Abstract
The rice Xa13 gene, whose promoter harbors a UPT (up-regulated by transcription activator-like [TAL] effector) box, UPTPthXo1, plays a pivotal role in the race-specific pathogenicity caused by Xanthomonas oryzae pv. oryzae (Xoo) strain PXO99. PXO99 causes rice disease by inducing Xa13. It is unknown, however, whether the UPTPthXo1 box is the only PXO99-responsive cis-regulating elements in the activation of Xa13 expression. We analyzed the expression of a series of end- and site-truncated and site-mutated Xa13 promoters in rice and the binding of PXO99 protein to the intact, partial, or site-mutated UPTPthXo1 boxes. In the Xa13 promoter, UPTPthXo1 box is the only Xoo-responsive cis-acting element that results in PXO99-induced Xa13 expression. The 5'-terminal second, third, and fourth nucleotides of the box are important for bacterial protein binding and gene activation; mutation of any one of these sites abolished PXO99-induced gene expression. Furthermore, the 3'-half of the UPTPthXo1 box is also required for protein binding and gene activation. These findings will enhance our understanding of the molecular mechanism of the interaction of rice and Xoo via UPT boxes and TAL effectors.
Keywords: Bacterial blight, disease, Oryza sativa, UPT box, Xanthomonas oryzae
INTRODUCTION
Bacteria blight, which is caused by Xanthomonas oryzae pv. oryzae (Xoo), is one of the most devastating diseases restricting rice production. More than 30 disease resistance (R) genes that mediate race-specific resistance to Xoo have been identified, and six of them have been characterized (Chu and Wang, 2007).
The recessive xa13 is a new type of R gene that confers resistance to Philippine Xoo strain PXO99 (Chu et al., 2006). PXO99 infection does not influence the expression of recessive xa13, but it induces the expression of its dominant (susceptible) allele Xa13; suppressing the expression of Xa13 can result in the same level of resistance to PXO99 as conferred by xa13 in rice (Chu et al., 2006). Sequence analysis of a series of rice lines carrying either dominant Xa13 or recessive xa13 revealed a set of recessive alleles of xa13, whose encoding proteins are different from or identical to that encoded by dominant Xa13. However, all the recessive alleles had nucleotide substitutions, deletions, or insertions in a promoter region corresponding to the –86 to –69 region of the promoter of dominant Xa13, suggesting that promoter mutations may result in xa13-mediated disease resistance (Chu et al., 2006). Promoter swap analyses further confirmed that the expressional non-reaction to PXO99 infection caused by promoter mutation, not its protein composition, is the key factor for xa13-mediated resistance (Yuan et al., 2009). Thus, the dominant Xa13 is a race-specific susceptibility gene. Activation of Xa13 is required for the development of disease caused by PXO99.
A recent study has revealed that PXO99 is sensitive to copper (Yuan et al., 2010), which is an essential micronutrient of plants and an important element for a number of pesticides in agriculture. PXO99 overcomes rice defense by regulating Xa13, which incorporates with another two genes to remove copper from the xylem vessels, where Xoo multiplies and spreads to cause disease.
The dominant Xa13 is also known as Os8N3, whose expression can be induced by a transcription activator-like (TAL) effector PthXo1 injected into rice through the type III secretion system of Xoo strain PXO99 (Yang et al., 2006). Effectors secreted by pathogenic bacteria play an essential role in promoting diseases in plants (Kay and Bonas, 2009). Several studies have demonstrated that pathogen TAL effectors can transcriptionally activate host genes by directly interacting with the cis-regulating elements, named UPT (up-regulated by TAL effectors) boxes, in the promoters of corresponding host susceptibility genes to promote diseases or in R genes to induce defense responses (Kay et al., 2007, 2009; Römer et al., 2007, 2009a, 2009b). This interaction is determined by the specific pairing of the repeat-variable diresidues (RVDs) of the repeat domain of a TAL effector and the nucleotides of a UPT box, with one RVD pairing to one specific nucleotide in the UPT box (Boch et al., 2009; Moscou and Bogdanove, 2009). Thus, based on the pairing codes of a given TAL effector, one can predict a putative UPT box that may interact with this TAL effector. A recent study revealed that the TAL effector PthXo1 from Xoo strain PXO99 directly binds to a UPT box, UPTPthXo1 (consisting of 25 nucleotides), in the Xa13 promoter to mediate gene activation (Römer et al., 2010). However, it is unknown whether the UPTPthXo1 box is the only cis-regulating element that is responsible for Xoo-induced activation of Xa13.
In the present study, we analyzed the expression of truncated and mutated Xa13 promoters in rice. We also examined the binding of PXO99 total proteins to the intact, incomplete, or site-mutated UPTPthXo1 box and corresponding DNA fragments from recessive xa13 alleles. Our results suggest that, in the Xa13 promoter, UPTPthXo1 box is the only cis-acting element that results in PXO99-induced Xa13 expression. The important nucleotide sites of the UPTPthXo1 box and the neighboring sites of this box needed for efficient activation of the gene are discussed.
RESULTS
Identification of the Pathogen-Responsive Region of the Xa13 Promoter
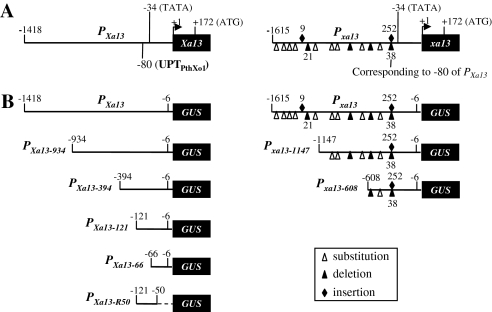
Sequence analysis suggested that the putative TATA boxes, which provide the binding sites for RNA polymerase, were at –34 for the promoters of both the dominant Xa13 gene (PXa13) and recessive xa13 gene (Pxa13) (Figure 1A). Based on the putative locations of TATA boxes, previous identification of the promoter regions of Xa13 and xa13 genes (Yuan et al., 2009), and the location of UPT box for PthXo1 effector binding (Römer et al., 2010), the DNA fragment located at the –1418 to –6 region of Xa13 and the corresponding DNA fragment located at the –1615 to –6 region of xa13 were fused with the reporter gene β-glucuronidase (GUS) to detect the pathogen-responsive regions; two truncated promoters PXa13-934 and PXa13-394 for PXa13 and Pxa13-1147 and Pxa13-608 for Pxa13 were also fused with GUS (Figure 1B). These constructs were transferred separately into rice, and each construct generated approximately 20 independent positive transgenic plants. Each transgenic plant was divided into two parts by separating the tillers at the tillering stage: one part for inoculation with Xoo strain PXO99 and another part for mock-inoculation at the booting (panicle development) stage.
Figure 1.
The Structures of Promoters PXa13 and Pxa13 from IR24 (Carrying Xa13) and IRBB13 (Carrying xa13), Respectively, and Truncated Promoters.
IR24 and IRBB13 are near-isogenic rice lines.
(A) The predicted basal regulatory element TATA in PXa13 and Pxa13. The transcription initiation site is indicated as +1. The nucleotide substitution, deletion, or insertion in Pxa13 compared with PXa13 were based on a previous report (Chu et al., 2006); the figure with a sign of deletion or insertion indicates the numbers of nucleotides inserted or deleted and the sign without a figure represents single-nucleotide deletion or insertion. ATG, translation start codon.
(B) The full-length and truncated promoters.
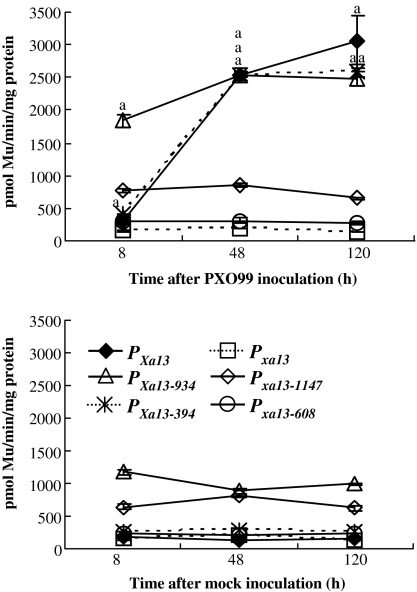
Because Xa13 expression was markedly induced at 1–5 d after PXO99 infection (Chu et al., 2006; Yuan et al., 2009, 2010), the expression level of marker gene in the transgenic plants was examine from 8 h to 5 d after infection. The expression of GUS in the transgenic plants carrying PXa13:GUS was strongly induced at 2 d and increased continually until 5 d after PXO99 infection compared with mock-inoculated control plants (Figure 2). Neither PXO99 infection nor mock-inoculation influenced GUS expression in the transgenic plants carrying Pxa13:GUS (Figure 2). The GUS level in the plants carrying PXa13:GUS at 5 d after PXO99 infection was 23-fold higher than that in the same plants at 5 d after mock-inoculation. Plants carrying PXa13-934 and those carrying PXa13-394 showed a similar induced expression pattern of GUS to plants carrying PXa13:GUS after PXO99 infection (Figure 2). However, plants carrying PXa13-934 and those carrying Pxa13-1147 showed a significantly higher level (P < 0.01) of GUS than the plants carrying PXa13:GUS or Pxa13:GUS at 8 h after infection or even without pathogen infection, respectively. Compared with mock-inoculation, PXO99 infection induced approximately two and three-fold increases of GUS levels in plants carrying PXa13-934 at 8 h, 2, and 5 d after infection, respectively. PXO99 infection did not markedly influence GUS expression in plants carrying Pxa13-1147 compared with the same plants after mock-inoculation. Plants carrying PXa13-394 and those carrying Pxa13-608 showed similar levels of GUS to the plants carrying PXa13:GUS or Pxa13:GUS when without pathogen infection. PXO99 infection showed approximately nine- and 10-fold increases in GUS levels in plants carrying PXa13-394 as compared with the same plants after mock-inoculation at 2 and 5 d after infection, respectively (Figure 2). PXO99 infection did not markedly influence GUS levels in the plants carrying Pxa13-608 compared to the same plants after mock-inoculation. These results suggest the following possibilities. First, the –1418 to –935 region of PXa13 and the corresponding –1615 to –1148 region of Pxa13 may contain PXO99-independent cis-regulating element(s) that suppress gene expression. Second, the –934 to –395 region of PXa13 and the corresponding –1147 to –609 region of Pxa13 may contain PXO99-independent cis-regulating element(s) that stimulate gene expression. Last, only the –394 to –6 region of PXa13 may contain PXO99-responsive element(s) that induce gene expression.
Figure 2.
The Expression of GUS Driven by Native (PXa13 and Pxa13) or Truncated (PXa13-934, PXa13-394, Pxa13-1147, and Pxa13-608) Promoters after PXO99-Inoculation or Mock-Inoculation at the Booting Stage.
GUS activity was determined by measuring the amount of 4-methylumbelliferone (Mu) produced under the catalysis of GUS in 1 mg total protein per minute. Sixteen to 22 T0 transgenic plants carrying each construct were used for analyses. For each time point examined, leaf fragments were collected from all the plants carrying the same construct and mixed to prepare a sample. The GUS activity at each time point was the average of three measurements ± standard deviation. The ‘a’ indicates that a significant difference (P < 0.01) was detected between pathogen-inoculated and mock-inoculated plants in the same time point.
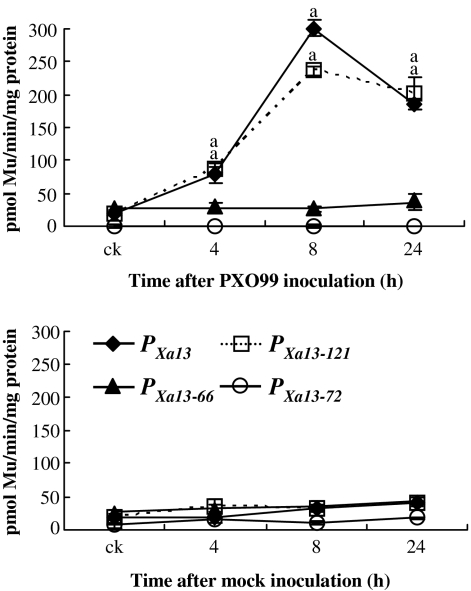
The –394 to –6 region of PXa13 harbors a PXO99-responsive element, the UPTPthXo1 box, located at –80 to –56 (Figure 1A; Römer et al., 2010). To determine whether this region may contain other PXO99-responsive elements in addition to the UPT box, two additional 5'-end truncated promoters, PXa13-121 and PXa13-66, and one 3'-end truncated promoter, PXa13-R50, of PXa13 were fused with GUS (Figure 1B). The three constructs as well as the construct carrying PXa13:GUS were transiently expressed in rice calli. The expression of GUS in the calli carrying PXa13-121:GUS was strongly induced by PXO99-inoculation compared with mock-inoculated control calli, as was the case in the calli carrying PXa13:GUS (Figure 3). PXO99-inoculation did not obviously influence GUS expression in the calli carrying PXa13-66:GUS or PXa13-R50:GUS compared with mock-inoculated calli. Because the PXa13-66:GUS construct only harbored incomplete UPTPthXo1 box, these results suggest that the –121 to –65 region of PXa13 may contain another PXO99-responsive element that induces gene expression or the –121 to –6 region may contain only one PXO99-responsive element, the UPTPthXo1 box. However, PXO99 infection did not obviously influenced GUS expression in the calli carrying PXa13-R50:GUS, which harbored the –121 to –50 region; this may be due to the lack of the basal transcriptional element, the TATA box. This assumption is supported by evidence that the GUS levels in the calli carrying PXa13-66:GUS were constantly approximately three-fold higher than those in the calli carrying PXa13-R50:GUS with either PXO99- or mock-inoculation.
Figure 3.
Transient Expression of GUS Driven by Native (PXa13) and Truncated (PXa13-121, PXa13-66, and PXa13-R50) Promoters in Rice Calli.
GUS activity was determined by measuring the amount of 4-methylumbelliferone (Mu) produced under the catalysis of GUS in 1 mg total protein per minute. Each sample was from 4.5 ml calli; all the calli were mixed to prepare a sample. The GUS activity of each sample was the average of three measurements ± standard deviation. ck, without pathogen- or mock-inoculation. The ‘a’ indicates that a significant difference (P < 0.01) was detected between pathogen-inoculated and mock-inoculated plants in the same time point. This experiment was biologically repeated twice and similar results were obtained.
More Xoo Proteins Bind to the Promoter Fragment of Xa13 than to the Promoter Fragment of xa13
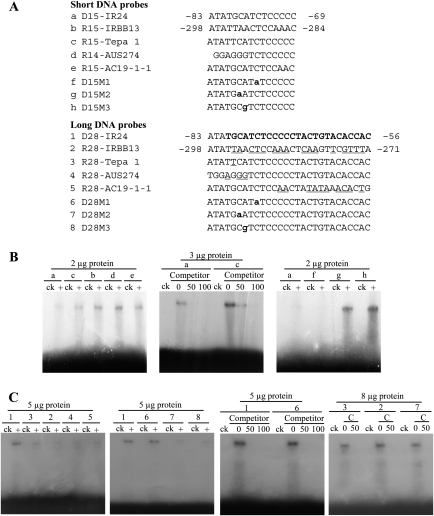
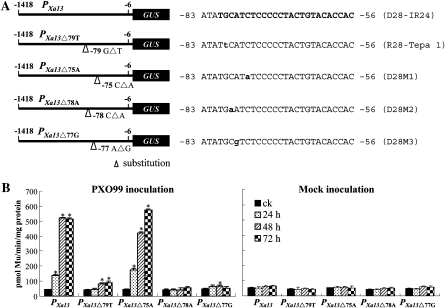
Although our findings indicated that the –121 to –66 region may harbor a PXO99-responsive element, a previous study showed that a region corresponding to the –86 to –69 region of PXa13 might be responsible for PXO99-induced expression of dominant Xa13, based on the comparison of the promoter sequences of seven rice lines carrying dominant Xa13 and 11 rice lines carrying recessive xa13 or its recessive alleles (Chu et al., 2006). To determine whether the protein(s) of PXO99 could directly bind to the putative PXO99-responsive element of PXa13, 14- to 15-nt DNA probes from the –86 to –69 region of PXa13, which harbored the 5'-half of the UPTPthXo1 box, and the corresponding regions of some promoters of recessive xa13 and its recessive alleles were designed based on sequence-specific analysis and used for protein binding analyses (Figure 4A).
Figure 4.
The Binding Ability of the Promoter Probes of Dominant Xa13 and Recessive xa13 to the Total Proteins of Xoo Strain PXO99 Analyzed by EMSA.
The binding assays were biologically repeated twice, with similar results ((B) and (C)).
(A) Probe sequences. Rice line IR24 carries dominant Xa13. Rice lines IRBB13, Tepa 1, AUS274, and AC19-1–1 carry different alleles of recessive xa13; another five rice lines, Chinsurah Boro2 (11484), Chinsurah Boro2 (11760), Chinsurah Boro2 (50930), Long Grain (64950), and BJ1 that carry different xa13 alleles from that of Tepa 1, have the same DNA sequence as Tepa 1 in the region corresponding to the –86 to –69 region of PXa13 (Chu et al., 2006). Probe D28-IR24 harbors the UPTPthXo1 box of PXa13 (bold italic letters). Compared with D28-IR24, the substitution sites of the probes from the promoters of xa13 and its recessive alleles are underlined. Mutation sites are shown in lower-case letters.
(B) The binding ability of PXO99 total proteins to different short (14- or 15-nt) probes. The labeled probes plus 50- or 100-fold unlabelled competitor probes or without plus competitor (0) were used for the binding assays. ck, without PXO99 proteins; +, with PXO99 proteins.
(C) The binding ability of PXO99 total proteins to different long (28-nt) probes. The labeled probes plus 50-fold unlabelled competitor (C) probes or without plus competitor (0) were used for the binding assays.
Because the TAL effector PthXo1 of PXO99 transcriptionally activates Xa13 (Yang et al., 2006), we expected that PXO99 proteins would bind more intensively to the probe from PXa13 than the probes from Pxa13, if there was protein binding. Unexpectedly, an electrophoretic mobility shift assay (EMSA) showed that PXO99 proteins bound more intensely to the four probes, R15-Tepa 1, R15-IRBB13, R14-AUS274, and R15-AC19-1–1, from the promoters of recessive xa13 and its recessive alleles than to probe D15-IR24 from PXa13 (Figure 4B). The protein-binding of D15-IR24 and R15-Tepa 1 was reduced or abolished by the competition of unlabelled probes, indicating specificity of the binding. To determine which nucleotide influenced the binding of PXO99 proteins, three site-mutated short DNA probes (D15M1, D15M2, and D15M3) were used for analysis of protein binding (Figure 4A). By comparing the protein binding intensity of D15M1, D15M2, and D15M3 with that of D15-IR24, substitution of C with A or A with G in probes D15M2 and D15M3, respectively, appeared to be important for protein binding (Figure 4B).
A recent study reported that the UPTPthXo1 box of PXa13 consists of 25 nucleotides (Figure 4A; Römer et al., 2010). Thus, we designed a new set of long DNA probes that harbored the full-length UPTPthXo1 box from Xa13 or the corresponding regions from xa13 or its recessive alleles (Figure 4A). Among these long probes, R28-IRBB13, R28-Tepa 1, R28-AUS274, and R28-AC19-1–1 from the promoters of recessive xa13 (IRBB13) and its recessive alleles (Tepa 1, AUS274, and AC19-1–1), respectively, harbored DNA fragments that had 1-, 3-, 10-, or 16-nucleotide differences from the UPTPthXo1 box. EMSA showed that PXO99 proteins bound intensively to probe D28-IR24 harboring the UPTPthXo1 box, but only very weakly to probes D28-IRBB13, D28-Tepa 1, D28-AUS274, and D28-AC19-1–1 (Figure 4C). Substitution of the third C nucleotide of the UPTPthXo1 box with A (probe D28M2) or the fourth A nucleotide with G (D28M3) markedly reduced the binding (Figure 4C). However, substitution of the sixth C of the UPTPthXo1 box with A (probe D28M1) did not influence the binding affinity of PXO99 proteins. The protein-binding signals of the probes were abolished by the competition of unlabelled probes, indicating specificity of the binding (Figure 4C). These results suggest that an intact UPTPthXo1 box is essential for bacterial protein binding. Furthermore, at least the second, third, and fourth nucleotides of the box are important for bacterial protein binding, whereas the sixth nucleotide of this box may not be important for protein binding.
Mutation of UPTPthXo1 Box Influences PXO99-Induced Transcriptional Activity
To ascertain whether the differential binding activities of the various probes to bacterial proteins affected transcriptional regulation of the promoter, we constructed four site-mutated promoters (PXa13Δ79T, PXa13Δ78A, PXa13Δ77G, and PXa13Δ75A) of PXa13 harboring the UPTPthXo1 box, in which the second, third, fourth, and sixth nucleotides were mutated, respectively (Figure 5A). These mutated promoters were fused with GUS and transferred separately into rice. Each construct generated approximately 20 independent positive transgenic plants. Each plant was divided into two parts by separating the tillers at the tillering stage for inoculation with Xoo strain PXO99 and mock-inoculation, respectively. Transgenic plants carrying different promoter constructs showed the similar levels of GUS expression when mock-inoculated. The PXO99-induced expression of GUS in the transgenic plants carrying PXa13Δ79T, PXa13Δ78A, or PXa13Δ77G was completely or partially suppressed compared to the expression of GUS in the plants carrying PXa13:GUS (Figure 5B). However, plants carrying PXa13Δ75A showed a similar level of PXO99-induced GUS expression to the plants carrying PXa13:GUS. The mutated UPTPthXo1 boxes in the promoters PXa13Δ79T, PXa13Δ78A, and PXa13Δ77G corresponded to DNA probes R28-Tepa1, D28M2, and D28M3, respectively, which showed only weak binding of PXO99 proteins (Figure 4C). The mutated UPTPthXo1 box in PXa13Δ75A was consistent with probe D28M1, which showed strong binding to PXO99 proteins (Figure 4C). These results suggest that the suppression or loss of PXO99-induced transcriptional activation in the transgenic plants carrying PXa13Δ79T, PXa13Δ78A, or PXa13Δ77G may have resulted from the unavailability or low affinity of binding bacterial protein to the promoters due to the mutation of the key sites of the UPTPthXo1 box. These results also suggest that the UPTPthXo1 box is the only PXO99-responsive element in the promoter of Xa13.
Figure 5.
Expression of GUS Driven by Native (PXa13) or Mutated (PXa13Δ79T, PXa13Δ75A, PXa13Δ78A, and PXa13Δ77G) Promoters after PXO99 Infection or Mock-Inoculation at the Booting Stage.
(A) The structures of mutated promoters and the sites of mutations. The UPTPthXo1 box of PXa13 is shown with bold italic letters. The figure with a sign of substitution indicates the site of nucleotides substitution.
(B) Expression of GUS in transgenic plants. GUS activity was determined by measuring the amount of 4-methylumbelliferone (Mu) produced under the catalysis of GUS in 1 mg total protein per minute. Twenty to 36 T0 transgenic plants carrying each construct were used for analyses. For each time point examined, leaf fragments were collected from all the plants carrying the same construct and mixed for preparing sample. The GUS activity at each time point was the average of three measurements ± standard deviation. ck, without pathogen- or mock-inoculation. The asterisk (*) indicates that a significant difference (P < 0.01) was detected between PXO99-inoculated plants and non-inoculated control (ck) plants carrying the same promoter.
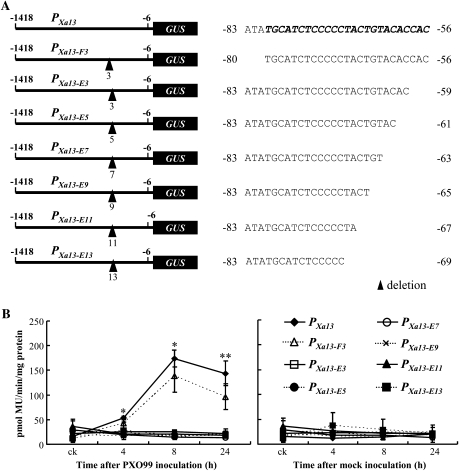
We also constructed seven site-truncated promoters. Three to 11 nucleotides either in front of the UPTPthXo1 box (PXa13-F3) or at 3'-terminal of the UPTPthXo1 box (PXa13-E3, PXa13-E5, PXa13-E7, PXa13-E9, PXa13-E11, and PXa13-E13) were deleted from PXa13 promoter (Figure 6A). These site-truncated constructs and the construct carrying PXa13:GUS were transiently expressed in separate rice calli. The expression of GUS in the calli carrying only PXa13:GUS or PXa13-F3:GUS but not other constructs was significantly induced (P < 0.01) by PXO99-inoculation compared to mock-inoculation (Figure 6B). However, GUS level in the calli carrying PXa13-F3:GUS was significantly lower than that in the calli carrying PXa13:GUS at 4–24 h after infection (Figure 6B). These results suggest that both the flanking sequence and the 3'-terminal part of UPTPthXo1 box may be important for Xoo-induced expression.
Figure 6.
Transient Expression of GUS Driven by Native (PXa13) and Site-Truncated (PXa13-F3, PXa13-E3, PXa13-E5, PXa13-E7, PXa13-E9, PXa13-E11, and PXa13-E13) Promoters in Rice Calli.
(A) The structures of site-truncated promoters and the sites of deletion. The UPTPthXo1 box of PXa13 is shown with bold italic letters. The figure with a sign of deletion indicates the numbers of nucleotides deleted.
(B) Expression of GUS driven by different promoters in rice calli. GUS activity was determined by measuring the amount of 4-methylumbelliferone (Mu) produced under the catalysis of GUS in 1 mg total protein per minute. Each sample was from 4.5 ml calli; all the calli were mixed to prepare a sample. The GUS activity of each sample was the average of three measurements ± standard deviation. One (*) or two (**) asterisks indicate that a significant difference between plants carrying PXa13:GUS and PXa13-F3:GUS in the same time point was detected at P < 0.05 or P < 0.01, respectively. ck, without pathogen- or mock-inoculation. This experiment was repeated twice biologically and similar results were obtained.
DISCUSSION
Xoo-induced Xa13 expression is critical for Xa13-facilited susceptibility (Yuan et al., 2009, 2010). The PthXo1 effector of Xoo strain PXO99 transcriptionally activates Xa13 by binding to a cis-acting element, the UPTPthXo1 box, in the gene's promoter (Yang et al., 2006; Römer et al., 2010). We analyzed a series of end- and site-truncated and site-mutated promoters, and our findings suggest that the UPTPthXo1 box is the only PXO99-responsive element in the Xa13 promoter. Thus, interaction of PthXo1 effector and UPTPthXo1 box results in the susceptibility of rice to PXO99. The promoters of recessive R gene xa13 and its recessive alleles carry mutated UPTPthXo1 boxes, which is the key reason for xa13-mediated resistance.
Although the present study used the total proteins of PXO99 instead of PthXo1 for DNA-protein binding analyses, the proteins bound to the DNA probe harboring the UPTPthXo1 box are likely mainly the PthXo1 effector, as supported by the following evidence. First, EMSA showed that PXO99 proteins bound intensively to the DNA probe harboring the full-length UPTPthXo1 box but weakly to the probes harboring mutated UPTPthXo1 boxes from the promoters of recessive xa13 and its recessive alleles (Figure 4C). These results agree with a recent report that His:PthXo1 fusion protein binds strongly to the DNA fragment harboring the UPTPthXo1 box but to a lesser extent to the corresponding promoter region of recessive xa13 (Römer et al., 2010). Second, replacement of the second G nucleotide of the UPTPthXo1 box with T in a natural mutation (the recessive allele of xa13 from rice variety Tepa 1) markedly reduced protein binding (Figure 4C). Likewise, substitution of the second nucleotide of the UPTPthXo1 box with A, C, or T significantly reduced or almost complete negated PthXo1-mediated promoter activation (Römer et al., 2010). Finally, the same mutations in the UPTPthXo1 box that reduced PXO99 protein binding abolished PXO99-induced Xa13 expression (Figure 5), which was similar to the knockout of PthXo1 in PXO99 (Yang et al., 2006).
Recently, several UPT boxes have been predicted by using the TAL effector code, the RVDs that are the hypervariable residues 12 and 13 in each repeat unit of the repeat domain of TAL effectors (Boch et al., 2009; Moscou and Bogdanove, 2009). Although about half of the identified RVDs of TAL effectors have degenerated pairing nucleotides based on the identified and predicted UPT boxes, with some RVDs codes varying between two to four nucleotides (Boch et al., 2009; Moscou and Bogdanove, 2009), this degeneracy appears to be influenced by the position of a RVD in TAL effectors or/and by the position of a nucleotide in a UPT box. For example, the RVD NN appears to recognize four types of nucleotide in UPT boxes (Boch et al., 2009; Moscou and Bogdanove, 2009). However, matching the NN-type RVD of PthXo1 to the 5'-end second G nucleotide but not the A, C, or T nucleotide of the UPTPthXo1 box is crucial for the interaction of the PthXo1 and UPTPthXo1 box (Römer et al., 2010). Our present results also showed that substitution of the second G to T in a natural mutation markedly reduced PXO99 protein binding and abolished PXO99-induced gene expression. Furthermore, the 5'-end third and forth nucleotides of the UPTPthXo1 box are also important for PXO99 protein binding and gene transcriptional activation. Both the RVDs pairing to the third and sixth nucleotides are HD-type (Römer et al., 2010), which is suggested to have degenerated pairing nucleotides in the UPT boxes (Boch et al., 2009; Moscou and Bogdanove, 2009). Interestingly, substitution of the third C to A in the UPTPthXo1 box abolished PXO99-induced gene expression and substitution of the sixth C to A did not influence gene activation (Figure 5). Similar results were observed in another study. The RVDs of TAL effector AvrBs3 pairing the 14th and 15th nucleotides of the UPAAvrBs3 box (also a UPT box) in the promoter of pepper R gene Bs3 are HD-type, but substitution of the 15th nucleotide C to A in the UPABs3 box abolished the Bs3-mediated hypersensitive response, while substitution of the 14th nucleotide C to A did not influence Bs3 function (Römer et al., 2009b). These results suggest that the three-dimensional positions of some RVDs may influence the interaction of a TAL effector and UPT box.
One RVD pairs with one nucleotide in a UPT box in the host–bacterium interaction; thus, the number of RVDs in a TAL effector determines the size of the corresponding UPT box (Boch et al., 2009; Moscou and Bogdanove, 2009). However, the length of a functional UPT box is not always as long as that predicted using the TAL effector code. The nucleotides in the 3'-end of some UPT boxes appear to not be required for gene activation. The AvrXa27 effector contains 17 RVDs (Moscou and Bogdanove, 2009). Omission of the last three nucleotides of the predicted UPTAvrXa27 box can still trigger hypersensitive response by AvrXa27 (Römer et al., 2009a). The AvrBs3Δrep16 effector contains 14 RVDs (Römer et al., 2010). Omission of the last two nucleotides of the predicted UPAAvrBs3Δrep16 box (also a UPT box) can also trigger hypersensitive response by AvrBs3Δrep16 (Römer et al., 2009b). A recent study reported that at least 6.5 RVDs were required to recognize the target DNA box and to activate gene expression, and 10.5 or more RVDs could efficiently activate gene expression (Boch et al., 2009). Our present results showed that the 12 5'-terminal nucleotides of the UPTPthXo1 box could not provide correct pathogen protein binding (Figure 4). Even deletion of the three nucleotides at the 3'-end of the UPTPthXo1 box abolished PXO99-induced gene expression (Figure 6). Likewise, an incomplete UPAAvrBs3Δrep16 box (lacking the first nucleotide) had a lower affinity for the AvrBs3Δrep16 effector than the complete UPAAvrBs3 box (Römer et al., 2007). However, the complete UPAAvrBs3Δrep16 box showed a higher affinity for AvrBs3Δrep16 than the complete UPAAvrBs3 box (Römer et al., 2009b). These results suggest that in some cases, an incomplete UPT box may result in non-specific protein binding.
The 5'-terminal T of UPT boxes has been reported to be crucial to transcriptional activation by TAL effectors (Boch et al., 2009; Römer et al., 2009b, 2010). Interestingly, the flanking sequence of the UPTPthXo1 box appears to influence PXO99-induced gene expression. The deletion of the three nucleotides flanking the 5'-end of the UPTPthXo1 significantly reduced the expression level of the gene activated by PXO99 (Figure 6). The neighboring nucleotides of at least some plant cis-acting elements also contribute to high-affinity binding of regulating proteins (Ciolkowski et al., 2008). It remains to be demonstrated whether it is a common feature that neighboring nucleotides of UPT boxes contribute to binding affinity of TAL effectors. In conclusion, the molecular mechanism of the specific interactions of TAL effectors and UPT boxes still needs to be refined. Investigating the binding of TAL effectors to UPT boxes at the three-dimensional level may help to clarify these issues.
METHODS
Constructing End-Truncated Promoters
Truncated promoters of dominant Xa13 gene (PXa13) and recessive xa13 gene (Pxa13) were obtained by PCR amplification from the 5'-end using the intermediate vector containing PXa13 and Pxa13 (Yuan et al., 2009), respectively, as a template. The 5'-end primers of Xa13P-F (5'-GGATCCGATGTTGAGCTTTAGGATTAGCGGGTT-3'), 5'-deletion-1147 (5'-GGATCCTCCCTTTCTTCAAGTTACCTCTCTC-3'), and 5'-deletion-608 (5'-GGATCCGCATCATTGTCCATGGTTGT-3'), which were specific to both PXa13 and Pxa13 and contained the BamHI digestion site (underlined), as well as primers XP-90F (5'-GGATCCGAAATATCAAGCACAAG-3') and XP-60F (5'-GGATCCCTGTACACCACCAAAAG-3'), which were specific to PXa13, were used in combination with the 3'-end primer X13-promR (5'-GGCAAGCTTGGCCTTGGCCATGGCTCAGT-3'), which was specific to both PXa13 and Pxa13 and contained the HindIII digestion site (underlined). The 5'-end primer XP-90F was also used in combination with the 3'-end primer XP-60R (5'-AAGCTTCTTTTGGTGGTGTACAG-3'), which was specific to PXa13 and contained the HindIII digestion site (underlined). The PCR products were ligated to vector pGEM-T (Promega Corporation, Madison, WI, USA) for sequencing, then the truncated promoter fragments were digested with BamHI and HindIII and ligated to vector pCAMBIA1381 to form truncated promoters fused with reporter gene GUS.
Rice Transformation
The promoter constructs were transferred into rice variety Mudanjiang 8 (Oryza sativa L. ssp. japonica) by Agrobacterium-mediated transformation (Lin and Zhang, 2005). A pair of PCR primers, GusF (5′-CCAGGCAGTTTTAACGATCAGTTCGC-3′) and GusR (5′-GAGTGAAGATCCCTTTCTTGTTACC), designed according to the sequence of GUS was used for detecting positive transgenic plants.
Pathogen Inoculation
Rice plants were inoculated with Xoo strain PXO99 by the leaf-clipping method at the booting (panicle development) stage (Sun et al., 2004). Mock-inoculated plants were treated under the same conditions except that the Xoo suspension was replaced with water.
Analysis of GUS Activity
Leaf fragments about 1 cm long right next to the inoculation sites were used for analysis of GUS expression. Quantitative analyses of GUS activity were conducted as described previously (Cai et al., 2007). Total protein concentration in the supernatant was quantified with the Bradford assay (Bradford, 1976). GUS protein in the supernatant was determined fluorometrically with an INFINITE 200 (Tecan Austria Gmbh, Ltd, Grodig, Austria).
Agrobacterium-Mediated Transient Expression
The calli of rice variety Zhonghua 11 (O. sativa ssp. japonica) were prepared as described previously (Lin and Zhang, 2005). Agrobacteria containing different transformation construct were co-cultured with the calli for over 16 h in a MS medium. The calli were then washed 10 times using autoclaved water and dried in a laminar flow cabinet. The calli were treated with Xoo strain PXO99 at 109 cfu ml−1 or mock-treated with autoclaved water for a certain period of time.
Site-Directed Mutation and Truncation
The GeneTailor Site-Directed Mutagenesis System (Invitrogen Life Technologies, Carlsbad, CA, USA) was used for site-directed mutation of Xa13 promoter as described previously (Cai et al., 2007). The pUC19 plasmid that contained the native promoter (PXa13) used as a PCR template was methylated before use. The mutagenic primer pair, in which the regulatory element was mutated, was used to amplify the promoter-containing target mutation. Primer pairs Xa13PM1F (5'-AAAGCAAAGGTTAGATATTCATCTCCCCCT-3')/Xa13PM1R (5'-ATATCTAACCTTTGCTTTTTTTTTTC-3'), Xa13PM2F (5'-CAAAGGTTAGATATGCATATCCCCCTACTG-3')/Xa13PM2R (5'-ATGCATATCTAACCTTTGCTTTTTTTTTTC-3'), Xa13PM4F (5'-AAGCAAAGGTTAGATATGAATCTCCCCCT-3')/Xa13PM4R (5'-CATATCTAACCTTTGCTTTTTTTTTTC-3'), and Xa13PM5F (5'-AGCAAAGGTTAGATATGCGTCTCCCCCTAC-3')/Xa13PM5R (5'-GCATATCTAACCTTTGCTTTTTTTTTTC-3') were used for site-directed mutation of PXa13, in which the mutation points were underlined. To help select the mutated construct, the PCR product was transferred into Escherichia coli strain DH5a-T1, in which the methylated plasmid could not replicate.
The site-directed truncation of Xa13 promoter was performed by nested PCR amplification using the intermediate vector containing PXa13 (Yuan et al., 2009) as a template. First, the forward primer BXa13(IR24)M17F (5'-TAGATATGCATCTCCCCCTACAAAAGTGGAG-3'), which contained a deletion in the target site, was used in combination with the reverse primer X13-promR to amplify the downstream of the deletion site. Simultaneously, the reverse primer BXa13(IR24)M17R (5'-GGGGGAGATGCATATCTAACCTTTGCTTTT-3'), which was overlapped with BXa13(IR24)M17F and also contained the same deletion as in BXa13(IR24)M17F, was paired with the forward primer Xa13P-F to amplify the upstream of the deletion site. The amplification products from the two PCR reactions were purified and mixed and used as the template for the second round of PCR using primers Xa13P-F and X13-promR. Another six pairs of primers, BXa13(IR24)M15F (5'-TAGATATGCATCTCCCCCCAAAAGTGGAGGG-3')/BXa13(IR24)M15R (5'-GGGAGATGCATATCTAACCTTTGCTTTTTT-3'), BXa13(IR24)M19F (5'-GATATGCATCTCCCCCTACTCAAAAGTGGAG-3')/BXa13(IR24)M19R (5'-TAGGGGGAGATGCATATCTAACCTTTGCTT-3'), BXa13(IR24)M21F (5'-TATGCATCTCCCCCTACTGTCAAAAGTGGAG-3')/BXa13(IR24)M21R (5'-AGTAGGGGGAGATGCATATCTAACCTTTGC-3'), BXa13(IR24)M23F (5'-TGCATCTCCCCCTACTGTACCAAAAGTGGAG-3')/BXa13(IR24)M23R (5'-ACAGTAGGGGGAGATGCATATCTAACCTTT-3'), BXa13(IR24)M25F (5'-CATCTCCCCCTACTGTACACCAAAAGTGGAG-3')/BXa13(IR24)M25R (5'-GTACAGTAGGGGGAGATGCATATCTAACCT-3'), and BXa13(IR24)M-3F (5'-GAAAAAAAAAAGCAAAGGTTAGTGCATCTCCCC-3')/BXa13(IR24)M-3R (5'-AACCTTTGCTTTTTTTTTTCTTGTGCTTGA-3'), were also used to construct other site-directed truncated promoters in the same way as described above.
Electrophoretic Mobility Shift Assay
To isolate the total proteins of Xoo strain PXO99, the fresh bacteria were ground with liquid nitrogen and suspended in buffer (10 mM Tris-HCl at pH 8.0, 1 mM DTT, 200 μM phenylmethanesulfonyl fluoride) and homogenized completely. The mixture was centrifuged at 4°C at 1000 g to collect the supernatant. The protein content in the nuclear extract and in the total proteins of Xoo strain PXO99 was quantified using the Bradford assay. EMSA was applied as described previously (Qiu et al., 2007).
Promoter Sequence Analysis
The TATA boxes of promoters PXa13 and Pxa13 were predicted using the computer programs TSSP provided at the Softberry website (www.softberry.com) and PROSCAN (http://bimas.dcrt.nih.gov/molbio/proscan).
Statistical Analysis
The significant differences between control and treatment of the samples were analyzed by the pair-wise t-test installed in the Microsoft Office Excel program.
FUNDING
This work was supported by grants from the National Natural Science Foundation of China (30930063 and 30921091) and the National Program of Transgenic Variety Development of China (2008ZX08009-004). No conflict of interest declared.
References
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cai M, Wei J, Li X, Xu C, Wang S. A rice promoter containing both novel positive and negative cis-elements for regulating green-tissue-specific gene expression in transgenic plants. Plant Biotechnol. J. 2007;5:664–674. doi: 10.1111/j.1467-7652.2007.00271.x. [DOI] [PubMed] [Google Scholar]
- Chu Z, Wang S. Isolation, structure, function relationship, and molecular evolution of disease resistance genes. In: Zhang Q, editor. Genetics and Improvement of Resistance to Bacterial Blight in Rice. Beijing: Science Press; 2007. pp. 349–377. [Google Scholar]
- Chu Z, et al. Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev. 2006;20:1250–1255. doi: 10.1101/gad.1416306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciolkowski I, Wanke D, Birkenbihl RP, Somssich IE. Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 2008;68:81–92. doi: 10.1007/s11103-008-9353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay S, Bonas U. How Xanthomonas type III effectors manipulate the host plant. Curr. Opin. Microb. 2009;12:37–43. doi: 10.1016/j.mib.2008.12.006. [DOI] [PubMed] [Google Scholar]
- Kay S, Hahn S, Marols E, Hause G, Bonas U. A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science. 2007;318:648–651. doi: 10.1126/science.1144956. [DOI] [PubMed] [Google Scholar]
- Kay S, Hahn S, Marols E, Wieduwild R, Bonas U. Detailed analysis of the DNA recognition motifs of the Xanthomonas type III effectors AvrBs3 and AvrBs3Δrep16. Plant J. 2009;59:859–871. doi: 10.1111/j.1365-313X.2009.03922.x. [DOI] [PubMed] [Google Scholar]
- Lin YJ, Zhang Q. Optimising the tissue culture conditions for high efficiency transformation of indica rice. Plant Cell Rep. 2005;23:540–547. doi: 10.1007/s00299-004-0843-6. [DOI] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- Qiu D, Xiao J, Ding X, Xiong M, Cai M, Cao Y, Li X, Xu C, Wang S. OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol. Plant Microbe Interact. 2007;20:492–499. doi: 10.1094/MPMI-20-5-0492. [DOI] [PubMed] [Google Scholar]
- Römer P, Fecht S, Straub T, Elsasser J, Schornack S, Boch J, Wang S, Lahaye T. Promoter elements of rice susceptibility genes are bound and activated by specific TAL effectors from the bacterial blight pathogen, Xanthomonas oryzae pv. oryzae. New Phytologist. 2010;187:1048–1057. doi: 10.1111/j.1469-8137.2010.03217.x. [DOI] [PubMed] [Google Scholar]
- Römer P, Hahn S, Jordan T, Strauss T, Bonas U, Lahaye T. Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science. 2007;318:645–648. doi: 10.1126/science.1144958. [DOI] [PubMed] [Google Scholar]
- Römer P, Recht S, Lahaye T. A single plant resistance gene promoter engineered to recognize multiple TAL effectors from disparate pathogens. Proc. Natl Acad. Sci. U S A. 2009a;106:20526–20531. doi: 10.1073/pnas.0908812106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römer P, Strauss T, Hahn S, Scholze H, Morbitzer R, Grau J, Bonas U, Lahaye T. Recognition of AvrBs3-like proteins is mediated by specific binding to promoters of matching pepper Bs3 alleles. Plant Physiol. 2009b;150:1697–1712. doi: 10.1104/pp.109.139931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Cao Y, Yang Z, Xu C, Li X, Wang S, Zhang Q. Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J. 2004;37:517–527. doi: 10.1046/j.1365-313x.2003.01976.x. [DOI] [PubMed] [Google Scholar]
- Yang B, Sugio A, White FF. Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc. Natl Acad. Sci. U S A. 2006;103:10503–10508. doi: 10.1073/pnas.0604088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Chu Z, Li X, Xu C, Wang S. Pathogen-induced expressional loss of function is the key factor in race-specific bacterial resistance conferred by a recessive R gene xa13 in rice. Plant Cell Physiol. 2009;50:947–955. doi: 10.1093/pcp/pcp046. [DOI] [PubMed] [Google Scholar]
- Yuan M, Chu Z, Li X, Xu C, Wang S. The bacterial pathogen Xanthomonas oryzae overcomes rice defenses by regulating host copper redistribution. Plant Cell. 2010;22:3164–2176. doi: 10.1105/tpc.110.078022. [DOI] [PMC free article] [PubMed] [Google Scholar]