Abstract
Human milk contains an unexpected abundance and diversity of complex oligosaccharides apparently indigestible by the developing infant and instead targeted to its cognate gastrointestinal microbiota. Recent advances in mass spectrometry-based tools have provided a view of the oligosaccharide structures produced in milk across stages of lactation and among human mothers. One postulated function for these oligosaccharides is to enrich a specific “healthy” microbiota containing bifidobacteria, a genus commonly observed in the feces of breast-fed infants. Isolated culture studies indeed show selective growth of infant-borne bifidobacteria on milk oligosaccharides or core components therein. Parallel glycoprofiling documented that numerous Bifidobacterium longum subsp. infantis strains preferentially consume small mass oligosaccharides that are abundant early in the lactation cycle. Genome sequencing of numerous B. longum subsp. infantis strains shows a bias toward genes required to use mammalian-derived carbohydrates by comparison with adult-borne bifidobacteria. This intriguing strategy of mammalian lactation to selectively nourish genetically compatible bacteria in infants with a complex array of free oligosaccharides serves as a model of how to influence the human supraorganismal system, which includes the gastrointestinal microbiota.
Keywords: glycoprofiling, human milk oligosaccharides, infant microbiota, Bifidobacterium, diet
The interaction of humans with microorganisms remains one of the most important relationships to both acute survival and long-term health. Humans emerged into a microbial world, and the microbial world continues to shape human evolutionary progress. For example, successes such as the discovery and application of small-molecule antibiotics have not only saved lives, but by intervening in the fundamental relationships between humans and microbes they have imposed selection pressures on the evolution of microorganisms. Understanding how to manage microbial biology in the future will require more sophisticated tools aimed at modifying microbial populations and functions toward human health benefits other than simply preventing pathogenic infection. Insights into how to guide human/microbial interactions to be net favorable for both are needed.
The connection between human breast milk and infants’ growth, development, and health exemplifies this link. Human milk is the culmination of 200 million years of Darwinian pressure on mammalian lactation as the sole source of early infant nourishment. Human milk components not only nourish the infant, they provide myriad bioactive compounds for the offspring that influence the growth, stimulation, and modulation of the immune system, cognitive development, protection from toxins and pathogenic diseases, and perhaps most remarkably, the establishment of the intestinal microbiota (1–3). Considerable efforts made to understand the biology of human milk and its effects on the infant (4) are beginning to elucidate the structure/function properties and benefits that milk provides.
The constant evolutionary pressure on milk as the sole source of nourishment for mammalian infants has resulted in a remarkable model for how diet affects all aspects of development and health. Maternal investment, specifically the composition of breast milk, has been shaped by natural selection acting on both the infant and the mother, maximizing infant survival, growth, and activity while minimizing the costs of lactation for the mother (5). One of the most remarkable apparent functions of breast milk is the selective colonization and support of a protective microbiota. How milk has been able to attain the goal of guiding the evolutionary emergence of specific strains of bacteria in infants for their mutual health benefit is precisely the kind of question that science needs to understand to achieve similar successes for a wide range of human/microbial interactions.
Human milk/colostrum contains between 5 and 23 g/L (6, 7) of oligosaccharides containing a lactose-reducing end elongated with fucosylated and/or sialylated N-acetyllactosamine units (8). This translates to over 200 different human milk oligosaccharide (HMO) structures that differ in their size, charge, and sequence (8).
Despite the fact that oligosaccharides are the third most abundant components in milk (after lactose and lipids; Fig. 1), they were long thought to have no biological significance. It is now known that oligosaccharides in milk are important for the healthy growth of infants (9–11). Certain HMOs derived from the mammary epithelial cells of the mother also share common structural motifs with glycans on the infant's intestinal epithelia known to be receptors for pathogens. The presence of such structures in milk implies a defensive strategy, with glycans acting as decoys to prevent binding of pathogens to epithelial cells, thereby protecting infants from disease (11). Consistent with these multiple functions, human milk is comprised of a complex mixture of oligosaccharides that differ in size, charge, and abundance (8). Several of the characteristic HMO structures and their isomers found in pooled samples of human milk are shown in Fig. 2. A direct mechanistic link between specific HMO structures and bifidobacterial growth has recently been established and will be discussed in this review (12–19).
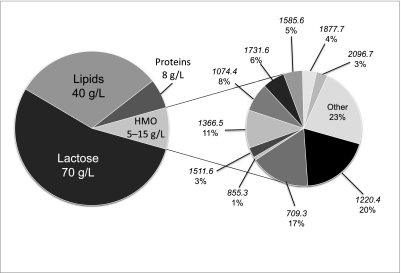
Fig. 1.
Human milk composition. (Left) Macronutrient composition of pooled human milk, with lactose being the most abundant component at 70 g/L, followed by lipids at 40 g/L. The third most abundant component is HMO at an estimated 5–15 g/L, followed by protein at 8 g/L. (Right) Pull-out pie chart showing composition of the most abundant HMOs. Masses of individual HMO structures are shown, along with their relative abundances, which were calculated from peak intensities presented in Ninonuevo et al. (21).
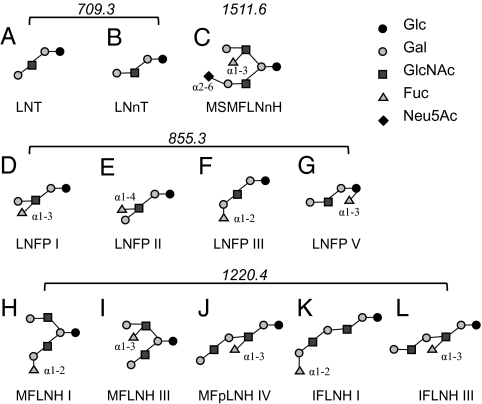
Fig. 2.
Characteristic HMO structures. The structures of several characteristic HMO structures found across human milk samples, along with their isomers, are shown. Two isomers with mass 709.3: (A) LNT, lacto-N-tetraose and (B) LNnT, lacto-N-neotetraose; (C) mass 1511.6: MSMFLNnH, monofucosylmonosialyllacto-N-hexaose; four isomers with mass 855.3: (D) LNFP I, lacto-N-fucopentaose, (E) LNFP II, (F) LNFP III, and (G) LNFP V; and five isomers with mass 1220.4: (H) MFLNH I, monofucosyllacto-N-hexaose, (I) MFLNH III, (J) MFpLNH IV, monofucosyl-paralacto-N-hexaose, (K) IFLNH I, isomer 1 fucosyl-paralacto-N-hexaose, and (L) IFLNH III, isomer 3 fucosyl-paralacto-N-hexaose. Glc, D-glucose; Gal, D-galactose; GlcNAc, N-acetylglucosamine; Fuc, L-fucose; Neu5Ac, N-acetyl-neuraminic acid.
Novel Analytical Tools: Structural Determination of HMO
The basic structure of HMOs includes a lactose core at the reducing end, which is elongated by N-acetyllactosamine units, with greater structural diversity provided by extensive fucosylation and/or sialylation wherein fucose and sialic acid residues are added at the terminal positions. Lactose, the most abundant component of milk, is a disaccharide composed of a galactose β-linked to the 4 position of glucose (Galβ1–4Glu) (6). HMOs are composed of both neutral and anionic species with building blocks of five monosaccharides: D-glucose, D-galactose, N-acetylglucosamine, L-fucose, and N-acetylneuraminic acid (Fig. 2). The estimated number of oligosaccharides ranges from a few hundred to the thousands (20); however, a recent study showed that a few of the most abundant HMO species accounted for as much as 80% of the total peak intensity (21).
Key to understanding the structure-function relationships of oligosaccharides has been the development of new and sensitive tools for structural analysis. Current analytical methods to characterize oligosaccharides in human milk include HPLC, high-pH anion exchange chromatography (HPAEC), capillary electrophoresis (CE), NMR, and MS (22–30). MS is evolving as the preferred method for elucidating HMO composition because it provides high sensitivity with structural information. A unique analytical strategy to rapidly profile oligosaccharides in human milk using HPLC-Chip/time-of-flight (TOF) MS technology has recently been developed (8, 31). This analytical technique uses an integrated microfluidic chip coupled with a high mass accuracy TOF mass analyzer. The HPLC-Chip/MS system replaces the traditional column and regular fittings in standard LC/MS systems with an integrated microfluidic chip, providing effective separation with significantly improved sensitivity and reproducibility.
Due to the nature of chemical structures of the oligosaccharides, a single atomic composition (or a single mass) can be made up of several structural isomers (Fig. 2). Variations in structures of isomers can range from linkage isomers (α1–2 vs. β1–2 or vs. α1–4) to variations in branching and positional isomers. The number of isomers can be large—>10 in some cases—with many of the isomers in relatively equal abundances. The extent and roles of this oligosaccharide heterogeneity in milk, or in any biological mixture, are still not well understood (32), primarily because the complexity of the mixture makes it difficult to monitor individual isomers and elucidate their functions.
A systematic method to elucidate the HMO structures has been developed (8). The HMO are first separated by standard HPLC into several fractions, and each fraction is then examined by both matrix-assisted laser desorption/ionization (MALDI) Fourier transform ion cyclotron resonance (FT-ICR) MS and HPLC-Chip/TOF MS to determine the number of structures in each fraction as well as their HPLC-Chip/TOF MS retention times. Tandem mass spectra of each component are obtained with either infrared multiphoton dissociation (IRMPD) or collision-induced dissociation (CID). The fragmentation patterns are used to elucidate the sequences of unknown structures. The exact linkages between each residue and the identity of the monosaccharide residue are then sequentially determined using a suite of biologically relevant exoglycosidases in a stepwise manner followed by MS after each step to evaluate the course of the reaction. Using this process, a complete library of structures found in human milk is being constructed. The combination of retention times, accurate masses, and tandem MS are being compiled to develop a rapid identification of milk oligosaccharides.
Structural Diversity and Functional Implications
An emerging feature of the structural analysis of oligosaccharides in human milk is the diversity among and within women. Approximately 200 molecular species have been identified in pooled human milk samples. These structures consist of neutral and acidic oligosaccharides containing a high degree of fucosylation, and to a lesser extent, sialylation (8). HMOs are terminated by fucose and sialic acid, and even in this aspect vary in the range of 50–70% fucosylated and 5–15% sialylated. Daily profiles of HMOs reflect the fluctuations between and within lactation in individual mothers (8). These results were supported by reports from other laboratories that showed similarly large heterogeneity in milk oligosaccharides using other methods, including HPLC and HPAEC methods but monitoring less than 20 oligosaccharide components (33, 34). This description of the basic composition of HMOs in humans will need to be broadened to an equally detailed understanding of the relationship between the levels of specific milk oligosaccharides and the specific functions these biomolecules contribute to maternal and infant health and development.
Several characteristic oligosaccharides (Fig. 2) illustrate the variation among individual mothers. For some mothers, the most dominant component lacto-N-neotetraose (LNnT; mass 709.3) can be 10× more intense than the next most abundant lacto-N-fucopentaose I/V (LNFP I/V; mass 855.3). For others, the three most abundant components—LNnT, lacto-N-tetraose (LNT; mass 709.3), and LNFP I/V—make up over 50% of the total. Among all samples analyzed to date, a neutral oligosaccharide with neutral mass 709.3 Da (3Hex, 1HexNAc;LNnT) is the most prominent. Importantly, this specific molecule was found to be preferentially consumed by several bifidobacterial strains (13). The next most common structures consist of fucosylated oligosaccharides with masses of 855.3 Da and 1220.4 Da (found in all five donors), and 1511.6 Da, a fucosylated species with a sialic acid residue. These specific HMOs are likely candidates for the important biological roles of milk in maintaining a healthy gut microbiota and in the prevention of pathogenic diseases among infants, as previously reported (6, 11, 18, 35).
The reasons for variation in HMOs among women have not yet been fully explored, and could be related to diet, lifestyle, ethnicity, and other factors. One factor that appears to be related to the variation among mothers is secretor status. Secretors have oligosaccharides that are more common to other secretors and different from those that are nonsecretors. Secretors produce Fuc1-2 motifs in secreted oligosaccharides. Nonsecretors do not have the gene that produces this motif. Although the secretor status of mothers has not been associated with infant outcomes, the ability of individuals to resist viral and bacterial infection has been correlated with their secretor status (36, 37). Kobata suggested a close relationship between the structures of milk oligosaccharides and the Lewis blood group systems (38). Thurl et al. (39) further observed that HMOs can be used to separate the subjects into four human milk groups corresponding to the presence of Lewis A and B. Lewis group blood types are named because of the fucose-containing glycans found on the surfaces of erythrocytes and in glycoproteins of secreted fluids. Lewis motifs are found on the surfaces of cells and have been implicated in many diseases, from cancer to infection. It has been shown, for example, that Lewis B exhibits preferential binding to pathogens, specifically Helicobacter pylori (40). Lewis-related antigens have been shown to have prognostic value for diseases as diverse as cancer and celiac disease (41). The Lewis structures are not synthesized in erythroblasts but are believed to be produced in the gut epithelium where they are shed into the digestive tract, digested, reabsorbed, and transported as glycolipids into the plasma where they are absorbed onto red blood cells (42). Lewis groups are not present on the erythrocytes of newborns, but appear between 3 and 6 mo and stabilize in concentration between 3 and 6 y (41). It is perhaps no coincidence that the mother administers large doses of Lewis structures to the infant during breast-feeding.
Fucosylation and sialylation are involved in structural motifs such as those belonging to the Lewis groups, specifically A, B, X, Y, and sialyl Lewis X. Many of these motifs are found in HMOs. Fucose and sialic acids both have distinct masses that can be readily obtained from the accurate mass. A mass profile of the mixture therefore provides a rapid method for determining the extent of fucosylation in HMO. The possibility that these differences relate to variations in the consequences of different human milks to the colonization, development, and net health properties of individual infants’ microbiota is compelling from these data. With these new analytical techniques in hand it will be possible to address important biological questions related to the fundamental functions of HMO: How do they affect the basic microbial ecology of the human infant intestine?
Milk Oligosaccharide Interactions with Bacteria
Oligosaccharides are known to interact directly with the surfaces of bacteria. Because the mechanism of milk oligosaccharide production by mammary cells involves the same enzymes as those for glycoproteins and glycolipids (10), it is believed that HMOs contain the same structural moieties as cell surface glycoconjugates. Bacterial surfaces have oligosaccharide binding proteins, including lectins and other target receptors such as Toll-like receptors (43). Though oligosaccharide binding to the surfaces of commensal bacteria is not well investigated, there is a larger body of work studying glycan binding with pathogenic bacteria. Various oligosaccharides and glycoconjugates in milk are believed to inhibit the binding of pathogenic bacteria and toxins, presumably by acting as decoys and binding to the bacterial surface, thus inhibiting their ability to bind to target oligosaccharides on the surface of epithelial cells (10, 11). Antiadhesive activity of free HMOs has been described for Streptococcus pneumoniae (44), enterpathogenic E. coli (45, 46), Listeria monocytogenes (47), Vibrio cholerae (48), Salmonella fyris (48), and HIV (49). Milk glycolipids or glycoproteins have also been implicated in protective mechanisms against pathogens such as Pseudomonas aeruginosa (50), Noroviruses (36), Cholera and Shiga toxins (51), and Rotovirus (52).
It is the impressive repertoire of structural diversity present in the aggregate HMO pool that likely contributes to the diverse protective functions against a range of pathogens or toxins. Several studies have shown that discrete fractions of oligosaccharides in human milk can differentially inhibit pathogen adhesion. Newburg and coworkers (35) have shown that fucosylated HMOs inhibit binding of Campylobactor jejuni-cultured HEp-2 intestinal cells and to human intestinal mucosa, and also reduce C. jejuni colonization of mice. Others have shown that sialylated milk oligosaccharides can block adhesion of enterotoxic and uropathogenic E. coli to human erythrocytes (53).
In addition to deflection of pathogens, milk oligosaccharides are also believed to enrich a beneficial microbiota containing bifidobacteria, a species commonly found in the feces of breast-fed infants (54–56)—an observation that dates back over 100 y (57). A prominent bifidobacterial population in the feces of breast-fed infants has been reported from numerous culture-based and non–culture-based studies (58). More recent deep-sequencing approaches have challenged the notion of strict bifidobacterial predominance (59, 60). Regardless, bifidobacteria are often overrepresented in the breast-fed infant microbiome by comparison with their appearance in adults.
If milk oligosaccharides act as prebiotic substrates that help shape the microbial content of the infant gastrointestinal tract, one would predict that the commensal bacteria normally enriched in a breast-fed infant would possess the capacity to grow on these complex structures. This concept is not new. Gyorgy and coworkers (61) first identified N-acetyl-glucosamine containing oligosaccharides as the “bifidus factor” over 50 y ago. More recently we have shown that, among a limited number of gut-related bacteria tested (including Lactobacillus, Clostridium, Eubacterium E. coli, Veillonella, and Enterococcus isolates), only Bifidobacterium and Bacteriodes species were able to consume HMOs as a sole carbon source and achieve high cell densities (14, 19). However, the ability to vigorously grow on HMOs as a sole carbon source is variable and, importantly, does not extend to all bifidobacterial isolates (12, 13, 19). This capability to grow vigorously on HMOs appears to be most common among B. bifidum and B. longum subsp. infantis strains, whereas isolates of B. longum subsp. longum and B. breve show more moderate growth, and other strains of B. adolescentis and B. animales lack this ability altogether (12).
Other studies have shown a similar restricted growth phenotype of specific HMO components. Xiao et al. (62) showed that lacto-N-biose (LNB), a core HMO component in type 1 glycans, supported the growth of B. bifidum, B. breve, and B. longum (subsp. infantis and longum) but did not support the growth of B. adolescentis, B. animalis, B. catenulatum, B. dentium, B. angulatum, and B. pseudolongum. Moreover, LNB did not significantly enable the growth of other gastrointestinal tract isolates, including species of Clostridium, Bacteroides, Eubacterium, Lactobacillus, Ruminococcus, and Propionibacterium (63). In aggregate, these data show the selective nature of HMOs as a growth substrate and provide a conceptual basis for their selective and bifidogenic activity in situ.
Though only specific bifidobacteria are able to consume HMOs, different species appear to have developed different strategies for using HMOs as a growth substrate. Glycoprofiling of HMO consumption has revealed that B. longum subsp. infantis ATCC15697 preferentially consumed oligosaccharides with a degree of polymerization (DP) 7 or less (13). These lower-DP oligosaccharides represent the most abundant species of HMO isomers in pooled human milk (15), indicating a selective correspondence between what the mother secretes and what this bacterium consumes. Conversely, B. longum subsp. longum DJO10A and B. breve ATCC15700 consumed only a portion of a single, nonfucosylated/nonsialylated HMO species, LNnT. Although LNnT is an abundant HMO in breast milk, the amount consumed by B. longum subsp. longum and B. breve represents only a small portion of the overall HMO pool (13). B. breve ATCC15700 was not able to readily consume the bulk of HMO structures, but did grow on all of the monomer constituents of HMO (19), suggesting a possible cross-feeding capacity in the gastrointestinal tract via liberated monosaccharides.
The catabolic capacity of these bacteria toward HMO can also be measured by monitoring the sialidase and fucosidase activities required to deconstruct these complex glycan structures. Enzymatic assays showed that B. longum subsp. infantis has a higher sialidase activity when grown on lactose as compared with B. longum subsp. longum and B. breve, respectively (13). Even though we cannot exclude a minimal/nonspecific sialidase activity in B. longum subsp. longum and B. breve, these data suggest that B. longum subsp. infantis has an inherent and constitutive ability to process sialylated compounds. Furthermore, among the three strains tested, fucosidase activity was only present in B. longum subsp. infantis and was only detected upon growth on HMOs (13).
A different mode of catalytic activity toward HMO consumption is illustrated by B. bifidum, which exports a 1,2-α-fucosidase (AfcA) and a 1–3/4-α-fucosidase (AfcB) that defucosylate HMO structures (64, 65). An extracellular lacto-N-biosidase then liberates LNB from HMO species lacking fucosylated and sialylated residues. Wada et al. (66) showed this lacto-N-biosidase activity is minimally conserved in bifidobacteria, being present in select B. bifidum and B. longum subsp. longum strains. Upon release, LNB is transported into B. bifidum via an ABC transporter and an associated LNB-specific solute-binding lipoprotein (67, 68) whereby it is further processed and fed into the central metabolic pathway.
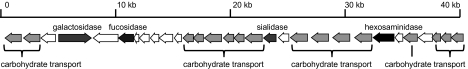
The recent genome sequencing of B. longum subsp. infantis ATCC15697, a prototypical HMO consumer, has greatly advanced understanding of the genetic underpinnings of this unusual phenotype (69). Strain ATCC15697 possesses a host of HMO-related genes clustered into four loci, including one large locus, HMO cluster 1, which contains all of the glycosidases (sialidase, fucosidase, galactosidase, and hexosaminidase) and transporters necessary for importing and metabolizing HMO (Fig. 3). Sequencing of several more isolates confirms this common HMO-related genomic architecture among B. longum subsp. infantis isolates and clearly provides a genetic rationale for the vigorous growth of this clade on HMOs. Strain ATCC15697 possesses both sialidase and fucosidase activities when grown on HMOs (13), and these results were corroborated by expression of the fucosidase and sialidase genes, Blon_2336 and Blon_2348, respectively, as observed via proteomics (17).
Fig. 3.
HMO-related gene cluster 1 from B. longum subsp. infantis ATCC15697. HMO gene cluster 1, shown here, contains all of the necessary glycosidases (sialidase, fucosidase, galactosidase, and hexosaminidase) and carbohydrate transporters necessary for importing and metabolizing HMOs.
A particularly interesting aspect of the large HMO cluster (Fig. 3) is the extensive repertoire of extracellular solute-binding proteins (SBP; pfam 01547) predicted to bind oligosaccharides. Six of these cluster I lipoproteins exhibit a pronounced evolutionary divergence relative to other SBP Family 1 proteins in bifidobacteria (17), suggesting a possible relationship with milk oligosaccharides. Interestingly, the B. longum subsp. infantis genome contains a total of 21 Family 1 SBP, roughly twice as many as observed in the B. longum subsp. longum or B. adolescentis genomes (17). Proteomics of HMO-grown B. longum subsp. infantis revealed expression of several Family 1 SBP from the large HMO cluster as well as two additional SBP that are located elsewhere on the genome.
Although the four HMO-related clusters are shared among B. longum subsp. infantis isolates, they are notably absent in other sequenced bifidobacteria, such as B. longum subsp. longum DJO10A (70) and B. adolescentis ATCC15703 (GenBank accession no. AP009256), which grow weakly or not all (respectively) on HMOs (12). Interestingly, one possible HMO-related gene set shared between ATCC15697 and DJO10A is the seven-gene operon responsible for LNB metabolism (71). Given that DJO10A is able to weakly grow on HMO, and glycoprofiling indicated a small consumption of LNnT, it is tempting to speculate that this operon is linked to consumption of that particular HMO moiety.
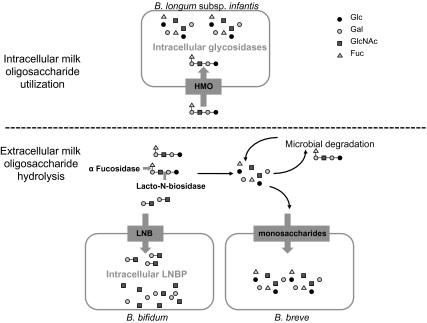
Although it is very hard to generalize the mechanisms of HMO catabolism across bifidobacteria because of strain heterogeneity and taxonomic confusion (72) within the genera, several important trends have emerged. The most common infant-borne bifidobacteria, B. bifidum, B. longum subsp. infantis, B. longum subsp. longum, and B. breve, possess different modes for consumption of HMO (Fig. 4). B. longum subsp. infantis likely imports the lower molecular-weight oligosaccharides via an army of dedicated SBP and ABC transporters. Once inside the cell, these oligosaccharides are catabolized by a complement of glycosidases before entry of the monosaccharides into central metabolic pathways. In contrast, B. bifidum exports fucosidases and lacto-N-biosidase to remove LNB from the HMO structure (leaving the free fucose behind) (19), internalize the free LNB, and catabolize it intracellularly. Both B. breve and B. longum subsp. longum are able to consume free LNnT from an HMO pool, whereas B. breve can also grow on the various monomer constituents of HMOs (13, 19). These different strategies suggest a possible mechanism for niche partitioning among the different bifidobacterial species within the developing infant gastrointestinal tract microbiota.
Fig. 4.
Strain-specific strategies for HMO import and catabolism. The most common infant-borne bifidobacteria, B. bifidum, B. longum subsp. infantis, and B. breve, possess different modes for consumption of HMO. B. longum subsp. infantis imports lower molecular-weight HMO via specific soluble binding proteins and transporters, followed by intracellular catabolism by a complement of glycosidases before entry of the monosaccharides into central metabolic pathways. B. bifidum exports fucosidases and lacto-N-biosidase for extracellular hydrolysis to remove lacto-N-biose (LNB) from the HMO structure, internalizes the free LNB, and catabolizes it intracellularly. B. breve consumes the various monomer constituents of HMO, imports them as monosaccharides, followed by intracellular catabolism. Glc, D-glucose; Gal, D-galactose; GlcNAc, N-acetylglucosamine; Fuc, L-fucose.
Conclusion
Microbial ecosystems are guided by a complex interplay of genetics and environment. Even the most narrowly defined ecosystem contains a complex array of microorganisms competing to maintain their respective niches. The human intestine is one such ecosystem in which the host represents an unusual dimension of the environment. The host dictates many of the conditions under which different bacteria compete. So, too, in an interesting sense is the microbiome part of each individual human's overall genotype. The concept of this mutually interactive community as a supraorganismal system is compelling (73). As scientists gain an increasingly detailed description of this supraorganismal system and its influence on human health, the opportunities to influence it will become increasingly attractive. However, two questions will be central to these opportunities. What direction should it change, and how? The answers to these questions will not be easy. Persistent, even minor, changes in a mature, established microbiota are difficult to achieve even with aggressive antibiotic treatments (74). When achieved, it is not clear whether they are in the long term net beneficial to the host's overall health. Human milk emerged from 200 million years of relentless Darwinian pressure to be nourishing to infants. The pervasive influence of bacteria and other microorganisms on the immunologically naïve infant was clearly a vital element of the evolutionary pressure on lactation. The evidence that is assembling from a wide variety of scientific disciplines points to oligosaccharide structures and their abundance as being a key element in evolution's strategy to establish and guide the human infant microbiome. Interestingly, weaning results in an immediate change in the infant microbiome (75). Weaning is also associated with an increased risk of a variety of intestine-related diseases (76). This basic strategy of selectively nourishing genetically compatible bacteria with a complex array of free oligosaccharides may well be the most immediate route to designing diets that are capable of influencing the human supraorganismal system for net health benefit.
Acknowledgments
We thank all the students, postdocs, and staff in the University of California–Davis Milk Bioactives Program (http://mbp.ucdavis.edu/) and Functional Glycobiology Program (http://fgp.ucdavis.edu/) for their hard work and inspiration. We specifically acknowledge David Sela and Mariana Barboza for their assistance in preparing this manuscript. This publication was made possible in part by support from the University of California Discovery Grant Program, the California Dairy Research Foundation, US Department of Agriculture National Research Initiative Cooperative State Research Education, and Extension Service Award 2008-35200-18776, National Institute on Environmental Health Sciences Superfund P42 ES02710, the Childhood Autism Risks from Genetics and the Environment Study Grant P01 ES11269, and National Institutes of Health–National Institute of Child Health and Human Development Awards 5R01HD059127 and 1R01HD061923.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Microbes and Health,” held November 2–3, 2009, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS Web site at http://www.nasonline.org/SACKLER_Microbes_and_Health.
This article is a PNAS Direct Submission.
References
- 1.Daniels MC, Adair LS. Breast-feeding influences cognitive development in Filipino children. J Nutr. 2005;135:2589–2595. doi: 10.1093/jn/135.11.2589. [DOI] [PubMed] [Google Scholar]
- 2.German JB, Dillard CJ, Ward RE. Bioactive components in milk. Curr Opin Clin Nutr Metab Care. 2002;5:653–658. doi: 10.1097/00075197-200211000-00007. [DOI] [PubMed] [Google Scholar]
- 3.Harmsen HJM, et al. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr. 2000;30:61–67. doi: 10.1097/00005176-200001000-00019. [DOI] [PubMed] [Google Scholar]
- 4.Schack-Nielsen L, Michaelsen KF. Advances in our understanding of the biology of human milk and its effects on the offspring. J Nutr. 2007;137:503S–510S. doi: 10.1093/jn/137.2.503S. [DOI] [PubMed] [Google Scholar]
- 5.Trivers RL. Parent-offspring conflict. Am Zool. 1974;14:249–264. [Google Scholar]
- 6.Kunz C, Rudloff S, Baier W, Klein N, Strobel S. Oligosaccharides in human milk: Structural, functional, and metabolic aspects. Annu Rev Nutr. 2000;20:699–722. doi: 10.1146/annurev.nutr.20.1.699. [DOI] [PubMed] [Google Scholar]
- 7.Coppa GV, et al. Changes in carbohydrate composition in human milk over 4 months of lactation. Pediatrics. 1993;91:637–641. [PubMed] [Google Scholar]
- 8.Ninonuevo MR, et al. A strategy for annotating the human milk glycome. J Agric Food Chem. 2006;54:7471–7480. doi: 10.1021/jf0615810. [DOI] [PubMed] [Google Scholar]
- 9.Bode L. Recent advances on structure, metabolism, and function of human milk oligosaccharides. J Nutr. 2006;136:2127–2130. doi: 10.1093/jn/136.8.2127. [DOI] [PubMed] [Google Scholar]
- 10.Newburg DS. Oligosaccharides in human milk and bacterial colonization. J Pediatr Gastroenterol Nutr. 2000;30(Suppl 2):S8–S17. [PubMed] [Google Scholar]
- 11.Newburg DS, Ruiz-Palacios GM, Morrow AL. Human milk glycans protect infants against enteric pathogens. Annu Rev Nutr. 2005;25:37–58. doi: 10.1146/annurev.nutr.25.050304.092553. [DOI] [PubMed] [Google Scholar]
- 12.LoCascio RG, et al. A versatile and scalable strategy for glycoprofiling bifidobacterial consumption of human milk oligosaccharides. Microb Biotechnol. 2009;2:333–342. doi: 10.1111/j.1751-7915.2008.00072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LoCascio RG, et al. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J Agric Food Chem. 2007;55:8914–8919. doi: 10.1021/jf0710480. [DOI] [PubMed] [Google Scholar]
- 14.Marcobal A, et al. Consumption of human milk oligosaccharides by gut-related microbes. J Agric Food Chem. 2010;58:5334–5340. doi: 10.1021/jf9044205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niñonuevo MR, et al. Daily variations in oligosaccharides of human milk determined by microfluidic chips and mass spectrometry. J Agric Food Chem. 2008;56:618–626. doi: 10.1021/jf071972u. [DOI] [PubMed] [Google Scholar]
- 16.Ninonuevo MR, et al. Methods for the quantitation of human milk oligosaccharides in bacterial fermentation by mass spectrometry. Anal Biochem. 2007;361:15–23. doi: 10.1016/j.ab.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 17.Sela DA, et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci USA. 2008;105:18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ward RE, Niñonuevo M, Mills DA, Lebrilla CB, German JB. In vitro fermentation of breast milk oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Appl Environ Microbiol. 2006;72:4497–4499. doi: 10.1128/AEM.02515-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward RE, Niñonuevo M, Mills DA, Lebrilla CB, German JB. In vitro fermentability of human milk oligosaccharides by several strains of bifidobacteria. Mol Nutr Food Res. 2007;51:1398–1405. doi: 10.1002/mnfr.200700150. [DOI] [PubMed] [Google Scholar]
- 20.Stahl B, et al. Oligosaccharides from human milk as revealed by matrix-assisted laser desorption/ionization mass spectrometry. Anal Biochem. 1994;223:218–226. doi: 10.1006/abio.1994.1577. [DOI] [PubMed] [Google Scholar]
- 21.Ninonuevo MR, et al. A strategy for annotating the human milk glycome. J Agric Food Chem. 2006;54:7471–7480. doi: 10.1021/jf0615810. [DOI] [PubMed] [Google Scholar]
- 22.Charlwood J, Tolson D, Dwek M, Camilleri P. A detailed analysis of neutral and acidic carbohydrates in human milk. Anal Biochem. 1999;273:261–277. doi: 10.1006/abio.1999.4232. [DOI] [PubMed] [Google Scholar]
- 23.Chaturvedi P, Warren CD, Ruiz-Palacios GM, Pickering LK, Newburg DS. Milk oligosaccharide profiles by reversed-phase HPLC of their perbenzoylated derivatives. Anal Biochem. 1997;251:89–97. doi: 10.1006/abio.1997.2250. [DOI] [PubMed] [Google Scholar]
- 24.Nakhla T, Fu D, Zopf D, Brodsky NL, Hurt H. Neutral oligosaccharide content of preterm human milk. Br J Nutr. 1999;82:361–367. doi: 10.1017/s0007114599001609. [DOI] [PubMed] [Google Scholar]
- 25.Pfenninger A, Karas M, Finke B, Stahl B. Structural analysis of underivatized neutral human milk oligosaccharides in the negative ion mode by nano-electrospray MSn (part 1: methodology) J Am Soc Mass Spectrom. 2002;13:1331–1340. doi: 10.1016/S1044-0305(02)00645-1. [DOI] [PubMed] [Google Scholar]
- 26.Pfenninger A, Karas M, Finke B, Stahl B. Structural analysis of underivatized neutral human milk oligosaccharides in the negative ion mode by nano-electrospray MSn (part 2: application to isomeric mixtures) J Am Soc Mass Spectrom. 2002;13:1341–1348. doi: 10.1016/S1044-0305(02)00646-3. [DOI] [PubMed] [Google Scholar]
- 27.Shen ZJ, Warren CD, Newburg DS. High-performance capillary electrophoresis of sialylated oligosaccharides of human milk. Anal Biochem. 2000;279:37–45. doi: 10.1006/abio.1999.4448. [DOI] [PubMed] [Google Scholar]
- 28.Sumiyoshi W, et al. Determination of each neutral oligosaccharide in the milk of Japanese women during the course of lactation. Br J Nutr. 2003;89:61–69. doi: 10.1079/BJN2002746. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki M, Suzuki A. Structural characterization of fucose-containing oligosaccharides by high-performance liquid chromatography and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Biol Chem. 2001;382:251–257. doi: 10.1515/BC.2001.032. [DOI] [PubMed] [Google Scholar]
- 30.Thurl S, Muller-Werner B, Sawatzki G. Quantification of individual oligosaccharide compounds from human milk using high-pH anion-exchange chromatography. Anal Biochem. 1996;235:202–206. doi: 10.1006/abio.1996.0113. [DOI] [PubMed] [Google Scholar]
- 31.Niñonuevo M, et al. Nanoliquid chromatography-mass spectrometry of oligosaccharides employing graphitized carbon chromatography on microchip with a high-accuracy mass analyzer. Electrophoresis. 2005;26:3641–3649. doi: 10.1002/elps.200500246. [DOI] [PubMed] [Google Scholar]
- 32.Varki A. Nothing in glycobiology makes sense, except in the light of evolution. Cell. 2006;126:841–845. doi: 10.1016/j.cell.2006.08.022. [DOI] [PubMed] [Google Scholar]
- 33.Chaturvedi P, et al. Fucosylated human milk oligosaccharides vary between individuals and over the course of lactation. Glycobiology. 2001;11:365–372. doi: 10.1093/glycob/11.5.365. [DOI] [PubMed] [Google Scholar]
- 34.Musumeci M, Simpore J, D'Agata A, Sotgiu S, Musumeci S. Oligosaccharides in colostrum of Italian and Burkinabe women. J Pediatr Gastroenterol Nutr. 2006;43:372–378. doi: 10.1097/01.mpg.0000228125.70971.af. [DOI] [PubMed] [Google Scholar]
- 35.Ruiz-Palacios GM, Cervantes LE, Ramos P, Chavez-Munguia B, Newburg DS. Campylobacter jejuni binds intestinal H(O) antigen (Fuc alpha 1, 2Gal beta 1, 4GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. J Biol Chem. 2003;278:14112–14120. doi: 10.1074/jbc.M207744200. [DOI] [PubMed] [Google Scholar]
- 36.Jiang X, et al. Human milk contains elements that block binding of noroviruses to human histo-blood group antigens in saliva. J Infect Dis. 2004;190:1850–1859. doi: 10.1086/425159. [DOI] [PubMed] [Google Scholar]
- 37.Newburg DS, et al. Human milk alphal,2-linked fucosylated oligosaccharides decrease risk of diarrhea due to stable toxin of E. coli in breastfed infants. Adv Exp Med Biol. 2004;554:457–461. doi: 10.1007/978-1-4757-4242-8_64. [DOI] [PubMed] [Google Scholar]
- 38.Kobata A. A journey to the world of glycobiology. Glycoconj J. 2000;17:443–464. doi: 10.1023/a:1011006122704. [DOI] [PubMed] [Google Scholar]
- 39.Thurl S, Henker J, Siegel M, Tovar K, Sawatzki G. Detection of four human milk groups with respect to Lewis blood group dependent oligosaccharides. Glycoconj J. 1997;14:795–799. doi: 10.1023/a:1018529703106. [DOI] [PubMed] [Google Scholar]
- 40.Borén T, Falk P, Roth KA, Larson G, Normark S. Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science. 1993;262:1892–1895. doi: 10.1126/science.8018146. [DOI] [PubMed] [Google Scholar]
- 41.Brooks SA, Dwek MV, Schumacher U. Functional and Molecular Glycobiology. New York: Garland; 2002. [Google Scholar]
- 42.Rothenbacher D, et al. Role of Lewis A and Lewis B blood group antigens in Helicobacter pylori infection. Helicobacter. 2004;9:324–329. doi: 10.1111/j.1083-4389.2004.00236.x. [DOI] [PubMed] [Google Scholar]
- 43.Boehm G, Moro G. Structural and functional aspects of prebiotics used in infant nutrition. J Nutr. 2008;138:1818S–1828S. doi: 10.1093/jn/138.9.1818S. [DOI] [PubMed] [Google Scholar]
- 44.Andersson B, Porras O, Hanson LA, Lagergård T, Svanborg-Edén C. Inhibition of attachment of Streptococcus pneumoniae and Haemophilus influenzae by human milk and receptor oligosaccharides. J Infect Dis. 1986;153:232–237. doi: 10.1093/infdis/153.2.232. [DOI] [PubMed] [Google Scholar]
- 45.Angeloni S, et al. Glycoprofiling with micro-arrays of glycoconjugates and lectins. Glycobiology. 2005;15:31–41. doi: 10.1093/glycob/cwh143. [DOI] [PubMed] [Google Scholar]
- 46.Coppa GV, et al. Human milk oligosaccharides inhibit the adhesion to Caco-2 cells of diarrheal pathogens: Escherichia coli, Vibrio cholerae, and Salmonella fyris. Pediatr Res. 2006;59:377–382. doi: 10.1203/01.pdr.0000200805.45593.17. [DOI] [PubMed] [Google Scholar]
- 47.Coppa GV, et al. Oligosaccharides of human milk inhibit the adhesion of Listeria monocytogenes to Caco-2 cells. Ital J Pediatr. 2003;29:61–68. [Google Scholar]
- 48.Coppa GV, Zampini L, Galeazzi T, Gabrielli O. Prebiotics in human milk: A review. Dig Liver Dis. 2006;38(Suppl 2):S291–S294. doi: 10.1016/S1590-8658(07)60013-9. [DOI] [PubMed] [Google Scholar]
- 49.Hong P, Ninonuevo MR, Lee B, Lebrilla C, Bode L. Human milk oligosaccharides reduce HIV-1-gp120 binding to dendritic cell-specific ICAM3-grabbing non-integrin (DC-SIGN) Br J Nutr. 2009;101:482–486. doi: 10.1017/s0007114508025804. [DOI] [PubMed] [Google Scholar]
- 50.Lesman-Movshovich E, Lerrer B, Gilboa-Garber N. Blocking of Pseudomonas aeruginosa lectins by human milk glycans. Can J Microbiol. 2003;49:230–235. doi: 10.1139/w03-027. [DOI] [PubMed] [Google Scholar]
- 51.Idota T, Kawakami H, Murakami Y, Sugawara M. Inhibition of cholera toxin by human milk fractions and sialyllactose. Biosci Biotechnol Biochem. 1995;59:417–419. doi: 10.1271/bbb.59.417. [DOI] [PubMed] [Google Scholar]
- 52.Yolken RH, et al. Human milk mucin inhibits rotavirus replication and prevents experimental gastroenteritis. J Clin Invest. 1992;90:1984–1991. doi: 10.1172/JCI116078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martín-Sosa S, Martín MJ, Hueso P. The sialylated fraction of milk oligosaccharides is partially responsible for binding to enterotoxigenic and uropathogenic Escherichia coli human strains. J Nutr. 2002;132:3067–3072. doi: 10.1093/jn/131.10.3067. [DOI] [PubMed] [Google Scholar]
- 54.Biavati B, Mattarelli P. The family Bifidobacteriaceae. Prokaryotes. 2006;3:322–382. doi: 10.1159/000092237. [DOI] [PubMed] [Google Scholar]
- 55.Haarman M, Knol J. Quantitative real-time PCR assays to identify and quantify fecal Bifidobacterium species in infants receiving a prebiotic infant formula. Appl Environ Microbiol. 2005;71:2318–2324. doi: 10.1128/AEM.71.5.2318-2324.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matsuki T, Watanabe K, Tanaka R, Fukuda M, Oyaizu H. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl Environ Microbiol. 1999;65:4506–4512. doi: 10.1128/aem.65.10.4506-4512.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moro E. A contribution to the knowledge of the normal intestinal bacteria of infants (Translated from German) Jahrb Kinderheilk. 1900;52:38–55. [Google Scholar]
- 58.Mariat D, et al. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009;9:123. doi: 10.1186/1471-2180-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kurokawa K, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gauhe A, et al. Bifidus factor. IV. Preparations obtained from human milk. Arch Biochem Biophys. 1954;48:214–224. doi: 10.1016/0003-9861(54)90326-4. [DOI] [PubMed] [Google Scholar]
- 62.Xiao JZ, et al. Distribution of in vitro fermentation ability of lacto-N-biose I, a major building block of human milk oligosaccharides, in bifidobacterial strains. Appl Environ Microbiol. 2010;76:54–59. doi: 10.1128/AEM.01683-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kiyohara M, et al. Prebiotic effect of lacto-N-biose I on bifidobacterial growth. Biosci Biotechnol Biochem. 2009;73:1175–1179. doi: 10.1271/bbb.80697. [DOI] [PubMed] [Google Scholar]
- 64.Katayama T, et al. Molecular cloning and characterization of Bifidobacterium bifidum 1,2-alpha-L-fucosidase (AfcA), a novel inverting glycosidase (glycoside hydrolase family 95) J Bacteriol. 2004;186:4885–4893. doi: 10.1128/JB.186.15.4885-4893.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nagae M, et al. Structural basis of the catalytic reaction mechanism of novel 1,2-alpha-L-fucosidase from Bifidobacterium bifidum. J Biol Chem. 2007;282:18497–18509. doi: 10.1074/jbc.M702246200. [DOI] [PubMed] [Google Scholar]
- 66.Wada J, et al. Bifidobacterium bifidum lacto-N-biosidase, a critical enzyme for the degradation of human milk oligosaccharides with a type 1 structure. Appl Environ Microbiol. 2008;74:3996–4004. doi: 10.1128/AEM.00149-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suzuki R, et al. Structural and thermodynamic analyses of solute-binding Protein from Bifidobacterium longum specific for core 1 disaccharide and lacto-N-biose I. J Biol Chem. 2008;283:13165–13173. doi: 10.1074/jbc.M709777200. [DOI] [PubMed] [Google Scholar]
- 68.Wada J, et al. Purification, crystallization and preliminary X-ray analysis of the galacto-N-biose-/lacto-N-biose I-binding protein (GL-BP) of the ABC transporter from Bifidobacterium longum JCM1217. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63:751–753. doi: 10.1107/S1744309107036263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Seibel J, et al. In utero and postnatal exposure to a phytoestrogen-enriched diet increases parameters of acute inflammation in a rat model of TNBS-induced colitis. Arch Toxicol. 2008;82:941–950. doi: 10.1007/s00204-008-0309-7. [DOI] [PubMed] [Google Scholar]
- 70.Lee JH, et al. Comparative genomic analysis of the gut bacterium Bifidobacterium longum reveals loci susceptible to deletion during pure culture growth. BMC Genomics. 2008;9:247. doi: 10.1186/1471-2164-9-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kitaoka M, Tian J, Nishimoto M. Novel putative galactose operon involving lacto-N-biose phosphorylase in Bifidobacterium longum. Appl Environ Microbiol. 2005;71:3158–3162. doi: 10.1128/AEM.71.6.3158-3162.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mattarelli P, Bonaparte C, Pot B, Biavati B. Proposal to reclassify the three biotypes of Bifidobacterium longum as three subspecies: Bifidobacterium longum subsp. longum subsp. nov., Bifidobacterium longum subsp. infantis comb. nov. and Bifidobacterium longum subsp. suis comb. nov. Int J Syst Evol Microbiol. 2008;58:767–772. doi: 10.1099/ijs.0.65319-0. [DOI] [PubMed] [Google Scholar]
- 73.Turnbaugh PJ, et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Edwards CA, Parrett AM. Intestinal flora during the first months of life: New perspectives. Br J Nutr. 2002;88(Suppl 1):S11–S18. doi: 10.1079/BJN2002625. [DOI] [PubMed] [Google Scholar]
- 76.Victora CG, et al. Evidence for protection by breast-feeding against infant deaths from infectious diseases in Brazil. Lancet. 1987;2:319–322. doi: 10.1016/s0140-6736(87)90902-0. [DOI] [PubMed] [Google Scholar]