Abstract
Human cystathionine β-synthase (CBS), a pivotal enzyme in the metabolism of homocysteine, is a pyridoxal-5′-phosphate-dependent enzyme that also contains heme, a second cofactor whose function is still unclear. One strategy for elucidation of heme function is its replacement with different metalloporphyrins or with porphyrins containing different substituent groups. This paper describes a novel expression approach and purification of cobalt CBS (CoCBS), which results in a high yield of fully active, high purity enzyme, in which heme is substituted by Co-protoporphyrin IX (CoPPIX). Metal content analysis showed that the enzyme contained 92% cobalt and 8% iron. CoCBS was indistinguishable from wild-type FeCBS in its activity, tetrameric oligomerization, PLP saturation and responsiveness to the allosteric activator, S-adenosyl-L-methionine. The observed biochemical and spectral characteristics of CoCBS provide further support for the suggestion that heme is involved in structural integrity and folding of this unusual enzyme.
Keywords: Cystathionine beta-synthase, heme, cobalt protoporphyrin IX, protein expression, S-adenosyl-L-methionine, heme replacement
1. Introduction
Cystathionine β-synthase (CBS) is the first enzyme in the transsulfuration pathway in which the toxic intermediate homocysteine is converted to cysteine and ultimately to glutathione. A deficiency in CBS activity, primarily due to the presence of missense mutations, is the most common cause of homocystinuria, an inherited metabolic disease characterized by elevated plasma homocysteine and methionine levels and by symptoms such as dislocated optic lenses, vascular manifestations, skeletal deformities and mental retardation [1].
Human CBS is a homotetramer composed of 63 kDa subunits each 551 amino acids in length. Each subunit has a modular structure consisting of three domains: an N-terminal heme binding domain, a highly conserved pyridoxal-5′-phosphate (PLP)-binding catalytic core, and a S-adenosyl-L-methionine (AdoMet)-binding C-terminal regulatory domain. PLP serves in the catalytic chemistry of CBS via a well-established mechanism [2–4]. AdoMet allosterically activates the enzyme up to 8-fold. Removal of the C-terminal regulatory domain leads to a change in oligomeric status from tetramer to dimer and loss of response to AdoMet. Binding of AdoMet causes a conformational change with displacement of an autoinhibitory region from the catalytic site [5, 6]. As a consequence, the 45 kDa C-terminal truncated enzyme has comparable activity to the AdoMet-stimulated full-length CBS.
The role of heme in CBS is not clear; however, our previous results demonstrate that heme does not function in redox sensing, ligand binding or the CBS catalytic step [7–13]. The presence of heme in the CBS enzyme seems to be limited to higher eukaryotes. The crystal structure of the dimeric 45 kDa truncated human CBS shows that the heme cofactor is relatively surface-exposed and axially coordinated by Cys52 and His65, both located on the loop within the N-terminus, in which the heme is nested [8, 14]. The iron is 6-coordinate and low spin in both reduced and oxidized states [15]. In oxidized CBS, the ferric heme does not bind exogenous ligands and appears to be a poor target for small molecule-mediated regulation. However, in reduced CBS, the ferrous heme binds CO, NO, CN− and various isonitriles. Binding affects CBS activity and generally leads to inactivation by disruption of thiolate coordination [13, 16, 17]. The redox properties of CBS heme and its effect on CBS activity suggested that the heme might function as a redox sensor [14, 15, 18, 19]. However, this theory is undermined by the complex redox behavior of CBS, which exhibits pH and temperature dependence incompatible with simple redox regulation [10–12]. Taken together, the complex behavior of the CBS heme iron suggests that allosteric regulation by small molecule ligand binding or changing heme redox state in CBS is unlikely.
A plausible alternative hypothesis is that heme plays a role in the structural stability of CBS. When expressed in a heme biosynthesis-deficient S. cerevisiae strain, the CBS enzyme does not accumulate in the absence of exogenous heme. Remarkably, heme may be replaced by the metal free protoporphyrin (PPIX) or with various alternate metalloporphyrins to yield accumulation of the CBS protein. The expression and activity of CBS are almost completely restored by inclusion of the chemical chaperone trimethylamine-N-oxide (TMAO) in the growth medium of the heme-requiring yeast cells, which implicates a folding defect in the absence of heme [20]. When expressed in heme biosynthesis-deficient E. coli RP523 cells, CBS accumulation was similarly dependent on exogenous porphyrins and CBS proteins containing MnPPIX and CoPPIX were isolated. These proteins were functional; however, the yields and activity of CoCBS and MnCBS anaerobically expressed in the presence of CoPPIX and MnPPIX, respectively, were lower than for FeCBS isolated from wild type E. coli cells in the presence of δ-aminolevulinic acid. In order to maximize the yield of metalloporphyrin-substituted protein, we have developed a new method which can be conveniently utilized for preparation of CoPPIX-substituted heme proteins [21]. In this paper, we present purification and characterization of cobalt-substituted human CBS (CoCBS) expressed in E. coli.
2. Material and Methods
2.1 Chemicals
Unless stated otherwise, all chemicals were purchased from Sigma or Fisher Scientific. Protoporphyrins were purchased from Frontier Scientific. L-[14C(U)]-serine was obtained from PerkinElmer Life Sciences.
2.2 Expression and purification of CoCBS
The construct pGEX-6P1-hCBS WT [22] was transformed into an improved bacterial host, E. coli Rosetta 2 (DE3) (Novagen), for expression. Cells were grown in M9 minimal medium. Media were always supplemented with 100 μg/ml ampicillin and 30 μg/ml chloramphenicol for selection of the CBS expression plasmid and pRARE2 plasmid, respectively. After overnight cultivation, cells were inoculated into fresh M9 minimal medium supplemented with CoCl2 at a final concentration of 150 μM and grown overnight. The cells were passaged seven times (7×) or twelve times (12×) in this medium, and the last culture served as an inoculum for the large-scale expression. The expression M9 minimal medium (pH 7.4) was supplemented with 0.5% glucose, 0.4% Casamino acids, 2 mM MgSO4, 100 μM CaCl2, 0.001% thiamine-HCl, 300 μM δ-aminolevulinic acid and 150 μM CoCl2. Cells were grown at 30°C to an A600 ~ 1.0, then protein expression was induced with 500 μM IPTG and carried out overnight.
The CBS containing CoPPIX was purified as described previously for wild type FeCBS [22] with several modifications. Briefly, the cells were harvested in the stationary phase of growth for preparation of CBS. After cleavage of the fusion protein with PreScission protease, the GST tag was removed chromatographically on DEAE Sepharose Fast Flow (GE Healthcare). The column was equilibrated in 15 mM potassium phosphate pH 7.2, 1 mM EDTA, 1 mM DTT and 10% ethylene glycol. Under these conditions, both GST tag and hCBS proteins bind to the DEAE Sepharose resin. The separation of the GST tag from CBS was achieved by elution with a linear gradient from 15 mM to 300 mM potassium phosphate pH 7.2, 1 mM EDTA, 1 mM DTT and 10% ethylene glycol. Protein-containing fractions were analyzed by electrophoresis on 9% SDS-PAGE. The CBS-rich fractions were pooled and subsequently concentrated on YM-100 membrane (Millipore). The buffer was changed by pressure dialysis for 20 mM HEPES pH 7.4, 1 mM TCEP and 0.01% Tween 20.
2.3 Pyridine hemochromogen assays
The pyridine hemochromogen assay was performed as described elsewhere [20, 21] using a HP diode array model 8453 spectrophotometer. For difference pyridine hemochromogen spectra of membrane-bound hemoproteins, the insoluble fractions of the cell lysates were washed twice with 120 volumes of Tris-saline buffer, pH 8.6. Difference spectra (i.e. reduced minus oxidized) were recorded from 650 to 380 nm with a Shimadzu 2401PC spectrophotometer.
2.4 Metal content determination
Metal content analysis was performed by inductively coupled plasma optical emission spectroscopy (ICP-OES) using a Perkin Elmer Optima 2000 DV in axial-view with an integrated AS-90 autosampler. The relevant measurement parameters were: plasma flow 15 L/min, auxiliary flow 0.2 L/min, nebulizer flow 0.6 L/min, power 1500 W, sample uptake 1 mL/min, single measurement mode with peak integration and high-resolution readout, background correction with manually selected 3-point interpolation, 10 s measurement time, 5 replicate measurements, 60 s wash between samples, 60 s read delay and 15 s flush time. Cobalt and iron were determined at commonly used analytical transitions of the atomic spectrum (228.616 nm for Co, 238.204 nm for Fe). Sample analysis was randomized to eliminate systematic bias in the measurement. Each sample was analyzed 3 times allowing precise measurement with high signal to noise ratios. Data processing was performed automatically with the integrated instrument software.
2.5 Spectroscopic characterization
Electronic absorption spectra were measured on double-beam Varian Cary 4 Bio spectrophotometer set to a spectral bandwidth of 0.5 nm at 25°C. Solutions containing 10 μM of enzyme were in 500 mM CHES pH 9.0, 100 mM NaCl. CBS is isolated in the oxidized form i.e. with the metal ion in protoporphyrin cofactor oxidized (either Co(III)PPIX or Fe(III)PPIX). Samples were kept under argon atmosphere in septum-sealed cuvettes. Reduction of oxidized samples was carried out by adding an anaerobically prepared stock solution of sodium dithionite to a final concentration of 5 mM.
2.6 Activity assay
The CBS activity in the classical reaction was determined by a previously described radioisotope assay using [14C] L-serine as the labeled substrate [23]. CBS enzyme (420 ng) was assayed in a 100 μL reaction mixture for 30 min at 37°C. The reaction mixture contained 100 mM Tris-HCl pH 8.6, 10 mM L-serine, 0.3 μCi L-[14C(U)]-serine and 0.5 mg/mL BSA. The extent of saturation of the enzyme with PLP was tested by running the activity assay in the presence or absence of 0.5 mM PLP. The reaction was performed in the presence or absence of CBS allosteric activator AdoMet in a final concentration of 0.3 mM. The reaction mixture with enzyme was incubated at 37°C for 5 min and the reaction was initiated by addition of L-homocysteine to final concentration of 10 mM. The colorimetric assay for the hydrogen sulfide-producing activity of CBS was essentially performed as described by Kayastha and Miles [24]. CBS enzyme (5 μg) was assayed in a 200 μL volume for 6 min at 37°C. The reaction mixture contained 50 mM Bicine pH 7.8, 40 mM L-cysteine, 50 mM β-mercaptoethanol and 0.5 mg/ml BSA. One unit of activity is defined as the amount of CBS that catalyzes the formation of 1 μmol of product in 1 h at 37°C under standard assay conditions.
3 Results
3.1 Purification of CoCBS
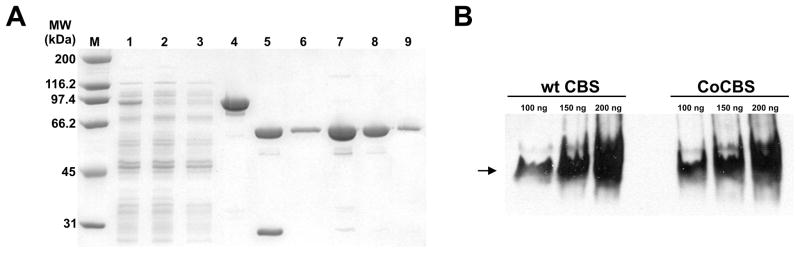
CoCBS was expressed as a GST fusion protein from a pGEX-6P1-hCBS WT construct transformed into the E. coli Rosetta 2 (DE3) expression strain. Expression of a GST-CBS fusion protein, where the fusion partner is specifically cleaved by PreScission protease leaving an extra glycine residue at the N-terminus, enabled us to isolate rapidly large amounts of highly purified CBS protein in a convenient two-step procedure. Unlike our previous procedure [22], we utilized a DEAE Sepharose column for the separation of the cleaved GST tag from CBS. After analysis of the fractions on 9% SDS-PAGE gels, the CBS-rich fractions were pooled and concentrated. To enhance solubility and prevent protein aggregation during concentration, we added a non-ionic detergent Tween 20 to a final concentration of 0.01%. We were typically able to recover 15 – 40 mg of highly purified CBS (> 97% pure according to inspection of the SDS-PAGE gel; Fig. 1A) from 12 L of overnight culture with a 44% recovery from the crude extract (Table 1). CoCBS under native conditions migrated predominantly as a tetramer, as did wild type FeCBS, with some higher oligomeric forms detectable (Fig. 1B).
Figure 1. SDS-PAGE analysis of fractions from CoCBS purification (A) and native Western blot of purified CoCBS (B).
(A) Each protein purification fraction was separated on a 9% SDS-PAGE gel and stained using Simply Blue Safe stain (Invitrogen). Lanes: M – Molecular weight marker (Biorad), 1 – crude extract (10 μg), 2 – Glutathione Sepharose flow-through (10 μg), 3 – Glutathione Sepharose wash (10 μg), 4 – GST-CBS fusion protein (10 μg), 5 – Fusion protein cleaved with PreScission protease (10 μg), 6 – DEAE Sepharose pool (2.5 μg), 7, 8, 9 – CoCBS (10 μg, 5 μg and 1 μg, respectively). (B) CoCBS and WT FeCBS were separated in 4–12% Tris-HCl native gel, transferred to PVDF membrane and probed with monoclonal anti-CBS antibody (Abnova).
Table 1.
CoCBS 12× purification table.
| Fraction | Total activity (U) | Total Protein (mg) | Specific activity (U/mg of protein) | Recovery yield (%) |
|---|---|---|---|---|
| Crude extract | 4520 | 6955 | 0.65 | 100 |
| Glutathione Sepharose flow-through | 1013 | 5279 | 0.19 | 22 |
| Glutathione Sepharose wash | 259 | 1140 | 0.23 | 6 |
| GST-CBS fusion protein | 2815 | 70 | 40.2 | 62 |
| Cleaved fusion protein | 3472 | 90 | 38.7 | 77 |
| DEAE pool | 1396 | 70 | 19.9 | 31 |
| CoCBS 12× | 2000 | 19 | 105.2 | 44 |
3.2 Spectral properties of CoCBS
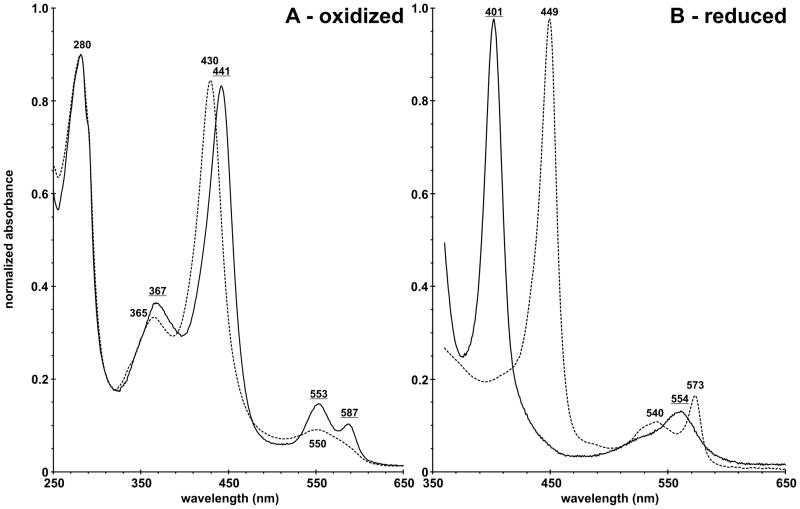
The visible spectrum of purified CoCBS was compared to the spectrum of wild type FeCBS. The A442/A280 ratio equal to 0.93 suggests that the purified CoCBS is fully saturated with CoPPIX. For comparison, the A429/A280 ratio of wild type FeCBS is equal to 0.94. The band shapes and positions of the peaks corresponding to the electronic absorption spectrum of CoCBS are distinct from those of wild type FeCBS and are indicative of a cobalt porphyrin in the +3 oxidation state [25, 26]. The Soret peak of Co(III)CBS was red-shifted by 11 nm to 441 nm, relative to that of Fe(III)CBS. The well-resolved α/β-region revealed two discrete peaks at 553 nm and 587 nm, which are different from the broad absorption envelope of Fe(III)CBS around 500 nm (Fig. 2A). The electronic absorption spectrum of Co(III)CBS also displays a distinct and well-resolved δ band at 367 nm compared to 365 nm for Fe(III)CBS. The combination of these spectral characteristics make Co(III)CBS unique among the family of cobalt-substituted heme proteins [27–31]. The reduction of Co(III)CBS was much slower compared to wild type Fe(III)CBS. The Soret peak of Co(II)CBS was blue-shifted by 48 nm to 401 nm, relative to that of Fe(II)CBS. The broad adsorption envelope of Co(II)CBS around 554 nm with blue-shifted shoulder is different from the well-resolved α/β region of Fe(II)CBS revealing two discrete peaks at 540 nm and 573 nm (Fig. 2B).
Figure 2. Electronic absorption spectrum of oxidized (A) and reduced (B) purified human CoCBS 12× (solid) compared to that of wild type FeCBS (dashed).
Enzyme (10 μM) was in 500 mM CHES pH 9.0, 100 mM NaCl at room temperature. Reduction was performed under anaerobic conditions by using sodium dithionite as a reductant. Underlined values correspond to CoCBS.
3.3 Metalloporphyrin analysis
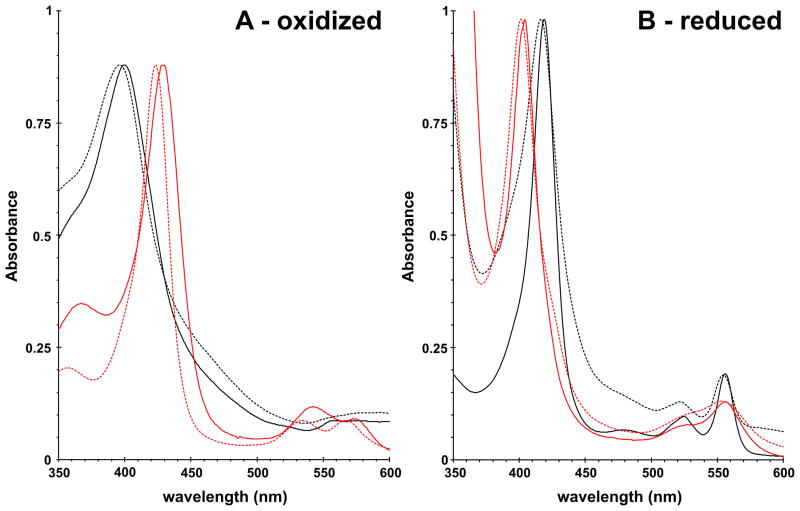
To confirm the presence of CoPPIX in the isolated CBS enzyme, we characterized the bis-pyridine adducts of the metalloporphyrin contained within the protein. The spectra of pyridine hemochromogen adducts were recorded in the oxidized and reduced states and compared with appropriate standards, i.e. oxidized or reduced FePPIX and CoPPIX, respectively (Fig. 3). The pyridine hemochromogen spectra of the protoporphyrins released from CoCBS closely resemble the spectra of the CoPPIX standard (Fig. 3, Table 2). Similarly, the pyridine hemochromogen spectra of wild type FeCBS were almost identical with the spectra of the FePPIX standard (Fig. 3, Table 2). The presence of iron and cobalt, respectively, in FeCBS and CoCBS is clearly visible from the positions of the Soret, α and β peaks of the oxidized (Fig. 3A) and reduced (Fig. 3B) pyridine hemochromogen spectra (Table 2).
Figure 3. Pyridine hemochromogen spectra of oxidized (A) and reduced (B) protoporphyrins released from wild type FeCBS and CoCBS.
Solid lines correspond to the spectra of metalloporphyrins released from the enzyme (black for wild-type FeCBS, red for CoCBS). Dashed lines represent the corresponding metalloporphyrin standard (black for FePPIX, red for CoPPIX).
Table 2.
Pyridine hemochromogen spectral peak positions.
| Sample | Soret (nm) | α/β peaks (nm) |
|---|---|---|
| Fe(III)PPIX standard | 396 | ND* |
| Oxidized PPIX released from wild type FeCBS | 399 | ND* |
| Co(III)PPIX standard | 423 | 534, 567 |
| Oxidized PPIX released from CoCBS | 429 | 542, 572 |
| Fe(II)PPIX standard | 416 | 522, 555 |
| Reduced PPIX released from wild type FeCBS | 419 | 524, 556 |
| Co(II)PPIX standard | 401 | 554 |
| Reduced PPIX released from CoCBS | 404 | 556 |
ND: not detected; bands were broad and indistinct with a maximum beyond the spectral range recorded
3.4 Metal content analysis
Quantitative determination of iron and cobalt content in CoCBS was carried out using ICP-OES analysis (Table 3). The CoCBS 7× protein was expressed in cells subjected to seven passages in cobalt-supplemented minimal medium prior to the induction of CBS expression. This enzyme contained 86.7% cobalt and 13.1% iron. The cobalt content increased with five additional passages in cobalt-containing medium, such that the CoCBS 12× protein contained 92.4% cobalt and 7.6% iron. For comparison, wild type FeCBS expressed in cells similarly grown in the presence of 150 μM FeCl3 contained only small amounts of cobalt. This minimal amount of cobalt may be attributed to cobalt in the growth medium; direct measurement of the minimal medium before supplementation with FeCl3 by ICP-OES revealed 1.38 ppb Co and 2.15 ppb Fe.
Table 3. Metal content analysis of wild type FeCBS and CoCBS using ICP-OES.
For the CoCBS samples, 7× and 12× denotes number of cell growth passages in minimal medium supplemented with 150 μM CoCl2 prior induction of CBS expression. Metal content is reported as percentage of total metal content.
| Sample | Iron content (μM) | Iron content (%) | Cobalt content (μM) | Cobalt content (%) |
|---|---|---|---|---|
| WT FeCBS | 1.3 ± 0.1 | 97.1 ± 0.7 | 0.036 ± 0.006 | 2.9 ± 0.2 |
| CoCBS 7× | 0.061 ± 0.002 | 13.1 ± 0.1 | 0.39 ± 0.04 | 86.7 ± 0.3 |
| CoCBS 12× | 0.083 ± 0.003 | 7.6 ± 0.4 | 0.96 ± 0.06 | 92.4 ± 0.8 |
3.5 The CoCBS activity measurements
Our previous results suggested that CoCBS prepared from cells grown under anaerobic conditions had 60% of the activity of FeCBS prepared from similarly cultivated cells. However, the activity of the wild type FeCBS isolated from anaerobically grown cells was substantially reduced compared to the wild type FeCBS isolated from aerobically grown cells [20]. CoCBS, isolated after 12 passages of cells through CoCl2-containing minimal medium, showed similar activity to wild type FeCBS in two different catalytic assays: the classical condensation of L-serine and homocysteine to L-cystathionine, and an alternative condensation of L-cysteine and β-mercaptoethanol to yield S-hydroxyethyl-L-cysteine and hydrogen sulfide (Table 4). The CoCBS remains fully responsive to the allosteric activator S-adenosyl-L-methionine. Furthermore, we conclude that the presence of the different metalloporphyrin in CBS did not affect the saturation of the enzyme with PLP, because similar increases in activity were observed for both CoCBS and wild type FeCBS when additional exogenous PLP was added.
Table 4. Enzymatic activity of CoCBS and wild type FeCBS.
The CBS activity was determined in two reactions. In the classical reaction, homocysteine and L-serine were used to generate cystathionine. In the alternative reaction, hydrogen sulfide was generated by condensing L-cysteine and β-mercaptoethanol. The extent of saturation of enzyme with the PLP was tested by running the activity assays in the presence or absence of 0.5 mM PLP. The reactions were performed in the presence or absence of CBS allosteric activator AdoMet in final concentration of 0.3 mM.
| Activity assay | PLP | AdoMet | CoCBS | FeCBS |
|---|---|---|---|---|
| Classical L-cystathionine-generation (U/mg of protein) | − | − | 75 ± 5 | 77 ± 9 |
| − | + | 204 ± 13 | 217 ± 15 | |
| + | − | 95 ± 3 | 101 ± 4 | |
| + | + | 372 ± 19 | 404 ± 25 | |
| Alternative H2S-generation (U/mg of protein) | − | − | 91 ± 6 | 112 ± 10 |
| − | + | 318 ± 21 | 329 ± 25 | |
| + | − | 124 ± 11 | 139 ± 6 | |
| + | + | 409 ± 24 | 341 ± 17 | |
4 Discussion
Heme replacement with another metalloporphyrin or a porphyrin with a different peripheral side chains represents a powerful approach for elucidating heme function in proteins. Typically, reconstitution of apoenzymes with heme analogs is an effective method for preparation of substituted hemoproteins (reviewed in [32]); however, this approach has been limited to proteins that can withstand heme removal and its replacement under partially denaturing conditions. Therefore, a different approach should be utilized for heme replacement in heme proteins sensitive to partial denaturation and/or simultaneous removal of other cofactors. Cystathionine β–synthase (CBS) of higher organisms is the only PLP-dependent enzyme known so far that also contains a heme (FePPIX) moiety. CBS condenses serine and homocysteine to form cystathionine, via a classic β–replacement reaction attributed to the PLP cofactor [3, 7, 8, 33, 34]. The heme in CBS is not essential for the catalytic reaction, nor is the heme involved in redox regulation or ligand binding [7–13]. Thus, the important question that remains unresolved is why CBS enzymes of higher organisms contain a heme (FePPIX) moiety.
Our previous attempt at heme replacement in CBS employed an E. coli strain deficient in heme biosynthesis [20]. Growing this strain anaerobically eliminated the need for heme. The medium was supplemented with heme analogs such as MnPPIX or CoPPIX. CBS expressed and purified under these conditions had significantly lower specific activity. The yield of the enzyme was also low thus preventing detailed characterization. In this communication, we report expression, purification and characterization of an aerobically expressed CoCBS in iron-depleted cells in the presence of CoCl2. This procedure yielded large amounts of CoCBS indistinguishable in activity from wild type FeCBS.
Recently, we showed that repeated growing of E. coli cells in a minimal medium supplemented with cobalt chloride resulted in the introduction of cobalt into protoporphyrin IX and subsequently in the incorporation of CoPPIX into heme proteins [21]. This approach allowed for more convenient cell cultivation and protein expression using a standard or an engineered E. coli expression strain, such as Rosetta 2 (DE3) employed in the present study [20, 35]. The presence of pRARE2 plasmid in E. coli Rosetta 2 (DE3), which supplies tRNAs for 7 rare codons and thus enhances the expression of eukaryotic proteins, appeared to have a beneficial influence on two aspects of CBS expression and purification: (i) increased abundance of full-length polypeptide compared to the most likely prematurely terminated polypeptides and (ii) concomitantly decreased presence of CBS antibody positive material in the insoluble fraction.
The CoCBS was expressed as a fusion protein with a GST partner at the N-terminus, which was subsequently cleaved off with PreScission protease leaving a single extra amino acid residue, glycine, at the N-terminus of CBS [22]. The DEAE Sepharose step was subsequently used for the separation of GST and CoCBS. The aggregative tendencies of the human full length CBS often led to protein precipitation during concentration and thus resulting in a great variability in yields [5, 15, 22]. The presence of non-ionic detergent (0.01% Tween 20) in the protein exchange buffer decreased the aggregation of wild type FeCBS as well as CoCBS during concentration.
CoCBS, with CoPPIX replacing heme, was isolated on expression in cobalt-supplemented minimal medium. The results of spectral analysis of CoCBS protein and pyridine hemochromogen assay correlate very well with our previous report on CoCBS prepared by an approach employing heme-deficient E. coli [20]. The metal analysis by using ICP-OES confirmed high-level substitution of iron by cobalt in CoCBS, wherein iron content was minimized to 13.1% in CoCBS 7× and further decreased to only 7.6% in CoCBS 12×. The analytical determination of cobalt content in these CoCBS enzymes correlates with spectrophotometric estimates of CoPPIX and FePPIX content reported previously [21]. The efficiency of present metalloporphyrin replacement method correlates well with previously employed total porphyrin replacement method utilizing a heme-deficient E. coli strain: the iNOSheme expressed in the presence of MnPPIX contained ≤ 5% of iron contamination [35].
The most striking difference between CoCBS described here and the one prepared by employing CBS expression in heme biosynthesis-deficient E. coli in the presence of CoPPIX [20] is the enzymatic activity data. We found CoCBS practically indistinguishable from wild type FeCBS for two different enzymatic activities. Importantly, the protein used in these assays is pure and contains >90% Co; clearly, the high activity is not due to residual FeCBS. The low activity of CoCBS reported previously must have been a consequence of the method employed to obtain the protein. The high activity of CoCBS reported herein demonstrates not only that heme (FePPIX) is not essential for the catalytic activity of CBS, but also that it can be replaced by other metalloporphyrin (such as CoPPIX) without any decrease of enzymatic activity. The fact that a non-native metal enables full activity supports the notion that porphyrin moiety in CBS performs a structural role.
Acknowledgments
This work was supported by Postdoctoral Fellowship 0920079G from the American Heart Association (to TM), by National Institutes of Health Grant HL065217 (to JPK and JNB), by American Heart Association Grant In-Aid 09GRNT2110159 and by a grant from the Jerome Lejeune Foundation (to JPK).
The authors thank Dr. Robert L. McClain (University of Wisconsin, Madison) for assistance with the ICP-OES instrument.
Abbreviations
- δ-ALA
δ-aminolevulinic acid
- AdoMet
S-adenosyl-L-methionine
- BSA
bovine serum albumin
- CBS
cystathionine β-synthase
- Co
cobalt
- Fe
iron
- GST
glutathione S-transferase
- ICP-OES
inductively coupled plasma optical emission spectroscopy
- IPTG
isopropyl-β-D-1-thiogalactopyranoside
- PLP
pyridoxal-5′-phosphate
- PPIX
protoporphyrin IX
- SEM
standard error of the mean
- WT
wild type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mudd SH, Levy HL, Kraus JP. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Kinzler K, Vogelstein B, editors. McGraw-Hill; New York: 2001. pp. 2007–2056. [Google Scholar]
- 2.Miles EW. In: Vitamin B6. Pyridoxal phosphate. Chemical, Biochemical, and Medical Aspects, Part B. Dolphin D, Poulson R, Avramovic O, editors. John Wiley & Sons; New York: 1986. pp. 253–310. [Google Scholar]
- 3.Kery V, Poneleit L, Meyer JD, Manning MC, Kraus JP. Biochemistry. 1999;38:2716–2724. doi: 10.1021/bi981808n. [DOI] [PubMed] [Google Scholar]
- 4.Banerjee R, Evande R, Kabil O, Ojha S, Taoka S. Biochim Biophys Acta. 2003;1647:30–35. doi: 10.1016/s1570-9639(03)00044-x. [DOI] [PubMed] [Google Scholar]
- 5.Janosik M, Kery V, Gaustadnes M, Maclean KN, Kraus JP. Biochemistry. 2001;40:10625–10633. doi: 10.1021/bi010711p. [DOI] [PubMed] [Google Scholar]
- 6.Shan X, Kruger WD. Nature Genet. 1998;19:91–93. doi: 10.1038/ng0598-91. [DOI] [PubMed] [Google Scholar]
- 7.Bruno S, Schiaretti F, Burkhard P, Kraus JP, Janosik M, Mozzarelli A. J Biol Chem. 2001;276:16–19. doi: 10.1074/jbc.C000588200. [DOI] [PubMed] [Google Scholar]
- 8.Meier M, Janosik M, Kery V, Kraus JP, Burkhard P. Embo J. 2001;20:3910–3916. doi: 10.1093/emboj/20.15.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oliveriusova J, Kery V, Maclean KN, Kraus JP. J Biol Chem. 2002;277:48386–48394. doi: 10.1074/jbc.M207087200. [DOI] [PubMed] [Google Scholar]
- 10.Pazicni S, Lukat-Rodgers GS, Oliveriusova J, Rees KA, Parks RB, Clark RW, Rodgers KR, Kraus JP, Burstyn JN. Biochemistry. 2004;43:14684–14695. doi: 10.1021/bi0488496. [DOI] [PubMed] [Google Scholar]
- 11.Pazicni S, Cherney MM, Lukat-Rodgers GS, Oliveriusova J, Rodgers KR, Kraus JP, Burstyn JN. Biochemistry. 2005;44:16785–16795. doi: 10.1021/bi051305z. [DOI] [PubMed] [Google Scholar]
- 12.Cherney MM, Pazicni S, Frank N, Marvin KA, Kraus JP, Burstyn JN. Biochemistry. 2007;46:13199–13210. doi: 10.1021/bi701159y. [DOI] [PubMed] [Google Scholar]
- 13.Vadon-Le Goff S, Delaforge M, Boucher JL, Janosik M, Kraus JP, Mansuy D. Biochem Biophys Res Commun. 2001;283:487–492. doi: 10.1006/bbrc.2001.4807. [DOI] [PubMed] [Google Scholar]
- 14.Taoka S, Lepore BW, Kabil O, Ojha S, Ringe D, Banerjee R. Biochemistry. 2002;41:10454–10461. doi: 10.1021/bi026052d. [DOI] [PubMed] [Google Scholar]
- 15.Taoka S, Ojha S, Shan X, Kruger WD, Banerjee R. J Biol Chem. 1998;273:25179–25184. doi: 10.1074/jbc.273.39.25179. [DOI] [PubMed] [Google Scholar]
- 16.Taoka S, Banerjee R. J Inorg Biochem. 2001;87:245–251. doi: 10.1016/s0162-0134(01)00335-x. [DOI] [PubMed] [Google Scholar]
- 17.Taoka S, Green EL, Loehr TM, Banerjee R. J Inorg Biochem. 2001;87:253–259. doi: 10.1016/s0162-0134(01)00336-1. [DOI] [PubMed] [Google Scholar]
- 18.Carballal S, Madzelan P, Zinola CF, Grana M, Radi R, Banerjee R, Alvarez B. Biochemistry. 2008;47:3194–3201. doi: 10.1021/bi700912k. [DOI] [PubMed] [Google Scholar]
- 19.Singh S, Madzelan P, Stasser J, Weeks CL, Becker D, Spiro TG, Penner-Hahn J, Banerjee R. J Inorg Biochem. 2009;103:689–697. doi: 10.1016/j.jinorgbio.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majtan T, Singh LR, Wang L, Kruger WD, Kraus JP. J Biol Chem. 2008;283:34588–34595. doi: 10.1074/jbc.M805928200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Majtan T, Frerman FE, Kraus JP. Biometals. 2010 doi: 10.1007/s10534-010-9400-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frank N, Kent JO, Meier M, Kraus JP. Arch Biochem Biophys. 2008;470:64–72. doi: 10.1016/j.abb.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kraus JP. Methods Enzymol. 1987;143:388–394. doi: 10.1016/0076-6879(87)43068-1. [DOI] [PubMed] [Google Scholar]
- 24.Kayastha AM, Miles EW. Anal Biochem. 1991;193:200–203. doi: 10.1016/0003-2697(91)90009-i. [DOI] [PubMed] [Google Scholar]
- 25.Falk JE. Porphyrins and Metalloporphyrins: Their General, Physical, and Coordination Chemistry, and Laboratory Methods. Elsevier; Amsterdam: 1964. [Google Scholar]
- 26.Dickinson LC, Chien JC. Biochemistry. 1975;14:3526–3534. doi: 10.1021/bi00687a003. [DOI] [PubMed] [Google Scholar]
- 27.Yonetani T, Yamamoto H, Woodrow GV., 3rd J Biol Chem. 1974;249:682–690. [PubMed] [Google Scholar]
- 28.Yonetani T, Yamamoto H, Iizuka T. J Biol Chem. 1974;249:2168–2174. [PubMed] [Google Scholar]
- 29.Wang MY, Hoffman BM, Hollenberg PF. J Biol Chem. 1977;252:6268–6275. [PubMed] [Google Scholar]
- 30.Dickinson LC, Chien JC. Biochemistry. 1975;14:3534–3542. doi: 10.1021/bi00687a004. [DOI] [PubMed] [Google Scholar]
- 31.Wagner GC, Gunsalus IC, Wang MY, Hoffman BM. J Biol Chem. 1981;256:6266–6273. [PubMed] [Google Scholar]
- 32.Fruk L, Kuo CH, Torres E, Niemeyer CM. Angew Chem Int Ed Engl. 2009;48:1550–1574. doi: 10.1002/anie.200803098. [DOI] [PubMed] [Google Scholar]
- 33.Braunstein AE, Goryachenkova EV. Adv Enzymol Relat Areas Mol Biol. 1984;56:1–89. doi: 10.1002/9780470123027.ch1. [DOI] [PubMed] [Google Scholar]
- 34.Evande R, Ojha S, Banerjee R. Arch Biochem Biophys. 2004;427:188–196. doi: 10.1016/j.abb.2004.04.027. [DOI] [PubMed] [Google Scholar]
- 35.Woodward JJ, Martin NI, Marletta MA. Nat Methods. 2007;4:43–45. doi: 10.1038/nmeth984. [DOI] [PubMed] [Google Scholar]