Abstract
The means by which vaginal microbiomes help prevent urogenital diseases in women and maintain health are poorly understood. To gain insight into this, the vaginal bacterial communities of 396 asymptomatic North American women who represented four ethnic groups (white, black, Hispanic, and Asian) were sampled and the species composition characterized by pyrosequencing of barcoded 16S rRNA genes. The communities clustered into five groups: four were dominated by Lactobacillus iners, L. crispatus, L. gasseri, or L. jensenii, whereas the fifth had lower proportions of lactic acid bacteria and higher proportions of strictly anaerobic organisms, indicating that a potential key ecological function, the production of lactic acid, seems to be conserved in all communities. The proportions of each community group varied among the four ethnic groups, and these differences were statistically significant [χ2(10) = 36.8, P < 0.0001]. Moreover, the vaginal pH of women in different ethnic groups also differed and was higher in Hispanic (pH 5.0 ± 0.59) and black (pH 4.7 ± 1.04) women as compared with Asian (pH 4.4 ± 0.59) and white (pH 4.2 ± 0.3) women. Phylotypes with correlated relative abundances were found in all communities, and these patterns were associated with either high or low Nugent scores, which are used as a factor for the diagnosis of bacterial vaginosis. The inherent differences within and between women in different ethnic groups strongly argues for a more refined definition of the kinds of bacterial communities normally found in healthy women and the need to appreciate differences between individuals so they can be taken into account in risk assessment and disease diagnosis.
Keywords: microbial communities, ecology, human microbiome, women’s health, bacterial vaginosis
The human body harbors microorganisms that inhabit surfaces and cavities exposed or connected to the external environment. Each body site includes ecological communities of microbial species that exist in a mutualistic relationship with the host. The kinds of organisms present are highly dependent on the prevailing environmental conditions and host factors and hence vary from site to site. Moreover, they vary between individuals and over time (1). The human vaginal microbiota seem to play a key role in preventing a number of urogenital diseases, such as bacterial vaginosis, yeast infections, sexually transmitted infections, urinary tract infections (2–9), and HIV infection (10, 11). Common wisdom attributes this to lactic acid–producing bacteria, mainly Lactobacillus sp., that commonly inhabit the vagina. These species are thought to play key protective roles by lowering the environmental pH through lactic acid production (12, 13), by producing various bacteriostatic and bacteriocidal compounds, or through competitive exclusion (13–16). The advent of culture-independent molecular approaches based on the cloning and sequencing of 16S rRNA genes has furthered our understanding of the vaginal microbiota by identifying taxa that had not been cultured (17–24). However, this technique is limited by high cost and low throughput, hence only small numbers of samples have usually been analyzed, and the depth of sample analysis was not great.
In this study we sought to develop an in-depth and accurate understanding of the composition and ecology of the vagina microbial ecosystem in asymptomatic women using a high-throughput method based on pyrosequencing of barcoded 16S rRNA genes. The data obtained are an essential prerequisite for comprehending the role and ultimately the function of vaginal microbiota in reducing the risk of acquiring diseases and identifying factors that determine disease susceptibility. Specifically we sought to characterize the vaginal microbial communities in a cohort of 396 North American women equally representing four ethnic backgrounds (Asian, white, black, and Hispanic) and further address three aims. The first was to establish whether there were correlations between community composition and vaginal pH because these would be indicative of community performance. The second was to explore how the species composition of vaginal communities was reflected in Nugent scores (25), a diagnostic factor commonly used to identify women with bacterial vaginosis (26). Finally, the third aim was to identify patterns in the relative abundances of different species because these might reflect antagonistic or cooperative interspecies interactions.
Results and Discussion
We characterized the vaginal microbiota and vaginal pH of 396 asymptomatic, sexually active women who fairly equally represented four self-reported ethnic groups: white (n = 98), black (n = 104), Asian (n = 97), and Hispanic (n = 97). The demographics and other characteristics of the women are given in Table S1. Each woman used two swabs to self-collect midvaginal samples. One swab was used to evaluate the vaginal microbiota on the basis of the Nugent criteria used for the diagnosis of bacterial vaginosis (25), and the second was used in procedures to determine the species composition and structure of the resident bacterial communities (27). The latter was accomplished by phylogenetic analysis of 16S rRNA gene sequences (28). Whole-genomic DNA was extracted from each swab, and variable regions 1 and 2 (V1–V2) of 16S rRNA genes were PCR amplified using the barcoded universal primers listed in Table S2 and pyrosequenced using a Roche 454 FLX instrument. This produced a dataset consisting of 897,345 high-quality sequences with an average length of 240 bp and ≈2,200 reads per sample that were classified using the Ribosomal Database Project (RDP) Naïve Bayesian Classifier (29). Species-level taxonomic assignments of Lactobacillus sp. were done using a bioinformatics algorithm based on a combination of species-level hidden Markov models and clustering as described in SI Materials and Methods. Overall, a total of 282 taxa were observed in the vaginal microbiota of these women (Table S3).
The taxonomic assignments of vaginal bacterial community members and the associated metadata for each subject are shown in Table S4. The depth of coverage for each community was sufficient to detect taxa that constitute ≈0.1% of the community. Although taxa present at less than this level are often referred to as low-abundance or “rare” taxa, they are only rare in the context of sampling depth. If a vaginal bacterial community has ≈108 cells per milliliter of vaginal secretion, then high numbers (105 cells/mL) of “rare” members are present in the community, whereas phylotypes present at densities of <105 cells per milliliter would remain undetected. These “rare” taxa could play major roles in the ecology of a community, whereas undetected members may constitute a “seed bank” of species whose numbers increase under conditions that favor their growth.
Vaginal Bacterial Community Composition and Structure.
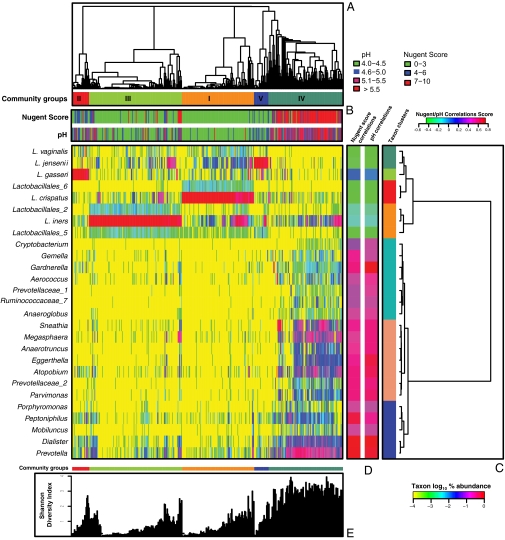
The vaginal bacterial communities were grouped according to community composition (Fig. 1A and Table S5), and the phylotypes were clustered according to their correlation profiles as explained in SI Materials and Methods (Fig. 1C). The heatmap in Fig. 1 shows the results obtained using log10-transformed percentage abundance of each taxon. It highlights the diversity found in all vaginal bacterial communities, even those where the phylotype abundance is highly skewed and dominated by a single phylotype, and identifies taxa with similar correlation profiles. By way of comparison, Fig. S1 shows the clustering of communities according to bacterial composition and abundance done as described above but based on only the 25 most abundant taxa.
Fig. 1.
Heatmap of log10-transformed proportions of microbial taxa found in the vaginal bacterial communities of 394 women of reproductive age (color key is indicated in the lower right corner). (A) Complete linkage clustering of samples based on the species composition and abundance of vaginal bacterial communities that define community groups I to V. (B) Nugent scores and pH measurements for each of the 394 community samples (color key is indicated above C). (C) Complete linkage clustering of taxa based on Spearman's correlation coefficient profiles, which were defined as the set of Spearman's correlation coefficients calculated between one taxon and all of the other taxa (SI Materials and Methods). (D) Spearman's correlation coefficients between the presence of a taxon and the Nugent score or pH of a sample. (E) Shannon diversity indices calculated for 394 vaginal communities (two singletons were excluded).
The analysis revealed five major groups of microbial communities (Fig. 1 and Fig. S1), which is reminiscent of previously published studies on microbial diversity in the human vagina (18). The five groups, designated I, II, III, IV, and V, contained 104, 25, 135, 108, and 21 taxa, respectively (Table S3). The most diverse communities were those of group IV, and this was reflected in the Shannon diversity indices of the communities (Fig. 1 and Fig. S1). In addition to these groupings there were two singletons: one was dominated by Lactobacillus_3 (99% of the community), and another was dominated by members of the genus Enterococcus (76% of the community).
Unlike any other anatomical site on the human body, most vaginal communities (73%) were dominated by one or more species of Lactobacillus that constitute >50% of all sequences obtained (Fig. S1 and Tables S4–S6). Communities in group I, which occurred in 26.2% of the women sampled, were dominated by L. crispatus, whereas groups II (6.3%), III (34.1%), and V (5.3%) were dominated by L. gasseri, L. iners, and L. jensenii, respectively. As shown in Table 1, communities belonging to group I have the lowest median pH (4.0 ± 0.3), whereas communities in group IV had the highest median pH (5.3 ± 0.6). Interestingly, communities dominated by species of Lactobacillus other than L. crispatus have slightly higher pH, ranging from 4.4 (group III) to 5.0 (group II), indicating that these communities as a whole might not produce as much lactic acid as those of group I or might have different buffering capabilities.
Table 1.
pH of vaginal community groups in women of different ethnicities
| Community groups* |
||||||||||||
| I (L. crispatus) |
II (L. gasseri) |
III (L. iners) |
IV (Diversity group) |
V (L. jensenii) |
All groups |
|||||||
| Ethnic groups | Subjects† | pH‡ | Subjects† | pH‡ | Subjects† | pH‡ | Subjects† | pH‡ | Subjects† | pH‡ | Subjects† | pH‡ |
| Asian | 24 | 4.4 ± 0.52 | 5 | 4.4 ± 0.44 | 41 | 4.0 ± 0.0 | 19 | 5.5 ± 0.44 | 7 | 5.0 ± 0.89 | 96 | 4.4 ± 0.59 |
| White | 44 | 4.0 ± 0.0 | 8 | 4.7 ± 0.44 | 26 | 4.3 ± 0.30 | 10 | 5.5 ± 0.74 | 9 | 4.85 ± 0.22 | 97 | 4.2 ± 0.30 |
| Black | 23 | 4.0 ± 0.0 | 5 | 5.0 ± 0.0 | 33 | 4.0 ± 0.0 | 42 | 5.3 ± 0.44 | 1 | 4.7 ± 0.44 | 104 | 4.7 ± 1.04 |
| Hispanic | 14 | 4.0 ± 0.0 | 7 | 4.7 ± 0.22 | 35 | 4.4 ± 0.59 | 37 | 5.3 ± 0.44 | 4 | 5.0 ± 0.59 | 97 | 5.0 ± 074 |
| All ethnic groups | 105 | 4.0 ± 0.3 | 25 | 5.0 ± 0.7 | 135 | 4.4 ± 0.6 | 108 | 5.3 ± 0.6 | 21 | 4.7 ± 0.4 | 394 | 4.4 ± 0.7 |
*Community groups are defined as in Fig. 1.
†Total number of subjects within a community group (two singleton clusters were identified and are not included).
‡pH values expressed as median ± median absolute deviation.
The skewed rank abundances of species in these communities leave the impression that these communities are species poor, but this may not be the case because there are an unknown number of “rare” species. The remaining communities found in 27% of the women formed a large heterogeneous group (IV) and were typified by higher proportions of strictly anaerobic bacteria, including Prevotella, Dialister, Atopobium, Gardnerella, Megasphaera, Peptoniphilus, Sneathia, Eggerthella, Aerococcus, Finegoldia, and Mobiluncus. This finding is consistent with those of previous studies wherein the species composition of vaginal communities was investigated by cloning and sequencing of 16S rRNA genes (17–24). It should be pointed out that although communities of group IV seem to be diverse relative to the other groups (Fig. S1), it could simply reflect greater species evenness. Resolution of this will require more intensive analysis by deep sequencing of 16S rRNA genes (>100,000 reads) of communities within all these groups.
Although communities in group IV were not dominated by Lactobacillus sp., L. iners and L. crispatus were detected in 78.7% and 51.9% of group IV communities (Table S6), respectively. Only four subjects in group IV lacked detectable Lactobacillus sp. in their vaginas, and those communities were dominated by Prevotella, Sneathia, Megasphaera, or Streptococcus. Interestingly, all communities contained members that have been assigned to genera known to produce lactic acid, including Lactobacillus, Megasphaera, Streptococcus, and Atopobium. This suggests that an important catabolic function, namely the production of lactic acid, may be conserved among communities despite differences in the species composition.
By clustering taxa on the basis of their correlation profiles, three subgroups of communities in group IV were identified (Fig. 1C). The first of these was defined by the cooccurrence of seven taxa, including Cryptobacterium, Gemella, Gardnerella, Aerococcus, Prevotellaceae_1, and Ruminococcaceae_7 and Anaeroglobus. The second cluster was also defined by the cooccurrence of seven taxa, including Sneathia, Megasphaera, Anaerotruncus, Eggerthella, Atopobium, Prevotellaceae_2, and Parvimonas. The third cluster was marked by the cooccurrence of five taxa, including Porphyromonas, Peptoniphilus, Mobiluncus, Dialister, and Prevotella. The significance of these associations is unknown, but they could reflect meaningful ecological interactions that should not be overlooked in considering differences among individuals.
Somewhat unexpectedly, Prevotella sp. were detected in 68.5% of the samples, with abundance ranging from a few percent to more than 45%. Although Prevotella sp. have been previously demonstrated to be members of vaginal communities, their prevalence may have been underappreciated, and their role in the community is unknown. However, it should be noted that Prevotella sp. have been shown to positively affect the growth of Gardnerella vaginalis and Peptostreptococcus anaerobius by producing key nutrients for these species, such as ammonia and amino acids. Both of these species have been linked to bacterial vaginosis, hence the wide distribution of Prevotella sp. in the vaginal microbiota might be a factor that facilitates bacterial vaginosis (4, 30, 31).
Core Microbiomes of the Human Vagina.
One objective of studies on the human microbiome is to determine whether there is a core set of microbial species associated with the bodies of all humans. It is postulated that changes to this “core microbiome” may be correlated with changes in human health or risk to disease. The results from this study suggest that for the human vagina there is no single core microbiome. Instead, it seems there are multiple core microbiomes that can be defined by the community groups I–V depicted in Fig. 1. As noted above, these groups can be readily distinguished on the basis of two criteria: (i) whether the constituent communities are dominated by Lactobacillus, and (ii) the particular species of Lactobacillus present. The vast majority of communities in groups I, II, III, and V had more than one phylotype of lactic acid bacteria, suggesting a degree of functional redundancy, but they differed widely in abundance. For example, a positive association between L. crispatus and Lactobacillales_6 was observed, with these taxa cooccurring in 99.1% of the communities in group I (Table S6), but the latter was much less abundant than the former. Likewise, in community group III, Lactobacillales_2 and Lactobacillales_5 cooccurred with L. iners in 97.8% and 97% of the samples, respectively (Table S6). Although we noted these strong positive associations, we cannot explain their existence. Aside from species of Lactobacillales no taxa were so consistently found together in any community group (Fig. 1). Although no core microbiome can be identified on the basis of the taxa found in these communities, we posit that core functions are conserved among communities despite differences in their species composition and that functional redundancy would be associated with increased community reliability in the face of environmental changes (32).
Correlation Profiles of Taxa and Nugent Scores.
The Nugent criteria (25) are widely used to diagnose bacterial vaginosis according to the proportions of different cellular morphologies seen in gram-stained smears of vaginal samples. The weighted score computed using these criteria is thought to reflect the relative abundance of the following morphotypes: lactobacilli, Gardnerella vaginalis or Bacteroides (small gram-variable rods or Gram-negative rods), and curved gram-variable rods. The resulting scores range from 0 to 10, with those of 7 and higher considered to be indicative of bacterial vaginosis, whereas scores of 4–6 and 3 or less are considered intermediate and normal, respectively. In this study communities with high Nugent scores were most often associated with communities in group IV but were also observed in communities belonging to other groups (Fig. 1).
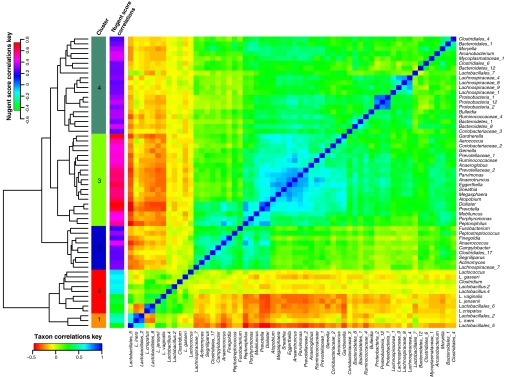
Because Nugent scores are based on weighted counts of different cellular morphotypes, we sought to better define what phylotypes actually form the basis of Nugent scores and to determine the kinds of communities associated with high Nugent scores. Phylotypes whose presence or absence is measured by Nugent scores were identified by computing the Pearson correlation coefficients for the relative abundances of all phylotypes and the corresponding Nugent scores, which were categorized as low (Nugent scores 0–3), medium (Nugent scores 4–6), and high (Nugent scores 7–10). We found that 119 phylotypes were either positively or negatively correlated with Nugent score. Of these, 59 had modest correlation coefficients in the range of 0.0–0.2 that were not statistically significant (P > 0.001; Fig. S2C). These were excluded in subsequent calculations. The phylotypes with highly significant correlations are listed in Table S7. A correlogram depicting the correlation profiles of the remaining 60 taxa that were significantly correlated to Nugent scores is shown in Fig. 2. In this figure phylotypes with similar correlation profiles were placed near each other using complete linkage hierarchical clustering based on Euclidean distances between correlation profiles. Clusters 3, 4, and 5 included taxa with high positive coefficients of correlation to Nugent score (Fig. 2). Of these, cluster 3 was most correlated to high Nugent scores, and it included the following phylotypes: Aerococcus, Anaeroglobus, Anaerotruncus, Atopobium, Coriobacteriaceae_2, Dialister, Eggerthella, Gardnerella, Gemella, Megasphaera, Mobiluncus, Parvimonas, Peptoiphilus, Prevotella, Porphyomonas, Prevotellaceae_1, Prevotellaceae_2, Ruminococcaceae, and Snethia. On the other hand, taxa whose correlation profiles were associated with low Nugent scores (clusters 1 and 2 in Fig. 2) included mainly phylotypes of Lactobacillus that were commonly found in community groups I, II, III, and V. The fact that phylotypes in clusters 1 and 2 are commonly found together is readily apparent, as is the cooccurrence of phylotypes of clusters 3, 4 and 5. Conversely, it is evident that phylotypes in clusters 1 and 2 tend not to occur with phylotypes in clusters 3–5. This may reflect antagonistic interactions or competitive exclusion among these groups of phylotypes.
Fig. 2.
Correlogram of 60 microbial taxa with negative or positive correlation to Nugent scores. Microbial taxa with the highest negative or positive correlation with Nugent scores were selected as described in SI Materials and Methods. The Spearman's correlation coefficients between each taxon and all other taxa were used to build the correlogram that illustrates the cooccurrence of taxa in communities. Spearman's correlation coefficients between taxa and Nugent scores are also indicated.
A comparable analysis was done based on pairwise interactions between Spearman's rank correlation coefficients based on the relative abundances of phylotype pairs for all 282 bacterial phylotypes found in the vaginal communities analyzed in this study, and the results are shown in Fig. S2B.
Differences in Vaginal Microbiomes of Ethnic Groups.
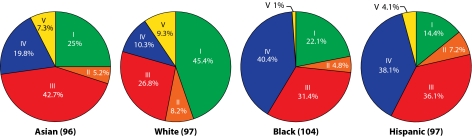
The study cohort consisted of roughly equal numbers of four self-described ethnicities (white, Asian, black, and Hispanic), and this offered the opportunity to assess the relationship of ethnic background on vaginal bacterial community composition. The proportions of each community group varied among the four ethnic groups (Fig. 3 and Fig. S3A), and these differences were statistically significant [χ2(10) = 36.8, P < 0.0001]. No statistically significant associations were observed between age and community types within or across ethnic groups.
Fig. 3.
Representation of vaginal bacterial community groups within each ethnic group of women. The number of women from each ethnic group is in parentheses.
Vaginal bacterial communities dominated by species of Lactobacillus (groups I, II, III, and V) were found in 80.2% and 89.7% of Asian and white women, respectively, but in only 59.6% and 61.9% of Hispanic and black women, respectively. The higher median pH values in Hispanic (pH 5.0 ± 0.59) and black (pH 4.7 ± 1.04) women reflects the higher prevalence of communities not dominated by Lactobacillus sp. (cluster IV) in these two ethnic groups when compared with Asian (pH 4.4 ± 0.59) and white (pH 4.2 ± 0.3) women (Table 1). This is significant because the occurrence of high numbers of lactobacilli and pH <4.5 have become synonymous with “healthy”. If accepted at face value, this common wisdom suggests that although most Asian and white women are “healthy”, a significant proportion of asymptomatic Hispanic and black women are “unhealthy”—a notion that seems implausible. It also begs the question of what kinds of bacterial communities should be considered “normal” in Hispanic and black women. We found that community group IV (diverse group) was overrepresented in Hispanic (34.3%) and black (38.9%) women as compared with Asian (17.6%) and white (9.3%) women (Fig. S3B). From these data we conclude that vaginal bacterial communities not dominated by species of Lactobacillus are common and appear normal in black and Hispanic women. The data from this study are in accordance with the results of Zhou et al. (17, 18), who studied the vaginal bacterial communities of white, black, and Japanese women. The reasons for these differences among ethnic groups are unknown, but it is tempting to speculate that the species composition of vaginal communities could be governed by genetically determined differences between hosts. These might include differences in innate and adaptive immune systems, the composition and quantity of vaginal secretions, and ligands on epithelial cell surfaces, among others. Although these may be key to shaping vaginal communities, previous studies have also shown that human habits and practices, including personal hygiene, methods of birth control, and sexual behaviors, also exert strong influences (33).
The small number of different kinds of vaginal communities is somewhat surprising given that these communities are probably assembled independently after birth. The repeatability of community assembly suggests that a host exerts strong selection for a rather limited number of different kinds of bacteria. This is especially evident in the limited number of Lactobacillus phylotypes and other lactic acid–producing bacteria that are abundant in these communities. The prominence of these populations and their important role in modulating vaginal pH suggests they might be drivers in these communities and thought of in terms of Walker's driver–passenger model (34, 35). This model posits that ecological function resides in “driver” species or in functional groups of such species that have key ecological functions that significantly structure ecosystems, whereas “passenger” species are those that have minor ecological impact. Studies done to tease out the influence of these various factors on vaginal community ecology will be important to understanding community stability, resistance, and resilience so that strategies can be developed to maintain human vaginal health and prevent disease.
Vaginal Community Space.
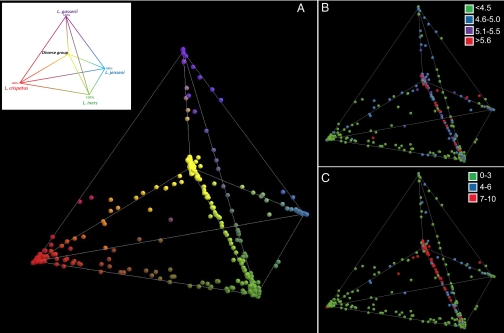
The relationships among communities were visualized by principal component analysis and displayed in 3D space. The three principle components explained 82% of the variance. Each point in Fig. 4 represents the vaginal community of an individual. Communities dominated by species of Lactobacillus and representing groups I, II, III, and V are shown at each of the four outer vertices of the tetrahedron, with communities of group IV at the inner vertex. Communities found on the edges joining two vertices are mixtures of the two Lactobacillus species that dominate the communities found at the corresponding vertices, with an equal proportion of each species at the midpoint of the edge. We refer to each location in this 3D space as a community state, and one can consider the entire space to represent the plausible alternative community states, or vaginal bacterial community space.
Fig. 4.
Relationships among vaginal bacterial communities visualized by principal component analysis in which the relative abundances are expressed as proportions of the total community and displayed in 3D space. Communities dominated by species of Lactobacillus and representing community groups I, II, III, and V are shown at each of the four outer vertices of the tetrahedron, with communities of group IV at the inner vertex and shown in the Inset. (A) Each point corresponds to a single subject and was colored according to the proportions of phylotypes in each community. (B) pH of each vaginal community shown in A. (C) Nugent score category of each vaginal community shown in A.
The cross-sectional design of this study with only one sample from each subject precludes knowing whether the locations of these communities in vaginal community space vary over time. Nonetheless, at this stage we can propose four distinct conceptual models for the variation of community composition over time. The first is the “dynamic equilibrium hypothesis”, in which the composition of a community is comparatively invariant over time and exists in a single dynamic equilibrium. A second “community space hypothesis” is the opposite of the first, and each community can and does occupy any position in community space over time and throughout a woman's lifetime. These changes are postulated to occur in response to hormonal cycles, an individual's habits and practices, changes in diet, or some other ecological force. A third model is an “alternative equilibrium states hypothesis”, wherein a woman's community can change over time, but the number of alternative states are limited in number and governed by unknown factors. A fourth possibility is a “community resilience hypothesis”, in which a community normally resides in a single region of space. Under this scenario the composition and structure of a vaginal community can change to a transitional state in response to disturbance, but the resistance and resilience of a community determine the extent and duration of a change, whereas homeostatic mechanisms drive communities back to their “ground state”. We expect that no single hypothesis will explain the dynamics of all communities. Each of these hypotheses can only be formally evaluated once time-series data on vaginal microbial community dynamics are available along with extensive metadata on subject's behaviors, habits and practices, health history, and other information. Currently the short-term temporal dynamics of vaginal communities are unknown because no studies have been done in which the same individuals are frequently sampled and variation in community composition assessed over time using cultivation-independent methods.
The pH and Nugent scores of each community are depicted in the 3D community space on Figs. 4 B and C. The figures (and Fig. S4) show a strong correlation between high pH and high Nugent scores. As presented in Table 1 and depicted here, the lowest pH values were associated with community states dominated by L. iners and L. crispatus, and the highest pH values were associated with community states not dominated by species of Lactobacillus. Both Nugent scores and pH values increased as the proportion of non-Lactobacillus sp. increased. This was most readily seen in communities that contained decreasing proportions of L. iners. Interestingly, elevated pH and high Nugent scores were observed in some communities that have high proportions of Lactobacillus species (also shown in Fig. 1), suggesting that these metrics cannot be predicted with absolute certainty solely on the basis of the proportion of Lactobacillus in a community. Clearly additional research is needed to understand the various factors that govern vaginal pH.
By analogy with other biological communities, it is reasonable to assume that vaginal microbial communities exist in a state of dynamic equilibrium and that homeostatic mechanisms exist to provide resilience. Given the fundamental differences in the species composition of these communities, one can speculate that they will differ in terms of number and strength of interspecies interactions. This will in turn have implications for the relative resistance and resilience of each community type to disturbances. If that is the case then invasive species, including both opportunistic and overt pathogens, are more likely to become established in communities that exhibit low stability, and the converse will also be true (36). This has direct implications for the assessment of susceptibility to infectious diseases. Importantly, it also suggests that differences in vaginal bacterial community composition should be taken into account in the estimation of disease risks. This would constitute the first step toward personalized medicine for women's reproductive health, wherein differences between the vaginal microbiomes of individuals would be taken into account in risk assessment and for disease diagnosis and treatment.
Materials and Methods
Sample Collection and Study Design.
Women were recruited at three clinical sites: two in Baltimore at the University of Maryland School of Medicine and one in Atlanta, at Emory University. Enrollment occurred between June 2008 and January 2009. All women were not pregnant, of reproductive age, ranging from 12 to 45 years (mean 30.6 ± 7.32 years), regularly menstruating (25- to 35-day menstrual cycles), with a history of sexual activity, and had not taken any antibiotic or antimycotic compounds in the past 30 days. Women were asked to refrain from sexual activity in the 48 h before the visit. Women were excluded from the study if they had used douches, vaginal medications or suppositories, feminine sprays, genital wipes or contraceptive spermicides, or had reported vaginal discharge in the past 48 h. Further, women who were menstruating or were currently using contraceptives that are directly delivered to the vaginal mucosa, such as NuvaRing, were also excluded. After obtaining informed consent, each participant completed a questionnaire on sexual health and behaviors.
Participants obtained two self-collected swabs using the Elution-swab system (Copan). The first swabs were stored in 1 mL of Amies transport medium (Copan) and frozen upright on dry ice until transported to the laboratory, where they were stored at −80 °C. The second swabs were rolled by the clinical nurse onto a microscope glass slide that was air-dried, gram-stained, and then scored using Nugent criteria (4). The second swab was then stored in 1 mL of Amies transport medium and frozen upright on dry ice until being transported to the laboratory, where they were archived at −80 °C. Two technicians independently scored the gram-stained slides, and a third technician evaluated discrepancies. Those with a score of 0–3 were reported as having a low score, whereas those with scores of 4–6 and 7–10 were categorized as intermediate and high, respectively.
Vaginal pH was measured using the VpH glove (Inverness Medical) and scored by the clinical nurse according to the manufacturer's instructions using a scale ranging from 4.0 to 7.7.
The institutional review boards at Emory University School of Medicine, Grady Memorial Hospital and the University of Maryland School of Medicine approved the protocol. Guidelines of the universities were followed in the conduct of the clinical research. The study was registered at clinicaltrials.gov under ID NCT00576797.
Whole-Genomic DNA Extraction from Vaginal Swabs.
The swabs were thawed on ice before analysis and vortexed vigorously for 5 min to resuspend the cells. A 0.5 mL-aliquot was transferred to a sterile 2.0 mL tube and stored on ice. Cell lysis was initiated by adding 50 μL of lyzosyme (10 mg/mL), 6 μL of mutanolysin (25,000 U/mL; Sigma- Aldrich), 3 μL of lysostaphin (4,000 U/mL in sodium acetate; Sigma- Aldrich), and 41 μL of TE50 buffer (10 mM Tris·HCL and 50 mM EDTA, pH 8.0). After a 1-h incubation at 37 °C the cells were disrupted by bead beating, which was performed with bleached and rinsed 0.1-mm-diameter zirconia/silica beads (BioSpec Products), for 1 min at room temperature, with 36 oscillations per second (2,100 rpm) in a Mini-Beadbeater-96 (BioSpec Products). The resulting crude lysate was processed using QIAamp DNA Mini Kit (Qiagen) according to the manufacturer's recommendation for crude lysates. The samples were eluted with 2 × 200 μL of AE buffer (10 mM Tris-Cl, 0.5 mM EDTA; pH 9.0) into separate tubes. The DNA concentrations in the samples were measured using the Quant-iT PicoGreen dsDNA assay kit from Molecular Probes (Invitrogen).
Pyrosequencing of Barcoded 16S rRNA Gene Amplicons.
Universal primers 27F and 338R were used for PCR amplification of the V1–V2 hypervariable regions of 16S rRNA genes (3). The 338R primer included a unique sequence tag to barcode each sample. The primers were as follows: 27F-5′-GCCTTGCCAGCCCGCTCAGTCAGAGTTTGATCCTGGCTCAG-3′ and 338R-5′-GCCTCCCTCGCGCCATCAGNNNNNNNNCATGCTGCCTCCCGTAGGAGT-3′, where the underlined sequences are the 454 Life Sciences FLX sequencing primers B and A in 27F and 338R, respectively, and the bold letters denote the universal 16S rRNA primers 27F and 338R. The 8-bp barcode within primer 338R is denoted by 8 Ns. Using 96 barcoded 338R primers (Table S2) the V1–V2 regions of 16S rRNA genes were amplified in 96-well microtiter plates using AmpliTaq Gold DNA polymerase (Applied Biosystems) and 50 ng of template DNA in a total reaction volume of 50 μL. Reactions were run in a PTC-100 thermal controller (MJ Research) using the following cycling parameters: 5 min of denaturation at 95 °C, followed by 20 cycles of 30 s at 95 °C (denaturing), 30 s at 56 °C (annealing), and 90 s at 72 °C (elongation), with a final extension at 72 °C for 7 min. Negative controls without a template were included for each barcoded primer pair. The presence of amplicons was confirmed by gel electrophoresis on a 2% agarose gel and staining with SYBRGreen. PCR products were quantified using a GelDoc quantification system (BioRad) and the Quant-iT PicoGreen dsDNA assay. Equimolar amounts (100 ng) of the PCR amplicons were mixed in a single tube. Amplification primers and reaction buffer were removed from each sample using the AMPure Kit (Agencourt). The purified amplicon mixtures were sequenced by 454 FLX pyrosequencing using 454 Life Sciences primer A by the Genomics Resource Center at the Institute for Genome Sciences, University of Maryland School of Medicine, using protocols recommended by the manufacturer as amended by the Center.
Sequence Read Binning.
Sequences were binned by samples using the sample-specific barcode sequences and trimmed by removal of the barcode and primer sequences. Criteria previously described (28) were used to assess the quality of sequence reads. To pass, a sequence read (i) included a perfect match to the sequence tag (barcode) and the 16S rRNA gene primer; (ii) was at least 200 bp in length; (iii) had no more than two undetermined bases; and (iv) had a least 60% match to a previously determined 16S rRNA gene sequence. On average 4.8% of the sequence reads did not pass this quality-control step.
Each processed 16S rRNA gene sequence was classified using the RDP Naïve Bayesian Classifier (7). RDP classifier quality score filtering was not used, and all reads were classified to the genus or species level as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Jane Schwebke and Charles Rivers (University of Alabama, Birmingham) for performing the Nugent scoring and Inverness Medical for graciously providing the Careplan VpH gloves for the study. This research was supported by Grant 1UO1AI070921 from the National Institute of Allergy and Infectious Diseases of the National Institutes of Heath, Grant UH2AI083264 funded under the Human Microbiome Project RoadMap Initiative from the National Institutes of Health, and in part by Public Health Service Grant (UL1 RR025008, KL2 RR025009, or TL1 RR025010) from the Clinical and Translational Science Award program, National Institutes of Health, National Center for Research Resources.
Footnotes
The authors declare no conflict of interest.
Data deposition: The bacterial 16S rRNA gene sequences have been deposited in the National Center for Biotechnology Information Short Read Archive (SRA022855).
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Microbes and Health” held November 2–3, 2009, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS Web site at http://www.nasonline.org/SACKLER_Microbes_and_Health.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1002611107/-/DCSupplemental.
References
- 1.Costello EK, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donders GG, et al. Pathogenesis of abnormal vaginal bacterial flora. Am J Obstet Gynecol. 2000;182:872–878. doi: 10.1016/s0002-9378(00)70338-3. [DOI] [PubMed] [Google Scholar]
- 3.Gupta K, et al. Inverse association of H2O2-producing lactobacilli and vaginal Escherichia coli colonization in women with recurrent urinary tract infections. J Infect Dis. 1998;178:446–450. doi: 10.1086/515635. [DOI] [PubMed] [Google Scholar]
- 4.Pybus V, Onderdonk AB. Microbial interactions in the vaginal ecosystem, with emphasis on the pathogenesis of bacterial vaginosis. Microbes Infect. 1999;1:285–292. doi: 10.1016/s1286-4579(99)80024-0. [DOI] [PubMed] [Google Scholar]
- 5.Cherpes TL, Meyn LA, Krohn MA, Lurie JG, Hillier SL. Association between acquisition of herpes simplex virus type 2 in women and bacterial vaginosis. Clin Infect Dis. 2003;37:319–325. doi: 10.1086/375819. [DOI] [PubMed] [Google Scholar]
- 6.Martin HL, et al. Vaginal lactobacilli, microbial flora, and risk of human immunodeficiency virus type 1 and sexually transmitted disease acquisition. J Infect Dis. 1999;180:1863–1868. doi: 10.1086/315127. [DOI] [PubMed] [Google Scholar]
- 7.Sobel JD. Is there a protective role for vaginal flora? Curr Infect Dis Rep. 1999;1:379–383. doi: 10.1007/s11908-999-0045-z. [DOI] [PubMed] [Google Scholar]
- 8.Watts DH, et al. Effects of bacterial vaginosis and other genital infections on the natural history of human papillomavirus infection in HIV-1-infected and high-risk HIV-1-uninfected women. J Infect Dis. 2005;191:1129–1139. doi: 10.1086/427777. [DOI] [PubMed] [Google Scholar]
- 9.Wiesenfeld HC, Hillier SL, Krohn MA, Landers DV, Sweet RL. Bacterial vaginosis is a strong predictor of Neisseria gonorrhoeae and Chlamydia trachomatis infection. Clinical Infect Dis. 2003;36:663–668. doi: 10.1086/367658. [DOI] [PubMed] [Google Scholar]
- 10.Lai SK, et al. Human immunodeficiency virus type 1 is trapped by acidic but not by neutralized human cervicovaginal mucus. J Virol. 2009;83:11196–11200. doi: 10.1128/JVI.01899-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taha TE, et al. Bacterial vaginosis and disturbances of vaginal flora: Association with increased acquisition of HIV. AIDS. 1998;12:1699–1706. doi: 10.1097/00002030-199813000-00019. [DOI] [PubMed] [Google Scholar]
- 12.Boskey ER. Baltimore: The Johns Hopkins University; 2000. Vaginal acidity is produced by vaginal bacteria. PhD thesis. [Google Scholar]
- 13.Boskey ER, Cone RA, Whaley KJ, Moench TR. Origins of vaginal acidity: High D/L lactate ratio is consistent with bacteria being the primary source. Hum Reprod. 2001;16:1809–1813. doi: 10.1093/humrep/16.9.1809. [DOI] [PubMed] [Google Scholar]
- 14.Kaewsrichan J, Peeyananjarassri K, Kongprasertkit J. Selection and identification of anaerobic lactobacilli producing inhibitory compounds against vaginal pathogens. FEMS Immunol Med Microbiol. 2006;48:75–83. doi: 10.1111/j.1574-695X.2006.00124.x. [DOI] [PubMed] [Google Scholar]
- 15.Klebanoff SJ, Hillier SL, Eschenbach DA, Waltersdorph AM. Control of the microbial flora of the vagina by H2O2-generating lactobacilli. J Infect Dis. 1991;164:94–100. doi: 10.1093/infdis/164.1.94. [DOI] [PubMed] [Google Scholar]
- 16.Voravuthikunchai SP, Bilasoi S, Supamala O. Antagonistic activity against pathogenic bacteria by human vaginal lactobacilli. Anaerobe. 2006;12:221–226. doi: 10.1016/j.anaerobe.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 17.Zhou X, et al. Differences in the composition of vaginal microbial communities found in healthy Caucasian and black women. ISME J. 2007;1:121–133. doi: 10.1038/ismej.2007.12. [DOI] [PubMed] [Google Scholar]
- 18.Zhou X, et al. The vaginal bacterial communities of Japanese women resemble those of women in other racial groups. FEMS Immunol Med Microbiol. 2009;58:169–181. doi: 10.1111/j.1574-695X.2009.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fredricks DN, Fiedler TL, Marrazzo JM. Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med. 2005;353:1899–1911. doi: 10.1056/NEJMoa043802. [DOI] [PubMed] [Google Scholar]
- 20.Srinivasan S, Fredricks DN. The human vaginal bacterial biota and bacterial vaginosis. Interdiscip Perspect Infect Dis. 2008;2008:750479. doi: 10.1155/2008/750479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferris MJ, et al. Association of Atopobium vaginae, a recently described metronidazole resistant anaerobe, with bacterial vaginosis. BMC Infect Dis. 2004;4:5. doi: 10.1186/1471-2334-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferris MJ, Norori J, Zozaya-Hinchliffe M, Martin DH. Cultivation-independent analysis of changes in bacterial vaginosis flora following metronidazole treatment. J Clin Microbiol. 2007;45:1016–1018. doi: 10.1128/JCM.02085-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verhelst R, et al. Cloning of 16S rRNA genes amplified from normal and disturbed vaginal microflora suggests a strong association between Atopobium vaginae, Gardnerella vaginalis and bacterial vaginosis. BMC Microbiol. 2004;4:16. doi: 10.1186/1471-2180-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verstraelen H, Verhelst R, Claeys G, Temmerman M, Vaneechoutte M. Culture-independent analysis of vaginal microflora: The unrecognized association of Atopobium vaginae with bacterial vaginosis. Am J Obstet Gynecol. 2004;191:1130–1132. doi: 10.1016/j.ajog.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 25.Nugent RP, Krohn MA, Hillier SL. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of gram stain interpretation. J Clin Microbiol. 1991;29:297–301. doi: 10.1128/jcm.29.2.297-301.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Centers for Disease Control and Prevention. Workowski KA, Berman SM. Sexually transmitted diseases treatment guidelines, 2006. MMWR Recomm Rep. 2006;55(RR-11):1–94. [PubMed] [Google Scholar]
- 27.Forney L, et al. Comparison of self-collected and physician-collected vaginal swabs for microbiome analysis. J Clin Microbiol. 2010;48:1741–1748. doi: 10.1128/JCM.01710-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pybus V, Onderdonk AB. Evidence for a commensal, symbiotic relationship between Gardnerella vaginalis and Prevotella bivia involving ammonia: Potential significance for bacterial vaginosis. J Infect Dis. 1997;175:406–413. doi: 10.1093/infdis/175.2.406. [DOI] [PubMed] [Google Scholar]
- 31.Pybus V, Onderdonk AB. A commensal symbiosis between Prevotella bivia and Peptostreptococcus anaerobius involves amino acids: Potential significance to the pathogenesis of bacterial vaginosis. FEMS Immunol Med Microbiol. 1998;22:317–327. doi: 10.1111/j.1574-695X.1998.tb01221.x. [DOI] [PubMed] [Google Scholar]
- 32.Konopka A. What is microbial community ecology? ISME J. 2009;3:1223–1230. doi: 10.1038/ismej.2009.88. [DOI] [PubMed] [Google Scholar]
- 33.Schwebke JR. New concepts in the etiology of bacterial vaginosis. Curr Infect Dis Rep. 2009;11:143–147. doi: 10.1007/s11908-009-0021-7. [DOI] [PubMed] [Google Scholar]
- 34.Peterson G, Allen CR, Holling CS. Ecological resilience, biodiversity, and scale. Ecosystems. 1998;1:6–18. [Google Scholar]
- 35.Walker BH. Biodiversity and ecological redundancy. Conserv Biol. 1992;6:18–23. [Google Scholar]
- 36.Hobbs RJ, Huenneke LF. Disturbance, diversity, and invasion: Implications for conservation. Conserv Biol. 1992;6:324–337. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.