Abstract
Cold-induced changes of gene expression and metabolism are critical for plants to survive freezing. Largely by changing gene expression, exposure to a period of non-freezing low temperatures increases plant tolerance to freezing—a phenomenon known as cold acclimation. Cold also induces rapid metabolic changes, which provide instant protection before temperature drops below freezing point. The molecular mechanisms for such rapid metabolic responses to cold remain largely unknown. Here, we use two-dimensional difference gel electrophoresis (2-D DIGE) analysis of sub-cellular fractions of Arabidopsis thaliana proteome coupled with spot identification by tandem mass spectrometry to identify early cold-responsive proteins in Arabidopsis. These proteins include four enzymes involved in starch degradation, three HSP100 proteins, several proteins in the tricarboxylic acid cycle, and sucrose metabolism. Upon cold treatment, the Disproportionating Enzyme 2 (DPE2), a cytosolic transglucosidase metabolizing maltose to glucose, increased rapidly in the centrifugation pellet fraction and decreased in the soluble fraction. Consistent with cold-induced inactivation of DPE2 enzymatic activity, the dpe2 mutant showed increased freezing tolerance without affecting the C-repeat binding transcription factor (CBF) transcriptional pathway. These results support a model that cold-induced inactivation of DPE2 leads to rapid accumulation of maltose, which is a cold-induced compatible solute that protects cells from freezing damage. This study provides evidence for a key role of rapid post-translational regulation of carbohydrate metabolic enzymes in plant protection against sudden temperature drop.
Keywords: 2-D DIGE, Arabidopsis, Cold response, freezing tolerance, heat shock protein, starch metabolism
INTRODUCTION
Plant survival and productivity are negatively affected by various environmental stresses, such as heat, cold, drought, and salinity. Understanding how plants perceive and respond to these signals is crucial for engineering stress-resistant plants (Xiong and Zhu, 2001; Salekdeh and Komatsu, 2007). Low temperature is one of the most important environmental factors that limit the yield, quality, post-harvest life, and geographic distribution of crop plants. In nature, plants can be exposed to gradual temperature drop during fall-to-winter transition or to sudden temperature drop in an early frost. Exposure to a period of non-freezing cold temperature activates transcriptional and metabolic reprogramming that increases tolerance to freezing temperatures—a phenomenon known as cold acclimation (Thomashow, 1999). Cold also induces rapid metabolic changes that help plants survive sudden temperature drop in an early frost. Genetic analysis in Arabidopsis thaliana has revealed much of the mechanism underlying transcriptional acclimation (Chinnusamy et al., 2007). However, little is known about how cold induces rapid changes of metabolite accumulation, some of which are too fast to be due to transcriptional regulation.
Cold responses involve complex molecular, biochemical and physiological changes, such as alteration in membrane composition and structure (Wang et al., 2006), and reprogramming of gene expression and metabolism (Thomashow, 1999; Xiong and Zhu, 2001; Chinnusamy et al., 2007). Studies of cold response using microarray profiling have identified over 3300 cold-responsive genes in Arabidopsis, including over 300 transcriptional factors (Fowler and Thomashow, 2002; Hannah et al., 2005). A number of transcription factors have been identified as key regulators of cold-induced gene expression and adaption, including the C-repeat binding transcription factors (CBF) (Yamaguchi-Shinozaki and Shinozaki, 2006; Chinnusamy et al., 2007), Inducer of CBF expression 1 (ICE1) (Chinnusamy et al., 2003), High Expression of OSmotically Responsive Genes 9 (HOS9) (Zhu et al., 2004), and a zinc finger protein ZAT12 (Vogel et al., 2005). Increase in freezing tolerance was observed in CBFs overexpression lines (Liu et al., 1998; Jaglo-Ottosen et al., 1998; Kasuga et al., 1999), in correlation with alteration of cold-responsive transcriptome and metabolome (Fowler and Thomashow, 2002; Cook et al., 2004; Gilmour et al., 2004), which indicates a prominent role of CBFs in cold acclimation. The mechanisms of primary cold sensing and signal transduction that control these transcription factors, however, remain unknown.
There has been a large body of evidence for post-translational regulation of metabolism by cold. Cold induces the accumulation of a large number of metabolites, including amino acid proline, glutamine, and low-molecular-weight carbohydrates such as fructose, glucose, maltose, and raffinose (Gilmour et al., 2000; Taji et al., 2002; Cook et al., 2004). Many of these metabolites play protective roles in freezing tolerance (Strauss and Hauser, 1986; Nanjo et al., 1999; Kaplan and Guy, 2004). Simultaneously monitoring changes in metabolites and associated gene expression showed that some metabolic changes, such as gamma-aminobutyric acid metabolism, correlate with changes in the transcripts of the corresponding metabolic enzymes, while changes in other metabolic processes, such as proline accumulation, seem to be independent of transcriptional regulation (Kaplan et al., 2007). Furthermore, some metabolites change more rapidly than the RNAs encoding for their metabolic enzymes. Rapid accumulation of maltose during cold acclimation has been documented in a number of detailed studies (Kaplan and Guy, 2004; Kaplan et al., 2004; Kaplan and Guy, 2005; Yano et al., 2005; Kaplan et al., 2007). For example, the level of maltose increases over 100-fold within 1 h of cold treatment (Kaplan and Guy, 2004), and such increase of maltose, which is a compatible solute, is believed to provide cellular protection against cryo-damage (Kaplan and Guy, 2004; Kaplan et al., 2007). These observations suggest that post-translational mechanisms are involved in cold responses.
The primary effects of low temperature on proteins that lead to changes of gene expression or enzyme activities remain unclear. Proteomic studies have identified cold-responsive proteins in several plant species, including rice (Imin et al., 2004; Cui et al., 2005; Imin et al., 2006; Yan et al., 2006; Hashimoto and Komatsu, 2007; Lee et al., 2009), Arabidopsis (Bae et al., 2003; Kawamura and Uemura, 2003; Amme et al., 2006), and poplar (Renaut et al., 2004). However, these studies used long time treatments, which led to the identification of late cold-responsive proteins that are most likely regulated through transcriptional changes (Bae et al., 2003; Yan et al., 2006). Two-dimensional difference gel electrophoresis (2-D DIGE) is a sensitive and quantitative method for proteomics profiling (Unlu et al., 1997; Tonge et al., 2001). We have recently shown that combining 2-D DIGE with sub-cellular fractionation can identify early response proteins involved in primary signal transduction processes (Deng et al., 2007; Tang et al., 2008a, 2008b).
In this study, protein fractionations followed by 2-D DIGE were used to profile Arabidopsis proteins that respond rapidly to cold treatment. Among many identified cold-responsive proteins, Disproportionating Enzyme 2 (DPE2) increased in the centrifugation pellet and decreased in the soluble fraction, resulting in a decrease of enzymatic activity of soluble DPE2 upon cold treatment. A dpe2 T-DNA knockout mutant showed increased freezing tolerance, which correlated with increased maltose content in this mutant reported previously (Chia et al., 2004). Our results reveal a post-translational mechanism for cold-induced rapid DPE2 inactivation, which plays an important role in freezing tolerance.
RESULTS
Our previous studies have demonstrated that pre-fractionation followed by 2-D DIGE analysis can identify proteins involved in signal perception or signal transduction (Deng et al., 2007; Tang et al., 2008a, 2008b). 2-D DIGE analysis of sub-fractions of Arabidopsis proteome was performed to identify early cold-responsive proteins. To achieve rapid cold treatment without other perturbations, we grew Arabidopsis seedlings in liquid suspension culture for 6 d when seedlings were still well separated from each other (Deng et al., 2007; Tang et al., 2008a, 2008b). Half of the medium was moved to a new flask and chilled on ice to 2°C, and then half of the cultured seedlings were moved into the cold medium to start cold treatment, while the other half of the seedlings were left at room temperature as untreated control (22°C). After 2 h of shaking, the seedlings were harvested and frozen immediately in liquid nitrogen. The soluble proteins, proteins extracted from microsomal fractions by sodium carbonate, and Triton-insoluble proteins were prepared from the tissues and analyzed by 2-D DIGE. Cold-responsive protein spots were identified by Decyder software analysis, excised from the 2-DE gels and analyzed by tandem mass spectrometry (MS/MS) after trypsin in-gel digestion to identify the proteins. A total of about 80 spots were detected as cold-responsive protein spots in different fractions and 50 of them were successfully identified by MS/MS and reported here.
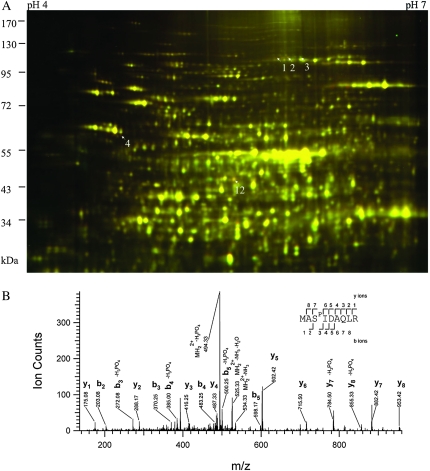
In the soluble fraction, only a few early cold-responsive proteins were identified, including phosphoenolpyruvate carboxylase 2 (PEPC2), initiation factor 4A-1 (EIF4A-1), and chaperonin-60 alpha (Figure 1A and Table 1). PEPC2 showed putative protein phosphorylation after cold treatment, as the spots shifted to more acidic side on the 2-DE gel. Tandem mass spectrometry analysis of the PEPC2 spots identified a phosphopeptide (9-MASPIDAQLR-17) (Figure 1B) only in the more acidic spots (spots 1 and 2, Figure 1A), but not in the more basic spot (spot 3, Figure 1A). This phosphoserine site was conserved in all plant PEPCs (Vidal and Chollet, 1997; Nimmo, 2003). The chaperonin-60 alpha is essential for chloroplast development (Apuya et al., 2001), and its increase is consistent with the observation that photosynthetic system is very sensitive to cold (Strand et al., 1999; Stitt and Hurry, 2002).
Figure 1.
2-D DIGE Analysis of Cold Response in the Soluble Fraction of Arabidopsis Proteome (A) and a MS Spectrum of the PEPC2 Phosphopeptide MASPIDAQLR (B). (A) Seedlings were treated at 2 or 22°C for 2 h, and the soluble protein fractions were compared by 2-D DIGE. In this image, proteins induced by cold treatment appear red, down-regulated appear green, while those unchanged show yellow. Arrows point to cold-responsive spots identified by tandem mass spectrometry.
(B) A spectrum of PEPC2 spots (spots 1 and 2 in (A)), corresponding to a phosphopeptide spanning Met-9 to Arg-17. Phosphorylated residue (Ser-11) is indicated in the peptide sequence as SP. The expected and theoretic m/z for the doubly charged peptide was 542.7461 and 542.7465, respectively, and the Error was 0.7 ppm and E-value 0.02.
Table 1.
Cold-Responsive Proteins Identified from the Soluble Fraction.
| Spot | Gene locus | Protein name | Abundance ratio | p-value (t-test) | Unique peptides | Sequence coverage (%) | Best expectation value |
| 1 | At2g42600 | PEPC2 | 1.42 | 1.1e–3 | 24 | 27.6 | 1.0e–6 |
| 2 | At2g42600 | PEPC2 | 1.32 | 3.3e–2 | 26 | 25.8 | 1.5e–7 |
| 3 | At2g42600 | PEPC2 | –1.52 | 5.2e–3 | 25 | 22.8 | 2.4e–7 |
| 4 | At2g28000 | Chaperonin-60 alpha | 1.64 | 3.5e–2 | 16 | 30.2 | 1.3e–5 |
| 12 | At3g13920 | EIF4A-1 | 1.34 | 8.2e–3 | 17 | 48.7 | 1.9e–7 |
The expression ratios of cold-treated to untreated (positive numbers) or untreated to treated (negative numbers) and the p-values of the Student's t-test of the comparison were calculated from three biological repeats. For the MS/MS identification, number of unique peptides, percentage of sequence coverage, and best expectation value (Protein Prospector) are listed.
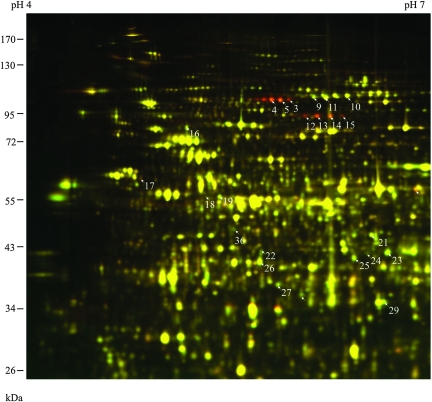
Sodium carbonate treatment reduces the complexity of the microsomal fraction, by releasing the peripheral and content proteins (Fujiki et al., 1982). In the soluble microsomal fraction (Figure 2), a number of protein spots showed more than three-fold increase after cold treatment, and tandem mass spectrometry analysis identified these spots as different forms of Disproportionating Enzyme 2 (DPE2) and alpha-Glucan Phosphorylase 2 (PHS2) (Table 2), two enzymes involved in maltose metabolism (Chia et al., 2004; Lu and Sharkey, 2004; Lu et al., 2006). Interestingly, several proteins involved in the tricarboxylic acid (TCA) cycle were down-regulated in this fraction, which include Aconitate Hydratase 2, Aconitate Hydratase 3, Isocitrate Dehydrogenase subunit 2, Malate Dehydrogenase 1, Succinyl-CoA Synthetase (beta chain), and Citrate Synthase 4 (Table 2), suggesting coordinated regulation of TCA enzymes during early cold response.
Figure 2.
2-D DIGE Analysis of Cold Response in the Sodium Carbonate-Soluble Microsomal Fraction of Arabidopsis. Seedlings were treated as in Figure 1, and the microsomal proteins solubilized by 0.1 M sodium carbonate were compared. The cold-induced proteins appear red, and the cold-repressed appear green, while those unchanged show yellow. Arrows point to MS-identified cold-responsive spots.
Table 2.
Cold-Responsive Proteins Identified from the Soluble-Microsomal Fraction.
| Spot | Gene locus | Protein name | Abundance ratio | p-value | Unique peptides | Sequence coverage (%) | Best expectation value |
| 3 | At2g40840 | DPE2 | 5.44 | 3.7e–4 | 57 | 49.9 | 1.4e–9 |
| 4 | At2g40840 | DPE2 | 4.88 | 1.3e–3 | 66 | 59.8 | 8.0e–11 |
| 5 | At2g40840 | DPE2 | 5.29 | 8.4e–4 | 74 | 69.6 | 1.9e–12 |
| 9 | At4g26970 | Aconitate hydratase 2 | –1.53 | 3.5e–2 | 83 | 53.3 | 7.0e–13 |
| 10 | At2g05710 | Aconitate hydratase 3 | –1.51 | 2.9e–2 | 81 | 62.9 | 1.2e–11 |
| 11 | At4g26970 | Aconitate hydratase 2 | –1.55 | 2.8e–2 | 87 | 59.9 | 3.1e–11 |
| 12 | At3g46970 | PHS2 | 3.50 | 1.6e–2 | 50 | 52.7 | 1.8e–8 |
| 13 | At3g46970 | PHS2 | 3.80 | 3.9e–3 | 75 | 61.6 | 4.2e–1 |
| 14 | At3g46970 | PHS2 | 2.74 | 7.6e–3 | 24 | 27.3 | 4.6e–9 |
| 15 | At3g46970 | PHS2 | 3.05 | 2.3e–3 | 81 | 66.1 | 4.7e–11 |
| 16 | At3g52200 | Dihydrolipoamide S-acetyltransferase | –1.51 | 3.7e–2 | 39 | 52.4 | 5.6e–11 |
| 17 | At2g28000 | Chaperonin-60 alpha | 1.52 | 4.5e–2 | 20 | 36.7 | 4.9e–8 |
| 18 | At5g08670 | Alanine aminotransferase | –1.68 | 4.9e–2 | 27 | 43.3 | 3.1e–1 |
| 19 | At5g08670 | Alanine aminotransferase | –1.49 | 3.0e–2 | 44 | 54.5 | 1.7e–11 |
| 21 | At2g44350 | Citrate synthase 4 | –1.49 | 4.9e–2 | 41 | 60.1 | 6.7e–11 |
| 22 | At2g17130 | Isocitrate dehydrogenase subunit 2 | –1.59 | 3.8e–2 | 23 | 48.8 | 1.5e–11 |
| 24 | At5g07440 | Glutamate dehydrogenase 2 | –1.55 | 9.5e–3 | 29 | 58.4 | 2.2e–11 |
| 25 | At4g02930 | Elongation factor Tu | –1.51 | 3.3e–2 | 66 | 86.1 | 6.7e–11 |
| 26 | At2g20420 | Succinyl–CoA synthetase, beta chain | –1.52 | 4.6e–2 | 57 | 74.1 | 7.6e–13 |
| 27 | At3g59760 | Cysteine synthase | –1.46 | 3.7e–2 | 29 | 58.8 | 5.2e–10 |
| 29 | At1g53240 | Malate dehydrogenase 1 | –1.60 | 3.7e–2 | 27 | 66 | 3.2e–12 |
| 36 | At3g13920 | EIF4A-1 | –1.34 | 1.4e–2 | 16 | 44.4 | 1.3e–7 |
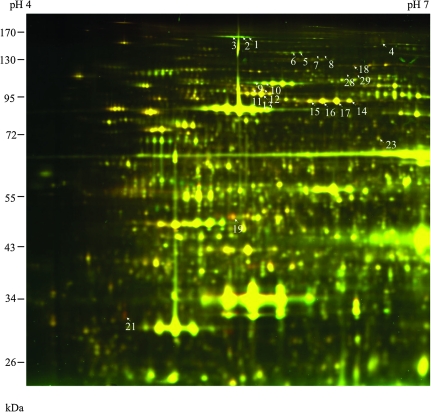
The triton-insoluble fraction is enriched with nuclei (Gampala et al., 2007), cell debris, and starch granules that are insoluble in Triton X-100 and sediment at 5000 g, but devoid of membranes of chloroplasts and mitochondria, although some macromolecular structures have been identified as Triton-insoluble fraction from the chloroplast (Phinney and Thelen, 2005). DPE2 increased in abundance while PHS2 decreased in abundance in this fraction (Figure 3 and Table 3). Two other proteins involved in starch degradation—alpha-Glucan Water Dikinase 1 (GWD1) (also named Starch Excess 1) (Yu et al., 2001) and Phosphoglucan Water Dikinase (GWD3) (Baunsgaard et al., 2005)—increased in abundance in this fraction. GWD1 has been previously shown to regulate freezing tolerance (Yano et al., 2005). Two proteins involved in sucrose biosynthesis—Sucrose Phosphate Synthase 1F and Phosphoglucomutase—decreased in abundance after cold treatment in this fraction. There are eight HSP100 proteins in Arabidopsis (Agarwal et al., 2001). Three of them, encoded by At5g15450, At5g50920, and At1g74310, were cold-repressed in the Triton-insoluble fraction (Figure 3 and Table 3). The HSP100 encoded by At1g74310 is essential for thermo-tolerance in Arabidopsis (Hong and Vierling, 2000; Queitsch et al., 2000). While the Triton-insoluble fraction contains a mixture of organelles, many of the proteins identified in this fraction were predicted to be plastid-localized (Heazlewood et al., 2007), indicating that the cold affects many proteins associated with plastids or starch.
Figure 3.
2-D DIGE Analysis of Cold Response Proteins in the Triton-Insoluble Fraction.
In this image, cold-induced protein spots appear green, and cold-repressed proteins spots appear red.
Table 3.
Cold-Responsive Proteins Identified from the Triton-Insoluble Fraction.
| Spot | Gene locus | Protein name | Abundance ratio | p-value | Unique peptides | Sequence coverage (%) | Best expectation value |
| 1 | At1g10760 | GWD1 | 2.13 | 5.4e–3 | 16 | 11.1 | 4.3e–6 |
| 2 | At1g10760 | GWD1 | 2.21 | 2.4e–2 | 14 | 9.4 | 5.4e–8 |
| 3 | At1g10760 | GWD1 | 2.11 | 1.1e–3 | 45 | 31.2 | 3.7e–9 |
| 4 | At3g09260 | beta-Glucosidase 23 | 1.75 | 2.2e–2 | 10 | 24.6 | 4.3e–8 |
| 5 | At5g26570 | GWD3 | 1.47 | 3.3e–2 | 35 | 34.1 | 1.6e–1 |
| 6 | At5g26570 | GWD3 | 1.37 | 1.4e–2 | 7 | 6.7 | 8.0e–5 |
| 7 | At5g20280 | Sucrose phosphate synthase 1F | –1.30 | 1.1e–2 | 4 | 4.3 | 1.2e–3 |
| 8 | At5g20280 | Sucrose phosphate synthase 1F | –1.39 | 1.6e–2 | 5 | 5.0 | 4.0e–6 |
| 9 | At2g40840 | DPE2 | 1.55 | 3.3e–7 | 58 | 55.9 | 8.5e–12 |
| 10 | At2g40840 | DPE2 | 1.72 | 3.3e–5 | 72 | 58.3 | 5.6e–11 |
| 11 | At5g15450 | HSP100 | –1.40 | 7.1e–3 | 70 | 50 | 2.5e–1 |
| 12 | At5g15450 | HSP100 | –1.43 | 1.5e–3 | 82 | 57.5 | 5.9e–1 |
| 13 | At5g50920 | HSP100 | –1.43 | 7.8e–6 | 95 | 67.4 | 8.9e–11 |
| 14 | At3g46970 | PHS2 | –1.39 | 4.7e–2 | 68 | 60.6 | 8.7e–12 |
| 15 | At3g46970 | PHS2 | –1.49 | 5.6e–3 | 73 | 72.4 | 1.1e–11 |
| 16 | At3g46970 | PHS2 | –1.56 | 7.8e–3 | 99 | 80.4 | 8.9e–12 |
| 17 | At3g46970 | PHS2 | –1.51 | 1.2e–2 | 102 | 83.9 | 2.9e–11 |
| 18 | At5g61780 | Tudor domain-containing protein | –1.43 | 1.4e–3 | 37 | 36.3 | 2.8e–9 |
| 19 | At3g13920 | EIF4A-1 | –2.27 | 5.7e–8 | 53 | 66.7 | 5.1e–1 |
| 21 | At2g41100 | Calmodulin-like protein 4 | –2.17 | 7.7e–4 | 8 | 23.5 | 5.4e–6 |
| 23 | At1g23190 | Phosphoglucomutase | –1.51 | 4.9e–5 | 22 | 38.9 | 3.0e–8 |
| 28 | At1g74310 | HSP100 | –1.77 | 4.1e–2 | 6 | 6.3 | 1.4e–7 |
| 29 | At1g74310 | HSP100 | –1.72 | 1.7e–2 | 20 | 28.3 | 1.3e–9 |
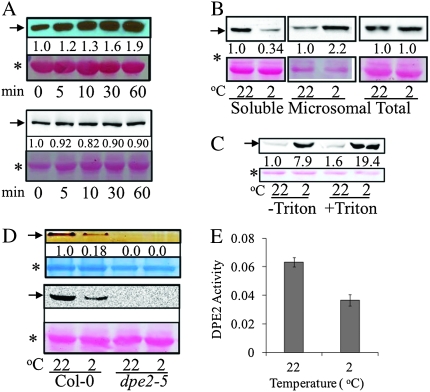
In 2-D DIGE analysis results, DPE2 showed a rapid and drastic increase in the protein extracted from microsomal fraction by sodium carbonate in the cold-treated samples. DPE2 is a cytosolic glucosyl transferase (Chia et al., 2004), and was proposed to use maltose as the donor, and use polyglucan, likely heteroglucan, as the receptor in vivo, and release another glucose unit (Fettke et al., 2006). The dpe2 loss-of-function mutant accumulates high levels of maltose (Chia et al., 2004). DPE2 is distinct from DPE1, which is a plastidial α-1,4-glucanotransferase responsible for maltotriose metabolism (Critchley et al., 2001). Immunoblotting with DPE2 antiserum (Chia et al., 2004) showed that DPE2 increased rapidly in the microsomal (100 000-g pellet) fraction within 5 min of cold treatment (Figure 4A). Interestingly, although DPE2 increased in the microsomal (100 000-g pellet) fraction, it decreased in the soluble fraction (Figure 4B), and its total amount in the tissue did not increase as determined in the SDS-extractable fraction (Figure 4A, bottom panel, and Figure 4B). To determine whether the accumulation of DPE2 in the 100 000-g pellet after the cold treatment was due to its association with membranes or aggregation into other insoluble structures, the microsomal pellet from both control and cold-treated (2°C for 2 h) samples were re-suspended in microsomal extraction buffer only, or extraction buffer containing 1% (v/v) Triton X-100, and then centrifuged at 100 000 g for 1 h. After the centrifugation, the pellets were dissolved in SDS buffer and analyzed by anti-DPE2 immunoblot. The results show that the DPE2 level in the pellet from both 22 and 2°C samples showed no decrease after Trion X-100 treatment compared with extraction-buffer-only treatment (Fig. 4C), suggesting that the addition of detergent can not increase solubilization of DPE2. These results suggest that sedimentation of DPE2 in ultracentrifugation after cold treatment was not due to association with membranes, but might be due to aggregation or association with other insoluble structures. The enzymatic activity of soluble DPE2 in the supernatant of 15 000-g centrifugation was significantly reduced after cold treatment, as assayed by the capacity of DPE2 to transfer one glucosyl unit of maltose to glycogen and releasing the other as free glucose (Chia et al., 2004) (Figure 4D and 4E). The amount of DPE2 protein also decreased in the soluble fraction (Figure 4D), suggesting the reduced enzymatic activity was most likely due to decreased protein solubility. Consistent with the results of Chia et al. (2004), the DPE2 activity was undetectable in the dpe2-5 knockout mutant (Figure 4D).
Figure 4.
Cold Induces Rapid Accumulation of DPE2 in the Centrifugation Pellet and Decrease of DPE2 Activity in Soluble Cell Extracts. (A) Cold induced a rapid increase of DPE2 in the microsomal (100 000-g pellets) fraction (top panel) but not in the total protein fraction (bottom panel). Arabidopsis seedlings were treated at 2°C for different time duration, and microsomal and total proteins were separated by SDS–PAGE and immunoblotted using the anti-DPE2 antibody. The DPE2 gel bands were quantified using ImageQuant 5.2 software (GE Healthcare) and normalized against the levels of Rubisco large subunit in each sample. Representative images of two repeat experiments performed with different batches of samples are shown. In this figure, the arrows point to DPE2 protein, the stars mark the Ponceau stained band of Rubisco large subunit as the loading control, and the numbers below the gel bands show the normalized signal intensity.
(B) DPE2 became less soluble after cold treatment. Seedlings were treated at 2°C for 2 h. Soluble, microsomal, and total proteins were extracted, separated by SDS–PAGE, and immunoblotted using the anti-DPE2 antibody.
(C) DPE2 is not directly associated with membranes. Microsome (100 000-g pellets) from both control and cold-treated (2°C for 2 h) samples were re-suspended in microsomal extraction buffer only, or extraction buffer containing 1% (v/v) Triton X-100, and then centrifuged at 100 000 g for 1 h; the pellets were dissolved in SDS buffer and analyzed by anti-DPE2 immunoblot.
(D, E) Soluble DPE2 activity was reduced after cold treatment (2°C for 2 h) as shown on native PAGE in-gel assay (D) or assayed in solution (E). (D) Top panel, soluble protein (15 000 g, 10 min) was first separated on native PAGE gel containing 1% oyster glycogen, and then incubated in 5 mM maltose solution, and finally stained with I2-KI solution. The arrow pointed to the band indicates transglucosyl activity, which is absent in the dep2-5 mutant samples. The lower panel shows anti-DPE2 immunoblot of the same protein extracts. (E) DPE2 activity (μmol min−1 g−1 fresh weight) measured in soluble fraction (15 000 g, 10 min) is presented as average of three biological repeats ± standard deviation.
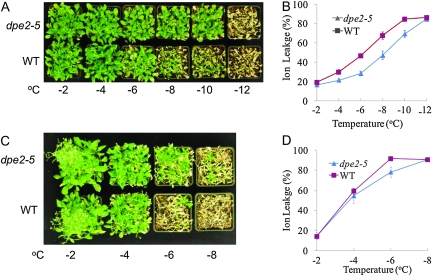
Previous studies have shown that dpe2 knockout lines contain dramatically higher levels of maltose and increased amounts of glucose and fructose, but reduced sucrose content compared to wild-type (Chia et al., 2004; Lu and Sharkey, 2004). Since maltose has been shown to function as a compatible solute that protects proteins, membranes, and photosynthesis apparatus from freezing stress (Kaplan and Guy, 2004), we tested whether the dpe2 mutant has altered freezing tolerance. As shown in Figure 5A, cold-acclimated (4°C for 7 d) dpe2-5 plants survived –8 and –10°C, but wild-type plants were severely damaged at both temperatures, with most of the rosette leaves dead. The dpe2-5 mutant also showed a much lower percentage of ion leakage than wild-type after the freezing treatment (Figure 5B), suggesting that plasma membranes were less damaged in the mutant during the freezing treatment. Without cold adaptation, the dpe2-5 also showed higher basal freezing tolerance than the wild-type plants (Figure 5C and 5D).
Figure 5.
The dpe2-5 Mutant Is More Tolerant to Freezing Damage. (A) Wild-type (WT) and dpe2 plants were grown at 22°C for 4 weeks, then 4°C for 1 week, and then treated at the indicated temperatures for 1 h and then grown for 1 week at 22°C. (B) Electrolyte leakage in wild-type and dpe2-5 after the 1-h freezing treatments in panel (A). The error bars indicate standard errors (n = 12). (C) Wild-type and dpe2 plants were grown at 22°C for 4 weeks, and then treated at the indicated temperatures for 1 h and then grown for 1 week at 22°C. (D) Electrolyte leakage after 1 h at various temperatures as in (C). The error bars represent standard errors (n = 8). The ion leakage percentage at –6°C was significantly lower in dpe2-5 compared to the wild-type (p-value of t-test = 0.0034).
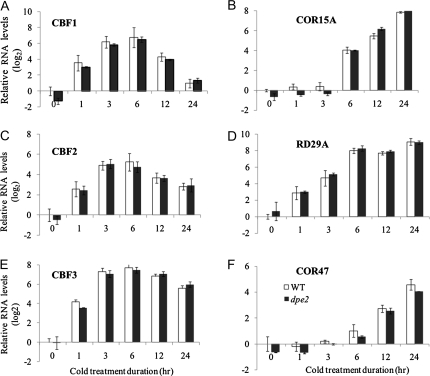
Since the dpe2-5 mutant showed increased freezing tolerance, we analyzed the expression of CBFs and their downstream targets to see whether dpe2-5 affects CBF-regulated gene expression (Figure 6). The basal expression levels of CBFs and their target genes, including COR15A, COR47, and RD29A (Jaglo-Ottosen et al., 1998), were comparable between wild-type and the dpe2-5 mutant. During cold treatment, CBF1, CBF2, and CBF3 showed rapid increases of transcript levels within 1 h, and peaked at approximately 6 h, in both wild-type and dpe2-5. The cold-induced expression of CBF downstream genes, including COR15A, COR47, and RD29A, was also similar between dpe2-5 and wild-type. These results indicate that the increased freezing tolerance of dpe2-5 was not because of increased expression of CBFs and their target genes, and thus was likely due to a direct effect of the accumulation of maltose.
Figure 6.
The CBFs and their Downstream Genes Show Similar Kinetics of Response to Cold in Wild-Type and dpe2-5 Plants. Seedlings were treated in ice-cold medium for the indicated time, and expression of individual genes was analyzed by real-time quantitative RT–PCR. The expression of each gene was first normalized to PP2A (At1g13320), and then to the levels of untreated wild-type samples. Data are means ± SD from three biological replicates.
DISCUSSION
Molecular genetics and transcriptional profiling have illustrated the transcriptional network that mediates cold acclimation in plants. But little is known about the direct effects of low temperature on the proteome. Rapid metabolic changes suggest post-translational regulation of metabolic enzymes is involved in early responses that protect plant cells from freezing damage (Stitt and Hurry, 2002; Kaplan and Guy, 2005). Using a combination of sub-cellular fractionation and 2-D DIGE followed by protein identification by tandem mass spectrometry, we have identified a number of early cold-responsive proteins. Some of these proteins are involved in starch metabolism, TCA cycle, and sucrose metabolism, in agreement with a prominent role of central carbohydrate metabolism in cold protection suggested by metabolic studies (Guy et al., 2008). In particular, our study reveals a post-translational mechanism for rapid inactivation of DPE2 by cold. We propose this inactivation of DPE2 could be responsible for the rapid accumulation of maltose, which protects cells from freezing damage.
Cold acclimation was accompanied by an increase of most TCA cycle intermediates, including α-ketoglutarate, fumarate, malate, and citrate (Cook et al., 2004; Kaplan et al., 2004, 2007). The biological implication of such metabolic changes is unclear, as these changes can reflect flux into or from the TCA cycle for the purpose of either energy production or biosynthetic processes coupled to the TCA cycle (Korn et al., 2010). Some TCA cycle intermediates are associated with heterosis in freezing tolerance in Arabidopsis, such as fumarate and succinate (Korn et al., 2010). The molecular mechanism underlying the accumulation of intermediates is unknown, as these metabolites increased rapidly in the early stage (within 4 h), but the transcript levels of their corresponding biosynthetic genes responded more slowly (increased during 12–24 h and sustained) (Kaplan et al., 2007). In our study, six proteins involved in the TCA cycle, including Aconitate Hydratase 2, Aconitate Hydratase 3, Isocitrate Dehydrogenase subunit 2, Malate Dehydrogenase 1, Succinyl-CoA Synthetase (beta chain), and Citrate Synthase 4, were affected by cold treatment. They were down-regulated in the soluble-microsomal fraction. TCA cycle enzymes are mostly localized in the mitochondrial matrix, and sodium carbonate treatment leads to their release into the soluble-microsomal fraction. It is of particular interest to further characterize how these proteins are coordinately regulated and whether their enzymatic activities are also coordinately controlled by cold treatment.
Another cold-responsive protein associated with the TCA cycle is PEPC2. PEPC catalyzes the reaction of β-carboxylation of phosphoenolpyruvate to yield oxaloacetate and phosphate—a reaction involved in several metabolic pathways (Nimmo, 2003). It has a conserved N-terminal phosphoserine residue in plants, which was also identified in PEPC2 by mass spectrometry in our study. Cold appears to increase PEPC2 phosphorylation, since its acidic forms increased in abundance while its basic form decreased in abundance upon cold treatment. The phosphopeptide was only observed in the acidic PEPC2 spots. Phosphorylation of this residue increases its sensitivity to activation by glucose-6-phosphate, and several lines of evidence support that phosphorylation of PEPC correlates with increased flux through this enzyme in vivo (Carter et al., 1991; Jiao et al., 1991; Bakrim et al., 1993). Since the levels of several TCA cycle intermediates increase during cold shock, phosphorylation of PEPC2 is likely to channel more oxaloacetate for the TCA cycle.
Four starch-degrading proteins were identified in this study as cold-responsive proteins, including GWD1, GWD3, DPE2, and PHS2, supporting the hypothesis that activation of starch degradation plays an important role in increased freezing tolerance in early cold acclimation (Yano et al., 2005; Kaplan et al., 2007). GWD1 was thought to phosphorylate amylopectin starch, and thus provide better accessibility by other starch-degrading enzymes (Yu et al., 2001). T-DNA knockout lines gwd1 were unable to accumulate sucrose, fructose, glucose, and soluble intermediates of starch-degradation products, and showed impaired freezing tolerance (Yano et al., 2005). GWD3 primarily catalyzes phosphorylation at the C-3 position of the glucose unit of preferably pre-phosphorylated amylopectin substrate with long side chains (Baunsgaard et al., 2005). PHS2 was proposed to phosphorylate the glucosyl residues of soluble heteroglycan, resulting in glucose 1-phosphate, and was involved in maltose metabolism in Arabidopsis (Lu et al., 2006). Absence of PHS2 gene expression in the T-DNA knockout line did not cause obvious growth phenotype, but resulted in increased nighttime maltose levels. Transcript profiling indicated that starch-degrading genes, such as beta-amylase and glucan-water dikinase (GWD1 and GWD3), were up-regulated in 1 or 4 d under cold conditions (Maruyama et al., 2009). However, GWD1 enzyme activity did not show a significant change for up to 12 h at 2°C (Yano et al., 2005), and it was proposed that components downstream of GWD1 are responsible for accumulation of starch-degradation products during early cold acclimation (Yano et al., 2005). In this study, GWD1 and GWD3 showed rapid increase in the Triton-insoluble fraction, and we hypothesize that GWDs may play an important role in cold activation of starch degradation through increased binding to starch.
Metabolite profiling has shown that maltose is the first carbohydrate to increase upon cold treatment (Kaplan et al., 2004, 2007). The levels of maltose increases over 100-fold with 1 h of cold treatment and reaches peak levels at 4 h after the onset of cold treatment (Kaplan et al., 2007). Previous studies have shown transcriptional regulation of beta-amylase 8, which is responsible for maltose biosynthesis during starch degradation (Kaplan and Guy, 2004). However, the rapid change of maltose levels is most likely due to post-translational rather than transcriptional regulation (Kaplan and Guy, 2004, 2005). Our results strongly suggest that cold-induced inactivation of DPE2 is one of the primary mechanisms leading to rapid accumulation of maltose.
Rapid and drastic increase of DPE2 in the centrifugation pellet is the most prominent change observed in this proteomics study. Since 1% Triton X-100 treatment did not increase solublization of DPE2 in the 100 000-g pellet (Figure 4C), DPE2 seems not directly associated with membrane structures. Thus, our results suggest that cold induces self-aggregation of DPE2 or association with insoluble cell structures. Such association occurs very rapidly and is detectable within 5 min of cold treatment. This leads to a reduction in the protein levels and enzyme activity of soluble DPE2. Reduced DPE2 activity will most likely contribute to cold-induced rapid accumulation of maltose (Kaplan and Guy, 2004; Kaplan et al., 2004; Kaplan and Guy, 2005; Yano et al., 2005; Kaplan et al., 2007), because the dpe2-5 mutant has been shown to accumulate high levels of maltose (Chia et al., 2004). Since maltose has been shown to act as a compatible solute and protects proteins, membranes, and thylakoids from temperature extremes (Kaplan and Guy, 2004), cold-induced inactivation of DPE2 and subsequent maltose accumulation should increase freeze tolerance. Indeed, we found that the dep2-5 mutant has significantly increased freezing tolerance (Figure 5). In addition to the role of maltose as a compatible solute, it is also possible that accumulation of maltose and other sugars increases freezing tolerance by regulating hormone biosynthesis and signaling. A wealth of evidence supports the link between sugar levels and hormonal (especially ABA and ethylene) production and signaling (Hare et al., 1998; Loreti et al., 2001; Leon and Sheen, 2003; Rolland et al., 2006). ABA is implicated in cold signaling (Thomashow, 1999), and reduced ABA levels impair freezing tolerance in Arabidopsis (Gilmour and Thomashow, 1991). Together, our proteomic and genetic data support a model that cold quickly inactivates DPE2 by removing it from soluble phase of the cytoplasm, leading to rapid accumulation of maltose, which protects plant cells against freezing damage.
METHODS
Plant Materials and Cold Treatment
Arabidopsis thaliana seedlings used for 2-D DIGE analysis were grown in liquid half-strength Murashige and Skoog medium containing 1.5% sucrose for 6 d under constant light conditions at 22°C. Half of the medium was transferred into a new container and chilled on ice to 2°C. Half of the cultured seedlings were then transferred into the chilled medium to start the cold treatment, while the other half remained at room temperature. At the end of cold treatments, seedlings were harvested and frozen in liquid nitrogen for proteomics analyses. Three biological repeats of 2-h cold-treated samples and control samples were used in each 2-D DIGE analysis. About 1.0–2.0 g of tissues were used for extraction of 2-D DIGE protein samples.
Freezing Tolerance Assays
Seedlings were first grown on Petri dishes for 10 d, and then on soil for 3 weeks, under long day (16-h light/8-h dark) at 22°C. Seedlings were treated with subfreezing temperatures in a bench-top temperature chamber (Tenney Model Jr, Tenney Engineering, Inc., Union, NJ, USA) for 1 h. For freezing tolerance after cold acclimation, 1-month-old plants were treated for 1 week at 4°C (cold acclimation) before freezing treatment. The freezing treatment was performed as described previously (Zhu et al., 2004). The temperature in the chamber containing the plants was reduced to –2 from 4°C in 30 min and held at –2°C for 1 h, and then the temperature was gradually reduced to –4, –6, –8, –10, and –12°C at a ramp rate of 2°C/30 min and 1 h hold at each temperature point. Ion leakage test was determined right after the freezing treatment. Approximately two-thirds of rosette leaves of each plant were taken, shaken in 10 ml de-ionized water overnight, and the conductivity of the solution was determined and compared with the conductivity after the leaves were autoclaved. To evaluate survival rate, freezing-treated plants were placed at 4°C overnight, and then grown under the long-day condition at 22°C for 1 week. At least two independent experiments were performed, with four seedlings in each assay.
2-D DIGE Protein Sample Preparation
Microsomal and soluble proteins were extracted as described previously (Deng et al., 2007; Tang et al., 2008b) with slight modifications. Liquid nitrogen-ground tissue powder was mixed with 3 vol. of microsomal extraction buffer (50 mM HEPES, pH 7.5, 0.33 M sucrose, 5% glycerol, 3 mM EDTA, 5 mM DTT) with phosphatase and protease inhibitors (10 mM sodium fluoride, 10 mM sodium molybdate, 10 mM imidazole, 1 mM activated sodium vanadate, 7 μM E–64, 1.5 μM bestatin, 2 μM pepstatin, 4 μM antipain, 1 mM PMSF), and then centrifuged at 10 000 g for 20 min to remove cell debris. The supernatant was further separated into soluble and microsomal (100 000-g pellet) fractions after 100 000 g for 1 h. The microsomal pellet was dissolved in the SDS extraction buffer (100 mM Tris-HCl, pH 8.0, 2% SDS, 1% β-mercaptoethanol, 5 mM EGTA, 10 mM EDTA), and then the proteins from both the microsomal pellet and soluble fractions were precipitated following a modified phenol-methanol protocol as described by Deng et al. (2007). The protein extract was dissolved in 2-D DIGE buffer (6 M urea, 2 M thiourea, 4% CHAPS) and quantified with the Bradford protein assay (BioRad).
Extraction of sodium-carbonate soluble microsomal protein was modified from Fujiki et al. (1982). The microsomal pellet obtained after 100 000-g centrifugation as described above was dissolved in 0.1 M sodium carbonate, and kept on ice for half an hour, then separated by centrifugation at 100 000 g for 60 min. The supernatant was mixed with equal amounts of phenol, and the protein was precipitated by the modified phenol-methanol protocol (Deng et al., 2007).
Triton-insoluble fraction was extracted as described previously (Gampala et al., 2007). Liquid nitrogen-ground tissue was mixed with 3 vol. of extraction buffer (20 mM HEPES, pH 7.5, 40 mM KCl, 1 mM EDTA, protease and phosphatase inhibitors), filtered through one layer of miracloth (Calbiochem, San Diego, CA, USA) and a 40-μm nylon mesh to obtain total extract. The total extract was treated with 1% Triton X-100 and 10 mM MgCl2 for 10 min, and then centrifuged at 5000 g for 5 min, and the pellet (Triton-insoluble fraction) was dissolved in the SDS extraction buffer and protein was precipitated using the phenol-methanol protocol as described above (Deng et al., 2007).
2-D DIGE Protocol and Image Analysis
2-D DIGE was performed as described (Deng et al., 2007), with minor modifications. Twenty micrograms of protein (pH 8.5) from control and cold-treated samples, respectively, was mixed with 80 pmol of Cy3 or Cy5 minimal dyes, and incubated on ice for at least 2 h in the dark. The labeling reaction was terminated by adding 0.5 μl of 10 mM lysine. Then, Cy3- and Cy5-labeled protein was combined and used for iso-electric focusing (IEF). IEF was performed on 24-cm IPG strips, pH 4–7 (GE Healthcare). The running conditions were as follows: rehydration for 2 h, 50 V for 10 h, step and hold at 500 V, 1000 V for 1 h each, gradient to 8000 V in 3 h, and then at 8000 V, until reaching a total of 74 000 V-h. Second-dimension electrophoresis was performed using 10% SDS-polyacrylamide gels, with molecular mass markers (170–26 kDa). The electrophoresis was performed at 40 V for 2 h and then at 120 V until the bromophenol blue front reached the end of the gel. Cy3- and Cy5-labeled images were acquired using a Typhoon Trio scanner (GE Healthcare, Piscataway, NJ, USA).
The DIGE images were analyzed using DeCyder 6.5 software (GE Healthcare). Differential in-gel analysis module with an estimated spot number 5000 was used for spot detection, and biological variation analysis module was employed to identify spots differentially expressed between cold-treated and control samples with statistically significant differences (p-value < 0.05) and minimal abundance changes of over 30% (Deng et al., 2007). At least three biological repeats were examined in each comparison, and spots of interests were manually checked to confirm spot matching between different gels and to remove artifacts.
Protein Identification Using Mass Spectrometry
Spot picking and reverse-phase liquid chromatography–electrospray tandem mass spectrometry (LC–MS/MS) analyses were performed as follows. Approximately 320 μg of protein were labeled with 40 pmol of Cy5 dye and separated by 2-DE. 2-DE gels were stained with deep-purple dye (GE Healthcare) after electrophoresis. After scanning the gels with Typhoon Trio, spots of interest were selected with DeCyder software and picked by an Ettan spot picker (GE Healthcare). The specific protein spots were washed twice with 50% acetonitrile in 25 mM ammonium bicarbonate (NH4HCO3) and vacuum-dried, re-hydrated in 10 μl of digestion buffer (10 ng μl−1 trypsin in 25 mM NH4HCO3), and covered with a minimum volume of NH4HCO3. After overnight digestion at 37°C, peptides were extracted twice with a solution containing 50% acetonitrile and 5% formic acid. The extracted digests were vacuum-dried and re-suspended in 10 μl of 0.1% formic acid in water, and then analyzed by LC–MS/MS. The digests were separated by nanoflow liquid chromatography using a 100-μm × 150-mm reverse-phase Ultra 120-μm C18Q column (Peeke Scientific, Redwood City, CA, USA) at a flow rate of 350 nl min−1 in an Agilent 1100 high-performance liquid chromatography system (Agilent Technologies, Inc., Santa Clara, CA, USA). Mobile phase A was 0.1% formic acid in water and mobile phase B was 0.1% formic acid in acetonitrile. Following equilibration of the column in 2% solvent B, one-half of each digest (5 μl) was injected, and then the organic content of the mobile phase was increased linearly to 40% over 30 min and then to 50% in 3 min. The liquid chromatography eluate was coupled to the mass spectrometer. Some samples were analyzed in hybrid linear ion trap–Fourier transform mass spectrometer (LTQ–FT, Thermo Scientific, San Jose, CA, USA) equipped with a nanoelectrospray ion source. Spraying was from an uncoated 15-μm-inner-diameter spraying needle (New Objective, Woburn, MA, USA). Peptides were analyzed in positive ion mode and in information-dependent acquisition mode to automatically switch between MS and MS/MS acquisition. MS spectra were acquired in profile mode using the ICR analyzer in the mass per charge (m/z) range between 310 and 1600. For each MS spectrum, the five most intense multiple charged ions over a threshold of 200 counts were selected. Product ions were analyzed on the linear ion trap in profile mode; the collision energy was automatically set to 35%. A dynamic exclusion window of 1 Da was applied that prevented the same m/z from being selected for 60 s after its acquisition. Peak lists were generated using Mascot Distiller version 2.1.0.0 (Matrix Science, Boston, MA, USA). Parameters for MS processing were set as follows: peak half-width, 0.02; data points per Da, 100. Parameters for MS/MS data were set as follows: peak half-width, 0.02; data points per Da, 100.
In some other cases, mass spectrometry was performed on either a QSTAR Pulsar or a QSTAR Elite mass spectrometer (AB SCIEX, Foster City, CA, USA). Peptides were analyzed in positive ion mode. MS spectra were acquired for 1 s in the m/z range between 310 and 1400. MS acquisitions were followed by two CID experiments in information-dependent acquisition mode. For each MS spectrum, the most intense multiply charged peaks over a threshold of 30 counts were selected for CID analysis. A dynamic exclusion window was applied to prevent the same m/z from being selected for 60 s after its acquisition. The data were analyzed with Analyst QS 1.1 software (AB SCIEX) and fragmentation data were converted to peaklists using the mascot.dll script (Mascot.dlll 1.6b19, AB SCIEX).
In all cases, the peak lists were searched in in-house ProteinProspector version 5.4.2 (Chalkley et al., 2005) (public version at http://prospector.ucsf.edu). Enzyme specificity was set to trypsin, and the maximum number of missed enzyme cleavages per peptide was set at one. The number of modifications was limited to two per peptide. Carbamidomethylation of cysteine was included as a fixed modification; N-acetylation of the N-terminus of the protein, oxidation of methionine, formation of pyro-Glu from N-terminal Glutamine, and phosphorylation of serine, threonine, or tyrosine were all allowed as variable modifications. In searches of LTQ–FT data, mass tolerance was 30 ppm for precursor and 0.8 Da for fragment ions. For QStar data, a precursor mass tolerance of 100 ppm and a fragment mass error tolerance of 0.2 Da were allowed for the data search.
The peak lists were searched against a subset of the UniprotKB database as of 15 December 2009, containing all entries for Arabidopsis (53 624 entries searched). In all protein identifications, a minimal protein score of 22, a peptide score of 15, and a minimal discriminate score threshold of 0.0 were used for initial identification criteria. Maximum expectation value threshold (number of different peptides with scores equivalent to or better than the result reported that are expected to occur in the database search by chance) was set to 0.05 for accepting individual spectra and 0.01 for accepting individual protein. When several accession numbers in the database matched the same set of peptides identified, the entries with the most descriptive name were reported. Individual isoforms of proteins were reported according to the detection of peptides unique to their sequences. If several isoforms shared the same set of peptides identified, they were all reported. Only proteins with at least two peptides identified were further considered and reported. In order to assign the modification site for peptides containing post-translational modifications, the MS/MS spectrum was reinterpreted manually by matching the observed fragment ions to a theoretical fragmentation obtained using MS Product (Protein Prospector).
DPE2 Protein Immunoblot and Activity Assay
DPE2 protein was detected in immunoblots using an anti-DPE2 antibody described previously (Chia et al., 2004). This antibody specifically detects DPE2 but not the chloroplast DPE1 (Chia et al., 2004). Total protein was extracted by mixing 0.1 g of frozen tissue powder with 300 μl of the SDS extraction buffer (100 mM Tris-HCl, pH 8.0, 2% SDS, 1% β-mercaptoethanol, 5 mM EGTA, 10 mM EDTA), followed by heating for 10 min at 65°C, and then centrifugation at 20 000 g for 20 min. Soluble and microsomal (100 000-g pellet) proteins in Figure 4A and 4B were extracted as described for 2-D DIGE samples. For the insoluble fraction in Figure 4C, microsomal pellet was further re-suspended in the microsomal extraction buffer only, or the microsomal buffer containing 1% Trion X-100, and then centrifuged at 100 000 g for 1 h. The insoluble proteins in the pellets were separated in SDS–PAGE gels and gel blots were probed using the anti-DPE2 antibody. Immunoblotting was performed as described (Deng et al., 2007). The blots were stained by Ponceau S to determine equal loading.
DPE2 activity was determined from extracts of whole seedlings prepared by 10-min 15 000-g centrifugation as described previously (Chia et al., 2004) except that the glucose content was determined using the glucose assay reagent (Sigma, St Louis, MO, USA). In-gel DPE2 activity assay was performed following a protocol modified from the one previously described for DPE1 (Critchley et al., 2001). The native resolving gel contained 7.5% (w/v) acrylamide (30:0.8 acrylamide/bisacrylamide), 375 mM Tris-HCl, pH 8.8, and 1% (w/v) oyster glycogen. The stacking gel contained 3.3% (w/v) acrylamide and 63 mM Tris, pH 6.8. A total of 30 μg of soluble protein was separated per lane. After electrophoresis at 12 mA and 4°C, the gel was washed twice with 40 ml of 100 mM Tris-HCl, pH 7.0, 1 mM MgCl2, 1 mM EDTA, and 1 mM DTT for 15 min then incubated for 2 h at 37°C in 20 ml of this medium plus 5 mM maltose. The gel was stained with 0.67% (w/v) I2 and 3.33% (w/v) KI. DPE2 activity assay and in-gel activity assay were performed three times with independent samples.
RNA Analysis
Total RNA extraction, first-strand cDNA synthesis, and real-time PCR were performed as described (Deng et al., 2007), except that the PCR conditions were as follows: 95°C for 8 min, then 45 cycles of 94°C for 10 s, 63°C for 15 s, and 72°C for 15 s. The primers used for each gene are listed in Table 4. For each data point, three biological repeats were used for total RNA extraction and subsequent analysis.
Table 4.
Quantitative RT–PCR Primers Used in this Study.
| Gene name | Locus | Forward primers | Reverse primers |
| CBF1 | AT4G25490 | 5'-GGAGACAATG TTTGGGATGCCGAC-3' | 5'-CTCCAAAGCGACACG TCACCATCTC-3' |
| CBF2 | AT4G25470 | 5'-CGGAATCAACCT GTGCCAAGGAAA-3' | 5'-AGACCATGAGCATCC GTCGTCATA-3' |
| CBF3 | AT2G42540 | 5'-GGCTTGGAGACT CCGAATCCCG-3' | 5'-AGCCTCCACCAACGTC TCCTCC-3' |
| COR15A | AT2G42540 | 5'-GCTGCGGCGTAT GTGGAGGAGAA-3' | 5'-GGTAAGACCCTACTTT GTGGCATCC-3' |
| RD29A | AT5G52310 | 5'-ACCAGCTACTAG GTCTAAAGTGGGT-3' | 5'-ACGATAGAGACAACA CCTCAACAAGTC-3' |
| COR47 | AT1G20440 | 5'-ACAAGCCTAGT GTCATCGAAAAGC-3' | 5'-TCTTCATCGCTCGAA GAGGAAG-3' |
| PP2A | AT1G13320 | 5'-TTCTCGCTCCAG TAATGGGATCCGA-3' | 5'-GTTCTCCACA ACCGCTTGGTCGACT-3' |
FUNDING
R.W. and T.L. were supported by the Carnegie Institution for Science and the China Scholarship Council. The UCSF Mass Spectrometry Facility (A.L. Burlingame, Director) is supported by the Biomedical Research Technology Program of the National Center for Research Resources, NIH NCRR P41RR001614, P41RR019934, and RR012961.
Acknowledgments
We thank Dr Alison M. Smith (John Innes Center, UK) for providing the anti-DPE2 antibody. Research was supported by grants from the US Department of Energy (DE-FG02-08ER15973), National Key Project (2009ZX08009–017B), and Carnegie Institution for Science. T-DNA line dpe2-5 was generated by Dr Joe Ecker at the Salk Institute and provided by the Arabidopsis Stock Center. No conflict of interest declared.
References
- Agarwal M, Katiyar-Agarwal S, Sahi C, Gallie DR, Grover A. Arabidopsis thaliana Hsp100 proteins: kith and kin. Cell Stress Chaperones. 2001;6:219–224. doi: 10.1379/1466-1268(2001)006<0219:athpka>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amme S, Matros A, Schlesier B, Mock HP. Proteome analysis of cold stress response in Arabidopsis thaliana using DIGE-technology. J. Exp. Bot. 2006;57:1537–1546. doi: 10.1093/jxb/erj129. [DOI] [PubMed] [Google Scholar]
- Apuya NR, Yadegari R, Fischer RL, Harada JJ, Zimmerman JL, Goldberg RB. The Arabidopsis embryo mutant schlepperless has a defect in the chaperonin-60alpha gene. Plant Physiol. 2001;126:717–730. doi: 10.1104/pp.126.2.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae MS, Cho EJ, Choi EY, Park OK. Analysis of the Arabidopsis nuclear proteome and its response to cold stress. Plant J. 2003;36:652–663. doi: 10.1046/j.1365-313x.2003.01907.x. [DOI] [PubMed] [Google Scholar]
- Bakrim N, et al. Regulatory phosphorylation of C4 phosphoenolpyruvate carboxylase (a cardinal event influencing the photosynthesis rate in sorghum and maize) Plant Physiol. 1993;101:891–897. doi: 10.1104/pp.101.3.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baunsgaard L, Lutken H, Mikkelsen R, Glaring MA, Pham TT, Blennow A. A novel isoform of glucan, water dikinase phosphorylates pre-phosphorylated alpha-glucans and is involved in starch degradation in Arabidopsis. Plant J. 2005;41:595–605. doi: 10.1111/j.1365-313X.2004.02322.x. [DOI] [PubMed] [Google Scholar]
- Carter PJ, Nimmo HG, Fewson CA, Wilkins MB. Circadian rhythms in the activity of a plant protein kinase. EMBO J. 1991;10:2063–2068. doi: 10.1002/j.1460-2075.1991.tb07737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalkley RJ, et al. Comprehensive analysis of a multidimensional liquid chromatography mass spectrometry dataset acquired on a quadrupole selecting, quadrupole collision cell, time-of-flight mass spectrometer: II. New developments in Protein Prospector allow for reliable and comprehensive automatic analysis of large datasets. Mol. Cell Proteomics. 2005;4:1194–1204. doi: 10.1074/mcp.D500002-MCP200. [DOI] [PubMed] [Google Scholar]
- Chia T, et al. A cytosolic glucosyltransferase is required for conversion of starch to sucrose in Arabidopsis leaves at night. Plant J. 2004;37:853–863. doi: 10.1111/j.1365-313x.2003.02012.x. [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, et al. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003;17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu J, Zhu JK. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007;12:444–451. doi: 10.1016/j.tplants.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Cook D, Fowler S, Fiehn O, Thomashow MF. A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proc. Natl Acad. Sci. U S A. 2004;101:15243–15248. doi: 10.1073/pnas.0406069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchley JH, Zeeman SC, Takaha T, Smith AM, Smith SM. A critical role for disproportionating enzyme in starch breakdown is revealed by a knock-out mutation in Arabidopsis. Plant J. 2001;26:89–100. doi: 10.1046/j.1365-313x.2001.01012.x. [DOI] [PubMed] [Google Scholar]
- Cui S, Huang F, Wang J, Ma X, Cheng Y, Liu J. A proteomic analysis of cold stress responses in rice seedlings. Proteomics. 2005;5:3162–3172. doi: 10.1002/pmic.200401148. [DOI] [PubMed] [Google Scholar]
- Deng Z, et al. A proteomics study of brassinosteroid response in Arabidopsis. Mol. Cell Proteomics. 2007;6:2058–2071. doi: 10.1074/mcp.M700123-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fettke J, Chia T, Eckermann N, Smith A, Steup M. A transglucosidase necessary for starch degradation and maltose metabolism in leaves at night acts on cytosolic heteroglycans (SHG) Plant J. 2006;46:668–684. doi: 10.1111/j.1365-313X.2006.02732.x. [DOI] [PubMed] [Google Scholar]
- Fowler S, Thomashow MF. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell. 2002;14:1675–1690. doi: 10.1105/tpc.003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y, Hubbard AL, Fowler S, Lazarow PB. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gampala SS, et al. An essential role for 14–3–3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell. 2007;13:177–189. doi: 10.1016/j.devcel.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Thomashow MF. Cold acclimation and cold-regulated gene expression in ABA mutants of Arabidopsis thaliana. Plant Mol. Biol. 1991;17:1233–1240. doi: 10.1007/BF00028738. [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Fowler SG, Thomashow MF. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 2004;54:767–781. doi: 10.1023/B:PLAN.0000040902.06881.d4. [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 2000;124:1854–1865. doi: 10.1104/pp.124.4.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy C, Kaplan F, Kopka J, Selbig J, Hincha DK. Metabolomics of temperature stress. Physiol. Plant. 2008;132:220–235. doi: 10.1111/j.1399-3054.2007.00999.x. [DOI] [PubMed] [Google Scholar]
- Hannah MA, Heyer AG, Hincha DK. A global survey of gene regulation during cold acclimation in Arabidopsis thaliana. PLoS Genet. 2005;1:e26. doi: 10.1371/journal.pgen.0010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare PD, Cress WA, Van Staden J. Dissecting the roles of osmolyte accumulation during stress. Plant. Cell & Environment. 1998;21:535–553. [Google Scholar]
- Hashimoto M, Komatsu S. Proteomic analysis of rice seedlings during cold stress. Proteomics. 2007;7:1293–1302. doi: 10.1002/pmic.200600921. [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Verboom RE, Tonti-Filippini J, Small I, Millar AH. SUBA: the Arabidopsis Subcellular Database. Nucleic Acids Res. 2007;35:D213–D218. doi: 10.1093/nar/gkl863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SW, Vierling E. Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc. Natl Acad. Sci. U S A. 2000;97:4392–4397. doi: 10.1073/pnas.97.8.4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imin N, Kerim T, Rolfe BG, Weinman JJ. Effect of early cold stress on the maturation of rice anthers. Proteomics. 2004;4:1873–1882. doi: 10.1002/pmic.200300738. [DOI] [PubMed] [Google Scholar]
- Imin N, Kerim T, Weinman JJ, Rolfe BG. Low temperature treatment at the young microspore stage induces protein changes in rice anthers. Mol. Cell Proteomics. 2006;5:274–292. doi: 10.1074/mcp.M500242-MCP200. [DOI] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science. 1998;280:104–106. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- Jiao J, Echevarria C, Vidal J, Chollet R. Protein turnover as a component in the light/dark regulation of phosphoenolpyruvate carboxylase protein-serine kinase activity in C4 plants. Proc. Natl Acad. Sci. U S A. 1991;88:2712–2715. doi: 10.1073/pnas.88.7.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F, Guy CL. beta-Amylase induction and the protective role of maltose during temperature shock. Plant Physiol. 2004;135:1674–1684. doi: 10.1104/pp.104.040808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F, Guy CL. RNA interference of Arabidopsis beta-amylase8 prevents maltose accumulation upon cold shock and increases sensitivity of PSII photochemical efficiency to freezing stress. Plant J. 2005;44:730–743. doi: 10.1111/j.1365-313X.2005.02565.x. [DOI] [PubMed] [Google Scholar]
- Kaplan F, et al. Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol. 2004;136:4159–4168. doi: 10.1104/pp.104.052142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F, et al. Transcript and metabolite profiling during cold acclimation of Arabidopsis reveals an intricate relationship of cold-regulated gene expression with modifications in metabolite content. Plant J. 2007;50:967–981. doi: 10.1111/j.1365-313X.2007.03100.x. [DOI] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 1999;17:287–291. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- Kawamura Y, Uemura M. Mass spectrometric approach for identifying putative plasma membrane proteins of Arabidopsis leaves associated with cold acclimation. Plant J. 2003;36:141–154. doi: 10.1046/j.1365-313x.2003.01864.x. [DOI] [PubMed] [Google Scholar]
- Korn M, Gartner T, Erban A, Kopka J, Selbig J, Hincha DK. Predicting Arabidopsis freezing tolerance and heterosis in freezing tolerance from metabolite composition. Mol. Plant. 2010;3:224–235. doi: 10.1093/mp/ssp105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DG, et al. Chilling stress-induced proteomic changes in rice roots. J. Plant Physiol. 2009;166:1–11. doi: 10.1016/j.jplph.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Leon P, Sheen J. Sugar and hormone connections. Trends Plant Sci. 2003;8:110–116. doi: 10.1016/S1360-1385(03)00011-6. [DOI] [PubMed] [Google Scholar]
- Liu Q, et al. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E, Bellis LD, Alpi A, Perata P. Why and how do plant cells sense sugars? Ann. Bot. 2001;88:803–812. [Google Scholar]
- Lu Y, Sharkey TD. The role of amylomaltase in maltose metabolism in the cytosol of photosynthetic cells. Planta. 2004;218:466–473. doi: 10.1007/s00425-003-1127-z. [DOI] [PubMed] [Google Scholar]
- Lu Y, Steichen JM, Yao J, Sharkey TD. The role of cytosolic alpha-glucan phosphorylase in maltose metabolism and the comparison of amylomaltase in Arabidopsis and Escherichia coli. Plant Physiol. 2006;142:878–889. doi: 10.1104/pp.106.086850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K, et al. Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol. 2009;150:1972–1980. doi: 10.1104/pp.109.135327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanjo T, Kobayashi M, Yoshiba Y, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K. Antisense suppression of proline degradation improves tolerance to freezing and salinity in Arabidopsis thaliana. FEBS Lett. 1999;461:205–210. doi: 10.1016/s0014-5793(99)01451-9. [DOI] [PubMed] [Google Scholar]
- Nimmo HG. Control of the phosphorylation of phosphoenolpyruvate carboxylase in higher plants. Arch. Biochem. Biophys. 2003;414:189–196. doi: 10.1016/s0003-9861(03)00115-2. [DOI] [PubMed] [Google Scholar]
- Phinney BS, Thelen JJ. Proteomic characterization of a triton-insoluble fraction from chloroplasts defines a novel group of proteins associated with macromolecular structures. J. Proteome Res. 2005;4:497–506. doi: 10.1021/pr049791k. [DOI] [PubMed] [Google Scholar]
- Queitsch C, Hong SW, Vierling E, Lindquist S. Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell. 2000;12:479–492. doi: 10.1105/tpc.12.4.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaut J, Lutts S, Hoffmann L, Hausman JF. Responses of poplar to chilling temperatures: proteomic and physiological aspects. Plant Biol. (Stuttg.) 2004;6:81–90. doi: 10.1055/s-2004-815733. [DOI] [PubMed] [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J. Sugar sensing and signaling in plants: conserved and novel mechanisms. Ann. Rev. Plant Biol. 2006;57:675–709. doi: 10.1146/annurev.arplant.57.032905.105441. [DOI] [PubMed] [Google Scholar]
- Salekdeh GH, Komatsu S. Crop proteomics: aim at sustainable agriculture of tomorrow. Proteomics. 2007;7:2976–2996. doi: 10.1002/pmic.200700181. [DOI] [PubMed] [Google Scholar]
- Stitt M, Hurry V. A plant for all seasons: alterations in photosynthetic carbon metabolism during cold acclimation in Arabidopsis. Curr. Opin. Plant Biol. 2002;5:199–206. doi: 10.1016/s1369-5266(02)00258-3. [DOI] [PubMed] [Google Scholar]
- Strand A, et al. Acclimation of Arabidopsis leaves developing at low temperatures: increasing cytoplasmic volume accompanies increased activities of enzymes in the Calvin cycle and in the sucrose–biosynthesis pathway. Plant Physiol. 1999;119:1387–1398. doi: 10.1104/pp.119.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss G, Hauser H. Stabilization of lipid bilayer vesicles by sucrose during freezing. Proc. Natl Acad. Sci. U S A. 1986;83:2422–2426. doi: 10.1073/pnas.83.8.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taji T, et al. Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J. 2002;29:417–426. doi: 10.1046/j.0960-7412.2001.01227.x. [DOI] [PubMed] [Google Scholar]
- Tang W, et al. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science. 2008a;321:557–560. doi: 10.1126/science.1156973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, et al. Proteomics studies of brassinosteroid signal transduction using prefractionation and two-dimensional DIGE. Mol. Cell Proteomics. 2008b;7:728–738. doi: 10.1074/mcp.M700358-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- Tonge R, et al. Validation and development of fluorescence two-dimensional differential gel electrophoresis proteomics technology. Proteomics. 2001;1:377–396. doi: 10.1002/1615-9861(200103)1:3<377::AID-PROT377>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Unlu M, Morgan ME, Minden JS. Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis. 1997;18:2071–2077. doi: 10.1002/elps.1150181133. [DOI] [PubMed] [Google Scholar]
- Vidal J, Chollet R. Regulatory phosphorylation of C4 PEP carboxylase. Trends Plant Sci. 1997;2:230–237. [Google Scholar]
- Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF. Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 2005;41:195–211. doi: 10.1111/j.1365-313X.2004.02288.x. [DOI] [PubMed] [Google Scholar]
- Wang X, Li W, Li M, Welti R. Profiling lipid changes in plant response to low temperatures. Physiologia Plantarum. 2006;126:90–96. [Google Scholar]
- Xiong L, Zhu JK. Abiotic stress signal transduction in plants: molecular and genetic perspectives. Physiol. Plant. 2001;112:152–166. doi: 10.1034/j.1399-3054.2001.1120202.x. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- Yan SP, Zhang QY, Tang ZC, Su WA, Sun WN. Comparative proteomic analysis provides new insights into chilling stress responses in rice. Mol. Cell Proteomics. 2006;5:484–496. doi: 10.1074/mcp.M500251-MCP200. [DOI] [PubMed] [Google Scholar]
- Yano R, Nakamura M, Yoneyama T, Nishida I. Starch-related alpha-glucan/water dikinase is involved in the cold-induced development of freezing tolerance in Arabidopsis. Plant Physiol. 2005;138:837–846. doi: 10.1104/pp.104.056374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TS, et al. The Arabidopsis sex1 mutant is defective in the R1 protein, a general regulator of starch degradation in plants, and not in the chloroplast hexose transporter. Plant Cell. 2001;13:1907–1918. doi: 10.1105/TPC.010091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, et al. An Arabidopsis homeodomain transcription factor gene, HOS9, mediates cold tolerance through a CBF-independent pathway. Proc. Natl Acad. Sci. U S A. 2004;101:9873–9878. doi: 10.1073/pnas.0403166101. [DOI] [PMC free article] [PubMed] [Google Scholar]