Abstract
Genetic association studies for binary diseases are designed as case-control studies: the cases are those affected with the primary disease and the controls are free of the disease. At the time of case-control collection, information about secondary phenotypes is also collected. Association studies of secondary phenotype and genetic variants have received a great deal of interest recently. To study the secondary phenotypes, investigators use standard regression approaches, where individuals with secondary phenotypes are coded as cases and those without secondary phenotypes are coded as controls. However, using the secondary phenotype as an outcome variable in a case-control study might lead to a biased estimate of odds ratios (ORs) for genetic variants. The secondary phenotype is associated with the primary disease; therefore, individuals with and without the secondary phenotype are not sampled following the principles of a case-control study. In this article, we demonstrate that such analyses will lead to a biased estimate of OR and propose new approaches to provide more accurate OR estimates of genetic variants associated with the secondary phenotype for both unmatched and frequency-matched (with respect to the secondary phenotype) case-control studies. We also propose a bootstrapping method to estimate the empirical confidence intervals for the corrected ORs. Using simulation studies and analysis of lung cancer data for single-nucleotide polymorphism associated with smoking quantity, we compared our new approaches to standard logistic regression and to an extended version of the inverse-probability-of-sampling-weighted regression. The proposed approaches provide more accurate estimation of the true OR.
Keywords: Odds ratio, bias, secondary phenotype, un-matched and frequency-matched study, SNP, genome-wide association study
1. Introduction
Genome-wide association (GWA) study has recently become a popular approach for detecting genetic variants for common diseases without prior knowledge of the variant's location or function. Typically, GWA studies were originally designed as case-control studies of the primary disease of interest, such as lung cancer, diabetes, or breast cancer (cases are those affected with the primary disease and controls are free of the disease). At the time of case-control collection, information about secondary phenotypes, which we define as traits associated with the primary disease of interest, such as smoking behavior and body mass index (BMI), are also collected. Thus, GWA studies provide a large number of datasets that can be used in studies of the association between secondary phenotypes and genetic variants. For example, in lung cancer GWA studies, data about smoking behavior and chronic obstructive pulmonary disease (COPD) are also available; both are secondary traits that are highly associated with lung cancer risk. The association studies between smoking and single-nucleotide polymorphisms (SNPs) have received a great deal of interest [Amos et al., 2008; Hung et al., 2008; Spitz et al., 2008; Thorgeirsson et al., 2008; Wang et al., 2010]. Similarly, in the GWA studies of type 2 diabetes, investigators also collected data for secondary phenotypes such as BMI and physical activity and were interested in the association between these secondary phenotypes and genetic variants [Frayling et al., 2007]. Similarly, in the GWA studies of breast cancer, the associations between genetic variants and different secondary phenotypes, such as ages of menarche and puberty, have been studied [He et al., 2009; Ong et al., 2009].
To study the secondary phenotypes, investigators have used the standard regression approaches, such as logistic regression, where individuals with secondary phenotypes are coded as cases and those without secondary phenotypes are coded as controls. However, using the secondary phenotype as an outcome variable in a case-control study might lead to a biased estimate of odds ratios (ORs) for genetic variants. This is because the secondary phenotype is associated with the primary disease of interest; therefore, individuals with (case subject) and without (control subject) the secondary phenotype are not sampled following the principle of a case-control study design, where cases are randomly ascertained from the group of individuals in the general population who have the specified phenotype and matched with a group of controls who do not have the phenotype. The commonly used solution to this problem is to utilize primary disease status as a covariate in the regression analyses. However, through simulation (see below) we showed that in many situations adjusting for the primary disease status will still result in a biased estimate of the OR. Alternative approaches for estimating ORs of genetic variants associated with secondary phenotypes include using only cases or only controls with respect to the primary disease. However, these approaches were similarly biased. Richardson et al. [2007] used stratum-weighted logistic regression to assess associations between the explanatory variables and the secondary phenotype for a nested case-control study within a prospective cohort, where the inverses of the sampling fractions were used as the weights. Monsees et al. [2009] investigated different scenarios for the problem of marker-secondary trait association in nested case-control samples using the inverse-probability-of-sampling-weighted (IPW) regression method proposed by Richardson et al. [2007] to estimate genotype-secondary trait association when the sampling fractions are available for case-control studies nested within a prospective cohort. Lin and Zeng [2009] proposed a likelihood-based approach for the analysis of a secondary phenotype. They considered several different scenarios involving a rare or not rare disease and a known or unknown disease rate, and for each scenario, they maximized the corresponding log-likelihood function via the Newton-Raphson algorithm. Li et al. [2010] focused on the situation when the primary disease is rare. They found that, when the primary disease is rare, some standard approaches (e.g. controls only or cases only) would still result in biased estimates if the secondary phenotype and the genetic variant have an interaction effect on the primary disease; and proposed an adaptively weighted method using both cases and controls for the study of association between secondary phenotype and the genetic variant. None of these authors considered commonly-used frequency-matched case-control design in their studies.
In this study, we focused on the scenario in which both the genetic variant and the secondary phenotype are associated with the primary disease. Under this scenario, it has been shown that the bias in the estimates of marker-secondary trait association is present under both the null and alternative hypotheses [Monsees et al., 2009]. Using a simulated example, we showed that either adjusting or not adjusting for the primary disease status resulted in a biased estimate of the OR of the genetic variant associated with the secondary phenotype and further demonstrated that the magnitude of the bias depended on the prevalence of the primary disease and, to a lesser extent, on the prevalence of the secondary phenotype. Therefore, we propose an approach to reduce the bias in OR estimation of the genetic variants associated with the secondary phenotype that accounts for the prevalences of the primary disease and secondary phenotype. The corrected ORs were obtained by solving non-linear equations involving prevalences iteratively. We also propose a bootstrapping method to estimate the empirical confidence intervals for the corrected ORs. We extended the IPW regression approach [Richardson et al., 2007] from the original nested case-control studies within a prospective cohort to retrospective case-control studies. We compared our proposed approach to the standard logistic regression method as well as to the extended IPW regression method. The performance of our approach was demonstrated via simulation studies as well as a real data analysis of SNPs associated with smoking quantity using lung cancer GWA data.
Frequency-matching is an important and commonly used study design for known risk confounders and has been widely used in case-control studies [Rothman and Greenland, 1998]. For example, in lung cancer studies, because smoking is a well-known risk confounder for the association between lung cancer and other risk factors, controls are typically frequency matched to cases with respect to smoking behavior. In this situation, both the logistic and IPW regression methods will result in a biased estimate of the OR of marker-secondary phenotype association. Therefore, we also propose a bias correction approach for the studies of marker-secondary phenotype associations in which the secondary phenotype in controls is frequency matched to that in primary disease cases. The results of this matched study design using logistic regression, the extended IPW regression, and our approach are reported.
2. Demonstration of Bias using a Simulated Example
2.1 Simulation Approach
We used the following simulation approach to demonstrate the bias of the OR estimates in studies of the association between genetic variants and secondary phenotypes using the logistic regression approach. First, denote two alleles at a SNP locus by A and a. Let A be the deleterious allele and a be the normal allele. We used a categorical random variable, X = {0,1, 2}, to denote the three genotypes, (a,a), (A,a), and (A,A). This coding assumes an additive genetic model, where the values of the random variable correspond to the number of copies of the A allele. When the dominant or recessive genetic model was assumed, we used a binary random variable, X = {0,1}, to denote the three genotypes. For the dominant genetic model, 0 represents genotype(a,a), and 1 represents genotypes (A,a) and (A,A). For the recessive genetic model, 0 represents genotypes (a,a) and (A,a), and 1 represents genotype (A,A). We defined another categorical random variable, Y = {0,1}, to indicate the case-control status of primary disease, with 0 representing individuals in the control group and 1 representing individuals in the case group. We defined the status of the secondary phenotype, also as a binary random variable, T = {0, 1}, with 0 representing individuals without the secondary phenotype and 1 representing individuals with the secondary phenotype.
In this article, we considered the simulation scenario as shown in Figure S1(A) in the Supplementary Materials. The simple network structure in Figure S1(A) represents the associations among the SNP (X), the secondary phenotype (T), and the primary disease (Y), where the SNP is associated with both the secondary phenotype and the primary disease and the secondary phenotype is associated with the primary disease.
First, genotypes of the SNP X were generated with the use of the genotype frequencies assuming the SNP is in Hardy-Weinberg proportion. We assumed a common SNP with a minor allele frequency (MAF) of 0.4. Therefore, the genotype frequencies were 0.36, 0.48, and 0.16 for the three genotypes (a,a), (A,a), and (A,A), respectively. Given the dataset of realizations of SNP X, secondary phenotype T was generated using logistic model Logit (Pr(T = 1| X)) = α0 + α1X and conditioned on the values of X and T, disease outcome Y was generated using the following logistic model: Logit (Pr(Y = 1|T, X)) = β0 + β1X + β2T.
In this way, we simulated a large amount of data on the population of interest and then randomly sampled 1,000 cases (individuals with the primary disease) along with 1,000 normal controls (individuals without the primary disease) from the population. When a frequency-matched study based on the secondary phenotype was considered, the cases were still sampled randomly. However, the controls were sampled so that the proportion of the presence of the secondary trait in the controls was approximately equal to that in the cases [Rothman and Greenland, 1998]. We assumed that the difference between the proportions of the presence of the secondary trait in cases and controls was ±2% with equal probability.
2.2 The Demonstration of Bias in OR Estimation as a Function of Primary Disease Prevalence
We assumed an additive genetic model for the SNP. We setα1 = β1 = 0.4055, which corresponds to an OR of 1.5 for the SNP association with primary and secondary phenotypes, and β2 = 1.9086, which corresponds to an OR of 3 for the association between secondary phenotype and primary trait. The intercept coefficients α0 and β0 were set from −6 to 3.5, with a 0.5 interval, for a total of 300 pairs of values of (α0,β0). Each pair of (α0,β0) values corresponds to specific prevalences of the secondary phenotype and the primary disease in the general population, denoted by fT and fD, respectively. This setting can cover a wide range of fD from ~0.5% to ~99% and of fT from ~3% to ~96%. Although, in reality, a disease prevalence of greater than 80% might not be realistic, many complex diseases, such as cancers and autoimmune disorders, will have an fD of less than 10% (nonetheless, we simulated the entire range of prevalence for the sake of completeness). For each pair of specific intercept coefficients, we simulated 1,000 replicates, each with 1,000 cases and 1,000 controls. Given a dataset of observations of random variables X, Y, and T, the OR of the SNP X associated with the secondary phenotype T for each replicate was determined by logistic regression. The ORs were estimated with and without using the primary disease status as a covariate. Medians of the ORs based on 1,000 replicates are shown in Figure S1 (B1, without adjusting for primary disease status; B2, with adjusting for primary disease status) as functions of the prevalence of disease fD. Meanwhile, we also estimated the percentages of replicates for which 95% confidence intervals include the pre-specified OR (OR = 1.5), based on 1,000 replicates. The results are shown in Figure S1 (C1, without adjusting for primary disease status; C2, with adjusting for primary disease status). The relationship between the median of the ORs and the prevalence of the secondary phenotype fT was also investigated but is not reported in this article because the impact of fT on bias in the estimation of OR is low.
Our results show that the bias exists for both logistic regression approaches, and the magnitude of the bias depends on the prevalence values of the primary disease and the secondary phenotype. Furthermore, it is clear that estimating ORs by simply adjusting for primary disease status in the logistic regression is not a uniformly better strategy than estimating ORs without adjustment. Similarly, ignoring primary disease status is also not a good strategy and leads to biased estimates of OR depending upon the prevalence values. If investigators decide to use one of these two strategies, it is necessary to define a threshold for disease prevalence values. That is, based on whether the disease is rare, very common, or moderately common, one chooses to adjust for the primary disease status as a covariate in the regression analysis or not. However, choosing threshold values for the prevalence of disease is not straightforward because the threshold value differs according to the true underlying OR, which is unknown. In addition, near the threshold value, both approaches will result in biased estimation of ORs. Therefore, developing a uniformly better approach for reducing the bias of OR estimates will result in a robust tool for case-control studies of secondary phenotypes and genetic variants. In this article, we have proposed a novel approach that uses information about the prevalence of both the primary disease and secondary phenotype to provide an accurate estimate of OR.
3. Model and Methods
3.1 OR Estimation of SNP Associated with Secondary Phenotype
Recall that we assumed binary random variables for primary disease and secondary phenotype, denoted as Y = {0, 1} and T = {0, 1}, respectively. We assumed a sample with N individuals, N = N0 + N1, where N0 is the number of controls and N1 is the number of cases with respect to the primary disease. For ease of presentation, we first present the dominant or recessive genetic model, so the SNP variable is denoted as X = {0,1}. The more complicated additive genetic model will be discussed later. According to the network structure shown in Figure S1(A), we can express the dependency of each random variable using the conditional probabilities as
| (1) |
for i, j, k = 0, 1. The conditional probabilities are explained by the logistic regression models. Note that the OR of the SNP's association with the secondary phenotype is a function of the regression coefficient α1 : OR = exp(α1). The OR of the SNP can be estimated as
where nki is the number of individuals in the sample with the secondary phenotype random variable T = k and the SNP random variable X = i. For example, n11 is the number of individuals in the sample for which the secondary phenotype is present and the SNP coding variable X is 1. Conditional on N1 and N0, the expected numbers of individuals nki can be obtained as
| (2) |
The conditional probabilities of pki|j in the above equation can be written as
Note that the conditional probabilities on the right hand side of the above equation are functions of parameters as shown in Equation (1). The probabilities pi, i = 0, 1, are related to the genotypic frequencies of the SNP of interest. When the dominant genetic model was assumed, p1 = p2 +2p(1−p); when the recessive genetic model was assumed, p1 = p2, where p is the MAF. The probability q1 is the prevalence of the disease (fD) in the general population, and q0 is calculated as 1 − fD.
Therefore, the OR of the SNP associated with the secondary phenotype can be written using the expected numbers of individuals Eki,
| (3) |
The conditional probabilities were given in Equation (1) expressed by logistic regression models. Recall that the true OR of the association between the secondary phenotype and the SNP is exp(α1), which is embedded in the conditional probabilities of pj|ki and pk|i, for i, j, k = 0, 1. It is easy to verify that the right-hand side of Equation (3) equals to the estimated from the data if we substituted all the regression coefficients assessed by logistic regressions, including , , , and , and also when is estimated using these regression coefficients. However, if we employed the estimated prevalence of the disease obtained from the literature, the right-hand side of the equation would fail to equal to the estimated . This is because, generally, for the logistic regression models, the estimated values of intercept coefficients, such as and in our models, are biased because not every explanatory variable is modeled in the logistic regression. The bias also arises because the proportion of cases in a case-control study is much higher than the prevalence of the cases in the population. Therefore, the estimated intercept coefficients and will not reflect the true prevalence of the disease and the secondary phenotype; therefore, the use of them directly in the conditional probabilities in Equation (3) will lead to a biased estimator for the OR. In other words, when we substituted the estimated intercept coefficients and to estimate the prevalence value, , and estimate conditional probabilities, and , we also introduced bias to these estimates. This, in turn, introduces bias into the estimated value of the genetic variant from Equation (3). Therefore, in this study, we put constraints on prevalence values when assessing the estimated using the right-hand side of Equation (3). We incorporated information about the true prevalences of the disease and secondary phenotype on the estimated values of the intercept coefficients α0 and β0 (as described in the next section) to estimate the intercept coefficients more accurately. We then estimated the OR as a function of the regression coefficients α0, β0 and α1, where the intercept coefficients α0 and β0 incorporate the information about the known prevalence values.
3.2 Prevalences of the Primary Disease and the Secondary Phenotype
Once again consider the network structure shown in Figure S1(A), which represents the dependent relationship among the three random variables. We can write the estimated prevalences of the primary disease and the secondary phenotype as
| (4) |
| (5) |
The conditional probabilities were given in Equation (1), and pi was defined above. Equations (4) and (5) show that the prevalence of the primary disease fD is a function of regression coefficients of β0, α0, and α1, and the prevalence of the secondary phenotype fT is a function of regression coefficients of α0 and α1.
3.3 Correction Approach for Estimating the OR for a SNP Associated with the Secondary Phenotype
Given a sample with N independent individuals for a case-control study of the primary disease of interest, one can estimate the regression coefficients and , as well as the biased for the SNP, using logistic regression as described above. Meanwhile, the genotype frequencies of the SNP , i = 0, 1, can also be estimated from the data set. Moreover, the estimated prevalences of the primary disease and the secondary phenotype in the general population can be obtained from the literature. Therefore, the Equations (3), (4), and (5) are a system of nonlinear equations with three unknown variables, β0, α0, and α1. The solution to this nonlinear equation system will give us the corrected OR for the SNP associated with the secondary phenotype. We denote the corrected OR as , where is the solution from the system of nonlinear equations (3), (4), and (5). We employed the `fsolve' function in Matlab [Mathworks, 2002] to solve the nonlinear equation system with the use of default settings. By default, the `fsolve' function uses the trust-region dogleg algorithm, which is a variant of the Powell dogleg method [Powell, 1970]. From now on, we will use to represent the biased OR for SNPs obtained using logistic regression without adjusting for the primary disease status and to refer to the corrected OR obtained by solving the system of nonlinear equations (3), (4), and (5). Furthermore, we will use to represent the biased OR obtained using logistic regression after adjusting for the primary disease status.
3.4 Additive Genetic Model
When an additive genetic model was assumed for the SNP, we used a categorical random variable, X = {0, 1, 2}, to denote the three genotypes (a,a), (A,a), and (A,A). The primary disease status and secondary phenotype status were denoted by binary random variables, Y = {0,1} and T = {0, 1}, as in the previous sections. In this situation, the biased obtained using logistic regression is given by the per-allele odds ratio, which corresponds to the odds ratio with respect to each copy of the deleterious allele. There are different ways to assess the per-allele odds ratio. We applied two approaches to evaluate the per-allele estimated OR, and therefore, obtained two corrected s. The final corrected was calculated as the average of the two per-allele s. First, the per-allele OR can be estimated as the odds ratio of X = 1 versus X = 0, so the equation will be the same as Equation (3). Therefore, using the system of nonlinear equations (3), (4), and (5), we can obtain a corrected per-allele . Meanwhile, it is known that the natural logarithm of the per-allele odds ratio can be evaluated as half of the natural logarithm of the odds ratio of X = 2 versus X = 0, as shown in the following equation
| (6) |
where the conditional probabilities were given in Equation (1), and pi, i = 0, 1, and 2, are the frequencies for the three genotypes, which equal to (1 − p)2, 2p(1 − p), and p2, respectively, under Hardy-Weinberg proportion, where p is the MAF. The second corrected per-allele can be obtained by solving the system of nonlinear equations (4), (5), and (6). Therefore, the final corrected for an additive genetic model can be estimated as the average of the two .
3.5 Confidence Intervals: Bootstrapping
We provided the empirical confidence intervals, using a resampling-based method [Efron and Tibshirani, 1993]. Given regression coefficient estimate , the empirical confidence interval of corrected was obtained by the following steps:
Take B samples from the normal distribution with mean and variance , where is the standard error of estimate . Denote the bootstrap samples as , u = 1, 2, …, B. The bootstrap is then estimated as , u = 1, 2, …, B.
For each , calculate the bootstrap corrected , u = 1, 2, …, B by solving the system of nonlinear equations as described in the previous sections. For the dominant or recessive genetic model, the equations (3) ~ (5) are employed; for the additive genetic mode, the equations (3) ~ (6) are employed.
Let be the uth ordered bootstrap estimate. Then 100(1− γ)% confidence interval of corrected is given as , .
3.6 Extended IPW Regression Approach for Retrospective Case-Control Studies
The IPW regression approach was first proposed by Richardson et al. to evaluate the OR estimate of secondary trait–genotype association in a nested case-control study within a prospective cohort [Richardson et al., 2007]. The idea is to use different weights for cases and controls in the logistic regression model, where the weights are given by the inverses of the sampling fractions of cases and controls in a study base or a prospective cohort. However, in a retrospective case-control study, the sampling fractions are usually unknown. In this article, we extended the IPW regression approach to the retrospective case-control study by introducing new weights based on use of the primary disease prevalence value similar to our proposed approach.
We assumed that there are M individuals in the finite general population. Given the prevalence of the primary disease fD, the number of cases with respect to the primary disease is fD × M, and the number of controls free of the primary disease is (1 − fD) × M. Considering a retrospective case-control study of N1 cases and N0 controls, the corresponding sampling fractions can be estimated as N1 /(fD × M) for cases and N0 /((1 − fD) × M) for controls. The weights for cases and controls are given as the reciprocals of the sample fractions. Moreover, if we use weight 1 for cases, then the weight for controls can be given as a ratio of (N1(1 − fD))/(N0fD). The size of the general population M is cancelled in the ratio formula; therefore, it does not have an impact on the weights. However, the prevalence of the primary disease still plays a major role in estimating the weights in the IPW regression approach. The IPW regression can be performed by using the SAS or R software packages [Monsees et al., 2009; Richardson et al., 2007]. We used ORIPW to represent the OR of the marker-secondary phenotype association obtained by using this extended IPW regression approach.
3.7 Bias Correction Approach for Frequency-Matching Study Design with Respect to the Secondary Phenotype
In many case-control studies, controls are frequency-matched to cases in order to reduce the effect of a known confounding factor. In the study of frequency-matched case-control design with respect to the secondary phenotype, in addition to the reasons we describe above, frequency matching also contributes to bias in the estimate of the OR for genetic variants associated with the secondary phenotype. The IPW regression approach and the proposed bias correction approach will not perform well unless accounted for in the frequency-matched study design. Currently, it is not clear to us how the IPW approach can be extended to frequency-matched case-control studies. However, we have adapted our proposed bias correction approach to frequency-matched case-control data analysis.
Note that the expected numbers of individuals Eki for T = k and X = i were calculated as the summation of the expected numbers of individuals from cases (Eki|1) and from controls (Eki|0) (see Equation (2)). When the frequency-matching design with respect to the secondary phenotype of interest is employed, the distribution of the secondary trait in controls should be the same as that in cases. That is, the expected numbers of individuals from cases are still the same as those in the unmatched case-control studies. However, the expected numbers of individuals from controls will be different from those in the unmatched case-control studies due to the frequency-matching design. Therefore, Equation (2) can be modified as follows:
| (7) |
The conditional probabilities of pki|1 were given in Section 3.1. After some manipulations, the conditional probabilities hi|k0hk|1 can be written as
The probabilities on the right-hand side of the above equation were given in Section 3.1. Equations (3) and (6) could then be modified accordingly using the new expected numbers of individuals Eki as in Equation (7).
Recall that the estimated regression coefficient (corresponding to the disease-secondary phenotype association) in our approach was evaluated from the sample data using logistic regression. In the frequency-matching case-control studies, the estimated value of is non-significant and could not represent the true association between the secondary phenotype and the primary disease. However, because the matching design considers the known risk-confounding factor at the study design phase, we typically know the associated risk for the primary disease and the secondary phenotype before the phase of analysis. Therefore, for the frequency-matching case-control studies, we added one more constraint on the value of , which is fixed as the risk coefficient estimated in the related unmatched case-control studies. The correct OR for the frequency-matching study was denoted as .
4. Results
4.1 Simulation Results
We examined the performance of the proposed approach by performing simulation studies. The details of the simulation approach have been presented in Section 2.1. Here we simulated different genetic models when simulating values for the SNP, including additive, dominant, and recessive models. We used the same specific parameters as those in Section 2.2, where α1 = β1 = 0.4055 and β2 = 1.9086. The same 300 pairs of the intercept coefficients α0 and β0 were used. The results of the simulation studies were based on 1,000 replicates, each replicate with 1,000 cases and 1,000 controls according to primary disease. We compared the corrected to the biased and obtained without or with adjusting for the primary disease status, respectively, as well as ORIPW obtained by the extended IPW regression.
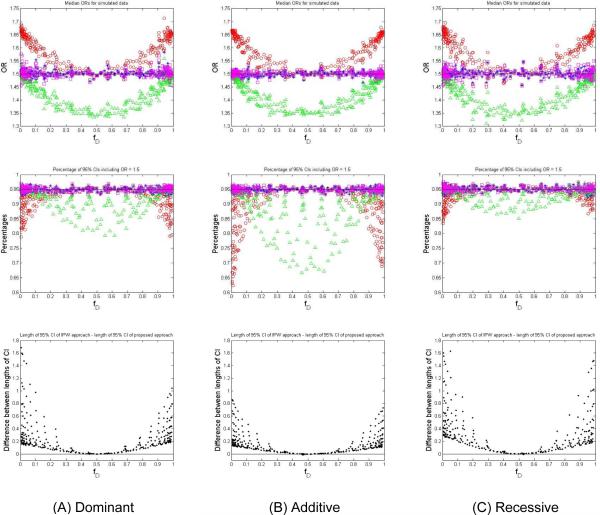
The results of the simulation studies were grouped into three columns with respect to three different genetic models: dominant, additive, and recessive (Figure 1). Within each column, the upper panel shows the median ORs based on 1,000 replicates obtained with the use of the different approaches: logistic regression without adjusting for the primary disease status (red symbol “◯”), logistic regression with adjusting for the primary disease status (green symbol “△”), the extended IPW regression (purple symbol “□”), and the bias correction approach proposed in this article (blue symbol“*”). For all three genetic models, the medians of ORIPW and our corrected were close to the pre-specified OR = 1.5 for the entire range of the prevalence of the primary disease. The other two logistic regression approaches, however, provided either upward- or downward-biased estimates of ORs. The middle panel in each column of Figure 1 represents the percentages of 95% confidence intervals for the 1,000 replicates covering the pre-specified OR of 1.5. These results also showed that both the extended IPW approach and the bias correction approach provide confidence intervals with accurate coverage probabilities for the underlying true OR. For example, considering the additive model, when , the median was 1.60, which overestimated the true OR, the median was 1.43, which underestimated the true OR, and the ORIPW and the corrected were 1.50; the corresponding percentages of 95% confidence intervals that included the true OR (OR = 1.5) were 85.1%, 90.2%, 94.6% and 94.5%, respectively. In another example with rare prevalence of disease, when , we obtained a median estimated of 1.66 and , ORIPW, and of 1.50. The corresponding percentages of 95% confidence intervals that included the true OR were 68.4%, 96.3%, 95.5%, and 96.3%, respectively. Compared to , the extended ORIPW, the corrected , and reduced the bias in risk estimation by 16% and estimated the true OR accurately.
Figure 1. Simulation results for three genetic models with risk secondary phenotype, based on 1,000 replicates, each with 1,000 cases and 1,000 controls. The true OR of the SNP associated with secondary phenotype was 1.5.
OR = odds ratio fD = estimated prevalence of primary disease
CI = confidence interval
Symbol “○” represents the results obtained by using logistic regression without adjusting for the primary disease status. Symbol “Δ” represents the results obtained by using logistic regression with adjusting for the primary disease status. Symbol “□” represents the results obtained by using the extended IPW regression approach. Symbol “*” represents the results obtained by using the bias correction approach proposed in this article. (A) Dominant genetic model. (B) Additive genetic model. (C) Recessive genetic model.
Overall, both the extended IPW regression and our bias correction approaches performed well when estimating the ORs for the genetic variants associated with the secondary phenotype. However, the confidence intervals (CI) for true OR based on our approach were always smaller than or equal to those based on the IPW regression at a fixed level of confidence coefficient. In the lower panel of each column of Figure 1, we show that the differences evaluated by subtracting the median lengths of 95% CIs based on our approach from those based on the extended IPW approach are always non-negative for the dominant, additive, and recessive model. In other words, the median lengths of the 95% CIs of the bias correction approach are always smaller than or equal to those of the IPW approach. For example, when in the additive genetic model, the median length of 95% CI of the bias correction approach is 0.38, compared with 0.51 for the IPW regression approach. Given Pr(real OR ε CI based on the IPW regression) ≈ Pr(real OR ε CI based on the proposed approach) at a fixed confidence coefficient then a desirable property would be to have a smaller length of CI. Because Length(CI based on the proposed approach) ≤ Length(CI based on the IPW regression), our proposed approach is preferred.
In addition, we also performed simulation studies where the secondary phenotype was the protective factor for the primary disease and the SNP was a risk factor for both the primary disease and the secondary phenotype. We used a set of specific parameters of the logistic models, where α1 = β1 = 0.4055 corresponded to OR = 1.5 and β2 = −0.6931 corresponded to OR = 0.5. Three genetic models were studied. The median , , ORIPW and corrected values, as well as the percentages of 95% confidence intervals covering OR = 1.5 for different approaches, were calculated (showed in Figure S2 in the Supplementary Materials). As expected, in the simulation studies in which the secondary phenotype was the protective factor for the primary disease, the median values of and were biased in a manner similar to those obtained when the secondary phenotype was a risk factor for the primary disease, as discussed previously. Except in these studies, the downward bias arose when using logistic regression without adjusting for the primary disease status, while the upward bias arose when using logistic regression with adjusting for the primary disease status. However, our proposed corrected estimated the underlying OR more accurately than both logistic regression approaches. Therefore, the bias correction approach proposed here and the extended IPW approach would perform well for estimating the OR, whether the secondary phenotype (or the SNP) is a risk or a protective factor to the primary disease. However, our bias correction approach is favored because it leads to smaller confidence intervals with the same confidence coefficient.
4.2 Sensitivity Analysis
In the simulation studies, we assumed that the prevalences of the primary disease and secondary phenotype were known. However, in reality, it cannot be known with certainty that prevalences obtained from the literature are accurate. Here, we assessed the sensitivity of the corrected to the estimated prevalence of the primary disease and the secondary phenotype . We considered several simulation scenarios under the assumption of an additive genetic model, and the real prevalence of the primary disease fD values used to simulate the data were 1.6%, 10.5%, and 54.3% (from rare disease to common disease), and the prevalence of the secondary phenotype fT was set as a fixed value of 34%. We evaluated the corrected using a range of prevalence values centered on the true prevalence values ([fD −△D, fD +△D] and [fT −△T, fT +△T]). The error term △D was defined differently with respect to different fD. We defined △T as 3% in all situations. The specific parameters and the results are listed in Table 1. All the results were very similar to those obtained using the real prevalences. For example, when the true prevalence of the primary disease and the secondary phenotype were 1.6% and 34%, respectively, the corrected was estimated as 1.4983 using real prevalence values, 1.4973 when and , 1.4969 when and , 1.4999 when and , and 1.4993 when and .
Table 1.
Results of sensitivity analysis when true OR = 1.5, based on 1,000 cases and 1,000 controls.
| fD | fT | Δ D | Δ T |
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| [fD, fT] | [fD−ΔD, fT− ΔT] | [fD−ΔD, fT + ΔT] | [fD+ΔD, fT−ΔT] | [fD+ΔD, fT + ΔT] | |||||||
| 1 | 1.6579 | 1.4877 | 1.6% | 34% | 0.2% | 3% | 1.4983 | 1.4973 | 1.4969 | 1.4999 | 1.4993 |
| 2 | 1.6009 | 1.4341 | 10.5% | 34% | 2% | 3% | 1.4983 | 1.4885 | 1.4853 | 1.5114 | 1.5073 |
| 3 | 1.4988 | 1.3475 | 54.3% | 34% | 4% | 3% | 1.4951 | 1.4986 | 1.4987 | 1.4896 | 1.4909 |
4.3 Frequency-Matched Case-Control Studies with Respect to the Secondary Phenotype
To investigate the performance of the proposed bias correction approach in frequency-matched case-control studies with respect to the secondary phenotype of interest, we performed simulations. We assumed a dominant genetic model. Without loss of generality, we considered different scenarios involving different prevalences of the primary disease (10%, 20%, 40%, and 60%), different prevalences of the secondary trait (5%, 15%, 40%, 60%, and 80%), and different ORs of genotype-secondary trait association (1.5, 2, and 3). All the other parameters were the same as those in Section 4.1. We simulated 1,000 replicates, each with 1,000 randomly sampled cases and 1,000 frequency-matched controls, with respect to the secondary phenotype. Median ORs obtained with the use of different approaches are reported in Table 2, including logistic regression without adjusting for the primary disease status (), the extended IPW regression (ORIPW), and the proposed bias correction approach (). Among all the scenarios with frequency-matching case-control data, both and ORIPW were biased. The extended IPW regression performed very similarly to the logistic regression. We observed that the magnitude of the bias in and ORIPW varied with respect to prevalence values of both the primary disease and secondary trait. However, the performed very well for estimating the true OR in all scenarios. For example, when the true OR of the genotype-secondary phenotype association was 2, and , we found that , ORIPW = 1.90, and = 2.00. Both and ORIPW had about a 10% bias in risk estimation, while estimated the real OR accurately.
Table 2.
Median ORs using different approaches for frequency-matched case-control studies with respect to secondary phenotype, using different prevalence values of primary disease and secondary phenotype, based on 1,000 replicates, each with 1,000 cases and 1,000 frequency-matched controls with respect to secondary phenotype.
|
fD = 10% |
fD = 20% |
fD = 40% |
fD = 60% |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ORt | fT | ORIPW | ORIPW | ORIPW | ORIPW | ||||||||
| 1.5 | 5% | 1.43 | 1.42 | 1.51 | 1.38 | 1.38 | 1.51 | 1.35 | 1.35 | 1.51 | 1.37 | 1.37 | 1.50 |
| 15% | 1.42 | 1.43 | 1.50 | 1.39 | 1.39 | 1.51 | 1.34 | 1.35 | 1.51 | 1.37 | 1.36 | 1.50 | |
| 40% | 1.44 | 1.45 | 1.50 | 1.40 | 1.39 | 1.50 | 1.34 | 1.34 | 1.49 | 1.36 | 1.36 | 1.51 | |
| 60% | 1.46 | 1.45 | 1.51 | 1.41 | 1.40 | 1.50 | 1.36 | 1.36 | 1.50 | 1.35 | 1.34 | 1.50 | |
| 80% | 1.45 | 1.46 | 1.50 | 1.42 | 1.42 | 1.50 | 1.36 | 1.36 | 1.50 | 1.34 | 1.34 | 1.49 | |
| 2.0 | 5% | 1.89 | 1.90 | 2.00 | 1.82 | 1.84 | 2.00 | 1.79 | 1.79 | 2.01 | 1.81 | 1.81 | 2.00 |
| 15% | 1.90 | 1.90 | 2.00 | 1.83 | 1.83 | 2.00 | 1.79 | 1.80 | 2.01 | 1.81 | 1.81 | 2.00 | |
| 40% | 1.91 | 1.93 | 2.00 | 1.86 | 1.85 | 1.99 | 1.80 | 1.80 | 2.01 | 1.80 | 1.80 | 2.00 | |
| 60% | 1.91 | 1.93 | 1.98 | 1.86 | 1.88 | 1.99 | 1.82 | 1.81 | 2.01 | 1.79 | 1.78 | 2.00 | |
| 80% | 1.94 | 1.94 | 2.00 | 1.91 | 1.91 | 2.02 | 1.81 | 1.80 | 1.98 | 1.79 | 1.79 | 2.02 | |
| 3.0 | 5% | 2.81 | 2.84 | 3.00 | 2.70 | 2.74 | 2.97 | 2.68 | 2.69 | 3.00 | 2.74 | 2.73 | 3.02 |
| 15% | 2.84 | 2.85 | 3.00 | 2.75 | 2.76 | 3.00 | 2.69 | 2.69 | 3.02 | 2.71 | 2.72 | 3.02 | |
| 40% | 2.85 | 2.85 | 2.98 | 2.77 | 2.78 | 2.99 | 2.68 | 2.67 | 3.00 | 2.68 | 2.68 | 2.99 | |
| 60% | 2.89 | 2.92 | 3.02 | 2.81 | 2.83 | 3.02 | 2.70 | 2.70 | 3.01 | 2.68 | 2.68 | 3.03 | |
| 80% | 2.91 | 2.93 | 3.01 | 2.81 | 2.80 | 2.99 | 2.72 | 2.71 | 3.01 | 2.68 | 2.68 | 3.00 | |
ORt = the true OR of the genetic variant associated with the secondary phenotype
5. Real Data Analysis
We next applied our approach to the case-control association study of smoking behavior and the CHRNA5-A3 region SNP, rs1051730, using lung cancer GWA study data [Amos et al., 2008; Spitz et al., 2008; Wang et al., 2010]. This analysis included N1 = 1153 lung cancer case subjects who were current or former smokers and N0 = 1137 control subjects frequency-matched to the cases by age, sex, and smoking status. All the case and control subjects were Caucasian. Lung cancer cases were accrued at The University of Texas MD Anderson Cancer Center and were histologically confirmed. Controls were ascertained through a multi-specialty physician practice from the same area. Questionnaire data were obtained by personal interview. This study was approved by the institutional review board at MD Anderson Cancer Center, and all participants provided written informed consent. We selected the number of cigarettes per day, or daily smoking quantity (SQ), a commonly used measurement of smoking intensity. Typically, the SQ measure is categorized into two levels: SQ<25, light smokers (coded as 0); SQ≥25, heavy smokers (coded as 1) [CDC, 2005]. The genetic variant (rs1051730) was coded assuming dominant, additive and recessive genetic models. In the original case-control association study of lung cancer, the lung cancer controls are frequency-matched to the cases by smoking status. Therefore, we employed the proposed bias correction approach for frequency-matching study design with respect to secondary phenotype to obtain the corrected . The ORIPW obtained by using the extended IPW regression was also reported.
Our aim here was to evaluate the performance of the proposed approach with real data. We first estimated the regression coefficients required to be substituted into the system of nonlinear equations, as well as the biased for rs1051730 associated with SQ, by applying logistic regression to the lung cancer case-control data. The regression coefficients and the biased s for different genetic models are reported in Table 3. For example, when the recessive genetic model was assumed, , (95% confidence interval (CI) = (1.2492, 2.0269)) and (95% CI = (1.2223, 1.9866)). The MAF was also estimated from the data as 37%, and therefore under Hardy-Weinberg proportion, the genotyping frequencies , i = 0, 1, and 2, were calculated as 0.40, 0.46, and 0.14, respectively. The prevalences of lung cancer () and heavy smokers () in ever smokers were obtained from the literature as 14% and 12%, respectively [CDC, 2005; Villeneuve and Mao, 1994]. We further assumed the OR of association between SQ and lung cancer as 1.86 as reported by Peto et al. [2000], therefore, for all models. The estimatedORIPW values were slightly higher than those from logistic regressions, and always had the widest 95% CIs among all the estimators. For the example of recessive genetic model, ORIPW = 1.6026 (95% CI = (1.1737, 2.1882). The corrected values of the SNP rs1051730 were: 1.3416 for dominant genetic model (95% CI = (1.1353, 1.5883)), 1.3214 for additive genetic model (95% CI = (1.1711, 1.4914)), and 1.6831 for recessive genetic model (95% CI = (1.3213, 2.1489). Therefore, the corrected could reduce the bias in risk estimation by certain percentages according to different genetic models. For example, when the recessive genetic model was assumed, , and ORIPW concluded that the individuals with one copy of the deleterious allele are about 59.1%, 56.0% or 60.3%, respectively, more likely to be heavy smokers than those with no deleterious allele, whereas the corrected reduced the bias in risk estimation by about 8~12% and suggested that individuals with one copy of the deleterious allele are 68.3% more likely to be heavy smokers. Moreover, the 95% CIs based on our approach are much smaller than those based on IPW regression, which suggests that our approach provides a more accurate estimation of the OR of SNP rs1051730 associated with smoker behavior.
Table 3.
OR estimators and corresponding 95% confidence intervals from real data analysis of genetic variant rs1051730 associated with the number of cigarettes smoked per day based on lung cancer case-control study data.
| Odds Ratio |
95% Confidence Intervals |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Genetic Model | ORIPW | Logistic (without adjustment) | Logistic (with adjustment) | IPW | Our approach | ||||
| Dominant | 0.1997 | 1.3071 | 1.2883 | 1.3245 | 1.3416 | (1.1055, 1.5453) | (1.0890, 1.5240) | (1.0787, 1.6287) | (1.1353, 1.5883) |
| Additive | 0.1883 | 1.2894 | 1.2739 | 1.3004 | 1.3214 | (1.1430, 1.4545) | (1.1286, 1.4378) | (1.1173, 1.5106) | (1.1711, 1.4914) |
| Recessive | 0.1951 | 1.5912 | 1.5583 | 1.6026 | 1.6831 | (1.2492, 2.0269) | (1.2223, 1.9866) | (1.1737, 2.1882) | (1.3213, 2.1489) |
6. Discussion
In genetic association studies, cases and controls were ascertained with respect to the primary diseases of interest. Other traits associated with the primary diseases were also collected. Recently, in the GWA studies, the same data is now used to identify SNPs that are associated with these secondary phenotypes. However, using logistic regression to study the secondary phenotype is problematic, since the data associated with secondary phenotype is not sampled according to the principals of case-control study design. In this article, we found that the odds ratios for genetic variants associated with secondary phenotypes can be biased. Generally, investigators assume that their results will not be biased if the odds ratios are adjusted for the primary disease status as a covariate in the association study. We show, however, that adjusting for primary disease status will still result in a biased estimate of the OR in many situations.
In our study, we further found that the magnitude of bias in the OR estimate of the genetic variant depends on the prevalence values of the primary disease and secondary phenotype in the general population. Moreover, the prevalence of the primary disease is most important in the determination of bias when estimating the OR. Therefore, we proposed an approach to provide a more accurate estimate of the OR, which incorporates information about the true prevalence values. The corrected obtained using the new approach was shown in our simulation studies to be a more accurate estimator of OR. In addition to the prevalence values of the primary disease and secondary phenotype, several other parameters also have an impact on the magnitude of bias in the OR estimate, including the correlation between the genetic variant and primary disease and the correlation between the secondary phenotype and primary disease. However, these parameters would not affect the performance of our approaches, since our approaches account for these parameters.
The major advantage of our approach is that there is no need to decide whether the primary disease is rare or common. Given a case-control sample and corresponding prevalences of disease and secondary phenotype, the corrected can be assessed by solving a nonlinear equation system with the use of iterative algorithms. Therefore, the estimates of the two prevalence values are the most important parameters for the performance of our approach. Generally, the true prevalence values in a population are not known with certainty, and we showed, by using a sensitivity analysis, that the misspecification of primary disease and secondary phenotype prevalence would not have a large impact on the estimate of the corrected . It should also be noted that in our theoretical model, the prevalences were defined as the proportion of individuals with the disease in the general population. Also, in this study, we investigated issues related to OR estimate for binary secondary phenotypes. Issues related to continuous secondary phenotypes will be considered in the future.
We also extended the IPW regression method originally proposed by Richardson et al. for prospective case-control studies [Richardson et al., 2007] to retrospective case-control studies based on our philosophy of using the prevalence of the primary disease. We compared the performance of our bias correction approach proposed in this article with that of the IPW approach. Overall, the two approaches performed similarly well, and both could provide more accurate estimations for the OR of genotype-secondary phenotype than the standard logistic regressions. However, our approach has a few advantages over IPW regression. First, our proposed approach always provides smaller or equal CIs at a fixed confidence coefficient, which is a desirable attribute. Furthermore, in addition to the prevalence of the primary disease, our approach can also account for the prevalence of the secondary phenotype. Although the prevalence of the secondary phenotype has less impact on OR estimation than the prevalence of the primary disease, it still introduces a bias in the estimation of true OR. Finally, our approach is robust even if the prevalence of the disease and the secondary trait are rare, whereas the approach based on the IPW method is sensitive to rare diseases. In this study, we also considered more complex association structures in which multiple covariates were included in the analysis. The proposed approach still resulted in accurate estimators; while interestingly, the IPW approach was biased for some scenarios (simulation results not reported).
Frequency matching has been widely applied in case-control association studies. When the secondary trait of interest is the risk-confounding factor in a frequency-matched case-control study, neither the standard logistic regression nor the IPW regression approach could estimate the OR of the genotype-secondary trait correctly. In this article, we also proposed a bias correction approach for the frequency-matched case-control study with respect to the secondary trait of interest. The simulation studies were performed to show that the corrected obtained using our approach is a more accurate estimator of OR than both standard logistic and IPW regressions. We also applied this proposed approach to a real data analysis of genetic variant (rs1051730) associated with number of cigarettes per day using lung cancer case-control study data.
In conclusion, to estimate the OR for genetic variants associated with a secondary phenotype, we propose a new approach that incorporates information about the prevalences of the primary disease and secondary phenotype. The proposed approach is more accurate and robust than the standard logistic regression and the extended IPW regression approaches, and the coverage probability of corresponding bootstrap confidence intervals is higher than that of the standard approach. We also propose a new approach for estimating the OR for genotype-secondary trait association in a frequency-matched case-control study with respect to the secondary trait of interest, which also provides an accurate estimation of the true OR.
Supplementary Material
Acknowledgments
We appreciate the two anonymous reviewers for their insightful and constructive comments. This research was supported by National Institutes of Health grant 1R01CA131324. This study makes use of lung cancer data generated by support from NIH grants R01CA55769 and R01CA121197.
References
- Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–22. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC Cigarette smoking among adults — United States, 2004. MMWR. 2005;54:1121–4. [PubMed] [Google Scholar]
- Efron B, Tibshirani RJ. An introduction to the bootstrap. Chapman and Hall; New York: 1993. [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–94. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Kraft P, Chen C, Buring JE, Pare G, Hankinson SE, Chanock SJ, Ridker PM, Hunter DJ, Chasman DI. Genome-wide association studies identify loci associated with age at menarche and age at natural menopause. Nat Genet. 2009;41:724–8. doi: 10.1038/ng.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–7. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- Li H, Gail MH, Berndt S, Chatterjee N. Using cases to strengthen inference on the association between single nucleotide polymorphisms and a secondary phenotype in genome-wide association studies. Genet Epidemiol. 2010;34:427–33. doi: 10.1002/gepi.20495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DY, Zeng D. Proper analysis of secondary phenotype data in case-control association studies. Genet Epidemiol. 2009;33:256–65. doi: 10.1002/gepi.20377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathworks . Matlab. Mathworks; Cambridge, MA: 2002. [Google Scholar]
- Monsees GM, Tamimi RM, Kraft P. Genome-wide association scans for secondary traits using case-control samples. Genet Epidemiol. 2009;33:717–28. doi: 10.1002/gepi.20424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong KK, Elks CE, Li S, Zhao JH, Luan J, Andersen LB, Bingham SA, Brage S, Smith GD, Ekelund U, et al. Genetic variation in LIN28B is associated with the timing of puberty. Nat Genet. 2009;41:729–33. doi: 10.1038/ng.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peto R, Darby S, Deo H, Silcocks P, Whitley E, Doll R. Smoking, smoking cessation, and lung cancer in the UK since 1950: combination of national statistics with two case-control studies. BMJ. 2000;321:323–9. doi: 10.1136/bmj.321.7257.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell MJD. A fortran subroutine for solving systems of nonlinear algebraic equations. In: Rabinowitz P, editor. Numerical methods for nonlinear algebraic equations. Gordon and Breach; 1970. pp. 115–61. [Google Scholar]
- Richardson DB, Rzehak P, Klenk J, Weiland SK. Analyses of case-control data for additional outcomes. Epidemiology. 2007;18:441–5. doi: 10.1097/EDE.0b013e318060d25c. [DOI] [PubMed] [Google Scholar]
- Rothman KJ, Greenland S. Modern epidemiology. Lippincott Williams & Wilkins; Philadelphia, PA: 1998. [Google Scholar]
- Spitz MR, Amos CI, Dong Q, Lin J, Wu X. The CHRNA5-A3 region on chromosome 15q24-25.1 is a risk factor both for nicotine dependence and for lung cancer. J Natl Cancer Inst. 2008;100:1552–6. doi: 10.1093/jnci/djn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Tobacco and Genetics Consortium Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42:441–7. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, Manolescu A, Thorleifsson G, Stefansson H, Ingason A, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–42. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve PJ, Mao Y. Lifetime probability of developing lung cancer, by smoking status, Canada. Can J Public Health. 1994;85:385–8. [PubMed] [Google Scholar]
- Wang J, Spitz MR, Amos CI, Wilkinson AV, Wu X, Shete S. Mediating effects of smoking and chronic obstructive pulmonary disease on the relation between the CHRNA5-A3 genetic locus and lung cancer risk. Cancer. 2010;116:3458–62. doi: 10.1002/cncr.25085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.