Abstract
We elucidate the detailed effects of gut microbial depletion on the bile acid sub-metabolome of multiple body compartments (liver, kidney, heart, and blood plasma) in rats. We use a targeted ultra-performance liquid chromatography with time of flight mass-spectrometry assay to characterize the differential primary and secondary bile acid profiles in each tissue and show a major increase in the proportion of taurine-conjugated bile acids in germ-free (GF) and antibiotic (streptomycin/penicillin)-treated rats. Although conjugated bile acids dominate the hepatic profile (97.0 ± 1.5%) of conventional animals, unconjugated bile acids comprise the largest proportion of the total measured bile acid profile in kidney (60.0 ± 10.4%) and heart (53.0 ± 18.5%) tissues. In contrast, in the GF animal, taurine-conjugated bile acids (especially taurocholic acid and tauro-β-muricholic acid) dominated the bile acid profiles (liver: 96.0 ± 14.5%; kidney: 96 ± 1%; heart: 93 ± 1%; plasma: 93.0 ± 2.3%), with unconjugated and glycine-conjugated species representing a small proportion of the profile. Higher free taurine levels were found in GF livers compared with the conventional liver (5.1-fold; P < 0.001). Bile acid diversity was also lower in GF and antibiotic-treated tissues compared with conventional animals. Because bile acids perform important signaling functions, it is clear that these chemical communication networks are strongly influenced by microbial activities or modulation, as evidenced by farnesoid X receptor-regulated pathway transcripts. The presence of specific microbial bile acid co-metabolite patterns in peripheral tissues (including heart and kidney) implies a broader signaling role for these compounds and emphasizes the extent of symbiotic microbial influences in mammalian homeostasis.
Keywords: farnesoid X receptor, gut microbiota, TGR5, ultra-performance liquid chromatography mass spectrometry, G protein-coupled bile acid receptor 1
The importance of gut microbiome variation in relation to human health and diverse diseases is now well-recognized (1–4). The microbiome is a virtual organ that performs many digestive and metabolic functions for the host, including enhanced calorific recovery from ingested foods and degradation of complex plant polysaccharides. Microbial communities have coevolved with man and show remarkable diversity dependent on topographical location and interperson variability (5). Co-evolution has refined the microbiome of organisms to a state where metabolic complementarity exists within the microbiota (6), and important biosynthetic/metabolic pathways are provided for the host that significantly extend host metabolic capacity (3). As such, the mammalian host can be considered a superorganism (7), whose metabolism is the sum of that of both the host and the collective microbial community. The enterohepatic circulation provides a vehicle for this transgenomic metabolism, and bile acids, whose functional role in the global mammalian system is multifaceted, are an important class of metabolites that undergo extensive enterohepatic recycling and gut microbial modification. Bile acids are cholesterol derivatives synthesized in the liver and then conjugated with glycine or taurine before secretion into the bile and small intestine. In the intestine, bile acids restrict bacterial proliferation and overgrowth, whereas bacterial enzymes modify primary bile acids through deconjugation, dehydrogenation, dehydroxylation, and sulfation reactions (8) to produce secondary bile acids, which are reabsorbed and returned to the liver for further processing.
Historically, bile acids have been primarily viewed as detergent molecules important for the absorption of dietary fats and lipid-soluble vitamins in the small intestine and the maintenance of cholesterol homeostasis in the liver. However, their role in the mammalian system is much broader than this, and they are now recognized as important signaling molecules with systemic endocrine functions. Bile acids are natural ligands for the nuclear receptor, farnesoid X receptor (FXR), and the plasma membrane-bound bile acid receptor TGR5 [also known as G protein-coupled bile acid receptor 1 (Gpbar1); membrane-type receptor for bile acids (M-BAR)]. Through activation of these receptors bile acids regulate lipid (9–11), glucose (12–16), and energy homeostasis (17) in addition to regulating their own synthesis (18), conjugation (19), transport (20–22), and detoxification (19, 23, 24). The global signaling capacity of bile acids is currently unclear; however, the expression of bile acid receptors FXR and TGR5 in tissues outside of the enterohepatic circulation, including the kidney (25) and heart (26, 27), suggests a greater role throughout the body. Expression of bile acid transporters in renal tubular cells [Asbt (28), organic anion transporting polypeptide 1 (OATP1) (29), and kidney specific organic anion transporter (OATK2) (30)] and cardiomyocytes (31, 32) further supports this proposition. Moreover, the involvement of bile acids in the regulation of glucose homeostasis is consistent with the expression of FXR in pancreatic β-cells, which has been shown to play an essential role in the regulation of insulin transcription and secretion induced by glucose (33).
The bile acid signature is heavily dependent on microbial activities and so, here we have explored the impact of the microbiota on the bile acid profiles of different tissues (liver, kidney, and heart) and in the plasma using two models of gut microbial modulation. We contrasted a germ-free (GF) rat model against the conventional (CV) rat model to assess the effect of life-long bacterial absence on the bile acid profile, and subsequently, we evaluated the impact of a more subtle microbiotal perturbation by adopting an antibiotic-treated (AB) rat model. Here, we compare tissue bile acid profiles from rats undergoing bacterial suppression achieved by 8 d of oral administration of antibiotics (penicillin and streptomycin) with those of control rats and characterize bile acid profiles across multiple tissue compartments using a targeted ultra performance liquid chromatography mass spectrometry (UPLC-MS) approach.
Results
Compartmental Bile Acid Profiles.
We detected a panel of bile acids in the liver, kidney, heart, and plasma of conventional animals using UPLC-MS (Fig. 1), which included a range of unconjugated bile acids in addition to those bile salt species conjugated with glycine and taurine. We found the bile acid intensity profile to be tissue-specific; for example, the hepatic bile acid profile was dominated by glycine and taurine conjugates, with glycocholic acid (GCA), glyco-β-muricholic acid (GβMCA), taurocholic acid (TCA), and tauro-β-muricholic (TβMCA) present at the highest intensities, consistent with extant literature (34). In the kidney, those species with the highest intensities were primary, secondary, and tertiary unconjugated bile acids. Glycine conjugates were present at a lower intensity to the unconjugated bile acids in the kidney, and taurine conjugates were the least intense. The total relative bile acid intensity of the kidney was ∼20.7-fold lower than that of the liver based on the summed signal intensity of the bile acids measured. The bile acid profile of the heart followed a similar, but not identical, trend to the kidney, with unconjugated bile acids the most intense followed by glycine and then taurine conjugates. We detected primary, secondary, and tertiary bile acids, but overall, fewer bile acid species were present compared with the liver and kidney, with α-muricholic acid (α-MCA) and its conjugated forms being absent. The total relative intensity of the heart bile acid pool was ∼47.4-fold lower than the summed signal intensity of the liver. In the case of the blood plasma, we found that the most intense signals derived from unconjugated bile acids, specifically cholic acid (CA) and β-muricholic acid (β-MCA). As in the kidney and heart, the levels of glycine conjugates were greater than taurine conjugates, although there were fewer species of glycine-conjugated bile acids detected than in the tissues, with GCA, GβMCA, and glycine-conjugated Δ22ω-MCA accounting for the majority of the total plasma bile acid signal intensity. The total relative bile acid signal in plasma was approximately 2-fold lower than the liver. Tissue bile acid profiles were different in composition to the plasma profiles, and several bile acids were present in the tissues that were absent in the plasma. Therefore, we are confident that the bile acid signals measured in tissues predominantly derive from tissue bile acids and not from residual blood contained within the tissue.
Fig. 1.
Relative bile acid profiles of the liver, plasma, kidney, and heart of conventional rats. Values are mean intensity values + SEM (scaled loge) measured using UPLC-MS. Abbreviations for the measured bile acids with additional structural information are given in the table. The prefixes G and T indicate conjugation with either glycine or taurine, respectively. For the bile acids labeled with the following scheme, C24Ox-yΔ, x is the number of oxygens on the bile acid skeleton, and y is the number of double bonds in the structure. 22Δ indicates a double bond on carbon 22.
Effects of Microbial Presence on Bile Acid Profiles.
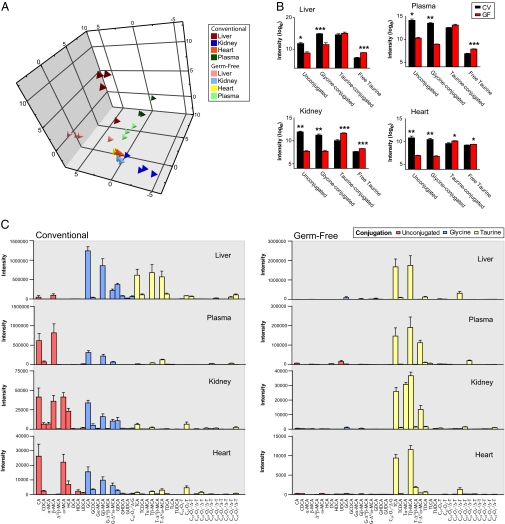
We established that the bile acid profiles of GF animals were markedly different from their CV equivalents across all sample types through principal components analysis (PCA) of the UPLC-MS spectral data (Fig. 2A). The bile acid signatures of all GF samples were dominated by taurine-conjugated bile acids with relatively lower unconjugated and glycine-conjugated bile acids compared with CV samples (Fig. 2C). From the UPLC-MS profiles, we found free taurine to be 5.1-fold higher in the GF liver with respect to the CV liver (P < 0.001) and observed a similar trend in the GF kidney, heart, and plasma, where free taurine was 1.9- (P < 0.001), 1.2- (P = 0.018), and 2.7-fold (P < 0.001) higher, respectively, than in CV animals (Fig. 2B). PCA scores plots constructed from the bile acid data of the individual tissues showed tight clustering of the GF animals with clear separation from the CV animals, whose bile acid profiles showed a larger degree of variation (Fig. S1). We showed that plasma bile acid signatures also differed between the two groups, although intragroup variation was comparable between GF and CV groups. Across all samples, we found that the GF profile was dominated by the primary taurine-conjugated bile acid, TCA, with TβMCA also dominant in terms of signal intensity in the plasma, kidney, and heart (Fig. 3A). GF tissues contained a significantly lower proportion of secondary and tertiary unconjugated and glycine-conjugated bile acids compared with CV animals. We found that the glycine conjugates of bile acids, GCA, glyco-chenodeoxycholic acid (GCDCA), GβMCA, and glyco-deoxycholic acid (GDCA), were lower in all GF tissues, including the plasma, whereas the secondary and tertiary glycine conjugates, glyco-hyodeoxycholic acid (GHDCA) and glyco-ursodeoxycholic acid (GUDCA), were lower in all tissues but not the plasma. Compared with the CV animals, the total intensity of the GF hepatic bile acid pool was 1.3-fold lower (P = 0.35), and those of the kidney and heart were 2.1- (P = 0.012) and 4.1-fold (P = 0.007) lower, respectively. The total intensity of the plasma bile acid pool was 4.7-fold lower (P < 0.021) in the GF with respect to the CV animals. Although the summed bile acid intensity was lower in all GF samples compared with the CV samples, the total taurine-conjugated bile acid intensity was higher in GF animals in all tissues, although this trend was only statistically significant in the kidney (P < 0.001) and heart (P = 0.031).
Fig. 2.
Effects of microbial absence on bile acid profiles. (A) PCA scores plot of the bile acid signatures in all sample matrices of GF and CV animals. (B) Total relative signals of the bile acids based on conjugation state are compared for the GF and CV animals in the liver, kidney, heart, and plasma. Free taurine is also compared. Values are means of the scaled (loge) data + SEM. Significant difference by Student t test. *P < 0.05; **P < 0.01; ***P < 0.001. (C) Intensity values for the measured bile acid species in all sample matrices of CV and GF rats. Values are mean intensities + SEM measured using UPLC-MS.
Fig. 3.
Effects of microbial absence and antibiotic treatment on bile acid profiles. (A) Intensity differences of individual bile acid species in GF rats in relation to CV rats in the liver, plasma, kidney, and heart. Bile acid species shown are those found to be statistically different (using a Student t test; P < 0.05) from those measured in CV rats; (B) Bile acid species found to be statistically different between control rats and those treated with antibiotics in the liver, kidney, and heart. Also shown is the relative intensity from the free taurine in the liver. Values are means of the scaled (loge) data ± SEM. Statistical significance was determined by Student t test. *P < 0.05; **P < 0.01; ***P < 0.001.
The analytical approach that we adopted here uses a targeted assay to allow the relative quantification of the major and some of the minor bile acid species present in the tissues; however, it is possible that some minor bile acid species may not have been measured. Such considerations should be taken into account when interpreting total bile acid comparisons. Nevertheless, the study allows an accurate direct comparison of the individual bile acid species measured.
Temporal Effects of Antibiotic Administration on Bile Acid Profiles.
We found that antibiotic-induced suppression of the microbiota caused a subtle perturbation of the bile acid profiles of the tissues, but no effect was observed in the plasma (Fig. 3B). PCA scores plots are shown in Fig. S2. The trend for this bile acid modulation was similar to the GF animals, with the profile skewed to increased presence of taurine-conjugated bile acids and lower abundance of unconjugated and glycine-conjugated bile acids compared with the control animals. In the liver, we show that microbial attenuation increased TCA, TβMCA, and an unsaturated taurine-conjugated bile acid (C24O4) and reduced the unconjugated species, αMCA and βMCA. We also detected an increase in hepatic taurine after bacterial suppression, as in GF animals (1.7-fold; P = 0.014). The kidney profiles of AB-treated animals had higher TCA and C24O2-1Δ-taurine and lower chenodeoxycholic acid (CDCA), hyocholic acid (HCA), GDCA, and C24O2-1Δ-taurine, whereas the heart profile of the AB animals contained significantly reduced GCDCA and C24O2-1Δ-taurine compared with the control animals.
Hepatic Metabolic and Signaling Pathways Modulated by the Gut Microbiota.
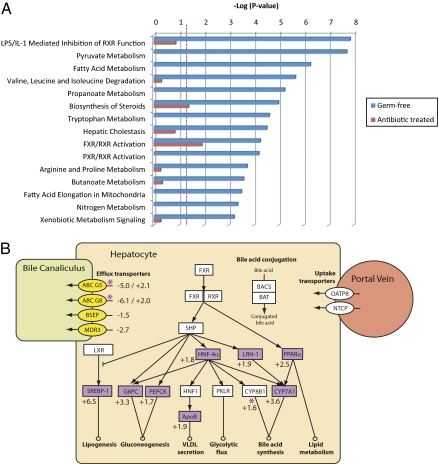
We identified differentially expressed genes modified in response to microbiotal deprivation in the livers of GF and AB rats relative to their respective controls. A greater number of transcripts were modulated by >1.5-fold (P < 0.05) in GF rats compared with AB rats (1,676 vs. 295, respectively) (Dataset S1). Canonical pathways altered in GF relative to CV rat livers were identified using Ingenuity Software (Ingenuity Systems, Inc.) (Fig. 4), and a large proportion of these were associated with metabolic and signaling pathways involved in bile acid metabolism, including FXR/retinoid X receptor (RXR) activation and the biosynthesis of steroids. The same pathway analysis method was carried out on the genes, with expression altered in response to AB treatment. Across both animal models of gut-microbial deprivation, two pathways were consistently different from their respective controls, and both pathways were regulated by bile acids (Fig. 4A). These were biosynthesis of steroids and FXR/RXR-activation pathways. Consistent with the bile acid profiles, these differences were more pronounced in the GF animals. The most significant pathways altered in the AB animals related to signaling events associated with axon guidance and cancer (Fig. S3).
Fig. 4.
Hepatic metabolic and signaling pathways modulated by the gut microbiota. (A) Significantly regulated canonical gene expression pathways in GF rats (blue bars) and pathways common to AB rats (red bars). Scale is a log-transformed P value calculated by Fisher's exact test that a pathway is over-represented within the altered genes. Dotted red lines indicate threshold value (P < 0.05) for pathway change to be considered statistically significant. (B) Relationship between FXR/RXR-regulated genes and bile acid metabolic pathways. Purple shapes indicate up-regulated genes, and yellow shapes indicate down-regulated genes in GF rats with respect to conventional rats. Purple stars denote up-regulated genes in AB rats with respect to control rats. White shapes indicate genes that were present on the microarray but were unchanged in GF rats. Values indicate fold change in gene transcription in GF rats with respect to conventional rats and in AB rats with respect to control rats. ApoB, apolipoprotein B; G6PC, glucose-6-phosphatase; HNF, hepatocyte nuclear factor; LRH-1, liver receptor homolog-1; LXR, liver X receptor; PEPCK, phosphoenolpyruvate carboxykinase; PKLR, pyruvate kinase; PPARα, peroxisome proliferator-activated receptor α; RXR, retinoid X receptor; SHP, short heterodimer partner; SREBP-1, sterol regulatory element binding protein-1.
The genes within the FXR/RXR activation pathway were related to the functional role of their encoded proteins in bile acid metabolism (Fig. 4B). In general, genes for bile acid synthetic enzymes [cytochrome P450 (CYP) 7A1 and CYP8B1] and efflux transporters [bile salt export pump (BSEP), multidrug resistance-associated protein 2 (MRP2), multidrug resistance protein 3 (MDR3), and ATP-binding cassette (ABC) G5/G8] were modulated in one or both regimens of reduced gut microbiota. In contrast, genes for bile acid conjugation enzymes [bile acid CoA synthetase (BACS) and bile acid-CoA:amino acid N-acyltransferase (BAT)] or uptake transporters [OATP8 and sodium-taurocholate cotransporting polypeptide (NTCP)] were not altered in either GF or AB rats. Genes encoding enzymes involved in glucose and lipid metabolism were also disrupted in the GF animals; these included genes that can be regulated by bile acids such as phosphoenolpyruvate carboxykinase (PEPCK), glucose-6-phosphatase (G6PC), and sterol regulatory element binding protein 1 (SREBP-1).
Discussion
We show that the gut microbiota impact significantly on the relative composition and abundance of bile acids across multiple tissue compartments. In the absence of gut microbiota, bile acid signatures of the liver, kidney, heart, and plasma were strikingly different to the conventional animals. The bile acid profiles for all tissues in the GF model were dominated by a small subset of taurine-conjugated bile acids, with glycine-conjugated and unconjugated bile acids present in negligible amounts. Short-term attenuation of the gut microbiota by antibiotics induced a similar, albeit less pronounced, trend of alterations in the tissue bile acid profiles. The global transcript profiles of the liver tissues revealed two pathways under the regulation of bile acids to be significantly different from the control animals in both GF and AB models. As with the bile acid profiles, these differences were exaggerated in the GF animals. The presence of bile acids in peripheral tissues, in addition to bile acid transporters and receptors, suggests a global signaling role for these transgenomic metabolites and may represent another mechanism through which the gut microbiome interacts with the mammalian host.
As expected we found that bile acids were most abundant in the liver, the site of bile acid synthesis, followed by the plasma, kidney, and heart. We show that the variation in tissue bile acid signatures reflects their position within and external to the enterohepatic circulation. Thus, unconjugated bile acids comprise the majority of the plasma, kidney, and heart signal but represent the smallest fraction of the total bile acid signal measured in the liver. The accumulation of unconjugated bile acids in the kidney and heart is consistent with the fact that, for these tissues, the bile acid pool has traversed the intestinal tract. Therefore, the majority of bile acids that returned to the liver and subsequently spilled over into systemic circulation are the microbially modified unconjugated forms. Additionally, it is known that the hepatic extraction of free bile acids is also less efficient compared with the conjugated forms (35), and this further explains the greater proportion of unconjugated bile acids in the systemic circulation and consequently, the kidney and heart. At physiological pH, conjugated bile acids are negatively charged and require carrier-mediated transport across tissue membranes; therefore, it is likely that these organ-specific profiles arise from selective uptake caused by differential expression of bile acid transporters in the renal tubular cells and cardiomyocytes (28–32).
In a previous study, Dupont et al. (36) showed organ-specific differences in the ratios of glycine- and taurine-conjugated bile acids in a range of tissues from miniature swine using gas liquid chromatography. In this swine model, one-half of the liver bile acids were shown to be conjugated with taurine, and one-third were conjugated with glycine; the heart profile was composed of equal proportions of glycine and taurine conjugates, and the kidney profile comprised almost exclusively of taurine-conjugated bile acids. These findings differ from ours and are most likely attributable to host-species differences. Additionally, because only the conjugation substrate was known and not the identity of the individual bile acid species, it is difficult to make a direct comparison.
Although the biological significance of bile acids in the heart remains to be elucidated, given their known function as signaling molecules and the presence of the FXR and TGR5 receptors in addition to bile acid transporters in the heart (26, 27, 31, 32), it is feasible that bile acids serve a regulatory role in cardiomyocyte metabolism. A minor route of bile acid elimination is urinary excretion, removing excess bile acids from systemic circulation. Therefore, the detection of bile acids in the kidneys is not unexpected (36). Bile acid transporters in the kidneys serve an important role in maintaining bile acid homeostasis, limiting urinary excretion while minimizing toxic accumulation (37). In this study, because we found negligible concentrations of bile acids in the urine of control or AB rats using UPLC-MS analysis but found diverse species of bile acids in the kidney, we assume that efficient bile acid reabsorption occurred. FXR is known to be involved in bile acid elimination through the regulation organic solute transporter (Ost)-α/β and multidrug resistance-associated protein 2 (Mrp2) transporters on the basolateral surface of renal tubular cells; however, the expression of the receptor TGR5 in this tissue suggests that bile acids may also serve a physiological role in the kidney.
Collectively, these data suggest that bile acids may have a signaling function outside of the enterohepatic circulation, possibly involved in glucose, lipid, and energy homeostasis through FXR and TGR5. Consistent with this notion is the fact that serum bile acid concentrations are known to increase by severalfold postprandially (38), which may represent a global signaling pathway to synchronize host metabolism with dietary (substrate) intake. Bile acids can increase energy expenditure in brown adipose tissue and muscle through activation of TGR5 and subsequent induction of the enzyme 2-iodothyronine deiodinase (17). This enzyme converts inactive thyroxine (T4) to tri-iodothyronine (T3), resulting in an increase in metabolic rate and energy expenditure (17). Through this mechanism, bile acids have been found to mitigate diet-induced obesity (39). Differential activation of TGR5 by variations in the bile acid pool between GF and CV rats may manifest in differences in energy expenditure. Indeed, GF animals are characterized by a lower basal metabolic rate compared with their CV equivalents (40). Recent work has confirmed an association between microbial composition and obesity (2, 4) mediated through several mechanisms such as increased fat storage through fasting-induced adipocyte factor (fiaf) suppression (41), increased energy harvest from the diet, and reduced fatty acid oxidation (42). Microbial influence over bile acid processing and conjugation patterns may represent another mechanism through which the microbiota can impact on host lipid metabolism and obesity, and in principle, the GF bile acid pool has a higher capacity for lipid emulsification and absorption. The importance of bile acids in the uptake and storage of dietary fats is well-known, and taurine-conjugated bile acids have a reduced hydrophobic:hydrophilic ratio compared with glycine-conjugated and unconjugated bile acids, increasing their solubility and lipid-emulsification properties.
Previously, we and others have identified higher concentrations of taurine-conjugated bile acids in the livers and intestines of GF mice (43) and rats (44) and in those colonized by human baby microbiota (45) compared with CV animals. Here, we show that, in the rat, gut microbial absence skews the bile acid profile of the liver to one dominated by the taurine-conjugated species, TCA, with glycine-conjugated and unconjugated species being markedly reduced. Although the global intensity of tauro-conjugated bile acids was higher in GF rats, the diversity of the taurine-conjugated species was notably reduced, which can be clearly seen in the PCA scores plots (Fig. S1), a similar finding to the ileal flushes of GF mice colonized with human baby flora (45). These observations were translated across the kidney, heart, and plasma profiles, with TβMCA also accounting for a large proportion of the total tauro-conjugated bile acid signal that increased in these matrices. Although the conjugation of bile acids with either taurine or glycine has been found to have minimal effect on the activation of FXR (46), the conjugates of CDCA, deoxycholic acid (DCA), and lithocholic acid (LCA) induced a stronger activation of FXR than conjugated forms of CA, and no activity was observed with tauro-MCA (47). TβMCA constituted a large proportion of the GF bile acid profile (34.5% of the GF plasma profile and 1.8% of the CV plasma profile) and suggests lower activation of FXR in the GF animal. Bacterial suppression through antibiotic treatment induced a similar shift in the tissue bile acid profiles to those seen in the GF animals, although this modulation was less pronounced and did not manifest in a difference in the plasma bile acid composition. Interestingly, microarray analysis identified FXR-related pathway changes in the liver with both methods of gut microbiotal deprivation. As with the microbially modulated bile acid signatures, these pathway changes were more pronounced in the GF rats compared with the AB rats. Consistent expression differences across both animal models indicate that bacterial loss rather than developmental consequences of the GF state underlie these FXR-related variations. In the GF animals, these FXR-related pathway changes included genes encoding enzymes involved in glucose and lipid metabolism.
The dominance of taurine-conjugated bile acids and diminished unconjugated and glycine-conjugated bile acids in GF and AB animals is consistent with the higher concentration of hepatic taurine in the absence or attenuation of the gut microbiota. Hardison and Proffitt (48) first showed that the bile salt glycine:taurine ratio is dependent on hepatic taurine concentration, even in the presence of high glycine concentrations (49). Conjugation of bile acids is catalyzed by two sequential enzymatic reactions involving BACS and BAT, with both glycine and taurine conjugation catalyzed by BAT in rats and humans (50). In rats, BAT has a much greater affinity for taurine than for glycine, which implies that taurine conjugation dominates at physiological concentrations (51). In the current study, no difference was observed in the transcription of either BACS or BAT enzymes between the GF and CV animals, and it remained unchanged in the AB animals. Considering that BAT is the enzyme for the conjugation of bile acids with either glycine or taurine, this supports the notion that the higher taurine pools in the GF and AB rats resulted in the dominance of taurine-conjugated bile acids in these animals. Higher taurine pools are likely to arise from the absence or attenuation of microbial taurine degradation. Taurine can be biosynthesized in hepatocytes, but the majority is obtained from the diet. Gut microbiota have been shown to have a high metabolic activity for degrading taurine to inorganic sulfate (52) or other organic sulfate catabolites (53), and therefore, reduced microbial activity could increase taurine bioavailability (54). Previous work by our group noted consistently higher concentrations of free taurine in the livers of GF mice (43) and the plasma of acclimatizing GF rats (55).
Although the functional role of the bile acids in the heart remains unclear, the different profiles between GF and CV animals may underlie the different morphological phenotypes of these two strains. GF animals have smaller hearts, livers, and lungs compared with CV animals, with a significantly reduced metabolic rate, lower oxygen consumption, and reduced cardiac output (40). Recent work by Crawford et al. (56) implicated myocardial ketone body metabolism in this reduced myocardial-mass phenotype observed in GF mice. It is feasible that microbial modulation of bile acid signaling through TGR5 may also contribute to this GF phenotype, because raised T3, as well as increasing metabolic rate, also increases cardiac output through elevations in heart rate and force of contraction. Additionally, TCA, one of the bile acids dominating the GF bile acid profiles, has been shown to reduce the rate and amplitude of contraction in rat neonatal and adult cardiomyocytes (57, 58). The effect on cardiomyocytes by glycine-conjugated bile acids, characteristic of the conventional animals, was less marked than TCA and unlike TCA, was shown to be completely reversible at higher concentrations (59). Therefore, the higher concentrations of TCA and TβMCA in the hearts of GF animals may directly impact on cardiac development. Interestingly, TCA has been identified to cause dysarrhythmia in cardiomyocytes, raising the potential for differences in microbial composition to impact on heart function through modification to the bile acid profile. In addition to the microbial impact exerted through influence on taurine-conjugated bile acids, the cardioprotective tertiary bile acid ursodeoxycholic acid (UDCA) formed through host and microbial cometabolism is also present in lower amounts in the GF hearts (60, 61). The trend in bile acid conjugation patterns in the AB animals was similar to that observed in the GF animals, although the effect was not statistically significant after 8 d of treatment. Further investigation is required to establish whether prolonged use of antibiotics would affect the equilibrium of bile acid composition in cardiac tissue with consequent impact on human health.
Conclusions
We have profiled a panel of bile acids in the liver, kidney, heart, and plasma of rats at steady state and found characteristic profiles to be specific to each tissue. We propose that the panel of bile acids measured in the heart may serve a regulatory/signaling function in this tissue, because both transporters and receptors specific to bile acids are expressed here. Life-long gut bacterial absence had a profound effect on the bile acid profiles of the liver, kidney, and heart, shifting the balance to almost exclusive taurine-conjugated species. Short-term perturbation of the gut microbiota by antibiotic treatment modulated the bile acid profiles in a similar manner, but the effect was less pronounced. These results suggest that the bile acids act as signaling molecules outside of the enterohepatic circulation, and consequently, modification of the bile acid composition by the gut microbiota may represent another mechanism by which the gut microbiota influence host metabolism.
Materials and Methods
Animal Handling and Sample Collection.
The animal studies were planned in accordance with the standards of animal care and ethics described in Guidance on the Operations of the Animal (Scientific Procedures) Act 1986 issued by the UK Home Office. Livers, kidneys, and hearts were obtained from four GF and four CV male Sprague-Dawley rats aged 7–8 wk (Charles River). All rats were fasted before necropsy and euthanized by halothane inhalation. Blood was also collected at necropsy. For the antibiotic-treated study, male Han-Wistar–derived rats aged ∼10 wk were separated into control (n = 6) and AB (n = 6) groups. The antibiotic dose (4 mg/mL streptomycin and 2 mg/mL penicillin) was provided in the drinking water of AB animals for 8 d, whereas the control group received antibiotic-free drinking water. After 8 d, we euthanized all animals by halothane inhalation and collected blood, liver, kidney, and heart tissues.
Reagents.
All solvents were HPLC grade. Water, acetonitrile (ACN), methanol, dichloromethane, leucine enkephalin, sodium formate, formic acid, and the taurine standard were obtained from Sigma-Aldrich. Bile acid standards were obtained from Steraloids.
Preparation of Extracts and Standards.
Tissue samples.
Thirty milligrams of tissue were obtained and washed with water to remove any residual blood to minimize the contribution of blood bile acids to the measured tissue profile. Washed tissue samples were then extracted with 300 μL chloroform:methanol (2:1), and the samples were homogenized for 8 min at 25 Hz in the TissueLyser (Qiagen). Vials were vortexed after the addition of 300 μL water and spun down at 10,000 × g for 10 min; the upper (aqueous) phase was subsequently separated into a clean tube. We repeated the extraction procedure with the remaining pellet and combined the aqueous phases to improve recovery. Extracted samples were snap frozen in liquid nitrogen and freeze dried. All samples were kept at −40 °C until reconstitution with 100 μL water and transferred to glass vials (Waters Corp.) before analysis.
Plasma samples.
Plasma samples were prepared for UPLC-MS analysis by methanol protein precipitation where ice-cold methanol (150 μL) was added to 50 μL plasma and vortexed for 30 s. The samples were then incubated at −20 °C for 20 min and centrifuged at 10,000 × g for 10 min, and the supernatant was removed to a clean tube. This supernatant was dried down in a Savant vacuum evaporator (Jencons), reconstituted in 100 μL water, and transferred into 96-well 350-μL plates.
Standard solutions.
Bile acid (1 mg/mL) stock solutions were prepared in water:methanol (80:20 vol/vol) and diluted in water to obtain the desired working solutions. A taurine solution was prepared in 10 μg/mL water.
UPLC-MS Analysis.
Tissue and plasma samples (5 μL) were injected onto a 2.1 × 100 mm (1.7 μm) HSS T3 Acquity column (Waters Corp.). Tissue extracts were eluted using a 25-min gradient of 100% A to 100% B (A, water, 0.1% formic acid; B, acetonitrile, 0.1% formic acid) at a flow rate of 500 μL/min and column temperature of 40 °C. Plasma samples were eluted using a 20-min gradient of 100% A to 100% B (A, water, 0.1% formic acid; B, methanol, 0.1% formic acid) at a flow rate of 400 μL/min and column temperature of 50 °C. Samples were analyzed using an Acquity UPLC system (Waters Ltd.) coupled online to an LCT Premier mass spectrometer (Waters MS Technologies, Ltd.) in negative electrospray mode with a scan range of 50–1,000 m/z. Bile acids ionize strongly in negative mode, producing a prominent [M-H]− ion. Capillary voltage was 2.4 Kv, sample cone was 35 V, desolvation temperature was 350 °C, source temperature was 120 °C, and desolvation gas flow was 900 L/h. Mass-spectrometric conditions were optimized through direct infusion of bile acid standards. The LCT Premier was operated in V optics mode, with a data-acquisition rate of 0.1 s and a 0.01-s interscan delay. Leucine enkephalin (m/z 556.2771) was used as the lockmass; a solution of 200 pg/μL (50:50 ACN:H2O) was infused into the instrument at 3 μL/min through an auxillary sprayer. Data were collected in centroid mode with a scan range of 50–1,000 m/z, with lockmass scans collected every 15 s and averaged over three scans to perform mass correction.
Data Analysis.
The UPLC-MS datasets were preprocessed through peak picking and alignment before multivariate statistical analysis using the freeware package XCMS (62). Marker tables were generated comprising m/z, retention time (RT), and intensity (peak area) values for each variable in every sample, which were exported into SIMCA-P 11.5 (Umetrics) for further multivariate analysis. Here, PCA was performed on all data after scaling to unit variance. Differences in individual bile acid abundance between CV and GF animals and between control and AB-treated animals were evaluated using a two-tailed unpaired t test. Authentic bile acid standards, where available, and a taurine standard were analyzed in negative ionization mode to identify the retention time and [M-H]− of these metabolites. The intensities (area under curve) of targeted metabolites were calculated from the extracted ion chromatograms using XCMS, and values were compared across study groups.
Method Validation.
Standard curves for a range of primary, secondary, and tertiary bile acids, in addition to unconjugated, glycine-conjugated, and taurine-conjugated bile acids, were calculated over a dynamic range of 0–80 μg/mL (Table S1). All bile acids were found to be linear over the measured range.
Gene-Expression Analysis.
RNA was extracted from liver samples, and gene-expression microarray analysis was carried out on Affymetrix rat 230 v2 gene arrays as described previously using Microarray Suite 5 normalization (63). Gene-expression analysis was performed on three animals from each (CV, GF, non-AB control, and AB) group. The microarray data discussed in this publication have been deposited in National Center for Biotechnology Information's Gene Expression Omnibus (64) and are accessible through GEO Series accession number GSE21937 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE21937). Genes were considered differentially expressed if they showed a fold change (FC) of greater than 1.5 with a P value less than 0.05 (by unpaired t test) and an absolute call of at least two treated samples for up-regulated genes or at least two control samples for down-regulated genes. Canonical metabolic and signaling pathways associated with the differentially expressed genes were identified using Ingenuity Pathway Analysis Software Version 8 (IPA; Ingenuity Systems). Statistically overrepresented pathways were identified by Fisher's exact test (P < 0.05).
Supplementary Material
Acknowledgments
The authors are grateful to the scientific and laboratory personnel of Safety Assessment (Alderley Park, AstraZeneca) for the excellent conduct of the animal studies. We thank John Shockcor for superb technical assistance. E.J.W. acknowledges Waters Corp. for funding.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Microbes and Health” held November 2–3, 2009, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS Web site at http://www.nasonline.org/SACKLER_Microbes_and_Health.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE21937).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006734107/-/DCSupplemental.
References
- 1.Dumas ME, et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Natl Acad Sci USA. 2006;103:12511–12516. doi: 10.1073/pnas.0601056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: Human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 3.Nicholson JK, Holmes E, Wilson ID. Gut microorganisms, mammalian metabolism and personalized health care. Nat Rev Microbiol. 2005;3:431–438. doi: 10.1038/nrmicro1152. [DOI] [PubMed] [Google Scholar]
- 4.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 5.Costello EK, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu D, et al. Metabolic complementarity and genomics of the dual bacterial symbiosis of sharpshooters. PLoS Biol. 2006;4:e188. doi: 10.1371/journal.pbio.0040188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lederberg J. Infectious history. Science. 2000;288:287–293. doi: 10.1126/science.288.5464.287. [DOI] [PubMed] [Google Scholar]
- 8.Midtvedt T. Microbial bile acid transformation. Am J Clin Nutr. 1974;27:1341–1347. doi: 10.1093/ajcn/27.11.1341. [DOI] [PubMed] [Google Scholar]
- 9.Hirokane H, Nakahara M, Tachibana S, Shimizu M, Sato R. Bile acid reduces the secretion of very low density lipoprotein by repressing microsomal triglyceride transfer protein gene expression mediated by hepatocyte nuclear factor-4. J Biol Chem. 2004;279:45685–45692. doi: 10.1074/jbc.M404255200. [DOI] [PubMed] [Google Scholar]
- 10.Kast HR, et al. Farnesoid X-activated receptor induces apolipoprotein C-II transcription: A molecular mechanism linking plasma triglyceride levels to bile acids. Mol Endocrinol. 2001;15:1720–1728. doi: 10.1210/mend.15.10.0712. [DOI] [PubMed] [Google Scholar]
- 11.Watanabe M, et al. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J Clin Invest. 2004;113:1408–1418. doi: 10.1172/JCI21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baquet A, Hue L, Meijer AJ, van Woerkom GM, Plomp PJ. Swelling of rat hepatocytes stimulates glycogen synthesis. J Biol Chem. 1990;265:955–959. [PubMed] [Google Scholar]
- 13.Häussinger D, Hallbrucker C, Saha N, Lang F, Gerok W. Cell volume and bile acid excretion. Biochem J. 1992;288:681–689. doi: 10.1042/bj2880681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katsuma S, Hirasawa A, Tsujimoto G. Bile acids promote glucagon-like peptide-1 secretion through TGR5 in a murine enteroendocrine cell line STC-1. Biochem Biophys Res Commun. 2005;329:386–390. doi: 10.1016/j.bbrc.2005.01.139. [DOI] [PubMed] [Google Scholar]
- 15.Saha N, Stoll B, Lang F, Häussinger D. Effect of anisotonic cell-volume modulation on glutathione-S-conjugate release, t-butylhydroperoxide metabolism and the pentose-phosphate shunt in perfused rat liver. Eur J Biochem. 1992;209:437–444. doi: 10.1111/j.1432-1033.1992.tb17307.x. [DOI] [PubMed] [Google Scholar]
- 16.Stayrook KR, et al. Regulation of carbohydrate metabolism by the farnesoid X receptor. Endocrinology. 2005;146:984–991. doi: 10.1210/en.2004-0965. [DOI] [PubMed] [Google Scholar]
- 17.Watanabe M, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439:484–489. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- 18.Goodwin B, et al. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell. 2000;6:517–526. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 19.Pircher PC, et al. Farnesoid X receptor regulates bile acid-amino acid conjugation. J Biol Chem. 2003;278:27703–27711. doi: 10.1074/jbc.M302128200. [DOI] [PubMed] [Google Scholar]
- 20.Ananthanarayanan M, Balasubramanian N, Makishima M, Mangelsdorf DJ, Suchy FJ. Human bile salt export pump promoter is transactivated by the farnesoid X receptor/bile acid receptor. J Biol Chem. 2001;276:28857–28865. doi: 10.1074/jbc.M011610200. [DOI] [PubMed] [Google Scholar]
- 21.Denson LA, et al. The orphan nuclear receptor, shp, mediates bile acid-induced inhibition of the rat bile acid transporter, ntcp. Gastroenterology. 2001;121:140–147. doi: 10.1053/gast.2001.25503. [DOI] [PubMed] [Google Scholar]
- 22.Neimark E, Chen F, Li X, Shneider BL. Bile acid-induced negative feedback regulation of the human ileal bile acid transporter. Hepatology. 2004;40:149–156. doi: 10.1002/hep.20295. [DOI] [PubMed] [Google Scholar]
- 23.Barbier O, et al. FXR induces the UGT2B4 enzyme in hepatocytes: A potential mechanism of negative feedback control of FXR activity. Gastroenterology. 2003;124:1926–1940. doi: 10.1016/s0016-5085(03)00388-3. [DOI] [PubMed] [Google Scholar]
- 24.Song CS, et al. Dehydroepiandrosterone sulfotransferase gene induction by bile acid activated farnesoid X receptor. J Biol Chem. 2001;276:42549–42556. doi: 10.1074/jbc.M107557200. [DOI] [PubMed] [Google Scholar]
- 25.Forman BM, et al. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell. 1995;81:687–693. doi: 10.1016/0092-8674(95)90530-8. [DOI] [PubMed] [Google Scholar]
- 26.Kawamata Y, et al. A G protein-coupled receptor responsive to bile acids. J Biol Chem. 2003;278:9435–9440. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Kast-Woelbern HR, Edwards PA. Natural structural variants of the nuclear receptor farnesoid X receptor affect transcriptional activation. J Biol Chem. 2003;278:104–110. doi: 10.1074/jbc.M209505200. [DOI] [PubMed] [Google Scholar]
- 28.Christie DM, Dawson PA, Thevananther S, Shneider BL. Comparative analysis of the ontogeny of a sodium-dependent bile acid transporter in rat kidney and ileum. Am J Physiol. 1996;271:G377–G385. doi: 10.1152/ajpgi.1996.271.2.G377. [DOI] [PubMed] [Google Scholar]
- 29.Bergwerk AJ, et al. Immunologic distribution of an organic anion transport protein in rat liver and kidney. Am J Physiol. 1996;271:G231–G238. doi: 10.1152/ajpgi.1996.271.2.G231. [DOI] [PubMed] [Google Scholar]
- 30.Takeuchi A, Masuda S, Saito H, Abe T, Inui K. Multispecific substrate recognition of kidney-specific organic anion transporters OAT-K1 and OAT-K2. J Pharmacol Exp Ther. 2001;299:261–267. [PubMed] [Google Scholar]
- 31.Couture L, Nash JA, Turgeon J. The ATP-binding cassette transporters and their implication in drug disposition: A special look at the heart. Pharmacol Rev. 2006;58:244–258. doi: 10.1124/pr.58.2.7. [DOI] [PubMed] [Google Scholar]
- 32.Gorelik J, et al. Genes encoding bile acid, phospholipid and anion transporters are expressed in a human fetal cardiomyocyte culture. BJOG. 2006;113:552–558. doi: 10.1111/j.1471-0528.2006.00918.x. [DOI] [PubMed] [Google Scholar]
- 33.Renga B, Mencarelli A, Vavassori P, Brancaleone V, Fiorucci S. The bile acid sensor FXR regulates insulin transcription and secretion. Biochim Biophys Acta. 2010;1802:363–372. doi: 10.1016/j.bbadis.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 34.Sakakura H, et al. Simultaneous determination of bile acids in rat liver tissue by high-performance liquid chromatography. J Chromatogr B Biomed Sci Appl. 1998;718:33–40. doi: 10.1016/s0378-4347(98)00342-9. [DOI] [PubMed] [Google Scholar]
- 35.Iga T, Klaassen CD. Hepatic extraction of bile acids in rats. Biochem Pharmacol. 1982;31:205–209. [PubMed] [Google Scholar]
- 36.Dupont J, Oh SY, O'Deen LA, Geller S. Cholanoic (bile) acids in hepatic and nonhepatic tissues of miniature swine. Lipids. 1974;9:294–297. doi: 10.1007/BF02532210. [DOI] [PubMed] [Google Scholar]
- 37.Zollner G, Marschall HU, Wagner M, Trauner M. Role of nuclear receptors in the adaptive response to bile acids and cholestasis: Pathogenetic and therapeutic considerations. Mol Pharm. 2006;3:231–251. doi: 10.1021/mp060010s. [DOI] [PubMed] [Google Scholar]
- 38.Pennington CR, Ross PE, Bouchier IA. Fasting and postprandial serum bile acid concentrations in normal persons using an improved GLC method. Digestion. 1978;17:56–62. doi: 10.1159/000198094. [DOI] [PubMed] [Google Scholar]
- 39.Thomas C, et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009;10:167–177. doi: 10.1016/j.cmet.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wostmann BS. The germfree animal in nutritional studies. Annu Rev Nutr. 1981;1:257–279. doi: 10.1146/annurev.nu.01.070181.001353. [DOI] [PubMed] [Google Scholar]
- 41.Bäckhed F, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci USA. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Claus SP, et al. Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Mol Syst Biol. 2008;4:1–14. doi: 10.1038/msb.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wostmann BS. Intestinal bile acids and cholesterol absorption in the germfree rat. J Nutr. 1973;103:982–990. doi: 10.1093/jn/103.7.982. [DOI] [PubMed] [Google Scholar]
- 45.Martin FP, et al. A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Mol Syst Biol. 2007;3:1–16. doi: 10.1038/msb4100153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reschly EJ, et al. Evolution of the bile salt nuclear receptor FXR in vertebrates. J Lipid Res. 2008;49:1577–1587. doi: 10.1194/jlr.M800138-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parks DJ, et al. Bile acids: Natural ligands for an orphan nuclear receptor. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- 48.Hardison WG, Proffitt JH. Influence of hepatic taurine concentration on bile acid conjugation with taurine. Am J Physiol. 1977;232:E75–E79. doi: 10.1152/ajpendo.1977.232.1.E75. [DOI] [PubMed] [Google Scholar]
- 49.Sweeny DJ, Barnes S, Diasio RB. Bile acid conjugation pattern in the isolated perfused rat liver during infusion of an amino acid formulation. JPEN J Parenter Enteral Nutr. 1991;15:303–306. doi: 10.1177/0148607191015003303. [DOI] [PubMed] [Google Scholar]
- 50.Falany CN, Johnson MR, Barnes S, Diasio RB. Glycine and taurine conjugation of bile acids by a single enzyme. Molecular cloning and expression of human liver bile acid CoA:amino acid N-acyltransferase. J Biol Chem. 1994;269:19375–19379. [PubMed] [Google Scholar]
- 51.Killenberg PG, Jordan JT. Purification and characterization of bile acid-CoA:amino acid N-acyltransferase from rat liver. J Biol Chem. 1978;253:1005–1010. [PubMed] [Google Scholar]
- 52.Hepner GW, Sturman JA, Hofmann AF, Thomas PJ. Metabolism of steroid and amino acid moieties of conjugated bile acids in man. 3. Cholyltaurine (taurocholic acid) J Clin Invest. 1973;52:433–440. doi: 10.1172/JCI107200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cook AM, Denger K. Dissimilation of the C2 sulfonates. Arch Microbiol. 2002;179:1–6. doi: 10.1007/s00203-002-0497-0. [DOI] [PubMed] [Google Scholar]
- 54.Kim SW, Rogers QR, Morris JG. Dietary antibiotics decrease taurine loss in cats fed a canned heat-processed diet. J Nutr. 1996;126:509–515. doi: 10.1093/jn/126.2.509. [DOI] [PubMed] [Google Scholar]
- 55.Swann J, et al. Gut microbiome modulates the toxicity of hydrazine: A metabonomic study. Mol Biosyst. 2009;5:351–355. doi: 10.1039/b811468d. [DOI] [PubMed] [Google Scholar]
- 56.Crawford PA, et al. Regulation of myocardial ketone body metabolism by the gut microbiota during nutrient deprivation. Proc Natl Acad Sci USA. 2009;106:11276–11281. doi: 10.1073/pnas.0902366106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gorelik J, et al. Taurocholate induces changes in rat cardiomyocyte contraction and calcium dynamics. Clin Sci (Lond) 2002;103:191–200. doi: 10.1042/cs1030191. [DOI] [PubMed] [Google Scholar]
- 58.Williamson C, et al. The bile acid taurocholate impairs rat cardiomyocyte function: A proposed mechanism for intra-uterine fetal death in obstetric cholestasis. Clin Sci (Lond) 2001;100:363–369. [PubMed] [Google Scholar]
- 59.Gorelik J, et al. Comparison of the arrhythmogenic effects of tauro- and glycoconjugates of cholic acid in an in vitro study of rat cardiomyocytes. BJOG. 2004;111:867–870. doi: 10.1111/j.1471-0528.2004.00166.x. [DOI] [PubMed] [Google Scholar]
- 60.Gorelik J, et al. Dexamethasone and ursodeoxycholic acid protect against the arrhythmogenic effect of taurocholate in an in vitro study of rat cardiomyocytes. BJOG. 2003;110:467–474. [PubMed] [Google Scholar]
- 61.Lee WY, Han SH, Cho TS, Yoo YH, Lee SM. Effect of ursodeoxycholic acid on ischemia/reperfusion injury in isolated rat heart. Arch Pharm Res. 1999;22:479–484. doi: 10.1007/BF02979156. [DOI] [PubMed] [Google Scholar]
- 62.Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 63.Craig A, et al. Systems toxicology: Integrated genomic, proteomic and metabonomic analysis of methapyrilene induced hepatotoxicity in the rat. J Proteome Res. 2006;5:1586–1601. doi: 10.1021/pr0503376. [DOI] [PubMed] [Google Scholar]
- 64.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.