Abstract
The human gut microbiota harbors three main groups of H2-consuming microbes: methanogens including the dominant archaeon, Methanobrevibacter smithii, a polyphyletic group of acetogens, and sulfate-reducing bacteria. Defining their roles in the gut is important for understanding how hydrogen metabolism affects the efficiency of fermentation of dietary components. We quantified methanogens in fecal samples from 40 healthy adult female monozygotic (MZ) and 28 dizygotic (DZ) twin pairs, analyzed bacterial 16S rRNA datasets generated from their fecal samples to identify taxa that co-occur with methanogens, sequenced the genomes of 20 M. smithii strains isolated from families of MZ and DZ twins, and performed RNA-Seq of a subset of strains to identify their responses to varied formate concentrations. The concordance rate for methanogen carriage was significantly higher for MZ versus DZ twin pairs. Co-occurrence analysis revealed 22 bacterial species-level taxa positively correlated with methanogens: all but two were members of the Clostridiales, with several being, or related to, known hydrogen-producing and -consuming bacteria. The M. smithii pan-genome contains 987 genes conserved in all strains, and 1,860 variably represented genes. Strains from MZ and DZ twin pairs had a similar degree of shared genes and SNPs, and were significantly more similar than strains isolated from mothers or members of other families. The 101 adhesin-like proteins (ALPs) in the pan-genome (45 ± 6 per strain) exhibit strain-specific differences in expression and responsiveness to formate. We hypothesize that M. smithii strains use their different repertoires of ALPs to create diversity in their metabolic niches, by allowing them to establish syntrophic relationships with bacterial partners with differing metabolic capabilities and patterns of co-occurrence.
Keywords: hydrogen-consuming microbes, metagenomics, microbial genome evolution, horizontal gene tranfer
Human microbiome projects seek to determine how microbial communities are assembled, maintained, and operate within our various body habitats as a function of our different cultural and socioeconomic conditions, family structures, stages of life, genotypes, and physiologies. Culture-independent metagenomic surveys have revealed that microbial communities cluster according to body habitat but with considerable interpersonal variation in bacterial species content (1), although differences are smaller within rather than between families (2). The gut harbors our largest collection of microbes, spanning all three domains of life. Bacteria dominate, specifically members of the phyla Bacteroidetes and Firmicutes (2–5).
Monozygotic (MZ) and dizygotic (DZ) twin pairs provide an attractive study paradigm for dissecting the relative contributions of host genotype and environmental exposures to shaping the microbial and viral landscape of our gut microbiota (2, 6). To date, bacterial 16S rRNA datasets indicate that adult MZ co-twins share no more similarity in their fecal bacterial communities than DZ co-twins, suggesting that shared environmental exposures likely play key roles in determining gut microbial community composition (2).
Here, we extend these twin studies to examine Methanobrevibacter smithii, the dominant archaeon in the human gut microbiota (3). We address several general questions. First, to what extent is the representation of Archaea influenced by host genotype versus shared environmental exposures? Second, what bacterial species, if any, co-occur with this hydrogen-consuming methanogen? Third, how does the genome of M. smithii vary within a co-twin, between co-twins, and across families?
Answers to these questions may have therapeutic implications. Accumulation of hydrogen from microbial fermentation inhibits bacterial NADH dehydrogenases, reducing the yield of ATP obtained by primary fermenters and the short chain fatty acid end-products of fermentation that can be absorbed by the host (7, 8). Thus, manipulation of the abundance of hydrogen consumers or their metabolic activities could affect the efficiency of energy harvest by the host. The human gut contains three major groups of organisms capable of consuming hydrogen that could be targeted: methanogenic archaea, acetogenic bacteria, and sulfate-reducing bacteria (SRB). M. smithii is an attractive therapeutic target not only because of its prominence among human gut-associated Archaea, but because it has a lower H2-utilization threshold than acetogens and, thus, is likely to be more efficient at depleting H2 from the gut environment. Acetogens are a metabolically diverse group of microbes that are distributed among a number of bacterial phyla; many consume H2 or formate and CO2 to generate acetate and ATP via the Wood–Ljungdahl pathway. Sulfate reducers can also use H2 as an electron donor to generate hydrogen sulfide (H2S) through anaerobic sulfate respiration. Desulfovibrio piger, a member of the δ-Proteobacteria, appears to be the dominant SRB present in the human gut microbiota (9). To consider the guild of H2 consumers as a therapeutic target, we must first identify which of their genetic and metabolic features are conserved within and between individual hosts and are therefore critical for fitness in the human gut. Unfortunately, in the case of M. smithii, tractable genetic systems are not yet available for genome-wide screens of fitness determinants.
Thirty to fifty percent of humans are positive for methanogens in the gut as measured either by breath tests (10, 11) or PCR assays targeting mcrA (methyl coenzyme M reductase subunit A, conserved in the methanogenesis pathway) (12). However, whether and how host physiologic factors, such as gastrointestinal transit time (13, 14) and BMI (15, 16), influence interpersonal variation in the representation of methanogens remains unclear. Methane excretion phenotyping of 274 Australian families containing adolescent twin pairs indicated that environmental exposures play a deterministic role in methanogen carriage, with similar concordance and correlations between MZ and DZ co-twins, and less concordance between parents and their offspring (17). As in other studies (18), more females than males had positive methane breath tests. Studies of intergenerational transfer of a positive methane excretion phenotype in rats demonstrated the critical effects of environmental factors during the weaning period; however, colonization through adulthood varied between strains of rats (17), suggesting that host genetic factors affect carriage.
In this report, we have characterized the representation of M. smithii in adult rather than adolescent female MZ and DZ twin pairs living in the United States, identified bacteria that co-occur with this methanogen, compared and contrasted the genomes of 20 sequenced M. smithii isolates recovered from the frozen fecal microbiota of MZ and DZ co-twins, and used RNA-Seq to perform whole genome transcriptional profiling of a subset of sequenced isolates under different growth conditions. The results provide an expanded view of the diversity and adaptations of this dominant archaeon to life in the human gut.
Results and Discussion
MZ Twins Have Higher Concordance for Gut Methanogens than DZ Twins.
We used a quantitative PCR (qPCR) assay of the mcrA gene to measure methanogens present in single fecal samples collected from 40 female MZ and 28 adult female DZ twin pairs (age 21–31 y). All were born in Missouri, although at the time they provided samples, only 29% were living in the same home and some lived >800 km apart (2). Based on a health questionnaire, all were healthy and none had a history of gastrointestinal disease including irritable bowel syndrome. Sixty-one percent were obese (BMI ≥ 30) and 7% overweight (BMI 25–30) at the time of sampling (2).
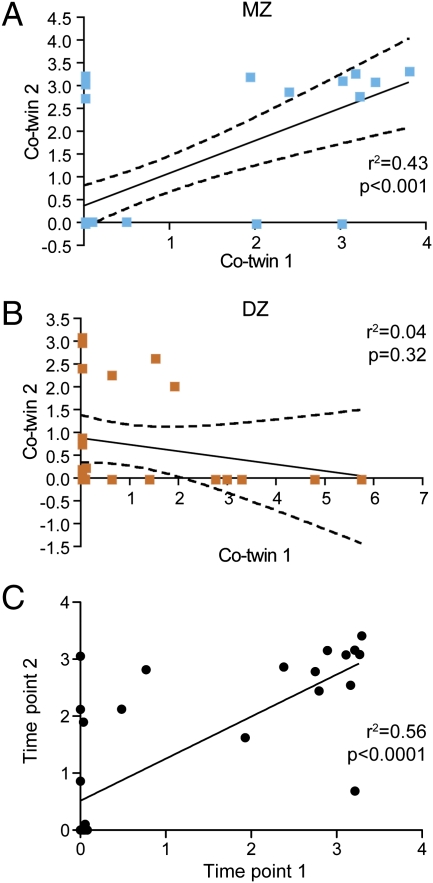
Thirty-two of the 136 individuals (23%) had levels of methanogens above our threshold for confidently calling the fecal sample “positive” (i.e., ≥4 × 107 genome equivalents per mg of total fecal DNA), and this proportion did not vary significantly by zygosity group (P = 0.59). The MZ twin pair concordance rate for carriage of methanogens was 74%, a value significantly higher than the DZ pair concordance rate (15%; P = 0.009 by Breslow-Day test). In addition, there was a significantly higher degree of correlation of methanogen levels between MZ pairs by linear regression (r2 = 0.43, P < 0.0001) than DZ pairs (r2 = 0.04, P = 0.32), (Fig. 1 A and B). Fecal samples were also collected from 23 of the MZ twin pairs and 12 of the DZ pairs 2 mo after the initial time point. Linear regression showed that time point 1 and time point 2 samples were highly correlated for both the presence of methanogens (r2 = 0.54, P < 0.0001; Fig. 1C) and their levels. Neither carriage nor levels of methanogens was significantly correlated with being overweight or obese in this study population (P = 0.37 and 0.38, respectively).
Fig. 1.
Correlation of methanogen levels in the fecal microbiota of MZ and DZ co-twins. The presence and levels of fecal methanogens were defined by qPCR assay that targeted the mcrA gene in samples obtained from MZ twin pairs (A) (n = 40) and DZ twin pairs (B) (n = 28). Dashed lines represent 95% confidence intervals for linear regression. (C) Correlation between mcrA levels in fecal samples collected at two time points per individual (2-mo interval between sampling). All axes in A–C are log10 (genome equivalents per ng total DNA +1).
Thirteen samples from the initial timepoint representing 4 MZ twin pairs, 1 DZ twin pair, plus 3 other unrelated individuals that were positive for mcrA were chosen for sequencing of amplicons generated by using the mcrA primers and previously described archaeal 16S rRNA primers (n = 5–10 amplicon subclones/primer set/fecal DNA sample). In 12 of the 13 samples, M. smithii was the only sequence detected by mcrA or 16S rRNA-directed PCR. In one MZ co-twin (TS17 in Dataset S1, Table S1), 2 of 6 16S rRNA amplicons and 2 of 8 mcrA amplicons matched to Methanosphaera stadtmanae, a mesophilic euryarchaeota known to be present in the gut microbiota of some humans (19); the remaining amplicons generated from her fecal DNA matched to M. smithii. Her co-twin (TS16) had no detectable methanogens.
We also examined fecal samples from 51 mothers in this study for presence of methanogens and found a similar overall degree of methanogen carriage in this population as found in their daughters (31% and 25%, respectively). Concordance for carriage of methanogens between mother and daughter (i.e., the probability that the daughter of a methanogen carrier was also a carrier, 32%) was nonsignificant (P = 0.33).
Co-Occurrence Between M. smithii and Bacterial Taxa.
Our qPCR results suggest that host genetic factors, including factors that influence the representation of potential syntrophic partners, may play a role in carriage of methanogens. In contrast, the study of Florin et al. (17), which used methane breath tests, showed no significant differences in concordance between young adolescent Australian MZ and DZ twin pairs. The difference could be explained if environmental factors play a dominant role in determining whether methanogens are acquired early in life, whereas persistent carriage in later life is determined by a variety of host factors. Such factors range from human genotype to the presence or absence of bacterial taxa that can collaborate or compete with the methanogens.
A role for host factors in determining carriage of methanogens is supported by previous studies of nonhuman primates. Methanogens were present in the gut microbiota of some primate phylogenetic lineages but not others; however, these patterns did not follow any identifiable features of gut physiology or morphology, nor behavior or diet (20). Another study that examined the distribution of methanogens within the guts of 253 vertebrate species found “methanogenic branches” of the host phylogenetic tree [i.e., branches containing ruminants (bovidae, cervidae, giraffidae) and “nonmethanogenic” branches (felidae, canidae, and ursidae)]. As with the primate study, the methane-producing groups could not be distinguished from the methane-negative groups based on their diets or features of their gut structure/physiology (21).
To understand whether methanogen carriage might be determined, in part, by the presence or absence of bacterial taxa that can collaborate or compete with the methanogens, we investigated co-occurrence patterns between methanogens and sulfate-reducing bacteria (SRB). SRB, which can use H2 as an electron donor to generate hydrogen sulfide (H2S) through anaerobic sulfate respiration, may show positive associations with methanogens if a hydrogen economy is more important in some individuals than others, or negative associations due to competition for H2. Positive associations between SRB and methanogens might also occur because of syntrophy, because some methanogens and SRB can grow syntrophically on lactate, with the methanogen removing H2 generated by the SRB (22, 23). Therefore, we determined whether SRB and methanogens had nonrandom codistribution patterns by SRB-directed qPCR assays of 87 fecal samples from the MZ and DZ twin pairs. The aps gene encodes adenosine-5′-phosphosulfate reductase, a key enzyme that catalyzes activation and then reduction of sulfate to sulfite (24). We chose aps as a target for a qPCR assay that used previously described and validated primers (25). Forty-five percent of the samples were positive for SRB (threshold of detection defined as ≈4 × 107 genome equivalents per mg of fecal DNA). The concordance rate for sulfate reducers was not significant for either MZ or DZ co-twins (31% and 27%, Dataset S1, Table S1). A logistic regression was performed to determine whether a higher level of mcrA is predictive of the presence of aps or vice versa. No statistically significant relationship was identified in either comparison (P = 0.10 and 0.07).
We also performed a general search for bacterial Operational Taxonomic Units (OTUs) that had positive or negative associations with M. smithii, using sequences generated from multiplex pyrosequencing of the V2 variable region of bacterial 16S rRNA genes from these same fecal samples (2). The raw sequences from this prior study were now processed by using the PyroNoise algorithm to remove sequencing noise (26), as implemented in QIIME (27). Using UCLUST (28), the denoised sequences were further divided into OTUs that each shared ≥96% nucleotide sequence identity (a value slightly more permissive than the 97% ID threshold typically used to denote a microbial species). The most abundant sequence within each of the resulting 12,833 OTUs was then selected as a representative of that OTU. Because some of the individuals in the study were sampled multiple times, we randomly selected one sample per individual. For each of the 607 OTUs that were found in at least 10 of the samples for which we had mcrA qPCR data, an ANOVA was performed to determine whether the OTU relative abundance was significantly different in methanogen-positive and -negative individuals. We also checked for associated presence/absence patterns by using the G-test of independence (an OTU was scored as present if it was observed one or more times). The resulting P values were corrected for multiple comparisons by using the Bonferroni correction (multiplied by 607; the number of comparisons) and the false discovery rate (FDR) method (multiplied by the number of comparisons divided by the P value rank).
Twenty-two OTUs had significantly different relative abundances in mcrA-positive versus negative individuals (P < 0.05 using ANOVA with the FDR correction). Of these 22 OTUs, 21 were more abundant in samples where methanogens were present, whereas one OTU was less abundant. The G-test identified five significant OTUs (P < 0.05 with FDR correction), and 4 of these 5 were also significant as judged by ANOVA. All G-test–identified associations were positive. Thus, the two statistical tests together identified 22 positively associated OTUs (Dataset S1, Table S2) and one negatively associated OTU.
To investigate the phylogenetic relationships of these OTUs to each other, and to bacterial isolates and lineages with known biological properties, we used parsimony insertion to add a representative sequence for each significant OTU into the Greengenes coreset tree (29) in the Arb software package (30). Because the closest relatives of the OTUs were mostly from other culture-independent metagenomic studies, we also inserted 16S rRNA sequences into the tree that were from well-characterized bacteria, including 16S rRNAs from fully sequenced genomes deposited in KEGG or sequenced through the Human Gut Microbiome Initiative (HGMI; http://genome.wustl.edu/genomes/list/human_gut_microbiome/), and 16S rRNA sequences from related organisms with known properties that were identified by using BLAST searches against the National Center for Biotechnology Information nonredundant database. To look for evidence of whether relatives of the OTUs were capable of growing in pure culture, we also BLASTed the 16S rRNA sequences against sequences in the RDP (31) that were marked as being from cultured bacterial isolates.
Remarkably, 20 of the 22 positively associated OTUs were members of the class Clostridiales (Firmicutes phylum). These 20 OTUs binned into five broad groups that were scattered throughout the class, including members of the three main clusters found in the human gut (clusters I, IV, and XIVa).
The group most positively associated with M. smithii was a lineage within Clostridia cluster IV that contains members of the genera Oscillospira and Sporobacter (Dataset S1, Table S2; note that this group had the four most significant OTUs according to the ANOVA test). Two of these OTUs are highly related to Oscillospira guilliermondii, an as yet uncultured, large, and morphologically conspicuous organism found in ruminants (32, 33). The most closely related cultured isolate that we could find for any of these OTUs is Sporobacter termitidis, a hydrogen-consuming acetogen from the termite gut (34).
Two of the positively associated OTUs are members of Clostridia cluster XIVa. The closest isolate with a sequenced genome was Blautia hydrogenotrophica, a hydrogen-consuming homoacetogen from the human gut, although the percent identity across the lanemasked V2 region was low (89–93%) and more closely related organisms to B. hydrogenotrophica are known not to be acetogens. Whether the Sporobacter and B. hydrogenotrophica-related OTUs are acetogens cannot be determined by using 16S rRNA sequences alone, because acetogenesis is only inconsistently associated with 16S rRNA-defined phylotypes (35). However, the relationship suggests that some OTUs may co-occur with methanogens because they are homoacetogens and have a shared preference for hydrogen. Nonetheless, the OTU most related to B. hydrogenotrophica in this analysis (99%ID) did not show significant co-occurrence with M. smithii (uncorrected P value = 0.38), indicating that not all homoacetogens in the human co-occur with M. smithii because of this preference for hydrogen.
Because members of the SRB can produce and consume H2, we were specifically interested in OTUs in the dataset that were in this group. Eighty-two of 281 fecal samples (29%) from the 16S rRNA analysis of these twin pairs (including additional fecal samples for which we did not obtain mcrA data) (2) had OTUs that were within the SRB clade (Fig. S1B). The actual prevalence of SRB is likely higher, because the samples were not exhaustively sequenced. Phylogenetic comparison indicated that these OTUs represented Desulfovibrio piger in 41 (14.6%) of the samples, Desulfovibrio desulfuricans in 10 samples (3.6%), and an additional taxon (1908) in 38 samples (13.5%) that was only distantly related to cultured isolates (Dataset S1, Table S2 and Fig. S1). Although significant associations were not detected with the SRB-specific qPCR, OTU 1908 showed a significantly positive association with methanogens (Dataset S1, Table S2). The abundant OTU representing D. piger (OTU 12050) did not have statistically significant co-occurrence with methanogens (Fig. S1), and the three different types of SRB did not significantly co-occur with each other. The differing distribution patterns of the three different SRB species, coupled with the smaller number of fecal samples for which we had aps compared with mcrA qPCR data, likely contributed to our inability to detect a significant association between methanogens and SRB with the aps qPCR assay.
The concentration of H2 in the gut lumen can vary over a wide range in healthy individuals (from 0.17% to 49% in a study of 11 subjects; ref. 36). Levels of H2 in the distal gut reflect the dynamic interplay between microbial production and consumption. One of the co-occurring groups within the Clostridiales may produce abundant amounts of hydrogen. Specifically, two of the positively associated OTUs in the Clostridiales family mapped to a clade that included isolate Rennanqilyf3, which was recovered from activated sludge by using a procedure designed to retrieve bacteria with particularly high yields of hydrogen (37). This isolate performs ethanol-type fermentation with glucose as an optimal carbon source for hydrogen production; however, its hydrogen production capacity varies with hydrogen concentration and pH. Thus, methanogen (M. smithii) abundance may be in part regulated by the presence of bacterial lineages that are efficient hydrogen producers. To our knowledge, no cultured isolates are available for members of this lineage from the gut.
Some of the OTUs that are positively associated with methanogens are quite distant from any cultured relatives (ribotypes): This observation is intriguing, because it suggests that syntrophic relationships may inhibit them from growing in monoculture. For example, four OTUs grouped in a clade of the Clostridiales family that is dominated by relatives identified in culture-independent studies of cellulose-degrading gut environments where methanogens also reside (e.g., termite gut and cow rumen) (Gut Clone Group; Dataset S1, Table S2 and Fig. S1A). The closest organism with a sequenced genome was only very distantly related, with a 78–86%ID over the lanemasked V2 region of rRNA. A BLAST search against the cultured component of the RDP revealed one successful attempt to culture a relative of one of these four OTUs (95%ID) from the forestomach of the kangaroo (38). However, this cultured isolate was much more distant from the other three co-occurring OTUs in this clade, and there are no reported cultured relatives for any of these four OTUs from the human gut. Three co-occurring OTUs fell within the Catabacter lineage. The closest cultured isolate, Catabacter sp. YIT12065, is only 82–92% identical to these co-occurring OTUs; very little is known about this isolate's biology. The presence of obligate syntrophs for methanogens in the human gut would not be surprising, because they are known to exist in other environments, such as sludge (39, 40).
Unfortunately, the lack of cultured relatives for these OTUs limits our ability to more fully interpret the co-occurrence results, because we lack knowledge about their biological properties. Targeted attempts to culture gut bacteria in the presence of M. smithii as well as targeted attempts to obtain and sequence their genomes from mixed populations should help to elucidate their functional relationships with human gut methanogens.
Analysis of the Pan-Genome of M. smithii.
We reasoned that one approach for further characterizing factors that affect M. smithii colonization of the human gut would be to develop a method for isolating strains from frozen fecal samples obtained from twins and their mothers, sequencing their genomes, and performing RNA-Seq to evaluate strain-level variations in patterns of gene expression during growth under varying levels of hydrogen and formate.
The method we developed for recovering M. smithii from frozen fecal samples is described in SI Methods. A total of 20 strains were isolated from two families: one consisting of a MZ twin pair and their mother and the other a DZ twin pair and their mother (n = 2–5 strains isolated and sequenced per individual). Deep draft genome assemblies were generated by using reads produced by Illumina GA-IIx and 454 sequencers. Dataset S1, Table S3 describes the details of genome coverage and of the assembly statistics. Assembled genomes were aligned by using Mauve (41), which iteratively reordered contigs based on the finished genome sequence of the M. smithii type strain PS (42). Dataset S1, Table S3 also provides information about previously generated, deep draft assemblies of the genomes of two other M. smithii type strains obtained from culture collections (42).
On average, any two strains shared 92.96 ± 6.5% of their single nucleotide polymorphisms (SNPs) [129,112 ± 6,322 (mean ± SD)]. A binary table of the presence or absence of a SNP was subsequently generated, a distance matrix was calculated, and a principal components analysis (PCA) was performed (Fig. S2 A and C). The PCA showed that strains from the same individual and strains from co-twins clustered together. Both MZ and DZ co-twins shared significantly more SNPs in their strains than with strains from their mothers or unrelated individuals (Fig. S2B).
Genes were identified by using Glimmer (v3.02) trained on contigs >500 bp in each of the 20 sequenced M. smithii isolate genomes, plus the PS type strain and the two other M. smithii isolates we had sequenced. Genes in all 23 genomes were binned by using the program CD-HIT and its default parameters (>90% nucleotide sequence identity over of the length of the shorter gene in each pairwise comparison; Fig. S3) into “operational gene units” (OGUs), a term we use in a way that is analogous to OTUs. If any predicted gene from an assembled genome was present in a given OGU bin, that OGU was called “present” within that genome (43). Functions were assigned to predicted proteins encoded by each gene by using the KEGG and STRING databases; Pfam and TIGRFAM annotations were also made. Note that all predicted protein-coding sequences <300 nt were filtered out and not considered in the analyses reported below.
Rarefaction analysis to determine the rate at which sequencing the genes of new strains revealed new OGUs showed that the number of new or unique OGUs identified begins to plateau by the time ≈6 strains were sequenced (≈10,000 genes) (Fig. S4 A and B). A total of 987 OGUs were present in all 23 strains (34.7% of 2,847 identified OGUs), whereas 1,532 (53.8%) were found in more than one strain but not all, and 328 (11.5%) in only a single strain (Fig. S3 A and B).
PCA of OGU assignments showed clustering of strains based on family of origin: Strains from MZ family members (TS94-96) generally clustered together, whereas strains from the DZ family (TS145-147) split into two groups (Fig. S3C). Further pairwise comparisons of the degree of sharing of OGUs in strains showed that strains within an individual and within MZ and DZ co-twins shared significantly more OGUs than strains from the co-twin's mother or from unrelated individuals. Moreover, the degree of sharing of OGUs was not significantly different between MZ and DZ twin pairs (Fig. S3D). As noted above, MZ twins have greater concordance for carriage and levels of methanogens in their fecal microbiota than DZ twins. The fact that the sequenced strains are no more similar between MZ co-twins than DZ twins suggests that although shared environmental exposures to methanogens direct which strains are found in an individual's gut, long-term persistence is influenced by a combination of host and microbial genetic factors.
We used KEGG to assign enzyme commission (EC) numbers to genes in all of the isolates’ genomes. A total of 412 ECs were identified: 349 were shared by all strains, 63 were variably represented, and 18 had significant differences in their representation between strains as judged by binomial test (Fig. S5D). These discriminatory ECs include (i) several restriction enzymes, (ii) two peptidases [a serine protease known as Do, HtrA, or DegP (44) that may protect against heat-stress and unfolded proteins and endopeptidase La (45)], both of which may be related to quality control in protein folding, and (iii) tRNA-guanine transglycosylases (involved in the anti-codon modification of tRNAs specific for Asn, Asp, His, and Tyr) (Fig. S5 B–D).
Genes assigned to COG M (cell envelope biogenesis/outer membrane) were prominently represented in the variable component of the pan-genome (Fig. S5A). Variability in surface proteins may directly impact the fitness of M. smithii strains in vivo, including their ability to adhere to host structures, or to interact with syntrophic partners. For example, all of the M. smithii strains contain the six genes involved in synthesis of pseudaminic acid structures related to sialic acid molecules expressed on host cell surfaces. The resulting surface epitopes are thought to play a role in the adaptation of M. smithii to the gut environment by mimicking the sialic acids that decorate the surfaces of host epithelial cells (46). Adhesin-like proteins (ALPs) are a novel class of proteins with homology to bacterial adhesins that were first identified in the M. smithii type strain. They are also hypothesized to play a role in adaptation to the gut environment (42). The 23 sequenced strains contain a total of 101 ALP OGUs (average 45 ± 6 ALP genes per strain): Only six were present in all strains. ALP sequences are quite divergent in terms of their domain structure: e.g., many have intimin domains, which in Escherichia coli mediate binding to intestinal epithelial cells; others have pectate lyase domains and/or parallel β-helix repeats that are often found in enzymes with polysaccharide substrates.
To better understand genomic differences among M. smithii strains, we searched the M. smithii pan-genome for evidence of horizontal gene transfer (HGT) (see SI Methods for details of compositional- and phylogenetic-based analyses of HGT). The results, described in SI Results and summarized in Dataset S1, Table S4C show that HGT has contributed to both the core and variable elements of the M. smithii pan-genome. They include core genes involved in methanogenesis and folate biosynthesis; e.g., both compositional- and phylogenetic-based methods revealed transfer of genes encoding THMP methyltransferase C subunit (EC 2.1.1.86), formate dehydrogenase (EC 1.2.1.2), and formylmethanofuran dehydrogenase subunit F (E.C. 1.2.99.5) (Dataset S1, Table S4B). Note that the early steps in synthesis of methanopterin, a C1 carrier coenzyme involved in the methanogenesis pathway (Fig. S6), are the same as those used for generation of folate (Dataset S1, Table S4B). In addition, between 52% and 65% ALPs show evidence of transfer: Large-scale HGT of ALPs would be consistent with their variability among strains (Dataset S1, Table S6A).
Expression Profiling of M. smithii Strains by RNA-Seq.
We used RNA-Seq to profile the transcriptomes of five of the M. smithii isolates: One from each member of the MZ family, one from each of the DZ co-twins, plus the PS type strain. The five strains from the two families were chosen because SNP, OGU, and EC analyses indicated that these isolates were representative of the strains from their human hosts, and because they exhibited consistent patterns of growth on MBC medium containing 2.8 or 44.1 mM formate, a substrate for the first enzyme involved in the methanogenesis pathway, formate dehydrogenase (EC 1.2.1.2 in Fig. S6B). Triplicate cultures were grown to midlog phase in medium with either low or high formate concentrations under an atmosphere that contained 80% hydrogen. Total RNA was extracted, structural RNAs were depleted (SI Methods), and double-stranded cDNA was synthesized and sequenced with an Illumina GA-IIx instrument (36-nt reads; 3–4 million reads per sample, with each biological triplicate sequenced twice as technical replicates). Reads were normalized to reads per kilobase per million (RPKM) and mapped back to each strain's own reference genome. At midlog phase, the number of protein-coding genes with ≥10 mapped mRNA-derived reads varied from 1,594 to 1,782 (89–97% of all CDS) among the 5 strains (Dataset S1, Table S5A). When we compared the 987 OGUs that comprise the conserved core of the M. smithii pan-genome to 31 sequenced methanogens associated with the human gut (M. stadmanae), cow rumen (M. ruminantium) or various environmental habitats, 55 OGUs were identified as unique to M. smithii (Blastp threshold E < 10−10), of which 42 encoded predicted conserved hypothetical or hypothetical proteins (Dataset S1, Table S5C). At the depth of sequencing achieved, RNA-Seq indicated that 34 of these 42 hypothetical genes were expressed in midlog phase in the PS type strain (Dataset S1, Table S5C).
We subsequently compared the phenotypes of strains based on normalized expression of each gene encoding each EC. Examining the gene expression data across functional groups allowed us to compare strains: The results revealed that no gene family was consistently regulated by formate across all strains. To identify genes significantly regulated by formate in each strain, we first analyzed normalized reads with CyberT. We used two criteria for determining significance in regulation: a posterior probability of differential expression (PPDE) threshold ≥0.97, and a ≥2-fold difference in expression (either direction) when a given strain was incubated in low versus high levels of formate (Dataset S1, Table S7).
All of the genes in the methanogenesis pathway illustrated in Fig. S6B were expressed in all six strains. Nonetheless, several of the genes in this pathway exhibited strain-specific differences in their levels of expression including EC 1.5.99.9 (F420-dependent methylene tetrahydromethanopterin dehydrogenase) and EC 1.5.99.11 (5,10-methylenetetrahydromethanopterin reductase). Cobalt, an important cofactor for some of the enzymes in the methanogenesis pathway, is translocated by an ABC transporter: Components of the transporter exhibited formate-responsive behavior in the PS type strain and in the strain from one of the DZ co-twins (TS145) but not in the strains from her sister or mother (Dataset S1, Table S7).
Looking beyond the methanogenesis pathway, none of the genes encoding ECs in the M. smithii pan-genome satisfied our criteria for being responsive to differences in formate levels in the medium at midlog phase in all strains. However, as with components of the methanogenesis pathway, some exhibited strain-specific differences in formate sensitivity e.g., in strain METSMITS145B (from DZ co-twin 1) genes encoding the subunits of MtrH (EC 2.1.1.86; tetrahydromethanopterin S-methyltransferase) were up-regulated in high formate, whereas in strain METSMITS146E (from the sister of DZ co-twin 1) they were down-regulated (see Dataset S1, Table S7 for additional examples).
M. smithii uses ammonia as a nitrogen source via an energy-dependent glutamine synthetase-glutamate synthase pathway, which has high affinity for ammonia, and a ATP-independent pathway with lower affinity (Fig. 2A). Both pathways are expressed in all strains, with 0.4–1.21% of reads mapping to enzymes involved in assimilation of ammonia. The energy-dependent GlnA pathway is generally expressed at a much higher level than the low affinity pathway, although strain-specific differences in levels expression were noted. With few exceptions, such as the genes encoding EC 1.4.1.4 and EC 1.4.1.13 in strains METSMITS145B and METSMITS96A, components of both pathways failed to exhibit a significant difference in their levels of expression in any of the strains as a function of formate concentration. Another exception was the ammonium transporter (AmtB) (Fig. 2 B and C and Dataset S1, Table S7).
Fig. 2.
Normalized RNA-Seq reads assigned to the gene encoding an ammonium transporter (AmtB) and ECs involved in ammonia assimilation. (A) Overview of the two pathways of M. smithii for assimilating ammonia: The energy-dependent glutamine synthetase-glutamate synthase pathway has high affinity for ammonia (red arrow); an ATP-independent pathway has lower affinity (orange). (B) Strain-specific differences in the relative expression of components of the high affinity Gln pathway and the energy-independent low affinity pathway for ammonia assimilation. Mean values ± SEM are plotted. Colors represent components of the two pathways shown in A; color codes are coordinated between A and B. (C) Strain-specific differences in levels of expression of amtB. P < 0.0001 by one-way ANOVA.
Using our threshold criteria for formate-responsive expression, four of the six strains were defined as having genes that were sensitive to levels of this compound. Dataset S1, Table S7 lists the 9 genes present in type strain PS, the 340 genes in the strain recovered from the mother of the DZ co-twins (TS145), the 23 genes in the strain isolated from one of her daughters (TS146), and the 81 genes in the strain from the mother of the MZ twins (TS96). Intriguingly, no genes were identified in strains from MZ twins of this mother (TS94, TS95) that exhibited significant formate responsiveness. The core component of M. smithii’s pan-genome contained no genes that met our criteria for formate-responsive behavior in every isolate.
The utility of using formate to identify strain-specific phenotypes is best illustrated by ALPs. As noted above, each sequenced strain contained a distinctive repertoire of genes encoding ALPs, with only 6 ALP OGUs shared by all isolates. ALP OGUs 112, 208, 412, and 827 are encoded by genes present in 4–6 of the strains: None of the genes are formate-responsive but members of each OGU exhibit strain-specific differences in their levels of expression (levels of expression are also notably different between ALP OGUs). OGUs 18, 37, 133, and 226 show strain-specific differences in their representation, strain-specific differences in their levels of expression, plus within-OGU differences in their formate sensitivity (Fig. 3).
Fig. 3.
Differential expression of M. smithii adhesin-like proteins (ALPs). Members of selected ALP OGUs with strain-specific differences in their expression profiles (A) and strain-specific, as well as OGU-associated, differences in their sensitivity to levels of formate during midlog phase growth (B). OGUs 112, 412, 827, and 208 exhibit strain-specific differences in their expression irrespective of formate concentration (one-way ANOVA, P < 0.0001), whereas OGUs 226, 287, 18, 133, and 37 contain at least one representative that is significantly regulated by formate concentration. Mean values ± SEM are plotted (n = 6 replicates per condition). * indicates a ≥2-fold difference, PPDE ≥ 0.97 (Dataset S1, Table S7).
Prospectus.
These results lead us to hypothesize that M. smithii strains use their different repertoires of ALPs and the different sensitivities of ALP genes to formate to create diversity in their physical locations and/or their metabolic niches within the gut. Stated another way, these variations in expressed ALP repertoires could have important effects on the ability of different strains to establish syntrophic relationships with bacterial partners that have different abilities to generate formate or other substrates, or that have differing patterns of co-occurrence within an individual over time and between individuals. To further explore this notion, it will be important to define the structures of representative members of different ALP clusters through an M. smithii-directed structural genomics effort: Selection of ALPs could be guided by a number of criteria, including their strain distribution and their patterns of expression, both in vitro in monoculture in the presence of a variety of potential substrates for their metabolic networks, and in vivo in gnotobiotic mice containing various collections of sequenced M. smithii isolates and available cultured co-occurring bacterial taxa. The interactions between isolates and co-occurring bacterial species can also be explored in vitro if cocolonization of gnotobiotic mice proves to be problematic either because of difficulty in identifying suitable host diets or strains that are fit in the mouse gut (e.g., we have not yet been able to achieve persistent colonization of gnotobiotic mice with any of the five strains characterized in vitro by RNA-Seq after inoculating all of them together with a consortium of human gut-derived members of the Firmicutes, Bacteroidetes, and Proteobacteria that include saccharolytic bacteria and hydrogen producers and consumers). A complementary approach will be to select taxa for these in vitro and in vivo studies by predicting potential syntrophic relationships through in silico metabolic reconstructions of the metabolic networks of sequenced co-occurring species and M. smithii isolates, using methods described by Borenstein et al. (47).
Methods
Genome Sequencing.
M. smithii strains are isolated and grown by using the procedure detailed in SI Methods. Genomic DNA was sequenced with an Illumina Genome Analyzer IIx instrument (36 base read lengths; 3.5–29 million reads per strain), and a 454 pyrosequencer (Titanium chemistry; 27,844–449,545 reads per strain). Reads were assembled using Velvet (48) for Illumina reads and Newbler v2.3 (Roche) for 454 reads. Hybrid assemblies were generated by using AMOS minimus2 (http://sourceforge.net/apps/mediawiki/amos/index.php?title=Minimus2), producing draft assemblies with on average 55 contigs, N50 contig lengths of 103,633 nucleotides, and total genome size of 1.9 Mb (Dataset S1, Table S3). Procedures used for genome annotation are described in SI Methods.
Other Methods.
Details of the experimental and computational approaches used for qPCR, co-occurrence, comparative genomic, HGT (Fig. S7), and microbial RNA-Seq analyses, plus how M. smithii RNA-Seq datasets compare with custom M. smithii GeneChip datasets, are provided in SI Methods and Figs. S8 and S9. Analyses of familial concordance or correlation for methanogen carriage or levels, and of their associations with overweight/obesity, were conducted by using logistic or linear regression, a robust variance estimator to adjust for the nonindependence of observations on family members.
Supplementary Material
Acknowledgments
We thank Sabrina Wagoner and Jill Manchester for superb technical assistance, Daniela Puiu and Steven Salzberg for generous help with genome assembly software, Stacey Marion and Deborah Hopper for assistance in obtaining fecal samples, plus Nicholas Griffin, Jeremiah Faith, Nathan McNulty, Ansel Hsiao, and Alejandro Reyes for many very helpful suggestions during the course of the study. This work was supported in part by National Institutes of Health Grants DK78669, DK30292, DK70977, and AA09022, The Crohn's and Colitis Foundation of America, and Howard Hughes Medical Institute. E.E.H. is a member of Washington University's Medical Scientist Training Program (NIH T32 GM07200-31).
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Microbes and Health” held November 2–3, 2009, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS Web site at http://www.nasonline.org/SACKLER_Microbes_and_Health.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported for M. smithii strains in this paper have been deposited in the GenBank database [accession nos. AEKU00000000 (for strain TS145A); AELL00000000 (TS145B); AELM00000000 (TS146A); AELN00000000 (TS146B); AELO00000000 (TS146C); AELP00000000 (TS146D); AELQ00000000 (TS146E); AELR00000000 (TS147A); AELS00000000 (TS147B); AELT00000000 (TS147C); AELU00000000 (TS94A); AELV00000000 (TS94B); AELW00000000 (TS94C); AELX00000000 (TS95A); AELY00000000 (TS95B); AELZ00000000 (TS95C); AEMA00000000 (TS95D); AEMB00000000 (TS96A); AEMC00000000 (TS96B); and AEMD0000000 (TS96C)]. RNA-Seq and GeneChip data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo [accession nos. GSE25408 (RNA-Seq) and GSE25535 (GeneChip)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000071108/-/DCSupplemental.
References
- 1.Costello EK, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qin J, et al. MetaHIT Consortium A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reyes A, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolin MJ, Miller TL. Interactions of microbial populations in cellulose fermentation. Fed Proc. 1983;42:109–113. [PubMed] [Google Scholar]
- 8.McNeil NI. The contribution of the large intestine to energy supplies in man. Am J Clin Nutr. 1984;39:338–342. doi: 10.1093/ajcn/39.2.338. [DOI] [PubMed] [Google Scholar]
- 9.Scanlan PD, Shanahan F, Marchesi JR. Culture-independent analysis of desulfovibrios in the human distal colon of healthy, colorectal cancer and polypectomized individuals. FEMS Microbiol Ecol. 2009;69:213–221. doi: 10.1111/j.1574-6941.2009.00709.x. [DOI] [PubMed] [Google Scholar]
- 10.Bond JH, Jr, Engel RR, Levitt MD. Factors influencing pulmonary methane excretion in man. An indirect method of studying the in situ metabolism of the methane-producing colonic bacteria. J Exp Med. 1971;133:572–588. doi: 10.1084/jem.133.3.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levitt MD, Furne JK, Kuskowski M, Ruddy J. Stability of human methanogenic flora over 35 years and a review of insights obtained from breath methane measurements. Clin Gastroenterol Hepatol. 2006;4:123–129. doi: 10.1016/j.cgh.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 12.Scanlan PD, Shanahan F, Marchesi JR. Human methanogen diversity and incidence in healthy and diseased colonic groups using mcrA gene analysis. BMC Microbiol. 2008;8:79. doi: 10.1186/1471-2180-8-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Attaluri A, Jackson M, Valestin J, Rao SSC. Methanogenic flora is associated with altered colonic transit but not stool characteristics in constipation without IBS. Am J Gastroenterol. 2010;105:1407–1411. doi: 10.1038/ajg.2009.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pimentel M, et al. Methane, a gas produced by enteric bacteria, slows intestinal transit and augments small intestinal contractile activity. Am J Physiol Gastrointest Liver Physiol. 2006;290:G1089–G1095. doi: 10.1152/ajpgi.00574.2004. [DOI] [PubMed] [Google Scholar]
- 15.Armougom F, Henry M, Vialettes B, Raccah D, Raoult D. Monitoring bacterial community of human gut microbiota reveals an increase in Lactobacillus in obese patients and methanogens in anorexic patients. PLoS ONE. 2009;4:e7125. doi: 10.1371/journal.pone.0007125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H, et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci USA. 2009;106:2365–2370. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Florin TH, Zhu G, Kirk KM, Martin NG. Shared and unique environmental factors determine the ecology of methanogens in humans and rats. Am J Gastroenterol. 2000;95:2872–2879. doi: 10.1111/j.1572-0241.2000.02319.x. [DOI] [PubMed] [Google Scholar]
- 18.Pitt P, de Bruijn KM, Beeching MF, Goldberg E, Blendis LM. Studies on breath methane: The effect of ethnic origins and lactulose. Gut. 1980;21:951–954. doi: 10.1136/gut.21.11.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fricke WF, et al. The genome sequence of Methanosphaera stadtmanae reveals why this human intestinal archaeon is restricted to methanol and H2 for methane formation and ATP synthesis. J Bacteriol. 2006;188:642–658. doi: 10.1128/JB.188.2.642-658.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hackstein JHP, Van Alen TA, Op Den Camp H, Smits A, Mariman E. Intestinal methanogenesis in primates—a genetic and evolutionary approach. Dtsch Tierarztl Wochenschr. 1995;102:152–154. [PubMed] [Google Scholar]
- 21.Hackstein JHP, et al. Fecal methanogens and vertebrate evolution. Evolution. 1996;50:559–572. doi: 10.1111/j.1558-5646.1996.tb03868.x. [DOI] [PubMed] [Google Scholar]
- 22.Scholten JC, Culley DE, Brockman FJ, Wu G, Zhang W. Evolution of the syntrophic interaction between Desulfovibrio vulgaris and Methanosarcina barkeri: Involvement of an ancient horizontal gene transfer. Biochem Biophys Res Commun. 2007;352:48–54. doi: 10.1016/j.bbrc.2006.10.164. [DOI] [PubMed] [Google Scholar]
- 23.Plugge CM, et al. Global transcriptomics analysis of the Desulfovibrio vulgaris change from syntrophic growth with Methanosarcina barkeri to sulfidogenic metabolism. Microbiology. 2010;156:2746–2756. doi: 10.1099/mic.0.038539-0. [DOI] [PubMed] [Google Scholar]
- 24.Friedrich MW. Phylogenetic analysis reveals multiple lateral transfers of adenosine-5′-phosphosulfate reductase genes among sulfate-reducing microorganisms. J Bacteriol. 2002;184:278–289. doi: 10.1128/JB.184.1.278-289.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stewart JA, Chadwick VS, Murray A. Carriage, quantification, and predominance of methanogens and sulfate-reducing bacteria in faecal samples. Lett Appl Microbiol. 2006;43:58–63. doi: 10.1111/j.1472-765X.2006.01906.x. [DOI] [PubMed] [Google Scholar]
- 26.Quince C, et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods. 2009;6:639–641. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- 27.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 29.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ludwig W, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cole JR, et al. The Ribosomal Database Project (RDP-II): Sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005;33(Database issue):D294–D296. doi: 10.1093/nar/gki038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mackie RI, et al. Ecology of uncultivated Oscillospira species in the rumen of cattle, sheep, and reindeer as assessed by microscopy and molecular approaches. Appl Environ Microbiol. 2003;69:6808–6815. doi: 10.1128/AEM.69.11.6808-6815.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yanagita K, et al. Flow cytometric sorting, phylogenetic analysis and in situ detection of Oscillospira guillermondii, a large, morphologically conspicuous but uncultured ruminal bacterium. Int J Syst Evol Microbiol. 2003;53:1609–1614. doi: 10.1099/ijs.0.02541-0. [DOI] [PubMed] [Google Scholar]
- 34.Grech-Mora I, et al. Isolation and characterization of Sporobacter termitidis gen nov sp nov, from the digestive tract of the wood-feeding termite Nasutitermes lujae. Int J Syst Bacteriol. 1996;46:512–518. [Google Scholar]
- 35.Drake HL, Gössner AS, Daniel SL. Old acetogens, new light. Ann N Y Acad Sci. 2008;1125:100–128. doi: 10.1196/annals.1419.016. [DOI] [PubMed] [Google Scholar]
- 36.Levitt MD. Volume and composition of human intestinal gas determined by means of an intestinal washout technic. N Engl J Med. 1971;284:1394–1398. doi: 10.1056/NEJM197106242842502. [DOI] [PubMed] [Google Scholar]
- 37.Li YF, et al. Molecular characterization and hydrogen production of a new species of anaerobe. J Environ Sci Health A Tox Hazard Subst Environ Eng. 2005;40:1929–1938. doi: 10.1080/10934520500184483. [DOI] [PubMed] [Google Scholar]
- 38.Ouwerkerk D, Klieve AV, Forster RJ, Templeton JM, Maguire AJ. Characterization of culturable anaerobic bacteria from the forestomach of an eastern grey kangaroo, Macropus giganteus. Lett Appl Microbiol. 2005;41:327–333. doi: 10.1111/j.1472-765X.2005.01774.x. [DOI] [PubMed] [Google Scholar]
- 39.Kosaka T, et al. The genome of Pelotomaculum thermopropionicum reveals niche-associated evolution in anaerobic microbiota. Genome Res. 2008;18:442–448. doi: 10.1101/gr.7136508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McInerney MJ, et al. The genome of Syntrophus aciditrophicus: Life at the thermodynamic limit of microbial growth. Proc Natl Acad Sci USA. 2007;104:7600–7605. doi: 10.1073/pnas.0610456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Darling AC, Mau B, Blattner FR, Perna NT. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Samuel BS, et al. Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proc Natl Acad Sci USA. 2007;104:10643–10648. doi: 10.1073/pnas.0704189104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giannakis M, et al. Response of gastric epithelial progenitors to Helicobacter pylori Isolates obtained from Swedish patients with chronic atrophic gastritis. J Biol Chem. 2009;284:30383–30394. doi: 10.1074/jbc.M109.052738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lipinska B, Zylicz M, Georgopoulos C. The HtrA (DegP) protein, essential for Escherichia coli survival at high temperatures, is an endopeptidase. J Bacteriol. 1990;172:1791–1797. doi: 10.1128/jb.172.4.1791-1797.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee I, Berdis AJ, Suzuki CK. Recent developments in the mechanistic enzymology of the ATP-dependent Lon protease from Escherichia coli: Highlights from kinetic studies. Mol Biosyst. 2006;2:477–483. doi: 10.1039/b609936j. [DOI] [PubMed] [Google Scholar]
- 46.Lewis AL, et al. Innovations in host and microbial sialic acid biosynthesis revealed by phylogenomic prediction of nonulosonic acid structure. Proc Natl Acad Sci USA. 2009;106:13552–13557. doi: 10.1073/pnas.0902431106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borenstein E, Kupiec M, Feldman MW, Ruppin E. Large-scale reconstruction and phylogenetic analysis of metabolic environments. Proc Natl Acad Sci USA. 2008;105:14482–14487. doi: 10.1073/pnas.0806162105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.