Abstract
Lactobacilli have long been regarded as important constituents of the healthy human vagina. Lactobacillus iners is the most frequently detected bacterial species in the vagina, but little is known about its characteristics. We report a description of the whole-genome sequence of L. iners AB-1 along with comparative analysis of published genomes of closely related strains of lactobacilli. The genome is the smallest Lactobacillus reported to date, with a 1.3-Mbp single chromosome. The genome seems to have undergone one or more rapid evolution events that resulted in large-scale gene loss and horizontal acquisition of a number of genes for survival in the vagina. L. iners may exhibit specialized adaptation mechanisms to the vaginal environment, such as an iron–sulfur cluster assembly system, and several unique σ factors to regulate gene transcription in this fluctuating environment. A potentially highly expressed homolog of a cholesterol-binding lysin may also contribute to host cell adhesion or act as a defense mechanism against other microbes. Notably, there is a lack of apparent adhesion proteins, but several cell-anchor proteins were identified and may be important for interaction with the host mucosal tissues. L. iners is widely present in healthy females as well as those suffering from bacterial vaginosis or who have undergone antimicrobial therapy, suggesting that it is an important indigenous species of the vagina.
Keywords: vaginal microbiota, cholesterol-dependent cytolysin, sortase-dependent protein
The vaginal microbiota of premenopausal women with no symptoms or signs of disease are comprised primarily of Lactobacillus species. The consistency of this finding is quite remarkable, given the several hundred other microbial species that have been detected in the vagina (1–5) and the accessibility of even more from the rectal–anal skin and through bathing and sexual contact. For decades, lactobacilli have been regarded as beneficial to the vagina by preventing infections through production of organic acids, hydrogen peroxide (H2O2), and other antimicrobial substances and by the fact that depletion of Lactobacillus populations often coincides with bacterial disease onset.
The fastidious L. iners is consistently the most common Lactobacillus sp. in the vagina (1, 3, 5–7), but little is known about its characteristics. The detection of L. iners in women with, and recovering from, bacterial vaginosis (BV) has led to the suggestion that it is not protective against disease (8). However, unlike other species that seem to be easily displaced by pathogens and infectious conditions, the ability of L. iners to persist (5–9) may prove it to be important in recovery of the microbiota, postdisease resolution. Interestingly, through 16s microbial community profiling, the L. iners sequences recovered from different women show homogeneity and suggest a lack of strain diversity in this species (10). Here, we report the genome sequence and annotation of a representative of the species, L. iners AB-1, and through comparative genomics, show that it is the smallest lactobacilli discovered to date (11), differing markedly from intestinal and other urogenital species.
Results and Discussion
Phylogeny.
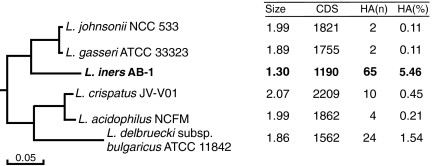
A phylogenetic tree was constructed from concatenated ribosomal subunit protein sequences of several Lactobacillus species available from the National Center for Biotechnology Information (NCBI) database with the addition of sequence for L. iners AB-1. The relationship between the species is in agreement with previous predictions (12) and places L. iners in the group known as the acidophilus complex (13) (Fig. 1 and SI Appendix, Fig. S1). This clade contains two other Lactobacillus species, L. gasseri and L. crispatus, that colonize the vaginal tract, the predominantly gut microbes L. johnsonii and L. acidophilus, and one dairy culture, L. delbrueckii subsp. bulgaricus (referred to here as L. bulgaricus). L. gasseri and L. johnsonii are sister species to L. iners within this clade, although L. iners is markedly more distantly related, which indicates a divergent evolutionary step. This small clade of organisms formed the basis for genomic comparisons with L. iners because of sequence similarity and in the case of L. crispatus and L. gasseri, the shared vaginal environment.
Fig. 1.
Subset of a phylogenetic tree constructed from concatenated ribosomal subunit protein sequences representing the L. iners clade, also part of the acidophilus complex (13) (full figure in SI Appendix, Fig. S1). Key genome features are summarized for each species. Sizes are in millions of base pairs (Mbp). CDS, coding sequences; HA(n), number horizontally acquired genes; HA(%), percent of CDS horizontally acquired.
General Genomic Features.
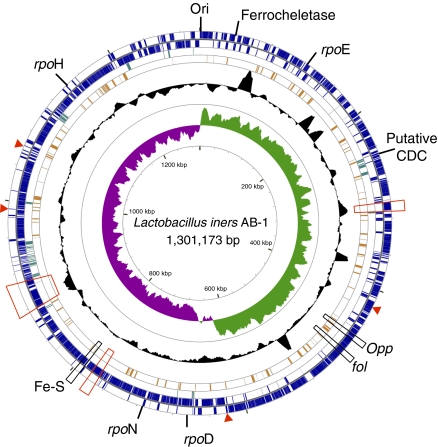
The L. iners AB-1 genome is the smallest Lactobacillus genome reported to date (11). Optical mapping showed a circular chromosome of 1.304 Mbp, a size which agreed with our assembled genome scaffold of 1.301 Mbp with no plasmids (Fig. 2 and SI Appendix, Figs. S2 and S3). Comparison of the optical map and the assembled scaffold suggests that there are 3 kb separating the two scaffolded contigs. The size of the L. iners genome places it within the range of several obligate symbionts and parasites (14, 15). In comparison, the smallest genome of any free-living organism is that of Pelagibacter ubique at 1,308,759 bp. The L. iners genome has a low cytosine + guanine (CG) content of 32.7%, which is similar to the most closely related species L. johnsonii (34.0%) and L. gasseri (35.3%) (16). Surprisingly, the small genome still contains six rRNA gene operons, which likely provide a competitive mechanism for responding to environmental pressures and resource availability (17). The L. bulgaricus genome also contains an exceptionally high number of rRNA operons relative to genome size. This was attributed to a recent phase of genome reduction (18). Because L. johnsonii (19) and L. gasseri (20) each have six rRNA operons, the L. iners genome may have undergone a large genome reduction phase after splitting from the L. johnsonii and L. gasseri lineage, but the rRNA clusters were retained.
Fig. 2.
Genomic atlas of L. iners AB-1. From the outer circle inward, coding regions are marked on the first two rings: outside the dividing line if encoded on the positive strand and inside the dividing line if encoded on the negative strand. The third ring (dark green) marks ORFs predicted to be horizontally acquired. The fourth ring (orange) shows ORFs predicted to be among the top 10% most highly expressed based on CAI. The fifth ring shows local CG content measured in a sliding window as a black plot. The innermost graph shows the CG skew, with sharp changes in skew occurring at the origin and terminus of replication. Genes of interest, as described in the text, are marked on the outside of the atlas. Regions containing adhesins are marked with red boxes, and single adhesin genes are maked with a red arrow. The atlas was constructed using the CGView Server (70), and a large-scale version can be found in SI Appendix, Fig. S2.
The L. iners genome was predicted by GeneMark (21) and Glimmer (22) to encode 1,190 ORFs greater than 100 nt, with an average length of 998 nt. A large fraction of the genes (85.7%) had one or more homologs with an E value of 1e-20 or lower in the NCBI nonredundant database. Functional classification of the 1,190 predicted genes by Clusters of Orthologous Genes (COGs) (23) showed that 956 (80.3%) were homologous to known gene families, including 201 poorly characterized genes. The number of pseudogenes was estimated by identifying ORFs that were truncated by at least 10% compared with the full length of the best protein match in the NCBI database (SI Appendix, Table S1). To account for gene miscalls by GeneMark, the truncated ORFs were verified with Glimmer predictions and corrected, where appropriate, when Glimmer predictions produced better alignments with predicted proteins in the NCBI nrdb. The analysis resulted in 95 possible pseudogenes, with approximately one-half predicted as hypothetical proteins or phage-related (SI Appendix, Table S2). Notable horizontally acquired genes are described in the following relevant sections.
Comparative Genomics.
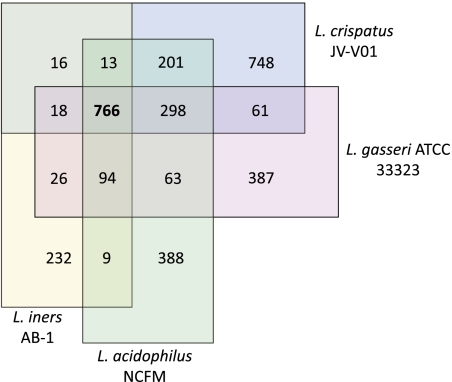
The InParanoid tool (24) was used to identify groups of orthologous proteins between L. iners and three other species of the clade (Fig. 3). There were a predicted 766 genes shared between all four species, representing a large proportion of the L. iners coding sequence (64%) compared with L. crispatus (37%), L. acidophilus (44%), and L. gasseri (44%). This illustrates the unique nature of L. iners, because most of its reduced genome contains core genes of the clade. Similar predictions were obtained when L. johnsonii was substituted for L. gasseri (SI Appendix, Fig. S4).
Fig. 3.
Venn diagram representing orthologous proteins between select species of the L. iners clade. Values are the number of orthologous genes between overlapping species as predicted by InParanoid (24). The 766 genes conserved between the four species represent a relatively large proportion of the total gene content of L. iners (64%).
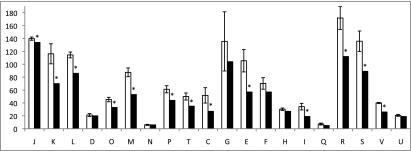
The distribution of genes with a COG functional category was compared between L. iners and the four other species within the clade for which there were COG functional predictions available from the NCBI database (Fig. 4). L. iners was considered to be significantly different within a category if the value was not within 2 SDs of the mean of the other four organisms. L. iners has significantly fewer genes in 13 of 20 COG functional categories, and it has the fewest number of genes of all five organisms in 15 categories, tying for lowest in two additional categories.
Fig. 4.
Comparison of the distribution of genes by COG functional category. The white bar represents the mean and SD of the number of genes in each COG functional category for four species of the L. iners clade (L. johnsonii, L. gasseri, L. acidophilus, and L. bulgaricus). The values for L. iners are plotted in the black bar. Marked with asterisks are values in L. iners that are at least 2 SDs away from the mean of the other organisms in the same COG category. COG functional categories: J, translation, ribosomal structure, and biogenesis; K, transcription; L, DNA replication, recombination, and repair; D, cell division and chromosome partitioning; O, posttranslational modification, protein turnover, and chaperones; M, cell envelope biogenesis, outer membrane; N, cell motility and secretion; P, inorganic ion transport and metabolism; T, signal transduction mechanisms; C, energy production and conversion; G, carbohydrate transport and metabolism; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; H, coenzyme metabolism; I, lipid metabolism; Q, secondary metabolites biosynthesis, transport, and catabolism; R, general function prediction only; S, function unknown; V, defense mechanisms; U, intracellular trafficking and secretion.
As observed previously (18), L. bulgaricus had approximately one-half the number of carbohydrate transport and metabolism (COG category G) genes as the mean of the remaining three lactobacilli (excluding L. iners), creating an unusually high variance in this category. The severely reduced number of genes in L. bulgaricus is because of its long use as a milk starter culture and its adaptation to this specific nutrient-rich but carbohydrate-limited environment. The authors noted many instances of loss or inactivation of carbohydrate-metabolizing enzymes other than lactose in this rapidly evolving organism. The vaginal environment is much more complex because of fluctuation of hormones affecting mucus and glycogen production, pH, and microbial species, and although L. iners also exhibited gene loss related to carbohydrate transport and metabolism, it was not to the extent of L. bulgaricus.
Horizontal Gene Transfer.
Genes in L. iners and the other five lactobacilli of the same clade (Fig. 1) were identified as foreign if the first three significant hits (E value of 1e-20 or less) to the NCBI nr database were to a genus other than Lactobacillus. L. iners has a striking number of predicted genes likely acquired from organisms outside its genus, with 65 genes identified, accounting for 5.5% of protein-coding genes (Fig. 1 and SI Appendix, Table S3). The next highest number was 24 foreign genes in the L. bulgaricus genome, accounting for 1.5% of protein-coding genes. For the other organisms in the group, less than 1% of their protein-coding genes are of putative foreign origin. Even more striking, 26 of the foreign genes in the L. iners genome have at least 80% amino acid identity to a non-Lactobacillus organism, indicating a recent large-scale introduction of these genes. Many of the horizontally acquired genes are most similar to a gene found in an organism that shares the vaginal environment, possibly indicating the origin of the L. iners gene (SI Appendix, Table S3). Interestingly, of the six members of the acidophilus group, L. iners and its sister species, L. johnsonii and L. gasseri, lack both clustered regularly interspaced short palindromic repeats (CRISPR) regions and the associated cas genes (25). Because CRISPR loci are a defense mechanism that excludes foreign DNA, this may explain, in part, why L. iners was able to acquire a large number of foreign genes. Several competence-related proteins were identified (LINAB1_1090 ComEB, LINAB1_0944 MecA, LINAB1_0943 CoiA, LINAB1_0502 ComEC, LINAB1_0501 ComEA, putative ComGC protein LINAB1_0373, LINAB1_0372 PulF, and LINAB1_0371 PulE), suggesting that L. iners has components of the machinery for natural competence.
It is not uncommon to find subjects colonized by one of either L. crispatus or L. iners (4, 5, 8), suggesting that there may be competition between the organisms. Pressure to compete with other organisms, including lactobacilli, may have been a reason for L. iners to acquire foreign genes, such as the adhesins and cytolysin described below. Interestingly, instillation of probiotic L. rhamnosus GR-1 with L. reuteri RC-14 or candidate probiotic L. crispatus CTV05 on its own was not inhibited by indigenous L. iners nor did they displace the L. iners from the vagina, indicating in vivo persistence of indigenous L. iners strains (1, 26).
Cholesterol-Dependent Cytolysin.
The L. iners AB-1 genome contains a cluster of related genes making up a complete horizontally acquired type I restriction modification (RM) system: restriction subunit (LINAB1_0222), methyltransferase subunit (LINAB1_0217), and three specificity proteins (LINAB1_0218, LINAB1_0220, and LINAB1_0221). There is one additional isolated restriction endonuclease encoded in the genome (LINAB1_1155). Neighboring the RM system is a putative integrase and a predicted cholesterol-dependent cytolysin (LINAB1_0216) that belongs to the family of Gram-positive pore-forming cholesterol-dependent cytolysins (CDC), formerly known as thiol-activated cytolysins. These proteins are typically found in pathogenic bacteria but are not found in other lactobacilli. When expressed and secreted, CDC proteins use cholesterol in host cell membranes as a receptor for binding. On binding, a conformational change in the protein causes cell lysis by forming large pores in the membrane (27).
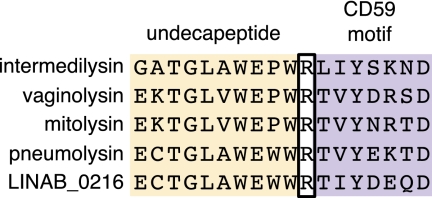
LINAB1_0216 is 519 aa long and has 55% amino acid identity to vaginolysin of Gardnerella vaginalis. A neighbor-joining tree derived from a multiple sequence alignment of representative CDCs shows that the putative L. iners AB-1 CDC is most closely related to the CD59-dependent intermedilysin class (SI Appendix, Fig. S5). CDC proteins have a well-conserved undecapeptide in the last domain of this four-domain protein that is important for host interaction and pore formation. Hughes et al. (28) recently showed in Streptococcus that a short peptide sequence adjacent to the undecapeptide was responsible for specific interaction to the complement regulatory protein, CD59. Fig. 5 shows an alignment of the undecapeptide and the CD59 interaction motif for LINAB1_0216 and its most closely related proteins. The putative cholesterol-dependent lysin was found to contain the consensus undecapeptide motif for cholesterol binding but to lack the CD59 motif. Thus, although intermedilysin and vaginolysin use the human complement factor CD59 as a dependent cofactor with cholesterol to mediate binding (29), the sequence differences in the L. iners AB-1 lysin indicate that the protein does not have the specificity to bind to CD59. We noted that domain 4 of LINAB1_0216, the membrane interaction domain, was more closely related to tetanolysins than to intermedilysins (SI Appendix, Fig. S6), supporting the conclusion that LINAB1_0216 is not dependent on CD59 for membrane binding. This putative lysin is predicted to be highly expressed by L. iners AB-1 based on a codon adaptation index (CAI) that ranks in the top 10% of this predicted measure of gene expression (SI Appendix, Table S4).
Fig. 5.
Alignment of conserved motifs in the putative cholesterol-dependent cytolysin (LINAB1_0216) with related cytolysins. The cholesterol-binding undecapeptide extends for the first 11 residues. The residues beginning at the R in intermedilysin compose the CD59 motif. Intermedilysin has a fourfold higher affinity to CD59 compared to mitolysin due to the SKN residues present in the CD59 motif compared to the NRT residues present in mitolysin (28). LINAB1_0216 has one or more differences from the intermedilysin in each of the two motifs, the most drastic being the SKN to DEQ difference that changes a polar–basic–polar trio to an acidic–acidic–polar trio.
L. iners is the predominant component of the vaginal microbiota in a significant proportion of healthy women (5, 6, 30, 31). There is no clinical evidence to date that the presence of L. iners is associated with induction of discharge, inflammation, or discomfort. Therefore, we speculate that the cytolysin could be used to acquire nutrients from the host in a symbiotic way or may exert antimicrobial activity against eukaryotic organisms in the environment, such as Candida species; additionally, it may contribute to attachment to host cell membranes. However, the exact function and role of LINAB1_0216 remains to be determined.
Carbohydrate Transport and Metabolic Capabilities.
Perinatally and until menopause, increased estrogen induces anaerobic metabolism of the large amounts of glycogen on the vaginal epithelium, causing an acidic environment in which the major unbound carbon sources are glucose, mannose, and glucosamine (32), all of which L. iners AB-1 is predicted to metabolize. The vaginal mucosa consists primarily of mucin glycoproteins composed of monosaccharide chains of L-fructose, N-acetylneuraminic acid (sialic acid), galactose, N-acetyl-galactosamine, and N-acetylglucosamine (33). Growth assays (SI Appendix, Fig. S7) determined that L. iners AB-1’s preferred carbon source is mucin, with maximum growth occurring within 24–48 h of initial inoculation. Lactose, glucose, and maltose also supported growth but peaked after a longer incubation of 72–96 h. We speculate that L. iners is adapted for an easily accessible component of the mucin polysaccharide, which after being depleted in culture, results in loss of viability. In the natural vaginal environment, mucin turnover (34) would provide new moieties of carbohydrates for L. iners to access. No growth was observed in fructose or mannose.
Complete phosphotransferase systems (PTSs) were found for lactose (LINAB1_0229 and LINAB1_0230 plus LINAB1_0228: LacI regulator), galactitol (LINAB1_0173, LINAB1_0174, and LINAB1_0175), glucose (LINAB1_00303, LINAB1_00304, and LINAB1_00306), and fructose (LINAB1_0669). The lactose-specific PTS has greater than 75% protein identity to several Streptococcus strains, while being less than 50% identical to other lactobacilli. There were two complete PTSs members of the mannose family of PTS transporters (LINAB1_0058 to LINAB1_0060 and LINAB1_0223 to LINAB1_0226) and one system predicted to be a mannose/fructose transporter with the most protein identity to Streptococcus spp. (LINAB1_0647 to LINAB1_0650). However, the mannose class of PTS transporters is structurally and functionally distinct from other PTS transporters, most notably for its ability to transport a wide variety of sugars, including mannose, glucose, glucosamine, fructose, galactosamine, and N-acetylgalactosamine (35). Since L. iners AB-1 did not grow in mannose supplemented media, we suspect these transporters import other carbohydrates. Two incomplete PTS operons containing only one transport unit have predicted specificity to mannose/fructose (LINAB1_1126) and glucitol/sorbitol (LINAB1_0935). Clusters of ATP-binding casette (ABC) transport genes composing complete transport machinery for general sugar transport (LINAB1_0031, LINAB1_0032, LINAB1_0033, LINAB1_0035, LINAB1_0036 and LINAB1_0341, LINAB1_0339, LINAB1_0338, and LINAB1_0337) and specifically, for maltose (LINAB1_0077 and LINAB1_0080) were found in addition to the carbohydrate PTSs.
Complete metabolic pathways are present to convert each transported sugar to glucose for entry into glycolysis, with the exception of galactitol. The enzyme galactitol-1-phosphate-5-dehydrogenase is missing in L. iners and related species L. johnsonii and L. gasseri, despite predicted galactitol transport ability. Unlike L. johnsonii and L. gasseri, L. iners is lacking PTSs and enzymes for sucrose and cellobiose metabolism. A predicted exported O-sialoglycoprotein endopeptidase (LINAB1_0262) contains a conserved glycoprotease motif (Pfam: Peptidase_M22), and the ORF protein sequence has greater than 80% identity to protein sequences in other vaginally associated lactobacilli. Four ORFs initially annotated as pullulanases (LINAB1_0206, LINAB1_0331, LINAB1_0631, and LINAB1_0626) have predicted glycosidase activity caused by several carbohydrate-binding and glycosidase domains. These findings of predicted carbohydrate metabolism and transport suggest that L. iners is adapted to hydrolyzing and extracting the carbohydrates of the vaginal mucosa.
All glycolytic enzymes are present to convert glucose to pyruvate and from pyruvate exclusively into L-lactate but not into other metabolites such as D-lactate, oxaloacetate, Acetyl-CoA, or acetate. L. iners has an incomplete citric acid cycle containing only fumarate reductase. Like most other lactobacilli, the organism is unable to synthesize or metabolize fatty acids.
Other Transport and Metabolism.
Consistent with a limited metabolic capability and other members of the lactic acid bacteria, L. iners AB-1 dedicates a large proportion of its genome [186 (15.6%) of protein-encoding genes] to transport (SI Appendix, Table S5). The closely related L. johnsonii is known for its auxotrophic nature, but in comparison, only 10.7% of protein-coding genes are related to transport. Other lactobacilli in the same clade range from 4.0% for L. bulgaricus to 8.0% and 9.1% for L. gasseri and L. acidophilus, respectively. Unique to L. iners compared with L. johnsonii and L. gasseri is a complete metal ion transport system likely transporting zinc or manganese. The system seems to be horizontally acquired, and each component has from 56% to 73% amino acid identity to Anaerococcus and Finegoldia species based on homology to proteins in the NCBI database. Manganese has long been known as an important protectant for lactobacilli against oxidative stress and has been shown as a required component for growth of several Lactobacillus species (36, 37).
L. iners AB-1 lacks enzymes for the biosynthesis of nearly all cofactors and vitamins. However, unlike a number of other lactobacilli including L. johnsonii and L. gasseri, L. iners AB-1 seems to have the complete biosynthesis pathway for folate, with a number of the enzymes clustered together in the genome (SI Appendix, Fig. S8). It remains to be determined if folate is produced and secreted by these bacteria into the vagina.
L. iners AB-1 has the complete pathway for de novo synthesis and salvage of pyrimidines. A partial purine metabolism pathway is present that contains the enzymes required for de novo synthesis of 5-phosphoribosyl-α-pyrophosphate (PRPP) but no subsequent enzymes for conversion into IMP. All genes are present for salvage of purines.
Amino Acid Biosynthesis.
The strain is unable to synthesize any amino acids de novo, with the possible exception of serine from pyruvate using a L-serine dehydratase (LINAB1_0514 and LINAB1_0513). Two genes allow for the interconversion of L-aspartate and L-aspargine: asparagine synthetase (asnB) and aspartate-ammonia ligase (asnA). Also present is a glutamine synthetase (LINAB1_0635) that can convert glutamate to glutamine in the presence of NH3. Three enzymatic steps are present to catabolize L-methionine into L-homocysteine and ultimately, into O-succinyl-L-homoserine, a process unique to L. iners compared with L. johnsonii and L. gasseri. This allows L. iners to form the methyl-donating coenzyme S-adenosyl-L-methionine (SAM) from methionine or homocysteine. It is unknown what additional role the met pathway plays in L. iners. Compensating for the inability of L. iners to synthesize most amino acids, several peptidases are predicted to be highly expressed based on CAI (SI Appendix, Table S4). There are also several genes involved in peptide transport, including a predicted highly expressed oligopeptide (Opp) transport system (LINAB1_0408 to LINAB1_0412).
Adherence and Host Interaction.
The ability of lactobacilli to adhere to vaginal epithelial surfaces is believed to be important to allow colonization and host interaction and exclude pathogens (38). The BV-associated microbes, G. vaginalis and Atopobium vaginae, are able to form dense biofilms recalcitrant to standard metronidazole treatment (39, 40). However, L. iners AB-1 can interfere with, and displace, G. vaginalis biofilms in vitro, suggesting a role in restoration of a healthy vagina post-BV (41).
Examination of the genome for potential adhesion factors surprisingly showed the L. iners genome lacks most of the known adhesion factors and conserved adhesion domains commonly found in the lactobacilli (Table 1 and SI Appendix, Table S6). One fibronectin-binding protein (LINAB1_0564) and one fibrinogen-binding protein (LINAB1_0798) were found, but no mucus-binding proteins common to lactobacilli that colonize the gastrointestinal tract were identified (42, 43), thus suggesting that L. iners uses different adherence mechanisms in the vagina.
Table 1.
Predicted adhesion-related proteins of L. iners AB-1
| Gene ID | Annotation | Size (aa) | Average CAI | TMD | SPP | Anchor motif | Conserved domains* | |
| LINAB1_0273 | Putative cell surface protein containing Gram-positive anchor | 1,457 | 0.493 | 0 | 0.000 | LPNTG | PF08428 and PF00746 | |
| LINAB1_0370 | Surface protein containing YSIRK and Gram-positive anchor | 101 | 0.515 | 1 | 0.000 | LPQTG | PF04650 and PF00746 | |
| LINAB1_0701 | Putative cell surface protein containing YSIRK and Gram-positive anchor | 3,006 | 0.651 | 1 | 0.807 | LPQTG | PF04650 and PF00746 | |
| LINAB1_0800 | Putative cell surface protein containing YSIRK and Gram-positive anchor | 1,804 | 0.610 | 2 | 1.000 | LPNTG | PF04650 and PF00746 | |
| LINAB1_0795 | Putative cell surface protein containing YSIRK and Gram-positive anchor | 2,258 | 0.616 | 0 | 0.793 | LPQTG | PF04650, PF00746, and PF05345 | |
| LINAB1_0796 | Putative cell surface protein containing YSIRK and Gram-positive anchor | 3,793 | 0.508 | 2 | 1.000 | LPQTG | PF04650, PF00746, and PF08428 | |
| LINAB1_0564 | Fibronectin/fibrinogen-binding protein | 565 | 0.658 | 0 | 0.000 | none | PF05833 and PF05670 | |
| LINAB1_0272 | Putative cell surface protein containing YSIRK motif | 1,559 | 0.547 | 1 | 0.966 | none | PF04650 | |
| LINAB1_0950 | Putative cell surface protein containing YSIRK motif | 448 | 0.756 | 1 | 1.000 | none | PF04650 | |
| LINAB1_0798 | Putative fibrinogen-binding protein | 778 | 0.682 | 1 | 1.000 | none | PF10425 | |
| LINAB1_0886 | Putative aggregation-promoting protein | 186 | 0.755 | 1 | 1.000 | none | none |
TMD, transmembrane domains (predicted by TMHMM); SPP, signal peptide probability (predicted by SignalP).
*Pfam database identification number.
The predicted ORFs were evaluated for cell wall anchor motifs that indicate that the protein is exposed on the surface of the cell. Sortase-dependent proteins (SDPs) often have a role in bacterial–host interactions and adhesion (44), and many Gram-positive cell wall-anchored proteins contain a conserved C-terminal LPXTG motif that is cleaved by the sortase A enzyme between the threonine and glycine residues. The cleaved protein is then covalently linked to the peptidoglycan of the cell wall (45). Recently, the probiotic L. rhamnosus GG was shown to have genes for three secreted LPXTG-like pilins (spaCBA) and a pilin-dedicated sortase that mediates binding to mucus (46).
Bereft of pili (Fig. 6), 1 ORF of L. iners was identified as sortase A (LINAB1_0482), and Table 1 shows that 6 ORFs were identified as possible SDPs containing the sortase cleavage motif LPXTG (Pfam ID PF00746). Two of the proteins (LINAB1_0273 and LINAB1_0796) have several tandemly repeated Rib domains (PF08428), which may function as adhesion and biofilm factors to human epithelial cells (47, 48). All of the SDPs have multiple repeated domains of unknown function within the protein that we speculate are involved in adhesion.
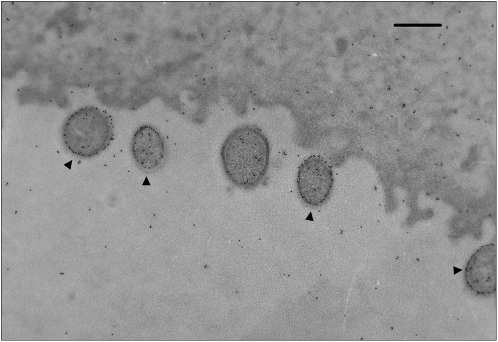
Fig. 6.
Transmission EM of immunogold-labeled L. iners AB-1 (black arrows) in association with human vaginal epithelial cells. Vaginal swab samples collected from healthy women were prepared for EM by thin sectioning. The L. iners cells were labeled with a polyclonal antiserum raised in rabbits against formalin-fixed whole cells of L. iners followed by a secondary goat anti-rabbit gold-conjugated antibody (10-nm gold particles). An even surface distribution of gold particles is expected for L. iners (marked by arrow) but not other bacteria because antibodies not specific to L. iners AB-1 were absorbed out of the serum as described in SI Appendix, SI Text. (Scale bar: 500 nm.)
A large fraction of Streptococcus and Staphylococcus SDP surface proteins with the LPXTG anchor motif also contain the YSIRK signal motif (49, 50). Deletion and substitution mutations of the YSIRK motif in cell wall-anchored protein A of Staphylococcus reduced secretion and sorting of the protein. Except for LINAB1_0273, all of the LPXTG proteins identified (Table 1) in L. iners also contained the YSIRK motif (PF04650), and another two proteins had the N-terminal YSIRK-type signal motif without an associated LPXTG motif (LINAB1_0272 and LINAB1_0950). The combinational signal and anchor motifs in these proteins suggest that they are under tight regulation to be properly secreted and anchored to the cell surface.
In addition, several of the putative adhesion factors show weak similarity to adhesion proteins. Two of the five SDPs have a significant hit (E value of 1e-20 or less) to the NCBI nr database: LINAB1_0273 and LINAB1_0701 are similar (26% and 47%, respectively) to a possible biofilm-associated protein of BV pathogen A. vaginae. Six putative binding proteins had significant hits to the nr database; however, only two of these had greater than 50% amino acid identity. LINAB1_0795 had 65% identity to a possible mucus binding protein precursor of L. jensenii. However, LINAB1_0795 itself does not contain a mucus-binding domain, and there is no significant similarity to any other mucus-binding protein in the database, suggesting that the L. jensenii protein is misannotated.
To determine if any putative cell surface proteins were expressed, cell wall protein extract from L. iners was separated by SDS/PAGE electrophoresis and analyzed by Western blot with antisera raised against L. iners AB-1. Bands were isolated for MS peptide mass fingerprinting. Isolated proteins were putatively identified based on predicted size in kilodaltons (SI Appendix, Fig. S10) and the peptide mass coverage. One band (marked with a black arrowhead in SI Appendix, Fig. S10) was predicted to be LINAB1_0950 based on six matched peptides and 15% peptide mass coverage. When search options included variable lysine acetylation, 16 peptides covered 25% of the protein. The 48,294 Da predicted mass of this protein is consistent with the size of the band. The protein sequence contains a YSIRK signal sequence supporting its predicted location outside the cell but has no significant similarity to proteins in the NCBI nonredundant database.
To further confirm the localization of the isolated proteins to the cell surface, transmission electron microscopy (TEM) of L. iners monoculture was prepared with a rabbit polyclonal antisera raised against formalin-fixed whole cells of L. iners and labeled with IgG conjugated to 10-nm gold particles. This showed specific labeling distributed around the cell membrane (Fig. 6 and SI Appendix, Fig. S9). Vaginal swab samples collected from healthy women were confirmed by PCR to contain L. iners (51), and TEM and immunogold labeling using the L. iners antisera showed L. iners to be consistently closely associated with vaginal epithelial cell surfaces (Fig. 6).
The lack of proteins containing adhesion domains and several cell-anchored proteins lacking sequence conservation to known proteins suggest that L. iners has a unique and unknown method of adherence. In support of this assertion, 7 of 14 putative adhesion proteins have a CAI greater than the mean CAI; two are predicted to have gene expression levels among the highest 10% of all genes. Taken together, these observations suggest that these genes may provide a competitive advantage in its specialized environment, but further characterization is required.
Stress Tolerance and Environmental Response.
L. iners AB-1 has the ability to survive both the low pH (3.6–4.5) conditions of a healthy vagina and the higher pH (greater than 4.5) conditions associated with BV (52, 53). The genome contains two alkaline-shock proteins (LINAB1_0403 and LINAB1_0434) also present in L. crispatus, which may aid in pH tolerance of the vaginal environment. The genome contains a heat-shock operon with a heat-inducible transcriptional repressor, HrcA, and molecular chaperones GrpE, DnaK, and DnaJ, plus a second stress-induced operon controlled by a redox-sensing transcriptional repressor and the chaperonins GroES and GroEL. Two universal stress proteins (LINAB1_0760 and LINAB1_0227) and a cold-shock protein (LINAB1_0419) were identified. Collectively, these may aid in allowing the organism to tolerate different pHs, mucus changes, and stress associated with infection.
RNA polymerase contains four standard subunits and an additional σ factor that directs the enzyme to a specific promoter. Alternative σ factors are active under different stress conditions to regulate the transcription of various stress response genes. In addition to the typical RpoD (LINAB1_0602) σ factor, L. iners has three alternative σ factors identified as RpoE (LINAB1_0946), RpoH (LINAB1_1028), and RpoN (LINAB1_0645). Interestingly, RpoE is used by the opportunistic intracellular pathogen Burkholderia cenocepacia for growth under stress (54). Another intracellular Gram-negative pathogen Brucella melitensis uses RpoH and RpoN to cope with stress from elevated temperature and exposure to H2O2 (55). These factors may contribute to the ability of L. iners to survive in the presence of H2O2-producing strains in the vagina and oxidative stresses associated with infection. LINAB1_0645 (RpoN) is adjacent to a complete mannose/fructose PTS with high amino acid identity to strains of Streptococcus pyogenes. Upstream is an unknown conserved protein (LINAB1_0651) with an RpoN interaction domain. Recent work by Stevens et al. (56) characterized an RpoN-dependent mannose PTS of L. plantarum with a similar operon structure, and RpoN acted as a major regulator of carbohydrate uptake.
There were four complete two-component systems identified, each consisting of a histidine kinase and a response regulator. Three of the paired systems have high similarity to proteins from other lactobacilli, whereas one system (LINAB1_0792 and LINAB1_0791) seems to be horizontally acquired, with 50–60% amino acid identity to proteins from either Anaerococcus prevotii or Oribacterium sinus. In addition to the complete coupled systems, three more response regulators without a corresponding histidine kinase were found (LINAB1_0366, LINAB1_0143, and LINAB1_0785).
We noted that a cluster of genes shared between L. iners, L. crispatus, and L. johnsonii encodes an iron–sulfur (Fe–S) cluster assembly system (LINAB1_0712 to LINAB1_0717). We examined homologs in the NCBI database and found these proteins to be most similar to those lactobacilli known to colonize the vaginal tract, but the complete system was rare in lactobacilli inhabiting other niches. Three of the proteins of L. iners in the cluster (LINAB1_0712, LINAB1_0714, and LINAB1_0716) did not match any of the proteins in the NCBI nonredundant database at greater than 60% protein identity, suggesting that they have a specified or unique function in L. iners.
The Fe–S clusters were first characterized as proteins needed to carry iron and sulfur to appropriate areas of the cell and electron transfer molecules (57). The so-called Suf system in Escherichia coli is activated under conditions of limiting iron or oxidative stress, such as the presence of H2O2 (58). Because of the sensitivity of a Fe–S cluster to oxygen, they can act as environmental sensors and regulate a response by the organism (59). This could be an important mechanism for resistance to oxidative stress in L. iners, where an abundance of H2O2 is produced by other lactobacilli in the vagina (60). Although iron is a required element for nearly all living organisms, it has commonly been accepted that lactobacilli do not require iron for growth and instead, use manganese and cobalt as cofactors for biological processes (36, 61). However, Elli et al. (62) found that depleting iron from a nucleotide-limited growth media resulted in no growth of certain Lactobacillus species, whereas Duhutrel et al. (63) reported the acquisition and use of iron in L. sakei, an organism especially well-adapted to meat. There were no apparent iron uptake systems in L. iners, but a ferrochelatase (LINAB1_0037) was detected that is not present in the other species of the clade. This enzyme plus an oxygen-independent coproporphyrinogen III oxidase (hemN) could break down heme for transport (64), although no transport system has yet been characterized and the hemN protein is lacking in L. iners. The potential for L. iners to sequester iron in the Fe–S complex could limit the availability of iron to vaginal pathogens and provide a competitive advantage against organisms unable to use iron in this iron-rich environment.
Finally, we noted that all steps in the oxidative branch of the pentose phosphate pathway are present for the production of NADPH, an important molecule for the prevention of oxidative stress.
Defense Mechanisms.
The vagina harbors a highly diverse community of competing microbes. Bacteriocins are produced by vaginal lactobacilli and enterococci (65, 66), and the ability to resist their antimicrobial effects would be advantageous for L. iners survival. No complete bacteriocin synthesis genes were found in the L. iners AB-1 genome, but three genes were found to encode putative bacteriocin immunity proteins. LINAB1_0852, identified by The RAST annotation server as a microcin self-immunity protein, has 91.5% amino acid identity to a putative bacteriocin immunity protein of Finegoldia magna and greater than 60% identity to MccC microcin immunity family protein from various strains of Streptococcus pneumoniae. Resistance to microcin may be significant, because it is an antimicrobial peptide produced by uropathogenic E. coli associated with aerobic vaginitis and urinary tract infection (67). This, along with a bacitracin resistance protein (LINAB1_0937) and putative enterocin A immunity protein, supports the assertion that L. iners incorporates foreign genes that may contribute to its ability to resist vaginal eradication by these pathogens. Of note, L. iners AB-1 has a putative pore-forming hemolysin gene (LINAB1_0545) of Lactobacillus origin in addition to the horizontally acquired cholesterol-dependent cytolysin described previously. Three other proteins contain domains of the hemolytic-related Tly family (68, 69) (LINAB1_0440, LINAB1_0684, and LINAB1_0698). The hemolytic potential of L. iners speculatively may be used for defense or may play a role in providing nutrients and surviving through menstruation. Interestingly, L. iners AB-1 exhibits clearing of sheep's red blood cells in culture (SI Appendix, Fig. S11), but it is unknown whether the putative hemolysins or cholesterol-dependent cytolysin are responsible.
Concluding Remarks.
This study describes the smallest Lactobacillus genome reported to date, with a genome that seems highly adapted for a specialized niche environment. The genome contains the largest fraction of genes composed of conserved proteins within the clade, and it also has the largest fraction and absolute number of genes predicted to be acquired from foreign sources. The conserved genes are largely core metabolic proteins shared between lactobacilli. L. iners is present in the vagina that is deemed healthy, infected with BV, or has just been subjected to antimicrobial therapy (5, 7). Recent quantification of bacterial numbers in these different stages of vaginal health has shown that the abundance of L. iners remains relatively constant, despite fluctuating environmental conditions (9). This remarkable ability to survive under a range of conditions suggests that, rather than L. iners being somehow associated with an aberrant microbiota, it may be an important member of the host's defenses by being a persistent mutualistic lactobacilli involved in restoration and maintenance of the normal microbiota. Further analysis of the functional characteristics associated with the L. iners genome may uncover desirable characteristics of a microbe that could contribute to the maintenance, and potentially restoration, of a healthy and stable vaginal microbiota.
Materials and Methods
DNA was isolated from L. iners AB-1 and submitted separately for Illumina and 454 sequencing (Next-Gen Sequencing Facility). Details of the sequence assembly, scaffolding, gap closure, and annotation are described in SI Appendix. Optical mapping (OpGen, Inc.) was used to validate the genome assembly and size.
Additional methods describing immunogold labeling and EM, cell wall protein isolation, MS, and growth assays are contained in SI Appendix. The University of Western Ontario (UWO) Research Ethics Board approved swab sample collection from volunteer human subjects, and each volunteer gave their signed informed consent before participation.
This Whole-Genome Shotgun project has been deposited in DNA Data Base in Japan (DDBJ)/European Molecular Biology Laboratory (EMBL)/GenBank under the accession ADHG00000000. The version described in this paper is the second version, ADHG02000000.
Supplementary Material
Acknowledgments
We thank Susan Koval and Judy Sholdice (UWO Transmission Electron Microscopy facility) for their work on preparing the TEMs and Kristina Jurcic (UWO Functional Proteomics Facility) for sample preparation and technical assistance of the MS analysis. Thanks to Kevin Chen and Michael Punnose for assistance in the genome assembly. The input and helpful discussions from Wayne Miller, Kate Crowley, Richard Gardiner, Jordon Bisanz, Ruben Hummelen, Rod MacPhee, Marc Monachese, Andrew Fernandes, and Russ Dickson are greatly appreciated. This work was supported by the Natural Sciences and Engineering Research Council of Canada.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Microbes and Health” held November 2–3, 2009, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS Web site at http://www.nasonline.org/SACKLER_Microbes_and_Health.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. ADHG00000000).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000086107/-/DCSupplemental.
References
- 1.Burton JP, Cadieux PA, Reid G. Improved understanding of the bacterial vaginal microbiota of women before and after probiotic instillation. Appl Environ Microbiol. 2003;69:97–101. doi: 10.1128/AEM.69.1.97-101.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fredricks DN, Fiedler TL, Marrazzo JM. Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med. 2005;353:1899–1911. doi: 10.1056/NEJMoa043802. [DOI] [PubMed] [Google Scholar]
- 3.Zhou X, et al. The vaginal bacterial communities of Japanese women resemble those of women in other racial groups. FEMS Immunol Med Microbiol. 2010;58:169–181. doi: 10.1111/j.1574-695X.2009.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ravel J, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci USA. 2010:4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hummelen R, et al. Deep sequencing of the vaginal microbiota of women with HIV. PLoS ONE. 2010;5:e12078. doi: 10.1371/journal.pone.0012078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi Y, Chen L, Tong J, Xu C. Preliminary characterization of vaginal microbiota in healthy Chinese women using cultivation-independent methods. J Obstet Gynaecol Res. 2009;35:525–532. doi: 10.1111/j.1447-0756.2008.00971.x. [DOI] [PubMed] [Google Scholar]
- 7.Ferris MJ, Norori J, Zozaya-Hinchliffe M, Martin DH. Cultivation-independent analysis of changes in bacterial vaginosis flora following metronidazole treatment. J Clin Microbiol. 2007;45:1016–1018. doi: 10.1128/JCM.02085-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verstraelen H, et al. Longitudinal analysis of the vaginal microflora in pregnancy suggests that L. crispatus promotes the stability of the normal vaginal microflora and that L. gasseri and/or L. iners are more conducive to the occurrence of abnormal vaginal microflora. BMC Microbiol. 2009;9:116. doi: 10.1186/1471-2180-9-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zozaya-Hinchliffe M, Lillis R, Martin DH, Ferris MJ. Quantitative PCR assessments of bacterial species in women with and without bacterial vaginosis. J Clin Microbiol. 2010;48:1812–1819. doi: 10.1128/JCM.00851-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou X, et al. Differences in the composition of vaginal microbial communities found in healthy Caucasian and black women. ISME J. 2007;1:121–133. doi: 10.1038/ismej.2007.12. [DOI] [PubMed] [Google Scholar]
- 11.Ventura M, et al. Genome-scale analyses of health-promoting bacteria: Probiogenomics. Nat Rev Microbiol. 2009;7:61–71. doi: 10.1038/nrmicro2047. [DOI] [PubMed] [Google Scholar]
- 12.Canchaya C, Claesson MJ, Fitzgerald GF, van Sinderen D, O'Toole PW. Diversity of the genus Lactobacillus revealed by comparative genomics of five species. Microbiology. 2006;152:3185–3196. doi: 10.1099/mic.0.29140-0. [DOI] [PubMed] [Google Scholar]
- 13.Kullen MJ, Sanozky-Dawes RB, Crowell DC, Klaenhammer TR. Use of the DNA sequence of variable regions of the 16S rRNA gene for rapid and accurate identification of bacteria in the Lactobacillus acidophilus complex. J Appl Microbiol. 2000;89:511–516. doi: 10.1046/j.1365-2672.2000.01146.x. [DOI] [PubMed] [Google Scholar]
- 14.Baker BJ, et al. Enigmatic, ultrasmall, uncultivated Archaea. Proc Natl Acad Sci USA. 2010;107:8806–8811. doi: 10.1073/pnas.0914470107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giovannoni SJ, et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- 16.Makarova K, et al. Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci USA. 2006;103:15611–15616. doi: 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klappenbach JA, Dunbar JM, Schmidt TM. rRNA operon copy number reflects ecological strategies of bacteria. Appl Environ Microbiol. 2000;66:1328–1333. doi: 10.1128/aem.66.4.1328-1333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van de Guchte M, et al. The complete genome sequence of Lactobacillus bulgaricus reveals extensive and ongoing reductive evolution. Proc Natl Acad Sci USA. 2006;103:9274–9279. doi: 10.1073/pnas.0603024103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Vries MC, et al. Comparative and functional analysis of the rRNA-operons and their tRNA gene complement in different lactic acid bacteria. Syst Appl Microbiol. 2006;29:358–367. doi: 10.1016/j.syapm.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 20.Berger B, et al. Similarity and differences in the Lactobacillus acidophilus group identified by polyphasic analysis and comparative genomics. J Bacteriol. 2007;189:1311–1321. doi: 10.1128/JB.01393-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isono K, McIninch JD, Borodovsky M. Characteristic features of the nucleotide sequences of yeast mitochondrial ribosomal protein genes as analyzed by computer program GeneMark. DNA Res. 1994;1:263–269. doi: 10.1093/dnares/1.6.263. [DOI] [PubMed] [Google Scholar]
- 22.Salzberg SL, Delcher AL, Kasif S, White O. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 1998;26:544–548. doi: 10.1093/nar/26.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tatusov RL, et al. The COG database: An updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Brien KP, Remm M, Sonnhammer ELL. Inparanoid: A comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 2005;33:D476–D480. doi: 10.1093/nar/gki107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 26.Antonio MAD, Meyn LA, Murray PJ, Busse B, Hillier SL. Vaginal colonization by probiotic Lactobacillus crispatus CTV-05 is decreased by sexual activity and endogenous lactobacilli. J Infect Dis. 2009;199:1506–1513. doi: 10.1086/598686. [DOI] [PubMed] [Google Scholar]
- 27.Tweten RK. Cholesterol-dependent cytolysins, a family of versatile pore-forming toxins. Infect Immun. 2005;73:6199–6209. doi: 10.1128/IAI.73.10.6199-6209.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughes TR, et al. Identification of the high affinity binding site in the Streptococcus intermedius toxin intermedilysin for its membrane receptor, the human complement regulator CD59. Mol Immunol. 2009;46:1561–1567. doi: 10.1016/j.molimm.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gelber SE, Aguilar JL, Lewis KLT, Ratner AJ. Functional and phylogenetic characterization of Vaginolysin, the human-specific cytolysin from Gardnerella vaginalis. J Bacteriol. 2008;190:3896–3903. doi: 10.1128/JB.01965-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vásquez A, Jakobsson T, Ahrné S, Forsum U, Molin G. Vaginal Lactobacillus flora of healthy Swedish women. J Clin Microbiol. 2002;40:2746–2749. doi: 10.1128/JCM.40.8.2746-2749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou X, et al. Characterization of vaginal microbial communities in adult healthy women using cultivation-independent methods. Microbiology. 2004;150:2565–2573. doi: 10.1099/mic.0.26905-0. [DOI] [PubMed] [Google Scholar]
- 32.Boskey ER, Telsch KM, Whaley KJ, Moench TR, Cone RA. Acid production by vaginal flora in vitro is consistent with the rate and extent of vaginal acidification. Infect Immun. 1999;67:5170–5175. doi: 10.1128/iai.67.10.5170-5175.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiggins R, Hicks SJ, Soothill PW, Millar MR, Corfield AP. Mucinases and sialidases: Their role in the pathogenesis of sexually transmitted infections in the female genital tract. Sex Transm Infect. 2001;77:402–408. doi: 10.1136/sti.77.6.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gipson IK. Mucins of the human endocervix. Front Biosci. 2001;6:D1245–D1255. doi: 10.2741/gipson. [DOI] [PubMed] [Google Scholar]
- 35.Saier MH, Hvorup RN, Barabote RD. Evolution of the bacterial phosphotransferase system: From carriers and enzymes to group translocators. Biochem Soc Trans. 2005;33:220–224. doi: 10.1042/BST0330220. [DOI] [PubMed] [Google Scholar]
- 36.Imbert M, Blondeau R. On the iron requirement of lactobacilli grown in chemically defined medium. Curr Microbiol. 1998;37:64–66. doi: 10.1007/s002849900339. [DOI] [PubMed] [Google Scholar]
- 37.Archibald FS, Fridovich I. Manganese and defenses against oxygen toxicity in Lactobacillus plantarum. J Bacteriol. 1981;145:442–451. doi: 10.1128/jb.145.1.442-451.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osset J, Bartolomé RM, García E, Andreu A. Assessment of the capacity of Lactobacillus to inhibit the growth of uropathogens and block their adhesion to vaginal epithelial cells. J Infect Dis. 2001;183:485–491. doi: 10.1086/318070. [DOI] [PubMed] [Google Scholar]
- 39.Swidsinski A, et al. Adherent biofilms in bacterial vaginosis. Obstet Gynecol. 2005;106:1013–1023. doi: 10.1097/01.AOG.0000183594.45524.d2. [DOI] [PubMed] [Google Scholar]
- 40.Swidsinski A, et al. An adherent Gardnerella vaginalis biofilm persists on the vaginal epithelium after standard therapy with oral metronidazole. Am J Obstet Gynecol. 2008;198:97.e1–97.e6. doi: 10.1016/j.ajog.2007.06.039. [DOI] [PubMed] [Google Scholar]
- 41.Saunders S, Bocking A, Challis J, Reid G. Effect of Lactobacillus challenge on Gardnerella vaginalis biofilms. Colloids Surf B Biointerfaces. 2007;55:138–142. doi: 10.1016/j.colsurfb.2006.11.040. [DOI] [PubMed] [Google Scholar]
- 42.Azcarate-Peril MA, et al. Analysis of the genome sequence of Lactobacillus gasseri ATCC 33323 reveals the molecular basis of an autochthonous intestinal organism. Appl Environ Microbiol. 2008;74:4610–4625. doi: 10.1128/AEM.00054-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boekhorst J, Helmer Q, Kleerebezem M, Siezen RJ. Comparative analysis of proteins with a mucus-binding domain found exclusively in lactic acid bacteria. Microbiology. 2006;152:273–280. doi: 10.1099/mic.0.28415-0. [DOI] [PubMed] [Google Scholar]
- 44.van Pijkeren JP, et al. Comparative and functional analysis of sortase-dependent proteins in the predicted secretome of Lactobacillus salivarius UCC118. Appl Environ Microbiol. 2006;72:4143–4153. doi: 10.1128/AEM.03023-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Navarre WW, Schneewind O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev. 1999;63:174–229. doi: 10.1128/mmbr.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kankainen M, et al. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human- mucus binding protein. Proc Natl Acad Sci USA. 2009;106:17193–17198. doi: 10.1073/pnas.0908876106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lindahl G, Stålhammar-Carlemalm M, Areschoug T. Surface proteins of Streptococcus agalactiae and related proteins in other bacterial pathogens. Clin Microbiol Rev. 2005;18:102–127. doi: 10.1128/CMR.18.1.102-127.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Conrady DG, et al. A zinc-dependent adhesion module is responsible for intercellular adhesion in staphylococcal biofilms. Proc Natl Acad Sci USA. 2008;105:19456–19461. doi: 10.1073/pnas.0807717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bae T, Schneewind O. The YSIRK-G/S motif of staphylococcal protein A and its role in efficiency of signal peptide processing. J Bacteriol. 2003;185:2910–2919. doi: 10.1128/JB.185.9.2910-2919.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tettelin H, et al. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science. 2001;293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 51.Alqumber MA, Burton JP, Devenish C, Tagg JR. A species-specific PCR for Lactobacillus iners demonstrates a relative specificity of this species for vaginal colonization. Microb Ecol Health Dis. 2008;20:135–139. [Google Scholar]
- 52.Simoes JA, et al. Clinical diagnosis of bacterial vaginosis. Int J Gynaecol Obstet. 2006;94:28–32. doi: 10.1016/j.ijgo.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 53.Tamrakar R, et al. Association between Lactobacillus species and bacterial vaginosis-related bacteria, and bacterial vaginosis scores in pregnant Japanese women. BMC Infect Dis. 2007;7:128. doi: 10.1186/1471-2334-7-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Flannagan RS, Valvano MA. Burkholderia cenocepacia requires RpoE for growth under stress conditions and delay of phagolysosomal fusion in macrophages. Microbiology. 2008;154:643–653. doi: 10.1099/mic.0.2007/013714-0. [DOI] [PubMed] [Google Scholar]
- 55.Delory M, Hallez R, Letesson JJ, De Bolle X. An RpoH-like heat shock sigma factor is involved in stress response and virulence in Brucella melitensis 16M. J Bacteriol. 2006;188:7707–7710. doi: 10.1128/JB.00644-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stevens MJ, Molenaar D, de Jong A, De Vos WM, Kleerebezem M. sigma54-Mediated control of the mannose phosphotransferase sytem in Lactobacillus plantarum impacts on carbohydrate metabolism. Microbiology. 2010;156:695–707. doi: 10.1099/mic.0.034165-0. [DOI] [PubMed] [Google Scholar]
- 57.Lill R. Function and biogenesis of iron-sulphur proteins. Nature. 2009;460:831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- 58.Outten FW, Djaman O, Storz G. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol Microbiol. 2004;52:861–872. doi: 10.1111/j.1365-2958.2004.04025.x. [DOI] [PubMed] [Google Scholar]
- 59.Beinert H, Kiley PJ. Fe-S proteins in sensing and regulatory functions. Curr Opin Chem Biol. 1999;3:152–157. doi: 10.1016/S1367-5931(99)80027-1. [DOI] [PubMed] [Google Scholar]
- 60.Martinez RC, et al. Analysis of vaginal lactobacilli from healthy and infected Brazilian women. Appl Environ Microbiol. 2008;74:4539–4542. doi: 10.1128/AEM.00284-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weinberg ED. The Lactobacillus anomaly: Total iron abstinence. Perspect Biol Med. 1997;40:578–583. doi: 10.1353/pbm.1997.0072. [DOI] [PubMed] [Google Scholar]
- 62.Elli M, Zink R, Rytz A, Reniero R, Morelli L. Iron requirement of Lactobacillus spp. in completely chemically defined growth media. J Appl Microbiol. 2000;88:695–703. doi: 10.1046/j.1365-2672.2000.01013.x. [DOI] [PubMed] [Google Scholar]
- 63.Duhutrel P, et al. Iron sources used by the nonpathogenic lactic acid bacterium Lactobacillus sakei as revealed by electron energy loss spectroscopy and secondary-ion mass spectrometry. Appl Environ Microbiol. 2010;76:560–565. doi: 10.1128/AEM.02205-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cavallaro G, Decaria L, Rosato A. Genome-based analysis of heme biosynthesis and uptake in prokaryotic systems. J Proteome Res. 2008;7:4946–4954. doi: 10.1021/pr8004309. [DOI] [PubMed] [Google Scholar]
- 65.Dezwaan DC, et al. Purification and characterization of enterocin 62-6, a two-peptide bacteriocin produced by a vaginal strain of Enterococcus faecium: Potential significance in bacterial vaginosis. Microb Ecol Health Dis. 2007;19:241–250. doi: 10.1080/08910600701538240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dover SE, Aroutcheva AA, Faro S, Chikindas ML. Safety study of an antimicrobial peptide lactocin 160, produced by the vaginal Lactobacillus rhamnosus. Infect Dis Obstet Gynecol. 2007;2007 doi: 10.1155/2007/78248. Article no. 78248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Azpiroz MF, Poey ME, Laviña M. Microcins and urovirulence in Escherichia coli. Microb Pathog. 2009;47:274–280. doi: 10.1016/j.micpath.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 68.Hsu T, Hutto DL, Minion FC, Zuerner RL, Wannemuehler MJ. Cloning of a beta-hemolysin gene of Brachyspira (Serpulina) hyodysenteriae and its expression in Escherichia coli. Infect Immun. 2001;69:706–711. doi: 10.1128/IAI.69.2.706-711.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.ter Huurne AA, et al. Characterization of three putative Serpulina hyodysenteriae hemolysins. Microb Pathog. 1994;16:269–282. doi: 10.1006/mpat.1994.1028. [DOI] [PubMed] [Google Scholar]
- 70.Grant JR, Stothard P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36:W181–W184. doi: 10.1093/nar/gkn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.