INTRODUCTION
Poly (ADP-ribose) polymerase (PARP) inhibitors have raised recent excitement because of the activity reported in triple negative breast cancer (TNBC) with iniparib (BSI 201) [1] and BRCA 1 or 2 associated ovarian or breast cancer with olaparib (AZ 2281) [2]. This class of agents is thought to augment cytotoxic therapy without increasing side effects and to kill cancer cells with DNA repair defects as a single agent. The genomic instability of some tumor cells allows PARP inhibitors to have selectivity for the tumor cells over normal cells.
DNA damages result from errors in replication, production of reactive oxygen species, and exposure to ultraviolet rays and ionizing radiation. These lesions that result from these noxious events include point mutations, single strand breaks (SSBs), double strand breaks (DSBs), intrastrand and interstrand cross-links. Cells employ multiple types of DNA repair mechanisms: base excision repair (BER), nucleic acid excision repair (NER), homologous recombination(HR), single strand annealing (SSA), Mismatch Repair (MMR), and non-homologous end joining (NHEJ) to repair these damages on a regular basis. As a result of DNA repair, injured cells can survive, which is optimal for normal cells, but exactly the opposite of the goal for tumor cells that undergo DNA damage in response to chemotherapy or radiation. In addition, errors can occur in the repair process especially with NHEJ that can lead to new abnormalities and dysfunction of the cells. Certain genetic disorders, such as BRCA1 and BRCA2 mutations, as well as other genetic anomalies that prevent DNA repair are associated with increased risk of malignancies. [3]
PARP is a family of proteins with enzymatic properties, scaffolding properties, and recruiting ability for other necessary DNA repair proteins. [4] PARP 1 and PARP 2 are the best known of these proteins and are critical for the function of BER. BER repairs single strand DNA breaks and inhibition of BER may ultimately lead to cell death. This makes PARP proteins ideal targets for anticancer therapy. PARP inhibitors interfere with BER and therefore DNA repair. By this route, PARP inhibitors can affect death of tumor cells. PARP inhibitors currently under clinical development are targeted to PARP 1 and PARP 2 proteins. They include Pfizer’s PF 01367338 (AG014699), AstraZeneca’s olaparib (AZD2281, KU-0059436), sanofi-aventis’ iniparib (BSI 201), Abbott Laboratories’ veliparib (ABT 888), Merck’s MK 4827, and Cephalon’s CEP 9722. Biomarin’s BMN673 (LT-673) and BiPar Science’s BSI 401 are in preclinical development.
Like with so many other therapies, resistance has been reported with PARP inhibitors. Resistance can develop via reversion of BRCA deficiency from the mutational reading frame to a reading frame that produces a wild type BRCA protein. This occurs through a second mutation, compensatory mutations, or crossovers.[5] Up-regulating the p-glycoprotein efflux pump and turning off 53BP1 have been shown as a possible mechanisms of resistance. [6–7] Also, resistance has been shown in tumors with increased tumor expression of PARP. Overcoming this resistance can be achieved through a mutation that converts the cell back to the mutated form, another mutation that inhibits HR, a proteosome inhibitor downregulating the P-glycoprotein pump, or up-regulation of 53BP1. Recently 6-Thioguanine has been shown to be active in cells resistant to PARP inhibitors in BRCA2 deficient tumors. [8]
The multiple areas of exploration of PARP inhibitors include the biology of the PARP inhibitors, DNA repair mechanisms, genetic defects of DNA repair, exploration of the clinical efficacy and toxicity, biomarkers for identifying target tumors, possibility of inducing tumors to be more sensitive to PARP inhibitors, development of new agents, and overcoming PARP inhibitor resistance (PIR). This review article will discuss these areas with focus on PARP inhibitors in the treatment of breast and ovarian cancers.
DNA REPAIR MECHANISMS
Genomic Instability of Tumors
Tumor cells, because of the frequency of their replication and their genomic susceptibility, have increased frequency of mutations which make them resilient against normal cell death, but may at the same time provide targets for antitumor therapy. Genomic instability can be in the form of mutational instability, consisting of point mutations and small deletions, and chromosomal instability, including gross rearrangements, such as loss or gains of whole chromosomes or fragments, amplifications, and fusion of genes.
BER, MMR and NER of SSB
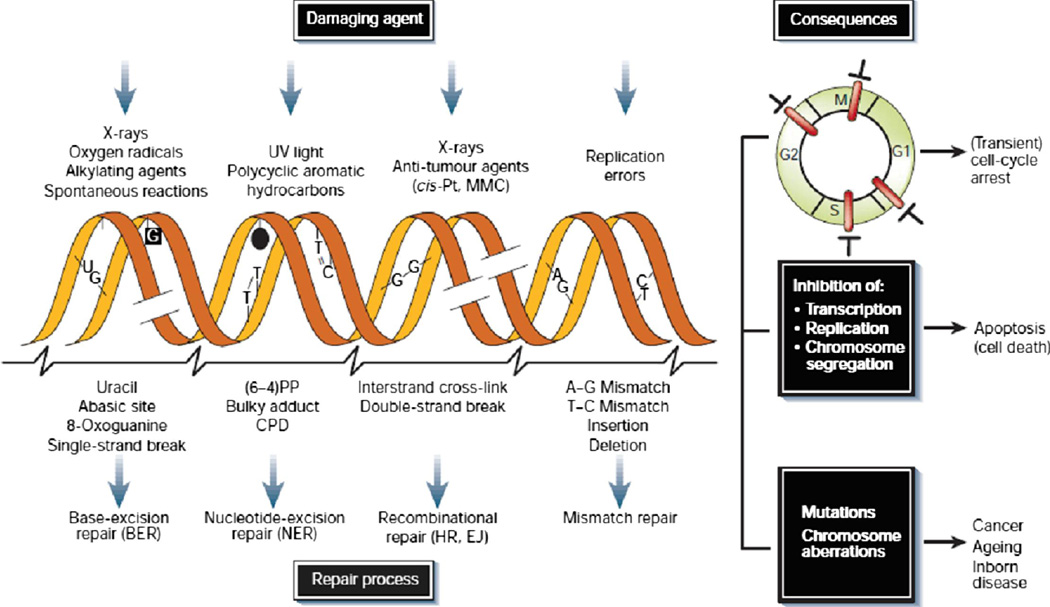
DNA repair mechanisms correct single strand breaks (SSBs) and double strand breaks (DSBs). (Fig. 1). In SSBs the complementary DNA strand is used as template; in DSBs the complementary strand is not readily available. There are estimated 10 4 SSBs daily. [10] The process for SSB repair is accomplished by BER and MMR and NER. BER involves removing a damaged base by a DNA glycosylase. BER is involved in the repair of damages from radiation and alkylating agents. BER is the defect involved in xeroderma pigmentosa, which increases UV sensitivity and skin cancer. PARP1 and 2 are integrally involved in BER. MMR corrects mismatched bases that may occur during the replication process. It involves genes MSH 2 and MLH1. Fifteen percent of colon cancer has MMR resulting in microsatellite instability. These tumors behave differently from other colon cancer and respond differently to treatment. [11] Knowing the genetic and molecular characteristics of tumors allows for differential treatment of individual patient’s tumors and subsequently personalized medicine. Nucleotide Excision Repair (NER) removes large patches of nucleotides around the “bad” base in its repair. It corrects damages from UV light and hydrocarbons.
Figure 1. Causes of DNA damage, Types of DNA Repair, Cellular Consequences.
Reprinted by permission from Macmillan Publishers Ltd: Nature 22;362(6422):709–15, copyright 2001. [9]
HR and NHEJ Repair of DSB
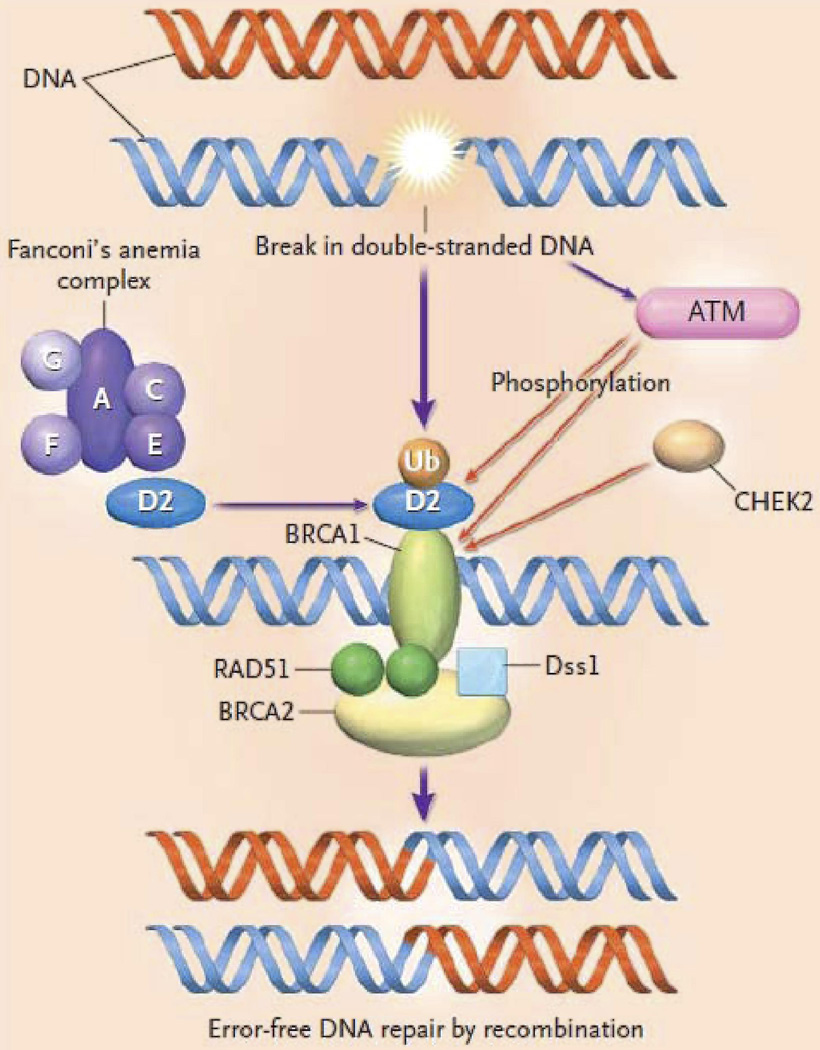
HR and NHEJ) work to correct DSBs. When DSBs occur, ATM and CHEK2 kinases mobilize proteins such as BRCA 1 protein. BRCA 2 carries Rad51, the recombination enzyme, to the DSB site. A complex of Fanconi anemia proteins, A, C, D2, E, F, and G, cause the ubiquitinization of D2 protein and the subsequent association of D2 with BRCA 1. (Fig. 2) All of this ultimately leads to repair the DSB with minimal error in the DNA. If there is a defect in BRCA 1 or BRCA 2, then the DSB repair is carried out by error prone mechanisms, such as NHEJ, increasing the risk of chromosomal aberrations.
Fig. 2. Proteins involved in Homologous Recombination.
When DSBs occur, ATM and CHEK2 kinases mobilize proteins such as BRCA 1 protein. BRCA 2 carries Rad51, the recombination enzyme, to the DSB site. A complex of Fanconi anemia proteins, A, C, D2, E, F, and G, cause the ubiquitinization of D2 protein and the subsequent association of D2 with BRCA 1. All of this ultimately leads to repair the DSB with minimal error in the DNA. Reprinted with permission [12], Copyright © 2003 Massachusetts Medical Society. All rights reserved.
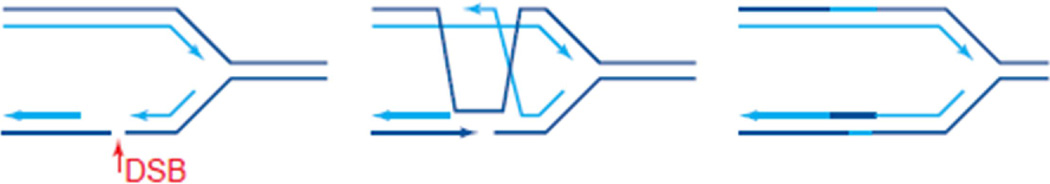
There are two processes of HR, gene conversion, which uses the homologous sequence, usually within the sister chromatid, as the template, and thereby usually repairs without error. (Fig. 3) It is this process that relies on RAD51 as the recombinase. It must be understood, however, that an “error-free” mechanism can result in loss of heterozygosity which can bring about inactivation of a tumor suppressor gene or activation of an oncogene, still leading the carcinogenesis of tissue. The other HR process is single strand annealing (SSA). SSA uses the homologous strand, but this time without RAD51, and frequently results in changes in the DNA sequence.
Fig. 3. Gene Conversion Homologous Recombination.
Gene conversion uses the homologous sequence, usually within the sister chromatid, as the template, and thereby usually repairs without error. Reprinted from Trends in Biochemical Sciences, 24(7), Haber JE, “DNA recombination: the replication connection,” 271–5, Copyright 1999, with permission from Elsevier. [13]
NHEJ is another mechanism that repairs DSBs. It is error prone, frequently resulting in gross chromosomal rearrangements, including translocations. NHEJ does not involve Rad 51. When HR is defective, NHEJ serves to repair DSBs, resulting in increased frequency of new mutations. [14]
Mutations in genes that result in defective HR, increase risk of developing breast and ovarian cancers. BRCA 1 and BRCA 2 are tumor suppressor genes involved in the HR pathway. Fanconi’s anemia, which predisposes to acute myeloid leukemia and squamous-cell carcinomas, also is associated with breast cancer. Six of the 8 subtypes of Fanconi’s anemia have a known germline mutation. Since the proteins in the Fanconi’s anemia complex are responsible for making D2 attach to BRCA 1 in the HR pathway, a defect in any of the subunits of the complex will cause chromosomal defects similar to defective BRCA 1 and BRCA 2 proteins. BRCA 2 itself is mutated in a small number of patients with Fanconi’s anemia, usually in Fanconi’s anemia type D1, and possibly also in type B. [12, 15] Defects in DSB repair are involved in ataxia telangiectasia (AT) and Nijmegen breakage syndrome (NBS). Ataxia telangiectasia, which is autosomal recessive with mutation at 11q22–q23 and results in a defect in the ataxia telangiectasia mutated (ATM) gene, predisposes patients to malignancy. The ATM protein is involved in the HR pathway and DSB repair. Ataxia telangiectasia patients have 16 fold greater risk of developing breast cancer over the general population. The heterozygous state is responsible for 7% of all breast cancers. Ataxia telangiectasia patients also have an increase in the risk of GI, lung and lymphoid cancers. The disorder makes the patient more sensitive to the effects of radiation. AT and NBS are just 2 examples of defects in DNA repair predisposing to the development of malignancies. [16–18]
DSBs can be quantitated which can be exploited as a biomarker for measuring the efficacy in anticancer treatment. As a reaction to DSBs, H2AX become phosphorylated and form γ-H2AX foci, which can be measured. Recently, NCI had developed an assay for measuring γ-H2AX in circulating tumor cells. [19] Many proteins involved in HR are contained in the Rad51 foci, which can also be measured in the laboratory. HR deficient cells do not form Rad51 foci in response to DNA damage. PARP 1 is not associated w/ Rad51 foci. Rad51 foci form in PARP −/− cells in response to hydroxyurea. PARP 1 inhibited cells respond to DSB with HR and show increased Rad51 foci. This shows that PARP 1 is not directly involved in HR. [20].
Poly-(Adenoribosyl)phoshatase, PARP
Poly-(Adenoribosyl)phoshatase, PARP, is in an enzyme that is involved in BER. This enzyme has been known since 1963 when Chambon, Weill, and Mandel wrote Nicotinamide Mononucleotide Activation of New DNA-Dependent Polyadenylic Acid Synthesizing Nuclear Enzyme in the journal, .Biochemical and Biophysical. Research Communications. [21] Shall, Durkacz, and Omidiji in 1980 proposed that modulating PARP 1 might augment the effect of alkylator chemotherapy. [22]
The PARP family consists of 17 proteins if based on structural similarity, but only 6 proteins if based on function. [23] The three groups consist of the following:
Group 1 containing PARP 1, PARP 2, and PARP 3,
Group 2 containing PARP 4, also called vault PARP, and
Group 3 containing tankyrase (TNKS) and TNKS2 [24]
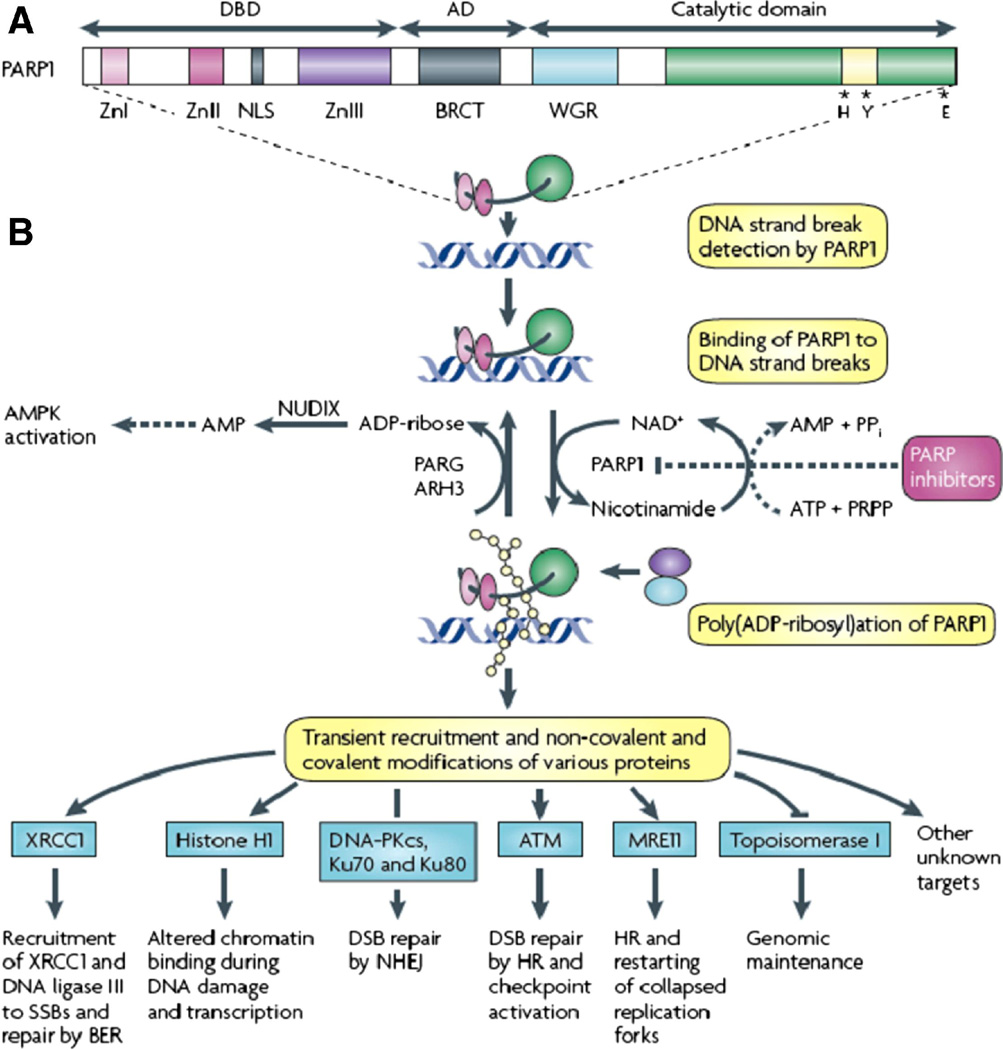
PARP proteins are composed of two ribose moieties and two phosphates per unit polymer. PARP 1 is the protein that is best understood. It is active in its homodimeric form. It has 3 functional domains, the DNA-binding domain (DBD), the automodification domain (AD), and the catalytic domain. The amino-terminal DBD is 42 kDa. It contains 3 zinc fingers, two that bind PARP 1 to DNA breaks, and a third that couples DNA damage-induced changes in the DBD to catalytic activity. The 16 kDa AD portion contains glutamate and lysine amino acids that accept ADP-ribose units, resulting in self poly (ADP-ribosyl)ation. The AD unit houses the BRCA 1 carboxyl-terminal (BRCT) repeat motif, similar to DNA sequences in other proteins involved in DNA repair. The C-terminal catalytic domain is 55 KDa. It holds the signature sequence containing the most conserved sequence of the PARP family. In this domain ADP-ribose transferase facilitates the transfer of ADP-ribose from nicotinamide adenine dinucleotide (NAD+) to protein acceptors within this domain.
PARP proteins are activated by DNA strand breaks. These proteins carry vital roles for survival of cells and organisms. Mice can survive without PARP 1 or PARP 2, but not without both. [25–26] Similarly, they can survive without TNKS or TNKS 2, but not without both. PARPs by definition must be able to transfer ADP-ribose from NAD+ to an acceptor protein and add multiple subunits to the PAR. [27] It is not known at this time if PARP 3 and PARP 4 form the multiple subunits of PAR, so these proteins may not be true PARPs. [4] PARP 3 might activate PARP 1 without DNA breaks. [28] PARP must be at least 90% inhibited to suppress DNA repair. [29]
PARP 1 is active in a homodimeric state. It detects and binds to sites of single strand DNA damage via the DNA-binding domain. It then synthesizes poly(ADP) ribose (pADPr, PAR) and transfers it to acceptor proteins. The acceptor proteins can be located on PARP 1 itself or on other proteins involved in DNA repair. The negative charge of PAR causes PARP 1 to lose its affinity for DNA. PAR recruits other repair proteins to the damaged DNA site. Poly(ADP-ribose) glycohydrolase (PARG) and possibly ADP-ribose hydrolase 3 (ARH3) break pADPr into ADP-ribose molecules, which are metabolized further to AMP. The increased AMP:ATP ratio triggers the metabolic sensor AMP-activated protein kinase (AMPK). MTORC1 is thereby inhibited, inducing autophagy. [30] Thus cellular energy homeostasis is regulated. In the process of making PAR, NAD+ is converted to nicotinamide. To replenish the NAD+ from nicotinamide, phosphoribosylpyrophosphate (PRPP) and ATP are converted to AMP and pyrophosphate. In the case of extreme DNA damage, as with ischemia, PARP 1 hyperactivation results in depletion of NAD+ and ATP, resulting in cell death by necrosis or apoptosis.
PAR covalently and noncovalently binds proteins that work in the repair of DNA or works on these proteins via pADPr-binding proteins. The largest amount of PAR stays attached to PARP 1. PAR binds XRCC1, the scaffolding protein. PAR regulates histone H1’s binding to chromatin, allowing the chromatin to relax. PARP is involved in methylation and transcription of genes coding for cell cycle and stress response, including p53. Experiments with PARP−/− mice [31] and human breast cancer cells that have PARP 1 downregulated by PARP 1 short-hairpin RNAs (shRNAs) showed alteration of these genes. [32]. PAR attaches DNA polymerase β to site to replace the missing bases. Finally PAR combines with DNA ligase III to seal the DNA.
PAR is involved in DSB repair as well. It attaches to DNA-protein kinase catalytic subunit (DNA-PKs), Ku 70 and Ku80, to allow DSB repair through NHEJ DNA ligase. [33] PAR recruits ATM, MRE11, and topoisomerase 1, all involved n DSB repair. [34–35] (Fig. 4)
Fig. 4. Structure and activity of PARP.
PARP 1 is active in its homodimeric form. It has 3 functional domains, the DNA-binding domain (DBD), the automodification domain (AD), and the catalytic domain. The amino-terminal DBD is 42 kDa. It contains 3 zinc fingers, two that bind PARP 1 to DNA breaks, and a third that couples DNA damage-induced changes in the DBD to catalytic activity. The 16 kDa AD portion contains glutamate and lysine amino acids that accept ADP-ribose units, resulting in self poly (ADP-ribosyl)ation. The AD unit houses the BRCA 1 carboxyl-terminal (BRCT) repeat motif, similar to DNA sequences in other proteins involved in DNA repair. The C-terminal catalytic domain is 55 KDa. It holds the signature sequence containing the most conserved sequence of the PARP family. In this domain ADP-ribose transferase facilitates the transfer of ADP-ribose from nicotinamide adenine dinucleotide (NAD+) to protein acceptors within this domain. PARP proteins are activated by DNA strand breaks. PARP 1 is active in a homodimeric state. It then synthesizes poly(ADP) ribose (pADPr, PAR) and transfers it to acceptor proteins. The acceptor proteins can be located on PARP 1 itself or on other proteins involved in DNA repair. The negative charge of PAR causes PARP 1 to lose its affinity for DNA. PAR recruits other repair proteins to the damaged DNA site. Poly(ADP-ribose) glycohydrolase (PARG) and possibly ADP-ribose hyrolase 3 (ARH3) break pADPr into ADP-ribose molecules, which are metabolized further to AMP. The increased AMP:ATP ratio triggers the metabolic sensor AMP-activated protein kinase (AMPK). MTORC1 is thereby inhibited, inducing autophagy. [30] Thus cellular energy homeostasis is regulated. In the process of making PAR, NAD+ is converted to nicotinamide. To replenish the NAD+ from nicotinamide, phosphoribosylpyrophosphate (PRPP) and ATP are converted to AMP and pyrophosphate. In the case of extreme DNA damage, as with ischemia, PARP 1 hyperactivation results in depletion of NAD+ and ATP, resulting in cell death by necrosis or apoptosis. PAR covalently and noncovalently binds proteins that work in the repair of DNA or works on these proteins via pADPr-binding proteins. Reprinted by permission from Macmillan Publishers Ltd: Nature Reviews Cancer, 10(4):293–301, copyright 2010. [4]
The half-life of PAR is seconds to minutes. However, it initiates activities of DNA repair that last longer.
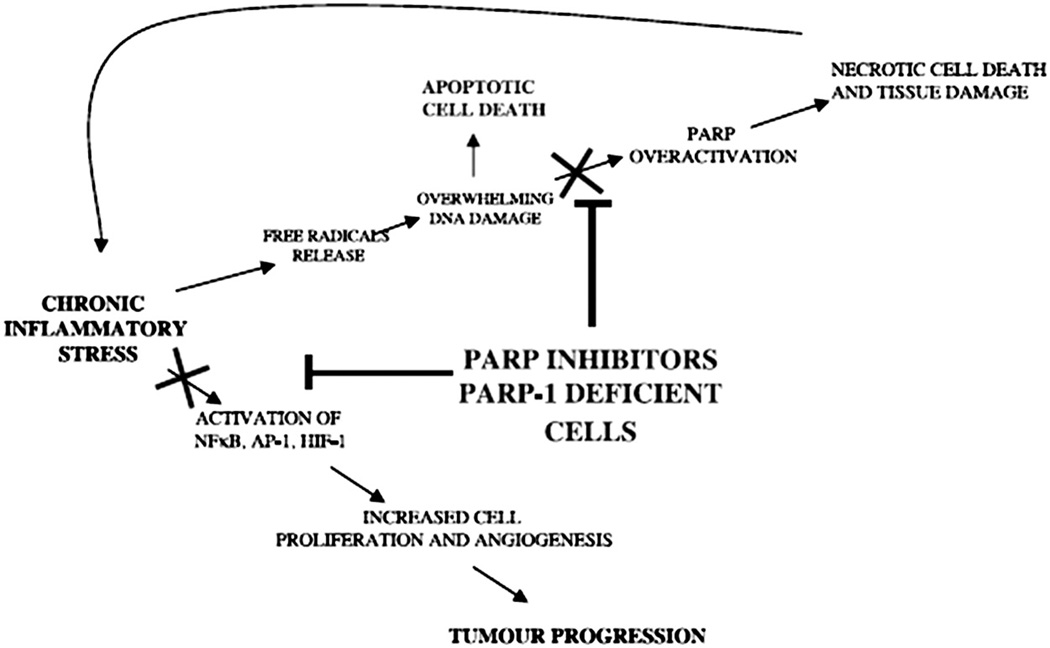
PARP 1 also activates genes in a more direct manner in addition to correcting DNA damage. It activates NF-κB, a stress-inducible transcription complex, which is part of the immune system, and which inhibits apoptosis and promotes proliferation. NF-κB exhibits increased expression in cancers. It is constitutively activated in breast cancer, particularly in those that are hormone refractory and those with worse prognoses. NF-κB correlates with progression of disease. It is also activated by XRT and chemotherapy. Inhibition of NF-κB sensitizes cells to XRT and chemotherapy. [36]
PARP 1 has some responsibility in the activation of HIF-1α. When PARP 1 was inhibited chemically or by knock-out genes in a mouse experiment, there was decreased tumor growth and tumor vasculature. There was also decreased expression of HIF-1α, activation protein-1 (AP-1), and NF-κB, and other genes involved in carcinogenesis and inflammation. [37] (Fig. 5)
Fig. 5. Multiple mechanisms of action of PARP Inhibitors.
PARP 1 inhibitors decreased tumor growth and tumor vasculature through decreased expression of HIF-1α, activation protein-1 (AP-1), and NF-κB, and other genes involved in carcinogenesis and inflammation.
Reprinted thanks to Cancer Research, AACR 2006. [37]
PARP 1 −/− cells in mice have shown increased sensitivity to DNA damaging chemicals, such as alkylating agents, and radiation. This finding makes sense since those cells have decreased ability to repair the DNA damage from those exposures. [38]
PARP 2 is also activated by DNA damage and synthesizes PAR; however, it is only responsible for 15% of the cell’s PAR production. [39–41] In PARP 1 −/− mice, where PARP2 responds to DNA damage, less NAD+ is consumed, and there is also less necrosis in the tissues compared with normal mice. PARP 2−/− mice have increased sensitivity to alkylators and radiation, increased genomic instability, abnormal spermatogenesis, abnormal adipogenesis, and abnormal T cell development. Defects in spermatogenesis, adipogenesis, and T cell development are not seen in PARP 1 −/− mice. [42]
PARP 3 and PARP 4 may not be true PARP family members. PARP 3 makes mono-ADP ribose moieties, rather than poly-ADP ribose. Whether PARP 4, a possible tumor suppressor, makes PAR at all is unknown. Interestingly, PARP 4 deficiency is associated with a higher incidence of colon cancer. [43]
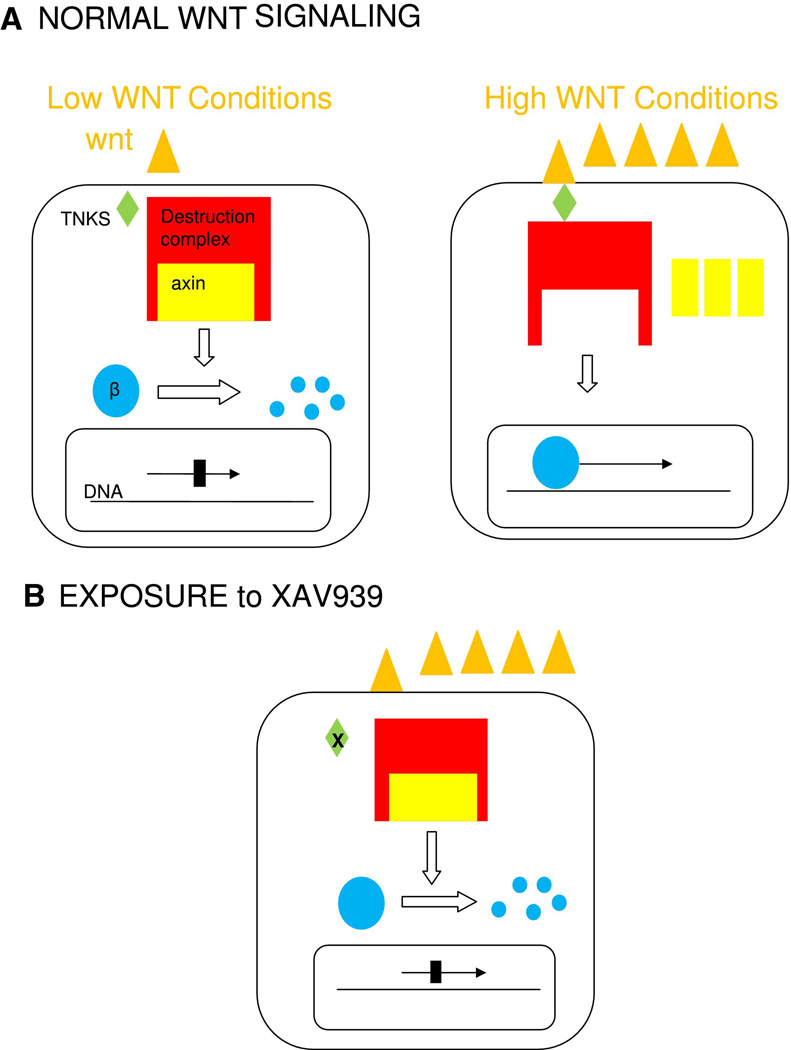
TNKS and TNKS 2 maintain telomere lengths via poly(ADP-ribosyl)ation in human cell lines, but not in mice. The structure of the protein differs between species. In human cells, TNKS is involved in mitotic spindle formation, but TNKD 2 is not. [44] TNKS and TNKS 2 are also involved in Wnt signaling. In the normal Wnt signaling pathway, Wnt binds to a cell surface receptor signaling beta-catenin to enter the nucleus and promote gene expression. If Wnt is not present, the beta-catenin is degraded by the beta-catenin destruction complex. The Wnt pathway is deregulated in many cancers. TNKS and TNKS 2 cause axin, the concentration-limiting piece of the destruction complex, to degrade through the ubiquitin-proteasome pathway. XAV939, a small molecule found through the high throughput process, inhibits TNKS and TNKS 2, thereby allowing axin to persist and destruction of Beta-catenin, inhibiting transcription. [45] (Fig. 6)
Fig. 6.
a. Normal Wnt signaling. b. exposure to TNKS inhibitor
SYNTHETIC LETHALITY
Synthetic lethality is when two conditions that independently would not cause cell death in combination with each other are lethal.
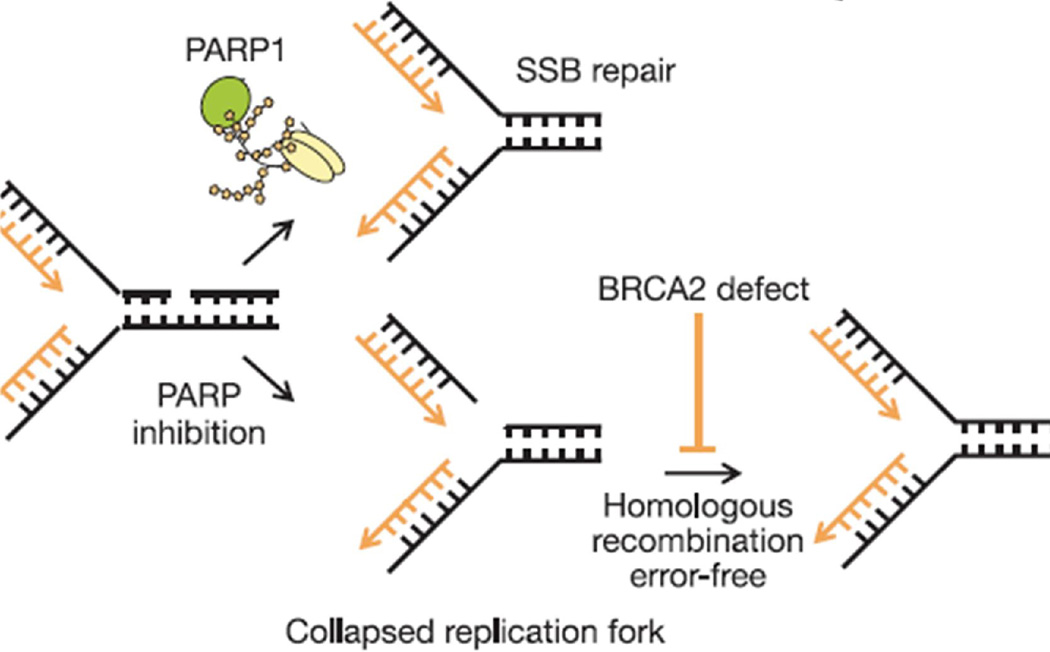
PARP 1 −/− mice are viable and fertile and do not develop tumors prematurely. PARP inhibitors bring about increase in γ-H2AX and RAD51 foci formation, indicating that DSB form and HR is initiated after exposure to PARP inhibitors. Without PARP 1, SSBs collapse replication forks and initiate HR. If the PARP 1 deficient cell is also deficient in BRCA 2 HR cannot occur. (Fig. 7), and the cell then dies or undergoes error prone recombination.
Fig. 7. PARP 1 inhibition leads to collapsed replication forks and need for homologous repair for cell survival.
Reprinted by permission from Macmillan Publishers Ltd: Nature, 14;434(7035):913–7, copyright 2005. [46]
In mouse experiments using siRNA to knock out specific genes, the following findings were noted:
Cells depleted of both PARP 1 + BRCA 2 had reduced survival
Cells depleted of both PARP 2 + BRCA 2 did not have reduced survival
Cells depleted of PARP 1+ PARP 2 + BRCA 2 had survival similar to the PARP 1 + BRCA 2 only depleted cells.
These results show that PARP 1, rather than PARP 2, is the PARP primarily responsible for DNA repair. [46] [47]
Proteins other than BRCA could cause synthetic lethality in combination with PARP inhibitors. Phosphatase and tensin homolog (PTEN) is a prevalent tumor suppressor gene involved in expression of RAD51, and thus involved in HR. As another example of synthetic lethality, PTEN-deficient cells are sensitive to PARP inhibitors in vitro and in vivo. Clinical trials are underway evaluating activity of PARP inhibitors in patients with decreased PTEN expression, common in endometrial cancer and glioblastoma, as well as in malignant melanoma, prostate, breast, lung and colorectal cancers. [48]
Mutated Fanconi’s anemia proteins, which also render HR ineffective, could also signal cells that might be susceptible to synthetic lethality when exposed to agents that inhibit PARP. [13]
There are two great advantages of exploiting synthetic lethality. One, inhibiting PARP 1 alone may be sufficient to cause tumor cell death and avoid the toxic effects of chemotherapy and radiation. Two, it is possible that the therapy can target only the tumor tissue and avoid normal tissue. Most people with BRCA mutations are heterozygous for the abnormality. As will be discussed later, in rare instances people have a double or triple heterozygous pattern. The homozygous pattern in a germline is nonviable. The genotype of the tumor, on the other hand, can be homozygous after a second hit occurs. Theoretically it makes sense that if the tumor, containing the homozygous pattern, resulted in defective HR, and normal tissue carries the heterozygous pattern and normal HR, then exposure to the PARP inhibitor would provide synthetic lethality selective only for the tumor.
Interestingly, however, a study done by King showed that whereas ovarian tumors demonstrate a homozygous pattern for BRCA mutations, breast cancers can demonstrate a heterozygous pattern in the tumor and there can even be loss of heterozygosity (LOH) of the mutant gene, leaving the wild type active. These findings confuse our theory of synthetic lethality in breast cancer. It is possible that PARP inhibitors are working by the other mechanisms. Alternatively, the study may have come to the wrong conclusion based on possible sampling error since not all parts of the tumor were sampled, or perhaps HR is defective in these cells based on epigenetic influences on the BRCA gene rather than changes in the genes themselves. [49]
BREAST CANCER
In U.S. in 2010 there will be 200,000 female and 2000 male new cases and 40,000 female and 400 male deaths from breast cancer. Breast cancer is a heterogeneous disease from the stand point of the tissue it affects and its genetic and proteomic characteristics. Ductal carcinoma occurs in the tubules that in a normal and lactating state would carry the milk from the lobules to the nipple; lobular carcinoma stems from the milk-producing glands. [50] Based on gene expression there are 5 types of breast cancer, including luminal, luminal B, human epidermal growth factor receptor-2 (HER-2) overexpressing, basal-like, and normal-like. HER2-over expressing tumors and basal breast cancers have the worst prognoses. [51] The basal-like phenotype is ER negative, PR, negative, HER2-negative, and either EGFR or cytokeratin 5/6 positive. Approximately 17% of breast cancers are triple negative (ER negative, PR negative and HER-2 negative), and 15% are basal-like. The basal-like tumor is prevalent in premenopausal African American women, which may account for the worse prognosis in that population. Basal-like tumors are mitotically active, high grade, invasive, and associated with younger age. Eighty to ninety percent of hereditary BRCA 1 breast carcinoma have features similar to basal-like tumors. [52] Basal-like breast cancers are basically triple negative breast cancers with one additional feature. Triple negative breast cancers express upregulation of PARP 1. [53] In fact most breast cancers that are classified as triple negative are also basal-like. The upregulation of PARP 1 in TNBC would make TNBC potentially sensitive to PARP inhibitors.
OVARIAN CANCER
In the U.S. there are 22,000 new cases of ovarian cancer and 14,000 deaths per year due to this diagnosis. Ovarian cancer accounts for 6% of cancers in women. Most ovarian tumors are either epithelial carcinomas, beginning from non-ova tissue or malignant germ cell tumors, from the ova tissues themselves. The epithelial tumors are further divided into type 1, including low grade and endometrioid, mucinous, and low grade serous, and type 2, including high-grade serous. If caught early ovarian cancer is 90% curable; however, the symptoms are nonspecific and the cancer is usually detected in advance stages, when the chance of cure is usually less than 20%. Ninety percent of cases are sporadic and ten percent have a predisposing genetic defect. [54] Ninety percent of patients with familial predisposition carry the BRCA defect. BRCA mutations are not usually seen in low grade serous ovarian cancer. [55]. Three syndromes predispose to the development of ovarian cancer, hereditary breast –ovarian cancer syndrome, hereditary non-polyposis colorectal cancer (Lynch syndrome), and hereditary site-specific ovarian cancer. Hereditary breast-ovarian cancer syndrome is associated with BRCA 1 and BRCA 2; hereditary site-specific ovarian cancer is associated with BRCA 1. Lynch Syndrome is associated with germline mutations of mismatch repair genes. Familial ovarian cancers usually occur in at a younger age than sporadic. Patients with BRCA mutations develop ovarian cancer at an average age of about 50 years old. A large percent of the familial ovarian cancers also have over-expression of Her2. [54]
Serous ovarian carcinomas account for 60–80% of ovarian tumors. Ninety percent of serous ovarian tumors are high grade. [56] They are the more aggressive subtype. BRCA 1 and BRCA 2 mutations are present in 23% of high grade serous cancers. [3] The prognosis with the genetic form of high grade serous ovarian carcinoma that is associated with BRCA mutation seems to be better than the sporadic cases. [54] [57]. [58] The improved prognosis of BRCA mutated tumors may be secondary to the impaired DNA repair in these cells that lead to greater sensitivity to treatment. [59] [60]
BRCA
Mutations of BRCA 1 and BRCA 2, two tumor suppressor genes, predispose cells to increased risk and malignancies. BRCA 1, on chromosome 17, and BRCA 2, on chromosome 13, both increase the risk of breast and ovarian cancers. BRCA2, however, contributes less to the risk of ovarian cancer, but more to the risk of male breast cancer and pancreatic cancer. [54, 61] The BRCA mutations are most prevalent in people of Ashkenazi Jewish (Jews from Eastern European) descent. In that population the predominant mutations are 185delAG and 5382insC in BRCA 1 and 6174delT in BRCA 2. Although these mutations occur only in 2.5% of the Ashkenazi population, they account for 70–85% of germline mutations in patients with heritable breast cancer and ovarian cancer. Only 5–10% of breast cancers and 10% of ovarian cancers are associated with a known genetic mutation, so BRCA mutations are not that common. Having the mutation increases the risk of contracting ovarian cancer up to 63% and breast cancer up to 87%. [62]
Usually the germline is heterozygous, not homozygous, for the mutation; however, double and even triple heterozygous combinations have been identified in rare instances. The incidence of a double heterozygous state is 0.22–0.87% in BRCA carriers and up to 1.8% in Ashkenazi BRCA carriers. Those genotypes have no worse features, in terms of age of onset, lifetime risks, and numbers of tumors, compared with the single heterozygous state. It important to evaluate for a multiple heterozygous state for genetic counseling and so others within the family are aware of their risks. [63]
BRCA testing is available commercially. Scales have been developed to provide guidance on the probability of having a mutation in BRCA 1 or 2, including FHAT, Manchester Score, Frank, Couch and Bayesian Probabilistic Model (BRCAPRO). The most accurate one is the BRCAPRO. [64] These scales are based on family history of breast and ovarian cancers, early age of onset of breast and ovarian cancer in patient and/or family members, multiple tumors in a single patient, breast cancer in men, cancer-free survival in first- or second degree relatives, and ethnicity.
“BRCAness”
BRCAness is a profile of tumor cells that share traits with BRCA 1 or BRCA 2 mutated tumors. Cells with BRCAness have defective HR and improved response and survival with exposure to platinum agents. The cells with BRCAness may or may not contain known BRCA 1 or BRCA 2 germline mutations. [65]
BRCA 1 mutations are rare in sporadic cancers, but reduced expression of normal BRCA 1 might still be an important feature in non-inherited breast cancer and ovarian cancer. BRCA1expression is decreased in high grade ductal carcinoma revealing that BRCA 1 expression might be reduced even in sporadic breast and ovarian carcinomas. The reduction in expression might be due to allelic loss of chromosome 17q which houses BRCA 1, a phenomena which is known to occur [66] [67]. In this situation, however, a wild type copy of BRCA 1 would still be present, accounting for at least some normal BRCA 1 expression. Hypermethylation of the BRCA1 regulatory region, which could inhibit transcription, has also been found in sporadic breast and ovarian cancers. [68] [69] [70] [71]
Sporadic breast cancers have been found to have two times less messenger RNA expression and 9 times higher level of BRCA 1 negative regulator, ID4. [72] Sixty three percent of metaplastic breast cancer, a rare form of basal-like breast cancer, had BRCA1 promoter methylation compared with 12% of controls. [73] Microarrays also show similar gene patterns between familial BRCA 1 breast cancers and basal-like sporadic breast cancers. [51, 73–74] So triple negative breast cancer, basal-like breast cancers, and BRCA 1 germline breast cancers might all harbor the same tumorgenesis mechanism through BRCA 1 dysfunction. For that reason, all of these types of breast cancer are being explored as possible histologies that might benefit from PARP inhibitors through the synthetic lethality concept. Sporadic ovarian tumors also showed gene profiles that were either BRCA 1-like or BRCA-2 like. [75] Twenty-five percent of BRCA mutations in the ovarian tumors were not germline. [76] Few breast and ovarian cancers have BRCA germline mutations; somatic BRCA mutations account for up to 20% of high-grade ovarian tumors. The presence of the sporadic mutation increases the population that could benefit from PARP inhibitor therapy. [3]
In addition, other mutations, [77–78] [3] and epigenetic effects have been found to impart BRCAness to the cells. Hypermethylation of the BRCA 1 promoter [71] and loss of function mutations in other genes that affect the HR pathway [79] have been found to endow BRCAness to cells.
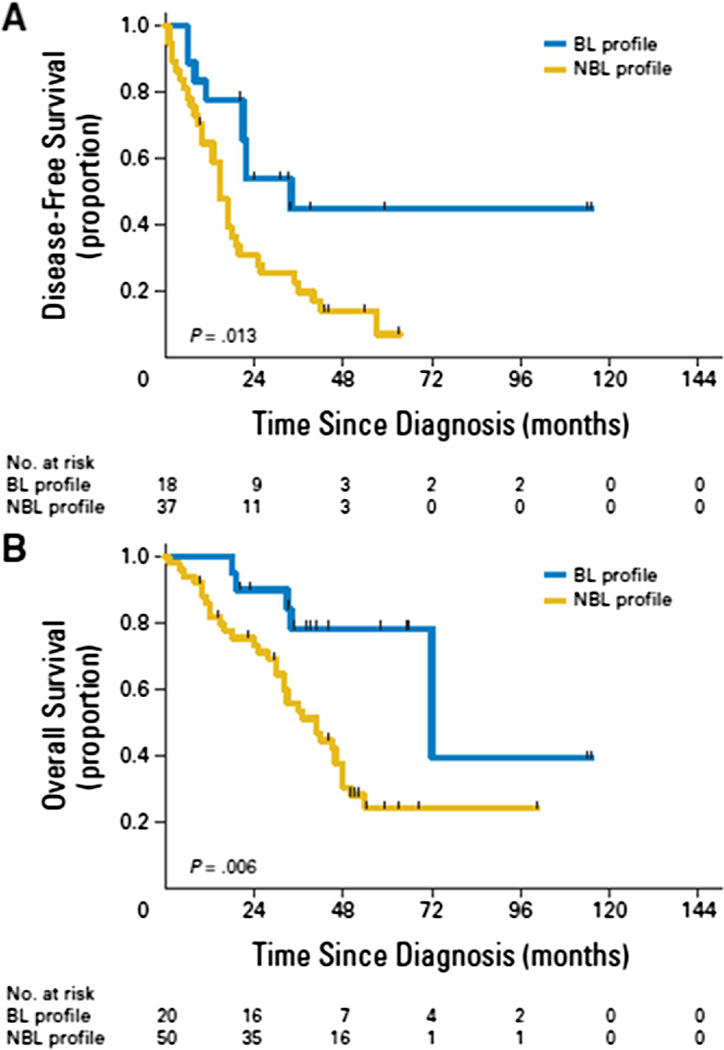
To find the genetic profile for cells with BRCAness, studies were carried out with microarrays of epithelial ovarian tumors with known germline mutations, a pancreatic tumor cell line known to have BRCA mutation, and sporadic epithelial ovarian tumors. A heat map showing the gene profile of tumors with and without the BRCA mutation differentiated specific patterns for BRCA like (BL) and not BRCA like (NBL) tumors with 94% accuracy in 61 patients.
In 6 patients with ovarian cancer with BRCA 1 and BRCA 2 germline mutations, biopsies were taken prior to cisplatin therapy and in 4 of those patients biopsies were taken after therapy. Eight of the ten biopsies showed correlation between BL and cisplatin sensitivity and between NBL and cisplatin resistance. In 2 of the 3 tumors that became resistant during the therapy, the microarray profile changed from BL to NBL. The correlation between BRCAness and RAD51 foci, as a marker of HR, was examined in the BRCA 2 mutated pancreatic cell line. Of 12 platinum resistant clones, seven clones formed RAD51 foci after receiving ionizing radiation. The profiles of 6 of those 7 clones showed reversion to functional BRCA 2 because of secondary BRCA 2 mutation that canceled the effect of the inherited BRCA 2 mutation. The 5 clones that did not exhibit RAD51 foci retained the nonfunctional BRCA2 mutation. In addition, the BRCAness profile accurately predicted PARP sensitivity in the clones. The restoration of BRCA function through a secondary mutation is a potential resistance mechanism to the PARP inhibitors. Evaluation of patients with epithelial ovarian cancer who did not harbor the BRCA mutation gene was evaluated for the BRCAness genetic profile. Those with BL profile had median DFS of 34 months compared with the NBL profile with median DFS of 15 months. The median OS for BL and NBL profiles were 72 and 41 months, respectively. The BRCAness profile had independent prognostic significance in multivariate analysis including age, stage, grade, histology, and debulking status (Fig. 8). [80]
Fig 8. Disease-Free Survival and Overall Survival in BRCA-like profile and Non-BRCA-like profile.
Reprinted with permission. © 2010 American Society of Clinical Oncology. All rights reserved. Konstantinopoulos PA, et al. J Clin Oncol 28(22), 2010: 3555–61. [80]
Some possible mechanisms of developing BRCAness, aside from inherent or sporadic BRCA mutations, include the following:
Increased expression of proteins that inactivate BRCA, including EMSY [81]
Defects in other Fanconi’s anemia complex proteins [12]
Decreased expression of other proteins involved in HR, including RAD51, ATR, ATM, CHK2. [85]
PTEN deficiency [48]
There are likely undiscovered mechanisms of accruing BRCAness as well.
The area of BRCAness is still undergoing multiple lines of investigation. TNBC is now being studied to see if the profiles for BRCAness in ovarian and pancreatic cancers apply. That TNBC shows some response to platinum agents would lead one to believe that the profile would apply. Clinical testing for PARP inhibitors for patients with sporadic tumors with the BRCAness profile is being planned.
PARP INHIBITORS
The rationale for PARP inhibitors is that by inhibiting BER, these agents can prevent repair that occurs after cytotoxic chemotherapy that causes SSBs, and also they can work by creating synthetic lethality in cells with underlying HR defects. PARP inhibitors compete with NAD+ at the enzyme’s active site. Because this site is present in other enzymes, PARP inhibitors might act in a nonspecific manner. PARP must be inhibited by at least 90% to obstruct DNA repair. [86] All PARP inhibitors are thought to inhibit both PARP 1 and PARP 2.
In 1971 nicotinamide was found to be a weak inhibitor of PARP. [87–89] The first generation of inhibitors included nicotinamide analogues. The first widely tested agent, 3 aminobenzamide developed in 1980 [22], was not as selective and 1000 times less potent compared with newer inhibitors. The second generation, including PD128763, NU1025, was 50 times more potent than 3-aminobenzamide. Current PARP inhibitors in development are the third generation PARP inhibitors and have greater potency and specificity for PARP. See Table 1. These inhibitors are primarily based on benzamide or purine structures. The specificity may allow for less off target effect of PARP inhibitors during treatment and less toxicities. [4]
Table 1.
Third Generation PARP Inhibitors
| PARP inhibitor |
Company | Clinical development |
Indications | Route of admin |
|---|---|---|---|---|
| AG014699 | Pfizer | Phase II | Melanoma, Breast | IV |
| Veliparib (ABT 888) | Abbott | Phase II | Melanoma, Breast, Glioblastoma, Ovarian | PO |
| Olaparib (AZ 2281, KU59436) | Astra Zeneca | Phase II | Breast, Ovarian, Melanoma | PO |
| Iniparib (BSI 201) | BiPar Sciences | Phase III | Breast, NSCLC | IV |
| BSI 401 | BiPar Sciences | Preclinical | PO | |
| MK4827 | Merck | Phase I | BRCA ovarian | PO |
| CEP 9722 | Cephalon | Phase I | PO | |
| BMN-673 | LEAD Parma | Preclinical |
PARP inhibitors in combination with cytotoxic therapy
DNA methylating agents, including dacarbazine and temozolomide, were discovered to activate PARP 1. The methylating agents caused SSB that required BER. PARP 1 elicited resistance to methylating agents. [90] However, if PARP inhibitors are utilized to disable BER, the SSBs caused by the methylating agents could not be repaired. Subsequently these SSB lead to DSBs. If HR is overwhelmed by the addition of SSB, cell death occurs.
Loss of mismatch repair (MMR) also caused cellular resistance to temozolomide. In wild type cells, MMR would either correct errors in replication or cause replication arrest or cell death; in MMR-deficient cells, survival prevails with abnormal DNA. [91] MMR-deficient cells have poor response to temozolomide. Defects in MMR are associated with colon and ovarian cancers. 3 aminobenzamide augmented the efficacy of temozolomide in MMR deficient and MMR proficient cells. [92–93] In a later experiment, AG14361, another PARP inhibitor, enhanced the effect of temozolomide in MMR-deficient cells more than in MMR-proficient cells, eliminating the resistance incurred by the MMR-deficient state. [94] Only tumor cells are deficient in MMR, providing selective killing of tumor cells by the combination of PARP inhibitors with methylating agents. [23, 90] In an orthotopic rat glioma model the combination of veliparib in combination with temozolomide significantly slowed tumor progression; whereas, temozolomide as single agent had no significant effect. [95]
The cytotoxicity of camptothecins, topoisomerase I inhibitors, is also enhanced by PARP inhibitors. Topoisomerase I cleaves, unwinds and decreases torsional strain of DNA. Topoisomerase I inhibitors promote DNA breakage. Studies in hamster ovary cells show that topoisomerase I inhibitors have a stronger effect on cell killing in BER defective cells compared with BER competent cells. However, when AG 14361 is added to topoisomerase inhibitors, there is a more significant decrease on the LC 50 in the BER competent cells. The PARP inhibitor seems to overcome the resistance to topoisomerase inhibition in BER competent cells. Resistance to camptothecins due to XRCC1 overexpression can be reversed by PARP inhibitors since the PARP inhibitor interferes with the attraction of XRCC1 to the break site. In vivo, PARP inhibitors improve irinotecan’s effect on human colon mouse xenografts. [96]
In a mouse BRCA mutated breast xenograft, veliparib enhanced the activity of cisplatin and carboplatin. [95] Older studies, too, showed the effect of PARP inhibitors on platinum agents. In 1993, nicotinamide in combination with cisplatin increased the survival of a cisplatin-resistant ovarian cancer xenograft model. [97] CEP-6800 in a xenograft of non-small cell lung cancer showed enhancement of the cytotoxic effect of cisplatin. [98] In addition the alkylating agent cyclophosphamide is potentiated by veliparib. [95] [90]
PARP inhibitors in combination with ionizing radiation
PARP inhibitors potentiate ionizing radiation, via the inhibition of BER, as well as possibly through inhibition of NFκB and other inflammatory proteins and altered regulation of cellular metabolism through AMP/ATP. PARP inhibitors preferentially sensitize cells in S-phase. In cells that have defective PARP ability and are exposed to radiation, there is accumulation of DSBs, revealing the conversion of SSBs to DSBs resulting from the collapse of replication forks. PARP inhibitors increase the sensitivity of cells in growth arrest that are otherwise resistant to radiation. One experiment showed that the latent cells after XRT improved the growth inhibition by 73% when exposed to AG 14361. [99] In addition to the accepted role of inhibiting SSBs, PARP inhibitors also inhibit DSBs. DSBs activate PARP more potently than SSBs. Whereas 2 Zn fingers are required to attach PARP to a SSB, only one Zn finger is required for a DSB to attach to PARP 1. Nu 1025 inhibited the repair of DSBs after radiation, by inhibiting NHEJ. DNA-PK, a protein active in NHEJ, may be stimulated by PARP 1. PARP 1 inhibition caused decrease in DNA-PK activity. Recent studies show synergism between PARP inhibition and DNA-PK inhibition when cells exposed to both were irradiated. When the NHEJ pathway is defective, PARP 1 is recruited for DSB repair. Cells with NHEJ defectiveness that are exposed to radiation, therefore, have heightened cell killing with PARP inhibitors. PARP inhibitors might also enhance radiation by impairment of NF-κB. [33, 100]
PARP inhibitors also augmented the effect of irradiation in vivo. in a mouse colon cancer xenograft, the addition of veliparib to irradiation extended survival from 23 days with radiation alone to 36 days with the combination. There was even one mouse that had a CR. [95]
PARP inhibitors as single agent
PARP inhibition effectiveness as a single agent was first described in 2005. PARP inhibitors killed BRCA 2 deficient cells at doses that were nontoxic to normal cells in vitro and in xenograft models. [46, 101] BRCA 2 deficient cells were found to be 90 times more sensitive to PARP inhibition than wild type cells. [102] PARP inhibition in BRCA defective cells leads to NHEJ or SSA, which cause cell arrest in G2 or M phases of the cell cycle and then apoptosis. PARP inhibition is 3 times more potent than cisplatin cytotoxicity in BRCA deficient cells. The PARP inhibitor Ku0058684 inhibited tumor formation in mice injected with a BRCA 2 deficient cell line, but not in mice injected with wild type cells. [101] P53 (17p13.1), the tumor suppressor gene that is thought to work downstream of PARP 1 after DNA damage, does not interfere in the cell killing by PARP inhibitors, nor does p53 mutation interfere with the effect of PARP inhibitors. [46, 103] Tankyrase I inhibitor causes chromosomal instability in BRCA1 and BRCA 2 cells, indicating that even non traditional PARP inhibitors could be beneficial in tumor killing. [104]
CLINICAL DEVELOPMENT OF PARP INHIBITORS
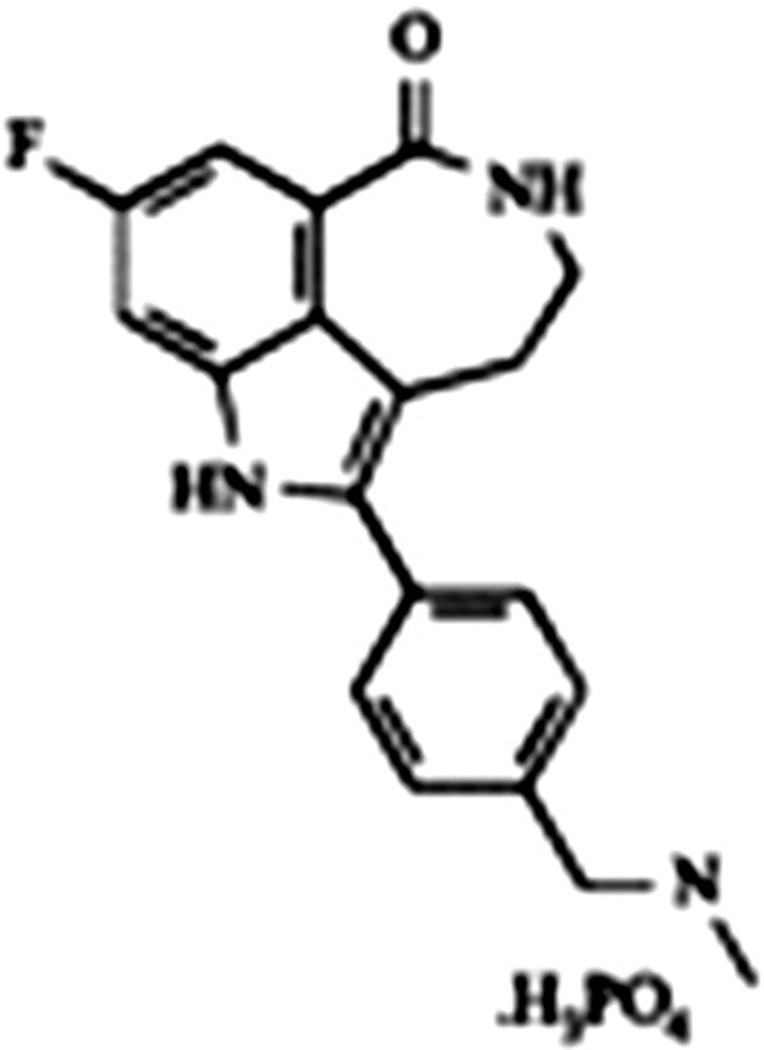
AG 014699 (PF01367338) Pfizer
AG 014699 (the phosphate salt of AG 14361) was the first agent studied clinically in 2003 (Fig. 9). Recently at EORTC, Ihnen reported preclinical data of AGO 14699 in ovarian cancer cell lines. The investigators evaluated 39 ovarian cancer cell lines with various molecular expressions either with single agent AGO14699 or in combination with carboplatin, doxorubicin, gemcitabine, paclitaxel or topotecan. They used combination index/isobologram analysis for multiple drug effect analysis. Concentration dependent efficacy was noted across the different cell lines to different degrees. The greatest impact appears to be in combination with carboplatin, topotecan and doxorubicin. The BRCA status of these cell lines was not reported. The study does suggest activity of AG014699 in ovarian cancer and not confined to BRCA deficient tumor or high grade serous cancer. [106]
Figure 9. AG 014699 (PF01367338) Pfizer.
(8-fluoro-2-(4-methylaminomethylphenyl)-1,3,4,5-tetrahydroazepino[5,4,3-cd]indol-6-one). [105]
Plummer conducted the initial phase I report with temozolomide in solid tumors and a subsequent phase II study was reported in melanoma patients. Overall, there was modest activity with significant myelosuppression. AG014699 was combined with temozolomide and both agents were given for 5 days in 28 day cycles. Temozolomide (TMZ) was started at ½ standard dose (100mg/m2) and AG014699 was escalated to PARP inhibitory dose (PID) as evaluated from PBMCs. PID was defined as > 50% decrease in PARP activity 24 hours after dosing. In the 17 patients in this first part of the study, no dose limiting toxicity was seen. The PID was determined to be 12 mg/m2. At this dose there was 74–97% inhibition of PBMC PARP. The mean terminal half life was 7.4–11.7 hours. There was linear pharmacokinetics. The kidneys were not a major route of excretion. AG014699 did not affect the PK for TMZ, nor did TMZ affect the PK of AG014699. The serum concentration varied 20 fold between patients and did not correlate with concentration in the biopsies. The PARP inhibition in the tumor correlated with dose. The drug remained in all tumor biopsies at 5 hours after administration. PARP was inhibited for over 24 hours so the cells were affected by the PARP inhibitor through the time they were exposed to TMZ. The PARP in the PBMCs recovered at least 50% function by 72 hours after dosing. Four patients were discovered to be homozygous for CYP2D6 G186A allele, known as CYP2D6 *4, which is expected to decrease the metabolism of AG 014699. The AUC was similar for patients with the CYP mutation compared with wild type. The toxicities in the part 1 portion of this phase I study were mild. There was 1 case of grade 3 toxicities for fatigue, infection, hypophosphatemia, and lymphopenia. Myelosuppression was the DLT for the highest dose level tested of 18mg/m2 in combination with standard dose TMZ. There was a partial response in a melanoma and a GIST patient, lasting more than 9 and 15 months. Seven patients had stable disease at least 6 months. Four of the patients with stable disease had melanoma, one had prostate cancer, one had pancreatic cancer, and one had leiomyosarcoma. [107]
In part 2 of the phase I study, the AG014699 dose was given at the PID of 12mg/m2 and TMZ was escalated to either MTD or standard dose of 200 mg/m2 in patients with metastatic melanoma. Again no DLT was experienced in the15 patients. Greater than 90% PARP inhibition was noted in PBMC. One patient with melanoma with CYP mutation had a CR and another with melanoma and the CYP mutation had a PR. A partial response was seen in a desmoid tumor patient previously treated. Seven additional patients had prolonged disease stabilization (>6 months), 4 with melanoma and one each with prostate cancer, pancreatic cancer, and leiomyosarcoma. [107]
The phase II study evaluated the efficacy of AG014699 at 12mg/m2 plus TMZ at 200 mg/m2 in 40 chemotherapy-naïve patients with advanced multiple melanoma. Myelosuppression was more significant in the phase II trial than seen in the phase I trial. There were 12% grade 4 thrombocytopenia, 15% neutropenia and one death from febrile neutropenia. Twelve patients required a dose reduction of TMZ to 150 mg/m2 and one patient required a reduction to 100mg/m2. Fatigue and nausea also occurred. There were 4 partial responses, 4 prolonged stable diseases, and 10 patients were too early to evaluate at the time of the report. [108]
There is an ongoing study evaluating AG 014699 in combination with various agents, including carboplatin, carboplatin and paclitaxel, cisplatin and premetrexate and epirubicin and cyclophosphamide. [109]
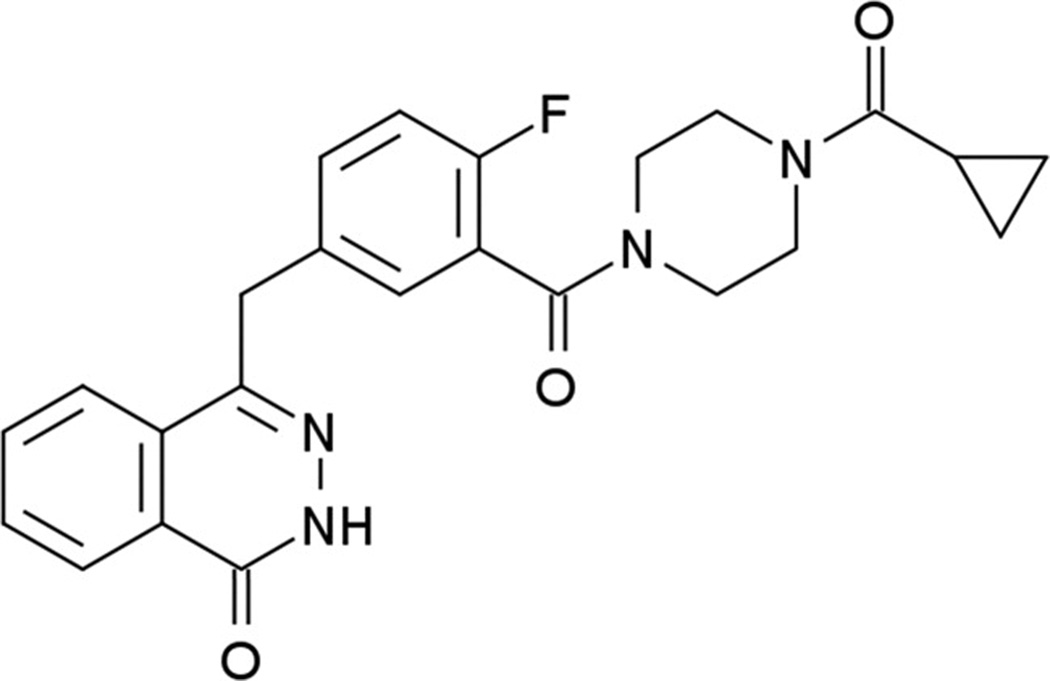
Olaparib (AZD 2281, KU-0059436) AstraZenieca
Olaparib is an oral PARP inhibitor with IC50 of 4.9 nM for PARP-1. It has been extensively evaluated in BRCA tumors (Fig. 10). It is the first PARP inhibitor to show activity in BRCA-related ovarian and breast cancers. It is been tested in combination with DNA damaging agents, such as topotecan, doxorubicin, carboplatin, carboplatin and paclitaxel, irinotecan, dacarbazine, and gemcitabine and cisplatin as well as with antiangiogenesis agentsand as a single agent.
Figure 10. Olaparib (AZD 2281, KU-0059436) AstraZeneca.
4-[3-(4-Cyclopropanecarbonylpiperazine-1-carbonyl)-4-fluorobenzyl]-2H-phthalazin-1-one C24H23FN4O3 [110]
One of the concerns regarding the addition of PARP inhibitors to chemotherapy had been the potential of enhancing toxicity. This is suggested with the addition of olaparib to gemcitabine and cisplatin. In this phase I trial, olaparib was given days 1–4, cisplatin on day 3 and gemcitabine on days 3 and 10 every 21 days. Five out of six patients experienced grade 3 or 4 thrombocytopenia. After de-escalation to dose level -1 (cisplatin 50 mg/m2 and gemcitabine of 400 mg/m2) and still experiencing significant myelosuppression in patients, the schedule was amended to only day 1 of olaparib. On this schedule two out of six patients still experienced grade 3 or 4 thrombocytopenia. Two PRs were reported in one pancreatic cancer and one NSCLC patient. The MTD was determined to be 100 mg BID of olaparib on day 1, cisplatin 60 mg/m2 on day 1 and gemcitabine of 500 mg/m2 on day 1 and 8 on a 21 day cycle. [111]
BRCA-related Ovarian Cancer and Olaparib
A phase I study of single agent olaparib, reported by Fong, enrolled 60 patients with solid tumors, including 22 BRCA mutation patients (and another patient who refused testing but had a strong family history). This study supported the synthetic lethality concept. Patients were treated at escalating doses and duration. Doses of 10 mg qd 2/3 weeks to 600 mg BID continuously were evaluated. The initial cohort were not restricted to BRCA deficient patients but was enriched for this population. In the expansion cohort, patients had to have BRCA mutation to enroll and were treated at 200 mg BID continuously. All of the DLTs were reversible. They included mood alterations and fatigue in 1 of 8 patients receiving 400 mg BID and recurred when that patient was treated with 200 mg BID. One patient out of 5 at the 600 mg BID dose level experience grade 4 thrombocytopenia with single agent olaparib. Lastly, grade 3 somnolence was seen in 1 patient on 600mg BID. The MTD was determined to be 400mg BID. Other adverse events were nausea, vomiting, diarrhea, dyspepsia, dysgeusia, stomatitis, altered taste, anorexia, dizziness, and anemia. There were no increased adverse effects in BRCA carriers compared to non BRCA carriers. Eight PRs by RECIST were seen out of the 15 patients with BRCA mutation-related advanced ovarian cancer group.All of the responses in ovarian cancer were seen in BRCA mutated tumors. One of the 3 patients with BRCA 2 breast cancer, who had progressed while receiving anthacyclines, had a CR for over a year after receiving olaparib. Another breast cancer patient had shrinkage of multiple subcentimeter brain lesions that had not been treated with radiation or steroids. [2]
The pharmacokinetics of olaparib was measured. The concentration was linear. The peak concentration was reached in 1–3 hours. The half life was 5–7 hours. Pharmacodynamics was also evaluated. PARP inhibition in PBMCs, hair follicles, and tumor were all measured. In PBMCs PARP was inhibited by 90% in patients treated with ≥ 60 mg BID. Immunoblots of tumor showed PAR inhibition also. Hair follicles had an increase in γH2AX, measuring DSBs, at all dose levels six hours after dosing. γH2AX remained elevated through the entire cycle in doses ≥ 200 mg BID. [2]
Fong reported in 2010 a follow-up of the ovarian cancer patients on this study. A total of 50 ovarian cancer patients with BRCA mutations were enrolled. Thirty-nine were treated at the expansion cohort dose of 200 mg BID. The rest were treated at various doses from 40 mg q d 2/3 weeks to 600 mg BID continuously. Twenty patients (40%) had CR or PR by RECIST and additional 3 patients had SD > 4 months (clinical benefit rate of 46%). The median duration of response was 28 months. Seventeen patients were on treatment for greater than 6 months including a patient who had received the drug for over 2 years. The most common drug related toxicities were mild GI symptoms and fatigue. A post ad hoc analysis of the study showed a statistically significant relationship of response to olaparib to the period of platinum-free interval. The response in platinum sensitive, resistant and refractory population as determined by RECIST or GCIG was 61%, 42% and 15%, respectively. There were no responses seen by RECIST in the refractory population vs. 46% and 33% in the platinum sensitive and resistant populations, respectively. No differences were noted in the duration of response or time to progression between the three platinum response groups. [112] These findings suggest that resistance to platinum decreases sensitivity to this PARP inhibitor. This correlation in tumor response to both platinum and PARP inhibitors can be explained by resistant disease being resistant to multiple therapies. Alternatively, the explanation might be a restoration of the HR through a second mutation to the BRCA gene, restoring it to wildtype, and thus decreasing the sensitivity that the mutated state imparts to platinum and PARP inhibitors. (see section Acquired Resistance to PARP Inhibitors). Though decreased, there was significant response to olaparib even in tumors that were resistant or refractory to platinum.
In a phase II international trial in women with confirmed BRCA 1 and BRCA 2 mutation with recurrent and incurable ovarian cancer (ICEBERG 2), olaparib was given daily in 28 day cycle to 33 patients at 400 mg BID and to a subsequent cohort of 24 patients at 100mg BID, a dose that had been shown previously to inhibit PARP. The RECIST ORR for the patients receiving the 400mg dose was 33% and for the patients receiving the 100mg dose was 12.5%. Two patients in the 400 mg cohort had complete responses and none in the lower dose group. The clinical benefit rate (ORR and/or decrease in CA125 by ≥ 50% and SD > 4 months) was 57.6% for the 400mg patients and 16.7% for the 100mg patients. Toxicity was mild with only grade 3 nausea in 7% and leucopenia in 5%. This study would imply that there might be other mechanisms causing tumor response by PARP inhibitors since the dose that was shown to inhibit PARP was not as effective as a higher dose. [113]
At 35th ESMO this year, Kaye presented a randomized phase II trial of two doses of olaparib (200 mg and 400 mg BID) vs. pegylated liposomal doxorubicin (PLD) in patients with BRCA mutation ovarian cancer who had recurred within 12 months of platinum-based chemotherapy. Ninety-seven patients were accrued. The PFS were 6.5 vs. 8.8 vs. 7.1 months for 200 mg, 400 mg and PDL arm, respectively. Olaparib did not reach the primary objective of improving PFS partly due to a better PFS seen in the PLD arm than expected. PRs were seen in 8/32 in the 200 mg, 10/31 in the 400 mg, and 6/33 patients in the PLD arms. No difference in OS is seen at this time. Twice as many grade 3+ toxicities were seen in the PLD arm. Though reported as a negative study, this trial still shows consistent response and decreased toxicitywith the use of single agent olaparib in BRCA mutation ovarian cancer patients. [114]
High Grade Serous Ovarian Cancer and Olaparib
Gelmon reported response of high grade serous ovarian cancer (HGSOC) in 2010 ASCO. In a multicenter study in Canada, patients with unknown BRCA mutation status with HGSOC were given olaparib. Biopsies were taken prior to starting treatment, after 2 cycles and at the time of progression. Fifty-five patients were enrolled and treated with continuous dosing of olaparib 400 mg BID. All patients agreed to BRCA testing prior to enrolling on study. There were 14 PRs out of 53 patients (26.4 %) in the unknown group. After BRCA testing 7 patients in the unknown group had BRCA mutations. In the 46 patients with negative BRCA mutation a response rate of 23.9 % (11/46) was reported. There were 3/7 responses in the BRCA mutated group. Progression free survival was 219 days in the ovarian group (17 BRCA mutations and 46 without). The toxicities profiles were mild. Grade 3 toxicities were reported for fatigue, anemia, diarrhea, in more than one patient. [115]
BRCA-related Breast Cancer and Olaprib
A proof of concept study was reported by Tutt with single agent olaparib in patients with BRCA1 or BRCA 2 mutation breast cancer patients. Patients all had at least one prior line of treatment for their breast cancer. Fifty-four patients were enrolled. Patients were randomized to either 100 mg BID or 400 mg BID of olaparib. ORR were 22% vs 41% and PFS were 3.8 vs. 5.7 months in the 100 mg vs. 400 mg cohorts, respectively. . Median duration of response were similar in the two cohorts, ~140 days. During an interim analysis, a difference of 2.5 months was noted in the median time to dose withdraw or dose escalation between the 100 mg and 400 mg arm. Patients in the 100 mg cohort were offered the option of having their doses escalated to the 400 mg level. There were more grade 3+ nausea, vomiting and fatigue associated with the higher dose cohort. This study again confirms the activity seen in the phase I trial of olaparib as a single agent in the treatment of BRCA mutation tumors. [116]
Olaparib and TNBC
There was also a phase I/II combination study with paclitaxel in TNBC. Nineteen patients were treated on the study. Fifteen of those had had prior taxane therapy. They were given 200mg daily of Olaparib orally along with Paclitaxel 90mg/m2 IV weekly for 3 out of 4 weeks. Thirty-seven percent of patients had confirmed PRs. Neutropenia was the primary adverse event even with GCSF support. Given the prior taxane exposure there is suggestion that the use of olaparib may overcome resistance to taxane. [117]
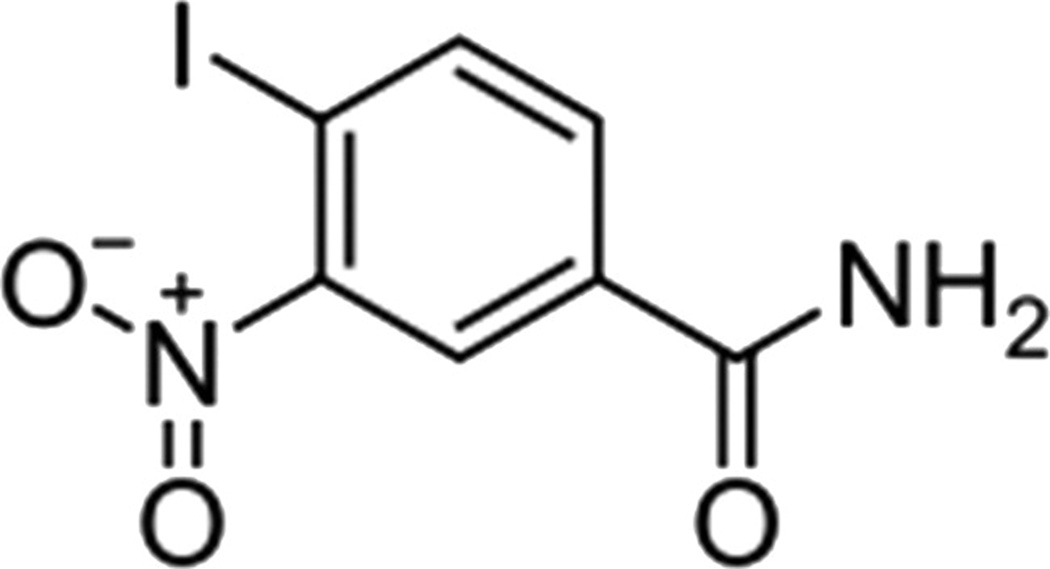
Iniparib (BSI 201, NSC-746045; IND-71677) sanofi-aventis
http://en.wikipedia.org/wiki/File:Iniparib.svg
Iniparib, also known as BSI 201, 4-iodo-3-nitrobenzamide, is an irreversible inhibitor of PARP1 (Fig. 11). It is a prodrug with 4-minute half life. Data on its active metabolite is unknown at this time though a nitroso metabolite might be one of the active metabolites. Iniparib is given intravenously twice a week. It is the first PARP inhibitor to show survival advantage with TNBC and has entered phase III testing.
Figure 11.
Iniparib (BSI 201, NSC-746045; IND-71677) sanofi-aventis
The phase I study included 23 patients with solid tumors. The patients were escalated through 7 dose levels up to 8 mg/kg without reaching MTD. The 1.4 mg/kg dose resulted in a Cmax ≥ 400 ng/ml. The concentration that brought about efficacy in preclinical models was 20–30ng/ml, so achievable levels were well over the preclinical efficacious levels. The 2.8 mg/kg dose caused PARP inhibition in PBMCs by more than 50% with the first dose. Subsequent dosing increased the amount of PARP inhibition to more than 80%. Six of the 23 heavily pretreated patients had stable disease for at least 2 months (up to over 9 months in one patient). The adverse events were mostly gastrointestinal. DLT was not seen. [119]
In another study patients with solid tumors were assigned to one of four combinations of iniparib with a cytotoxic agent, topotecan, gemcitabine, temozolomide, or carboplatin with taxol. Assignment to each of the 4 regimens was not randomized, but based on physician preference. Iniparib was given on days 1 and 4 of each week. The dose of iniparib was escalated to 8mg/kg. 55 patients have been treated on the study at the time of the report. All regimens were reported to be well tolerated. There were no SAEs attributed to the drug. A patient with ovarian cancer achieved a CR that lasted at least 6 months. Five patients, with breast cancer, uterine cancer, renal cancer, and a sarcoma, achieved PRs. Nineteen patients had SD for ≥ 2 months. [120]
TNBC and iniparib
In a phase II study conducted by O'Shaughnessy focusing on TNBC randomized patients to gemcitabine with carboplatin vs. gemcitabine with carboplatin plus iniparib. The dosing schedule was gemcitabine 1000mg/m2 and carboplatin at AUC=2 days 1 and 8 and iniparib 5.6 mg/kg days 1, 4,8,11 every 21days. One hundred and sixteen patients were treated on the study. Updated results were reported at the 35th ESMO this year. The clinical benefit rate (CBR) was defined as CR+PR+SD ≥ 6 months. CBR of 55.7% vs. 33.9% (p=0.015), overall response rate (ORR) of 52.5% vs. 32.5% (p=0.023), median PFS 5.9months vs. 3.6 months (p=0.012), and OS 12.3 months vs. 7.7 months (p = 0.014) in the iniparib combination arm vs. chemotherapy arm, respectively. The p values were not adjusted for several interim analyses. Adverse events were similar in the 2 groups. [121]The regimen of gemcitabine and carboplatin was not a standard regimen used in treatment of TNBC prior to this report. The carboplatin dose was lower than standard dosing. The use of an uncommon regimen may lead to a less effective control arm and magnify the advantage of PARP inhibitor; however, the difference in OS does support the role of PARP inhibitor with chemotherapy. A phase III study evaluating this combination as first to third line treatment for metastatic TNBC has completed accrual. Food and Drug Administration (FDA) recently approved an Expanded Access Protocol (EAP) for iniparib, in metastatic triple negative breast cancer (mTNBC).
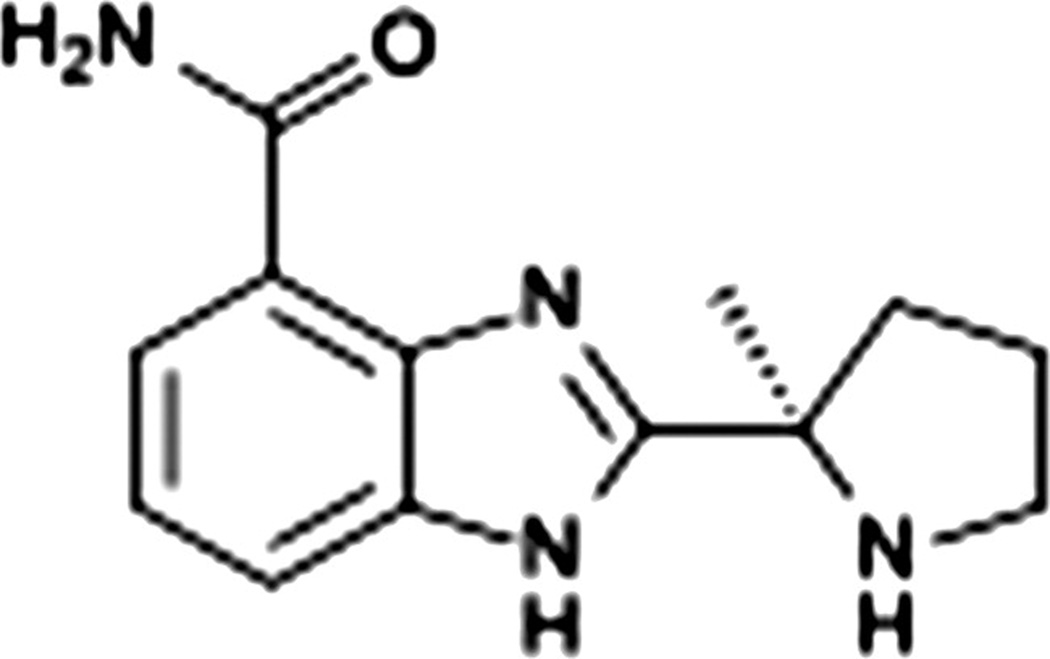
Veliparib (ABT888) Abbott Laboratories
Preclinically veliparib was shown to be a potent inhibitor of PARP. It had good bioavailability (Fig. 12). It crossed the blood brain barrier as seen with the PKs done in rat brains. The addition of temozolomide did not change the PKs of veliparib. Veliparib potentiated temozolomide, platinum agents, cyclophosphamide, and radiation in syngeneic and xenograft tumor models. [95]
Figure 12. Veliparib (ABT888).
Abbott Laboratories. 2-((R)-2-Methylpyrrolidin-2-yl)-1H-benzimidazole-4-carboxamide. C13H16N4O [122]
http://upload.wikimedia.org/wikipedia/commons/d/da/Veliparib_skeletal.svg
The first phase 0 study under FDA’s new Exploratory IND was performed by Kummar with veliparib, ABT 888. Phase 0 studies are new mechanisms for expediting drug development. Veliparib was chosen because it has a wide therapeutic index and a validated pharmacodynamic assay. The pharmacokinetics and pharmacodynamics are evaluated over a short time period after single dose of veliparib that is thought to be nontoxic. Three dose levels were tested, 10, 25, 50mg, each with three patients. The 10 mg starting dose was based on 1/50 of the no observed adverse effect level (NOAEL) in the most sensitive species, dog. The study showed that peak plasma levels occurred between 30 minutes and 1.5 hours after dosing. The target concentration based on PARP inhibition concentration in animals was exceeded even in the patients receiving the lowest dose. The drug was primarily renally secreted.
PAR levels were assessed in tumors and PBMCs. Statistically significant decreases in PAR were defined to be 55% in PBMCs and 95% in tumors. Statistically significant reduction in PAR for both tumor and PBMC was seen at 25 and 50 mg at 3–6 hours. The higher doses were, therefore, not evaluated. However, three additional subjects at the 50 mg dose underwent tumor biopsy around 24 hours after the administration of veliparib. PAR levels in the tumor at 24 hours were still at least 49% below baseline levels, but only in 1 of the 3 patients was the reduction significant.
One patient at the 50 mg dose did not experience a decrease in PAR. No abnormalities in PARP, nor poly(ADP-ribose) glycohydrolase (PARG), the enzyme responsible for degradation of PAR, were found to explain this phenomenon. This finding raises the possibility of evaluating for resistance to PARP inhibitors by ex vivo screening of PBMCs. [123]
Veliparib in combination with topotecan also showed significant myelosuppression. The original schedule was topotean days -8 and 2–5 at 1.2 mg/m2 and veliparib 10 mg BID days 1–7. The schedule was changed to days 1–5 of topotecan when 0.9 mg/m2 of topotecan was not tolerated. The final schedule found to be tolerable was topotecan 0.6 mg/m2 days 1–5 and veliparib 10 mg BID on day 1 only. Six out of ten patients at the higher dose levels had significant increases in γH2AX. γH2AX was not seen with the lower doses of topotecan alone. There was a correlation of γH2AX upregulation with PARP inhibition. [124](Personal communication with Dr. Kummar)
There are multiple ongoing phase I and II trials with Veliparib as single agent and in various chemotherapy combinations.
Ovarian Cancer and veliparib
A phase I study of veliparib in combination with metronomic cyclophosphamide in patients with refractory solid tumors and lymphomas registered 18 patients in 6 dose levels. Adverse events have been grade 3 and 4 lymphopenia in 3 patients and grade 2 neutropenia in 2 patients. PBMC PAR level reductions of ≥ 50% were seen in 16 of 18 patients. Two patients exhibited >95% reduction in PAR within the tumors. [125] Two patients with BRCA 2 ovarian cancer achieved PR. Both patients achieved PRs in the second dose level consisting of oral cyclophosphamide at 50 mg q d days 1–21 and veliparib 30 mg q day × 7 days on a 21 day cycle. A phase II randomized trial evaluating the role of veliparib in combination with oral cyclophosphamide in ovarian cancer patients with BRCA mutation or high grade serous ovarian cancer will be activated in the near future.
Breast Cancer and Veliparib
Kummar reported a PR in the phase I trial of veliparib with oral cyclophosphamide in an ER + with BRCA 2 mutation breast cancer patient. The patient was treated with 50 mg q d of oral cyclophosphamide and veliparib 60 mg q d oral continuous dosing. Patient was previously treated with doxorubicin/cyclophosphamide, letrozole, fulvestrant, gemcitabine, traztuzemab and bevacizumab. A phase II randomized trial evaluating metronomic cyclophosphamide with or without veliparib in TNBC will be starting soon.
Veliparib in combination with temozolomide was evaluated in metastatic breast cancer. Forty-one patients were treated with 40 mg PO BID veliparib days 1–7 and temozolomide 150 mg/m2 days 1–5 every 28 days. The initial schedule was revised due to higher than expected grade 4 thrombocytopenia. Veliparib was reduced to 30 mg PO BID day 1–7. Fifteen patients had TNBC. One CR and 2 PRs were reported in 24 evaluable patients. [126]
MK 4827 Merck
MK 4827 is an oral PARP 1 and 2 inhibitor with IC50 of 3.8 nM for PARP 1. Preclincial data showed single agent anti-tumor activity against BRCA mutant cell lines in culture and xenograft models. Also, MK4827 showed activity in combination with DNA damaging agents in cell cultures and xenograft models. It is currently being tested in phase 1 development as a single agent, in advanced solid tumors, ovarian tumors and prostate tumors, and as combination therapy in patients with advanced solid tumors in combination with carboplatin,carboplatin with paclitaxel, and carboplatin with liposomal doxorubicin. [127]
MK4827 in Ovarian Cancer
At 2010 ASCO, Sandhu presented the phase I trial of MK4827 as a single agent enriched with BRCA 1 or 2 mutation patients. An expansion cohort of high grade serous ovarian cancer patients were added at the MTD. Seven dose levels ranging from 30–400 mg of MK4827 were given daily for 21 days out of 28 days initially and then on a continuous basis. Patients with prior PARP inhibitor exposure were excluded. The preliminary PK showed a half-life of 40 hours. PARP inhibition was shown in PBMC of patients treated with doses higher than 110 mg. The MTD was determined to be 300 mg qd. The DLT was thrombocytopenia. Eleven patients had BRCA mutations. Nineteen ovarian cancer patients were treated in the phase I trial. Six ovarian cancer patients achieved PR; 5 of the six patients had BRCA mutations. Responses were seen in all dose levels. The expansion cohort in high grade serous ovarian cancer is ongoing. [128]
CEP 9722 and CEP 8983 Cephalon
CEP 9722 is the prodrug to CEP 8983. Preclinical evaluation of CEP 8983 in chemoresistant glioblastoma (RG2), rhabdomyosarcoma (RH18), neuroblastoma (NB1691) and colon carcinoma (HT29) showed the agent sensitized these cells to temozolomide and camptothecins. In addition, using a granulocyte-macrophage colony-forming assay, CEP-8983 did not potentiate myelotoxicity in the presence of temozolomide or topotecan. These cytotoxic agents have been associated with significant myelosuppression when combined with other PARP inhibitors. In animal studies, CEP 8983 increased the sensitivity of chemoresistant tumor cells to temozolomide and irinotecan without exacerbating myelotoxicity. CEP 9722 is being studied in a phase 1 study in solid tumors with and without combination with temozolomide. [129]
BMN 673 Biomarin
LT 673 is an oral agent which the company reports to be the most potent of the PARP inhibitors to date. It is still in preclinical development, but shows promise in mouse xenograft models. [130]
BSI 401 sanofi aventis
BSI-401 is a derivative of 6-iodo-5-amino-1,2-benzopyrone, a noncovalently binding PARP-1 inhibitor. It is an oral PARP inhibitor that may be entering clinic in the near future. It has shown activity in orthotopic nude mouse models bearing pancreatic cells both as single agent and in combination with oxaloplatin. Also, the study reported BSI 401 protected the animals from oxaliplatin-induced neurotoxicity. [131]
ACQUIRED RESISTANCE to PARP INHIBITORS
As stated above, BRCA 2 mutated tumor cells are sensitive to platinum agents. With time, however, they become resistant to platinum chemotherapy. Resistance has been found to result from a secondary mutation in BRCA 2 that corrected the frameshift of the original mutation. This finding has been confirmed to occur in patients. [132] Multiple clinical disorders have been found to have the potential to revert inherited mutations to a normal state, including, Bloom’s syndrome, epidermolysis bullosa, severe combined immunodeficiency, tyrosinemia, Wiscott-Aldrich syndrome, and Fanconi anemia. The mechanisms found for reversion to wild type in these genetic disorders were secondary mutations that alter the reading frame to wild type, compensatory mutations, and crossovers.[5]
Both a DNA damaging agent, such as cisplatin, and the BRCA 2 mutated cells, that are defective in DNA repair, could increase the incidence of a secondary mutation. The mutational event that converts the BRCA 2 mutation status to wild type also converts the cells to being resistant not only to platinum agents, but also to PARP inhibitors due to restoration of the HR pathway. BRCA 2 mutated tumors that happen to be platinum resistant, but not on the basis of a secondary mutation converting the cells to wild type, would be expected to retain responsiveness to PARP inhibitors. For cells that have been converted to wild type on the basis of a second mutation, it is possible that resensitization to PARP inhibitors could occur if another part of the HR pathway was blocked. Because proteasome inhibitors inhibit FANCD2, 53BP1, phospho-ATM, NBS1, BRCA1, FANCD2, and RAD51, which are all necessary in HR, proteasome inhibitors might be another option to create “BRCAness”. [132–133]
Another mechanism of resistance to PARP inhibitors had been reported with AZ 2281. In a genetically engineered mouse model for BRCA 1 associated breast cancer, AZ2281 was administered over long term until resistance developed. The resistance was secondary to up-regulation of Abcb1a/b gene encoding for P-glycoprotein efflux pump. With the coadministration of tariquidar, P-glycoprotein inhibitor, the resistance was reversed. [134]
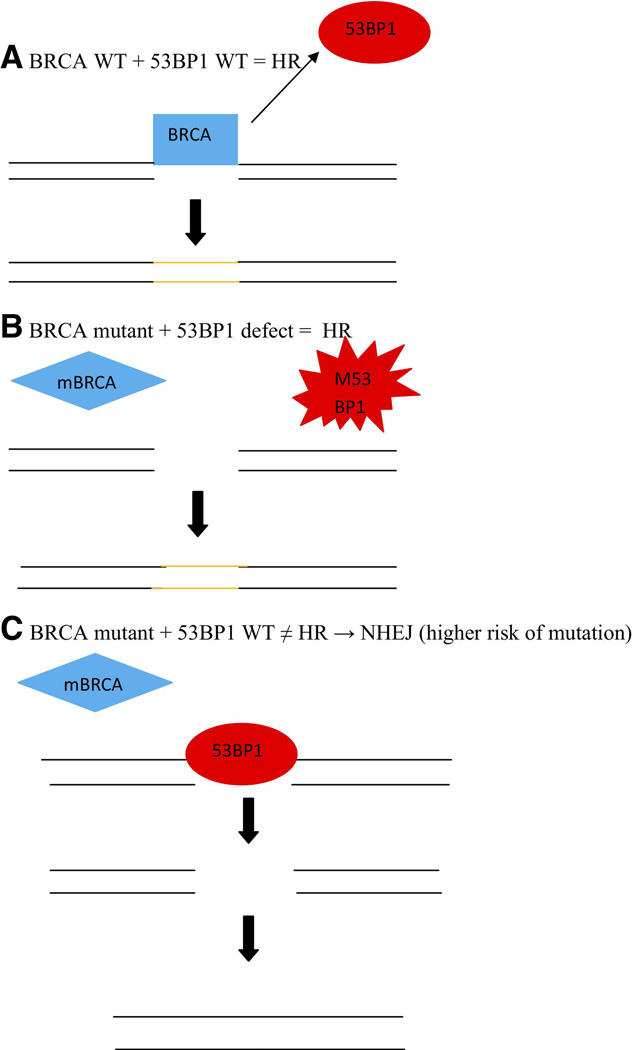
Adding another layer of complication into this system, it has recently been found that 53BP1 regulates the repair mechanism in BRCA deficient cells and, therefore, could play a role in PARP inhibition. In a cell with normal BRCA 1 function, after a DSB, BRCA 1 displaces 53BP1 and HR repair is completed. In the case of BRCA1 mutation and absence of 53BP1, downstream proteins of HR are still initiated and HR is still activated. It is only when BRCA 1 is mutated and 53BP1 is present that 53BP1 sticks to the site of the DSB preventing HR, but allowing for error prone NHEJ to takeover. In BRCA-deficient mice bearing mammary tumor, loss of 53BP1 reduced tumorgenesis. The presence of 53BP1 and BRCA 1 changes the balance between HR and NHEJ. This would suggest PARP inhibitors would be expected to have activity primarily in cells retaining normal 53BP1, with a greater dependence on HR to repair DSB. Absence of 53BP1 would confer PARP inhibitor resistance. In addition, an inhibitor of 53BP1 might decrease the tumor risk in carriers of BRCA mutations. However, this protein is also necessary for B cell immunoglobulin switching. In addition, ATM inhibitors might prevent BRCA mutated cells that have defective 53BP1 from maintaining HR capability and restore synthetic lethality in the presence of PARP inhibitors to cells that otherwise might be resistant to PARP inhibitors (Fig. 13). [135]
Figure 13. Effect of 53BP1 on HR.
Recently 6-thioguanine (6-TG) has been found to be active in cells resistant to PARP inhibitors. In a screen for drugs that selectively kill BRCA 2 – deficient cells, 6TG stood out. 6-TG induces DSBs that cannot be repaired by the BRCA mutated cells. 6-TG was as efficient as a PARP inhibitor in selectively killing BRCA 2 mutated tumor cells in a xenograft model. 6-TG was effective against BRCA 1 defective, PARP inhibitor resistant (PIR) tumors. The resistance was thought to be mediated through increased p-glycoprotein. 6 TG is not a substrate for p-glycoprotein so could overcome the PIR mechanism of increased p-glycoprotein. In BRCA 2 deficient tumor cells with PIR through reactivation of HR, the reactivated HR is not sufficient to repair the 6TG induced lesions to allow tumor cell survival. 6 TG forms both mismatch repair-dependent and independent HR lesions. 6-TG appears to be active in BRCA mutated cells that have acquired platinum and/or PARP inhibitor resistance [7]
Discussion regarding other possible mechanisms that might allow for pathways for killing of tumor in PARP inhibitor resistant situations have been discussed above, including triggering other pathways that obstruct HR, inhibiting p-glycoprotein efflux, and up-regulating 53BP1. Proteosome inhibitors, which would be expected to downregulate p-glycoprotein, also cause degradation of 53BP1, which would allow HR to proceed in DSB repair and, by that mechanism might not overcome resistance to PARP inhibition. [133]
FUTURE DIRECTIONS
PARP inhibitors are an exciting new class of agents that have shown efficacy, especially for BRCA-related ovarian and breast cancer. Also, activity has been seen in both TNBC and high grade serous ovarian cancer. There are many PARP inhibitors in development. They vary in their route of administration, toxicity profile, efficacy and resistance mechanism. Currently it is not clear how close the different PARP inhibitors behave clinically since few are studied in similar conditions. To date trials have varied with respect to the type of tumors eligible, as well as the combination with cytotoxic chemotherapy.
PARP inhibitors also work as single agents in tumors with certain DNA repair defects, using synthetic lethality. Multiple preclinical studies have shown activity of the different PARP inhibitors in BRCA 1 and BRCA 2 mutated tumors. Also, single agent activity has been shown clinically in BRCA mutated tumors with olaparib and MK4827. There is interest in exploring the activity of PARP inhibitors in cells with other DNA repair abnormalities in the HR pathway besides BRCA 1 or 2 germline mutation,. Some of the possible HR abnormalities include PTEN defects, Fanconi’s anemia protein defects, ATM abnormalities, RAD51 dysfunction, EMSY defects, and TNKS abnormalities. There is a search for biomarkers to identify tumors that are more likely to respond to PARP inhibitors, for example the abnormalities causing HR dysfunction and gene profiling, including microarrays that identify BRCAness. Genetic manipulation of the BRCA tumor suppressor gene might affect prevention of tumor formation and might also augment targeted therapy. Also screening for resistance to PARP inhibitors, for example by measuring PAR in PBMCs ex vivo or by genome analysis, might select for patients with more susceptibility to PARP inhibitor treatment. More investigation needs to be done regarding overcoming resistance.
The other area of significant research is the combination of PARP inhibitor with DNA damaging chemotherapy especially those that causes SSB. The most common chemotherapy combinations are with temozolomide, topotecan, irinotecan, and carboplatin. Also, radiation is another area of interest since it also depends on the BER for repair. With olaparib and veliparib, the myelosuppression seen with cisplatin/gemcitabine and topotecan, respectively, are significantly potentiated with the addition of the PARP inhibitors. It is not clear at this time if same dose of PARP inhibitor could be combined with different regimens of chemotherapy. With BSI 201, same dose of BSI 201 had been used with various chemotherapy. With veliparib this had not been the case. Also, aside from myelosuppression is it not clear if PARP inhibitors potentiate other toxicities. Data with BSI 401 suggest some toxicities may be improved with the addition of PARP inhibitors. This area would also benefit from additional research. An agent, like BSI 401, that will alleviate neuropathy and potentially increase cytotoxicity, would be a great addition to the oncologic therapeutic arena. This will need to be evaluated clinically.
Additional areas of research include other enzymes PARP inhibitors might affect by their action on the NAD+ site, which is present in other enzymes. In the olaparib single agent study, 100 mg was not as efficacious as the 400 mg dose. From the report 100 mg of olaparib inhibited PARP. This finding suggests that other mechanisms of action may be responsible for the difference in activity between the two dose levels. Also, PARP 1 leads to down regulation of mTORC and to autophagy as part of the cellular metabolic regulation. PARP inhibitors may cause decreased autophagy and overall cellular dysregulation. This nonspecific aspect of PARP inhibitors could be a road to untoward side effects not yet recognized.
As this class of agents moves into the (neo)adjuvant and prevention setting, caution should be raised regarding secondary malignancy. PARP repairs DNA mutations which occur daily. By inhibiting PARP long term, inability to repair a normal process may lead to malignancy similar to a BRCA carrier. The late effects of being treated with PARP inhibitors will only become known over time.
This is an exciting new class of agents that has many areas of development and potential. Over the next few years, current studies answering many of the above questions will mature leading to better understanding of the PARP inhibitors and additional use for these agents. It may also lead to FDA approval of one or more of these agents for use in the treatment of breast, ovarian and other cancers.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.O'Shaughnessy J. Efficacy of BSI-201, a poly (ADP-ribose) polymerase-1 (PARP1) inhibitor, in combination with gemcitabine/carboplatin (G/C) in patients with metastatic triple-negative breast cancer (TNBC): Results of a randomized phase II trial. J Clin Oncol. 2009;27(18s) ASCO abstr 3. [Google Scholar]
- 2.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009 Jul 9;361(2):123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 3.Hennessy BT, Timms KM, Carey MS, Gutin A, Meyer LA, Flake DD, 2nd, et al. Somatic mutations in BRCA1 and BRCA2 could expand the number of patients that benefit from poly (ADP ribose) polymerase inhibitors in ovarian cancer. J Clin Oncol. 2010 Aug 1;28(22):3570–3576. doi: 10.1200/JCO.2009.27.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010 Apr;10(4):293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirschhorn R. In vivo reversion to normal of inherited mutations in humans. J Med Genet. 2003 Oct;40(10):721–728. doi: 10.1136/jmg.40.10.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amin DN, Masocha W, Ngan'dwe K, Rottenberg M, Kristensson K. Suramin and minocycline treatment of experimental African trypanososmiasis at an early stage of parasite brain invasion. Acta Trop. 2008 Apr;106(1):72–74. doi: 10.1016/j.actatropica.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 7.Abbadie C, McManus OB, Sun SY, Bugianesi RM, Dai G, Haedo RJ, et al. Analgesic effects of a substituted N-triazole oxindole (TROX-1), a state-dependent, voltage-gated calcium channel 2 blocker. J Pharmacol Exp Ther. 2010 Aug;334(2):545–555. doi: 10.1124/jpet.110.166363. [DOI] [PubMed] [Google Scholar]
- 8.Issaeva N, Thomas HD, Djurenovic T, Jaspers JE, Stoimenov I, Kyle S, et al. 6-Thioguanine Selectively Kills BRCA2-Defective Tumors and Overcomes PARP Inhibitor Resistance. Cancer Res. 2010 Aug 1;70(15):6268–6276. doi: 10.1158/0008-5472.CAN-09-3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001 May 17;411(6835):366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 10.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993 Apr 22;362(6422):709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 11.Sargent DJ, Marsoni S, Monges G, Thibodeau SN, Labianca R, Hamilton SR, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010 Jul 10;28(20):3219–3226. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venkitaraman AR. A growing network of cancer-susceptibility genes. N Engl J Med. 2003 May 8;348(19):1917–1919. doi: 10.1056/NEJMcibr023150. [DOI] [PubMed] [Google Scholar]
- 13.Haber JE. DNA recombination: the replication connection. Trends Biochem Sci. 1999 Jul;24(7):271–275. doi: 10.1016/s0968-0004(99)01413-9. [DOI] [PubMed] [Google Scholar]
- 14.Bennardo N. Alternative-NHEJ Is a Mechanistically Distinct Pathway of Mammalian Chromosome Break Repair. PLoS Genet. 2008 doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002 Jul 26;297(5581):606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 16.Easton D, Ford D, Peto J. Inherited susceptibility to breast cancer. Cancer Surv. 1993;18:95–113. [PubMed] [Google Scholar]
- 17.Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol. 2008 Oct;9(10):759–769. doi: 10.1038/nrm2514. [DOI] [PubMed] [Google Scholar]
- 18.Taylor AM, Edwards MJ. Malignancy, DNA damage and chromosomal aberrations in ataxia telangiectasia. IARC Sci Publ. 1982 39;:119–126. [PubMed] [Google Scholar]
- 19.Wang LH, Pfister TD, Parchment RE, Kummar S, Rubinstein L, Evrard YA, et al. Monitoring drug-induced gammaH2AX as a pharmacodynamic biomarker in individual circulating tumor cells. Clin Cancer Res. 2010 Feb 1;16(3):1073–1084. doi: 10.1158/1078-0432.CCR-09-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schultz N, Lopez E, Saleh-Gohari N, Helleday T. Poly(ADP-ribose) polymerase (PARP-1) has a controlling role in homologous recombination. Nucleic Acids Res. 2003 Sep 1;31(17):4959–4964. doi: 10.1093/nar/gkg703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chambon P, Weill JD, Mandel P. Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem Biophys Res Commun. 1963 Apr 2;11:39–43. doi: 10.1016/0006-291x(63)90024-x. [DOI] [PubMed] [Google Scholar]
- 22.Durkacz BW, Omidiji O, Gray DA, Shall S. (ADP-ribose)n participates in DNA excision repair. Nature. 1980 Feb 7;283(5747):593–596. doi: 10.1038/283593a0. [DOI] [PubMed] [Google Scholar]
- 23.Zaremba T, Curtin NJ. PARP inhibitor development for systemic cancer targeting. Anticancer Agents Med Chem. 2007 Sep;7(5):515–523. doi: 10.2174/187152007781668715. [DOI] [PubMed] [Google Scholar]
- 24.Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci. 2008;13:3046–3082. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- 25.Menissier de Murcia J, Ricoul M, Tartier L, Niedergang C, Huber A, Dantzer F, et al. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 2003 May 1;22(9):2255–2263. doi: 10.1093/emboj/cdg206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Murcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci U S A. 1997 Jul 8;94(14):7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiang YJ, Hsiao SJ, Yver D, Cushman SW, Tessarollo L, Smith S, et al. Tankyrase 1 and tankyrase 2 are essential but redundant for mouse embryonic development. PLoS One. 2008;3(7):e2639. doi: 10.1371/journal.pone.0002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loseva O, Jemth AS, Bryant HE, Schuler H, Lehtio L, Karlberg T, et al. PARP-3 is a mono-ADP-ribosylase that activates PARP-1 in the absence of DNA. J Biol Chem. 2010 Mar 12;285(11):8054–8060. doi: 10.1074/jbc.M109.077834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farzaneh F, Zalin R, Brill D, Shall S. DNA strand breaks and ADP-ribosyl transferase activation during cell differentiation. Nature. 1982 Nov 25;300(5890):362–366. doi: 10.1038/300362a0. [DOI] [PubMed] [Google Scholar]
- 30.Hoyer-Hansen M, Jaattela M. AMP-activated protein kinase: a universal regulator of autophagy? Autophagy. 2007 Jul–Aug;3(4):381–383. doi: 10.4161/auto.4240. [DOI] [PubMed] [Google Scholar]
- 31.Simbulan-Rosenthal CM, Ly DH, Rosenthal DS, Konopka G, Luo R, Wang ZQ, et al. Misregulation of gene expression in primary fibroblasts lacking poly(ADP-ribose) polymerase. Proc Natl Acad Sci U S A. 2000 Oct 10;97(21):11274–11279. doi: 10.1073/pnas.200285797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frizzell KM, Gamble MJ, Berrocal JG, Zhang T, Krishnakumar R, Cen Y, et al. Global analysis of transcriptional regulation by poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase in MCF-7 human breast cancer cells. J Biol Chem. 2009 Dec 4;284(49):33926–33938. doi: 10.1074/jbc.M109.023879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang M, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34(21):6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haince JF, Kozlov S, Dawson VL, Dawson TM, Hendzel MJ, Lavin MF, et al. Ataxia telangiectasia mutated (ATM) signaling network is modulated by a novel poly(ADP-ribose)-dependent pathway in the early response to DNA-damaging agents. J Biol Chem. 2007 Jun 1;282(22):16441–16453. doi: 10.1074/jbc.M608406200. [DOI] [PubMed] [Google Scholar]
- 35.Haince JF, McDonald D, Rodrigue A, Dery U, Masson JY, Hendzel MJ, et al. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem. 2008 Jan 11;283(2):1197–1208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- 36.Veuger SJ, Hunter JE, Durkacz BW. Ionizing radiation-induced NF-kappaB activation requires PARP-1 function to confer radioresistance. Oncogene. 2009 Feb 12;28(6):832–842. doi: 10.1038/onc.2008.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin-Oliva D, Aguilar-Quesada R, O'Valle F, Munoz-Gamez JA, Martinez-Romero R, Garcia Del Moral R, et al. Inhibition of poly(ADP-ribose) polymerase modulates tumor-related gene expression, including hypoxia-inducible factor-1 activation, during skin carcinogenesis. Cancer Res. 2006 Jun 1;66(11):5744–5756. doi: 10.1158/0008-5472.CAN-05-3050. [DOI] [PubMed] [Google Scholar]
- 38.Shall S, de Murcia G. Poly(ADP-ribose) polymerase-1: what have we learned from the deficient mouse model? Mutat Res. 2000 Jun 30;460(1):1–15. doi: 10.1016/s0921-8777(00)00016-1. [DOI] [PubMed] [Google Scholar]
- 39.Ame JC, Rolli V, Schreiber V, Niedergang C, Apiou F, Decker P, et al. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J Biol Chem. 1999 Jun 18;274(25):17860–17868. doi: 10.1074/jbc.274.25.17860. [DOI] [PubMed] [Google Scholar]
- 40.Shieh WM, Ame JC, Wilson MV, Wang ZQ, Koh DW, Jacobson MK, et al. Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J Biol Chem. 1998 Nov 13;273(46):30069–30072. doi: 10.1074/jbc.273.46.30069. [DOI] [PubMed] [Google Scholar]
- 41.Blum W, Faigle JW, Pfaar U, Sallmann A. Characterization of a novel diclofenac metabolite in human urine by capillary gas chromatography-negative chemical ionization mass spectrometry. J Chromatogr B Biomed Appl. 1996 Oct 25;685(2):251–263. doi: 10.1016/s0378-4347(96)00198-3. [DOI] [PubMed] [Google Scholar]
- 42.Yelamos J, Schreiber V, Dantzer F. Toward specific functions of poly(ADP-ribose) polymerase-2. Trends Mol Med. 2008 Apr;14(4):169–178. doi: 10.1016/j.molmed.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 43.Raval-Fernandes S, Kickhoefer VA, Kitchen C, Rome LH. Increased susceptibility of vault poly(ADP-ribose) polymerase-deficient mice to carcinogen-induced tumorigenesis. Cancer Res. 2005 Oct 1;65(19):8846–8852. doi: 10.1158/0008-5472.CAN-05-0770. [DOI] [PubMed] [Google Scholar]
- 44.Hsiao SJ, Smith S. Tankyrase function at telomeres, spindle poles, and beyond. Biochimie. 2008 Jan;90(1):83–92. doi: 10.1016/j.biochi.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 45.Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009 Oct 1;461(7264):614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 46.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005 Apr 14;434(7035):913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 47.Bryant HE, Petermann E, Schultz N, Jemth AS, Loseva O, Issaeva N, et al. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009 Sep 2;28(17):2601–2615. doi: 10.1038/emboj.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mendes-Pereira AM, Martin SA, Brough R, McCarthy A, Taylor JR, Kim JS, et al. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol Med. 2009 Sep;1(6–7):315–322. doi: 10.1002/emmm.200900041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alazawi W, Pett M, Strauss S, Moseley R, Gray J, Stanley M, et al. Genomic imbalances in 70 snap-frozen cervical squamous intraepithelial lesions: associations with lesion grade, state of the HPV16 E2 gene and clinical outcome. Br J Cancer. 2004 Dec 13;91(12):2063–2070. doi: 10.1038/sj.bjc.6602237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.National Institutes of Health. Breast Cancer. cancer gov. 2010
- 51.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003 Jul 8;100(14):8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheang MC, Voduc D, Bajdik C, Leung S, McKinney S, Chia SK, et al. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res. 2008 Mar 1;14(5):1368–1376. doi: 10.1158/1078-0432.CCR-07-1658. [DOI] [PubMed] [Google Scholar]
- 53.O'Shaughnessy J. Triple Negative Metastatic Breast Cancer: A Phase 2, Multi-Center, Open-Label, Randomized Trial of Gemcitabine/Carboplatin (G/C) With or Without BSI-201, A PARP Inhibitor. Poster Presentation at San Antonio Breast Cancer Symposium 2008.2008. [Google Scholar]
- 54.Johns Hopkins Pathology. Johns Hopkins Pathology. Ovarian Cancer. Available from: http://www.ovariancancer.jhmi.edu/hereditary.cfm. [Google Scholar]
- 55.Banerjee S. Ovarian Cancer: Making the Best of PARP Inhibitors : BRCA1/BRA Mutations in Ovarian Cancer. Nat Rev Clin Oncol. 2010;7(9) doi: 10.1038/nrclinonc.2010.116. [DOI] [PubMed] [Google Scholar]
- 56.Schmeler KM, Gershenson DM. Low-grade serous ovarian cancer: a unique disease. Curr Oncol Rep. 2008 Nov;10(6):519–523. doi: 10.1007/s11912-008-0078-8. [DOI] [PubMed] [Google Scholar]
- 57.Levanon K, Crum C, Drapkin R. New insights into the pathogenesis of serous ovarian cancer and its clinical impact. J Clin Oncol. 2008 Nov 10;26(32):5284–5293. doi: 10.1200/JCO.2008.18.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tan DS, Rothermundt C, Thomas K, Bancroft E, Eeles R, Shanley S, et al. "BRCAness" syndrome in ovarian cancer: a case-control study describing the clinical features and outcome of patients with epithelial ovarian cancer associated with BRCA1 and BRCA2 mutations. J Clin Oncol. 2008 Dec 1;26(34):5530–5536. doi: 10.1200/JCO.2008.16.1703. [DOI] [PubMed] [Google Scholar]
- 59.Cass I, Baldwin RL, Varkey T, Moslehi R, Narod SA, Karlan BY. Improved survival in women with BRCA-associated ovarian carcinoma. Cancer. 2003 May 1;97(9):2187–2195. doi: 10.1002/cncr.11310. [DOI] [PubMed] [Google Scholar]
- 60.Cannistra SA. Cancer of the ovary. N Engl J Med. 2004 Dec 9;351(24):2519–2529. doi: 10.1056/NEJMra041842. [DOI] [PubMed] [Google Scholar]
- 61.Basham VM, Lipscombe JM, Ward JM, Gayther SA, Ponder BA, Easton DF, et al. BRCA1 and BRCA2 mutations in a population-based study of male breast cancer. Breast Cancer Res. 2002;4(1):R2. doi: 10.1186/bcr419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wooster R, Neuhausen SL, Mangion J, Quirk Y, Ford D, Collins N, et al. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13. Science. 1994 Sep 30;265(5181):2088–2090. doi: 10.1126/science.8091231. [DOI] [PubMed] [Google Scholar]
- 63.Leegte B, van der Hout AH, Deffenbaugh AM, Bakker MK, Mulder IM, ten Berge A, et al. Phenotypic expression of double heterozygosity for BRCA1 and BRCA2 germline mutations. J Med Genet. 2005 Mar;42(3):e20. doi: 10.1136/jmg.2004.027243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.James PA, Doherty R, Harris M, Mukesh BN, Milner A, Young MA, et al. Optimal selection of individuals for BRCA mutation testing: a comparison of available methods. J Clin Oncol. 2006 Feb 1;24(4):707–715. doi: 10.1200/JCO.2005.01.9737. [DOI] [PubMed] [Google Scholar]
- 65.Turner N, Tutt A, Ashworth A. Hallmarks of 'BRCAness' in sporadic cancers. Nat Rev Cancer. 2004 Oct;4(10):814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 66.Futreal PA, Soderkvist P, Marks JR, Iglehart JD, Cochran C, Barrett JC, et al. Detection of frequent allelic loss on proximal chromosome 17q in sporadic breast carcinoma using microsatellite length polymorphisms. Cancer Res. 1992 May 1;52(9):2624–2627. [PubMed] [Google Scholar]
- 67.Russell SE, Hickey GI, Lowry WS, White P, Atkinson RJ. Allele loss from chromosome 17 in ovarian cancer. Oncogene. 1990 Oct;5(10):1581–1583. [PubMed] [Google Scholar]
- 68.Dobrovic A, Simpfendorfer D. Methylation of the BRCA1 gene in sporadic breast cancer. Cancer Res. 1997 Aug 15;57(16):3347–3350. [PubMed] [Google Scholar]
- 69.Mancini DN, Rodenhiser DI, Ainsworth PJ, O'Malley FP, Singh SM, Xing W, et al. CpG methylation within the 5' regulatory region of the BRCA1 gene is tumor specific and includes a putative CREB binding site. Oncogene. 1998 Mar 5;16(9):1161–1169. doi: 10.1038/sj.onc.1201630. [DOI] [PubMed] [Google Scholar]
- 70.Wilson CA, Ramos L, Villasenor MR, Anders KH, Press MF, Clarke K, et al. Localization of human BRCA1 and its loss in high-grade, non-inherited breast carcinomas. Nat Genet. 1999 Feb;21(2):236–240. doi: 10.1038/6029. [DOI] [PubMed] [Google Scholar]
- 71.Esteller M, Silva JM, Dominguez G, Bonilla F, Matias-Guiu X, Lerma E, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 2000 Apr 5;92(7):564–569. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- 72.Umetani N, Mori T, Koyanagi K, Shinozaki M, Kim J, Giuliano AE, et al. Aberrant hypermethylation of ID4 gene promoter region increases risk of lymph node metastasis in T1 breast cancer. Oncogene. 2005 Jul 7;24(29):4721–4727. doi: 10.1038/sj.onc.1208538. [DOI] [PubMed] [Google Scholar]
- 73.Turner NC, Reis-Filho JS, Russell AM, Springall RJ, Ryder K, Steele D, et al. BRCA1 dysfunction in sporadic basal-like breast cancer. Oncogene. 2007 Mar 29;26(14):2126–2132. doi: 10.1038/sj.onc.1210014. [DOI] [PubMed] [Google Scholar]
- 74.Lamers E. Microarray based expression profi ling of BRCA1 mutated human tumours using a breast-specifi c platform to identify a profile of BRCA1 deficiency. Breast Cancer Res. 2010;12 Suppl 1:poster 44. [Google Scholar]
- 75.Jazaeri AA, Yee CJ, Sotiriou C, Brantley KR, Boyd J, Liu ET. Gene expression profiles of BRCA1-linked, BRCA2-linked, and sporadic ovarian cancers. J Natl Cancer Inst. 2002 Jul 3;94(13):990–1000. doi: 10.1093/jnci/94.13.990. [DOI] [PubMed] [Google Scholar]
- 76.Hennessy BT. Somatic BRCA status in ovarian tumors. Journal of Clinical Oncology. 2009 May 20;27(155) Supplement:5528. [abstract] [Google Scholar]
- 77.Geisler JP, Hatterman-Zogg MA, Rathe JA, Buller RE. Frequency of BRCA1 dysfunction in ovarian cancer. J Natl Cancer Inst. 2002 Jan 2;94(1):61–67. doi: 10.1093/jnci/94.1.61. [DOI] [PubMed] [Google Scholar]
- 78.Hilton JL, Geisler JP, Rathe JA, Hattermann-Zogg MA, DeYoung B, Buller RE. Inactivation of BRCA1 and BRCA2 in ovarian cancer. J Natl Cancer Inst. 2002 Sep 18;94(18):1396–1406. doi: 10.1093/jnci/94.18.1396. [DOI] [PubMed] [Google Scholar]
- 79.Turner NC, Lord CJ, Iorns E, Brough R, Swift S, Elliott R, et al. A synthetic lethal siRNA screen identifying genes mediating sensitivity to a PARP inhibitor. EMBO J. 2008 May 7;27(9):1368–1377. doi: 10.1038/emboj.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Konstantinopoulos PA, Spentzos D, Karlan BY, Taniguchi T, Fountzilas E, Francoeur N, et al. Gene expression profile of BRCAness that correlates with responsiveness to chemotherapy and with outcome in patients with epithelial ovarian cancer. J Clin Oncol. 2010 Aug 1;28(22):3555–3561. doi: 10.1200/JCO.2009.27.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hughes-Davies L, Huntsman D, Ruas M, Fuks F, Bye J, Chin SF, et al. EMSY links the BRCA2 pathway to sporadic breast and ovarian cancer. Cell. 2003 Nov 26;115(5):523–535. doi: 10.1016/s0092-8674(03)00930-9. [DOI] [PubMed] [Google Scholar]
- 82.Baldwin RL, Nemeth E, Tran H, Shvartsman H, Cass I, Narod S, et al. BRCA1 promoter region hypermethylation in ovarian carcinoma: a population-based study. Cancer Res. 2000 Oct 1;60(19):5329–5333. [PubMed] [Google Scholar]
- 83.D'Andrea AD. The Fanconi Anemia/BRCA signaling pathway: disruption in cisplatin-sensitive ovarian cancers. Cell Cycle. 2003 Jul–Aug;2(4):290–292. [PubMed] [Google Scholar]
- 84.Taniguchi T, Tischkowitz M, Ameziane N, Hodgson SV, Mathew CG, Joenje H, et al. Disruption of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian tumors. Nat Med. 2003 May;9(5):568–574. doi: 10.1038/nm852. [DOI] [PubMed] [Google Scholar]
- 85.McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift S, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006 Aug 15;66(16):8109–8115. doi: 10.1158/0008-5472.CAN-06-0140. [DOI] [PubMed] [Google Scholar]
- 86.Satoh MS, Poirier GG, Lindahl T. Dual function for poly(ADP-ribose) synthesis in response to DNA strand breakage. Biochemistry. 1994 Jun 14;33(23):7099–7106. doi: 10.1021/bi00189a012. [DOI] [PubMed] [Google Scholar]
- 87.Clark JB, Ferris GM, Pinder S. Inhibition of nuclear NAD nucleosidase and poly ADP-ribose polymerase activity from rat liver by nicotinamide and 5'-methyl nicotinamide. Biochim Biophys Acta. 1971 Apr 29;238(1):82–85. doi: 10.1016/0005-2787(71)90012-8. [DOI] [PubMed] [Google Scholar]
- 88.Zaremba T, Thomas H, Cole M, Plummer ER, Curtin NJ. Doxorubicin-induced suppression of poly(ADP-ribose) polymerase-1 (PARP-1) activity and expression and its implication for PARP inhibitors in clinical trials. Cancer Chemother Pharmacol. 2010 Sep;66(4):807–812. doi: 10.1007/s00280-010-1359-0. [DOI] [PubMed] [Google Scholar]
- 89.Underhill C, Toulmonde M, Bonnefoi H. A review of PARP inhibitors: from bench to bedside. Ann Oncol. 2010 Jul 19; doi: 10.1093/annonc/mdq322. [DOI] [PubMed] [Google Scholar]
- 90.Tentori L, Graziani G. Chemopotentiation by PARP inhibitors in cancer therapy. Pharmacol Res. 2005 Jul;52(1):25–33. doi: 10.1016/j.phrs.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 91.Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006 May;7(5):335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 92.Wedge SR, Porteus JK, May BL, Newlands ES. Potentiation of temozolomide and BCNU cytotoxicity by O(6)-benzylguanine: a comparative study in vitro. Br J Cancer. 1996 Feb;73(4):482–490. doi: 10.1038/bjc.1996.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu X, Shi Y, Guan R, Donawho C, Luo Y, Palma J, et al. Potentiation of temozolomide cytotoxicity by poly(ADP)ribose polymerase inhibitor ABT-888 requires a conversion of single-stranded DNA damages to double-stranded DNA breaks. Mol Cancer Res. 2008 Oct;6(10):1621–1629. doi: 10.1158/1541-7786.MCR-08-0240. [DOI] [PubMed] [Google Scholar]
- 94.Curtin NJ, Wang LZ, Yiakouvaki A, Kyle S, Arris CA, Canan-Koch S, et al. Novel poly(ADP-ribose) polymerase-1 inhibitor, AG14361, restores sensitivity to temozolomide in mismatch repair-deficient cells. Clin Cancer Res. 2004 Feb 1;10(3):881–889. doi: 10.1158/1078-0432.ccr-1144-3. [DOI] [PubMed] [Google Scholar]
- 95.Donawho CK, Luo Y, Penning TD, Bauch JL, Bouska JJ, Bontcheva-Diaz VD, et al. ABT-888, an orally active poly(ADP-ribose) polymerase inhibitor that potentiates DNA-damaging agents in preclinical tumor models. Clin Cancer Res. 2007 May 1;13(9):2728–2737. doi: 10.1158/1078-0432.CCR-06-3039. [DOI] [PubMed] [Google Scholar]
- 96.Smith LM, Willmore E, Austin CA, Curtin NJ. The novel poly(ADP-Ribose) polymerase inhibitor, AG14361, sensitizes cells to topoisomerase I poisons by increasing the persistence of DNA strand breaks. Clin Cancer Res. 2005 Dec 1;11(23):8449–8457. doi: 10.1158/1078-0432.CCR-05-1224. [DOI] [PubMed] [Google Scholar]
- 97.Burkle A, Chen G, Kupper JH, Grube K, Zeller WJ. Increased poly(ADP-ribosyl)ation in intact cells by cisplatin treatment. Carcinogenesis. 1993 Apr;14(4):559–561. doi: 10.1093/carcin/14.4.559. [DOI] [PubMed] [Google Scholar]
- 98.Miknyoczki SJ, Jones-Bolin S, Pritchard S, Hunter K, Zhao H, Wan W, et al. Chemopotentiation of temozolomide, irinotecan, and cisplatin activity by CEP-6800, a poly(ADP-ribose) polymerase inhibitor. Mol Cancer Ther. 2003 Apr;2(4):371–382. [PubMed] [Google Scholar]
- 99.Calabrese CR, Almassy R, Barton S, Batey MA, Calvert AH, Canan-Koch S, et al. Anticancer chemosensitization and radiosensitization by the novel poly(ADP-ribose) polymerase-1 inhibitor AG14361. J Natl Cancer Inst. 2004 Jan 7;96(1):56–67. doi: 10.1093/jnci/djh005. [DOI] [PubMed] [Google Scholar]
- 100.Boulton S, Kyle S, Durkacz BW. Interactive effects of inhibitors of poly(ADP-ribose) polymerase and DNA-dependent protein kinase on cellular responses to DNA damage. Carcinogenesis. 1999 Feb;20(2):199–203. doi: 10.1093/carcin/20.2.199. [DOI] [PubMed] [Google Scholar]
- 101.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005 Apr 14;434(7035):917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 102.Evers B, Drost R, Schut E, de Bruin M, van der Burg E, Derksen PW, et al. Selective inhibition of BRCA2-deficient mammary tumor cell growth by AZD2281 and cisplatin. Clin Cancer Res. 2008 Jun 15;14(12):3916–3925. doi: 10.1158/1078-0432.CCR-07-4953. [DOI] [PubMed] [Google Scholar]
- 103.Munoz-Gamez JA, Martin-Oliva D, Aguilar-Quesada R, Canuelo A, Nunez MI, Valenzuela MT, et al. PARP inhibition sensitizes p53-deficient breast cancer cells to doxorubicin-induced apoptosis. Biochem J. 2005 Feb 15;386(Pt 1):119–125. doi: 10.1042/BJ20040776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McCabe N, Cerone MA, Ohishi T, Seimiya H, Lord CJ, Ashworth A. Targeting Tankyrase 1 as a therapeutic strategy for BRCA-associated cancer. Oncogene. 2009 Mar 19;28(11):1465–1470. doi: 10.1038/onc.2008.483. [DOI] [PubMed] [Google Scholar]
- 105. http://clincancerres.aacrjournals.org/content/14/23/7917/F1.small.gif. AG 014699 Structure. [Google Scholar]
- 106.Ihnen M. Therapeutic Advantage of Chemotherapy Drugs in Combination with PARP Inhibitor PF-01367338 (AG-014699) in Human Ovarian Cancer Cells. EORTC. 2010 [abstract] [Google Scholar]
- 107. http://www.lookchem.com/chemical-dictionary/en/product_a/459868-92-9/. AG 014699 Structure.
- 108.Plummer R. First and final report of a phase II study of the poly(ADP-ribose) polymerase (PARP) inhibitor, AG014699, in combination with temozolomide (TMZ) in patients with metastatic malignant melanoma (MM) J Clin Oncol. 2006;24(18s) ASCO abstr 8013. [Google Scholar]
- 109.ClinicalTrials.gov. A Study Of Poly (ADP-Ribose) Polymerase Inhibitor PF-01367338. Combination With Several Chemotherapeutic Regimens. 2010
- 110.Olaparib, Wikipedia. http://en.wikipedia.org/wiki/File:Olaparib.png. [Google Scholar]
- 111.Rajan A. A Phase I Combination Study of Olaparib (AZD 2208; KU-0059436) and Cisplatin Plus Gemcitabine in Adults With Solid Tumors. TAT. 2010 doi: 10.1158/1078-0432.CCR-11-2425. [abstract] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fong PC, Yap TA, Boss DS, Carden CP, Mergui-Roelvink M, Gourley C, et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J Clin Oncol. 2010 May 20;28(15):2512–2519. doi: 10.1200/JCO.2009.26.9589. [DOI] [PubMed] [Google Scholar]
- 113.Audeh MW, Carmichael J, Penson RT, Friedlander M, Powell B, Bell-McGuinn KM, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010 Jul 24;376(9737):245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- 114.Kaye S, et al. Phase II study of the oral PARP inhibitor olaparib (AZD2281) versus liposomal doxorubicin in ovarian cancer patients with BRCA1 and/or BRCA2 mutations. ESMO. 2010:2010. [abstract] [Google Scholar]
- 115.Gelman K. Can we define tumors that will respond to PARP inhibitors? A phase II correlative study of olaparib in advanced serous ovarian cancer and triple negative breast cancer. Journal of Clinical Oncology. 2010;28(15 supplement) [abstract], abstract 3002. [Google Scholar]
- 116.Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010 Jul 24;376(9737):235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 117.Dent R. Safety and efficacy of the oral PARP inhibitor olaparib (AZD2281) in combination with paclitaxel for the first- or second-line treatment of patients with metastatic triple-negative breast cancer: Results from the safety cohort of a phase I/II multicenter trial. J Clin Oncol. 2010;28(15s) doi: 10.1186/bcr3484. ASCO abstr 1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Iniparib, wikipedia. http://en.wikipedia.org/wiki/Iniparib. [Google Scholar]
- 119.Kopetz S. First in human phase I study of BSI-201, a small molecule inhibitor of poly ADP-ribose polymerase (PARP) in subjects with advanced solid tumors. J Clin Oncol. 2008 May 20;26 suppl ASCO abstr 3577. [Google Scholar]
- 120.Mahany J. A phase IB study evaluating BSI-201 in combination with chemotherapy in subjects with advanced solid tumors. J Clin Oncol. 2008 May 20;26 suppl ASCO abstr 3579. [Google Scholar]
- 121.O'Shaughnessy J, et al. Final efficacy and safety results of a randomized phase II study of the parp inhibitor iniparib (BSI-201) in combination with gamcitabine/carboplatin in metastatic triple negative breast cancer. ESMO. 2010:2010. [abstract] [Google Scholar]
- 122.Veliparib skeletal, wikipedia. http://en.wikipedia.org/wiki/File:Veliparib_skeletal.svg. [Google Scholar]
- 123.Kummar S, Kinders R, Gutierrez ME, Rubinstein L, Parchment RE, Phillips LR, et al. Phase 0 clinical trial of the poly (ADP-ribose) polymerase inhibitor ABT-888 in patients with advanced malignancies. J Clin Oncol. 2009 Jun 1;27(16):2705–2711. doi: 10.1200/JCO.2008.19.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kummar S, et al. Pharmacodynamic response in phase I combination study of ABT-888 and topotecan in adults with refractory solid tumors and lymphomas. ASCO. 2010:2010. [abstract] [Google Scholar]
- 125.Kummar S. A phase I study of ABT-888 (A) in combination with metronomic cyclophosphamide (C) in adults with refractory solid tumors and lymphomas. J Clin Oncol. 2010;28(15s) ASCO abstr 2605. [Google Scholar]
- 126.Isakoff J, et al. Phase II trial of the PARP inhibitor veliparib (ABT888) and temozolomide for metastatic breast cancerS. ASCO. 2010 [abstract]. 2010:abstract 1019. [Google Scholar]
- 127.ClinicalTrials.gov. MK 4827. clinicaltrialsgov. 2010
- 128.Sandhu SK, et al. First-in-human trial of a poly(ADP-ribose) polymerase (PARP) inhibitor MK-4827 in advanced cancer patients (pts) with antitumor activity in BRCA-deficient and sporadic ovarian cancers. ASCO. 2010 [abstract]. 2010:abstract 3001. [Google Scholar]
- 129.Miknyoczki S, Chang H, Grobelny J, Pritchard S, Worrell C, McGann N, et al. The selective poly(ADP-ribose) polymerase-1(2) inhibitor, CEP-8983, increases the sensitivity of chemoresistant tumor cells to temozolomide and irinotecan but does not potentiate myelotoxicity. Mol Cancer Ther. 2007 Aug;6(8):2290–2302. doi: 10.1158/1535-7163.MCT-07-0062. [DOI] [PubMed] [Google Scholar]
- 130.The Medical News. LEAD Therapeutics discovers orally available PARP inhibitor. The Medical News. 2009 [Google Scholar]
- 131.Melisi D, Ossovskaya V, Zhu C, Rosa R, Ling J, Dougherty PM, et al. Oral poly(ADP-ribose) polymerase-1 inhibitor BSI-401 has antitumor activity and synergizes with oxaliplatin against pancreatic cancer, preventing acute neurotoxicity. Clin Cancer Res. 2009 Oct 15;15(20):6367–6377. doi: 10.1158/1078-0432.CCR-09-0910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sakai W, Swisher EM, Karlan BY, Agarwal MK, Higgins J, Friedman C, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008 Feb 28;451(7182):1116–1120. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jacquemont C, Taniguchi T. Proteasome function is required for DNA damage response and fanconi anemia pathway activation. Cancer Res. 2007 Aug 1;67(15):7395–7405. doi: 10.1158/0008-5472.CAN-07-1015. [DOI] [PubMed] [Google Scholar]
- 134.Rottenberg S, Jaspers JE, Kersbergen A, van der Burg E, Nygren AO, Zander SA, et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc Natl Acad Sci U S A. 2008 Nov 4;105(44):17079–17084. doi: 10.1073/pnas.0806092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010 Apr 16;141(2):243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]