Abstract
Vancomycin, metronidazole, and the bacteriocin lacticin 3147 are active against a wide range of bacterial species, including Clostridium difficile. We demonstrate that, in a human distal colon model, the addition of each of the three antimicrobials resulted in a significant decrease in numbers of C. difficile. However, their therapeutic use in the gastrointestinal tract may be compromised by their broad spectrum of activity, which would be expected to significantly impact on other members of the human gut microbiota. We used high-throughput pyrosequencing to compare the effect of each antimicrobial on the composition of the microbiota. All three treatments resulted in a decrease in the proportion of sequences assigned to the phyla Firmicutes and Bacteroidetes, with a corresponding increase in those assigned to members of the Proteobacteria. One possible means of avoiding such “collateral damage” would involve the application of a narrow-spectrum antimicrobial with specific anti-C. difficile activity. We tested this hypothesis using thuricin CD, a narrow-spectrum bacteriocin produced by Bacillus thuringiensis, which is active against C. difficile. The results demonstrated that this bacteriocin was equally effective at killing C. difficile in the distal colon model but had no significant impact on the composition of the microbiota. This offers the possibility of developing a targeted approach to eliminating C. difficile in the colon, without collateral damage.
Keywords: gut microbiota, pyrosequencing, bacteriocin, antibiotic, thuricin
Antibiotics have revolutionized the treatment of infections since the middle of the last century and are one of the most important factors in the extended life expectancy of modern humans. However, antibiotics may also play a role in initiating infections, such as Clostridium difficile-associated diarrhea (CDAD), mainly through the disruption of the microbial diversity of the gastrointestinal (GI) tract. The mammalian gut microbiota has been described as a virtual organ, playing a role not only in food digestion and the provision of nutrients, but also in host metabolism, neural development, and the development of a fully functional immune system. An intact, fully developed GI tract microbiota also protects the host against invasion by pathogenic microorganisms through a highly complex set of events known as “colonization resistance” (1). The consequences of disturbing the normally mutualistic association between the gut microbiota and the host can be extreme (2). Indeed it is thought that the pathogenesis of some chronic GI diseases, such as inflammatory bowel disease, may also be associated with disturbances in the normal flora of the host (3–5).
Broad-spectrum antibiotics bring about a particularly severe perturbation of the gut microbiota, primarily in the form of dysbiosis. Such dysbiosis can create opportunities for the overgrowth of bacteria normally restricted by microbial competition in an undisrupted flora. When originally isolated by Hall and O'Toole in 1935 (6), C. difficile was classified as a harmless inhabitant of the GI tract of infants; only 40 y later was its potential as a pathogen fully recognized (7). It is now accepted that C. difficile is an opportunistic pathogen with most infections arising as a direct consequence of dysbiosis after antibiotic use. C. difficile is the causative agent of between 20% and 25% of all cases of antibiotic-associated diarrhea but is responsible for the majority (90–100%) of cases of the serious complication, pseudomembranous colitis (8). Current antibiotics of choice for the treatment of CDAD are the broad-spectrum antibiotics vancomycin or metronidazole, but treatment failures and recurrence of infection are common at 2–38% and 8–50%, respectively (9). The emergence of strains with increased resistance to these antibiotics has also been reported (10), and it is estimated that within the European Union alone the annual cost of hospital treatment of patients with CDAD amounts to €3 billion (11).
The appearance worldwide of a more virulent strain of C. difficile (PCR ribotype 027/NAP-1) poses additional problems for health care providers. This strain is characterized by fluoroquinolone resistance, binary toxin production, and a mutation of the tcdC gene that results in increased toxin production (12). Although vancomycin remains effective against C. difficile, its use creates the potential for the selection of resistant strains of clostridia and other species, such as vancomycin-resistant enterococci, and could facilitate the spread of resistance determinants within the hospital environment (13). As a consequence, there is a resurgence of interest in the search for novel antimicrobial compounds effective against C. difficile. One avenue currently being explored is ribosomally synthesized antimicrobial peptides (AMPs), which are produced by many plants, animals, and bacteria. Bacterial AMPs, or bacteriocins, may have potential as therapeutics in the treatment of GI disease. This has been demonstrated in vivo by Corr et al. (14), who showed that the feeding of mice with the probiotic Lactobacillus salivarius UCC118 provided bacteriocin-mediated protection to the animals against subsequent oral infection with Listeria monocytogenes.
We propose that to provide a significant advantage over existing therapies, a novel treatment for CDAD should have most or all of the following characteristics. It should (i) be better than, or of comparable efficacy to, current therapies, (ii) have minimal impact on the diversity of the gut microbiota, (iii) present a reduced likelihood of resistance development among target and nontarget species, and (iv) obviously, be nontoxic to the host. We have previously shown that a broad-spectrum bacteriocin, lacticin 3147, is effective in eliminating C. difficile in a model fecal environment, but we also established that there was an associated negative impact on populations of Lactobacillus and Bifidobacterium (15). Screening of the human gut microbiota for novel bacteriocin producers resulted in the identification of a strain of B. thuringiensis, DPC 6431, which produces a unique two-component narrow-spectrum bacteriocin with potent anti-C. difficile properties, which we have named thuricin CD (16). In this study we investigated the efficacy of vancomycin, metronidazole, lacticin 3147, and thuricin CD on the viability of C. difficile in a model colonic environment and also performed a high-throughput pyrosequence-based assessment of the impact of these antimicrobials on the diversity of the native microbiota. On the basis of the outcomes of these studies, we conclude that thuricin CD meets all of the characteristics required of an improved therapy for the treatment of CDAD.
Results
Comparison of the Efficacy of Vancomycin, Metronidazole, and Lacticin 3147 Against C. difficile.
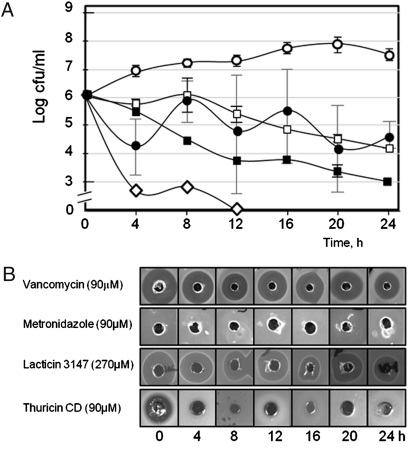
Although vancomycin and metronidazole (both antibiotics) and lacticin 3147 (a member of the lantibiotic class I bacteriocins) are all active against C. difficile (15, 17, 18), their relative antimicrobial activity has not been compared to date. To address this issue, the ability of these three broad-spectrum antimicrobials to control C. difficile ribotype 001 was assessed using a model of the distal colon. Initially, it was established that, after its introduction into the model system at a level of 106 cfu/mL, C. difficile 001 numbers remained relatively stable and even increased slightly over a 24-h period (Fig. 1A). Vancomycin, metronidazole, and lacticin 3147 were introduced at a concentration of 90 μM at 8-h intervals (0, 8, and 16 h). The addition of vancomycin resulted in a consistent reduction in pathogen levels, with a 3.9 log reduction after 24 h (Fig. 1A). However, in the corresponding metronidazole treatment the C. difficile numbers decreased in each 4-h period after antibiotic addition but then increased during the subsequent 4-h period until the antibiotic was introduced again. In contrast to both vancomycin and metronidazole, lacticin 3147 at 90 μM had no effect on the viability of C. difficile. However, when lacticin 3147 concentrations were increased 3-fold (270 μM), C. difficile was successfully inhibited to the extent that the pathogen could not be detected after 12 h. Antibiotic bioavailability analysis of extracted samples revealed that vancomycin and lacticin 3147 (at both 90-μM and 270-μM concentrations) could be detected throughout the 24 h period but that metronidazole was only detected immediately after its introduction at 8 and 16 h (Fig. 1B). This inability to recover metronidazole has been previously noted (18), and it has been suggested that inactivation of metronidazole by human gut microbiota such as enterococci may be responsible for the lack of activity (19, 20).
Fig. 1.
Effect of antimicrobials on levels of C. difficile in a model of the human distal colon. (A) Antimicrobials were administered at three time points (0, 8, and 16 h) [control, open circles; vancomycin (90 μM), black squares; metronidazole (90 μM), black circles; lacticin 3147 (270 μM), open diamonds; thuricin (90 μM), open squares]. (B) Activity (measured as zones of inhibition from a fecal cell-free supernatant in a lawn of C. difficile) recovered from colonic model at indicated time points.
Comparison of the Impact of Vancomycin, Metronidazole, and Lacticin 3147 on the Human Gut Microbiota.
An inevitable consequence of the use of broad-spectrum antimicrobials to treat C. difficile, including vancomycin, metronidazole, and lacticin 3147, is the potential for impact on the other members of the gut microbiota. Although the impact of these three antimicrobials on the gut microbial composition has been assessed to some extent previously (15, 18, 21), none of these assessments has benefitted from the detailed analysis of bacterial populations that becomes possible through massively parallel sequencing of variable regions of the 16s rRNA gene. To facilitate such an analysis, samples were taken at 24 h from the simulated human distal colon experiments already described, and total metagenomic DNA was extracted. Although a number of variable regions of the 16s rRNA gene have been the focus of high-throughput sequencing studies to investigate complex bacterial communities, such as those present in deep-sea vents (variable region 6, V6) (1, 2), human (V1, V2, V6, and V3) (3–6), and soil samples (V9) (7), V4 was selected for our study because it was shown to most efficiently identify populations of microbes present in samples with high levels of sequence diversity (22). A set of primers that match perfectly to 94.6% of sequences present in Ribosomal Database Project (RDP) release 9.53 (http://pyro.cme.msu.edu/pyro/help.jsp) was used to amplify the V4 regions from the extracted metagenomic DNA samples, and a total of almost 31,000 sequence reads (vancomycin-treated, 5,488 reads; lacticin 3147–treated (270 μM), 8,793 reads; metronidazole-treated, 6,553 reads; antibiotic control, 9,986 reads) were generated by DNA sequencing on a Roche 454 GS-FLX sequencer. V4 reads from all samples were BLAST searched against the Small Subunit (SSU) rRNA database compiled by Urich et al. (23), and assignment to the National Center for Biotechnology Information (NCBI) taxonomy was performed using the lowest common ancestor algorithm. MEGAN uses the BLAST bit-score to assign taxonomy. A bit-score of 86 was used as a cutoff for MEGAN, as recommended for 16s rRNA sequences. Reads that were not assigned were those for which there was a hit that fell below the 86 bit-score criteria for assigning taxonomy or those for which there was no hit in the database. Species richness, coverage, and diversity estimations were calculated for each data set (Table 1). At the 97% similarity level the Shannon index, a metric for community diversity, revealed a high level of overall biodiversity within all fermentation vessels, with values exceeding 2.5. Good's coverage is an estimator of sampling completeness and calculates the probability that a randomly selected amplicon sequence from a sample has already been sequenced. The Good's coverage at the 97% similarity level ranged between 97% and 98.8% for the control, metronidazole-, vancomycin-, and purified lacticin–containing vessels, indicating that between 30 and 80 additional reads [1/(1 − “Good's coverage”)] would need to be sequenced before detecting a new phylotype.
Table 1.
Estimations of diversity within each data set
| Data set | Control | Metronidazole | Vancomycin | Lacticin 3147 | Thuricin control | Thuricin | ||||||
| Similarity | 97% | 98% | 97% | 98% | 97% | 98% | 97% | 98% | 97% | 98% | 97% | 98% |
| Chao1 richness estimation | 616 | 859 | 475 | 701 | 582 | 898 | 445 | 645 | 439 | 577 | 566 | 774 |
| Shannon's index for diversity | 4.5 | 4.8 | 2.7 | 3.5 | 3.8 | 4.5 | 3.6 | 4.1 | 4.0 | 4.2 | 4.4 | 4.8 |
| Good's coverage | 98.5 | 97.8 | 98.2 | 97.4 | 97.1 | 96.0 | 98.8 | 98.2 | 96.7 | 95.7 | 96.5 | 95.3 |
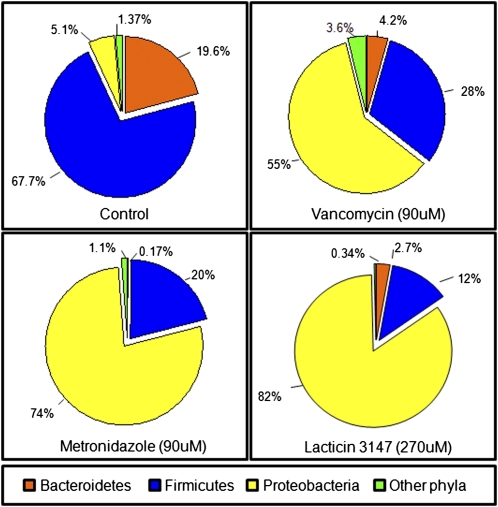
Rarefaction curves for each dataset were parallel or approaching parallel with the x axis, indicating that the total bacterial diversity present within these samples is well represented and that additional sampling would yield a limited increase in species richness (Fig. S1). High-throughput sequencing revealed that of the assigned sequence reads from the control (no antimicrobial) community, 67.7% corresponded to Firmicutes, 19.6% Bacteroidetes, and 5.1% Proteobacteria, with Actinobacteria, Spirochaetes, Lentisphaerae, and Tenericutes collectively constituting 1.37%. This composition is well within the normal range found in healthy adults (24). The introduction of vancomycin, metronidazole, or lacticin 3147 resulted in a major disturbance in the overall fecal microbial composition. The respective proportions of the three major phyla were altered dramatically with respect to each other, with a large reduction in Firmicutes and Bacteroidetes and a dramatic increase in the Proteobacteria. More specifically, the Firmicutes decreased from 67.7% to 28% (vancomycin), 20% (metronidazole), and 12% (lacticin 3147), whereas the corresponding shift in Bacteroidetes was from 19.6% to 4.2% (vancomycin), 0.17% (metronidazole), and 2.7% (lacticin 3147). Proteobacteria assignments increased from just 5.1% of the total sequences in the control to 74% (metronidazole), 55% (vancomycin), and 82% (lacticin 3147) (Fig. 2). The population shifts in the antimicrobial treated samples were shown to be highly significant compared with the control when Pearson correlation coefficients were calculated (Table S1). Further analysis of reads that could be assigned at the family level revealed that members of the Enterobacteriaceae (Proteobacteria subgroup) accounted for more than 50% of the total assignments in the metronidazole-containing vessel and increased to 71% in the vancomycin-containing samples and 80.3% in the lacticin 3147–containing vessel. In contrast, the corresponding Enterobacteriaceae assignments in the non–antimicrobial-containing control sample corresponded to just 1.4% of the total assignable sequence reads (Fig. S2).
Fig. 2.
Phylum level diversity of gut communities in a model of the distal colon, expressed as percentage of total population of assignable tags. Other phyla: Actinobacteria, Spirochaetes, Lentisphaerae, and Tenericutes.
Efficacy of a Narrow-Spectrum Bacteriocin, Thuricin CD, Against C. difficile in the Human Distal Colon Model.
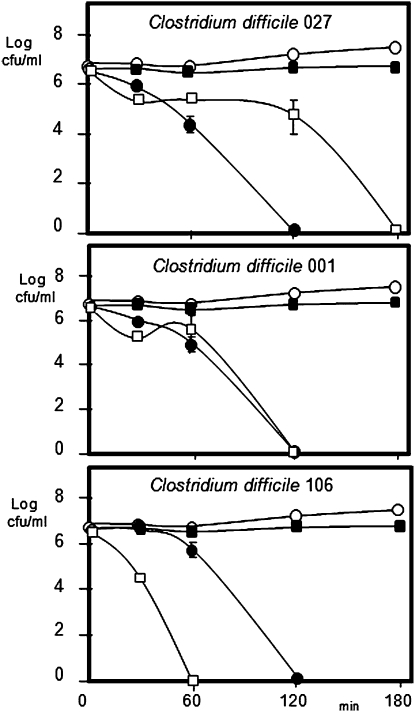
Thuricin CD is a narrow-spectrum bacteriocin produced by Bacillus thuringiensis that exhibits potent activity against C. difficile (16). Indeed, when assessed in broth-based assays with strains representing ribotypes 001, 027, and 106 of the pathogen, it is apparent that this potency is consistently greater than that of vancomycin and, in some cases, is also greater than that of metronidazole (Fig. 3). In contrast, thuricin CD does not exhibit antimicrobial activity against any of a selection of commercially available probiotic strains tested, confirming its relatively narrow host range (probiotics tested included Lactobacillus rhamnosus GG, Lactobacillus johnsonii LA1, Lactobacillus casei Shirota (Yakult), Lactobacillus casei DN-114001 (Actimel), and Bifidobacterium lactis Bb12). The effect of thuricin CD on C. difficile numbers in the distal colon model are presented in Fig. 1A, where a 3.0 log decrease in pathogen numbers compares favorably with a 3.9 log reduction for vancomycin over 24 h. With respect to bioavailability, although detected immediately after each addition, thuricin CD activity was not detected 4 h later (Fig. 1B). Although this could be as a consequence of degradation by the microbiota (as suggested for metronidazole), the consistent reduction in C. difficile numbers suggests that the antimicrobial continues to be active. An alternative explanation may be that thuricin CD is no longer detectable as a consequence of binding to target cells. It should also be noted in this regard that the use of well diffusion assays (WDAs) can only give a crude estimation of actual antimicrobial bioavailability.
Fig. 3.
Effect of thuricin CD (white squares), vancomycin (black squares), and metronidazole (black circles) on C. difficile ribotypes in RCM at 37 °C (control, open circles).
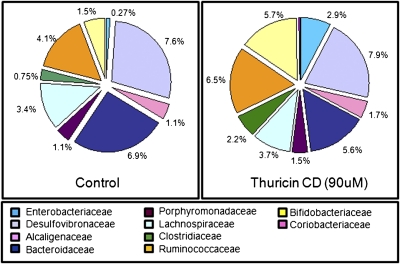
The impact of thuricin CD on the composition of the gut microbiota was assessed by the same high-throughput sequencing strategy as before. This yielded 3,990 sequence reads for the thuricin CD–treated sample and 3,614 for the corresponding control. Assignment of these reads at the phylum and family level revealed that in contrast to the major alterations in microbial composition that results from the utilization of vancomycin, metronidazole, and lacticin 3147, there was no significant difference between the thuricin CD–containing sample and the corresponding control (we present the family data owing to the limited taxonomic assignment of classification programs to the genus level) (Fig. 4 and Table S1). LIBSHUFF (a computer program designed to compare two libraries of 16S rRNA gene sequences) confirmed that there is no difference between the two control vessels, and the Student t test confirmed no significant difference at the phylum (P = 0.45), order (P = 0.45), family (P = 0.31), or genus (P = 0.27) level (Fig. S3). This indicates that the 24-h time point is directly comparable between vessels.
Fig. 4.
Family-level taxonomic distribution of the microbial communities present in model of the distal colon, expressed as percentage of total assignable sequences.
Discussion
Although often an indispensable tool for combating infection, broad-spectrum antimicrobials can also have a very significant impact on the resident microbiota of treated individuals. A detailed analysis of the consequence of antibiotic treatment on the gut microbiota has only become possible recently through the advent of powerful parallel sequencing technologies. Two studies have used this approach to investigate the impact on the gut microbiota of exposure to ciprofloxacin and cefoperazone. It was established that, although previously reported to have lower side effects than some other broad-spectrum antibiotics, ciprofloxacin affected the abundance of ≈30% of taxa across three individuals (25), whereas the cephalosporin cefoperazone caused a dramatic reduction in representatives of the phylum Bacteroidetes (26). Even before these technologies became available, valuable data were generated with both traditional and molecular-based methods, which have provided us with insight into the impact of vancomycin on the gut microflora. Sekirov et al. (27) used FISH to reveal that vancomycin treatment of mice resulted in the proliferation of Enterobacteriaceae at the expense of other Gram-negative bacteria (Cytophaga-Flavobacterium-Bacteroidetes) and of Firmicutes. This decrease in the relative abundance of the Firmicutes was also apparent when a denaturing gradient gel electrophoresis–based approach demonstrated a decrease of populations of Clostridium leptum, Clostridium coccoides, Clostridium symbiosum, and Photorabdus luminescens in mouse stool samples 1 d after vancomycin treatment (28). Similarly, with respect to lacticin 3147, previous culture-based investigations have revealed the negative impact of lacticin 3147 on the Lactobacillus and Bifidobacterium populations of the gut microbiota (15).
Here we investigated the impact of each of four antimicrobials on the human gut microbiota using a model of the human distal colon. All four agents resulted in a decrease of C. difficile levels in the model, albeit that lacticin required a higher concentration. Both thuricin CD and metronidazole seem to be less effective against C. difficile against the fecal background than they are when tested against the same strain in pure culture. This may reflect issues related to bioavailability, although it is notable that vancomycin gives the opposite effect, working more effectively in the fecal fermentation. The impact of administering the three broad-spectrum antimicrobials, vancomycin, metronidazole, and lacticin 3147, is very dramatic, with a consistent observation being a shift in the sequence assignments at the phylum level from the Firmicutes to the Proteobacteria, with a major reduction in overall diversity due to a massive proportionate increase in the Enterobacteriaceae. Exposure to these agents also brought about a decrease in the proportion of reads assigned to the Bacteroidetes. Metronidazole brought about a greater decrease in Bacteroidetes proportions than vancomycin, which is not surprising given the greater activity of the former against representatives of this phylum (29). Lacticin 3147 also impacted on the microorganisms within the Bacteroidetes. It is not apparent whether this impact is direct or arises as a consequence of a change in relative proportions resulting from the activity of lacticin 3147 against other targets. There are a number of similarities between the general patterns identified here and that observed in another study, in which mice were fed a combination of amoxicillin, metronidazole, and bismuth (26). This study also reported a dramatic increase in Proteobacteria, and Enterobacteriaceae in particular, and a decrease in Firmicutes. Although the Bacteroidetes numbers also decreased, this reduction was not as dramatic as that observed in the present study. Some similarities also exist with the trends observed by Hill et al. (30), who treated mice with a combination of ampicillin, gentamicin, metronidazole, neomycin, and vancomycin through oral gavage, in that Proteobacteria and Firmicute reads increased and decreased, respectively. However, this study did differ in that a massive outgrowth of Bacteroidetes was observed, with reads representative of this phylum representing 95% of hits from samples collected from antibiotic-treated mice, 1 d after treatment.
Although vancomycin and metronidazole possess potent anti-C. difficile activity, their application is frequently followed by a recurrence of infection. Enhanced susceptibility to infection has frequently been associated with the negative impacts of broad-spectrum antimicrobials on the gut microbiota (21, 27, 31). With respect to CDAD and antibiotic treatment, the strategies that have been used to address this issue include supplementation of the diet with probiotics (26, 32) and fecal transplantation (33). However, a more desirable strategy would be one in which the negative impact on the gut microbiota is prevented or limited, rather than attempting to regenerate diversity after therapy. Such an alternate strategy requires the utilization of narrow-spectrum antimicrobials. Indeed, the fact that the novel macrocycle OPT-80 eliminates C. difficile without impacting on members of the Bacteroides species (Bacteroides fragilis group) was regarded as noteworthy in a recent study (34). Culture-based techniques confirmed the narrow-spectrum nature of thuricin CD, in that it was not effective against a number of commercially important probiotic strains. However, the utilization of powerful high-throughput sequencing-based strategies allowed us to assess the effect of a narrow-spectrum antimicrobial on both the culturable and unculturable gut microbiota. In this respect thuricin CD was shown to cause no significant shift in the relative proportions of the dominant populations when compared with the control. Although narrow-spectrum activity is one of the key desirable features in novel anti-C. difficile antimicrobials, the fact that thuricin CD also exhibited potency that is comparable to that of its broad-spectrum equivalents makes it a plausible alternative to conventional antibiotic therapies. We also show that although thuricin CD is effective in controlling the pathogen, it is not detectable in the model colonic system within hours of administration. This may be a positive attribute, in that gut microbiota would not be exposed to the antimicrobial for prolonged periods, which would be expected to reduce the pressure on target species to develop thuricin CD resistance. This study also demonstrates the value of using the distal gut system as a model to investigate interventions that target gut flora populations.
In conclusion, we have shown that the three broad-spectrum antimicrobials, vanomcycin, metronidazole, and lacticin 3147, cause dramatic shifts in the relative proportions of the dominant phyla of the human microbiota in a distal colon model. These changes included population shifts from Firmicutes to Proteobacteria in the presence of the antimicrobials. In contrast, the narrow-spectrum bacteriocin thuricin CD seems to cause negligible changes in gut flora but was at least as effective as the two antibiotics at killing C. difficile. We thus demonstrate the potential of narrow-spectrum antimicrobials to target particular pathogens in complex microbial settings such as the gut while the surrounding commensal flora remain relatively unaffected. This study also provides a basis for the development of combined therapies involving coadministration of thuricin CD and probiotics to help to eliminate the pathogen and reduce the dysbiosis associated with currently used antibiotic therapies.
Materials and Methods
Strains Used in This Study.
C. difficile ATCC 43596, Lactococcus lactis HP, and L. lactis HP pMRC01were used to determine the biological activity of the antimicrobial compounds using the WDA, as previously described (15, 35). L. lactis MG1363(pOM02)(pMRC01) and Bacillus thuringiensis 6431 was used to produce the purified lacticin 3147 and thuricin CD, respectively. B. thuringiensis 6431 was grown from a spore isolated from a human fecal sample after ethanol shock. It was the only strain recovered that produces thuricin CD, which is active against clostridia, including C. difficile, but has an otherwise narrow spectrum of activity (16). Neither the strain nor the bacteriocin is in current commercial use. C. difficile DPC 6537 (ribotype 027), DPC 6538 (ribotype 001), and DPC 6539 (ribotype106) were used in this study.
Preparation of Lacticin 3147 and Thuricin CD.
Lacticin 3147 was prepared as described by Rea et al. (16), whereas thuricin CD was purified as follows. Brain heart infusion (BHI) broth was clarified, before autoclaving at 121 °C for 15 min, by passing through a column containing propan-2-ol washed XAD beads (Sigma-Aldrich). B. thuringiensis DPC 6431 was subcultured twice in BHI broth at 37 °C before use. One liter of BHI broth was inoculated with the culture at 0.1% and incubated shaking at 37 °C overnight. The culture was centrifuged at 8,280 × g for 15 min. The cell pellet and supernatant were retained. The cells were resuspended in 200 mL of 70% propan-2-ol (pH 2.0) per liter of broth and stirred at 4 °C for 4 h. The culture supernatant was passed through XAD beads and prewashed with 1 L of distilled H20. The column was washed with 500 mL of 30% ethanol and the inhibitory activity eluted in 400 mL of 70% propan-2-ol (pH 2.0) and retained (S1). The cells that had been resuspended in 70% propan-2-ol (pH 2.0) were centrifuged at 8,280 × g for 15 min and the supernatant (S2) retained; S1 and S2 were combined. The propan-2-ol was evaporated using a rotary evaporator (Buchi) and the sample applied to a 6 g (20 mL) Phenomenex C-18 column preequilibrated with methanol and water. The column was washed with two column volumes of 30% ethanol, and the inhibitory activity was eluted in 1.5 column volumes of 70% propan-2-ol (pH 2.0). This preparation was concentrated using rotary evaporation before separation of peptides using HPLC as follows: aliquots of ≈2 mL were applied to a Phenomenex C18 reverse phase–HPLC column (Primesphere 10 μ C18-MC 30, 250 × 10.0 mm, 10 μm) previously equilibrated with 45% acetonitrile and 0.1% trifluoroacetric acid (TFA). The column was subsequently developed in a gradient of 45% acetonitrile containing 0.1% TFA to 65% acetonitrile containing 0.1% TFA from 4 to 40 min at a flow rate of 9.9 mL/min. Biologically active fractions were identified using C. difficile as target organism in WDA. Fractions containing the active peptides were pooled, freeze-dried, and reconstituted at the required concentration in 70% propan-2-ol (pH 2.0) and frozen at −20 °C until use. Subsequent dilutions were made in sterile 50 mM phosphate buffer (pH 6.5). The C18 SPE eluent from step 2 was added to the colonic model system, and HPLC analysis confirmed that the preparation was at least 90% pure.
Comparison of Thuricin CD, Metronidazole, and Vancomycin on Viability of C. difficile in Pure Culture.
The effect of thuricin, metronidazole, and vancomycin on C. difficile R027 NAP1, R001, and R106 was determined in reinforced Clostridium medium (RCM; Merck). Three independent cultures were prepared for each strain and grown overnight at 37 °C. Three replicate 1-mL volumes of sterile double-strength broth medium was prepared for each strain and inoculated with the test organisms to give initial cell numbers of 106 cfu/mL Thuricin, vancomycin, and metronidazole were added to give the required concentration and the volume made up to 2 mL with sterile distilled water. The antimicrobial was omitted from the control and volume substituted with sterile water. Samples were removed at intervals, serially diluted, and plated on reinforced Clostridium agar incubated anaerobically at 37 °C.
Susceptibility of Probiotic Strains to Thuricin CD.
The sensitivity of probiotic strains to thuricin CD (90 μM) was assayed as follows: L. casei, (Actimel), L. casei (Yakult), L. rhamnosus, L. johnsonii LA1, and B. lactis Bb12 were subcultured twice in de Man, Rogosa, and Sharpe (MRS) medium containing 0.05% cysteine (mMRS) at 37 °C. mMRS agar plates were seeded with 0.5% of the overnight culture and 50 μL of 90 μM thuricin added to well in agar. Plates were incubated at 37 °C overnight and observed for zones of inhibition. Seeded C. difficile agar plates were prepared as described previously (15).
Distal Colon Model.
Fecal medium was prepared according to Fooks et al. (36). The medium (160 mL) was added to each vessel of the MultiFors fermentation system (Infors UK) and autoclaved at 121 °C for 15 min. Once cool the pH of the medium was adjusted to 6.8, and the medium was sparged with oxygen-free N2 for at least 120 min to ensure anaerobic conditions were established. Throughout the experiment the pH was maintained at 6.8.
A fresh composite fecal sample was obtained from at least two donors who were healthy and between 25 and 60 y old (who work in the laboratory but who had tested negative for carriage of thuricin-producing B. thuringiensis) and who had not taken antibiotics for the previous 3 mo. The sample was collected on the morning of the experiment. Just before inoculation the fecal sample was weighed into a sterile filtered stomacher bag and mixed for 1 min in the stomacher with maximum recovery diluent containing 0.05% cysteine, adjusted to pH 7 (which had been previously boiled and cooled in the anaerobic cabinet) to give a 20% solution. Immediately after stomaching, 35 mL of the sample was introduced into the fermentation vessel, which had been sparged with nitrogen for at least the previous 120 min. The vessels were inoculated with 35 mL of the fecal slurry, 2 mL of the overnight culture, and 2 mL of each antimicrobial separately to give the desired concentration and a final volume in the vessel of 200 mL. C. difficile ribotype 001 was added to achieve final cell numbers of 106 cfu/mL. The control vessel contained C. difficile in the absence of any antimicrobial.
At 4-h intervals samples were withdrawn from the vessels, and viable counts of C. difficile were enumerated on CCEY agar (Lab M) incubated at 37 °C for 48 h. The presence of thuricin CD, vancomycin, and metronidazole was determined using C. difficile ATCC 6539 as the target strain, and lacticin 3147 was detected using L. lactis HP using the WDA. Two milliliters of sample was also centrifuged at 20,800 × g for 5 min and the DNA extracted for high-throughput pyrosequencing, as described below.
At 8 and 16 h an additional aliquot of antimicrobial (90 μM) was added to the test vessels, and the level of antimicrobial present in the vessel was sampled before and after addition.
Preparation of DNA for High-Throughput Pyrosequencing.
Samples were collected at the 24-h time point and frozen at −20 °C upon collection. DNA extraction was performed using the QIAamp DNA Stool Mini Kit (Qiagen) coupled with an initial bead-beating step (30 s × 3). Universal 16s primers, designed to amplify from highly conserved regions corresponding to those flanking the V4 region [i.e., the forward primer F1 (5′-AYTGGGYDTAAAGNG) and a combination of four reverse primers R1 (5′-TACCRGGGTHTCTAATCC), R2 (5′-TACCAGAGTATCTAATTC), R3 (5′-CTACDSRGGTMTCTAATC), and R4 (5′-TACNVGGGTATCTAATC)] (RDP's Pyrosequencing Pipeline: http://pyro.cme.msu.edu/pyro/help.jsp) were used for Taq-based PCR amplification. Molecular identifier tags were attached between the 454 adaptor sequence and the forward primers. Amplicons were cleaned using the AMPure purification system (Agencourt) and sequenced on a 454 Genome Sequencer FLX platform (Roche Diagnostics) according to 454 protocols.
Analysis of Sequencing Data.
Raw sequencing reads was quality trimmed using a locally installed version of the RDP Pyrosequencing Pipeline applying the following criteria: (i) exact matches to primer sequences and barcode tags; (ii) no ambiguous bases (Ns); and (iii) read lengths not shorter than the main distribution (150 bp for V4). Clustering and statistical analysis of sequence data were performed using the MOTHUR software package (37). Trimmed FASTA sequences were then BLASTed (38) against a previously published 16s-specific database using default parameters. The resulting BLAST output was parsed using MEGAN (39). MEGAN assigns reads to NCBI taxonomies by using the lowest common ancestor algorithm. Bit-scores were used from within MEGAN for filtering the results before tree construction and summarization. A bit-score of 86 was selected as previously used for 16s ribosomal sequence data (23). To determine their similarity or difference, individual libraries of 16S rRNA gene sequences were compared using LIBSHUFF (http://whitman.myweb.uga.edu/libshuff.html).
Supplementary Material
Acknowledgments
We thank Glenn Gibson (University of Reading, United Kingdom) for his assistance in establishing the human distal colon model. This work was supported by the Science Foundation of Ireland-funded Centre for Science, Engineering and Technology, the Alimentary Pharmabiotic Centre.
Footnotes
Conflict of interest statement: B.K. is Chief Executive Officer of Alimentary Health Ltd., a company that has obtained rights to the thuricin antimicrobial from the research institutes of the other authors involved in this study.
This article is a PNAS Direct Submission.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Microbes and Health,” held November 2–3, 2009, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS Web site at http://www.nasonline.org/SACKLER_Microbes_and_Health.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1001224107/-/DCSupplemental.
References
- 1.Vollaard EJ, Clasener HA. Colonization resistance. Antimicrob Agents Chemother. 1994;38:409–414. doi: 10.1128/aac.38.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donskey CJ. The role of the intestinal tract as a reservoir and source for transmission of nosocomial pathogens. Clin Infect Dis. 2004;39:219–226. doi: 10.1086/422002. [DOI] [PubMed] [Google Scholar]
- 3.Frank DN, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shanahan F. The host-microbe interface within the gut. Best Pract Res Clin Gastroenterol. 2002;16:915–931. doi: 10.1053/bega.2002.0342. [DOI] [PubMed] [Google Scholar]
- 6.Hall IC, O'Toole E. Intestinal flora in new-born infants with a description of a new pathogenic anaerobe, Bacillus difficilis. Am J Dis Child. 1935;49:390–402. [Google Scholar]
- 7.George RH, et al. Identification of Clostridium difficile as a cause of pseudomembranous colitis. Br Med J. 1978;1:695. doi: 10.1136/bmj.1.6114.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cramer JP, Burchard GD, Lohse AW. [Old dogmas and new perspectives in antibiotic-associated diarrhea] Med Klin (Munich) 2008;103:325–338. doi: 10.1007/s00063-008-1040-0. quiz 339–340. [DOI] [PubMed] [Google Scholar]
- 9.Aslam S, Hamill RJ, Musher DM. Treatment of Clostridium difficile-associated disease: Old therapies and new strategies. Lancet Infect Dis. 2005;5:549–557. doi: 10.1016/S1473-3099(05)70215-2. [DOI] [PubMed] [Google Scholar]
- 10.Peláez T, et al. Reassessment of Clostridium difficile susceptibility to metronidazole and vancomycin. Antimicrob Agents Chemother. 2002;46:1647–1650. doi: 10.1128/AAC.46.6.1647-1650.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anonymous. Clostridium difficile infection. Available at: http://ecdc.europa.eu/EN/HEALTHTOPICS/Pages/Clostridium_Difficile_Infection.aspx. Accessed May 1, 2010.
- 12.Kuijper EJ, et al. Update of Clostridium difficile infection due to PCR ribotype 027 in Europe, 2008. Euro Surveill. 2008;13:1–3. [PubMed] [Google Scholar]
- 13.Sood S, Malhotra M, Das BK, Kapil A. Enterococcal infections & antimicrobial resistance. Indian J Med Res. 2008;128:111–121. [PubMed] [Google Scholar]
- 14.Corr SC, et al. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc Natl Acad Sci USA. 2007;104:7617–7621. doi: 10.1073/pnas.0700440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rea MC, et al. Antimicrobial activity of lacticin 3,147 against clinical Clostridium difficile strains. J Med Microbiol. 2007;56:940–946. doi: 10.1099/jmm.0.47085-0. [DOI] [PubMed] [Google Scholar]
- 16.Rea MC, et al. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc Natl Acad Sci USA. 2010;107:9352–9357. doi: 10.1073/pnas.0913554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baines SD, O'Connor R, Saxton K, Freeman J, Wilcox MH. Comparison of oritavancin versus vancomycin as treatments for clindamycin-induced Clostridium difficile PCR ribotype 027 infection in a human gut model. J Antimicrob Chemother. 2008;62:1078–1085. doi: 10.1093/jac/dkn358. [DOI] [PubMed] [Google Scholar]
- 18.Freeman J, Baines SD, Saxton K, Wilcox MH. Effect of metronidazole on growth and toxin production by epidemic Clostridium difficile PCR ribotypes 001 and 027 in a human gut model. J Antimicrob Chemother. 2007;60:83–91. doi: 10.1093/jac/dkm113. [DOI] [PubMed] [Google Scholar]
- 19.Nagy E, Földes J. Inactivation of metronidazole by Enterococcus faecalis. J Antimicrob Chemother. 1991;27:63–70. doi: 10.1093/jac/27.1.63. [DOI] [PubMed] [Google Scholar]
- 20.Rafii F, Wynne R, Heinze TM, Paine DD. Mechanism of metronidazole-resistance by isolates of nitroreductase-producing Enterococcus gallinarum and Enterococcus casseliflavus from the human intestinal tract. FEMS Microbiol Lett. 2003;225:195–200. doi: 10.1016/S0378-1097(03)00513-5. [DOI] [PubMed] [Google Scholar]
- 21.Croswell A, Amir E, Teggatz P, Barman M, Salzman NH. Prolonged impact of antibiotics on intestinal microbial ecology and susceptibility to enteric Salmonella infection. Infect Immun. 2009;77:2741–2753. doi: 10.1128/IAI.00006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Claesson MJ, et al. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS ONE. 2009;4:e6669. doi: 10.1371/journal.pone.0006669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Urich T, et al. Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS ONE. 2008;3:e2527. doi: 10.1371/journal.pone.0002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mariat D, et al. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009;9:123. doi: 10.1186/1471-2180-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Antonopoulos DA, et al. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun. 2009;77:2367–2375. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sekirov I, et al. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect Immun. 2008;76:4726–4736. doi: 10.1128/IAI.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yap IKS, et al. Metabonomic and microbiological analysis of the dynamic effect of vancomycin-induced gut microbiota modification in the mouse. J Proteome Res. 2008;7:3718–3728. doi: 10.1021/pr700864x. [DOI] [PubMed] [Google Scholar]
- 29.Citron DM, et al. In vitro activities of ramoplanin, teicoplanin, vancomycin, linezolid, bacitracin, and four other antimicrobials against intestinal anaerobic bacteria. Antimicrob Agents Chemother. 2003;47:2334–2338. doi: 10.1128/AAC.47.7.2334-2338.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hill DA, et al. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2009;3:148–158. doi: 10.1038/mi.2009.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garner CD, et al. Perturbation of the small intestine microbial ecology by streptomycin alters pathology in a Salmonella enterica serovar typhimurium murine model of infection. Infect Immun. 2009;77:2691–2702. doi: 10.1128/IAI.01570-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McFarland LV. Evidence-based review of probiotics for antibiotic-associated diarrhea and Clostridium difficile infections. Anaerobe. 2009;15:274–280. doi: 10.1016/j.anaerobe.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 33.Khoruts A, Dicksved J, Jansson JK, Sadowsky MJ. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile-associated diarrhea. J Clin Gastroenterol. 2010;44:354–360. doi: 10.1097/MCG.0b013e3181c87e02. [DOI] [PubMed] [Google Scholar]
- 34.Louie TJ, Emery J, Krulicki W, Byrne B, Mah M. OPT-80 eliminates Clostridium difficile and is sparing of bacteroides species during treatment of C. difficile infection. Antimicrob Agents Chemother. 2009;53:261–263. doi: 10.1128/AAC.01443-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ryan MP, Rea MC, Hill C, Ross RP. An application in cheddar cheese manufacture for a strain of Lactococcus lactis producing a novel broad-spectrum bacteriocin, lacticin 3147. Appl Environ Microbiol. 1996;62:612–619. doi: 10.1128/aem.62.2.612-619.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fooks LJ, Gibson GR. Mixed culture fermentation studies on the effects of synbiotics on the human intestinal pathogens Campylobacter jejuni and Escherichia coli. Anaerobe. 2003;9:231–242. doi: 10.1016/S1075-9964(03)00043-X. [DOI] [PubMed] [Google Scholar]
- 37.Schloss PD, Handelsman J. A statistical toolbox for metagenomics: assessing functional diversity in microbial communities. BMC Bioinformatics. 2008;9:34. doi: 10.1186/1471-2105-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.