SUMMARY
The developmental potential of human pluripotent stem cells suggests that they can produce disease-relevant cell types for biomedical research. However, substantial variation has been reported among pluripotent cell lines, which could affect their utility and clinical safety. Such cell-line-specific differences must be better understood before one can confidently use embryonic stem (ES) or induced pluripotent stem (iPS) cells in translational research. Toward this goal we have established genome-wide reference maps of DNA methylation and gene expression for 20 previously derived human ES lines and 12 human iPS cell lines, and we have measured the in vitro differentiation propensity of these cell lines. This resource enabled us to assess the epigenetic and transcriptional similarity of ES and iPS cells and to predict the differentiation efficiency of individual cell lines. The combination of assays yields a scorecard for quick and comprehensive characterization of pluripotent cell lines.
INTRODUCTION
Human embryonic stem (ES) cell lines can be cultured and expanded for many passages in vitro, without losing their ability to differentiate into all three embryonic germ layers (Thomson et al., 1998). The same is true for induced pluripotent stem (iPS) cell lines, which are obtained by reprogramming somatic cells using ectopic expression of the transcription factors OCT4, SOX2, KLF4, and C-MYC (Takahashi et al., 2007) or alternative reprogramming cocktails (reviewed in Stadtfeld and Hochedlinger, 2010). Both ES and iPS cell lines are powerful research tools and could provide substantial quantities of disease-relevant cells for biomedical research. Several groups have already used human pluripotent cell lines as a model system for dissecting the cellular basis of monogenic diseases, and the range of diseases under investigation is rapidly expanding (reviewed in Colman and Dreesen, 2009). Future applications of human pluripotent stem cell lines could include the study of complex diseases that emerge from a mixture of genetic and environmental effects; cell-based drug screening in disease-relevant cell types; and the use of pluripotent cells as a renewable source for transplantation medicine (Colman and Dreesen, 2009; Daley, 2010; Rubin, 2008).
All of these applications require the selection and characterization of cell lines that reliably, efficiently, and stably differentiate into disease-relevant cell types. However, significant variation has been observed for the differentiation efficiency of various human ES cell lines (Di Giorgio et al., 2008; Osafune et al., 2008), and further concerns have been raised about the equivalence of human ES and iPS cell lines. For example, it has been reported that human iPS cells collectively deviate from ES cells in the expression of hundreds of genes (Chin et al., 2009), in their genome-wide DNA methylation patterns (Doi et al., 2009), and in their neural differentiation properties (Hu et al., 2010). Such differences must be better understood before human ES and iPS cell lines can be confidently used for translational research. In particular, it is necessary to establish genome-wide reference maps for patterns of gene expression and DNA methylation in a large collection of pluripotent cell lines, providing a baseline against which comparisons of epigenetic and transcriptional properties of new ES and iPS cell lines can be made. Previous research has shown that human pluripotent cells exhibit highly characteristic patterns of DNA methylation and gene expression (Guenther et al., 2010; Hawkins et al., 2010; Lister et al., 2009; Müller et al., 2008). However, these studies focused on few cell lines and therefore could not systematically investigate the role of epigenetic and transcriptional variation.
In order to firmly establish the nature and magnitude of epigenetic variation that exists among human pluripotent stem cell lines, three genomic assays were applied to 20 established ES cell lines (Chen et al., 2009; Cowan et al., 2004; Thomson et al., 1998) and 12 iPS cell lines that were recently derived and functionally characterized (Boulting et al., 2011). The assays performed on each cell line included DNA methylation mapping by genome-scale bisulfite sequencing, gene expression profiling using microarrays, and a novel quantitative differentiation assay that utilizes high-throughput transcript counting of 500 lineage marker genes in embryoid bodies (EBs). Collectively, our data provide a reference of variation among human pluripotent cell lines. This reference enabled us to perform a systematic comparison between ES and iPS cell lines, to identify cell-line-specific outlier genes, and to predict each cell line's differentiation propensity into the three germ layers. Finally, we show that the differentiation propensities that we report here are highly predictive of the efficiencies by which Boulting and colleagues could direct the differentiation of the 12 iPS cell lines into motor neurons (Boulting et al., 2011).
In summary, we found that epigenetic and transcriptional variation is common among human pluripotent cell lines and that this variation can have significant impact on a cell line's utility. Our observation applies to both ES and iPS cell lines, underlining the need to carefully characterize each cell line, regardless of how it was derived. As a step toward lowering the experimental burden of comprehensive cell line characterization and to improve the accuracy over existing assays, we have combined our three genomic assays into a bioinformatic scorecard. This scorecard enables high-throughput prediction of the quality and utility of any pluripotent cell line.
RESULTS
A Reference of DNA Methylation and Gene Expression in Human ES Cell Lines
Human ES cell lines are subject to many factors of influence that could contribute to epigenetic and transcriptional variation, such as their genetic background, differences between derivation protocols, and varying cell culture conditions. To establish a baseline of variation among high-quality pluripotent cell lines, we obtained low-passage freezes of 20 well-characterized and widely used human ES cell lines (Table S1). These cell lines were cultured for several passages under standardized conditions, and we confirmed the expression of pluripotency markers by immunostainings (Figure S1A) before collecting material for genomic analysis of DNA methylation and gene expression.
DNA methylation profiling was performed by reduced-representation bisulfite sequencing (RRBS) as described previously (Gu et al., 2010; Meissner et al., 2008) and resulted in DNA methylation measurements for approximately four million individual CpG dinucleotides per cell line. The genomic coverage was sufficient to determine DNA methylation levels for three quarters of all gene promoters, the majority of CpG islands and many other genomic elements (Figures S1B and S1C). Gene expression profiling was performed using commercially available Affymetrix microarrays and gave rise to expression levels for a total of 15,210 Ensembl genes. All data are publicly available for visual browsing and download (http://scorecard.computational-epigenetics.org/).
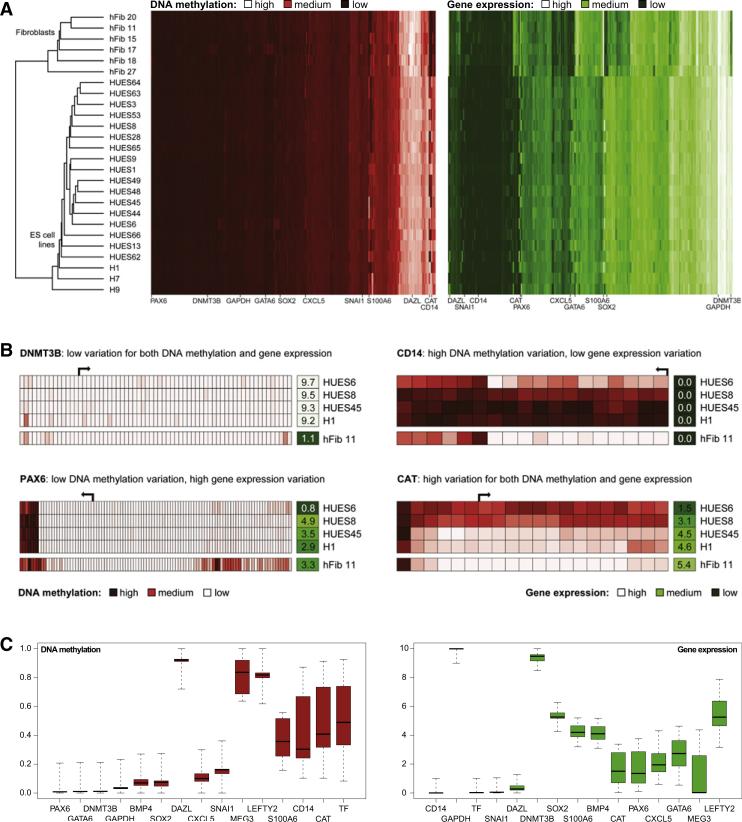
To determine whether the global patterns of DNA methylation and gene expression would segregate ES cell lines into subclasses, we performed hierarchical clustering (Figure 1A, Table S2), which also included data from the 6 primary fibroblast cell lines as nonpluripotent controls. Two well-separated clusters emerged, one comprising all ES cell lines and the other comprising all fibroblast cell lines. Within the ES cell cluster, there was some indication that cell lines derived at the same institution cluster together (HUES cell lines versus H1, H7, and H9), which is consistent with a prior study of marker gene expression in human ES cell lines (Adewumi et al., 2007). However, this trend was mild compared to the difference between pluripotent and nonpluripotent cells and did not significantly influence the results reported below.
Figure 1. DNA Methylation and Gene Expression Profiles Quantify Variation among Human ES Cell Lines.
(A) Joint hierarchical clustering of DNA methylation and gene expression in 20 human ES cell lines (“HUESx,” “Hx”) and 6 primary fibroblast cell lines (“hFibx”). Light colors indicate high levels of DNA methylation (red) or gene expression (green), and dark colors indicate low levels. Joint DNA methylation and gene expression data are available from Table S2.
(B) High-resolution view of DNA methylation and gene expression at four selected genes. DNA methylation patterns are shown for the promoter regions (–5kb to +1 kb) of representative Ensembl-annotated transcripts. Each box on the left represents a single CpG dinucleotide (dark red: high methylation, light red: little or no methylation). The single boxes on the right visualize the normalized expression levels of each gene (dark green: little or no expression, light green: high expression). The DNA methylation patterns are not drawn to scale.
(C) Boxplots of gene-specific DNA methylation (left) and gene expression levels (right) among 20 low-passage human ES cell lines, illustrating the concept of an epigenetic/transcriptional reference corridor. Boxplot boxes correspond to center quartiles, the median is marked by a black bar, and whiskers indicate the width of the reference corridor as defined in the Extended Experimental Procedures (i.e., value of the most extreme data point that is no more than 1.5 times the interquartile range from the box if the distance from the median exceeds a minimum threshold of 0.2 for DNA methylation and 1 for gene expression; otherwise these thresholds—which correspond to 20 percentage points for DNA methylation and a 2-fold change for gene expression—define the reference corridor). Data points that fall outside the whiskers are flagged as outliers and are suppressed in this figure; their position relative to the reference corridor is shown in Figure 4A.
Consistent with the overall similarity among all 20 ES cell lines, the majority of genetic loci exhibit similar DNA methylation and gene expression levels between different ES cell lines, as exemplified by the DNA methyltransferase gene DNMT3B (Figure 1B). However, a moderate number of genes show variable DNA methylation and/or gene expression levels. For example, the antioxidant gene CAT exhibits substantial and correlated variation of DNA methylation and gene expression; the developmental regulator PAX6 exhibits gene expression variation and a consistently unmethylated gene promoter; and the macrophage/granulocyte surface marker CD14 exhibits DNA methylation variation while not being expressed in any of the 20 ES cell lines (Figure 1B). Importantly, cell-line-specific differences were maintained when we collected biological replicates from different passages of the same cell line (Figure S1D).
To investigate the variation observed among human ES cell lines in a more quantitative manner, we calculated, for each gene, the distribution of DNA methylation and gene expression among the 20 ES cell lines (Table S3). The resulting “reference corridor” quantifies the range of DNA methylation and gene expression values for a given gene (or genomic region) in a reference set of pluripotent cell lines. Any measurement that falls outside of this corridor is regarded as an outlier and could potentially affect that cell line's functional properties. We illustrate the concept of the reference corridor using boxplots (Figure 1C), which display the median and range of observed DNA methylation/expression levels for representative genes with different degrees of variability. For each gene (or genomic region), these plots impose upper and lower thresholds between which the DNA methylation/expression levels must fall to be considered “within the range of the current ES cell reference.” With this reference in hand, it becomes possible to determine the number and identity of deviations in any pluripotent cell line by using a statistical outlier filter (Tukey, 1977) and to investigate the causes and potential consequences of this variation.
Causes and Consequences of Epigenetic and Transcriptional Variation among Human ES Cell Lines
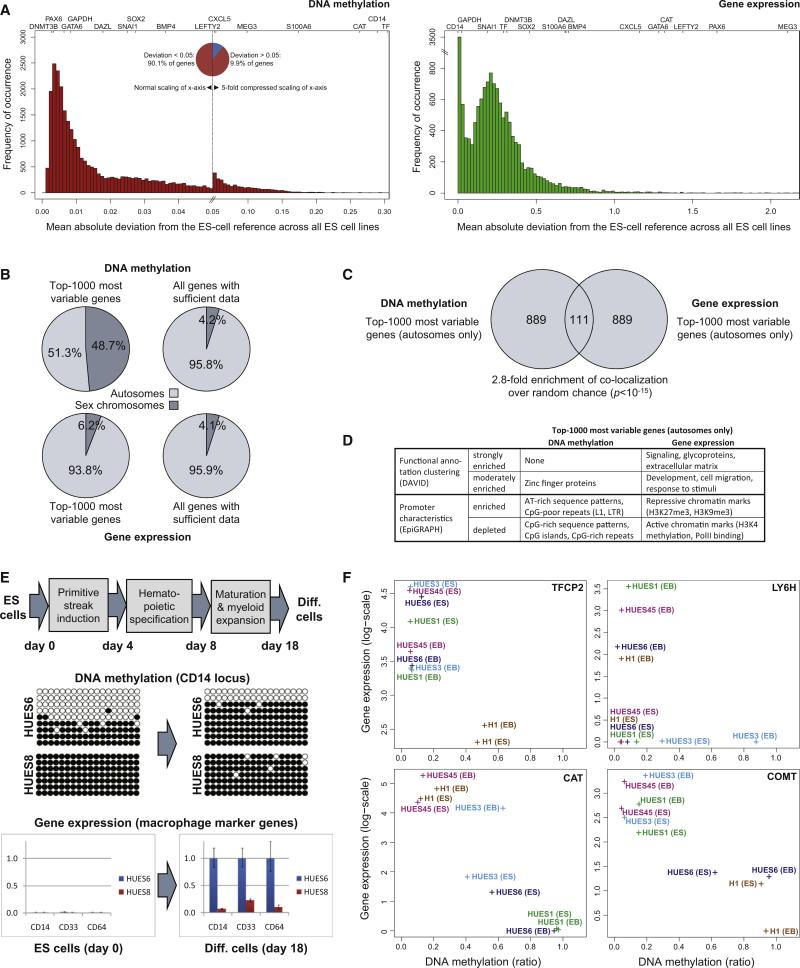
Plotting the deviation from the ES cell reference maps for all genes confirmed our initial observation that epigenetic and transcriptional variation focuses on a subset of genes, whereas most genes exhibit little deviation from the reference in any of the ES cell lines (Figure 2A). Specifically, 13% of genes account for half of the total DNA methylation variation, and 20% of genes account for half of the total gene expression variation (Table S3). As one might have expected, housekeeping genes such as GAPDH were among the least variable genes between the cell lines. Similarly, we observed relatively low variation among several genes that are highly expressed in pluripotent cell lines, including SOX2 and DNMT3B. In contrast, moderate to high levels of variation were found for several genes that regulate embryonic development and are induced upon ES cell differentiation, including GATA6, LEFTY2, and PAX6. Finally, a small number of loci exhibited highly variable DNA methylation levels between cell lines, ranging from close to 0% methylation in some cell lines to almost 100% methylation in other cell lines. The most prominent cases were CAT and CD14 (both shown in Figure 1B) and the transferrin-encoding gene TF.
Figure 2. Epigenetic and Transcriptional Variation Targets Specific Genes and Influences Cellular Differentiation.
(A) Distribution of cell-line-specific variation in terms of DNA methylation and gene expression. The histogram shows the number of genes (y axis) that fall into each interval when calculating the mean absolute deviation of individual ES cell lines relative to the reference of all other ES cell lines (x axis). The position of selected genes within each histogram is highlighted on top. Note that the DNA methylation histogram (left) is extremely skewed; for better representation the x axis has been compressed 5-fold for the right half of the diagram, which gives rise to an artificial peak in the center of the histogram. The gene expression histogram (right) is characterized by a strong peak at zero, due to a large number of genes with zero expression and zero variation in all ES cell lines. Variation data for all genes are available from Table S3.
(B) Chromosomal distribution of the 1000 most variable genes in terms of DNA methylation (top left) and gene expression (bottom left). For comparison, the diagram also shows the chromosomal distribution of all genes with sufficient DNA methylation (top right) or gene expression data (bottom right).
(C) Comparison of the 1000 most variable genes in terms of DNA methylation (left) and gene expression (right). To prevent bias due to the chromosomal differences of male versus female cell lines, all X-linked and Y-linked genes were excluded. Significance of overlap was confirmed by Fisher's exact test.
(D) Functional and structural characteristics of the 1000 most variable genes in terms of DNA methylation (left) and gene expression (right). Functional annotation clustering was performed with the DAVID software (Huang et al., 2007), and the promoter characteristics were analyzed by the EpiGRAPH web service (Bock et al., 2009). This panel provides a summary of the results; the full results tables are available online http://scorecard.computational-epigenetics.org/.
(E) Epigenetic and transcriptional differences between two ES cell lines (HUES6 and HUES8) subjected to a defined hematopoietic differentiation protocol. DNA methylation levels were measured by clonal bisulfite sequencing at day 0 and day 18 of the differentiation protocol. White beads correspond to unmethylated CpGs, and black beads correspond to methylated CpGs. Rows correspond to individual clones, and columns correspond to specific CpGs in the promoter region of CD14. Similarly, gene expression of CD14 and two additional macrophage marker genes (CD33 and CD64) was measured by qPCR in two independent experiments (shown are three technical replicates) at day 0 and day 18 of the differentiation protocol. Error bars indicate ± one standard deviation.
(F) Cell-line-specific DNA methylation and gene expression levels at four genes with a known role in hematopoiesis (TFCP2, LY6H) and neural processes (COMT, CAT). Each data point denotes the combined DNA methylation (x axis) and gene expression (y axis) levels of an ES cell lines (“ES”) or the corresponding 16 day embryoid body (“EB”).
To gain insight into potential causes of the differences in variation, we bioinformatically compared the top 1000 most variable genes with all other genes that were covered by our dataset. We detected a striking enrichment of DNA methylation-variable genes located on the sex chromosomes (Figure 2B), which is—at least partially—due to the fact that we included both male and female cell lines in our comparison. The measured levels of Y-linked methylation and transcription vary between cell lines because the Y chromosome is absent in female lines. Similarly, DNA methylation measurements vary between cell lines because female ES cell lines often exhibit high levels of DNA methylation on the inactivated X chromosome, which is not observed in male cell lines (Lengner et al., 2010). As X-linked and Y-linked genes were such a significant source of variation, we were concerned that they might interfere with our ability to identify gene features that more subtly influence transcriptional or epigenetic variability. We therefore excluded all loci that map to the sex chromosomes from subsequent analyses.
We also found significant overlap between the sets of genes that showed the greatest epigenetic and transcriptional variability (Figure 2C). This observation suggests that DNA methylation may play a regulatory role for a subset of the most transcriptionally variable genes. Furthermore, bioinformatic analysis identified significant enrichment of specific gene functions and promoter patterns that characterize highly variable genes (Figure 2D). The most variably expressed genes were strongly enriched for Gene Ontology categories related to cellular signaling, development, and the response to external stimuli. In contrast, genes with variable DNA methylation levels showed little evidence of enrichment for any particular function. Instead, we found that the promoters of these genes shared common structural characteristics. Most notably, these promoters were relatively depleted in CpG dinucleotides compared to the promoters of nonvariable genes (most of which are CpG island promoters). Comparatively low CpG frequency is a known characteristic of genomic regions that are susceptible to variation in DNA methylation (Bock et al., 2008; Keshet et al., 2006; Meissner et al., 2008). These observations suggest that variation among ES cell lines is not random but follows certain biological principles.
To test whether epigenetic variation has consequences for ES cell function, we compared the differentiation potential of ES cell lines that differed in the DNA methylation level at the CD14 locus, which stood out as one of the most epigenetically variable genes in our dataset (Figure 2A). CD14 encodes a well-characterized macrophage/granulocyte surface marker and is functionally important for the innate immune response to bacterial lipopolysaccharide (Kitchens, 2000). Epigenetic defects at this locus could therefore compromise the ability of ES cell lines to differentiate into CD14-positive macrophages. We selected two ES cell lines that differed in their DNA methylation at the CD14 locus (HUES6: partial methylation, HUES8: complete methylation) and performed directed differentiation toward the hematopoietic lineage (Figure 2E). We found that HUES6 was able to upregulate CD14 expression, whereas the CD14 locus remained fully methylated and silent during hematopoietic differentiation of the HUES8 cell line. Two additional macrophage marker genes (CD33 and CD64) were also more highly expressed in hematopoietic cells derived from HUES6 compared to HUES8, indicating that the latter cell line is compromised in its ability to produce macrophage-like cells in vitro.
To identify further examples of cell-line-specific DNA methylation defects that might interfere with differentiation, we compared the DNA methylation and gene expression levels of five ES cell lines with their corresponding day 16 EBs (Figure 2F). Among the most interesting cases were two additional genes with a known role in hematopoietic cells: the alpha-globin transcription factor TFCP2 is hypermethylated and lowly expressed in the H1 ES cell line, indicating that this cell line may be less suitable for studying erythrocyte differentiation; and the lymphocyte antigene LY6H is hypermethylated and silenced specifically in the HUES3 cell line. We also found that the widely varying DNA methylation and gene expression levels of CAT (Figure 1B) were maintained during EB differentiation (Figure 2F). Given the central role of CAT in the response to oxidative stress, these differences could be relevant for a wide range of cell types including hematopoietic and neural cells. COMT is another example with potential relevance for neural cells. It is hypermethylated and downregulated in HUES6 and H1, suggesting that these two cell lines may produce neural cells that are defective in their ability to inactivate neurotransmitters, which is an important function of the COMT gene. All of these cases highlight the relevance of monitoring DNA methylation and gene expression to prospectively identify cell-line-specific defects that could interfere with their differentiation or the functional properties of derived cell types.
Comparison of DNA Methylation and Gene Expression Variation between Human ES and iPS Cell Lines
The reference maps of variation among ES cell lines enabled us to systematically address the contentious issue of epigenetic and transcriptional differences between human ES and iPS cell lines (Chin et al., 2009, 2010; Doi et al., 2009; Guenther et al., 2010; Newman and Cooper, 2010). Four technical aspects distinguish our comparison from previous studies: (1) we compare both DNA methylation and gene expression in the same cell lines; (2) we use a relatively large sample size of 20 ES and 12 iPS cell lines; (3) all cell lines were maintained under the same standardized culture conditions; (4) we compare each ES or iPS cell line individually against the ES cell reference, rather than comparing the set of all ES cell lines against the set of all iPS cell lines (this approach increases the robustness toward a small number of strong outliers that could easily skew a group-wise comparison).
The iPS cell lines were derived by Boulting and colleagues using retroviral transduction of OCT4, SOX2, and KLF4 of fibroblasts obtained from six unrelated donors. This “test set” of 12 well-characterized human iPS cell lines is available as an independent resource via the Harvard Stem Cell Institute's iPS Cell Core Facility (Boulting et al., 2011). To match the passage numbers of the 20 ES cell lines and to avoid increased noise in extremely low-passage iPS cell lines (Chin et al., 2009; Polo et al., 2010), we focused our analysis on iPS cells in the range of passage 15 to passage 30 (Table S1). DNA methylation and gene expression profiling were performed on these iPS cell lines in the same way as for the ES cell lines.
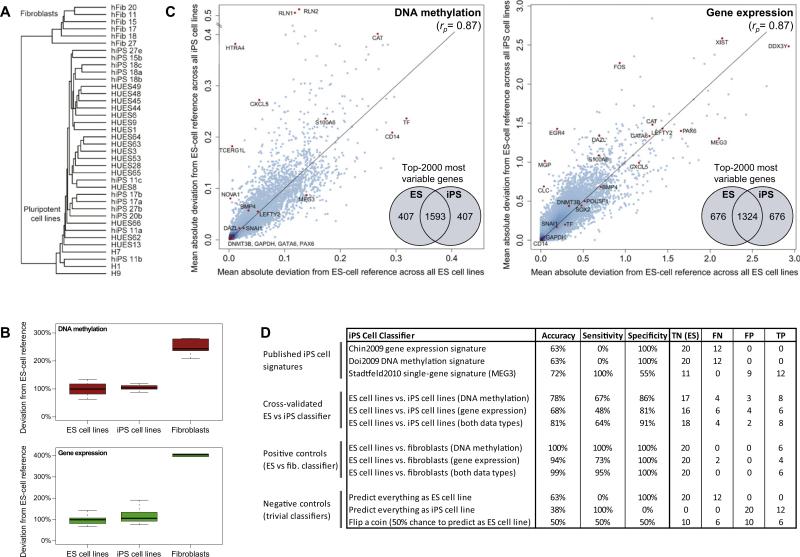
Hierarchical clustering confirmed that all iPS cell lines grouped with the ES cell lines, rather than with the fibroblasts (Figure 3A and Figure S2A). No clear-cut separation between ES and iPS cell lines was observed, indicating that their global DNA methylation and gene expression profiles are highly similar. However, hierarchical clustering has known limitations (Allison et al., 2006), which may prevent it from picking up subtle differences between these ES and iPS cell lines.
Figure 3. Cell-Line-Specific Deviation from the Reference Is Slightly Higher in iPS than in ES Cell Lines.
(A) Joint hierarchical clustering of 12 iPS cell lines (“hiPSx”), 20 ES cell lines (“HUESx,” “Hx”), and 6 primary fibroblast cell lines (“hFibx”). An extended version that includes heatmaps is available from Figure S2A. The numbers of the iPS cell lines connect them to the fibroblasts from which they were derived (e.g., hFib 18 was used to generate hiPS 18a, 18b, and 18c).
(B) Boxplots of the cell-line-specific deviation from the ES cell reference, averaged over all genes and scaled such that the mean deviation of the 20 ES cell lines is equal to 100%.
(C) Scatterplots comparing the gene-specific deviation of 20 ES cell lines (x axis) with the gene-specific deviation of 12 iPS cell lines (y axis), in both cases measured relative to the ES cell reference and averaged over all ES or iPS cell lines, respectively. To prevent comparing cell lines to themselves, each ES cell line was temporarily removed from the ES cell reference when it was compared to the reference. Selected genes are highlighted in orange, rp refers to Pearson's correlation coefficient, and the inset Venn diagrams visualize the overlap between the 2000 most deviating genes in ES versus iPS cell lines. The reprogramming factors OCT4, SOX2, and KLF4 were excluded from the DNA methylation analysis because transgene silencing gives rise to spurious hypermethylation among the iPS cell lines (Figure 4A and Figure S2C).
(D) Performance table summarizing the predictive power of three previously published iPS cell signatures and three newly derived classifiers for distinguishing between ES and iPS cell lines. For comparison, the table also lists the performance of three newly derived classifiers for distinguishing between ES cell lines and fibroblasts (positive controls) and the performance of three trivial classifiers (negative controls). Shown are the prediction accuracy, sensitivity, and specificity for identifying iPS cell lines (true positives, TP) among ES cell lines (true negatives, TN), while minimizing the number of cell lines that are incorrectly predicted as iPS cell lines (false positives, FP) or incorrectly predicted as ES cell lines (false negatives, FN). To increase the robustness of the results, all values were averaged over 100 randomized repetitions of the cross-validation. Minor numerical inconsistencies in the table are due to rounding all values to whole numbers.
For a more quantitative comparison, we calculated—for each cell line—the mean deviation from the ES cell reference over all genes (Figure 3B). The results indicate that many iPS cell lines fall well within the range of global deviation that is also observed among ES cell lines, although the average deviation is somewhat higher among the iPS cell lines compared to the ES cell lines. We also investigated whether there are any specific marker genes that reproducibly distinguish ES and iPS cell lines. To that end, we calculated—for each gene—the mean deviation from the ES cell reference separately in all ES and iPS cell lines and plotted these gene-specific deviations against each other (Figure 3C). The vast majority of genes exhibit similar deviation averages in ES cell lines and in iPS cell lines. This is true for highly variable genes (e.g., CAT) as well as for genes that exhibit little variation (e.g., GAPDH). This global concordance is also reflected in high correlation between deviation in ES and iPS cell lines (Pearson's r = 0.87). However, for a small number of genes we observed substantially increased deviation from the ES cell reference among the iPS cell lines (Figure 3C). Some of these genes were hypermethylated in a subset of iPS lines, for example the protease HTRA4 (9 out of 12 iPS cell lines) and the relaxin hormones RLN1/2 (9 out of 12 iPS cell lines; although also hypermethylated in one ES cell line). Others were transcribed at higher levels in some iPS cell lines, such as the transcription factor EGR4 (6 out of 12 iPS cell lines) and the matrix Gla protein MGP (3 out of 12 iPS cell lines).
The HTRA4 gene, which is most frequently hypermethylated in iPS cell lines compared to ES cell lines (9 out of 12 iPS versus 0 out of 20 ES cell lines), is also hypermethylated in all six fibroblast cell lines. This observation suggests that somatic cell memory (i.e., incomplete reprogramming of DNA methylation at genes that are methylated in fibroblasts) might provide a potential explanation for the deviation in some iPS cell lines. To address this point in a quantitative way, we built a statistical model that estimates the relative contribution of epigenetic memory to the DNA methylation levels observed in iPS cell lines. Specifically, we asked how much better we can predict each gene's average DNA methylation in iPS cell lines if we know its DNA methylation state in both fibroblasts and ES cells, compared to knowing only its DNA methylation state in ES cells. This question can be addressed by comparing the predictive power of linear models that implement both explanations. The results were highly conclusive: including the DNA methylation in fibroblasts led to a significantly more accurate model (p < 10–8), but the increase in accuracy was extremely low (Δr2 < 10–5). Similar results were also obtained for somatic memory of gene expression (p < 10–8, Δr2 ≈ 10–4), indicating that somatic memory cannot explain more than a very small fraction (0.01% to 0.001%) of the DNA methylation and gene expression observed in human iPS cell lines.
Performance Evaluation of Classifiers for Distinguishing between ES and iPS Cell Lines
The analysis described above identified small but significant differences between the ES cell lines and iPS cell lines. Two alternative models could explain these observations. On the one hand, these differences could systematically affect all iPS cell lines; on the other hand, they could be specific to a subset of “deviant” iPS cell lines. To quantitatively address this issue, we reframed them as a classification problem: Can we use DNA methylation and gene expression profiles to accurately predict whether a specific cell line is an ES or iPS cell line?
Several gene signatures of differences between human ES and iPS cell lines have been reported in the literature (Chin et al., 2009; Doi et al., 2009; Stadtfeld et al., 2010). We started our prediction efforts by evaluating the predictive power of three published signatures on the current sample set. (1) The iPS-specific transcription signature reported by Chin et al. exhibits significant overlap with the set of genes that are more highly expressed in iPS than in ES cell lines in the current study (odds ratio = 1.54, p = 0.01, Table S4). However, the overlap is low in absolute terms and insufficient for correctly identifying individual iPS cell lines (Figure 3D). (2) The iPS-specific DNA methylation signature reported by Doi et al. also shows a trend toward being consistent with the current sample set (odds ratio = 1.58, p = 0.73, Table S4), but this trend was not significant and also insufficient for correctly identifying individual iPS cell lines (Figure 3D). Importantly, a much higher concordance was observed for the fibroblast-specific DNA methylation signature from the same study (odds ratio = 152.03, p < 10–15, Table S4), suggesting that the low concordance for the iPS-specific DNA methylation signature cannot be explained by the different DNA methylation assays used. (3) The Gtl2/MEG3 single-gene signature that was reported by Stadtfeld et al. in mouse exhibited 100% sensitivity but only moderate specificity in our sample set (Figure 3D). Almost half of the ES cell lines were MEG3 negative and thus incorrectly classified as iPS cell lines (Figure S2A). It is not possible to test whether absence of MEG3 expression has the same consequences as reported in mouse, namely to interfere with normal development in the tetraploid embryo complementation assay (Stadtfeld et al., 2010). However, we found no evidence that would speak against using MEG3-negative ES or iPS cell lines in biomedical research. Specifically, MEG3-negative ES cell lines exhibit similar levels of variation in DNA methylation and gene expression as did MEG3-positive ES cell lines (Figure S2B), and several MEG3-negative ES cell lines have been widely and productively used for in vitro studies.
Finally, we tested whether we could use the current dataset to develop a more accurate classifier for distinguishing between ES and iPS cell lines. To minimize the risk of overfitting to the training data, or overestimating the prediction accuracy of our classifier, we employed a stringent statistical learning approach (Hastie et al., 2001). Specifically, we abstained from any manual parameter optimization or feature selection (which are notorious for inflating accuracies when used incorrectly), and we assessed the performance of the trained classifiers only on test cases that were not included in the training dataset. The best classifier—a support vector machine trained on DNA methylation and gene expression data—achieved an overall accuracy of 81%, which outperformed all three previously reported iPS gene signatures. The classifier's specificity was high (91%), indicating that few ES cell lines were incorrectly predicted to be iPS cell lines. However, it achieved only moderate sensitivity (64%), hence there were more iPS cell lines predicted to be ES cell lines than vice versa. In summary, these results indicate that most, but not all, iPS cell lines exhibit characteristic DNA methylation and/or gene expression profiles by which they can be distinguished from ES cell lines.
A Scorecard for Evaluating the Quality and Utility of Human Pluripotent Cell Lines
It has become clear from our analysis so far that human pluripotent cell lines vary in their DNA methylation and gene expression (Figure 1), which can have functional implications (Figure 2) and affects both ES and iPS cell lines (Figure 3). These results indicate that all ES and iPS cell lines should be carefully monitored for DNA methylation or gene expression alterations that could interfere with an intended application or confound biological interpretations. To provide an informative and practically useful method for high-throughput cell-line characterization, we bioinformatically integrated several genomic assays into a scorecard that measures the quality and utility of any human pluripotent cell line.
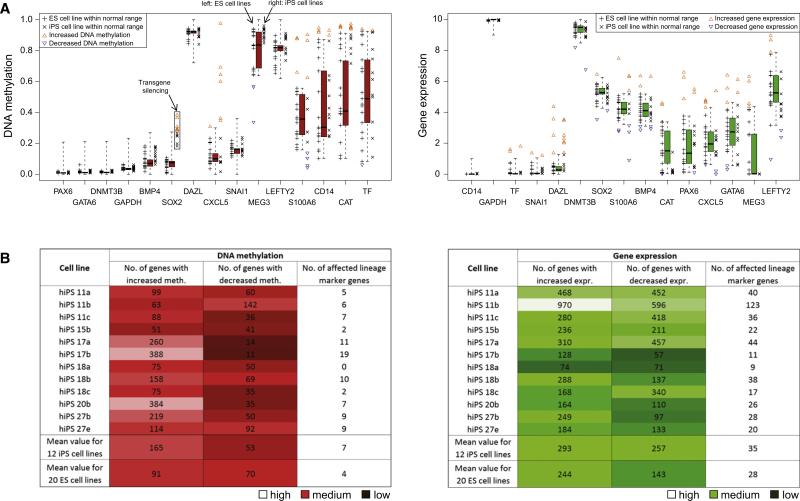
In a first step, we compared the DNA methylation and gene expression profiles of the 12 iPS cell lines with the ES cell reference, in order to identify iPS cell lines with epigenetic or transcriptional defects that might interfere with motor neuron differentiation. The hierarchical clustering already yields important information (Figure 3A): All 12 iPS cell lines globally cluster with the ES cell lines, confirming that no partially reprogrammed or grossly abnormal cell lines were included in our study. Next, we tested for each gene whether or not its DNA methylation and gene expression levels fall within the range observed among ES cell lines (Figure 4A). Genes outside of this range were flagged, and the number and identity of these outlier genes were tracked for each iPS cell line. The results of the outlier detection were summarized as a “deviation scorecard” (Figure 4B). It is apparent from this scorecard that individual iPS cell lines can harbor several hundred outlier genes. Importantly, this was also true for the ES cell lines we studied, and it is likely that not all outliers will have detectable functional consequences. We manually inspected the extended version of the deviation scorecard (Table S5), searching for known genes that might specifically interfere with neural differentiation or motor neuron function. One cell line (hiPS 17a) was flagged because it exhibits significantly increased DNA methylation at the glutamate receptor gene GRM1, a gene that is important for motor neuron function and survival (Nistri et al., 2006). In contrast, if we were studying pancreatic differentiation rather than motor neuron function, we might have kept hiPS 17a but avoided hiPS 27b due to hypermethylation at the pancreatic transcription factor PAX4.
Figure 4. Comparison with the Reference Corridor Identifies Cell-Line-Specific Outlier Genes.
(A) Distribution of gene-specific DNA methylation (left) and gene expression levels (right) among 20 ES cell lines and 12 iPS cell lines, plotted against the ES cell reference corridor (cf. Figure 1C). ES or iPS cell lines that fall outside of the corridor are highlighted by colored triangles.
(B) Deviation scorecard summarizing the cell-line-specific number of outliers relative to the ES cell reference, in terms of DNA methylation (left) and gene expression (right). As an additional indication of a cell line's quality, the scorecard lists the number of affected lineage marker genes. The table also shows the mean number of deviating genes in the 20 low-passage ES cell lines (bottom row), providing an indication of what numbers are within a range that is also observed among low-passage ES cell lines. A more comprehensive version of this scorecard is available from Table S5.
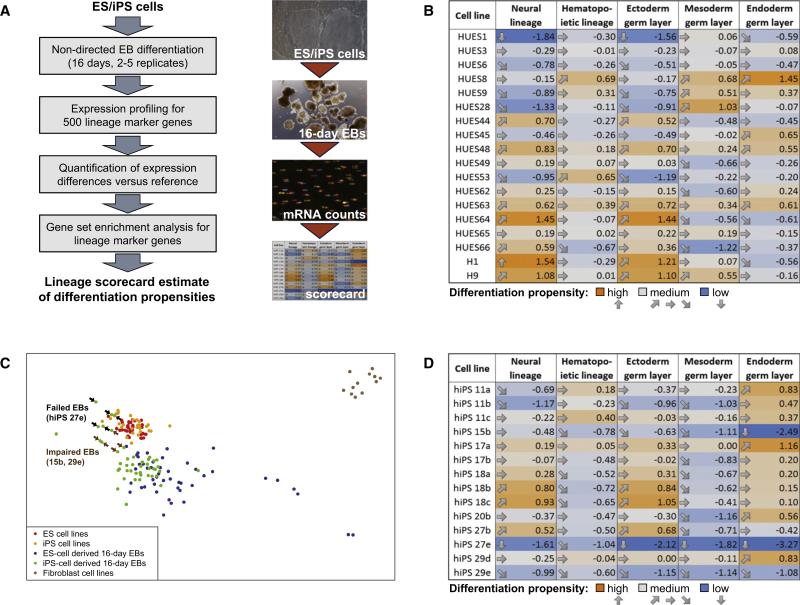
In summary, DNA methylation and gene expression profiling in combination with bioinformatic comparison to an ES cell reference provide a quick and comprehensive method for excluding cell lines that could be problematic for an intended application. However, there may be other characteristics of a cell line that we cannot readily predict from epigenetic and transcriptional profiles, for example its specific genetic background or the presence of acquired mutations in key developmental genes. To overcome these limitations, we sought to complement the “deviation scorecard” with a “lineage scorecard” that directly reflects a cell line's in vitro differentiation potential. To be practically useful, such a lineage scorecard cannot rely on expensive and time-consuming directed differentiation protocols. Instead we chose a simple nondirected EB differentiation assay and combined it with highly quantitative gene expression profiling and a bioinformatic algorithm that quantifies a cell line's differentiation propensity for multiple lineages. The experimental and bioinformatic protocol of this quantitative differentiation assay is outlined in Figure 5A and described in more detail in Figure S4 and in the Extended Experimental Procedures.
Figure 5. A Quantitative Differentiation Assay Measures Cell-Line-Specific Differentiation Propensities.
(A) Outline of the lineage scorecard assay for quantifying cell-line-specific differentiation propensities using a combination of nondirected EB differentiation, highly quantitative expression profiling, and bioinformatic analysis of lineage marker gene enrichment.
(B) Lineage scorecard summarizing cell-line-specific differentiation propensities of a set of low-passage human ES cell lines. The numbers indicate relative enrichment (positive values) or depletion (negative values) of lineage marker expression in the EBs derived from each cell line. An ES cell line will exhibit a differentiation propensity of zero if it differentiates just like the average of all other ES cell lines that were used to calibrate the assay. Values should be interpreted relative to each other, with higher numbers indicating higher differentiation propensities and lower values indicating lower differentiation propensities, while the absolute values have no measurement unit and no direct biological interpretation. Gene lists, expression values, and gene-specific enrichment values are available from Table S6.
(C) Multidimensional scaling map of the transcriptional similarity between ES and iPS cell lines, ES-derived and iPS-derived EBs, and primary fibroblast cell lines. Each point corresponds to a single biological replicate. Cell lines that were impaired or unable to form normal EBs are highlighted by arrows.
(D) Lineage scorecard summarizing cell-line-specific differentiation propensities of a set of human iPS cell lines. The scorecard was derived in the same way as Figure 5B, and all values were normalized relative to the ES cell reference. The scores were calculated across all biological replicates that were available for each cell line. Further details on single biological replicates and the reproducibility of the lineage scorecard are available from Table S6G.
To test and calibrate the lineage scorecard for pluripotent cells, we initially applied it to our reference set of 20 ES cell lines. Embryoid bodies were obtained in biological duplicate for each ES cell line, RNA was collected and profiled for the expression levels of 500 marker genes, and the cell-line-specific differentiation propensities were estimated for each of the three germ layers as well as for the neural and hematopoietic lineages (Figure 5B, Table S6). The resulting lineage scorecard pinpoints quantitative differences among the cell-line-specific differentiation propensities. For example, HUES8 showed the greatest propensity for endoderm differentiation, corroborating previous results showing that this cell line performs well in directed endoderm differentiation (Osafune et al., 2008). This result may also explain why HUES8 is frequently used for directed endoderm differentiation (Borowiak et al., 2009). In contrast, H1 and H9 received high scores for neural lineage differentiation (Figure 5B), suggesting that they might be an excellent choice for applications in the study or treatment of neural degeneration. These cell lines indeed performed well in a recent report of directed differentiation into motor neurons (Hu et al., 2010).
We performed several additional validations of the lineage scorecard, in order to establish its utility for quantifying cell-line-specific differentiation propensities. First, we compared the differentiation propensities determined by the lineage scorecard with the expression levels of five widely used lineage marker genes (NES, TUBB3, KDR, ACTA2, AFP) and found good qualitative agreement (Figure S3A). Second, we subjected four ES cell lines to two differentiation protocols that were biasing cells toward ectoderm and mesoderm, respectively. Cell lines that were cultured in the presence of Noggin and an ALK inhibitor (SB431542) to promote ectoderm differentiation exhibited substantially increased ectoderm scores and lower mesoderm scores, compared to cell lines that were cultured in the presence of Activin A and BMP4 to promote mesoderm differentiation (Figure S3B). These validation data suggested that the lineage scorecard accurately detects differences in the differentiation propensity of human pluripotent cell lines.
We next performed nondirected differentiation of 14 iPS cell lines into EBs and profiled the expression levels of the 500 marker genes after 16 days of EB differentiation. To globally assess the similarity between ES cell- and iPS cell-derived EBs, we calculated a two-dimensional similarity map of all biological replicates (Figure 5C). The results were consistent with Figure 3, indicating that most, but not all, EBs can be identified as ES cell or iPS cell derived. Furthermore, the scorecard predicted that three iPS cell lines had an impaired ability to differentiate (hiPS 15b, hiPS 27e, and hiPS 29e), which might limit their usefulness for many applications. Indeed, the lineage scorecard indicates that the neural differentiation propensity of hiPS 27e and hiPS 29e is very low, whereas the predicted neural differentiation propensity of hiPS 15b is only marginally reduced relative to an average human ES cell line (Figure 5D). This prediction is consistent with observations by Boulting and colleagues, who showed that lines 27e and 29e are impaired in motor neuron-directed differentiation, whereas line 15b differentiated relatively well (Boulting et al., 2011). In addition, line 27e seemed to be impaired in its ability to differentiate into any germ layer. To confirm this prediction for an additional germ layer, we performed flow cytometry to analyze the percentage of cells that expressed the endodermal marker gene AFP in dissociated EBs (Figure S3C). The percentage was substantially lower in hiPS 27e as compared to hiPS 17a (which we used as a control), providing further confirmation of the lineage scorecard's ability to detect cell-line-specific differences in the differentiation propensities.
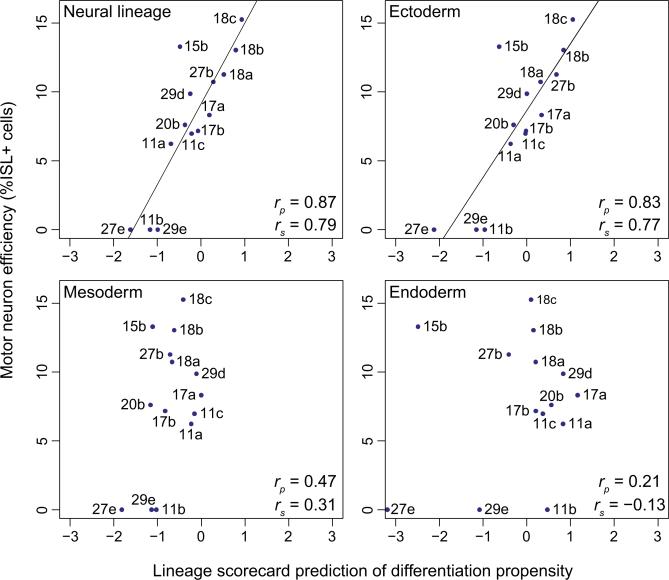
Based on the results of the lineage scorecard, hiPS 18b, hiPS 18c, and hiPS 27b appear to be well-suited for studying neural function in vitro, as these cell lines obtained high scores for ectoderm and neural differentiation propensity. Independent results obtained in the study by Boulting et al. provide an opportunity to quantitatively test these predictions. They used the test set of iPS cell lines, applied a 32 day motor neuron-directed differentiation protocol (Di Giorgio et al., 2008), and then quantified the efficiency with which each cell line could be differentiated into motor neurons. When we compared the scorecard predictions for neural differentiation for each iPS cell line with the actual motor neuron differentiation efficiency they observed (Figure 6, Table S7), we found a remarkably high correlation (Pearson's r = 0.87). Notably, the three iPS cell lines that were predicted to behave optimally by our scorecard (hiPS 18b, hiPS 18c, and hiPS 27b) were all among the cell lines they found to differentiate best into motor neurons. The high correlation between the lineage scorecard predictions and the experimentally determined differentiation efficiencies was specific for the ectoderm germ layer and did not extend to mesoderm or endoderm (Figure 6). This final observation shows that the scorecard can detect lineage-specific differences in the differentiation propensities of a given cell line, rather than merely measuring the overall recalcitrance or amenability of a cell line toward differentiation into any sort of cell.
Figure 6. The Lineage Scorecard Predicts Cell-Line-Specific Differences in the Efficiency of Motor Neuron Differentiation.
Correlation between the lineage scorecard estimates for the neural lineage and three germ layers versus the cell-line-specific efficiency of directed differentiation into motor neurons (rp, Pearson's correlation coefficient; rs, Spearman's correlation coefficient). Motor neuron efficiencies were measured by the percentage of ISL1-positive cells at the end point of a 32 day neural differentiation protocol. Further details including biological replicates and standard errors are available from Table S7.
In summary, we have described how a “deviation scorecard” derived from genome-wide maps of DNA methylation and gene expression can have utility for predicting which iPS cell lines should be avoided for a given application. In addition, we developed a “lineage scorecard” that combines simple nondirected differentiation with RNA counting, which could predict the efficiency with which iPS cell lines made motor neurons in an independent study (Boulting et al., 2011). Together, these scorecards enabled us to predict the quality and utility of more than 30 pluripotent cell lines for a broad range of applications.
DISCUSSION
To better understand the causes and consequences of variation among human pluripotent cell lines, we used genomic methods to characterize a panel of 20 ES cell lines and 12 iPS cell lines. All cell lines exhibited similar DNA methylation and gene expression levels, which clearly denoted them as pluripotent and set them apart from somatic cells. Despite their global similarity, we could identify in each cell line a number of genes that deviated from the DNA methylation or gene expression levels of the other cell lines. These cell-line-specific outliers were relatively stable over time, and our dataset suggests that some may have functional consequences, for example by interfering with differentiation into certain cell types. Cell-line-specific outliers were slightly more prevalent among iPS cell lines than among ES cell lines, but we could not find any epigenetic or transcriptional deviation that was unique to and shared by all iPS cell lines. This observation was confirmed by developing bioinformatic classifiers, which could correctly identify most but not all iPS cell lines in our dataset based on their DNA methylation and gene expression profiles.
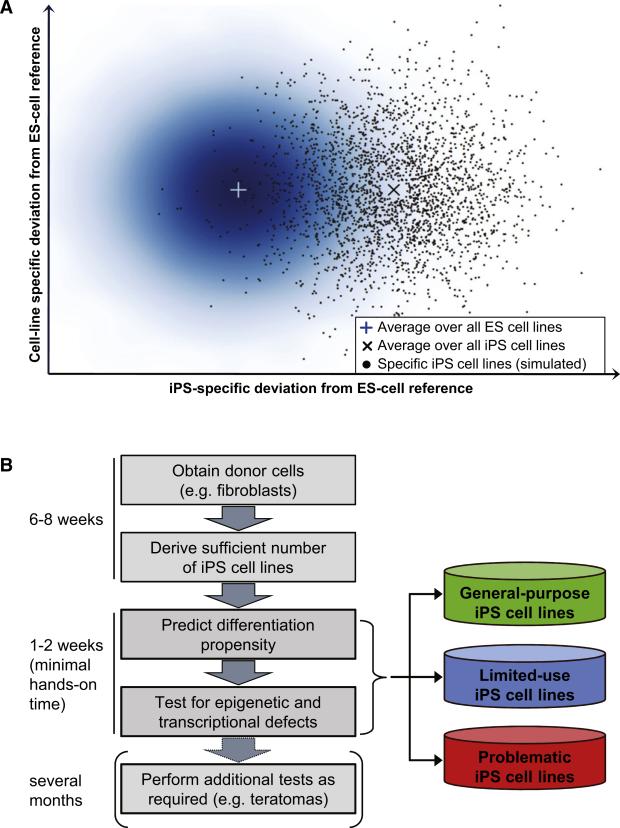
These results suggest that ES and iPS cells should not be regarded as one or two well-defined points in the cellular space but rather as two partially overlapping point clouds with inherent variability among both ES and iPS cell lines (Figure 7A). In this model, a single iPS cell line can be indistinguishable from ES cell lines, even though there is a difference in our current dataset between the average ES cell line and the average iPS cell line (denoted by the two crosses in Figure 7A).
Figure 7. The Scorecard Enables Quick and Comprehensive Characterization of Human Pluripotent Cell Lines.
(A) Schematic illustration of the similarity between ES and iPS cell lines in the epigenetic and transcriptional space. The density plot on the left depicts the variation observed among human ES cells. The two crosses indicate the (hypothetical) average of all ES and iPS cell lines, which this study approximated by profiling 20 human ES cell lines and 12 human iPS cell lines. The scatterplot on the right simulates the distribution of a large number of human iPS cell lines, taking into account their moderately increased variation (Figure 3B) as well as the observation that a minority of iPS cell lines were indistinguishable from ES cell lines (Figure 3D). Gaussians were used to simulate the ES cell and iPS cell distribution in silico.
(B) Outline of a workflow for high-throughput characterization of human pluripotent cell lines. Cell line characterization is performed in an iterative fashion, starting with the quantitative differentiation assay and performing additional characterizations only on those cell lines that the lineage scorecard identifies as useful for the application of interest.
These observations have important practical implications. On the one hand, equivalence to ES cell lines is unlikely to be a sufficient indicator of an iPS cell line's utility for a specific application, given that cell-line-specific outliers were prevalent even among ES cell lines. On the other hand, no single cell line may be equally powerful for deriving all cell types in vitro, implying that researchers would benefit from identifying the best cell lines specifically for each application. Unfortunately, the teratoma assay (Daley et al., 2009) does not provide the level of specificity and detail that would support application-specific selection of the most suitable cell lines (cf. Boulting et al., 2011). Teratomas are also too time consuming and expensive to be feasible for validating a large cohort of iPS cell lines, highlighting the demand for more informative and efficient assays that can be used to validate human pluripotent cell lines.
We sought to address the need for better validation assays by developing a genomic scorecard of pluripotent cell line quality and utility. The cell-line-specific outliers detected by DNA methylation and gene expression profiling were aggregated into a deviation scorecard (Figure 4 and Table S5), which enables researchers to quickly identify defects at known genes that are relevant for the intended application. This gene-specific view was complemented by the lineage scorecard, which provides a systems-level assay for quantifying how well each cell line can be differentiated into the neural and hematopoietic lineages, and into the three germ layers (Figures 5B and 5D). We tested the practical utility of this scorecard by comparing its results with independently derived motor neuron differentiation efficiencies and showed that it was highly predictive (Boulting et al., 2011).
Because the scorecard does not involve any labor-intensive steps, it becomes feasible to quickly screen through a large number of iPS cell lines in order to find the most appropriate cell lines for an intended application (Figure 7B). Furthermore, the scorecard provides a substantially more detailed characterization than for example the teratoma assay, and it therefore seems plausible that genomic scorecards could over time supersede the teratoma assay as the gold standard for validating human pluripotent cell lines. To assist researchers who want to use the scorecard on their own cell lines, we provide an extended technical note in the Extended Experimental Procedures. The scorecard can readily be adapted to other protocols for DNA methylation and gene expression profiling, and it is easy to incorporate new cell types in the prediction of the lineage scorecard. In the future, it will be necessary to validate the predictiveness for additional directed differentiation protocols, and it may occasionally be necessary to recalibrate the scorecard (e.g., for directed differentiation protocols that do not involve an EB step). The scorecard could also provide a useful readout when optimizing cell culture conditions, developing new reprogramming protocols, or continuously monitoring cell line quality in large-scale production facilities. For example, it will be interesting to measure whether the use of integration-free methods for reprogramming (Soldner et al., 2009; Warren et al., 2010) has an effect on the differentiation propensities of iPS cell lines.
In conclusion, the discovery of human pluripotent cells and the reprogramming methods to produce them from selected patient populations has revolutionized the way we think about studying and treating human disease. However, if we are to efficiently and effectively use these discoveries to improve the lives of patients, we must continue to develop tools (such as the scorecard described herein) that optimize and streamline the selection and monitoring of pluripotent cell lines and their differentiating progeny.
EXPERIMENTAL PROCEDURES
Cells Lines
A total of 20 human ES cell lines, 14 human iPS cell lines, and 6 primary fibroblast cell lines were included in the study (Table S1). The ES cell lines were obtained from the Human Embryonic Stem Cell Facility of the Harvard Stem Cell Institute (17 ES cell lines) and from the WiCell Research Institute's WISC Bank (3 ES cell lines). The iPS cell lines were derived by retroviral transduction of OCT4, SOX2, and KLF4 in dermal fibroblasts (Boulting et al., 2011). All pluripotent cell lines have been characterized by conventional methods (Chen et al., 2009; Cowan et al., 2004) and were grown under standardized conditions as described in the Extended Experimental Procedures.
DNA Methylation Mapping
RRBS was performed according to a previously published protocol (Smith et al., 2009) with some optimizations for small cell numbers (Gu et al., 2010). Using Maq's bisulfite alignment mode (Li et al., 2008), the raw sequencing reads were aligned to a human genome sequence that had been MspI-digested and size-selected in silico. DNA methylation calling was performed using custom software (Gu et al., 2010). Next, we calculated mean DNA methylation levels for all gene promoters that were covered by a minimal number of DNA methylation measurements (Bock et al., 2010). Gene promoters were defined as the –5 kb to +1 kb sequence window surrounding the annotated transcription start site of Ensembl-annoted genes (Hubbard et al., 2009). Data processing was performed by custom Python (http://python.org/) and R(http://www.r-project.org/) scripts.
Gene Expression Profiling
Microarray analysis was performed by the microarray core facility at the Broad Institute. Affymetrix GeneChip HT HG-U133A microarrays were used throughout. The microarray intensity data were normalized using Bioconductor's gcRMA package (Gentleman et al., 2004) and quality-controlled using arrayQualityMetrics (Kauffmann et al., 2009). Data analysis was performed with the R statistics package (http://www.r-project.org/).
Quantitative Embryoid Body Assay
EB differentiation was performed as described in the Extended Experimental Procedures. On day 16, cells were lysed and total RNA was extracted using Trizol (Invitrogen), followed by column clean-up using the RNeasy kit (QIAGEN). Subsequently, 300 ng to 500 ng of RNA was profiled on the Nano-String nCounter system according to manufacturer's instructions. A custom nCounter codeset was used, which covers 500 genes that were selected for their ability to monitor cell state, pluripotency, and differentiation (Table S6). Because the nCounter system has been introduced only recently, no best practices exist for normalizing the expression values. We tested several different procedures and found that a combination of spike-in normalization using positive controls and the VSN algorithm (Huber et al., 2002) produced best results. Data analysis was performed with the R statistics package (http://www.r-project.org/).
Scorecard Calculation and Bioinformatics
The deviation scorecard is based on Tukey's outlier filter (Tukey, 1977), denoting all genes as putative outliers whose DNA methylation or gene expression levels fall by more than 1.5 times the interquartile range outside of the center quartiles. The lineage scorecard performs a parametric gene set enrichment analysis on t scores obtained from a pairwise comparison between all replicates of the cell line of interest and the reference of ES cell-derived EBs. A more detailed description of the bioinformatic methods is available in Figure S4 and in the Extended Experimental Procedures.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Christopher Henderson and Hynek Wichterle (Columbia University Motor Neuron Center, Project A.L.S./Jenifer Estess Laboratory for Stem Cell Research) for sharing fibroblast and iPS cell lines prior to publication and for many helpful discussions. We would also like to thank Tarjei Mikkelsen, Eleni Tomazou, Chad Cowan, Leon Plastek, and Tim Ahfeldt for their involvement in early pilot experiments; Patrick Boyle for technical assistance; the Harvard Research Computing team for support and computation time on the Odyssey cluster; and the members of the Eggan and Meissner labs for their comments on the manuscript. Furthermore, we acknowledge the members of the Broad Institute's Genome Sequencing Platform, Genetic Analysis Platform, and Epigenomics Initiative, including Ido Amit, Fontina Kelley, Kathleen Tibbetts, Tim Fennell, Supriya Gupta, Andrew Crenshaw, Charles Epstein, and Brad Bernstein. C.B. is supported by a Feodor Lynen Fellowship from the Alexander von Humboldt Foundation. E.K. is a fellow of the European Molecular Biology Organization. K.E. is an assistant investigator of the Stowers Medical Institute, a Howard Hughes Medical Institute early career scientist, and fellow of the MacArthur Foundation. The described work was funded by the NIH Roadmap Initiative on Epigenomics (U01ES017155), the Massachusetts Life Science Center (MLSC), and the Pew Charitable Trusts. A patent has been filed for the scorecard described in the manuscript.
C.B., E.K., G.V., K.E., and A.M. designed the study and interpreted the results. C.B. developed the scorecard and analyzed the data; E.K. and G.V. performed the experiments; H.G., Z.S., and A.G. performed DNA methylation profiling; G.B. contributed data on motor neuron differentiation efficiencies; M.Z. contributed bioinformatic tools; G.C., M.A., and D.O. derived human fibroblasts and contributed data on motor neuron differentiation efficiencies; K.E. co-supervised the project; A.M. supervised the project. C.B., E.K., G.V., K.E., and A.M. wrote the paper with assistance from the other authors.
Footnotes
ACCESSION NUMBERS
Microarray data have been submitted to the Gene Expression Omnibus (GEO) and are available under accession number GSE25970.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, four figures, and seven tables and can be found with this article online at doi:10.1016/j.cell.2010.12.032.
REFERENCES
- Adewumi O, Aflatoonian B, Ahrlund-Richter L, Amit M, Andrews PW, Beighton G, Bello PA, Benvenisty N, Berry LS, Bevan S, et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat. Biotechnol. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nat. Rev. Genet. 2006;7:55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- Bock C, Walter J, Paulsen M, Lengauer T. Inter-individual variation of DNA methylation and its implications for large-scale epigenome mapping. Nucleic Acids Res. 2008;36:e55. doi: 10.1093/nar/gkn122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C, Halachev K, Büch J, Lengauer T. EpiGRAPH: User-friendly software for statistical analysis and prediction of (epi-) genomic data. Genome Biol. 2009;10:R14. doi: 10.1186/gb-2009-10-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C, Tomazou EM, Brinkman AB, Muller F, Simmer F, Gu H, Jager N, Gnirke A, Stunnenberg HG, Meissner A. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat. Biotechnol. 2010;28:1106–1114. doi: 10.1038/nbt.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiak M, Maehr R, Chen S, Chen AE, Tang W, Fox JL, Schreiber SL, Melton DA. Small molecules efficiently direct endodermal differentiation of mouse and human embryonic stem cells. Cell Stem Cell. 2009;4:348–358. doi: 10.1016/j.stem.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulting GL, Kiskinis E, Croft GF, Amoroso MW, Oakley DH, Wainger BJ, Williams DJ, Kahler DJ, Yamaki M, Davidow L, et al. A functionally characterized test set of human induced pluripotent stem cells. Nat. Biotechnol. 2011 doi: 10.1038/nbt.1783. Published online February 3, 2011. 10.1038/nbt.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AE, Egli D, Niakan K, Deng J, Akutsu H, Yamaki M, Cowan C, Fitz-Gerald C, Zhang K, Melton DA, et al. Optimal timing of inner cell mass isolation increases the efficiency of human embryonic stem cell derivation and allows generation of sibling cell lines. Cell Stem Cell. 2009;4:103–106. doi: 10.1016/j.stem.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, Ambartsumyan G, Aimiuwu O, Richter L, Zhang J, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin MH, Pellegrini M, Plath K, Lowry WE. Molecular analyses of human induced pluripotent stem cells and embryonic stem cells. Cell Stem Cell. 2010;7:263–269. doi: 10.1016/j.stem.2010.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman A, Dreesen O. Pluripotent stem cells and disease modeling. Cell Stem Cell. 2009;5:244–247. doi: 10.1016/j.stem.2009.08.010. [DOI] [PubMed] [Google Scholar]
- Cowan CA, Klimanskaya I, McMahon J, Atienza J, Witmyer J, Zucker JP, Wang S, Morton CC, McMahon AP, Powers D, et al. Derivation of embryonic stem-cell lines from human blastocysts. N. Engl. J. Med. 2004;350:1353–1356. doi: 10.1056/NEJMsr040330. [DOI] [PubMed] [Google Scholar]
- Daley GQ. Stem cells: roadmap to the clinic. J. Clin. Invest. 2010;120:8–10. doi: 10.1172/JCI41801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley GQ, Lensch MW, Jaenisch R, Meissner A, Plath K, Yamanaka S. Broader implications of defining standards for the pluripotency of iPSCs. Cell Stem Cell. 2009;4:200–201. doi: 10.1016/j.stem.2009.02.009. author reply 202. [DOI] [PubMed] [Google Scholar]
- Di Giorgio FP, Boulting GL, Bobrowicz S, Eggan KC. Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. Cell Stem Cell. 2008;3:637–648. doi: 10.1016/j.stem.2008.09.017. [DOI] [PubMed] [Google Scholar]
- Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat. Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Bock C, Mikkelsen TS, Jager N, Smith ZD, Tomazou E, Gnirke A, Lander ES, Meissner A. Genome-scale DNA methylation mapping of clinical samples at single-nucleotide resolution. Nat. Methods. 2010;7:133–136. doi: 10.1038/nmeth.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Frampton GM, Soldner F, Hockemeyer D, Mitalipova M, Jaenisch R, Young RA. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell. 2010;7:249–257. doi: 10.1016/j.stem.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, Friedman JH. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. Springer; New York: 2001. [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu BY, Weick JP, Yu J, Ma LX, Zhang XQ, Thomson JA, Zhang SC. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc. Natl. Acad. Sci. USA. 2010;107:4335–4340. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(Suppl 1):S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- Kauffmann A, Gentleman R, Huber W. arrayQualityMetrics–a bioconductor package for quality assessment of microarray data. Bioinformatics. 2009;25:415–416. doi: 10.1093/bioinformatics/btn647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshet I, Schlesinger Y, Farkash S, Rand E, Hecht M, Segal E, Pikarski E, Young RA, Niveleau A, Cedar H, et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat. Genet. 2006;38:149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- Kitchens RL. Role of CD14 in cellular recognition of bacterial lipopolysaccharides. Chem. Immunol. 2000;74:61–82. doi: 10.1159/000058750. [DOI] [PubMed] [Google Scholar]
- Lengner CJ, Gimelbrant AA, Erwin JA, Cheng AW, Guenther MG, Welstead GG, Alagappan R, Frampton GM, Xu P, Muffat J, et al. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell. 2010;141:872–883. doi: 10.1016/j.cell.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller FJ, Laurent LC, Kostka D, Ulitsky I, Williams R, Lu C, Park IH, Rao MS, Shamir R, Schwartz PH, et al. Regulatory networks define phenotypic classes of human stem cell lines. Nature. 2008;455:401–405. doi: 10.1038/nature07213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AM, Cooper JB. Lab-specific gene expression signatures in pluripotent stem cells. Cell Stem Cell. 2010;7:258–262. doi: 10.1016/j.stem.2010.06.016. [DOI] [PubMed] [Google Scholar]
- Nistri A, Ostroumov K, Sharifullina E, Taccola G. Tuning and playing a motor rhythm: how metabotropic glutamate receptors orchestrate generation of motor patterns in the mammalian central nervous system. J. Physiol. 2006;572:323–334. doi: 10.1113/jphysiol.2005.100610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osafune K, Caron L, Borowiak M, Martinez RJ, Fitz-Gerald CS, Sato Y, Cowan CA, Chien KR, Melton DA. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat. Biotechnol. 2008;26:313–315. doi: 10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin LL. Stem cells and drug discovery: the beginning of a new era? Cell. 2008;132:549–552. doi: 10.1016/j.cell.2008.02.010. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Gu H, Bock C, Gnirke A, Meissner A. High-throughput bisulfite sequencing in mammalian genomes. Methods. 2009;48:226–232. doi: 10.1016/j.ymeth.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, Hargus G, Blak A, Cooper O, Mitalipova M, et al. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev. 2010;24:2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Tukey JW. Exploratory Data Analysis. Addison-Wesley Pub. Co.; Reading, MA: 1977. [Google Scholar]
- Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.