Abstract
Objective
Bax-Interacting Factor-1 (Bif-1) protein plays a critical role in apoptosis, mitochondrial morphogenesis and autophagy, and its loss promotes tumorigenesis. The role of Bif-1 in pancreatic cancer has not been studied.
Methods
We determined Bif-1 expression in 82 human pancreatic ductal adenocarcinomas (PDC) and in 82 non-malignant pancreatic specimens (NMP), using immunohistochemistry and tissue microarray. Bif-1 immunostain was semiquantitatively scored on a scale of 0 to 9.
Results
Bif-1 scores in NMP were either 6 or 9, with lower scores in only 19 of 82 NMP (23.2%). Low Bif-1 expression (score < 6) was found in 37 of 82 PDC (45.1%), a proportion significantly greater than that found in NMP (p = 0.005). Bif-1 expression was twice as likely to be low in PDC as in NMP (RR = 1.95, 95% CI: 1.23, 3.09). Kaplan-Meier survival estimates showed no difference in survival between patients with low and high Bif-1 expression (p = 0.21, log-rank test).
Conclusions
Bif-1 expression is down-regulated in a subset of PDC. This novel finding is in agreement with the tumor suppressor function of Bif-1. The lack of association with Bif-1 expression with patient survival may be best explained by the complexity of carcinogenesis.
Keywords: Bif-1, pancreatic ductal adenocarcinoma, tissue microarray, immunohistochemistry, outcomes
INTRODUCTION
The annual incidence of pancreatic cancer has risen over the last decade, with more than 42,000 new cases expected in 2009 (1–3). Pancreatic cancer is the fourth most-common cause of death from cancer in men and women, with the great majority of these being ductal adenocarcinomas. Furthermore, the 5-year survival of patients with ductal pancreatic cancer is less than 5% and it has shown no improvement over the past 3 decades (1, 3).
It is well documented that acquired resistance to programmed cell death or apoptosis is essential for tumor development, progression and metastasis (4). For instance, aberrant expression of Bcl-2 family proteins, growth factors, and cell receptors has been described in pancreatic cancer (5–9). Mutations in p53 have been reported in 37–63% of pancreatic ductal adenocarcinomas (10,11) and oncogenic K-ras mutations have been observed in 80% of such cancers (12). CD44 variants 6 and 2 are over-expressed on pancreatic tumor cells, and predict a decreased survival (13,14). Moreover, lysosomal cathepsins and laminin receptors, as well as urokinase plasminogen activator (uPA) and its receptor (uPAR), have been linked to the metastatic potential of pancreatic cancer cells (15–17).
Bif-1 (also known as endophilin B1 and SH3GLB1) was originally discovered by yeast two-hybrid screenings for proteins that bind the pro-apoptotic Bcl-2 family protein Bax (18,19). While Bif-1 over-expression promotes Bax activation and apoptosis (18), inhibition of Bif-1 expression suppresses Bax-dependent apoptosis in response to intrinsic death signals (20). Bif-1 has also been shown to regulate mitochondrial morphogenesis (21). Moreover, Bif-1 is involved in endocytic trafficking of growth factor receptors [22–24] and autophagy (25). Bif-1 interacts with Beclin 1 through UVRAG and plays a role in autophagosome formation (25). Importantly, suppression of Bif-1 expression promotes xenograft tumor growth and the development of spontaneous tumors in mice (25).
While apoptotic evasion represents one of the true hallmarks of cancer, accumulating evidence suggests that defects in autophagy also contribute to tumor development and progression (26–29). Autophagy is an intracellular catabolic process that plays a vital role in maintaining cellular homeostasis. It has been shown that the rate of autophagy is reduced during chemical carcinogenesis in rats (13–15). Oncogenes, such as mTOR, Akt and Bcl-2, suppress autophagy, whereas tumor suppressors, such as DAPK, p53 and PTEN, stimulate autophagy (5,6,16,17). Moreover, mutation of autophagy genes, such as beclin 1, is frequently detected in human cancer (9).
In this study we evaluated Bif-1 protein expression in 82 pancreatic ductal adenocarcinomas using semi-quantitative immunohistochemistry and tissue microarray technology. We then sought an association between level of Bif-1 expression and clinical outcomes.
MATERIALS AND METHODS
The present study is focused on 82 patients with stage IA – IIB ductal adenocarcinoma of the pancreas who underwent pancreatectomy with curative intent, between 1987 and 2006 at the Moffitt Cancer Center, and survived at least 30 days. The histologic diagnosis in each case was confirmed by expert pathology review (DC, MG) of sections cut from fixed surgical specimens that had been stained with hematoxylin and eosin. Data from the Moffitt Cancer Registry, the patient’s medical record, the Anatomic Pathology database (CoPath), and the Social Security Death Index were reviewed to determine the patient’s clinicopathologic characteristics, American Joint Committee on Cancer (AJCC) anatomic staging, survival of each patient after surgery, whether the patient was deceased as of most recent knowledge, and if there was evidence of disease at that time. The study was approved by the Institutional Review Board of the University of South Florida, Tampa.
Selection of Cases and Construction of Tissue Microarrays
For immunohistochemical study we used stage-oriented human pancreatic cancer tissue microarrays (TMAs) prepared in the Histology Laboratory of the Moffitt Cancer Center Tissue Core Facility. TMAs were constructed using specimens of both pancreatic ductal adenocarcinoma and non-malignant pancreas taken from adjacent to the cancer. Two separate cores, 0.6 cm in diameter, were obtained for each pancreatic cancer specimen, as well as for the non-malignant pancreas specimens taken from adjacent to the cancers. All specimens used in constructing TMAs had been preserved in 10% buffered formalin prior to embedding in paraffin.
Immunohistochemistry
The tissues were stained using the avidin-biotin-peroxidase method and anti-Bif-1 mouse monoclonal antibody (Imgenex, San Diego, CA). TMA slides were dewaxed by heating at 55° C for 30 minutes and by three washes, five minutes each, with xylene. Tissues were rehydrated by a series of five-minute washes in 100%, 95%, and 80% ethanol, and distilled water. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide for 20 minutes. After blocking with universal blocking serum (Ventana Medical Systems, Inc., Tucson, Arizona OMNIMAP) for 30 minutes, the samples were incubated with anti-Bif-1 mouse monoclonal antibody (Imgenex, dilution 1:2500) at 4° C overnight. The samples were then incubated with biotin-labeled secondary antibody and streptavidin-horseradish peroxidase for 30 minutes each (Ventana Medical Systems). The slides were developed with 3, 3’-diaminobenzidine tetrahydrochloride substrate (Ventana Medical Systems Inc. Tucson, Arizona) and counterstained with hematoxylin (Ventana Medical Systems Inc. Tucson, Arizona). The tissue samples were dehydrated and coversliped. Standard cell conditioning following Ventana proprietary recommendations was used for antigen retrieval. The specificity of the anti-Bif-1 monoclonal antibody was confirmed by immunoblot analysis of Bif-1-positive and -negative cells (20,30). A negative control was included by omitting Bif-1 antibody during the primary antibody incubation step.
Analytical and Statistical Methods
A semi-quantitative measure of Bif-1 expression was determined by two pathologists (DC, MG) who independently scored stained tissue sections for intensity of Bif-1 immunostain and percent of cells stained, with consensus scores determined for each section. Intensity of immunostain was subjectively ranked on a scale of increasing intensity: 0, 1+, 2+, and 3+. The percent of cells stained for Bif-1 was estimated on a scale of 0 to 3: 0 (0%), 1 (1–33%), 2 (34 66%), and 3 (67–100%). Bif-1 expression score was calculated as the product of the intensity and percent-stained scores. Thus, the Bif-1 score can take on only the values 0, 1, 2, 3, 4, 6, and 9.
Association between variables was assessed by Fisher’s exact test. Relative risk (RR) was used to compare the probability (risk) of Bif-1 expression being low between two groups of patients. Disease-specific survival was estimated by the Kaplan-Meier method, using death with evidence of disease as the endpoint. The log-rank test for equality of survival functions was used to compare Kaplan-Meier survival curves.
RESULTS
A total of 82 patients who underwent resection of stage IA–IIB ductal adenocarcinoma of the pancreas had fixed surgical cancer specimens immunostained for Bif-1. Adjacent non-malignant pancreas specimens were stained in 75 of these 82 patients. In 7 additional pancreatic cancer patients only non-malignant pancreas specimens taken from adjacent to the cancer were stained, making a total of 82 non-malignant pancreas specimens stained. Clinical features, AJCC anatomic staging, and histologic characteristics for the 82 patients whose cancer specimens were stained for Bif-1 are shown in Table 1.
TABLE 1.
Clinical features, AJCC anatomic staging and histologic characteristics of 82 patients who underwent resection of stage IA–IIB ductal pancreatic cancer with curative intent and survived at least 30 days after surgery
| n (%) | |
|---|---|
| Gender | 50 female : 32 male |
| Median Age | 66 yr, range (25, 87) |
| Stage Group | |
| IA–tumor ≤ 2 cm, confined to pancreas, no positive nodes | 5 (6%) |
| IB–tumor > 2 cm, confined to pancreas, no positive nodes | 13 (16%) |
| IIA–local extrapancreatic extension without celiac axis or SMA involvement, no positive nodes | 29 (35%) |
| IIB–regional node metastasis | 35 (43%) |
| T Classification | |
| T 1–tumor ≤ 2 cm, confined to pancreas | 9 (11%) |
| T 2–tumor > 2cm, confined to pancreas | 16 (19%) |
| T 3–local extrapancreatic extension without celiac axis or SMA involvement | 57 (70%) |
| N Classification–regional node metastases (N 1) | 36 (44%) |
| R Classification–microscopically positive resection margin (R 1) | 23 (28%) |
| Histologic Differentiation | |
| well | 19 (23% of 82) |
| moderate | 49 (60% of 82) |
| poor & undifferentiated | 14 (17% of 82) |
| Lymphatic invasion present | 55 (67% of 82) |
| Venous invasion present | 36 (44% of 82) |
| Perineural invasion present | 72 (88% of 82) |
Staging as per American Joint Committee on Cancer (AJCC) Staging Manual, 6th edition
Immunohistochemical Observations
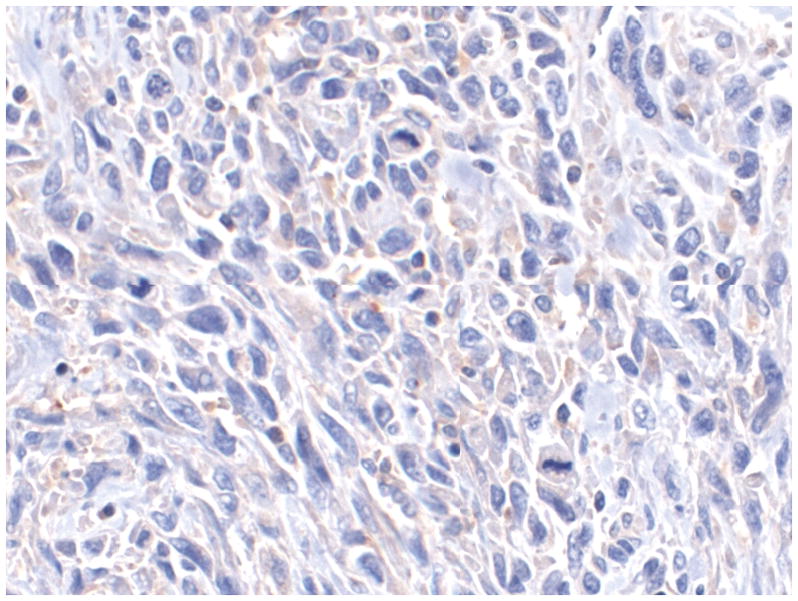
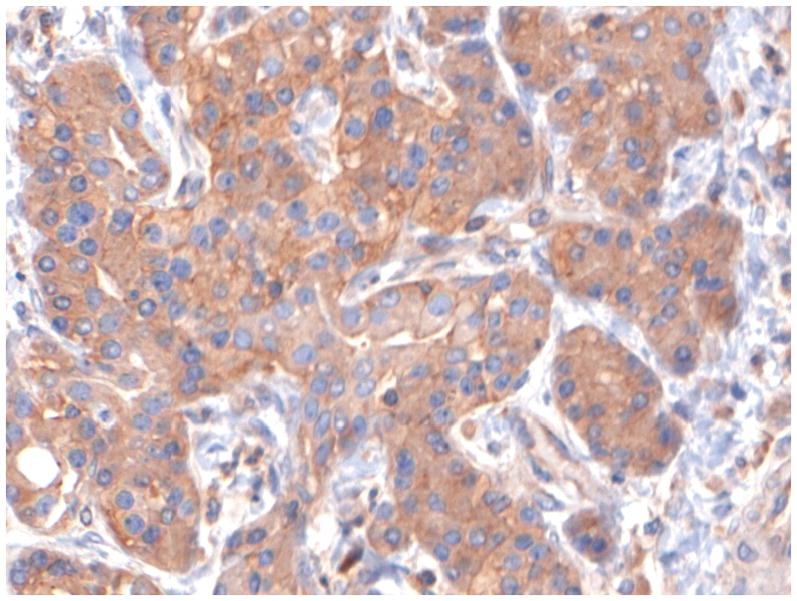
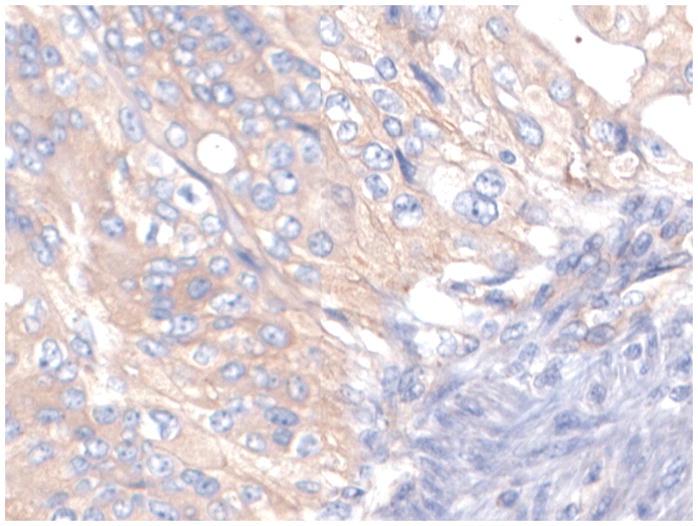
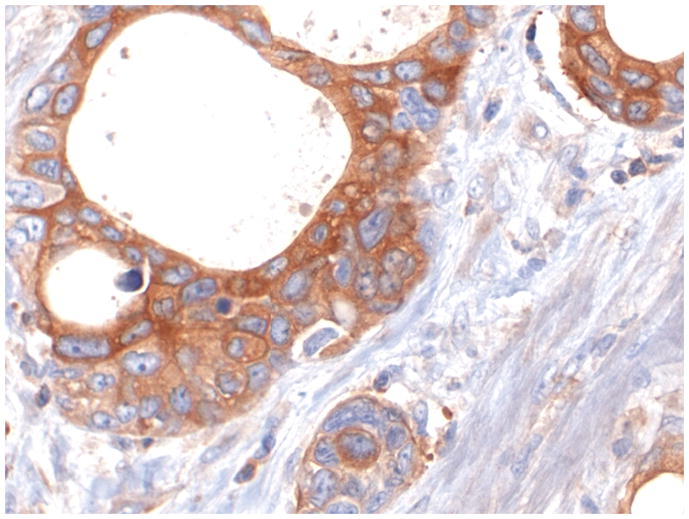
Figure 1 shows representative examples of cancer and non-malignant pancreas immunohistochemically-stained for Bif-1, along with their respective Bif-1 expression scores. In all cases expressing Bif-1 the staining was cytoplasmic and diffusely granular. In some cases there was variation in intensity within the same lesion. In such event the intensity score was assigned based on the predominant staining intensity and the percent of positive cells was calculated based on the amount of lesion showing the predominant intensity.
Figure 1. Distribution of Big-1 stain in pancreatic cellular components and in pancreatic adenocarcinoma.
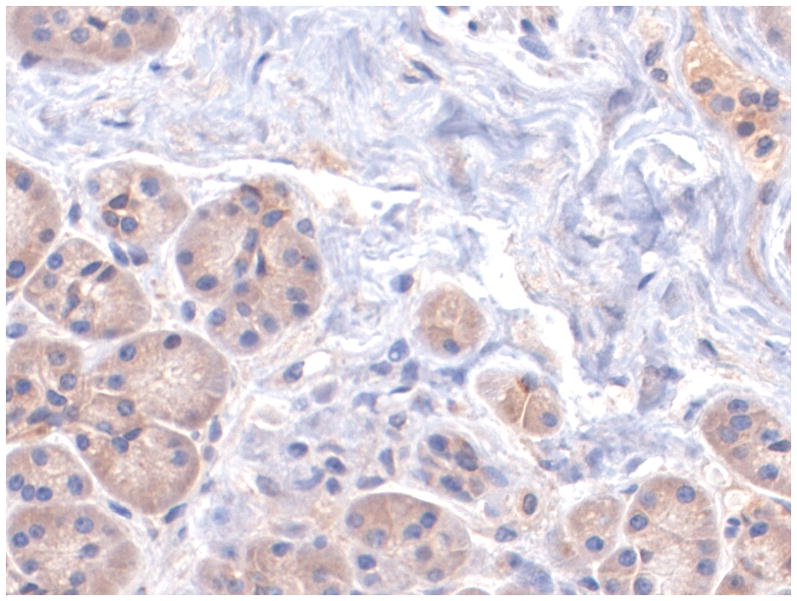
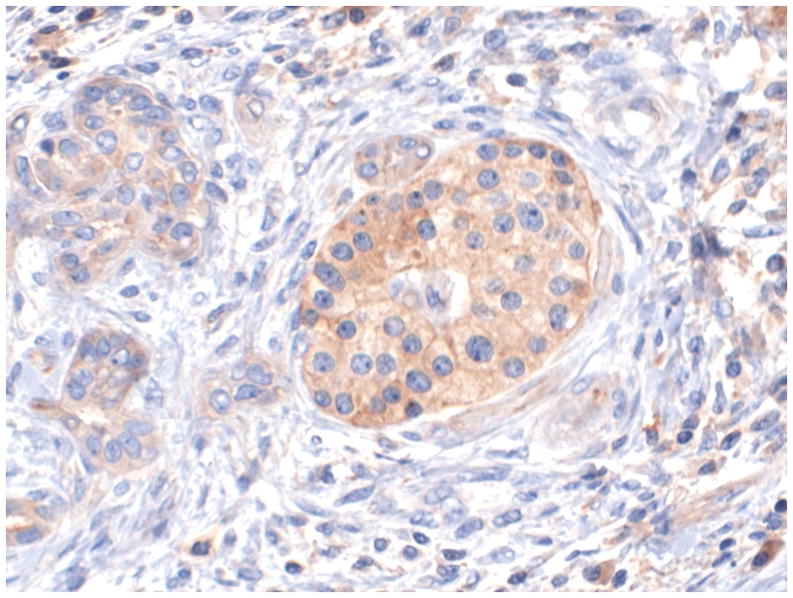
A: Bif-1 staining the non neoplastic pancreatic acinar cells. Case representing IHC score 3.
B: Bif-1 staining the non neoplastic pancreatic ductal cells. Case representing IHC score 6.
C: Bif-1 staining non neoplastic pancreatic ductal cells (left) and islet cells (right)
D: A Bif-1 negative pancreatic ductal adenocarcinoma (PDA). Case representing IHC score 0.
E: A Pancreatic ductal adenocarcinoma weakly immunoreactive with Bif-1 antibody. Case representing IHC score 3.
F: A Bif-1 positive pancreatic ductal adenocarcinoma (PDA). Case representing IHC score 9.
All pictures X200
Bif-1 Expression in Cancer and Non-Malignant Pancreas
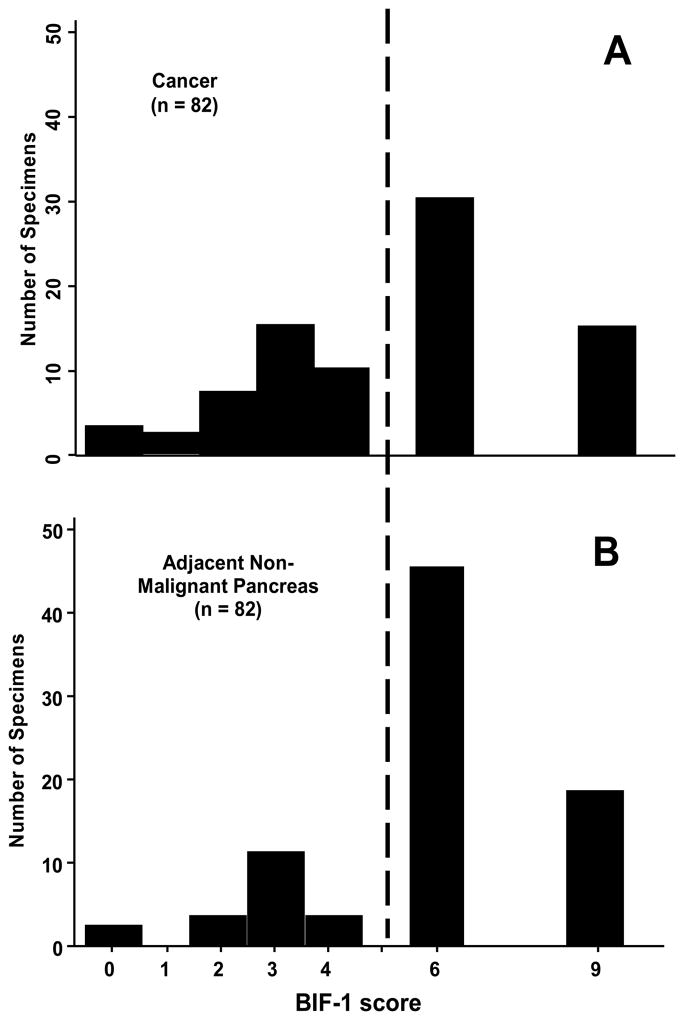
The stain was localized to the pancreatic duct epithelium, but also in the acinar cells and in the neuroendocrine cells of the islets of Langerhans. We first considered the expression of Bif-1 in non-malignant pancreas by inspection of the frequency distribution of expression scores in 82 non-malignant pancreas specimens (Figure 2A). The great majority of Bif-1 scores in nonmalignant pancreas were either 6 or 9, with a lower Bif-1 expression being found in only 19 of 82 non-malignant specimens (23.2%). It is this normally-low percentage of non-malignant pancreas specimens with low Bif-1 expression to which we compared Bif-1 expression in cancer to determine if Bif-1 expression was lost in pancreatic cancer.
Figure 2.
Frequency distribution of BIF-1 scores in A) 82 non-malignant pancreatic specimens taken from adjacent to pancreatic cancer and B) 82 pancreatic cancer specimens. The normally-high level of BIF-1 expression in non-malignant pancreas is indicated by BIF-1 scores to the right of the dashed line.
The frequency distribution of Bif-1 expression scores for the 82 pancreatic cancer specimens is shown in Figure 2B. Bif-1 expression scores were low in 37 of 82 cancer specimens (45.1%), which was significantly greater than the proportion of non-malignant specimens (23.2%) with an expression score of less than 6 (p = 0.005). Bif-1 expression was about twice as likely to be low (score less than 6) in pancreatic cancers than in non-malignant pancreas (RR = 1.95, 95% CI: 1.23, 3.09).
Bif-1 Expression in Cancer and Clinical Outcomes
Kaplan-Meier survival estimates by Bif-1 score were done for the 82 patients who underwent pancreatectomy for cancer and had their tumor evaluated for Bif-1 expression. Survival for patients with low Bif-1 expression did not differ significantly from those with high Bif-1 expression (p = 0.21).
We considered whether low Bif-1 expression (score less than 6) in pancreatic cancer might be associated with the presence of histopathologic characteristics regarded as portending an adverse clinical outcome. Table 2 shows the RR of low Bif-1 expression by the presence of potentially-adverse histopathologic characteristics. RR was calculated as the ratio of the probability (risk) of Bif-1 expression being low in patients with the specified characteristic being present to the probability in patients without that characteristic. No RR differed significantly from 1.00, indicating that Bif-1 expression was independent of the presence of each histopathologic characteristic (p > 0.50 in all instances).
Table 2.
Association between potentially-adverse histolopathologic characteristics and low Bif-1 expression in 82 patients who underwent resection of stage IA-IIB ductal pancreatic cancer with curative intent
| Characteristic | Present | Bif-1 Expression |
Association p-value | RR | 95% CI | |
|---|---|---|---|---|---|---|
| Low | High | |||||
| Local Extrapancreatic Extension (T3 classification) | yes | 25 | 32 | 0.81 | 0.91 | (0.55, 1.51) |
| no | 12 | 13 | ||||
| Regional Node Metastases (N1 classification) | yes | 18 | 18 | 0.51 | 1.21 | (0.75, 1.95) |
| no | 19 | 27 | ||||
| Positive Resection Margin (R1 classification) | yes | 10 | 13 | 1.00 | 0.95 | (0.55, 1.63) |
| no | 27 | 32 | ||||
| Poorly-Differentiated | Yes | 5 | 9 | 0.56 | 0.76 | (0.36, 1.60) |
| no | 32 | 36 | ||||
| Lymphatic Invasion | yes | 24 | 31 | 0.81 | 0.91 | (0.55, 1.48) |
| no | 13 | 14 | ||||
| Perivenous Invasion | yes | 15 | 21 | 0.66 | 0.87 | (0.53, 1.42) |
| no | 22 | 24 | ||||
| Perineural Invasion | yes | 32 | 40 | 0.75 | 0.89 | (0.45, 1.74) |
| no | 5 | 5 | ||||
Relative Risk (RR) is the ratio of the probability of Bif-1 expression being low in patients with the specified characteristic to the probability in patients without the characteristic; 95% Confidence Interval (CI) for the RR
DISCUSSION
Evidence of a role for loss of Bif-1 tumor suppressor activity in carcinogenesis has been found at the chromosomal level, in transcription, and in the protein product for a number of different cancers. The human Bif-1 gene is mapped to chromosome 1p22, a chromosome in which loss of heterozygosity (LOH) is frequently found in many tumor types (31–39). As an example, a homozygous deletion of Bif-1 has been reported in mantle cell lymphomas (31). Moreover, it has been shown that Bif-1 mRNA levels are down-regulated in lung carcinoma (40) and colon cancer (41). At the protein level, several reports described findings similar to ours, with subsets of tumors retaining Bif-1 expression, and others loosing such expression. For example, approximately 60% of gastric carcinoma [42], 21% of prostate cancer [43], 23% of colorectal adenocarcinoma [44], and 15% of invasive bladder cancer [45] are Bif-1 negative as determined by immunohistochemistry. Moreover, Bif-1 expression is down-regulated during clinical progression of breast carcinoma in situ to invasive and metastatic carcinomas [46]. However, somatic mutation of bif-1 gene is rare in common human cancers [47].
In this study, we found for the first time that Bif-1 was under-expressed in about 45% of pancreatic cancers, in contrast to the high levels normally expressed in nonmalignant pancreas. This novel finding is consistent with the function of Bif-1 as a tumor suppressor. However, it remains to be determined whether loss of Bif-1 tumor suppressor activity in pancreatic cancer would occur through reduced Bax activation with resultant decrease in apoptosis, inhibition of autophagy, or both.
As already noted, a number of the biochemical and molecular changes that occur in pancreatic cancer have been shown to be associated with differences in tumor behavior and clinical outcome. We found, however, that survival after pancreatectomy in patients who had low Bif-1 expression did not differ from survival in those who had the high level of Bif-1 expression most often found in non-malignant pancreas. Having failed to find that survival differed by Bif-1 expression, we considered that various histopathologic characteristics have been reported to be individually predictive of survival after pancreatectomy with curative intent. These characteristics include, but are not limited to, anatomic tumor extent as defined by AJCC staging, and tumor histologic characteristics. We recently reported that a simple combination of histopathologic characteristics reflecting aggressiveness of tumor biology (histologic differentiation, lymphatic invasion within the tumor, and local extrapancreatic tumor extension) was predictive of survival after pancreatectomy (48). When we considered whether loss of Bif-1 expression might be associated with these and other potentially adverse histopathologic characteristics, we were unable to find an association.
Loss of Bif-1 expression in a subset of patients with ductal pancreatic cancer is presumptive evidence of a role for loss of Bif-1 tumor suppressor activity in pancreatic carcinogenesis. Yet, we were unable to demonstrate an association between loss of Bif-1 expression and clinical outcome, a finding that may be best explained by the complexity of carcinogenesis. For example, one explanation might be that loss of Bif-1 tumor suppressor activity may be more important in earlier stages of carcinogenesis, while factors other than loss of Bif-1 tumor suppressor activity may dominate the outcome after pancreatectomy. Nevertheless, finding a loss of Bif-1 expression in pancreatic adenocarcinoma is an important step in understanding the biology of the disease, and one which may help design new targeted molecular strategies for treatment of a cancer that has been resistant to therapy thus far.
Acknowledgments
We thank the Histology Section of the Tissue Core and the Analytic Microscopy Core at the Moffitt Cancer Center and Research Institute for the support in performing the immunohistochemical stains and providing the gallery images necessary to appropriately evaluate the immunostains.
None of the authors has a personal or financial conflict of interest.
Sources of Financial Support: This study was supported by a grant (CA82197) from the National Cancer Institute and a grant (RSG-05-244-01-CCG) from the American Cancer Society.
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Thomas A, Murray T, et al. Cancer statistics, 2002. CA Cancer J Clin. 2002;52:23–47. doi: 10.3322/canjclin.52.1.23. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 5.Miyamoto Y, Hosotani R, Wada M, et al. Immunohistochemical analysis of Bcl-2, Bax, Bcl-X, and Mcl-1 expression in pancreatic cancers. Oncology. 1999;56:73–82. doi: 10.1159/000011933. [DOI] [PubMed] [Google Scholar]
- 6.Wilentz RE, Iacobuzio-Donahue CA, Argani P, et al. Loss of expression of Dpc4 in pancreatic intraepithelial neoplasia: evidence that DPC4 inactivation occurs late in neoplastic progression. Cancer Res. 2000;60:2002–6. [PubMed] [Google Scholar]
- 7.Barton CM, Hall PA, Hughes CM, et al. Transforming growth factor alpha and epidermal growth factor in human pancreatic cancer. J Pathol. 1991;163:111–6. doi: 10.1002/path.1711630206. [DOI] [PubMed] [Google Scholar]
- 8.Korc M, Meltzer P, Trent J. Enhanced expression of epidermal growth factor receptor correlates with alterations of chromosome 7 in human pancreatic cancer. Proc Natl Acad Sci U S A. 1986;83:5141–4. doi: 10.1073/pnas.83.14.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szepeshazi K, Halmos G, Schally AV, et al. Growth inhibition of experimental pancreatic cancers and sustained reduction in epidermal growth factor receptors during therapy with hormonal peptide analogs. J Cancer Res Clin Oncol. 1999;125:444–52. doi: 10.1007/s004320050301. [DOI] [PubMed] [Google Scholar]
- 10.DiGiuseppe JA, Hruban RH, Goodman SN, et al. Overexpression of p53 protein in adenocarcinoma of the pancreas. Am J Clin Pathol. 1994;101:684–8. doi: 10.1093/ajcp/101.6.684. [DOI] [PubMed] [Google Scholar]
- 11.Lundin J, Nordling S, von Boguslawsky K, et al. Prognostic value of immunohistochemical expression of p53 in patients with pancreatic cancer. Oncology. 1996;53:104–11. doi: 10.1159/000227545. [DOI] [PubMed] [Google Scholar]
- 12.Almoguera C, Shibata D, Forrester K, et al. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53:549–54. doi: 10.1016/0092-8674(88)90571-5. [DOI] [PubMed] [Google Scholar]
- 13.Gotoda T, Matsumura Y, Kondo H, et al. Expression of CD44 variants and its association with survival in pancreatic cancer. Jpn J Cancer Res. 1998;89:1033–40. doi: 10.1111/j.1349-7006.1998.tb00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castella EM, Ariza A, Ojanguren I, et al. Differential expression of CD44v6 in adenocarcinoma of the pancreas: an immunohistochemical study. Virchows Arch. 1996;429:191–5. doi: 10.1007/BF00198333. [DOI] [PubMed] [Google Scholar]
- 15.Tumminello FM, Leto G, Pizzolanti G, et al. Cathepsin D, B and L circulating levels as prognostic markers of malignant progression. Anticancer Res. 1996;16:2315–9. [PubMed] [Google Scholar]
- 16.Pelosi G, Pasini F, Bresaola E, et al. High-affinity monomeric 67-kD laminin receptors and prognosis in pancreatic endocrine tumours. J Pathol. 1997;183:62–9. doi: 10.1002/(SICI)1096-9896(199709)183:1<62::AID-PATH1095>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 17.Cantero D, Friess H, Deflorin J, et al. Enhanced expression of urokinase plasminogen activator and its receptor in pancreatic carcinoma. Br J Cancer. 1997;75:388–95. doi: 10.1038/bjc.1997.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cuddeback SM, Yamaguchi H, Komatsu K, et al. Molecular cloning and characterization of Bif-1. A novel Src homology 3 domain-containing protein that associates with Bax. J Biol Chem. 2001;276:20559–65. doi: 10.1074/jbc.M101527200. [DOI] [PubMed] [Google Scholar]
- 19.Farsad K, Ringstad N, Takei K, et al. Generation of high curvature membranes mediated by direct endophilin bilayer interactions. J Cell Biol. 2001;155:193–200. doi: 10.1083/jcb.200107075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takahashi Y, Karbowski M, Yamaguchi H, et al. Loss of Bif-1 suppresses Bax/Bak conformational change and mitochondrial apoptosis. Mol Cell Biol. 2005;25:9369–82. doi: 10.1128/MCB.25.21.9369-9382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karbowski M, Jeong SY, Youle RJ. Endophilin B1 is required for the maintenance of mitochondrial morphology. J Cell Biol. 2004;166:1027–39. doi: 10.1083/jcb.200407046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahashi Y, Meyerkord CL, Wang HG. Bif-1/endophilin B1: a candidate for crescent driving force in autophagy. Cell Death Differ. 2009;16:947–955. doi: 10.1038/cdd.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thoresen SB, Pedersen NM, Liestol K, Stenmark H. A phosphatidylinositol 3-kinase class III sub-complex containing VPS15, VPS34, Beclin 1, UVRAG and BIF-1 regulates cytokinesis and degradative endocytic traffic. Exp Cell Res. 2010 doi: 10.1016/j.yexcr.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 24.Wan J, Cheung AY, Fu WY, Wu C, Zhang M, et al. ndophilin B1 as a novel regulator of nerve growth factor/ TrkA trafficking and neurite outgrowth. J Neurosci. 2008;28:9002–9012. doi: 10.1523/JNEUROSCI.0767-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takahashi Y, Coppola D, Matsushita N, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–51. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gozuacik D, Kimchi A. Autophagy as a cell death and tumor suppressor mechanism. Oncogene. 2004;23:2891–906. doi: 10.1038/sj.onc.1207521. [DOI] [PubMed] [Google Scholar]
- 27.Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–5. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mathew R, White E. Why sick cells produce tumors: the protective role of autophagy. Autophagy. 2007;3:502–5. doi: 10.4161/auto.4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Green DR, Evan GI. A matter of life and death. Cancer Cell. 2002;1:19– 30. doi: 10.1016/s1535-6108(02)00024-7. [DOI] [PubMed] [Google Scholar]
- 30.Takahashi Y, Coppola D, Matsushita N, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–51. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balakrishnan A, von Neuhoff N, Rudolph C, et al. Quantitative microsatellite analysis to delineate the commonly deleted region 1p22.3 in mantle cell lymphomas. Genes Chromosomes Cancer. 2006;45:883–92. doi: 10.1002/gcc.20352. [DOI] [PubMed] [Google Scholar]
- 32.Walker GJ, Indsto JO, Sood R, et al. Deletion mapping suggests that the 1p22 melanoma susceptibility gene is a tumor suppressor localized to a 9-Mb interval. Genes Chromosomes Cancer. 2004;41:56–64. doi: 10.1002/gcc.20056. [DOI] [PubMed] [Google Scholar]
- 33.Lee WC, Balsara B, Liu Z, et al. Loss of heterozygosity analysis defines a critical region in chromosome 1p22 commonly deleted in human malignant mesothelioma. Cancer Res. 1996;56:4297–301. [PubMed] [Google Scholar]
- 34.Roberts T, Chernova O, Cowell JK. Molecular characterization of the 1p22 breakpoint region spanning the constitutional translocation breakpoint in a neuroblastoma patient with a t(1;10)(p22;q21) Cancer Genet Cytogenet. 1998;100:10–20. doi: 10.1016/s0165-4608(97)00013-7. [DOI] [PubMed] [Google Scholar]
- 35.Kakinuma H, Habuchi T, Ito T, et al. BCL10 is not a major target for frequent loss of 1p in testicular germ cell tumors. Cancer Genet Cytogenet. 2001;126:134–8. doi: 10.1016/s0165-4608(00)00405-2. [DOI] [PubMed] [Google Scholar]
- 36.Mora J, Cheung NK, Kushner BH, et al. Clinical categories of neuroblastoma are associated with different patterns of loss of heterozygosity on chromosome arm 1p. J Mol Diagn. 2000;2:37–46. doi: 10.1016/S1525-1578(10)60613-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emi M, Matsumoto S, Iida A, et al. Correlation of Allelic Losses and Clinicopathological Factors in Primary Breast Cancers. Breast Cancer. 1997;4:243–6. doi: 10.1007/BF02966514. [DOI] [PubMed] [Google Scholar]
- 38.Bello MJ, de Campos JM, Vaquero J, et al. High-resolution analysis of chromosome arm 1p alterations in meningioma. Cancer Genet Cytogenet. 2000;120:30–6. doi: 10.1016/s0165-4608(99)00249-6. [DOI] [PubMed] [Google Scholar]
- 39.Knosel T, Petersen S, Schwabe H, et al. Incidence of chromosomal imbalances in advanced colorectal carcinomas and their metastases. Virchows Arch. 2002;440:187–94. doi: 10.1007/s004280100493. [DOI] [PubMed] [Google Scholar]
- 40.Bonner AE, Lemon WJ, Devereux TR, et al. Molecular profiling of mouse lung tumors: association with tumor progression, lung development, and human lung adenocarcinomas. Oncogene. 2004;23:1166–76. doi: 10.1038/sj.onc.1207234. [DOI] [PubMed] [Google Scholar]
- 41.Ho J, Kong JW, Choong LY, et al. Novel breast cancer metastasis associated proteins. J Proteome Res. 2009;8:583–94. doi: 10.1021/pr8007368. [DOI] [PubMed] [Google Scholar]
- 42.Lee JW, Jeong EG, Soung YH, et al. Decreased expression of tumour suppressor Bax-interacting factor-1 (Bif-1), a Bax activator, in gastric carcinomas. Pathology. 2006;38:312–315. doi: 10.1080/00313020600820880. [DOI] [PubMed] [Google Scholar]
- 43.Coppola D, Oliveri C, Sayegh Z, et al. Bax-interacting factor-1 (Bif-1) expression in prostate cancer. Clin Genitourin Cancer. 2008 Sep;6(2):117–21. doi: 10.3816/CGC.2008.n.018. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coppola D, Khalil F, Eschrich S, et al. Down regulation of Bax- interacting factor-1 (Bif-1) in colon cancer. Cancer. 2008 doi: 10.1002/cncr.23892. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim SY, Oh YL, Kim KMJ, et al. Decreased expression of Bax-interacting factor-1 (Bif-1) in invasive urinary bladder and gallbladder cancers. Pathology. 2008;40:553–557. doi: 10.1080/00313020802320440. [DOI] [PubMed] [Google Scholar]
- 46.Ho J, Kong JW, Choong LY, et al. Novel breast cancer metastasis-associated proteins. J Proteome Res. 2009;8:583–594. doi: 10.1021/pr8007368. [DOI] [PubMed] [Google Scholar]
- 47.Kim MS, Yoo NJ, Lee SH. Somatic mutation of pro-cell death Bif-1 gene is rare in common human cancers. APMIS. 2008;116:939–940. doi: 10.1111/j.1600-0463.2008.01091.x. [DOI] [PubMed] [Google Scholar]
- 48.Helm J, Centeno BA, Coppola D, et al. Histologic characteeristics enhance predictive value of American Joint Committee on Cancer staging in resectable pancreas cancer. Cancer. 2009;115:4080–9. doi: 10.1002/cncr.24503. [DOI] [PubMed] [Google Scholar]