Abstract
Background
Hereditary breast cancer runs in families where several members in different generations are affected. Most of these breast cancers are caused by mutations in the high penetrance genes BRCA1 and BRCA2 accounting for about 5% of all breast cancers. Other genes that include CHEK2, PTEN, TP53, ATM, STK11/LKB1, CDH1, NBS1, RAD50, BRIP1 and PALB2 have been described to be high or moderate penetrance breast cancer susceptibility genes, all contributing to the hereditary breast cancer spectrum. However, in still a part of familial hereditary breast cancers no relationship to any of these breast cancer susceptibility genes can be found. Research on new susceptibility genes is therefore ongoing.
Design
In this review we will describe the function of the today known high or moderate penetrance breast cancer susceptibility genes and the consequences of their mutated status. Furthermore, we will focus on the histology, the immunophenotype and genotype of breast cancers caused by mutations in BRCA1 and BRCA2 genes and the other high or moderate penetrance breast cancer susceptibility genes. Finally, an overview of the clinical implications of hereditary breast cancer patients will be provided.
Conclusion
This information leads to a better understanding of the morphological, immunohistochemical and molecular characteristics of different types of hereditary breast cancers. Further, these characteristics offer clues for diagnosis and new therapeutic approaches.
Keywords: Hereditary breast cancer, BRCA1, BRCA2, Pathology, Genetics
Introduction
In 1866, Paul Broca was the first to describe a family with a high prevalence of carcinoma of the breast. His wife suffered from early onset of breast cancer and when he made a pedigree of her family, four generations with breast cancer could be identified [24]. The “Broca” report is the first of many that pointed out that breast cancer can be inherited, passing through from one generation to the other. Family history of breast cancer is now an established risk factor for the development of the disease. In fact, among those variables that have been shown to bear a causal relationship with breast cancer, the highest increased risk, after age, is a positive family history of breast cancer [31]. With the knowledge of today, only in about 5% of all the breast cancer cases, the disease will occur as part of a hereditary cancer susceptibility syndrome, caused by mutations in high penetrance susceptibility genes. A substantial proportion of hereditary breast cancers, about 16% [2, 138], can be attributed to germline mutations in either of the BRCA (breast cancer 1 and 2) early onset genes. Since the identification of the BRCA1 and BRCA2 genes in 1994, several studies have been undertaken to find other high penetrance breast cancer susceptibility genes than BRCA1 and BRCA2, with less spectacular results so far [124].
Nevertheless, various other genes conferring an increased risk of breast cancer involved in hereditary cancer syndromes have been identified, including CHEK2, PTEN, TP53, ATM, STK11/LKB1, CDH1, NBS1, RAD50, BRIP1 and PALB2. Some of these genes are involved in multiple cancer syndromes like Li-Fraumeni (TP53), Peutz-Jeghers (STK11/LKB1) and Cowden syndrome (PTEN) [49, 69, 94, 125, 161]. In Table 1, an overview of the hereditary cancer susceptibility syndrome genes is shown, including their chromosomal location, which syndrome is involved and the clinical features of these syndromes.
Table 1.
Summary of the syndromes associated with hereditary breast cancer, adapted from Tan et al. [170]
| Gene involved | Cytoband | Breast cancer risk | Syndrome | Clinical features |
|---|---|---|---|---|
| BRCA1 | 17q21 | High | Hereditary breast cancer and ovarian syndrome | Breast cancer, ovarian cancer |
| BRCA2 | 13q12.3 | High | Hereditary breast cancer and ovarian syndrome | Breast cancer, ovarian cancer, prostate cancer, pancreatic cancer, melanoma |
| TP53 | 17p13.1 | High | Li-Fraumeni syndrome | Breast cancer Sarcomas Brain tumors |
| ATM | 11q22.3 | Intermediate | Louis-Bar syndrome | Lymphoma, cerebellar ataxia, immune deficiency, glioma, medulloblastoma, breast cancer |
| CDH1 | 16q22.1 | Intermediate | Familial diffuse gastric cancer syndrome | Gastric cancer, lobular breast cancer |
| PTEN | 10q23.31 | Intermediate | Cowden syndrome | Increased risk of neoplasms: breast cancer, thyroid cancer, endometrial carcinomas, hamartomatous polyps of the gastrointestinal tract |
| Bannayan-Riley-Rivalcaba syndrome | Breast cancer, meningioma | |||
| STK1 | 19p13.3 | Intermediate | Peutz-Jeghers syndrome | Melanocytic macules of the lips and others multiple gastrointestinal hamartomatous polyps increased risk of neoplasms; breast, testis, pancreas and cervix |
| NBS1 | 8q21 | Intermediate | Nijmegen breakage syndrome | Microcephaly,growth retardation, immunodeficiency and a marked susceptibility to cancer |
| Moderate risk of breast cancer | ||||
| BRIP/FANCJ | 17q22 | Intermediate | Fanconi anemia | Developmental anomalies affecting the skeleton (absent or abnormal thumbs and radii), kidneys, heart or any other major organ system |
| PALB2/FANCN | 16p12 | Intermediate | Aplastic anaemia, acute myeloid leukaemia and squamous cell carcinoma, breast cancer | |
| FANCA | 16q24.3 | Low | ||
| FANCE | 6p22-p21 | Low | ||
| MSH2 | 2p22-p21 | Low | Lynch cancer family syndrome | Endometrial cancer, colorectal cancer, breast cancer, ovarian cancer, genitourinary cancer, sarcomas, glioblastomas and leukaemia (often multiple) |
| MSH3 | 5q11-q12 | Low | ||
| MSH6 | 2p16 | Low | ||
| MLH1 | 3p21.3 | Low | ||
| PMS1 | 2q31-q33 | Low | ||
| PMS2 | 7p22 | Low |
In this paper, we will mainly focus on the hereditary breast cancer syndromes caused by germline mutations in the BRCA1 and BRCA2 genes since these have been well studied for their pathological features. Thereafter, we will briefly discuss the rarer hereditary cancer susceptibility syndrome genes mentioned earlier where yet little is known on tumor pathology [8, 140, 174].
The BRCA1 and BRCA2 genes
Discovery
The BRCA1 and BRCA2 genes were discovered in the nineties, starting in 1990 where BRCA1 was for the first time linked to breast cancer using a large group of early onset breast cancer families and linkage analysis. The BRCA1 gene was mapped to chromosome 17q21 [57]. In 1994, the BRCA1 gene was cloned and truncating mutations were identified in the coding sequence of the BRCA1 gene in families with multiple cases of breast cancer [118]. Search for more genes that might be involved in these hereditary susceptibility breast cancer families led in 1995 to the discovery of the BRCA2 gene. The BRCA2 gene is located on chromosome 13q12.3, and was discovered also by linking analysis and positional cloning using familial breast cancer pedigrees in successive generations [201, 202]. At the same time, families with high frequencies of male breast cancer were found to carry the BRCA2 mutation [167].
Carriers of the BRCA1 and BRCA2 mutations do not only develop breast cancer and ovarian cancer but also bear an increased risk for developing Fallopian tube, colon, melanoma, prostate and pancreatic cancer [56, 83, 93, 123, 128, 131, 206].
Structure
Both BRCA genes bear rather complex genomic structures. BRCA1 is composed of 24 exons and BRCA2 of 27 exons. They both encode very large proteins: BRCA1 consists of 1,863 amino acids and BRCA2 of 3,418 amino acids. In both genes, exon 1 is non-coding and exon 11 is unusually large [163, 201, 202] (Fig. 1). BRCA1 has a highly conserved zinc-binding RING finger domain which is located close to the amino-terminus. RING finger domain proteins are recognized as E3 ligase enzymes that participate in ubiquitination [110]. Mutations in the RING finger domain inactivate BRCA E3 ligase and have an effect on the other tumor suppressor activities of BRCA1 [152]. Towards the carboxyl terminus of BRCA1 two tandem copies of the same motif are found, designated the BRCT domains. These BRCT domains are regions reported to activate transcription when fused to a DNA binding domain [27]. BRCA2 contains a number of recognizable motifs, the eight copies of a 20–30 amino acid repeat, termed BRC repeats and the ssDNA binding region. Their function is to bind to RAD51 to regulate DNA repair (Fig. 1) [95, 198].
Fig. 1.
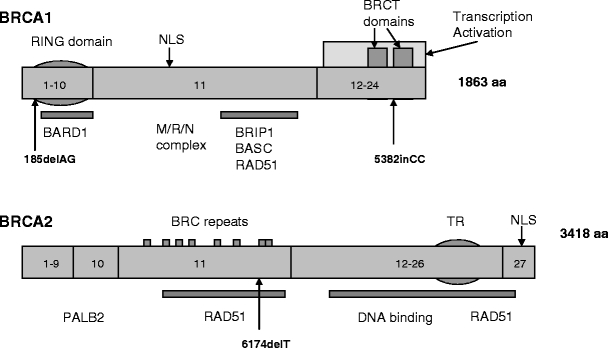
Schematic representation of BRCA1 and BRCA2 functional domains and selected binding partners, partially adapted from Narod et al. [124]. NLS = Nuclear Localization signal. Some of the proteins interacting with BRCA1 or BRCA2 are marked below the site of interaction
Function
Both BRCA genes are involved in DNA repair. They form complexes that will activate the repair of double strand breaks (DSBs) and initiate homologous recombination (HR). RAD51 is the key component of this mechanism. Co-localization of BRCA1 and BRCA2 with RAD51 at the site of recombination and DNA damaged induced foci strongly suggest that they are involved in the detection and the repair of DSBs. The roles played by BRCA1 and BRCA2 in this process appear to differ. Small ubiquitin-like modifier ligases are essential for localization of BRCA1 at the sites of DNA damage and sumoylated BRCA1 itself, together with BARD1 acts as E3 ligase and further ubiquitinates local proteins [47, 121]. BRCA1 will associate with RAD51 upon DNA damage and subsequently gets phosphorylated, but the nature of interaction with RAD51 is yet unknown [156]. BRCA2 has a more direct role through its strict interaction with RAD51 via the BRC repeats [200]. In addition, RAD51 also interacts with the C-terminal region of BRCA2, TR2 [119, 160]. This part of BRCA2 is thought to serve a regulatory role in recombinational repair. Phosphorylation of this part of BRCA2 can provide a dual function, resulting in inhibition or activation during HR [36, 40]. BRCA2 also has a role in the HR in meiosis via an interaction with RAD51 and DMC1. Given the fact that they have distinct non-overlapping binding sites, it has been suggested that there might well be a BRCA2-RAD51-DMC1 complex. However, more data has to be obtained to confirm this. It does suggest that BRCA2 not only plays a role in carcinogenesis but in addition contributes to fertility problems in affected carriers [176].
Cells that are defective for BRCA1 and BRCA2 are hypersensitive for crosslinking agents that will produce double strand breaks like mitomycin c and cisplatin [122, 135, 172]. Also, ionizing radiation will produce these same breaks and both will be resolved by error-prone repair, such as non homologous end joining [204, 205]. The levels of expression of BRCA1, BRCA2 and RAD51 increase in cells when they enter the S phase, indicating that they function during or after DNA replication. This means that BRCA1 and BRCA2 function in a common pathway that is responsible for the integrity of the genome and the maintenance of chromosomal stability [157]. BRCA1 is part of the BRCA1 associated genome surveillance complex (BASC) This complex includes MSH2, MSH6, MLH1, ATM, BLM , the RAD50-MRE11-NBS1 complex and the DNA replication factor C. All the members of this complex have roles in recognition of abnormal or damaged DNA, suggesting that the BASC may serve as a sensor for DNA damage and as a regulator of the post-replication repair process. BRCA1 functions also as a checkpoint control, playing an essential role in cell survival by preventing the propagation of DNA damage through cell cycle progression before DNA repair has taken place [197]. Taken together, BRCA1 is an integral part of the DNA damage signalling cascade; downstream of ATM and ATR kinases and both downstream and upstream of the checkpoint protein kinases CHEK1 and CHEK2 suggesting that there is a positive feedback loop to increase the magnitude of DNA damage response. In addition, BRCA1 regulates the expression of additional G2M cell cycle checkpoint proteins thereby preventing unscheduled transition into mitosis at multiple levels of regulation. Ubiquitination is the process by which proteins are tagged for degradation by the proteasome. BRCA1 functions with BARD1 in this ubiquitination process [144]. It has been suggested that BRCA1 plays a role in both transcription coupled repair [103] and global genome repair [59]. So, in conclusion, both BRCA genes are involved in DNA repair and both function in a common pathway that is responsible for the integrity of the genome and the maintenance of chromosomal stability.
Mutations
The Breast Cancer Information Core (BIC) database has recorded 1,639 and 1,853 distinct mutations, polymorphisms and variants in the BRCA1 and BRCA2 genes, respectively (data 2010). Mutations appear to be reasonably evenly distributed across the coding sequences, with no obvious “mutation hot spots”. Most mutations found in the breast and ovarian cancer families are predicted to truncate the protein product, which will lead to shortened and non-functional BRCA1 and BRCA2 proteins. The most common types of mutations are small frameshift insertions or deletions, non-sense mutation or mutations affecting splice sites, resulting in deletion of complete or partial exons or insertion of intronic sequences. These mutations will cover approximately 70% of the BRCA1 mutations and 90% of the BRCA2 mutations in linked families, as estimated by the Breast Cancer Linkage Consortium (BCLC) [174]. Large-scale rearrangements including insertions, deletions or duplications of more than 500 kb of DNA have also been identified. There have been reports of at least 19 distinct large genomic rearrangements in BRCA1 and two genomic rearrangements in BRCA2, identified using multiplex ligation dependent probe amplification (MLPA). The majority of the rearrangements are deletions of one or more exons [73, 120]. These mutations can be all classified with reasonable confidence but classification of rare missense changes is still a challenge. According to the BIC database, approximately half of the unique BRCA1 and BRCA2 variants detected (excluding common polymorphisms) are missense variants of unknown pathogenic potential, termed “unclassified variants”. Note of concern here is that the BIC database did not take into account the frequency in which these variants were found in the population undergoing testing. Furthermore, the clinical relevance of only a few unclassified variants has been established. For the others, the subtle alteration might not alter the function of the protein and there might also be insufficient information about the family history to classify these unclassified variants as cancer predisposing changes. However, these alterations can provide indications to do further functional and family studies [30, 104]. It has been stated that a new approach is needed to improve the association between these unclassified variants and breast cancer risk, and that using histopathology data of tumors from carries of an unclassified variant could be helpful [165].
Loss of heterozygosity (LOH) of the wildtype allele has been robustly demonstrated for BRCA linked breast cancer. Although some of the studies mentioning a role of LOH in approximately 80% of the cases included in the studies, for the rest of the cases LOH affecting the BRCA gene could not be detected [32, 33, 54, 127]. This might be caused by the practical and conceptual problems associated with LOH studies [178]. Furthermore, LOH of the wild type allele is not required for BRCA linked breast tumorigenesis and when it occurs it is probably a late event [92]. Another consideration is whether a second somatic mutation or methylation dependent silencing affecting the wild type allele accounts for these findings. However, no evidence of a second somatic mutation in BRCA linked breast cancer has been found so far. BRCA promoter hypermethylation as a gene silencing mechanism has been reported in 11%–30% and 42%–51% of sporadic breast cancer and non-BRCA1/BRCA2 related hereditary breast cancers, respectively [19, 42, 76, 171]. BRCA1 and BRCA2 related breast tumours only rarely showed BRCA promoter hypermethylation [37, 41, 169, 171]
Population specific occurrence
The majority of all the mutation described above are found throughout the population. However, certain mutations in BRCA1 and BRCA2 have been observed to be common in specific populations. Such founder mutations in BRCA1 and BRCA2 have been described in French Canadian [162], Swedes [86], Icelandic women [175], Norwegians [7], Finns [80], Dutch women [136, 139], Russians [50], Japanese women [82] and African Americans [48]. Three founder mutations are very commonly found in the Ashkenazi Jewish population, the 185delAG [129, 168] and 5382insC in BRCA1 and 6147delT in BRCA2 [9, 126]. The 185delAG is prevalent in 1% of all Ashkenazi Jews but has also been reported in other Jewish groups [16]. The 5382insC mutation found in 0.1% of the Ashkenazi Jews is described to occur more widespread, being common in Poland, Russia, and other parts of Eastern Europe and occurring in most European populations. In Ashkenazi Jewish women with breast cancer, the 185delAG mutation in BRCA1 is found in 20% [130]. The 6147delT mutation in BRCA2 is found to be present in 8% of the Ashkenazi Jews with breast cancer [126, 179]. In Iceland, a single BRCA2 mutation 999del5 has been identified and is present in the majority of familial breast cancer cases in this population [55, 175].
Pathology of BRCA1 related breast cancer
Histology
The histology of BRCA1 associated breast cancers differs from the histopathological features of sporadic breast cancers in various aspects. The majority of the BRCA1 associated tumors are invasive ductal adenocarcinomas (74%). However, compared to sporadic breast cancer, a significantly higher frequency of the BRCA1 associated tumors are classified as medullary like carcinomas, 2% versus 13% respectively [1, 99]. The remaining histological types of breast cancer occur about equally in BRCA1 mutation associated tumors and in sporadic breast cancer [1]. With regard to other histopathological characteristics it is observed that BRCA1 tumors are more frequently poorly differentiated (grade 3), have a high mitotic count and show an high frequency of necrotic areas [186]. Tubule formation is decreased, but a higher degree of pleimorphism is observed, all aspects pointing at a more aggressive phenotype [10, 75, 99, 112]. In addition, tumors are often well demarcated and show a remarkable degree of lymphoplasmocytic infiltration, and a high frequency of lymphovascular invasion [66].
When considering the age of onset of these BRCA1 mutation carriers, less than 50 years of age compared to age above 50 years, significant differences in grade (higher) and in percentage of medullary type (more cases), of breast cancer are seen in the younger population [38]. With regard to pre-invasive breast lesions, it has initially been reported that ductal carcinoma in situ (DCIS) and lobular carcinoma in situ (LCIS) are seen less frequently in BRCA1 mutation carriers, being 41% and 2% respectively versus 56% and 6% in non-carriers. These concerned however pre-invasive lesions in cases where invasive breast cancer was seen [1, 13]. Studies investigating the occurrence of premalignant lesions in prophylactic mastectomies of BRCA1 mutation carriers usually showed more frequent occurrence of premalignant lesions. These premalignant lesions concern DCIS [79, 84, 90], LCIS [79], atypical ductal (ADH) [71, 79, 84, 90] and atypical lobular hyperplasia (ALH) [71, 79, 84, 90], usual ductal hyperplasia [71], columnar cell lesions [90] and fibroadenoma [71, 97]. Interestingly, the remarkable lymphocytoplasmic infiltrate described in invasive BRCA1 related cancers has also been described in DCIS lesions and even the normal breast shows T-cell lobulitis [72].
Immunophenotype
The immunophenotype of the BRCA1 mutation related breast cancers (Table 2) is first of all characterized by a low expression of the estrogen receptor alpha (ERα). In 1997 the first reports about the low expression of ERα in BRCA1 tumors compared to sporadic tumors were described. Subsequent reports confirmed this observation and in addition described a significant relationship between low ERα on the one hand and high grade [11, 38, 44, 87, 88, 101, 111, 134] and an earlier age of onset [38, 44, 192] on the other. In contrast, overexpression of estrogen receptor beta (ERβ) is seen in breast cancers of BRCA1 mutation carriers [109]. Similar low expression of the progesterone receptor (PR) has been reported [11, 38, 101, 111, 134].
Table 2.
Chromosomal loci showing significant differences in frequency of gain or loss by array comparative genomic hybridization between BRCA1 and BRCA2 related and sporadic breast cancers [185]
| Locus | Frequency | p-value | ||||
|---|---|---|---|---|---|---|
| BRCA1 | BRCA2 | Sporadic | BRCA1 vs sporadic | BRCA2 vs sporadic | BRCA1 vs BRCA2 | |
| Gains | ||||||
| 1cen-p13 | 89 | 68 | 87 | 0.054 | ||
| 3pter-p22 | 33 | 16 | 0 | 0.006 | ||
| 3q13-q27 | 67 | 56 | 13 | 0.000 | 0.073 | |
| 8p12-cent | 11 | 16 | 47 | 0.012 | ||
| 9p | 33 | 16 | 3 | 0.078 | ||
| 9q22-q34 | 0 | 32 | 3 | 0.013 | ||
| 10pter-p12 | 50 | 20 | 7 | 0.000 | ||
| 10p12-q21 | 36 | 4 | 3 | 0.089 | ||
| 13q3 | 25 | 8 | 0 | 0.059 | ||
| 16p | 17 | 24 | 57 | 0.019 | ||
| 18p | 28 | 16 | 3 | 0.025 | ||
| Losses | ||||||
| 5cent-q23 | 72 | 40 | 27 | 0.025 | ||
| 14q1-q2 | 39 | 8 | 10 | 0.048 | ||
Overall, the expression of the human epidermal growth receptor 2 (HER-2/neu) is low in BRCA1 related breast cancers when compared with controls [11, 75, 101]. Furthermore, HER-2/neu amplifications among BRCA1 tumors have only rarely been reported. One explanation could be that in the background of a BRCA1 germline mutation, HER-2/neu is lost during loss of heterozygosity (LOH) at the BRCA1 locus since HER-2/neu is localized close to BRCA1 on chromosome 17 [3, 53, 134].
In contrast to HER-2/neu, overexpression of the epidermal growth factor receptor (EGFR) has been strongly associated with BRCA1 associated breast cancers [45, 100, 187, 188, 191].
BRCA1 related breast cancers often lack cyclin D1 (CCND1) expression. Also the expression of p27Kip1 is very low in BRCA1 related breast cancers and this is seen together with high levels of cyclin E [44]. Mutations in the TP53 gene are seen in 30%–77% of BRCA1 tumors whereas they are only present in about 20% of sporadic controls. As a consequence, accumulation of p53 is often seen in BRCA1 related breast cancer. Furthermore, the distribution of the TP53 mutations might be influenced by the BRCA1 and BRCA2 genes [11, 34, 52, 74, 141].
Evaluating the expression of several basal markers in BRCA1 related breast cancers it was observed that most of these tumors are positive for cytokeratins CK5/CK6 and CK14 [45, 100], caveolin 1[143], vimentin, laminin [151] and p-cadherin [12].
Expression of the apoptosis related proteins BAX and BCL2 in BRCA1 related breast cancers is lower compared to sporadic breast cancers is reported [46, 133, 134]. In contrast, high levels of active caspase 3 were observed in BRCA1 tumors [133]. Hypoxia inducible factor-1α (HIF-1α) is the key regulator of the hypoxia response. HIF-1α is overexpressed during sporadic breast carcinogenesis [22] and correlated with poor prognosis [21, 195]. It appears to be involved in BRCA1 related breast cancers, where HIF-1α is overexpressed in most of these tumors [186]. Expression of the stem cell marker ALDH1 appeared to be higher in BRCA1 related breast cancers compared to sporadic cancers [64] , indicating that these cancers bear an increased cancer stem cell compartment. Intrestingly, there was no difference between normal breast tissue of BRCA1 mutation carriers and controls [65]
Altogether, this immunophenotype indicates that BRCA1 related invasive breast cancer largely shows the immunophenotype of progenitor cells of the breast, indicating that they initially may (in contrast to BRCA2 related cancers, see below) derive from these cells. While the immunophenotype of invasive BRCA1 related cancers has been well studied little is yet known on the immunophenotype of pre-invasive lesions from the BRCA1 carcinogenetic spectrum. We recently showed that the immunophenotype of DCIS in BRCA1 carriers is similar to that of their accompanying invasive cancers [190].
Genetic profile
Gene expression profile analysis has provided a tool to distinguish distinct subtypes of breast cancers [137, 164]. Based on these data BRCA1 associated breast cancers are classified as basal. The gene expression profile of BRCA1 associated tumors involves genes that were found to have functions in proliferation, angiogenesis, cell motility, cell adhesion, transcription and DNA repair. As mentioned above, BRCA1 related breast tumors express basal markers like CK 5/6, CK14, EGFR, P-cadherin and caveolin 1, vimentin and laminin, thereby confirming the basal subtype as established by immunohistochemistry [45, 100, 151]. These data further underline that carcinogenesis in BRCA1 germline mutation carriers very often occurs within the “basal” progression route.
Promotor hypermethylation of tumor suppressor genes has been shown to be somewhat less abundant in BRCA1 germline mutation related breast cancers [169], although it is still clearly higher than in normal tissue.
As to copy number changes, a different pattern of chromosomal copy-number gains and losses compared to sporadic controls has been found. Copy number changes frequently occurring in BRCA1 related breast cancers are gains of 3q, 7p, 8q 10p, 12p, 16p and 17q and loss of 2q, 3p, 4p, 4q, 5q, 12q, 16p and 18q. This only partly overlaps with copy number changes found in sporadic and BRCA2 germline mutation related breast cancers, see Table 3 [177, 185, 199].
Table 3.
Chromosomal loci showing significant differences in frequency of gain or loss by array comparative genomic hybridization between BRCA1 and BRCA2 related and sporadic breast cancers [185]
| Locus | Frequency | p-value | ||||
|---|---|---|---|---|---|---|
| BRCA1 | BRCA2 | Sporadic | BRCA1 vs sporadic | BRCA2 vs sporadic | BRCA1 vs BRCA2 | |
| Gains | ||||||
| 1cen-p13 | 89 | 68 | 87 | 0.054 | ||
| 3pter-p22 | 33 | 16 | 0 | 0.006 | ||
| 3q13-q27 | 67 | 56 | 13 | 0.000 | 0.073 | |
| 8p12-cent | 11 | 16 | 47 | 0.012 | ||
| 9p | 33 | 16 | 3 | 0.078 | ||
| 9q22-q34 | 0 | 32 | 3 | 0.013 | ||
| 10pter-p12 | 50 | 20 | 7 | 0.000 | ||
| 10p12-q21 | 36 | 4 | 3 | 0.089 | ||
| 13q3 | 25 | 8 | 0 | 0.059 | ||
| 16p | 17 | 24 | 57 | 0.019 | ||
| 18p | 28 | 16 | 3 | 0.025 | ||
| Losses | ||||||
| 5cent-q23 | 72 | 40 | 27 | 0.025 | ||
| 14q1-q2 | 39 | 8 | 10 | 0.048 | ||
Prognosis
In BRCA1 associated tumors, a lower rate of bone metastases and a higher frequency of lung and brain metastases have been described [96]. Investigating overall survival in BRCA1 associated breast cancer versus age matched sporadic breast cancer patients have yielded contradictive results with some studies describing a worse survival and others a similar survival rate [23, 58, 107, 147, 150, 194].
Pathology of BRCA2 related breast cancer
Histology
Similar to BRCA1 related breast cancers, the most common histological type in BRCA2 tumors is invasive ductal carcinoma (76%) [1]. Reports of a higher incidence of tumors belonging to invasive (pleiomorphic) lobular, tubular and cribiform carcinomas in BRCA2 related breast cancers compared to sporadic breast cancer have been published [4, 10, 14, 99, 112, 113]. BRCA2 tumors are more frequently moderately or poorly differentiated carcinomas (grade 2 and 3) [4, 99, 111, 134] due to less tubule formation [1] more nuclear pleiomorphism and higher mitotic rates compared to controls [4, 14]. BRCA2 related breast cancers have, as BRCA1 related cancers, a higher proportion of continuous pushing margins in comparison to sporadic breast cancers [14, 99]. With regard to pre-invasive breast lesions it has been described that DCIS and LCIS in BRCA2 mutation carriers occur in the same frequency, 52% and 3% respectively compared to 56% and 6% in control individuals [1, 13, 78]. The occurrence of premalignant lesions in prophylactic mastectomies of BRCA2 mutation carriers show different results, similar to what has been observed in BRCA1 mutation carriers, ranging from no differences to the more frequent occurrence of premalignant lesions in prophylactic mastectomies of BRCA2 mutation carriers like DCIS [79, 90], LCIS [79, 84], ADH [71, 79, 84, 90], ALH [71, 79, 84, 90] and columnar cell lesions [90]. Interestingly, the remarkable lymphocytoplasmic infiltrate described in invasive BRCA2, and in BRCA1 as mentioned before, related cancers has also been described in DCIS lesions, and even the normal breast shows T-cell lobulitis [72].
Immunophenotype
The immunophenotype of BRCA2 related breast cancers is similar to the immunophenotype of sporadic breast cancers (Table 2). As a consequence, most BRCA2 tumors show a different immunophenotype compared to BRCA1 related breast tumors as discussed above. BRCA2 cancers show more frequently expression of ERα and PR [4, 10, 14, 38, 45]. Furthermore, these ER positive BRCA2 related breast cancers decrease in frequency with increasing age [44]. In BRCA2 related breast cancer different studies report no or low expression of HER-2/neu compared to sporadic breast cancer and only in rare cases a HER-2/neu amplification was found [14, 101, 132, 134]. Furthermore, a more recent study described that BRCA2 related breast cancers are characterized by a higher expression of fibroblast growth factor 1 (FGF1) and fibroblast growth factor receptor 2 (FGFR2) compared to BRCA1 related breast cancers. This could help to distinguish BRCA2 related breast cancers from other breast cancers [15]. The BRCA2 related breast cancers usually express only “luminal” cytokeratins like CK8 and CK18 and not CK5/6 and CK14 [133]. In BRCA2 related breast cancers no expression of caveolin1 has been described in contrast to the expression of caveolin in BRCA1 related tumors [143]. No differences or even lower levels of the incidence of p53 have been reported for BRCA2 related breast cancers in comparison with BRCA1 related breast cancers [133]. Higher expression of cyclin D1, BAX and BCL2 in BRCA2 related breast cancers compared to BRCA1 and non-BRCA carriers have been described [133, 134]. However, anecdotic data suggest that EGFR expression is high in BRCA2 related cancers [188]. While BRCA1 related cancers have been described to be frequently positive for P-cadherin, vimentin and HIF-1α, no such data are yet available for BRCA2 related cancers. No data on ALDH1 expression in BRCA2 related cancers are available.
While the immunophenotype of invasive BRCA2 related cancers has been well studied, little is yet known on the immunophenotype of pre-invasive lesions from the BRCA2 carcinogenetic spectrum. We recently showed that the immunophenotype of DCIS in BRCA2 carriers is similar to that of their accompanying invasive cancers [190].
In conclusion, most of the BRCA2 related breast cancers are of the so called luminal type with overexpression of ER, PR, CK8 and CK18. This is clearly different from the observations in BRCA1 related breast cancers [101, 134], pointing to a different origin from the luminal cells of the breast rather than the progenitor cells as in BRCA1 related breast cancer.
Genetic profile
In a recent study using gene expression analysis to distinguish BRCA2 associated tumors, discriminating genes were those related to transcription, signal transduction, cell proliferation, cell adhesion and extracellular matrix remodelling. In this study, a relative high expression of FGF1 and FGFR2 was observed and this was confirmed by immunohistochemistry as stated above [15, 61, 184, 199]. When using the gene expression profile mentioned before, most of the BRCA2 related breast cancers were classified as luminal [137, 164]. Looking more specifically at the molecular genetics, BRCA2 related breast cancers show patterns of chromosomal copy-number gains and losses that are not found in sporadic controls. Copy number changes more frequently occurring in BRCA2 related breast cancers are gains of 8q, 17q22-q24 and 20q13 and loss of 8p, 6q, 11q and 13q [177, 185], see Table 3.
Prognosis
In women with BRCA2 associated breast cancer, bone and soft tissue metastases are observed more frequently likely associated with their more frequent ER positivity [96]. As is the case in BRCA1 patients, for BRCA2 patients conflicting data with regard to outcome have been presented [23, 58, 107, 147, 150, 194].
Other hereditary breast cancer genes
TP53
TP53 (tumor protein p53) is a tumor suppressor gene located on chromosome 17p13.1 encoding a nuclear phosphoprotein (p53). TP53 acts as a transcription factor involved in the control of cell cycle progression, repair of DNA damage, genomic stability, and apoptosis [196]. TP53 is constitutionally mutated in the Li-Fraumeni syndrome, an autosomal dominant predisposition to breast cancer and other forms of cancer (see Table 1). Most mutations are point mutations leading to proteins defective for sequence-specific DNA binding and activation of p53 responsive genes [49, 94, 161]. The TP53 gene is more commonly altered in BRCA1 (56%–100%) and BRCA2 (29%) related breast cancer in comparison with non-BRCA related breast [29, 74]. In BRCA1 or BRCA2 deficient cells changes were seen at TP53 codons that are not the mutation hotspots. Structural modelling showed that most of these p53 non-hot spot aminoacids are distributed in a region of the protein on the opposite side of the p53 DNA-binding surface in these BRCA1 or BRCA2 deficient cells [52]. Breast cancers with these TP53 non-hot spot mutations were associated with a significantly better prognosis when compared with TP53 mutations in conserved or structural domains [6]. Preliminary data suggest that BRCA1 or BRCA2 mutations influence the distribution of the TP53 mutations and the way of carcinogenesis, but additional studies must be performed to support this [52].
CHEK2
The CHEK2 (checkpoint kinase 2) gene is located on chromosome 22q12.1 and encodes a cell cycle checkpoint kinase which is a key mediator in DNA damage response [115, 203]. Mutations in CHEK2 were originally thought to result in the Li-Fraumeni syndrome or in a Li-Fraumeni-like syndrome (mentioned above and described in Table 1), since the first CHEK2 mutations were found in these Li-Fraumeni families [18]. More recent studies question this association, following the identification of the 1000delC and 1157T CHEK2 germline variants among breast cancer patients that otherwise show no signs of Li-Fraumeni like features [5, 183]. The CHEK2 gene has been proposed to be a low penetrance breast cancer susceptibility gene. The 1000delC variant results in an approximately two fold risk of breast cancer in women and a tenfold risk in men. In these cases, there is no mention of co-existence of BRCA1 and BRCA2 mutations [117, 182]. So far, beside the 1000delC and 1157T mutations, no additional CHEK2 mutations have been found [155].
ATM
The ATM (ataxia teleangiectasia mutated) gene is located on chromosome 11q22.3 and encodes a checkpoint kinase that plays a role in DNA repair. Biallelic mutations in this gene are linked to the rare human autosomal recessive disorder called ataxia teleangiectasia (AT) [153], causing a variety of somatic disorders as described in Table 1. A heterozygous mutation of ATM does not lead to the AT phenotype but carriers have a two to five fold risk of breast cancer [148, 173] (Table 1).
CDH1
CDH1 (Cadherin 1, E-cadherin) is a gene located on chromosome 16q22.1 encoding E-cadherin, a calcium dependent cell adhesion glycoprotein, which is important for cell-to-cell adhesion [17]. Familial diffuse gastric cancer, an autosomal dominant cancer syndrome is caused by mutations in the CDH1 gene and affected women are predisposed to lobular breast cancer. Patient with a familial diffuse gastric cancer have a risk of about 50% of getting breast cancer [91, 154] (Table 1).
PTEN
PTEN (phosphatase and tensin homolog), is a tumor suppressor gene located on chromosome 10q23.3. PTEN encodes for the protein phosphatidylinositol phosphate phosphatase and has multiple and as yet incompletely understood roles in cellular regulation[106, 166]. Germline mutations in PTEN can lead to a rare autosomal dominant inherited cancer syndrome, Cowden disease, characterized by a high risk of breast-, thyroid- and endometrial carcinomas and hamartomas [125]. Mutations in PTEN also cause the related syndrome, Bannayan-Riley-Rivalcaba syndrome [114], see for more details Table 1.
STK11
STK11 (LKB1) (Serine/theronine kinase 11) is a gene located on chromosome 19p13.3 that encodes a serine/threonine kinase and functions mainly through inhibition of the mTOR pathway. STK11 is mutated in the autosomal dominant condition Peutz-Jeghers syndrome, characterized by perioral pigmentation and hamartomatous polyposis [69]. Patients with this syndrome have a 30%–50% risk of developing breast cancer [51, 60, 108] (Table 1).
NBS1
NBS1 is a gene located on chromosome 8q21 and involved in the Nijmegen breakage syndrome, a chromosome instability syndrome. Proteins of the gene NSB together with proteins of the genes RAD50 and MRE11, form the so called MRN complex. The MRN complex is involved in the recognition and repair of DNA double strand breaks [102]. The estimated prevalence of the most common mutation is very low and the breast cancer risk conferred by a NBS1 mutation is estimated to be low [20].
FANCONI
A rare recessive repair defect disorder called Fanconi anaemia (FA) is linked to a number of genes, in total 12 so far, that, together with BRCA1, are involved in homologous recombination DNA repair mechanisms [35, 85, 193]. Mutations in FANCJ (=BRIP1) and FANCN (= PALB2) are associated with a two fold increased risk of breast cancer [145, 158]. The remaining ten FA genes may likewise be involved in the carcinogenesis of breast cancer but their role has not been elucidated yet. It has been suggested that the remaining FA genes are inactivated through epigenetic/transcriptional mechanisms. For example, the FANCD2 protein is down regulated in sporadic and in hereditary breast carcinomas [189].
Mismatch repair
Postreplication mismatch repair (MMR) is a critical mechanism for maintaining microsatellite stability through the correction of base substitution mismatches and insertion/deletion events. The mismatch repair genes (MMR), MLH1, MSH2 and MSH6, play a role in hereditary non-polyposis colorectal cancer, the Lynch-syndrome. In a few of these families, breast cancer is part of this syndrome, which seems to be related to the absence of the MLH1and MSH2 proteins [159]. Furthermore, a causative role of MSH6 in the occurrence of breast cancer has been suggested but only one case has been reported so far [70].
Together with BRCA1 and BRCA2, the above described genes account for most, but not all, hereditary breast cancers (Fig. 2). Obviously, the search for other genes involved in hereditary breast cancer is still continuing [149].
Fig. 2.
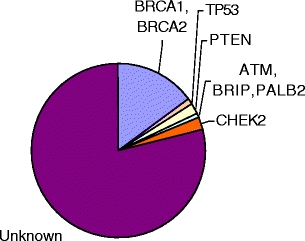
Affected genes in hereditary breast cancer
Pathology of non-BRCA1 or non-BRCA2 related breast cancers
Phenotypic characteristics of cancers developing in patients with a strong family history without a BRCA1 or BRCA2 germline mutation are various. These breast cancers develop as a consequence of mutations in different moderate to low penetrance genes, like the genes mentioned earlier (see Table 1), or in genes yet to be discovered. It has been established that these tumors have even a lower grade compared to sporadic breast cancers. Furthermore, the immunophenotype is more or less the same as shown in sporadic breast disease [76, 98]. One study describing a gene expression profile of non-BRCA related breast cancer was able to classify these tumors into two homogenous subsets, ribosomal genes were more represented in one of these groups compared to the other based on a 60 gene set. Additional experiments should be done on these non-BRCA related tumors to further describe their molecular characteristics [62].
CHEK2
Morphologic and immunophenotypic studies of breast cancer in patients with a CHEK2 mutation has yielded conflicting results, largely due to the limited cases of breast cancers that have been found being related to this mutation. Studies on ER and PR expression have reported contradictory results, ranging from similar to overexpression of ER and PR. Breast cancer in patients with a U157T mutation has been associated with an increased incidence of lobular carcinomas as has been described earlier [77, 81].
PALB2
A recent study describes for the first time some tumors characteristics of PALB2, 1592delT, mutation carriers. Most of these breast tumors exhibited a phenotype of high grade mostly of ductal type and ER, PR and HER-2/neu receptor negativity. They were mostly CK5/6, CK14 and CK17 negative, showed high expression of Ki67 and low expression of Cyclin D1 as compared with other familial and sporadic patients [67]
In conclusion, the pathology of hereditary breast cancers not related to a BRCA1 or BRCA2 mutation has not been studied extensively and so far does not seem very specific.
Clinical relevance
Surgical options for surveillance include prophylactic bilateral mastectomy and prophylactic bilateral salpingo-oophorectomy (BSO) [146]. Prophylactic bilateral mastectomy reduces the risk of breast cancer by almost 100% in mutation carriers [63, 116]. In view of the additional high lifetime risk of ovarian cancer, especially in BRCA1 mutation carriers, these women are strongly advised to undergo BSO including the removal of the Fallopian tubes at the completion of childbearing [89, 105, 142].
Preliminary results from one study suggests that the use of hormone therapy in postmenopausal women with a BRCA1 mutation was associated with a decreased risk of breast cancer. It is important to confirm this in a larger study including different populations and a longer study period [39].
The association of negativity ER/PR/HER-2/neu status classifies many of BRCA1 related cancers in the “triple negative” category, which is clinically under scrutiny as these cancers may require an alternate chemotherapeutic approach. Due to the important role of the BRCA genes in DNA repair it could be expected that DNA cross linking agents, like cisplatin and mitomycine-c, would have an effect especially in those diseases that occur as a consequence of mutated and therefore dysfunctional BRCA1 and BRCA2 genes [181]. Higher tumor responses to platinum based chemotherapy have indeed been observed in patients with BRCA1 mutated ovarian cancers when compared with the effects observed in non hereditary ovarian cancer [26, 28].
A potentially new strategy that has emerged for treatment of BRCA1 and BRCA2 related tumors is the use of poly(ADP-ribose) polymerase 1 (PARP1) inhibitors. BRCA1 and BRCA2 are both involved in DNA double strand break repair, as mentioned before. PARP1 is involved in base excision repair, a key pathway in the repair of DNA single strand break. The absence of PARP leads to spontaneous single strand breaks which collapse replication forks into double strand breaks, triggering homologous recombination for repair. However, with the loss of functional BRCA1 or BRCA2, cells will be sensitized to inhibit PARP activity, apparently leading to the persistence of the DNA lesions which are usually repaired by homologous recombination. When both pathways are defect this will result in chromosomal instability, cell cycle arrest and finally apoptosis. Cell survival assays show that cell lines lacking wildtype BRCA1 or BRCA2 were extremely sensitive to PARP inhibitors compared to heterozygous mutant or the wildtype cells [43]. Similar results were obtained using non embryonic cells deficient for BRCA2. These results suggest the potential use of PARP inhibitors in the treatment of BRCA1 and BRCA2 related breast cancer. This is presently evaluated in various clinical trials in BRCA carriers suffering from breast and/or ovarian cancer [25, 43, 68, 180].
Conclusions
BRCA1 related breast cancers are very well characterized by morphological, immunohistochemical and molecular features that clearly help to differentiate them from sporadic tumors and identify high risk patients for mutation testing. BRCA2 related breast cancers on the other hand, offer yet only a few morphological, immunohistochemical or molecular features to separate them from sporadic controls.
Finally, although numbers studied so far are small, breast cancers caused by other breast cancer susceptibility genes do not, as in BRCA2 related disease, seem to differ significantly from sporadic breast cancers.
More studies should be performed on morphological, immunohistochemical and molecular characterization of BRCA2 related breast cancers, breast cancers caused by unclassified variants of BRCA1 and BRCA2, and breast cancers caused by other breast cancer susceptibility genes to gain insight into the development of these breast cancers, and to subsequently be able to offer clues for diagnosis and new therapeutic approaches.
Acknowledgments
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Supported by a grant of PinkRibbon, The Netherlands
References
- 1.Pathology of familial breast cancer: differences between breast cancers in carriers of BRCA1 or BRCA2 mutations and sporadic cases. Breast Cancer Linkage Consortium. Lancet 349, 1505–1510 (1997) [PubMed]
- 2.Prevalence and penetrance of BRCA1 and BRCA2 mutations in a population-based series of breast cancer cases. Anglian Breast Cancer Study Group. Br. J. Cancer 83, 1301–1308 (2000) [DOI] [PMC free article] [PubMed]
- 3.Adem C, Soderberg CL, Hafner K, Reynolds C, Slezak JM, Sinclair CS, Sellers TA, Schaid DJ, Couch F, Hartmann LC, Jenkins RB. ERBB2, TBX2, RPS6KB1, and MYC alterations in breast tissues of BRCA1 and BRCA2 mutation carriers. Genes Chromosom. Cancer. 2004;41:1–11. doi: 10.1002/gcc.20057. [DOI] [PubMed] [Google Scholar]
- 4.Agnarsson BA, Jonasson JG, Bjornsdottir IB, Barkardottir RB, Egilsson V, Sigurdsson H. Inherited BRCA2 mutation associated with high grade breast cancer. Breast Cancer Res. Treat. 1998;47:121–127. doi: 10.1023/a:1005853022804. [DOI] [PubMed] [Google Scholar]
- 5.Allinen M, Huusko P, Mantyniemi S, Launonen V, Winqvist R. Mutation analysis of the CHK2 gene in families with hereditary breast cancer. Br. J. Cancer. 2001;85:209–212. doi: 10.1054/bjoc.2001.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alsner J, Yilmaz M, Guldberg P, Hansen LL, Overgaard J. Heterogeneity in the clinical phenotype of TP53 mutations in breast cancer patients. Clin. Cancer Res. 2000;6:3923–3931. doi: 10.1186/bcr109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersen TI, Borresen AL, Moller P. A common BRCA1 mutation in Norwegian breast and ovarian cancer families? Am. J. Hum. Genet. 1996;59:486–487. [PMC free article] [PubMed] [Google Scholar]
- 8.Antoniou AC, Easton DF. Models of genetic susceptibility to breast cancer. Oncogene. 2006;25:5898–5905. doi: 10.1038/sj.onc.1209879. [DOI] [PubMed] [Google Scholar]
- 9.Antoniou AC, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, Olsson H, Johannsson O, Borg A, Pasini B, Radice P, Manoukian S, Eccles DM, Tang N, Olah E, Anton-Culver H, Warner E, Lubinski J, Gronwald J, Gorski B, Tulinius H, Thorlacius S, Eerola H, Nevanlinna H, Syrjakoski K, Kallioniemi OP, Thompson D, Evans C, Peto J, Lalloo F, Evans DG, Easton DF. Breast and ovarian cancer risks to carriers of the BRCA1 5382insC and 185delAG and BRCA2 6174delT mutations: a combined analysis of 22 population based studies. J. Med. Genet. 2005;42:602–603. doi: 10.1136/jmg.2004.024133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Armes JE, Egan AJ, Southey MC, Dite GS, McCredie MR, Giles GG, Hopper JL, Venter DJ. The histologic phenotypes of breast carcinoma occurring before age 40 years in women with and without BRCA1 or BRCA2 germline mutations: a population-based study. Cancer. 1998;83:2335–2345. [PubMed] [Google Scholar]
- 11.Armes JE, Trute L, White D, Southey MC, Hammet F, Tesoriero A, Hutchins AM, Dite GS, McCredie MR, Giles GG, Hopper JL, Venter DJ. Distinct molecular pathogeneses of early-onset breast cancers in BRCA1 and BRCA2 mutation carriers: a population-based study. Cancer Res. 1999;59:2011–2017. [PubMed] [Google Scholar]
- 12.Arnes JB, Brunet JS, Stefansson I, Begin LR, Wong N, Chappuis PO, Akslen LA, Foulkes WD. Placental cadherin and the basal epithelial phenotype of BRCA1-related breast cancer. Clin. Cancer Res. 2005;11:4003–4011. doi: 10.1158/1078-0432.CCR-04-2064. [DOI] [PubMed] [Google Scholar]
- 13.Arun B, Vogel KJ, Lopez A, Hernandez M, Atchley D, Broglio KR, Amos CI, Meric-Bernstam F, Kuerer H, Hortobagyi GN, Albarracin CT. High prevalence of preinvasive lesions adjacent to BRCA1/2-associated breast cancers. Cancer Prev. Res. (Phila. Pa.) 2009;2:122–127. doi: 10.1158/1940-6207.CAPR-08-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bane AL, Beck JC, Bleiweiss I, Buys SS, Catalano E, Daly MB, Giles G, Godwin AK, Hibshoosh H, Hopper JL, John EM, Layfield L, Longacre T, Miron A, Senie R, Southey MC, West DW, Whittemore AS, Wu H, Andrulis IL, O’Malley FP. BRCA2 mutation-associated breast cancers exhibit a distinguishing phenotype based on morphology and molecular profiles from tissue microarrays. Am. J. Surg. Pathol. 2007;31:121–128. doi: 10.1097/01.pas.0000213351.49767.0f. [DOI] [PubMed] [Google Scholar]
- 15.A.L. Bane, D. Pinnaduwage, S. Colby, S.B. Bull, F.P. O’Malley, I.L. Andrulis, Expression profiling of familial breast cancers demonstrates higher expression of FGFR2 in BRCA2-associated tumors. Breast Cancer Res. Treat. (2008) [DOI] [PMC free article] [PubMed]
- 16.Bar-Sade RB, Kruglikova A, Modan B, Gak E, Hirsh-Yechezkel G, Theodor L, Novikov I, Gershoni-Baruch R, Risel S, Papa MZ, Ben-Baruch G, Friedman E. The 185delAG BRCA1 mutation originated before the dispersion of Jews in the diaspora and is not limited to Ashkenazim. Hum. Mol. Genet. 1998;7:801–805. doi: 10.1093/hmg/7.5.801. [DOI] [PubMed] [Google Scholar]
- 17.Becker KF, Atkinson MJ, Reich U, Becker I, Nekarda H, Siewert JR, Hofler H. E-cadherin gene mutations provide clues to diffuse type gastric carcinomas. Cancer Res. 1994;54:3845–3852. [PubMed] [Google Scholar]
- 18.Bell DW, Varley JM, Szydlo TE, Kang DH, Wahrer DC, Shannon KE, Lubratovich M, Verselis SJ, Isselbacher KJ, Fraumeni JF, Birch JM, Li FP, Garber JE, Haber DA. Heterozygous germ line hCHK2 mutations in Li-Fraumeni syndrome. Science. 1999;286:2528–2531. doi: 10.1126/science.286.5449.2528. [DOI] [PubMed] [Google Scholar]
- 19.Birgisdottir V, Stefansson OA, Bodvarsdottir SK, Hilmarsdottir H, Jonasson JG, Eyfjord JE. Epigenetic silencing and deletion of the BRCA1 gene in sporadic breast cancer. Breast Cancer Res. 2006;8:R38. doi: 10.1186/bcr1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bogdanova N, Schurmann P, Waltes R, Feshchenko S, Zalutsky IV, Bremer M, Dork T. NBS1 variant I171V and breast cancer risk. Breast Cancer Res. Treat. 2008;112:75–79. doi: 10.1007/s10549-007-9820-4. [DOI] [PubMed] [Google Scholar]
- 21.Bos R, van der Groep P, Greijer AE, Shvarts A, Meijer S, Pinedo HM, Semenza GL, van Diest PJ, van der Wall E. Levels of hypoxia-inducible factor-1alpha independently predict prognosis in patients with lymph node negative breast carcinoma. Cancer. 2003;97:1573–1581. doi: 10.1002/cncr.11246. [DOI] [PubMed] [Google Scholar]
- 22.Bos R, Zhong H, Hanrahan CF, Mommers EC, Semenza GL, Pinedo HM, Abeloff MD, Simons JW, van Diest PJ, van der Wall E. Levels of hypoxia-inducible factor-1 alpha during breast carcinogenesis. J. Natl. Cancer Inst. 2001;93:309–314. doi: 10.1093/jnci/93.4.309. [DOI] [PubMed] [Google Scholar]
- 23.Brekelmans CT, Seynaeve C, Menke-Pluymers M, Bruggenwirth HT, Tilanus-Linthorst MM, Bartels CC, Kriege M, van Geel AN, Crepin CM, Blom JC, Meijers-Heijboer H, Klijn JG. Survival and prognostic factors in BRCA1-associated breast cancer. Ann. Oncol. 2006;17:391–400. doi: 10.1093/annonc/mdj095. [DOI] [PubMed] [Google Scholar]
- 24.Broca, Traite des tumeurs (1866)
- 25.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 26.T. Byrski, T. Huzarski, R. Dent, J. Gronwald, D. Zuziak, C. Cybulski, J. Kladny, B. Gorski, J. Lubinski, S.A. Narod, Response to neoadjuvant therapy with cisplatin in BRCA1-positive breast cancer patients. Breast Cancer Res. Treat. (2008) [DOI] [PubMed]
- 27.Callebaut I, Mornon JP. From BRCA1 to RAP1: a widespread BRCT module closely associated with DNA repair. FEBS Lett. 1997;400:25–30. doi: 10.1016/s0014-5793(96)01312-9. [DOI] [PubMed] [Google Scholar]
- 28.Cass I, Baldwin RL, Varkey T, Moslehi R, Narod SA, Karlan BY. Improved survival in women with BRCA-associated ovarian carcinoma. Cancer. 2003;97:2187–2195. doi: 10.1002/cncr.11310. [DOI] [PubMed] [Google Scholar]
- 29.Chappuis PO, Nethercot V, Foulkes WD. Clinico-pathological characteristics of BRCA1- and BRCA2-related breast cancer. Semin. Surg. Oncol. 2000;18:287–295. doi: 10.1002/(sici)1098-2388(200006)18:4<287::aid-ssu3>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 30.Chenevix-Trench G, Healey S, Lakhani S, Waring P, Cummings M, Brinkworth R, Deffenbaugh AM, Burbidge LA, Pruss D, Judkins T, Scholl T, Bekessy A, Marsh A, Lovelock P, Wong M, Tesoriero A, Renard H, Southey M, Hopper JL, Yannoukakos K, Brown M, Easton D, Tavtigian SV, Goldgar D, Spurdle AB. Genetic and histopathologic evaluation of BRCA1 and BRCA2 DNA sequence variants of unknown clinical significance. Cancer Res. 2006;66:2019–2027. doi: 10.1158/0008-5472.CAN-05-3546. [DOI] [PubMed] [Google Scholar]
- 31.Claus EB, Schildkraut J, Iversen ES, Jr, Berry D, Parmigiani G. Effect of BRCA1 and BRCA2 on the association between breast cancer risk and family history. J. Natl. Cancer Inst. 1998;90:1824–1829. doi: 10.1093/jnci/90.23.1824. [DOI] [PubMed] [Google Scholar]
- 32.Collins N, McManus R, Wooster R, Mangion J, Seal S, Lakhani SR, Ormiston W, Daly PA, Ford D, Easton DF, et al. Consistent loss of the wild type allele in breast cancers from a family linked to the BRCA2 gene on chromosome 13q12-13. Oncogene. 1995;10:1673–1675. [PubMed] [Google Scholar]
- 33.Cornelis RS, Neuhausen SL, Johansson O, Arason A, Kelsell D, Ponder BA, Tonin P, Hamann U, Lindblom A, Lalle P, et al. High allele loss rates at 17q12-q21 in breast and ovarian tumors from BRCAl-linked families. The Breast Cancer Linkage Consortium. Genes Chromosom. Cancer. 1995;13:203–210. doi: 10.1002/gcc.2870130310. [DOI] [PubMed] [Google Scholar]
- 34.Crook T, Brooks LA, Crossland S, Osin P, Barker KT, Waller J, Philp E, Smith PD, Yulug I, Peto J, Parker G, Allday MJ, Crompton MR, Gusterson BA. p53 mutation with frequent novel condons but not a mutator phenotype in BRCA1- and BRCA2-associated breast tumours. Oncogene. 1998;17:1681–1689. doi: 10.1038/sj.onc.1202106. [DOI] [PubMed] [Google Scholar]
- 35.D’Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nat. Rev. Cancer. 2003;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 36.Davies OR, Pellegrini L. Interaction with the BRCA2 C terminus protects RAD51-DNA filaments from disassembly by BRC repeats. Nat. Struct. Mol. Biol. 2007;14:475–483. doi: 10.1038/nsmb1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dworkin AM, Spearman AD, Tseng SY, Sweet K, Toland AE. Methylation not a frequent “second hit” in tumors with germline BRCA mutations. Fam. Cancer. 2009;8:339–346. doi: 10.1007/s10689-009-9240-1. [DOI] [PubMed] [Google Scholar]
- 38.Eerola H, Heikkila P, Tamminen A, Aittomaki K, Blomqvist C, Nevanlinna H. Relationship of patients’ age to histopathological features of breast tumours in BRCA1 and BRCA2 and mutation-negative breast cancer families. Breast Cancer Res. 2005;7:R465–R469. doi: 10.1186/bcr1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eisen A, Lubinski J, Gronwald J, Moller P, Lynch HT, Klijn J, Kim-Sing C, Neuhausen SL, Gilbert L, Ghadirian P, Manoukian S, Rennert G, Friedman E, Isaacs C, Rosen E, Rosen B, Daly M, Sun P, Narod SA. Hormone therapy and the risk of breast cancer in BRCA1 mutation carriers. J. Natl. Cancer Inst. 2008;100:1361–1367. doi: 10.1093/jnci/djn313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Esashi F, Galkin VE, Yu X, Egelman EH, West SC. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat. Struct. Mol. Biol. 2007;14:468–474. doi: 10.1038/nsmb1245. [DOI] [PubMed] [Google Scholar]
- 41.Esteller M, Fraga MF, Guo M, Garcia-Foncillas J, Hedenfalk I, Godwin AK, Trojan J, Vaurs-Barriere C, Bignon YJ, Ramus S, Benitez J, Caldes T, Akiyama Y, Yuasa Y, Launonen V, Canal MJ, Rodriguez R, Capella G, Peinado MA, Borg A, Aaltonen LA, Ponder BA, Baylin SB, Herman JG. DNA methylation patterns in hereditary human cancers mimic sporadic tumorigenesis. Hum. Mol. Genet. 2001;10:3001–3007. doi: 10.1093/hmg/10.26.3001. [DOI] [PubMed] [Google Scholar]
- 42.Esteller M, Silva JM, Dominguez G, Bonilla F, Matias-Guiu X, Lerma E, Bussaglia E, Prat J, Harkes IC, Repasky EA, Gabrielson E, Schutte M, Baylin SB, Herman JG. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J. Natl. Cancer Inst. 2000;92:564–569. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- 43.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 44.Foulkes WD, Brunet JS, Stefansson IM, Straume O, Chappuis PO, Begin LR, Hamel N, Goffin JR, Wong N, Trudel M, Kapusta L, Porter P, Akslen LA. The prognostic implication of the basal-like (cyclin E high/p27 low/p53+/glomeruloid-microvascular-proliferation+) phenotype of BRCA1-related breast cancer. Cancer Res. 2004;64:830–835. doi: 10.1158/0008-5472.can-03-2970. [DOI] [PubMed] [Google Scholar]
- 45.Foulkes WD, Stefansson IM, Chappuis PO, Begin LR, Goffin JR, Wong N, Trudel M, Akslen LA. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J. Natl. Cancer Inst. 2003;95:1482–1485. doi: 10.1093/jnci/djg050. [DOI] [PubMed] [Google Scholar]
- 46.Freneaux P, Stoppa-Lyonnet D, Mouret E, Kambouchner M, Nicolas A, Zafrani B, Vincent-Salomon A, Fourquet A, Magdelenat H, Sastre-Garau X. Low expression of bcl-2 in Brca1-associated breast cancers. Br. J. Cancer. 2000;83:1318–1322. doi: 10.1054/bjoc.2000.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 2009;462:935–939. doi: 10.1038/nature08657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao Q, Neuhausen S, Cummings S, Luce M, Olopade OI. Recurrent germ-line BRCA1 mutations in extended African American families with early-onset breast cancer. Am. J. Hum. Genet. 1997;60:1233–1236. [PMC free article] [PubMed] [Google Scholar]
- 49.Gasco M, Yulug IG, Crook T. TP53 mutations in familial breast cancer: functional aspects. Hum. Mutat. 2003;21:301–306. doi: 10.1002/humu.10173. [DOI] [PubMed] [Google Scholar]
- 50.Gayther SA, Harrington P, Russell P, Kharkevich G, Garkavtseva RF, Ponder BA. Frequently occurring germ-line mutations of the BRCA1 gene in ovarian cancer families from Russia. Am. J. Hum. Genet. 1997;60:1239–1242. [PMC free article] [PubMed] [Google Scholar]
- 51.Giardiello FM, Brensinger JD, Tersmette AC, Goodman SN, Petersen GM, Booker SV, Cruz-Correa M, Offerhaus JA. Very high risk of cancer in familial Peutz-Jeghers syndrome. Gastroenterology. 2000;119:1447–1453. doi: 10.1053/gast.2000.20228. [DOI] [PubMed] [Google Scholar]
- 52.Greenblatt MS, Chappuis PO, Bond JP, Hamel N, Foulkes WD. TP53 mutations in breast cancer associated with BRCA1 or BRCA2 germ-line mutations: distinctive spectrum and structural distribution. Cancer Res. 2001;61:4092–4097. [PubMed] [Google Scholar]
- 53.Grushko TA, Blackwood MA, Schumm PL, Hagos FG, Adeyanju MO, Feldman MD, Sanders MO, Weber BL, Olopade OI. Molecular-cytogenetic analysis of HER-2/neu gene in BRCA1-associated breast cancers. Cancer Res. 2002;62:1481–1488. [PubMed] [Google Scholar]
- 54.Gudmundsson J, Barkardottir RB, Eiriksdottir G, Baldursson T, Arason A, Egilsson V, Ingvarsson S. Loss of heterozygosity at chromosome 11 in breast cancer: association of prognostic factors with genetic alterations. Br. J. Cancer. 1995;72:696–701. doi: 10.1038/bjc.1995.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gudmundsson J, Johannesdottir G, Arason A, Bergthorsson JT, Ingvarsson S, Egilsson V, Barkardottir RB. Frequent occurrence of BRCA2 linkage in Icelandic breast cancer families and segregation of a common BRCA2 haplotype. Am. J. Hum. Genet. 1996;58:749–756. [PMC free article] [PubMed] [Google Scholar]
- 56.Hahn SA, Greenhalf B, Ellis I, Sina-Frey M, Rieder H, Korte B, Gerdes B, Kress R, Ziegler A, Raeburn JA, Campra D, Grutzmann R, Rehder H, Rothmund M, Schmiegel W, Neoptolemos JP, Bartsch DK. BRCA2 germline mutations in familial pancreatic carcinoma. J. Natl. Cancer Inst. 2003;95:214–221. doi: 10.1093/jnci/95.3.214. [DOI] [PubMed] [Google Scholar]
- 57.Hall JM, Lee MK, Newman B, Morrow JE, Anderson LA, Huey B, King MC. Linkage of early-onset familial breast cancer to chromosome 17q21. Science. 1990;250:1684–1689. doi: 10.1126/science.2270482. [DOI] [PubMed] [Google Scholar]
- 58.Hamann U. Hereditary breast cancer: high risk genes, genetic testing and clinical implications. Clin. Lab. 2000;46:447–461. [PubMed] [Google Scholar]
- 59.Hartman AR, Ford JM. BRCA1 induces DNA damage recognition factors and enhances nucleotide excision repair. Nat. Genet. 2002;32:180–184. doi: 10.1038/ng953. [DOI] [PubMed] [Google Scholar]
- 60.Hearle N, Schumacher V, Menko FH, Olschwang S, Boardman LA, Gille JJ, Keller JJ, Westerman AM, Scott RJ, Lim W, Trimbath JD, Giardiello FM, Gruber SB, Offerhaus GJ, de Rooij FW, Wilson JH, Hansmann A, Moslein G, Royer-Pokora B, Vogel T, Phillips RK, Spigelman AD, Houlston RS. Frequency and spectrum of cancers in the Peutz-Jeghers syndrome. Clin. Cancer Res. 2006;12:3209–3215. doi: 10.1158/1078-0432.CCR-06-0083. [DOI] [PubMed] [Google Scholar]
- 61.Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, Simon R, Meltzer P, Gusterson B, Esteller M, Kallioniemi OP, Wilfond B, Borg A, Trent J, Raffeld M, Yakhini Z, Ben-Dor A, Dougherty E, Kononen J, Bubendorf L, Fehrle W, Pittaluga S, Gruvberger S, Loman N, Johannsson O, Olsson H, Sauter G. Gene-expression profiles in hereditary breast cancer. N. Engl. J. Med. 2001;344:539–548. doi: 10.1056/NEJM200102223440801. [DOI] [PubMed] [Google Scholar]
- 62.Hedenfalk I, Ringner M, Ben-Dor A, Yakhini Z, Chen Y, Chebil G, Ach R, Loman N, Olsson H, Meltzer P, Borg A, Trent J. Molecular classification of familial non-BRCA1/BRCA2 breast cancer. Proc. Natl. Acad. Sci. USA. 2003;100:2532–2537. doi: 10.1073/pnas.0533805100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heemskerk-Gerritsen BA, Brekelmans CT, Menke-Pluymers MB, van Geel AN, Tilanus-Linthorst MM, Bartels CC, Tan M, Meijers-Heijboer HE, Klijn JG, Seynaeve C. Prophylactic mastectomy in BRCA1/2 mutation carriers and women at risk of hereditary breast cancer: long-term experiences at the Rotterdam Family Cancer Clinic. Ann. Surg. Oncol. 2007;14:3335–3344. doi: 10.1245/s10434-007-9449-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.M.R. Heerma van Voss, P. van der Groep, J. Bart, E. van der Wall, P.J. van Diest, Expression of the stem cell marker ALDH1 in BRCA1 related breast cancer, In preparation [DOI] [PMC free article] [PubMed]
- 65.M.R. Heerma van Voss, P. van der Groep, J. Bart, E. van der Wall, P.J. van Diest, Expression of the stem cell marker ALDH1 in the normal breast of BRCA1 mutation carriers. Breast Cancer Res. Treat. [DOI] [PubMed]
- 66.M.R. Heerma van Voss, P. van der Groep, J. Bart, E. van der Wall, P.J. van Diest, Lympho-vascular invasion in BRCA related breast cancer compared to sporadic controls. BMC Cancer 10, 145 [DOI] [PMC free article] [PubMed]
- 67.Heikkinen T, Karkkainen H, Aaltonen K, Milne RL, Heikkila P, Aittomaki K, Blomqvist C, Nevanlinna H. The breast cancer susceptibility mutation PALB2 1592delT is associated with an aggressive tumor phenotype. Clin. Cancer Res. 2009;15:3214–3222. doi: 10.1158/1078-0432.CCR-08-3128. [DOI] [PubMed] [Google Scholar]
- 68.Helleday T, Bryant HE, Schultz N. Poly(ADP-ribose) polymerase (PARP-1) in homologous recombination and as a target for cancer therapy. Cell Cycle. 2005;4:1176–1178. doi: 10.4161/cc.4.9.2031. [DOI] [PubMed] [Google Scholar]
- 69.Hemminki A, Markie D, Tomlinson I, Avizienyte E, Roth S, Loukola A, Bignell G, Warren W, Aminoff M, Hoglund P, Jarvinen H, Kristo P, Pelin K, Ridanpaa M, Salovaara R, Toro T, Bodmer W, Olschwang S, Olsen AS, Stratton MR, de la Chapelle A, Aaltonen LA. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature. 1998;391:184–187. doi: 10.1038/34432. [DOI] [PubMed] [Google Scholar]
- 70.Hendriks YM, Wagner A, Morreau H, Menko F, Stormorken A, Quehenberger F, Sandkuijl L, Moller P, Genuardi M, Van Houwelingen H, Tops C, Van Puijenbroek M, Verkuijlen P, Kenter G, Van Mil A, Meijers-Heijboer H, Tan GB, Breuning MH, Fodde R, Wijnen JT, Brocker-Vriends AH, Vasen H. Cancer risk in hereditary nonpolyposis colorectal cancer due to MSH6 mutations: impact on counseling and surveillance. Gastroenterology. 2004;127:17–25. doi: 10.1053/j.gastro.2004.03.068. [DOI] [PubMed] [Google Scholar]
- 71.Hermsen BB, van Diest PJ, Berkhof J, Menko FH, Gille JJ, Piek JM, Meijer S, Winters HA, Kenemans P, Mensdorff-Pouilly S, Verheijen RH. Low prevalence of (pre) malignant lesions in the breast and high prevalence in the ovary and Fallopian tube in women at hereditary high risk of breast and ovarian cancer. Int. J. Cancer. 2006;119:1412–1418. doi: 10.1002/ijc.21988. [DOI] [PubMed] [Google Scholar]
- 72.Hermsen BB, von Mensdorff-Pouilly S, Fabry HF, Winters HA, Kenemans P, Verheijen RH, van Diest PJ. Lobulitis is a frequent finding in prophylactically removed breast tissue from women at hereditary high risk of breast cancer. J. Pathol. 2005;206:220–223. doi: 10.1002/path.1774. [DOI] [PubMed] [Google Scholar]
- 73.Hogervorst FB, Nederlof PM, Gille JJ, McElgunn CJ, Grippeling M, Pruntel R, Regnerus R, van Welsem T, van Spaendonk R, Menko FH, Kluijt I, Dommering C, Verhoef S, Schouten JP, van’t Veer LJ, Pals G. Large genomic deletions and duplications in the BRCA1 gene identified by a novel quantitative method. Cancer Res. 2003;63:1449–1453. [PubMed] [Google Scholar]
- 74.Holstege H, Joosse SA, van Oostrom CT, Nederlof PM, de Vries A, Jonkers J. High incidence of protein-truncating TP53 mutations in BRCA1-related breast cancer. Cancer Res. 2009;69:3625–3633. doi: 10.1158/0008-5472.CAN-08-3426. [DOI] [PubMed] [Google Scholar]
- 75.Honrado E, Benitez J, Palacios J. The molecular pathology of hereditary breast cancer: genetic testing and therapeutic implications. Mod. Pathol. 2005;18:1305–1320. doi: 10.1038/modpathol.3800453. [DOI] [PubMed] [Google Scholar]
- 76.Honrado E, Osorio A, Milne RL, Paz MF, Melchor L, Cascon A, Urioste M, Cazorla A, Diez O, Lerma E, Esteller M, Palacios J, Benitez J. Immunohistochemical classification of non-BRCA1/2 tumors identifies different groups that demonstrate the heterogeneity of BRCAX families. Mod. Pathol. 2007;20:1298–1306. doi: 10.1038/modpathol.3800969. [DOI] [PubMed] [Google Scholar]
- 77.Honrado E, Osorio A, Palacios J, Benitez J. Pathology and gene expression of hereditary breast tumors associated with BRCA1, BRCA2 and CHEK2 gene mutations. Oncogene. 2006;25:5837–5845. doi: 10.1038/sj.onc.1209875. [DOI] [PubMed] [Google Scholar]
- 78.Hoogerbrugge N, Bult P, Bonenkamp JJ, Ligtenberg MJ, Kiemeney LA, de Hullu JA, Boetes C, Niermeijer MF, Brunner HG. Numerous high-risk epithelial lesions in familial breast cancer. Eur. J. Cancer. 2006;42:2492–2498. doi: 10.1016/j.ejca.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 79.Hoogerbrugge N, Bult P, de Widt-Levert LM, Beex LV, Kiemeney LA, Ligtenberg MJ, Massuger LF, Boetes C, Manders P, Brunner HG. High prevalence of premalignant lesions in prophylactically removed breasts from women at hereditary risk for breast cancer. J. Clin. Oncol. 2003;21:41–45. doi: 10.1200/JCO.2003.02.137. [DOI] [PubMed] [Google Scholar]
- 80.Huusko P, Paakkonen K, Launonen V, Poyhonen M, Blanco G, Kauppila A, Puistola U, Kiviniemi H, Kujala M, Leisti J, Winqvist R. Evidence of founder mutations in Finnish BRCA1 and BRCA2 families. Am. J. Hum. Genet. 1998;62:1544–1548. doi: 10.1086/301880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huzarski T, Cybulski C, Domagala W, Gronwald J, Byrski T, Szwiec M, Woyke S, Narod SA, Lubinski J. Pathology of breast cancer in women with constitutional CHEK2 mutations. Breast Cancer Res. Treat. 2005;90:187–189. doi: 10.1007/s10549-004-3778-2. [DOI] [PubMed] [Google Scholar]
- 82.Inoue R, Fukutomi T, Ushijima T, Matsumoto Y, Sugimura T, Nagao M. Germline mutation of BRCA1 in Japanese breast cancer families. Cancer Res. 1995;55:3521–3524. [PubMed] [Google Scholar]
- 83.Iscovich J, Abdulrazik M, Cour C, Fischbein A, Pe’er J, Goldgar DE. Prevalence of the BRCA2 6174 del T mutation in Israeli uveal melanoma patients. Int. J. Cancer. 2002;98:42–44. doi: 10.1002/ijc.10155. [DOI] [PubMed] [Google Scholar]
- 84.Isern AE, Loman N, Malina J, Olsson H, Ringberg A. Histopathological findings and follow-up after prophylactic mastectomy and immediate breast reconstruction in 100 women from families with hereditary breast cancer. Eur. J. Surg. Oncol. 2008;34:1148–1154. doi: 10.1016/j.ejso.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 85.Joenje H, Arwert F. Connecting Fanconi anemia to BRCA1. Nat. Med. 2001;7:406–407. doi: 10.1038/86458. [DOI] [PubMed] [Google Scholar]
- 86.Johannsson O, Ostermeyer EA, Hakansson S, Friedman LS, Johansson U, Sellberg G, Brondum-Nielsen K, Sele V, Olsson H, King MC, Borg A. Founding BRCA1 mutations in hereditary breast and ovarian cancer in southern Sweden. Am. J. Hum. Genet. 1996;58:441–450. [PMC free article] [PubMed] [Google Scholar]
- 87.Johannsson OT, Idvall I, Anderson C, Borg A, Barkardottir RB, Egilsson V, Olsson H. Tumour biological features of BRCA1-induced breast and ovarian cancer. Eur. J. Cancer. 1997;33:362–371. doi: 10.1016/s0959-8049(97)89007-7. [DOI] [PubMed] [Google Scholar]
- 88.Karp SE, Tonin PN, Begin LR, Martinez JJ, Zhang JC, Pollak MN, Foulkes WD. Influence of BRCA1 mutations on nuclear grade and estrogen receptor status of breast carcinoma in Ashkenazi Jewish women. Cancer. 1997;80:435–441. doi: 10.1002/(sici)1097-0142(19970801)80:3<435::aid-cncr11>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 89.Kauff ND, Barakat RR. Risk-reducing salpingo-oophorectomy in patients with germline mutations in BRCA1 or BRCA2. J. Clin. Oncol. 2007;25:2921–2927. doi: 10.1200/JCO.2007.11.3449. [DOI] [PubMed] [Google Scholar]
- 90.Kauff ND, Brogi E, Scheuer L, Pathak DR, Borgen PI, Hudis CA, Offit K, Robson ME. Epithelial lesions in prophylactic mastectomy specimens from women with BRCA mutations. Cancer. 2003;97:1601–1608. doi: 10.1002/cncr.11225. [DOI] [PubMed] [Google Scholar]
- 91.Keller G, Vogelsang H, Becker I, Hutter J, Ott K, Candidus S, Grundei T, Becker KF, Mueller J, Siewert JR, Hofler H. Diffuse type gastric and lobular breast carcinoma in a familial gastric cancer patient with an E-cadherin germline mutation. Am. J. Pathol. 1999;155:337–342. doi: 10.1016/S0002-9440(10)65129-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.King TA, Li W, Brogi E, Yee CJ, Gemignani ML, Olvera N, Levine DA, Norton L, Robson ME, Offit K, Borgen PI, Boyd J. Heterogenic loss of the wild-type BRCA allele in human breast tumorigenesis. Ann. Surg. Oncol. 2007;14:2510–2518. doi: 10.1245/s10434-007-9372-1. [DOI] [PubMed] [Google Scholar]
- 93.Kirchhoff T, Kauff ND, Mitra N, Nafa K, Huang H, Palmer C, Gulati T, Wadsworth E, Donat S, Robson ME, Ellis NA, Offit K. BRCA mutations and risk of prostate cancer in Ashkenazi Jews. Clin. Cancer Res. 2004;10:2918–2921. doi: 10.1158/1078-0432.ccr-03-0604. [DOI] [PubMed] [Google Scholar]
- 94.Ko LJ, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 95.Koonin EV, Altschul SF, Bork P. BRCA1 protein products. Functional motifs. Nat. Genet. 1996;13:266–268. doi: 10.1038/ng0796-266. [DOI] [PubMed] [Google Scholar]
- 96.Kriege M, Seynaeve C, Meijers-Heijboer H, Collee JM, Menke-Pluymers MB, Bartels CC, Tilanus-Linthorst MM, van den Ouweland A, van Geel B, Brekelmans CT, Klijn JG. Distant disease-free interval, site of first relapse and post-relapse survival in BRCA1- and BRCA2-associated compared to sporadic breast cancer patients. Breast Cancer Res. Treat. 2008;111:303–311. doi: 10.1007/s10549-007-9781-7. [DOI] [PubMed] [Google Scholar]
- 97.Kuijper A, Preisler-Adams SS, Rahusen FD, Gille JJ, van der Wall E, van Diest PJ. Multiple fibroadenomas harbouring carcinoma in situ in a woman with a family history of breast/ovarian cancer. J. Clin. Pathol. 2002;55:795–797. doi: 10.1136/jcp.55.10.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lakhani SR, Gusterson BA, Jacquemier J, Sloane JP, Anderson TJ, van de Vijver MJ, Venter D, Freeman A, Antoniou A, McGuffog L, Smyth E, Steel CM, Haites N, Scott RJ, Goldgar D, Neuhausen S, Daly PA, Ormiston W, McManus R, Scherneck S, Ponder BA, Futreal PA, Peto J, Stoppa-Lyonnet D, Bignon YJ, Stratton MR. The pathology of familial breast cancer: histological features of cancers in families not attributable to mutations in BRCA1 or BRCA2. Clin. Cancer Res. 2000;6:782–789. [PubMed] [Google Scholar]
- 99.Lakhani SR, Jacquemier J, Sloane JP, Gusterson BA, Anderson TJ, van de Vijver MJ, Farid LM, Venter D, Antoniou A, Storfer-Isser A, Smyth E, Steel CM, Haites N, Scott RJ, Goldgar D, Neuhausen S, Daly PA, Ormiston W, McManus R, Scherneck S, Ponder BA, Ford D, Peto J, Stoppa-Lyonnet D, Bignon YJ, Struewing JP, Spurr NK, Bishop DT, Klijn JG, Devilee P, Cornelisse CJ, Lasset C, Lenoir G, Barkardottir RB, Egilsson V, Hamann U, Chang-Claude J, Sobol H, Weber B, Stratton MR, Easton DF. Multifactorial analysis of differences between sporadic breast cancers and cancers involving BRCA1 and BRCA2 mutations. J. Natl. Cancer Inst. 1998;90:1138–1145. doi: 10.1093/jnci/90.15.1138. [DOI] [PubMed] [Google Scholar]
- 100.Lakhani SR, Reis-Filho JS, Fulford L, Penault-Llorca F, van der Vijver M, Parry S, Bishop T, Benitez J, Rivas C, Bignon YJ, Chang-Claude J, Hamann U, Cornelisse CJ, Devilee P, Beckmann MW, Nestle-Kramling C, Daly PA, Haites N, Varley J, Lalloo F, Evans G, Maugard C, Meijers-Heijboer H, Klijn JG, Olah E, Gusterson BA, Pilotti S, Radice P, Scherneck S, Sobol H, Jacquemier J, Wagner T, Peto J, Stratton MR, McGuffog L, Easton DF. Prediction of BRCA1 status in patients with breast cancer using estrogen receptor and basal phenotype. Clin. Cancer Res. 2005;11:5175–5180. doi: 10.1158/1078-0432.CCR-04-2424. [DOI] [PubMed] [Google Scholar]
- 101.Lakhani SR, Van De Vijver MJ, Jacquemier J, Anderson TJ, Osin PP, McGuffog L, Easton DF. The pathology of familial breast cancer: predictive value of immunohistochemical markers estrogen receptor, progesterone receptor, HER-2, and p53 in patients with mutations in BRCA1 and BRCA2. J. Clin. Oncol. 2002;20:2310–2318. doi: 10.1200/JCO.2002.09.023. [DOI] [PubMed] [Google Scholar]
- 102.Lavin MF. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene. 2007;26:7749–7758. doi: 10.1038/sj.onc.1210880. [DOI] [PubMed] [Google Scholar]
- 103.Le Page F, Randrianarison V, Marot D, Cabannes J, Perricaudet M, Feunteun J, Sarasin A. BRCA1 and BRCA2 are necessary for the transcription-coupled repair of the oxidative 8-oxoguanine lesion in human cells. Cancer Res. 2000;60:5548–5552. [PubMed] [Google Scholar]
- 104.Lee E, McKean-Cowdin R, Ma H, Chen Z, Van Den Berg D, Henderson BE, Bernstein L, Ursin G. Evaluation of unclassified variants in the breast cancer susceptibility genes BRCA1 and BRCA2 using five methods: results from a population-based study of young breast cancer patients. Breast Cancer Res. 2008;10:R19. doi: 10.1186/bcr1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Leeper K, Garcia R, Swisher E, Goff B, Greer B, Paley P. Pathologic findings in prophylactic oophorectomy specimens in high-risk women. Gynecol. Oncol. 2002;87:52–56. doi: 10.1006/gyno.2002.6779. [DOI] [PubMed] [Google Scholar]
- 106.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner SH, Giovanella BC, Ittmann M, Tycko B, Hibshoosh H, Wigler MH, Parsons R. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 107.Liebens FP, Carly B, Pastijn A, Rozenberg S. Management of BRCA1/2 associated breast cancer: a systematic qualitative review of the state of knowledge in 2006. Eur. J. Cancer. 2007;43:238–257. doi: 10.1016/j.ejca.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 108.Lim W, Olschwang S, Keller JJ, Westerman AM, Menko FH, Boardman LA, Scott RJ, Trimbath J, Giardiello FM, Gruber SB, Gille JJ, Offerhaus GJ, de Rooij FW, Wilson JH, Spigelman AD, Phillips RK, Houlston RS. Relative frequency and morphology of cancers in STK11 mutation carriers. Gastroenterology. 2004;126:1788–1794. doi: 10.1053/j.gastro.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 109.Litwiniuk MM, Roznowski K, Filas V, Godlewski DD, Stawicka M, Kaleta R, Breborowicz J. Expression of estrogen receptor beta in the breast carcinoma of BRCA1 mutation carriers. BMC Cancer. 2008;8:100. doi: 10.1186/1471-2407-8-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lynch BJ, Holden JA, Buys SS, Neuhausen SL, Gaffney DK. Pathobiologic characteristics of hereditary breast cancer. Hum. Pathol. 1998;29:1140–1144. doi: 10.1016/s0046-8177(98)90427-0. [DOI] [PubMed] [Google Scholar]
- 112.Marcus JN, Watson P, Page DL, Narod SA, Lenoir GM, Tonin P, Linder-Stephenson L, Salerno G, Conway TA, Lynch HT. Hereditary breast cancer: pathobiology, prognosis, and BRCA1 and BRCA2 gene linkage. Cancer. 1996;77:697–709. doi: 10.1002/(sici)1097-0142(19960215)77:4<697::aid-cncr16>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 113.Marcus JN, Watson P, Page DL, Narod SA, Tonin P, Lenoir GM, Serova O, Lynch HT. BRCA2 hereditary breast cancer pathophenotype. Breast Cancer Res. Treat. 1997;44:275–277. doi: 10.1023/a:1005830230664. [DOI] [PubMed] [Google Scholar]
- 114.Marsh DJ, Kum JB, Lunetta KL, Bennett MJ, Gorlin RJ, Ahmed SF, Bodurtha J, Crowe C, Curtis MA, Dasouki M, Dunn T, Feit H, Geraghty MT, Graham JM, Jr, Hodgson SV, Hunter A, Korf BR, Manchester D, Miesfeldt S, Murday VA, Nathanson KL, Parisi M, Pober B, Romano C, Eng C, et al. PTEN mutation spectrum and genotype-phenotype correlations in Bannayan-Riley-Ruvalcaba syndrome suggest a single entity with Cowden syndrome. Hum. Mol. Genet. 1999;8:1461–1472. doi: 10.1093/hmg/8.8.1461. [DOI] [PubMed] [Google Scholar]
- 115.Matsuoka S, Huang M, Elledge SJ. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science. 1998;282:1893–1897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- 116.McGaughey A. Body image after bilateral prophylactic mastectomy: an integrative literature review. J. Midwifery Womens Health. 2006;51:e45–e49. doi: 10.1016/j.jmwh.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 117.Meijers-Heijboer H, van den Ouweland A, Klijn J, Wasielewski M, de Snoo A, Oldenburg R, Hollestelle A, Houben M, Crepin E, van Veghel-Plandsoen M, Elstrodt F, van Duijn C, Bartels C, Meijers C, Schutte M, McGuffog L, Thompson D, Easton D, Sodha N, Seal S, Barfoot R, Mangion J, Chang-Claude J, Eccles D, Eeles R, Evans DG, Houlston R, Murday V, Narod S, Peretz T, Peto J, Phelan C, Zhang HX, Szabo C, Devilee P, Goldgar D, Futreal PA, Nathanson KL, Weber B, Rahman N, Stratton MR. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat. Genet. 2002;31:55–59. doi: 10.1038/ng879. [DOI] [PubMed] [Google Scholar]
- 118.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 119.Mizuta R, LaSalle JM, Cheng HL, Shinohara A, Ogawa H, Copeland N, Jenkins NA, Lalande M, Alt FW. RAB22 and RAB163/mouse BRCA2: proteins that specifically interact with the RAD51 protein. Proc. Natl. Acad. Sci. USA. 1997;94:6927–6932. doi: 10.1073/pnas.94.13.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Montagna M, Dalla Palma M, Menin C, Agata S, De Nicolo A, Chieco-Bianchi L, D’Andrea E. Genomic rearrangements account for more than one-third of the BRCA1 mutations in northern Italian breast/ovarian cancer families. Hum. Mol. Genet. 2003;12:1055–1061. doi: 10.1093/hmg/ddg120. [DOI] [PubMed] [Google Scholar]
- 121.Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A, Butler L, Galanty Y, Pangon L, Kiuchi T, Ng T, Solomon E. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462:886–890. doi: 10.1038/nature08593. [DOI] [PubMed] [Google Scholar]
- 122.Moynahan ME, Cui TY, Jasin M. Homology-directed dna repair, mitomycin-c resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer Res. 2001;61:4842–4850. [PubMed] [Google Scholar]
- 123.Narod SA, Feunteun J, Lynch HT, Watson P, Conway T, Lynch J, Lenoir GM. Familial breast-ovarian cancer locus on chromosome 17q12-q23. Lancet. 1991;338:82–83. doi: 10.1016/0140-6736(91)90076-2. [DOI] [PubMed] [Google Scholar]
- 124.Narod SA, Foulkes WD. BRCA1 and BRCA2: 1994 and beyond. Nat. Rev. Cancer. 2004;4:665–676. doi: 10.1038/nrc1431. [DOI] [PubMed] [Google Scholar]
- 125.Nelen MR, Padberg GW, Peeters EA, Lin AY, van den Helm B, Frants RR, Coulon V, Goldstein AM, van Reen MM, Easton DF, Eeles RA, Hodgsen S, Mulvihill JJ, Murday VA, Tucker MA, Mariman EC, Starink TM, Ponder BA, Ropers HH, Kremer H, Longy M, Eng C. Localization of the gene for Cowden disease to chromosome 10q22-23. Nat. Genet. 1996;13:114–116. doi: 10.1038/ng0596-114. [DOI] [PubMed] [Google Scholar]
- 126.Neuhausen S, Gilewski T, Norton L, Tran T, McGuire P, Swensen J, Hampel H, Borgen P, Brown K, Skolnick M, Shattuck-Eidens D, Jhanwar S, Goldgar D, Offit K. Recurrent BRCA2 6174delT mutations in Ashkenazi Jewish women affected by breast cancer. Nat. Genet. 1996;13:126–128. doi: 10.1038/ng0596-126. [DOI] [PubMed] [Google Scholar]
- 127.Neuhausen SL, Marshall CJ. Loss of heterozygosity in familial tumors from three BRCA1-linked kindreds. Cancer Res. 1994;54:6069–6072. [PubMed] [Google Scholar]
- 128.Niell BL, Rennert G, Bonner JD, Almog R, Tomsho LP, Gruber SB. BRCA1 and BRCA2 founder mutations and the risk of colorectal cancer. J. Natl. Cancer Inst. 2004;96:15–21. doi: 10.1093/jnci/djh008. [DOI] [PubMed] [Google Scholar]
- 129.Oddoux C, Struewing JP, Clayton CM, Neuhausen S, Brody LC, Kaback M, Haas B, Norton L, Borgen P, Jhanwar S, Goldgar D, Ostrer H, Offit K. The carrier frequency of the BRCA2 6174delT mutation among Ashkenazi Jewish individuals is approximately 1% Nat. Genet. 1996;14:188–190. doi: 10.1038/ng1096-188. [DOI] [PubMed] [Google Scholar]
- 130.Offit K, Gilewski T, McGuire P, Schluger A, Hampel H, Brown K, Swensen J, Neuhausen S, Skolnick M, Norton L, Goldgar D. Germline BRCA1 185delAG mutations in Jewish women with breast cancer. Lancet. 1996;347:1643–1645. doi: 10.1016/s0140-6736(96)91484-1. [DOI] [PubMed] [Google Scholar]
- 131.Ozcelik H, Schmocker B, Di Nicola N, Shi XH, Langer B, Moore M, Taylor BR, Narod SA, Darlington G, Andrulis IL, Gallinger S, Redston M. Germline BRCA2 6174delT mutations in Ashkenazi Jewish pancreatic cancer patients. Nat. Genet. 1997;16:17–18. doi: 10.1038/ng0597-17. [DOI] [PubMed] [Google Scholar]
- 132.Palacios J, Honrado E, Cigudosa JC, Benitez J. ERBB2 and MYC alterations in BRCA1- and BRCA2-associated cancers. Genes Chromosom. Cancer. 2005;42:204–205. doi: 10.1002/gcc.20125. [DOI] [PubMed] [Google Scholar]
- 133.Palacios J, Honrado E, Osorio A, Cazorla A, Sarrio D, Barroso A, Rodriguez S, Cigudosa JC, Diez O, Alonso C, Lerma E, Dopazo J, Rivas C, Benitez J. Phenotypic characterization of BRCA1 and BRCA2 tumors based in a tissue microarray study with 37 immunohistochemical markers. Breast Cancer Res. Treat. 2005;90:5–14. doi: 10.1007/s10549-004-1536-0. [DOI] [PubMed] [Google Scholar]
- 134.Palacios J, Honrado E, Osorio A, Cazorla A, Sarrio D, Barroso A, Rodriguez S, Cigudosa JC, Diez O, Alonso C, Lerma E, Sanchez L, Rivas C, Benitez J. Immunohistochemical characteristics defined by tissue microarray of hereditary breast cancer not attributable to BRCA1 or BRCA2 mutations: differences from breast carcinomas arising in BRCA1 and BRCA2 mutation carriers. Clin. Cancer Res. 2003;9:3606–3614. [PubMed] [Google Scholar]
- 135.Patel KJ, Yu VP, Lee H, Corcoran A, Thistlethwaite FC, Evans MJ, Colledge WH, Friedman LS, Ponder BA, Venkitaraman AR. Involvement of Brca2 in DNA repair. Mol. Cell. 1998;1:347–357. doi: 10.1016/s1097-2765(00)80035-0. [DOI] [PubMed] [Google Scholar]
- 136.Peelen T, van Vliet M, Petrij-Bosch A, Mieremet R, Szabo C, van den Ouweland AM, Hogervorst F, Brohet R, Ligtenberg MJ, Teugels E, van der Luijt R, van der Hout AH, Gille JJ, Pals G, Jedema I, Olmer R, van Leeuwen I, Newman B, Plandsoen M, van der Est M, Brink G, Hageman S, Arts PJ, Bakker MM, Devilee P, et al. A high proportion of novel mutations in BRCA1 with strong founder effects among Dutch and Belgian hereditary breast and ovarian cancer families. Am. J. Hum. Genet. 1997;60:1041–1049. [PMC free article] [PubMed] [Google Scholar]
- 137.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 138.Peto J, Collins N, Barfoot R, Seal S, Warren W, Rahman N, Easton DF, Evans C, Deacon J, Stratton MR. Prevalence of BRCA1 and BRCA2 gene mutations in patients with early-onset breast cancer. J. Natl. Cancer Inst. 1999;91:943–949. doi: 10.1093/jnci/91.11.943. [DOI] [PubMed] [Google Scholar]
- 139.Petrij-Bosch A, Peelen T, van Vliet M, van Eijk R, Olmer R, Drusedau M, Hogervorst FB, Hageman S, Arts PJ, Ligtenberg MJ, Meijers-Heijboer H, Klijn JG, Vasen HF, Cornelisse CJ, van’t Veer LJ, Bakker E, van Ommen GJ, Devilee P. BRCA1 genomic deletions are major founder mutations in Dutch breast cancer patients. Nat. Genet. 1997;17:341–345. doi: 10.1038/ng1197-341. [DOI] [PubMed] [Google Scholar]
- 140.Pharoah PD, Antoniou AC, Easton DF, Ponder BA. Polygenes, risk prediction, and targeted prevention of breast cancer. N. Engl. J. Med. 2008;358:2796–2803. doi: 10.1056/NEJMsa0708739. [DOI] [PubMed] [Google Scholar]
- 141.Phillips KA, Nichol K, Ozcelik H, Knight J, Done SJ, Goodwin PJ, Andrulis IL. Frequency of p53 mutations in breast carcinomas from Ashkenazi Jewish carriers of BRCA1 mutations. J. Natl. Cancer Inst. 1999;91:469–473. doi: 10.1093/jnci/91.5.469. [DOI] [PubMed] [Google Scholar]
- 142.Piek JM, van Diest PJ, Zweemer RP, Kenemans P, Verheijen RH. Tubal ligation and risk of ovarian cancer. Lancet. 2001;358:844. doi: 10.1016/S0140-6736(01)05992-X. [DOI] [PubMed] [Google Scholar]
- 143.Pinilla SM, Honrado E, Hardisson D, Benitez J, Palacios J. Caveolin-1 expression is associated with a basal-like phenotype in sporadic and hereditary breast cancer. Breast Cancer Res. Treat. 2006;99:85–90. doi: 10.1007/s10549-006-9184-1. [DOI] [PubMed] [Google Scholar]
- 144.Polanowska J, Martin JS, Garcia-Muse T, Petalcorin MI, Boulton SJ. A conserved pathway to activate BRCA1-dependent ubiquitylation at DNA damage sites. EMBO J. 2006;25:2178–2188. doi: 10.1038/sj.emboj.7601102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rahman N, Seal S, Thompson D, Kelly P, Renwick A, Elliott A, Reid S, Spanova K, Barfoot R, Chagtai T, Jayatilake H, McGuffog L, Hanks S, Evans DG, Eccles D, Easton DF, Stratton MR. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat. Genet. 2007;39:165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rebbeck TR. Prophylactic oophorectomy in BRCA1 and BRCA2 mutation carriers. Eur. J. Cancer. 2002;38(Suppl 6):S15–S17. doi: 10.1016/s0959-8049(02)00269-1. [DOI] [PubMed] [Google Scholar]
- 147.Rennert G, Bisland-Naggan S, Barnett-Griness O, Bar-Joseph N, Zhang S, Rennert HS, Narod SA. Clinical outcomes of breast cancer in carriers of BRCA1 and BRCA2 mutations. N. Engl. J. Med. 2007;357:115–123. doi: 10.1056/NEJMoa070608. [DOI] [PubMed] [Google Scholar]
- 148.Renwick A, Thompson D, Seal S, Kelly P, Chagtai T, Ahmed M, North B, Jayatilake H, Barfoot R, Spanova K, McGuffog L, Evans DG, Eccles D, Easton DF, Stratton MR, Rahman N. ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat. Genet. 2006;38:873–875. doi: 10.1038/ng1837. [DOI] [PubMed] [Google Scholar]
- 149.T. Ripperger, D. Gadzicki, A. Meindl, B. Schlegelberger, Breast cancer susceptibility: current knowledge and implications for genetic counselling. Eur. J. Hum. Genet. (2008) [DOI] [PMC free article] [PubMed]
- 150.Robson ME, Offit K. Breast MRI for women with hereditary cancer risk. JAMA. 2004;292:1368–1370. doi: 10.1001/jama.292.11.1368. [DOI] [PubMed] [Google Scholar]
- 151.Rodriguez-Pinilla SM, Sarrio D, Honrado E, Moreno-Bueno G, Hardisson D, Calero F, Benitez J, Palacios J. Vimentin and laminin expression is associated with basal-like phenotype in both sporadic and BRCA1-associated breast carcinomas. J. Clin. Pathol. 2007;60:1006–1012. doi: 10.1136/jcp.2006.042143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ruffner H, Joazeiro CA, Hemmati D, Hunter T, Verma IM. Cancer-predisposing mutations within the RING domain of BRCA1: loss of ubiquitin protein ligase activity and protection from radiation hypersensitivity. Proc. Natl. Acad. Sci. USA. 2001;98:5134–5139. doi: 10.1073/pnas.081068398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle DA, Smith S, Uziel T, Sfez S, Ashkenazi M, Pecker I, Frydman M, Harnik R, Patanjali SR, Simmons A, Clines GA, Sartiel A, Gatti RA, Chessa L, Sanal O, Lavin MF, Jaspers NG, Taylor AM, Arlett CF, Miki T, Weissman SM, Lovett M, Collins FS, Shiloh Y. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 154.Schrader KA, Masciari S, Boyd N, Wiyrick S, Kaurah P, Senz J, Burke W, Lynch HT, Garber JE, Huntsman DG. Hereditary diffuse gastric cancer: association with lobular breast cancer. Fam Cancer. 2008;7:73–82. doi: 10.1007/s10689-007-9172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Schutte M, Seal S, Barfoot R, Meijers-Heijboer H, Wasielewski M, Evans DG, Eccles D, Meijers C, Lohman F, Klijn J, van den Ouweland A, Futreal PA, Nathanson KL, Weber BL, Easton DF, Stratton MR, Rahman N. Variants in CHEK2 other than 1100delC do not make a major contribution to breast cancer susceptibility. Am. J. Hum. Genet. 2003;72:1023–1028. doi: 10.1086/373965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston DM. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 157.Scully R, Livingston DM. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature. 2000;408:429–432. doi: 10.1038/35044000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Seal S, Thompson D, Renwick A, Elliott A, Kelly P, Barfoot R, Chagtai T, Jayatilake H, Ahmed M, Spanova K, North B, McGuffog L, Evans DG, Eccles D, Easton DF, Stratton MR, Rahman N. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat. Genet. 2006;38:1239–1241. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- 159.S. Shanley, C. Fung, J. Milliken, J. Leary, R. Barnetson, M. Schnitzler, J. Kirk, Breast cancer immunohistochemistry can be useful in triage of some HNPCC families. Fam. Cancer (2009) [DOI] [PubMed]
- 160.Sharan SK, Morimatsu M, Albrecht U, Lim DS, Regel E, Dinh C, Sands A, Eichele G, Hasty P, Bradley A. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature. 1997;386:804–810. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- 161.Sigal A, Rotter V. Oncogenic mutations of the p53 tumor suppressor: the demons of the guardian of the genome. Cancer Res. 2000;60:6788–6793. [PubMed] [Google Scholar]
- 162.Simard J, Tonin P, Durocher F, Morgan K, Rommens J, Gingras S, Samson C, Leblanc JF, Belanger C, Dion F, et al. Common origins of BRCA1 mutations in Canadian breast and ovarian cancer families. Nat. Genet. 1994;8:392–398. doi: 10.1038/ng1294-392. [DOI] [PubMed] [Google Scholar]
- 163.Smith TM, Lee MK, Szabo CI, Jerome N, McEuen M, Taylor M, Hood L, King MC. Complete genomic sequence and analysis of 117 kb of human DNA containing the gene BRCA1. Genome Res. 1996;6:1029–1049. doi: 10.1101/gr.6.11.1029. [DOI] [PubMed] [Google Scholar]
- 164.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lonning PE, Brown PO, Borresen-Dale AL, Botstein D. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc. Natl. Acad. Sci. USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Spearman AD, Sweet K, Zhou XP, McLennan J, Couch FJ, Toland AE. Clinically applicable models to characterize BRCA1 and BRCA2 variants of uncertain significance. J. Clin. Oncol. 2008;26:5393–5400. doi: 10.1200/JCO.2008.17.8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T, Frye C, Hu R, Swedlund B, Teng DH, Tavtigian SV. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat. Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 167.Stratton MR, Ford D, Neuhasen S, Seal S, Wooster R, Friedman LS, King MC, Egilsson V, Devilee P, McManus R, et al. Familial male breast cancer is not linked to the BRCA1 locus on chromosome 17q. Nat. Genet. 1994;7:103–107. doi: 10.1038/ng0594-103. [DOI] [PubMed] [Google Scholar]
- 168.Struewing JP, Abeliovich D, Peretz T, Avishai N, Kaback MM, Collins FS, Brody LC. The carrier frequency of the BRCA1 185delAG mutation is approximately 1 percent in Ashkenazi Jewish individuals. Nat. Genet. 1995;11:198–200. doi: 10.1038/ng1095-198. [DOI] [PubMed] [Google Scholar]
- 169.Suijkerbuijk KP, Fackler MJ, Sukumar S, van Gils CH, van Laar T, van der Wall E, Vooijs M, van Diest PJ. Methylation is less abundant in BRCA1-associated compared with sporadic breast cancer. Ann. Oncol. 2008;19:1870–1874. doi: 10.1093/annonc/mdn409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tan DS, Marchio C, Reis-Filho JS. Hereditary breast cancer: from molecular pathology to tailored therapies. J. Clin. Pathol. 2008;61:1073–1082. doi: 10.1136/jcp.2008.057950. [DOI] [PubMed] [Google Scholar]
- 171.Tapia T, Smalley SV, Kohen P, Munoz A, Solis LM, Corvalan A, Faundez P, Devoto L, Camus M, Alvarez M, Carvallo P. Promoter hypermethylation of BRCA1 correlates with absence of expression in hereditary breast cancer tumors. Epigenetics. 2008;3:157–163. doi: 10.4161/epi.3.3.6387. [DOI] [PubMed] [Google Scholar]
- 172.Tassone P, Tagliaferri P, Perricelli A, Blotta S, Quaresima B, Martelli ML, Goel A, Barbieri V, Costanzo F, Boland CR, Venuta S. BRCA1 expression modulates chemosensitivity of BRCA1-defective HCC1937 human breast cancer cells. Br. J. Cancer. 2003;88:1285–1291. doi: 10.1038/sj.bjc.6600859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Thompson D, Duedal S, Kirner J, McGuffog L, Last J, Reiman A, Byrd P, Taylor M, Easton DF. Cancer risks and mortality in heterozygous ATM mutation carriers. J. Natl. Cancer Inst. 2005;97:813–822. doi: 10.1093/jnci/dji141. [DOI] [PubMed] [Google Scholar]
- 174.Thompson D, Easton D. The genetic epidemiology of breast cancer genes. J. Mammary Gland Biol. Neoplasia. 2004;9:221–236. doi: 10.1023/B:JOMG.0000048770.90334.3b. [DOI] [PubMed] [Google Scholar]
- 175.Thorlacius S, Olafsdottir G, Tryggvadottir L, Neuhausen S, Jonasson JG, Tavtigian SV, Tulinius H, Ogmundsdottir HM, Eyfjord JE. A single BRCA2 mutation in male and female breast cancer families from Iceland with varied cancer phenotypes. Nat. Genet. 1996;13:117–119. doi: 10.1038/ng0596-117. [DOI] [PubMed] [Google Scholar]
- 176.Thorslund T, West SC. BRCA2: a universal recombinase regulator. Oncogene. 2007;26:7720–7730. doi: 10.1038/sj.onc.1210870. [DOI] [PubMed] [Google Scholar]
- 177.Tirkkonen M, Johannsson O, Agnarsson BA, Olsson H, Ingvarsson S, Karhu R, Tanner M, Isola J, Barkardottir RB, Borg A, Kallioniemi OP. Distinct somatic genetic changes associated with tumor progression in carriers of BRCA1 and BRCA2 germ-line mutations. Cancer Res. 1997;57:1222–1227. [PubMed] [Google Scholar]
- 178.Tomlinson IP, Lambros MB, Roylance RR. Loss of heterozygosity analysis: practically and conceptually flawed? Genes Chromosom. Cancer. 2002;34:349–353. doi: 10.1002/gcc.10085. [DOI] [PubMed] [Google Scholar]
- 179.Tonin P, Weber B, Offit K, Couch F, Rebbeck TR, Neuhausen S, Godwin AK, Daly M, Wagner-Costalos J, Berman D, Grana G, Fox E, Kane MF, Kolodner RD, Krainer M, Haber DA, Struewing JP, Warner E, Rosen B, Lerman C, Peshkin B, Norton L, Serova O, Foulkes WD, Garber JE, et al. Frequency of recurrent BRCA1 and BRCA2 mutations in Ashkenazi Jewish breast cancer families. Nat. Med. 1996;2:1179–1183. doi: 10.1038/nm1196-1179. [DOI] [PubMed] [Google Scholar]
- 180.R.S. Tuma, Combining carefully selected drug, patient genetics may lead to total tumor death. J. Natl. Cancer Inst. 99, 1505-1506, 1509 (2007) [DOI] [PubMed]
- 181.Turner N, Tutt A, Ashworth A. Targeting the DNA repair defect of BRCA tumours. Curr. Opin. Pharmacol. 2005;5:388–393. doi: 10.1016/j.coph.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 182.Vahteristo P, Bartkova J, Eerola H, Syrjakoski K, Ojala S, Kilpivaara O, Tamminen A, Kononen J, Aittomaki K, Heikkila P, Holli K, Blomqvist C, Bartek J, Kallioniemi OP, Nevanlinna H. A CHEK2 genetic variant contributing to a substantial fraction of familial breast cancer. Am. J. Hum. Genet. 2002;71:432–438. doi: 10.1086/341943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Vahteristo P, Tamminen A, Karvinen P, Eerola H, Eklund C, Aaltonen LA, Blomqvist C, Aittomaki K, Nevanlinna H. p53, CHK2, and CHK1 genes in Finnish families with Li-Fraumeni syndrome: further evidence of CHK2 in inherited cancer predisposition. Cancer Res. 2001;61:5718–5722. [PubMed] [Google Scholar]
- 184.van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 185.van Beers EH, van Welsem T, Wessels LF, Li Y, Oldenburg RA, Devilee P, Cornelisse CJ, Verhoef S, Hogervorst FB, van’t Veer LJ, Nederlof PM. Comparative genomic hybridization profiles in human BRCA1 and BRCA2 breast tumors highlight differential sets of genomic aberrations. Cancer Res. 2005;65:822–827. [PubMed] [Google Scholar]
- 186.van der Groep P, Bouter A, Menko FH, van der Wall E, van Diest PJ. High frequency of HIF-1alpha overexpression in BRCA1 related breast cancer. Breast Cancer Res. Treat. 2008;111:475–480. doi: 10.1007/s10549-007-9817-z. [DOI] [PubMed] [Google Scholar]
- 187.van der Groep P, Bouter A, van der Zanden R, Menko FH, Buerger H, Verheijen RH, van der Wall E, van Diest PJ. Re: Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst. 2004;96:712–713. doi: 10.1093/jnci/djh114. [DOI] [PubMed] [Google Scholar]
- 188.van der Groep P, Bouter A, van der Zanden R, Siccama I, Menko FH, Gille JJ, van Kalken C, van der Wall E, Verheijen RH, van Diest PJ. Distinction between hereditary and sporadic breast cancer on the basis of clinicopathological data. J. Clin. Pathol. 2006;59:611–617. doi: 10.1136/jcp.2005.032151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.van der Groep P, Hoelzel M, Buerger H, Joenje H, de Winter JP, van Diest PJ. Loss of expression of FANCD2 protein in sporadic and hereditary breast cancer. Breast Cancer Res. Treat. 2008;107:41–47. doi: 10.1007/s10549-007-9534-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.van der Groep P, van Diest PJ, Menko FH, Bart J, de Vries EG, van der Wall E. Molecular profile of ductal carcinoma in situ of the breast in BRCA1 and BRCA2 germline mutation carriers. J. Clin. Pathol. 2009;62:926–930. doi: 10.1136/jcp.2009.065524. [DOI] [PubMed] [Google Scholar]
- 191.van Diest PJ, van der Groep P, van der Wall E. EGFR expression predicts BRCA1 status in patients with breast cancer. Clin Cancer Res. 2006;12:670. doi: 10.1158/1078-0432.CCR-05-2098. [DOI] [PubMed] [Google Scholar]
- 192.Vaziri SA, Krumroy LM, Elson P, Budd GT, Darlington G, Myles J, Tubbs RR, Casey G. Breast tumor immunophenotype of BRCA1-mutation carriers is influenced by age at diagnosis. Clin. Cancer Res. 2001;7:1937–1945. [PubMed] [Google Scholar]
- 193.Venkitaraman AR. Tracing the network connecting BRCA and Fanconi anaemia proteins. Nat. Rev. Cancer. 2004;4:266–276. doi: 10.1038/nrc1321. [DOI] [PubMed] [Google Scholar]
- 194.Verhoog LC, Brekelmans CT, Seynaeve C, van den Bosch LM, Dahmen G, van Geel AN, Tilanus-Linthorst MM, Bartels CC, Wagner A, van den Ouweland A, Devilee P, Meijers-Heijboer EJ, Klijn JG. Survival and tumour characteristics of breast-cancer patients with germline mutations of BRCA1. Lancet. 1998;351:316–321. doi: 10.1016/s0140-6736(97)07065-7. [DOI] [PubMed] [Google Scholar]
- 195.Vleugel MM, Greijer AE, Shvarts A, van der Groep P, van Berkel M, Aarbodem Y, van Tinteren H, Harris AL, van Diest PJ, van der Wall E. Differential prognostic impact of hypoxia induced and diffuse HIF-1alpha expression in invasive breast cancer. J. Clin. Pathol. 2005;58:172–177. doi: 10.1136/jcp.2004.019885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 197.Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- 198.Welcsh PL, King MC. BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Hum. Mol. Genet. 2001;10:705–713. doi: 10.1093/hmg/10.7.705. [DOI] [PubMed] [Google Scholar]
- 199.Wessels LF, van Welsem T, Hart AA, van’t Veer LJ, Reinders MJ, Nederlof PM. Molecular classification of breast carcinomas by comparative genomic hybridization: a specific somatic genetic profile for BRCA1 tumors. Cancer Res. 2002;62:7110–7117. [PubMed] [Google Scholar]
- 200.Wong AK, Pero R, Ormonde PA, Tavtigian SV, Bartel PL. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J. Biol. Chem. 1997;272:31941–31944. doi: 10.1074/jbc.272.51.31941. [DOI] [PubMed] [Google Scholar]
- 201.Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 202.Wooster R, Neuhausen SL, Mangion J, Quirk Y, Ford D, Collins N, Nguyen K, Seal S, Tran T, Averill D, et al. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13. Science. 1994;265:2088–2090. doi: 10.1126/science.8091231. [DOI] [PubMed] [Google Scholar]
- 203.Wu X, Webster SR, Chen J. Characterization of tumor-associated Chk2 mutations. J. Biol. Chem. 2001;276:2971–2974. doi: 10.1074/jbc.M009727200. [DOI] [PubMed] [Google Scholar]
- 204.Yuan SS, Lee SY, Chen G, Song M, Tomlinson GE, Lee EY. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 1999;59:3547–3551. [PubMed] [Google Scholar]
- 205.Zhong Q, Chen CF, Li S, Chen Y, Wang CC, Xiao J, Chen PL, Sharp ZD, Lee WH. Association of BRCA1 with the hRad50-hMre11-p95 complex and the DNA damage response. Science. 1999;285:747–750. doi: 10.1126/science.285.5428.747. [DOI] [PubMed] [Google Scholar]
- 206.Zweemer RP, van Diest PJ, Verheijen RH, Ryan A, Gille JJ, Sijmons RH, Jacobs IJ, Menko FH, Kenemans P. Molecular evidence linking primary cancer of the fallopian tube to BRCA1 germline mutations. Gynecol. Oncol. 2000;76:45–50. doi: 10.1006/gyno.1999.5623. [DOI] [PubMed] [Google Scholar]


