Abstract
Roseburia inulinivorans is a recently identified motile representative of the Firmicutes that contributes to butyrate formation from a variety of dietary polysaccharide substrates in the human large intestine. Microarray analysis was used here to investigate substrate-driven gene-expression changes in R. inulinivorans A2-194. A cluster of fructo-oligosaccharide/inulin utilization genes induced during growth on inulin included one encoding a β-fructofuranosidase protein that was prominent in the proteome of inulin-grown cells. This cluster also included a 6-phosphofructokinase and an ABC transport system, whereas a distinct inulin-induced 1-phosphofructokinase was linked to a fructose-specific phosphoenolpyruvate-dependent sugar phosphotransferase system (PTS II transport enzyme). Real-time PCR analysis showed that the β-fructofuranosidase and adjacent ABC transport protein showed greatest induction during growth on inulin, whereas the 1-phosphofructokinase enzyme and linked sugar phosphotransferase transport system were most strongly up-regulated during growth on fructose, indicating that these two clusters play distinct roles in the use of inulin. The R. inulinivorans β-fructofuranosidase was overexpressed in Escherichia coli and shown to hydrolyze fructans ranging from inulin down to sucrose, with greatest activity on fructo-oligosaccharides. Genes induced on starch included the major extracellular α-amylase and two distinct α-glucanotransferases together with a gene encoding a flagellin protein. The latter response may be concerned with improving bacterial access to insoluble starch particles.
Keywords: anaerobic gut bacteria, differential gene expression, fructo-oligosaccharides, butyrate, prebiotic
Plant cell wall polysaccharides, storage polysaccharides such as resistant starch and inulin, and many oligosaccharides of plant origin remain undigested in the upper gastrointestinal tract and become important substrates for the growth of colonic bacteria. Prebiotics are defined as dietary substrates that reach the colon where they selectively stimulate the growth of beneficial gut bacteria (1), with the most widely used prebiotics being the fructans inulin and fructo-oligosaccharides (FOS). Inulin is a linear polymer [degree of polymerization (DP) = 3–60] of β-2,1-linked fructose monomers with a terminal glucose residue, whereas FOS have the same backbone but a maximal chain length of 8 monomeric units. The increasing use of prebiotics to enhance gut health is driving research into their mode of action. Many studies have shown that both inulin and FOS selectively stimulate the growth of potentially beneficial gut bacteria such as bifidobacteria and lactobacilli (reviewed in ref. 2). However, only a few studies have considered the potential for stimulation of other bacterial groups that may also be beneficial (3–6).
The bifidogenic dose of inulin has been defined as 5–8 g/d (7), yet the effect of such supplementation on other beneficial groups of gut bacteria and the differences relating to different compositions of inulin have received little attention. For example, the stimulation of the Bifidobacterium genus by inulin and FOS prebiotics does not explain why levels of butyrate are enhanced by inulin and FOS (4). This may be caused by bacterial cross-feeding (5, 8), the direct stimulation of butyrate-producing bacteria by the prebiotics (3, 4) or the butyrogenic effect of reduced pH (9). Rossi et al. (4) established that only 8 of 55 bifidobacterial strains tested were able to grow on inulin in pure culture, although virtually all of the 55 strains used FOS for growth. They concluded that the elevated butyrate production observed in mixed fecal cultures indicated that other bacterial groups were able to degrade inulin and that the bifidobacteria were able to use the mono- and oligosaccharide (FOS) products released by the primary inulin degraders (4). Kleessen et al. (10) used fluorescent in situ hybridization (FISH) to investigate changes in specific bacterial groups in human flora-associated rats fed diets containing various mixtures of short- and long-chain fructans. A mix of oligofructose with long-chain inulin or inulin alone enhanced the numbers of the Clostridium coccoides/Eubacterium rectale group. This bacterial group includes Roseburia inulinivorans and other butyrate-producing bacteria (11) and was unaffected by FOS supplementation. In contrast, numbers of bifidobacteria were only stimulated by FOS (10). In a human study, numbers of the butyrate-producing species Faecalibacterium prausnitzii were significantly increased after inulin–oligofructose consumption (12). The chain length of the fructan molecule seems to be critical in defining the stimulatory effect. Longer chain molecules have a greater and longer-lasting effect on bacterial fermentation in in vitro models (13).
The simple linear structure of FOS and inulin means that only a few enzymes are required for their degradation. The β-fructosidase superfamily includes all enzymes involved in the hydrolysis of nonreducing β-D,fructosidic bonds to release fructose (14). Despite the diversity in nomenclature of this group of enzymes (β-fructosidases, β-fructofuranosidases, sucrose-6-phosphate hydrolases, endo-inulinases, exo-inulinases, invertases, and saccharases), they all contain highly conserved residues and motifs and are members of glycosyl hydrolase (GH) families 32 or 68. This superfamily also includes fungal enzymes (15, 16). Exo-acting β-fructofuranosidases cleave the terminal β,2–1 bonds linking the fructose monomers, but most enzymes also have some invertase activity against the β,2–6 bonds between the terminal glucose–fructose units (17). Thus, activities of specific enzymes against sucrose, FOS, and inulin vary (18). β-fructofuranosidases have been identified in the genome sequences of Bifidobacterium longum (19), Bi. breve (20), Bi. lactis (21, 22), and Lactobacillus plantarum WCFS1 (23). The enzyme in Bi. breve had greatest activity against β,2–1 glucose–fructose links and no activity against β,2–1 fructose–fructose links (20). Few published enzymes have considerable activity against long-chain inulin molecules.
R. inulinivorans, a butyrate producing low G+C Firmicute bacterium (3, 24), is a Clostridium cluster XIVa bacterium that can comprise up to 2% of the gut microbiota (25) and is able to grow on some sugars (24) and the complex substrates inulin and starch in pure culture (3, 26). Inulin also enhanced the survival of R. inulinivorans A2-194 against a background of total fecal bacteria after introduction into a fermentor system designed to simulate the colon (3). In the present study, we identified specific genes involved in inulin and starch utilization by R. inulinivorans A2-194 using microarray analysis and were able to show the induction of certain genes and proteins during growth on different dietary polysaccharide substrates. Expression and purification of a highly regulated β-fructofuranosidase confirmed its role in using both mixed-chain length FOS and long-chain inulin.
Results
Microarray Detection of Genes Differentially Expressed During Growth on Inulin Compared with Starch.
A shotgun genomic microarray of random R. inulinivorans A2-194 DNA fragments (27) was used to assess changes in gene expression during growth on inulin compared with starch. Bacterial mRNA was purified from exponentially growing cells [optical density (OD)650 = 0.4] and hybridized as described previously (27). Spots that were up-regulated on either substrate were ranked based on the log2 normalized data (Materials and Methods). Only eight clones were induced more than 5-fold on inulin compared with starch (and 12 induced >3-fold), whereas 22 clones were up-regulated more than 5-fold on starch compared with inulin. These 30 clones were sequenced, and the gene functions based on the closest database matches are shown in Table 1 with full descriptions in Table S1.
Table 1.
Summary of the ORFs and main contigs identified through microarray analysis
| Strain A2-194 protein | ||
| Contig or clone | ORF | Predicted function |
| Clones induced >5-fold on inulin compared with starch | ||
| Contig I1 β-fructosidase cluster (7,858 nt) | 5 (318) | 6-phosphofructokinase (6-pfk) |
| 6 (505) | β-fructofuranosidase | |
| 7 (556) | ABC sugar-binding protein | |
| 8 (298 | ABC sugar-transport system | |
| 9 (312) | ABC sugar-transport system | |
| 10 (341) | Repressor protein | |
| Clone I2 ferredoxin (428nt) | 1 (143 tr) | Oxidoreductase |
| Contig I3 (2,211nt) | 1 (539) | PTS I |
| Contig I4 (3,077 nt) | 1 (309) | 1-phosphofructokinase (1-pfk) |
| 2 (638) | PTS II | |
| Contig I5 (1,364 nt) | 1 | Transcription regulator |
| Clones induced >5-fold on starch compared with inulin | ||
| Contig S1 (3,075 nt) | 1 (510) | 4-α-glucanotransferase |
| 2 (429) | ABC transport-binding protein | |
| Contig S2 (3,607 nt) | 1 (98) | Regulator |
| 3 (142) | Regulator | |
| 4 (94) | Regulator | |
| 5 (392) | Flagellin (FlaA) | |
| 7 (180tr) | Regulator | |
| Contig S3 | Amylopullulanase (AM055811) | |
| Contig S4 (1,538 nt) | 1 (437tr) | 4-α-glucanotransferase |
| Single clones | ||
| 10.e6 | 1 (108tr) | Membrane proteins |
| 2 (312 tr) | ||
| 6.o9 (4,496 nt) | 1 (354 tr) | Arginosuccinate lyase |
| 2 (736) | Glycogen phosphorylase | |
| 3 (260) | AraC transcriptional regulator | |
| 12.g10 | (371 tr) | α-amylase |
| 7.b10 | (189 tr) | tRNA synthetase |
| 2.j15 | (275tr) | Transposase |
| 8.c12 | (306tr) | Membrane protein |
| 10.f5 | (232 tr) | DNA topoisomerase |
Identification and assembly into functional clusters of those clones induced >5-fold during growth on inulin or starch. The function of each ORF within a contig is based on the closest database match. ORF length in amino acids is indicated in parentheses. Tr, truncated ORF.
An additional microarray analysis was done to compare genes expressed during growth on fructose, short-chain FOS (scFOS; P95), and inulin to distinguish those required for degradation of long- and short-chain fructan substrates. In this case, a three-way comparison of hybridization intensities was done. In general, the strongest induction was observed during growth on inulin compared with either of the other two substrates, with the expression of only three clones changed >5-fold on scFOS compared with fructose (Fig. S1). Several clones matched those identified in the starch:inulin comparison, and any additional clones that were up- or down-regulated more than 7-fold on at least two substrates were sequenced; these data are also included in Table S1.
Genes Induced During Growth on Starch Relative to Inulin.
Three overlapping clones with identity to α-amylases were induced 3- to 5-fold during growth on starch. These represent the major extracellular multidomain α-amylase/amylopullulanase described previously (accession number AM055811) (26) that is postulated to be anchored to the cell wall. An ABC transport protein located adjacent to this gene presumably functions in transporting the small monomeric/oligomeric breakdown products into the cell. Another starch-induced clone contained the catalytic domain of a different α-amylase, with only 22% amino acid sequence identity to the first clone.
Six of the clones induced on starch overlapped to form a 3-kb sequence (Table 1, contig S1) encoding a 4-α-glucanotransferase and an extracellular solute binding protein component of an ABC transport system with a signal peptide. One function of 4-α-glucanotransferases (EC 2.4.1.25) is to transfer single or multiple glucose residues from long chains of glucose molecules to acceptor sugars such as maltose (28). A second 4-α-glucanotransferase was also detected (Table 1, contig S4) that showed ∼4-fold induction on starch compared with inulin and was also strongly induced on inulin compared with FOS (94- or 62-fold) and fructose (16- and 20-fold). This suggests that this enzyme might have broad substrate specificity. These two GH family 77 4-α-glucanotransferase enzymes (encoded in S1 and S4) had only 44% amino acid identity. Both had strongly hydrophobic sequences at the C termini, with that on S1 also having a terminal basic region, but neither possessed an obvious N-terminal signal peptide.
Interestingly, the remaining group of four overlapping clones significantly up-regulated (8- to 9-fold) on starch compared with inulin (Table 1, contig S2) encoded the flagellin protein FlaA and several ORFs with identity (>40%) to transcriptional regulators. These clones were not induced during growth on any of the fructan substrates.
Genes Induced During Growth on Inulin or FOS.
Four of six clones most strongly induced on inulin overlapped, and the sequence of this contig was extended to 7.8 kb by genome walking to reveal a β-fructofuranosidase, a 6-phosphofructokinase, an ABC transport system, and a LacI family regulatory gene belonging to the type 1 periplasmic binding-protein superfamily. These genes together form a fructan utilization cluster, illustrated diagrammatically in Fig. 1.
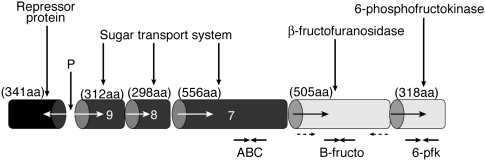
Fig. 1.
Diagram showing the organization of the ORFs in the fructan utilization cluster in R. inulinivorans A2-194. The number of amino acids (aa) encoded by each complete ORF is given. The promoter region in the 257-nt gap between the genes encoding the repressor and the ABC transport proteins is shown (P). The solid arrows indicate the positions of the primer pairs used for the RT-Q-PCR. The hashed arrows indicate the positions of the primers used to clone the active β-fructofuranosidase gene (∼1,500 nt) into the pET vector.
The regulatory protein is separated from the divergently transcribed structural genes by a noncoding gap of 257 nucleotides, within which are two classic promoter sequences, one on each strand, and an operator region capable of forming a strong secondary structure (ΔG = −28.2). The β-fructofuranosidase enzyme and the three components of the ABC transport system share 48–71% identity (Table S1) with similar proteins encoded in an identical cluster structure by C. beijerinckii NCIMB 8052 (genome sequence accession number CP000721). These β-fructofuranosidases contain the three crucial active-site residues, D/D/E(C) within the conserved domains D1, N3, and N5 (16). No characteristic signal peptide (SignalP predict program) or membrane-anchoring domain could be identified on the R. inulinivorans enzyme.
The secondary structure of each protein component of the R. inulinivorans A2-194 ABC transport system is consistent with their proposed functions. The putative extracellular solute binding protein (ORF 7) is predicted to extrude into the extracellular matrix where it can selectively bind the specific substrate, in this case inulin or FOS, and deliver it to the gated translocation pathway formed by the transmembrane components encoded by ORFs 8 and 9. These ORFs showed the classic hydrophobicity plots of membrane-bound proteins. In addition, ORF8 contains a motif that specifically acts as the interface between the transmembrane gates and the ATPase, which provides the energy for the transport system.
Two different phosphoenolpyruvate-dependent sugar phosphotransferase system (PTS) transport enzymes were also induced on inulin (Table 1). One is a PTS enzyme I, with 64% identity to the homolog from Bi. longum (contig I3). The other is a PTS enzyme IIbc transport protein encoded by a gene adjacent to a 1-phosphofructokinase sequence (contig I4). The two phosphofructokinase (pfk) enzymes detected in the R. inulinivorans A2-194 genome are unrelated with different catalytic specificities (6-pfk and 1-pfk; 19% amino acid identity).
In the three-way comparison (growth on inulin, FOS, or fructose), PTS transporters were strongly induced (>40-fold) on fructose compared with scFOS (Fig. S1). The β-fructofuranosidase was up-regulated 71-fold during growth on inulin and 7-fold on FOS compared with fructose. It was also up-regulated 10-fold on inulin compared with scFOS, suggesting that it is an essential enzyme required to degrade the long fructose chains in inulin (Table S1).
Two overlapping clones induced on inulin compared with starch encoded a partial protein with considerable identity (76%) to iron-sulfur oxidoreductases of the aldo/keto reductase family from Clostridia spp. (Table 1, contig I2). Bacterial iron-sulfur ferredoxins perform essential roles in anaerobic electron transport chains, mediating the transfer of electrons between molecules in metabolic reactions. The same gene was also identified in the three-way comparison, induced on inulin compared with both scFOS (6-fold) and fructose (10-fold) (Fig. S1).
Comparison of Gene Induction on Different Substrates Using Reverse Transcription–Quantitative PCR.
The expression of genes shown by microarray analysis to be up-regulated during growth on inulin, FOS, or starch was confirmed using quantitative PCR (Q-PCR) analysis. Specific PCR primer pairs were designed targeting genes encoding the β-fructofuranosidase, the solute binding protein, and the 6-pfk in the fructan cluster (contig I1) (Fig. 1), the second phosphofructokinase enzyme (1-pfk) and PTSII in contig I4, and the PTSI enzyme on contig I3. These primers were used in reverse transcription–Q-PCR (RT-Q-PCR) to amplify RNA purified from exponentially growing R. inulinivorans cells cultured on inulin, sucrose, scFOS, fructose, and glucose. The results illustrated the differential induction of the genes relative to the glucose control (Fig. 2).
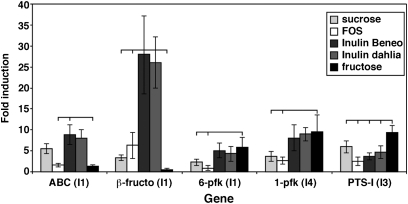
Fig. 2.
Results of RT-Q-PCR amplification of selected genes encoded on three inulin contigs (Fig. 1), which were detected as up-regulated on the inulin:starch microarray. Primer pairs were used to amplify mRNA extracted from cells during exponential growth (OD650 = 0.4) on glucose, fructose, sucrose, scFOS, and two types of long-chain inulin—Dahlia inulin (Sigma) and HP Inulin-Beneo (Orafti). The fold induction of the data converted from logarithmic to linear scale (x = 2−Ct) for each gene was calculated relative to the glucose standard. Results are the means of triplicate results obtained from two biological replicates. Significant differences between gene expression on different substrates (P < 0.05) are indicated by horizontal lines.
The largest changes in expression were for the β-fructofuranosidase (Fig. 2). Expression of the gene encoding this enzyme was elevated 25-fold on two long-chain inulin substrates compared with 6-fold on scFOS and only 3-fold on sucrose. Expression on fructose was virtually the same as on glucose. The 6-pfk located on the fructan cluster was not induced as strongly as the 1-pfk enzyme, which was significantly induced on fructose compared with FOS (Fig. 2). The gene encoding the fructofuranosidase-linked ABC transport protein was induced strongly during growth on longer-chain inulin substrates, whereas the PTSI had maximum induction on fructose, reflecting the different transport capabilities of these two systems (29).
Specific up-regulation of the flagellin gene was also confirmed by RT-Q-PCR, illustrating that the gene was induced 24-fold on starch compared with inulin, in a direct comparison. In contrast, expression of this gene on maltose and inulin was virtually the same as the glucose control (2- and 3.8-fold up-regulation, respectively) (Fig. S2).
Activity of R. inulinivorans β-Fructofuranosidase Expressed in Escherichia coli.
The β-fructofuranosidase coding region (Fig. 1) was cloned into the pET-30 expression vector incorporating a 6HIS-tag at both the N and C termini. The activity of the purified protein was confirmed in a colorimetric microtitre plate assay with several fructose-based substrates of varying chain length. After the optimal assay conditions were established (Materials and Methods), the specific activity of the enzyme was tested against a range of substrates, including the short-chain oligofructose substrates kestose (GF2), nystose (GF3), and fructofuranosylnystose (GF4), the fructo-oligosaccharides P95 scFOS and Synergy1, and inulin. The end products of the reaction were visualized by TLC after 2 and 24 h, and a range of degradation products was visible for all substrates (Fig. S3).
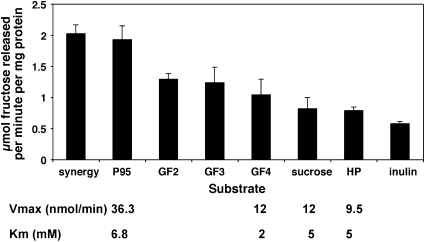
Glucose and fructose monomers were not distinguishable by TLC, but the release of fructose was calculated using an enzymatic assay. Similar amounts of fructose were released from each of the defined fructo-oligosaccharides (GF2, GF3, and GF4) (Fig. 3), with less from both sucrose and the complex long-chain inulin substrates. The enzyme was most active (in terms of release of fructose) against intermediate chain-length FOS substrates such as P95 and Synergy1. The Vmax values (nmol/min) of the purified recombinant enzyme were calculated for four substrates and were greatest for P95, whereas the Km values ranged from 2mM for GF4 to 6.8 mM for P95. These results indicate that this enzyme can release fructose residues from the nonreducing ends of fructan chains of variable length.
Fig. 3.
Activity of the cloned enzyme with specific substrates (at 10 mM) after 2-h incubation. The stated amount of fructose (μmol) released per milligram of protein per minute is the average of four independent experimental values. The specific enzyme activities (Vmax and Km), calculated from four independent experiments, are shown for four substrates.
Proteomic Analysis of R. inulinivorans A2-194.
To compare the gene-expression profiles with protein expression under the same growth conditions, the R. inulinivorans proteome was resolved using 2D gel electrophoresis. Pair-wise comparisons of the proteomes of bacteria grown on either inulin or starch revealed few differences in protein-expression patterns, with about 86% of spots conserved and the densities of only about 14% (80 of 560 spots identified) differing by a factor greater than 2. More proteins were overexpressed during growth on starch, with the densities of 10 spots increased at least 5-fold (one 12-fold), whereas of four spots elevated 5-fold on inulin, two were >10-fold (Fig. 4).
Fig. 4.
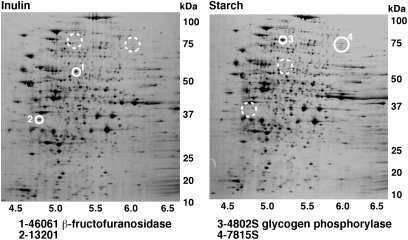
Representative 2D PAGE gels of bacterial proteomes of R. inulinivorans A2-194 cells grown on either inulin or starch. The protein spot excised and found to be the β-fructofuranosidase is marked 1 on the inulin gel, and the complementary position of this missing spot is indicated on the starch gel. The positions of the other three spots that were excised for sequencing are numbered (2–4), and the corresponding “empty” regions are indicated. Molecular weight size markers are given to the right of each gel (kDa), and the pH gradient is shown along the bottom.
Four of the most distinct differentially expressed protein spots were excised from the gels and peptide sequences derived from liquid chromatography-tandem mass spectrometry (LC-MS/MS) fragmentation spectra compared with the sequence data obtained after microarray analysis. Two of four spots corresponded to proteins encoded by clones identified on the microarray (Table S2). The protein most strongly expressed during growth on inulin and absent on starch corresponded to the β-fructofuranosidase. One of the proteins up-regulated on starch corresponded to the glycogen phosphorylase (Table 1, clone 6.o9).
Potential for Bacterial Cross-Feeding in Coculture.
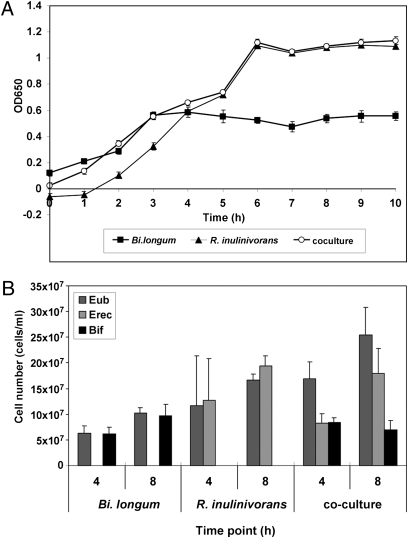
Bi. longum 20219 cannot use long-chain inulin for growth, although it can grow on FOS (Fig. S4). Because R. inulinivorans A2-194 possesses an enzyme capable of degrading long-chain inulin, these two bacteria were incubated together on either long-chain inulin or the mixed-chain length Synergy1 (consisting of inulin and FOS in a 1:1 ratio) to ascertain whether growth of Bi. longum improved in the presence of R. inulinivorans A2-194. Aliquots were removed from the culture tubes at selected time points, and numbers of each bacterial species were enumerated using FISH (9). After 10 h growth on inulin, ∼99% of the bacterial cells present corresponded to R. inulinivorans, with low numbers of Bi. longum cells detected (Fig. S4). During growth on Synergy1, both bacteria competed equally for the substrate in the first 4 h, but after 8 h, there were two times as many R. inulinivorans cells (Fig. 5). This was consistent with the growth-curve data where Bi. longum seemed to stop growing after 3–4 h, presumably when all of the short-chain FOS was depleted, but R. inulinivorans continued to grow for up to 8 h on the long-chain inulin component of the substrate (Fig. 5A). Thus, R. inulinivorans A2-194 was better adapted than Bi. longum 20219 to use inulin for growth under these conditions.
Fig. 5.
Data from the coculture experiment after growth of R. inulinivorans A2-194 and Bi. longum 20219 on Synergy1. (A) Growth curve showing the increasing OD650 at hourly intervals of Bi. longum (■), R. inulinivorans (▲), and coculture (○). (B) Number of bacterial cells at 4-h and 8-h time points enumerated by FISH using the eubacterial probe (Eub338) and probes to specifically detect R. inulinivorans (Erec482) and Bi. longum (Bif164).
R. inulinivorans A2-194 cultures grown overnight on long-chain inulin were fractionated to investigate the distribution of fructanosidase activity on a range of fructan substrates. No activity was observed for the concentrated supernatant fractions on inulin, Synergy1, P95, or sucrose, whereas both the cell pellet and sonicated cells were active on all substrates (Fig. S5).
Discussion
Investigation of substrate-driven changes in gene expression in R. inulinivorans A2-194 has provided insights into the mechanisms that allow this bacterium to use polysaccharide substrates. The identification of the GH family 32 β-fructofuranosidase as the most strongly induced gene during growth on long-chain inulin indicated that it has an important role in the use of fructan substrates. Gene expression was greater on inulin than FOS substrates, and the enzyme was also detected as a prominent up-regulated protein in the bacterial proteome from cells grown on inulin. Analysis of the purified enzyme after expression in E. coli shows that it is capable of degrading fructan molecules of variable chain length. The absence of a signal peptide suggests that this β-fructofuranosidase is probably intracellular, acting on fructans that enter the cell through the inulin-induced ABC transport system that is encoded by the same gene cluster. C. beijerinckii NCIMB 8052 encodes a distinct sucrose hydrolase on a separate cluster linked to a fructokinase and a PTSII transporter (30). This C. beijerinckii sucrose hydrolase has greatest activity against sucrose (30) and is not closely related to the β-fructofuranosidase identified in R. inulinivorans A2-194.
Growth of R. inulinivorans A2-194 on inulin and scFOS was accompanied by increased expression of a second gene cluster encoding a 1-phosphofructokinase and a fructose-specific PTSII transport system. The Bacillus subtilis genome encodes 15 PTS proteins with different sugar specificities (31), one of which is also adjacent to a 1-pfk enzyme. PTS transport systems are linked with uptake of monomeric substrates, whereas ABC transport systems are required for the transport of larger substrates, as shown for L. acidophilus (29). Sucrose-use clusters, including PTSII transporter systems that are up-regulated during growth on sucrose or FOS, have been identified in C. beijerinckii (30), C. acetobutylicum (32), and L. plantarum (23).
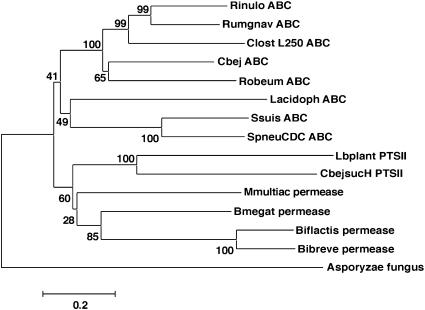
In the present study, expression of the ABC transport system in R. inulinivorans was most strongly up-regulated in response to inulin, whereas the PTS system was most strongly induced by the monosaccharide fructose. Interestingly, phylogenetic analysis of selected GH family 32 β-fructofuranosidase enzymes from a wide range of bacteria indicated that those that are adjacent to ABC transport systems cluster together, independent of the phylogenetic relationships of the bacteria carrying them, whereas those associated with sugar permeases or PTS transport enzymes form separate branches on the phylogenetic tree (Fig. 6). It seems likely that other β-fructofuranosidases whose preference is for longer-chain substrates, like that from R. inulinivorans, will be encoded by genes adjacent to ABC transport systems, whereas enzymes specific for shorter substrates such as sucrose may be encoded alongside PTS transporters. Thus the type of transport molecule adjacent to β-fructofuranosidases is indicative of the enzyme function.
Fig. 6.
Phylogenetic tree showing the relationship between selected β-fructofuranosidase sequences in the database and indicating their linkage to ABC transport systems (ABC), permeases (permease), and PTS transport systems (PTS II). Bootstrapping values are given, and the scale bar indicates amino acid substitutions per position. The sequences extracted from the database have the following accession numbers: Rinulo ABC, R. inulinivorans (this study; GU591787); Rumgnav ABC (Ruminicoccus gnavus EDN78469); ClostL250 ABC (Clostridium sp. EDO57496); Cbej ABC (C. beijerinckii ABR35991); Robeum ABC (R. obeum EDM87838); Lacidoph ABC (L. acidophilus AY172019); Ssuis ABC (Streptococcus suis EAP40155); SpneuCDC ABC (S. pneumoniae EDT91128); Lbplant PTSII (L. plantarum Q88ZV8); CbejsucH PTSII (C. beijerinckii AF059741); Mmultiac permease (Mitsuokella multiacidus EEC92082); Bmegat permease (Bacillus megaterium AAM19071); Biflactis permease (Bi. lactis AJ437479); Bibreve permease (Bi. breve AY549965); Asporyzae fungus (Aspergillus oryzae A8D1N0).
Although we cannot exclude the possibility that R. inulinivorans encodes an additional extracellular enzyme to specifically degrade long inulin molecules, such an enzyme was not detected during our analysis of inulin-inducible genes. The only protein with identity to β-fructosidases within the newly available genome sequence of R. inulinivorans (accession number ACFY01000156) was the β-fructofuranosidase identified here. Interestingly, a strain of Bi. longum that uses fructose and scFOS was unable to grow in coculture with R. inulinivorans A2-194, indicating that there was no extracellular accumulation of monomeric units during degradation of inulin by R. inulinivorans. This is in contrast to coculture studies with B. thetaiotaomicron and various Bifidobacterium strains (33). In these cases, B. thetaiotaomicron does release short-chain oligofructose molecules into the medium following extracellular inulin degradation, which different bifidobacterial species have varying abilities to use (33). Analysis of the activity of fractionated R. inulinivorans cells to degrade fructans did not indicate significant extracellular activity, although the cell pellets and sonicated material were active. These findings strongly suggest that, within R. inulinivorans A2-194, the β-fructofuranosidase enzyme identified here is responsible for inulin and FOS degradation. Because this enzyme seems to be intracellular, the ABC transport system may be able to internalize longer-chain inulin substrates to facilitate bacterial growth.
Advances in molecular-profiling methods have enabled the composition of the gut microbiota to be reassessed, and it is now clear that bifidobacteria comprise <5% of the total, with other groups of obligate anaerobes being equally or more abundant (25). The two major groups of butyrate-producing bacteria are more abundant in healthy adults, with the Roseburia and E. rectale group estimated at 7–10% of the total population (34) and F. prausnitzii forming up to 15% of the total population (35). The results presented here indicate that, within the Roseburia genus, R. inulinivorans is able to use scFOS and inulin directly for growth.
Genome-sequence information is becoming available for an increasing number of commensal gut bacteria, and the most recent database searches with the R. inulinivorans β-fructofuranosidase sequence have identified hypothetical proteins in many other commensal gut anaerobes that have significant amino acid sequence homology to the R. inulinivorans β-fructofuranosidase (Table S3). Further work investigating the growth of these bacteria on specific FOS and inulin substrates will be required to ascertain the actual function of these enzymes in vivo. One problem that has been encountered in determining the FOS/inulin specificities of specific bacterial groups and enzymes in vitro is that it is difficult to obtain pure long-chain substrate preparations that do not include a small percentage of low molecular-weight FOS molecules that the bacteria can use for growth, perhaps preferentially.
Inulin and scFOS are widely used as prebiotics to promote the growth of beneficial gut bacteria. In the past, evidence to support this has focused on the increase in numbers of bifidobacterial species. The resident population of bifidobacteria varies markedly between individuals, both in terms of abundance and the species present (36, 37), and it is now clear that few bifidobacterial species can grow directly on long-chain inulin, although many can use scFOS (4). The results presented here clearly show that other specific members of the gut microbiota are capable of using prebiotics as growth substrates. Some of these, including R. inulinivorans itself, produce butyrate as fermentation products, indicating that stimulation of this group of bacteria could, at least in part, explain increases in fecal butyrate concentration after prebiotic consumption (4, 38). Although the bifidogenic dose of inulin has been determined to be 5–8 g/d (7), a lower concentration equivalent to 1.5 g/d was sufficient to stimulate numbers of R. inulinivorans in a model system (3). This illustrates the ability of this bacterial species to compete for inulin against a background of total gut microbiota. Numbers of the butyrate-producer F. prausnitzii have also been shown to be significantly increased during supplementation with an inulin/oligofructose mix (12). Very few studies to date have monitored the response of all of these inulin-using groups of bacteria in vivo, and further work will, therefore, be needed to define the competitive interactions between them.
Two genes encoding amylase activity were among those induced on starch. One of these was the major cell surface-anchored α-amylase/neopullulanase identified previously (26). The second amylase catalytic domain identified had only 22% identity to the characterized neopullulanase. A separate starch-induced gene cluster encoded an α-glucanotransferase and the substrate binding protein of an ABC transport system, suggesting that there may be parallel systems for the processing of malto- and fructo-oligosaccharides in R. inulinivorans.
The observation that flagellin synthesis was up-regulated during growth on starch, but not on inulin, is intriguing. One possible explanation is that most of the starch surviving into the large intestine is likely to exist in insoluble food particles. Chemotaxis and motility may, therefore, be particularly important in colonizing this substrate but may not be required to access the more soluble inulin molecules. The considerable energy input required for flagellin synthesis indicates that it must be of considerable benefit to the bacterial cell to produce them when starch is available.
The human gut represents a challenging environment for bacterial survival, in part because of the sudden influxes of dietary substrates followed by periods of relative starvation. In addition, the composition of the available food varies considerably, both daily and weekly. To cope with these factors, commensal gut bacteria have evolved with the ability to degrade many different substrates, but conserve energy by tightly regulating the expression of genes required for specific metabolic pathways. In conclusion, this analysis of the global transcriptional response to growth on alternative dietary sources has provided valuable insights into substrate use in an important group of human colonic anaerobes. In particular, it indicates an important role for inducible fructofuranosidases linked to ABC transport systems in the processing of long-chain fructan molecules. More unexpectedly, the up-regulation of flagellin genes in response to starch implies a motility response to access certain growth substrates. Together with previous data describing the ability of R. inulinivorans to use fucose as a growth substrate (27), these results indicate that this colonic bacterium has considerable ability to respond to the availability of alternative dietary substrates, an attribute that undoubtedly enhances its survival in the highly competitive microbial ecosystem of the human large intestine.
Materials and Methods
Additional details on various methods, including gene-expression analyses, proteomic studies, and TLC and FISH methodologies, are provided in SI Materials and Methods.
Bacteria and Growth Conditions.
Routine anaerobic culturing of R. inulinivorans A2-194 was in M2GSC medium (39). Growth on single carbon sources used basal YCFA medium (YCFA - yeast extract-casein hydrolysate-fatty acids) (3) supplemented with 0.5% wt/vol of the specific substrate (glucose, FOS, Dahlia inulin, fructose or amylopectin starch from Sigma). FOS P95 (DP 2–8), Synergy1 (DP 2–60), and Beneo HP (chicory inulin DP > 5) were provided by Beneo-Orafti. Defined fructo-oligosaccharides (GFn) were obtained from Wako Chemicals (GmbH). Bacterial growth was determined spectrophotometrically by following changes in OD at 650 nm (OD650). Bi. longum was cultured in the same way on M2GSC or supplemented YCFA medium. In the coculture experiment, equal cell numbers of overnight cultures in M2GSC were subinoculated into basal YCFA containing 0.5% inulin or Synergy1 and growth monitored (OD650) for up to 10 h, withdrawing samples for analysis by FISH at selected time points (SI Materials and Methods).
RNA Purification and Microarray Hybridization.
Microarray slides on which the total genome of R. inulinivorans was arrayed were prepared in house, and all RNA purifications, hybridizations, and analyses were carried out as described previously (27). Briefly, RNA was purified from midexponential phase (OD650 = 0.4) cultures on basal YCFA supplemented with a single substrate using the RNeasy RNA purification kit (Qiagen), and the mRNA component was enriched using the MICROBExpress system (Ambion). The purified RNA (1 μg) was labeled by reverse transcription (Amersham) using random nonamer extension incorporating either dCTP-Cy3 or dCTP-Cy5 dyes. To ensure reproducibility and obtain statistically significant results, the dye labeling was swapped for a second hybridization. RNA purified from a separate biological replicate was labeled and hybridized two times in the same way. The fluorescence of each spot was measured in two channels using a GeneTAC LS IV (Genomic Solutions) with GTLS software. The microarray data were log2 transformed and normalized by Loess normalization to remove intensity dependent dye effects. P values were calculated by applying a one-sample t test to the log ratios from the replicate experiments. A threshold of at least 5-fold induction was used to select genes, because the P values were based on only two biological replicates. The whole experiment conformed to the MIAME criteria for the documentation of microarray experiments. Data have been deposited in the GEO database with accession number GSE22245. The DNA sequences reported in this paper have been deposited in the GenBank database with accession numbers GU591772–GU591787.
RT-Q-PCR.
Primers were designed and checked using Netprimer to specifically amplify fragments of 80–200 bp from selected clones. The optimal annealing temperature for each primer pair was determined in a standard PCR amplification, using the appropriate clone DNA as a template. The same RNA purification (10 ng) used to hybridize to the array slides was reverse-transcribed using the iScript cDNA synthesis kit (Bio-Rad). Aliquots of the resulting cDNA (2 μL) were used in triplicate in real-time quantitative PCR amplifications, as detailed in SI Materials and Methods. Gene expression on the different substrates was calculated relative to a glucose control by comparing the fluorescence threshold cycles (40). The log-transformed data were analyzed statistically to determine significant differences in gene induction.
β-Fructosidase Assays.
Primers were designed to amplify the full-length β-fructofuranosidase gene, incorporating histidine tags at the N and C termini. The PCR amplicon was cloned into the pET-30Ek/LIC expression vector (Novagen) following the manufacturer's guidelines. The cloned enzyme was overexpressed and purified using Sigma NiCam Hc Resin N3158. Initial activity tests on microtitre plates showed that the enzyme was active against inulin and Beneo HP as well as sucrose. Optimal activity of the enzyme occurred at pH 6 and 37 °C, and subsequent assays were done using these parameters. The relative activity of the cloned enzyme against various substrates was established using the fructose assay kit (Sigma) following the manufacturer's instructions. This assay determines the amount of free fructose released into solution after incubation by specifically monitoring the conversion of any free fructose to 6-phosphogluconate over 15 min in a reaction generating NADH. Fructose residues preexisting in the reaction mixture are subtracted with the background so that only enzymatically released fructose is measured.
Analysis of the Bacterial Proteome.
This method is fully described in SI Materials and Methods. Briefly, bacterial cell pellets harvested from exponentially growing cells (OD650 = 0.4) were washed in buffer before sonicating to disrupt the bacterial cells. The protein concentration in the sonicated extracts was measured (41), and 150 μg of total protein was analyzed. The proteins were separated in two dimensions, first using isoelectric focusing followed by polyacrylamide gel electrophoresis. After staining with Coomassie blue (0.1%) (42), gels were scanned, and the digitized images were analyzed using PDQuest version 7.0.1 software, incorporating total spot normalization (43). In total, five biological replicates were run in triplicate for each growth condition, and the resolved proteomes were compared. The four protein spots displaying the greatest increases in their apparent intracellular abundance in the proteomes of R. inulinivorans A2-194 cells grown in the presence of either inulin or starch were chosen for subsequent LC-MS/MS analysis to generate peptide fingerprints.
Supplementary Material
Acknowledgments
We thank Martin Reid and Gary Duncan for excellent technical assistance and Grietje Holtrop (Biomathematics and Statistics Scotland, Rowett Institute) for statistical analysis of Q-PCR data. The Rowett Institute of Nutrition and Health, University of Aberdeen, and Biomathematics and Statistics Scotland receive support from the Scottish Government Rural and Environment Research and Analysis Directorate (SG-RERAD).
Footnotes
The authors declare no conflict of interest.
Data deposition: The DNA sequences reported in this paper have been deposited in the GenBank database (accession nos. GU591772M–GU591787). The microarray data reported in this paper have been deposited in the NCBI GEO repository (accession no. GSE22245).
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Microbes and Health” held November 2–3, 2009, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS Web site at http://www.nasonline.org/SACKLER_Microbes_and_Health.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000091107/-/DCSupplemental.
References
- 1.Gibson GR, Probert HM, Loo JV, Rastall RA, Roberfroid MB. Dietary modulation of the human colonic microbiota: Updating the concept of prebiotics. Nutr Res Rev. 2004;17:259–275. doi: 10.1079/NRR200479. [DOI] [PubMed] [Google Scholar]
- 2.Van Loo JA. The specificity of the interaction with intestinal bacterial fermentation by prebiotics determines their physiological efficacy. Nutr Res Rev. 2004;17:89–98. doi: 10.1079/NRR200377. [DOI] [PubMed] [Google Scholar]
- 3.Duncan SH, et al. Effects of alternative dietary substrates on competition between human colonic bacteria in an anaerobic fermentor system. Appl Environ Microbiol. 2003;69:1136–1142. doi: 10.1128/AEM.69.2.1136-1142.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rossi M, et al. Fermentation of fructooligosaccharides and inulin by bifidobacteria: A comparative study of pure and fecal cultures. Appl Environ Microbiol. 2005;71:6150–6158. doi: 10.1128/AEM.71.10.6150-6158.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Falony G, Vlachou A, Verbrugghe K, De Vuyst L. Cross-feeding between Bifidobacterium longum BB536 and acetate-converting, butyrate-producing colon bacteria during growth on oligofructose. Appl Environ Microbiol. 2006;72:7835–7841. doi: 10.1128/AEM.01296-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flint HJ, Duncan SH, Scott KP, Louis P. Interactions and competition within the microbial community of the human colon: Links between diet and health. Environ Microbiol. 2007;9:1101–1111. doi: 10.1111/j.1462-2920.2007.01281.x. [DOI] [PubMed] [Google Scholar]
- 7.Kolida S, Meyer D, Gibson GR. A double-blind placebo-controlled study to establish the bifidogenic dose of inulin in healthy humans. Eur J Clin Nutr. 2007;61:1189–1195. doi: 10.1038/sj.ejcn.1602636. [DOI] [PubMed] [Google Scholar]
- 8.Belenguer A, et al. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol. 2006;72:3593–3599. doi: 10.1128/AEM.72.5.3593-3599.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker AW, Duncan SH, McWilliam Leitch EC, Child MW, Flint HJ. pH and peptide supply can radically alter bacterial populations and short-chain fatty acid ratios within microbial communities from the human colon. Appl Environ Microbiol. 2005;71:3692–3700. doi: 10.1128/AEM.71.7.3692-3700.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleessen B, Hartmann L, Blaut M. Oligofructose and long-chain inulin: Influence on the gut microbial ecology of rats associated with a human faecal flora. Br J Nutr. 2001;86:291–300. doi: 10.1079/bjn2001403. [DOI] [PubMed] [Google Scholar]
- 11.Barcenilla A, et al. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl Environ Microbiol. 2000;66:1654–1661. doi: 10.1128/aem.66.4.1654-1661.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramirez-Farias C, et al. Effect of inulin on the human gut microbiota: Stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br J Nutr. 2009;101:541–550. doi: 10.1017/S0007114508019880. [DOI] [PubMed] [Google Scholar]
- 13.van de Wiele T, Boon N, Possemiers S, Jacobs H, Verstraete W. Inulin-type fructans of longer degree of polymerization exert more pronounced in vitro prebiotic effects. J Appl Microbiol. 2007;102:452–460. doi: 10.1111/j.1365-2672.2006.03084.x. [DOI] [PubMed] [Google Scholar]
- 14.Barrangou R, Altermann E, Hutkins R, Cano R, Klaenhammer TR. Functional and comparative genomic analyses of an operon involved in fructooligosaccharide utilization by Lactobacillus acidophilus. Proc Natl Acad Sci USA. 2003;100:8957–8962. doi: 10.1073/pnas.1332765100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan XL, et al. Database mining and transcriptional analysis of genes encoding inulin-modifying enzymes of Aspergillus niger. Microbiology. 2006;152:3061–3073. doi: 10.1099/mic.0.29051-0. [DOI] [PubMed] [Google Scholar]
- 16.Naumoff DG. β-fructosidase superfamily: Homology with some α-L-arabinases and β-D-xylosidases. Proteins. 2001;42:66–76. doi: 10.1002/1097-0134(20010101)42:1<66::aid-prot70>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 17.Ricca E, Calabrò V, Curcio S, Iorio G. The state of the art in the production of fructose from inulin enzymatic hydrolysis. Crit Rev Biotechnol. 2007;27:129–145. doi: 10.1080/07388550701503477. [DOI] [PubMed] [Google Scholar]
- 18.Warchol M, Perrin S, Grill JP, Schneider F. Characterization of a purified β-fructofuranosidase from Bifidobacterium infantis ATCC 15697. Lett Appl Microbiol. 2002;35:462–467. doi: 10.1046/j.1472-765x.2002.01224.x. [DOI] [PubMed] [Google Scholar]
- 19.Schell MA, et al. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc Natl Acad Sci USA. 2002;99:14422–14427. doi: 10.1073/pnas.212527599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryan SM, Fitzgerald GF, van Sinderen D. Transcriptional regulation and characterization of a novel β-fructofuranosidase-encoding gene from Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2005;71:3475–3482. doi: 10.1128/AEM.71.7.3475-3482.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janer C, et al. Hydrolysis of oligofructoses by the recombinant β-fructofuranosidase from Bifidobacterium lactis. Syst Appl Microbiol. 2004;27:279–285. doi: 10.1078/0723-2020-00274. [DOI] [PubMed] [Google Scholar]
- 22.Ehrmann MA, Korakli M, Vogel RF. Identification of the gene for β-fructofuranosidase of Bifidobacterium lactis DSM10140(T) and characterization of the enzyme expressed in Escherichia coli. Curr Microbiol. 2003;46:391–397. doi: 10.1007/s00284-002-3908-1. [DOI] [PubMed] [Google Scholar]
- 23.Saulnier DMA, Molenaar D, de Vos WM, Gibson GR, Kolida S. Identification of prebiotic fructooligosaccharide metabolism in Lactobacillus plantarum WCFS1 through microarrays. Appl Environ Microbiol. 2007;73:1753–1765. doi: 10.1128/AEM.01151-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duncan SH, et al. Proposal of Roseburia faecis sp. nov., Roseburia hominis sp. nov. and Roseburia inulinivorans sp. nov., based on isolates from human faeces. Int J Syst Evol Microbiol. 2006;56:2437–2441. doi: 10.1099/ijs.0.64098-0. [DOI] [PubMed] [Google Scholar]
- 25.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramsay AG, Scott KP, Martin JC, Rincon MT, Flint HJ. Cell-associated α-amylases of butyrate-producing Firmicute bacteria from the human colon. Microbiology. 2006;152:3281–3290. doi: 10.1099/mic.0.29233-0. [DOI] [PubMed] [Google Scholar]
- 27.Scott KP, Martin JC, Campbell G, Mayer CD, Flint HJ. Whole-genome transcription profiling reveals genes up-regulated by growth on fucose in the human gut bacterium “Roseburia inulinivorans.”. J Bacteriol. 2006;188:4340–4349. doi: 10.1128/JB.00137-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goda SK, Eissa O, Akhtar M, Minton NP. Molecular analysis of a Clostridium butyricum NCIMB 7423 gene encoding 4-α-glucanotransferase and characterization of the recombinant enzyme produced in Escherichia coli. Microbiology. 1997;143:3287–3294. doi: 10.1099/00221287-143-10-3287. [DOI] [PubMed] [Google Scholar]
- 29.Barrangou R, et al. Global analysis of carbohydrate utilization by Lactobacillus acidophilus using cDNA microarrays. Proc Natl Acad Sci USA. 2006;103:3816–3821. doi: 10.1073/pnas.0511287103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reid SJ, Rafudeen MS, Leat NG. The genes controlling sucrose utilization in Clostridium beijerinckii NCIMB 8052 constitute an operon. Microbiology. 1999;145:1461–1472. doi: 10.1099/13500872-145-6-1461. [DOI] [PubMed] [Google Scholar]
- 31.Reizer J, et al. Novel phosphotransferase system genes revealed by genome analysis—the complete complement of PTS proteins encoded within the genome of Bacillus subtilis. Microbiology. 1999;145:3419–3429. doi: 10.1099/00221287-145-12-3419. [DOI] [PubMed] [Google Scholar]
- 32.Tangney M, Mitchell WJ. Analysis of a catabolic operon for sucrose transport and metabolism in Clostridium acetobutylicum ATCC 824. J Mol Microbiol Biotechnol. 2000;2:71–80. [PubMed] [Google Scholar]
- 33.Falony G, Calmeyn T, Leroy F, De Vuyst L. Coculture fermentations of Bifidobacterium species and Bacteroides thetaiotaomicron reveal a mechanistic insight into the prebiotic effect of inulin-type fructans. Appl Environ Microbiol. 2009;75:2312–2319. doi: 10.1128/AEM.02649-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aminov RI, et al. Molecular diversity, cultivation, and improved detection by fluorescent in situ hybridization of a dominant group of human gut bacteria related to Roseburia spp. or Eubacterium rectale. Appl Environ Microbiol. 2006;72:6371–6376. doi: 10.1128/AEM.00701-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suau A, et al. Fusobacterium prausnitzii and related species represent a dominant group within the human fecal flora. Syst Appl Microbiol. 2001;24:139–145. doi: 10.1078/0723-2020-00015. [DOI] [PubMed] [Google Scholar]
- 36.Turroni F, et al. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl Environ Microbiol. 2009;75:1534–1545. doi: 10.1128/AEM.02216-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fuller R, Gibson GR. Modification of the intestinal microflora using probiotics and prebiotics. Scand J Gastroenterol Suppl. 1997;222:28–31. doi: 10.1080/00365521.1997.11720714. [DOI] [PubMed] [Google Scholar]
- 38.Gibson GR. Fibre and effects on probiotics (the prebiotic concept) Clin Nutr Suppl. 2004;1:25–31. [Google Scholar]
- 39.Miyazaki K, Martin JC, Marinsek-Logar R, Flint HJ. Degradation and utilization of xylans by the rumen anaerobe Prevotella bryantii (formerly P. ruminicola subsp. brevis) B(1)4. Anaerobe. 1997;3:373–381. doi: 10.1006/anae.1997.0125. [DOI] [PubMed] [Google Scholar]
- 40.Hancock LE, Shepard BD, Gilmore MS. Molecular analysis of the Enterococcus faecalis serotype 2 polysaccharide determinant. J Bacteriol. 2003;185:4393–4401. doi: 10.1128/JB.185.15.4393-4401.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 42.Anderson NL. Two Dimensional Gel Electrophoresis: Operation of the ISO-DALT System. 2nd Ed. Rockville, MD: Large Scale Biology Press; 1991. [Google Scholar]
- 43.Choe LH, Lee KH. Quantitative and qualitative measure of intralaboratory two-dimensional protein gel reproducibility and the effects of sample preparation, sample load, and image analysis. Electrophoresis. 2003;24:3500–3507. doi: 10.1002/elps.200305614. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.