Abstract
Establishing a diagnosis in patients with a suspected mitochondrial disorder is often a challenge. Both knowledge of the clinical spectrum of mitochondrial disorders and the number of identified disease-causing molecular genetic defects are continuously expanding. The diagnostic examination of patients requires a multi-disciplinary clinical and laboratory evaluation in which the biochemical examination of the mitochondrial functional state often plays a central role. In most cases, a muscle biopsy provides the best opportunity to examine mitochondrial function. In addition to activity measurements of individual oxidative phosphorylation enzymes, analysis of mitochondrial respiration, substrate oxidation, and ATP production rates is performed to obtain a detailed picture of the mitochondrial energy-generating system. On the basis of the compilation of clinical, biochemical, and other laboratory test results, candidate genes are selected for molecular genetic testing. In patients in whom an unknown genetic variant is identified, a compatible biochemical phenotype is often required to firmly establish the diagnosis. In addition to the current role of the biochemical analysis in the diagnostic examination of patients with a suspected mitochondria disorder, this report gives a future perspective on the biochemical diagnosis in view of both the expanding genotypes of mitochondrial disorders and the possibilities for high throughput molecular genetic diagnosis.
Introduction
Mitochondrial disorders are a clinically heterogeneous group of disorders that are caused by defects in the mitochondrial ATP production system. The previously published guidelines and diagnostic criteria for mitochondrial disorders illustrate the challenges of diagnosing patients with a suspected mitochondrial disorder (Bernier et al. 2002; Haas et al. 2008; Morava et al. 2006; Thorburn and Smeitink 2001; Walker et al. 1996; Wolf and Smeitink 2002). Given the extremely broad clinical spectrum and the relatively poor genotype–phenotype correlation of mitochondrial genetic defects, it is usually preferred to perform a complete laboratory diagnostic work-up, and to evaluate the results in the context of the clinical phenotype and family history of the patient (Haas et al. 2007; Kirkman et al. 2008; Zeviani and Di Donato 2004). Laboratory diagnostic analysis of patients with a suspected mitochondrial disorder is a multi-disciplinary approach involving a broad spectrum of laboratory tests, including metabolite analysis, enzymatic measurements, and molecular genetic analysis (Haas et al. 2008). In a minority of cases, those with a well-recognized phenotype, it might be possible to shortcut the diagnostic process and go directly to molecular genetic analysis (Finsterer et al. 2009). Although time- and cost-effective when successful, there are drawbacks to this approach. Importantly, there is a chance of finding a mutation that is irrelevant to the pathogenesis of the disease of the patient. It has been shown that the prevalence of pathogenic mtDNA mutations is at least 1 in 200, (Elliott et al. 2008), while the prevalence of mtDNA diseases or of being at risk to develop an mtDNA disease has been estimated to be around 1 in 10,000 and 1 in 6,000, respectively (Schaefer et al. 2008). Establishing the biochemical phenotype is not only important for candidate gene selection but also provides important information that may be required to interpret genetic test results. For these and other reasons, as outlined below, the biochemical examination of a muscle biopsy to evaluate the functional state of mitochondria is still regarded as the cornerstone of the diagnostic examination of patients with a suspected mitochondrial disease.
Metabolite analysis
Prior to the biochemical examination of a muscle biopsy, metabolite analysis in blood and urine is usually performed. The results often provide important clues for the presence of a mitochondrial defect and, in some cases, can even give some indications for the location of the primary cause of the disease. Defects in the mitochondrial energy-generating system may lead to high lactate levels in blood, urine, and/or CSF due to reduced pyruvate utilization by the mitochondria. In the case of a respiratory chain defect, the lactate/pyruvate ratio in blood will increase because of a shift in the mitochondrial redox state (Trijbels et al. 1988). This will also affect the ratio of the ketone bodies 3-OH-butyrate and acetoacetate. In the case of a pyruvate dehydrogenase defect, the ratio between lactate and pyruvate may be normal while both metabolites are elevated. However, these features are neither very specific nor sensitive for use as a diagnostic test. Amino acid analysis can reveal elevated alanine, as a product of the transamination of pyruvate by alanine aminotransferase. Elevated levels of the branched chain amino acids could be an indication for an E3 deficiency; an enzyme module shared by the branched chain amino acid dehydrogenase, 2-oxoglutarate dehydrogenase, and pyruvate dehydrogenase. Generalized amino aciduria is sometimes seen as a result of renal tubulopathy caused by mitochondrial defects. Urine organic acids of mitochondrial patients may show increased lactate excretion. In addition, increased levels of TCA cycle intermediates, such as malate, succinate, 2-oxoglutarate, and fumarate can be found. A high level of fumarate excretion is an indication for fumarase deficiency. Elevated 2-oxoglutarate excretion is seen in patients with a 2-oxoglutarate deficiency, but also in other mitochondrial defects. Elevated levels of several other metabolites can be found in mitochondrial patients, including ethylmalonate, sometimes seen in respiratory chain deficient patients but also in SCAD, MADD, and in ETHE1 defects, methylmalonate, e.g., in succinate-CoA ligase deficiency (Carrozzo et al. 2007), and 3-methylglutaconate (3-MG). Four types of 3-MG acidurias can be discriminated, including the classical 3-MGA hydratase deficiency (type I MGA), Barth syndrome (type II; TAZ defect), Costeff syndrome (type III; OPA1 defect), and the group of type IV MGAs which includes several mitochondrial disorders, such as those due to genetic defects in POLG, SUCLA2, and TMEM70 (Wortmann et al. 2009). In addition, this group contains patients with MEGDEL (3-methylglutaconic aciduria, sensory-neural deafness, encephalopathy, and Leigh-like syndrome) association (Wortmann et al. 2006).
Enzyme measurements
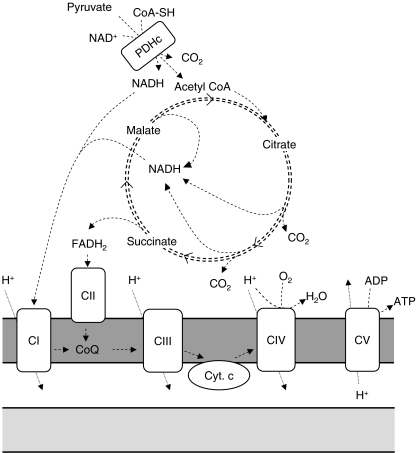
Biochemical diagnostic examination of tissue and cell samples from mitochondrial patients includes measurements of enzyme activities of the oxidative phosphorylation (OXPHOS) system, consisting of complex I (EC 1.6.5.3), complex II (EC 1.3.5.1), complex III (EC 1.10.2.2), complex IV (EC 1.9.3.1), and complex V (EC 3.6.1.3) (Fig. 1). Assays to quantify OXPHOS enzyme activities are usually based on spectrophotometry (Benit et al. 2006; Janssen et al. 2007; Kirby et al. 2007; Rustin et al. 1994). Table 1 gives an overview of the assays that are currently operational in our center. Alternatively, or in addition, assays that determine the amounts of OXPHOS complexes are performed, such as Blue Native gel electrophoresis followed by western blot analysis (Schagger and von Jagow 1991). A drawback of the latter approach could be that catalytic defects can be missed, although for complex I it has been shown that there is often a correlation between residual enzyme activities and the amounts of holo-complex I (Koopman et al. 2008). Another way to examine OXPHOS enzymes is by colorimetric in-gel measurements of enzyme activities after blue-native gel electrophoresis, or other colorimetric assays based on a similar principle (Van Coster et al. 2001; Willis et al. 2009; Wittig et al. 2007; Zerbetto et al. 1997). For complex I, these assays makes use of an electron acceptor that changes color upon reduction (e.g., nitrotetrazolium blue), instead of analogues of the natural substrate coenzyme Q that are used in spectrophotometric complex I assays (Benit et al. 2006; Janssen et al. 2007; Kirby et al. 2007). This means that the colorimetric assay measures only the activity of the NADH dehydrogenase module of complex I, instead of the entire NADH:ubiquinone oxidoreductase activity that is measured in spectrophotometric assays. Moreover, in-gel activity assays are not designed to provide highly quantitative information, and therefore mild deficiencies of OXPHOS enzymes may be difficult to detect. This also holds true for other gel-based assays to quantify enzyme activities or amounts, such as western blot assays. Therefore, most diagnostic centers prefer to make use of spectrophotometric enzyme assays. At this moment, there is no universal assay design for spectrophotometric quantification of OXPHOS enzyme activities. Different assay protocols are in use in different laboratories. Assays differ with respect to buffer conditions, reaction temperatures, substrate concentrations, and other assay ingredients, such as bovine serum albumin or salts. Nevertheless, for most of these assays, the principles on which the different protocols are based are similar. The use of different assay protocols can make an inter-laboratory comparison of test results difficult. For both ethical and practical reasons, a quality control scheme involving the analysis of patient-derived material, including tissue samples with OXPHOS deficiencies, seems impossible. Indeed, attempts to perform a laboratory quality assessment have been hampered by the limited availability of patient-derived muscle samples, and therefore have been done with animal muscle tissue (Gellerich et al. 2004; Medja et al. 2009). Despite the different assay protocols, as each laboratory has established its own reference ranges to which diagnostic test results are related, it is questionable if the use of different assay protocols could be a major cause for possible misdiagnoses.
Fig. 1.
Schematic representation of the mitochondrial energy generating system. Substrates are shuttled into mitochondria and are metabolized in the matrix in the TCA cycle, during which reduction equivalents NADH and FADH2 are formed. These are oxidized by complex I (CI) and II (CII) of the respiratory chain, and electrons coming from NADH and FADH2 are shuttled through complexes I or II, coenzyme Q, complex III (CIII), cytochrome c, and complex IV (CIV) to molecular oxygen. During the electron transport process, protons are pumped out of the mitochondrial matrix to the intermembrane space by complexes I, III, and IV. The electrochemical gradient thus formed is used by complex V (CV; F1/F0 ATPase) to convert ADP into ATP. The intramitochondrial ATP/ADP ratio is balanced by shuttling ADP and ATP in and out of the mitochondrial matrix by the adenosine nucleotide transporter ANT (not shown here). For diagnostic purposes, the mitochondrial energy generating system can be analyzed in several ways. By using 14C labeled pyruvate, malate, or succinate, the conversion rates of these substrates can be determined by measuring the amounts of released 14CO2 as parameters for the overall capacity of the mitochondrial energy-generating system (Janssen et al 2006). A similar parameter is the oxygen consumption rate in the presence of different substrates, e.g., pyruvate + malate, and which can be measured by respirometry or by fluorescent probes (Jonckheere et al 2010; Rustin et al 1994). The rate of synthesis of the end product ATP in the presence of different mitochondrial substrates is also representative for the capacity of the mitochondrial energy-generating system (Janssen et al 2006). In addition to these flux-parameters of the mitochondrial energy-generating system, individual enzymes can be determined by spectrophotometric and radiochemical assays
Table 1.
Overview of the assays currently in use in the Nijmegen Center for mitochondrial disorders
| Enzyme | Substrate | Assay conditions | Read-out |
|---|---|---|---|
| Complex I | 0.2 mmol/L NADH | 25 mM phosphate buffer, pH 7.6 | 1 µmol/L rotenone-sensitive DCIP reduction at 600 nm |
| 70 µmol /L coenzyme Q1 | 0.35% BSA | ||
| 60 µmol/L DCIP | |||
| Complex II | 10 mmol/L succinate | 80 mM phosphate buffer, pH 7.8 | 5 mmol/L malonate-sensitive DCIP reduction at 600 nm |
| 80 µmol/L decylubiquinone | 0.2 % BSA | ||
| 0.2 mmol/L ATP | |||
| 80 µmol/L DCIP | |||
| 2.0 mmol/L EDTA | |||
| 0.3 mmol/L sodium azide | |||
| Complex III | 300 µmol/L decylubiquinol | 50 mM phosphate buffer, pH 7.8 | Cytochrome c reduction at 550 nm |
| 50 µmol/L cytochrome c | 1.0 mmol/L EDTA | ||
| 3.0 mmol/L sodium azide | |||
| 0.04% Tween20 | |||
| Complex IV | 70 µmol/L reduced cytochrome c | 30 mM phosphate buffer, pH 7.4 | Cytochrome c oxidation at 550 nm |
| Complex V | 3 mmol/L ATP | 25 mM phosphate buffer, pH 8.0 | 10 µmol/L oligomycin-sensitive NADH oxidation at 340 nm |
| 0.2 mmol/L EGTA | |||
| 5 mg/L Ap5A | |||
| 0.3% BSA | |||
| 250 mmol/L sucrose | |||
| 7.5 mmol/L MgCl2 | |||
| 50 mmol/L KCl | |||
| 0.1 mmol/L phosphoenolpyruvate | |||
| 2.5 U/ml lactate dehydrogenase | |||
| 1.5 U/ml pyruvate kinase |
The assays are performed at 37°C. The samples that are tested with these assays include isolated mitochondria from muscle and fibroblasts. In addition, more crude preparations from muscle, such as 10% homogenates in SETH-buffer (Janssen et al 2006), and the 600-g supernatants of these homogenates, are measured in this way. Extracts from heart, liver, brain, and chorionic villi are examined in a similar manner. These assays can be performed on any spectrophotometric device that has sufficient sensitivity.
The interpretation of enzymatic measurements is not as straightforward as for many other metabolic enzyme disorders. For the OXPHOS enzymes, there is a only a small margin between patient ranges and control ranges (Rustin et al. 1994). Therefore, activities around the lowest reference value cannot always be considered as definite proof of either a normal or deficient enzyme activity, and, as mentioned before, additional observations (mtDNA sequence, metabolites, clinical features, etc.) should be included in the evaluation of the enzyme test results. It can be hypothesized that one of the reasons for this problem is the complexity of the OXPHOS enzymes, which are multi-subunit enzymes encoded by both mitochondrial and nuclear genomes, except for complex II, which exclusively consists of nuclear encoded subunits. It seems likely that the genetic make-up of the entire OXPHOS system and of the individual complexes could modulate the effect of single subunit mutations. As there is no universal correlation between the severity of the enzyme deficiency and the severity of the clinical phenotype, detection of even very mild deficiencies is very important. Ratios between mitochondrial enzymes give a much narrower range of normal values compared to activities expressed on the basis of protein content of the sample (Rustin et al. 1991). Often, citrate synthase is used as a reference enzyme, although ratios between OXPHOS enzymes and tricarboxylic acid (TCA) cycle enzymes are less constant compared to ratios within the OXPHOS system (Rustin et al. 1991). Furthermore, a moderate increase of the activity of citrate synthase is frequently observed in tissues with an OXPHOS deficiency. For neonates, results should be interpreted very cautiously as it has been shown that there are age-dependent differences in OXPHOS enzyme activities in very early childhood (Honzik et al. 2008; Sperl et al. 1992). Table 2 gives an overview of the results of the biochemical diagnostics in Nijmegen for 2009. This shows that complex I and IV deficiencies are the most frequently encountered OXPHOS deficiencies. In addition to single OXPHOS enzyme measurements, combinations of enzymes can be measured. For complex I, an additional measurement of complex I + complex III (NADH:cytochrome c oxidoreductase) can be performed. This activity is reduced in the case of a complex I deficiency. In addition, as this is a coenzyme Q-dependent reaction, this activity is also reduced in the case of a CoQ biosynthesis defect. Similarly, the activity of complex II + III is also coenzyme Q-dependent and reduced in the case of a CoQ deficiency. Further indications for a CoQ deficiency can be obtained by addition of a CoQ analogue to the complex II + III enzyme assay, which will result in normalization of the activity in the case of a CoQ deficiency (Lopez et al. 2006; Rahman et al. 2001). Both the combined complex I + III activity and the II + III activity are less sensitive to detect complex III deficiencies, so these assays are not a good substitute for complex III measurements and should only be used to obtain additional evidence for deficiencies of complex I, II, or coenzyme Q (Taylor et al. 1993). Coenzyme Q can also be measured directly by HPLC in muscle, blood, or other sample types (Duncan et al. 2005). In addition to the OXPHOS enzymes, activities of pyruvate dehydrogenase or TCA cycle enzymes can be considered in the case of a suspected mitochondrial disorder (Bonnefont et al. 1992; Carrozzo et al. 2007; Reisch and Elpeleg 2007; Zinn et al. 1986). A particular feature of defects in the TCA cycle enzyme succinate-CoA ligase is depletion of mtDNA (Carrozzo et al. 2007; Ostergaard et al. 2007; Reisch and Elpeleg 2007). Patients with a defect in this enzyme often display a characteristic combination of clinical features, including early hypotonia, dystonia, and sensori-neuronal deafness (Morava et al. 2009; Ostergaard et al. 2010). The mechanism leading to mtDNA depletion in succinate-CoA ligase deficiency is poorly understood. It is assumed that it involves the mitochondrial deoxynucleotide salvage enzyme nucleoside diphosphate kinase, which has been shown to physically interact with succinate-CoA ligase (Kavanaugh-Black et al. 1994). As a consequence of mtDNA depletion, patients with mutations in the succinate:CoA ligase α-subunit SUCLA2 or β-subunit SUCLG1 can show deficiencies of OXPHOS enzymes complexes I, III, and IV in muscle tissue.
Table 2.
Overview of the results of mitochondrial biochemical diagnostics in Nijmegen in the years 2005–2009
| Parameter | % reduced |
|---|---|
| ATP production | 39 |
| Complex I | 8 |
| Complex II | 2 |
| Complex III | 3 |
| Complex IV | 5 |
| Complex V | 1 |
| Combination of enzymes | 7 |
| ATP production (normal OXPHOS) | 13 |
The table gives the percentage of fresh muscle samples that showed an activity below the lowest control value. Results from frozen muscle samples are not included in this table. In total, 1,406 fresh muscle samples were examined in Nijmegen in 2005–2009. Of these, 39% showed a reduced rate of ATP production from the oxidation of pyruvate and malate. Approximately 2/3 of the samples with a reduced ATP production rate also showed a reduced activity of one or more OXPHOS enzymes. The remaining 1/3 showed normal OXPHOS enzyme activities. Approximately 5% of this latter group had a reduced PDHc activity. The figures presented here are similar to those reported in 2000, only in that case complex V and ATP production were not included (Loeffen et al. 2000). The reason for a reduced ATP production rate in the remaining 30% of the patients with normal OXPHOS and PDHc enzyme activities is either a secondary mitochondrial dysfunction or an unknown mitochondrial defect, such as enzymes and transporters for which a diagnostic assay is not (yet) available. The possibility of unknown defects is quite likely, given the total number of proteins/genes involved in mitochondrial energy metabolism and given the fact that new mitochondrial defects are still being discovered very regularly. This table should not be regarded as illustrative for the total group of patients with a suspected mitochondrial disorder, as not all patients have been tested for enzyme activities in muscle. Moreover, samples were received from many different hospitals in different countries, and the indications for doing a muscle biopsy are not the same in each hospital.
Which tissue to investigate?
In the majority of mitochondrial patients, skeletal muscle is affected. Muscle is mitochondria-rich tissue and has a high energy demand. Therefore, a muscle biopsy provides an optimal opportunity to detect possible aberrations in the functional state of mitochondria. The biochemical analysis of muscle mitochondria can be performed on frozen or on freshly collected muscle tissue. It should be noted that analysis of complex V in frozen muscle tissue is considered to be less reliable than in fresh muscle tissue (Kirby et al. 2007). For optimal preservation of mitochondrial enzymes, it is important to snap-freeze the sample immediately after collection. A fresh muscle sample needs to be collected in cooled (not frozen) transport buffer and should be processed within 2–3 h after collection (Janssen et al. 2003). Fresh muscle has the advantage that, in addition to the measurement of individual mitochondrial enzymes, measurements on the entire mitochondrial energy-generating system (MEGS) can be performed (Fig. 1). These measurements can include the analysis of mitochondrial oxygen consumption in the presence of different combinations of mitochondrial substrates, such as pyruvate, malate, and glutamate (Rustin et al. 1994; Will et al. 2006). Alternatively, the oxidation rates of radio-labeled substrates can be determined, such as the decarboxylation of radio-labeled pyruvate in the presence of non-labeled co-substrates (Janssen et al. 2006). In addition, the ATP production rate in intact mitochondria can be determined as a measure for the MEGS capacity (Janssen et al. 2006). By examining the MEGS in the presence of different combinations of substrates, co-substrates, co-factors, and inhibitors, a detailed picture of the MEGS is obtained, providing important clues for the location of the primary mitochondrial defect. For example, a reduced pyruvate oxidation rate in the presence of malate, combined with a normal to mildly reduced pyruvate oxidation rate in the presence of carnitine is an indication for an OXPHOS defect (Hoefs et al. 2008; Jonckheere et al. 2008), while equally reduced rates of both types of pyruvate oxidations are an indication for a pyruvate dehydrogenase complex (PDHc) deficiency (Janssen et al. 2006). This is not trivial, as in several patients with a complex I deficiency in muscle tissue, a mild, secondary PDHc deficiency has been observed (unpublished observations, and Loeffen et al. 1998; van den Heuvel et al. 1998). Reduced pyruvate oxidation rates have also been observed in a mitochondrial pyruvate transport defect (Brivet et al. 2003). Reduced glutamate + malate oxidation rates have been seen in muscle of patients with glutamate carrier deficiencies (Molinari et al. 2005; Wibom et al. 2009). A reduced pyruvate oxidation rate that is normalized by addition of an uncoupler, such as FCCP, indicates a defect in complex V, the adenine nucleotide translocator, or the phosphate transporter (Jonckheere et al. 2008; Mayr et al. 2007). The MEGS assays described above not only provide in-depth information on the mitochondrial functional state, it has also been shown that these assays are more sensitive to detect mitochondrial defects, compared to measurements of individual OXPHOS enzyme activities (Janssen et al. 2008). In the case of a patient with a clinical phenotype with profound involvement of tissues other than muscle, a biopsy of the affected tissue should be considered. Many cases have been described in which the diagnostic examination of a heart or liver biopsy has lead to the diagnosis (Garcia-Cazorla et al. 2006; Taylor et al. 2003). For example, for the mtDNA depletion syndromes with predominant liver involvement, e.g., due to defects in MPV17, DGUOK, or POLG (Mandel et al. 2001; Spinazzola et al. 2006; Van Goethem et al. 2004), a liver biopsy is often more informative than a muscle biopsy.
Cultured fibroblasts
When a tissue biopsy is performed, it is important to collect a skin biopsy at the same time, to obtain fibroblasts for possible additional biochemical and genetic studies. In addition to the added value of fibroblasts in the diagnostic evaluation of patients, fibroblasts can be stored and retested when desired, e.g., when new diagnostic possibilities emerge in the future. There are several other benefits of fibroblast testing in the diagnostic evaluation of patients with a suspected mitochondrial disorder (Cameron et al. 2004; van den Heuvel et al. 2004). Fibroblast OXPHOS enzyme activities provide information that is helpful for the selection of candidate genes for molecular genetic analysis, even if enzyme activities are normal. For example, in patients with reduced OXPHOS enzymes in muscle and normal enzyme activities in fibroblasts, a genetic defect might be present in one of the nuclear genes causing mtDNA depletion, for example POLG (de Vries et al. 2007). It should be noted that normal enzyme activities have also been observed in muscle of several POLG patients (de Vries et al. 2008; Horvath et al. 2006; Van Goethem et al. 2004). Biochemical observations in fibroblasts of patients with mtDNA mutations are variable. The main reason for this is tissue/cell-type-specific differences in heteroplasmy of mtDNA mutations. Although many nuclear defects will give rise to reduced enzyme activities in fibroblasts, there are several exceptions. In addition to POLG, other nuclear genetic defects causing mtDNA depletion syndromes, such as DGUOK and MPV17, often show normal enzyme activities in fibroblasts (Mandel et al. 2001; Spinazzola et al. 2006). The combination of complex I, III, IV, and/or V deficiency in muscle and normal OXPHOS enzymes in fibroblasts is suggestive for involvement of mtDNA depletion, deletions/rearrangements, or point mutations (van den Heuvel et al. 2004). For mutations in two aminoacyl-tRNA synthetase genes, DARS2 and RARS2, normal or mild OXPHOS deficiencies in fibroblasts, respectively, have been reported, while for RARS2, it has been shown that OXPHOS enzyme activities in muscle vary from normal to severely reduced (Edvardson et al. 2007; Scheper et al. 2007). Fibroblast enzyme deficiencies are common in nuclear defects of genes encoding structural components and assembly factors of the OXPHOS system, and also for defects in genes encoding proteins involved in mitochondrial translation (Coenen et al. 2004; Miller et al. 2004; Saada et al. 2007; Smeitink et al. 2006). It should be noted that a limited number of cases have been reported in which a nuclear genetic defect of a structural respiratory chain building block did not result in an enzyme deficiency of the corresponding enzyme, e.g., complex I (Benit et al. 2001), complex II (Taylor et al. 1996), and complex IV (Vesela et al. 2004), although according to the experiences we have had in Nijmegen, these appear to be quite exceptional cases. Occasionally, patients with normal enzyme activities in muscle show reduced enzyme activities in cultured fibroblasts, in particular for complex I and PDHc (unpublished observations). Confirmation of enzyme deficiencies in fibroblasts of the OXPHOS-deficient index case in families in which the pathogenic genetic defect has not (yet) been uncovered, is required before reliable prenatal testing by measuring OXPHOS enzymes in chorionic villi or amniocytes can be performed (Niers et al. 2003). In Nijmegen, prenatal diagnosis is performed by measuring respiratory chain or PDHc enzyme activities in chorionic villi, provided that the diagnostic examination of the index case has revealed a clearly detectable enzyme deficiency in fibroblasts and muscle (or other tissue), and that mtDNA mutations have been excluded in mtDNA from a sample in which the enzyme deficiency has been detected (Niers et al. 2003; Niers et al. 2001). When these prerequisites are met, reliable prenatal diagnosis can be performed. In addition to the diagnostic applications, fibroblasts provide a model system to study the pathogenesis of novel genetic defects, for example by performing complementation experiments. Several assays have been described to examine MEGS activities in cultured fibroblasts. These are based on the same principles as the MEGS assays for fresh muscle. ATP synthesis, oxygen consumption, and substrate oxidation rates can all be measured in fibroblasts (Cameron et al. 2004; Rustin et al. 1994; Wanders et al. 1993). In many of these assays, cells are permeabilized with digitonin, which allows substrates, ADP, and other assay ingredients to reach the mitochondria and enter the matrix via the mitochondrial transporters. In this way, mitochondrial ATP production, substrate oxidation and respiration rates can be examined in a controlled manner, providing additional information on the biochemical phenotype of patients.
Avoiding potential pitfalls
Accurate diagnostic biochemical testing of tissue and cell samples can be hampered by many different circumstances. As with many other types of diagnostic laboratory tests, a frequent cause for problems is sample collection and handling, for example muscle samples that were thawed during transport, or needle muscle biopsies contaminated with lidocaine. In experienced diagnostic centers where muscle biopsies are performed and analyzed on a daily basis, these problems are not likely to occur. In addition to these practical issues, there are also several other reasons for difficulties in reaching a diagnosis by muscle biochemistry. These can include secondary mitochondrial dysfunction arising from other metabolic defects, e.g., due to toxicity of accumulating toxic metabolites, such as propionyl-CoA (Schwab et al. 2006) or sulfide (Tiranti et al. 2009), or due to drug effects, such as valproate (Haas et al. 1981). A diagnosis can be missed when relying solely on a single diagnostic test, e.g., biochemical testing of a muscle biopsy in patients with hepatocerebral mtDNA depletion syndromes, as outlined in the previous paragraph. Also, in the case of mtDNA disorders, a normal muscle biochemistry can sometimes be observed. This is usually due to a relatively low percentage of heteroplasmy of the pathogenic mtDNA mutation. For the m.3243G>A “MELAS” mutation, we have shown that there are different thresholds for different biochemical defects, such as complex I deficiency and reduced ATP production rate (Janssen et al. 2008). In practice, mtDNA analysis and muscle biochemistry are often performed in parallel, as a normal biochemistry does not exclude a mtDNA mutation, and vice versa, and, in addition, as biochemical observations can be very helpful in the interpretation of mtDNA sequence analysis results, especially when unknown mtDNA variants are encountered. In general, most of the potential pitfalls mentioned here can be avoided by performing a complete laboratory diagnostic examination, including analysis of blood and urine metabolites, muscle and fibroblast biochemistry, muscle histology, mtDNA, and other candidate genes. The combined results of these laboratory tests should be interpreted in the context of the clinical phenotype of the patient (Haas et al. 2008).
Outlook
The number of mitochondrial proteins in mammals has been estimated to be between 1,130 and 1,500 (Meisinger et al. 2008; Pagliarini et al. 2008). To date, pathogenic defects have been found in only a fraction of these proteins. Given the number of genes encoding mitochondrial proteins and the considerable subgroup of patients with a mitochondrial disease phenotype in whom, despite new developments in diagnostic testing, it is still not possible to establish a definite diagnosis (Kirkman et al. 2008), it is likely that more defects leading to a mitochondrial disorder will be found in the mitochondrial proteome in the future. The analysis of fresh muscle biopsies has led to the identification of several genetic defects that are outside the classical OXPHOS genetic defects. These include defects in mitochondrial carriers, e.g., glutamate and phosphate carriers (Mayr et al. 2007; Molinari et al. 2005; Wibom et al. 2009). These studies have clearly demonstrated the added value of fresh muscle sample analysis, as it has guided the search for the specific genetic defects that caused the observed biochemical abnormalities and clinical phenotypes of the patients. It is likely that additional pathogenic mitochondrial carrier defects will be found in the future. To date, more than 20 different mitochondrial functional carrier systems have been identified, and there are many other carriers of which the exact function remains to be established (Palmieri 2008). Genes encoding proteins with a role in mitochondrial transcription and translation are another likely group of candidate genes. These include the mitochondrial aminoacyl-tRNA synthetases, in which only two defects have been found thus far (Edvardson et al. 2007; Scheper et al. 2007). Several other groups of candidate genes can be identified or postulated, including those involved in mtDNA maintenance (Copeland 2008), tRNA modification (Bykhovskaya et al. 2004), mitochondrial ribosomal assembly (Miller et al. 2004), nucleoid formation (Hoffmann et al. 2009), enzymes regulating the activity of key enzymes in the MEGS (Cameron et al. 2009; Phillips et al. 2009), and several other cell biological processes required for mitochondrial ATP production. For all these mitochondrial defects, functional tests could have an important added value in the biochemical diagnosis of mitochondrial disorders in the future.
In the near future, developments in next generation sequencing technologies (Mardis 2008) will make it possible to include high throughput sequence analysis in the diagnostic work-ups of mitochondrial patients. The first applications of this technology in mitochondrial diagnostics have already been published (Vasta et al. 2009). In the case of a suspected mitochondrial disorder, the number of candidate genes that could be examined is quite large. For example, for complex I there are more than 50 different nuclear genes encoding for structural subunits and assembly proteins, and many other genes of which it could be postulated that a defect might lead to complex I deficiency. Screening of this many genes using conventional sequencing techniques is an expensive and time-consuming effort. Therefore, there is no doubt that high throughput molecular genetic assays will increase the possibilities for diagnostic testing of mitochondrial disorders in the near future. The developments in genetic testing may change the role of biochemical diagnosis. It can be expected that there will be a greater demand for more detailed functional studies to confirm the pathogenicity of new molecular genetic variants. This is already the case for patients in with unknown mtDNA variants detected by sequence analysis of the mitochondrial genome, e.g., by mtDNA sequencing arrays (van Eijsden et al. 2006). Nowadays, the complete sequence analysis of the 16.5-kb mitochondrial genome is routinely being performed in many diagnostic centers. Often, the prediction of the pathogenicity of unknown genetic variants is not possible on the basis of sequence information alone. Usually, the level of heteroplasmy of the mtDNA variant is checked in different tissues of the patient and, in addition, in family members in the maternal lineage. Should this not provide a clear answer to the question on pathogenicity of mtDNA variants, functional testing of mtDNA variants is possible by creating transmitochondrial cybrids (King and Attardi 1989), in which the observed mitochondrial function can provide additional diagnostic information. It is expected that this problem of uncharacterized genetic variants will be even greater for results of high throughput nuclear genomic sequencing data. In a recent study in which 362 genes were sequenced in two patients (Vasta et al. 2009), it was found that, after extensive bioinformatical analysis of all genetic variants that were identified, two to three genetic variants remained for which it was not possible to predict possible pathogenicity without follow-up functional testing. It is likely that these figures will improve in time, as bioinformatical tools will improve in time and databases are continuously being filled with new genetic variants. Nevertheless, there will be an increasing demand for a more detailed functional analysis of individual genetic variants, in order to confirm the diagnosis in individual patients. The combination of a sophisticated bioinformatical approach with detailed functional analysis of individual proteins and pathways will be required to unravel the mechanisms behind protein malfunction and pathogenesis of mitochondrial disorders (Huynen et al. 2009). In addition, detailed information on the biochemical phenotype prior to high throughput genetic testing will be of benefit to the selection of groups of candidate genes and to bioinformatical evaluation of genetic variants. For example, in the case of a proven complex I deficiency, the high-throughput genetic examination could focus on complex I genes. Therefore, the biochemical evaluation of mitochondrial functional state will continue to play an important role in the diagnostic examination of mitochondrial patients in the foreseeable future.
Acknowledgments
The author would like to thank the colleagues of the NCMD tissue culture and muscle laboratories of the Laboratory for Genetic, Endocrine, and Metabolic disorders for excellent technical assistance, and Dr. Leo Kluijtmans for critical reading of the manuscript and helpful suggestions. This work has been made possible thanks to grants from Metakids, the Prinses Beatrix Fonds, and Energy4All.
Footnotes
Competing interests: None declared.
References
- Benit P, Chretien D, Kadhom N, et al. Large-scale deletion and point mutations of the nuclear NDUFV1 and NDUFS1 genes in mitochondrial complex I deficiency. Am J Hum Genet. 2001;68:1344–1352. doi: 10.1086/320603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benit P, Goncalves S, Philippe DE, Briere JJ, Martin G, Rustin P. Three spectrophotometric assays for the measurement of the five respiratory chain complexes in minuscule biological samples. Clin Chim Acta. 2006;374:81–86. doi: 10.1016/j.cca.2006.05.034. [DOI] [PubMed] [Google Scholar]
- Bernier FP, Boneh A, Dennett X, Chow CW, Cleary MA, Thorburn DR. Diagnostic criteria for respiratory chain disorders in adults and children. Neurology. 2002;59:1406–1411. doi: 10.1212/01.wnl.0000033795.17156.00. [DOI] [PubMed] [Google Scholar]
- Bonnefont JP, Chretien D, Rustin P, et al. Alpha-ketoglutarate dehydrogenase deficiency presenting as congenital lactic acidosis. J Pediatr. 1992;121:255–258. doi: 10.1016/s0022-3476(05)81199-0. [DOI] [PubMed] [Google Scholar]
- Brivet M, Garcia-Cazorla A, Lyonnet S, et al. Impaired mitochondrial pyruvate importation in a patient and a fetus at risk. Mol Genet Metab. 2003;78:186–192. doi: 10.1016/s1096-7192(03)00016-7. [DOI] [PubMed] [Google Scholar]
- Bykhovskaya Y, Casas K, Mengesha E, Inbal A, Fischel-Ghodsian N. Missense mutation in pseudouridine synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic anemia (MLASA) Am J Hum Genet. 2004;74:1303–1308. doi: 10.1086/421530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron JM, Levandovskiy V, MacKay N, Robinson BH. Respiratory chain analysis of skin fibroblasts in mitochondrial disease. Mitochondrion. 2004;4:387–394. doi: 10.1016/j.mito.2004.07.039. [DOI] [PubMed] [Google Scholar]
- Cameron JM, Maj M, Levandovskiy V, et al. Pyruvate dehydrogenase phosphatase 1 (PDP1) null mutation produces a lethal infantile phenotype. Hum Genet. 2009;125:319–326. doi: 10.1007/s00439-009-0629-6. [DOI] [PubMed] [Google Scholar]
- Carrozzo R, Dionisi-Vici C, Steuerwald U, et al. SUCLA2 mutations are associated with mild methylmalonic aciduria, Leigh-like encephalomyopathy, dystonia and deafness. Brain. 2007;130:862–874. doi: 10.1093/brain/awl389. [DOI] [PubMed] [Google Scholar]
- Coenen MJ, Antonicka H, Ugalde C, et al. Mutant mitochondrial elongation factor G1 and combined oxidative phosphorylation deficiency. N Engl J Med. 2004;351:2080–2086. doi: 10.1056/NEJMoa041878. [DOI] [PubMed] [Google Scholar]
- Copeland WC. Inherited mitochondrial diseases of DNA replication. Annu Rev Med. 2008;59:131–146. doi: 10.1146/annurev.med.59.053006.104646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vries MC, Rodenburg RJ, Morava E, et al. Multiple oxidative phosphorylation deficiencies in severe childhood multi-system disorders due to polymerase gamma (POLG1) mutations. Eur J Pediatr. 2007;166:229–234. doi: 10.1007/s00431-006-0234-9. [DOI] [PubMed] [Google Scholar]
- Vries MC, Rodenburg RJ, Morava E, et al. Normal biochemical analysis of the oxidative phosphorylation (OXPHOS) system in a child with POLG mutations: a cautionary note. J Inherit Metab Dis. 2008;31:547–547. doi: 10.1007/s10545-008-0871-4. [DOI] [PubMed] [Google Scholar]
- Duncan AJ, Heales SJ, Mills K, Eaton S, Land JM, Hargreaves IP. Determination of coenzyme Q10 status in blood mononuclear cells, skeletal muscle, and plasma by HPLC with di-propoxy-coenzyme Q10 as an internal standard. Clin Chem. 2005;51:2380–2382. doi: 10.1373/clinchem.2005.054643. [DOI] [PubMed] [Google Scholar]
- Edvardson S, Shaag A, Kolesnikova O, et al. Deleterious mutation in the mitochondrial arginyl-transfer RNA synthetase gene is associated with pontocerebellar hypoplasia. Am J Hum Genet. 2007;81:857–862. doi: 10.1086/521227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott HR, Samuels DC, Eden JA, Relton CL, Chinnery PF. Pathogenic mitochondrial DNA mutations are common in the general population. Am J Hum Genet. 2008;83:254–260. doi: 10.1016/j.ajhg.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finsterer J, Harbo HF, Baets J, et al. EFNS guidelines on the molecular diagnosis of mitochondrial disorders. Eur J Neurol. 2009;16:1255–1264. doi: 10.1111/j.1468-1331.2009.02811.x. [DOI] [PubMed] [Google Scholar]
- Garcia-Cazorla A, Lonlay P, Rustin P, et al. Mitochondrial respiratory chain deficiencies expressing the enzymatic deficiency in the hepatic tissue: a study of 31 patients. J Pediatr. 2006;149:401–405. doi: 10.1016/j.jpeds.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Gellerich FN, Mayr JA, Reuter S, Sperl W, Zierz S. The problem of interlab variation in methods for mitochondrial disease diagnosis: enzymatic measurement of respiratory chain complexes. Mitochondrion. 2004;4:427–439. doi: 10.1016/j.mito.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Haas R, Stumpf DA, Parks JK, Eguren L. Inhibitory effects of sodium valproate on oxidative phosphorylation. Neurology. 1981;31:1473–1476. doi: 10.1212/wnl.31.11.1473. [DOI] [PubMed] [Google Scholar]
- Haas RH, Parikh S, Falk MJ, et al. Mitochondrial disease: a practical approach for primary care physicians. Pediatrics. 2007;120:1326–1333. doi: 10.1542/peds.2007-0391. [DOI] [PubMed] [Google Scholar]
- Haas RH, Parikh S, Falk MJ, et al. The in-depth evaluation of suspected mitochondrial disease. Mol Genet Metab. 2008;94:16–37. doi: 10.1016/j.ymgme.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoefs SJ, Dieteren CE, Distelmaier F, et al. NDUFA2 complex I mutation leads to Leigh disease. Am J Hum Genet. 2008;82:1306–1315. doi: 10.1016/j.ajhg.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Bellance N, Rossignol R, et al. C. elegans ATAD-3 is essential for mitochondrial activity and development. PLoS ONE. 2009;4:e7644–e7644. doi: 10.1371/journal.pone.0007644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honzik T, Wenchich L, Bohm M, et al. Activities of respiratory chain complexes and pyruvate dehydrogenase in isolated muscle mitochondria in premature neonates. Early Hum Dev. 2008;84:269–276. doi: 10.1016/j.earlhumdev.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Horvath R, Hudson G, Ferrari G, et al. Phenotypic spectrum associated with mutations of the mitochondrial polymerase gamma gene. Brain. 2006;129:1674–1684. doi: 10.1093/brain/awl088. [DOI] [PubMed] [Google Scholar]
- Huynen MA, de HM, Szklarczyk R. Mitochondrial proteome evolution and genetic disease. Biochim Biophys Acta. 2009;1792:1122–1129. doi: 10.1016/j.bbadis.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Janssen AJ, Smeitink JA, Heuvel LP. Some practical aspects of providing a diagnostic service for respiratory chain defects. Ann Clin Biochem. 2003;40:3–8. doi: 10.1258/000456303321016114. [DOI] [PubMed] [Google Scholar]
- Janssen AJ, Trijbels FJ, Sengers RC, et al. Measurement of the energy-generating capacity of human muscle mitochondria: diagnostic procedure and application to human pathology. Clin Chem. 2006;52:860–871. doi: 10.1373/clinchem.2005.062414. [DOI] [PubMed] [Google Scholar]
- Janssen AJ, Trijbels FJ, Sengers RC, et al. Spectrophotometric assay for complex I of the respiratory chain in tissue samples and cultured fibroblasts. Clin Chem. 2007;53:729–734. doi: 10.1373/clinchem.2006.078873. [DOI] [PubMed] [Google Scholar]
- Janssen AJ, Schuelke M, Smeitink JA, et al. Muscle 3243A−>G mutation load and capacity of the mitochondrial energy-generating system. Ann Neurol. 2008;63:473–481. doi: 10.1002/ana.21328. [DOI] [PubMed] [Google Scholar]
- Jonckheere AI, Hogeveen M, Nijtmans LG, et al. A novel mitochondrial ATP8 gene mutation in a patient with apical hypertrophic cardiomyopathy and neuropathy. J Med Genet. 2008;45:129–133. doi: 10.1136/jmg.2007.052084. [DOI] [PubMed] [Google Scholar]
- Jonckheere AI, Huigsloot M, Janssen AJ, Kappen AJ, Smeitink JA, Rodenburg RJ (2010) High-Throughput Assay to Measure Oxygen Consumption in Digitonin-Permeabilized Cells of Patients with Mitochondrial Disorders. Clin Chem 56:424–431 [DOI] [PubMed]
- Kavanaugh-Black A, Connolly DM, Chugani SA, Chakrabarty AM. Characterization of nucleoside-diphosphate kinase from Pseudomonas aeruginosa: complex formation with succinyl-CoA synthetase. Proc Natl Acad Sci USA. 1994;91:5883–5887. doi: 10.1073/pnas.91.13.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MP, Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- Kirby DM, Thorburn DR, Turnbull DM, Taylor RW. Biochemical assays of respiratory chain complex activity. Methods Cell Biol. 2007;80:93–119. doi: 10.1016/S0091-679X(06)80004-X. [DOI] [PubMed] [Google Scholar]
- Kirkman MA, Yu-Wai-Man P, Chinnery PF. The clinical spectrum of mitochondrial genetic disorders. Clin Med. 2008;8:601–606. doi: 10.7861/clinmedicine.8-6-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman WJ, Verkaart S, Emst-de Vries SE, et al. Mitigation of NADH: ubiquinone oxidoreductase deficiency by chronic Trolox treatment. Biochim Biophys Acta. 2008;1777:853–859. doi: 10.1016/j.bbabio.2008.03.028. [DOI] [PubMed] [Google Scholar]
- Loeffen J, Smeitink J, Triepels R, et al. The first nuclear-encoded complex I mutation in a patient with Leigh syndrome. Am J Hum Genet. 1998;63:1598–1608. doi: 10.1086/302154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffen JL, Smeitink JA, Trijbels JM, et al. Isolated complex I deficiency in children: clinical, biochemical and genetic aspects. Hum Mutat. 2000;15:123–134. doi: 10.1002/(SICI)1098-1004(200002)15:2<123::AID-HUMU1>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Lopez LC, Schuelke M, Quinzii CM, et al. Leigh syndrome with nephropathy and CoQ10 deficiency due to decaprenyl diphosphate synthase subunit 2 (PDSS2) mutations. Am J Hum Genet. 2006;79:1125–1129. doi: 10.1086/510023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel H, Szargel R, Labay V, et al. The deoxyguanosine kinase gene is mutated in individuals with depleted hepatocerebral mitochondrial DNA. Nat Genet. 2001;29:337–341. doi: 10.1038/ng746. [DOI] [PubMed] [Google Scholar]
- Mardis ER. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- Mayr JA, Merkel O, Kohlwein SD, et al. Mitochondrial phosphate-carrier deficiency: a novel disorder of oxidative phosphorylation. Am J Hum Genet. 2007;80:478–484. doi: 10.1086/511788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medja F, Allouche S, Frachon P, et al. Development and implementation of standardized respiratory chain spectrophotometric assays for clinical diagnosis. Mitochondrion. 2009;9:331–339. doi: 10.1016/j.mito.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Meisinger C, Sickmann A, Pfanner N. The mitochondrial proteome: from inventory to function. Cell. 2008;134:22–24. doi: 10.1016/j.cell.2008.06.043. [DOI] [PubMed] [Google Scholar]
- Miller C, Saada A, Shaul N, et al. Defective mitochondrial translation caused by a ribosomal protein (MRPS16) mutation. Ann Neurol. 2004;56:734–738. doi: 10.1002/ana.20282. [DOI] [PubMed] [Google Scholar]
- Molinari F, Raas-Rothschild A, Rio M, et al. Impaired mitochondrial glutamate transport in autosomal recessive neonatal myoclonic epilepsy. Am J Hum Genet. 2005;76:334–339. doi: 10.1086/427564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morava E, Heuvel L, Hol F, et al. Mitochondrial disease criteria: diagnostic applications in children. Neurology. 2006;67:1823–1826. doi: 10.1212/01.wnl.0000244435.27645.54. [DOI] [PubMed] [Google Scholar]
- Morava E, Steuerwald U, Carrozzo R, et al. Dystonia and deafness due to SUCLA2 defect; clinical course and biochemical markers in 16 children. Mitochondrion. 2009;9:438–442. doi: 10.1016/j.mito.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Niers LE, Smeitink JA, Trijbels JM, Sengers RC, Janssen AJ, Heuvel LP. Prenatal diagnosis of NADH:ubiquinone oxidoreductase deficiency. Prenat Diagn. 2001;21:871–880. doi: 10.1002/pd.162. [DOI] [PubMed] [Google Scholar]
- Niers L, Heuvel L, Trijbels F, Sengers R, Smeitink J. Prerequisites and strategies for prenatal diagnosis of respiratory chain deficiency in chorionic villi. J Inherit Metab Dis. 2003;26:647–658. doi: 10.1023/b:boli.0000005605.57420.b4. [DOI] [PubMed] [Google Scholar]
- Ostergaard E, Christensen E, Kristensen E, et al. Deficiency of the alpha subunit of succinate-coenzyme A ligase causes fatal infantile lactic acidosis with mitochondrial DNA depletion. Am J Hum Genet. 2007;81:383–387. doi: 10.1086/519222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostergaard E, Schwartz M, Batbayli M, et al. A novel missense mutation in SUCLG1 associated with mitochondrial DNA depletion, encephalomyopathic form, with methylmalonic aciduria. Eur J Pediatr. 2010;169:201–205. doi: 10.1007/s00431-009-1007-z. [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri F. Diseases caused by defects of mitochondrial carriers: a review. Biochim Biophys Acta. 2008;1777:564–578. doi: 10.1016/j.bbabio.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Phillips D, Aponte AM, French SA, Chess DJ, Balaban RS. Succinyl-CoA synthetase is a phosphate target for the activation of mitochondrial metabolism. Biochemistry. 2009;48:7140–7149. doi: 10.1021/bi900725c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S, Hargreaves I, Clayton P, Heales S. Neonatal presentation of coenzyme Q10 deficiency. J Pediatr. 2001;139:456–458. doi: 10.1067/mpd.2001.117575. [DOI] [PubMed] [Google Scholar]
- Reisch AS, Elpeleg O. Biochemical assays for mitochondrial activity: assays of TCA cycle enzymes and PDHc. Methods Cell Biol. 2007;80:199–222. doi: 10.1016/S0091-679X(06)80010-5. [DOI] [PubMed] [Google Scholar]
- Rustin P, Chretien D, Bourgeron T, et al. Assessment of the mitochondrial respiratory chain. Lancet. 1991;338:60–60. doi: 10.1016/0140-6736(91)90057-v. [DOI] [PubMed] [Google Scholar]
- Rustin P, Chretien D, Bourgeron T, et al. Biochemical and molecular investigations in respiratory chain deficiencies. Clin Chim Acta. 1994;228:35–51. doi: 10.1016/0009-8981(94)90055-8. [DOI] [PubMed] [Google Scholar]
- Saada A, Shaag A, Arnon S, et al. Antenatal mitochondrial disease caused by mitochondrial ribosomal protein (MRPS22) mutation. J Med Genet. 2007;44:784–786. doi: 10.1136/jmg.2007.053116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer AM, McFarland R, Blakely EL, et al. Prevalence of mitochondrial DNA disease in adults. Ann Neurol. 2008;63:35–39. doi: 10.1002/ana.21217. [DOI] [PubMed] [Google Scholar]
- Schagger H, Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- Scheper GC, van der KT, van Andel RJ et al (2007) Mitochondrial aspartyl-tRNA synthetase deficiency causes leukoencephalopathy with brain stem and spinal cord involvement and lactate elevation. Nat Genet 39:534–539 [DOI] [PubMed]
- Schwab MA, Sauer SW, Okun JG, et al. Secondary mitochondrial dysfunction in propionic aciduria: a pathogenic role for endogenous mitochondrial toxins. Biochem J. 2006;398:107–112. doi: 10.1042/BJ20060221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeitink JA, Elpeleg O, Antonicka H, et al. Distinct clinical phenotypes associated with a mutation in the mitochondrial translation elongation factor EFTs. Am J Hum Genet. 2006;79:869–877. doi: 10.1086/508434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperl W, Sengers RC, Trijbels JM, et al. Enzyme activities of the mitochondrial energy generating system in skeletal muscle tissue of preterm and fullterm neonates. Ann Clin Biochem. 1992;29:638–645. doi: 10.1177/000456329202900604. [DOI] [PubMed] [Google Scholar]
- Spinazzola A, Viscomi C, Fernandez-Vizarra E, et al. MPV17 encodes an inner mitochondrial membrane protein and is mutated in infantile hepatic mitochondrial DNA depletion. Nat Genet. 2006;38:570–575. doi: 10.1038/ng1765. [DOI] [PubMed] [Google Scholar]
- Taylor RW, Birch-Machin MA, Bartlett K, Turnbull DM. Succinate-cytochrome c reductase: assessment of its value in the investigation of defects of the respiratory chain. Biochim Biophys Acta. 1993;1181:261–265. doi: 10.1016/0925-4439(93)90030-5. [DOI] [PubMed] [Google Scholar]
- Taylor RW, Birch-Machin MA, Schaefer J, et al. Deficiency of complex II of the mitochondrial respiratory chain in late-onset optic atrophy and ataxia. Ann Neurol. 1996;39:224–232. doi: 10.1002/ana.410390212. [DOI] [PubMed] [Google Scholar]
- Taylor RW, Giordano C, Davidson MM, et al. A homoplasmic mitochondrial transfer ribonucleic acid mutation as a cause of maternally inherited hypertrophic cardiomyopathy. J Am Coll Cardiol. 2003;41:1786–1796. doi: 10.1016/s0735-1097(03)00300-0. [DOI] [PubMed] [Google Scholar]
- Thorburn DR, Smeitink J. Diagnosis of mitochondrial disorders: clinical and biochemical approach. J Inherit Metab Dis. 2001;24:312–316. doi: 10.1023/a:1010347808082. [DOI] [PubMed] [Google Scholar]
- Tiranti V, Viscomi C, Hildebrandt T, et al. Loss of ETHE1, a mitochondrial dioxygenase, causes fatal sulfide toxicity in ethylmalonic encephalopathy. Nat Med. 2009;15:200–205. doi: 10.1038/nm.1907. [DOI] [PubMed] [Google Scholar]
- Trijbels JM, Sengers RC, Ruitenbeek W, Fischer JC, Bakkeren JA, Janssen AJ. Disorders of the mitochondrial respiratory chain: clinical manifestations and diagnostic approach. Eur J Pediatr. 1988;148:92–97. doi: 10.1007/BF00445910. [DOI] [PubMed] [Google Scholar]
- Coster R, Smet J, George E, et al. Blue native polyacrylamide gel electrophoresis: a powerful tool in diagnosis of oxidative phosphorylation defects. Pediatr Res. 2001;50:658–665. doi: 10.1203/00006450-200111000-00020. [DOI] [PubMed] [Google Scholar]
- Heuvel L, Ruitenbeek W, Smeets R, et al. Demonstration of a new pathogenic mutation in human complex I deficiency: a 5-bp duplication in the nuclear gene encoding the 18-kD (AQDQ) subunit. Am J Hum Genet. 1998;62:262–268. doi: 10.1086/301716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuvel LP, Smeitink JA, Rodenburg RJ. Biochemical examination of fibroblasts in the diagnosis and research of oxidative phosphorylation (OXPHOS) defects. Mitochondrion. 2004;4:395–401. doi: 10.1016/j.mito.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Eijsden RG, Gerards M, Eijssen LM, et al. Chip-based mtDNA mutation screening enables fast and reliable genetic diagnosis of OXPHOS patients. Genet Med. 2006;8:620–627. doi: 10.1097/01.gim.0000237782.94878.05. [DOI] [PubMed] [Google Scholar]
- Goethem G, Luoma P, Rantamaki M, et al. POLG mutations in neurodegenerative disorders with ataxia but no muscle involvement. Neurology. 2004;63:1251–1257. doi: 10.1212/01.wnl.0000140494.58732.83. [DOI] [PubMed] [Google Scholar]
- Vasta V, Ng SB, Turner EH, Shendure J, Hahn SH. Next generation sequence analysis for mitochondrial disorders. Genome Med. 2009;1:100–100. doi: 10.1186/gm100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesela K, Hansikova H, Tesarova M, et al. Clinical, biochemical and molecular analyses of six patients with isolated cytochrome c oxidase deficiency due to mutations in the SCO2 gene. Acta Paediatr. 2004;93:1312–1317. doi: 10.1080/08035250410008761. [DOI] [PubMed] [Google Scholar]
- Walker UA, Collins S, Byrne E. Respiratory chain encephalomyopathies: a diagnostic classification. Eur Neurol. 1996;36:260–267. doi: 10.1159/000117269. [DOI] [PubMed] [Google Scholar]
- Wanders RJ, Ruiter JP, Wijburg FA. Studies on mitochondrial oxidative phosphorylation in permeabilized human skin fibroblasts: application to mitochondrial encephalomyopathies. Biochim Biophys Acta. 1993;1181:219–222. doi: 10.1016/0925-4439(93)90024-u. [DOI] [PubMed] [Google Scholar]
- Wibom R, Lasorsa FM, Tohonen V, et al. AGC1 deficiency associated with global cerebral hypomyelination. N Engl J Med. 2009;361:489–495. doi: 10.1056/NEJMoa0900591. [DOI] [PubMed] [Google Scholar]
- Will Y, Hynes J, Ogurtsov VI, Papkovsky DB. Analysis of mitochondrial function using phosphorescent oxygen-sensitive probes. Nat Protoc. 2006;1:2563–2572. doi: 10.1038/nprot.2006.351. [DOI] [PubMed] [Google Scholar]
- Willis JH, Capaldi RA, Huigsloot M, Rodenburg RJ, Smeitink J, Marusich MF. Isolated deficiencies of OXPHOS complexes I and IV are identified accurately and quickly by simple enzyme activity immunocapture assays. Biochim Biophys Acta. 2009;1787:533–538. doi: 10.1016/j.bbabio.2008.10.009. [DOI] [PubMed] [Google Scholar]
- Wittig I, Carrozzo R, Santorelli FM, Schagger H. Functional assays in high-resolution clear native gels to quantify mitochondrial complexes in human biopsies and cell lines. Electrophoresis. 2007;28:3811–3820. doi: 10.1002/elps.200700367. [DOI] [PubMed] [Google Scholar]
- Wolf NI, Smeitink JA. Mitochondrial disorders: a proposal for consensus diagnostic criteria in infants and children. Neurology. 2002;59:1402–1405. doi: 10.1212/01.wnl.0000031795.91814.d8. [DOI] [PubMed] [Google Scholar]
- Wortmann S, Rodenburg RJ, Huizing M, et al. Association of 3-methylglutaconic aciduria with sensori-neural deafness, encephalopathy, and Leigh-like syndrome (MEGDEL association) in four patients with a disorder of the oxidative phosphorylation. Mol Genet Metab. 2006;88:47–52. doi: 10.1016/j.ymgme.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Wortmann SB, Rodenburg RJ, Jonckheere A, et al. Biochemical and genetic analysis of 3-methylglutaconic aciduria type IV: a diagnostic strategy. Brain. 2009;132:136–146. doi: 10.1093/brain/awn296. [DOI] [PubMed] [Google Scholar]
- Zerbetto E, Vergani L, Dabbeni-Sala F. Quantification of muscle mitochondrial oxidative phosphorylation enzymes via histochemical staining of blue native polyacrylamide gels. Electrophoresis. 1997;18:2059–2064. doi: 10.1002/elps.1150181131. [DOI] [PubMed] [Google Scholar]
- Zeviani M, Donato S. Mitochondrial disorders. Brain. 2004;127:2153–2172. doi: 10.1093/brain/awh259. [DOI] [PubMed] [Google Scholar]
- Zinn AB, Kerr DS, Hoppel CL. Fumarase deficiency: a new cause of mitochondrial encephalomyopathy. N Engl J Med. 1986;315:469–475. doi: 10.1056/NEJM198608213150801. [DOI] [PubMed] [Google Scholar]