;19:2270–2279. doi: 10.1093/emboj/19.10.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buck J, Sinclair M L, Schapal L, Cann M J, Levin L R. Proc Natl Acad Sci USA. 1999;96:79–84. doi: 10.1073/pnas.96.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lewis J M, Woolkalis M J, Gerton G L, Smith R M, Jarett L, Manning D R. Cell Regul. 1991;2:1097–1113. doi: 10.1091/mbc.2.12.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]