Summary
Human PUMILIO1 (PUM1) and PUMILIO2 (PUM2) are members of the PUMILIO/FBF (PUF) family that regulate specific target mRNAs posttranscriptionally. Recent studies have identified mRNA targets associated with human PUM1 and PUM2. Here we explore the structural basis of natural target RNA recognition by human PUF proteins through crystal structures of the RNA-binding domains of PUM1 and PUM2 in complex with four cognate RNA sequences including sequences from p38α and erk2 MAP kinase mRNAs. We observe three distinct modes of RNA binding around the 5th RNA base, two of which are different from the prototypical 1 repeat:1 RNA base binding mode previously identified with model RNA sequences. RNA-binding affinities of PUM1 and PUM2 are not affected dramatically by the different binding modes in vitro. However, these modes of binding create structurally variable recognition surfaces that suggest a mechanism in vivo for recruitment of downstream effector proteins defined by the PUF:RNA complex.
INTRODUCTION
RNA-binding proteins play critical roles in gene expression through regulation of RNA splicing, localization, translation, and decay. Members of the PUF family, named after the two founding members Drosophila melanogaster Pumilio (PUM) and Caenorhabditis elegans fem-3 binding factor (FBF), are RNA-binding proteins that regulate gene expression posttranscriptionally. They induce mRNA decay or repress translation (Wickens et al., 2002) and have recently been shown to activate translation of some mRNA targets (Kaye et al., 2009; Suh et al., 2009). PUF proteins exist exclusively in eukaryotes and bind sequence specifically to regulatory sequences in the 3′ UTRs of their target mRNAs. All PUF proteins share a highly conserved RNA-binding domain known as the Pumilio-homology domain (PUM-HD) or PUF domain (Wharton et al., 1998; Zamore et al., 1997; Zhang et al., 1997). Structural studies of different PUF proteins with RNA have revealed the details of their RNA recognition schemes (Gupta et al., 2008; Miller et al., 2008; Wang et al., 2002; Wang et al., 2009b; Zhu et al., 2009).
Crystal structures of the PUM-HD of human PUMILIO1 (PUM1) bound to the Nanos Response Element (NRE) sequences in D. melanogaster hunchback (hb) mRNA provide a prototypical model of modular RNA recognition (Wang et al., 2002). The PUM-HD comprises eight α-helical PUM repeats and two pseudo-repeats at the N and C termini, which together adopt a crescent shape. The inner concave surface binds target RNAs in an ‘anti-parallel’ orientation with the N -terminal end of the protein binding to the 3′ end of the RNA. Each PUM repeat recognizes one RNA base using three side chains at specific positions in the repeat. Thus 8 RNA bases are recognized by 8 PUM repeats. We refer to this as a 1:1 binding mode. Two side chains contact the Watson-Crick edge of the base and a third side chain stacks with the same base and/or preceding base. Certain combinations of side chains recognize particular RNA bases. Mutation of these conserved combinations of residues allows design of the RNA recognition specificity of PUM-HDs (Cheong and Hall, 2006; Furman et al., 2010; Koh et al., 2009; Opperman et al., 2005; Ozawa et al., 2007; Stumpf et al., 2008; Tilsner et al., 2009; Wang et al., 2002; Wang et al., 2009a).
In contrast to other RNA-binding protein families with hundreds of different family members per organism, the PUF protein family is small. Humans and other mammals express two PUF proteins, D. melanogaster express one, Saccharomyces cerevisiae express six, C. elegans express nine, and Arabidopsis thaliana encode up to 26. Each organism expresses at least one PUF family member closely related to human PUM1 and Drosophila PUM, which contains a PUM-HD that binds to the recognition sequence found in hb mRNA, 5′-UGUANAUA-3′. All PUM-HDs bind to sequences containing a 5′ UGU sequence.
The identification of mRNA targets of PUF proteins has revealed more variability in mRNA sequence recognition than expected based on the 1:1 binding mode observed in crystal structures of human PUM1 with hb RNA and the high conservation of RNA recognition side chains among PUF proteins. Yeast Puf4p and Puf5p use 8 PUM repeats to bind to sequences containing, respectively, 9 or 10 bases starting from the 5′ UGU (Gerber et al., 2004). Similarly, worm PUF proteins with 8-repeat PUM-HDs recognize longer RNA sequences (Koh et al., 2009; Opperman et al., 2005; Stumpf et al., 2008). Crystal structures of yeast Puf4p and worm FBF-2 demonstrate that additional bases can be accommodated by direct stacking of bases or flipping bases away from the RNA-binding surface, influenced by changes in curvature of the RNA-binding surfaces of these proteins (Miller et al., 2008; Wang et al., 2009b). Additional specificity of a PUM-HD is achieved by a specialized binding pocket at the C-terminal end of the domain of Puf3p, which specifically recognizes a cytosine two bases upstream of the 5′ UGU motif (Zhu et al., 2009). A cytosine at this (−2) position is required for in vivo target recognition. Hence, PUF proteins utilize binding modes in addition to 1 repeat:1 RNA base to recognize RNA targets. As our understanding of the structures and RNA target recognition by specific PUF proteins grows along with corresponding knowledge of downstream effects, computational prediction of binding modes and biological effects may be possible.
Mammalian cells express two PUF proteins, PUM1 and PUM2, whose RNA-binding domains are highly similar to D. melanogaster PUM (80% and 78% amino acid positions identical to PUM, respectively). Until recently, little was known about target mRNAs of human PUM1 and PUM2. Several studies in the past few years have identified mRNA targets associated with human PUM1 and PUM2 and revealed the same consensus recognition sequence as that of fly PUM, 5′-UGUANAUA-3′, where N is A, U, or C (Galgano et al., 2008; Gerber et al., 2006; Hafner et al., 2010; Morris et al., 2008).
The primordial function of PUF proteins appears to be regulating germline stem cell differentiation (Wickens et al., 2002). mRNAs of mitogen-activated protein (MAP) kinases have been shown to be targets of PUF proteins in stem cells (Lee et al., 2007). FBF regulates mapk/erk2 mRNAs in C. elegans germline cells. Human PUM2 has been shown to down-regulate the expression of MAP kinase homolog mRNAs, p38α and erk2, in human embryonic stem cells (Lee et al., 2007). These two mRNAs contain sequences similar to the PUM consensus sequence, and mutation of the UGU motifs in these sequences results in reporter mRNAs refractory to PUM2 regulation.
These advances in the identification of native mRNA targets of human PUF proteins prompted us to revisit how cognate RNA target sequences are recognized and further examine human PUF protein substrate specificities. We determined crystal structures of PUM1-HD and PUM2-HD in complex with four different RNA sequences including three cognate target sequences from MAPK homolog mRNAs. We also analyzed biochemically the affinity and specificity of binding to these RNAs by PUM1 and PUM2. We observe three different modes of binding to RNAs around the 5th RNA base, which varies in the consensus sequences. The different modes of binding do not appear to affect binding affinity in vitro, but in vivo the protein:RNA complexes may present alternative recognition surfaces that could direct downstream effector complex formation.
RESULTS
Structural overview of human PUM1-HD and PUM2-HD in complex with cognate RNAs
To examine recognition of natural mRNA target sequences by human PUF proteins, we determined crystal structures of human PUM1-HD and PUM2-HD bound to 8-nt recognition sequences from the 3′-UTRs of p38α mRNA NREa (5′-UGUAAAUA-3′) and NREb (5′-UGUAGAUA-3′) and erk2 mRNA NRE (5′-UGUACAUC-3′) (Tables 1 and S1). We also determined the crystal structure of PUM1-HD in complex with the Drosophila hb mRNA NRE1 (5′-UGUAUAUA-3′). These RNA targets share the PUM consensus recognition sequence: U1G2U3A4N5A6U7A8, where N is any base. Per convention, PUM recognition sequences are numbered beginning with the conserved 5′ UGU sequence.
Table 1.
Refinement resolution and R values
| PUM1-HD | PUM2-HD | |||||||
|---|---|---|---|---|---|---|---|---|
| RNA |
p38α NREa |
p38α NREb |
erk2 NRE |
erk2 NRE |
hb NRE1 |
p38α NREa |
p38α NREb |
erk2 NRE |
| PDB ID | 3Q0L | 3Q0M | 3Q0N | 3Q0O | 3Q0P | 3Q0Q | 3Q0R | 3Q0S |
| Resolution, Å | 50-2.5 | 50-2.7 | 50-2.4 | 50-2.8 | 50-2.6 | 50-2.0 | 50-2.0 | 50-2.0 |
| Completeness, % | 99.6 | 79.0 | 99.2 | 94.8 | 96.6 | 93.4 | 93.1 | 99.2 |
| Rwork / Rfree ,% | 19.2/ 25.9 | 21.2/ 29.8 | 20.2/ 26.0 | 22.4/ 29.7 | 19.3/ 26.1 | 19.4/ 25.7 | 20.7/ 25.7 | 20.7/ 24.9 |
See supplemental Table S1 for complete statistics
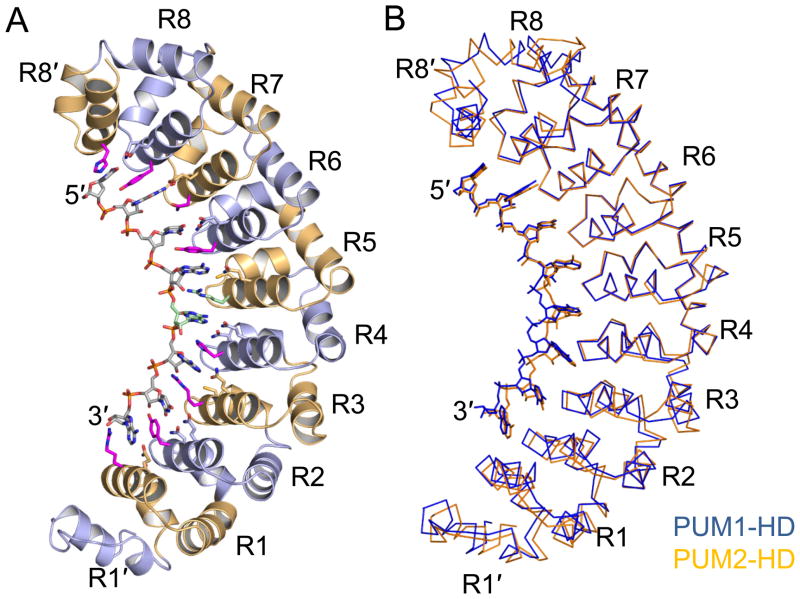
The crystal structures of PUM1-HD and PUM2-HD in complex with p38α NREa are used here to describe the general structural features of these two homology domains (Figure S1). PUM1-HD and PUM2-HD adopt similar folds, in agreement with their high amino acid sequence conservation (94% of amino acid residues are identical). As noted previously for PUM1-HD, the eight PUM repeats (R1-R8) in PUM2-HD form a crescent shape flanked by two pseudo repeats (R1′ and R8′), one at each terminus (Figure 1A).
Figure 1.
Crystal structures of human PUM1-HD and PUM2-HD in complex with p38α NREa RNA. (A) Ribbon diagram of PUM2-HD in complex with p38α NREa (UGUAAAUA). PUM repeats are colored alternately light blue and orange. The RNA is colored according to atom type (gray, carbon; red, oxygen; blue, nitrogen; orange, phosphorus). The 5th RNA base and the side chain of Arg888 are colored cyan. Protein side chains that contact the RNA bases are shown. (B) Superposition of Cα traces of PUM1-HD:p38α NREa (blue) and PUM2-HD:p38α NREa (orange). The two structures are aligned over PUM repeats R4-R8′. The figures were prepared with PyMOL (DeLano, 2002). See also Figure S1 for a stereo view of a PUM2-HD Cα trace.
Structural alignments of PUM1-HD and PUM2-HD reveal a subtle difference in their overall curvatures (Figure 1B). The root mean square deviation (RMSD) over 342 Cα atoms in PUM1-HD and PUM2-HD is 4.4 Å, but the RMSD decreases to 1.7 Å and 1.3 Å, respectively, by aligning separately the two corresponding regions R1′-R3 (133 Cα atoms) and R4-R8′ (209 Cα atoms) in PUM1-HD and PUM2-HD. Aligning other corresponding fragments does not reduce the RMSD. Thus a shift to a flatter curvature in PUM2-HD is centered near the transition from repeat 3 to repeat 4. The flatter curvature is also observed in mouse PUM2-HD (Jenkins et al., 2009), with an RMSD of 2.1 Å over 322 Cα atoms when compared to human PUM2-HD (99% of amino acid residues are identical). As with human PUM1-HD, it appears that the overall structure of PUM2-HD does not change upon RNA binding.
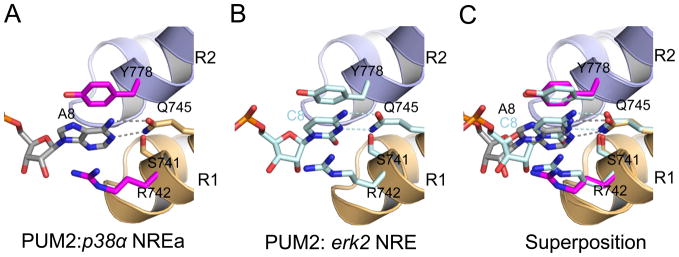
As for all PUF proteins, the inner concave surfaces of PUM1 and PUM2 serve as the platforms for RNA binding. Recognition of the conserved 5′ U1G2U3A4 and 3′ A6U7 regions is identical in all structures to that observed previously for PUM1-HD. Similarly, recognition of A8 in the p38α and hb sequences is conserved in PUM1 and PUM2. However, the 8th base in the PUM2 target erk2 mRNA is C8 instead of A8, which is defined in consensus sequences. The frequency of appearance of C8 in PUM1 RNA-binding sequences is less than 5% (Morris et al., 2008). Nevertheless, recognition of C8 in the erk2 mRNA sequence by PUM1-HD and PUM2-HD resembles that of A8 (Figure 2). A glutamine in PUM repeat 1 (Gln867 in PUM1 or Gln745 in PUM2) contacts the Watson-Crick edge of A8 or C8, which is stacked between arginine (Arg864 in PUM1 or Arg742 in PUM2) and tyrosine (Tyr900 in PUM1 or Tyr778 in PUM2) residues. Similarly, the 1st repeat of C. elegans FBF-2 can recognize different RNA bases using identical side chains (Wang et al., 2009b), although A is preferred in selection experiments and other residues appear in natural target sequences (Bernstein et al., 2005; Opperman et al., 2005).
Figure 2.
Recognition of the 8th RNA base by PUM2. Interaction of PUM repeat 1 of PUM2 with (A) A8 in p38α NREa and (B) C8 in erk2 NRE. (C) Superposition of panels A and B. Structures are shown as in Figure 1 with the carbon atoms in p38α NREa colored grey and erk2 NRE colored light cyan. Hydrogen bonds are indicated by dashed lines.
Crystal structures of PUM1-HD and PUM2-HD in complex with natural target RNA sequences from p38α and erk2 reveal how different bases are accommodated at the 5th position. These natural target RNAs contain A5, G5, or C5, and consensus RNA recognition sequences indicate that PUM1-HD and PUM2-HD bind to RNAs with different bases at the 5th position (Galgano et al., 2008; Hafner et al., 2010; Morris et al., 2008).
Recognition of the Hoogsteen edge of the 5th RNA base
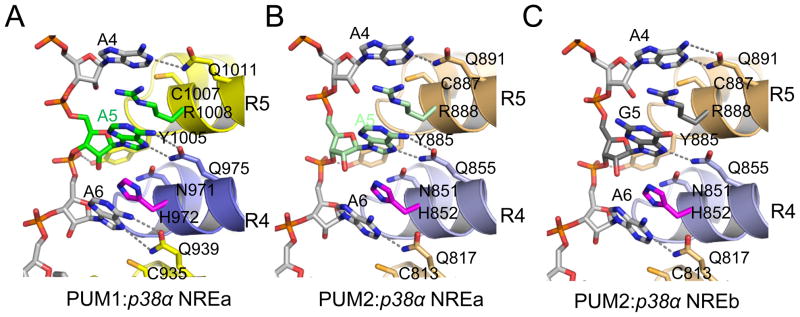
When the 5th RNA base is A (as in p38α NREa), both PUM1-HD and PUM2-HD recognize the Hoogsteen edge of A5 with a conserved glutamine residue (Gln975 in PUM1 and Gln855 in PUM2) (Figure 3A&B). In order to present the Hoogsteen edge of A5, the p38α NREa RNA backbone in the PUM1-HD or PUM2-HD structure adopts a different conformation from that observed in the previously determined PUM1-HD structures in which the Watson-Crick edge of the 5th RNA base is recognized (Figure S2) (Wang et al., 2002). The ribose group of A5 is in a C2′-endo conformation, and Tyr1005 of PUM1 (Tyr885 of PUM2) forms a hydrogen bond with the phosphate group between A5 and A6. PUM1 and PUM2 adopt different strategies for recognizing A6. PUM1 contacts the Hoogsteen edges of both A5 and A6, while PUM2 recognizes the Watson-Crick edge of A6 instead. Similar changes in the RNA backbone conformation and presentation of the Hoogsteen edge of adenosine were also observed for the Puf3 protein of S. cerevisiae (Zhu et al., 2009). Puf3p recognizes the Hoogsteen edges of A5 and A6 in the COX17 Puf3p binding site A, and Tyr695, equivalent to PUM1 Tyr1005 and PUM2 Tyr885, also makes a hydrogen bond with the phosphate group between A5 and A6. Thus, this feature of PUF protein RNA recognition is shared across species.
Figure 3.
Recognition of purine bases at the 5th position by PUM1 and PUM2. The Hoogsteen edges of the 5th purine bases are recognized by PUM1 and PUM2, as in (A) PUM1:p38α NREa, (B) PUM2:p38α NREa, and (C) PUM2:p38α NREb.
We saw a difference in recognition of A5 for the second PUM1-HD:p38α RNA complex in the asymmetric unit. In this complex PUM1 instead recognizes the Watson-Crick edge of A5. This difference is influenced by crystal packing. Tyr1005 in PUM1, which would have supported an RNA backbone conformation change to present the Hoogsteen edge, forms a hydrogen bond with the side chain of Glu912 from a symmetry-related molecule. This crystal contact prohibits Tyr1005 from stabilizing the rearrangement of the backbone of p38α NREa between A5 and A6.
When the 5th RNA base is G (as in p38α NREb), both PUM1 and PUM2 recognize the Hoogsteen edge of G5 instead of the Watson-Crick edge. As with A5, the ribose group of G5 is in a C2′-endo conformation, and a tyrosine residue (Tyr1005 in PUM1 or Tyr885 in PUM2) makes a hydrogen bond with the phosphate group between G5 and A6, stabilizing this conformation (Figure 3C). Thus it appears that a general mechanism for accommodating purine bases at the 5th position is to present the Hoogsteen edge, and PUM1 and PUM2 can recognize either the Watson-Crick or Hoogsteen edges of bases.
A distinct base-omission binding mode at the 5th RNA base
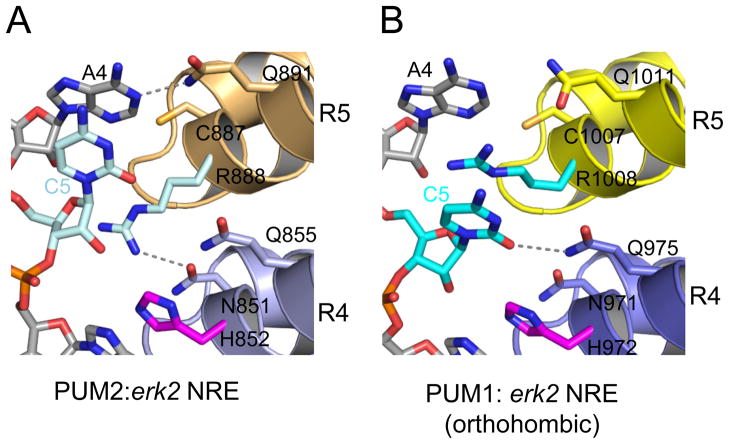
When the 5th RNA base is C (as in erk2 NRE), we observe a third RNA recognition mode by PUM1 and PUM2, which we refer to as the base-omission mode (Figure 4A). In structures of PUM1-HD and PUM2-HD in complex with RNA, the side chain of a conserved arginine in repeat 4 (Arg1008 in PUM1 or Arg888 in PUM2) is typically inserted between the 4th and 5th bases of the RNA, forming stacking interactions with both bases. In the structures of PUM1-HD:erk2 NRE and PUM2-HD:erk2 NRE (monoclinic C2 crystal form), however, C5 stacks directly with A4, and the base is not contacted by repeat 4 of PUM1-HD or PUM2-HD. The arginine side chain that would have stacked between bases 5 and 6 of the erk2 NRE instead interacts with the RNA backbone of bases 5 and 6.
Figure 4.
Base-omission mode of recognition of the 5th RNA base. PUM1 and PUM2 can recognize the C5 RNA base with both (A) the base-omission mode in PUM2:erk2 NRE and (B) the 1:1 binding mode in PUM1: erk2 NRE. See also Figure S2 illustrating the recognition of U5 and C5 by PUM1.
Alternatively, recognition of the Watson-Crick edge of C5 by repeat 4 can occur. In another crystal form of PUM1-HD:erk2 NRE reported here (orthorhombic p212121) and in the previously described crystal structure of PUM1-HD with hb NRE2, Gln975 in repeat 4 recognizes the Watson-Crick edge of C5 and Arg1008 stacks with both A4 and C5 (Figure 4B). Thus PUM1-HD, and possibly PUM2-HD, can bind erk2 NRE in both 1:1 binding and base-omission modes.
RNA-binding preferences of PUM1-HD and PUM2-HD
Given the three observed binding modes to the 5th base of natural target RNA sequences (1:1 Watson-Crick or Hoogsteen binding modes and base-omission mode), we wished to examine whether these changes in structure correlate with changes in RNA-binding affinity. We used electrophoretic mobility shift assays to measure binding affinities of PUM1-HD and PUM2-HD for 8-nt PUM recognition sequences in natural target mRNAs. Both PUM1-HD and PUM2-HD have strong affinities for the cognate 8-nt sequences with Kds for PUM1-HD of 0.63-19 nM and for PUM2-HD of 0.08-11.1 nM (Table 2, Figure S3). For both PUM1-HD and PUM2-HD the weakest binding RNA was the erk2 NRE sequence, which bears a C5 and a C8.
Table 2.
RNA-binding analyses of PUM1 and PUM2
| RNA | RNA sequence | Kd (nM) | |
|---|---|---|---|
| 1 2 3 4 5 6 7 8a | PUM1 | PUM2 | |
| p38α NREa | UGUAAAUA | 0.91 ± 0.08 | 0.17 ± 0.02 |
| p38α NREb | UGUAGAUA | 0.63 ± 0.01 | 0.08 ± 0.01 |
| erk2 | UGUACAUC | 19.02 ± 2.09 | 11.07 ± 1.15 |
| hb NRE1 | UGUAUAUA | 0.40 ± 0.01 | 0.36 ± 0.02 |
| hb NRE2 | UGUACAUA | 1.01 ± 0.05 | 0.07 ± 0.01 |
Numbers indicate positions in the core recognition sequence.
Boldface letters indicate substitutions relative to the p38α NREa sequence. See Figure S3 for representative data.
Since the consensus PUM recognition sequences from genomic screening suggest PUM1-HD and PUM2-HD can bind to any base at position 5, we tested binding to hb NRE1 (5′-UGUAUAUA-3′) and NRE2 (5′-UGUACAUA-3′), which contain U5 and C5, respectively, and are bound by PUM1-HD in 1:1 Watson-Crick binding mode (Figure S2) (Wang et al., 2002). PUM1-HD and PUM2-HD bind tightly to hb NRE1 (U5), hb NRE2 (C5), p38α NREa (A5), and p38α NREb (G5). Thus neither the identity of the 5th base nor the binding mode appears to affect binding affinity dramatically. In addition, the number of observed hydrogen bonds to the 5th base (0-2 in the crystal structures) does not appear to correlate with affinity, likely due to the larger binding energy associated with the stacking interactions. Consequently, C5 in erk2 NRE and the base-omission mode of binding are unlikely to cause the weaker binding of this RNA. However, by comparing binding to erk2 NRE vs. hb NRE2, we conclude that the affinity difference is due to the C at the 8th position in the erk2 NRE sequence. This preference is consistent with the low frequency of appearance of C at position 8 in mRNAs associated with PUM1 (Morris et al., 2008).
DISCUSSION
Although human PUM1 has been a prototypical model for RNA recognition by PUF proteins, recognition of cognate RNAs by human Pumilio proteins remained unexplored due to little information about their natural mRNA targets. Our study here has identified distinct binding modes by PUM1-HD and PUM2-HD near the 5th RNA base in cognate mRNA targets. In the 1:1 binding mode, the Watson-Crick or Hoogsteen edge of the 5th RNA base is recognized by the 4th PUM repeat. Alternatively, in the base-omission mode, a C5 RNA base stacks directly with the preceding A4 and is not contacted by the protein. This flexibility of human PUMILIO proteins allows a broader range of RNA target sequences to be recognized. In general, PUM1-HD and PUM2-HD adopt identical RNA-binding modes and have similar binding affinities with cognate RNA sequences, consistent with the finding that they share the majority of their natural RNA substrates (Galgano et al., 2008).
Our discovery of the alternative recognition modes used by PUM1-HD and PUM2-HD suggests they are general mechanisms to broaden specific RNA recognition by PUF proteins. Similar features have been observed in crystal structures of other PUF proteins. For example, Puf4p from S. cerevisiae and FBF-2 from C. elegans both utilize the base-omission mode with the 4th and 5th bases of target RNAs directly stacking. For Puf4p and FBF-2, the direct stacking of the 4th and 5th bases appears to be part of RNA conformational changes that may be necessary for binding of 9 RNA bases by the 8-repeat proteins. This does not seem to be the case for PUM1 and PUM2, since the experimentally-determined consensus sequences suggest binding to a well-conserved 8-base sequence. Non-cognate 9-nt RNA sequences can be accommodated in vitro by PUM1-HD, but the base-omission mode is not observed (Gupta et al., 2008).
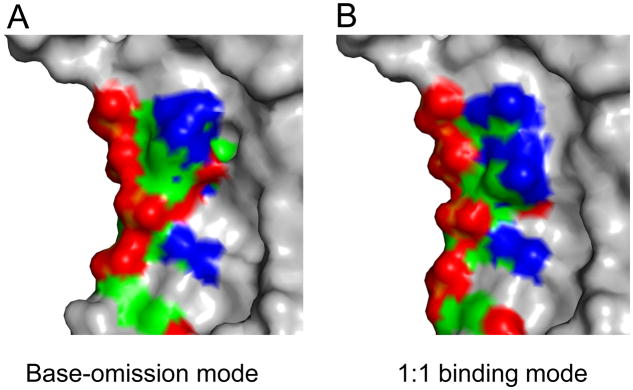
The distinct binding modes do not appear to alter binding affinity to RNA target sequences. Yet they do present differing recognition surfaces that could be specific for downstream factors. For example, PUM1 is able to bind to erk2 NRE with both the base-omission and 1:1 binding modes. The charge distribution on the protein surface differs for the two binding modes, which also have different shape complementarities for potential effector proteins (Figure 5). Currently, little is known about additional factors in human PUF protein effector complexes, but in Drosophila, the interaction of NANOS and BRAIN TUMOR proteins with DmPUM requires hb mRNA (Sonoda and Wharton, 1999). The conformation of the bound RNA could influence the interaction with components of effector complexes. Similarly, flipped bases observed in crystal structures of yeast Puf4p and worm FBF can vary the recognition surfaces of PUF protein:RNA complexes presented to other molecules. Such differences could explain the different outcomes of PUF protein:RNA target interaction such as mRNA decay (Wickens et al., 2002), translational repression (Chritton and Wickens, 2010; Wickens et al., 2002), or translational activation (Kaye et al., 2009; Suh et al., 2009).
Figure 5.
Different modes of RNA binding may present alternative recognition surfaces for downstream effector proteins. Surface representation of PUM1-HD:erk2 NRE (UGUACAUC) in (A) the base-omission mode and (B) the canonical 1:1 mode. RNA bases and protein residues with different conformations in both modes are colored by atom type (green, carbon; red, oxygen; blue, nitrogen; orange, phosphorus), and the rest of the structure is colored gray.
EXPERIMENTAL PROCEDURES
Preparation of human PUM1-HD and PUM2-HD and protein:RNA complexes
PUM1-HD was expressed and purified as described previously (Wang et al., 2001). A cDNA encoding human PUM2-HD (Gly706-Gly1056) was amplified from a pOTB7 vector containing a partial human PUM2 cDNA (Open Biosystems) and inserted into the pDEST527 plasmid encoding an N-terminal six-histidine tag using the Gateway TOPO cloning method (Invitrogen). His-PUM2-HD was expressed in Rosetta2 (DE3) Escherichia coli cells (Novagen), which were induced with 0.2 mM IPTG at 16°C overnight. Cell pellets were resuspended in cell lysis buffer (20 mM Tris-HCl pH 7.5, 100 mM NaCl, and 5 mM beta-mercaptoethanol) and subsequently lysed by sonication. His-PUM2-HD was purified from the soluble fraction using Ni-NTA resin (Qiagen). The resin was washed extensively with lysis buffer and with lysis buffer containing 1 M NaCl. The fusion protein was eluted from the Ni-NTA resin with lysis buffer containing 250 mM imidazole. The eluate was then dialyzed against lysis buffer at 4°C to remove the imidazole, and 1% (w/w) TEV protease was added to remove the N-terminal six-histidine tag from the fusion protein. PUM2-HD was further purified on a Heparin HiTrap column (Buffer A: 20 mM Tris-HCl pH 7.5 and 5 mM beta-mercaptoethanol; Buffer B: 20 mM Tris-HCl pH 7.5, 1M NaCl, and 5 mM beta-mercaptoethanol) followed by a Superdex 200 10/300 column (GE Healthcare) pre-equilibrated with lysis buffer. Purified PUM2-HD was concentrated to 3 mg/ml in lysis buffer.
RNA oligonucleotides were obtained from Dharmacon (Lafayette, CO). PUM1-HD or PUM2-HD was incubated with cognate RNA substrate at a molar ratio of 1:1.1 overnight at 4°C. Protein:RNA mixtures were purified with a Superdex 200 10/300 column run with lysis buffer. Fractions containing protein:RNA complex were pooled and concentrated to ~3 mg/ml.
Crystallization and Structure Determination
Both monoclinic and orthorhombic crystals of PUM1-HD:RNA complexes were obtained by hanging drop vapor diffusion at 20 °C. One microliter of PUM1-HD:RNA solution was mixed with 1 μl of well solution containing 15–20% (w/v) PEG 3350, 100 mM Li2SO4, and 100 mM Na3Citrate pH 5.5-6.0. Crystals were transferred into a cyroprotectant solution containing 22% (w/v) PEG 3350, 100 mM Li2SO4, 100 mM Na3Citrate pH 5.5, and 15% (v/v) ethylene glycol and flash-cooled in liquid nitrogen. Both triclinic and monoclinic crystals of PUM2-HD:RNA complexes were crystallized by vapor diffusion in either hanging or sitting drops at 20°C. Three microliters of PUM2-HD:RNA solution were mixed with 1.5 μl of well solution containing 17–20% (w/v) PEG 3000, 100 mM Na3Citrate pH 5.0–6.0. PUM2-HD:RNA complexes were flash cooled in liquid nitrogen after incubation in a cyroprotectant solution with 22% (w/v) PEG 3000, 100 mM Na3Citrate, and 15% (v/v) ethylene glycol. Diffraction data were collected at −180°C with a Rigaku microMAX 007 X-ray generator equipped with a Saturn92 CCD detector. All data sets were indexed, integrated, and scaled with the HKL2000 suite (Otwinowski and Minor, 1997).
The structures of PUM1-HD:RNA and PUM2-HD:RNA complexes were determined by molecular replacement using the structure of PUM1-HD (PDB ID: 1M8Y) as a search model with Phaser (McCoy et al., 2007) and AMoRe (Navaza, 1994). Iterative model building was carried out with O (Emsley and Cowtan, 2004; Jones et al., 1991) and COOT (Emsley and Cowtan, 2004). Torsion angle, positional, B-factor, and TLS refinement were performed in CNS (Brunger et al., 1998) and PHENIX (Adams et al.). All structures were evaluated with the program MolProbity (Davis et al., 2007) with no outliers detected in both protein and nucleic acid geometry.
Electrophoretic Mobility Shift Assays (EMSA)
Detailed procedures for the RNA-binding assay were described previously (Cheong and Hall, 2006; Wang et al., 2002; Wang et al., 2009b), except that PUM1-HD or PUM2-HD was incubated with 32P-radiolabeled RNAs for 16 hours at 4°C prior to the electrophoresis. All binding experiments were done at least in triplicate, and a representative experiment is shown in Figure S3. The percentage of active PUM1-HD (88%) or PUM2-HD (93%) in each protein preparation was determined by size-exclusion chromatography (Cheong and Hall, 2006). All dissociation constants were adjusted based on the percentage of active protein to allow direct comparison between different proteins.
Supplementary Material
Figure S1. Cα trace of PUM2 in complex with p38α NREa RNA, as shown in Figure 1. Every tenth Cα atom in PUM2, starting with residue 710, is indicated with a sphere.
Figure S2. Recognition by PUM1 of the 5th base in hb NRE1 and hb NRE2, related to Figure 4.
Figure S3. Representative data for determination of equilibrium binding affinities reported in Table 2. (A) Representative electrophoretic mobility shift assay of PUM2-HD binding to p38α NREa. (B) Curve-fitting analysis of the binding data in A.
Table S1. Complete data collection and refinement statistics for crystal structures summarized in Table I.
Acknowledgments
We are grateful to Drs. L. Pedersen and J. Krahn of the National Institute of Environmental Health Sciences for crystallographic and data collection support. We also thank our colleagues for fruitful discussion on the experiments and the manuscript. This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession codes
Coordinates and structure factor files have been deposited into the Protein Data Bank with the accession codes: 3Q0L, 3Q0M, 3Q0N, 3Q0O, 3Q0P, 3Q0Q, 3Q0R, 3Q0S.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein D, Hook B, Hajarnavis A, Opperman L, Wickens M. Binding specificity and mRNA targets of a C. elegans PUF protein, FBF-1. RNA. 2005;11:447–458. doi: 10.1261/rna.7255805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Cheong CG, Hall TM. Engineering RNA sequence specificity of Pumilio repeats. Proc Natl Acad Sci U S A. 2006;103:13635–13639. doi: 10.1073/pnas.0606294103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chritton JJ, Wickens M. Translational repression by PUF proteins in vitro. RNA. 2010;16:1217–1225. doi: 10.1261/rna.2070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Furman JL, Badran AH, Ajulo O, Porter JR, Stains CI, Segal DJ, Ghosh I. Toward a General Approach for RNA-Templated Hierarchical Assembly of Split-Proteins. J Am Chem Soc. 2010 doi: 10.1021/ja104395b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galgano A, Forrer M, Jaskiewicz L, Kanitz A, Zavolan M, Gerber AP. Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PLoS ONE. 2008;3:e3164. doi: 10.1371/journal.pone.0003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Luschnig S, Krasnow MA, Brown PO, Herschlag D. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2006;103:4487–4492. doi: 10.1073/pnas.0509260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta YK, Nair DT, Wharton RP, Aggarwal AK. Structures of human Pumilio with noncognate RNAs reveal molecular mechanisms for binding promiscuity. Structure. 2008;16:549–557. doi: 10.1016/j.str.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA–binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins HT, Baker-Wilding R, Edwards TA. Structure and RNA binding of the mouse Pumilio-2 Puf domain. J Struct Biol. 2009;167:271–276. doi: 10.1016/j.jsb.2009.06.007. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M. Improved methods for the building of protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kaye JA, Rose NC, Goldsworthy B, Goga A, L'Etoile ND. A 3'UTR pumilio-binding element directs translational activation in olfactory sensory neurons. Neuron. 2009;61:57–70. doi: 10.1016/j.neuron.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh YY, Opperman L, Stumpf C, Mandan A, Keles S, Wickens M. A single C. elegans PUF protein binds RNA in multiple modes. RNA. 2009;15:1090–1099. doi: 10.1261/rna.1545309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Hook B, Pan G, Kershner AM, Merritt C, Seydoux G, Thomson JA, Wickens M, Kimble J. Conserved regulation of MAP kinase expression by PUF RNA-binding proteins. PLoS Genet. 2007;3:e233. doi: 10.1371/journal.pgen.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MT, Higgin JJ, Hall TM. Basis of altered RNA-binding specificity by PUF proteins revealed by crystal structures of yeast Puf4p. Nat Struct Mol Biol. 2008;15:397–402. doi: 10.1038/nsmb.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AR, Mukherjee N, Keene JD. Ribonomic analysis of human Pum1 reveals cis-trans conservation across species despite evolution of diverse mRNA target sets. Mol Cell Biol. 2008;28:4093–4103. doi: 10.1128/MCB.00155-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallogr A. 1994;A50:157–163. [Google Scholar]
- Opperman L, Hook B, DeFino M, Bernstein DS, Wickens M. A single spacer nucleotide determines the specificities of two mRNA regulatory proteins. Nat Struct Mol Biol. 2005;12:945–951. doi: 10.1038/nsmb1010. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Ozawa T, Natori Y, Sato M, Umezawa Y. Imaging dynamics of endogenous mitochondrial RNA in single living cells. Nat Methods. 2007;4:413–419. doi: 10.1038/nmeth1030. [DOI] [PubMed] [Google Scholar]
- Sonoda J, Wharton RP. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev. 1999;13:2704–2712. doi: 10.1101/gad.13.20.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf CR, Kimble J, Wickens M. A Caenorhabditis elegans PUF protein family with distinct RNA binding specificity. RNA. 2008;14:1550–1557. doi: 10.1261/rna.1095908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh N, Crittenden SL, Goldstrohm A, Hook B, Thompson B, Wickens M, Kimble J. FBF and its dual control of gld-1 expression in the Caenorhabditis elegans germline. Genetics. 2009;181:1249–1260. doi: 10.1534/genetics.108.099440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilsner J, Linnik O, Christensen NM, Bell K, Roberts IM, Lacomme C, Oparka KJ. Live-cell imaging of viral RNA genomes using a Pumilio-based reporter. Plant J. 2009;57:758–770. doi: 10.1111/j.1365-313X.2008.03720.x. [DOI] [PubMed] [Google Scholar]
- Wang X, McLachlan J, Zamore PD, Hall TM. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110:501–512. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- Wang X, Zamore PD, Hall TM. Crystal structure of a Pumilio homology domain. Mol Cell. 2001;7:855–865. doi: 10.1016/s1097-2765(01)00229-5. [DOI] [PubMed] [Google Scholar]
- Wang Y, Cheong CG, Hall TM, Wang Z. Engineering splicing factors with designed specificities. Nat Methods. 2009a;6:825–830. doi: 10.1038/nmeth.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Opperman L, Wickens M, Hall TM. Structural basis for specific recognition of multiple mRNA targets by a PUF regulatory protein. Proc Natl Acad Sci U S A. 2009b;106:20186–20191. doi: 10.1073/pnas.0812076106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton RP, Sonoda J, Lee T, Patterson M, Murata Y. The Pumilio RNA-binding domain is also a translational regulator. Mol Cell. 1998;1:863–872. doi: 10.1016/s1097-2765(00)80085-4. [DOI] [PubMed] [Google Scholar]
- Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3'UTR regulation as a way of life. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Williamson JR, Lehmann R. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA. 1997;3:1421–1433. [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Gallegos M, Puoti A, Durkin E, Fields S, Kimble J, Wickens MP. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997;390:477–484. doi: 10.1038/37297. [DOI] [PubMed] [Google Scholar]
- Zhu D, Stumpf CR, Krahn JM, Wickens M, Hall TM. A 5' cytosine binding pocket in Puf3p specifies regulation of mitochondrial mRNAs. Proc Natl Acad Sci U S A. 2009;106:20192–20197. doi: 10.1073/pnas.0812079106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Cα trace of PUM2 in complex with p38α NREa RNA, as shown in Figure 1. Every tenth Cα atom in PUM2, starting with residue 710, is indicated with a sphere.
Figure S2. Recognition by PUM1 of the 5th base in hb NRE1 and hb NRE2, related to Figure 4.
Figure S3. Representative data for determination of equilibrium binding affinities reported in Table 2. (A) Representative electrophoretic mobility shift assay of PUM2-HD binding to p38α NREa. (B) Curve-fitting analysis of the binding data in A.
Table S1. Complete data collection and refinement statistics for crystal structures summarized in Table I.