Abstract
The human oropharynx is a reservoir for many potential pathogens, including streptococcal species that cause endocarditis. Although oropharyngeal microbes have been well described, viral communities are essentially uncharacterized. We conducted a metagenomic study to determine the composition of oropharyngeal DNA viral communities (both phage and eukaryotic viruses) in healthy individuals and to evaluate oropharyngeal swabs as a rapid method for viral detection. Viral DNA was extracted from 19 pooled oropharyngeal swabs and sequenced. Viral communities consisted almost exclusively of phage, and complete genomes of several phage were recovered, including Escherichia coli phage T3, Propionibacterium acnes phage PA6, and Streptococcus mitis phage SM1. Phage relative abundances changed dramatically depending on whether samples were chloroform treated or filtered to remove microbial contamination. pblA and pblB genes of phage SM1 were detected in the metagenomes. pblA and pblB mediate the attachment of S. mitis to platelets and play a significant role in S. mitis virulence in the endocardium, but have never previously been detected in the oral cavity. These genes were also identified in salivary metagenomes from three individuals at three time points and in individual saliva samples by PCR. Additionally, we demonstrate that phage SM1 can be induced by commonly ingested substances. Our results indicate that the oral cavity is a reservoir for pblA and pblB genes and for phage SM1 itself. Further studies will determine the association between pblA and pblB genes in the oral cavity and the risk of endocarditis.
Keywords: endocarditis, metagenomics, oropharyngeal viruses, phage-encoded virulence, streptococcal phage
The human oropharynx is constantly exposed to a wide variety of viruses and microbes from the environment—from both inhaled air and ingested food and water. The oropharynx serves as a niche for commensal bacteria, some of which (e.g., Streptococcus and Neisseria spp.) can be pathogenic when introduced into other body sites (1–3). In healthy individuals, these normal flora prevent colonization by invading organisms by changing the local pH, by producing bacteriocins, and by providing a mechanical barrier that prevents adherence to mucosal surfaces (2, 4, 5). The oropharynx is also a reservoir for several viruses, including HIV, as well as for papillomaviruses and Epstein-Barr virus, which are associated with oropharyngeal carcinomas (6–9). Although oropharyngeal and oral microbes in general have been studied extensively using culturing and 16S sequencing, little is known about viral communities in the oropharyngeal spaces of healthy individuals (1, 10–13). The advent of viral metagenomics—i.e., the culture-independent sequencing of viral nucleic acids—has made it possible to rapidly screen human samples for both known and previously undetected viruses (14–17). For example, a number of known pathogenic viruses, as well as previously unknown types, were detected in nasopharyngeal aspirates from patients with respiratory infections (15, 16).
Here, we present a description of oropharyngeal DNA viral communities in healthy individuals. The initial purpose of this study was to evaluate the feasibility of viral screening using metagenomics in asymptomatic human subjects. Characterization of viral communities in healthy individuals is critical because it establishes a baseline for comparison with samples from diseased individuals (18). However, in healthy individuals, viruses are likely to present in very small numbers, presenting a greater challenge for detection. We demonstrate that oropharyngeal swabs coupled with high-throughput sequencing are an effective method for sampling and characterizing oropharyngeal DNA viral communities, including both phage and eukaryotic viruses. Viral metagenomic sequences from a pool of 19 oropharyngeal samples provided complete coverage of several phage genomes and identified the oropharynx as a potential reservoir for enterobacteria phage T3. Additionally, phage-encoded platelet-binding factors associated with Streptococcus mitis virulence in the endocardium were detected in the oral cavity, providing a potential link between viral communities in the oropharynx and heart disease.
Results and Discussion
Metagenomic Detection of Oropharyngeal Viruses.
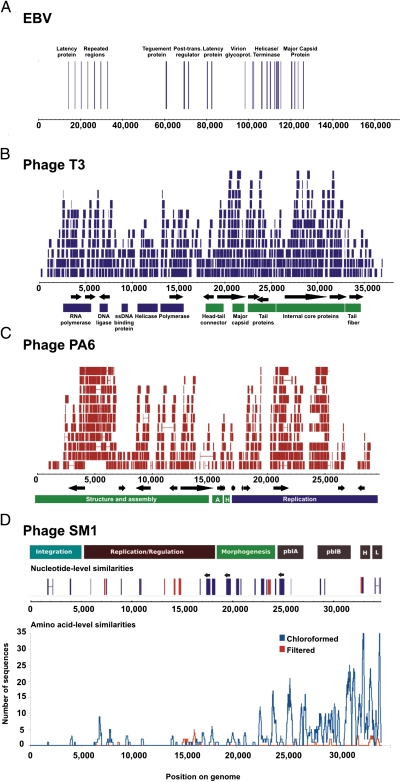
Metagenomic sequencing of oropharyngeal swabs detected both phage and eukaryotic viruses (Fig. 1). Taxonomy was assigned to metagenomic sequences on the basis of BLAST comparisons to the nonredundant database (e-value <10−5). BLASTn analysis identified 53 sequences that were nearly identical (>98% identity at the nucleotide level) to Epstein-Barr virus (EBV). The majority of these sequences aligned to ORFs in the EBV genome, including genes involved in viral replication and latency as well as virion structure (Fig. 1A). No additional sequences were recruited to the EBV genome using amino-acid-level searches (tBLASTx). EBV primarily infects epithelial cells in the oropharynx (19). EBV infection is generally controlled by the immune system in healthy individuals, but the virus remains latent in circulating B lymphocytes (19, 20). Viral reactivation can occur in seropositive-normal individuals, resulting in viral shedding in the oropharynx (21). Although it is estimated that 90% of the healthy adult population is seropositive for EBV, reactivation occurs in only 10–20% of individuals with latent EBV infections (20, 22). The incomplete coverage of EBV in the oropharyngeal metagenome was likely a reflection of the low prevalence of individuals actively shedding virus in the pooled sample population, as the metagenome was a composite from all 19 study subjects. Detection of EBV in a pooled sample indicates that metagenomic sequencing of oropharyngeal swabs has adequate sensitivity to serve as a rapid noninvasive screen for viruses in individuals.
Fig. 1.
Coverage of viral genomes by oropharyngeal metagenomic sequences: Epstein-Barr virus (A), E. coli phage T3 (B), P. acnes phage PA6 (C), and S. mitis phage SM1 (D). Similarities obtained from the chloroformed metagenome are shown in blue, and those from the filtered metagenome are shown in red. Nucleotide-level coverage (A, B, C, and top of D) was determined by alignment of metagenomic sequences to complete viral genome sequences obtained from the National Center for Biotechnology Information using BLAT. Amino acid level coverage (D, bottom) was plotted using significant tBLASTx (e-value <10−5) similarities to each genome. Contigs were assembled using the 454 gsAssembler and aligned to genomes using BLAT.
The complete genome of Escherichia coli phage T3 was recovered from oropharyngeal swabs (Fig. 1B). Over 500 sequences were at least 98% identical to the T3 genome at the nucleotide level. These sequences provided ≈3× the coverage of T3 and could be combined into contigs as large as 4 kb. Laboratory strains of phage T3 are widely used for experimental purposes; however, the origins of and natural reservoirs for T3 are largely unknown (23). A BLAST search of publicly available environmental metagenomes revealed a very low prevalence of sequences similar to T3, even in fecal samples that are considered to be a source of the phage (Table S1). Our results indicate that the oropharynx may be a previously undiscovered environmental reservoir for phage T3.
Metagenomic sequences provided high coverage of Propionibacterium acnes phage PA6 at the nucleotide level and of S. mitis phage SM1 at the amino acid level (Fig. 1 C and D). Contigs of up to 2 kb could be assembled and aligned to the PA6 genome; however, no contigs larger than 500 bp that were significantly similar to SM1 could be assembled . Phage PA6 is a lytic phage whose host, P. acnes, is highly abundant in the oral cavity (4). Phage SM1 is a temperate phage previously isolated from S. mitis SF100, an endocarditis strain (24). SM1 carries two genes, pblA and pblB, which contribute to S. mitis virulence in the endocardium (24–27). Although S. mitis is a ubiquitous member of the normal oral flora, the presence of phage SM1 has never previously been reported in the mouth or oropharynx (1). The lack of long contigs and discontinuous coverage at the nucleotide level suggests that the SM1 nucleotide sequence was highly variable between individuals, within individuals, or both. Temperate phage adopt the oligonucleotide usage patterns of their hosts, which can lead to sequence divergence at the nucleotide level if multiple different hosts are present (28, 29). Because the oropharyngeal metagenomes were constructed from pooled samples from 19 individuals, it is likely that phage with varied hosts and host ranges were sampled.
Sample Processing Methods Affect Metagenomic Composition.
The composition of the oropharyngeal metagenomes differed depending on which sample preparation method was used (Fig. 2 A and B). Before DNA extraction, the pooled oropharyngeal swab sample was split, and each half was treated to reduce microbial contamination either by the addition of chloroform or 0.22-μm filtration. The filtered metagenome contained a higher percentage of bacterial sequences, whereas the chloroformed metagenome was enriched in viral (including phage) sequences (Fig. 2A). Filtering at 0.22 μm should trap bacterial cells while allowing viral particles to pass through (30). However, some viral particles will stick to the filter, especially larger viruses. EBV was the only eukaryotic virus detected in the oropharyngeal metagenomes, and it was present only in the chloroformed metagenome. EBV virions range in diameter from 120 to 220 nm and thus may not have passed through the filter (31). Additionally, some bacterial cells are likely to have escaped filtration. These cells would have been lysed during viral DNA extraction, releasing chromosomal DNA and contaminating the viral metagenome (30). Chloroform treatment permeabilizes the membranes of bacterial cells, leading to cell death and the release of chromosomal DNA into the medium, where it can be digested with DNase I (30). This treatment also releases any intracellular viral particles, which may be fully assembled but have not yet induced host cell lysis. In general, viral capsids are resistant to chloroform and remain intact until lysis during DNA extraction.
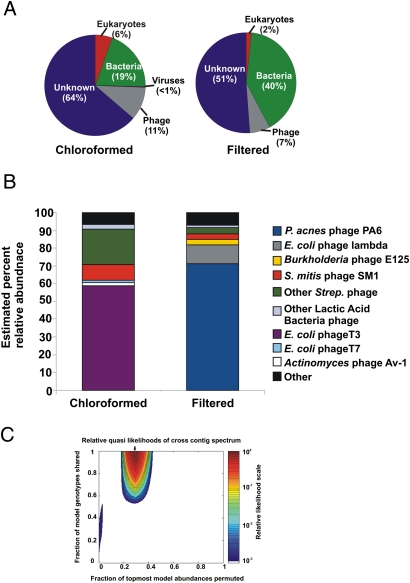
Fig. 2.
Taxonomic composition and diversity of the oropharyngeal metagenomes. (A) Composition of complete metagenomes as determined by best tBLASTx similarities to the nonredundant database (e-value <10−5). (B) Composition of viral communities. Viral relative abundances were determined by GAAS on the basis of tBLASTx similarities (e-value <10−5, percentage identity >30%, query coverage >80%) to a database containing all complete viral genomes currently available at the National Center for Biotechnology Information. (C) Monte Carlo analysis of cross-contig spectra for oropharyngeal metagenomes. The area of maximum likelihood is indicated by an arrow. The metagenomes were predicted to share more than 95% of genotypes with 30% of their relative abundances permuted.
Phage communities in the two oropharyngeal metagenomes shared many species, but in different relative abundances (Fig. 2B). Community composition was estimated using GAAS, which calculates relative abundances on the basis of all significant BLAST similarities (32). E. coli phage T3 was the most abundant phage in the chloroformed sample, yet comprised only 1.6% of the community in the filtered sample. Similarly, P. acnes phage PA6 was the most abundant phage in the filtered sample, yet appeared in extremely low abundance (<0.01%) in the chloroformed sample. P. acnes is ubiquitous in the healthy oral cavity, whereas Gram-negative bacteria such as E. coli are generally present in low abundance or not at all because they are rapidly cleared in healthy people (4, 33, 34). Abedon (35) demonstrated that phage with more abundant hosts tend to have shorter latent periods, i.e., a smaller lag time between adsorption and host lysis. Phage with less abundant hosts have longer latent periods and will produce more progeny before lysis, generating a larger burst size (35). The addition of chloroform would cause the release of progeny phage from host cells, whereas filtration would remove host cells and their intracellular phage. The shift in phage T3 abundance between the filtered and chloroformed samples is likely the result of the release of intracellular T3 phage during chloroform treatment. This may also account for the increased abundance of T7 in the chloroformed sample (1.3% versus 0.1% in the filtered sample). The enrichment of E. coli phage λ in the filtered metagenome (11.1% versus <0.01%) is seemingly in contrast to the long-latent-period hypothesis. However, λ is a temperate phage, and several λ sequences with flanking host sequences were detected in the metagenomes. This indicates that λ was present as a prophage element integrated into the host genome, not as a free phage particle. Therefore, the higher abundance of λ in the filtered metagenome was due to the higher level of bacterial DNA contamination.
Streptococcal phage were more abundant in the chloroformed sample than in the filtered sample, comprising 33% and 7% of the viral communities respectively (Fig. 2B). S. mitis phage SM1 was the most abundant streptococcal phage in both metagenomes. Phage of other lactic acid bacteria (LAB) were also more abundant in the chloroformed metagenome (2.5% versus 1.0% in filtered). All of the LAB and streptococcal phage detected were temperate phage. No flanking host sequences were detected adjacent to these phage sequences in the metagenomes, indicating the presence of free phage particles. Free phage would be enriched in the chloroformed sample due to the release of intracellular phage from host cells as described above. Streptococci are among the first bacteria to colonize the oral cavity and remain in the mouth and oropharynx at high population densities throughout an individual's life (2, 5). Lactobacilli and other lactic acid bacteria are also common constituents of the normal oral flora, but are present at lower abundances, as reflected by the lower abundances of their phage in the oropharyngeal viral communities (2, 5).
Diversity of Viruses in the Oropharynx.
Viral communities had the same predicted diversity, regardless of the sample preparation method. Viral diversity was estimated using the PHACCs program as described in ref. 36. The PHACCs method uses all metagenomic sequences, not just those with significant BLAST similarities (36). Viral communities in both filtered and choloroformed samples were predicted to follow a power law distribution. The estimated richness of viral communities in both samples was 236 species, which was similar to estimates for the human respiratory tract and low compared with the viral richness in marine environments (14, 37, 38). Estimates of microbial richness in the healthy oral cavity are similarly low; although over 700 microbial species have been identified, each individual is thought to harbor only 100–200 at any given time (1, 10). Despite the constant introduction of environmental microbes from food, water, and air, microbial and viral richness in the oropharynx is limited by several anatomical and biological mechanisms. Microbiota can be trapped in the mucosa before adherence, inhibited by chemicals in saliva such as lactoferrin, or cleared by the host immune system (2, 33). Additionally, normal flora prevent the adherence and growth of transient microbiota by producing bacteriocins and manipulating the pH of oral microenvironments (2, 33).
Viral communities in the filtered and chloroformed samples share many genotypes, but at different relative abundances. Taxonomic data indicated that the majority of viruses that could be identified using BLAST appeared in both metagenomes, but in different proportions. To test whether this was true for all viral genotypes, not just those with significant BLAST similarities, cross-contigs were generated between the metagenomes, and a Monte Carlo simulation was conducted as described in ref. 37. The simulation uses cross-contig spectra to estimate what proportion of genotypes are shared between communities and what proportion of these shared genotypes are permuted, i.e., present in different abundances. The filtered and chloroformed viral communities were predicted to share more than 95% of genotypes with 30% permuted (Fig. 2C). When each sample was compared with itself as a control, nearly all (>99%) of the sequences were shared; however, less than 0.1% were permuted (Fig. S1). The simulation results corroborated the BLAST-based comparisons, demonstrating that although the filtered and chloroformed samples shared the same population of viruses, sample preparation methods altered their relative abundances.
Phage-Encoded Platelet-Binding Factors in Oropharyngeal and Salivary Metagenomes.
Two genes of S. mitis phage SM1 that encode the platelet-adhesion factors pblA and pblB were detected in the oropharyngeal metagenomes (Fig. 1D and Fig. S2). Although few sequences similar to pblA and pblB at the nucleotide level were identified using BLASTn, coverage of pblA, pblB, and holin and lysin genes was much higher than for the rest of the SM1 genome at the amino acid level (Fig. 1D). pblA and pblB are integral phage tail proteins, and phage with pblA and pblB gene deletions have intact capsids but no tails (24, 25). pblA and pblB also mediate the attachment of S. mitis to platelets, which has been shown in a rabbit model to contribute significantly to the virulence of S. mitis in the endocardium (25–27, 39). The interaction between S. mitis cells and platelets requires phage induction for maximal release of intracellular pblA and pblB, but the soluble proteins can bind to choline residues on the surface of host or nonhost cells (27, 39). Theoretically, pblA and pblB genes inserted into any phage capable of host cell permeabilization or lysis would be sufficient to mediate S. mitis adhesion to platelets. pblA and pblB may be prime targets for horizontal gene transfer (HGT), as genes encoding phage tail proteins are especially labile regions and can be highly variable even in phage with nearly identical genomes (24, 40, 41). Phage are major agents of HGT in many streptococci and have been shown to mediate interspecies genetic exchange, indicating that streptococcal phage may have wide host ranges and undergo frequent recombination events (42–44). S. mitis and other streptococci have been shown to enter the blood stream from the oral cavity following tooth extractions, so the dissemination of pblA and pblB genes in oral phage and microbes could potentially translate into an increased risk of endocarditis (3).
Phage SM1-like pblA and pblB genes were also detected in salivary metagenomes from three individuals at three time points (Fig. 2A). Subsequent to our analysis of the oropharyngeal metagenomes, nine preexisting salivary metagenomes became available to us for screening for pblA and pblB genes. Sequences with significant tBLASTx similarities (e-value <10−5, identity >30%, query coverage >80%) to pblA were found in all individuals at all time points, although pblB was absent in subject 2 at the 30-d time point. Aas et al. demonstrated that although some microbes preferentially colonize particular sites, S. mitis is ubiquitous and can be detected throughout the oral cavity (1). The presence of phage SM1 pblA and pblB genes in both oropharyngeal and salivary metagenomes suggests that these genes, and most likely phage SM1 itself, have a similarly widespread distribution.
In the salivary metagenomes pblA sequences varied both between and within subjects (Fig. S2B). The sequence comparison tool cd-hit-est-2d was used to assess the degree of variability of pblA sequences at the nucleotide level (45). pblB sequences were not analyzed because at some time points fewer than five sequences were identified. pblA sequences with 90% identity at the nucleotide level were considered to be congruent. A dissimilarity matrix was constructed from cd-hit-est-2d results and used as an input to multidimensional scaling (MDS) (Table S2). A scatterplot of MDS coordinates showed that pblA sequences differed between individuals and within individuals at different time points (Fig. S2B). In all three individuals, sequences from closer time points appeared to be more similar than those from more distant time points (i.e., 1 versus 90 d). Sequences from subject 3 were extremely dissimilar from those from subjects 1 and 2 at all time points. To determine whether this divergence was driven by coverage differences, coverage of pblA genes was compared between metagenomes (Table S3). Coverage was not significantly correlated with sequence dissimilarities (Spearman's ρ = 0.26, P = 0.13). Similar to the results in the oropharyngeal metagenomes, this suggests that pblA genes are variable between individuals, and in addition, within individuals over time. These nucleotide-level changes may be indicative of the adaptation of phage sequences to host oligonucleotide usage, reflecting the movement of phage SM1 genes into different and potentially novel hosts either through host range expansion or lateral gene transfer (28, 46).
PCR Detection of pblA in Saliva Samples.
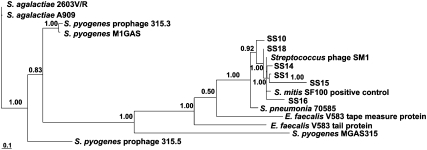
Gene fragments of pblA with high homology to phage SM1 pblA were detected in individual saliva samples from healthy individuals (Fig. 3, Figs. S3 and S4, and Table S4). Saliva samples were collected from 20 individuals and screened for the presence of an ≈750-bp region of pblA to confirm the presence of pblA in the oral cavity. This region, spanning nucleotides 1,456–2,222 of the pblA gene, was notably under-represented in both the oropharyngeal and the salivary metagenomes (Fig. S2A). The pblA gene fragment was detected in 6 of the 20 individuals tested and was sequenced, along with positive control DNA, from cultured S. mitis SF100. Negative PCR results in the other 14 individuals may have indicated the absence of pblA or possibly sequences that were highly divergent from the SM1 pblA gene sequence because the primers used in the assay were specific to SM1 pblA. At the nucleotide level, the divergence between saliva sequences and the reference sequence (phage SM1 pblA) ranged from 0.0 to 2.0% (Fig. S3). The positive control sequence, derived from the same strain of S. mitis as the reference sequence, was 3.2% divergent, which is less similar to the reference than the saliva sequences. At the amino acid level, the positive control sequence was 7.1% divergent from the reference, which is similar to the saliva sequences, which ranged from 4.5% and 37.5% divergence (Fig. S4). Phylogenetic analysis indicated that all saliva sequences and the positive control sequence were more closely related to the SM1 reference sequence than to any other pblA homolog (Fig. 3). These results confirm the presence of SM1-like pblA genes in the healthy human oral cavity.
Fig. 3.
Phylogenetic relationships between pblA sequences from saliva samples and reference genomes. The Bayes values show the proportion of sampled trees in which the sequences to the right of the branch point clustered together.
Induction of Phage SM1.
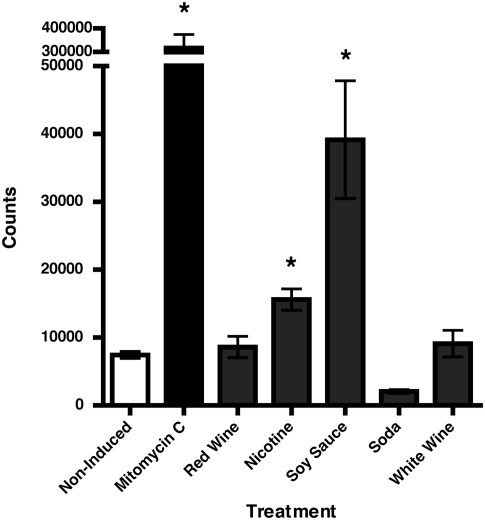
Phage SM1 was induced by commonly ingested substances, such as nicotine and soy sauce (Fig. 4; Fig. S5). To determine the relative amounts of phage induced, we used a flow cytometry method to enumerate phage in each sample (47). Cultures of S. mitis SF100 were treated with red wine, white wine, soda, solubilized nicotine, soy sauce, or mitomycin C for phage induction and compared with an untreated control culture. Nicotine and soy sauce treatments produced significantly more phage particles (P < 0.05) than the noninduced control, whereas red wine, white wine, and soda had no significant effect on phage production. Phage induction provides a vehicle for virulence genes to travel between bacterial species. Acquisition of toxin genes by group A and group C Streptococcus has been shown to occur by lysogenization following prophage induction, and it is likely that pblA and pblB genes could disseminate in the same manner (42). Mitchell et al. (27) demonstrated that even at low levels phage SM1 induction is sufficient to facilitate S. mitis binding to platelets. Induction of phage by food or beverages in individuals with severe periodontal disease could lead to an increased endocarditis risk, as they are highly prone to acquiring bacteremia from routine activities such as toothbrushing (48, 49).
Fig. 4.
Phage induction assay. Data are presented as induction treatment on the x axis versus the mean number of phage events counted using flow cytometry (±SEM) on the y axis (n = 3). White wine, nicotine, soda, and soy sauce treatments were diluted 1:10, and the red wine treatment was diluted 1:100. Asterisks indicate that nicotine, soy sauce, and mitomycin C caused a statistically significant increase in phage count (P < 0.05).
Additional Considerations.
The oropharyngeal viral community described here consisted almost exclusively of phage. With the exception of EBV, no eukaryotic viruses were detected. In healthy individuals, the absence of eukaryotic viruses may be characteristic of the nondiseased state. However, it is also possible that enveloped viruses were not efficiently isolated by the cesium chloride density gradient method due to their anomalous density (30). Additionally, the effects of the chloroform and filtration treatments on reducing microbial contamination were not evaluated quantitatively in this study. Future studies to test the efficacy of these two methods could use samples containing known amounts of viral and bacterial cells or quantitative PCR for bacterial marker genes as a proxy to measure bacterial DNA concentration before and after treatment.
Pooled oropharyngeal swab samples from 19 individuals were used to generate the oropharyngeal metagenomes in this study. Pooled samples have often been used in metagenomic studies to allow for sampling of a large group of individuals simultaneously. In microarray studies, pooling has been shown to decrease biological variation and enhance detection of features shared by members of the pool (50). Here, pooling allowed for the successful detection of phage SM1 and pblA and pblB genes, which may not have been present in all individuals and may not have been detected if only one or two subjects had been sampled. Future studies of oropharyngeal viruses should include contrasts between viral communities in nonpooled samples to better describe variation between individuals.
A caveat to this study was the use of multiple displacement amplification (MDA) with phi29 polymerase before 454 sequencing. Although MDA generally does not bias the representation of individual genomes in metagenomic samples, small circular and long linear genomes may be disproportionately amplified (51, 52). In any case, all metagenomes were amplified using the same reaction conditions, allowing for valid comparisons between samples even if bias were introduced.
Conclusions.
Metagenomics is a powerful tool for both characterization of environmental viral communities and discovery. In this study, we set out to evaluate the use of oropharyngeal swabs as a general screen for viruses and unexpectedly discovered phage-encoded virulence genes in oropharyngeal viral communities. Detection of the pblA and pblB genes in saliva as well as in oropharyngeal samples suggests that they are widely disseminated both in the oral cavity and in the human population at large. Within the metagenomes, pblA and pblB sequences varied significantly at the nucleotide level between individuals and within individuals over time. HGT and the host range expansion of phage SM1 could potentially be facilitated in the oral cavity by commonly ingested substances, as shown by our phage induction assay. Several studies have established a link between endocarditis and oral hygiene, demonstrating that streptococci readily enter the bloodstream during tooth extractions and following toothbrushing in individuals with periodontal disease (3, 48, 49). Future studies should include characterization of oral phage communities in individuals with endocarditis, as well as comparative studies between endocarditis and nonendocarditis individuals to determine the endocarditis risk associated with the presence of phage SM1 and/or pblA and pblB genes in the oral cavity.
Materials and Methods
Ethics Statement.
Subject recruitment for the oropharyngeal metagenomes and saliva PCR assay was approved by the San Diego State University Institutional Review Board (SDSU IRB 2121) and Environmental Health Services (BUA 06–02-062R). Subject recruitment for the saliva metagenomic study was approved by the Stanford University Administrative Panel on Human Subjects in Medical Research.
Viral Metagenome Preparation.
Viruses were concentrated from pooled oropharyngeal and individual saliva samples as described in ref. 30. The pooled oropharyngeal sample was split, and half was chloroform treated and half was 0.2-μm filtered. Saliva metagenomic samples were filtered only. Viral DNA was extracted using a cetyltrimethylammonium bromide/formamide protocol and submitted for 454 sequencing (53).
Bioinformatics for Metagenomic Sequences.
Metagenomic sequences were compared with the nonredundant database at GenBank for taxonomic assignment using BLAST (54). Sequences with best BLAST similarities to microbial and eukaryotic genomes were removed before further analysis. Nucleotide-level coverage of individual viral genomes was assessed with BLAT and visualized with the Integrated Genome Browser (55, 56). Viral community composition was determined using GAAS (32). Diversity of metagenomes was assessed using PHACCs, and comparisons between metagenomes were performed using a Monte Carlo simulation (36, 37). Saliva metagenomic sequences were clustered using cd-hit-2-est, and sequence dissimilarity was evaluated by multidimensional scaling (45). The pblA and pblB gene sequences were obtained from GenBank and coverage by the metagenomic sequences was compared by using the XIPE program (57).
Saliva PCR Assay.
Total DNA was extracted from 20 saliva samples from randomly selected healthy individuals (according to the criteria of the initial oropharyngeal study) as described in ref. 58. Positive and negative control DNA was extracted from overnight cultures of S. mitis SF100 and PS344, respectively. PCR primers and reaction conditions are provided in the SI Materials and Methods. PCR products were purified using the Accu-Prep PCR purification kit (Bioneer) and sequenced using an ABI Prism 3100 Genetic Analyzer. Sequences were deposited in GenBank under accession numbers GU586484, GU586485, GU586486, GU586487, GU5864848, GU586489, and GU586490. Sequences were translated using the online tool TranSeq and aligned using ClustalW2 (59, 60). A phylogenetic tree was constructed using MrBayes version 3.1 (61).
Phage Induction Assay.
One of five treatments, 0.2 mg/mL mitomycin C, or no treatment was added to overnight cultures of S. mitis SF100. Cultures were incubated for 3.5 h and then 0.45-μm filtered to remove remaining bacterial cells. Samples were fixed and stained and phage particles were counted using flow cytometry as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the Joint Genome Institute for sequencing the oropharyngeal metagenomes, Paul Sullam and Ho Seong Seo (University of California, San Francisco) for generously providing S. mitis strains SF100 and PS344, Brett Hilton and the San Diego State University Fluorescence Activated Cell Sorting core for assistance with flow cytometry, and Kelly Doran, Tom Gibson, Marlene DeMers, and Mike Gurney for their microbiology assistance. We also thank Gary C. Armitage and Peter Loomer for their assistance in the saliva metagenomic project and Linda Wegley for helpful discussions. This work was performed under the auspices of the US Department of Energy's Office of Science, Biological and Environmental Research Program; the University of California Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344; and by the University of California Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231 and Lawrence and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396. This work was also supported in part by grants from the Robert Wood Johnson Foundation, the United Negro College Fund--Merck Science Initiative, and the Burroughs Wellcome Fund (to D.T.P.); the National Institutes of Health Director's Pioneer Award DP1OD000964 (to D.A.R.); and a grant from Cystic Fibrosis Research Inc. (to F.L.R.). D.A.R. is supported by the Thomas C. and Joan M. Merigan Endowment at Stanford University.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Microbes and Health” held November 2–3, 2009, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS Web site at http://www.nasonline.org/SACKLER_Microbes_and_Health.
This article is a PNAS Direct Submission.
Data deposition: Oropharyngeal metagenomic sequences are publicly available from Metagenome Rapid Annotation using Subsystem Technology (http://metagenomics.nmpdr.org) under IDs 4444195.3 and 4444196.3. Metagenomic sequences were also deposited in Genbank as genome project 43627 (oropharyngeal metagenomes) and genome project 43629 (salivary metagenome subset). Saliva PCR sequences were deposited under accession nos. GU586484, GU586485, GU586486, GU586487, GU586488, and GU586489 and the positive control sequences were deposited under accession no. GU586490.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000089107/-/DCSupplemental.
References
- 1.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hull MW, Chow AW. Indigenous microflora and innate immunity of the head and neck. Infect Dis Clin North Am. 2007;21:265–282. doi: 10.1016/j.idc.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 3.Bahrani-Mougeot FK, et al. Diverse and novel oral bacterial species in blood following dental procedures. J Clin Microbiol. 2008;46:2129–2132. doi: 10.1128/JCM.02004-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hentges DJ. The anaerobic microflora of the human body. Clin Infect Dis. 1993;16(Suppl 4):S175–S180. doi: 10.1093/clinids/16.supplement_4.s175. [DOI] [PubMed] [Google Scholar]
- 5.Jenkinson HF, Lamont RJ. Oral microbial communities in sickness and in health. Trends Microbiol. 2005;13:589–595. doi: 10.1016/j.tim.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Moutsopoulos NM, Greenwell-Wild T, Wahl SM. Differential mucosal susceptibility in HIV-1 transmission and infection. Adv Dent Res. 2006;19:52–56. doi: 10.1177/154407370601900111. [DOI] [PubMed] [Google Scholar]
- 7.Moutsopoulos NM, et al. Tonsil epithelial factors may influence oropharyngeal human immunodeficiency virus transmission. Am J Pathol. 2007;171:571–579. doi: 10.2353/ajpath.2007.061006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shillitoe EJ. The role of viruses in squamous cell carcinoma of the oropharyngeal mucosa. Oral Oncol. 2009;45:351–355. doi: 10.1016/j.oraloncology.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 9.Szkaradkiewicz A, et al. Epstein-Barr virus and human papillomavirus infections and oropharyngeal squamous cell carcinomas. Clin Exp Med. 2002;2:137–141. doi: 10.1007/s102380200019. [DOI] [PubMed] [Google Scholar]
- 10.Paster BJ, Olsen I, Aas JA, Dewhirst FE. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol 2000. 2006;42:80–87. doi: 10.1111/j.1600-0757.2006.00174.x. [DOI] [PubMed] [Google Scholar]
- 11.Diaz PI, et al. Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl Environ Microbiol. 2006;72:2837–2848. doi: 10.1128/AEM.72.4.2837-2848.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kazor CE, et al. Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients. J Clin Microbiol. 2003;41:558–563. doi: 10.1128/JCM.41.2.558-563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nasidze I, Li J, Quinque D, Tang K, Stoneking M. Global diversity in the human salivary microbiome. Genome Res. 2009;19:636–643. doi: 10.1101/gr.084616.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willner D, et al. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS ONE. 2009;4:e7370. doi: 10.1371/journal.pone.0007370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allander T, et al. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci USA. 2005;102:12891–12896. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamura S, et al. Direct metagenomic detection of viral pathogens in nasal and fecal specimens using an unbiased high-throughput sequencing approach. PLoS ONE. 2009;4:e4219. doi: 10.1371/journal.pone.0004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breitbart M, Rohwer F. Method for discovering novel DNA viruses in blood using viral particle selection and shotgun sequencing. Biotechniques. 2005;39:729–736. doi: 10.2144/000112019. [DOI] [PubMed] [Google Scholar]
- 18.Turnbaugh PJ, et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fields BN, et al. Fields Virology. 3rd Ed. Philadelphia: Lippincott Williams & Wilkins; 1996. [Google Scholar]
- 20.Vetsika EK, Callan M. Infectious mononucleosis and Epstein-Barr virus. Expert Rev Mol Med. 2004;6:1–16. doi: 10.1017/S1462399404008440. [DOI] [PubMed] [Google Scholar]
- 21.Ling PD, et al. The dynamics of herpesvirus and polyomavirus reactivation and shedding in healthy adults: A 14-month longitudinal study. J Infect Dis. 2003;187:1571–1580. doi: 10.1086/374739. [DOI] [PubMed] [Google Scholar]
- 22.Pender MP. Infection of autoreactive B lymphocytes with EBV, causing chronic autoimmune diseases. Trends Immunol. 2003;24:584–588. doi: 10.1016/j.it.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 23.Abedon ST. The murky origin of Snow White and her T-even dwarfs. Genetics. 2000;155:481–486. doi: 10.1093/genetics/155.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siboo IR, Bensing BA, Sullam PM. Genomic organization and molecular characterization of SM1, a temperate bacteriophage of Streptococcus mitis. J Bacteriol. 2003;185:6968–6975. doi: 10.1128/JB.185.23.6968-6975.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bensing BA, Siboo IR, Sullam PM. Proteins PblA and PblB of Streptococcus mitis, which promote binding to human platelets, are encoded within a lysogenic bacteriophage. Infect Immun. 2001;69:6186–6192. doi: 10.1128/IAI.69.10.6186-6192.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bensing BA, Rubens CE, Sullam PM. Genetic loci of Streptococcus mitis that mediate binding to human platelets. Infect Immun. 2001;69:1373–1380. doi: 10.1128/IAI.69.3.1373-1380.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchell J, Siboo IR, Takamatsu D, Chambers HF, Sullam PM. Mechanism of cell surface expression of the Streptococcus mitis platelet binding proteins PblA and PblB. Mol Microbiol. 2007;64:844–857. doi: 10.1111/j.1365-2958.2007.05703.x. [DOI] [PubMed] [Google Scholar]
- 28.Blaisdell BE, Campbell AM, Karlin S. Similarities and dissimilarities of phage genomes. Proc Natl Acad Sci USA. 1996;93:5854–5859. doi: 10.1073/pnas.93.12.5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mrázek J, Karlin S. Distinctive features of large complex virus genomes and proteomes. Proc Natl Acad Sci USA. 2007;104:5127–5132. doi: 10.1073/pnas.0700429104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thurber RV, Haynes M, Breitbart M, Wegley L, Rohwer F. Laboratory procedures to generate viral metagenomes. Nat Protoc. 2009;4:470–483. doi: 10.1038/nprot.2009.10. [DOI] [PubMed] [Google Scholar]
- 31.Buchen-Osmond C. ICTVdB: The Universal Virus Database. New York: Columbia University; 2006. [Google Scholar]
- 32.Angly FE, et al. The GAAS metagenomic tool and its estimations of viral and microbial average genome size in four major biomes. PLOS Comput Biol. 2009;5:e1000593. doi: 10.1371/journal.pcbi.1000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mobbs KJ, van Saene HK, Sunderland D, Davies PD. Oropharyngeal Gram-negative bacillary carriage: A survey of 120 healthy individuals. Chest. 1999;115:1570–1575. doi: 10.1378/chest.115.6.1570. [DOI] [PubMed] [Google Scholar]
- 34.Van Saene HK, Stoutenbeek CP, Torres A. The abnormal oropharyngeal carrier state: Symptom or disease? Respir Med. 1992;86:183–186. doi: 10.1016/s0954-6111(06)80052-1. [DOI] [PubMed] [Google Scholar]
- 35.Abedon ST. Selection for bacteriophage latent period length by bacterial density: A theoretical examination. Microb Ecol. 1989;18:79–88. doi: 10.1007/BF02030117. [DOI] [PubMed] [Google Scholar]
- 36.Angly F, et al. PHACCS, an online tool for estimating the structure and diversity of uncultured viral communities using metagenomic information. BMC Bioinformatics. 2005;6:41. doi: 10.1186/1471-2105-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Angly FE, et al. The marine viromes of four oceanic regions. PLoS Biol. 2006;4:e368. doi: 10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marhaver KL, Edwards RA, Rohwer F. Viral communities associated with healthy and bleaching corals. Environ Microbiol. 2008;10:2277–2286. doi: 10.1111/j.1462-2920.2008.01652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitchell J, Sullam PM. Streptococcus mitis phage-encoded adhesins mediate attachment to alpha2-8-linked sialic acid residues on platelet membrane gangliosides. Infect Immun. 2009;77:3485–3490. doi: 10.1128/IAI.01573-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Angly F, et al. Genomic analysis of multiple Roseophage SIO1 strains. Environ Microbiol. 2009;11:2863–2873. doi: 10.1111/j.1462-2920.2009.02021.x. [DOI] [PubMed] [Google Scholar]
- 41.Lucchini S, Desiere F, Brüssow H. The structural gene module in Streptococcus thermophilus bacteriophage phi Sfi11 shows a hierarchy of relatedness to Siphoviridae from a wide range of bacterial hosts. Virology. 1998;246:63–73. doi: 10.1006/viro.1998.9190. [DOI] [PubMed] [Google Scholar]
- 42.Vojtek I, et al. Lysogenic transfer of group A Streptococcus superantigen gene among streptococci. J Infect Dis. 2008;197:225–234. doi: 10.1086/524687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davies MR, et al. Inter-species genetic movement may blur the epidemiology of streptococcal diseases in endemic regions. Microbes Infect. 2005;7:1128–1138. doi: 10.1016/j.micinf.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 44.Holden MTG, et al. Genomic evidence for the evolution of Streptococcus equi: Host restriction, increased virulence, and genetic exchange with human pathogens. PLoS Pathog. 2009;5:e1000346. doi: 10.1371/journal.ppat.1000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li W, Godzik A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 46.Pride DT, Wassenaar TM, Ghose C, Blaser MJ. Evidence of host-virus co-evolution in tetranucleotide usage patterns of bacteriophages and eukaryotic viruses. BMC Genomics. 2006;7:8. doi: 10.1186/1471-2164-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brussaard CPD. Enumeration of bacteriophages using flow cytometry. Methods Mol Biol. 2009;501:97–111. doi: 10.1007/978-1-60327-164-6_11. [DOI] [PubMed] [Google Scholar]
- 48.Lockhart PB, et al. Bacteremia associated with toothbrushing and dental extraction. Circulation. 2008;117:3118–3125. doi: 10.1161/CIRCULATIONAHA.107.758524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lockhart PB, et al. Poor oral hygiene as a risk factor for infective endocarditis-related bacteremia. J Am Dent Assoc. 2009;140:1238–1244. doi: 10.14219/jada.archive.2009.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kendziorski C, Irizarry RA, Chen K-S, Haag JD, Gould MN. On the utility of pooling biological samples in microarray experiments. Proc Natl Acad Sci USA. 2005;102:4252–4257. doi: 10.1073/pnas.0500607102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pinard R, et al. Assessment of whole genome amplification-induced bias through high-throughput, massively parallel whole genome sequencing. BMC Genomics. 2006;7:216. doi: 10.1186/1471-2164-7-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dean FB, et al. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci USA. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sambrook J. Molecular Cloning: A Laboratory Manual. 3rd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 54.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 55.Nicol JW, Helt GA, Blanchard SG, Jr, Raja A, Loraine AE. The Integrated Genome Browser: Free software for distribution and exploration of genome-scale datasets. Bioinformatics. 2009;25:2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kent WJ. BLAT: The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodriguez-Brito B, Rohwer F, Edwards RA. An application of statistics to comparative metagenomics. BMC Bioinformatics. 2006;7:162. doi: 10.1186/1471-2105-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Quinque D, Kittler R, Kayser M, Stoneking M, Nasidze I. Evaluation of saliva as a source of human DNA for population and association studies. Anal Biochem. 2006;353:272–277. doi: 10.1016/j.ab.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 59.Larkin MA, et al. ClustalW and ClustalX version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 60.Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 61.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.