Abstract
Dietary polyphenols are components of many foods such as tea, fruit, and vegetables and are associated with several beneficial health effects although, so far, largely based on epidemiological studies. The intact forms of complex dietary polyphenols have limited bioavailability, with low circulating levels in plasma. A major part of the polyphenols persists in the colon, where the resident microbiota produce metabolites that can undergo further metabolism upon entering systemic circulation. Unraveling the complex metabolic fate of polyphenols in this human superorganism requires joint deployment of in vitro and humanized mouse models and human intervention trials. Within these systems, the variation in diversity and functionality of the colonic microbiota can increasingly be captured by rapidly developing microbiomics and metabolomics technologies. Furthermore, metabolomics is coming to grips with the large biological variation superimposed on relatively subtle effects of dietary interventions. In particular when metabolomics is deployed in conjunction with a longitudinal study design, quantitative nutrikinetic signatures can be obtained. These signatures can be used to define nutritional phenotypes with different kinetic characteristics for the bioconversion capacity for polyphenols. Bottom-up as well as top-down approaches need to be pursued to link gut microbial diversity to functionality in nutritional phenotypes and, ultimately, to bioactivity of polyphenols. This approach will pave the way for personalization of nutrition based on gut microbial functionality of individuals or populations.
Keywords: polyphenol bioconversion, gut microbiota, metabolomics, metagenomics, microbiomics
Plant polyphenols are phytochemicals (1), which have been suggested to play a major role in the health enhancing effects of fruits and vegetables (2, 3). Polyphenols are secondary plant metabolites originating from the shikimic pathway and share at least one aromatic ring structure with one or more hydroxyl groups. Despite this simple common structural motif, a large number of polyphenol classes exist, including flavonoids, stilbenes, coumarins, lignans, lignins, cinnamic, and benzoic acids. Prospective and cross-sectional epidemiological studies have associated polyphenol consumption with reduced risk for cardiovascular disease (2) and cancer (3). Intervention studies in human and animals have provided further evidence for the protective effects of polyphenols in the direction of modulation of vascular and platelet function, blood pressure (4), and an improved plasma lipid profile (1–3, 5). Mechanistic studies have identified among others oxidative stress, inflammation, and endothelial function as important targets where polyphenols may exert their beneficial bioactive effects. Indeed, numerous studies with polyphenols in pure form, or as occurring in natural extracts, have been performed that confirm their antioxidant capacity by chemical assays or in cell models (6). However, the continual emergence of studies on the bioavailability of polyphenols has created increasing doubt about the biological relevance of such findings (7). It has become clear that the bioavailability of polyphenols, as they occur in dietary formats, is highly variable between individuals and generally far too low to explain direct antioxidant effects in vivo (8). Dietary flavonoids, for example, are mostly present as poorly absorbed glycosides, which require deglycosylation by mammalian β-glucosidases in the small intestine before being absorbed as aglycons (9). Levels of aglycones in the circulation vary widely but tend to be low. This property also holds for oxidized and conjugated flavonoid forms due to phase I and II metabolism. Other specific mechanisms of action have been proposed, yet also here the concern has been raised that bioavailability of polyphenols in the forms as they occur in the diet is probably very low (10–12). An important reason for this concern is that, depending on the glycosylation pattern and polymerization degree, a significant fraction of dietary polyphenols can persist to the colon, where they are exposed to the gut microbial community. Given its enormous gene pool, it is now becoming clear that the resident colonic microbiota can be regarded as a separate organ within the human host, which can perform many functions of which the human host is incapable. These strong and symbiotic microbiota–host interactions have led to the recognition of humans as superorganisms, in which the colon operates as a bioreactor with a virtually unlimited metabolic potential (13). The consideration of the highly individual gut microbial activity has been recognized as an essential part of personalized nutrition approaches (14). The recently introduced pharmaco-metabonomics approach demonstrated that the presence of a predose gut-mediated dietary metabolite in urine was predictive for the metabolic fate of a drug substance (15, 16). The gut-mediated nature of this interaction has been proposed as a route for modulation of drug efficacy (and safety). This concept may be well applicable to microbiota-mediated bioactivation of dietary polyphenols. This area exists as a largely uncharted territory because of the lack of experimental approaches to address the overwhelming complexity of cometabolome interactions, in particular when viewed from a nutritional context. The rapid emergence of functional genomics approaches for the holistic assessment of microbial communities and metabolism, in combination with adequate in vitro and in vivo models, opens new avenues for studying the human superorganism in an integrated approach.
Gut-Mediated Bioactivation of Polyphenols in the Human Superorganism
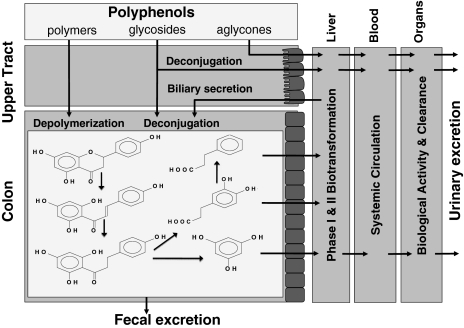
The human colon harbors a highly complex microbial ecosystem, at concentrations of 1012 microorganisms per gram of gut content. The gut microbiota composition of each individual is unique and is influenced through a legacy acquired at birth, genotype and physiological status of the host, diet, and lifestyle (17, 18). Fig. 1 depicts the cometabolome interactions between the colonic microbiota and the human host and the resulting complex metabolic fate of dietary polyphenols. Note that the colon not only receives dietary polyphenols, but also phase I and II metabolites that have been excreted back into the intestine through the enterohepatic cycle. Microbial polyphenol metabolism follows a very general pattern in which the extremely diverse group of natural polyphenols is funneled to a relatively small number of metabolites. Besides deglycosylation by bacterial enzymes, the microbiota can perform mild transformation such as dehydroxylation and demethylation and, in addition, catabolism of polyphenols into small fragments (19). As a consequence, a relatively small number of metabolites are formed in the colon from the extremely diverse group of natural polyphenols (20). With the flavonoid naringenin as an example (Fig. 1), bioconversion starts with isomerization of the flavanone's C ring at the hetero atom into the corresponding chalcone phloretin. After reduction into a dihydrochalcone, further splicing takes place at the carbonyl moiety, yielding phloroglucinol and 3-(4-hydroxyphenyl)propionic acid. The metabolite 3-(4-hydroxyphenyl)propionic acid may be dehydroxylated into 3-phenylpropionic acid, and the mixture can be absorbed from the colon. Both components are often recovered as such or in a conjugated form in urine (21), but may also be subjected to β-oxidation and glycination in the liver, yielding hippuric acid and 4-hydroxyhippuric acid (20). In contrast, phloroglucinol is hardly ever recovered as a final metabolite, because it can be catabolized into acetate, butyrate, and CO2 (22).
Fig. 1.
Schematic depiction of metabolic fate of dietary polyphenols in the human–microbial superorganism. Within the colonic compartment, the microbial bioconversion pathways of naringenin are depicted. Within the host, dietary polyphenols and their microbial bioconversion products successively undergo liver phase I and II metabolism, absorption in the systemic circulation, interaction with organs, and excretion in the urine.
Colonic bioconversion of polyphenols is most well described for flavonoids and is highly variable because of three main reasons. First, large interindividual differences have been noted in the bioconversion of specific flavonoids (17, 18, 23). This variability can be attributed to the individual colonic microbiota composition and has led to the recognition of low to high flavonoid converters (24, 25). Second, small differences in substitution pattern of flavonoids can lead to major changes in colonic bioconversion (24, 26). A third factor is the dietary context of the ingested polyphenols that can modulate polyphenol–microbiota interaction (27).
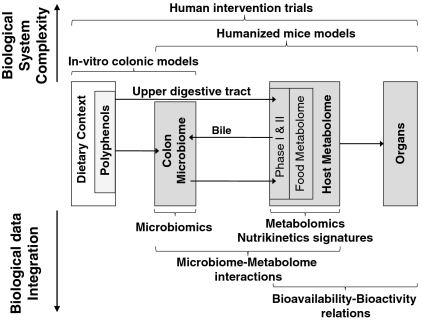
Besides bioconversion of active dietary compounds into less active metabolites, a number of bacterial transformations are also known to produce metabolites with increased biological activity. A good example are the phytoestrogens such as soy isoflavones, prenylflavonoids from hops, and lignans, for which the pseudoestrogenic activity is determined by intestinal bacterial activation followed by absorption of the microbial metabolites (28, 29). Establishing the role of the gut microbiota in the bioconversion of polyphenols into potentially bioactive species is challenging. Cometabolome interactions in the human superorganism are complex and experimentally difficult to capture. The holistic view provided by functional genomics can be overwhelming, thus we have adopted a strategic approach that is outlined in Fig. 2. Here, the upper part depicts our experimental approach to address biological complexity in the human superorganism by taking recourse to adequate in vitro and animal model systems. The lower part of Fig. 2 highlights microbiomics and metabolomics as the most relevant functional genomics technologies. We define microbiomics for the purpose of this review as the study of entire mixed microbial populations, their common genetic elements (metagenome), functionality (metatranscriptome and -proteome), and environmental interactions in a defined environment. These platforms are approaching full coverage of the wide variety in microbial metabolites and involved microorganisms, hence we will also discuss approaches for integrating the generated information. This integrated strategy will aid in further unraveling of metabolome–microbiome relations and properly assess the health-modulating potential of dietary polyphenols.
Fig. 2.
Experimental strategy for assessment of the metabolic fate of polyphenols in the human superorganism. Biological complexity can be addressed by combining In Vitro Colonic Models, Humanized Mice Models, and Human Intervention Trials. Microbiomics, Metabolomics, and Nutrikinetic Signatures need to be integrated to resolve Microbiome–Metabolome Interactions and establishing Bioavailability-Bioactivity Relations. Terms in italics correspond to section headers.
Human Intervention Trials
In humans one can study the full complexity of dietary polyphenol interaction with the microbiota–host cometabolome. Most human intervention studies on the absorption, distribution, metabolism, and excretion (ADME) of polyphenols focused on phase I and II metabolism in the small intestine and liver (30). Typically, intact polyphenols appear as aglycones or conjugates in plasma at low levels within a few hours after ingestion. The involvement of the gut microbiota in polyphenol bioconversion is typically reflected in delayed (6–8 h) appearance of metabolites in systemic circulation (31) and significant interindividual variation in absorption rates and levels. These kinetic features can serve as a functional measure for the polyphenol bioconverting capacity of the microbiota and, hence, need to be accounted for in the design of the studies (11, 32, 33). Considering that structurally rather diverse polyphenols are assumed to be bioconverted by the gut microbiota to a limited number of key metabolites, the polyphenol content of the background diet also needs to be accounted for in the study design (34, 35) and subsequent data analysis (31). Nonpolyphenol dietary components may interact with microbial bioconversion (27), hence the background diet also needs to be controlled and standardized. Consideration of the aforementioned constraints in the design of a human study will inevitably meet practical and ethical constraints. Hence, the small effects of dietary interventions in human intervention trials are commonly clouded by biological variation. The use of cross-over designs where volunteers serve as their own control allows for multilevel analysis schemes that increase power, but still require significant numbers of volunteers to allow for statistical significant multivariate models (36).
Many human intervention trials have aimed at establishing bioactivity of polyphenols, but only few took bioavailability into account. No clear relationship between bioavailability and bioactivity in humans has been established (37). However, polyphenol metabolites are most likely interacting with multiple molecular targets in a subtle manner (10). Global metabolic profiling in human polyphenol intervention studies primarily revealed effects on the gut microbiota-mediated metabolites (35, 38, 39). The endogenous effects observed in these NMR-based studies are subtle and may be related to tight homeostatic regulation in healthy humans. Hence, the use of homeostatic challenges has been advocated as a sensitive approach to assess regulation of inflammatory, oxidative, and metabolic stress as overarching drivers of health (40). Such an approach would also require MS-based profiling approaches focusing at specific submetabolomes and deployment of other functional genomics approaches (transcriptomics), but no published study has appeared yet. Given the many current restrictions of human intervention trials, animal and in vitro models have been developed to allow more experimental freedom in defining and modifying polyphenol doses, background diet, microbial diversity, longitudinal sampling of metabolic profiles, and assessment of bioactivity.
Humanized Mice Models
Numerous rodent studies have investigated the metabolism of polyphenols, especially for their impact on metabolic disorders (41, 42). Only a few of these studies have followed the dynamics and composition of the intestinal microbiota in association with polyphenol metabolites retrieved from the host (43, 44). These studies have demonstrated that polyphenols can modify the intestinal bacterial community diversity, which has meanwhile been supported by preliminary in vitro fecal incubation experiments and human intervention studies (45, 46). Therefore, studies demonstrating beneficial health effects of polyphenols must include gut microbiological analysis to link shifts in bacterial populations to polyphenol metabolites and beneficial effects. However, caution is required in extrapolating results to humans because culture-independent comparisons have revealed that most bacterial genera and species found in mice are not seen in humans, although the distal gut microbiota of mice and humans harbors the same bacterial phyla (47). Therefore, the use of germ-free mice inoculated with the fecal microbiota of a healthy human is of great relevance. Such humanized (gnotobiotic) mice provide a model system for controlling host genotype, gut microbiota composition, diet, and housing conditions. Demonstrating the applicability of the model, several studies involving germ-free and humanized rodents showed that the presence and, therefore, bioavailability of certain polyphenol derivatives depended on the presence of an intestinal microbiota (48–50). So far, no studies have followed the composition of the intestinal microbiota in humanized rodents fed a polyphenol-rich diet. Studies addressing this gap would be of relevance because shifts in specific bacterial groups could be the first step towards an improved description of microbial polyphenol bioconversion and may be linked to health effects. An important aspect in studies involved in determining the impact of polyphenols in vivo is their dietary level, which can vary from 0.1 to 4% (51). The level can have major repercussion in terms of intestinal bacterial abundance and composition because it is known that certain polyphenols and their microbial bioconversion products can influence the overall intestinal ecosystem, in particular due to antimicrobial properties (45).
In Vitro Colonic Models
Although in vivo human or animal intervention trials are physiologically most relevant to study the ADME of polyphenols, they are less suited to perform mechanistic research on how gut microbiota affect polyphenol bioavailability. Therefore, in vitro tools have been designed that simulate intestinal conditions. In combination with in vivo trials, in vitro experiments may elucidate to what extent bioconversion processes are mediated by the gut microbiota or by the host itself (28, 52, 53). The complexity of in vitro gut models is diverse, ranging from simple static models to advanced continuous models (54). A static model approach is primarily used when assessing the stability of polyphenols in the presence of human-derived gut microbiota and evaluating which environmental conditions favor or limit polyphenol bioconversion. Fecal batch incubations are of particular interest for a first assessment of gut microbial polyphenol metabolism, which is characterized by a high human interindividual variability due to differences in microbial community composition (53). This way, a first distinction can be made between high and low polyphenol-converting individuals. In addition, fecal incubations have been extensively used to characterize microbial fermentation products of dietary polyphenols (20, 55, 56).
In contrast to short-duration experiments with static gut models, longer-term experiments are required when the adaptation of the gut microbial community to dietary polyphenols needs to be assessed. To this end, dynamic in vitro gut models such as the Reading model (57), the Simulator of the Human Intestinal Microbial Ecosystem (SHIME) (58), and the TNO Intestinal Model 2 (TIM2) (59) have been developed where gut microbiota are cultured over a longer timeframe (days to weeks) in one or multiple connected, pH-controlled vessels representing different parts of the human colon. Validation of multistage culture systems by analyses of the colonic contents of sudden death victims demonstrated a good comparability (60). The SHIME has already been used to monitor the behavior of human gut microbiota with different polyphenol converting potency (61, 62), whereas an interesting feature of the TIM2 is the use of dialysis modules (59), whereby end products of microbial metabolism are removed, thus preventing end product inhibition.
In vitro gastrointestinal models are important tools in research leads for dietary polyphenols. First, the flexibility of in vitro studies can effectively build mechanistic knowledge around microbial polyphenol bioconversion (32). Linking metabolomic datasets with in-depth microbiome analyses may not only result in the discovery of new polyphenol metabolites with health-promoting potential, but also point out which microbial groups and species are involved in polyphenol bioconversion. In addition, dynamic in vitro gut models may even be used to evaluate strategies that target the enrichment of polyphenol-converting species such as Clostridium orbiscindens and Eubacterium ramulus (63, 64) with low doses of polyphenols. Such strategies offer possibilities to steer the bacterial community toward a composition with specific bioconversion properties. This strategy was successfully applied for steering isoflavone and prenylflavonoid conversion in vitro and in vivo (61, 65). The isolates of such consortia may be subsequently investigated at the genome level and, thereby, complement the information generated from other omics platforms. Second, in vitro gut models allow a more profound analysis of how polyphenols affect the gut microbial community. Antimicrobial effects of polyphenols have been described as well as stimulation of bacterial groups (56, 66). In this respect, in vitro dynamic ileal models and mucus adhesion modules (67), respectively, may be used to evaluate effects of polyphenols toward the activity of opportunistic pathogens or the mucus colonizing potency of gut microbiota.
Microbiomics
Thus far, relatively little is known regarding microbial species responsible for the polyphenol bioconversion in the human colon, but emerging technologies will enable breakthroughs in this area. Molecular approaches, mainly based on the 16S ribosomal RNA (rRNA) gene, have revolutionized the field of gut microbial ecology, bypassing tedious culturing of anaerobic gut microbes. Novel high-throughput diversity approaches, such as phylogenetic microarrays (68, 69) and barcoded “next generation” sequencing (70), may be applied to monitor and compare polyphenol-induced global gut community shifts with greater sensitivity than former fingerprinting methods (such as PCR-DGGE) (71). Complementary quantitative technologies such as fluorescence in situ hybridization (FISH), which is truly quantitative at the single-cell level (72), and real-time quantitative PCR (RT-qPCR) (73) can be used to confirm shifts in particular groups or species. Stable isotope probing (SIP) technology has been successfully applied to identify microbes that grow and incorporate a 13C label of a substrate into biomarker(s) (74). However, the nature of chemical conversions that take place during polyphenol conversion is not always suited to monitor with SIP; the energy gain of polyphenol metabolism is low, many polyphenols have antimicrobial effects, and polyphenol metabolism can be performed by microbial consortium, thereby further reducing the label signal. Thus, there are both biological and practical limitations to the deployment of SIP for identification of polyphenol bioconverting microbes.
Currently, the field of gut microbial ecology is witnessing a second revolution in molecular approaches by the rapid advances in next-generation DNA sequencing, which are dramatically reducing costs and markedly increasing capacity. This progress allows culture independent microbiomics methods to be readily deployed, not only to determine diversity but more critically the functionality of the gut microbiome (75–77). The use of metagenomic libraries for identifying gut bacterial genes from intestinal communities has recently been demonstrated and, notably, for a novel polyphenol oxidase gene from a metagenome expression library of bovine rumen (78, 79). Key to this approach is the creation of suitable screening assays for the relevant polyphenol family and, moreover, that the gene set coding for a specific functionality is clustered on the bacterial genome. The recent description of a gut microbial gene catalog of 3.3 million genes greatly enhances the annotation possibilities for omics approaches (80). This new catalog supports associated technologies such as metatranscriptomics, which uses gene expression patterns to understand the metabolic activities of microbial communities, or potentially emerging high-throughput RNA-seq approaches (81). Reproducible protocols for messenger RNA (mRNA) isolation from intestinal samples and procedures to reduce ribosomal RNA before cDNA sequencing are becoming available (82, 83). The sequencing of cDNA synthesized from the mRNA and further bioinformatics analysis largely depends on the progress and development of sequencing and analyzing capacities but is now feasible (83). Metatranscriptomics has been elegantly applied to assess gut microbial gene regulation upon dietary intervention in a mouse model, as well as a preliminary array-based approach for infant fecal bacteria (84, 85), but no application to polyphenols has been reported. Similar to metatranscriptomics, metaproteomics is becoming a feasible approach for identification of proteins involved in polyphenol metabolism for complex communities (86, 87). Metaproteomics has the advantage that because of the stability of the proteins, a more representative result of functional activity within the human intestine from a fecal sample may be obtained compared with that of metatranscriptomics. Initial knowledge on genetic determinants for polyphenol bioconversion may be obtained more readily from in vitro gut model experiments that can use higher polyphenol concentrations to amplify effects, although antimicrobial and stress effects also risk to be magnified. The use of fecal inocula from different individuals can be used to account for interindividual variation and generate a more comprehensive view on microbial determinants that drive polyphenol metabolism. The application of such targeted approaches, using in vitro models and humanized mice, is anticipated to identify microbial genomic pathways for polyphenolic bioconversion.
Metabolomics
The extensive bioconversion of polyphenols by colonic microbiota and the host produces a large range of metabolites that can potentially be taken up into systemic circulation. These metabolites are part of the so-called “food metabolome” (10, 88) that, in its turn, interacts with the endogenous metabolome of the host. Effects on food (89) and endogenous (90) metabolomes have typically been assessed by targeting a limited number of predefined (hypothesis-driven) metabolites. Such targeted approaches hold the risk of overlooking the complexity of phytochemical diversity in the diet and the complexity of their effect on endogenous metabolism (10). Advances in analytical instrumentation now allow for comprehensive and simultaneous profiling of metabolites in biological samples (91). Global metabolic profiling, which nonselectively captures metabolites in an unbiased manner, is typically performed by high-field (>600 MHz) NMR and has successfully been applied to describe both the food as well as endogenous metabolome in urine, plasma, and feces (92). Although NMR metabolomics is considered relatively insensitive, it still captures a significant part of the microbially mediated part of the food metabolome in urine. Thus, it played a key role in developing the concept of the human superorganism encompassing the symbiotic gut microbiota–host cross-talk (93). NMR-based profiling was also used to demonstrate the concept of pharmaco-metabonomics (15, 16). More sensitive and selective focused MS-based profiling approaches have been developed to capture less abundant food and endogenous metabolites. Sensitivity enhancement is typically achieved by defining submetabolomes in a wide-angle hypothesis-driven manner. An example is the focused profiling of phenolic acids by GC-MS (94) in plasma, urine, feces, and in vitro models, hence capturing a major part of the microbial bioconversion products of polyphenols. The disadvantage of these focused GC-MS profiling methods is the laborious sample pretreatment requiring deconjugation and subsequent derivatisation to increase volatility. Thus, most information on conjugative host metabolism is discarded, and only relatively small bioconversion products are observed. In comparison, LC-MS profiling platforms are less biased in their sample pretreatment, capture more and larger conjugated species, but encounter bottlenecks in identification of relevant metabolites (88). An intrinsic limitation of most MS-based profiling approaches is that they are mostly semiquantitative and not suitable for making mass balances between dietary input and metabolic output in urine, plasma, and feces.
In addition to technological developments, high demands are made on the statistical analysis to unambiguously characterize the usually subtle metabolic response induced by polyphenol intervention. Whereas univariate approaches neglect covariance effects and suffer from multiple testing issues, the multivariate approaches are fraught by the risk of overfitting (95). Hence, rigorous cross-validation approaches have been proposed (96). Multilevel approaches have been shown to be capable of distinguishing subtle polyphenol-induced effects in the presence of large phenotypic variation between subjects (31, 97).
Nutrikinetic Signatures
The timecourse of polyphenol metabolites contains information on their ADME characteristics between the different compartments of the human superorganism. Like other dietary constituents, the disposition of polyphenol metabolites mostly follows first-order kinetics across compartments and can be parameterized with appropriate nutrikinetic models (31). The estimated set of nutrikinetic model parameters can therefore be considered as a lumped, quantitative summary of a compartmental model. Hence, we investigated whether these nutrikinetic signatures can be used to map the highly individual interactions among polyphenol intake, the host metabolome, and the gut microbiome.
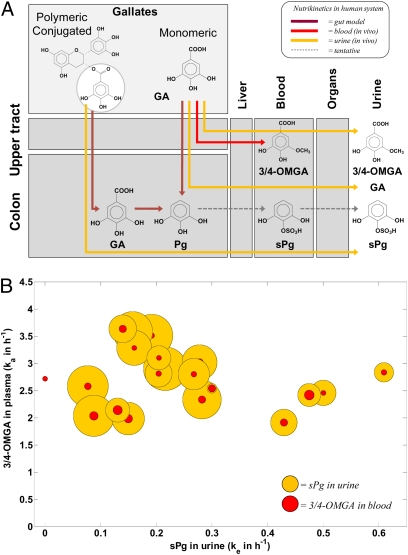
In a recently performed human intervention study, 20 healthy volunteers were given a bolus dose of polyphenols. Global [1H NMR (31)] and focused (GC-MS) (94)] urinary metabolic profiling was performed as well as targeted LC-MS analysis on selected phenolic compounds in plasma. Multilevel PLS-DA on the urinary profiles revealed the most discriminatory markers of treatment (36, 97). The concentration–time curves of the selected biomarkers were then fitted by using an adapted cumulative, one-compartmental first-order nutrikinetic model (31). The resulting nutrikinetic signatures showed large interindividual variation, which points toward nutritional phenotypes that differ in onset, rate, and extent of microbial bioconversion. The variation in nutrikinetic signatures therefore provides a functional measure for the interindividual differences in gut microbial composition, bioconversion, and/or bioactivity (23). This variation in signature can be illustrated by the metabolic fate of gallic acid (GA) as it occurs in the free and conjugated form in black tea. 3/4-O-methylgallic acid (3/4-OMGA) and sulfated pyrogallol (sPg) are considered as end metabolites of the GA pathway, which most likely progresses according to the evolution scheme in Fig. 3A. This figure also summarizes the sources of nutrikinetic information now available for us. In Fig. 3B, the interindividual variation in urinary excretion rate constant (ke) of sPg is plotted against the rate constant (ka) of 3/4-OMGA in blood. In this bubbleplot representation, the bubble-size is proportional to their estimated total molar levels (micromoles). High blood levels and fast (formation) rate constants of 3/4-OMGA may therefore directly point toward high and fast liver phase II metabolism (98). The nutrikinetic signatures of sPg on the other hand may be more indicative for the activity of gut microbial bioconversion. Hence, even though the population shares a common topology of the metabolic network, large interindividual variation of the nutrikinetic signatures can be observed. The interindividual variation in Fig. 3B is primarily created by differences in gut microbial metabolism (large range of sPg rates) rather than liver metabolism (more narrow range of 3/4-OMGA rates). The observed variation in liver metabolism and bioconversion capacity of the gut microbiota may serve to distinguish nutritional phenotypes within test populations (99). Such nutrikinetic signatures may also be used to strengthen the concept of pharmaco-metabonomics, which is based on a single predose metabolic snapshot (15).
Fig. 3.
(A) Proposed GA metabolic pathways in the human superorganism. The red, yellow, and brown lines, respectively, represent nutrikinetic information obtained from plasma, urine (in vivo in humans), and in vitro models. (B) Bubbleplot representing the nutrikinetics of sPg in urine (ke) and 3/4-OMGA in blood per individual (ka). Bubble diameters are proportional to the estimated total molar level of sPg in urine (0–124 μmol) and 3/4-OMGA in blood (5–21 μmol).
Microbiome–Metabolome Interactions
Establishing functional links between polyphenol metabolism and the human gut microbiome is lagging behind the rapid developments in metabolomics and microbiomics (100). Microbiome–metabolome correlations may be established in vivo, as was demonstrated in a small human cohort study, where several microbial species were found to correlate to metabolites that appear in polyphenol bioconversion pathways (101). The nutrikinetic phenotyping of the 20 human volunteers described in the prior section provided us with the opportunity to correlate the human metabolome with the phylogenetically (Human Intestinal Tract Chip, HITChip) defined gut microbiome. Robust Spearman correlations were calculated between the nutrikinetic parameters of a range of metabolites appearing in plasma and microbial species as observed by HITChip. Members of the Actinobacteria and the Clostridium clusters showed significant correlation with the plasma nutrikinetic parameters of 5-(3-methoxy-4-hydroxyphenyl)-γ-valerolactone, which results from the first microbial ring fission of catechin-type structures. To obtain more detailed insight in the functionality of the gut microbiota, further work should focus on establishing metabolome–microbiome correlations at the metagenomic or transcriptomic level. To go beyond the single-gene, single-metabolite correlation, penalized multivariate regression modeling (102) can be used. The penalization ensures that only few species or genes are selected to correlate with nutrikinetic signatures, but still rigorous validation will be required to prevent chance results. The established correlations will also need in vitro experiments for validation and establishing causal relationships between microbiota and bioconversion steps.
So far, nutrikinetics has been used to summarize the functional (bioconversion) capacity of the human gut microbiota. The established first-generation nutrikinetic models can be used to phenotype the functional (bioconversion) capacity of both host and microbiota, but do not cover the systems biology underlying the observed metabolic bioavailability. Therefore, we aim to develop second-generation nutrikinetic models that also summarize features of metabolic networks and cometabolome cross-talks (103, 104) within the mammalian superorganism. The complexity of such relationships cannot be unraveled by using merely in vivo measurements on the human superorganism. As depicted in Figs. 2 and 3A, combinations of in vitro and in vivo approaches are required to resolve bioconversion pathways and to identify the microbiota involved. The sources of information obtained from these additional tools can be used to build up a systems biology model in a bottom-up fashion, provided metabolic data are acquired in an absolute quantitative manner (105). Finally, the various sources of prior information have to be combined with the nutrikinetic signatures obtained from human interventions, for which we advocate the gray modeling approach. Gray modeling is a distribution free approach of Bayesian statistics where prior information is used and its confidence is expressed by a soft penalty (106). These gray models may be applied to find a relationship between the polyphenol intervention, the gut microbial levels, and the nutrikinetic signature of the individuals, in agreement with outcomes from in vitro and humanized mice experiments. Such models will enable more detailed phenotyping of human individuals than the current “snapshot” approaches of pharmaco-metabonomics (15), hence bringing us closer to opportunities in personalized nutrition.
Ultimately, knowledge of the genetic determinants for polyphenol bioconversion rather than phylogeny of the gut microbial species may be more appropriate for making microbiome–metabolome correlations. Because polyphenols are not considered an energy source for gut microbial growth (relative to polysaccharides), they may have modest impact on microbial growth and composition, although there will be metabolic activity that could be measured at the RNA and protein level. Microbial shifts may occur because of other effects of polyphenols, such as antimicrobial activity and knock-on effects. Importantly for human interventions, dietary polyphenol intake cannot be radically altered; a polyphenol-free diet for a prolonged period obviously would be unethical and, for safety reasons, there are limits to single polyphenol doses. Consequently, microbial community shifts in human interventions may be limited, thus requiring highly sensitive analysis, for example, a sequencing depth beyond what is realistically feasible with current next generation sequencing technologies (71). This demand especially holds true when analyzing the full gene content (metagenome) rather than the microbial composition (16S rRNA). Moreover, because of the functional redundancy of the human gut microbiome, the actual genes/proteins rather composition are probably more relevant. Consequently, metatranscriptome and metaproteome approaches, and possible future modifications of the RNA-seq approach (81), that focus on those genes (and proteins) impacted by polyphenols, are likely to be more powerful for use in correlation analysis.
Bioavailability–Bioactivity Relations
So far only a few microbial polyphenol bioconversion products have been associated with systemic biological effects in the host or at the local level at the gut wall (107). In vitro and human trials are revealing an increasing number of metabolites that appear at high levels in the colon and systemic circulation (33, 36). The biological relevance for most of these metabolites is unknown, and systematic approaches are required to address this. In a bottom-up approach, potential bioactivity of gut microbiota-mediated metabolites needs to be screened in relevant in vitro and animal models. In a recent screening exercise, gut-microbial polyphenol metabolites were indeed able to reduce the ex vivo response of peripheral blood mononuclear cells to an inflammatory challenge (108). When screening gut-mediated metabolites for biological activity, one should however take into account the different conjugation routes once they have entered systemic circulation (109).
In top-down systems biology approaches, we will need to address the challenge to link the microbiota-mediated metabolome with other functional genomics levels (110). A recent attempt in this area associated circulation levels of a single microbial polyphenol metabolite with differences in long-term bioactivity as defined by transcriptomics of leukocytes (111). We envisage that the introduction of nutrikinetic phenotypes will allow for stronger associations between nutritional phenotypes and bioactivity of polyphenols. The use of homeostatic challenge tests to obtain more sensitive readouts of long-term bioactivity will introduce a next level of complexity in data analyses. Another route is presented by numerous comprehensive human gut microbiome projects that are in progress around the globe (112, 113). These microbiomic surveys not only capture the gut microbial diversity associated with humans but will also promote investigations into the functional contributions that our microbes make to our physiologic phenotypes, health, and disease predispositions.
Conclusions
Deployment of in vitro gut models, humanized mouse models, and human intervention trials, in combination with deployment of metabolomics and microbiomics, is a prerequisite for unraveling the role of colonic microbiota in the bioconversion of polyphenols. Longitudinal nutrikinetic type interventions are useful for distinguishing human subpopulations that differ in their microbiota-mediated bioavailability. Bottom-up as well as top-down strategies need to be pursued for linking gut microbial diversity to nutritional phenotypes and bioactivity of polyphenols. Ultimately, this approach will realize opportunities and direct strategies for personalization of nutrition based on dietary modulation of gut microbial functionality of individuals or populations.
Acknowledgments
Martin Foltz (Unilever) is thanked for providing useful comments on the manuscript. We acknowledge the financial support of the European Community under the Framework 6 Marie-Curie Host Fellowships for the Transfer of Knowledge Industry-Academia Strategic Partnership scheme, specifically GUTSYSTEM Project MTKI-CT-2006-042786. Sam Possemiers and Tom Van de Wiele are postdoctoral research fellows of the Flemish Science Foundation (FWO-Vlaanderen). Part of this project was carried out within the research program of the Netherlands Metabolomics Centre, which is part of the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Microbes and Health” held November 2-3, 2009, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS Web site at http://www.nasonline.org/SACKLER_Microbes_and_Health.
This article is a PNAS Direct Submission.
References
- 1.Manach C, Scalbert A, Morand C, Rémésy C, Jiménez L. Polyphenols: Food sources and bioavailability. Am J Clin Nutr. 2004;79:727–747. doi: 10.1093/ajcn/79.5.727. [DOI] [PubMed] [Google Scholar]
- 2.Manach C, Mazur A, Scalbert A. Polyphenols and prevention of cardiovascular diseases. Curr Opin Lipidol. 2005;16:77–84. doi: 10.1097/00041433-200502000-00013. [DOI] [PubMed] [Google Scholar]
- 3.Arts IC, Hollman PC. Polyphenols and disease risk in epidemiologic studies. Am J Clin Nutr. 2005;81(1, Suppl):317S–325S. doi: 10.1093/ajcn/81.1.317S. [DOI] [PubMed] [Google Scholar]
- 4.Hodgson J, Kroft KD. Dietary flavonoids: Effects on endothelial function and blood pressure. J Sci Food Agric. 2006;86:2492–2498. [Google Scholar]
- 5.Scalbert A, Manach C, Morand C, Rémésy C, Jiménez L. Dietary polyphenols and the prevention of diseases. Crit Rev Food Sci Nutr. 2005;45:287–306. doi: 10.1080/1040869059096. [DOI] [PubMed] [Google Scholar]
- 6.Higdon JV, Frei B. Tea catechins and polyphenols: Health effects, metabolism, and antioxidant functions. Crit Rev Food Sci Nutr. 2003;43:89–143. doi: 10.1080/10408690390826464. [DOI] [PubMed] [Google Scholar]
- 7.Crozier A, Jaganath IB, Clifford MN. Dietary phenolics: Chemistry, bioavailability and effects on health. Nat Prod Rep. 2009;26:1001–1043. doi: 10.1039/b802662a. [DOI] [PubMed] [Google Scholar]
- 8.Lotito SB, Frei B. Consumption of flavonoid-rich foods and increased plasma antioxidant capacity in humans: Cause, consequence, or epiphenomenon? Free Radic Biol Med. 2006;41:1727–1746. doi: 10.1016/j.freeradbiomed.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 9.Walle T. Absorption and metabolism of flavonoids. Free Radic Biol Med. 2004;36:829–837. doi: 10.1016/j.freeradbiomed.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Manach C, Hubert J, Llorach R, Scalbert A. The complex links between dietary phytochemicals and human health deciphered by metabolomics. Mol Nutr Food Res. 2009;53:1303–1315. doi: 10.1002/mnfr.200800516. [DOI] [PubMed] [Google Scholar]
- 11.Forester SC, Waterhouse AL. Metabolites are key to understanding health effects of wine polyphenolics. J Nutr. 2009;139:1824S–1831S. doi: 10.3945/jn.109.107664. [DOI] [PubMed] [Google Scholar]
- 12.Scalbert A, Morand C, Manach C, Rémésy C. Absorption and metabolism of polyphenols in the gut and impact on health. Biomed Pharmacother. 2002;56:276–282. doi: 10.1016/s0753-3322(02)00205-6. [DOI] [PubMed] [Google Scholar]
- 13.Lederberg J. Infectious history. Science. 2000;288:287–293. doi: 10.1126/science.288.5464.287. [DOI] [PubMed] [Google Scholar]
- 14.Nicholson JK, Holmes E, Wilson ID. Gut microorganisms, mammalian metabolism and personalized health care. Nat Rev Microbiol. 2005;3:431–438. doi: 10.1038/nrmicro1152. [DOI] [PubMed] [Google Scholar]
- 15.Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci USA. 2009;106:14728–14733. doi: 10.1073/pnas.0904489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clayton TA, et al. Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature. 2006;440:1073–1077. doi: 10.1038/nature04648. [DOI] [PubMed] [Google Scholar]
- 17.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2008;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zoetendal EG, Vaughan EE, de Vos WM. A microbial world within us. Mol Microbiol. 2006;59:1639–1650. doi: 10.1111/j.1365-2958.2006.05056.x. [DOI] [PubMed] [Google Scholar]
- 19.Cermak R, Breves GM, Lupke M, Wolffram S. In vitro degradation of the flavonol quercetin and of quercetin glycosides in the porcine hindgut. Arch Anim Nutr. 2006;60:180–189. doi: 10.1080/17450390500467695. [DOI] [PubMed] [Google Scholar]
- 20.Rechner AR, et al. Colonic metabolism of dietary polyphenols: Influence of structure on microbial fermentation products. Free Radic Biol Med. 2004;36:212–225. doi: 10.1016/j.freeradbiomed.2003.09.022. [DOI] [PubMed] [Google Scholar]
- 21.Felgines C, et al. Bioavailability of the flavanone naringenin and its glycosides in rats. Am J Physiol Gastrointest Liver Physiol. 2000;279:G1148–G1154. doi: 10.1152/ajpgi.2000.279.6.G1148. [DOI] [PubMed] [Google Scholar]
- 22.Brune A, Schink B. Phloroglucinol pathway in the strictly anaerobic Pelobacter acidigalli: Fermentation of trihydroxybenzenes to acetate via triacetic acid. Arch Microbiol. 1992;157:417–424. [Google Scholar]
- 23.Blaut M, Clavel T. Metabolic diversity of the intestinal microbiota: Implications for health and disease. J Nutr. 2007;137(3, Suppl 2):751S–755S. doi: 10.1093/jn/137.3.751S. [DOI] [PubMed] [Google Scholar]
- 24.Simons AL, Renouf M, Hendrich S, Murphy PA. Human gut microbial degradation of flavonoids: Structure-function relationships. J Agric Food Chem. 2005;53:4258–4263. doi: 10.1021/jf0500177. [DOI] [PubMed] [Google Scholar]
- 25.Erlund I. Review of the flavonoids quercetin, hesperetin, and naringenin. Dietary sources, bioactivities, bioavailability, and epidemiology. Nutr Res. 2004;24:851–874. [Google Scholar]
- 26.Schoefer L, Mohan R, Braune A, Birringer M, Blaut M. Anaerobic C-ring cleavage of genistein and daidzein by Eubacterium ramulus. FEMS Microbiol Lett. 2002;208:197–202. doi: 10.1111/j.1574-6968.2002.tb11081.x. [DOI] [PubMed] [Google Scholar]
- 27.Roowi S, Mullen W, Edwards CA, Crozier A. Yoghurt impacts on the excretion of phenolic acids derived from colonic breakdown of orange juice flavanones in humans. Mol Nutr Food Res. 2009;53(Suppl 1):S68–S75. doi: 10.1002/mnfr.200800287. [DOI] [PubMed] [Google Scholar]
- 28.Possemiers S, et al. The prenylflavonoid isoxanthohumol from hops (Humulus lupulus L.) is activated into the potent phytoestrogen 8-prenylnaringenin in vitro and in the human intestine. J Nutr. 2006;136:1862–1867. doi: 10.1093/jn/136.7.1862. [DOI] [PubMed] [Google Scholar]
- 29.Rowland I, Wiseman H, Sanders T, Adlercreutz H, Bowey E. Metabolism of oestrogens and phytoestrogens: Role of the gut microflora. Biochem Soc Trans. 1999;27:304–308. doi: 10.1042/bst0270304. [DOI] [PubMed] [Google Scholar]
- 30.Spencer JP, Abd El Mohsen MM, Minihane AM, Mathers JC. Biomarkers of the intake of dietary polyphenols: Strengths, limitations and application in nutrition research. Br J Nutr. 2008;99:12–22. doi: 10.1017/S0007114507798938. [DOI] [PubMed] [Google Scholar]
- 31.van Velzen EJ, et al. Phenotyping tea consumers by nutrikinetic analysis of polyphenolic end-metabolites. J Proteome Res. 2009;8:3317–3330. doi: 10.1021/pr801071p. [DOI] [PubMed] [Google Scholar]
- 32.Selma MV, Espín JC, Tomás-Barberán FA. Interaction between phenolics and gut microbiota: Role in human health. J Agric Food Chem. 2009;57:6485–6501. doi: 10.1021/jf902107d. [DOI] [PubMed] [Google Scholar]
- 33.Van Dorsten FA, et al. The metabolic fate of red wine and grape juice polyphenols in humans assessed by metabolomics. Mol Nutr Food Res. 2009 doi: 10.1002/mnfr.200900212. 10.1002/mnfr.200900212. [DOI] [PubMed] [Google Scholar]
- 34.Winnike JH, Busby MG, Watkins PB, O'Connell TM. Effects of a prolonged standardized diet on normalizing the human metabolome. Am J Clin Nutr. 2009;90:1496–1501. doi: 10.3945/ajcn.2009.28234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walsh MC, et al. Influence of acute phytochemical intake on human urinary metabolomic profiles. Am J Clin Nutr. 2007;86:1687–1693. doi: 10.1093/ajcn/86.5.1687. [DOI] [PubMed] [Google Scholar]
- 36.van Velzen EJJ, et al. Multilevel data analysis of a crossover designed human nutritional intervention study. J Proteome Res. 2008;7:4483–4491. doi: 10.1021/pr800145j. [DOI] [PubMed] [Google Scholar]
- 37.Atkinson C, Frankenfeld CL, Lampe JW. Gut bacterial metabolism of the soy isoflavone daidzein: Exploring the relevance to human health. Exp Biol Med (Maywood) 2005;230:155–170. doi: 10.1177/153537020523000302. [DOI] [PubMed] [Google Scholar]
- 38.Solanky KS, et al. Biofluid 1H NMR-based metabonomic techniques in nutrition research—metabolic effects of dietary isoflavones in humans. J Nutr Biochem. 2005;16:236–244. doi: 10.1016/j.jnutbio.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 39.Van Dorsten FA, Daykin CA, Mulder TP, Van Duynhoven JP. Metabonomics approach to determine metabolic differences between green tea and black tea consumption. J Agric Food Chem. 2006;54:6929–6938. doi: 10.1021/jf061016x. [DOI] [PubMed] [Google Scholar]
- 40.van Ommen B, Keijer J, Heil SG, Kaput J. Challenging homeostasis to define biomarkers for nutrition related health. Mol Nutr Food Res. 2009;53:795–804. doi: 10.1002/mnfr.200800390. [DOI] [PubMed] [Google Scholar]
- 41.Bose M, et al. The major green tea polyphenol, (-)-epigallocatechin-3-gallate, inhibits obesity, metabolic syndrome, and fatty liver disease in high-fat-fed mice. J Nutr. 2008;138:1677–1683. doi: 10.1093/jn/138.9.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang CS, Wang X, Lu G, Picinich SC. Cancer prevention by tea: Animal studies, molecular mechanisms and human relevance. Nat Rev Cancer. 2009;9:429–439. doi: 10.1038/nrc2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dolara P, et al. Red wine polyphenols influence carcinogenesis, intestinal microflora, oxidative damage and gene expression profiles of colonic mucosa in F344 rats. Mutat Res. 2005;591:237–246. doi: 10.1016/j.mrfmmm.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 44.Smith AH, Mackie RI. Effect of condensed tannins on bacterial diversity and metabolic activity in the rat gastrointestinal tract. Appl Environ Microbiol. 2004;70:1104–1115. doi: 10.1128/AEM.70.2.1104-1115.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee HC, Jenner AM, Low CS, Lee YK. Effect of tea phenolics and their aromatic fecal bacterial metabolites on intestinal microbiota. Res Microbiol. 2006;157:876–884. doi: 10.1016/j.resmic.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 46.Clavel T, et al. Isoflavones and functional foods alter the dominant intestinal microbiota in postmenopausal women. J Nutr. 2005;135:2786–2792. doi: 10.1093/jn/135.12.2786. [DOI] [PubMed] [Google Scholar]
- 47.Ley RE, et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bowey E, Adlercreutz H, Rowland I. Metabolism of isoflavones and lignans by the gut microflora: A study in germ-free and human flora associated rats. Food Chem Toxicol. 2003;41:631–636. doi: 10.1016/s0278-6915(02)00324-1. [DOI] [PubMed] [Google Scholar]
- 49.Lhoste EF, et al. The human colonic microflora influences the alterations of xenobiotic-metabolizing enzymes by catechins in male F344 rats. Food Chem Toxicol. 2003;41:695–702. doi: 10.1016/s0278-6915(03)00010-3. [DOI] [PubMed] [Google Scholar]
- 50.Hanske L, Loh G, Sczesny S, Blaut M, Braune A. The bioavailability of apigenin-7-glucoside is influenced by human intestinal microbiota in rats. J Nutr. 2009;139:1095–1102. doi: 10.3945/jn.108.102814. [DOI] [PubMed] [Google Scholar]
- 51.Gonthier MP, et al. Microbial aromatic acid metabolites formed in the gut account for a major fraction of the polyphenols excreted in urine of rats fed red wine polyphenols. J Nutr. 2003;133:461–467. doi: 10.1093/jn/133.2.461. [DOI] [PubMed] [Google Scholar]
- 52.Jacobs DM, Gaudier E, van Duynhoven J, Vaughan EE. Non-digestible food ingredients, colonic microbiota and the impact on gut health and immunity: A role for metabolomics. Curr Drug Metab. 2009;10:41–54. doi: 10.2174/138920009787048383. [DOI] [PubMed] [Google Scholar]
- 53.Bolca S, et al. Microbial and dietary factors associated with the 8-prenylnaringenin producer phenotype: A dietary intervention trial with fifty healthy post-menopausal Caucasian women. Br J Nutr. 2007;98:950–959. doi: 10.1017/S0007114507749243. [DOI] [PubMed] [Google Scholar]
- 54.Macfarlane GT, Macfarlane S. Models for intestinal fermentation: Association between food components, delivery systems, bioavailability and functional interactions in the gut. Curr Opin Biotechnol. 2007;18:156–162. doi: 10.1016/j.copbio.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 55.Aura AM, et al. In vitro metabolism of anthocyanins by human gut microflora. Eur J Nutr. 2005;44:133–142. doi: 10.1007/s00394-004-0502-2. [DOI] [PubMed] [Google Scholar]
- 56.Tzounis X, et al. Flavanol monomer-induced changes to the human faecal microflora. Br J Nutr. 2008;99:782–792. doi: 10.1017/S0007114507853384. [DOI] [PubMed] [Google Scholar]
- 57.Gibson GR, Cummings JH, Macfarlane GT. Use of a three-stage continuous culture system to study the effect of mucin on dissimilatory sulfate reduction and methanogenesis by mixed populations of human gut bacteria. Appl Environ Microbiol. 1988;54:2750–2755. doi: 10.1128/aem.54.11.2750-2755.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Molly K, Vande Woestyne M, Verstraete W. Development of a 5-step multi-chamber reactor as a simulation of the human intestinal microbial ecosystem. Appl Microbiol Biotechnol. 1993;39:254–258. doi: 10.1007/BF00228615. [DOI] [PubMed] [Google Scholar]
- 59.Minekus M, et al. A computer-controlled system to simulate conditions of the large intestine with peristaltic mixing, water absorption and absorption of fermentation products. Appl Microbiol Biotechnol. 1999;53:108–114. doi: 10.1007/s002530051622. [DOI] [PubMed] [Google Scholar]
- 60.Macfarlane GT, Macfarlane S, Gibson GR. Validation of a three-stage compound continuous culture system for investigating the effect of retention time on the ecology and metabolism of bacteria in the human colon. Microb Ecol. 1998;35:180–187. doi: 10.1007/s002489900072. [DOI] [PubMed] [Google Scholar]
- 61.Possemiers S, et al. Eubacterium limosum activates isoxanthohumol from hops (Humulus lupulus L.) into the potent phytoestrogen 8-prenylnaringenin in vitro and in rat intestine. J Nutr. 2008;138:1310–1316. doi: 10.1093/jn/138.7.1310. [DOI] [PubMed] [Google Scholar]
- 62.Eeckhaut E, et al. Metabolism of the lignan macromolecule into enterolignans in the gastrointestinal lumen as determined in the simulator of the human intestinal microbial ecosystem. J Agric Food Chem. 2008;56:4806–4812. doi: 10.1021/jf800101s. [DOI] [PubMed] [Google Scholar]
- 63.Schoefer L, Mohan R, Schwiertz A, Braune A, Blaut M. Anaerobic degradation of flavonoids by Clostridium orbiscindens. Appl Environ Microbiol. 2003;69:5849–5854. doi: 10.1128/AEM.69.10.5849-5854.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Clavel T, Borrmann D, Braune A, Doré J, Blaut M. Occurrence and activity of human intestinal bacteria involved in the conversion of dietary lignans. Anaerobe. 2006;12:140–147. doi: 10.1016/j.anaerobe.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 65.Decroos K, Eeckhaut E, Possemiers S, Verstraete W. Administration of equol-producing bacteria alters the equol production status in the Simulator of the Gastrointestinal Microbial Ecosystem (SHIME) J Nutr. 2006;136:946–952. doi: 10.1093/jn/136.4.946. [DOI] [PubMed] [Google Scholar]
- 66.Fattouch S, et al. Comparative analysis of polyphenolic profiles and antioxidant and antimicrobial activities of tunisian pome fruit pulp and peel aqueous acetone extracts. J Agric Food Chem. 2008;56:1084–1090. doi: 10.1021/jf072409e. [DOI] [PubMed] [Google Scholar]
- 67.Van den Abbeele P, et al. In vitro model to study the modulation of the mucin-adhered bacterial community. Appl Microbiol Biotechnol. 2009;83:349–359. doi: 10.1007/s00253-009-1947-2. [DOI] [PubMed] [Google Scholar]
- 68.Palmer C, et al. Rapid quantitative profiling of complex microbial populations. Nucleic Acids Res. 2006;34:e5. doi: 10.1093/nar/gnj007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rajilić-Stojanović M, et al. Development and application of the human intestinal tract chip, a phylogenetic microarray: Analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environ Microbiol. 2009;11:1736–1751. doi: 10.1111/j.1462-2920.2009.01900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Andersson AF, et al. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS ONE. 2008;3:e2836. doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Metzker ML. Sequencing technologies—the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 72.Ben-Amor K, Vaughan EE. In: Gastrointestinal Microbiology. Ouwehand A, Vaughan EE, editors. New York: Taylor & Francis Group; 2006. pp. 1–23. [Google Scholar]
- 73.Carey CM, Kirk JL, Ojha S, Kostrzynska M. Current and future uses of real-time polymerase chain reaction and microarrays in the study of intestinal microbiota, and probiotic use and effectiveness. Can J Microbiol. 2007;53:537–550. doi: 10.1139/W07-039. [DOI] [PubMed] [Google Scholar]
- 74.Neufeld JD, Wagner M, Murrell JC. Who eats what, where and when? Isotope-labelling experiments are coming of age. ISME J. 2007;1:103–110. doi: 10.1038/ismej.2007.30. [DOI] [PubMed] [Google Scholar]
- 75.Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol. 2009;587:4153–4158. doi: 10.1113/jphysiol.2009.174136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hamady M, Knight R. Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Res. 2009;19:1141–1152. doi: 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kurokawa K, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Beloqui A, et al. Novel polyphenol oxidase mined from a metagenome expression library of bovine rumen: Biochemical properties, structural analysis, and phylogenetic relationships. J Biol Chem. 2006;281:22933–22942. doi: 10.1074/jbc.M600577200. [DOI] [PubMed] [Google Scholar]
- 79.Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci USA. 2008;105:13580–13585. doi: 10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Qin JJ, et al. MetaHIT Consortium. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Perkins TT, et al. A strand-specific RNA-Seq analysis of the transcriptome of the typhoid bacillus Salmonella typhi. PLoS Genet. 2009;5:e1000569. doi: 10.1371/journal.pgen.1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zoetendal EG, et al. Isolation of RNA from bacterial samples of the human gastrointestinal tract. Nat Protoc. 2006;1:954–959. doi: 10.1038/nprot.2006.143. [DOI] [PubMed] [Google Scholar]
- 83.Warnecke F, Hess M. A perspective: Metatranscriptomics as a tool for the discovery of novel biocatalysts. J Biotechnol. 2009;142:91–95. doi: 10.1016/j.jbiotec.2009.03.022. [DOI] [PubMed] [Google Scholar]
- 84.Turnbaugh PJ, et al. The effect of diet on the human gut microbiome: A metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Klaassens ES, et al. Mixed-species genomic microarray analysis of fecal samples reveals differential transcriptional responses of bifidobacteria in breast- and formula-fed infants. Appl Environ Microbiol. 2009;75:2668–2676. doi: 10.1128/AEM.02492-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.VerBerkmoes NC, et al. Shotgun metaproteomics of the human distal gut microbiota. ISME J. 2009;3:179–189. doi: 10.1038/ismej.2008.108. [DOI] [PubMed] [Google Scholar]
- 87.Rudney JD, Xie H, Rhodus NL, Ondrey FG, Griffin TJ. A metaproteomic analysis of the human salivary microbiota by three-dimensional peptide fractionation and tandem mass spectrometry. Mol Oral Microbiol. 2010;25:38–49. doi: 10.1111/j.2041-1014.2009.00558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fardet A, et al. Metabolomics provide new insight on the metabolism of dietary phytochemicals in rats. J Nutr. 2008;138:1282–1287. doi: 10.1093/jn/138.7.1282. [DOI] [PubMed] [Google Scholar]
- 89.Manach C, Williamson G, Morand C, Scalbert A, Rémésy C. Bioavailability and bioefficacy of polyphenols in humans. I. Review of 97 bioavailability studies. Am J Clin Nutr. 2005;81(1, Suppl):230S–242S. doi: 10.1093/ajcn/81.1.230S. [DOI] [PubMed] [Google Scholar]
- 90.Williamson G, Manach C. Bioavailability and bioefficacy of polyphenols in humans. II. Review of 93 intervention studies. Am J Clin Nutr. 2005;81(1, Suppl):243S–255S. doi: 10.1093/ajcn/81.1.243S. [DOI] [PubMed] [Google Scholar]
- 91.Lindon JC, Nicholson JK. Analytical technologies for metabonomics and metabolomics, and multi-omic information recovery. Trends Analyt Chem. 2008;27:194–204. [Google Scholar]
- 92.Jacobs DM, et al. (1)H NMR metabolite profiling of feces as a tool to assess the impact of nutrition on the human microbiome. NMR Biomed. 2008;21:615–626. doi: 10.1002/nbm.1233. [DOI] [PubMed] [Google Scholar]
- 93.Nicholson JK, Lindon JC. Systems biology: Metabonomics. Nature. 2008;455:1054–1056. doi: 10.1038/4551054a. [DOI] [PubMed] [Google Scholar]
- 94.Grün CH, et al. GC-MS methods for metabolic profiling of microbial fermentation products of dietary polyphenols in human and in vitro intervention studies. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;871:212–219. doi: 10.1016/j.jchromb.2008.04.039. [DOI] [PubMed] [Google Scholar]
- 95.Westerhuis JA, et al. Assessment of PLSDA cross validation. Metabolomics. 2008;4:81–89. [Google Scholar]
- 96.Westerhuis J, van Velzen EJJ, Hoefsloot HCJ, Smilde AK. Discriminant Q2 (DQ2) for improved discrimination in PLSDA models. Metabolomics. 2008;4:293–296. [Google Scholar]
- 97.Westerhuis JA, van Velzen EJ, Hoefsloot HC, Smilde AK. Multivariate paired data analysis: Multilevel PLSDA versus OPLSDA. Metabolomics. 2010;6:119–128. doi: 10.1007/s11306-009-0185-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hodgson JM, Morton LW, Puddey IB, Beilin LJ, Croft KD. Gallic acid metabolites are markers of black tea intake in humans. J Agric Food Chem. 2000;48:2276–2280. doi: 10.1021/jf000089s. [DOI] [PubMed] [Google Scholar]
- 99.van Ommen B, et al. The challenges for molecular nutrition research 2: Quantification of the nutritional phenotype. Genes Nutr. 2008;3:51–59. doi: 10.1007/s12263-008-0084-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Turnbaugh PJ, Gordon JI. An invitation to the marriage of metagenomics and metabolomics. Cell. 2008;134:708–713. doi: 10.1016/j.cell.2008.08.025. [DOI] [PubMed] [Google Scholar]
- 101.Li M, et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci USA. 2008;105:2117–2122. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chun H, Keleş S. Sparse partial least squares regression for simultaneous dimension reduction and variable selection. J R Stat Soc Series B Stat Methodol. 2010;72:3–25. doi: 10.1111/j.1467-9868.2009.00723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Claus SP, et al. Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Mol Syst Biol. 2008;4:219. doi: 10.1038/msb.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Martin FP, et al. Panorganismal gut microbiome-host metabolic crosstalk. J Proteome Res. 2009;8:2090–2105. doi: 10.1021/pr801068x. [DOI] [PubMed] [Google Scholar]
- 105.Duarte NC, et al. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci USA. 2007;104:1777–1782. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Westerhuis JA, Derks EPPA, Hoefsloot HCJ, Smilde AK. Grey component analysis. J Chemom. 2007;21:474–485. [Google Scholar]
- 107.Russell WR, Labat A, Scobbie L, Duncan SH. Availability of blueberry phenolics for microbial metabolism in the colon and the potential inflammatory implications. Mol Nutr Food Res. 2007;51:726–731. doi: 10.1002/mnfr.200700022. [DOI] [PubMed] [Google Scholar]
- 108.Monagas M, et al. Dihydroxylated phenolic acids derived from microbial metabolism reduce lipopolysaccharide-stimulated cytokine secretion by human peripheral blood mononuclear cells. Br J Nutr. 2009;102:201–206. doi: 10.1017/S0007114508162110. [DOI] [PubMed] [Google Scholar]
- 109.Kroon PA, et al. How should we assess the effects of exposure to dietary polyphenols in vitro? Am J Clin Nutr. 2004;80:15–21. doi: 10.1093/ajcn/80.1.15. [DOI] [PubMed] [Google Scholar]
- 110.García-Cañas V, Simó C, León C, Cifuentes A. Advances in Nutrigenomics research: Novel and future analytical approaches to investigate the biological activity of natural compounds and food functions. J Pharm Biomed Anal. 2010;51:290–304. doi: 10.1016/j.jpba.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 111.Niculescu MD, Pop EA, Fischer LM, Zeisel SH. Dietary isoflavones differentially induce gene expression changes in lymphocytes from postmenopausal women who form equol as compared with those who do not. J Nutr Biochem. 2007;18:380–390. doi: 10.1016/j.jnutbio.2006.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Turnbaugh PJ, et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mullard A. Microbiology: The inside story. Nature. 2008;453:578–580. doi: 10.1038/453578a. [DOI] [PubMed] [Google Scholar]