Summary
Roesleria subterranea causes root rot in grapevine and fruit trees. The fungus has long been underestimated as a weak parasite, but during the last years it has been reported to cause severe damages in German vineyards. Direct, observation-based detection of the parasite is time consuming and destructive, as large parts of the rootstocks have to be uprooted and screened for the tiny, stipitate, hypogeous ascomata of R. subterranea. To facilitate rapid detection in vineyards, protocols to extract DNA from soil samples and grapevine roots, and R.-subterranea-specific PCR primers were designed. Twelve DNA–extraction protocols for soil samples were tested in small-scale experiments, and selected parameters were optimised. A protocol based on ball-mill homogenization, DNA extraction with SDS, skim milk, chloroform, and isopropanol, and subsequent purification of the raw extracts with PVPP-spin-columns was most effective. This DNA extraction protocol was found to be suitable for a wide range of soil-types including clay, loam and humic-rich soils. For DNA extraction from grapevine roots a CTAB-based protocol was more reliable for various grapevine rootstock varieties. Roesleria-subterranea-specific primers for the ITS1–5.8S–ITS2 rDNA-region were developed and tested for their specificity to DNA extracts from eleven R. subterranea strains isolated from grapevine and fruit trees. No cross reactions were detected with DNA extracts from 44 different species of fungi isolated from vineyard soils. The sensitivity of the species-specific primers in combination with the DNA extraction method for soil was high: as little as 100 fg μl−1 R.-subterranea-DNA was sufficient for a detection in soil samples and plant material. Given that specific primers are available, the presented method will also allow quick and large-scale testing for other root pathogens.
Keywords: grapevine dieback, Vitis sp, soil-borne pathogen, DNA extraction, root rot
Introduction
In 1877 Thümen introduced the genus Roesleria with the single species Roesleria hypogaea Thümen & Passerini. The current name of this species is R. subterranea (Weinm.) Redhead (Redhead, 1984; Kirchmair et al., 2008). The species was originally described from roots of grapevine and recognized as a facultative root-rotting pathogen. But there are also records from different fruit trees, where Roesleria causes root damage and dieback (Beckwith, 1924; Véghelyi, 1987; Véghelyi, 1989). R. subterranea infections start with the colonisation of the root surface. Then hyphae grow from the cortex towards the vascular cylinder. Höfer (1993) demonstrated that the hyphae aggregate particularly in the xylem and constipate the water-transport vessels, and this eventually damages the roots and causes dieback. In contrast to earlier publications, Huber et al. (2006a, 2006c) pointed out that R. subterranea is a primary parasite, and not only a weak parasite.
In 2001, during field work on a project on soil-borne fungi in phylloxera-infested vineyards, ascomata of R. subterranea were found on the roots of stunted grapevines in the German winegrowing region Rheingau (Huber, 2007). According to the literature, a wide distribution of R. subterranea was not expected. But in autumn 2003 and 2004 masses of fruiting bodies were observed in different vineyards (Huber et al., 2006b). In August 2005, a vineyard with distinct signs of dieback and stunted growth was screened for R. subterranea. This vineyard was found to be heavily infested: in October ascomata of R. subterranea were detected on 89% of all grapevines (Huber et al., 2006c, 2006d). Consequently, twenty-three vineyards some with and some without signs of stunted growth were screened for R. subterranea in the Rheingau and the Mosel-Saar-Ruwer Region. Roesleria subterranea was found in all vineyards (Huber et al., 2006c). Usually, an aggregated spatial distribution of infected vines was observed. Above-ground symptoms typically did not appear before the roots had extensively decayed.
To date, the occurrence of R. subterranea can be assessed only by time-consuming, direct observation of ascomata on the roots down to a depth of 1.5 m. Large parts of the rootstock have to be uprooted and screened for the tiny ascomata. This destructive method is often not feasible in commercial vineyards. In addition, fruiting bodies are not obligatory formed on infected plants and it is hardly possible to detect the beginning of R. subterranea infections with traditional methods. Identification of root-rotting fungi is usually impossible until the infection has spread through the root system and severe dieback of the crop is obvious. For sustainable plant protection it is necessary to detect and reliably identify parasites before they can cause serious damage; in general, prophylaxis is much easier than acute therapy. With PCR assays, starting infections not yet visible to the naked eye can be detected. But applying PCR-based methods to detect phytopathogens is often difficult. To obtain reproducible high-quality DNA extracts from plants and soil is a well-known problem (Mumford et al., 2006): (i) Fungi are unevenly distributed in the soil; they can be bound to soil particles or aggregated around organic matter. (ii) PCR can be hampered by various impurities co-extracted with DNA, such as humic acids, metal ions, and polysaccharides (Wilson, 1997; Robe et al., 2003); the composition of these inhibitory compounds varies with the soil type and with the type of soil management (e.g. conventional or organic). (iii) Both vital and dead material is detected with PCR. (iv) The interpretation of the results is difficult, as threshold limits for a pathogen vary with soil type, plant cultivar, climate or season. (v) False positive results caused by cross contamination have to be avoided from the sampling to the final diagnostic PCR assay.
In this study highly sensitive DNA extraction methods for soil and root samples are presented. Together with the PCR assay described here, these methods allow a quick, reliable and high-resolution PCR-based detection of R. subterranea in vineyard soils.
Materials and methods
Fungal strains
The R. subterranea strains used in this study were: Germany, MJG–040832, IB 2005/506: Kiedrich; on Vitis berlandieri × V. riparia; leg. det. Huber, Hoffmann, Michaelis 2005. MJG–040836, IB 2005/504: Hochheim; on V. riparia 183G × V. cinerea Arnold; leg. det. Huber, Michaelis, Hoffmann 2005. MJG–040834, IB 2005/509: Wiltingen; on V. berlandieri × V. riparia; leg. det. Huber, Michaelis 2005. MJG–040837, IB 2005/508: Hochheim, on V. rupestris 193G × V. riparia 1G; leg. det. Huber, Michaelis, Hoffmann 2005. MJG–040833, IB 2005/507: Kiedrich; on V. berlandieri × V. riparia; leg. det. Huber, Hoffmann, Michaelis 2005. MJG–040835, IB 2005/505: Hattenheim; on V. berlandieri × V. riparia; leg. det. Huber, Michaelis, Hoffmann 2005. Austria: CBS 339.96, IB1995/966: Eberstein; twig gof deciduous shrub lying on the ground; leg. det. M. Kirchmair 1995. Netherlands: CBS 320.33: Nijmegen; Malus sylvestris root; leg. det. Diddens 1933. CBS 271.82: Oostelijk Flevoland, Bremerbergerbos; decayed wood, root of Populus sp.; leg. det. van der Aa 1982. Italy: CBS 407.51: V. vinifera root; leg. det. Ciferri 1951. USA: CBS 320.33: V. vinifera; leg. det. A.M. Beckwith, 1925.
Different strains of 44 fungal species isolated form the rhizosphere, the rhizoplane and the roots of a vineyard in the German Rheingau (Table 1) were used to determine the specificity of the R.-subterranea primers. All fungal isolates were cultured on PDA (potato dextrose agar, Oxoid, Basingstoke, UK).
Table 1.
Fungi isolated from a trial vineyard in Kiedrich (Germany) used to test the specificity of the designed primers.
| Species | Root free soil | Rhizoplane | Rhizosphere | Roots |
|---|---|---|---|---|
| Acremonium charticola (J. Lindau) W. Gams | + | + | ||
| Acremonium furcatum Moreau & R. Moreau ex Gams | + | + | ||
| Acremonium kiliense Grütz | + | + | ||
| Acremonium strictum W. Gams | + | + | ||
| Acrodontium crateriforme (J.F.H. Beyma) de Hoog | + | |||
| Alternaria alternata (Fr.) Keissl. | + | + | + | |
| Aspergillus ustus (Bainier) Thom & Church | + | + | ||
| Cladosporium cladosporioides (Fresen.) G.A. de Vries | + | + | + | |
| Cylindrocarpon magnusianum Wollenw. | + | |||
| Cylindrocarpon destructans (Zinssm.) Scholten | + | + | + | + |
| Fusarium equiseti (Corda) Sacc. | + | + | + | + |
| Fusarium merismoides var. merismoides Corda | + | + | + | |
| Fusarium oxysporum Schltdl. | + | + | + | + |
| Fusarium sacchari (E.J. Butler & Hafiz Khan) W. Gams | + | |||
| Fusarium sambucinum s.l. Fuckel | + | |||
| Fusarium semitectum Berk. & Ravenel | + | |||
| Fusarium solani (Mart.) Sacc. | + | + | + | |
| Fusarium tabacinum (J.F.H. Beyma) W. Gams | + | + | + | |
| Geotrichum candidum Link | + | |||
| Gliocladium catenolatum J.C. Gilman & E.V. Abbott | + | + | + | + |
| Gliocladium roseum Bainier | + | + | + | + |
| Leptosphaeria coniothyrium (Fuckel) Sacc. | + | + | + | |
| Mortierella alpina Peyronel | + | |||
| Mucor hiemalis Wehmer | + | |||
| Nectria inventa Pethybr. | + | + | + | |
| Penicillium aurantiogriseum Dierckx | + | + | ||
| Penicillium brevicompactum Dierckx | + | + | ||
| Penicillium chrysogenum Thom | + | + | + | |
| Penicillium citrinum Thom | + | + | ||
| Penicillium corylophilum Thom | + | |||
| Penicillium expansum Link | + | + | + | |
| Penicillium janthinellum Biourge | + | + | ||
| Penicillium miczynskii K.M. Zalessky | + | + | ||
| Penicillium restrictum C. Gilman & E.V. Abbott | + | + | + | |
| Penicillium simplicissimum (Oudem.) Thom | + | + | + | |
| Penicillium spinulosum Thom | + | + | ||
| Penicillium viridicatum Westling | + | |||
| Penicillium wakesmanni K.M. Zalessky | + | + | + | |
| Ramichloridium subulatum de Hoog | + | + | ||
| Rhizopus stolonifer (Ehrenb.) Vuill. | + | + | ||
| Roesleria subterranea (Weinm.) Redhead | + | |||
| Schizophyllum commune Fr. | + | + | ||
| Trichoderma koningii Oudem. | + | + | + | |
| Verticillium nigrescens Pethybr. | + | + | + |
Soil samples
Different types of vineyard and agricultural soils were used to develop the DNA-extraction method (Table 2). All soil samples were taken with a soil corer from 0–20 cm depth, put into plastic bags and transported to the laboratory. The samples were allowed to air-dry overnight, were passed trough a 2 mm mesh sieve and were subsequently stored at 4°C until use.
Table 2.
Soil samples used for the development of the DNA extraction protocol.
| Code | Crop | Origin | Soil-type |
|---|---|---|---|
| Kiedrich | Vitis sp. | Germany, Rheinland-Pfalz, Kiedrich | Loamy sand, high water- holding capacity, loess |
| Mosel | Vitis sp. | Germany, Rheinland-Pfalz, Brenkastel-Wehlen | Loam; high water holding capacity |
| Martell | Fragaria sp. | Italy, South Tyrol, Martell | Rich in humus loamy sand |
| Wachau | Vitis sp. | Austria, Lower Austria, Weiβenkirchen | Loamy sand |
| Weinviertel | Vitis sp. | Austria, Lower Austria, Groβ-Engersdorf | Chernosem |
| Ozora | Zea mays | Hungary, Tolna, Ozora | Chernosem |
| Gölle | Zea mays | Hungary, Somogy, Gölle | Chernosem |
| Bonyhad | Zea mays | Hungary, Tolna, Bonyhad | Chernosem |
Plant material
Root samples were taken with a spade 10–20 cm away from the grapevine trunk, underneath the row, and from the upper 25 cm soil horizon (Porten and Huber, 2003). Soil was removed as far as possible without damaging the roots. Vitis vinifera roots (1 sample), and different rootstock varieties (V. berlandieri × V. riparia: SO4, 2 samples, V. riparia × V. cinerea: Börner, 2 samples) were used to develop a DNA extraction method from the grapevine roots. Randomly selected root samples from different not further determined rootstock-varieties were used to validate the method (10 samples).
DNA extraction
DNA extraction from fungal cultures
All fungal cultures were grown on PDA at 20°C for 7–14 days. Approximately 1 cm2 mycelium was transferred into 1.5-ml tubes with 0.2 g of glassbeads (2 mm diameter) and 100 μl CTAB-buffer (2% CTAB, 1.4 M NaCl, 0.1 M Tris-HCl pH 7.5, 0.2 M EDTA). The mixture was homogenized in a Ball-mill (MM301 Retsch, Haan, Germany) at maximum speed for 30 seconds. Samples were then spun down, 400 μl CTAB-buffer were added, and the mixture was incubated at 65°C for one hour. Four hundred μl chloroform/isoamylalcohol (24:1) were added, the samples were vortexed and centrifuged for 5 min at 10000 ×g and the aqueous top layer was transferred to a new 1.5-ml tube. This step was repeated twice. Two hundred μl 5 M Ammonium-acetate were added and the samples were incubated at 4°C for at least 30 min and subsequently centrifuged (20 min, 4°C, 16000 ×g). DNA was precipitated with an equal volume of isopropanol at −20°C overnight. DNA was pelleted (15 min, 4°C, 16000 ×g), washed with ice-cold 70% ethanol (10 min, 4°C, 16000 ×g), air-dried and re-dissolved in 50 μl TE buffer (10 mM Tris-HCl, 10 mM EDTA, pH 8) containing 4.5 U RNase ml−1.
DNA extraction from plant material
Grapevine roots were put into a 2-ml tube with glass-beads of different sizes: 2–3 glass beads with 5 mm diameter; approx. 0.14 g of beads with 2 mm diameter, and approx. 0.07 g glass beads with 0.2 mm diameter. Samples were homogenised without any buffer in a ball-mill (Retsch MM301) at maximum speed for 2 minutes. One ml of CTAB-PVPP-extraction buffer (200 mM Tris-HCl pH 7.5; 20 mM EDTA; 1,4 M NaCl; 2% CTAB, 4.5% PVPP) was added, the samples were vortexed, and incubated with shaking at 60°C for 30 min. Samples were centrifuged for 5 min at 5000 ×g, and the supernatant was transferred to a new 1.5-ml tube. 500 μl chloroform/isoamylalcohol (24:1) were added, samples were mixed on a vortex, centrifuged (5 min, 16000 ×g), and the aqueous top layer recovered to a new 1.5-ml tube. This step was repeated twice. 300 μl 5 M ammonium acetate were added, samples were incubated at -20°C for 10 min, centrifuged (10 min, 4°C, 16000 ×g) and the supernatant was transferred to a new 1.5-ml tube. An equal volume of isopropanol was added and DNA precipitated at −20 °C for 30 minutes. The DNA was pelleted (5 min, 4°C, 16000 ×g), washed with ice-cold 70% ethanol (5 min, 4°C, 16000 ×g), air-dried and re-dissolved in 100 μl TE buffer (10 mM Tris, 10 mM EDTA, pH 8). No further cleaning of the DNA extracts was required.
Development of a DNA extraction method from soil samples
Based on a meta-study on soil-DNA extraction protocols (about 280 references) twelve different methods were compiled and tested. Fundamental differences between these methods are summarized in Table 3. All DNA extraction methods were based on homogenisation of the soil samples in a ball-mill (Retsch MM301), a step including detergent (SDS or CTAB) and salt, chloroform extraction, and alcohol precipitation of the DNA. A subsequent cleaning step with PVPP-spin columns was used to purify the DNA extracts. The most suitable DNA extraction protocol (Method 10), which is illustrated below, was optimised.
Table 3.
Differences between the DNA-extraction protocols tested. Varied concentrations (w/v) of CTAB, SDS and skim milk used in extraction buffers are indicated.
| Method | CTAB (%) | SDS (%) | Skim milk (%) | Time (d) | Additional treatment |
|---|---|---|---|---|---|
| 1 | 2 | − | − | 2 | Proteinase K |
| 2 | 2 | − | 2 | 2 | Proteinase K |
| 3 | 0.02 | − | − | 1 | − |
| 4 | 0.02 | − | 10 | 1 | − |
| 5 | 10 | − | − | 1 | Incubation (65°C) before homogenisation |
| 6 | 10 | − | − | 1 | Incubation (65°C) after homogenisation |
| 7 | − | 10 | − | 1 | − |
| 8 | − | 10 | 2 | 1 | − |
| 9 | 2 | − | − | 2 | − |
| 10 | − | 10 | 2 | 1 | − |
| 11 | − | 10 | 2 | 1 | Sodium-acetate + ethanol combined |
| 12 | 2 | − | 2 | 1 | Sodium-acetate + ethanol combined |
Soil (0.2 g) was transferred to a 1.5-ml tube with 0.2 g glass-beads (2 mm and 0.2 mm diameter, ratio 2:1), 300 μl SDS-buffer (10% SDS, 100 mM NaCl, 500 mM Tris-HCl pH 8), 300 μl skim milk (2% skim milk powder in aqueous solution), and 5 μl RNase (10 μg ml−1). Samples were homogenized in a Ball-mill (Retsch MM301) at maximum speed for 2 min, spun down, and incubated shaking at 60°C for 10 min. 400 μl chloroform/isoamylalcohol (24:1) were added, samples were vortexed, centrifuged (5 min, 10000 ×g), and the aqueous top layer was transferred into a new 1.5-ml tube. This step was repeated twice. 200 μl of 5 M ammonium-acetate were added, samples were incubated at -20°C for 20 min, centrifuged (20 min, 4°C, 16000 ×g) and the supernatant was transferred into a new 1.5-ml tube. An equal volume of isopropanol was added and DNA precipitated at −20°C for 30 minutes. DNA was pelleted (5 min, 4°C, 16000 ×g), washed with 400 μl ice-cold 70% ethanol (5 min, 4°C, 16000 ×g), air-dried and re-dissolved in 200 μl TE buffer (10 mM Tris-HCl, 10 mM EDTA, pH 8).
To find the most appropriate protocol, selected parameters of this DNA extraction method were varied. Different skim milk concentrations (0, 1, 2, 3, 4, 5, 8 and 10% skim milk powder in aqueous solution) were tested as well as different quantities of soil (0.1, 0.2, 0.3, 0.4 and 0.5 g). Ammonium-acetate (5 M) was substituted for 3 M sodium-acetate and isopropanol was substituted for ethanol (96%). DNA was also extracted from sieved (2 mm mesh diameter) and unsieved soil samples.
To remove remaining PCR inhibitors from the soil DNA raw extracts, PVPP columns were used according to Damm and Fourie (2005): with a blood lancet an opening (approximately 0.3 mm to 0.7 mm in diameter) was made into the bottom of a 0.5-ml tube. The tube was filled with a PVPP/TE suspension (0.4 g ml−1 PVPP, TE-buffer: 10 mM Tris, 1 mM EDTA, pH 8), placed in a 2-ml tube, and centrifuged for 1 min at 900 ×g. The flow-through was discarded, the column was replaced in the 2-ml tube and the previous step was repeated until the PVPP spin-column was approximately 15 mm high. The column was spindried for 3 min at 1400 ×g and placed in a fresh 1.5-ml tube. Fifty μl of DNA extract were transferred to the column, incubated for 5 min at room temperature, and spun down for 90 s at 1400 ×g. The cleaned DNA extracts were used without further dilution for the PCR reactions.
Evaluation of soil DNA extraction methods
To evaluate the soil DNA extracts, ethidium-bromide stained agarose gels were used. Each DNA extract was analysed using three agarose concentrations (0.8, 1.5 and 2%, in 1×TAE, w/v). One hundred ml agarose gel were supplemented with 2 μl ethidium-bromide solution (1% w/v). 4 μl DNA extract and 2 μl loading dye (1.6 mM Tris-HCl (pH 7.6), 0.005% bromophenol blue, 0.005% xylene cyanol FF, 30% glycerol, 10 mM EDTA) were loaded onto the gel and run in 1× TAE (40 mM Tris-HCl, 19 mM acetic acid, 1 mM EDTA, pH 8) at 100 V in a RunOne elecrophoresis system (Embi Tec, San Diego, CA, USA) for 10–15 minutes. The DNA bands were visualised with trans-illuminating UV light (254 nm wavelength) in an AlphaDigiDoc 1201 (Alpha Innotech, San Leandro, CA, USA).
All DNA extracts were rated visually. For an unbiased evaluation of the DNA extracts the fluorescence intensities of the DNA bands were analysed with the 1D-Multi analysis tool of the AlphaDigiDoc™ FC software (Alpha Innotech, San Leandro, CA, USA). The 1D-Multi tool converts digital images of ethidium-bromide stained agarose gels into line graphs, where the fluorescence intensity of the DNA bands is plotted on the ordinate. The height of the peaks was used to compare DNA extracts. Only one peak per lane of long DNA fragments was evaluated (Rf equal to or less than 0.25). The settings of the 1D-Multi analysis tool of the Alpha DigiDoc™ FC software were: grid controls: “width 25”; contrast adjustment: “linear”, “auto contrast”; baseline: “rubber band”. Two DNA ladders (peqGOLD 100 bp DNA-Leiter Plus, Peqlab, Erlangen, Germany; prepared according to manufacturer’s instructions) were added to every row on the gel (2 μl each). The 500 bp band was used to standardise the intensities: the mean of the intensities of the two 500 bp bands was defined as 100% for each row and the fluorescence intensities of the DNA extracts in each row were adapted to this value. To linearise the dataset, it was transformed by calculating the natural logarithm. Fluorescence intensity of bands was scored on a scale from one to five in which 1 denoted 81–100% fluorescence of the brightest band; 2 denoted 61–80%; 3 denoted 41–60%, 4 denoted 21–40% and 5 denoted 0-100% fluorescence of the brightest band. For each DNA extract the median from the six different ratings was calculated with Microsoft Office Excel 2003 and used for subsequent statistics. Data were analysed with SPSS 14 for Windows: median and the inter-quartile range (Q3–Q1) were analysed as box-and-whisker plots.
PCR
All PCR reactions were carried out with a Primus 96 advanced thermocycler (peqLab Biotechnology) in 200 μl reaction tubes. The final PCR reaction-mix comprised 0.2 mM of each dNTP, 1 mM of each primer, 10% of PCR reaction buffer, 3.5 mM MgCl2, 200 μg ml−1 BSA (bovine serum albumin), 3.75 U GoTaq® DNA polymerase (Promega, Mannheim, Germany) in 100 μl reaction mix. Two μl of DNA template were used for every 20 μl PCR reaction. For nested PCR 1 μl of the first PCR product served as template for the second PCR. Standard PCR conditions were: 120 s initial denaturation at 94°C; followed by 30 cycles with 30 s at 94°C, 45 s at primer annealing temperature, 60 s at 72°C; and a final slope for 10 min at 72°C. Primers used to amplify the fungal ITS1–5.8S–ITS2 rDNA region: V9G (5′–TTA CgT CCC TgC CCT TTg TA–3′) and LS266 (5′–gCA TTC CCA AAC AAC TCgACT C–3′) at an annealing temperature of 56°C (Gerrits van den Ende and De Hoog, 1999); ITS5 (5′–ggA AgT AAA AgT CgT AAC AAg g–3′) and ITS4 (5′–TCC TCC gCT TAT TgA TAT gC–3′) at 54°C (White et al., 1990). Positive (R. subterranea) and negative (sterile distilled water) controls were included in every PCR run.
Sequencing
Excess primers and dNTPs were removed with chromatography columns (Microspin S–300 HR, Amersham Biosciences). To sequence the ITS1–5.8S–ITS2 rDNA, primers V9G and LS266 were used at a concentration of 1.6 μM. Sequencing was carried out with the ABI PRISM BigDye™ Terminator Cycle Sequencing Kit (Applied Biosystems) according to the manufacturer’s instructions. The parameters for cycle sequencing were: 18 sec delay at 96°C, followed by 25 cycles with 18 sec at 96°C, 5 sec at 50°C and 4 min at 60°C. Sequences were analysed using an automated sequence analyzer (ABI PRISM 3130, Applied Biosystems) in conjunction with the ABI Prism Auto Assembler™ software (version 140, Applied Biosystems).
Design of R.-subterranea-specific PCR primers
Roesleria-subterranea-specific primers for the ITS1–5.8S–ITS2 rDNA region were designed. Sequences of R. subterranea isolates were compared with published sequences of selected ascomycetous fungi (GenBank). A 20–30 bp region uniform for all Roesleria isolates and distinct from all other ascomycetes examined was selected and blasted against the NCBI nucleotide database (blastn). Primers were synthesized from sequences with no hits on organisms other than R. subterranea (genXpress, Wiener Neudorf). These primers were tested with the R. subterranea isolates and with different soil fungi isolated from a vineyard in Germany (Table 1). To avoid false negative results, all fungal DNA extracts were tested with primers broadly targeting fungi (LS266/V9G, or ITS1/ITS5).
To determine the sensitivity of the PCR primers, autoclaved soil samples were used. Soil and root samples were extracted with the methods described. Roesleria subterranea DNA was used in decimal dilutions from 1 ng μl−1 to 1 fg μl−1 (i) directly in PCR or (ii) to spike the DNA extracts of the autoclaved soil or root samples (1 μl for every 20 μl PCR reaction). This method was used because spiking samples with spores of R. suberranea is not possible as no anamorph is known and no fruiting bodies are formed on agar medium. Soil and root DNA extracts were tested in PCR without the addition of R. subterranea DNA to ensure that the latter was absent.
Results
Species-specific PCR primers for R. subterranea
Species-specific primers were designed based on the ITS1–5.8S–ITS2 rDNA region. The primer pair amplifying specifically for R. subterranea was: Rs1R (5′–TCC ggA ACg TCT ATA gCg Agg AgA–3′) and Rs2F (5′–TCg Cgg gCA ACC ggC TCA CgC–3′) at an annealing temperature of 60°C. This primer pair tested positive with all R. subterranea isolates. No PCR product was obtained from DNA extracts of the soil fungi (Table 1) using the Rs1R/Rs2F primer pair. All fungal DNA extracts were processed with the fungal universal primers V9G/LS266 or ITS5/ITS4 to exclude false negative results; these PCRs were successful for all fungal isolates tested. The primers were tested with naturally infested field samples (soil and roots) from the trial site Kiedrich. The PCR products were sequenced to confirm the specificity of the Rs-primers. A BLAST search revealed 100% similarity with the published R. subterranea sequences.
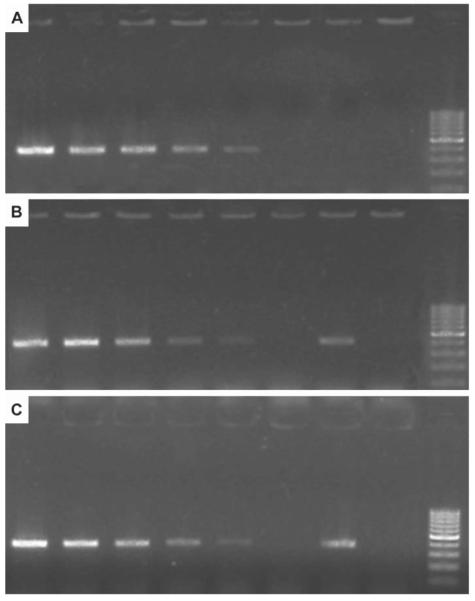
To estimate the sensitivity of the R. subterranea primers, autoclaved soil and root samples were extracted. When soil or root DNA extracts were spiked with serial dilutions of R. subterranea DNA, a detection limit of 100 fg DNA was determined. A weak inhibition of PCR by the purified soil or root extracts was observed when these PCR products were compared with PCR products of pure R. subterranea DNA. On agarose gel the fluorescence intensity of the PCR products from the spiked soil or root DNA extracts was lower than it was in the internal positive control, a pure DNA template of the same concentration (Fig. 1).
Fig. 1.
Primer sensitivity test with (A) pure R. subterranea DNA; (B) spiked soil extracts; (C) spiked grapevine root extracts. (A) PCR products of DNA extracts from R. subterranea. From left to right: 1 ng, 100 pg, 10 pg, 1 pg, 100 fg, 10 fg, 1 fg, negative control (a.d.) and DNA ladder. (B) PCR products of DNA extracts from sterile soil spiked with R. subterranea DNA. From left to right: 1 ng, 100 pg, 10 pg, 1 pg, 100 fg, pure soil extract, 1 pg DNA of R. subterranea (positive control), negative control (a.d.) and DNA ladder. (C) PCR products of DNA extracts from grapevine roots spiked with R. subterranea DNA. From left to right: 1 ng, 100 pg, 10 pg, 1 pg, 100 fg, pure root extract, 1 pg DNA of R. subterranea (positive control), negative control (a.d.) and DNA ladder.
DNA extraction from soil samples
A total of 839 soil-DNA extracts was rated in the study to optimise the DNA extraction protocol for vineyard and agricultural soils. Twelve DNA extraction protocols were tested and rated on the basis of their DNA contents. On the five-point-scale (1, best; 5, worst), the DNA extracts of methods 1, 9 and 10 were in category two. The extracts of all other methods were between categories four and five. Samples from all DNA-extraction methods were tested by PCR using a primer pair broadly targeting fungi (ITS1/ITS4 and/or V9G/LS266). Only DNA extracts of methods 9 and 10 yielded reproducible PCR products. The extraction and purification procedure lasted 28 h with method 9, but only three hours with method 10 which was therefore superior. Improvement processes were therefore accomplished with method 10.
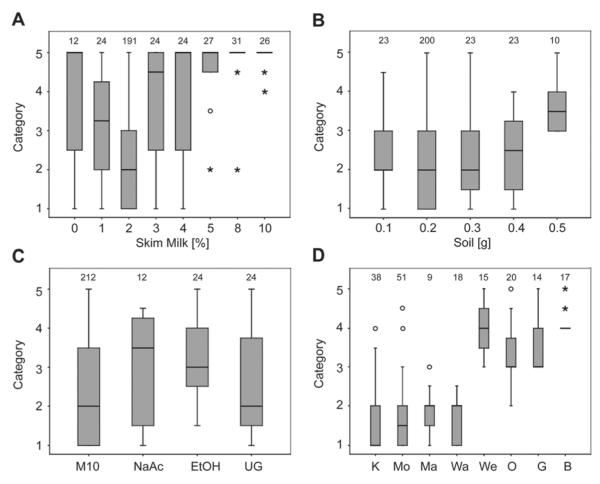
Different skim milk concentrations were used in combination with DNA extraction method 10 (Fig. 2A). Best results were obtained when 2% skim milk powder in aqueous solution was used: the DNA extracts were rated within category two. When 1% or 3% skim milk were used, the DNA extracts were rated as categories 3.25 and 4.50, respectively, and with skim milk concentrations of 0, 4, 5, 8, or 10%, the DNA extracts were rated in category 5. Different amounts of soil were extracted with method 10. When 0.1, 0.2 or 0.3 g of soil was extracted, the DNA extracts were in category two (Fig. 2B), when 0.4 g was extracted the DNA extract rating was 2.5, and when 0.5 g of soil was extracted it was 3.5. However with 0.4 and 0.5 g of soil the 1.5-ml tubes repeatedly ruptured during the first centrifugation step.
Fig. 2.
Ratings of DNA extracts. A five-point-scale was used, whereby category one was assigned to bands with the highest intensity. Data are expressed as a box-and-whisker plot showing median, inter-quartile range (IQR), and extreme values. Atypical outliers are indicated with an open dot (values 1.5–3×IQR), extreme outliners with an asterisk (values >3×IQR). Numbers above the boxes represent the number of DNA extracts examined for each treatment. (A) Method 10, with varying concentrations of skim milk used in the extraction buffer. Percentages indicate the concentration (w/v) of skim milk. (B) Amounts of soil extracted with method 10. (C) Method 10 compared with varying chemicals or soil treatments (M10, method 10; NaAc, sodium acetate instead of ammonium acetate; EtOH, ethanol instead of isopropanol; UG, unsieved soil). (D) Ratings of DNA extracts from different agricultural soils (K, Kiedrich; Mo, Mosel; Ma, Martell; Wa, Wachau; We, Weinviertel; O, Ozora; G, Gölle; B, Bonyhad).
Apart form different concentrations of skim milk and different amounts of soil, ammonium-acetate was replaced by sodium-acetate, and isopropanol by 96% ethanol. As a further experimental variable, some soil samples were also not sieved prior to DNA extraction (Fig. 2C). When ammonium-acetate was replaced by sodium-acetate, the DNA extracts were in category 3.5, and when ethanol was used the DNA extracts were rated in category 3. Both these replacements therefore gave results worse than those from the unmodified method 10 (category 2). The DNA extracts of unsieved samples were in the same category those of the sieved samples. Different agricultural soils were tested with method 10 (Fig. 2D). Relatively high yields of DNA could be extracted from loamy sand soils (Kiedrich, Mosel, Wachau) and rich in humus, loamy soils (Martell). Relatively little DNA was extracted form chernosem soils (Ozora, Gölle, Bonyhad, Weinviertel). Nevertheless, PCR was successful with DNA extracts from all soil samples (primer pairs ITS1/ITS4 and/or V9G/LS266).
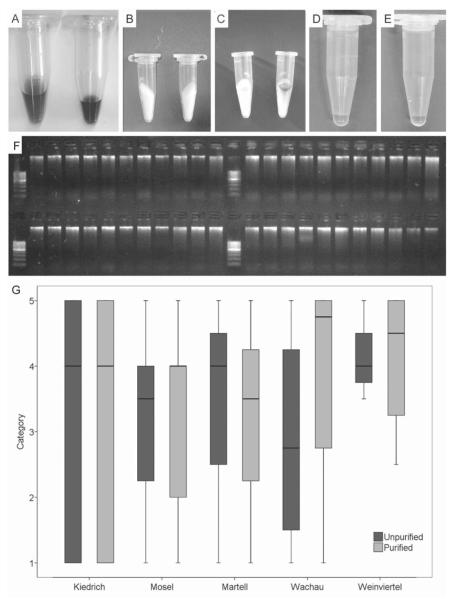
PCR was effective only, if DNA extracts were first purified with PVPP spin-columns. After this purification step the previously dark coloured soil extracts were cleared considerably (Fig. 3A–E). To examine possible DNA loss during purification, the DNA extracts were loaded on an agarose gel before and after purification. No excessive loss of DNA was observed (Fig. 3F). Random DNA extracts of all soil samples were purified (for every soil sample n=12). No or only very small differences in the DNA contents of the purified and un-purified DNA extracts were found with the soil samples Kiedrich, Mosel, Martell and Weinviertel; purified DNA extracts from the soil sample Wachau contained less DNA than the raw extracts (Fig. 3G).
Fig. 3.
DNA extract purification. (A) Unpurified DNA raw extracts. (B) PVPP spin-columns. (C) Dark substances of DNA extracts bound to the spin column. (D, E) Purified DNA extracts. (F) DNA raw extracts (upper row) and purified DNA extracts (lower row) on 1.5% agarose gel. (G) Ratings of unpurified and purified DNA extracts of different soil samples (n= 12 for each soil). Data are expressed as a box-and-whisker plot showing median, inter-quartile range (IQR), and extreme values. Atypical outliers are indicated with an open dot (values 1.5–3×IQR), extreme outliners with an asterisk (values >3×IQR). A five-point-scale was used, whereby category one was assigned to bands with the highest intensity.
With method 10, R. subterranea was detected in soil samples from all vineyards known to be infested with this fungus.
Discussion and conclusions
Fungi are unevenly distributed in soil particles. The composition of soil particles is a result of interactions between the soil microbial communities, the soil properties, the plants growing in the soil, and the history of the soil ecosystem under investigation (Tate, 2000; Herrera and Cockell, 2007). Because of this inhomogeneous distribution, more than one sub-sample should be taken from a vineyard to detect phytopathogenic fungi. We orientated our soil sampling strategy on a standard protocol for the detection of entomopathogenic fungi (Längle et al., 2005) where at least 15 sub-samples per hectare are randomly taken and mixed together. To release the fungal DNA, the soil aggregates as well as the fungal cell walls have to be destroyed. Many protocols to disrupt soil samples have been reported, including grinding in liquid nitrogen, lysozyme treatment, microwave heating, ultrasonication, or homogenising with ball-mills and bead beaters (Zhou et al., 1996; Miller et al., 1999; Lloyd-Jones and Hunter, 2001; Martin-Laurent et al., 2001; Schneegurt et al., 2003; Lakay et al., 2007). The highest DNA yields are consistently reported when soil samples are mechanically ruptured with ball-mills or bead beaters (Damm and Fourie, 2005; Lakay et al., 2007), a method also successfully applied in this study.
A drawback of ball-mill homogenisation is that more inhibitory compounds such as humic substances or polysaccharides are co-extracted with the DNA (Miller et al., 1999). Sorption of DNA to clay minerals, humic substances, and other high molecular weight compounds must be minimized (Wilson, 1997). Skim milk has been reported to limit these DNA binding effects in soil DNA extracts (Volossiouk et al., 1995; Takada Hoshino and Matsumoto, 2005). Skim milk was autoclaved, as the nucleases and proteins in un-autoclaved skim milk have been demonstrated to inhibit PCR (Wilson, 1997). In the present study it was demonstrated that moderate concentrations of skim milk in the SDS extraction buffer (1% final concentration) were essential for a high DNA yield and a successful PCR. Volossiouk et al. (1995) and Takada Hoshino and Matsumoto (2005) reported that DNA extraction and subsequent PCR were very effective when extraction buffers with a final concentration of 4% skim milk were used. However, when similar skim milk concentrations were used in the presented extraction method, soil DNA extracts were of poor quality and no PCR products were obtained.
Though the sorption of DNA to different soil compounds is minimised by the addition of skim milk, humic substances co-extracted with the DNA must be removed. These substances are powerful inhibitors of PCR reactions. As little as 100 pg μl−1 humic acids inhibit PCR amplification (Tsai and Olsen, 1992). To remove humic substances PVPP spin-columns are very effective, and they are easy to produce and handle (Berthelet et al., 1996; Cullen and Hisch, 1998; Damm and Fourie, 2005, Neuhauser et al., 2008). A highly efficient purification of soil DNA extracts with PVPP spin-columns was also demonstrated in this study. The application of PVPP in DNA extraction from grapevine roots very effectively removed PCR inhibitors such as tannins and other polyphenols produced by plants in response to stress (Winkel-Shirley, 2002). When DNA from grapevine roots was extracted the production of PVPP spin columns was omitted by adding the PVPP directly to the CTAB-extraction buffer. This strategy was however not successful with the soil DNA extracts.
DNA-based methods do not provide information about the physiological status of the organism under study: both vital and dead material can be detected (Ward et al., 2004; Levy-Booth et al., 2007). Complex processes at the molecular level can stabilise free DNA in the soil matrix (for reviews see Boyd and Mortland, 1990; Huang, 1990; Janvier et al., 2007). Amplifying DNA from dead cells is sometimes seen as a minor problem, as the rapid degeneration of DNA in soil is expected (Ruano-Rosa et al., 2007). However, there is good evidence that sorption of DNA to clay minerals or other soil particles can protect DNA from degradation (Ivarson et al., 1982; Paget et al., 1992; Levy-Booth et al., 2007). Nevertheless, when soil-borne pathogens such as root-rotting fungi are to be detected in soil samples from land under permanent crops, the problem of DNA sorption to soil particles is negligible, as it is highly unlikely that a well-established pathogen will disappear while its host is still present.
To verify an infection with a soil-borne pathogen such as R. subterranea, root samples should be tested along with soil samples. If the pathogen can be detected in the roots, the plant must be considered infected. Compared to the time consuming and expensive process of uprooting grapevine root-stocks that is required to detect R. subterranea by traditional methods, the high sensitivity PCR assay combined with the rapid soil or plant DNA extraction method enables R. subterranea to be detected in less than one day. Comprehensive testing and a risk assessment for R. subterranea infestations will therefore only be possible when detection in vineyard soils and detection in grapevine roots are combined. The early detection of R. subterranea will be of economic interest for vine growers, since it costs an estimated € 20,000–30,000 to replant one hectare vineyard in plains; on steep hills the cost is much higher. Previous studies gave evidence that a high percentage (60–80%) of infected plants die within the first three years (Véghelyi, 1989; Höfer, 1993). When R. subterranea is detected in the roots, the plant must be removed according to current guidelines for root pathogens (e.g. Pérez-Jiménez, 2006); when it is detected in the soil the procedures to be followed become more complex. R. subterranea can survive for many years in the soil (Höfer, 1993) although it does not necessarily infect plants. Soil management strategies may be crucial in determining whether pathogenic micro-organisms become established in agricultural land or not. Pathogen-suppressive properties of the soil may influence whether or not plants become infected by a parasite (Alabouvette et al., 1982; Janvier et al., 2007). But the mechanisms leading to soil suppressiveness are far from being understood. For this reason, benchmarks for R. subterranea in the soil are difficult to define and will require elaborate field trials for the future.
To facilitate high-throughput experiments with many parameters only small scale experiments were conducted in this study. As a consequence the DNA extraction method presented was developed for small amounts of soil (0.2 g) with samples taken from the upper soil layer. In view of the much greater depth at which fruiting bodies of R. subterranea have been found, strong sampling strategies will be required. For efficient field sampling it would be advisable to scale up DNA extraction to a more representative sample size (Mumford et al., 2006). The method presented here will serve as a basis to determine adequate guidelines for the detection of R. subterranea.
This work is intended to serve as basis for future field research on soil-borne grapevine pathogens. The PCR-based detection method permits the comprehensive testing of root pathogens such as R. subterranea in vineyards. A better knowledge of the abundance and distribution of R. subterranea will improve our general understanding of this devastating grapevine pest. The presented detection method will serve as basis to determine adequate guidelines for the detection of soil-borne fungi in vineyards.
Acknowledgements
The authors wish to thank M. Hoffmann and P. Michaelis for soil sampling and M. Flöck, F. Kraler, B. Laimgruber, M. Margreiter, V. Pauli, and S. Primig for technical assistance in the laboratory. They gratefully acknowledge W. Buzina, Medical University Graz, Austria for sequencing support. We are indebted to R. Pöder, University of Innsbruck, Austria for passing on his knowledge and expertise. This work was partially supported by the Hessian Ministry for Environment, the Feldbausch Foundation, and the Heinrich-Birk-Gesellschaft e.V., Geisenheim. S. Neuhauser was supported by a postgraduate grant of the Leopold Franzens University Innsbruck and the Austrian Science Fund (FWF; grant T379).
Abbreviations
- CTAB
cetyl trimethyl ammonium bromide
- EDTA
ethylene diamine tetraacetic acid
- PVPP
polyvinylpyrrolidone
- SDS
sodium dodecylsulfate
Literature cited
- Alabouvette C, Couteaudier Y, Louvet J. Comparaison de la réceptivité de différents sols et substrats de culture aux fusarioses vasculaires. Agronomie. 1982;2:1–6. [Google Scholar]
- Beckwith AM. The life history of the grape root rot fungus Roesleria hypogaea Thüm. et Pass. Journal of Agricultural Research. 1924;27:609–616. [Google Scholar]
- Berthelet M, Whyte LG, Greer CW. Rapid, direct extraction of DNA from soils for PCR analysis using polyvinylpolypyrrolidone spin columns. FEMS Microbiology Letters. 1996;138:17–22. doi: 10.1111/j.1574-6968.1996.tb08128.x. [DOI] [PubMed] [Google Scholar]
- Boyd SA, Mortland M. Enzyme interactions with clays and clay–organic matter complexes. In: Bollag JM, Strotzky G, editors. Soil Biochemistry. Vol. 6. Marcel Dekker, Inc.; New York, NY, USA: 1990. pp. 1–28. [Google Scholar]
- Cullen DW, Hirsch PR. Simple and rapid method for direct extraction of microbial DNA from soil for PCR. Soil Biology and Biochemistry. 1998;30:983–993. [Google Scholar]
- Damm U, Fourie P. A cost-effective protocol for molecular detection of fungal pathogens in soil. South African Journal of Science. 2005;101:135–139. [Google Scholar]
- Gerrits Van Den Ende AHG, De Hoog GS. Variability and molecular diagnostics of the neurotropic species Cladophialophora bantiana. Studies in Mycology. 1999;43:151–162. [Google Scholar]
- Herrera A, Cockell CS. Exploring microbial diversity in volcanic environments: a review of methods in DNA extraction. Journal of Microbiological Methods. 2007;70:1–12. doi: 10.1016/j.mimet.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Höfer M. Untersuchungen über Roesleria hypogaea Thym. & Pass. als Erreger des Wurzelschimmels der Weinrebe. Geisenheimer Berichte. 1993;13:135. [Google Scholar]
- Huang PM. Role of soil minerals in transformations of natural organics and xenobiotics in soil. In: Bollag JM, Stotzky G, editors. Soil Biochemistry. Vol. 6. Marcel Dekker; New York, NY, USA: 1990. pp. 29–115. [Google Scholar]
- Huber L. Johannes Gutenberg University; Mainz, Germany: 2007. Schaderreger im Wurzelraum von Reben (Vitis spp.), Vorkommen, Wirkung, Interaktionen - und Möglichkeiten zu deren Kontrolle durch Maβnahmen des Integrated Pest Management (IPM) p. 460. PhD thesis. [Google Scholar]
- Huber L, Eisenbeis G, Hoffmann M, Neuhauser S, Porten M, Rühl EH. Wurzelschimmel - verborgene Gefahr im Untergrund. Die Winzer-Zeitschrift. 2006a;4:34–35. [Google Scholar]
- Huber L, Eisenbeis G, Hoffmann M, Neuhauser S, Porten M, Rühl EH. Die Bekämpfung des Wurzelschimmels. Die Winzer-Zeitschrift. 2006b;5:36–37. [Google Scholar]
- Huber L, Eisenbeis G, Hoffmann M, Rühl EH, Neuhauser S, Porten M. Wurzelschimmelerreger Roesleria subterranea (Weinm.) Redhead: Absterbeerscheinungen und Kümmerwuchs - Gefahr im Verborgenen. Das Deutsche Weinmagazin. 2006c;6:25–31. [Google Scholar]
- Huber L, Eisenbeis G, Kirchmair M, Neuhauser S, Porten M, Rühl E, Forneck A. Rebensterben durch Wurzelschimmel. Der Winzer. 2006d;62(5):22–24. [Google Scholar]
- Ivarson KC, Schnitzer M, Cortez J. The biodegra-dability of nucleic acid bases adsorbed on inorganic and organic soil components. Plant and Soil. 1982;64:343–353. [Google Scholar]
- Janvier C, Villeneuve F, Alabouvette C, Edel-Hermann V, Mateille T, Steinberg C. Soil health through soil disease suppression: which strategy from descriptors to indicators? Soil Biology and Biochemistry. 2007;39:1–23. [Google Scholar]
- Kirchmair M, Neuhauser S, Buzina W, Huber L. The taxonomic position of Roesleria subterranea. Mycological Research. 2008;112:1210–1219. doi: 10.1016/j.mycres.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Lakay F, Botha A, Prior B. Comparative analysis of environmental DNA extraction and purification methods from different humic acid-rich soils. Journal of Applied Microbiology. 2007;102:265–273. doi: 10.1111/j.1365-2672.2006.03052.x. [DOI] [PubMed] [Google Scholar]
- Längle T, Pernfuss B, Seger C, Strasser H. Field efficacy evaluation of Beauveria brongniartii against Melolontha melolontha in potato cultures. Sydowia. 2005;57:54–93. [Google Scholar]
- Levy-Booth DJ, Campbell RG, Gulden RH, Hart MM, Powell JR, Klironomos JN, Pauls KP, Swanton CJ, Trevors JT, Dunfield KE. Cycling of extra-cellular DNA in the soil environment. Soil Biology and Biochemistry. 2007;39:2977–2991. [Google Scholar]
- Lloyd-Jones G, Hunter DWF. Comparison of rapid DNA extraction methods applied to contrasting New Zealand soils. Soil Biology and Biochemistry. 2001;33:2053–2059. [Google Scholar]
- Martin-Laurent F, Philippot L, Hallet S, Chaussod R, Germon JC, Soulas G, Catroux G. DNA extraction from soils: old bias for new microbial diversity analysis methods. Applied and Environmental Microbiology. 2001;67:2354–2359. doi: 10.1128/AEM.67.5.2354-2359.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DN, Bryant JE, Madsen EL, Ghiorse WC. Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Applied and Environmental Microbiology. 1999;65:4715–4724. doi: 10.1128/aem.65.11.4715-4724.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumford R, Boonham N, Tomlinson J, Barker I. Advances in molecular phytodiagnostics: new solutions for old problems. European Journal of Plant Pathology. 2006;116:1–19. doi: 10.1007/s10658-006-9037-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhauser S, Lass-Flörl C, Buzina W, Mayr A, Kirchmair M. A rapid DNA extraction protocol for a PCR-based detection of Histoplasma capsulatum in bat guano. Sydowia. 2008;60:123–130. [Google Scholar]
- Paget E, Monrozier LJ, Simonet P. Adsorption of DNA on clay minerals: protection against DNase I and influence on gene transfer. FEMS Microbiology Letters. 1992;97:31–39. [Google Scholar]
- Pérez-Jiménez RM. A review of the biology and pathogenicity of Rosellinia necatrix: the cause of white root rot disease of fruit trees and other plants. Journal of Phytopathology. 2006;154:257–266. [Google Scholar]
- Porten M, Huber L. An assessment method for the quantification of Daktulosphaira vitifoliae (Fitch; Hemiptera: Phylloxeridae) populations in the field. Journal of Applied Entomology. 2003;127:157–162. [Google Scholar]
- Redhead SA. Roeslerina gen. nov. (Caliciales, Caliciaceae), an ally of Roesleria and Coniocybe. Canadian Journal of Botany. 1984;62:2514–2519. [Google Scholar]
- Robe P, Nalin R, Capellano C, Vogel TM, Simonet P. Extraction of DNA from soil. European Journal of Soil Biology. 2003;39:183–190. [Google Scholar]
- Ruano-Rosa D, Schena L, Ippolito A, Lopez-Herrera CJ. Comparison of conventional and molecular methods for the detection of Rosellinia necatrix in avocado orchards in southern Spain. Plant Pathology. 2007;56:251–256. [Google Scholar]
- Schneegurt MA, Dore SY, Kulpa CF. Direct extraction of DNA from soils for studies in microbial ecology. Current Issues in Molecular Biology. 2003;5:1–8. [PubMed] [Google Scholar]
- Takada Hoshino Y, Matsumoto N. Skim milk drastically improves the efficacy of DNA extraction from Andisol, a vulcanic ash soil. Japan Agricultural Research Quaterly. 2005;39:247–252. [Google Scholar]
- Tate RL. Soil Microbiology. 2nd ed John Wiley & Sons, Inc.; New York, NY, USA: 2000. p. 508. [Google Scholar]
- Tsai Y–L, Olson BH. Rapid method for separation of bacterial DNA from humic substances in sediments for polymerase chain reaction. Applied and Environmental Microbiology. 1992;58:2292–2295. doi: 10.1128/aem.58.7.2292-2295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Véghelyi K. A Roesleria pallida (Fries) Sacc. szerpepe a gyümölcsfák fiatlalkori puszulásában. [The role of Roesleria pallida (Fries) Sacc. in the early death of fruit trees]. Növényvédelem. 1987;23(9):405–412. [Google Scholar]
- Véghelyi K. The isolation and characteristics of Roesleria hypogaea Thüm. et Pass. Acta Phytopathologica et Entomologica Hungarica. 1989;24:293–299. [Google Scholar]
- Volossiouk T, Robb EJ, Nazar RN. Direct DNA extraction for PCR–mediated assays of soil organisms. Applied and Environmental Microbiology. 1995;61:3972–3976. doi: 10.1128/aem.61.11.3972-3976.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward E, Foster SJ, Fraaije BA, McCartney HA. Plant pathogen diagnostics: immunological and nucleic acid-based approaches. Annals of Applied Biology. 2004;145:1–16. [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols: A Guide to Methods and Applications. Academic Press, Inc.; New York, NY, USA: 1990. pp. 315–322. [Google Scholar]
- Wilson IG. Inhibition and facilitation of nucleic acid amplification. Applied and Environmental Microbiology. 1997;63:3741–3751. doi: 10.1128/aem.63.10.3741-3751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. Biosynthesis of flavonoids and effects of stress. Current Opinion in Plant Biology. 2002;5:218–223. doi: 10.1016/s1369-5266(02)00256-x. [DOI] [PubMed] [Google Scholar]
- Zhou J, Bruns MA, Tiedje JM. DNA recovery from soils of diverse composition. Applied and Environmental Microbiology. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]