Abstract
The indigenous human microbiota is essential to the health of the host. Although the microbiota can be affected by many features of modern life, we know little about its responses to disturbance, especially repeated disturbances, and how these changes compare with baseline temporal variation. We examined the distal gut microbiota of three individuals over 10 mo that spanned two courses of the antibiotic ciprofloxacin, analyzing more than 1.7 million bacterial 16S rRNA hypervariable region sequences from 52 to 56 samples per subject. Interindividual variation was the major source of variability between samples. Day-to-day temporal variability was evident but constrained around an average community composition that was stable over several months in the absence of deliberate perturbation. The effect of ciprofloxacin on the gut microbiota was profound and rapid, with a loss of diversity and a shift in community composition occurring within 3–4 d of drug initiation. By 1 wk after the end of each course, communities began to return to their initial state, but the return was often incomplete. Although broadly similar, community changes after ciprofloxacin varied among subjects and between the two courses within subjects. In all subjects, the composition of the gut microbiota stabilized by the end of the experiment but was altered from its initial state. As with other ecosystems, the human distal gut microbiome at baseline is a dynamic regimen with a stable average state. Antibiotic perturbation may cause a shift to an alternative stable state, the full consequences of which remain unknown.
Keywords: human microbiome, microbial community resilience, alternative stable state, ecosystem, ciprofloxacin
All mammals have evolved in the presence of a complex and intimately associated microbial ecosystem, with the highest cell densities found in the gut. The vertebrate gut was no doubt a microbial habitat even before the origin of mammals; during this long coevolutionary history, distinct lineages of bacteria arose that are specialized for the gut habitat. Because the gut microbiota tends to be shared within families (1), these lineages can expand their habitat by enhancing the fitness of their hosts. Mammalian hosts have come to depend on microbial activities to assist digestion, provide vitamins, resist pathogens, and regulate metabolism and the immune system. Thus, there has been and continues to be selection for mutualism between the microbiota and its host (2, 3).
Many aspects of the modern world influence the interactions of humans with their microbiota in ways that are unprecedented in our evolutionary history. These factors include urbanization, rapid global mobility, highly processed diets, improved sanitation and hygiene, nonfamilial child care, and medical therapies, especially antibiotics (2). There are a number of concerns related to the impact of antibiotics on the human microbiota. One is the spread of antibiotic resistance among pathogens, which is facilitated by transfer from and between mutualists (4, 5). More generally, there is a concern that altering the composition of the microbiota will interfere with some of the human–microbe interactions that are integral to human physiology (6). Although most courses of antibiotics result in no immediate signs or symptoms, acute (7) and chronic (8, 9) health problems are associated with antibiotic use. The hygiene hypothesis asserts that increasing rates of autoimmune disorders in the developed world, such as asthma and inflammatory bowel disease, are related to the disruption of the normal interactions within and between the human microbiota and the host (10).
The dynamics of a single complex community over time can reveal more about interactions between community members than a collection of one-time snapshot samples from distinct communities in similar habitats. The interpersonal variation in the composition of the human microbiota implies that the same species may occupy somewhat different niches in different individuals and have different linkages to other taxa, thus displaying different responses to disturbance. However, averaging the effects of a disturbance across multiple individuals may inappropriately treat these diverse phenomena as a single, albeit noisy phenomenon. In contrast, measurements within an individual over time may reveal the range of variation possible in a system governed by the same set of interactions. Time series that span an experimental intervention in a complex community can be particularly useful, because the hypothesized relationships can be examined in potentially different states.
We present here a cultivation-independent survey through time of the composition of the distal gut microbiota of three individuals before, during, and after two exposures to the same antibiotic (in this case, ciprofloxacin). The findings reveal a dynamic ecological system with considerable resilience but also suggest that, in some cases, the system retains a memory of past disturbance; in all cases, repeated disturbance led to a persistent regime shift.
Results
We monitored bacterial communities in the distal gut of three subjects (D, E, and F) by collecting stool samples (52–56 per subject) over a 10-mo interval, during which time these subjects took two 5-d courses of the antibiotic ciprofloxacin (Cp) separated by 6 mo. Samples were collected daily for two 19-d periods surrounding each Cp course and weekly or monthly at other times (Table S1). Analysis of 18 samples before, during, and after the first Cp course in subjects A–C from the same study has been reported previously (11).
Colony Counts in the Presence of 0, 1, and 10 μg/mL Cp.
Aliquots of fresh stool samples from immediately before, immediately after, and 4 wk after each Cp course were diluted and plated in duplicate on aerobic trypticase soy agar medium containing no, low, or high levels of Cp (0, 1, and 10 μg/mL Cp). The density of cells capable of growth in these conditions ranged from 1.3 × 104 cfu/mL to 1.6 × 107 cfu/mL at times not associated with Cp and from 7.5 × 103 cfu/mL to 2.2 × 105 cfu/mL immediately after Cp (Table S2). The concentration of cultivable cells increased during the first Cp in subject D and then, fell over the next 4 wk. For the second Cp in D and both courses in the other two subjects, the cultivable population decreased by ∼1–2 log with Cp administration and increased again over the subsequent 4 wk. The proportion of cultivable cells that were capable of growth in 10 μg/mL Cp was increased by Cp and fell over the subsequent 4 wk in all instances. The same pattern was observed in most cases for cells capable of growth in 1 μg/mL Cp. For both courses in subjects D and F, the concentrations of bacteria capable of growth at both low and high Cp concentrations at 4-wk posttreatment were greater than they had been immediately before Cp exposure.
Sequencing Depth and Community Diversity.
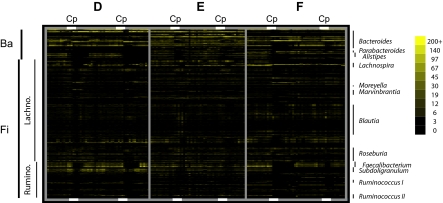
From PCR amplicons spanning the V1, V2, and V3 hypervariable regions of the 16S rRNA gene, 1.76 million pyrosequencing reads (5,901–41,901 per sample) (Table 1) were clustered into 2,582 reference operational taxonomic units (refOTUs; 1,610–2,019 per subject) (Table 1 and SI Text). Good's coverage (estimated probability that the next read will belong to a refOTU that has already been found) averaged 98.5% ± 0.5% (mean ± SD, range = 96.9–99.5%) for individual samples; coverage for each subject exceeded 99.99%. Fig. 1 depicts refOTU relative abundance as a heat map for the Bacteroidetes phylum and subsets of Lachnospiraceae and Ruminococcaceae in the Firmicutes phylum; a heat map with all 2,582 refOTUs is provided in Fig. S1. The same clades of related refOTUs tended to be abundant across all samples and subjects; most refOTUs declined in relative abundance immediately after Cp. On closer examination (Fig. 1 and Fig. S2), a variety of temporal patterns was evident, such as taxa that became more abundant after Cp (e.g., Moreyella and related refOTUs in the Lachnospiraceae), those that showed perturbations of different duration after the two courses (e.g., Faecalibacterium), those that recovered from the first Cp but not the second Cp (Alistipes in D), those that were no longer detected after the first Cp (e.g., Subdolilgranulum in F), and those that varied in abundance independently of Cp treatment (e.g., a clade within Lachnospira in E). Despite the traditional characterization of the gut microbiota as a stable community, few refOTUs maintained uniform abundance, even considering only the Cp-free intervals of the experiment.
Table 1.
Pyrosequencing reads, refOTUs, and coverage
| Subject | Reads (range per sample) | Observed refOTUs (range per sample) | Estimated refOTUs* ± SEM | Good's coverage (range per sample) | Phyla |
| D | 506,662 (5,901–16,415) | 1,732 (137–552) | 2,621 ± 163 | 0.9993 (0.969−0.994) | 8 |
| E | 672,947 (7,013–41,901) | 2,019 (336–796) | 2,787 ± 110 | 0.9995 (0.973−0.995) | 10 |
| F | 581,365 (6,426–14,583) | 1,610 (135–596) | 2,267 ± 73 | 0.9994 (0.981−0.995) | 7 |
| Total | 1,760,974 | 2,691 | 3,520 ± 139 | 0.9998 | 11 |
*Parametric estimate of total refOTU richness by best-fitting model of abundance distribution (12).
Fig. 1.
Heat map displaying the relative abundance of refOTUs in three prominent clades of bacteria. Relative abundance is based on the number of pyrosequencing reads clustering into each refOTU after normalizing the number of reads per sample. Clades are indicated on the left; Ba is all 174 refOTUs within the Bacteroidetes phylum. Within the Firmicutes phylum (Fi), the figure shows clades within the Lachnospiraceae (573 refOTUs) and the Ruminococcaceae (211 refOTUs) that contain the prominent genera named on the right. Each column corresponds to a sample in sequential order for each of subjects D, E, and F, with the times of Cp administration indicated by white bars at the top and bottom of the heat map. Each row corresponds to 1 refOTU, listed in phylogenetic order as defined by the Silva 100 reference tree. Color intensity is proportional to the logarithm of refOTU abundance from 0 to 200 reads as indicated by the scale; color is saturated for abundance of 200 or more.
Of the 2,582 refOTUs detected in the study (Dataset S1), 1,089 (42%) were found at least one time in each of the three subjects, and 150 (5.8%) were detected in at least one-half of the samples of each subject. Only 120 refOTUs (4.6%) were found at a relative abundance of at least 10−3 in at least one sample from each subject, and 4 refOTUs (0.4%) attained at least that abundance in at least one-half of the samples of all three subjects. A taxon at 10−3 relative abundance has about a 99% probability of being detected in the sample with the fewest pyrosequencing reads. For questions related to the core microbiota, it makes sense to consider only non-Cp samples rather than confounding the effects of a deliberate perturbation with comparisons of taxon representation across subjects. Individual non-Cp samples contained 91 ± 14, 96 ± 11, and 109 ± 21 refOTUs (mean ± SD) with relative abundance of at least 10−3 in subjects D, E, and F, respectively, with 309, 336, and 351 refOTUs attaining that abundance at least one time over all non-Cp time points from a single subject. There were 111 refOTUs that attained 10−3 relative abundance in at least one non-Cp sample of each subject. The list of abundant refOTUs (>10−3 relative abundance) in a single non-Cp sample typically contained about one-half or fewer of these 111 refOTUs that were shared among subjects and at least sometimes abundant (50%, 47%, and 43% on average for D, E, and F).
Between 49% and 56% of the reads in each subject were affiliated with the Bacteroidetes phylum (167 refOTUs), 37–48% with Firmicutes (2,266 refOTUs), 1.2–3.1% with Proteobacteria (90 refOTUs), 1.2–2.7% with Verrucomicrobia (3 refOTUs), and fewer than 0.1% with Actinobacteria (35 refOTUs) (Fig. S3). The proportion of reads classified to the same refOTU correlated well between technical replicates (mean Pearson correlation = 0.9905 ± 0.0159 SD, range = 0.9183–0.9991, n = 39 pairs of replicates) (Fig. S4 and Table S3). Additional discussion of the inferred community composition and the reproducibility of the results is found in SI Text.
The observed richness of refOTUs differed significantly between subjects (E > D > F, P < 10−6) when comparisons were made after rarefaction to the same number of reads (506,000/subject) to standardize sampling effort. Parametric estimation of total OTU richness per subject according to the best fitting of several models (12) showed the same relative order, although the difference between E and D was not significant (Table 1 and additional discussion in SI Text). The rank order was reversed for taxon evenness (F, 0.88; D, 0.85; E, 0.82; P < 10−6) and diversity (F, 6.47; D, 6.35; E, 6.22; P < 10−6) assessed through Shannon entropy. Within-subject (α) 1° diversity according to Jost (13) showed the same pattern (F, 644; D, 584; E, 502; P < 10−6), with an average of 577 effective refOTUs/subject (i.e., average diversity per subject was equivalent to a community with 577 equally common taxa). Between-subject (β) 1° diversity across all subjects was 1.84 effective communities [i.e., the 1° diversity in the three subjects combined (γ diversity) was 1,062 refOTUs, which was 1.84 times the average α diversity for a single subject].
Effects of Cp on Diversity.
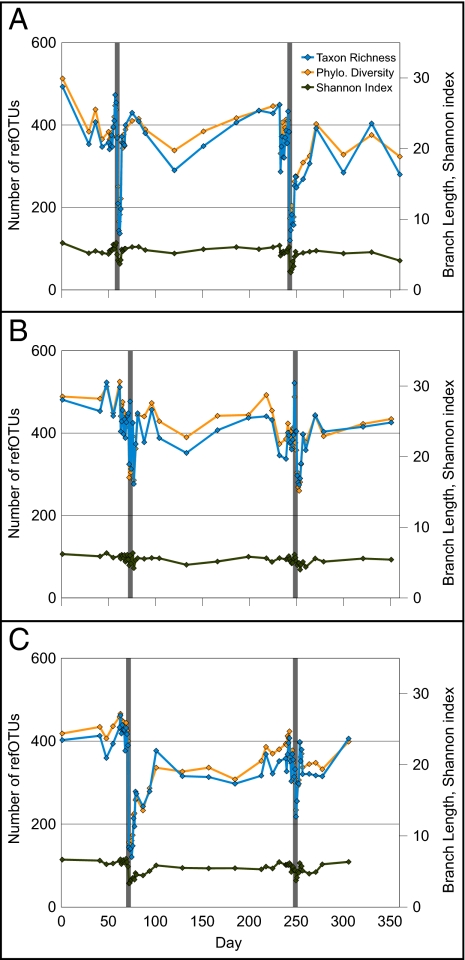
Cp had a marked effect on the distal gut microbiota, despite the absence of any gastrointestinal symptoms reported by subjects. The Cp-associated disturbances shared some features across all subjects and both Cp courses, as indicated by plots of refOTU richness, phylogenetic diversity, and Shannon entropy over time (Fig. 2). Richness and diversity (assessed after rarefaction to 5,900 reads/sample to standardize sampling intensity) plunged rapidly 3–4 d after the subjects began Cp; the lowest values of these parameters occurred on the last 2 d of Cp treatment or during the several days thereafter. Richness and diversity also rebounded rapidly, recovering much of their lost value in 1–2 d. Parametric estimation of total richness per treatment interval within each subject (12) showed a drop in richness in each Cp-perturbed interval relative to the intervals immediately before and after; the drop was statistically significant in all cases except the comparison of pre-Cp with first Cp in E (Fig. S5 and SI Text). In addition to the obvious similarities between each Cp perturbation, however, there were less obvious differences between subjects and between courses in the effect of Cp on richness and diversity. Displacement from the pre-Cp values of these parameters was strongest in D and weakest in E (P < 10−6), and although the magnitude of the first and second perturbations did not differ significantly in D and E (P > 0.05), the second perturbation was smaller than the first in F (P < 10−6). The differences between subjects in the effects of Cp on diversity and richness had no obvious relationship to the initial richness and diversity values. The response to the first Cp course in E was unique, with wide day-to-day fluctuations in the diversity of the microbiota (Fig. 2). A subset of taxa with daily oscillations in abundance was responsible for this phenomenon (e.g., refOTUs in the genera Blautia, Roseburia, and Dorea) (Fig. 1, Fig. S1, and SI Text).
Fig. 2.
Three measures of biological diversity for samples from subjects D (A), E (B), and F (C). Calculations were made after rarefying to an equal number of reads for all samples to control for unequal sampling effort. Narrow gray rectangles indicate the 5-d Cp courses; daily sampling around these times allowed visualization of daily fluctuations in diversity parameters that were not evident during less frequent sampling. RefOTU richness (number of refOTUs observed per sample) is shown on the left y axis; phylogenetic diversity (PD; total branch length of the phylogenetic tree relating all refOTUs in the sample) and the Shannon index of diversity are shown on the right y axis. The x axis reflects experiment day.
The short-duration, asymptomatic drop in richness and diversity of the microbiota after Cp was consistent with our previously reported results (11). However, the restricted set of samples in the previous analysis limited the temporal resolution of recovery time to 4 wk after the first course of Cp and did not permit comparison of the Cp perturbation with non–Cp-associated temporal fluctuations in the diversity of the gut microbiota at a range of time scales.
Effects of Cp on Community Membership.
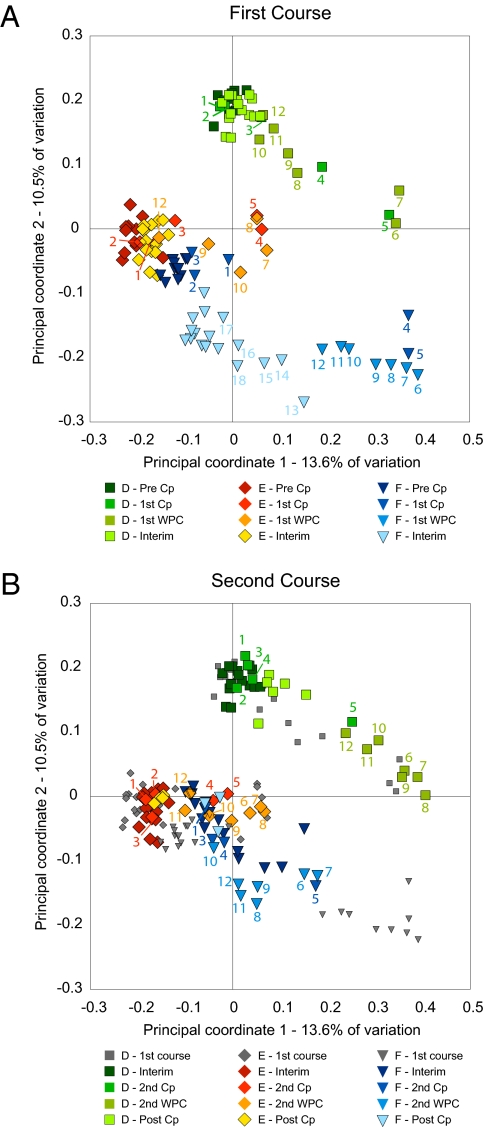
Similarities and differences between the membership of the distal gut microbial communities of the three subjects and between the first and second Cp responses are portrayed by principal coordinate analysis (PCoA) of unweighted UniFrac distances, a measure of community dissimilarity based on OTU presence/absence (community membership) that takes into account evolutionary relatedness among OTUs (14) (Fig. 3). As suggested by the diversity statistics (Fig. 2), the responses of the gut microbiota to Cp shared features across all subjects, despite interindividual community differences. On closer examination, differences in the Cp response among subjects and between the first and second Cp course in some subjects were also apparent.
Fig. 3.
PCoA of unweighted UniFrac distances, a phylogenetically aware measure of intersample (β) diversity. A covers the period encompassing the first Cp, with samples before Cp (pre-Cp), during the first Cp course (first Cp), during the first week post-Cp (first WPC), and in the interim period between Cp courses (interim) progressing from darker to lighter shades. B covers the second Cp period, with interim samples in the darkest shades and the second Cp course, second WPC, and post-Cp periods becoming progressively lighter. Data points from A are also shown on B with small gray symbols to facilitate comparisons between the two parts of the experiment. Numbers adjacent to some samples track the Cp perturbation by indicating the number of days elapsed since the start of Cp administration; missing numbers indicate that samples were not collected on those days.
The first principal coordinate (PC1) explains 13.6% of intersample variance and is driven primarily by the response to Cp, with maximally perturbed samples on the right clearly separated from clusters of non-Cp (pre-Cp, interim, and post-Cp) samples on the left. The community response to the first Cp in F is unique in that it required several months, not merely 1 wk, to approach a stable interim state; the prolonged transition represents slow directional change as opposed to prolonged instability. Both PC2 (10.5% of variance) and PC3 (9.7% of variance) (Fig. S6) reflect a mixture of the Cp response and interpersonal differences. Most samples of the three subjects are separated into distinct regions on PC2. The gut microbiota of subjects D and F responded to the first Cp in the same direction along PC2, but whereas samples from D eventually regained their pre-Cp values and then responded similarly to the second Cp, samples from F did not rebound along PC2 after the initial Cp perturbation and were not influenced on this axis by the second Cp. Samples from E showed no obvious trend along PC2 in response to Cp.
On PC3 (Fig. S6), non-Cp samples of D and F are offset only slightly from each other, and the Cp effects of both courses are reflected in vectors of similar direction and magnitude for these two subjects. The coordinates of non-Cp samples were lower and the Cp effect was smaller along PC3 for E than for the other subjects, but the direction of the effect was consistent. In contrast to the first three PCoA axes, PC4 (4.0%) captures an effect that differs in direction between the subjects. After each Cp course, samples from D shifted in a positive direction on PC4 and returned, whereas those from E did the opposite. The refOTUs that most differentiated the responses of D and E (SI Text and Dataset S1) were roughly evenly divided among the Bacteroidetes (particularly Bacteroides and Parabacteroides), the Lachnospiraceae (Blautia and uncharacterized), and the Ruminococcaceae (Faecalibacterium and several other genera.) The pattern was more complex in F and differed between the courses.
For comparison, previously reported results examining a single Cp course showed responses in the same direction for all subjects along the first ordination axis and in the opposite direction for one of three subjects along the second ordination axis, with the magnitude of the Cp response being much smaller in one of three subjects (11). Because only one or two Cp-affected samples were included in the earlier analysis, we were unable at that time to assess the dynamics of the response and whether it followed a linear trajectory to the perturbed state and back or some more complicated pattern.
Because the unweighted UniFrac distance between communities represents the absence of shared, related (or identical) taxa, samples with similar PCoA coordinates (i.e., low UniFrac distance between them) share a set of related taxa. A trajectory of changing coordinates over time represents the gain or loss of taxa, and therefore, Cp responses associated with vectors of similar direction on the first three axes indicate that most taxa that appeared or disappeared in response to Cp in one subject tended to behave similarly in other subjects, if they responded at all. The refOTUs that by their presence or absence most clearly differentiated the perturbed and nonperturbed samples across all subjects (Dataset S1) were dominated by taxa that were often present in non-Cp samples and absent in most Cp-perturbed samples (and not taxa that appeared only in perturbed samples). About one-half of these disturbance-differentiating taxa are affiliated with the Ruminococcaceae in the Firmicutes phylum, and about one-half of these Ruminococcaceae refOTUs can be assigned to the genus Faecalibacterium, a genus we found to be reduced or eliminated in response to Cp in other subjects as well (11). The second most common affiliation of refOTUs that differentiated non-Cp from Cp-perturbed samples by their presence or absence was uncultivated or uncharacterized Lachnospiraceae, highlighting the importance of continuing attempts to cultivate and characterize strains from this prominent family of intestinal microbes.
Effects of Cp on Community Composition.
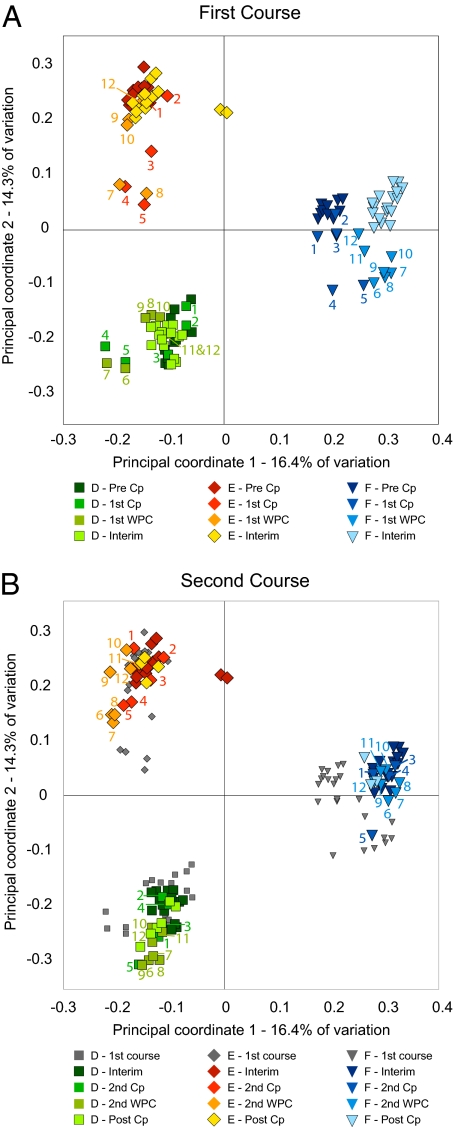
We assessed the changing community composition during different time periods of the experiment by using distance-based redundancy analysis (dbRDA) of Bray–Curtis intersample distances (BC). BC is a common metric of community dissimilarity because it makes the reasonable assumption that the shared absence of a taxon is not evidence of community similarity. dbRDA is a supervised ordination technique designed to handle ecologically meaningful but non-Euclidean measures of dissimilarity, such as BC (15). Taxon relative abundance was represented as the logarithm of the number of reads affiliated with each refOTU after rarefaction of the number of reads/sample to a consistent value to control for differences in sampling effort. Log weighting of counts is appropriate for organisms capable of exponential growth on the time scale of our sampling; comparisons of growth (or death) rates are measured by changes in the logarithm of organism abundance. Ordination followed by permutation testing confirmed that the composition of the microbiota varied between subjects and between the time intervals defined with respect to Cp administration (P < 0.01 for each) and that the effect of Cp was not the same in each subject (P < 0.01 for interaction term), with 58.7% of intersample variance explained by these terms. dbRDA of datasets containing only samples from Cp-free intervals of individual subjects indicated that the composition of the pre-Cp, interim, and post-Cp communities differed in every subject (P < 0.01, P = 0.04, and P < 0.01 for D, E, and F, respectively). Posthoc testing within each subject revealed that (i) for both courses in each subject, the composition of the Cp-perturbed microbiota differed from that of the preceding time interval, (ii) the maximally perturbed communities differed between the first and second courses for all subjects, and (iii) the composition of the pre-Cp and post-Cp communities was different in every subject, but in D, the interim community resembled the pre-Cp state, whereas in E and F, the interim communities resembled the post-Cp community.
Visualization of the dbRDA ordination of BC distances (Fig. 4) shows that the first two components (together explaining 30.7% of intersample variance) primarily separate samples according to subject but also shows a Cp effect that varies in direction for each subject. Relative to other samples from the same subject, the perturbed samples from D, E, and F tended to be displaced left, down, and right, respectively. In contrast, the third component explaining 9.4% of variance was driven almost entirely by a Cp response that was similar in all subjects (Fig S7). The fourth component (3.8%) captures the unique phenomenon of a prolonged directional shift in the community of F after the first Cp, which was also evident along the first principal coordinate of the unweighted Unifrac analysis. Taken together, the dbRDA data show that community composition is influenced primarily by subject identity and secondarily by Cp effects and that the latter exhibits shared and unique features in these subjects. Plots of the BC intersample distances over time in comparison with a reference sample (Fig S8), chosen as the last sample before the first Cp, also show the relative stability of communities during Cp-free intervals and the shifts in community state that can occur in these communities when they are perturbed by Cp.
Fig. 4.
Distance-based redundancy analysis (15) of Bray–Curtis intersample distances calculated with log2-transformed abundance data. As in Fig. 3, A and B show samples surrounding the first and second Cp course respectively, with interim samples appearing in both panels. Symbol color and numbering are as described for Fig. 3.
The taxa making the greatest contribution to the common Cp response among subjects (SI Text and Dataset S1) include 43 refOTUs that decrease and 11 refOTUs that increase in relative abundance after Cp administration. These taxa include 11 of 20 most abundant refOTUs over all subjects. Within this set of 54 shared Cp responders, those that decrease in abundance after Cp include eight refOTUs in the genus Faecalibacterium and similar numbers of refOTUs elsewhere in the Ruminococcaceae and the Lachnospiraceae; most of these refOTUs are closely related to butyrate-producing isolates from the gut. Seven of eleven shared Cp responders that increase in abundance after Cp are also affiliated with Lachnospiraceae, at least some of which are also closely related to known butyrate producers. The common Cp response also features representatives of the Bacteroidetes phylum, including some that decrease in abundance (seven refOTUs in the genus Bacteroides, four in the genus Alistipes, and two in the family Porphyromonadaceae) and some that increase (four Bacteroides refOTUs) as well as refOTUs affiliated with Sutterella (a β proteobacterium gut symbiont), Thalassospira (first described as an oligotrophic marine α-proteobacterium genus), and 4C0d-2, representing a little-known, deeply branching lineage related to Cyanobacteria, all of which decrease after Cp.
Of note, the diversity statistics (Fig. 2), unweighted UniFrac PCoA (Fig. 3 and Fig. S6), and dbRDA of BC distances (Fig. 4 and Fig. S7) all showed differences between the responses to the first and second courses of Cp in all subjects. For example, after the first Cp, the diversity of the gut microbiota in D recovered in essentially 1 d. In contrast, after the second Cp, there was a biphasic response in measurements of diversity in D. In F, it was the opposite case: a biphasic response after the first Cp and a single-day return after the second Cp. The subject-specific Cp effects displayed along the first two components of the BC ordination differed subtly between the first and second Cp in D and E; for these subjects, the direction of the Cp displacement shifted 30–45° between courses, implying that within each subject, the taxa responding to the two courses are not entirely the same. The difference between the two perturbations was not subtle in F; we have already noted the prolonged return to a stable state in F after the first Cp that is evident on the first UniFrac ordination axis and fourth BC ordination axis. Furthermore, a large proportion of the first Cp perturbation in F was changes from which no recovery was made; unidirectional displacement is found along the first dbRDA component and the second UniFrac component. After the second Cp, the community composition in F showed essentially no displacement along either of these axes. Another distinction between the first and second Cp in F is found along the third dbRDA component; the second perturbation has a similar direction but roughly one-half the magnitude of the first perturbation.
The taxa that differentiated the two Cp responses were typically found in low abundance in most samples and increased in abundance after Cp (SI Text and Dataset S1). Most such taxa increased dramatically with the first Cp course and less so, if at all, with the second. This pattern contrasts with that of the taxa that most effectively distinguished between Cp-associated and non–Cp-associated samples, which were typically abundant at most sampling times and then decreased in abundance after Cp. The most prominent taxa differentiating between the two Cp responses were refOTUs affiliated with Bacteroides dorei, Akkermansia municiphilia, and several Roseburia species (a genus of butyrate-producing microbes) (16, 17) as well as other known butyrate producers. Fewer taxa increased to a greater extent during the second Cp course than they did during the first, but among these refOTUs, there are also close relatives to the Roseburia species and to Bacteroides thetaiotaomicron. Intriguingly, this relatively small group of refOTUs included two in different phyla whose closest cultivated relative was named for the ability to degrade xylans (Bacteroides xylanisolvens and Clostridium xylanolyticum) and in general, included refOTUs more distant from their nearest cultivated relative than were the refOTUs showing a greater increase after the first Cp. For example, in addition to the C. xylanolyticum relative, the group included a refOTU clustered around a database clone sequence ∼95% (genus level) similar to a cultivated Oscillospira sp., another with ∼8–9% (family level) sequence divergence from Eubacterium rectale, and a number of uncharacterized butyrate-producing isolates.
We examined the matrix of BC distances for temporal autocorrelation, finding that, for the pre-Cp and interim periods, the composition of the microbiota was more similar on adjacent sampling days than it was on average over all pairs of samples over that Cp-free interval (P range from 0.001 to 0.043 for five of six comparisons; P = 0.054 for pre-Cp in E). The similarity between adjacent-day samples was significantly less than the similarity between technical replicates (P < 0.001). It is difficult to estimate the time required for autocorrelation of community composition to decay (i.e., the interval after which the community is, on average, as different from its starting composition as it ever becomes without a perturbation), both because of limited data for any given time interval between samples and because of variance in the BC data. Nonetheless, we took the average BC distance between samples in the same Cp-free interval but separated by at least 2 wk as an estimate of the expected dissimilarity between uncorrelated samples. For subject D, intersample BC distances averaged over a sliding 3-d window (i.e., samples separated by 1–3 d, 2–4 d, etc.) were within an SD of the uncorrelated BC distance after samples were separated by 4–6 d and 5–7 d for pre-Cp and interim samples, respectively. Comparable intervals for F are 3–5 d and 4–6 d, but for E, average BC distances for samples separated by 1–3 d were already within 1 SD of the average for uncorrelated samples during both the pre-Cp and interim intervals. Considering the human gut as a flow-through system with a retention time of about 1–2 d, the decay of autocorrelation over about three retention times is not surprising.
Discussion
The human distal gut is one of the most complex ecosystems on the planet. However, it may be a tractable and powerful system for the study of both basic ecological principles and health-related community interactions through the exploitation of disturbance. Daily sampling in each subject allowed us to compare day to day with longer term changes in the composition of the gut microbiota under both perturbed and nonperturbed conditions. We found that the composition of the gut microbiota is, on average, more similar on adjacent sampling days than on a random pair of days in the same disturbance-free interval and that this temporal autocorrelation decays over several days to a week, depending on the subject. Although most taxa changed in relative abundance from day to day, communities were no more different when compared at times separated by 2–5 mo, on average, than they were at times separated by more than about 1 wk.
The dynamic composition of the gut microbiota over time makes it more difficult to address the concept of the core microbiota. The core microbiota has been taken to mean those components (taxa or genes) common to all or the vast majority of humans (18), although others have used the term simply to mean those taxa present in a majority of their subjects (19). The incomplete characterization of these complex communities must be acknowledged when addressing questions about the core microbiota and especially, its absence (1); the problems are exacerbated if complete characterization requires sampling an individual over time. The apparent conflict between our finding of considerable overlap of refOTUs between subjects and the recent conclusion of Turnbaugh et al. (1) that a core microbiota may not exist with respect to phylogenetic groups (1) evaporates when one realizes that our comparison involves over 5 × 105 pyrosequencing reads per subject collected over many months from three subjects, whereas the Turnbaugh et al. (1) paper involved an average of ∼1.2 × 104 reads collected at a single time from each of 154 subjects.
The routine fluctuations in community composition indicate that the long-term stability of the distal gut community is not maintained by inertia, or resistance to change, but rather, by the action of restoring forces that maintains the state of this dynamic system within a certain range. One might imagine that such restoring forces would be strong enough to allow the community composition to resist change in the face of disturbance. This was not the case for exposure to Cp, despite the fact that Cp is generally believed to have minimal effects on the anaerobic microbiota of the gut. Within 3–4 d of initiating Cp in each subject (perhaps the soonest that might be expected given the time required for Cp concentrations to rise in the cecum and large intestine and the transit time required for material to leave the colon), the community composition made a dramatic shift to a different state. After Cp was discontinued and a lag that must at least partially be explained by these same factors, the community began to return to a state more similar to its pre-Cp state. This return occurred despite the fact that abundant taxa accounting for 25–50% of the community before Cp exposure in the three subjects were essentially wiped out after Cp exposure. For Bacteroides and Lachnospiraceae and to a lesser extent, for Ruminococcaceae (although not Faecalibaterium), taxa closely related to those that had been eliminated surged in abundance at these times but were then rapidly replaced by the original taxa after Cp was withdrawn. At this time, intersubject differences were apparent. In subject D, there was a complete return to pre-Cp conditions after the first perturbation. In E, the first return was largely complete, but the composition of the interim samples remained slightly closer to the perturbed samples. In F, the first Cp perturbation included some taxa that largely rebounded to pre-Cp values (cf dbRDA component 2) and others that made no return at all (cf dbRDA components 1 and 4). Furthermore, whereas 1 wk was sufficient time for samples of D and E to attain the composition that they maintained during the interim between courses, samples from F continued to show directional change in composition for about 2 mo.
The absence of any GI-related symptoms experienced by the subjects during these times supports the idea that the gut microbiota has functional redundancy among its constituent taxa (1, 20), at least for functions likely to generate symptoms within several days, such as the fermentation of various food- and host-derived resources entering the large intestine. Thus, the mechanisms responsible for the restoring forces on community composition would not seem to include deficiencies in substrate fermentation in the colon or overt intolerance of the altered communities by the host.
By the end of the study, the community composition in each subject was different from what it had been before the first course of antibiotic, and it seemed to be stable in the new state over the final 2 mo of the study. The contrast with the initial community was most evident in F, where even interim samples never regained the position of pre-Cp samples along the primary ordination axes. This discrepancy was more evident in the dbRDA analysis than in the unweighted UniFrac analysis, suggesting that the change was driven more by altered taxon abundance than by the gain or loss of community members. However, the recovery in F was more complete after the second Cp, with little change between the interim and post-Cp communities. In contrast, the microbiota of D made essentially a complete recovery from the first Cp but stabilized in a state distinct from the initial composition after the second Cp. The stability of community composition over 2 mo in all three subjects before Cp exposure offered no clue as to the different degrees of resilience in the microbiota of the subjects after Cp exposure. However, the existence of sudden regimen shifts in ecosystems, sometimes triggered by perturbations, is a familiar ecological phenomenon, and the return of external conditions to their former state may not reverse such changes in community composition (21). Repeated perturbations may be particularly likely to cause such shifts, even when the community seems to have recovered from the initial perturbation (22).
One potential ramification of the altered community is an enhanced carriage of antibiotic-resistance genes in the human population (5). The responses to Cp that we observed are likely to have included both direct effects because of intrinsic or acquired resistance of strains to Cp and indirect effects mediated through numerous ecological interactions among microbial populations (23). Although 16S rRNA surveys cannot track the spread of antibiotic resistance, the persistence of some changes in community composition that occurred at the time of Cp may mean that the proportion of resistant strains was increased. The proportion of cfu that were able to grow in the presence of 1 or 10 μg/mL Cp increased with Cp treatment, although the total number of cfu per mass of stool decreased in five of six cases for these three subjects. A higher proportion of Cp-resistant strains in the community is one possible explanation for an increased lag time between initiating Cp and community perturbation for the second Cp course in subjects D and F.
The functional consequences of the alterations that we observed in the composition of the gut microbiota are unclear. It seems likely that the ability of the community to ferment substrates in the colon was grossly unchanged, not only because of the absence of antibiotic-associated diarrhea (an osmotic effect after reduced colonic fermentation and thus, lower concentrations of short-chain fatty acids) (7) but also because the presence of an unused fermentable substrate in the colon is likely to stimulate the growth of strains that can use it. If the substrate is sufficiently abundant to attain a high concentration in the colon, there are likely to be many such strains. With respect to the use of growth substrates, competition among microbes is likely to ensure a rapid return to an efficient microbiota (2, 3) if the collective ability of the microbiota to use the available resources is ever diminished in the first place.
Other traits attributed to members of the gut microbiota, however, such as inhibiting the growth (24), attachment (25), or virulence (26) of particular pathogens, helping to regulate host immunity (21–23) or energy balance (24), or participating with host enzymes in cometabolism of specific substances (27, 28), may be restricted to a small subset of the community. Unlike carbon use, these traits are not essential to the microbes; variation in the trait between close relatives is predicted by evolutionary theory (29, 30) and observed in practice (24, 31, 32). If a trait is beneficial to the community as a whole (e.g., by keeping the host healthy), community-level selection acting over host generations can favor the mutualistic trait in the long term (33, 34). However, if expression of the trait incurs some cost for the microbe, cheater phenotypes lacking the trait will have a fitness advantage over altruist phenotypes within a host generation (29). If a cheater occupies the former niche of a mutualist that has been eliminated by an antibiotic, host health is diminished. Every course of antibiotics may represent another roll of the dice, potentially allowing displacement of a mutualist with a strain that may or may not provide the same benefit. Although it is possible for a mutualist to replace a cheater, the dice are loaded in the opposite direction for altruistic traits that impose a cost on the bearer. Furthermore, to the extent that antibiotic treatment weakens the fidelity of the association between lineages of microbes and hosts across generations, it weakens community-level selection for mutualistic traits in the microbiota by eliminating some strains that were inherited from kin and allowing outside strains to enter the community (33, 34).
The use of broad-spectrum antibiotics to treat acute infectious disease will undoubtedly continue because of immediate, undeniable benefits for human health. Nonetheless, the imminent and well-publicized threat of losing those benefits because of the spread of antibiotic-resistant microbes has led to constraints on antibiotic use. Even if the success of such resistance-control strategies could be assured, however, there would remain a less obvious but perhaps more important risk to antibiotic use. The antimicrobial agents that we deploy against pathogens also disrupt coevolved microbial communities that are integral to human health. Fortunately, our native microbiota can display considerable resilience as well as functional redundancy for at least some processes. However, because we have only a limited understanding of the ecosystem services provided to us by our resident microbiota, caution and additional research are warranted.
Materials and Methods
Participants and Sampling.
Healthy adult participants were recruited from Stanford and the surrounding community; exclusion criteria included antibiotics within the previous 12 mo, past reactions to fluoroquinolone antibiotics, pregnancy or nursing, and age under 18. Written informed consent was obtained; the study was approved by the Stanford University Institutional Review Board. Participants provided stool samples over ∼10 mo at frequencies varying from daily to monthly (Table S1). Daily samples were collected in the weeks before, during, and after each of two 5-d courses of Cp (500 mg orally two times daily), which took place at 2 mo and 8 mo into the 10-mo study. Subjects collected samples in sterile vials, which were frozen immediately in their home freezers (−20 °C), and brought them to the laboratory within several days for storage at −80 °C. Participants were requested to report any symptoms co-occurring with Cp administration, including mild gastrointestinal symptoms; none were reported.
Cultivation.
Unfrozen stool samples were collected by participants in tubes containing Cary-Blair medium (Medical Chemical) on six sampling dates: 1 d before, 1 d after, and 4 wk after each Cp course. Samples were transported to the lab and processed within no more than 6 h, most often within 2 h. Subsamples of ∼1 g stool (wet weight) were suspended and diluted in neutral Hepes-buffered saline and plated in duplicate at dilutions ranging from 10−3 to 10−7 on trypticase soy agar plates containing 5 g/L glucose and 0, 1, or 10 μg/mL Cp. Plates were incubated aerobically at 37 °C for 5 d, with colonies counted daily. Aerobic conditions were chosen because of the logistical constraints of rapidly transferring samples to anaerobic conditions. The intention was not to characterize the gut microbiota through cultivation but to examine the effect of Cp treatment on a consistent subset of the microbiota defined by the ability to grow under a particular set of conditions.
DNA Extraction, Amplification, and Pyrosequencing.
DNA extraction was as described in ref. 11. Briefly, the QIAamp DNA stool mini kit (Qiagen) was used as directed for extraction of bacterial DNA, with the addition of a bead-beating step (FastPrep machine for 45 s at setting 5; Bio 101), which took place in the lysis buffer immediately before the initial incubation at 95 °C. Controls treated identically but lacking fecal material uniformly failed to produce detectable bands after PCR and gel electrophoresis. PCR amplification of a region of the small subunit rRNA (16S rRNA) gene was performed with 50 ng or 5 ng template DNA (Nanodrop) in 50 μL reactions as described (Roche). Fusion primers adapted for the general sequencing kit of the GS FLX Titanium pyrosequencing platform (Roche) comprised of A linker–10-mer barcode–dinucleotide spacer–533–515R reverse primer (proximal primer) or B linker–dinucleotide spacer–8–27F forward primer (distal primer). (Forward and reverse refer to 16S rRNA orientation; sequencing was from the A-linked proximal toward the B-linked distal primer.) Spacers were chosen to match few or no dinucleotides adjacent to the priming site (with reference to public 16S rRNA databases) to interrupt chance complementarity with the barcode or linker. Primer sequences are listed in Table S4. PCR amplicon libraries were gel purified, quantified using PicoGreen (Invitrogen) in 96-well plates on a Typhoon scanner (GE Healthcare), pooled in equal ratios by mass, and submitted for pyrosequencing.
Data Analysis.
Approximately 5 million raw pyrosequencing reads were processed using mothur version 1.7 (33) to obtain 2.32 million filtered, quality-trimmed reads that were assigned unambiguously to a sample. Unique reads were clustered at a 5% genetic distance threshold using Uclust software (http://www.drive5.com/uclust) with nondefault settings resulting in thorough searches for the optimal clusters and cluster seeds provided by a high-quality subset of the Silva 100 reference database (http://www.arb-silva.de/). The database sequences were preclustered so that the 27,231 reference sequences were approximately uniformly spaced at 3% genetic distance in densely sampled regions of bacterial phylogeny. About 0.34 million pyrosequencing reads failing to cluster with a reference sequence at the 5% distance threshold were omitted from the analysis to minimize the effect of pyrosequencing errors (34, 35). Another 0.21 million reads were derived from samples that were not analyzed as part of the time series dataset (control experiments, aberrant replicates, and data from other experiments), leaving 1,760,974 reads in the current dataset (Table 1 and Dataset S1). Data-processing parameters are described in greater detail in SI Text.
Reads that clustered with the same reference sequence were defined as refOTUs (11). An abundance matrix of refOTUs by subject and sample (Dataset S1) and a phylogenetic tree of reference sequences obtained by pruning the Silva 100 reference tree (35) to the observed cluster seeds were the basis of subsequent analysis. Quantitative Insights Into Microbial Ecology (QIIME) 0.8 (http://qiime.sourceforge.net/) was used to perform rarefactions, calculate α and β diversity measures, and conduct PCoA of unweighted UniFrac distances between samples. Calculation of true diversity values as the effective number of taxa (α diversity) and effective number of communities (β diversity) according to Jost (13, 36) and simple tests of statistical significance were performed using NeoOffice spreadsheet software. dbRDA (15) was performed using the capscale command of the vegan package (1.15–4) of R statistical software (2.9.1; http://cran.r-project.org).
Supplementary Material
Acknowledgments
We thank Lisa Bukovnik of the Duke Institute for Genome Science and Policy Sequencing Core Facility for assistance with pyrosequencing, Robert Edgar for making Uclust available and for assistance with its use, members of the Knight laboratory for assistance with the QIIME pipeline, members of the D.A.R. laboratory for helpful discussion and support, and all of our study participants. This work was funded by the Doris Duke Charitable Trust (D.A.R.) and by a National Institutes of Health Pioneer Award DP1OD000964 (to D.A.R.). D.A.R. is supported by the Thomas C. and Joan M. Merigan Endowment at Stanford University.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Microbes and Health,” held November 2–3, 2009, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS Web site at http://www.nasonline.org/SACKLER_Microbes_and_Health.
This article is a PNAS Direct Submission.
Database deposition: The sequences reported in this paper have been deposited in the NCBI Short Read Archive database (accession no. SRA020961).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000087107/-/DCSupplemental.
References
- 1.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 4.Sommer MO, Dantas G, Church GM. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science. 2009;325:1128–1131. doi: 10.1126/science.1176950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salyers AA, Gupta A, Wang Y. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 2004;12:412–416. doi: 10.1016/j.tim.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 6.Blaser MJ, Falkow S. What are the consequences of the disappearing human microbiota? Nat Rev Microbiol. 2009;7:887–894. doi: 10.1038/nrmicro2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beaugerie L, Petit JC. Microbial-gut interactions in health and disease. Antibiotic-associated diarrhoea. Best Pract Res Clin Gastroenterol. 2004;18:337–352. doi: 10.1016/j.bpg.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Flöistrup H, et al. Allergic disease and sensitization in Steiner school children. J Allergy Clin Immunol. 2006;117:59–66. doi: 10.1016/j.jaci.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 9.Marra F, et al. Does antibiotic exposure during infancy lead to development of asthma?: A systematic review and metaanalysis. Chest. 2006;129:610–618. doi: 10.1378/chest.129.3.610. [DOI] [PubMed] [Google Scholar]
- 10.Guarner F, et al. Mechanisms of disease: The hygiene hypothesis revisited. Nat Clin Pract Gastroenterol Hepatol. 2006;3:275–284. doi: 10.1038/ncpgasthep0471. [DOI] [PubMed] [Google Scholar]
- 11.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bunge J, Barger K. Parametric models for estimating the number of classes. Biom J. 2008;50:971–982. doi: 10.1002/bimj.200810452. [DOI] [PubMed] [Google Scholar]
- 13.Jost L. Entropy and diversity. Oikos. 2006;113:363–375. [Google Scholar]
- 14.Lozupone C, Hamady M, Knight R. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Legendre P, Anderson MJ. Distance-based redundancy analysis: Testing multispecies responses in multifactorial ecological experiments. Ecol Monogr. 1999;69:1–24. [Google Scholar]
- 16.Duncan SH, et al. Proposal of Roseburia faecis sp. nov., Roseburia hominis sp. nov. and Roseburia inulinivorans sp. nov., based on isolates from human faeces. Int J Syst Evol Microbiol. 2006;56:2437–2441. doi: 10.1099/ijs.0.64098-0. [DOI] [PubMed] [Google Scholar]
- 17.Duncan SH, Hold GL, Barcenilla A, Stewart CS, Flint HJ. Roseburia intestinalis sp. nov., a novel saccharolytic, butyrate-producing bacterium from human faeces. Int J Syst Evol Microbiol. 2002;52:1615–1620. doi: 10.1099/00207713-52-5-1615. [DOI] [PubMed] [Google Scholar]
- 18.Turnbaugh PJ, et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tap J, et al. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol. 2009;11:2574–2584. doi: 10.1111/j.1462-2920.2009.01982.x. [DOI] [PubMed] [Google Scholar]
- 20.Kurokawa K, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheffer M, Carpenter SR. Catastrophic regime shifts in ecosystems: Linking theory to observation. Trends Ecol Evol. 2003;18:648–656. [Google Scholar]
- 22.Paine RT, Tegner MJ, Johnson EA. Compounded perturbations yield ecological surprises. Ecosystems. 1998;1:535–545. [Google Scholar]
- 23.Flint HJ, Duncan SH, Scott KP, Louis P. Interactions and competition within the microbial community of the human colon: Links between diet and health. Environ Microbiol. 2007;9:1101–1111. doi: 10.1111/j.1462-2920.2007.01281.x. [DOI] [PubMed] [Google Scholar]
- 24.Servin AL. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol Rev. 2004;28:405–440. doi: 10.1016/j.femsre.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 25.Ingrassia I, Leplingard A, Darfeuille-Michaud A. Lactobacillus casei DN-114 001 inhibits the ability of adherent-invasive Escherichia coli isolated from Crohn's disease patients to adhere to and to invade intestinal epithelial cells. Appl Environ Microbiol. 2005;71:2880–2887. doi: 10.1128/AEM.71.6.2880-2887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medellin-Peña MJ, Wang H, Johnson R, Anand S, Griffiths MW. Probiotics affect virulence-related gene expression in Escherichia coli O157:H7. Appl Environ Microbiol. 2007;73:4259–4267. doi: 10.1128/AEM.00159-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li M, et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci USA. 2008;105:2117–2122. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 29.Velicer GJ. Social strife in the microbial world. Trends Microbiol. 2003;11:330–337. doi: 10.1016/s0966-842x(03)00152-5. [DOI] [PubMed] [Google Scholar]
- 30.West SA, Griffin AS, Gardner A, Diggle SP. Social evolution theory for microorganisms. Nat Rev Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- 31.Reid G, Bruce AW. Probiotics to prevent urinary tract infections: The rationale and evidence. World J Urol. 2006;24:28–32. doi: 10.1007/s00345-005-0043-1. [DOI] [PubMed] [Google Scholar]
- 32.Peña JA, et al. Genotypic and phenotypic studies of murine intestinal lactobacilli: Species differences in mice with and without colitis. Appl Environ Microbiol. 2004;70:558–568. doi: 10.1128/AEM.70.1.558-568.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Foster KR, Wenseleers T. A general model for the evolution of mutualisms. J Evol Biol. 2006;19:1283–1293. doi: 10.1111/j.1420-9101.2005.01073.x. [DOI] [PubMed] [Google Scholar]
- 34.Sachs JL, Mueller UG, Wilcox TP, Bull JJ. The evolution of cooperation. Q Rev Biol. 2004;79:135–160. doi: 10.1086/383541. [DOI] [PubMed] [Google Scholar]
- 35.Pruesse E, et al. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jost L. Partitioning diversity into independent alpha and beta components. Ecology. 2007;88:2427–2439. doi: 10.1890/06-1736.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.