Abstract
BRCA1 and BRCA2 carriers are at increased risk for both breast and ovarian cancer, but estimates of lifetime risk vary widely, suggesting their penetrance is modified by other genetic and/or environmental factors. The BRCA1 and BRCA2 proteins function in DNA repair in conjunction with RAD51. A preliminary report suggested that a single nucleotide polymorphism in the 5′ untranslated region of RAD51 (135C/G) increases breast cancer risk in BRCA1 and BRCA2 carriers. To investigate this effect we studied 257 female Ashkenazi Jewish carriers of one of the common BRCA1 (185delAG, 5382insC) or BRCA2 (6174delT) mutations. Of this group, 164 were affected with breast and/or ovarian cancer and 93 were unaffected. RAD51 genotyping was performed on all subjects. Among BRCA1 carriers, RAD51-135C frequency was similar in healthy and affected women [6.1% (3 of 49) and 9.9% (12 of 121), respectively], and RAD-135C did not influence age of cancer diagnosis [Hazard ratio (HR) = 1.18 for disease in RAD51-135C heterozygotes, not significant]. However, in BRCA2 carriers, RAD51-135C heterozygote frequency in affected women was 17.4% (8 of 46) compared with 4.9% (2 of 41) in unaffected women (P = 0.07). Survival analysis in BRCA2 carriers showed RAD51-135C increased risk of breast and/or ovarian cancer with an HR of 4.0 [95% confidence interval 1.6–9.8, P = 0.003]. This effect was largely due to increased breast cancer risk with an HR of 3.46 (95% confidence interval 1.3–9.2, P = 0.01) for breast cancer in BRCA2 carriers who were RAD51-135C heterozygotes. RAD51 status did not affect ovarian cancer risk. These results show RAD51-135C is a clinically significant modifier of BRCA2 penetrance, specifically in raising breast cancer risk at younger ages.
Germ-line mutations in the BRCA1 and BRCA2 genes increase susceptibility for both breast and ovarian cancer. Penetrance of these mutations is incomplete and age-dependent, thus cancer risk in carriers continues to increase with age even though the mean age of cancer diagnosis is younger in BRCA1/BRCA2 carriers compared with noncarriers (1). Estimates of penetrance have varied widely, perhaps as a result of different ascertainment schemes and/or allelic effects. In families ascertained for multiple affected individuals suitable for linkage analysis, lifetime cancer risk (by age 70) was 85% for breast cancer in both BRCA1 and BRCA2 carriers (2, 3), 63% for ovarian cancer in BRCA1 carriers (2), and 27% for ovarian cancer in BRCA2 carriers (3). Significantly lower risk estimates were obtained in studies performed in less selected families or at the population level, with a 36–56% lifetime risk for breast cancer (4–7) and a 16% lifetime risk for ovarian cancer (5). These studies were performed in specific ethnic groups (Ashkenazi Jewish and the Iceland population) that harbor a limited number of specific mutations and could therefore be representative of these alleles, rather than reflect the general penetrance of BRCA1/BRCA2 mutations. Such differences suggest that penetrance of BRCA1/BRCA2 mutations is modified by other genetic and/or environmental factors. Identification of such modifiers has important implications, e.g., in facilitating more accurate risk assessment in carriers who face difficult clinical choices regarding prophylactic mastectomy and oophorectomy.
Candidate modifiers include genes whose products are known to interact with BRCA1 and BRCA2 (reviewed in ref. 8). RAD51 is a homologue of bacterial RecA, which is required for meiotic and mitotic recombination and for recombinational repair of double-strand DNA breaks. Both BRCA1 and BRCA2 have been shown to interact with RAD51 (9–11), and the phenotype of murine Brca1- and Brca2-knockout mice is similar to that of Rad51 knockouts (reviewed in ref. 8). A missense mutation in RAD51 (Arg-150–Glu) has been described in two Japanese patients with bilateral breast cancer (12), and Wang et al. orally presented evidence that a single nucleotide polymorphism (SNP) in the 5′ untranslated region (UTR) of RAD 51 is associated with increased breast cancer risk in BRCA1 and BRCA2 carriers but does not influence breast cancer risk in women who are not BRCA1/BRCA2 carriers.‖ This SNP, designated 135 g/c, is a substitution of G for C at position 135 of the human RAD51 cDNA (GenBank accession no. D14134). The aim of the present study was to investigate further the association between the RAD51 5′ UTR polymorphism and disease status in BRCA1 and BRCA2 carriers and to determine whether the RAD51-135C polymorphism is indeed a modifier of BRCA1 and/or BRCA2 penetrance.
Methods
Subjects and Clinical Data.
Participants included all 289 female Ashkenazi Jewish carriers, both healthy and affected, ascertained through Cancer Genetics clinics at two institutions in Israel: Shaare Zedek Medical Center (SZMC) in Jerusalem and Rambam Medical Center (RMC) in Haifa. Nineteen carriers at SZMC were identified through an ongoing study of all Ashkenazi Jewish women diagnosed with breast or ovarian cancer at SZMC since January 1995. All other carriers were identified through patients counseled for family history of breast and/or ovarian cancer history. Clinical data collected on each subject included type of malignancy (based on pathology reports), age at diagnosis or at last follow-up exam, and age at prophylactic surgery if any was performed. All women received genetic counseling and gave informed consent for genetic testing. The study was approved by the institutional review boards (Helsinki committees) at both SZMC and RMC. Samples from healthy Ashkenazi controls (SZMC) were anonymous DNA samples from unrelated healthy persons who reported all four grandparents as being Ashkenazi Jewish and who gave their consent for anonymous testing.
Molecular Analysis.
DNA extraction.
Genomic DNA was extracted from peripheral blood samples by using standard high-salt extraction (13).
Analysis of BRCA1 and BRCA2 mutations.
Genomic DNA was analyzed for the three founder mutations common in Ashkenazi Jews (BRCA1–185delAG, 5382insC, and BRCA2–6174delT) by using previously published methods (4, 14).
RAD51 genotyping.
RAD51 genotyping was performed by PCR amplification of a 157-bp region around nucleotide 135. This amplicon contains a single MvaI site that is abolished by the 135C polymorphism. Wild-type alleles are digested by MvaI resulting in 86- and 71-bp products. The 135C allele is not digested by MvaI, resulting in a single 157-bp product. PCR was performed by using the following primers: RAD51AF (5′-TGGGAACTGCAACTCATCTGG-3′) and RAD51RR (5′-GCGCTCCTCTCTCCAGCAG-3′) at a final Mg concentration of 1.5 mM and an annealing temperature of 53°C. After digestion with MvaI (Fermentas, Vilnius, Lithuania) for 4 h at 37°C, samples were run on a 3% agarose gel. Direct sequencing of the RAD51 amplicon (ABIprism 377, Perkin–Elmer) was performed on three samples heterozygous for the MvaI site, confirming the accuracy of the restriction-digest assay in identifying the RAD51-135C polymorphism.
Statistical Analysis.
RAD51 allele frequencies in affected vs. unaffected subjects were compared by using the χ2 test. The association of disease status in BRCA1/BRCA2 carriers and RAD51-135C was analyzed by using logistic regression, and analysis of disease-free survival was done by using Cox proportional hazard. Several outcomes were analyzed–breast and/or ovarian cancer, breast cancer only, and ovarian cancer only. In all analyses, healthy carriers were censored at the age of last follow-up exam or at the age of the relevant prophylactic surgery (i.e., prophylactic oophorectomy for ovarian cancer and prophylactic mastectomy for breast cancer). Outcomes in affected women were treated as follows: (i) for analysis of breast and/or ovarian cancer, outcome in affected women was the age at diagnosis of the first malignancy; (ii) for analysis of breast cancer only, the outcome in women affected with breast cancer was the age of breast cancer diagnosis; women with ovarian cancer were censored at the age of prophylactic mastectomy, last follow-up exam, or death (whichever came first); and (iii) for analysis of ovarian cancer only, the outcome in women affected with ovarian cancer was the age of ovarian cancer diagnosis, and women with breast cancer were censored at age of prophylactic oophorectomy, last follow-up exam, or death (whichever came first).
Results
There are 289 BRCA1 and BRCA2 carriers currently followed at both participating institutions. RAD51 mutation analysis could not be performed in 25 cases, and the age at diagnosis was not known in three cases, leaving a total of 261 subjects in which complete clinical data and genetic analyses were available. Two subjects were excluded because they were affected with cancers other than breast or ovarian (one with colorectal cancer and one with lymphoma). Two additional subjects were excluded because they were double heterozygotes (BRCA1–185delAG and BRCA2- 6174delT) and could not be assigned a single mutation status. Analysis therefore was performed on 257 carriers who are members of 205 unrelated families (141 segregating BRCA1 mutations and 64 segregating BRCA2 mutations).
Among all carriers, 93 were unaffected, and 164 were affected with breast and/or ovarian cancer. Of 170 BRCA1 carriers, 49 (29%) were unaffected, and of 87 BRCA2 carriers, 41 (47%) were unaffected. Mean age at diagnosis in affected carriers and mean age at last follow-up exam in healthy carriers were not significantly different (Table 1). No RAD51-135C homozygotes were identified among all carriers or among 73 healthy Ashkenazi controls. RAD51-135C heterozygote frequency among all healthy carriers was 5.5% (5 of 90) compared with 12% (20 of 167) among affected carriers. This difference was not significant (Table 1). In BRCA2 carriers, there was a trend to higher frequency of RAD51-135C among affected carriers (P = 0.07). RAD51-135C heterozygote frequency in 73 healthy Ashkenazi controls was 8.2% (6 of 73), intermediate between the frequencies observed in affected and unaffected BRCA1/BRCA2 carriers.
Table 1.
RAD51-135C frequency in BRCA1 and BRCA2 mutation carriers
| Disease status |
BRCA1
carriers
|
BRCA2 carriers
|
||
|---|---|---|---|---|
| RAD51-135C frequency | Mean age*, SD | RAD51-135C frequency** | Mean age*, SD | |
| Healthy | 3/49 (6.1%) | 47.8 (13.5) | 2/41 (4.9%) | 45.6 (10.7) |
| Total affected | 12/121 (9.9%) | 44.9 (10.3) | 8/46 (17.4%) | 49.9 (10.9) |
| Unilateral breast cancer | 6/64 (9.4%) | 43.0 (10.6) | 5/28 (17.9%) | 46.0 (9.2) |
| Bilateral breast cancer | 0/14 (0%) | 41.9 (7.8) | 1/5 (20%) | 45.6 (10.4) |
| Breast & ovarian cancer | 3/13 (23%) | 50.4 (7.4) | 1/1 (100%) | 62 |
| Ovarian cancer | 3/30 (10%) | 48.0 (10.6) | 1/12 (8.3%) | 61.2 (7.3) |
Mean age was not significantly different between affected and unaffected BRCA1 carriers. In BRCA2 carriers, P = 0.07.
In BRCA2 carriers χ2 = 11.1 for RAD51-135C frequency and disease status (4 df), P = 0.03. For RAD51-135C frequency in healthy vs. affected carriers, P = 0.07.
Initial analysis was performed only for unrelated cases. Results were similar to those presented below for all cases, but power was obviously lower because of the smaller sample size. Except for one mother/daughter pair, both of which were BRCA1 carriers and RAD51-135C heterozygotes, all other RAD51-135C heterozygotes were unrelated to each other. Furthermore, RAD51, BRCA1, and BRCA2 are located on different chromosomes and segregate independently, and thus information gained from multiple members of the same family is pertinent to the question of genetic interaction between BRCA1/BRCA2 and RAD51. Results therefore are presented for all cases (257 carriers from 205 families). We first analyzed the effect of RAD51-135C on diagnosis of any related malignancy (breast and/or ovarian cancer) in all carriers (BRCA1 and BRCA2 combined). RAD51 status did not influence total malignancy risk [Hazard ratio (HR) = 1.46 for RAD51-135C vs. normal homozygotes, not significant (NS)]. However, as a covariate, the specific BRCA gene was a significant predictor of disease risk [HR = 0.6, P = 0.01 for BRCA2 vs. BRCA1], and thus further analyses were performed separately for BRCA1 and BRCA2 carriers. This separate analysis is justified also on the grounds that any gene–gene interaction would not necessarily be similar for both BRCA1 and BRCA2.
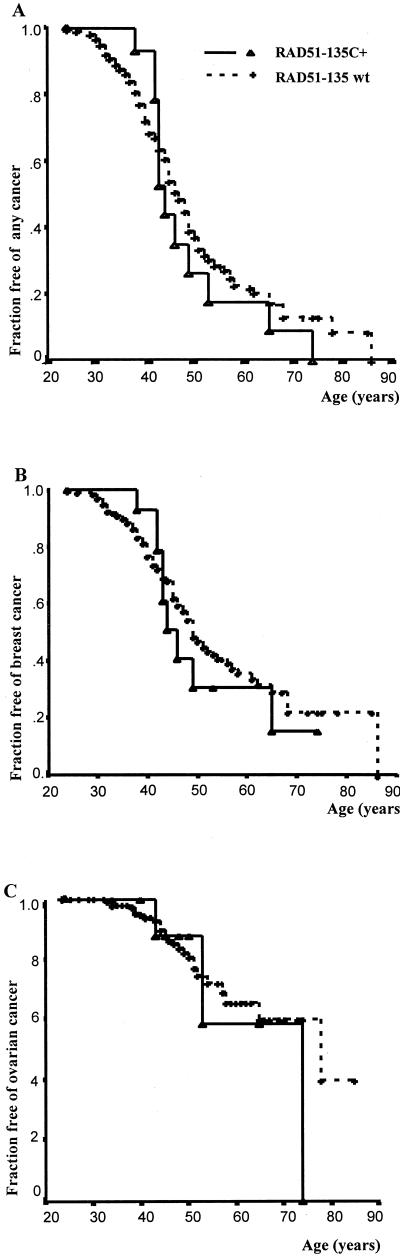
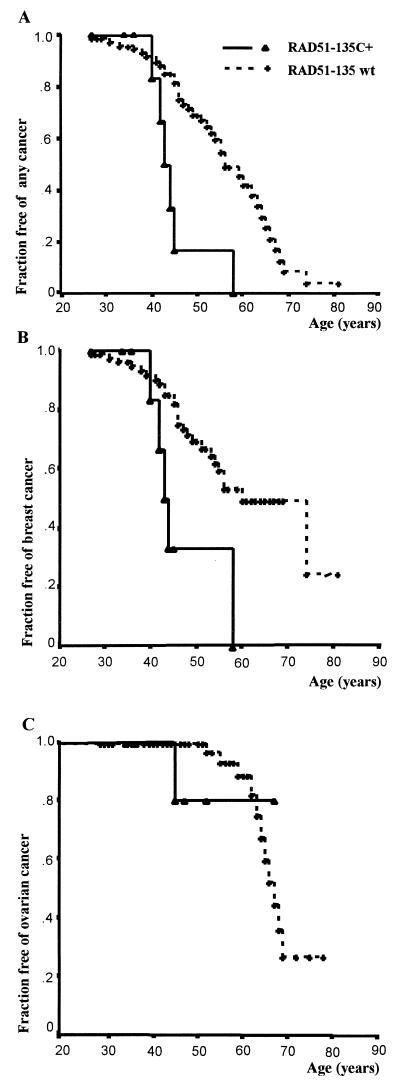
In BRCA1 carriers, RAD51-135C was not found to influence disease risk (Fig. 1). This lack of effect was true for both breast and ovarian cancer combined (HR = 1.18, NS; Fig. 1A), breast cancer only (HR = 1.16, NS; Fig. 1B), and ovarian cancer only (HR = 1.28, NS; Fig. 1C). However in BRCA2 carriers, RAD51-135C significantly increased cancer risk (Fig. 2). For both breast and ovarian cancer combined, the HR for BRCA2 carriers who were also RAD51-135C heterozygotes was 4.0 (95% CI 1.6–9.8, P = 0.003; Fig. 2A). As noted above, information gained from multiple family members is relevant to genetic interaction between independently segregating loci. However, if a substantial proportion of RAD51-135C heterozygotes among BRCA2 carriers were related closely, other unidentified familial factors could have confounded the observed RAD51–BRCA2 interaction. To exclude this possible bias rigorously (even though all RAD51-135C heterozygotes among BRCA2 carriers were unrelated), analysis in BRCA2 carriers was performed also by using only unrelated subjects. In unrelated BRCA2 carriers, for both breast and ovarian cancer combined, the HR for RAD51-135C heterozygotes was 3.5 (95% CI 1.4–8.9, P = 0.008). Notably, all BRCA2/RAD51-135C carriers became affected by age 58, whereas among BRCA2 carriers who were not RAD51-135C heterozygotes, 50.4% remained unaffected at the same age.
Figure 1.
Cancer risk in BRCA1 carriers–survival analysis according to RAD51 genotype. RAD51-135C +- heterozygotes for the RAD51-135C polymorphism. RAD51-135wt (wild type) − homozygous normal (GG) at nucleotide 135. (A) Fraction of BRCA1 carriers remaining free of both breast and ovarian cancer. For RAD51-135C heterozygotes, HR = 1.18 (NS). (B) Fraction of BRCA1 carriers remaining free of breast cancer. For RAD51-135C heterozygotes, HR = 1.16 (NS). (C) Fraction of BRCA1 carriers remaining free of ovarian cancer. For RAD51-135C heterozygotes, HR = 1.28 (NS).
Figure 2.
Cancer risk in BRCA2 carriers–survival analysis according to RAD51 genotype. RAD51-135C +-heterozygotes for the 135C polymorphism. RAD51-135wt (wild type) − homozygous normal (GG) at nucleotide 135. (A) Fraction of BRCA2 carriers remaining free of both breast and ovarian cancer. For RAD51-135C heterozygotes, HR = 4.0 [95% confidence interval (CI) 1.6–9.8, P = 0.003]. (B) Fraction of BRCA2 carriers remaining free of breast cancer. For RAD51-135C heterozygotes, HR = 3.46 (95% CI 1.3–9.2, P = 0.01). All BRCA2 carriers who also were RAD51-135C heterozygotes became affected by age 58, whereas 50.4% of those who were RAD51-135 normal homozygotes remained unaffected at the same age. (C) Fraction of BRCA2 carriers remaining free of ovarian cancer. For RAD51-135C heterozygotes, HR = 1.23 (NS).
To explore the RAD51–BRCA2 interaction further, it also was tested for breast and ovarian cancer as separate outcomes. RAD51-135C did not influence ovarian cancer risk in BRCA2 carriers [HR = 1.23 (NS), P = 0.85 for RAD51-135C heterozygotes vs. normal homozygotes], but the number of ovarian cancer outcomes in BRCA2 carriers was small (Fig. 2C). Breast cancer risk, however, was elevated significantly in BRCA2 carriers who were also RAD51-135C heterozygotes with an HR of 3.46 (95% CI 1.3–9.2, P = 0.01; Fig. 2B). Thus, most of the elevated cancer risk associated with the RAD51-135C polymorphism in BRCA2 carriers is explained by an increase in breast cancer risk. Similar results were obtained with analysis by logistic regression, in which the outcome is dichotomous (presence or absence of breast cancer diagnosis, regardless of age at diagnosis). In BRCA1 carriers, breast cancer risk was not elevated significantly in RAD51-135C heterozygotes (odds ratio = 1.1, NS), but in BRCA2 carriers, the odds ratio for breast cancer in RAD51-135C heterozygotes was 4.3 (P = 0.046). Notably, the magnitude of the RAD51-135C effect found by logistic regression was similar to that found with survival analysis by using Cox proportional hazard. The level of significance is lower because of reduced power when information from age at diagnosis is not taken into account.
Discussion
We observed that in BRCA2–6174delT carriers, presence of the RAD51-135C allele results in an approximately 4-fold increase in breast cancer risk (Fig. 2). This elevated risk was observed consistently by using different analyses (Cox proportional hazard and logistic regression) and in independent unrelated cases as well as in the entire study group. RAD51-135C did not influence breast or ovarian cancer risk in BRCA1–185delAG/5382insC carriers (Fig. 1). Because both genetic and environmental modifiers are more likely to influence low-penetrance mutations than high-penetrance mutations, these results are consistent with previous observations of lower penetrance of BRCA2 and more specifically the BRCA2–6174delT mutation. As noted above, studies in high-risk families found ovarian cancer risk is lower in BRCA2 carriers compared with BRCA1 carriers (3), and despite similar lifetime breast cancer risk, a large study found a suggestion of lower risk in BRCA2 carriers under 50 years of age (3). In Ashkenazi Jews, there is evidence that the BRCA2–6174delT mutation may be associated with lower cancer risk than the BRCA1 mutations common in this population (185delAG and 5382insC). BRCA2–6174delT is the most common mutation at the population level, with an estimated carrier frequency of 1.4%, compared with 1.1% for both BRCA1 mutations combined (15). However, among affected carriers, the frequency of BRCA2 mutations is approximately half that of BRCA1 mutations, suggesting that many BRCA2 carriers remain unaffected (ref. 15; Table 2). In this study, in which families were ascertained mostly on the basis of moderate family history (the mean number of affected relatives, excluding the proband, was 1.8), the BRCA2–6174delT mutation was approximately half as frequent as the BRCA1–185delAG and 5382insC mutations combined.
Table 2.
Relative frequency of the common BRCA1 and BRCA2 mutations in Ashkenazi Jewish subjects and families affected with breast/ovarian cancer
| Study subjects | Number | Affected BRCA1 carriers, % | Affected BRCA2 carriers, % | Ref. |
|---|---|---|---|---|
| Unselected living breast cancer cases | 412 | 34 (8.3%) | 15 (3.6%) | 23 |
| Unselected breast cancer cases, pathology slides | 268 | 10 (3.7%) | 8 (3%) | 6 |
| Unselected ovarian cancer cases | 208 | 57 (27%) | 29 (14%) | 24 |
| Selected breast cancer cases | 178 | 16 (9%) | 8 (4.5%) | 25 |
| Breast and breast/ovarian families | 220 | 91 (41%) | 9 (4%) | 26 |
| Families with known BRCA1/BRCA2 mutations | 205 | 141 (69%) | 64 (31%) | This study |
| Breast or ovarian cancer survivors among Jewish volunteers | 302 | 16 (5.3%) | 11 (3.6%) | 5 |
The biological effect of the RAD51-135C polymorphism is yet to be elucidated and will be important to investigate. It is located in the 5′ untranslated region of the RAD51 gene and theoretically could affect mRNA stability and/or translation efficiency, leading to altered RAD51 protein levels. Altered RAD51 levels could influence the activity of the multiprotein DNA-repair complex that includes BRCA1, BRCA2, and RAD51. Indeed, recent studies suggest altered RAD51 expression may play a role in breast cancer pathogenesis. In breast carcinomas, loss of heterozygosity at the RAD51 locus has been reported in 32% (41 of 127) (16), and reduced Rad51 protein levels have been found in 30% (54 of 179) (17). However, histological grading of sporadic invasive ductal carcinoma has been correlated with overexpression of wild-type Rad51 (18). In one case report, a missense mutation in RAD51 (Arg-150–Glu) has been associated with bilateral breast cancer in two Japanese patients (12), although it is unknown whether this mutation segregated with the disease in their families and whether they were also carriers of BRCA1 or BRCA2 mutations. In our study, RAD51-135C was not associated specifically with bilaterality of breast cancer, although this result could be due to the small number of bilateral breast cancer cases. Among BRCA2 carriers, the frequency of RAD51-135C heterozygotes in women with bilateral breast cancer was 1 in 5 (20%), similar to 17.9% (5 in 28) in unilateral cases (Table 1). Among BRCA1 carriers, none of the 14 women with bilateral breast cancer were RAD51-135C heterozygotes, compared with 9.4% (6 of 64) of those with unilateral breast cancer.
The differential effect of RAD135C on BRCA2 vs. BRCA1 may be related to the different pathways in which the BRCA proteins play a role. Although both BRCA1 and BRCA2 are involved in DNA double-strand break repair by means of homologous recombination (a process requiring RAD51), modulation of RAD51 may not be as crucial for BRCA1 compared with BRCA2-mediated repair (reviewed in ref. 19). Conversely, as suggested by the observation that some BRCA1 mutations are more severe than BRCA2 mutations, DNA repair may be so impaired in the presence of BRCA1 defects that more minor alterations in RAD51 have no detectable impact. Our results also raise the possibility that RAD51-135C may have a differential effect on breast vs. ovarian cancer risk. The lack of effect on ovarian cancer could be the result of the small number of ovarian cancer outcomes in BRCA2 carriers in this study. However, if it is confirmed in a larger series, it may be explained by differences in downstream targets in breast compared with ovarian tissues.
Current breast cancer prevention strategies in BRCA2 carriers include chemoprevention with tamoxifen, prophylactic mastectomy, and probably prophylactic oophorectomy (20–22). Tamoxifen is expected to reduce breast cancer risk by up to 49%, although its efficacy has not been demonstrated specifically in BRCA1 or BRCA2 carriers (20). Prophylactic mastectomy is effective in reducing breast cancer incidence by 90%, but this reduction is counterbalanced by the psychological impact of such surgery and the large number of mastectomies probably needed for each life saved (21). If the association of RAD51-135C with breast cancer risk in BRCA2 carriers is confirmed in additional studies, RAD51 status may be useful in differentiating those carriers most likely to benefit from aggressive prevention measures from those in whom more conservative management would be appropriate.
Acknowledgments
We thank Ms. Gaya Chicco for excellent technical assistance. This study was supported in part by a project grant from the Israel Cancer Research Fund (to E.L.L. and R.C.) and a gift from the Basker family, in loving memory of Eileen Basker.
Abbreviations
- HR
Hazard ratio
- NS
not significant
- CI
confidence interval
Footnotes
Wang, W., Tucker, M. A., Doody, M. M., Tarone, R. E. & Struewing, J. P. (1999) Am. J. Hum. Genet. 655, 22 (abstr.).
References
- 1.Brody L C, Biesecker B B. Medicine (Baltimore) 1998;77:208–226. doi: 10.1097/00005792-199805000-00006. [DOI] [PubMed] [Google Scholar]
- 2.Easton D F, Ford D, Bishop D T. Am J Hum Genet. 1995;56:265–271. [PMC free article] [PubMed] [Google Scholar]
- 3.Ford D, Easton D F, Stratton M, Narod S, Goldgar D, Devilee P, Bishop D T, Weber B, Lenoir G, Chang-Claude J, et al. Am J Hum Genet. 1998;62:676–689. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levy-Lahad E, Catane R, Eisenberg S, Kaufman B, Hornreich G, Lishinsky E, Shohat M, Weber B L, Beller U, Lahad A, Halle D. Am J Hum Genet. 1997;60:1059–1067. [PMC free article] [PubMed] [Google Scholar]
- 5.Struewing J P, Hartge P, Wacholder S, Baker S M, Berlin M, McAdams M, Timmerman M M, Brody L C, Tucker M A. N Engl J Med. 1997;336:1401–1408. doi: 10.1056/NEJM199705153362001. [DOI] [PubMed] [Google Scholar]
- 6.Fodor F H, Weston A, Bleiweiss I J, McCurdy L D, Walsh M M, Tartter P I, Brower S T, Eng C M. Am J Hum Genet. 1998;63:45–51. doi: 10.1086/301903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thorlacius S, Struewing J, Hartge P, Olafsdottir G H, Sigvaldason H, Tryggvadottir L, Wacholder S, Tulinius H, Eyfjord J E. Lancet. 1998;352:1337–1339. doi: 10.1016/s0140-6736(98)03300-5. [DOI] [PubMed] [Google Scholar]
- 8.Welcsh P L, Owens K N, King M C. Trends Genet. 2000;16:69–74. doi: 10.1016/s0168-9525(99)01930-7. [DOI] [PubMed] [Google Scholar]
- 9.Wong A K C, Pero R, Ormonde P A, Tavtigian S V, Bartel P L. J Biol Chem. 1997;272:31941–31943. doi: 10.1074/jbc.272.51.31941. [DOI] [PubMed] [Google Scholar]
- 10.Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston D M. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 11.Chen P L, Chen C F, Chen Y, Xiao J, Sharp Z D, Lee W H. Proc Natl Acad Sci USA. 1998;95:5287–5292. doi: 10.1073/pnas.95.9.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato M, Yano K, Matsuo F, Saito H, Katagiri T, Kurumizaka H, Yoshimoto M, Kasumi F, Akiyama F, Sakamoto G, et al. J Hum Genet. 2000;45:133–137. doi: 10.1007/s100380050199. [DOI] [PubMed] [Google Scholar]
- 13.Miller S A, Dykes D D, Polesky H F. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gershoni-Baruch R, Dagan E, Fried G, Kepten I, Robinson E. Eur J Hum Genet. 1999;7:833–836. doi: 10.1038/sj.ejhg.5200371. [DOI] [PubMed] [Google Scholar]
- 15.Roa B B, Boyd A A, Volcik K, Richards C S. Nat Genet. 1996;14:185–187. doi: 10.1038/ng1096-185. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez R, Silva J M, Dominguez G, Garcia J M, Martinez G, Vargas J, Provencio M, Espana P, Bonilla F. Br J Cancer. 1999;81:503–509. doi: 10.1038/sj.bjc.6690722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshikawa K, Ogawa T, Baer R, Hemmi H, Honda K, Yamauchi A, Inamoto T, Ko K, Yazumi S, Motoda H, et al. Int J Cancer. 2000;88:28–36. [PubMed] [Google Scholar]
- 18.Maacke H, Opitz S, Jost K, Hamdorf W, Henning W, Kruger S, Feller A C, Lopens A, Diedrich K, Schwinger E, Sturzbecher H W. Int J Cancer. 2000;88:907–913. doi: 10.1002/1097-0215(20001215)88:6<907::aid-ijc11>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 19.Karran P. Curr Opin Genet Dev. 2000;10:144–150. doi: 10.1016/s0959-437x(00)00069-1. [DOI] [PubMed] [Google Scholar]
- 20.Fisher B, Costantino J P, Wickerham D L, Redmond C K, Kavanah M, Cronin W M, Vogel V, Robidoux A, Dimitrov N, Atkins J, et al. J Natl Cancer Inst. 1998;90:1371–1388. doi: 10.1093/jnci/90.18.1371. [DOI] [PubMed] [Google Scholar]
- 21.Hartmann L C, Schaid D J, Woods J E, Crotty T P, Myers J L, Arnold P G, Petty P M, Sellers T A, Johnson J L, McDonnell S K, et al. N Engl J Med. 1999;340:77–84. doi: 10.1056/NEJM199901143400201. [DOI] [PubMed] [Google Scholar]
- 22.Rebbeck T R, Levin A M, Eisen A, Snyder C, Watson P, Cannon-Albright L, Isaacs C, Olopade O, Garber J E, Godwin A K, et al. J Natl Cancer Inst. 1999;91:1475–1479. doi: 10.1093/jnci/91.17.1475. [DOI] [PubMed] [Google Scholar]
- 23.Warner E, Foulkes W, Goodwin P, Meschino W, Blondal J, Paterson C, Ozcelik H, Goss P, Allingham-Hawkins D, Hamel N, et al. J Natl Cancer Inst. 1999;91:1241–1247. doi: 10.1093/jnci/91.14.1241. [DOI] [PubMed] [Google Scholar]
- 24.Moslehi R, Chu W, Karlan B, Fishman D, Risch H, Fields A, Smotkin D, Ben-David Y, Rosenblatt J, Russo D, et al. Am J Hum Genet. 2000;66:1259–1272. doi: 10.1086/302853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abeliovich D, Kaduri L, Lerer I, Weinberg N, Amir G, Sagi M, Zlotogora J, Heching N, Peretz T. Am J Hum Genet. 1997;60:505–514. [PMC free article] [PubMed] [Google Scholar]
- 26.Tonin P, Weber B, Offit K, Couch F, Rebbeck T R, Neuhausen S, Godwin A K, Daly M, Wagner-Costalos J, Berman D, et al. Nat. Med. 1996. 1179–1183. [DOI] [PubMed] [Google Scholar]